Abstract
Cancer immunotherapy has shown great potential as witnessed by an increasing number of immuno-oncology drug approvals in the past few years. Meanwhile, the field of nucleic acid therapeutics has made significant advancement. Nucleic acid therapeutics, such as plasmids, antisense oligonucleotides (ASO), small interfering RNA (siRNA) and microRNA, messenger RNA (mRNA), immunomodulatory DNA/RNA, and gene-editing guide RNA (gRNA) are attractive due to their versatile abilities to alter the expression of target endogenous genes or even synthetic genes, and modulate the immune responses. These abilities can play vital roles in the development of novel immunotherapy strategies. However, limited by the intrinsic physicochemical properties such as negative charges, hydrophilicity, as well as susceptibility to enzymatic degradation, the delivery of nucleic acid therapeutics faces multiple challenges. It is therefore pivotal to develop drug delivery systems that can carry, protect, and specifically deliver and release nucleic acid therapeutics to target tissues and cells. In this review, we attempted to summarize recent advances in nucleic acid therapeutics and the delivery systems for these therapeutics in cancer immunotherapy.
Keywords: Drug delivery, Nucleic acid therapeutics, Therapeutic vaccines, Immunostimulatory nucleic acids, mRNA drug, Cancer immunotherapy
1. Introduction
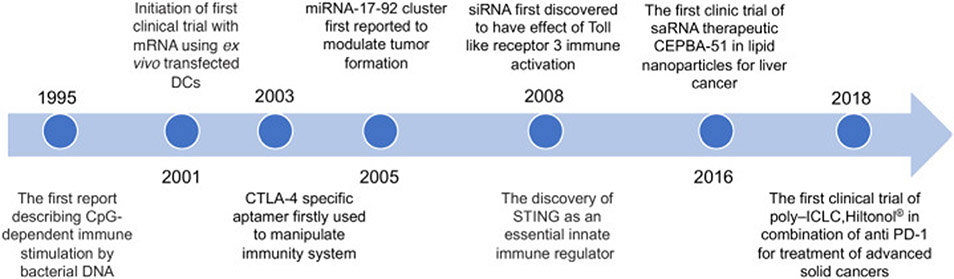
Nucleic acid therapeutics have emerged as promising candidates for cancer treatment, including immunotherapy [1]. Nucleic acid therapeutics are a diverse class of DNA or RNA such as plasmids, mRNA, ASO, siRNA, miRNA, small-activating RNA (saRNA), aptamers, gene-editing gRNA, as well as immunomodulatory DNA/RNA. Nucleic acid therapeutics have versatile functionalities ranging from altering (up- or down- regulating) gene expression [2], to modulating immune responses [3,4]. Research on immunomodulatory nucleic acids can date back to 1995 [5], and since then, a growing number of potential immunomodulatory nucleic acids have been discovered and tested for immunotherapy (Fig. 1).
Fig. 1.
Milestones for the development of nucleic acid therapeutics for immunotherapy.
The high specificity, versatile functionality, reproducible batch-to-batch manufacture, and tunable immunogenicity of nucleic acid therapeutics make them good candidates for cancer immunotherapy. In this article, we will mainly discuss five categories of nucleic acid therapeutics for cancer immunotherapy. Gene-regulating nucleic acid drugs such as siRNA and ASO can regulate post-transcriptional gene expression, and silence targeted genes, further regulating intracellular signaling pathway involved in cancer progression [6]; Nucleic acids immunostimulants such as unmethylated cytosine-guanosine deoxynucleotides (CpG), poly I:C, 5′-triphosphate RNA, as well as di-cyclic nucleotides that active stimulator interferon genes (STING) that induce or augment anticancer immune activation [7-9]. mRNA therapeutics and plasmid DNA (pDNA) can be tailor-designed to express proteins or peptides of interest, such as antigens or cancer immunotherapeutic proteins [10]. Aptamers, which are short single-stranded nucleic acids, have been studied as nucleic acid versions of antibodies for cancer immunotherapy [11]. Finally, genome editing-related nucleic acids such as gRNA have been recently implemented to precisely edit target genes and therefore modulate gene expression for cancer immunotherapy [12].
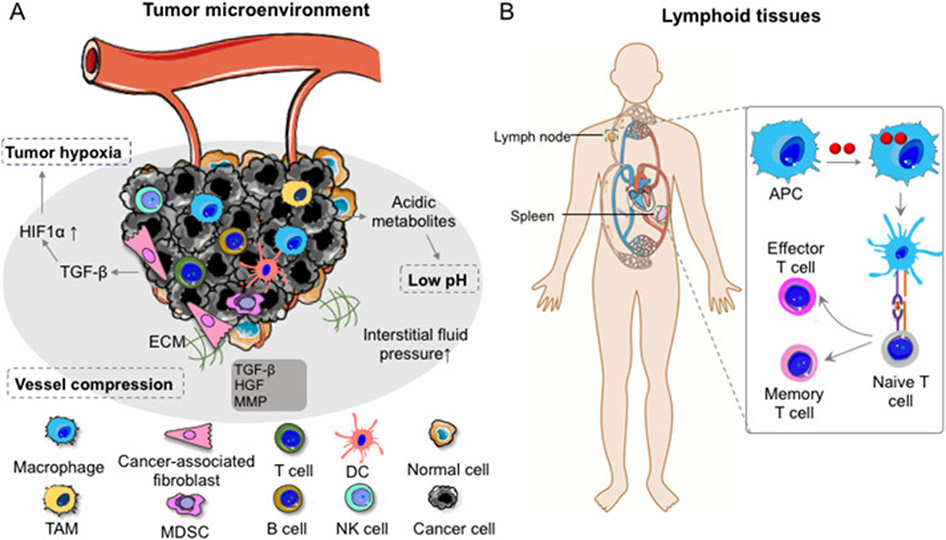
Wide application of nucleic acid therapeutics is hinged on effective and specific delivery of these drugs into target tissues (Fig. 2), cells, as well as subcellular locations. Complex tumor microenvironment (TME) makes drug delivery to tumor cells intractable, which is further aggravated by the immunosuppression in TME. For instance, TME has tumor hypoxia, acidosis as well as high interstitial fluid pressure. Further, stromal cells in TME, such as cancer-associated fibroblasts (CAFs) that overexpress TGF-β, HGF, and TGF-β, facilitate the malignant tumor evolution and reduce the delivery efficiency of nucleic acid therapeutics into TME [13] (Fig. 2A). Another important class of tissue targets for cancer immunotherapy is lymphoid tissues, where various immune cells reside and a myriad of immune responses are orchestrated (Fig. 2B). It is thus pivotal to understand the delivery targets and accordingly design delivery systems for nucleic acid therapeutics.
Fig. 2.
Tumor microenvironment and lymphoid tissues as two common tissue-level targets for the delivery of nucleic acid therapeutics in cancer immunotherapy. (A) The tumor microenvironment harbors a variety of cells including tumor cells, antigen presenting cells (DC, macrophage), lymphocytes (T cells, B cells and NK cells), and fibroblasts, which play a critical role in the development of tumor initiation, proliferation, as well as metastasis. (B) Lymphatic organs such as lymph nodes and spleen are among the desired sites for cancer therapeutics such as vaccines. DC: dendritic cells; TAM: tumor-associated macrophage; MDSC: myeloid-derived suppressor cells; NK cell: natural killer cell; APC: antigen-presenting cells; TGF-β: transforming growth factor-β; HGF: hepatocyte growth factor; MMP: matrix metalloproteinase.
Unlike traditional small molecule medicines or large biologics, the delivery of nucleic acid therapeutics can be generally impacted by their unique properties such as negative charges, hydrophilicity, susceptibility to enzyme degradation if not modified, potentially unwanted immunogenicity [14], as well as off-targeting effect [15]. Decades of research and development have devised a good number of strategies to address these issues and advance the development of nucleic acid therapeutics for disease treatment, such as cancer immunotherapy [10,16]. One commonly-used strategy to improve the biostability of nucleic acids is to chemically modify nucleic acids with, for example, 2’-F and 2’-O-methyl on the sugar, or phosphorothioate in the backbone [17,18]. Moreover, nanocarriers, which are widely used to load nucleic acid therapeutics, can also protect nucleic acids from nuclease degradation by generating steric hindrance for nucleases to access nucleic acids [19].
