Abstract
Many biologically active natural products are synthesized by nonribosomal peptide synthetases (NRPSs), polyketide synthases (PKSs) and their hybrids. These megasynthetases contain modules possessing distinct catalytic domains that allow for substrate initiation, chain extension, processing and termination. At the end of a module, a terminal domain, usually a thioesterase (TE), is responsible for catalyzing the release of the NRPS or PKS as a linear or cyclized product. In this review, we address the general cyclization mechanism of the TE domain, including oligomerization and the fungal C-C bond forming Claisen-like cyclases (CLCs). Additionally, we include examples of cyclization catalysts acting within or at the end of a module. Furthermore, condensation-like (CT) domains, terminal reductase (R) domains, reductase-like domains that catalyze Dieckmann condensation (RD), thioesterase-like Dieckmann cyclases, trans-acting TEs from the penicillin binding protein (PBP) enzyme family, product template (PT) domains and others will also be reviewed. The studies summarized here highlight the remarkable diversity of NRPS and PKS cyclization catalysts for the production of biologically relevant, complex cyclic natural products and related compounds.
Keywords: Enzymes, Natural Products, Biocatalysis, Polyketide Synthase, Nonribosomal Peptide Synthetase
1. Introduction
Nonribosomal peptide synthetases (NRPSs), polyketide synthases (PKSs) and hybrids (PKS/NRPS) thereof are responsible for the synthesis of numerous natural products in bacterial and fungal species.[1] Several of these compounds are clinically approved therapeutics to treat human diseases, including cancer (e.g. calicheamicin and bleomycin) autoimmune disorders (e.g. rapamycin), parasitic infections (e.g. ivermectin) and bacterial pathogens (e.g. erythromycin, tiacumicin B and vancomycin).[2] The multifunctional machinery of these megasynthases makes them particularly unique. Individual modules containing diverse domains incorporate specific substrates as they work together to assemble complex nonribosomal peptides or polyketides in a linear manner to generate a final product. NRPS and PKS products possess widely varied structures with linear, cyclic and branched elements.[3]
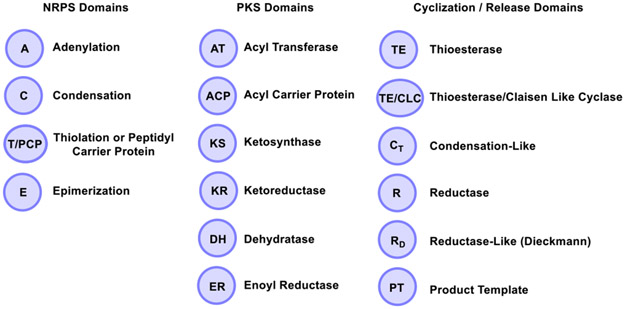
The NRPS and PKS megasynthases contain modules that mediate substrate initiation, chain extension, processing and termination. A module contains a series of domains that catalyze diverse reactions on a peptide or polyketide chain. The NRPS core module contains an adenylation (A) domain that selects and activates amino acid monomers, a condensation (C) domain which catalyzes the formation of peptide bonds, and a thiolation domain (T) or peptidyl carrier protein (PCP) with a phosphopantetheine (Ppant) group for transferring substrates into the catalytic site. NRPS modules might also contain accessory domains including an epimerization (E) domain for the conversion of L-amino acids to D-amino acids. The PKS core module contains an acyltransferase (AT) domain, which selects and transfers an extender unit, an acyl carrier protein (ACP) also with a Ppant group (similarly to the PCP in NRPS) for extender unit loading, and a ketosynthase (KS) domain for decarboxylative Claisen condensation.[4] In the case of PKS, the elective processing domains include a ketoreductase (KR), dehydratase (DH) and enoylreductase (ER) (Figure 1).[1] During the biosynthesis of a fully elongated and mature linear peptide or polyketide, a terminal domain, usually a thioesterase (TE) catalyzes the release of the peptide and polyketide as a linear or cyclized product.[4] In this review, we describe examples of biosynthetic cyclization catalysts found in NRPS and PKS systems. The general cyclization mechanism of the TE domain will be discussed including oligomerization and the fungal C-C bond forming Claisen-like cyclases (CLCs). Moreover, a number of unusual or rare termination domains have been identified including terminal condensation-like (CT) domains, reductase (R) domains, Dieckmann cyclases, trans-acting TEs from the penicillin binding protein (PBP) enzyme family, product template (PT) domains and select others. Although various detailed reviews on these topics have appeared in the literature previously,[1, 5] we focus on the current literature relating to fascinating NRPS and PKS cyclization catalysts responsible for the assembly of particularly unique natural products and related compounds.
Figure 1.
NRPS and PKS domains including the cyclization and release domains discussed throughout this review.
2. Thioesterases
As previously described, NRPSs and PKSs are organized into a series of modules that contain distinct catalytic domains. Utilizing peptidyl or acyl chain initiation, elongation and termination mechanisms, these enzymatic assembly lines are capable of generating a mature product. At the end of a biosynthetic pathway, a terminal TE domain usually located at the C-terminus of a PKS or the NRPS module is responsible for catalyzing the cyclization step during termination.[1, 6] Due to the importance and wide range of thioesterases found in nature, this topic has been reviewed with detailed discussions on PKS TEs, NRPS TEs, comparisons of their sequence as well as insights into their structure.[5a, 7] Herein, we provide an overview of the cyclization mechanisms employed by TEs and highlight recent examples of work in this area of natural product biosynthesis. Studies on the TE domain are particularly compelling due to their ability to catalyze diverse reactions, including cyclization via oligomerization, and through carbon-carbon bond formation, among others discussed below. In addition, we will cover thioesterase-like Dieckmann cyclases later in the review. Although an important feature of PKS and NRPS systems, the TEII class of thioesterases involved in editing of polyketide intermediates during assembly will not be discussed.[8]
2.1. Mechanistic Overview
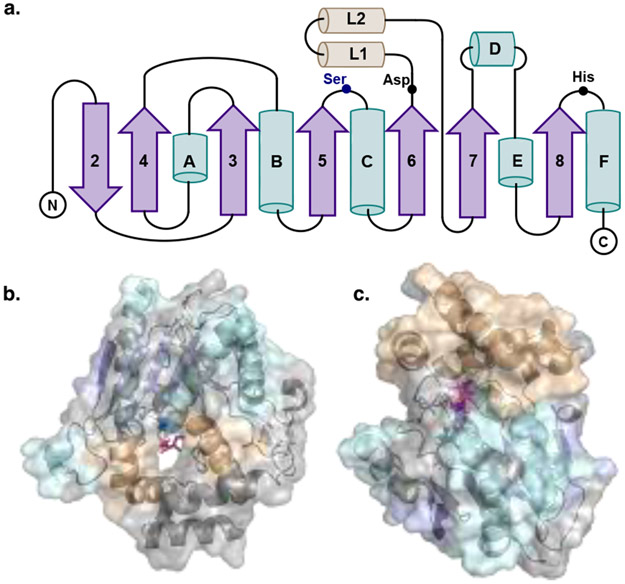
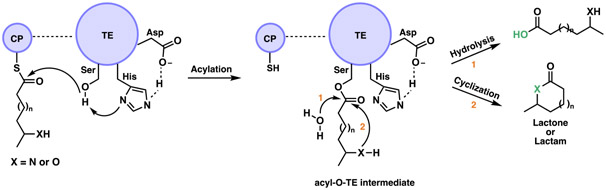
Thioesterases are prevalent in both NRPS and PKS pathways. These enzymes are broadly characterized as members of the α/β hydrolase superfamily, which also includes serine proteases (Figure 2). Macrocyclizing TEs, termed type I TEs are generally found at the end of the C-terminal module of NRPS and PKS pathways. In the active site, the TE features a conserved serine (Ser), histidine (His) and aspartate (Asp) catalytic triad (Figure 3). After the peptide or polyketide is fully extended and elongated, it is typically transferred from the upstream module PCP or ACP to the hydroxyl group of the Ser. The Ser residue acts as a catalytic nucleophile activated by the His and Asp dyad, which generates an acyl-O-TE covalent enzyme intermediate.[5a, 9] The Ppant group covalently attached to the ACP or PCP active site Ser residue mediates the approach and transfer of the thioester substrate to the TE. Then, an acyl hydroxyl or amino nucleophile adds into the carbonyl of the acyl-O-TE intermediate. An intermolecular nucleophile, generally water, leads to a linear hydrolyzed product, while an intramolecular nucleophile (amino or hydroxyl group) generates a lactam or lactone during cyclization, respectively (Figure 3).[1, 5a]
Figure 2.
Overall structure of PKS and NRPS thioesterases with the core α - helices depicted in teal, the lid α-helices depicted in yellow, β strands represented in purple. a. Topology diagram of TEs with the catalytic residues highlighted. b. Representation of PikTE (PDB: 2H7X) highlighting the substrate channel typically seen in PKS TEs and a triketide substrate seen in pink. c. Representation of VlmTE (PDB: 6ECE) depicting the buried, bowl shaped active site with the lid forming the top cap.
Figure 3.
Mechanism of the lactonization or hydrolysis of a CP-tethered peptide or polyketide intermediate. Highlighted in green throughout the review, the bond formed via the described enzyme.
Multiple macrocyclizing TE X-ray crystal structures have been solved including for the pikromycin (1) and erythromycin (2) PKS TEs as well as tyrocidine[10] and fengycin[11] NRPS TEs. These structures provide the basis for information about the similarity of the protein fold, nature of the substrate channel and catalytic mechanism for ring closure.[12] Despite nearly identical fold architecture (Figure 2a), PKS TEs are dimers and contain dimerization helices at the N-terminus as well as a hydrophobic substrate channel through the protein (Figure 2b). In contrast NRPS TEs are monomeric and do not contain these extra helices but sometimes a primary β-strand at the N-terminus. NRPS TEs also have a more bowl-shaped active site that is capped by a formal lid (Figure 2c). These differences are born out in the current understanding of molecular mechanisms employed by TEs to effect macrocyclization. In 2006, the Fecik, Sherman and Smith groups captured a substrate mimic in the active site of the pikromycin (Pik) TE leading to a new hypothesis for macrocyclization.[13] The structure revealed a previously unrecognized water barrier in the enzyme that appears to induce a curled substrate conformation to direct macrolactone ring formation.[13] Similar studies on NRPS domains cited both key interactions in the active site (especially apparent in the fengycin TE) as well as inherent preorganization of the linear peptides as the driving force behind cyclization.[9a, 11] Indeed, early studies on the tyrocidine TE demonstrated that it was capable of cyclizing substrates with widely differing amino acids compared to the native structure, lending biochemical evidence to preorganization of the linear intermediate as a driving force in the NRPS TE cyclization mechanism.[14] This new information provided insights for the design of enzymes capable of catalyzing regioselective macrocyclization of natural or synthetic substrates.[12b]
2.2. General Cyclization Reactions PKS/NRPS TEs
The canonical cyclization of PKSs and NRPSs refers to the TE mediated mechanism that includes the Ser-His-Asp catalytic triad and the acyl-O-TE intermediate, which facilitates the ring closing reaction.[15] A number of examples of cyclic structures can be found including lactones and lactams that are formed via canonical and non-canonical mechanisms. The different types of mechanisms will be discussed below. Moreover, chemoenzymatic applications and engineering of these systems for the synthesis of various natural product structural analogs will also be highlighted in the context of recent work involving TE domain mediated cyclization.
2.2.1. Lactones
Recently, the Wencewicz group studied the NRPS cis-monocyclic β-lactone antibiotic obafluorin (3).[16] Obafluorin (Obi) was isolated in 1984 from Pseudomonas fluorescens ATCC 39502.[17] The biosynthetic gene cluster (BGC) was subsequently identified, and the ObiF TE domain shown to play a direct role in β-lactone formation. The authors demonstrated that the TE catalyzes cyclization of an active site-tethered β-hydroxythioester during product release to generate the strained ring system. Primary sequence analysis revealed that there is a cysteine active site residue (Cys1141) instead of the expected serine for the ObiF TE. This work has the potential for future NRPS engineering to produce targeted peptide β-lactones.
In 2016, the Moore group reported a study of salinamide A (4), which is a cyclic depsipeptide possessing antibacterial activity (Figure 4).[18] Salinamides A and B are the first depsipeptides from this class isolated from marine actinomycete Streptomyces sp. CNB-091.[19] They found the sln9 gene encodes a tetradomain NRPS module with a C-terminal TE. Biochemical characterization of the Sln9 TE showed that it catalyzes an intermolecular transesterification of a serine residue of desmethylsalinamide E with acylated glycyl thioesters to give desmethylsalinamide C in salinamide biosynthesis. Lysobactin (5), is another example of a cyclic depsipeptide with high antibacterial activity against human pathogens (Figure 4). This molecule was originally isolated from Lysobacter sp. ATCC 53042 in 1988.[20] In 2011, the Marahiel group identified and characterized the gene cluster responsible for the lysobactin biosynthesis.[21] The lyb gene cluster codes for two NRPSs, LybA and LybB. The latter was found to contain an unusual tandem TE architecture. The authors characterized in vitro the two individual terminal TEs (TE1, TE2) using a thiophenol activated linear lysobactin analog. Their results demonstrated that only the penultimate thioesterase domain mediates cyclization and simultaneous release of lysobactin. TE1 exclusively catalyzes the formation of the macrocyclic structure, whereas TE2 mediates hydrolytic cleavage of the synthetic substrate.
Figure 4.
Chemical structures of the lactones pikromycin, erythromycin, obafluorin, salinamide A and lysobactin.
2.2.2. Lactams
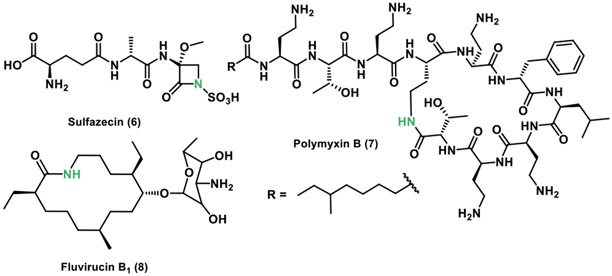
In 2018, the Townsend group revealed the biosynthetic routes for monobactam synthesis in sulfazecin (6).[22] This β-lactam antibiotic is produced by NRPS enzymes and was isolated from Pseudomonas acidophila G-6302 in 1981.[23] In the gene cluster, they identified an unusual TE domain possessing a cysteine active site, and described possible routes for β-lactam formation. The proposed mechanism involves an unknown NRPS TE reaction type in which an assembled tripeptide is N-sulfonated in-trans before β-lactam ring synthesis in the TE domain, which is distinct compared to other systems (Figure 5).
