Abstract
COVID-19 has caused a pandemic crisis that threatens the world in many areas, especially in public health. For the diagnosis of COVID-19, computed tomography has a prognostic role in the early diagnosis of COVID-19 as it provides both rapid and accurate results. This is crucial to assist clinicians in making decisions for rapid isolation and appropriate patient treatment. Therefore, many researchers have shown that the accuracy of COVID-19 patient detection from chest CT images using various deep learning systems is extremely optimistic. Deep learning networks such as convolutional neural networks (CNNs) require substantial training data. One of the biggest problems for researchers is accessing a significant amount of training data. In this work, we combine methods such as segmentation, data augmentation and generative adversarial network (GAN) to increase the effectiveness of deep learning models. We propose a method that generates synthetic chest CT images using the GAN method from a limited number of CT images. We test the performance of experiments (with and without GAN) on internal and external dataset. When the CNN is trained on real images and synthetic images, a slight increase in accuracy and other results are observed in the internal dataset, but between and in the external dataset. It is promising according to the performance results that the proposed method will accelerate the detection of COVID-19 and lead to more robust systems.
Keywords: COVID-19, Computed tomography, Deep learning, Generative adversarial network, Lung segmentation, Data augmentation
Introduction
COVID-19 is an infectious disease with a high mortality rate caused by the mutant SARS-CoV-2 virus. Following the identification of the first case in Wuhan, China, in December 2019, it quickly spread all over the world and was declared a pandemic by WHO on March 11, 2020. COVID-19 has caused major public health problems in the international community due to its rapid spread all over the world [1].
It is extremely important to be able to diagnose COVID-19 in an infected patient during the pandemic process. Although polymerase chain reaction (PCR) testing is the standard for confirming COVID-19 positive patients, medical imaging such as X-ray and non-contrast computed tomography (CT) plays an important role in the detection of COVID-19. Since COVID-19 can be detected in the early stages with the presence of lung ground-glass opacities in CT images, which are clearer and more accurate than X-ray images, the diagnosis of COVID-19 from CT will help clinicians make quick decisions for rapid isolation and appropriate patient treatment [2].
Recently, computer vision has led to extraordinary developments with the advancement of artificial intelligence technology and especially the development of convolutional neural networks (CNN). It is widely used in the medical field and provides support for medical diagnosis [3–5]. Additionally, due to the heavy workload of large numbers of infected patients and healthcare professionals during the pandemic crisis, the AI-based computer-aided system could speed up the diagnostic process. These systems can provide effective diagnosis of the disease in a shorter time in cases such as radiologist insufficiency and deficiency. It can also reduce the need for human surveillance and identify details invisible to the human eye. In this context, many deep learning models have also begun to be developed for the diagnosis of COVID-19.
There is currently a great deal of interest in data-driven approaches to diagnosing COVID-19. Many researchers have used deep convolutional neural networks (CNN) such as VGG, ResNet, Inception, Xception and DenseNet, with well-known and proven performance, to diagnose COVID-19. However, deep learning networks such as convolutional neural networks require large amounts of training data. In addition, because CNNs have a large number of parameters, overfitting can easily occur in small datasets. However, medical imaging data are scarce, expensive and fraught with legal concerns over patient privacy, so datasets are small in size. A quick and easy method, data augmentation, can be used to overcome this problem [6]. The dataset is artificially expanded using techniques such as image rotation, transformation, scaling, brightness or contrast enhancement. However, it cannot be said that completely different images are produced since the same data are displayed differently with this method. In addition, the dataset can be enlarged by generating unique data by producing synthetic data with GAN, which is an advanced technique [7, 8].
The main contributions of this article can be summarized as follows:
It is difficult to identify COVID-19 as findings on CT images differ in both position and shape in different patients due to infections caused by COVID-19. In addition, we present a method for generating synthetic CT images by developing a GAN-based model to overcome the problem of overfitting in CNNs. In addition, we demonstrate the effectiveness of GAN-generated synthetic images in improving CNN’s generalization ability for COVID-19 detection in the context of performance criteria in internal and external datasets.
We use a segmentation model based on ConvLSTMU-Net architecture and graph-cut image processing for segmentation of the relevant lung region from CT images.
We use data enlargement techniques such as random distortion, rotation, flip and zoom with an in-place / on-the-fly augmentation approach to increase the effectiveness of the model during model training.