2. Delivery of nucleic acid therapeutics in cancer immunotherapy
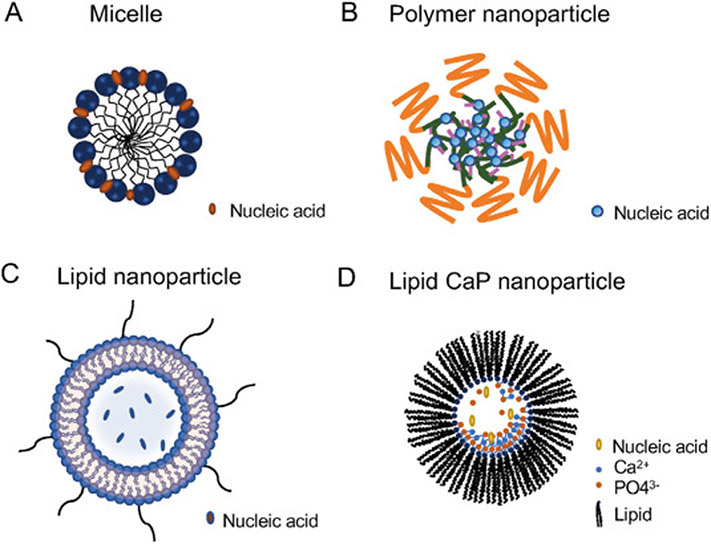
Nanodrug delivery systems have been developed to improve the efficacy of nucleic acid therapeutics by passive homing to target tissues such as tumor or lymph nodes [20]. Nanocarriers can be further modified with functional ligands for active targeted delivery and to improve cell endocytosis, via receptor-ligand interactions [21]. Lipid nanoparticles [22] and polymer nanoparticles [16] are among the most commonly used nanocarriers for nucleic acid delivery (Fig. 3). In addition, molecular bioconjugates of nucleic acid, such as N-Acetylgalactosamine (GalNAc), have emerged as a promising approach for nucleic acid delivery to the liver [23].
Fig. 3.
Representative nanoparticulate drug delivery systems for nucleic acid therapeutics. (A) Delivery of nucleic acid therapeutics using micelles comprise of amphiphilic molecules, such as amphiphilic block polymers. Micelles contain hydrophilic regions as well as hydrophobic regions. After reaching the critical micelle concentration, micelles can be formed with nucleic acid loading in hydrophilic side via mechanisms such as electrostatic interactions or Van der Waals interactions. (B) Delivery of nucleic acid therapeutics using polymer nanoparticles that can load nucleic acids through various mechanism, most commonly using cationic polymers, which can form nanoparticles with negative nucleic acid via electrostatic interactions. (C) Delivery of nucleic acid therapeutics using lipid nanoparticles comprise of lipids such as lecithin, DOPC, and DSPE. Polyethylene glycol (PEG) modification of lipids enhance the half-life of lipid nanoparticles. DOPC: 1,2-dioleoyl-sn-glycero-3-phosphocholine; DSPE: 1,2-distearoyl-sn-glycero-3-phosphorylethanolamine. (D) Delivery of nucleic acid therapeutics using lipid calcium phosphate (CaP) nanoparticles. CaP encapsulates nucleic acids due to the co-precipitation of the phosphate on nucleic acid backbones with calcium-phosphate. Lipids make CaP nanoparticles more stable and can facilitate cellular delivery.
2.1. Delivery of immunostimulatory nucleic acids
Immunostimulants can activate antitumor immune responses for cancer immunotherapy. Among such immunostimulants are nucleic acid of particular sequences, ranging from agonists for toll-like receptors (TLR), retinoic acid-inducible gene I (RIG-1) agonists, cyclic-GMP-AMP-synthase (cGAS) activators, to STING agonists. For instance, STING is a 379 amino-acid protein located in the endoplasmic reticulum in a variety of cells types, and is composed of several transmembrane regions [7]. Upon sensing intracellular dsDNA, cGAS cyclizes endogenous GTP and ATP to produce 2′3’-cyclic-GMP-AMP (cGAMP). cGAMP binds to and activates the endoplasmic reticulum (ER) transmembrane receptor STING, triggering the activation of STING. Activated STING then serves as an adaptor for kinase TBK1 and transcription factor IRF-3 and leads to IRF-3 phosphorylation and dimerization. Phosphorylated IRF3 dimers translocate to the nucleus and induce the production and secretion of type I interferons (IFNs) [24].
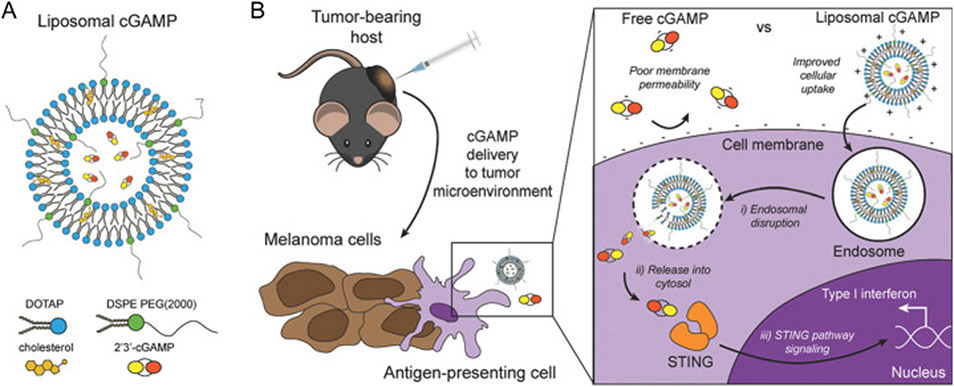
STING agonists have been studied in preclinical systems and recently in clinical trials for cancer immunotherapy [25]. STINGs can be activated by CDNs such as cGAMP [26], oligonucleotides (directly activate cGAS to produce cGAMP for STING activation) [27], and non-nucleic acid small molecules [28]. For instance, Ramanjulu et al. reported a small molecule STING agonist, diABZIs, which is a symmetric amidobenzimidazole (ABZI)-based compound that binds to human STING. diABZI activated CD8+ T cells in vivo after intravenous administration [28]. Nanoparticles, such as liposomes, that are loaded with STING agonists can accumulate in lymph nodes after local administration and mediate efficient cytosolic delivery of STING agonists [29-31]. For instance, Koshy et al. used cationic liposomes modified with PEG (Fig. 4) to deliver cGAMP. Such cGAMP-loaded liposome can promote the intracellular delivery of cGAMP and activate the STING signaling pathway, resulting in the secretion of type I IFNs into TME and improved tumor therapeutic efficacy in a metastatic melanoma model [32]. In another example, Shae et al. designed pH-sensitive polymersomes with the ability of membrane-destabilization for intracellular release of the loaded cGAMP. These STING-NPs not only enhanced the cellular uptake, but also promoted endosomal escape of cGAMP into cytosol. When administrated by intratumorally or intravenously, STING-NPs significantly promoted the therapeutic efficacy of STING agonists in mouse models [33]. Interestingly, it has also been reported by Luo et al. that polymeric nanoparticles, named PC7A NPs, per se stimulated the STING pathway to activate antitumor immunity [34], which provides a novel platform for nanovaccines based on the unique characteristics of the polymer.
Fig. 4.
Schematic illustration of cGAMP delivery using liposomes. (A) The structure of liposomal cGAMP. (B) Compared to free cGAMP, liposomal cGAMP improved cellular uptake by antigen-presenting cells due to the benefit of positive charge, and after cell endocytosis, liposomal cGAMP can facilitate cGAMP to release from endosomes to further bind STING located in cytosol, triggering STING signaling pathway. Adapted by permission from [32] Copyright 2017, WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Toll-like receptor (TLR) agonists are another class of immunostimulatory adjuvants that have been extensively explored in preclinical studies as well as clinical trials. Representative examples of immunostimulatory TLR agonists include unmethylated cytosine-phosphate-guanine-containing oligonucleotides (CpG) for TLR9, R848 for TLR7/8, flagellin for TLR5, and polyinosinic-polycytidylic acid (poly I:C) for TLR3. For example, via pattern recognition, specific DNA sequences with unmethylated cytosine-phosphate-guanine (CpG) can be recognized by TLR9 to induce innate and adaptive immune responses via the MyD88-dependent nuclear factor (NF-κB) and mitogen-activated protein kinase signaling pathways, promoting the secretion of proinflammatory cytokines such as IL-2, IL-6, IFN-α, IFN-γ [8]. The delivery carriers for CpG range from organic biomaterials (polymer [35], albumin [36] and lipid nanoparticles [37] as well as inorganic materials [38-40]. TLR3 agonist, poly I:C, is a synthetic immunostimulatory polymer agent that resembles dsRNA as it comprises long strands of inosine and cytidine, and is recognized mainly by TLR3 and MDA5 to work as adjuvants to induce antitumor immune response [9]. A vaccine named BiVax was developed, which mixed poly-IC with synthetic peptides, was shown to magnify the antigen-specific CD8+ T cell immune responses [41]. Poly I:C, as well as its formulation derivative poly-l-lysine and carboxymethylcellulose (poly-ICLC) are commonly used in clinical trials as immunostimulatory adjuvants [42,43].