Figure 5.
Chemical structures of the lactams sulfazecin, polymyxin B and fluvirucin B1.
In 2017, the Li and Velkov groups characterized the polymyxin B (7) TE to understand its mode of assembly (Figure 5).[15a] polymyxins remain one of the few antibiotics available for treating antibiotic resistant bacteria. In their study they found that the TE contains a catalytic cysteine in the active site similarly to the TE involved in the synthesis of sulfazecin noted above. They also demonstrated that the polymyxin B pathway TE is able to cyclize a linear N-acetylcysteamine (SNAc) polymyxin B analog. Additionally, homology model examinations of the TE substrate-binding cleft was found to be negatively charged, suggesting a potential mechanism for cyclization of the substrate. The authors also noted that charged interactions could play a role in the binding of the positively charged linear substrate to the negatively charged TE binding site facilitating cyclization.
The Schnarr group in 2013 identified and characterized the fluvirucin B1 (8) polyketide synthase (Figure 5).[24] This 14-membered macrolactam produced by Actinomadura vulgaris has shown antifungal and antiviral activity.[25] They found that the fluvirucin B1 PKS consists of five extender modules, flanked by an initial N-terminal loading ACP that incorporates the initial β-alanine and a C-terminal TE, which catalyzes ring closure of the linear polyketide substrate. This macrolactam forming TE was also found to include a cysteine in the active site instead of a serine.
2.2.3. Chemoenzymatic syntheses
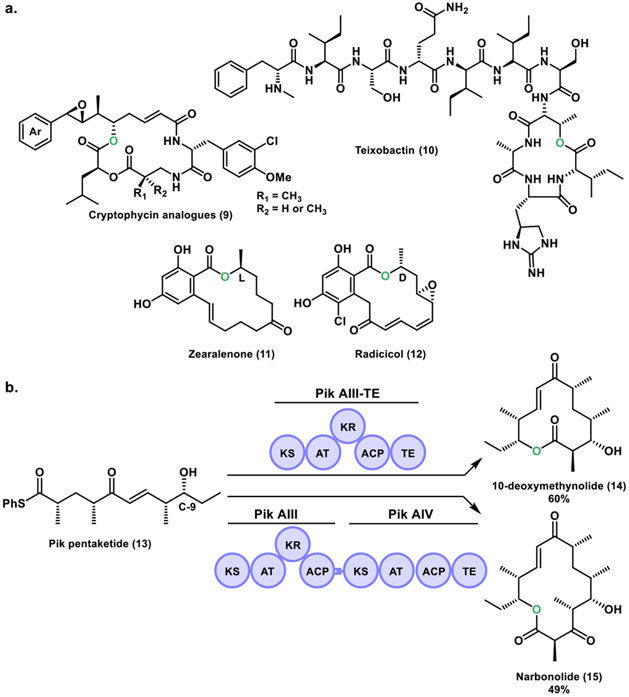
Recently, the Sherman group described the chemoenzymatic synthesis of anti-cancer cryptophycin analogs (9).[26] These macrocycles are a class of 16-membered ring depsipeptide natural products generated by hybrid PKS/NRPS system (Figure 6a). Cryptophycins were first isolated in Nostoc sp. ATCC 53789, and showed antifungal activity.[27] These compounds were rediscovered in Nostoc sp. GSV 224, as a potent antiproliferative microtubule binding agent.[28] The penultimate step in cryptophycin biosynthesis involves a macrocyclization catalyzed by the cryptophycin thioesterase (Crp TE).[29] In their work, a series of novel cryptophycin chain elongation intermediates were synthesized and the Crp TE demonstrated its utility as a biocatalyst through the formation of a library of cryptophycin analogs. Additionally, biological activity studies identified a low picomolar potency analog containing a styrene functionality that overcomes the need for a β-epoxide group. This work shed light into the production of complex bioactive molecules for the potential treatment of malignant diseases and may represent a promising candidate as an antibody-drug conjugate payload.[30]
Figure 6.
a. Chemical structures of cryptophycin analogs, teixobactin, zearalenone and radicicol. b. In vitro biocatalytic platform using the terminal two monomodules (PikAIII and PikAIV) of the pikromycin pathway.
In 2018, the Zhang group studied the teixobactin (10) TE domains.[31] Teixobactin is a depsipeptide isolated from a β-proteobacterium Eleftheria terrae, with antimicrobial activity against many notorious Gram-positive pathogens (Figure 6a).[32] The BGC encodes two large NRPSs, Txo1 and Txo2. In the Txo2 termination module two TE domains (TE1, TE2) were identified, similarly to the lysobactin biosynthetic system.[21] The Zhang group developed a TE based chemoenzymatic approach for the development of teixobactin analogs. Their work shows that the two TE domains operate together to generate a cyclized product and are functionally exchangeable. In contrast to lysobactin biosynthesis in which only the penultimate thioesterase domain (TE1) mediates cyclization while TE2 mediates hydrolytic cleavage, in teixobactin biosynthesis, the TE domains (TE1, TE2) appear to form a combined active site cavity that is essential for cyclization. Their work further demonstrates the impressive mechanistic diversity found in NRPS systems.
The Sherman group in 2013 reported an in vitro, scalable biocatalytic platform utilizing the final two modules (PikAIII and PikAIV) of the pikromycin biosynthetic pathway for the generation of 12- and 14-membered ring macrolactones (Figure 6b).[33] A thiophenol-activated Pik pentaketide (13) substrate was synthesized, and loaded onto PikAIII-TE or PikAIII-PikAIV enzyme systems to afford 10-deoxymethynolide (14) and narbonolide (15), respectively. Furthermore, these macrolactones were converted to their macrolide counterparts in vivo by direct appendage of D-desosamine and final P450-mediated C-H oxidation(s). In a follow-up study, a series of unnatural pentaketides were synthesized, which revealed the substrate tolerance of PikAIII-TE in vitro for production of novel 12-membered ring macrolactones.[34] However, interrogation of pentaketides with epimerized chiral centers in the C-9 hydroxyl group nucleophile revealed the inability of Pik TE to catalyze macrocycle formation. Instead, exclusive hydrolysis of the epimerized hexaketide substrates was observed, suggesting that the TE domain is a catalytic bottleneck in the processing of these chain elongation intermediates.
The Boddy group in 2014 reported work on the TE domains from the zearalenone (11) and radicicol (12) biosynthetic pathways (Figure 6a).[35] These natural products form part of the resorcylic acid lactone (RALs) class of macrocyclic fungal polyketides that contain a resorcylate (2,4-dihydroxybenzoate) within the 14-membered ring macrolactone. Interestingly, these compounds have opposite configuration in the lactone alcohol group, where zearalenone possesses a L (S) configuration, whereas radicicol contains a D (R) configuration. In their work, they show that the TE domains responsible for substrate cyclization are highly stereotolerant to D and L configurations contained in synthetic substrate analogs. Both zearalenone and radicicol TE have the ability to macrocyclize substrates with a different stereochemistry and low competing hydrolysis, in contrast to bacterial PKS TEs, which are highly stereoselective. They further demonstrated the ability of these RAL TEs in chemoenzymatic synthesis of 12- to 18-membered ring macrolactones and a 14-membered macrolactam.[36]
2.2.4. Engineering in these systems
In 2017, the Sherman group reported the engineering of the PikTE, where they mutated the serine 148 in the active site to a cysteine (Figure 7a).[37] The application of the engineered Pik TES148C into a hybrid protein PikAIII-TES148C enabled processing of the C-9 epimerized pentaketide (16) for the production of an epimeric 12-membered ring macrolactone (17). Comparison of quantum mechanics calculations for wild type Pik TE and the S148C mutant revealed that it significantly lowers the activation barriers to macrolactonization, resulting in a more efficient catalyst. In another recent study, the group created a series of hybrid PKS modules containing exchanged TE domains from heterologous pathways.[38] The modules selected were the terminal two modules of the pikromycin (Pik) and erythromycin (DEBS) pathways, and the penultimate module of the juvenimicin (Juv) system. These hybrid PKS modules were interrogated with native and non-native polyketide substrates. Their results showed that substrate TE pairing was a major driving factor in macrolactone production. These works highlight that the TE is often a critical catalytic gatekeeper for the processing of unnatural polyketide chain elongation intermediates. It also showed the substrate flexibility of PKS TE domains and their application in future engineering strategies for the production of polyketide natural product analogs.
Figure 7.
a. Engineering of the Pik TE for synthesis of novel macrolactones. b. Chemical structures of cilengitide and the tautomycetin polyketide natural products.
The Zhu group in 2019, reported a chemoenzymatic method to synthesize the cyclic peptide cilengitide (18) based on the activity of the TE domain from Microcystis aeruginosa microcystin synthetase C (Figure 7b).[39] Moreover, they generated a single S85C mutation in McyC TE, following the strategy used by the Sherman group for the Pik TE.[37] For their chemoenzymatic method using initially the wild type McyC TE domain, they synthesized a pentapeptide-BMT substrate. Enzymatic reactions showed that the McyC TE was able to recognize the linear pentapeptide chain and cyclized it into cilengitide. This was further improved when the McyC TES85C mutant was employed. Kinetic analysis revealed that the McyC TES85C mutation resulted into a superior cyclization catalyst for the pentapeptide-BMT substrate for the production of cilengitide with a 12-fold increase in the Kcat/Km for macrolactonization.
The Sherman and Kim groups in 2016 reported the polyketide tautomycetin (19) TE exchange with the pikromycin TE in Streptomyces sp. CK4412 (Figure 7b).[40] In this work, the PKS cluster in Streptomyces sp. CK4412 that produces tautomycetin (TMC) was engineered by replacing the TMC TE domain with the Pik TE, in order to test the ability of the constructed TMC-PikTE chimera to catalyze macrocyclization of the linear substrate. Their findings showed that indeed, the TMC-PikTE chimera produced not only the expected TMC linear product, but also a cyclized form of TMC. This work shows the potential to engineer PKS strains for in vivo production of new natural products by exchanging a hydrolyzing TE for a macrocyclizing TE.
2.3. TE catalyzed Oligomerization
In addition to canonical head to tail macrocyclization, TE domains are also known to catalyze oligomerization of various natural products during biosynthesis. Cereulide (20), valinomycin (21), elaiophylin (22) and conglobatin (23) are representative examples of both PKS and NRPS derived compounds all formulated by an oligomerization mechanism (Figure 8a). Initially, one monomer is covalently attached to the active site serine of the thioesterase and the second to the carrier protein directly upstream (ACP or PCP). From here two modes of dimerization have been proposed including a “forward transfer” model (originally hypothesized for the enterobactin biosynthesis[41]) that involves nucleophilic attachment of the TE bound monomer on the carrier protein monomer, functionally building the oligomerized substrate on the TE. The second model, termed the “backward transfer” shifts the TE bound monomer back to a nucleophilic functional group on the carrier protein bound monomer, and building the oligomer on the carrier protein. This dimerized (or oligomerized when this cycle is repeated more than once) is then transferred back to the TE active site serine for final cyclization. Evidence for this backward mechanism initially comes from in vitro work with the gramicidin S TE in which linear dimerized thioester intermediates were detected.[42] Recent structural work by the Schmeing and Chin groups on the valinomycin TE have further defined the mechanisms employed in this system.[43] Utilizing site-specific incorporation of a non-canonical amino acid 2,3-diaminopropionic acid (DAP) into recombinant TE, they demonstrated efficient capture of both the first and last acyl-enzyme intermediates. Their crystallographic data demonstrated that for this enzyme, the “backwards transfer” method is employed, further corroborating the gramicidin S TE data and indicating this is likely a global mechanism employed by this subset of TEs. Deeper investigations on the molecular mechanism of the cyclization function revealed a massive lid rearrangement associated with the dodecadepsipeptidyl bound Vlm TE. The terminal domain reorients the substrate from its position during oligomerization placing it into a pocket that entropically controls cyclization. Overall, this work provided an innovative approach to characterize and capture diverse acyl-enzyme complexes of transiently acylated proteins.
Figure 8.
a. Chemical structures of cereulide, valinomycin and elaiophylin. b. Proposed mechanism for TE oligomerization in conglobatin biosynthesis.
The Boddy group has also investigated the TE from the highly homologous cereulide pathway, which is also produced by two NRPS, CesA and CesB.[44] Cereulide is a depsidodecapeptide isolated in 1994 from Bacillus cereus.[45] Despite the similarities between the structures and BGCs of cereulide and the natural product valinomycin, it was previously suggested that cereulide was synthesized through a different mechanism.[46] However, the Boddy group demonstrated that cereulide is in fact produced in a similar manner to valinomycin where the Ces TE catalyzes the trimerization and macrocyclization of a tetradepsipeptide substrate. Interestingly, they also show the Ces TE can utilize related substrates like the valinomycin tetradepsipeptide and a cereulide-valinomycin hybrid tetradepsipeptide with the native substrate to generate natural product analogs. In 2015, the Leadlay group reported the C2-symmetrical macrodiolide elaiophylin produced by PKS enzymes.[47] They investigated the elaiophylin (Ela) TE domain mechanism of the diolide formation in vitro by using synthetic N-acetylcysteaminyl thioesters (SNAc thioesters) of tetraketide and pentaketide analogs of the natural octaketide monomers. It was determined that the Ela TE can catalyze homodimerization of the synthetic pentaketide to a novel 16-membered decaketide diolide. Iterative use of the Ela TE active site during ligation and cyclization is suggested since a linear dimeric thioester intermediate was identified in the ring formation. The authors also proposed that Ela TE must catalyze a total of two acylation and two deacylation reactions to form the diolide. Thus, in a similar manner to that determined for the NRPS dimerizing TEs, the Ela TE first ligates two monomers and then re-loads the linear dimer in the active site for cyclization. However, further studies are required to firmly establish the Ela TE dimerization mechanism.
In the same year, the Leadlay group also studied conglobatin biosynthesis by a PKS/NRPS hybrid (Figure 8b).[48] This compound is an unusual C2-symmetrical macrodiolide with antitumor activity isolated from Streptomyces conglobatus ATCC 31005.[49] The group utilized a single step in vitro cloning method to obtain the entire gene cluster that contains the five genes congA-E resulting in conglobatin production in a heterologous host. A model substrate was also utilized to mimic the conglobatin monomer, which revealed that the Cong TE acts iteratively. The Cong TE ligates two monomers and then re-binds the dimeric product and cyclizes it similarly to the Ela TE described above. Additional work showed that Cong TE is capable of combining two different monomers into hybrid linear dimers and trimers forming additional complex polyketides.