The rest of the paper is organized as follows. In Sect. 2, brief information about the studies related to our study is given. The materials and methods used in the study are presented in Sect. 3. Section 4 presents the results of different analyzes for different deep learning algorithms in the proposed frameworks on internal and external datasets. The comparisons with the other related studies and the discussion of the results are given in Sect. 5. Section 6 presents the conclusion of this study.
Related works
Recently, many studies have been conducted to perform data augmentation on medical images. Zhoe et al. [9] developed the forward and backward GAN (F&BGAN) method to create synthetic images of lung nodules in lung cancer and used the VGG16 network for benign and malignant classification. Chuquicusma et al. [10] produced realistic lung nodules using DCGAN for the training of radiologists. Frid-Adar et al. [11] proposed a magnification scheme for advanced liver lesion classification based on a combination of standard image perturbation and synthetic liver lesion generation using GAN. Guibas et al. [12] proposed a two-stage pipeline for generating synthetic medical images from a pair of GANs, which was tested in practice on retinal fundi images. Shin et al. [13] proposed a method to generate synthetic abnormal MRI images with brain tumors by training a GAN using two publicly available datasets of brain MRI.
Nowadays, there is intense interest in diagnosing COVID-19 using deep learning methods. Wu et al. [14] aimed to diagnose COVID-19 cases with the concept of multi-view fusion using the ResNet50 architecture. The datasets they used in their studies included 622 CT images collected from two different hospitals in China. They resized their images in the datasets to . The system they developed achieved accuracy, sensitivity and specificity. Xu et al. [15] collected the data they used in their study from 3 different hospitals in China. It achieved accuracy, sensitivity, accuracy and F1-score with pretrained ResNet18 architecture using 618 CT images. Jin et al. [16] used pretrained CNN models such as DPN-92, Inceptionv3, ResNet50 and Attention ResNet50 with 3D U-Net++ for the diagnosis of COVID-19. The datasets contain 850 COVID-19 positives and 541 negative COVID-19 images and were collected from 5 different hospitals in China. According to the performance criteria, their system accepted the 3D U-Net++ResNet50 model as the best model with accuracy, sensitivity and AUC values. Yousefzadeh et al. [17] introduced a deep learning framework using different CNN architectures DenseNet, ResNet, Xception and EfficientNetB0. The datasets contain a total of 2124 CT images. The system they proposed resulted in accuracy, sensitivity, specificity, F1 score and AUC. Ardakani et al. [18] proposed a system for detecting COVID-19 using CNN architectures such as AlexNet, VGG-16, VGG-19, SqueezeNet, GoogleNet, MobileNetv2, ResNet18, ResNet50, ResNet101 and Xception. The dataset they use contains 1020 CT images in total. Among the 10 networks, ResNet101 and Xception performed better than the others according to their performance values. Chen et al. [19] used the U-Net++ segmentation model together with the ResNet50 model to diagnose COVID-19 using a very large and non-public dataset of 106 patients from the Renmin Hospital of Wuhan University. The results of COVID-19 classification are accuracy, sensitivity and specificity. The U-Net and 3D CNN models are used for lung segmentation and diagnosing of COVID-19, respectively, in Ref. [20]. The results of the model are sensitivity, specificity and AUC. Most of the previous works on COVID-19 are given in Refs. [21–23].
Material and methods
Dataset
We use two different datasets in our study:
The dataset, which was collected from different open sources [24], contains 1607 COVID-19 and 1667 normal CT images. We randomly allocate 80% of the dataset for training and 20% for testing. We will refer to this dataset as the internal dataset throughout the study.
The external dataset in Ref. [25] contains 4001 COVID-19 and 9575 normal CT images. We use this dataset to evaluate the performance of the trained models.
Preprocessing
Since the internal dataset is collected from different sources, it contains CT images with a resolution of min - max . For these reasons, the images in the internal dataset were resized to resolution. In the external dataset, all CT images are high-resolution images. We also reduce the size of the CT images in this dataset to to be standard and perform central zooming.
Lung segmentation
Since distinctive information is in the lungs on CT images, we apply lung segmentation to obtain only the lung area. For segmentation, we applied a similar approach to BidirectionalConvLSTMU-Net (BDCU-Net) [26] architecture. A total of 1667 normal CT data and masks in the internal dataset were used for training the lung segmentation architecture. We reserve of the dataset for training and of the dataset for validation.