2.2. Delivery of mRNA therapeutics
Natural RNA has various functions ranging from encoding proteins, eliciting innate immune responses, modulating protein or RNA functions [44]. mRNA relay genomic information from DNA to ribosome that uses mRNA as templates to translate protein in the cytoplasm [10]. The ability of mRNA to express necessarily any proteins and peptides over a long duration without reliance on nuclear localization for gene expression [45] makes mRNA therapeutics of tremendous potential for versatile applications, including cancer immunotherapy. Compared to gene therapy that inserts genes into genome, mRNA therapeutics bypass the risk of permanent gene integration into genome that might lead to adverse side effects. Nowadays, nucleic acid chemistry has been developed to synthesize mRNA that can resist enzymatic degradation or alleviate the immunogenicity of mRNA. Poly A tail of endogenous mRNA is able to bind multiple poly A binding proteins (PABPs) in a tandem manner, which can improve mRNA stability, as well as stimulate mRNA translation [46]. Consistently, for mRNA vaccines, a poly-A tail measuring around 120 nucleotides enhanced mRNA stability and translational efficiency, providing a potential strategy for optimizing mRNA vaccines [47]. Other strategies of chemical modification include N1-methylpseudouridine, 5-methoxyuridine and pseudouridine modification of mRNA were proved to be effective to enhance mRNA stability and improve protein expression [48]. A recent study by Li et al. designed a translation initiation nanoplex, which comprised of an inherent molecular recognition between 7-methyguanosine-capped mRNA and eukaryotic initiation factor 4E (eIF4E) protein loaded in cationic carriers, polyamine. This ribonucleoproteins (RNPs) can mimick the first step of protein synthesis. They demonstrated that the nanoplex enhanced the mRNA transfection by improving the stability of mRNA, and increased activation of cytotoxic CD8 T cells after induction to murine dendritic cells [49]. Besides, mRNA purification can reduce the innate immune activation caused by the leftover during the generation of mRNA therapeutics [50], so high-performance liquid chromatography (HPLC) is applied to purify mRNA to reduce adverse immune response.
Further, mRNA can now be manufactured in vitro at large scales at a low cost. Specifically, mRNA can be reproducibly synthesized by in vitro transcription (IVT) using DNA templates, a T7, a T3 or an Sp6 phage RNA polymerase [44]. These technology advancements have altogether render mRNA therapeutics very promising for cancer immunotherapy, by versatile approaches such as ex vivo mRNA transfer for therapeutic adoptive cell engineering, and using cancer-specific antigen-encoding mRNA directly as cancer therapeutic vaccines [45].
The field of mRNA therapeutics has indeed made remarkable progress in the past decade. One example is RNActive® vaccines that was shown to be taken up by leukocytic cells and presented by APCs to trigger immune cell activation and adaptive immunity [51]. Meanwhile, the immune-stimulation was limited to the injection site and lymphoid site, and proinflammatory cytokine levels were not elevated by these vaccines in the serum of the immunized mice, suggesting the specificity of this mRNA vaccine.
Unmodified or unformulated mRNA can be degraded by ubiquitous RN-ases and can be difficult to be delivered to the cytoplasm. Lipid nanoparticles (LNP) can encapsulate mRNA, protect them from enzymatic degradation, and markedly enhance the potency of mRNA [52]. For example, LNPs that encapsulated self-amplifying mRNA encoding secreted alkaline phosphatase showed higher levels of transporter gene expression than naked RNA delivery [52]. To optimize the formulation of LNPs for mRNA vaccine delivery, Miao et al screened 1000 lipid formulations that were used for mRNA vaccination and found that the lipids with an unsaturated lipid tail, a dihydroimid-azole linker and cyclic amine head group performed best in melanoma and human papillomavirus (HPV) mouse tumor models. Moreover, cyclic amino head groups can activate the MyD88/RLR-independent STING pathway, eliciting a robust type I IFN activation and APC maturation, thus providing an additional mechanism for the mRNA carriers to potentiate immunostimulatory immune responses [53]. Moffett et al. developed a polymeric nanoparticle loaded mRNA that encoded a rare-cleaving megaTAL nuclease into lymphocytes (Fig. 5). Biodegradable poly (β-amino ester) (PBAE) was used to form nanoparticles to encapsulate mRNA, and PGA modified with targeting ligands,anti-CD3 and anti-CD8, was additionally added to balance the positive charge of PBAE mRNA nanoparticles [54]. These polymer nanoparticles showed a high transfection efficiency of mRNA, therefore efficiently reprogrammed naïve T cells to effector T cells.
Fig. 5.
T cell-targeting mRNA nanoparticles or in vivo T cell gene editing in cancer immunotherapy. (A) mRNA nanoparticles controllably deliver mRNA to lymphocytes or hematopoietic stem cells (HSCs) by merely mixing the mRNA nanoparticles with the cells in vitro. (B) Design of targeted mRNA-carrying nanoparticles. mRNA was complexed with a positively-charged PBAE polymer, which condenses mRNA into nanocomplexes. Antibody-functionalized polyglutamic acid (PGA) was added to shield the positive charge of the PBAE-mRNA particles and confer lymphocyte-targeting. Adapted by permission from reference [54]. Copyright, 2017, Springer Nature.
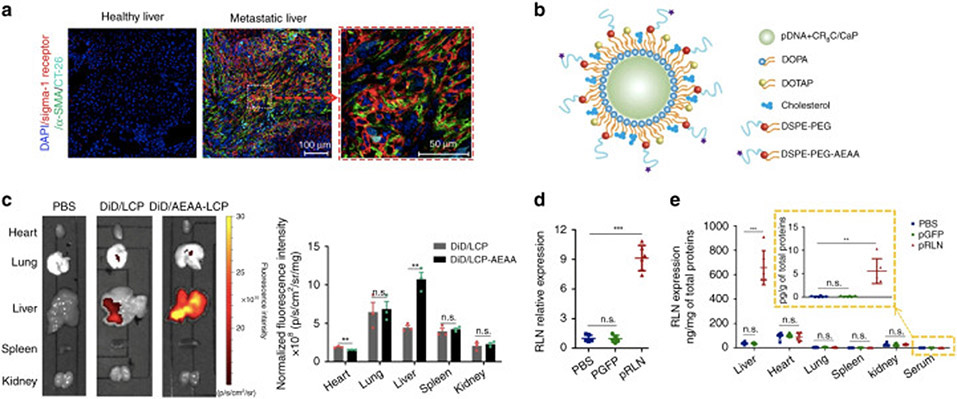
pDNA is double-stranded DNA (dsDNA) composed of genes of interest and can be used to express mRNA or proteins/peptides or interest for gene therapy [55]. pDNA can replicate independently from chromosomal DNA, and the gene fragment in pDNA can also be integrated into the chromosomal genome for permanent expression of protein or peptide products. pDNA has been studied to encode regulatory proteins [56-58] or express RNA to silence target genes for cancer immunotherapy. CaP NP is one of the most common strategies for pDNA delivery as it shows excellent biocompatibility, biodegradability, as well as pH-responsiveness [59]. Olton et al. reported that by optimizing the ratio of Ca/P and the mixing mode of Ca2+ and PO43− solutions, the CaP-pDNA nanoparticles yielded high transfection efficiencies [19]. CaP lipid nanoparticles were further developed and modified with targeting ligands for specific delivery to target cells in cancer immunotherapy [59,60]. Hu et al. prepared lipid-CaP nanoparticles modified with aminoethyl anisamide to deliver pDNA encoding relaxin, which reversed the stromal microenvironment and inhibit liver metastasis of colorectal, pancreatic, and breast cancer (Fig. 6). Furthermore, the combination of this relaxin-expression pDNA nanoparticles with programmed death-ligand 1(PD-L1) blockade further potentiated the immunotherapeutic efficacy in this cancer model [61].Polymer nanoparticles are also commonly used for pDNA delivery. Liu et al synthesized a copolymer named LA-PegPI, which is composed of PEI (molecular weight: ~800 Da) cross-linked by myo-inositol (INO) and conjugated with a galactose-grafted PEG chain, to deliver a pDNA that encodes interleukin-15 (IL-15). The resulting IL-15 activated CD8+ T cells and natural killer (NK) cells to upregulate the expression of antitumor cytokines such as interferons-γ (IFN-γ), tumor necrosis factor (TNF), and interleukin-12 (IL-12). As a result, LA-PegPI/pDNA effectively inhibited tumor growth in orthotopic hepatocellular carcinoma mouse model [62].