2.4. Fungal TE domains: C-C bond forming Claisen-Like Cyclases (CLCs)
Fungal nonreducing PKS (NR-PKS) are megasynthases that include the SAT (starter unit: ACP transacylase), KS (β-ketosynthase), MAT (malonyl CoA: ACP transacylase), PT (product template), ACP (acyl carrier protein) and TE/CLC (thioesterase/Claisen like cyclase) domains.[50] For the majority of fungal NR-PKS, the TE catalyzes the last step, which releases the product via a Claisen cyclization of a polyketide intermediate, leading to a C-C bond formation.[51] This process is accompanied by the cleavage of the thioester bond between the ACP and the polyketide to generate fused ring structures.[52] Similarly to the TE domains from bacterial PKSs, the TE/CLC domain also contains a Ser, His and Asp catalytic triad.[12a] In 2001, the Ebizuka group identified for the first time a TE with a Claisen cyclase function in the wA PKS gene cluster (Figure 9a) from Aspergillus nidulans, which produces the naphthopyrone YWA1 (24).[52a]
Figure 9.
a. WA TE/CLC mechanism for the production of YWA1. b. PksA TE/CLC domain catalyzes the Claisen cyclization (C-C bond formation) of noranthrone.
In 2008, the Kelleher and Townsend groups studied the NR-PKS PksA from the aflatoxin B1 (26) biosynthesis in Aspergillus parasiticus (Figure 9b).[50a] With respect to the TE/CLC domain, their results suggested that it performs a Claisen cyclization on the intermediate from the PT domain (discussed further below) leading to the formation of 25. In 2010, Townsend and Tsai groups reported a 1.7 Å crystal structure, which provided additional information related to the structure and function of the C-C bond forming TE/CLC domain.[15b] Their study revealed that this domain conducts a Claisen cyclization to release the product from the 4’-phosphopantetheine (Ppant)-tethered ACP affording noranthrone (25). Further biosynthetic processing steps results in formation of aflatoxin B1.
The Townsend group also reported studies on fungal PKS TE function involved in melanin biosynthesis.[53] In 1995, the Kubo group proposed that a polyketide synthase from the PKS1 gene in Colletotrichum lagenarium was involved in production of this pigment.[54] During the biosynthesis of these molecules, a 1,3,6,8-tetrahydroxynaphthalene (THN) intermediate undergoes two rounds of enzymatic reduction and dehydration forming 1,8-dihydroxynaphthalene (DHN), which ultimately affords melanin. THN is the major product from the NR-PKS Pks1 obtained from the heterologous expression in Aspergillus oryzae.[55] In their work, they performed an in vitro characterization of Pks1 from Colletotrichum lagenarium. The Townsend group showed that Pks1 TE is both a Claisen cyclase and deacetylase of an enzyme-bound monocyclic hexaketide precursor that generates THN. This rare dual function demonstrates the notable versatility of the α/β hydrolase fold family of enzymes.
Our final example is the TE/CLC domain found in the herqueinone biosynthesis from Penicillium herquei. In 2016, the Houk and Tang groups studied the NR-PKS PhnA that is responsible for biosynthesis of a heptaketide backbone as well as for its cyclization into a hemiketal-containing naphtho-γ-pyrone prephenalenone (27).[50b] They showed that the PhnA TE/CLC catalyzes a C1-C10 Claisen cyclization generating the corresponding naphthopyrone products (Figure 10a). Remarkably, when the TE/CLC domain was deleted in PhnA (TE0) shunt products 28 and 29 appeared, which are 8- and 10-membered ring macrolactones, respectively (Figure 10b). Their study revealed the basis for new and unexpected metabolites derived from polyketide cyclization.
Figure 10.
a. PhnA TE/CLC C1-C10 Claisen cyclization producing the naphthopyrone prephenalenone. b. Shunt products from the PhnA TE deletion (TE0).
3. Condensation Domains
3.1. Condensation-like (CT) domains
The minimal NRPS module is typically composed of a condensation (C), adenylation (A) and a carrier protein (CP) domain, also termed a thiolation (T) or a peptidyl-carrier-protein (PCP). In bacterial NRPSs, the final off-loading reaction is frequently catalyzed by a thioesterase (TE) domain responsible for the hydrolysis or cyclization of the resulting peptide.[56] However, fungal NRPSs have evolved to employ a different mechanism for macrocyclization, a C-terminal condensation-like (CT) domain, reserving TEs only for hydrolytic offloading.[1, 56b, 57] Genome sequencing data has shown that 60-90% of the fungal NRPSs have a terminal CT domains demonstrating the nearly universal adoption of this offloading mechanism in fungi.[56b] Understanding the basis of the CT domain catalysis in fungal NRPSs offers interesting avenues for biocatalysis and chemoenzymatic approaches for synthesis of numerous cyclic nonribosomal peptides.
3.2. Structural and Mechanistic Overview of CT domains
The macrocyclization mechanism of fungal NRPSs differs significantly from bacterial TE domains. The activity of CT domains was originally established by the Walsh and Tang groups through the in vitro reconstitution of the trimodular NRPS genes (TqaA) involved in formation of fumiquinazoline F (30), a 6,10 fused heterocyclic ring isolated from Aspergillus fumigatus.[56b] They confirmed that the CT domain was responsible for catalyzing the intramolecular attack of an amine nucleophile on the CP domain thioester linkage, and proposed the mechanistic basis for cyclization. Analysis of the fused ring structure generated two initial hypotheses regarding how this core is formed. First, the CT domain could catalyze an initial 10-membered ring and subsequent transannular closure followed by dehydration to generate the fused system (Figure 11a, pathway A). Second, the CT domain could form an initial diketopiperazine like intermediate, followed by nucleophilic attack of the aniline amino group (Figure 11a, pathway B) to produce the same scaffold. In order to distinguish between these two mechanisms, an N-methylated tripeptide analog that would preclude amine attack was synthesized and used as a mechanistic probe. The experiment showed buildup of the 10-membered ring product, suggesting initial macrocyclization is favored over DKP formation. Moreover, this mechanism is comparable to that proposed for the CT domains from the 6,11 bicycle in the asperlicin systems,[58] indicating the larger rings are likely formed first, followed by the intramolecular rearrangement to generate the smaller fused heterocyclic systems.
Figure 11.
a. CT cyclization routes. A and B represent two possible cyclization routes by the CT domain for the formation of fumiquinazoline F. b. Crystal structure of the holo T domain (gold) in conjunction with the CT domain (purple). The Ppant arm and the conserved histidine are drawn in pink. The salt bridge between E4019 (pink) of the β12 - β13 loop and R3652 (teal) of helix α2 is highlighted to show the closure of the solvent channel at the acceptor site.
Further efforts have highlighted the structural and mechanistic differences between CT domains, canonical NRPS condensation domains and bacterial TEs.[59] As previously discussed, bacterial NRPSs terminate by first transferring the growing peptide to the TE domain and then activating it for cyclization through a His-Asp-Ser catalytic triad.[5a] In contrast, CT domains, along with canonical C domains, complete their chemistry on T domain tethered substrates. Biochemical investigations have demonstrated that the CT domain is only active on substrates bound to the correctly paired T-domain partner and that peptidyl-SNAc or peptidyl-CoA substrates are not recognized by the CT domain (as they would be for a canonical bacterial TE domain).[56b] Both types of condensation domains also utilize a histidine in the active site that functions as a general base.[56b, 60] The catalytic histidine deprotonates the amine nucleophile, which then promotes attack on the thioester carbonyl for cyclization and further release in a single step. Recently, a crystal structure of the TqaA CT domain has demonstrated that despite low sequence homology (less than 20%), the overall CT domain fold compared to the canonical condensation domain VibH and SrfA-C is well conserved.[59] The most notable differences are that the N-terminal loop in the C domains is replaced with a helix (α1). This alpha helix causes a shift in the truncated α2 helix toward the acceptor site. A notable salt bridge between Glu4019 and Arg3652, effectively closes off the position where a downstream T domain would interact with the acceptor site.[59] Furthermore, this α2 helix is capable of making extensive contacts with the β12 - β13 loop, which obstructs the solvent channel generally found in C domains, preventing water and other nucleophiles from competing with the cyclization reaction (Figure 11b).
3.3. Examples of CT domains
3.3.1. Gliotoxin
Gliotoxin (31) is one of the most notable members of the epipolythiodiketopiperazines (ETPs), a family of toxic NRPS-derived diketopiperazines (DKPs) (Figure 12).[61] The biosynthesis of gliotoxin is mediated by the gli BGC in Aspergillus fumigatus as well as other related fungi. The gene cluster contains a core bimodular NRPS gliP gene, that has been characterized to convert L-Phe and L-Ser into the DKP cyclo(L-Phe-L-Ser).[62] Due to the lack of an apparent offloading domain, initial reports speculated that the core DKP was formed by a spontaneous cyclization. However, the presence of a conserved histidine within an SHXXXDXXS/T sequence suggested the presence of a CT domain. Notably, although GliP specifies attachment of two amino acids, it contains a third, terminal T domain that may play a role in product offloading in this pathway, a feature not identified in the aforementioned CT domain containing pathways. This prompted the Keller and Schroeder groups to investigate the formation of DKP by these specialized condensation domains and to elucidate its association with the second and third T domains.[62c] They first investigated the function of the GliP-CT and the GliP-T3 domains in vivo by generating two truncation mutants, GliP-ΔCTT3 and GliP-ΔT3 in A. fumigatus. Their results show a complete loss of 31-34 for the GliP-ΔCTT3 strain. Next, GliP-ΔT3 strain alone was investigated and again no 31 was detected or the shunt metabolites 32 or 33 for the GliP-ΔT3, only trace quantities of 34, the detoxification product of 31. The experiment was repeated by growing cultures supplemented with 31 since it has been shown that it serves as a positive feedback for its own production through regulation of gli cluster expression.[63] Again, similar results were obtained using LC-HRMS comparison of the extracts. No production of DKP containing metabolites were observed with the GliP-ΔCTT3 strain, whereas the GliP-ΔT3 strain produced small amounts of cyclized materials 32 and 33. These results demonstrate that for normal biosynthesis of 31, the T3 domain of GliP is required.[62c] Trace products observed with only the GliP-ΔT3 are potentially formed via spontaneous cyclization.
Figure 12.
Function of GliP and abbreviated biosynthesis of gliotoxin showing the most abundant intermediates or shunt metabolites (32) and (33), as well as the detoxification product (34).
Finally, heterologous expression of a GliP mutant protein was conducted with point mutations designed to inactivate the first and second T domains (GliP-ΔT1T2). In order to correct for potential cyclization caused by non-T3 tethered substrate, a second mutant protein with all three T domains inactivated was engineered (GliP-ΔT1T2T3). Utilizing a synthetic L-Phe-L-Ser-N-acetylcysteamine (L-Phe-L-Ser-SNAc) in conjunction with the heterologous expressed GliP-ΔT1T2 and GliP-ΔT1T2T3, in vitro cyclization was confirmed to be greatly enhanced by the presence of the activated T3 domain. This study, in conjunction with previous in vivo studies demonstrated that L-Phe-L-Ser dipeptide cyclization is not spontaneous as previously proposed.[62a, 64] Rather, it requires two additional GliP domains, the second condensation-like domain (CT) and the terminal (third) thiolation domain (T3) in which the nascent dipeptide appears to be transferred from the T2 domain to the T3 domain prior to CT mediated cyclization (Figure 12).[62c] This is in contrast to the previously discussed CT domains for tripeptide substrates in the fumiquinazoline pathway.
3.3.2. Nanangelenin A
Nanangelenin A (36) contains a 1-benzazepine core, which is rare among natural products (Figure 13a).[65] The Piggott and Chooi groups explored the biosynthesis of this alkaloid isolated from a novel Australian fungus, Aspergillus nanangensis.[66] Nanangelenin A contains a 3,4-dihydro-1-benzazepine-2,5-dione-N-prenyl-N-acetoxy-anthranilamide scaffold and its biosynthesis involves the NRPS nan gene cluster, showing a similar T3/CT domain as discussed above for gliotoxin.[66a]
Figure 13.
a. Initial steps in the biosynthesis of nanangelenin A. b. Biosynthesis of benzomalvins proceeds through formation of the Anth-NmPhe dipeptide covalently bound via a thioester bond to the second T-domain of BenZ. BenZ-C2 subsequently catalyzes formation of the Anth-NmPhe-Anth tripeptide covalently bound to BenY-T. BenYCT then catalyzes cyclization and cleavage of the thioester bond, leading to trans-annulation and production of benzomalvin A/D.
