Abstract
Global HIV-1 genetic diversity forms a major obstacle to the development of an HIV vaccine. It may be necessary to employ subtype-specific HIV-1 vaccines in individual countries according to their HIV-1 subtype distribution. We estimated the global and regional need for subtype-specific HIV-1 vaccines. We took into account the proportions of different HIV-1 variants circulating in each country, the genetic composition of HIV-1 recombinants, and the different genome segments (gag, pol, env) that may be incorporated into vaccines. We modeled different scenarios according to whether countries would employ subtype-specific HIV-1 vaccines against (1) the most common subtype; (2) subtypes contributing more than 5% of HIV infections; or (3) all circulating subtypes. For therapeutic vaccines targeting the most common HIV-1 subtype in each country, 16.5 million doses of subtype C vaccine were estimated globally, followed by subtypes A (14.3 million) and B (4.2 million). A vaccine based on env required 2.6 million subtype E doses, and a vaccine based on pol required 4.8 million subtype G doses. For prophylactic vaccines targeting the most common HIV-1 subtype in each country, 1.9 billion doses of subtype A vaccine were estimated globally, followed by subtype C (1.1 billion) and subtype B (1.0 billion). A vaccine based on env required 1.2 billion subtype E doses, and a vaccine based on pol required 0.3 billion subtype G doses. If subtype-specific HIV-1 vaccines are also directed against less common subtypes in each country, vaccines targeting subtypes D, F, H, and K are also needed and would require up to five times more vaccine doses in total. We conclude that to provide global coverage, subtype-specific HIV-1 vaccines need to be directed against subtypes A, B, and C. Vaccines targeting env also need to include subtype E and those targeting pol need to include subtype G.
Keywords: HIV, subtype, recombinant, CRF, URF, vaccine
Introduction
Thirty-eight million people globally were living with HIV in 2019 (UNAIDS, 2020). Despite the increased availability of antiretroviral therapy, there were 690,000 deaths and 1.7 million new infections in 2019 (UNAIDS, 2020). A globally effective preventative HIV vaccine is likely necessary to end the HIV pandemic (Fauci, 2017). Furthermore, a therapeutic vaccine that augments the immune system of HIV-infected individuals may reduce the need for antiretroviral therapy (Dorrell, 2005). However, a key stumbling block to the development of an HIV vaccine is the extensive global genetic diversity of HIV (Barouch, 2008; Hemelaar, 2012, 2013).
HIV has its origins in the zoonotic transmission of Simian Immunodeficiency Virus (SIV) from chimpanzees to humans a century ago (Gao et al., 1999). Subsequent to this, HIV-1 Group M diversified in Central Africa into multiple distinct subtypes: A, B, C, D, F, G, H, J, K, and L (Robertson et al., 2000; Worobey et al., 2008; Yamaguchi et al., 2020). The global spread of HIV throughout the second half of the twentieth century led to the differential global distribution of HIV-1 subtypes (Tebit and Arts, 2011; Hemelaar, 2012; Faria et al., 2014).
Genetic divergence between HIV-1 subtypes is substantial, with Env displaying a median difference of 25% (range 20–36%) at the amino acid level between strains from different subtypes, whereas the difference is 17% (15–22%) for Gag (Korber et al., 2001). Recombination between different HIV strains has led to further diversification of the HIV pandemic (Hemelaar, 2013). Recombinant forms are classified as circulating recombinant forms (CRFs) if they are found in three or more epidemiologically distinct individuals or unique recombinant forms (URFs) if there is no evidence of onward transmission (Robertson et al., 2000). To date, 106 distinct CRFs have been identified (Zhou et al., 2020), and collectively, these CRFs have been estimated to account for 16.7% of HIV-1 infections worldwide (Hemelaar et al., 2019).
The immune response to HIV is multifaceted, with antibodies mainly directed against the envelope component of the virus, whereas cytotoxic T lymphocyte responses are preferentially directed against Gag and/or Pol (Goulder et al., 2000). The large genetic divergence between HIV-1 subtypes makes it difficult to elicit immune responses that are sufficiently cross-reactive between HIV-1 subtypes (Korber et al., 2001). Given the variation between HIV-1 subtypes, it has been a common approach in HIV vaccine design to match the HIV-1 subtype(s) of the immunogen in the candidate vaccine to the HIV-1 subtype(s) circulating in the target population. To date, a number of different vaccine concepts, each using immunogen HIV-1 subtype(s) matched to circulating HIV-1 subtype(s), have been tested in large-scale efficacy trials.
Firstly, recombinant HIV-1 envelope proteins were used as immunogens aimed at generating broadly neutralizing antibodies. A bivalent subtype B/B recombinant glycoprotein gp120 vaccine was trialed in North America and The Netherlands, where subtype B dominates, and a bivalent subtype B/E recombinant gp120 vaccine was tested in Thailand, where subtype B and CRF01_AE cocirculate. Neither of these vaccines proved efficacious (HIV Vaccine Study Group et al., 2005; Pitisuttithum et al., 2006).
Next, viral vectors were used with the aim of eliciting cytotoxic T lymphocyte responses. The first such vaccine consisted of adenovirus type-5 (Ad5) vectors expressing subtype B Gag, Pol, and Nef proteins. This vaccine was tested in North America, the Caribbean, South America, and Australia, where subtype B is the predominant HIV-1 subtype, and in South Africa, where subtype C dominates. In both trials, the vaccine did not prevent HIV-1 infection or lower the viral-load setpoint (Buchbinder et al., 2008; Gray et al., 2011).
A subsequent approach aimed to elicit both antibody and T-cell responses. This strategy consisted of priming with DNA plasmids expressing subtype B Gag, Pol, and Nef and subtypes A, B, and C Env proteins, followed by a boost consisting of Ad5 vectors expressing a subtype B Gag-Pol fusion protein and Env glycoproteins of subtypes A, B, and C. When tested in MSM populations in the United States (mainly subtype B), the vaccine did not reduce the rate of HIV-1 acquisition or the viral-load set point (Hammer et al., 2013).
A further attempt aimed at eliciting both humoral and cell-mediated immunity used a combination of a canarypox vector expressing subtype B Env gp41TM, Gag and Pol and CRF01_AE Env gp120 followed by a boost with bivalent subtype B/E recombinant gp120 proteins, chosen to match the B and CRF01_AE strains circulating in Thailand. The RV144 trial showed modest efficacy of this vaccine (Rerks-Ngarm et al., 2009). This vaccine concept was then adapted for use in South Africa by replacing the B/CRF01_AE immunogens with subtype C immunogens to match HIV-1 subtype C endemic in South Africa (Bekker et al., 2018). However, the trial of this vaccine was recently halted due to lack of efficacy (Adepoju, 2020).
Given the genetic divergence between HIV-1 subtypes and their differential global spread, it may be necessary to employ subtype-specific HIV-1 vaccines in individual countries according to their HIV-1 subtype distribution. To aid prioritization of HIV-1 subtypes for vaccine development, we aimed to estimate the global and regional need for therapeutic and prophylactic vaccines specific for different HIV-1 subtypes, taking account of the proportions of different HIV-1 variants circulating in each country, the genetic composition of HIV-1 recombinants, and the different genome segments of HIV that may be incorporated into a vaccine.