Fig. 6.
Using CaP lipid nanoparticles for the delivery of relaxin-encoding pDNA for tumor immunotherapy. (A) High expression level of sigma-1 receptor (Sig-1R) on activated hepatic stellate cells (aHSCs). (B) Schematic illustration of CaP lipid nanoparticles. A Sig-1R-targeting ligand, aminoethyl anisamide (AEAA), was conjugated on liposome surface for active targeting. (C) Distribution of DiD-labeled CaP lipid nanoparticles and DiD-labeled AEAA-conjugated CaP lipid nanoparticles in the organs after injection in mice bearing liver metastatic CT26-FL3 for 13 days. (D) Relative relaxin mRNA levels after the injection of pDNA CaP lipid nanoparticles. (E) Relative relaxin expression levels after the injection of pDNA CaP lipid nanoparticles. Adapted by permission from reference [61]. Copyright 2019, Springer Nature.
2.3. Nucleic acid aptamer-based delivery of cancer immunotherapeutics
Aptamers are single-stranded nucleic acid ligands that can be isolated from a random pool of oligonucleotides in the process of systematic evolution of ligands by exponential enrichment (SELEX) [63]. Aptamers can be efficiently selected in vitro against virtually any targets, exhibit high specificity with high binding affinity to targets, and can be reproducibly manufactured [11], all of which make them attractive alternatives to antibody- or peptide-based target ligands [64]. The first aptamer drug approved by the US FDA in 2004 is a VEGF-targeting aptamer called Pegaptanib that treats age-related macular degeneration [64]. For cancer immunotherapy, McNamara et al. developed an aptamer that binds to 4-1 BB, which is a major costimulatory receptor expressed on APCs and plays an essential role in the activation of CD8+ T cells. They went on and showed that the corresponding multivalent aptamer co-stimulated T cell activation in vitro and mediated tumor rejection in mice 65]. Aptamers were also selected to block cytotoxic T-lymphocyte associated protein 4 (CTLA4) [66,67], T cell immunoglobulin-3 (TIM-3) [68], or act as an agonist for OX40 [69] for cancer immunotherapy. In addition, aptamers may be internalized into cells together with their receptors for the intracellular delivery of immune therapeutics. For example, IL-2, as an activation marker of shortlived effector cells has been reported that the downregulation of IL-2 signaling is beneficial to develop long-term memory T cells [70]. Rajagopalan et al. used a 4-1 BB-targeting aptamer conjugated to siRNA against CD25 (IL-2 receptor) to downregulate the IL-2 signaling in CD8+ T cells. The results showed that the aptamer-siRNA conjugate could activate circulating T cells, increase the differentiation to memory T cells, and further suppress the tumor growth in a breast carcinoma model [71].
2.4. Delivery of genome editing nucleic acids
Genome editing nucleases include zinc-finger nuclease [72], transcription activator-like effector nuclease (TALEN) [73] and clustered regularly interspaced short palindromic repeats/CRISPR associated protein (CRISPR/Cas) system [73,74]. Since the first application in mammalian cells in 2013, the CRISPR/Cas system, based on a RNA-guided nuclease, has revolutionized the precise genome manipulations [75]. Gene-editing has been employed to engineer T cells for the immunotherapy of diseases, such as acquired immunodeficiency syndrome (AIDS) [76] and cancers [77]. The CRISPR/Cas9 system is currently one of the most comprehensively studied tools because of its simple utilities, programmability, cost effectiveness, and most importantly, the highly efficient multiplex genome engineering capability [77]. For instance, CRISPR/Cas9 system contains two critical components, Cas9 nuclease, and a gRNA, the latter of which is a fusion of a crRNA and a constant tracrRNA [74]. Recently, a class II CRISPR system has been developed based on Cas12a, also called Cpf1, which is a single RNA-guided endonuclease lacking tracrRNA. Cas12a cleaves DNA, forming a staggered DNA double-stranded break, and the sticky-end mediated DNA repair would facilitate the gene modifications in non-dividing cells, in which homology-directed repair (HDR) is difficult to achieve [75]. Genomic editing has expanded the landscape of cancer immunotherapy approaches such as adoptive T cell engineering and therapy. Recent work on T cell engineering applications using CRISPR/Cas9 has attracted the focus on improving the efficacy and safety of TCR re-directed T cells and the pre-clinical development of off-the-shelf universal chimeric antigen receptor (CAR) T cells [78]. This propelled a clinical trial involving the first use of CRISPR/Cas9 for cancer treatment in humans in the USA [79]. Using multiplex CRISPR/Cas9 genomic editing, allogeneic universal CAR T cells that are deficient in the TCR beta chain, beta-2-microglobulin (β2M), PD-1 and CTLA-4 have been generated [80-82]. These CAR T cells maintain functions both in vitro and in vivo without causing graft-versus-host-disease (GVHD). Furthermore, with the additional disruption of PD-1, TCR and human leukocyte antigen (HLA) class I-negative CAR T cells have exhibited significantly improved in vivo anti-tumor activities [81]. CRISPR/Cas9 can also be used to target a CAR to the T-cell receptor α constant (TRAC) locus to generate universal CAR T cells. It has been demonstrated that directing a CD19-specific CAR to the TRAC locus not only resulted in uniform CAR expression in human peripheral blood T cells but also enhanced T-cell potency; these edited cells largely outperformed conventionally generated CAR T cells in a mouse model of acute lymphoblastic leukemia [83].
To overcome the physiological barriers against the delivery of gene-editing nucleic acid therapeutics [84], traditional physical approaches such as microinjection [85], electroporation [86], and viral vectors [87], as well as more recently non-viral carriers based on nanoparticles, have been explored [88-90]. Zuris et al. found that the engineered Cas9 nucleases with negative charged protein can be efficiently delivered by positive charged liposomal reagents [91]. Liu et al. synthesized a lipid nanoparticle integrated with disulfide bonds to efficiently deliver Cas9 mRNA and sgRNA into hepatocytes, and knocked down proprotein convertase subtili-sin/kexin type 9 in mouse serum down to 20% of that in non-treated mice [92]. When forming the Cas9 (ribonucleoproteins) RNPs, despite the positive charge of the nature nuclease proteins, the presence of a negatively charged gRNA molecule can result in a net negative charge, which facilitated loading of the RNP/gRNA complexes into cationic lipid nanoparticles.
2.5. Delivery of gene-regulating nucleic acid therapeutics
Gene-regulating nucleic acid therapeutics have long been studied to modulate gene expression for the treatment of genetic diseases. Such nucleic acid therapeutics include siRNA, miRNA, small-activating RNA (saRNA), antagomir, antisense oligonucleotides, decoy oligonucleotides, as well as gRNA involved in gene editing.
siRNA is double-strand RNA (dsRNA) fragments of about 21–23 nucleotides long. In the cytoplasm, siRNA binds to RNA-induced silencing complex (RISC), where the passenger strand of siRNA is cleaved. The guide RNA of siRNA in the RISC will selectively bind with and degrade target mRNA, whereby down-regulating the expression of cellular protein [93]. In 2018, the first siRNA drug named Patisiran was approved by FDA to treat hereditary transthyretin amyloidosis (hAATR) with polyneuropathy [94]. siRNA therapeutics have also been enthusiastically developed for cancer immunotherapy [95], using drug delivery carriers such as polymer nanoparticles [6,96,97] and lipid nanoparticles [98-100]. For instance, Warashinia, et al. used YSK12-C4, an ionizable-cationic lipid to form lipid nanoparticle (YSK12-MEND) to load siRNA that can silence the suppressor of cytokine signaling 1 (SOCS 1). YSK12-MEND resulted in a gene silencing efficiency higher than 90% in murine dendritic cells, showing a drastic enhancement in cytokine production and significant tumor suppression [99]. Song, et al. reported a polymer nanoparticle comprising of polyethylene glycol (PEG)- and mannose-modified trimethyl chitosan along with citraconic anhydride grafted poly (allylamine hydrochloride) (PC) to deliver siRNAs to silence genes encoding vascular endothelial growth factor (VEGF) and placental growth factor (PIGF). The nanoparticles efficiently accumulated in the tumor tissue and be internalized by M2 type-tumor associated macrophage (TAM) as well as breast cancer cells, exerting robust suppression of breast tumor growth and lung metastasis [101].
