Abstract
The herbicide active ingredient glyphosate can affect the growth of microorganisms, which rely on the shikimate pathway for aromatic amino acid biosynthesis. However, it is uncertain whether glyphosate exposure could lead to perturbations in the population of human gut microbiota. We have addressed this knowledge gap by analysing publicly available datasets to provide new insights into possible effects of glyphosate on the human gut microbiome. Comparison of the abundance of the shikimate pathway in 734 paired metagenomes and metatranscriptomes indicated that most gut bacteria do not possess a complete shikimate pathway, and that this pathway is mostly transcriptionally inactive in the human gut microbiome. This suggests that gut bacteria are mostly aromatic amino acid auxotrophs and thus relatively resistant to a potential growth inhibition by glyphosate. As glyphosate blocking of the shikimate pathway is via inhibition of EPSPS, we classified E. coli EPSPS enzyme homologues as class I (sensitive to glyphosate) and class II (resistant to glyphosate). Among 44 subspecies reference genomes, accounting for 72% of the total assigned microbial abundance in 2144 human faecal metagenomes, 9 subspecies have class II EPSPS. The study of publicly available gut metagenomes also indicated that glyphosate might be degraded by some Proteobacteria in the human gut microbiome using the carbon–phosphorus lyase pathway. Overall, there is limited experimental evidence available for the effects of glyphosate on the human gut microbiome. Further investigations using more advanced molecular profiling techniques are needed to ascertain whether glyphosate and glyphosate-based herbicides can alter the function of the gut microbiome with consequent health implications.
Keywords: Glyphosate, Gut microbiome, Metagenome, Pesticides, Shikimate
Graphical abstract
Highlights
-
•
It is debated whether glyphosate can perturb the human gut microbiota
-
•
Gut microbiome EPSPS enzymes are predicted to be sensitive to glyphosate
-
•
Shikimate pathway is mostly transcriptionally inactive in the human gut microbiome
-
•
Most gut microbiome bacteria do not possess a complete shikimate pathway
-
•
Investigations using more advanced techniques such as metabolomics are needed
1. Introduction
The human gastrointestinal tract is inhabited by a large collection of microorganisms including bacteria, fungi, viruses and small eukaryotes collectively known as the gut microbiota. Although diet plays a primary role in determining the status of the gut microbiota population (Carmody et al., 2015; Rothschild et al., 2018), several studies have suggested that it can also be affected by the exposure to environmental chemicals (Mesnage et al., 2018). Xenobiotic metabolism by gut microbiota has been shown to affect the therapeutic effects of common drugs (Clayton et al., 2009; Saha et al., 1983), be a source of toxic effects (Zheng et al., 2013), or even influence animal species evolution by providing them with the ability to feed on toxic plants (Kohl et al., 2014). In most cases, it is not clear whether these changes in gut microbiome composition will be pathological. There is no clear definition of an ‘healthy’ or ‘normal’ gut microbiome (Schmidt et al., 2018). However, since effects of chemicals on the gut microbiome are not systematically evaluated in the battery of pre-commercial toxicity tests conducted for regulatory approval purposes in accordance with OECD guidelines, concerns have been raised about the potential impacts of environmental contaminants frequently found in human foodstuff such as pesticides (Claus et al., 2016).
One pesticide that is of particular concern in this regard is glyphosate, the major herbicidal active ingredient used across the world (Benbrook, 2016). Glyphosate was patented as an herbicide active ingredient in 1971 (U.S. Patent No 3,799,758) and subsequently commercialised under the brand name Roundup by Monsanto Company. Glyphosate residues are frequently found in the food chain due to either being sprayed on cereals to accelerate ripening, facilitate more uniform drying of the grain and thus expedite harvesting of the crop, or to clear weeds during cultivation of Roundup-tolerant genetically modified crops (Rothschild et al., 2018). Glyphosate residues are routinely detected in human urine at levels around 1 μg/L, with some studies showing levels up to 10 μg/L in non-occupationally exposed populations (Mesnage et al., 2018). The mode of glyphosate herbicidal action in plants is to inhibit the binding of phosphoenolpyruvate (PEP) to the enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) a component of the shikimate pathway (Fig. 1A). This causes a shortage in aromatic amino acid synthesis and consequent death. The binding of glyphosate to EPSPS is highly specific and it is assumed that it is unlikely that this compound can bind to the PEP association site of other enzymes in mammals (Schonbrunn et al., 2001). Crucially, many strains of bacteria and fungi, including some present in the gut microbiome, also possess the shikimate pathway raising the possibility that glyphosate may also interfere with the viability of these microorganisms. Indeed, the patenting of glyphosate as an antibiotic was based on this possibility (US Patent No. 7,771,736 B2).
Fig. 1.
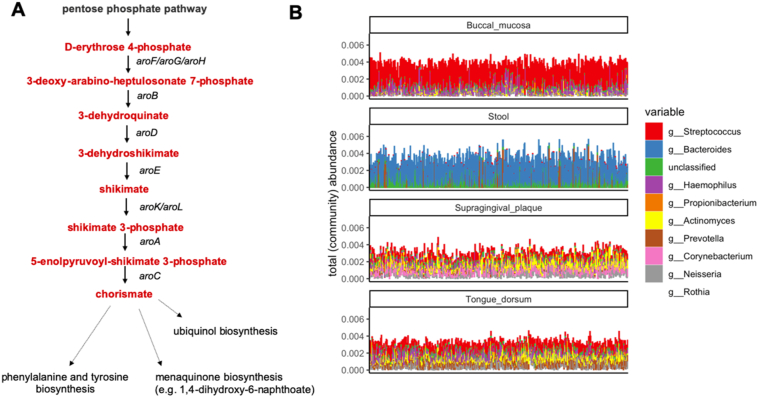
The shikimate pathway in the human digestive tract. A. the first part of the shikimate pathway resulting in the formation of chorismate from erythrose 4-phosphate. B. Shotgun metagenomics data generated by the human microbiome project for 4 major sites of the digestive tract was analysed to determine the abundance of the shikimate pathway (as MetaCyc superpathway of aromatic amino acid biosynthesis) and the contribution of bacterial genera to this abundance. Pathway bar plots display total (community) abundance and the contributions of the top seven genera proportionally scaled to the total abundance. The abundance data was sourced from the HMP portal (https://www.hmpdacc.org/hmp/hmmrc2/).
EPSPS enzymes have been classified into two categories: EPSPS class I enzymes are sensitive to glyphosate inhibition while EPSPS class II enzymes are not affected by glyphosate. The first genetic evidence showing that EPSPS was the biochemical target of glyphosate in enteric bacteria were presented in 1983, when a mutant aroA gene (coding for EPSPS) cloned from Salmonella typhimurium was shown to confer glyphosate resistance to E. coli (Sugata et al., 1991). The discovery of glyphosate tolerant EPSPS was seen as an opportunity to generate transgenic plants tolerant to this compound (Kishore et al., 1992). EPSPS enzymes isolated from bacteria naturally tolerant to glyphosate, such as from Agrobacterium sp. strain CP4, Achromobacter sp. strain LBAA and Pseudomonas sp. strain PG2982, were designated as class II EPSPS. All class I EPSPS show a remarkable degree of homology. For example, class I EPSPS from plants and bacteria are approximately 54% identical and with similarity as high as 80% (U.S. patent No. 6248876B1). In contrast, class II CP4 EPSPS present substantial differences in their amino acid sequences in comparison to class I EPSPS from E. coli, S. typhimurium, B. pertussis, P. hydriba, or S cerevisiae (similarity/identity = 50–53%/23–30%) (U.S. patent No. 6248876B1).