Utilizing bioinformatic analysis, the authors identified a putative biosynthetic cluster associated with the formation of this scaffold. Through sequential activation of the reconstituted gene cluster they found five biosynthetic intermediates, which established the role of each encoded enzyme towards assembly of the final product. Importantly, the NRPS nanA gene was found to incorporate anthranilic acid (Ant) and L-kynurenine (L-Kyn) leading to a dipeptide containing two potential cyclizing amines (Figure 13a). Heterologous reconstitution of nanA produced only a single product indicating the terminal CT/T domain pair effects regiospecific cyclization to form nanangelenin B (35).4 This was unequivocally established by constructing NanA carrying mutations in the CT and T3 active sites. Inactivation of the nanA CT domain (generating the NanAH2106A mutant protein) resulted in the loss of (35) production and instead accumulated small amounts of the linear Ant-L-Kyn dipeptide. In a similar manner, the NanA-T3 mutant (S2401A) was tested and resulted in low level production (<3% of WT) of (35) and even lower amounts of the linear Ant-L-Kyn. Expression of both NanA CT and T3 mutants together with NanC also yielded a small amount of the linear dipeptide.[66a]
Additionally, to further probe CT domain regioselectivity, the investigators synthesized the Ant-L-Kyn dipeptide SNAc thioester (Ant-L-Kyn-SNAc) to mimic the T domain Ppant-tethered Ant-L-Kyn, and expressed the stand alone CT domain in E. coli. The native reaction demonstrated that this CT domain was able to catalyze cyclization to 35 despite the lack of a carrier protein. Inactivation of the CT domain produced a new product. NMR studies revealed this to be a spontaneous cyclization event from the anthranilic acid amine, demonstrating the clear divergence of the enzymatically catalyzed cyclization from the kynurenine amine to the more favorable anthranilic amine.[66a]
3.3.3. Benzomalvin A/D
Benzomalvin A and D (37) are benzodiazepines, which were first isolated from a Penicillium species (Figure 13b). These compounds have been shown to antagonize the human NK1 receptor, inhibiting the effects of substance P.[67] The Wu, Keller and Kelleher groups described for the first time the BGC for benzomalvin A and D (ben).[68] The system was identified using fungal artificial chromosomes with a metabolomic scoring (FAC-MS) platform in Aspergillus terreus (ATCC 20542). The FAC-MS approach utilizes high-throughput capture of intact fungal BGCs from genomic DNA in an Escherichia coli-Aspergillus nidulans shuttle vector called a FAC.[69] This is followed by gene cluster heterologous expression in A. nidulans and detection and analysis of small molecule products by metabolomics and cheminformatics using high resolution mass spectrometry (HRMS).[69]
The ben BGC is composed of a benX gene that encodes a putative SAM-binding methyltransferase and two NRPS genes benY and benZ. Benzomalvin A and D biosynthesis proceeds through formation of an NRPS-tethered linear tripeptide (Anth-NmPhe-Anth) containing two anthranilic acid (Anth) residues and one N-methylphenylalanine (NmPhe) residue, which is later cyclized to form an 11-membered macrocyclic intermediate (38). Recently, these investigators deleted C-domains of benY and benZ from a FAC containing the entire benzomalvin gene cluster.[70] The results from the deletions were analyzed by heterologous expression in A. nidulans and targeted metabolomics was employed to determine the biosynthesis of benzomalvin in fungi and the role of the C domains. The results show the production of 37 and 38 by AtFAC9J20-ΔbenY-C, although at a lower level compared to AtFAC9J20 (without any deletions), suggesting that benY-C encodes the CT domain. The biosynthetic pathway for benzomalvin was also deciphered from these findings (Figure 13b). Further data indicated that BenZ-C2 acts as the second internal C-domain while BenY-C functions as the CT domain.[70]
This work suggests that BenY-CT could also mediate formation of the 7-membered ring, representing the first reported benzodiazepine synthase. This process might be occurring through a trans-annulation reaction using 38 as the substrate. This hypothesis was tested, and the group obtained results suggesting that BenY-CT could directly catalyze trans-annulation. However, the possibility that another enzyme was involved from the A. terreus strain or the A. nidulans heterologous host could not be ruled out. In general, the FAC-MS strategy was beneficial to identify the roles of the C domains in the biosynthesis of benzomalvin A and D, showing that there are two internal C-domains (BenZ-C1 and BenZ-C2) and the terminal cyclization domain BenY-CT.[70]
4. Reductive Cyclization
Typical TE domains catalyze offloading in PKS, NRPS or PKS/NRPS hybrids to generate carboxylic acids or cyclic macrocycles. Another release mechanism sometimes utilized to terminate biosynthetic assembly involves a terminal reductase (R) domain. Originally reported for lysine primary metabolism, [1, 5b, 71] these domains are part of the short-chain dehydrogenase/reductase (SDR) superfamily catalyzing NAD(P)H-dependent reductive release of acyl-S-polyketides or peptidyl-S-nonribosomal peptides. One of the most conserved features of the SDR enzymes is an α/β folding pattern with a central beta sheet flanked by two - three α-helices from each side, which is characteristic of a Rossmann-fold.[72] This key motif is characteristic of a NAD(P)H nucleotide binding site, which then facilitates recruiting of a NAD(P)H/NAD(P)+ redox cofactor.[73] The mechanism relies on a hydride and proton transfer that involves nicotinamide in conjunction with a tyrosine residue in the active site of the enzyme.[73a] R domains enable product release via a catalyzed two-electron or four-electron reduction, resulting in an aldehyde or alcohol functional group, respectively. Interestingly, after a two-electron reduction and subsequent aldehyde formation, the product could undergo a spontaneous cyclization process.[1, 73a] Even though the formal cyclization is generally nonenzymatic, the fact that a reductase catalyzes the release of the product as a reactive aldehyde, which then spontaneously cyclizes, makes these mechanism noteworthy. Many examples of these linear and cyclized compounds are also found in a recent review from the Kelleher and Thomson groups where they discussed compounds produced from reductase terminated BGCs.[5b] Herein, we will focus on the two-electron reduction and spontaneous cyclization catalyzed by these domains.
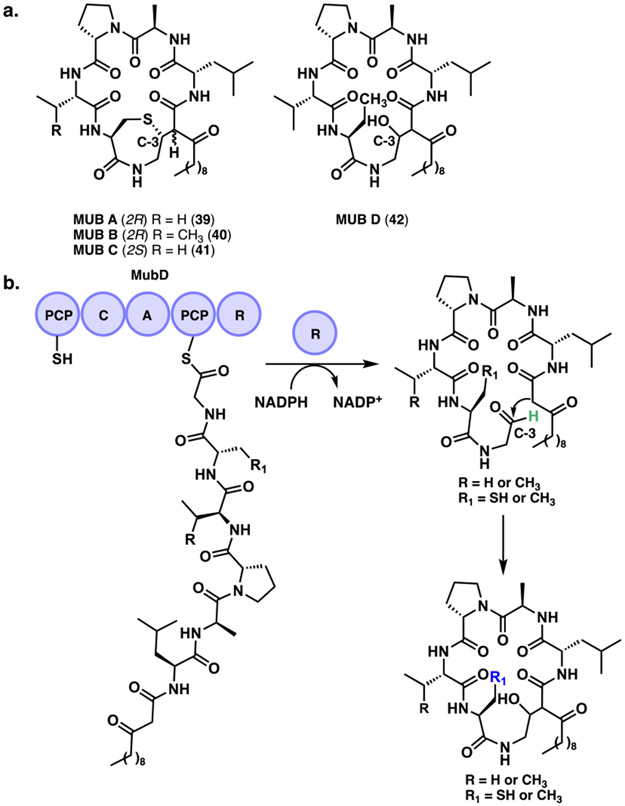
4.1. Mutanobactins
Recently, the Chen group studied the macrocyclic lipohexapeptides mutanobactins A-D (39-42) (MUBs) derived from a PKS/NRPS hybrid pathway (Figure 14a).[74] These compounds were isolated from the human oral bacterium Streptococcus mutans UA159.[75] The mub BGC found in many S. mutans strains is the primary causative agent of tooth decay.[76] The authors found that the C-terminal reductase domain of MubD initially releases the linear peptide as an aldehyde (Figure 14b). From there, a three-step spontaneous process can occur depending on the reactive functional groups leading to either macrocycles or linear derivatives. Initially, an intramolecular aldol addition mediates the C-C macrocyclization event. This intermediate can be further modified spontaneously via the presence of a reactive thiol group that can attack the C-3 position generating the 1,4-thiazepane rings of select MUBs. Finally, in the presence of a hemithioacetal, the macrocyclized β-thiodiketones can be converted to linear lipopeptides through a retro-aldol reaction. These molecules represent an interesting route used by bacteria that takes advantage of biosynthetically formed reactive intermediates to generate complex ring structures.
Figure 14.
a. Mutanobactins A – D. b. Reductase (R) domain mediated release of aldehyde precursor in the biosynthesis of mutanobactins.
4.2. Macrocyclic iminopeptides
Macrocyclic iminopeptides represent another great example of aldehyde offloading mediated by an R domain in which the linear product undergoes spontaneous cyclization.[73a] In 2001, the Moore group discovered the nostocyclopeptides, the first cyclic iminopeptides from the terrestrial cyanobacterium Nostoc sp. ATCC 53789.[77] Despite the biosynthetic cluster terminating in a reductase domain, no linear products were observed. Further studies into the macrocyclization of these compounds, demonstrated that they are offloaded as an aldehyde by the reductase domain (NcpB) and then spontaneously cyclize to form the imine. This was further supported by NMR studies in which the solution phase structure demonstrated the importance of preorganization for cyclization over polymerization and other side reactions.[78]
More recently, other members of this family have been discovered including the scytonemides (whose biosynthesis is currently unknown but is proposed to form via a similar aldehyde amine addition) and the koranimines.[79] In 2011, the Kelleher group discovered the cyclic heptapeptide koranimine (43), from a Bacillus sp. collected in Koran, Louisiana (Figure 15a).[79b] They utilized a scanning microbial proteome mining approach to identify expressed gene clusters. The koranimine structure was determined using tandem mass spectrometry, feeding studies with stable isotope tracers, NMR spectroscopy and in vitro enzyme reconstitution. The gene cluster encodes four NRPS proteins and five adenylation (A) domains. Interestingly, the natural product was determined to be a heptapeptide with an NRPS bearing only five A domains (suggesting it should incorporate only five amino acids) due to their absence from KorA and KorC. It was proposed that a single adenylation domain in KorA serves two carrier protein sites although the exact mechanism remains unknown. Another unusual finding is a domain structure containing a T-T-C tridomain and no linked A domain in KorC. Utilizing a Ppant ejection assay, the investigators determined that the thiolation domains in KorC are both charged by the A domain of KorD. Moreover, the cyclization process is initiated by the R domain on KorD, which performs a two-electron NADPH-dependent reduction of the C-terminal acyl-S-thioester acid to an aldehyde. This is followed by a nonenzymatic macrocyclization event that yields the imine product 43. In 2017, the Baran group confirmed the spontaneous macrocyclization producing koranimine by the synthesis of the aldehyde precursor.[80] An additional example is the novel peptide antibiotic lugdunin (44) that was reported in 2016 as the first compound of a new class of macrocyclic thiazolidine peptides produced by a NRPS system (Figure 15b).[81] This compound has potent antibacterial activity against major human pathogens, which have been analyzed in animal models. Lugdunin was isolated from Staphylococcus lugdunensis IVK28, which prevented S. aureus colonization in the human nasal cavity. Similar to koranimine, lugdunin is comprised of a heptapeptide derived from a five A domain containing NRPS. Their study suggests that in this pathway, the single adenylation domain of LugC is responsible for the iterative activation of three consecutive valine units with alternating L- and D-configurations. The R domain in LugC cleaves the thioester-bound peptide using a NAD(P)H cofactor. The heptapeptide is released from the enzyme complex with a C-terminal aldehyde (L-Val) and is subsequently cyclized by the N-terminal amine (L-Cys) forming a macrocyclic imine, similar to that of koranimine. However, in this example, further nucleophilic attack of the cysteine thiol group generates the unusual 5-membered thiazolidine heterocycle found in 44.
Figure 15.
a. Termination step mediated by the reductase (R) domain in the biosynthesis of koranimine B. b. Final step in the biosynthesis of lugdunin and nucleophilic attack of the cysteine thiol group forming the 5-membered thiazolidine heterocycle.
4.3. Pyrrolobenzodiazepines
Pyrrolobenzodiazepines containing natural products feature a tricyclic ring system formed by an anthranilate (A ring), a diazepine (B ring) and a hydropyrrole (C-ring).[5b, 82] Anthramycin (45) is the first known bacterially NRPS derived benzodiazepine alkaloid with potent antitumor and antibiotic activity (Figure 16). This compound is a pyrrolo [1,4]-benzodiazepine from Streptomyces refuineus sbsp. thermotolerans discovered during the 1960s.[83] During assembly, the dipeptide anthramycin precursor is released by a terminal R domain. Intramolecular addition of the anthranilate-derived arylamine then occurs by adding into the dehydroproline acrylamide aldehyde resulting in the formation of the 7-membered hemiaminal benzodiazepine skeleton of anthramycin.[84] Moreover, tomaymycin (46) is another pyrrolobenzodiazepine structurally similar to anthramycin. It is also released as an aldehyde by an R domain and spontaneously cyclized (Figure 16). This compound is an antitumor antibiotic produced by Streptomyces achromogenes.[85] In 2009, the Gerratana group identified and sequenced the BGC providing insights into A ring formation in tomaymycin.[86] Later, in 2017, the Müller group reported an in vitro reconstitution of the NRPS system including intact protein mass spectrometry analysis, which dissected every step in tomaymycin biogenesis.[87] Their method could be broadly applicable to diverse systems for dissection of the reaction steps in secondary metabolic pathways.
Figure 16.
Structures of anthramycin, tomaymycin, tilivalline and tilimycin.
Our last example of this class is tilivalline (47) produced by an NRPS was isolated from Klebsiella oxytoca found in the human gut microbiota, and represents a marker for diagnosis and treatment of colitis.[88] The Breinbauer and Zechner groups studied the biosynthesis of tilivalline, whose gene cluster did not appear to contain any genes associated with the characteristic indole functionality.[89] Genome analysis revealed a trypticase gene tnaA, which was subsequently inactivated. Cultures of this ΔtnaA K. oxytoca produced a new product, tilimycin (48), containing an alcohol functionality in place of the indole moiety. Further analysis of the biosynthesis of tilimycin shows that the dipeptide is reductively released as an aldehyde via an R domain followed by a spontaneous ring closure. From there, tilimycin was shown to react nonenzymatically with free indole (generated from tryptophan) to generate tilivalline (Figure 16).