ASO is single-stranded oligodeoxynucleotides that can target a specific mRNA to degrade the targeted complex by mechanisms such as endogenous cellular RNase H [5]. The first approved ASO, fomivirsen, is applied for the treatment of cytomegalovirus (CMV) retinitis in 1998 [102]. ASOs have been studied for the immunotherapy of cancer. For example, an ASO that target CD39 mRNA has been studied for cancer immunotherapy. CD39 is a protein which serves as ectonucleotidase to degrade ATP to immunosuppressive adenosine, and the knockdown of CD39 by ASOs can improve CD8+ T cell proliferation, whereby improving antitumor immune responses [103].
miRNA is endogenous small non-coding RNA of about 22 nt in length that can regulate target gene expression by a mechanism similar to siRNA [104]. One main challenge of miRNA delivery is to deliver them into tumor tissue with deep tissue penetration efficiently. Moreover, the complexation of tumor microenvironment also prevents miRNA from efficient intracellular delivery into target tumorcells [105]. In order to enhance the biostability, chemical modifications of miRNA have been used to avoid nuclease degradation [106-108]. Polymer-based nanoparticles such as PLGA-PEG nanoparticles [110] and PEI nanoparticles [111,112] were commonly used for miRNA therapy. For instance, Parayath, et al. used CD44-targeting hyaluronic acid-poly(ethylenimine)(HA-PEI)-based nanoparticles to deliver miRNA-125b, an miRNA that can reprogram TAMs into an antitumor/proinflammatory (M1) phenotype, and found that the HA-PEI nanoparticles can accumulate in the macrophage-abated lung tissues of both naïve and KRAS/p53 double mutant genetically engineered(KP-GEM) non-small cell lung cancer mouse model. Moreover, through transfection with miRNA-125b, a > 6-fold increase in the M1/M2 macrophage ratio and 300-fold increase in the iNOS (M1 marker)/Arg-1(M1 marker) ratio in TAMs were observed compared to the control group [113].
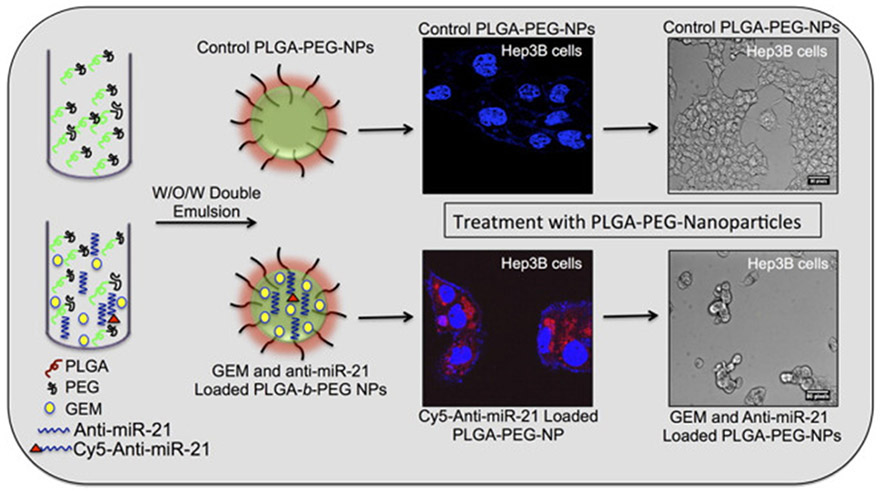
Antagomir, which is also called anti-micro RNA, prevents miRNA activity by irreversibly binding to the targeted miRNA [114]. For instance, as the upregulation of miRNA-21 results in HCC cell proliferation, Devulapally et al. used PLGA-b-PEG nanoparticles to encapsulate anti-miRNA-21 with gemcitabine to inhibit miRNA-21 (Fig. 7). The study demonstrates that down-regulation of endogenous miRNA-21 function with anti-miRNA-21 can reduce HCC cell proliferation by up-regulating miRNA-21 target proteins levels and can improve treatment efficiency in HCC cells in comparison to anti-miRNA-21 or GEM nanoparticles alone [109].
Fig. 7.
PEGylated-PLGA nanoparticles co-encapsulated with anti-miRNA-21 and GEM for HCC therapy. Anti-miRNA-21 and GEM co-encapsulated nanoparticles can improve treatment efficiency in HCC cells in comparison to nanoparticles treated with nanoparticles with equal concentrations of individually loaded anti-miRNA-21 and GEM. Adapted by permission from reference [109]. Copyright 2016, American Chemical Society.
saRNA is a class of noncoding dsRNA about 21 nt in length with 2 nt overhangs at both end [115]. Though it shares similar structure with siRNA but has the opposite mechanism of gene regulation, saRNA in the cytoplasm is specifically loaded to an AGO2 protein and this RNA-AGO2 complex is transported to the nucleus to induce targeted gene promoters for gene activation [116]. It has been reported that saRNA-AGO2 complex in the nucleus recruits essential protein for transcription initiation such as RNA helicase A, RNA polymerase-associated protein CTR9 homolog (CTR9) and RNA polymerase II-associated factor 1 homolog(PAF1) [117]. Due to its ability of gene upregulation, saRNA shows the potential for applications such as cancer immunotherapy. Voutila et al. developed an saRNA that can upregulate the transcription factor CCATT/enhancer binding protein alpha (CEBPA) and found that saRNA retains activation of CEBPA mRNA, leading to an increase in functional C/EBP protein and albumin, and inhibits growth of liver cancer in a rat model of hepatocellular carcinoma (HCC) [118].
3. Conclusion and outlook
Nucleic acid therapeutics have shown unprecedented potential for a wide variety of diseases. Recent advancements in the fields of nucleic acid therapeutics and immuno-oncology have garnered broad interest in applying nucleic acid therapeutics for cancer immunotherapy. Nucleic acid therapeutics have versatile functionalities. From the development of gene-regulating nucleic acid drugs (ASO, siRNA, gRNA, etc.) to pDNA and mRNA therapeutics, immunomodulatory nucleic acids, nuclei acid therapeutics take advantages of manipulating gene expression and hence regulating protein functions to modulate immune responses. Natural nucleic acids have characteristic negative charges, susceptibility to enzymatic degradation, as well as fast clearance from the body, all of which could hinder their clinical translation as therapeutics. Moreover, in order to achieve the efficient delivery of nucleic acid drugs to the desired tissues, cells, as well as subcellular locations, multiple physiological barriers present considerable challenges. For instance, systemically administered tumor-targeted nucleic acid drugs may need to overcome the endothelial barrier in blood vessels, and then penetrate the tumor microenvironment to be efficiently uptaken by cells in the tumor microenvironment. As for nucleic acid drugs that enhance the immune responses, nucleic acid drugs are desired to accumulate in the lymphoid tissues and immune cells to improve the ability of tumor-infiltrating lymphocytes. Lastly, how to improve endosome escape upon cell uptake by endocytosis is critical for nucleic acid therapeutics that execute in the cytosol or even in nuclei. For instance, mRNA therapeutics need to be delivered to the cytosol for protein or peptide translation, immunostimulatory adjuvants based on cytosolic nucleic acid sensing, such as STING agonists, are also expected to be delivered to the cytosol, while the subcellular destinations of pDNA and genome-editing nucleic acids are typically nuclei. To address these challenges, various nucleic acid chemical modifications and drug delivery systems have been developed and validated to improve their biostability, pharmacokinetics, as well as tissue and cell delivery. Nucleic acid chemistry has been leveraged to develop a wide range of chemical modifications on the backbone, sugar, and nucleosides in order to increase the biostability and therapeutic potency, or ameliorate the off-targeting and adverse side effects. Moreover, a variety of drug delivery systems have been reported for tissue and cell delivery of nucleic acid therapeutics. Representative delivery systems for nucleic acid therapeutics include lipid nanoparticles, polymer nanoparticles, bioconjugates, as well as viruses which were not in the scope of the discussion in this article. Recently, multiple siRNA have been approved by US FDA for the treatment of liver diseases using lipid nanoparticles or GalNAc bioconjugates, showcasing the potential for nucleic acid therapeutics. Future efforts are desired to develop nucleic acid therapeutics, including delivery systems, for extrahepatic organs and tissues for disease treatment such as cancer immunotherapy. Moreover, such achievements in ASO and siRNA therapeutics are expected to inspire the development of other oligonucleotide therapeutics, such as microRNA and gRNA. Lastly, the experience in the field of pDNA therapeutics should also benefit the development of other nucleic acid therapeutics, especially the often large mRNA. As more and more immuno-oncology mechanisms are dissected, it is expected that the development of nucleic acid therapeutics as well as their delivery systems would play a pivotal role to address issues (e.g., generally low response rate and resistance to current immune checkpoint blockade) associated with current immunotherapy approaches and improve treatment outcomes in the immunotherapy of cancer.