Materials and Methods
HIV-1 Molecular Epidemiology Data
Country-level HIV-1 molecular epidemiology data was collected by conducting a global survey and a comprehensive systematic review, as described previously (Hemelaar et al., 2019; Hemelaar et al., 2020). In total, 2,203 datasets with 383,519 samples were obtained from 116 countries across 1990–2015, with the data analyzed for four time periods: 2010–2015, 2005–2009, 2000–2004, and 1990–1999. In the current study, we used data from the latest time period (2010–2015) for analysis for most countries. For the small number of countries for which no data for 2010–2015 was available, data from the next most recent time period (2005–2009, 2000–2004, or 1990–1999) was used. These latter countries, as well as the time period for which data was used, are listed in Supplementary Material, p. 7.
Reassignment of CRFs to “Pure” HIV-1 Subtypes
Given the large number of different CRFs (106 distinct CRFs identified to date (Los Alamos National Laboratory; Zhou et al., 2020), it would be impractical to make a vaccine specific for each CRF. Hence, we determined which “pure” HIV-1 subtypes contributed most to each CRF, both to each genome segment (gag, pol, env) as well as the full-length genome (which also includes accessory genes (vif, vpu, vpr, nef), the regulatory genes (rev, tat), and the 5′ and 3′ long terminal repeat regions). Information on the genetic composition of individual CRFs was obtained from the Los Alamos National Laboratory (LANL) website (Los Alamos National Laboratory). CRFs were reassigned to “pure” HIV-1 subtypes according to the HIV-1 subtype making the largest contribution to the full-length genome as well as each genome segment. In situations where unclassified sequences made the largest contribution, the next largest contributing HIV-1 subtype was taken. If the subtype composition of certain genome regions was unclear from the LANL website, the original paper describing the CRF was examined. The full list of CRFs and their reassignment to the “pure” HIV-1 subtypes, for full length as well as each genome segment, can be found in Supplementary Material, pp. 4–6. Unfortunately, reassignment could not be performed for CRF30_0206, CRF75_01B, CRF77_cpx, CRF79_0107, CRF80_0107, CRF81_cpx, and CRF84_A1D due to lack of data on their subtype composition.
HIV-1 Subtype Proportions in Countries After Reassignment of CRFs to “Pure” HIV-1 Subtypes
Upon completion of the reassignment of CRFs to “pure” HIV-1 subtypes, the proportions of infections accounted for by each CRF in each country, as previously estimated (Hemelaar et al., 2020), were added to those of the relevant “pure” HIV-1 subtypes, thereby generating new proportions of infections that could be ascribed to each “pure” HIV-1 subtype for the full-length genome and each genome segment. Country-level “pure” HIV-1 subtype distributions were combined with UNAIDS data on the number of people living with HIV in each country in 2016 (UNAIDS, 2017) to generate estimates of regional and global “pure” HIV-1 subtype proportions (Supplementary Material, p. 2).
Estimation of Numbers of Doses for Subtype-Specific Therapeutic and Prophylactic HIV-1 Vaccines
Upon estimation of the proportions of “pure” HIV-1 subtypes in each country, estimates for the numbers of doses needed for either therapeutic or prophylactic subtype-specific HIV-1 vaccines were calculated. Calculations were performed for three different scenarios, each using a different cut-off for HIV-1 subtypes eligible for inclusion in vaccines for each country: (1) Vaccinating against only the most common subtype circulating in each country (“most common subtype” scenario), (2) Vaccinating against subtypes with a prevalence of >5% in people living with HIV in each country (“>5% prevalence” scenario), and (3) Vaccinating against all circulating subtypes in each country (“all circulating subtypes” scenario). A fourth scenario was assessed for therapeutic vaccines, in which each HIV-infected individual would be vaccinated with a vaccine based on the HIV-1 subtype they were infected with. All calculations for each scenario were conducted for the full-length genome and each genome region (gag, pol, env).
For therapeutic vaccines, the target population was all people living with HIV in 2016, as estimated by UNAIDS (UNAIDS, 2017). For prophylactic vaccines, the target population was 10–49-year-old men and women, chosen to include most of the sexually active population as well as other high-risk groups, using estimates of population numbers in 2015 reported by the United Nations (United Nations, 2017). For both the therapeutic and prophylactic HIV-1 vaccine analyses, the entire target population in each country was to be vaccinated against every subtype that made the cut-off in each scenario, i.e., the most common subtype, all subtypes which contributed >5% of HIV infections, or all circulating subtypes. The estimated number of subtype-specific HIV-1 vaccine doses per country were subsequently aggregated at both the regional and global levels.
The term “dose” in this study was used to describe a “course of vaccination” with a subtype-specific HIV-1 vaccine. It may, however, be that a course of vaccination may consist of multiple doses of the same vaccine or a combination of different types of vaccines in a “prime-boost” configuration. All calculations were performed in Microsoft Excel.
Results
Global and Regional Distribution of HIV-1 Subtypes
Given the difficulty in generating a vaccine for each individual HIV-1 recombinant, we first reassigned each CRF to a “pure” HIV-1 subtype (A-K) according to the HIV-1 subtype that contributed most to each CRF, both for the full length genome and the gag, pol, and env regions (Supplementary Material, pp. 4–6). Following reassignment of CRFs, we determined the global and regional proportions of infections caused by each of the “pure” HIV-1 subtypes, based on the most recent available HIV-1 subtype distribution data for each country (Figures 1, 2 and Supplementary Material, pp. 8–10).
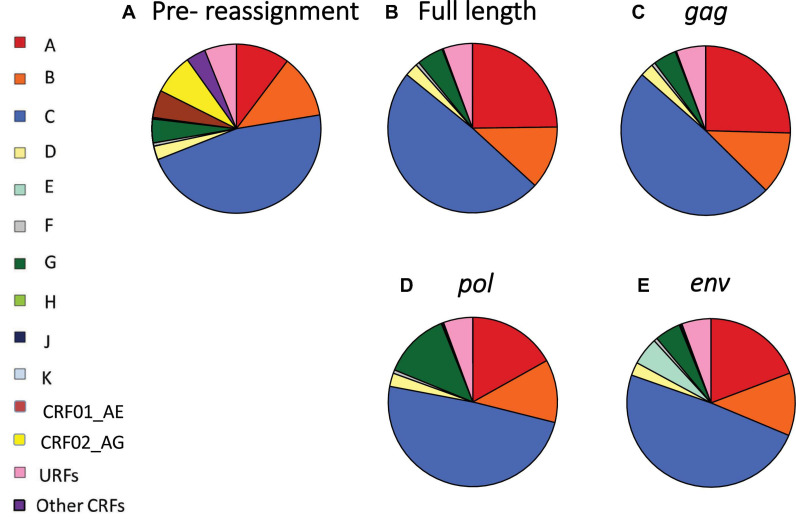
FIGURE 1.