Glyphosate can affect the growth of a large range of microorganisms, which rely on the shikimate pathway and EPSPS for aromatic amino acid biosynthesis (Tsiaoussis et al., 2019; Sviridov et al., 2015). However, it is debated as to whether glyphosate exposure could lead to perturbations in the gut microbiota by inhibiting EPSPS and thus impeding microorganism growth. Although no study has directly investigated the effects of glyphosate on the human gut microbiome, some studies have been performed both in vivo in rats (Nielsen et al., 2018; Mao et al., 2018; Lozano et al., 2018), honeys bees (Motta et al., 2018) and turtles (Kittle et al., 2018), and employing in vitro model systems simulating the bovine (Riede et al., 2016) and poultry (Shehata et al., 2013) digestive tracks. These studies present contradictory evidence and it is not clear to what extent they provide meaningful information regarding potential effects on the human gut microbiome. Since some intracellular parasites rely on the shikimate pathway for aromatic amino acid biosynthesis (Roberts et al., 1998), the use of glyphosate has been proposed to tackle pathogenic infections provoked by Toxoplasma gondii, Plasmodium falciparum and Cryptosporidium parvum (Roberts et al., 1998). We recently used a multi-omics strategy combining shotgun metagenomics and metabolomics to test whether glyphosate or the Roundup herbicide formulation MON 52276 has an effect on the rat gut microbiome in a 90-day subchronic toxicity test (Mesnage et al., 2019). Caecum metabolomics revealed that glyphosate inhibits EPSPS of the shikimate pathway as evidenced by an accumulation of shikimic acid and 3-dehydroshikimic acid, which would normally be metabolised by this enzyme. However, the rat gut microbiome is substantially different from that of humans, and it is thus not possible to predict if our findings will be of relevance for human physiology without investigating in detail the function of the shikimate pathway in the human gut microbiome.
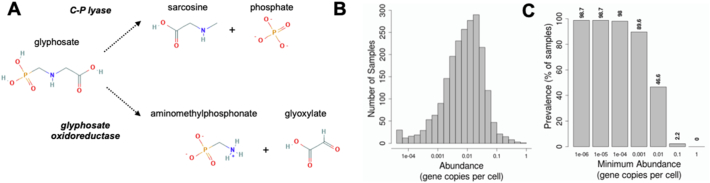
Another area of uncertainty is whether glyphosate can be metabolised by the gut microbiome. A large number of studies have shown that soil microorganisms can metabolise glyphosate using two major pathways (Sviridov et al., 2015); a cleavage of the C—P bond to phosphate and sarcosine (the C—P lyase pathway also called the sarcosine pathway), or a cleavage to AMPA and glyoxylate (the glycine oxidase pathway also known as the AMPA pathway) (Fig. 5A). The C—P lyase complex is the product of a 14 gene operon (phnCDEFGHIJKLMNOP) (Zhan et al., 2018). Glyphosate oxidoreductase (GOX) is another enzyme which can perform glyphosate degradation to AMPA by C—N bond cleavage. GOX genes have been identified in Ochrobactrum spp. (Zhan et al., 2018). A synthetic GOX has also been used as a transgene to produce glyphosate-resistant canola crops (Hadi et al., 2012). Another enzyme found to metabolise glyphosate is glycine oxidase from B. subtilis (Pedotti et al., 2009). The variety of bacteria and fungi isolated from environmental sources with the ability of degrading glyphosate is already well described (Zhan et al., 2018; Hove-Jensen et al., 2014).
Fig. 5.
Glyphosate metabolism by microorganisms. A. Illustration of the metabolism of glyphosate by microorganisms using either the C—P lyase or glycine oxidase pathways. However, the existence of these pathways in the human gut microbiome is unclear. We studied the distribution of genes related to E. coli alpha-d-ribose 1-methylphosphonate 5-triphosphate synthase (P16687|PHNI_ECOLI) in metagenomic data from MetaQuery. B. Abundance of the P16687|PHNI_ECOLI gene across samples. C. Prevalence of the P16687|PHNI_ECOLI gene across faecal metagenomes at different abundance thresholds.
In order to provide more insight into the possible effects of glyphosate on the human gut microbiome, we have studied the presence and activity of the shikimate pathway in diverse body sites in human populations by analysing data generated by the Human Microbiome Project (HMP) (Lloyd-Price et al., 2017). Comparative evaluation of shikimate pathway abundance in faecal metagenomes and faecal metatranscriptomes informed us on its transcriptional activity in faecal microbiomes. Since the potential effects of glyphosate on the gut microbiome also depends on its ability to affect EPSPS activity, we then predicted the classification of E. coli EPSPS homologues in the most frequently found bacterial strains in 2144 human faecal metagenomes by comparing amino acid composition for 5 motifs representative of class II enzymes (Yi et al., 2016; Li et al., 2009).
Overall, the data presented here have informed on the potential human health implications of findings from laboratory animal investigations such as our recent study showing that glyphosate inhibited EPSPS in the rat caecum microbiome causing an accumulation of shikimate pathway intermediates (Mesnage et al., 2019).
2. Material and methods
2.1. Transcriptional activity of the shikimate pathway in the human gut microbiome
The MetaCyc superpathway of aromatic amino acid biosynthesis (Fig. 1A) was considered as a good indicator of the shikimate pathway. This is because it includes all the steps leading to the synthesis of aromatic amino acids from the precursor D-erythrose 4-phosphate. We present the total (community) abundance for the 10 genera found to be the most involved in this pathway, for 4 body sites namely buccal mucosa, stool, supragingival plaque and tongue dorsum. We did not re-process the raw data from this project but used the output of the HUMAnN2 (Franzosa et al., 2018) analysis to quantify pathways representative of major biochemical reactions as included in the MetaCyc database (Caspi et al., 2018), a curated catalogue of experimentally elucidated pathways. In brief, firstly HUMAnN2 uses the results of the taxonomic binning and profiling program MetaPhlAn2 (Truong et al., 2015). In addition, it maps reads to the ChocoPhlAn pan-genome database, containing coding sequences from NCBI deposited genomes, using Bowtie2. These two steps allow an organism-specific functional profiling. Then, a translated search (i.e., using nucleotide information at the protein level) with sequences of unclassified organisms is performed against the UniRef protein database to identify protein families with known functions.