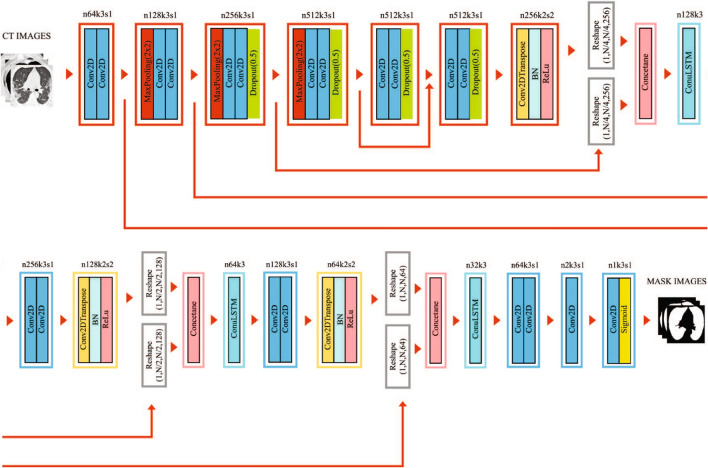
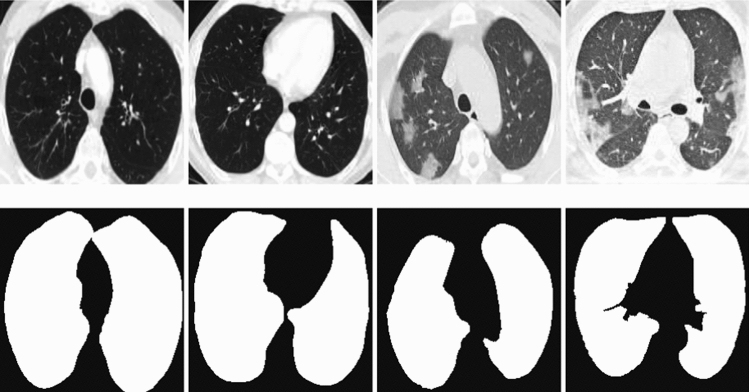
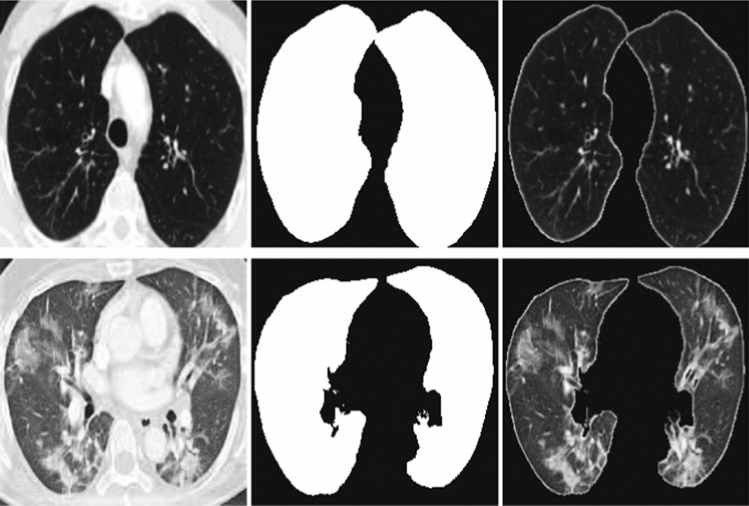
We initially started with learning rate and training epochs of 30. We dynamically reduced the learning rate when there was no improvement in validation error for 4 consecutive training periods. When there is no improvement in validation error for 8 consecutive training epochs, it is taken as the criterion for early stopping of training. The Adam stochastic optimization algorithm, which was developed as a solution to the disappearing gradient problem, was used for parameter optimization. At the end of the 26th epoch, an early stop occurs, and accuracy: 0.9675 is obtained. The architecture of obtaining the mask images from the CT images by using lung segmentation model is given in Fig. 1. The sample images and obtained mask images by using the lung segmentation model are given in Fig. 2. The example images for the relevant region obtained as a result of Graphcut image processing [27] are given in Fig. 3.
Fig. 1.
The architecture of obtaining the mask images from the CT images by using BDCU-Net model
Fig. 2.