Global distribution of HIV-1 variants before and after reassignment of CRFs to “pure” HIV-1 subtypes. (A) Global proportions of HIV-1 subtypes A–K, CRF01_AE, CRF02_AG, other CRFs and URFs, based on most recent data available for each country. (B–E) Global proportions of HIV-1 variants after reassignment of CRFs to “pure” HIV-1 subtypes, based on full-length sequence (B), gag (C), pol (D), and env (E). CRF, circulating recombinant form; URF, unique recombinant form. Data underlying this figure is displayed in Supplementary Material, pp. 8–10.
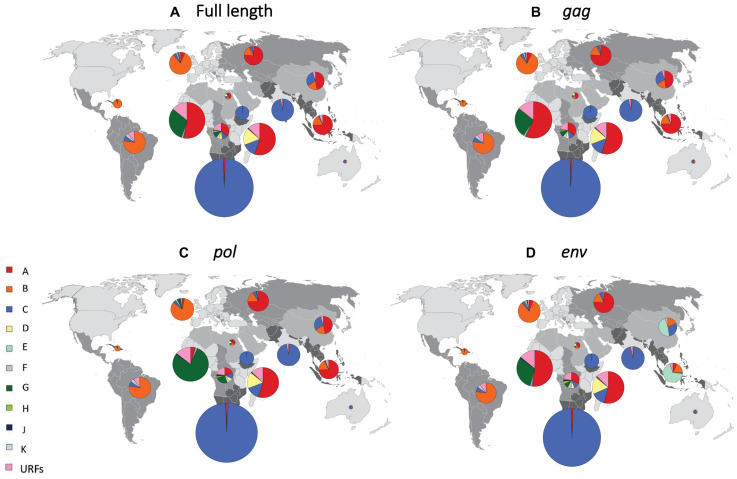
FIGURE 2.
Regional distribution of HIV-1 subtypes after reassignment of CRFs to “pure” HIV-1 subtypes. Regional proportions of HIV-1 subtypes after reassignment of CRFs to “pure” HIV-1 subtypes, based on full-length sequence (A), gag (B), pol (C), and env (D). We grouped all countries into 14 regions, as previously described (Hemelaar et al., 2019). Individual regions are shaded differently on the world map. The proportion of HIV infections attributed to each subtype in each region is shown in pie charts superimposed onto the regions. The sizes (surface area) of the pie charts for each region are proportional to the relative number of people living with HIV in each region. URF, unique recombinant form. Data underlying this figure is displayed in Supplementary Material, pp. 8–10.
After reassignment of CRFs based on the full-length genome, half of the global HIV infections were attributable to subtype C (49.1%), followed by subtype A (24.8%), B (12.0%), G (5.1%), D (2.5%), F (0.7%), and H, J, and K (0.1% each) (Figure 1B and Supplementary Material, pp. 8–10). Major changes in global HIV-1 subtype distribution resulting from reassignment of CRFs to “pure” HIV-1 subtypes were driven by the major recombinants CRF01_AE and CRF02_AG, as CRF01_AE is composed of subtype A in gag and pol, but subtype E in env, whereas CRF02_AG is composed of subtype A in gag and env and subtype G in pol (Figure 1 and Supplementary Material, pp. 4–6 and 8–10).
Subtype A contributed 25.4% of global HIV infections after CRF reassignment based on gag, 19.2% for env, and 16.9% for pol (Figures 1C–E and Supplementary Material, pp. 9–10). Subtype E contributed 5.3% of global infections after reassignment based on env, but none for gag and pol, as subtype E has never been identified for those genome segments. Subtype G constituted 12.7% of infections after reassignment based on pol, but only 5.0% for env and 4.4% for gag. The global contributions of other subtypes remained relatively stable following CRF reassignment (Figure 1 and Supplementary Material, pp. 8–10).
In South-East Asia, where CRF01_AE plays an important role, the proportion of infections attributable to subtype A was 74.4% after CRF reassignment based on the full-length analysis (Figure 2A and Supplementary Material, pp. 8–10). Subtype A also constituted 74.4 and 74.2% of infections for gag and pol in this region, whereas subtype E constituted 67.8% for env (Figures 2B–D and Supplementary Material, pp. 8–10). In East Asia, subtype A constituted 47.0% of infections for full length and gag, and 46.7% for pol. However, subtype E constituted 46.8% of infections for env in this region. In West Africa, where CRF02_AG plays a major role, subtype A constituted 52.6% of infections for full length and similar percentages for gag and env. However, subtype G constituted 78.7% for pol. In the other regions, which had fewer CRF infections, there was less change in HIV-1 subtype proportions following reassignment of CRFs to “pure” HIV-1 subtypes (Figure 2 and Supplementary Material, pp. 8–10).
Therapeutic HIV-1 Vaccines
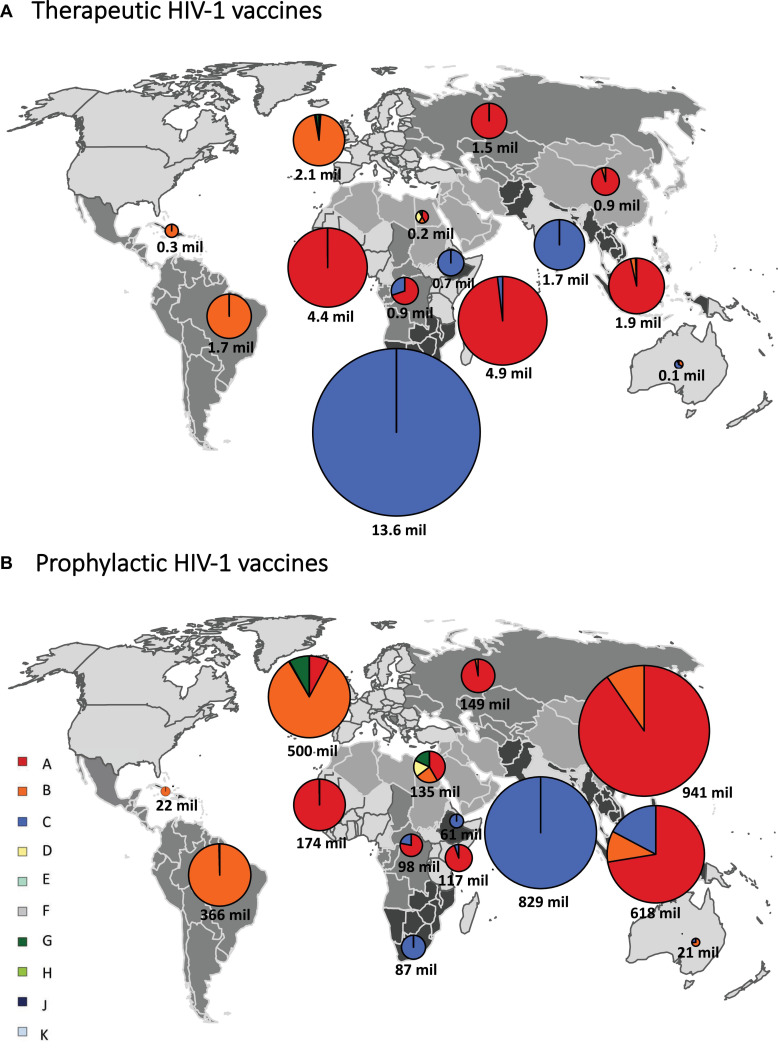
If HIV-infected people would be vaccinated against the most common subtype circulating in each country (“most common subtype” scenario), based on the full-length genome, 35.1 million vaccine doses would be required globally, of which 16.5 million were subtype C, 14.3 million subtype A, and 4.2 million subtype B, with much fewer doses for other subtypes (Figure 3A and Table 1). A vaccine based on env required 2.6 million subtype E doses, and a vaccine based on pol required 4.8 million subtype G doses (Figure 3A and Table 1). The global need for a therapeutic subtype C vaccine was largely driven by Southern Africa and South Asia (Figure 4A and Supplementary Material, p. 11). The need for a subtype A vaccine was largely driven by East and West Africa, as well as South-East Asia and Eastern Europe and Central Asia. Finally, the global need for a subtype B vaccine was driven by Western and Central Europe and North America and Latin America (Figure 4A and Supplementary Material, p. 11).