Files were downloaded from the HMP portal (https://www.hmpdacc.org/hmp/hmmrc2/, file name ‘hmp1-II_humann2_pathabundance-nrm-mtd-qcd’), generated using bioinformatics pipelines described in (Lloyd-Price et al., 2017), or from the Inflammatory Bowel Disease Multi'omics Database (IBDMDB) (https://ibdmdb.org/tunnel/public/summary.html, tables ‘pathabundances.tsv.gz’) for the paired metatranscriptomes and metagenomes described in (Lloyd-Price et al., 2019). In addition, in order to assess the completeness of the pathway in given organisms, we also analysed enzyme abundance levels by using the gene family level analysis aggregated at different enzyme commission (EC) number level (file ‘ecs.tsv.gz’ from the IBDMDB dabatase). Note that the two datasets from (Lloyd-Price et al., 2017; Lloyd-Price et al., 2019) were run with different parametrisations of the HUMAnN2 pipeline, with details described in the material and methods sections of the respective publications. Abundance tables were imported in R and the relative contribution of the different bacterial species to the abundance of the pathway COMPLETE-ARO-PWY plotted with the R package ggplot2.
2.2. Comparisons of class I and class II EPSPS conserved regions
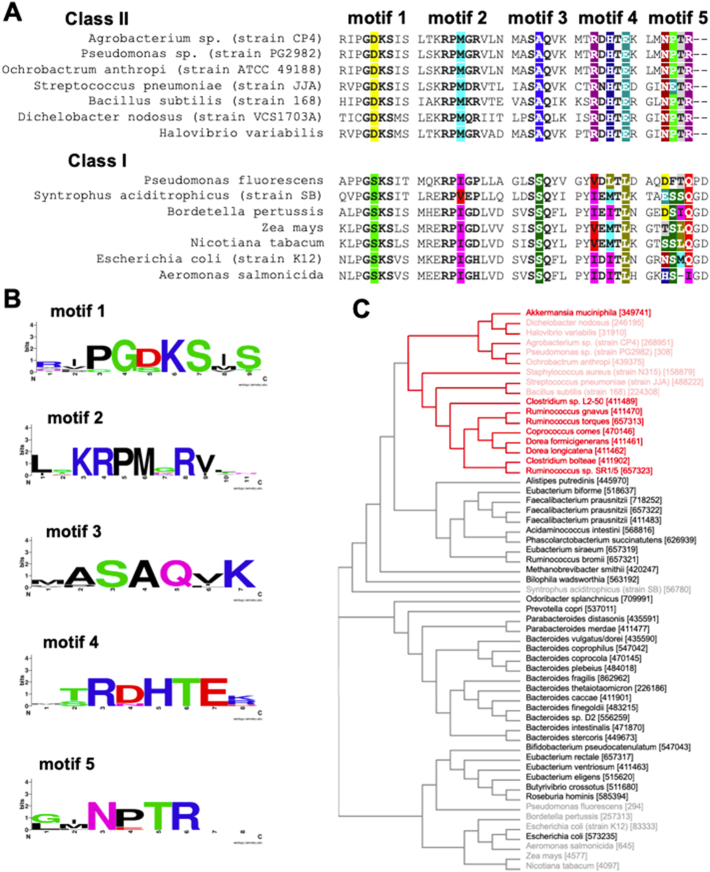
We studied the classification of EPSPS from the most highly abundant subspecies of bacteria in the human gut microbiome selecting 44 reference genomes from species accounting for 72% of the total assigned microbial abundance in 2144 human faecal metagenomes (Costea et al., 2017). EPSPS sequences were retrieved with the NCBI BLAST tool, by a BLAST of E. coli EPSPS to the reference genomes of the 44 target bacteria (Supplementary Material 1). In order to compare the different protein sequences, EPSPS sequences were aligned using T-Coffee with default parameters (Di Tommaso et al., 2011). The classification was visualised as a neighbour-joining tree without distance corrections performed with default parameters in ClustalO (Sievers et al., 2011). In order to make sure that we classified the different EPSPS appropriately, we also included sequences of different species with experimentally identified EPSPS class II or class I (Supplementary Material 1). Ultimately, classification was done by considering the 5 molecular motifs allowing EPSPS class II classification described above (Fig. 3, Supplementary Material 1).
Fig. 3.
Classification of EPSPS enzymes present in the human gut microbiome. A. Comparison of EPSPS enzyme amino acid sequences for their classification and the detection of molecular motifs. B. Graphical illustration of the 5 EPSPS class II motifs generated with WebLogo 3 (http://weblogo.berkeley.edu/). C. Classification of gut microbiome EPSPS variants by analysis of the EPSPS sequences from 44 subspecies, accounting for 72% of the total assigned bacterial abundance in 2144 human faecal metagenomes. The sequences of known class I (shaded black) and II (shaded red) EPSPS enzymes are used as a positive control. The classification was visualised as a neighbour-joining tree without distance corrections performed with default parameters in ClustalO.
Experimental evidence for the involvement of these molecular motifs in glyphosate resistance was retrieved from previous publications (Yi et al., 2016; Li et al., 2009). First, the aspartate residues in the class II EPSPS motif 1 (GDKX) is replaced by a serine in class I EPSPS. The lysine residue (K) facilitates PEP binding. The second motif RPMXR is the most recently described and was experimentally shown to be required for resistance to glyphosate (Li et al., 2009). The third motif is a triplet of amino acids (SAQ) in which the serine (S) and glutamine (Q) residues are important for productive shikimate-3-phosphate binding. The central alanine residue in this EPSPS class II motif is mostly replaced by a serine in class I EPSPS. Motif 4 has the sequence RXHXE. In motif 4, the positive charge of the guanidine group of the arginine (R) destabilizes glyphosate binding (Li et al., 2009). In the fifth motif (NXTR), the arginine residue (R) has also been shown to destabilize the binding of glyphosate (U.S. patent No. 6248876B1).
2.3. Re-analysis of glyphosate effects on mammalian faecal microbiomes
The gut microbiome study of Nielsen and colleagues (Nielsen et al., 2018) combined 16S rRNA gene sequencing with metabolomics to understand the effects of glyphosate and a glyphosate formulated product on the gut microbiota at different stages of the rat digestive tract (ileum, colon, cecum, feces). In this study, Sprague-Dawley rats were exposed to glyphosate (2.5 and 25 mg/kg/day) or Glyfonova® (25 mg/kg/day glyphosate acid equivalent) by oral gavage for two weeks. This corresponded to doses 5 times and 50 times respectively above the EU ADI. The gut microbiota of these animals was subjected to an analysis based on the amplificon of fragments from the 16S rRNA genes. The bioinformatics analysis was initially performed with QIIME. Although this analysis informs on taxonomic composition, the most recent developments for the analysis of PCR-amplified marker gene sequencing data includes the generation of functional predictions from amplicon sequences in a QIIME2 module called PICRUSt2 (Douglas et al., 2019).