Acknowledgment
This research was supported partially by the start-up funds from the Center for Pharmaceutical Engineering and Sciences of the VCU Department of Pharmaceutics, KL2 scholarship under an NCATS Clinical & Translational Science Award (CTSA) (UL1TR002649), and pilot grant from VCU Massey Cancer Center (P30 CA106059)); and American Cancer Society Institutional Research Grants (IRG-18-159-43).
Funding Source
This research was supported partially by the start-up funds from VCU Pharmaceutics, KL2 scholarship under an NCATS Clinical & Translational Science Award (CTSA) (UL1TR002649), and pilot grant from VCU Massey Cancer Center (P30 CA106059).
Footnotes
Declaration of competing interest
The authors declare that there are no conflicts of interest.
References
- [1].Opalinska JB, Gewirtz AM. Nucleic-acid therapeutics: basic principles and recent applications. Nat Rev Drug Discov. 2002;1(7):503–14. [DOI] [PubMed] [Google Scholar]
- [2].Pastor F, Berraondo P, Etxeberria I, Frederick J, Sahin U, Gilboa E, et al. An RNA tool-box for cancer immunotherapy. Nat Rev Drug Discov. 2018;17(10):751–67. [DOI] [PubMed] [Google Scholar]
- [3].Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, et al. Sequence- and target-independent angiogenesis suppression by SiRNA via TLR3. Nature. 2008;452(7187):591–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sridharan K, Gogtay NJ. Therapeutic nucleic acids: current clinical status. Br J Clin Pharmacol. 2016;82(3):659–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dahlman JE, Barnes C, Khan OF, Thiriot A, Jhunjunwala S, Shaw TE, et al. In vivo endothelial SiRNA delivery using polymeric nanoparticles with low molecular weight. Nature Nanotechnol. 2014;9(8):648–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Barber GN. STING: Infection, Inflammation and Cancer. Nat Rev Immunol. 2015;15(12):760–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Vollmer J, Krieg AM. Immunotherapeutic applications of CpG Oligodeoxynucleotide TLR9 agonists. Adv Drug DelivRev. 2009;61(3):195–204. [DOI] [PubMed] [Google Scholar]
- [9].Kyi C, Roudko V, Sabado R, Saenger Y, Loging W, Mandeli J, et al. Therapeutic immune modulation against solid cancers with intratumoral Poly-ICLC: a pilot trial. Clin Cancer Res. 2018;24(20):4937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hajj KA, Whitehead KA. Tools for translation: non-viral materials for therapeutic MRNA delivery. Nat Rev Mater. 2017;2(10):17056. [Google Scholar]
- [11].Gilboa E, Berezhnoy A, Schrand B. Reducing toxicity of immune therapy using Aptamer-targeted drug delivery. Cancer Immunol Res. 2015;3(11):1195–200. [DOI] [PubMed] [Google Scholar]
- [12].Yin H, Kauffman KJ, Anderson DG. Delivery technologies for genome editing. Nat Rev Drug Discov. 2017;16(6):387–99. [DOI] [PubMed] [Google Scholar]
- [13].Li H, Fan X, Houghton J. Tumor microenvironment: the role of the tumor stroma in cancer. J Cell Biochem. 2007;101(4):805–15. [DOI] [PubMed] [Google Scholar]
- [14].Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–93. [DOI] [PubMed] [Google Scholar]
- [15].Verma IM, Somia N. Gene therapy - promises, Problems and Prospects. Nature. 1997;389(6648):239–42. [DOI] [PubMed] [Google Scholar]
- [16].Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–93. [DOI] [PubMed] [Google Scholar]
- [17].Egli M, Manoharan M. Re-engineering RNA molecules into therapeutic agents. Acc Chem Res. 2019;52(4):1036–47. [DOI] [PubMed] [Google Scholar]
- [18].Brown JA, Suo Z. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry. 2011;50(7):1135–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Olton D, Li J, Wilson ME, Rogers T, Close J, Huang L, et al. Nanostructured calcium phosphates (NanoCaPs) for non-viral gene delivery: influence of the synthesis parameters on transfection efficiency. Biomaterials. 2007;28(6):1267–79. [DOI] [PubMed] [Google Scholar]
- [20].Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv Drug Deliv Rev. 2014;66:2–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Patel S, Kim J, Herrera M, Mukherjee A, Kabanov AV, Sahay G. Brief update on endocytosis of nanomedicines. Adv Drug Deliv Rev. 2019;144:90–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang Y, Miao L, Satterlee A, Huang L. Delivery of oligonucleotides with lipid nanoparticles. Adv Drug Deliv Rev. 2015;87:68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Huang Y Preclinical and clinical advances of GalNAc-decorated nucleic acid therapeutics. Mol Ther - Nucleic Acids. 2017;6:116–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ergun SL; Fernandez D; Weiss TM; Li L STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition. Cell 2019, 178 (2), 290–301. e10. [DOI] [PubMed] [Google Scholar]
- [25].Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 2015;11(7):1018–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wu J, Sun L, Chen X, Du F, Shi H, Chen C, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339(6121):826–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Herzner A-M, Hagmann CA, Goldeck M, Wolter S, Kübler K, Wittmann S, et al. Sequence-specific activation of the DNA sensor CGAS by Y-form DNA structures as found in primary HIV-1 CDNA. Nat Immunol. 2015;16(10):1025–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ramanjulu JM, Pesiridis GS, Yang J, Concha N, Singhaus R, Zhang S-Y, et al. Design of Amidobenzimidazole STING receptor agonists with systemic activity. Nature. 2018;564(7736):439–43. [DOI] [PubMed] [Google Scholar]
- [29].Hanson MC, Crespo MP, Abraham W, Moynihan KD, Szeto GL, Chen SH, et al. Nanoparticulate STING agonists are potent lymph node–targeted vaccine adjuvants. J Clin Invest. 2015;125(6):2532–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nakamura T, Miyabe H, Hyodo M, Sato Y, Hayakawa Y, Harashima H. Liposomes loaded with a STING pathway ligand, cyclic Di-GMP, enhance cancer immunotherapy against metastatic melanoma. J Control Release. 2015;216:149–57. [DOI] [PubMed] [Google Scholar]
- [31].Smith TT, Moffett HF, Stephan SB, Opel CF, Dumigan AG, Jiang X, et al. Biopolymers codelivering engineered T cells and STING agonists can eliminate heterogeneous tumors. J Clin Invest. 2017;127(6):2176–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Koshy ST, Cheung AS, Gu L, Graveline AR, Mooney DJ. Liposomal delivery enhances immune activation by STING agonists for Cancer immunotherapy. Adv Biosys. 2017;1(1–2):1600013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shae D, Becker KW, Christov P, Yun DS, Lytton-Jean AKR, Sevimli S, et al. Endosomolytic polymersomes increase the activity of cyclic dinucleotide STING agonists to enhance cancer immunotherapy. Nat Nanotechnol. 2019;14(3):269–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Luo M, Wang H, Wang Z, Cai H, Lu Z, Li Y, et al. A STING-activating nanovaccine for cancer immunotherapy. Nature Nanotechnol. 2017;12(7):648–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Casey LM, Kakade S, Decker JT, Rose JA, Deans K, Shea LD, et al. Cargo-less nanoparticles program innate immune cell responses to toll-like receptor activation. Biomaterials. 2019;218:119333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ai S-L, He X-Y, Liu B-Y, Zhuo R-X, Cheng S-X. Targeting delivery of Oligodeoxynucleotides to macrophages by mannosylated cationic albumin for immune stimulation in cancer treatment. Mol Pharm. 2019;16(6):2616–25. [DOI] [PubMed] [Google Scholar]
- [37].Yoshizaki Y, Yuba E, Sakaguchi N, Koiwai K, Harada A, Kono K. PH-sensitive polymer-modified liposome-based immunity-inducing system: effects of inclusion of cationic lipid and CpG-DNA. Biomaterials. 2017;141:272–83. [DOI] [PubMed] [Google Scholar]
- [38].Zhang H, Cheng T, Lai L, Deng S, Yu R, Qiu L, et al. BN Nanospheres functionalized with mesoporous silica for enhancing CpG oligodeoxynucleotide-mediated cancer immunotherapy. Nanoscale. 2018;10(30):14516–24. [DOI] [PubMed] [Google Scholar]