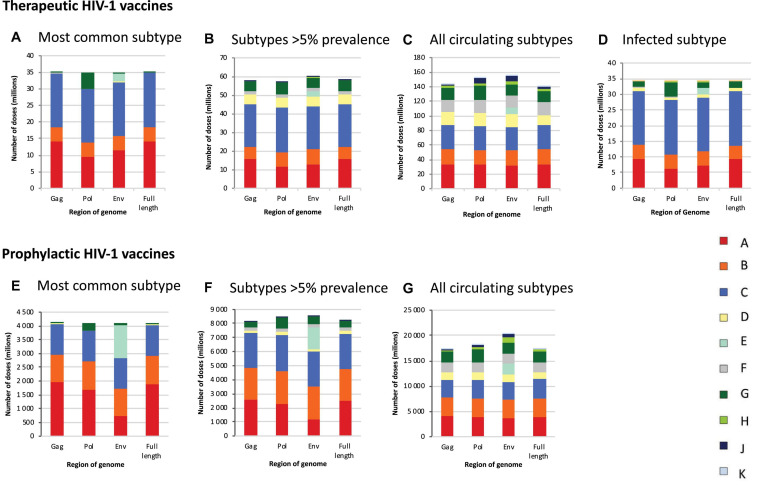
FIGURE 3.
Global estimates of the number of doses of subtype-specific therapeutic and prophylactic HIV-1 vaccines. Estimates of the number of doses of subtype-specific therapeutic (A–D) and prophylactic (E–G) HIV-1 vaccines. Estimates are stratified according to genome region (gag, pol, env, and full length) and the number of subtypes against which people in each country are vaccinated, according to different scenarios, i.e., vaccinating people against the most common subtype in each country (A,E), subtypes with a prevalence of >5% in people living with HIV in each country (B,F), all subtypes circulating in each country (C,G), and the HIV-1 subtype with which HIV-positive people are infected (D). Data underlying this figure is displayed in Table 1.
TABLE 1.
Global estimates of numbers of doses (millions) of subtype-specific therapeutic and prophylactic HIV-1 vaccines.
|
HIV-1 Subtypes |
|||||||||||||
| Vaccine Type | Scenario | Genome Region | A | B | C | D | E | F | G | H | J | K | Total |
| Therapeutic | Most common | gag | 14.28 | 4.21 | 16.21 | 0.06 | 0.00 | 0.02 | 0.04 | 0.00 | 0.00 | 0.00 | 34.81 |
| pol | 9.54 | 4.22 | 16.21 | 0.06 | 0.00 | 0.02 | 4.78 | 0.00 | 0.00 | 0.00 | 34.81 | ||
| env | 11.65 | 4.23 | 16.21 | 0.06 | 2.60 | 0.00 | 0.06 | 0.00 | 0.00 | 0.00 | 34.81 | ||
| Full length | 14.25 | 4.22 | 16.50 | 0.06 | 0.00 | 0.02 | 0.06 | 0.00 | 0.00 | 0.00 | 35.10 | ||
| >5% | gag | 15.94 | 6.47 | 22.73 | 5.34 | 0.00 | 1.62 | 5.11 | 0.28 | 0.20 | 0.00 | 57.69 | |
| pol | 11.67 | 7.75 | 23.89 | 5.34 | 0.00 | 1.62 | 6.53 | 0.28 | 0.32 | 0.00 | 57.38 | ||
| env | 13.00 | 7.75 | 23.14 | 5.28 | 2.92 | 1.62 | 5.70 | 0.31 | 0.32 | 0.00 | 60.02 | ||
| Full length | 15.94 | 6.52 | 22.74 | 5.28 | 0.00 | 1.63 | 5.63 | 0.28 | 0.20 | 0.00 | 58.24 | ||
| All subtypes | gag | 33.12 | 20.78 | 32.73 | 18.11 | 0.00 | 17.82 | 15.20 | 3.14 | 2.27 | 0.70 | 143.89 | |
| pol | 32.71 | 20.78 | 32.73 | 18.11 | 0.00 | 17.89 | 18.70 | 3.05 | 8.05 | 1.09 | 153.12 | ||
| env | 31.20 | 20.89 | 32.11 | 17.96 | 8.54 | 17.81 | 14.22 | 4.02 | 8.10 | 1.09 | 155.94 | ||
| Full length | 32.86 | 20.78 | 33.31 | 14.08 | 0.00 | 17.69 | 15.66 | 3.14 | 2.68 | 1.07 | 141.27 | ||
| Infected subtype | gag | 9.50 | 4.34 | 17.33 | 1.03 | 0.00 | 0.25 | 1.62 | 0.04 | 0.04 | 0.02 | 34.17 | |
| pol | 6.37 | 4.36 | 17.33 | 1.03 | 0.00 | 0.24 | 4.66 | 0.04 | 0.12 | 0.02 | 34.17 | ||
| env | 7.33 | 4.38 | 17.33 | 1.02 | 1.85 | 0.25 | 1.81 | 0.07 | 0.12 | 0.02 | 34.17 | ||
| Full length | 9.26 | 4.36 | 17.33 | 1.02 | 0.00 | 0.25 | 1.85 | 0.04 | 0.04 | 0.02 | 34.17 | ||
| Prophylactic | Most common | gag | 1,968.11 | 1,004.82 | 1,095.35 | 23.24 | 0.00 | 2.97 | 1.34 | 0.00 | 0.00 | 0.00 | 4,095.84 |
| pol | 1,702.24 | 1,010.52 | 1,095.35 | 23.24 | 0.00 | 2.97 | 261.52 | 0.00 | 0.00 | 0.00 | 4,095.84 | ||
| env | 718.02 | 1,012.00 | 1,095.35 | 23.24 | 1,180.43 | 1.50 | 65.31 | 0.00 | 0.00 | 0.00 | 4,095.84 | ||
| Full length | 1,898.45 | 1,010.52 | 1,116.87 | 23.24 | 0.00 | 2.97 | 65.31 | 0.00 | 0.00 | 0.00 | 4,117.36 | ||
| >5% | gag | 2,561.34 | 2,256.68 | 2,486.53 | 203.94 | 0.00 | 253.16 | 356.69 | 21.52 | 8.40 | 0.00 | 8,148.27 | |
| pol | 2,298.97 | 2,345.43 | 2,567.32 | 203.94 | 0.00 | 253.16 | 748.03 | 21.52 | 19.02 | 0.00 | 8,457.38 | ||
| env | 1,160.36 | 2,345.43 | 2,481.26 | 196.47 | 1,557.26 | 253.16 | 489.63 | 27.85 | 19.02 | 0.00 | 8,530.44 | ||
| Full length | 2,522.69 | 2,269.40 | 2,491.46 | 180.70 | 0.00 | 258.09 | 477.17 | 21.52 | 8.40 | 0.00 | 8,229.43 | ||
| All subtypes | gag | 4,033.34 | 3,627.84 | 3,660.23 | 1,443.14 | 2.46 | 1,952.32 | 2,096.18 | 349.77 | 84.54 | 71.15 | 17,320.97 | |
| pol | 3,907.84 | 3,627.84 | 3,660.23 | 1,443.14 | 2.46 | 2,007.39 | 2,685.46 | 345.61 | 457.35 | 158.67 | 18,296.00 | ||
| env | 3,643.27 | 3,637.83 | 3,604.44 | 1,407.96 | 2,112.43 | 1,950.97 | 2,190.60 | 1,200.43 | 457.57 | 157.98 | 20,363.47 | ||
| Full length | 3,942.57 | 3,627.84 | 3,814.19 | 1,274.02 | 2.46 | 1,945.75 | 2,243.83 | 362.80 | 151.96 | 129.63 | 17,495.05 | ||
Estimates are stratified according to genome region (gag, pol, env, and full length) and the number of subtypes against which people in each country are vaccinated, according to different scenarios, i.e., vaccinating people against the most common subtype in each country (“most common”), subtypes with a prevalence of > 5% in people living with HIV in each country (“>5%”), all subtypes circulating in each country (“all subtypes”), and the HIV-1 subtype with which HIV-positive people are infected (infected subtype).
FIGURE 4.