We therefore used PICRUSt2 to predict the function of 16S amplicon sequences from the study of Nielsen and colleagues (Nielsen et al., 2018). For this purpose, the sequencing data from this investigation (BioProject PRJNA412959) was re-analysed using QIIME2 2019.1. Data analysis was done with Rosalind, the Biomedical Research Centre/King's College London high-performance computing cluster. We used the read denoiser DADA2, the Silva 132 99% OTUs full-length sequence taxonomy classifier, a sequence placement using q2-fragment-insertion, and PICRUSt2 plugin for QIIME2, as described in the GitHub q2 picrust2 Tutorial (https://github.com/picrust/picrust2/wiki/q2-picrust2-Tutorial). Data was imported in R version 3.6.1 for statistical analysis. Variables with more than 20% missing values were discarded. In order to make the samples comparable, raw counts were normalised to the relative abundance of all bacteria observed. Data was then log-transformed before statistical comparisons in a two-way ANOVA taking into account the treatment (glyphosate 5 mg/kg/day, glyphosate 25 mg/kg/day, formulation Glyfonova) and the sample location (caecum, ileum, colon, feces). P-values for the factor treatment were then corrected for multiple comparison using the Benjamini–Hochberg procedure. Data was plotted with the ggplot2 and ggbeeswarm R packages.
2.4. Finding homologues of glyphosate metabolizing enzyme
We investigated the presence of genes related to E. coli alpha-d-ribose 1-methylphosphonate 5-triphosphate synthase (P16687|PHNI_ECOLI) in the human gut metagenome by using the MetaQuery database, which interrogates more than 2000 publicly available metagenomes for the presence and abundance of genes using BLAST (Pollard et al., 2015). This tool was used with default parameters. We used a similar strategy to investigate the presence of homologues of glycine oxidase thiO of B. subtilis (O31616) and of the glyphosate oxidase gox of O. anthropic (D2KI28) in human gut metagenome datasets.
3. Results
Although it is widely recognised that many microorganisms rely on the shikimate pathway for aromatic amino acid biosynthesis, the extent to which this pathway is active in the human gut microbiome is unknown. We show that the shikimate pathway is present in almost all individuals (Fig. 1B). The genus Bacteroides is the main contributor in the faecal microbiome, which reflects the composition of the gut microbiome since Bacteroides spp. are most common (Fig. 1B, panel Stool). The contribution to the shikimate pathway in the oral microbiome also reflects the abundance of the different species at this site since Streptococcus spp. are dominant (Fig. 1B, panel Buccal mucosa). In other compartments, a more varied composition of shikimate pathway positive bacterial species is evident (Fig. 1B, panels Supragingival plaque, Tongue dorsum).
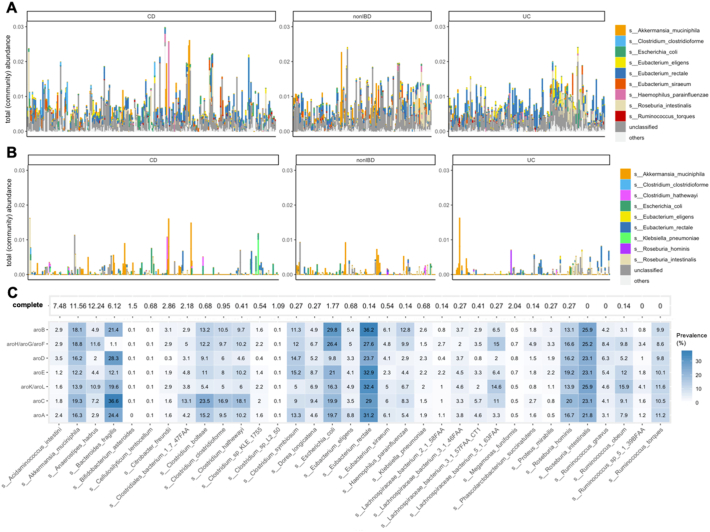
Gut bacteria that are highly abundant can also be relatively transcriptionally inactive (Lloyd-Price et al., 2019) . We therefore compared the abundance of the shikimate pathway in 734 paired metagenomes and metatranscriptomes from human faecal samples available in the Inflammatory Bowel Disease Multi'omics Database (IBDMDB). The metatranscriptome profiles were different from those of the metagenomes (Fig. 2). Although the bacterial species contributing to the shikimate pathway were comparable (Fig. 2A), abundance profiles suggests that this pathway is mostly transcriptionally inactive (Fig. 2B). Although the shikimate pathway was detected in 98% of the metagenome samples, it was only detected in 35% of the metatranscriptome samples. The total assigned abundance to the shikimate pathway was approximately 10 times lower for the metatranscriptome than for the metagenome. The shikimate pathway's transcript pool was dominated by Akkermansia muciniphila, which represented 29.4% of the gene expression assigned to the shikimate pathway although this species only represented 5% of the assigned abundance of at the genome level.
Fig. 2.
Analysis of paired metagenomes and metatranscriptomes suggests that the shikimate pathway is mostly transcriptionally inactive in the human gut microbiome. Paired metagenomics (a) and metatranscriptomics (B) samples from 734 patients with inflammatory bowel disease (CD, Crohn's disease; nonIBD, non-IBD control patients; UC, ulcerative. colitis). Abundance data for paired metatranscriptomes and metagenomes was downloaded from the inflammatory bowel disease Multi'omics database (https://ibdmdb.org/tunnel/public/summary.html). (C) Completeness of the shikimate pathway (first part leading to the synthesis of chorismic acid including the enzymes catalysing the reaction 1.1.1.25 (gene aroE), 2.5.1.19 (gene aroA, coding for the EPSPS enzyme), 2.5.1.54 (aroG/aroH/aroF), 2.7.1.71 (aroL/aroK), 4.2.3.4 (aroB), 4.2.3.5 (aroC), as well as 4.2.1.10 (aroD) in the 734 individuals having their metatranscriptome sequenced in the IBDMDB project. Numbers in the tiles indicate the percentage of individuals for which transcripts coding for these enzymes were detected. The ‘complete’ row indicates the percentage of individuals for which the complete set of genes have been found to be transcribed, for each gut bacterial species in which the shikimate pathway was found to be active.
Since our recent study provided evidence that glyphosate can cause an accumulation of shikimate and 3-dehydroshikimic acid by blocking the activity of EPSPS in the caecum microbiome in rats (Mesnage et al., 2019), we questioned if bacterial species found in the human gut microbiome harboured transcriptionally active genes allowing the synthesis of shikimate. For this purpose, we investigated the completeness of the shikimate pathway at the gene family level in order to understand if the 734 individuals from the IBDMDB project harbour the complete set of enzymes allowing the biosynthesis of chorismate from erythrose 4-phosphate. A small proportion of individuals had all the transcripts coded from the genes of the shikimate pathway (Fig. 2C). The bacteria Akkermansia muciniphila was found to have all the transcriptionally active genes necessary for the synthesis of chorismate in 11.6% of the individuals. Most other bacterial species had complete shikimate pathways in less than 1% of the individuals. This suggested that the shikimate is produced only by a small proportion of the gut bacteria, although we cannot assess if different species can cooperate and use each other's metabolic products to form shikimate. This also suggests that gut microbiome bacteria are mostly aromatic amino acid auxotrophs. However, it is not clear if this can be different in cases of malnourished individuals on an impoverished, low protein diet with a consequent shortage of aromatic amino acids.