- [39].Wang Z, Zhang Y, Liu Z, Dong K, Liu C, Ran X, et al. A bifunctional nanomodulator for boosting CpG-mediated cancer immunotherapy. Nanoscale. 2017;9(37):14236–47. [DOI] [PubMed] [Google Scholar]
- [40].Luo J, Cheng Y, He X-Y, Liu Y, Peng N, Gong Z-W, et al. Self-assembled CpG oligodeoxynucleotides conjugated hollow gold nanospheres to enhance cancer-associated Immunostimulation. Colloids Surf B Biointerfaces. 2019;175:248–55. [DOI] [PubMed] [Google Scholar]
- [41].Cho H-I, Barrios K, Lee Y-R, Linowski AK, Celis E. BiVax: a peptide/poly-IC subunit vaccine that mimics an acute infection elicits vast and effective anti-tumor CD8 T-cell responses. Cancer Immunol Immunother. 2013;62(4):787–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Salazar AM, Erlich RB, Mark A, Bhardwaj N, Herberman RB. Therapeutic in situ autovaccination against solid cancers with Intratumoral poly-ICLC: case report, hypothesis, and clinical trial. Cancer Immunol Res. 2014;2(8):720–4. [DOI] [PubMed] [Google Scholar]
- [43].Rodríguez-Ruiz ME; Perez-Gracia JL; Rodríguez I; Alfaro C; Oñate C; Pérez G; Gil-Bazo I; Benito A; Inogés S; López-Diaz de Cerio A; et al. Combined Immunotherapy Encompassing Intratumoral Poly-ICLC, Dendritic-Cell Vaccination and Radiotherapy in Advanced Cancer Patients. Ann Oncol 2018, 29 (5), 1312–1319. [DOI] [PubMed] [Google Scholar]
- [44].Pardi N, Hogan MJ, Porter FW, Weissman D. MRNA vaccines — a new era in vaccinology. Nat Rev Drug Discov. 2018;17(4):261–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sahin U, Karikó K, Türeci Ö. MRNA-based therapeutics — developing a new class of drugs. Nat Rev Drug Discov. 2014;13(10):759–80. [DOI] [PubMed] [Google Scholar]
- [46].Wang Z, Day N, Trifillis P, Kiledjian M. An MRNA stability complex functions with poly (a)-binding protein to stabilize MRNA in vitro. Mol Cell Biol. 1999;19(7):4552–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Holtkamp S, Kreiter S, Selmi A, Simon P, Koslowski M, Huber C, et al. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108(13):4009–17. [DOI] [PubMed] [Google Scholar]
- [48].Li B, Luo X, Dong Y. Effects of chemically modified messenger RNA on protein expression. Bioconjug Chem. 2016;27(3):849–53. [DOI] [PubMed] [Google Scholar]
- [49].Li J, Wang W, He Y, Li Y, Yan EZ, Zhang K, et al. Structurally programmed assembly of translation initiation nanoplex for superior MRNA delivery. ACS Nano. 2017;11(3):2531–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Karikó K, Muramatsu H, Ludwig J, Weissman D. Generating the optimal MRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified. Protein-Encoding MRNA Nucleic Acids Res. 2011;39(21):e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sebastian M, Papachristofilou A, Weiss C, Früh M, Cathomas R, Hilbe W, et al. Phase Ib study evaluating a self-adjuvanted MRNA cancer vaccine (RNActive®) combined with local radiation as consolidation and maintenance treatment for patients with stage IV non-small cell lung cancer. BMC Cancer. 2014;14(1):748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, Banerjee K, et al. Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci U S A. 2012;109(36):14604–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Miao L, Li L, Huang Y, Delcassian D, Chahal J, Han J, et al. Delivery of MRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat Biotechnol. 2019;37:1174–85. [DOI] [PubMed] [Google Scholar]
- [54].Moffett HF, Coon ME, Radtke S, Stephan SB, McKnight L, Lambert A, et al. Hit-and-run programming of therapeutic cytoreagents using MRNA nanocarriers. Nat Commun. 2017;8(1):389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Murakami T, Sunada Y. Plasmid DNA gene therapy by electroporation: principles and recent advances. CGT. 2011;11(6):447–56. [DOI] [PubMed] [Google Scholar]
- [56].Liu Q, Zhu H, Tiruthani K, Shen L, Chen F, Gao K, et al. Nanoparticle-mediated trap ping of Wnt family member 5A in tumor microenvironments enhances immunotherapy for B-Raf proto-oncogene mutant melanoma. ACS Nano. 2018;12(2):1250–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Song W, Shen L, Wang Y, Liu Q, Goodwin TJ, Li J, et al. Synergistic and low adverse effect cancer immunotherapy by immunogenic chemotherapy and locally expressed PD-L1 trap. Nat Commun. 2018;9(1):2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Luo C, Miao L, Zhao Y, Musetti S, Wang Y, Shi K, et al. A novel cationic lipid with intrinsic antitumor activity to facilitate gene therapy of TRAIL DNA. Biomaterials. 2016;102:239–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Qi C, Musetti S, Fu L-H, Zhu Y-J, Huang L. Biomolecule-assisted green synthesis of nanostructured calcium phosphates and their biomedical applications. Chem Soc Rev. 2019;48(10):2698–737. [DOI] [PubMed] [Google Scholar]
- [60].Song W, Musetti SN, Huang L. Nanomaterials for cancer immunotherapy. Biomaterials. 2017;148:16–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hu M, Wang Y, Xu L, An S, Tang Y, Zhou X, et al. Relaxin gene delivery mitigates liver metastasis and synergizes with check point therapy. Nat Commun. 2019;10(1):2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Liu L, Zong Z-M, Liu Q, Jiang S-S, Zhang Q, Cen L-Q, et al. A novel galactose-PEG-conjugated biodegradable copolymer is an efficient gene delivery vector for immunotherapy of hepatocellular carcinoma. Biomaterials. 2018;184:20–30. [DOI] [PubMed] [Google Scholar]
- [63].Zhu G, Chen X. Aptamer-based targeted therapy. Adv Drug Deliv Rev. 2018;134:65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ng EWM, Shima DT, Calias P, Cunningham ET, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2006;5(2):123–32. [DOI] [PubMed] [Google Scholar]
- [65].McNamara JO, Kolonias D, Pastor F, Mittler RS, Chen L, Giangrande PH, et al. Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J Clin Invest. 2008;118(1):376–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Santulli-Marotto S, Nair SK, Rusconi C, Sullenger B, Gilboa E. Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 2003;63(21):7483–9. [PubMed] [Google Scholar]
- [67].Herrmann A, Priceman SJ, Kujawski M, Xin H, Cherryholmes GA, Zhang W, et al. CTLA4 aptamer delivers STAT3 SiRNA to tumor-associated and malignant T cells. J Clin Invest. 2014;124(7):2977–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gefen T, Castro I, Muharemagic D, Puplampu-Dove Y, Patel S, Gilboa E. A TIM-3 oligonucleotide Aptamer enhances T cell functions and potentiates tumor immunity in mice. Mol Ther. 2017;25(10):2280–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Dollins CM, Nair S, Boczkowski D, Lee J, Layzer JM, Gilboa E, et al. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem Biol. 2008;15(7):675–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. Prolonged interleukin-2Rα expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32(1):91–103. [DOI] [PubMed] [Google Scholar]
- [71].Rajagopalan A, Berezhnoy A, Schrand B, Puplampu-Dove Y, Gilboa E. Aptamer-targeted attenuation of IL-2 signaling in CD8 + T cells enhances antitumor immunity. Mol Ther. 2017;25(1):54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–46. [DOI] [PubMed] [Google Scholar]
- [73].Maeder ML, Gersbach CA. Genome-editing technologies for gene and cell therapy. Mol Ther. 2016;24(3):430–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Li L, Hu S, Chen X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: challenges and opportunities. Biomaterials. 2018;171:207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Das AT, Binda CS, Berkhout B. Elimination of infectious HIV DNA by CRISPR-Cas9. Curr Opin Virol. 2019;38:81–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Yin H, Xue W, Anderson DG. CRISPR–Cas: a tool for cancer research and therapeutics. Nat Rev Clin Oncol. 2019;16(5):281–95. [DOI] [PubMed] [Google Scholar]