5. Dieckmann Cyclases
The family of Dieckmann cyclases were recently found in the biosynthesis of natural products containing tetramate or pyridone scaffolds.[5b, 90] The Dieckmann condensation is an intramolecular Claisen-type reaction of diesters or similar functionalities leading to stable 5- and 6-membered heterocyclic rings.[1] In fungal PKS/NRPS hybrids a reductase-type domain performs a non-redox, NAD(P)H independent Dieckmann condensation carrying out a cyclization. Similarly, in actinomycete-derived tetramic acid and pyridone natural products from bacterial PKS/NRPS hybrids, stand-alone Dieckmann cyclases have been identified as cyclization catalysts for the biosynthesis of these compounds.[90]
5.1. Reductase domain catalyzed Dieckmann Condensation (RD)
Reductase (R) domains have been found to catalyze a non-redox Dieckmann condensation instead of a reductive release of a polyketide or a peptide, as shown previously. In this review, we will refer to these non-redox R domains as RD. As noted previously for NRPS terminal R domains, they enable product release in an aldehyde form, which could then undergo a spontaneous cyclization. However, for these RD domains, the release of the polyketide or peptide occurs directly via a Dieckmann condensation.[1, 5b]
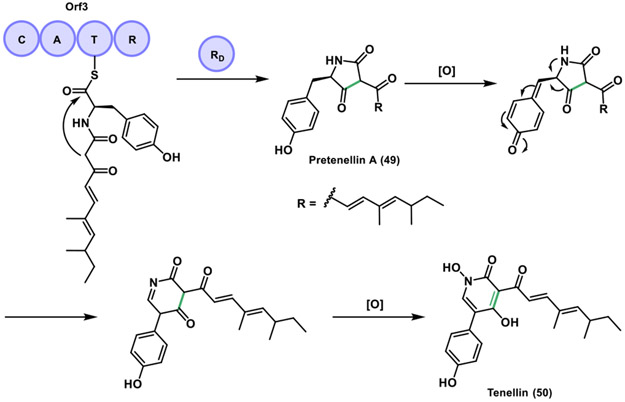
Tenellin (50) is an example of this category, isolated in 1977 from the fungus Beauveria bassiana and produced by a PKS/NRPS hybrid.[91] This compound contains a 2-pyridone ring that derives from a tetramic acid (pyrrolidine-2,4-dione) via a ring expansion. In 2007, it was proposed that the tetramate ring was produced through an R domain catalyzed reductive release to give an aldehyde, which then cyclized non-enzymatically generating pretenellin A (49). Then, this tetramate ring would be further oxidized by tailoring enzymes to afford the 2-pyridone in tenellin.[92] A year later, the tenS gene that encodes the PKS/NRPS tenellin synthetase (TENS) was expressed in the heterologous host Aspergillus oryzae M-2-3.[93] It was determined that the terminal R domain in the tenellin biosynthesis catalyzes a Dieckmann-type cyclization to generate the tetramic acid moiety in pretenellin A, as opposed to a spontaneous cyclization via an off-loaded aldehyde as originally proposed. This intermediate requires two additional oxidations by two BGC-encoded cytochrome P450s to be converted to tenellin through the 2-pyridone (Figure 17).[94]
Figure 17.
Dieckmann cyclization mediated by a reductase domain (RD) in the biosynthesis of tenellin.
Additional tetramic acid containing natural products can be formed from an RD domain such as equisetin (51) and cyclopiazonic acid (52) (Figure 18a). Equisetin was isolated from the fungus Fusarium heterosporum.[95] Later, it was found to be an HIV-1 integrase inhibitor and it also has shown antibacterial properties.[96] Recently, equisetin was shown to inhibit bacterial acetyl-CoA carboxylase (ACC), the first step in fatty acid synthesis.[97] This compound derives from the PKS/NRPS hybrid equisetin synthetase (EqiS) and involves an RD domain mediated cyclization.[98] It also bears a structural similarity with the cholesterol lowering agent lovastatin.[99] In 2008, the Schmidt group overexpressed the EqiS RD domain and studied its reactivity utilizing synthetic substrate analogs. Their results show that the EqiS RD domain catalyzes a Dieckmann condensation rather than a reductive release.[98] Subsequently, they showed that EqiS does not produce equisetin, but instead it produces a 2,4-pyrrolidinedione, fusaridione A (53). This newly identified compound leads to a spontaneous reverse-Dieckmann reaction due to the methylated 3-position of the compound scaffold (Figure 18b). A new gene cluster, eqx was described as responsible for the biosynthesis of equisetin.[100] Another example is cyclopiazonic acid, produced by the fungus Aspergillus flavus, which possesses nanomolar inhibitory activity against Ca2+− ATPase.[101] Similarly, cyclopiazonic acid follows a similar mechanism to equisetin where the CpaS R domain catalyzes a Dieckmann condensation producing the tetramate ring. Interestingly, the Walsh group determined that the CpaS RD domain lacks the Ser-Tyr-Lys catalytic triad found in the SDR superfamily.[102] Their results show a conserved aspartic acid residue (D3803), which may play an important role in the RD domain since mutations abolished the activity.
Figure 18.
a. RD domain in the biosynthesis of equisetin and fusaridione A. The activity of the RD domain results in release of the intermediate as the tetramate, trichosetin. Then, N-methylation is carried out to give equisetin. b. For the case of fusaridione A, the unstable pyrrolidinedione ring is opened through a reverse-Dieckmann reaction. c. Proposed biosynthetic pathway for burnettramic acids A and B.
Burnettramic acids A (54) and B (55) are a new class of antibiotics derived from a Dieckmann cyclization during their biosynthesis (Figure 18c). Recently, the Chooi and Piggott groups studied rare PKS/NRPS derived bolaamphiphilic pyrrolizidinediones, isolated from an Australian fungus Aspergillus burnettii.[103] They reported the discovery, biological investigation and biosynthetic studies of the burnettramic acids. In the proposed pathway, an RD domain releases the polyketide-peptide via a Dieckmann condensation to produce a tetramic acid fused to the hydroxyproline, generating the bicyclic pyrrolidinedione moiety. The C-terminal domain in BuaA was confirmed in their study as a Dieckmann condensation domain, containing a phenylalanine in place of a tyrosine found in the typical catalytic triad of the short chain SDR superfamily.[102, 104]
5.2. Thioesterase-like Dieckmann Cyclases
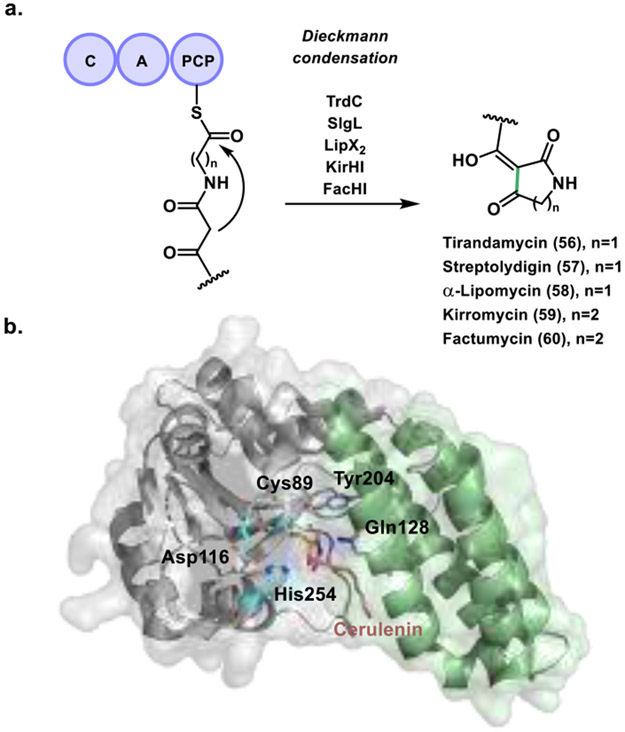
In 2015, the Ju group discovered a new family of Dieckmann cyclases related to actinomycete-derived tetramic acid and pyridone natural products (Figure 19a).[90a] The compounds in this study included tirandamycin B (56), streptolydigin (57), α-lipomycin (58), kirromycin (59), and factumycin (60). Although their biosynthetic assembly lines have been identified, none have been found to contain TE or R domains presumed to produce the characteristic tetramic acid or pyridine moiety. Intrigued by this, the Ju group studied the TrdC, SlgL, LipX2, KirHI, and FacHI group of homologous proteins. Their results showed that this highly conserved new family of enzymes catalyzes formation of tetramic acid and pyridone moieties via a Dieckmann cyclization mechanism. Further investigation into the closest homologs of TrdC revealed that it is most related to FAS, PKS and NRPS TEs. The conserved catalytic triad is also present in TrdC with the typical serine being replaced by a cysteine. Surprisingly, mutation of this catalytic cysteine to serine (C88S) retained cyclase activity. Finally, an analysis of GenBank found that these TE like Dieckmann cyclases are prevalent in bacterial secondary metabolism and may serve as effective genetic probes for identifying bacterially derived tetramic acids and 2-pyridone based natural products.
Figure 19.
Activity and structure of Dieckmann cyclases. a. Dieckmann cyclization reaction in the biosynthesis of five actinomycete-derived tetramic acid and pyridone natural products. b. NcmC-cerulenin complex with the α/β hydrolase subdomain highlighted in grey and the four-helix bundle in green. Catalytic residues important for function are depicted in blue and purple.
More recently, the Nair group identified a Dieckmann cyclase (NcmC) that installs the tetramate headgroup in the PKS/NRPS hybrid derived nocamycin peptide natural product.[90b] They identified a putative biosynthetic pathway for nocamycin I in Saccharothrix syringae NRRL B-16468. By testing a PCP-linked, and a SNAc mimic substrate, they measured NcmC activity in vitro, which revealed that the enzyme catalyzes a Dieckmann condensation reaction. They also reported a 1.6 Å resolution crystal structure of NcmC that provided mechanistic insights about this family of cyclases. Thus, NcmC was found to possess two subdomains, with the first consisting of the characteristic α/β hydrolase fold found in TEs and other hydrolases (Figure 19b, grey). In contrast, the second subdomain consists of a four-helix bundle inserted between strands β5 and β6, which is unique to these Dieckmann cyclases (Figure 19b, green). The α/β hydrolase subdomain contains the common active site catalytic triad with a cysteine residue present instead of a serine (Cys89, Asp116 and His254), a substitution observed in previously discussed TE containing PKS pathways. Although they were unable to capture a native substrate, cerulenin was employed and formed a covalent adduct with the active site, yielding insights into other important active site residues. Identification of Tyr204 and Gln128 located on the unique four helix bundle in conjunction with mutagenesis studies demonstrated that these residues play a crucial role in catalysis. Mutations at both positions showed a marked increase in hydrolytic activity and a corresponding decrease in cyclization. These results coupled with the crystallographic analysis led the authors to hypothesize that Gln128 (positioned next to the thioester bond) is responsible for shielding the thioester intermediate from water mediated hydrolysis. The Tyr204 residue appears to be highly conserved among similar proteins, and likely participates in stabilizing the enol tautomer necessary for nucleophilic attack and cyclization. This is supported by the hydrogen bonding network observed between the tyrosine side chain, a water molecule, and the cerulenin inhibitor.
6. Cyclization by Other Mechanisms
6.1. Penicillin Binding Protein (PBP) Cyclases
Surugamides A-E (61-65) are cyclic octapeptides with four D-amino acids originally isolated from the marine-derived actinomycete Streptomyces sp. JAMM992. These compounds have been shown to have cathepsin B inhibitory bioactivity.[105] Sequencing and genome mining identified the biosynthetic gene cluster responsible for the production of surugamides.[106] The authors reported a putative gene cluster containing four successive NRPS genes, surA, surB, surC and surD, with the surA and surD being responsible for the assembly of surugamides A-E (61-65) as well as surB and surC required to produce surugamide F (66), an unrelated linear decapeptide (Figure 20a).[106] Interestingly, these biosynthetic studies demonstrated a lack of chain termination domain such as a TE, R, or CT encoded by the surD or surC genes. Hence, the enzyme responsible for the chain termination remained unknown.
Figure 20.
a. The fully elongated peptide chains tethered to the last NRPS modules are presumed to be released by SurE to generate cyclopeptides 61–65 and the linear peptide 66.[110] b. Overall Structure of SurE with PBP domain depicted in light grey for the apo form and dark grey for the complexed form, lipocalin like domain in cyan and the linker between the two in purple. Active site residues are depicted in blue with the density for the visible leucine in pink. The loop rearrangement is highlighted in green for the ordered apo form. c. Active site of SurE including comparison of loop region His225 and Met226 in apo (green) and complex structure with D-Leu (grey).
In 2018, the Wakimoto group showed the total synthesis of surugamide B (62) and identified the chain termination enzyme.[107] Their work describes SurE as the candidate offloading enzyme, which is encoded just upstream of the NRPS genes and was originally annotated as a penicillin-binding protein (PBP).[108] Utilizing a recombinant SurE protein and SNAc chain elongation intermediate to mimicking the PCP-bound octapeptide substrate, the authors demonstrated not only that SurE catalyzes peptide cyclization in vitro, but also that it appeared to exhibit a preference for substrates bearing a C-terminal D-amino acid, contrary to the L orientation usually preferred by TEs. Further investigation including in vivo knockouts of the surE gene and testing with alternative substrates revealed that SurE is catalyzes offloading of the surA/surD specified products 61-65, and also the surB/surC product 66. [101, 102] Notably, unlike TEs that are appended to the terminal module of NRPS modules, these PBP like TEs are stand-alone proteins that act in-trans to offload as either cyclic or hydrolyzed products.
Recently, the Abe and Wakimoto groups reported interesting results about the substrate scope of SurE using synthetic substrate derivatives.[109] Their work confirmed a strict requirement for heterochirality between N- and C-terminal residues with L- and D-amino acids, respectively. They also revealed that macrolactamization by canonical cis-acting offloading domains such as type I TE and CT domains is completed via heterochiral coupling with the opposite configuration (D-amino acid at the N-terminus and L-amino acid at the C-terminus). Based on these findings, PBP-type TEs could be employed as a strategy for creating alternative heterochirality in the biosynthesis of cyclopeptides.
In order to gain mechanistic insights, the authors also disclosed a 2.2 Å resolution X-ray crystal structure for this enzyme.[109] This work revealed two subdomains linked by a long disordered loop region: an N-terminal PBP domain containing the typical α/β hydrolase fold seen in TEs and a C-terminal lipocalin like domain comprised of β strands. This C-terminal domain acts as a lid on the PBP subdomain, forming a cleft that houses the active site tetrad Ser63, Lys66, Asn156 and His305. To understand the basis for the unusual C-terminal selectivity, a model of the peptide-O-SurE complex using the residues located within 8 Å of the active-site serine (Ser63) was calculated. Their data suggests that in SurE the D-stereoselectivity in the C-terminus is governed by the oxyanion hole, a side chain-recognizing hydrophobic pocket (Leu231, Ala234, Gly235 and Val309), and the carbonyl-recognizing Arg446. In addition, substrate recognition studies were conducted using the crystal of apo-SurE protein soaked with a substrate that cannot be cyclized, which enabled capture of a peptide-O-SurE complex. Density for a single amino acid was well defined (with the rest weakly visible) attached to the active site serine. Analysis of the complex vs the apo structures revealed a conformational rearrangement of a flexible loop (Figure 20b, green loop) induced by the formation of the peptidyl-O-SurE complex due to interactions with the tethered peptide. This rearrangement however, caused a shift in His225 and Met226 (Figure 20c), which now occupy the presumed hydrophobic pocket that was identified in the simulations, contradicting that original hypothesis. Although the exact mechanism for the C-terminal recognition remains elusive, these studies overall show the potential of SurE has a compelling candidate for synthetic biology applications and developing new methodology towards the biocatalytic production of NRPS based macrocycles.[109]
6.2. Product Template Domains (PTs) in Fungi
The PT domain in fungal NR-PKS controls the aldol cyclization of poly-β-ketone intermediates. These oligoketides are assembled upstream in the β-ketoacyl synthase (KS) catalyzing intramolecular closure to cyclic and fused polycyclic compounds.[111] In 2008, the Kelleher and Townsend groups showed the PT domain present in the NR-PKS PksA from the aflatoxin B1 biosynthesis in Aspergillus parasiticus. Their results demonstrated that a PT domain unites with the KS and TE in the iterative PKS to assemble precisely seven malonyl-derived building blocks to a hexanoyl starter unit and mediates a specific cyclization cascade (Figure 21).[50a]
Figure 21.