Regional estimates of the number of doses of subtype-specific therapeutic and prophylactic HIV-1 vaccines. Regional estimates for therapeutic (A) and prophylactic (B) HIV-1 vaccines based on the most common subtype, based on the full-length sequence, in each country. We grouped all countries into 14 regions, as previously described (Hemelaar et al., 2019). Individual regions are shaded differently on the world map. The estimates for vaccines based on each HIV-1 subtype are shown in pie charts superimposed onto the regions. The sizes of the pie charts (surface area) correspond to the relative number of vaccine doses for each region and the total number of vaccine doses is shown below each pie chart. mil, million. Data underlying this figure is displayed in Supplementary Material, p. 11.
If HIV-infected people would be vaccinated against subtypes with a prevalence of >5% in people living with HIV in each country (“>5% prevalence” scenario), based on the full-length genome, 58.2 million vaccine doses were estimated, of which 22.7 million would be subtype C, 15.9 million subtype A, 6.5 million subtype B, 5.6 million subtype G, and 5.3 million subtype D (Figure 3B and Table 1). A vaccine based on env required 2.9 million subtype E doses.
If HIV-infected people would be vaccinated against all circulating subtypes in each country (“all circulating subtypes” scenario), 141.3 million doses of vaccine would be required, based on the full-length genome, of which 33.3 million would be subtype C, 32.9 million subtype A, 20.8 million subtype B, 17.7 million subtype F, 15.7 million subtype G, 14.1 million subtype D, 3.1 million subtype H, 2.7 million subtype J and 1.1 million subtype K (Figure 3C and Table 1). A vaccine based on env required 8.5 million subtype E doses.
In the final scenario in which each infected individual would be vaccinated only against the subtype with which they are already infected, 34.2 million vaccine doses were estimated, based on the full-length genome, of which 17.3 million would be subtype C, 9.3 million subtype A, 4.4 million subtype B, 1.9 million subtype G, and 1.0 million subtype D, reflecting the global distribution of HIV-1 variants (Figures 1, 3D, Table 1, and Supplementary Material, 8–10).
Prophylactic HIV-1 Vaccines
If all 10–49-year-old people would be vaccinated against the most common subtype circulating in each country (“most common subtype” scenario), based on the full-length genome, an estimated 4.1 billion doses of vaccines would be required globally, of which 1.9 billion were subtype A, 1.1 billion subtype C, and 1.0 billion subtype B (Figure 3E and Table 1). A vaccine based on env required 1.2 billion subtype E doses, and a vaccine based on pol required 262 million subtype G doses. The global need for a prophylactic subtype A vaccine was largely driven by East Asia and South-East Asia (Figure 4B and Supplementary Material, p. 11). This was due to their large populations as well as the endemic nature of CRF01_AE in these regions. The need for a subtype C vaccine was driven largely by South Asia, and the need for a subtype B vaccine was driven by Western and Central Europe and North America and Latin America.
In the “>5% prevalence” scenario, 8.2 billion doses of a vaccine based on the full-length genome were estimated, of which 2.5 billion would be subtype A, 2.5 billion subtype C, and 2.3 billion subtype B (Figure 3F and Table 1). A vaccine based on env required 1.6 billion subtype E doses.
Finally, in the “all circulating subtypes” scenario, for a vaccine based on the full-length genome, 17.5 billion vaccine doses would be required, of which 3.9 billion would be subtype A, 3.8 billion subtype C, 3.6 billion subtype B, 2.2 billion subtype G, 1.9 billion subtype F, 1.3 billion subtype D, and 0.4 billion for subtype H (Figure 3G and Table 1). A vaccine based on env required 2.1 billion subtype E doses.
Discussion
In this study, we estimated the global and regional need for subtype-specific therapeutic and prophylactic HIV-1 vaccines. When targeting the most common HIV-1 subtype in each country, we estimated the largest number of therapeutic vaccine doses were needed for a subtype C vaccine (16.5 million), followed by subtype A (14.3 million) and subtype B (4.2 million). A vaccine based on env required 2.6 million subtype E doses, and a vaccine based on pol required 4.8 million subtype G doses. The need for therapeutic subtype C vaccines was largely driven by the endemicity of subtype C in Southern Africa and South Asia, the need for subtype A vaccines by East and West Africa, and the need for subtype B vaccines by Western and Central Europe and North America and Latin America.
For prophylactic vaccines targeting the most common HIV-1 subtype in each country, 1.9 billion doses of subtype A vaccine were estimated, followed by subtype C (1.1 billion) and subtype B (1.0 billion). A vaccine based on env required 1.2 billion subtype E doses, and a vaccine based on pol required 0.3 billion subtype G doses. The need for prophylactic subtypes A and E vaccines was largely driven by East Asia and South-East Asia, owing to their large populations as well as prevalence of CRF01_AE. The need for a prophylactic subtype C vaccine was largely driven by South Asia.