- [78].Liu X, Zhao Y. CRISPR/Cas9 genome editing: fueling the revolution in cancer immunotherapy. Curr Res Transl Med. 2018;66(2):39–42. [DOI] [PubMed] [Google Scholar]
- [79].Cornel MC, Howard HC, Lim D, Bonham VL, Wartiovaara K. Moving towards a cure in genetics: what is needed to bring somatic gene therapy to the clinic? Eur J Hum Genet. 2019;27(3):484–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Ren J; Zhang X; Liu X; Fang C; Jiang S; June CH; Zhao Y A Versatile System for Rapid Multiplex Genome-Edited CAR T Cell Generation. Oncotarget 2017, 8 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ren J, Liu X, Fang C, Jiang S, June CH, Zhao Y. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin Cancer Res. 2017;23(9):2255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Liu X, Zhang Y, Cheng C, Cheng AW, Zhang X, Li N, et al. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res. 2017;27(1):154–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJC, Hamieh M, Cunanan KM, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543(7643):113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Yin H, Kauffman KJ, Anderson DG. Delivery technologies for genome editing. Nat Rev Drug Discov. 2017;16(6):387–99. [DOI] [PubMed] [Google Scholar]
- [85].Graessmann M, Graessmann A. Microinjection of tissue culture cells. Meth Enzymol. 1983;101:482–92. [DOI] [PubMed] [Google Scholar]
- [86].Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K, et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun. 2015;6(1):7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MDC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32(3):267–73. [DOI] [PubMed] [Google Scholar]
- [88].Alsaiari SK, Patil S, Alyami M, Alamoudi KO, Aleisa FA, Merzaban JS, et al. Endosomal escape and delivery of CRISPR/Cas9 genome editing machinery enabled by nanoscale zeolitic Imidazolate framework. J Am Chem Soc. 2018;140(1):143–6. [DOI] [PubMed] [Google Scholar]
- [89].Kretzmann JA, Ho D, Evans CW, Plani-Lam JHC, Garcia-Bloj B, Mohamed AE, et al. Synthetically controlling dendrimer flexibility improves delivery of large plasmid 0DNA. Chem Sci. 2017;8(4):2923–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Mout R, Ray M, Yesilbag Tonga G, Lee Y-W, Tay T, Sasaki K, et al. Direct cytosolic delivery of CRISPR/Cas9-Ribonucleoprotein for efficient gene editing. ACS Nano. 2017;11(3):2452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33(1):73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Liu J, Chang J, Jiang Y, Meng X, Sun T, Mao L, et al. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv Mater. 2019;31(33):1902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in SiRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Setten RL, Rossi JJ, Han S. The current state and future directions of RNAi-based therapeutics. Nat Rev Drug Discov. 2019;18(6):421–46. [DOI] [PubMed] [Google Scholar]
- [95].Zuckerman JE, Davis ME. Clinical experiences with systemically administered SiRNA-based therapeutics in cancer. Nat Rev Drug Discov. 2015;14(12):843–56. [DOI] [PubMed] [Google Scholar]
- [96].Ben David-Naim M, Grad E, Aizik G, Nordling-David MM, Moshel O, Granot Z, et al. Polymeric nanoparticles of SiRNA prepared by a double-emulsion solvent-diffusion technique: physicochemical properties, toxicity, biodistribution and efficacy in a mammary carcinoma mice model. Biomaterials. 2017;145:154–67. [DOI] [PubMed] [Google Scholar]
- [97].Naito M, Yoshinaga N, Ishii T, Matsumoto A, Miyahara Y, Miyata K, et al. Enhanced intracellular delivery of SiRNA by controlling ATP-responsivity of phenylboronic acid-functionalized polyion complex micelles. Macromol Biosci. 2018;18(1):1700357. [DOI] [PubMed] [Google Scholar]
- [98].Li J, Chen Y-C, Tseng Y-C, Mozumdar S, Huang L. Biodegradable calcium phosphate nanoparticle with lipid coating for systemic SiRNA delivery. J Control Release. 2010;142(3):416–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Warashina S, Nakamura T, Sato Y, Fujiwara Y, Hyodo M, Hatakeyama H, et al. A lipid nanoparticle for the efficient delivery of SiRNA to dendritic cells. J Control Release. 2016;225:183–91. [DOI] [PubMed] [Google Scholar]
- [100].Wayteck L, Dewitte H, De Backer L, Breckpot K, Demeester J, De Smedt SC, et al. Hitchhiking nanoparticles: reversible coupling oflipid-based nanoparticles to cytotoxic T lymphocytes. Biomaterials. 2016;77:243–54. [DOI] [PubMed] [Google Scholar]
- [101].Song Y, Tang C, Yin C. Combination antitumor immunotherapy with VEGF and PIGF SiRNA via systemic delivery of multi-functionalized nanoparticles to tumor-associated macrophages and breast cancer cells. Biomaterials. 2018;185:117–32. [DOI] [PubMed] [Google Scholar]
- [102].Orr RM. Technology evaluation: Fomivirsen, Isis pharmaceuticals Inc/CIBA vision. Curr Opin Mol Ther. 2001;3(3):288–94. [PubMed] [Google Scholar]
- [103].Kashyap AS, Thelemann T, Klar R, Kallert SM, Festag J, Buchi M, et al. Antisense oligonucleotide targeting CD39 improves anti-tumor T cell immunity. J Immunotherapy Cancer. 2019;7(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136(1):26–36. [DOI] [PubMed] [Google Scholar]
- [105].Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. MiRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6(3):235–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Davis S Improved targeting of MiRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34(8):2294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, et al. Antagonism of MicroRNA-122 in mice by systemically administered LNA-AntimiR leads to up-regulation of a large set of predicted target MRNAs in the liver. Nucleic Acids Res. 2008;36(4):1153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Peacock H, Fucini RV, Jayalath P, Ibarra-Soza JM, Haringsma HJ, Flanagan WM, et al. Nucleobase and ribose modifications control immunostimulation by a MicroRNA-122-mimetic RNA. J Am Chem Soc. 2011;133(24):9200–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Devulapally R, Foygel K, Sekar TV, Willmann JK, Paulmurugan R. Gemcitabine and antisense-MicroRNA co-encapsulated PLGA–PEG polymer nanoparticles for hepatocellular carcinoma therapy. ACS Appl Mater Interfaces. 2016;8(49):33412–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].van Vlerken LE, Vyas TK, Amiji MM. Poly(ethylene glycol)-modified nanocarriers for tumor-targeted and intracellular delivery. Pharm Res. 2007;24(8):1405–14. [DOI] [PubMed] [Google Scholar]
- [111].Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92(16):7297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7(5):657–63. [DOI] [PubMed] [Google Scholar]
- [113].Parayath NN, Parikh A, Amiji MM. Repolarization of tumor-associated macrophages in a genetically engineered nonsmall cell lung cancer model by Intraperitoneal administration of hyaluronic acid-based nanoparticles encapsulating MicroRNA-125b. Nano Lett. 2018;18(6):3571–9. [DOI] [PubMed] [Google Scholar]
- [114].Yue J MiRNA and vascular cell movement. Adv Drug Deliv Rev. 2011;63(8):616–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kwok A, Raulf N, Habib N. Developing small activating RNA as a therapeutic: current challenges and promises. Ther Deliv. 2019;10(3):151–64. [DOI] [PubMed] [Google Scholar]
- [116].Li L-C, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, et al. Small DsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A. 2006;103(46):17337–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Portnoy V, Lin SHS, Li KH, Burlingame A, Hu Z-H, Li H, et al. SaRNA-guided Ago2 targets the RITA complex to promoters to stimulate transcription. Cell Res. 2016;26(3):320–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Voutila J, Reebye V, Roberts TC, Protopapa P, Andrikakou P, Blakey DC, et al. Development and mechanism of small activating RNA targeting CEBPA, a novel therapeutic in clinical trials for liver cancer. Mol Ther. 2017;25(12):2705–14. [DOI] [PMC free article] [PubMed] [Google Scholar]