Sample CT images and mask images obtained as a result of BDCU-Net
Fig. 3.
Sample images for the region of interest obtained after applying Graphcut image processing
Generating synthetic images
Generative adversarial network (GAN)
The GAN framework is a widely used modern method for generating synthetic data from many domains [28, 29]. Examples of data generation with GANs include text-to-image synthesis, superimage resolution, style transfer and symbolic music production [30].
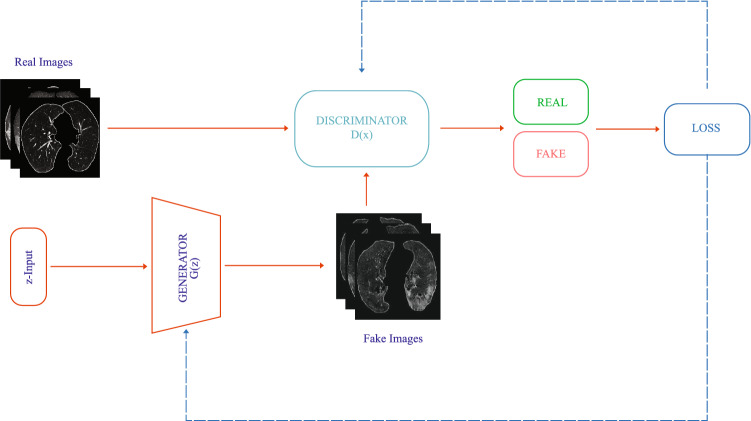
The working principle of GANs is simply; GAN is a min-max game between two hostile networks, generator G(z) and discriminator D(x). The generator tries to convert the random noise into observations that appear to be sampled from the original dataset, and the discriminator tries to guess whether an observation comes from the original dataset or is one of the generator’s fakes. At the start of the process, generator G(z) takes a z-point (random noise) from a latent space and outputs noisy images and tries to fool the discriminator D(x). If the discriminator is D(x), the generator tries to distinguish whether the images from are real or fake. The key of GANs lies in how we change the training of the two networks so that as the generator becomes more adept at deceiving the discriminator, the discriminator must adapt to maintain its ability to accurately identify which observations are fake. Thus, the generator tries to find new ways to fool the discriminator so that the loop continues between the two networks. The purpose of G(z) is to minimize the cost function V(D, G) and to maximize the D(x) by training both G(z) and D(x) at the same time:
| 1 |
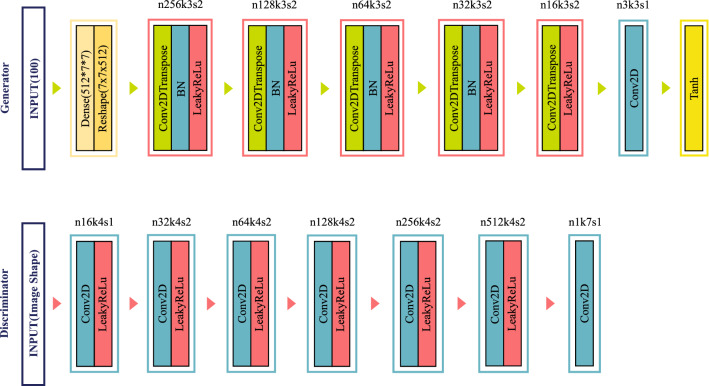
The architecture of GAN applied in the study is given in Fig. 4
Fig. 4.
The framework of GAN
GAN Train Procedure
Initially, the parameters of the discriminator network are set to non-trainable. The output of the generator network feeds the discriminator and the generator is updated through the discriminator, so the generator is stacked on the discriminator. The hyperparameter of LeakyReLu layers used in discriminator and generator networks is , and momentum value of batch normalization layer used in generator is 0.8. The detailed designs of the discriminator and generator network used in the study are given in Fig. 5.
Fig. 5.
The detailed designs of the discriminator and generator network used in the study
of the internal dataset reserved for training iz used in GAN training, i.e., 1286 COVID-19 and 1333 normal CT images. Test data are not included in the training. The pixel values of the CT images are normalized from [0, 255] to as a preprocessing. In the training of both networks, Adam optimization algorithm with learning rate and momentum value is used as hyperparameters. The least-squares function is used as the loss function. In addition, the number of training periods is determined as 40000 and the batch size is 64. The latent space dimension where the generator is fed, is taken as 100.