Employing vaccines against more than one HIV-1 subtype in each country, as estimated in the “>5% prevalence” and “all circulating subtypes” scenarios, dramatically increases the number of vaccine doses and number of different subtype-specific vaccines required for both therapeutic and prophylactic vaccines.
It is apparent that to provide global coverage against the most common HIV-1 subtype circulating in each country, subtype-specific therapeutic and prophylactic HIV-1 vaccines need to be directed against subtypes A, B, and C. Vaccines targeting the envelope protein would also need to include subtype E and those targeting Pol need to include subtype G. If subtype-specific vaccines are also directed against less common HIV-1 subtypes in each country, vaccines targeting subtypes D, F, H, and K also need to be considered and would require up to five times more vaccine doses in total.
This study has several strengths. We utilized the largest available global HIV-1 molecular epidemiology database and applied the novel approach of reassigning CRFs to “pure” HIV-1 subtypes based on their genetic composition. This enabled us to address the complexity posed by multiple distinct recombinants and to generate new estimates of the proportion of infections caused by each “pure” HIV-1 subtype. In addition, because HIV-1 vaccines could aim to elicit antibodies or T-cell responses or both, we generated estimates for subtype-specific HIV-1 vaccines based on env, gag, and pol as well as the full-length genome. Moreover, we conducted separate analyses for therapeutic and prophylactic vaccines. In addition, we examined a number of different scenarios according to the number of HIV-1 subtypes eligible for inclusion in a vaccine.
Our study also has some limitations. Although CRFs were reassigned to “pure” HIV-1 subtypes based on their genetic composition, we do not know the extent of their overlapping immunogenic properties. Although the target population for a therapeutic vaccine are people living with HIV, the target population for a prophylactic vaccine is less certain. We opted to estimate a one-off vaccination of all people aged 10–49 years old, to include most sexually active people and other risk groups (Marzetta et al., 2010). We made no distinction between routine and catch-up vaccinations (Marzetta et al., 2010). This comprehensive approach may have led to higher estimates of numbers of doses needed, but did allow us to gauge the relative importance of HIV-1 subtypes for regional and global vaccine development to enable prioritization of relevant HIV-1 subtypes. Vaccine efficacy may differ between subtype-specific vaccines and was not factored in Dimitrov et al. (2015). Moreover, a putative vaccine with high efficacy would likely be administered to larger populations whereas a vaccine with low/moderate efficacy would more likely be limited to high risk groups (Esparza et al., 2003). Duration of protection offered by a putative vaccine was not modeled and consequently revaccination was also not factored in. Furthermore, we aimed to estimate the need for subtype-specific vaccines and did not estimate the actual uptake or use, which depends on factors such as adoption time, accessibility and acceptability, which will vary by country (Marzetta et al., 2010). Lastly, cost and cost-effectiveness were not taken into account.
There are also limitations to the concept of subtype-specific HIV-1 vaccines. One issue is the need to generate multiple different vaccines for the different HIV-1 subtypes. This could be partially overcome by formulation of multivalent vaccines (“cocktails”) incorporating multiple subtype-specific preparations (Korber et al., 2017). Another limitation is that a subtype-specific HIV-1 vaccine would need to be matched to locally circulating strains, which requires availability of up-to-date HIV-1 diversity data and the relevant subtype-specific HIV-1 vaccines. Furthermore, protection would be limited to a certain geographical region and thereby limit travel to other regions, while at the same time leave vaccinated people vulnerable to infection by newly imported strains of HIV, as HIV-1 subtype distribution is very dynamic (Hemelaar et al., 2019).
A crucial outstanding limitation of subtype-specific HIV-1 vaccines is the issue of intrasubtype genetic diversity (Korber et al., 2001; Gaschen et al., 2002). The HIV-1 vaccine recently tested, and proven ineffective, in South Africa consisted of a recombinant canarypox vaccine, which contained a subtype C env gp120 isolate sequence (96ZM651 from Zambia), and a bivalent subtype C gp120 consisting of two distinct subtype C recombinant monomeric Env gp120 proteins (derived from isolates TV1.C from South Africa and 1086.C from Malawi) (Zambonelli et al., 2016; Bekker et al., 2018). Utilization of subtype C isolate sequences in the vaccine, matching subtype C dominating in South Africa, was hoped to lead to protective immunogenicity. However, intrasubtype diversity is considerable, with median percentage amino acid differences within HIV-1 subtypes estimated at 17% (range 4–30%) for Env and 8% (2–15%) for Gag (Korber et al., 2001), thereby limiting the potential for eliciting cross-reactive protective immune responses. One way to reduce the genetic distance between vaccine immunogens and circulating strains is the inclusion of artificial centralized sequences, such as consensus, ancestral or center-of tree sequences (Gaschen et al., 2002; Nickle et al., 2003). For example, subtype C isolate sequences are around 5–15% different to other subtype C isolate sequences, whereas, a subtype C consensus amino acid sequence is only around 3–8% different from individual subtype C isolates (Gaschen et al., 2002). Of note, a group M consensus sequence (i.e., a consensus of all subtype consensus sequences) would be around 5–15% different to individual circulating HIV-1 isolates and therefore not better than a subtype-matched isolate sequence (Gaschen et al., 2002). Indeed, the use of isolate sequences in all candidate HIV vaccines tested in phase 3 trials to date may have limited cross-reactivity and hence limited efficacy (HIV Vaccine Study Group et al., 2005; Pitisuttithum et al., 2006; Buchbinder et al., 2008; Rerks-Ngarm et al., 2009; Gray et al., 2011; Hammer et al., 2013; Bekker et al., 2018). Thus, the use of subtype consensus (or ancestral or center-of-tree) sequences may be a more successful approach in the future.
Ideally, a globally effective HIV vaccine will need to confer protection against all diverse HIV-1 subtypes and recombinants. There are multiple HIV vaccine efforts on-going, utilizing a number of different approaches to address HIV-1 diversity. One approach is to use mosaic vaccines, which have been shown to elicit increased breadth and depth of immune responses (Barouch et al., 2010). The HVTN 705/Imbokodo trial currently underway in southern Africa is evaluating a tetravalent vaccine composed of adenovirus serotype 26 vector expressing mosaic gag, pol and env inserts combined with subtype C gp140 Env protein, with the intention of eliciting responses against a wide range of HIV subtypes, but still matching subtype C predominant in the region. The HVTN 706/Mosaico trial taking place in North America, Western Europe, and Latin America evaluates a nearly identical mosaic vaccine, which also includes a mosaic gp140 glycoprotein (Baden et al., 2020). Other approaches in preclinical development include focussing on conserved or structurally important regions of HIV (Letourneau et al., 2007; Gaiha et al., 2019). For all HIV-1 vaccine approaches, it is crucial to have up-to-date knowledge of HIV-1 genetic diversity to allow prioritization and development of vaccine concepts that are likely to provide the greatest benefit to specific countries, regions, and the world.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by local ethics committees of contributing studies. The participants provided their written informed consent to participate in this study.