In order to understand if EPSPS enzymes in the human gut microbiome are likely to be inhibited by glyphosate, we attempted to classify them from the most highly abundant subspecies of bacteria in this body compartment (Fig. 1C, Supplementary Material 1). The 44 reference genomes were chosen because these species accounted for 72% of the total assigned microbial abundance in 2144 human faecal metagenomes (Costea et al., 2017). EPSPS enzymes have been classified as either class I (sensitive to glyphosate) or class II (resistant to glyphosate) based on a visual inspection of 5 amino acid sequence molecular motifs and based on the classification of EPSPS sequences in a neighbour-joining tree (Fig. 3). A total of 9 subspecies (17.7% of the assigned abundance) were predicted to possess a class II EPSPS (Fig. 3C, red annotation), which are thus most likely resistant to glyphosate. However, most of the bacteria inhabiting the human gut microbiome were classified by default as having class I EPSPS enzymes (Fig. 3C, black annotation), which would render them sensitive to inhibition by glyphosate.
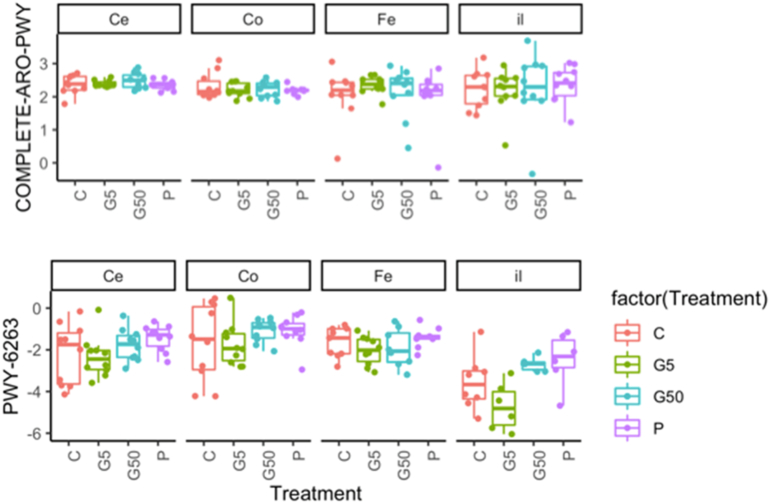
The fact that the shikimate pathway could be transcriptionally inactive in faecal samples may not be so surprising since bacteria inhabiting the digestive tract do not necessarily need to synthetize aromatic amino acids if they are provided by the diet. This is corroborated by an experimental study conducted by Nielsen and colleagues, which compared the effects of glyphosate and a commercial glyphosate based herbicide on the Sprague-Dawley rat gut microbiota (Nielsen et al., 2018). Rats administered with two doses of glyphosate, or a glyphosate formulation (Roundup), at 2.5 or 25 mg/kg bw/day presented little difference in gut microbiota composition. This observation seemed to indicate that sufficient quantities of aromatic amino acids were available in the rodent diets. Since this study did not include functional prediction at the pathway level, allowing the measurement of shikimate pathway abundance, we used PICRUSt2 to predict the function of 16S amplicon sequences detected from this study (Nielsen et al., 2018). The functional prediction analysis with PICRUSt2 suggested that the treatments have produced subtle changes in the function of the gut microbiome (Fig. 4; Supplementary Material 3). The shikimate pathway was not affected, and its abundance was comparable in the different sections of the gastrointestinal tract (ileum, colon and cecum) as well as in the faecal samples. However, it is noteworthy that the most significantly affected pathways (adjusted p-value for treatment effect = 0.03, Supplementary Material 3) were MetaCyc-7374 (1,4-dihydroxy-6-naphthoate biosynthesis I), METACYC-6263 (superpathway of menaquinol-8 biosynthesis II), and MetaCyc-7371 (MetaCyc 1,4-dihydroxy-6-naphthoate biosynthesis II). The pathways MetaCyc-7374 and MetaCyc-7371 are both part of the superpathway METACYC-6263. They all include chorismate, a central compound in the shikimate pathway, which can be expected to be affected by exposure to the glyphosate formulation. However, differences in pathway abundance between individual animals within a group were greater than the effect of test diets, which prevented a definitive conclusion on their potential reliability (Fig. 4).
Fig. 4.
Reanalysis of published data suggests that glyphosate may affect chorismate metabolism. The 16S rRNA gene sequence data from Nielsen et al. (Nielsen et al., 2018) was reanalysed using QIIME2 2019.1. Pathway abundance in the caecum (Ce), colon (Co), feces (Fe), and ileum (il) was compared for rats exposed to glyphosate (2.5 and 25 mg/kg/day, G5 (5× EU ADI) and G50 (50× EU ADI) or the commercial glyphosate herbicide formulation Glyfonova® (25 mg/kg/day glyphosate acid equivalent, P). Although the shikimate pathway was not affected (COMPLETE-ARO-PWY, upper panel), PICRUSt2 analysis suggested that treatment with the Glyfonova® might have produced some changes in the abundance of the METACYC-6263 pathway (lower panel) using chorismate as a precursor compound (superpathway of menaquinol-8 biosynthesis II).
Besides its potential interaction with the shikimate pathway, it is also not clear if glyphosate can be metabolised by gut microbiota in the same manner as it is by some environmental microorganisms. We investigated the presence of genes related to E. coli alpha-d-ribose 1-methylphosphonate 5-triphosphate synthase (P16687|PHNI_ECOLI) in the human gut metagenome by using the MetaQuery database. We found homologues of phnI in a large number of Proteobacteria (Escherichia, Enterobacter, Citrobacter, Klebsiella) frequently found in the human gut microbiome (Supplementary Material 4). They were found in almost all metagenomes surveyed (Fig. 5B and C), suggesting that the processing of phosphonates into usable phosphate may be a core function in the human gut microbiome. We used a similar strategy employing MetaQuery to investigate the presence of homologues of the glycine oxidase thiO of B. subtilis (O31616) and of the glyphosate oxidase gox of O. anthropic (D2KI28) in human gut metagenomes. No homologues were found using this bioinformatics tool.
4. Discussion
Whether the exposure to glyphosate is a source of health risk is a recurring subject of intense debate amplified by political and economic vested interests. Since the effects of chemicals on the gut microbiome are not systematically evaluated in the battery of pre-commercial toxicity tests conducted according to OECD guidelines, concerns have been raised about the potential impacts of glyphosate on the human gut microbiome.