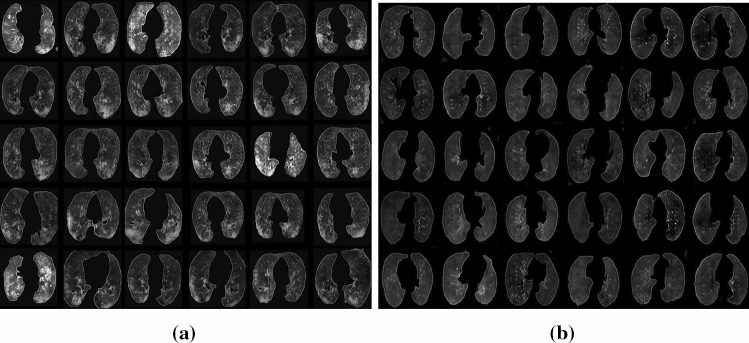
A total of 2314 normal and 2267 COVID-19 synthetic images are generated by GAN. The samples of the COVID-19 and normal synthetic images produced after the GAN training are given in Fig. 6.
Fig. 6.
a The samples of the COVID-19 synthetic images produced after the training with GAN b The samples of the normal synthetic images produced after the training with GAN
CNN architectures
Deep CNN architectures pretrained on 9 well-known ImageNet [31] datasets were used in this study: VGG16, VGG19, Xception, ResNet50, ResNet50v2, Inceptionv3, InceptionResNetv2, DenseNet121 and DenseNet169.
VGG16 was developed to win the ILSVRC2014 (Large Scale Visual Recognition Challenge 2014) in 2014 [32]. VGG16 is a convolutional neural network architecture named after the Visual Geometry Group of Oxford developed. VGG-16 is a convolutional neural network with a depth of 16 layers. The model loads a pretrained set of weights on ImageNet and reaches accuracy on ImageNet, a dataset of more than 14 million images belonging to 1000 classes. VGG-19 is the version of VGG with a depth of 19 layers. VGG16 has 138, 357, 544 parameters while VGG19 has 143, 667, 240 parameters.
He et al. [33] developed the architecture called residual network (ResNet) to solve the vanishing/exploding gradient problem. ResNet is created by adding the blocks that feed the residual values to the next layers into the model. This is also called residual learning. ResNet-50 has 16 residual blocks. The main difference between ResNet50 and ResNet50v2 is now the arrangement of layers in blocks. ResNet50 and ResNet50v2 achieved and accuracy on the ImageNet dataset, respectively.
Inception [34] and InceptionResNet [35] are developed by Google. The concept of Inception represents Inception modules within the network. This is a simple and powerful architectural unit that also allows the model to learn parallel filters of different sizes and multiple scales. The main difference between Inception and InceptionResNet is that InceptionResNet uses residual layers. Inceptionv3 using the Inception modules has 23, 851, 784 while InceptionResNet has 55, 873, 736. Inceptionv3 and InceptionResNetv2 achieved and accuracy on the ImageNet dataset, respectively.
Xception [36] is a deep convolutional neural network architecture that includes deeply separable convolutions and developed by Google researchers. Google presented an interpretation of the Initiation modules in convolutional neural networks as an intermediate step between regular convolution and deeply separable convolution processing. Xception has 22, 910, 480 parameters and achieved accuracy on the ImageNet dataset.
DenseNet [37] is a type of convolutional neural network that uses dense connections between layers through dense blocks that connect all layers directly. Each layer receives additional inputs from all previous layers and transmits its own feature maps to all subsequent layers. The difference between DenseNet121 and DenseNet169 is the number of layers. DenseNet121 has 8, 062, 504 parameters while DenseNet169 has 14, 307, 880 parameters. DenseNet121 and DenseNet169 achieved and accuracy on the ImageNet dataset, respectively.
Training of models and implementation details
In this study, two types of experimental studies (with GAN and without GAN) are conducted using the real images from the internal dataset and incorporating synthetic images generated by the GAN into the internal dataset. Details of the dataset used in the two studies are given in Table 1. The 1667 normal class images in the internal dataset used in the training of models are the same as the images used in the BConvLSTM U-Net training.
Table 1.