PT domain in the NR-PKS PksA for the biosynthesis of aflatoxin B1.
In 2009, the Tsai and Townsend groups solved the crystal structure of the PT domain at 1.8 Å resolution and also conducted mutational studies in the PT domain from PksA.[112] They found that the PT structure displays a "double hot dog" (DHD) fold and differs from those observed in the dehydratase (DH) domains in animal FASs and bacterial PKSs.[113] The DHD fold controls cyclization specificity, which is one of the key programmed steps in aromatic-polyketide biosynthesis.[50a, 114] Additional data provided some insights on the possible origins of the PT domains. Interestingly, native PAGE analysis of the oligomerization states coupled with previous phylogenetic results[115] suggested that the dimeric PT domains evolved from ancient DH domains in reducing PKSs and would be positioned at the DH locus in animal FAS.
Furthermore, docking and mutagenesis studies in the PT domain allowed identification of important residues for substrate binding and catalysis. It also identified the presence of a phosphopantetheine localization channel and a deep two-part interior binding pocket and reaction chamber. The proposed mechanism in this study suggested the ACP-bound linear substrate 67 binds to the PT substrate pocket in an extended conformation. His1345 acting together with Asp1543 (hydrogen bonded to Gln1547) functions as the key catalytic base deprotonating C4, leading to the enolate intermediate stabilized by the backbone N-H of Asn1568. The enolate subsequently collapses followed by C4 aldol addition to carbonyl C9 (C4-C9 cyclization event) leading to the first ring cyclization product 68 with a network of water molecules bound to Ser1356, Asp1543, Asn1568 and Thr1546 stabilizing the oxyanion. The second ring is formed in the same manner as the first to achieve C2-C11 closure in 70 (Figure 21).
In 2017, Townsend, Burkart and Tsai provided new insights about the PT domain function through crystallographic applications of "atom replacement" mimetics.[111] Isoxazole rings linked by thioethers were employed to mimic the alternating sites of carbonyls in the poly-β-ketone intermediates. In their work, they utilized 4’-phosphorylated and unphosphorylated atom-replaced mimetics, which were selected to represent substrate chain lengths of 8-16 carbon units of the linear and proposed monocyclic intermediates from aflatoxin biosynthesis. A 1.8 Å resolution co-crystal structure of the PksA PT domain was presented with a 4′-phosphopantetheinylated linear heptaketide mimetic. In summary, using structural biology, structure-based mutagenesis, and enzymatic assays the collaborative team identified key residues involved in substrate recognition that have been difficult to identify due to oligoketide substrate instability and inherent reactivity. They observed that the heptaketide C4 carbon is positioned proximal to the catalytic His1345, providing further evidence to the previously proposed mechanism of polyketide cyclization mediated by the PT domain. Also, the role of the protein-coordinated water network to selectively activate the C9 carbonyl for nucleophilic addition was shown as well as the importance of the 4’-phosphate distal to the pantetheine arm to deliver and anchor the heptaketide mimetic into the PT active site. Moreover, atom-replacement mimetics proved to be an alternative way to probe other PKS enzymes (e.g. bacterial type II PKS) together with highly reactive substrates/intermediates, which are challenging or impossible to capture using X-ray crystallography.
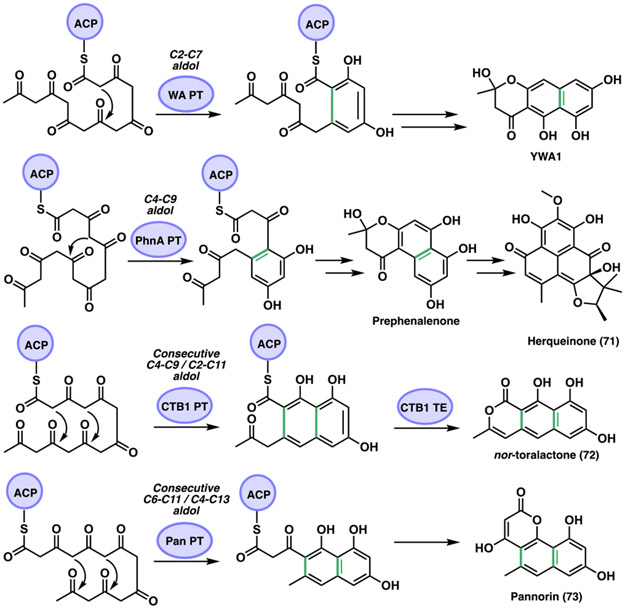
Another interesting study of the fungal PKS PT domains was reported by the Houk and Tang groups.[50b] They investigated the biosynthesis of herqueinone (71) isolated from Penicillium herquei (Figure 22). This compound forms part of the phenalenone family of natural products that contains a peri-fused tricyclic ring core system cyclized from a linear polyketide precursor. 71 is formed after the phenalenone is heavily derivatized and oxidized. The identified gene cluster is phn from P. herquei containing an encoded NR-PKS (PhnA), that contains SAT-KS-MAT-PT-ACP-ACP-TE/CLC domains. Additional genes encode a flavin-dependent monooxygenase (FMO, PhnB and PhnG), an O-methyltransferase (O-MT, PhnC), and a prenyltransferase (PrT, PhnF). In their work, the Houk and Tang team demonstrated that the NR-PKS PhnA is responsible for the synthesis of a heptaketide backbone as well as for its cyclization into a hemiketal-containing naphtho-γ-pyrone prephenalenone. They also found that the FMO (PhnB) catalyzes a C2 aromatic hydroxylation of the prephenalenone NR-PKS product converting it into phenalenone followed by a simultaneous ring opening of the γ-pyrone ring. Density functional theory (DFT) calculations were presented that supported their mechanistic hypothesis. In addition, they compared prephenalenone with different NR-PKS shown to synthesize the common heptaketide precursor that is cyclized into diverse final products due to PT domain regioselectivity (Figure 22). The NR-PKS derived naphthopyrone products in the analysis include 1) YWA1 from Aspergillus nidulans wA[52a, 116] (WA, NR-PKS) 2) nor-toralactone (72) from Cercospora nicotianae[117] (CTB1, NR-PKS) and 3) pannorin (73) from Chrysosporium pannorum[118] M10539 (Pan, NR-PKS). PT catalyzed cyclization of oligoketide substrates has been demonstrated to perform consecutive C4-C9 and C2-C11 intramolecular aldol condensations.[50a, 117a] In addition, they showed that the PT domain in PhnA catalyzes only the C4-C9 aldol condensation, while the TE/CLC catalyzes a C1-C10 cyclization. Both PhnA and WA produce a heptaketide compound and catalyze an intramolecular aldol condensation and a C1-C10 Claisen cyclization generating the corresponding naphthopyrone products. A difference between them is the PT regioselectivity, which for the case of WA a C2-C7 cyclization leads to YWA1 and for PhnA a C4-C9 cyclization results in prephenalenone. In the CTB1 system, the PT domain catalyzes tandem C4-C9 and C2-C11 aldol cyclization reactions that produce nor-toralactone (72),[117a] which is similar to the PT domain in PksA described above.[112] Lastly, the NR-PKS for pannorin (73) has not been identified, however, the PT domain has been suggested to catalyze consecutive C6-C11 and C4-C13 cyclization reactions.[119] In general, the PT domain in the NR-PKS PhnA is functionally distinct since it catalyzes only a C4-C9 cyclization compared to the corresponding PT homologs that catalyze C4-C9 or C2-C11 cyclizations.
Figure 22.
PT domain cyclization regioselectivities in NR-PKSs.
7. Summary and Outlook
In this review we describe the fascinating work reported on peptide and polyketide biosynthetic cyclization catalysts during the past decade. We presented several examples of new cyclic structures with interesting biological activities and summarized the recent findings on the diversity of enzymes that catalyze cyclization of linear peptides or polyketide chains. These enzymes possess structural and mechanistic diversity including cyclization mediated by thioesterase (TE) domains, oxidative, reductive, condensation (CT domains and Dieckmann cyclases) and other mechanisms.
Initial attention was focused on thioesterases (TEs), which have been studied extensively throughout the past two - three decades. Even though TEs are relatively well understood in comparison with other cyclization catalysts, additional NRPS and PKS BGCs were recently characterized, showing the production of cyclic lactone and lactam structures mediated by a TE domain.[15a, 16, 18, 21-22, 24] Examples of these TE cyclases are derived from obafluorin, salinamide A, lysobactin, sulfazecin, polymyxin B and fluvirucin B1. In some cases, a cysteine instead of a serine active site residue was found[15a, 16, 22, 24]; in addition to the unusual tandem TE architecture employed by lysobactin and teixobactin biosynthesis.[21, 31] Furthermore, chemoenzymatic syntheses[26, 31, 33-36] and pathway engineering in these systems[37-40] represent leading examples for the synthesis of novel compounds. TE domain oligomerization[43-44, 47-48, 120] and C-C bond forming Claisen-Like Cyclases[15b, 50, 53] (CLCs) were also described, which reflect the mechanistic diversity of these domains. The remarkable achievements toward understanding these systems show the potential of these cyclization biocatalysts. In the future, mechanistic, structural insights (X-ray crystallography and cryo-electron microscopy) and protein engineering will guide the path for deeper exploration of the capabilities of the variant TEs for the synthesis of numerous natural products and bioactive, high value analogs.
In addition to canonical TE domains, we also discussed functionally similar enzymes that perform a cyclization via a condensation mechanism (CT domains and R domain catalyzed Dieckmann condensation) or the release of a linear chain substrate as an aldehyde (terminal R domains), followed by spontaneous cyclization. For the fungal NRPS CT domains, phylogenetic analysis, mechanistic and structural investigations have provided significant information about these terminal domains.[56b, 59, 62c, 66, 70] However, only a few reports are available on CT domains, suggesting that further studies are needed on fungal NRPSs to explore their full biocatalytic potential. For the case of terminal R domains, remarkable work has been reported on these enzymes. However, it would be interesting to understand mechanistically how the initial product is compelled to cyclize upon reductive release,[5b, 74, 79b, 81, 87, 89] and similarly the initial product of a non-redox Dieckmann condensation[90a, 94, 98, 102-103] leading to cyclization. Structural elucidation of NRPS R domains could further elucidate linear off-loading or cyclization routes for these homologous SDR enzymes. Lastly, cyclization mediated by other mechanisms were also described. The PBP cyclases and product template (PT) domains are a pronounced example of additional enzymes in NRPS and PKS systems with cyclization chemistry. Recent crystal structures and mechanistic studies have demonstrated the impressive strategies resulting in cyclization of secondary metabolites.[50b, 109, 111-112] Future approaches using genome mining for the discovery of new biocatalysts, as well as protein engineering efforts on NRPS and PKS mega-enzyme systems are ripe with potential for the production of pharmaceutically relevant peptide and polyketide derived natural products.
Acknowledgements
We gratefully acknowledge NIH grant R35 GM118101, and the Hans W. Vahlteich Professorship (D.H.S.). We also thank Rajani Arora for the design of our frontispiece.
Biography

Maria L. Adrover-Castellano graduated from the University of Puerto Rico-Rio Piedras with a B.S. in chemistry in 2018. Soon after joining the Ph.D. Program in Chemical Biology at the University of Michigan-Ann Arbor, she started working under the supervision of Prof. David Sherman. Currently, she explores chemo-enzymatic synthesis of macrolactones, and macrolides utilizing polyketide synthases (PKSs) and applies enzyme bioengineering approaches for the synthesis of these complex molecules.

Jennifer J. Schmidt received her B.S. in Biochemistry and Molecular Biology at UC Santa Cruz followed by a Ph.D. in Medicinal Chemistry under the supervision of Prof. David Sherman. Her work focuses on the merging of synthetic chemistry and biocatalysis for the production of unnatural polyketide and nonribosomal peptide molecules of interest.

David H. Sherman received his B.A. in chemistry at UC Santa Cruz and Ph.D. at Columbia University. Following 13 years at the University of Minnesota, Prof. Sherman moved to the University of Michigan and is now the Hans W. Vahlteich Professor of Medicinal Chemistry, Professor of Chemistry, and Professor of Microbiology & Immunology. Sherman’s laboratory is in the U-M Life Sciences Institute where his research focuses on the discovery and analysis of bioactive natural products and their metabolic pathways from diverse bacteria and fungi.