Comparative abundance analysis of the shikimate pathway in 734 paired metagenomes and metatranscriptomes with HUMAnN2, suggested that this pathway is mostly transcriptionally inactive in the human gut microbiome. Further analyses at the gene family level showed that some bacteria species may have “lost” genes and do not possess a complete shikimate pathway. This is corroborated by a study of the presence of genes encoding shikimate pathway enzymes in 488 sequenced prokaryotes using Hidden Markov Model (HMM) profiles, which revealed that 31% of host associated bacteria do not possess a complete shikimate pathway (Zucko et al., 2010). Out of the 91 host-associated bacteria lacking a complete shikimate pathway, most (67/91) had lost five or more enzymes from this pathway, whereas nearly all of the remaining (20/91) have only lost a single enzyme (Zucko et al., 2010). Although it is likely that gut bacteria source amino acids from the host, especially from ingested food, they might also have a shared metabolic adaptation in mutualistic symbiosis with other gut bacteria. It is frequent for bacteria to cooperate and use each other's metabolic products to perform the biosynthesis of some compounds. It could thus be postulated that if a bacterium, which provides a crucial component of the shikimate pathway that is shared with other microorganisms, is negatively impacted by glyphosate, this could lead to gut dysbiosis. In addition, the transcriptional activity of bacteria residing in the gut microbiome depends on their microenvironment and can thus vary in different parts of the gastrointestinal tract (Lloyd-Price et al., 2019). Another limit of this analysis is that the analysis of metagenomes and metatranscriptomes is generally not complete for a given individual because of the incompleteness of the taxonomic classification, as well as the zero-inflation of metagenomic gene count data (Knight et al., 2012).
The study conducted by Nielsen and colleagues showed that an inhibition of the shikimate pathway by glyphosate in all likelihood would have minor consequences because the quantity of aromatic amino acids included in the diet should be adequate to prevent a change in essential amino acid levels. Although these findings are not sufficient to claim that glyphosate has an effect on chorismate metabolism, our pathway analysis with PICRUSt2 suggested that the commercial glyphosate herbicide formulation affected the synthesis of menaquinols as the abundance for this pathway was increased (Fig. 4). Even if the gut bacteria are not fully equipped to synthesise shikimate from its precursor D-erythrose 4-phosphate, glyphosate may block the conversion of dietary shikimate to menaquinols, which can have important health implications (Beulens et al., 2013). This highlights that further studies should not only look at aromatic amino acid synthesis, since the shikimate pathway is also involved in the synthesis of other phenolic compounds (Seigler, 1998) (Fig. 4B). However, menaquinol metabolism was probably not altered to a level which would result in toxic effects since menaquinones are critical for electron transport and a deficiency would result in a significant growth deficit of bacteria, which was not detected.
There is no evidence to suggest that glyphosate undergoes active metabolism upon absorption in mammals. Glyphosate is generally rapidly absorbed from the intestinal tract (~30% absorption rate), and rapidly eliminated in urine (99% eliminated 7 days after a single dose administration) (Brewster et al., 1991). The main metabolite of glyphosate is considered to be aminomethylphosphonic acid (AMPA) and glyoxylate. Although our analysis did not find indications for the presence of the degradation AMPA pathway in the human gut microbiome, glyphosate can be degraded to AMPA in the environment, or spontaneously without active metabolism in the human body. The degradation of glyphosate could be toxicologically relevant since one of its degradation products, glyoxylate, has been reported to not only conjugate with proteins but also increase liver triglyceride and cholesteryl ester levels (Ford et al., 2017). Our search for the presence of homologues of genes known to metabolise glyphosate suggested the C—P lyase pathway may be present in some Proteobacteria (Escherichia, Enterobacter, Citrobacter, Klebsiella). Although it is not possible to define bacterial species as either ‘good’ or ‘bad’, Proteobacteria are generally regarded to have a negative influence on human health (Shin et al., 2015). In a recent study using bioreactor systems inoculated with colonic bacteria from German Landrace pigs, glyphosate concentrations remained unchanged after a short two day exposure (Fritz-Wallace et al., 2020). However, it remains unclear as to whether glyphosate metabolic pathways exist in the human gut microbiome.
In this study, we explored a variety of mechanisms by which glyphosate may affect the human gut microbiome. However, their relevance to human health needs to take into account the typical daily intake of this pesticide. Typical herbicide exposure occurs through diet, dermal contact and inhalation. In the case of glyphosate, ingestion through the diet appears to be the main route of exposure. Glyphosate residues are frequently found in the food chain due to either being sprayed on cereals to facilitate more uniform drying of the grain and thus expedite harvesting of the crop, or to clear weeds during cultivation of glyphosate-tolerant genetically modified crops (EFSA, 2020).
In order to evaluate the probability of adverse health effects in humans, it is possible to estimate pesticide dietary exposure using standard daily intake for a variety of foodstuffs for which contaminating levels are known. International Estimates of Daily Intake (IEDI) for the WHO GEMS/Food 17 Cluster Diets have been estimated to range between 1.7 and 4.9 μg/kg bw/day in a recent analysis (Stephenson and Harris, 2016). In this case, the highest contributor to the total glyphosate intake was barley in the Irish diet. Barley in breakfast cereals was found to be contaminated with 5.85 mg/kg of glyphosate (Stephenson and Harris, 2016). This ingestion resulted in glyphosate detection in human urine at levels around 1 μg/L (Mesnage et al., 2018), which can be greater in higher use territories such as the US (Gillezeau et al., 2019). In order to perform an evaluation of risk, in current legislative practise human exposures are compared to toxicity thresholds derived from animal studies, such as the acceptable daily intake (ADI) or its US equivalent, the chronic reference dose (cRfD). In the case of glyphosate (EFSA, 2020), it is estimated that human dietary exposure represented between 0.06% (most conservative estimate) and 0.26% (least conservative estimate) of the ADI. Although our recent shotgun metagenomics analysis suggested an inhibition of the shikimate pathway at levels of exposure above the ADI (Mesnage et al., 2019), there is no experimental evidence showing effects of glyphosate on gut microbiome metabolism in real-world exposure scenarios. Most of the studies describing antimicrobial effects of glyphosate are using doses several orders of magnitude higher than current dietary levels of exposure.
5. Conclusion
We have assessed evidence for effects of glyphosate in the human gut microbiome. Bioinformatics analyses of the human gut metagenome and metatranscripome databases suggested that the presence of the shikimate biochemical pathway targeted by glyphosate is widespread in microorganisms inhabiting the gastrointestinal tract, but that this pathway is mostly transcriptionally inactive. Aromatic amino acid biosynthesis via the shikimate pathway is likely not essential for the gut microbiota, presumably because of the presence of sufficient amounts of these amino acids in the digestive tract entering from food consumption. Antimicrobial effects are mostly found at high doses, which are unlikely to cause adverse health outcomes in real-world exposure scenarios. However, glyphosate may affect shikimate metabolism without drastically affecting microorganism abundance and these effects might not be detectable with the most common gut microbiome profiling techniques. Overall, there is limited experimental evidence available for the effects of glyphosate on gut microbiome metabolism. Environmental epidemiological studies measuring shikimic acid levels as a biomarker for the inhibition of the shikimate pathway could be performed to understand if glyphosate can have biological effects in human populations.
The following are the supplementary data related to this article.
EPSPS classification.
Pathway table.
Pathway results.
Homologues metabolism.
CRediT authorship contribution statement
Robin Mesnage: Conceptualization, Methodology, Formal analysis, Writing - original draft. Michael N. Antoniou: Writing - review & editing.