Author Contributions
RE and MJ conducted the analyses, prepared the figures and tables, interpreted the data, and wrote the first draft of the manuscript. RE, JY, LD-T, SK, and JH collected the data. JH conceived, designed and coordinated the study, designed the analysis and figures, interpreted the data, and wrote the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Contributor Information
WHO-UNAIDS Network for HIV Isolation and Characterisation:
Alash’le G Abimiku, Simon Agwale, Chris Archibald, Boaz Avidor, María Gabriela Barbás, Francoise Barre-Sinoussi, Banson Barugahare, El Hadj Belabbes, Silvia Bertagnolio, Deborah Birx, Aleksei F Bobkov, James Brandful, Helba Bredell, Catherine A Brennan, James Brooks, Marie Bruckova, Luigi Buonaguro, Franco Buonaguro, Stefano Buttò, Anne Buvé, Mary Campbell, Jean Carr, Alex Carrera, Manuel Gómez Carrillo, Connie Celum, Beth Chaplin, Macarthur Charles, Dimitrios Chatzidimitriou, Zhiwei Chen, Katsumi Chijiwa, David Cooper, Philip Cunningham, Anoumou Dagnra, Cillian F de Gascun, Julia Del Amo, Elena Delgado, Ursula Dietrich, Dominic Dwyer, Dennis Ellenberger, Barbara Ensoli, Max Essex, Hervé Fleury, Peter N Fonjungo, Vincent Foulongne, Deepak A Gadkari, Feng Gao, Federico García, Roger Garsia, Guy Michel Gershy-Damet, Judith R Glynn, Ruth Goodall, Zehava Grossman, Monick Lindenmeyer Guimarães, Beatrice Hahn, Raph L Hamers, Osamah Hamouda, Ray Handema, Xiang He, Joshua Herbeck, David D Ho, Africa Holguin, Mina Hosseinipour, Gillian Hunt, Masahiko Ito, Mohamed Ali Bel Hadj Kacem, Erin Kahle, Pontiano Kaleebu, Marcia Kalish, Adeeba Kamarulzaman, Chun Kang, Phyllis Kanki, Edward Karamov, Jean-Claude Karasi, Kayitesi Kayitenkore, Tony Kelleher, Dwip Kitayaporn, Leondios G Kostrikis, Claudia Kucherer, Claudia Lara, Thomas Leitner, Kirsi Liitsola, Jai Lingappa, Marek Linka, Ivette Lorenzana de Rivera, Vladimir Lukashov, Shlomo Maayan, Luzia Mayr, Francine McCutchan, Nicolas Meda, Elisabeth Menu, Fred Mhalu, Doreen Mloka, John L Mokili, Brigitte Montes, Orna Mor, Mariza Morgado, Fausta Mosha, Awatef Moussi, James Mullins, Rafael Najera, Mejda Nasr, Nicaise Ndembi, Joel R Neilson, Vivek R Nerurkar, Florian Neuhann, Claudine Nolte, Vlad Novitsky, Philippe Nyambi, Marianna Ofner, Fem J Paladin, Anna Papa, Jean Pape, Neil Parkin, Chris Parry, Martine Peeters, Alexandra Pelletier, Lucía Pérez-Álvarez, Deenan Pillay, Angie Pinto, Trinh Duy Quang, Cecilia Rademeyer, Filimone Raikanikoda, Mark A. Rayfield, Jean-Marc Reynes, Tobias Rinke de Wit, Kenneth E Robbins, Morgane Rolland, Christine Rousseau, Jesus Salazar-Gonzales, Hanan Salem, Mika Salminen, Horacio Salomon, Paul Sandstrom, Mario L Santiago, Abdoulaye D Sarr, Bryan Schroeder, Michel Segondy, Philippe Selhorst, Sylvester Sempala, Jean Servais, Ansari Shaik, Yiming Shao, Amine Slim, Marcelo A Soares, Elijah Songok, Debbie Stewart, Julie Stokes, Shambavi Subbarao, Ruengpung Sutthent, Jun Takehisa, Amilcar Tanuri, Kok Keng Tee, Kiran Thapa, Michael Thomson, Tyna Tran, Willy Urassa, Hiroshi Ushijima, Philippe van de Perre, Guido van der Groen, Kristel van Laethem, Joep van Oosterhout, Ard van Sighem, Eric van Wijngaerden, Anne-Mieke Vandamme, Jurgen Vercauteren, Nicole Vidal, Lesley Wallace, Carolyn Williamson, Dawit Wolday, Jianqing Xu, Chunfu Yang, Linqi Zhang, and Rong Zhang
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.690647/full#supplementary-material
References
- Adepoju P. (2020). Moving on from the failed HIV vaccine clinical trial. Lancet HIV 7:e161. 10.1016/s2352-3018(20)30047-3 [DOI] [PubMed] [Google Scholar]
- Baden L. R., S9itieh D. J., Sarnecki M., Walsh S. R., Tomaras G. D., Kublin J. G., et al. (2020). Safety and immunogenicity of two heterologous HIV vaccine regimens in healthy, HIV-uninfected adults (TRAVERSE): a randomised, parallel-group, placebo-controlled, double-blind, phase 1/2a study. Lancet HIV 7 e688–e698. 10.1016/s2352-3018(20)30229-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch D. H. (2008). Challenges in the development of an HIV-1 vaccine. Nature 455 613–619. 10.3109/9781420060744-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch D. H., O’Brien K. L., Simmons N. L., King S. L., Abbink P., Maxfield L. F., et al. (2010). Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat. Med. 16 319–323. 10.1038/nm.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker L.-G., Moodie Z., Grunenberg N., Laher F., Tomaras G. D., Cohen K. W., et al. (2018). Subtype C ALVAC-HIV and bivalent subtype C gp120/MF59 HIV-1 vaccine in low-risk, HIV-uninfected, South African adults: a phase 1/2 trial. Lancet HIV 5 e366–e378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchbinder S. P., Buchbinder S. P., Mehrotra D. V., Duerr A., Fitzgerald D. W., Mogg R., et al. (2008). Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372 1881–1893. 10.1016/s0140-6736(08)61591-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D., Kublin J. G., Ramsey S., Corey L. (2015). Are clade specific HIV vaccines a necessity? An analysis based on mathematical models. EBioMedicine 2 2062–2069. 10.1016/j.ebiom.2015.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell L. (2005). Therapeutic immunization strategies for the control of HIV-1. Expert Rev. Vaccines 4 513–520. 10.1586/14760584.4.4.513 [DOI] [PubMed] [Google Scholar]
- Esparza J., Chang M. L., Widdus R., Madrid Y., Walker N., Ghys P. D. (2003). Estimation of “needs” and “probable uptake” for HIV/AIDS preventive vaccines based on possible policies and likely acceptance (a WHO/UNAIDS/IAVI study). Vaccine 21 2032–2041. 10.1016/s0264-410x(02)00775-2 [DOI] [PubMed] [Google Scholar]
- Faria N. R., Rambaut A., Suchard M. A., Baele G., Bedford T., Ward M. J., et al. (2014). The early spread and epidemic ignition of HIV-1 in human populations. Science 346 56–61. 10.1126/science.1256739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauci A. S. (2017). An HIV vaccine is essential for ending the HIV/AIDS pandemic. J. Am. Med. Assoc. 318 1535–1536. 10.1001/jama.2017.13505 [DOI] [PubMed] [Google Scholar]