The numbers of the real images without GAN and the real+synthetic images produced with GAN in training and test sets
| Framework | Types | Training size | Test size |
|---|---|---|---|
| Real images | COVID-19 | 1286 | 321 |
| Without GAN | Normal | 1333 | 334 |
| Real+synthetic images | COVID-19 | 3600 | 321 |
| Produced with GAN | Normal | 3600 | 334 |
All networks in both frameworks, the pixel values of the CT images, were normalized from [0, 255] to [0, 1] as a preprocessing. In both experiments, Adam optimization algorithm with learning rate and momentum value was used as hyperparameter. Binary cross-entropy function is used as loss function. In addition, the number of training epochs is determined as 40 and the batch size was 16. The learning rate is reduced when there is no improvement in validation error for 5 consecutive training periods. It was taken as the criterion for early discontinuation of training when there is no improvement in validation error for 10 consecutive training periods. A fivefold cross-validation is performed and tested on a randomly selected of the internal dataset and the external dataset.
In both experiments, pretrained deep CNN models VGG16, VGG19, Xception, ResNet50, ResNet50V2, DenseNet121, DenseNet169, InceptionV3 and InceptionResNetV2 were used and fine-tuned. Dense(512, relu), BatchNormalization (0.9), Dense(256, relu), Dense (1, Sigmoid) layers are used, respectively, in the fully connected layer for classification. The working principle of this study is given in Fig. 7
Fig. 7.
The flow chart of both experiments is given in Fig. 5
Data augmentation
The performance of deep learning neural networks is generally directly proportional to the amount of data available. Data augmentation is referred to as a technique of artificially deriving data from existing training data through much more image processing such as rotating, panning, zooming, flipping [6]. The aim is to increase the generalization performance of the model while expanding the training dataset with new examples. Thus, the trained model constantly sees new, different versions of the input data, and the model can learn more robust features.
There are three types of data augmentation methods: first, expanding the existing dataset. The problem with this approach is that it does not fully increase the generalization ability of the model. The second is in-place/on-the-fly data augmentation. The network is trained to see new variations of the data in each epoch by using this type of data augmentation. This method increases the generalization ability of the model. Finally, a hybrid approach combines the first and the second approaches.
In our study, we use only in-place/on-the-fly augmentation approach in models trained with real data; in the second experiment, we increase the size of the dataset with synthetic data produced with GAN, and we adopt a hybrid approach with in-place/on-the-fly augmentation during the training of models. The applied data augmentation processes and sample images are given in Table 2 and Fig. 8, respectively. Random distortion allows you to distort the image while maintaining elastically the image aspect ratio. The magnitude parameter determines how much it degrades. Flip left-right acts as a mirror on images. Rotation was used to rotate left and right a certain degree. Zoom was used to enlarge and reduce the image centrally.
Table 2.
Types of data augmentation
| Types | Parameters |
|---|---|
| Random distortion | probability=0.5 |
| Grid width=4 | |
| Grid height=4 | |
| Magnitude=10 | |
| Flip (left, right) | Probability=0.5 |
| Rotate | Probability=0.5 |
| Max left rotation=10 | |
| Max right rotation=10 | |
| Zoom | Probability=0.5 |
| Min factor=0.9 | |
| Max factor=1.20 |
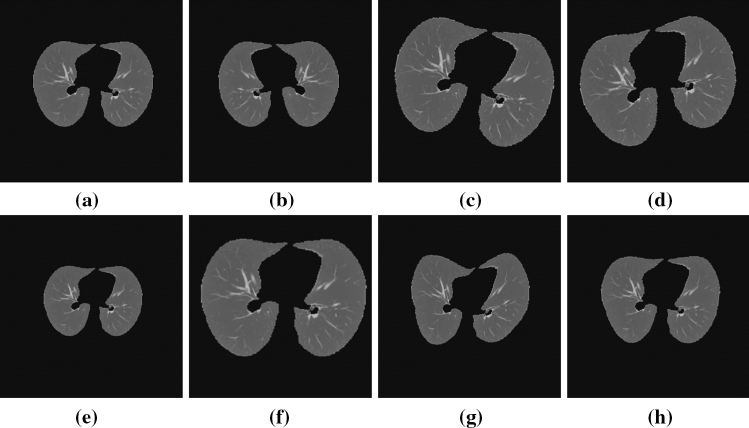
Fig. 8.
a Original image b result of the flip left-right c result of the right rotation d result of the left rotation e result of a 0.9 rate zoom f result of a 1.20 rate zoom. g and h results of random distortion
Results
All training and testing processes are performed using AMD Ryzen 3970X CPU with 128GB RAM and Nvidia RTX 3080 GPU with 10GB memory. The Keras [38] deep learning library is used for processes.