Declaration of competing interest
RM has served as a consultant on glyphosate risk assessment issues as part of litigation in the US over glyphosate health effects.
Acknowledgements
RM is supported by a grant from the Sustainable Food Alliance (USA).
References
- Benbrook C.M. Trends in glyphosate herbicide use in the United States and globally. Environ. Sci. Eur. 2016;28:3. doi: 10.1186/s12302-016-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beulens J.W., Booth S.L., van den Heuvel E.G., Stoecklin E., Baka A., Vermeer C. The role of menaquinones (vitamin K(2)) in human health. Br. J. Nutr. 2013;110:1357–1368. doi: 10.1017/S0007114513001013. [DOI] [PubMed] [Google Scholar]
- Brewster D.W., Warren J., Hopkins W.E., 2nd Metabolism of glyphosate in Sprague-Dawley rats: tissue distribution, identification, and quantitation of glyphosate-derived materials following a single oral dose. Fundam. Appl. Toxicol. 1991;17:43–51. doi: 10.1016/0272-0590(91)90237-x. [DOI] [PubMed] [Google Scholar]
- Carmody R.N., Gerber G.K., Luevano J.M., Jr., Gatti D.M., Somes L., Svenson K.L., Turnbaugh P.J. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R., Billington R., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Midford P.E., Ong Q., Ong W.K. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018;46:D633–D639. doi: 10.1093/nar/gkx935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus S.P., Guillou H., Ellero-Simatos S. The gut microbiota: a major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes. 2016;2:16003. doi: 10.1038/npjbiofilms.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton T.A., Baker D., Lindon J.C., Everett J.R., Nicholson J.K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costea P.I., Coelho L.P., Sunagawa S., Munch R., Huerta-Cepas J., Forslund K., Hildebrand F., Kushugulova A., Zeller G., Bork P. Subspecies in the global human gut microbiome. Mol. Syst. Biol. 2017;13:960. doi: 10.15252/msb.20177589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J.M., Taly J.F., Notredame C. T-coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:W13–W17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas G.M., Maffei V.J., Zaneveld J., Yurgel S.N., Brown J.R., Taylor C.M., Huttenhower C., Langille M.G.I. PICRUSt2: An improved and extensible approach for metagenome inference. bioRxiv. 2019 doi: 10.1101/672295. [DOI] [Google Scholar]
- EFSA The 2018 European Union report on pesticide residues in food. EFSA Journal. 2020;18(4):6057. doi: 10.2903/j.efsa.2020.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford B., Bateman L.A., Gutierrez-Palominos L., Park R., Nomura D.K. Mapping proteome-wide targets of glyphosate in mice. Cell Chem Biol. 2017;24:133–140. doi: 10.1016/j.chembiol.2016.12.013. [DOI] [PubMed] [Google Scholar]
- Franzosa E.A., McIver L.J., Rahnavard G., Thompson L.R., Schirmer M., Weingart G., Lipson K.S., Knight R., Caporaso J.G., Segata N. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods. 2018;15:962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Wallace K., Engelmann B., Krause J.L., Schape S.S., Poppe J., Herberth G., Rosler U., Jehmlich N., von Bergen M., Rolle-Kampczyk U. Quantification of glyphosate and AMPA from microbiome reactor fluids. Rapid Commun. Mass Spectrom. 2020;34(715) doi: 10.1002/rcm.8668. [DOI] [PubMed] [Google Scholar]
- Gillezeau C., van Gerwen M., Shaffer R.M., Rana I., Zhang L., Sheppard L., Taioli E. The evidence of human exposure to glyphosate: a review. Environ. Health. 2019;18:2. doi: 10.1186/s12940-018-0435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi F., Mousavi A., Salmanian A.H., Akbari Noghabi K. Glyphosate tolerance in transgenic canola by a modified glyphosate oxidoreductase (gox) gene. Progress in Biological Sciences. 2012;2:50–58. [Google Scholar]
- Hove-Jensen B., Zechel D.L., Jochimsen B. Utilization of glyphosate as phosphate source: biochemistry and genetics of bacterial carbon-phosphorus lyase. Microbiol. Mol. Biol. Rev. 2014;78:176–197. doi: 10.1128/MMBR.00040-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore G.M., Padgette S.R., Fraley R.T. History of herbicide-tolerant crops, methods of development and current state of the art: emphasis on glyphosate tolerance. Weed Technol. 1992;6:626–634. [Google Scholar]
- Kittle R.P., McDermid K.J., Muehlstein L., Balazs G.H. Effects of glyphosate herbicide on the gastrointestinal microflora of Hawaiian green turtles (Chelonia mydas) Linnaeus. Mar. Pollut. Bull. 2018;127:170–174. doi: 10.1016/j.marpolbul.2017.11.030. [DOI] [PubMed] [Google Scholar]
- Knight R., Jansson J., Field D., Fierer N., Desai N., Fuhrman J.A., Hugenholtz P., van der Lelie D., Meyer F., Stevens R. Unlocking the potential of metagenomics through replicated experimental design. Nat. Biotechnol. 2012;30:513–520. doi: 10.1038/nbt.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl K.D., Weiss R.B., Cox J., Dale C., Dearing M.D. Gut microbes of mammalian herbivores facilitate intake of plant toxins. Ecol. Lett. 2014;17:1238–1246. doi: 10.1111/ele.12329. [DOI] [PubMed] [Google Scholar]
- Li L., Lu W., Han Y., Ping S., Zhang W., Chen M., Zhao Z., Yan Y., Jiang Y., Lin M. A novel RPMXR motif among class II 5-enolpyruvylshikimate-3-phosphate synthases is required for enzymatic activity and glyphosate resistance. J. Biotechnol. 2009;144:330–336. doi: 10.1016/j.jbiotec.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J., Mahurkar A., Rahnavard G., Crabtree J., Orvis J., Hall A.B., Brady A., Creasy H.H., McCracken C., Giglio M.G. Strains, functions and dynamics in the expanded human microbiome project. Nature. 2017;550:61–66. doi: 10.1038/nature23889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J., Arze C., Ananthakrishnan A.N., Schirmer M., Avila-Pacheco J., Poon T.W., Andrews E., Ajami N.J., Bonham K.S., Brislawn C.J. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano V.L., Defarge N., Rocque L.M., Mesnage R., Hennequin D., Cassier R., de Vendomois J.S., Panoff J.M., Seralini G.E., Amiel C. Sex-dependent impact of roundup on the rat gut microbiome. Toxicol. Rep. 2018;5:96–107. doi: 10.1016/j.toxrep.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Q., Manservisi F., Panzacchi S., Mandrioli D., Menghetti I., Vornoli A., Bua L., Falcioni L., Lesseur C., Chen J. The Ramazzini institute 13-week pilot study on glyphosate and roundup administered at human-equivalent dose to Sprague Dawley rats: effects on the microbiome. Environ. Health. 2018;17:50. doi: 10.1186/s12940-018-0394-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage R., Antoniou M.N., Tsoukalas D., Goulielmos G.N., Tsatsakis A. Gut microbiome metagenomics to understand how xenobiotics impact human health. Current Opinion in Toxicology. 2018;11–12:51–58. [Google Scholar]