- Gaiha G. D., Rossin E. J., Urbach J., Landeros C., Collins D. R., Nwonu C., et al. (2019). Structural topology defines protective CD8(+) T cell epitopes in the HIV proteome. Science 364 480–484. 10.1126/science.aav5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Bailes E., Robertson D. L., Chen Y., Rodenburg C. M., Michael S. F., et al. (1999). Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397 436–441. 10.1038/17130 [DOI] [PubMed] [Google Scholar]
- Gaschen B., Taylor J., Yusim K., Foley B., Gao F., Lang D., et al. (2002). Diversity considerations in HIV-1 vaccine selection. Science 296 2354–2360. 10.1126/science.1070441 [DOI] [PubMed] [Google Scholar]
- Goulder P. J., Brander C., Annamalai K., Mngqundaniso N., Govender U., Tang Y., et al. (2000). Differential narrow focusing of immunodominant human immunodeficiency virus gag-specific cytotoxic T-lymphocyte responses in infected African and caucasoid adults and children. J. Virol. 74 5679–5690. 10.1128/jvi.74.12.5679-5690.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray G. E., Allen M., Moodie Z., Churchyard G., Bekker L. G., Nchabeleng M., et al. (2011). Safety and efficacy of the HVTN 503/Phambili study of a clade-B-based HIV-1 vaccine in South Africa: a double-blind, randomised, placebo-controlled test-of-concept phase 2b study. Lancet Infect. Dis. 11 507–515. 10.1016/s1473-3099(11)70098-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer S. M., Sobieszczyk M. E., Janes H., Karuna S. T., Mulligan M. J., Grove D., et al. (2013). Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N. Engl. J. Med. 369 2083–2092. 10.1056/NEJMoa1310566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelaar J. (2012). The origin and diversity of the HIV-1 pandemic. Trends Mol. Med. 18 182–192. 10.1016/j.molmed.2011.12.001 [DOI] [PubMed] [Google Scholar]
- Hemelaar J. (2013). Implications of HIV diversity for the HIV-1 pandemic. J. Infect. 66 391–400. 10.1016/j.jinf.2012.10.026 [DOI] [PubMed] [Google Scholar]
- Hemelaar J., Elangovan R., Yun J., Dickson-Tetteh L., Fleminger I., Kirtley S., et al. (2019). Global and regional molecular epidemiology of HIV-1, 1990–2015: a systematic review, global survey, and trend analysis. Lancet Infect. Dis. 19 143–155. [DOI] [PubMed] [Google Scholar]
- Hemelaar J., Elangovan R., Yun J., Dickson-Tetteh L., Kirtley S., Gouws-Williams E., et al. (2020). Global and regional epidemiology of HIV-1 recombinants in 1990-2015: a systematic review and global survey. Lancet HIV 7 e772–e781. [DOI] [PubMed] [Google Scholar]
- HIV Vaccine Study Group, Flynn N. M., Forthal D. N., Harro C. D., Judson F. N., Mayer K. H., et al. (2005). Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 191 654–665. 10.1086/428404 [DOI] [PubMed] [Google Scholar]
- Korber B., Gaschen B., Yusim K., Thakallapally R., Kesmir C., Detours V. (2001). Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 58 19–42. 10.1093/bmb/58.1.19 [DOI] [PubMed] [Google Scholar]
- Korber B., Hraber P., Wagh K., Hahn B. H. (2017). Polyvalent vaccine approaches to combat HIV-1 diversity. Immunol. Rev. 275 230–244. 10.1111/imr.12516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneau S., Im E. J., Mashishi T., Brereton C., Bridgeman A., Yang H., et al. (2007). Design and pre-clinical evaluation of a universal HIV-1 vaccine. PLoS One 2:e984. 10.1371/journal.pone.0000984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los Alamos National Laboratory Laboratory HIV Sequence Database. Los Alamos, NM: Los Alamos National Laboratory. Available online at: http://www.hiv.lanl.gov/ [Google Scholar]
- Marzetta C. A., Lee S. S., Wrobel S. J., Singh K. J., Russell N., Esparza J. (2010). The potential global market size and public health value of an HIV-1 vaccine in a complex global market. Vaccine 28 4786–4797. [DOI] [PubMed] [Google Scholar]
- Nickle D. C., Jensen M. A., Gottlieb G. S., Shriner D., Learn G. H., Rodrigo A. G., et al. (2003). Consensus and ancestral state HIV vaccines. Science 299 1515–1518. [DOI] [PubMed] [Google Scholar]
- Pitisuttithum P9., Gilbert P., Gurwith M., Heyward W., Martin M., van Griensven F., et al. (2006). Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J. Infect. Dis. 194 1661–1671. 10.1086/508748 [DOI] [PubMed] [Google Scholar]
- Rerks-Ngarm S., Pitisuttithum P., Nitayaphan S., Kaewkungwal J., Chiu J., Paris R., et al. (2009). Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361 2209–2220. [DOI] [PubMed] [Google Scholar]
- Robertson D. L., et al. (2000). HIV-1 nomenclature proposal. Science 288 55–56. [DOI] [PubMed] [Google Scholar]
- Tebit D. M., Arts E. J. (2011). Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infectious Diseases 11 45–56. 10.1016/s1473-3099(10)70186-9 [DOI] [PubMed] [Google Scholar]
- UNAIDS (2017). Global AIDS update 2017. Ending AIDS: Progress Towards the 90-90-90 Targets. Geneva: UNAIDS. [Google Scholar]
- UNAIDS (2020). Global AIDS Update. Geneva: UNAIDS. [Google Scholar]
- United Nations (2017). Department of Economic and Social Affairs, Population Division (2017): World Population Prospects 2017 – Data Booklet (ST/ESA/SER.A/401). New York, NY: United Nations. [Google Scholar]
- Worobey M., Gemmel M., Teuwen D. E., Haselkorn T., Kunstman K., Bunce M., et al. (2008). Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455 661–664. 10.1038/nature07390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi J., Vallari A., McArthur C., Sthreshley L., Cloherty G. A., Berg M. G., et al. (2020). Brief report: complete genome sequence of CG-0018a-01 establishes HIV-1 subtype L. J. Acquir. Immune Defic. Syndr. 83 319–322. 10.1097/qai.0000000000002246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambonelli C., Dey A. K., Hilt S., Stephenson S., Go E. P., Clark D. F., et al. (2016). Generation and characterization of a bivalent HIV-1 subtype C gp120 protein boost for proof-of-concept HIV vaccine efficacy trials in southern Africa. PLoS one 11:e0157391. 10.1371/journal.pone.0157391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li M., Min C., Ma Y., Shao Y., Xing H. (2020). Near full-length genomic characterization of a novel HIV-1 circulating recombinant form (CRF106_cpx) identifified among heterosexuals in China. AIDS Res. Hum. Retroviruses 36 875–880. 10.1089/aid.2020.0101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.