References
- [1].Du L, Lou L, Nat. Prod. Rep 2010, 27, 255–278. [DOI] [PubMed] [Google Scholar]
- [2].Williams GJ, Curr. Opin. Struc. Biol 2013, 23, 603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hwang S, Lee N, Cho S, Palsson B, Cho BK, Front. Mol. Biosci 2020, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ansari MZ, Yadav G, Gokhale RS, Mohanty D, Nucleic Acids Res. 2004, 32, W405–W413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].a) Horsman ME, Hari TPA, Boddy CN, Nat. Prod. Rep 2016, 33, 183–202 [DOI] [PubMed] [Google Scholar]; b) Mullowney MW, McClure RA, Robey MT, Kelleher NL, Thomson RJ, Nat. Prod. Rep 2018, 35, 847–878 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Tang MC, Zou Y, Watanabe K, Walsh CT, Tang Y, Chem. Rev 2017, 117, 5226–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Weissman KJ, Nat. Prod. Rep 2016, 33, 203–230. [DOI] [PubMed] [Google Scholar]
- [7].Cantu DC, Chen Y, Reilly PJ, Protein Sci 2010, 19, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kotowska M, Pawlik K, Appl. Microbiol. Biotechnol 2014, 98, 7735–7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Bruner SD, Weber T, Kohli RM, Schwarzer D, Marahiel MA, Walsh CT, Stubbs MT, Structure 2002, 10, 301–310 [DOI] [PubMed] [Google Scholar]; b) Miller BR, Gulick AM, in Nonribosomal Peptide and Polyketide Biosynthesis: Methods in Molecular Biology Vol. 1401 (Ed.: Evans B), Humana Press, New York, 2016, pp. 3–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Loll PJ, Upton EC, Nahoum V, Economou NJ, Cocklin S, Biochim Biophys Acta 2014, 1838, 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Samel SA, Wagner B, Marahiel MA, Essen LO, J Mol Biol 2006, 359, 876–889. [DOI] [PubMed] [Google Scholar]
- [12].a) Tsai SC, Miercke LJ, Krucinski J, Gokhale R, Chen JC, Foster PG, Cane DE, Khosla C, Stroud RM, Proc. Natl. Acad Sci. U.S.A 2001, 98, 14808–14813 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Tsai SC, Lu H, Cane DE, Khosla C, Stroud RM, Biochemistry 2002, 41, 12598–12606. [DOI] [PubMed] [Google Scholar]
- [13].Akey DL, Kittendorf JD, Giraldes JW, Fecik RA, Sherman DH, Smith JL, Nat. Chem. Biol. 2006, 2, 537–542. [DOI] [PubMed] [Google Scholar]
- [14].Trauger JW, Kohli RM, Walsh CT, Biochemistry 2001, 40, 7092–7098. [DOI] [PubMed] [Google Scholar]
- [15].a) Galea CA, Roberts KD, Zhu Y, Thompson PE, Li J, Velkov T, Biochemistry 2017, 56, 657–668 [DOI] [PubMed] [Google Scholar]; b) Korman TP, Crawford JM, Labonte JW, Newman AG, Wong J, Townsend CA, Tsai SC, Proc. Natl. Acad. Sci. U.S.A 2010, 107, 6246–6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Schaffer JE, Reck MR, Prasad NK, Wencewicz TA, Nat. Chem. Biol 2017, 13, 737–744. [DOI] [PubMed] [Google Scholar]
- [17].a) Wells JS, Trejo WH, Principe PA, Sykes RB, J. Antibiot. (Tokyo) 1984, 37, 802–803 [DOI] [PubMed] [Google Scholar]; b) Tymiak AA, Culver CA, Malley MF, Gougoutas JZ, J. Org. Chem 1985, 50, 5491–5495. [Google Scholar]
- [18].Ray L, Yamanaka K, Moore BS, Angew. Chem. Int. Ed 2016, 55, 364–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Trischman JA, Tapiolas DM, Jensen PR, Dwight R, Fenical W, Mckee TC, Ireland CM, Stout TJ, Clardy J, J. Am. Chem. Soc 1994, 116, 757–758. [Google Scholar]
- [20].a) Bonner DP, Osullivan J, Tanaka SK, Clark JM, Whitney RR, J. Antibiot. (Tokyo) 1988, 41, 1745–1751 [DOI] [PubMed] [Google Scholar]; b) Osullivan J, Mccullough JE, Tymiak AA, Kirsch DR, Trejo WH, Principe PA, J. Antibiot. (Tokyo) 1988, 41, 1740–1744. [DOI] [PubMed] [Google Scholar]
- [21].Hou J, Robbel L, Marahiel MA, Chem. Biol 2011, 18, 655–664. [DOI] [PubMed] [Google Scholar]
- [22].Oliver RA, Li RF, Townsend CA, Nat. Chem. Biol 2018, 14, 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Asai M, Haibara K, Muroi M, Kintaka K, Kishi T, J. Antibiot. (Tokyo) 1981, 34, 621–627. [DOI] [PubMed] [Google Scholar]
- [24].Lin TY, Borketey LS, Prasad G, Waters SA, Schnarr NA, ACS Synth. Biol 2013, 2, 635–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].a) Naruse N, Konishi M, Oki T, Inouye Y, Kakisawa H, J. Antibiot. (Tokyo) 1991, 44, 756–761 [DOI] [PubMed] [Google Scholar]; b) Naruse N, Tenmyo O, Kawano K, Tomita K, Ohgusa N, Miyaki T, Konishi M, Oki T, J. Antibiot. (Tokyo) 1991, 44, 733–740 [DOI] [PubMed] [Google Scholar]; c) Naruse N, Tsuno T, Sawada Y, Konishi M, Oki T, J. Antibiot. (Tokyo) 1991, 44, 741–755. [DOI] [PubMed] [Google Scholar]
- [26].Schmidt JJ, Khatri Y, Brody SI, Zhu C, Pietraszkiewicz H, Valeriote FA, Sherman DH, ACS Chem. Biol 2020, 15, 524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Schwartz RE, Hirsch CF, Sesin DF, Flor JE, Chartrain M, Fromtling RE, Harris GH, Salvatore MJ, Liesch JM, Yudin K, J. Ind. Microbiol 1990, 5, 113–123. [Google Scholar]
- [28].Smith CD, Zhang XQ, Mooberry SL, Patterson GML, Moore RE, Cancer Res. 1994, 54, 3779–3784. [PubMed] [Google Scholar]
- [29].Magarvey NA, Beck ZQ, Golakoti T, Ding Y, Huber U, Hemscheidt TK, Abelson D, Moore RE, Sherman DH, ACS Chem. Biol 2006, 1, 766–779. [DOI] [PubMed] [Google Scholar]
- [30].Verma VA, Pillow TH, DePalatis L, Li G, Phillips GL, Polson AG, Raab HE, Spencer S, Zheng B, Bioorg. Med. Chem. Lett 2015, 25, 864–868. [DOI] [PubMed] [Google Scholar]
- [31].Mandalapu D, Ji X, Chen J, Guo C, Liu WQ, Ding W, Zhou J, Zhang Q, J. Org. Chem 2018, 83, 7271–7275. [DOI] [PubMed] [Google Scholar]
- [32].Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schaberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K, Nature 2015, 517, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hansen DA, Rath CM, Eisman EB, Narayan AR, Kittendorf JD, Mortison JD, Yoon YJ, Sherman DH, J. Am. Chem. Soc 2013, 135, 11232–11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hansen DA, Koch AA, Sherman DH, J. Am. Chem. Soc 2017, 139, 13450–13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Heberlig GW, Wirz M, Wang M, Boddy CN, Org. Lett 2014, 16, 5858–5861. [DOI] [PubMed] [Google Scholar]
- [36].Heberlig GW, Brown JTC, Simard RD, Wirz M, Zhang W, Wang M, Susser LI, Horsman ME, Boddy CN, Org. Biomol. Chem 2018, 16, 5771–5779. [DOI] [PubMed] [Google Scholar]
- [37].Koch AA, Hansen DA, Shende VV, Furan LR, Houk KN, Jimenez-Oses G, Sherman DH, J. Am. Chem. Soc 2017, 139, 13456–13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Koch AA, Schmidt JJ, Lowell AN, Hansen DA, Coburn KM, Chemler JA, Sherman DH, Angew. Chem. Int. Ed 2020, 59, 13575–13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Qiao L, Fang J, Zhu P, Huang H, Dang C, Pang J, Gao W, Qiu X, Huang L, Li Y, Protein J 2019, 38, 658–666. [DOI] [PubMed] [Google Scholar]
- [40].Tripathi A, Choi SS, Sherman DH, Kim ES, J. Ind. Microbiol. Biotechnol 2016, 43, 1189–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gehring AM, Mori I, Walsh CT, Biochemistry 1998, 37, 2648–2659. [DOI] [PubMed] [Google Scholar]
- [42].Hoyer KM, Mahlert C, Marahiel MA, Chem Biol 2007, 14, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Huguenin-Dezot N, Alonzo DA, Heberlig GW, Mahesh M, Nguyen DP, Dornan MH, Boddy CN, Schmeing TM, Chin JW, Nature 2019, 565, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Heberlig GW, Boddy CN, J. Nat. Prod 2020, 83, 1990–1997. [DOI] [PubMed] [Google Scholar]
- [45].a) Agata N, Mori M, Ohta M, Suwan S, Ohtani I, Isobe M, FEMS Microbiol. Lett 1994, 121, 31–34 [DOI] [PubMed] [Google Scholar]; b) Agata N, Ohta M, Mori M, Isobe M, FEMS Microbiol. Lett 1995, 129, 17–19. [DOI] [PubMed] [Google Scholar]
- [46].Marxen S, Stark TD, Rutschle A, Lucking G, Frenzel E, Scherer S, Ehling-Schulz M, Hofmann T, Sci. Rep 2015, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhou YJ, Prediger P, Dias LC, Murphy AC, Leadlay PF, Angew. Chem 2015, 54, 5232–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhou YJ, Murphy AC, Samborskyy M, Prediger P, Dias LC, Leadlay PF, Chem. Biol 2015, 22, 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Westley JW, Liu CM, Evans RH, Blount JF, J. Antibiot. (Tokyo) 1979, 32, 874–877. [DOI] [PubMed] [Google Scholar]
- [50].a) Crawford JM, Thomas PM, Scheerer JR, Vagstad AL, Kelleher NL, Townsend CA, Science 2008, 320, 243–246 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gao SS, Duan A, Xu W, Yu P, Hang L, Houk KN, Tang Y, J. Am. Chem. Soc 2016, 138, 4249–4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhou H, Li Y, Tang Y, Nat. Prod. Rep 2010, 27, 839–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].a) Fujii I, Watanabe A, Sankawa U, Ebizuka Y, Chem. Biol 2001, 8, 189–197 [DOI] [PubMed] [Google Scholar]; b) Awakawa T, Yokota K, Funa N, Doi F, Mori N, Watanabe H, Horinouchi S, Chem. Biol 2009, 16, 613–623. [DOI] [PubMed] [Google Scholar]
- [53].Vagstad AL, Hill EA, Labonte JW, Townsend CA, Chem. Biol 2012, 19, 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Takano Y, Kubo Y, Shimizu K, Mise K, Okuno T, Furusawa I, Mol. Gen. Genet 1995, 249, 162–167. [DOI] [PubMed] [Google Scholar]
- [55].a) Fujii I, Mori Y, Watanabe A, Kubo Y, Tsuji G, Ebizuka Y, Biochemistry 2000, 39, 8853–8858 [DOI] [PubMed] [Google Scholar]; b) Fujii I, Mori Y, Watanabe A, Kubo Y, Tsuji G, Ebizuka Y, Biosci. Biotechnol. Biochem 1999, 63, 1445–1452. [DOI] [PubMed] [Google Scholar]
- [56].a) McErlean M, Overbay J, Van Lanen S, J. Ind. Microbiol. Biotechnol 2019, 46, 493–513 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gao X, Haynes SW, Ames BD, Wang P, Vien LP, Walsh CT, Tang Y, Nat. Chem. Biol 2012, 8, 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Sieber SA, Marahiel MA, Chem. Rev 2005, 105, 715–738 [DOI] [PubMed] [Google Scholar]; d) Fischbach MA, Walsh CT, Chem. Rev 2006, 106, 3468–3496. [DOI] [PubMed] [Google Scholar]
- [57].a) Berry D, Mace W, Grage K, Wesche F, Gore S, Schardl CL, Young CA, Dijkwel PP, Leuchtmann A, Bode HB, Scott B, Proc. Natl. Acad. Sci. U.S.A 2019, 116, 25614–25623 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kopp F, Marahiel MA, Nat. Prod. Rep 2007, 24, 735–749 [DOI] [PubMed] [Google Scholar]; c) von Döhren H, Fungal Genet. Biol 2009, 46 Suppl 1, S45–S52. [DOI] [PubMed] [Google Scholar]
- [58].Haynes SW, Gao X, Tang Y, Walsh CT, J. Am. Chem. Soc 2012, 134, 17444–17447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhang J, Liu N, Cacho RA, Gong Z, Liu Z, Qin W, Tang C, Tang Y, Zhou J, Nat. Chem. Biol 2016, 12, 1001–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].a) Bergendahl V, Linne U, Marahiel MA, Eur. J. Biochem 2002, 269, 620–629 [DOI] [PubMed] [Google Scholar]; b) Keating TA, Marshall CG, Walsh CT, Keating AE, Nat. Struct. Biol 2002, 9, 522–526. [DOI] [PubMed] [Google Scholar]
- [61].a) Cui CB, Kakeya H, Okada G, Onose R, Osada H, J. Antibiot. (Tokyo) 1996, 49, 527–533 [DOI] [PubMed] [Google Scholar]; b) Wang Y, Li ZL, Bai J, Zhang LM, Wu X, Zhang L, Pei YH, Jing YK, Hua HM, Chem. Biodivers 2012, 9, 385–393 [DOI] [PubMed] [Google Scholar]; c) Scharf DH, Brakhage AA, Mukherjee PK, Environ. Microbiol 2016, 18, 1096–1109 [DOI] [PubMed] [Google Scholar]; d) Wang X, Li Y, Zhang X, Lai D, Zhou L, Molecules 2017, 22, 2026. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Nilov DK, Yashina KI, Gushchina IV, Zakharenko AL, Sukhanova MV, Lavrik OI, Svedas VK, Biochemistry (Moscow) 2018, 83, 152–158. [DOI] [PubMed] [Google Scholar]