- Mesnage R., Teixeira M., Mandrioli D., Falcioni L., Ducarmon Q.R., Zwittink R.D., Amiel C., Panoff J.-M., Belpoggi F., Antoniou M.N. Shotgun metagenomics and metabolomics reveal glyphosate alters the gut microbiome of Sprague-Dawley rats by inhibiting the shikimate pathway. bioRxiv. 2019 doi: 10.1101/870105. [DOI] [Google Scholar]
- Motta E.V.S., Raymann K., Moran N.A. Glyphosate perturbs the gut microbiota of honey bees. Proc Natl Acad Sci U S A. 2018;115(41):10305–10310. doi: 10.1073/pnas.1803880115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen L.N., Roager H.M., Casas M.E., Frandsen H.L., Gosewinkel U., Bester K., Licht T.R., Hendriksen N.B., Bahl M.I. Glyphosate has limited short-term effects on commensal bacterial community composition in the gut environment due to sufficient aromatic amino acid levels. Environ. Pollut. 2018;233:364–376. doi: 10.1016/j.envpol.2017.10.016. [DOI] [PubMed] [Google Scholar]
- Pedotti M., Rosini E., Molla G., Moschetti T., Savino C., Vallone B., Pollegioni L. Glyphosate resistance by engineering the flavoenzyme glycine oxidase. J. Biol. Chem. 2009;284:36415–36423. doi: 10.1074/jbc.M109.051631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K.S., Nayfach S., Fischbach M.A. MetaQuery: a web server for rapid annotation and quantitative analysis of specific genes in the human gut microbiome. Bioinformatics. 2015;31:3368–3370. doi: 10.1093/bioinformatics/btv382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riede S., Toboldt A., Breves G., Metzner M., Kohler B., Braunig J., Schafft H., Lahrssen-Wiederholt M., Niemann L. Investigations on the possible impact of a glyphosate-containing herbicide on ruminal metabolism and bacteria in vitro by means of the ‘Rumen Simulation Technique’. J. Appl. Microbiol. 2016;121:644–656. doi: 10.1111/jam.13190. [DOI] [PubMed] [Google Scholar]
- Roberts F., Roberts C.W., Johnson J.J., Kyle D.E., Krell T., Coggins J.R., Coombs G.H., Milhous W.K., Tzipori S., Ferguson D.J. Evidence for the shikimate pathway in apicomplexan parasites. Nature. 1998;393:801–805. doi: 10.1038/31723. [DOI] [PubMed] [Google Scholar]
- Rothschild D., Weissbrod O., Barkan E., Kurilshikov A., Korem T., Zeevi D., Costea P.I., Godneva A., Kalka I.N., Bar N. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- Saha J.R., Butler V.P., Jr., Neu H.C., Lindenbaum J. Digoxin-inactivating bacteria: identification in human gut flora. Science. 1983;220:325–327. doi: 10.1126/science.6836275. [DOI] [PubMed] [Google Scholar]
- Schmidt T.S.B., Raes J., Bork P. The human gut microbiome: from association to modulation. Cell. 2018;172:1198–1215. doi: 10.1016/j.cell.2018.02.044. [DOI] [PubMed] [Google Scholar]
- Schonbrunn E., Eschenburg S., Shuttleworth W.A., Schloss J.V., Amrhein N., Evans J.N., Kabsch W. Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1376–1380. doi: 10.1073/pnas.98.4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigler D.S. Shikimic acid pathway. In: Seigler D.S., editor. Plant Secondary Metabolism. Springer; US: 1998. pp. 94–105. [Google Scholar]
- Shehata A.A., Schrodl W., Aldin A.A., Hafez H.M., Kruger M. The effect of glyphosate on potential pathogens and beneficial members of poultry microbiota in vitro. Curr. Microbiol. 2013;66:350–358. doi: 10.1007/s00284-012-0277-2. [DOI] [PubMed] [Google Scholar]
- Shin N.R., Whon T.W., Bae J.W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson C.L., Harris C.A. An assessment of dietary exposure to glyphosate using refined deterministic and probabilistic methods. Food Chem. Toxicol. 2016;95:28–41. doi: 10.1016/j.fct.2016.06.026. [DOI] [PubMed] [Google Scholar]
- Sugata F., Aoki N., Shioda T., Hayashi T., Shimada K., Mitamura K., Shibuta H. Immune response of mice infected with recombinant vaccinia viruses carrying the HIV gag gene. Microbiol. Immunol. 1991;35:849–861. doi: 10.1111/j.1348-0421.1991.tb02025.x. [DOI] [PubMed] [Google Scholar]
- Sviridov A.V., Shushkova T.V., Ermakova I.T., Ivanova E.V., Epiktetov D.O. Leont’evskii AA: [microbial degradation of glyphosate herbicides (review)] Prikl. Biokhim. Mikrobiol. 2015;51:183–190. doi: 10.7868/s0555109915020221. [DOI] [PubMed] [Google Scholar]
- Truong D.T., Franzosa E.A., Tickle T.L., Scholz M., Weingart G., Pasolli E., Tett A., Huttenhower C., Segata N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- Tsiaoussis J., Antoniou M.N., Koliarakis I., Mesnage R., Vardavas C.I., Izotov B.N., Psaroulaki A., Tsatsakis A. Effects of single and combined toxic exposures on the gut microbiome: current knowledge and future directions. Toxicol. Lett. 2019;312:72–97. doi: 10.1016/j.toxlet.2019.04.014. [DOI] [PubMed] [Google Scholar]
- Yi S.Y., Cui Y., Zhao Y., Liu Z.D., Lin Y.J., Zhou F. A novel naturally occurring class I 5-enolpyruvylshikimate-3-phosphate synthase from Janibacter sp. Confers High Glyphosate Tolerance to Rice. Sci Rep. 2016;6:19104. doi: 10.1038/srep19104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan H., Feng Y., Fan X., Chen S. Recent advances in glyphosate biodegradation. Appl. Microbiol. Biotechnol. 2018;102:5033–5043. doi: 10.1007/s00253-018-9035-0. [DOI] [PubMed] [Google Scholar]
- Zheng X., Zhao A., Xie G., Chi Y., Zhao L., Li H., Wang C., Bao Y., Jia W., Luther M. Melamine-induced renal toxicity is mediated by the gut microbiota. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3005114. (172ra122) [DOI] [PubMed] [Google Scholar]
- Zucko J., Dunlap W.C., Shick J.M., Cullum J., Cercelet F., Amin B., Hammen L., Lau T., Williams J., Hranueli D. Global genome analysis of the shikimic acid pathway reveals greater gene loss in host-associated than in free-living bacteria. BMC Genomics. 2010;11:628. doi: 10.1186/1471-2164-11-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EPSPS classification.
Pathway table.
Pathway results.
Homologues metabolism.