- [62].a) Scharf DH, Heinekamp T, Remme N, Hortschansky P, Brakhage AA, Hertweck C, Appl. Microbiol. Biotechnol 2012, 93, 467–472 [DOI] [PubMed] [Google Scholar]; b) Balibar CJ, Walsh CT, Biochemistry 2006, 45, 15029–15038 [DOI] [PubMed] [Google Scholar]; c) Baccile JA, Le HH, Pfannenstiel BT, Bok JW, Gomez C, Brandenburger E, Hoffmeister D, Keller NP, Schroeder FC, Angew. Chem 2019, 58, 14589–14593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Cramer RA Jr., Gamcsik MP, Brooking RM, Najvar LK, Kirkpatrick WR, Patterson TF, Balibar CJ, Graybill JR, Perfect JR, Abraham SN, Steinbach WJ, Eukaryot. Cell 2006, 5, 972–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].a) Scharf DH, Remme N, Habel A, Chankhamjon P, Scherlach K, Heinekamp T, Hortschansky P, Brakhage AA, Hertweck C, J. Am. Chem. Soc 2011, 133, 12322–12325 [DOI] [PubMed] [Google Scholar]; b) Scharf DH, Chankhamjon P, Scherlach K, Heinekamp T, Willing K, Brakhage AA, Hertweck C, Angew. Chem. Int. Ed 2013, 52, 11092–11095 [DOI] [PubMed] [Google Scholar]; c) Dolan SK, O'Keeffe G, Jones GW, Doyle S, Trends Microbiol. 2015, 23, 419–428 [DOI] [PubMed] [Google Scholar]; d) Chang SL, Chiang YM, Yeh HH, Wu TK, Wang CC, Bioorg. Med. Chem. Lett 2013, 23, 2155–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].a) Chiaroni A, Randriambola L, Riche C, Husson HP, J. Am. Chem. Soc 1980, 102, 5920–5921 [Google Scholar]; b) Hu XL, Bian XQ, Wu X, Li JY, Hua HM, Pei YH, Han AH, Bai J, Tetrahedron Lett. 2014, 55, 3864–3867 [Google Scholar]; c) Kitajima M, Nakazawa M, Wu YQ, Kogure N, Zhang RP, Takayama H, Tetrahedron 2016, 72, 6692–6696. [Google Scholar]
- [66].a) Li H, Gilchrist CLM, Phan CS, Lacey HJ, Vuong D, Moggach SA, Lacey E, Piggott AM, Chooi YH, J. Am. Chem. Soc 2020, 142, 7145–7152 [DOI] [PubMed] [Google Scholar]; b) Lacey HJ, Gilchrist CLM, Crombie A, Kalaitzis JA, Vuong D, Rutledge PJ, Turner P, Pitt JI, Lacey E, Chooi YH, Piggott AM, Beilstein J. Org. Chem 2019, 15, 2631–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sun HH, Barrow CJ, Sedlock DM, Gillum AM, Cooper R, J. Antibiot. (Tokyo) 1994, 47, 515–522. [DOI] [PubMed] [Google Scholar]
- [68].Clevenger KD, Bok JW, Ye R, Miley GP, Verdan MH, Velk T, Chen C, Yang K, Robey MT, Gao P, Lamprecht M, Thomas PM, Islam MN, Palmer JM, Wu CC, Keller NP, Kelleher NL, Nat. Chem. Biol 2017, 13, 895–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Bok JW, Ye R, Clevenger KD, Mead D, Wagner M, Krerowicz A, Albright JC, Goering AW, Thomas PM, Kelleher NL, Keller NP, Wu CC, BMC Genomics 2015, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Clevenger KD, Ye R, Bok JW, Thomas PM, Islam MN, Miley GP, Robey MT, Chen C, Yang K, Swyers M, Wu E, Gao P, Wu CC, Keller NP, Kelleher NL, Biochemistry 2018, 57, 3237–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ehmann DE, Gehring AM, Walsh CT, Biochemistry 1999, 38, 6171–6177. [DOI] [PubMed] [Google Scholar]
- [72].Kavanagh KL, Jornvall H, Persson B, Oppermann U, Cell. Mol. Life Sci 2008, 65, 3895–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].a) Kavanagh K, Jornvall H, Persson B, Oppermann U, Cell. Mol. Life Sci 2008, 65, 3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Selles Vidal L, Kelly CL, Mordaka PM, Heap JT, Biochim. Biophys. Acta Proteins Proteomics 2018, 1866, 327–347. [DOI] [PubMed] [Google Scholar]
- [74].Wang M, Xie Z, Tang S, Chang EL, Tang Y, Guo Z, Cui Y, Wu B, Ye T, Chen Y, Org. Lett 2020, 22, 960–964. [DOI] [PubMed] [Google Scholar]
- [75].a) Joyner PM, Liu J, Zhang Z, Merritt J, Qi F, Cichewicz RH, Org. Biomol. Chem 2010, 8, 5486–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang X, Du L, You J, King JB, Cichewicz RH, Org. Biomol. Chem 2012, 10, 2044–2050. [DOI] [PubMed] [Google Scholar]
- [76].a) Wu C, Cichewicz R, Li Y, Liu J, Roe B, Ferretti J, Merritt J, Qi F, Appl. Environ. Microbiol 2010, 76, 5815–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mousa WK, Athar B, Merwin NJ, Magarvey NA, Nat. Prod. Rep 2017, 34, 1302–1331. [DOI] [PubMed] [Google Scholar]
- [77].Golakoti T, Yoshida WY, Chaganty S, Moore RE, J. Nat. Prod 2001, 64, 54–59. [DOI] [PubMed] [Google Scholar]
- [78].a) Becker JE, Moore RE, Moore BS, Gene 2004, 325, 35–42 [DOI] [PubMed] [Google Scholar]; b) Kopp F, Mahlert C, Grunewald J, Marahiel MA, J. Am. Chem. Soc 2006, 128, 16478–16479 [DOI] [PubMed] [Google Scholar]; c) Enck S, Kopp F, Marahiel MA, Geyer A, ChemBioChem 2008, 9, 2597–2601. [DOI] [PubMed] [Google Scholar]
- [79].a) Krunic A, Vallat A, Mo S, Lantvit DD, Swanson SM, Orjala J, J. Nat. Prod 2010, 73, 1927–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Evans BS, Ntai I, Chen Y, Robinson SJ, Kelleher NL, J. Am. Chem. Soc 2011, 133, 7316–7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Malins LR, deGruyter JN, Robbins KJ, Scola PM, Eastgate MD, Ghadiri MR, Baran PS, J. Am. Chem. Soc 2017, 139, 5233–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, Burian M, Schilling NA, Slavetinsky C, Marschal M, Willmann M, Kalbacher H, Schittek B, Brotz-Oesterhelt H, Grond S, Peschel A, Krismer B, Nature 2016, 535, 511–516. [DOI] [PubMed] [Google Scholar]
- [82].Huang T, Wang X, Guo W, Lin S, in Comprehensive Natural Products III (Eds.: Liu HW, Begley TP), Elsevier, 2020, pp. 393–445. [Google Scholar]
- [83].a) Tendler MD, Korman S, Nature 1963, 199, 501. [DOI] [PubMed] [Google Scholar]; b) Leimgruber W, Stefanovic V, Schenker F, Karr A, Berger J, J. Am. Chem. Soc 1965, 87, 5791–5793 [DOI] [PubMed] [Google Scholar]; c) Leimgruber W, Batcho AD, Schenker F, J. Am. Chem. Soc 1965, 87, 5793–5795. [DOI] [PubMed] [Google Scholar]
- [84].Hu Y, Phelan V, Ntai I, Farnet CM, Zazopoulos E, Bachmann BO, Chem. Biol 2007, 14, 691–701. [DOI] [PubMed] [Google Scholar]
- [85].Arima K, Kosaka M, Tamura G, Imanaka H, Sakai H, J. Antibiot. (Tokyo) 1972, 25, 437–444. [DOI] [PubMed] [Google Scholar]
- [86].Li W, Chou S, Khullar A, Gerratana B, Appl. Environ. Microbiol 2009, 75, 2958–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].von Tesmar A, Hoffmann M, Pippel J, Fayad AA, Dausend-Werner S, Bauer A, Blankenfeldt W, Muller R, Cell Chem. Biol 2017, 24, 1216–1227 e1218. [DOI] [PubMed] [Google Scholar]
- [88].Schneditz G, Rentner J, Roier S, Pletz J, Herzog KA, Bucker R, Troeger H, Schild S, Weber H, Breinbauer R, Gorkiewicz G, Hogenauer C, Zechner EL, Proc. Natl. Acad. Sci. U.S.A 2014, 111, 13181–13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Dornisch E, Pletz J, Glabonjat RA, Martin F, Lembacher-Fadum C, Neger M, Hogenauer C, Francesconi K, Kroutil W, Zangger K, Breinbauer R, Zechner EL, Angew. Chem. Int. Ed 2017, 56, 14753–14757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].a) Gui C, Li Q, Mo X, Qin X, Ma J, Ju J, Org. Lett 2015, 17, 628–631 [DOI] [PubMed] [Google Scholar]; b) Cogan DP, Ly J, Nair SK, ACS Chem. Biol 2020, 15, 2783–2791. [DOI] [PubMed] [Google Scholar]
- [91].Wat CK, Mcinnes AG, Smith DG, Wright JLC, Vining LC, Can. J. Chem 1977, 55, 4090–4098. [Google Scholar]
- [92].Eley KL, Halo LM, Song Z, Powles H, Cox RJ, Bailey AM, Lazarus CM, Simpson TJ, ChemBioChem 2007, 8, 289–297. [DOI] [PubMed] [Google Scholar]
- [93].Halo LM, Marshall JW, Yakasai AA, Song Z, Butts CP, Crump MP, Heneghan M, Bailey AM, Simpson TJ, Lazarus CM, Cox RJ, ChemBioChem 2008, 9, 585–594. [DOI] [PubMed] [Google Scholar]
- [94].Halo LM, Heneghan MN, Yakasai AA, Song Z, Williams K, Bailey AM, Cox RJ, Lazarus CM, Simpson TJ, J. Am. Chem. Soc 2008, 130, 17988–17996. [DOI] [PubMed] [Google Scholar]
- [95].a) Vesonder RF, Tjarks LW, Rohwedder WK, Burmeister HR, Laugal JA, J. Antibiot. (Tokyo) 1979, 32, 759–761 [DOI] [PubMed] [Google Scholar]; b) Singh SB, Zink DL, Goetz MA, Dombrowski AW, Polishook JD, Hazuda DJ, Tetrahedron Lett. 1998, 39, 2243–2246. [Google Scholar]
- [96].a) Phillips NJ, Goodwin JT, Fraiman A, Cole RJ, Lynn DG, J. Am. Chem. Soc 1989, 111, 8223–8231 [Google Scholar]; b) Jeong YC, Moloney MG, Future Med. Chem 2015, 7, 1861–1877. [DOI] [PubMed] [Google Scholar]
- [97].Larson EC, Lim AL, Pond CD, Craft M, Cavuzic M, Waldrop GL, Schmidt EW, Barrows LR, PLoS One 2020, 15, e0233485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Sims JW, Schmidt EW, J. Am. Chem. Soc 2008, 130, 11149–11155. [DOI] [PubMed] [Google Scholar]
- [99].Campbell CD, Vederas JC, Biopolymers 2010, 93, 755–763. [DOI] [PubMed] [Google Scholar]
- [100].Kakule TB, Sardar D, Lin Z, Schmidt EW, ACS Chem. Biol 2013, 8, 1549–1557. [DOI] [PubMed] [Google Scholar]
- [101].a) Chang PK, Ehrlich KC, Fujii I, Toxins 2009, 1, 74–99 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Liu X, Walsh CT, Biochemistry 2009, 48, 8746–8757 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Seidler NW, Jona I, Vegh M, Martonosi A, J. Biol. Chem 1989, 264, 17816–17823. [PubMed] [Google Scholar]
- [102].Liu X, Walsh CT, Biochemistry 2009, 48, 8746–8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Li H, Gilchrist CLM, Lacey HJ, Crombie A, Vuong D, Pitt JI, Lacey E, Chooi YH, Piggott AM, Org. Lett 2019, 21, 1287–1291. [DOI] [PubMed] [Google Scholar]
- [104].Ladenstein R, Winberg JO, Benach J, Cell. Mol. Life Sci 2008, 65, 3918–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Takada K, Ninomiya A, Naruse M, Sun Y, Miyazaki M, Nogi Y, Okada S, Matsunaga S, J. Org. Chem 2013, 78, 6746–6750. [DOI] [PubMed] [Google Scholar]
- [106].Ninomiya A, Katsuyama Y, Kuranaga T, Miyazaki M, Nogi Y, Okada S, Wakimoto T, Ohnishi Y, Matsunaga S, Takada K, ChemBioChem 2016, 17, 1709–1712. [DOI] [PubMed] [Google Scholar]
- [107].Kuranaga T, Matsuda K, Sano A, Kobayashi M, Ninomiya A, Takada K, Matsunaga S, Wakimoto T, Angew. Chem. Int. Ed 2018, 57, 9447–9451. [DOI] [PubMed] [Google Scholar]
- [108].Pratt RF, Cell. Mol. Life Sci 2008, 65, 2138–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Matsuda K, Zhai R, Mori T, Kobayashi M, Sano A, Abe I, Wakimoto T, Nat. Catal 2020, 507–515. [Google Scholar]
- [110].Zhou YJ, Lin X, Xu CM, Shen YY, Wang SP, Liao HZ, Li L, Deng H, Lin HW, Cell Chem. Biol 2019, 26, 737–744. [DOI] [PubMed] [Google Scholar]
- [111].Barajas JF, Shakya G, Moreno G, Rivera H, Jackson DR, Topper CL, Vagstad AL, La Clair JJ, Townsend CA, Burkart MD, Tsai SC, Proc. Natl. Acad. Sci. U.S.A 2017, 114, E4142–E4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Crawford JM, Korman TP, Labonte JW, Vagstad AL, Hill EA, Kamari-Bidkorpeh O, Tsai SC, Townsend CA, Nature 2009, 461, 1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].a) Maier T, Leibundgut M, Ban N, Science 2008, 321, 1315–1322 [DOI] [PubMed] [Google Scholar]; b) Keatinge-Clay A, J. Mol. Biol 2008, 384, 941–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Crawford JM, Townsend CA, Nat. Rev. Microbiol 2010, 8, 879–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG, Proc. Natl. Acad. Sci. U.S.A 2003, 100, 15670–15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].a) Mayorga ME, Timberlake WE, Genetics 1990, 126, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mayorga ME, Timberlake WE, Mol. Gen. Genet 1992, 235, 205–212 [DOI] [PubMed] [Google Scholar]; c) Rugbjerg P, Naesby M, Mortensen UH, Frandsen RJ, Microb. Cell Fact 2013, 12, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].a) Newman AG, Vagstad AL, Belecki K, Scheerer JR, Townsend CA, Chem. Commun 2012, 48, 11772–11774 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Newman AG, Townsend CA, J. Am. Chem. Soc 2016, 138, 4219–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Ogawa H, Hasumi K, Sakai K, Murakawa S, Endo A, J. Antibiot. (Tokyo) 1991, 44, 762–767. [DOI] [PubMed] [Google Scholar]
- [119].Vagstad AL, Newman AG, Storm PA, Belecki K, Crawford JM, Townsend CA, Angew. Chem. Int. Ed 2013, 52, 1718–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Kudo F, Asou Y, Watanabe M, Kitayama T, Eguchi T, Synlett 2012, 1843–1846. [Google Scholar]