Estimation performances of the methods in this study are measured with metrics such as accuracy, precision, recall and F1-score. The confusion matrix used to explain the performance of the classification model consists of true positive (), true negative (TN), false positive (FP) and false negatives (FN).
Dividing the number of correctly classified cases into the total number of test images shows the accuracy and is calculated as follows.
| 2 |
where is the number of instances that correctly predicted, is the number of instances that incorrectly predicted, is the number of negative instances that correctly predicted, is the number of negative instances that incorrectly predicted.
The recall is used to measure correctly classified COVID-19 cases. Recall is calculated as follows.
| 3 |
The percentage of correctly classified labels in truly positive patients is defined as the precision and is calculated as follows.
| 4 |
Specificity is the proportion of people who test negatively among those who actually do not have the disease and is defined as:
| 5 |
The F1-score is defined as the weighted average of precision and recall combining both precision and recall and is calculated as follows.
| 6 |
The same real images are used in the tests of the both experiments on both internal and external datasets. Information on test sets size is given in Table 3. The comparison of performance results in internal and external datasets for both experiments is given in Tables 4 and 5, respectively. Confusion values and performance results for each model are given in Section Appendix.
Table 3.
Information on the size of the test sets
| Dataset | Types | Testing set |
|---|---|---|
| Internal | COVID-19 | 321 |
| Normal | 334 | |
| External | COVID-19 | 9545 |
| Normal | 4001 |
Table 4.
Comparison performance results of models without GAN and with GAN on internal dataset
| Model | Method | Accuracy | Precision | Recall | Specificity | F1-score |
|---|---|---|---|---|---|---|
| VGG16 | without GAN | 0.9814 | 0.9898 | 0.9720 | 0.9904 | 0.9808 |
| with GAN | 0.9872 | 0.9949 | 0.9788 | 0.9952 | 0.9868 | |
| VGG19 | without GAN | 0.9841 | 0.9924 | 0.9751 | 0.9928 | 0.9837 |
| with GAN | 0.9875 | 0.9894 | 0.9850 | 0.9898 | 0.9872 | |
| Xception | without GAN | 0.9902 | 0.9900 | 0.9900 | 0.9904 | 0.9900 |
| with GAN | 0.9927 | 0.9913 | 0.9938 | 0.9916 | 0.9925 | |
| ResNet50 | without GAN | 0.9798 | 0.9819 | 0.9769 | 0.9826 | 0.9794 |
| with GAN | 0.9911 | 0.9969 | 0.9850 | 0.9970 | 0.9909 | |
| ResNet50v2 | without GAN | 0.9860 | 0.9931 | 0.9782 | 0.9934 | 0.9856 |
| with GAN | 0.9942 | 0.9975 | 0.9907 | 0.9976 | 0.9941 | |
| InceptionV3 | without GAN | 0.9881 | 0.9949 | 0.9807 | 0.9952 | 0.9878 |
| with GAN | 0.9951 | 0.9949 | 0.9913 | 0.9952 | 0.9950 | |
| InceptionResNetV2 | without GAN | 0.9893 | 0.9931 | 0.9850 | 0.9934 | 0.9890 |
| with GAN | 0.9893 | 0.9962 | 0.9913 | 0.9964 | 0.9938 | |
| DenseNet121 | without GAN | 0.9893 | 0.9919 | 0.9863 | 0.9922 | 0.9891 |
| with GAN | 0.9921 | 0.9919 | 0.9919 | 0.9922 | 0.9919 | |
| DenseNet169 | withoutGAN | 0.9911 | 0.9944 | 0.9875 | 0.9946 | 0.9909 |
| with GAN | 0.9927 | 0.9956 | 0.9894 | 0.9958 | 0.9925 |
Table 5.
Comparison performance results of models without GAN and with GAN on external dataset
| Model | Method | Accuracy | Precision | Recall | Specificity | F1-score |
|---|---|---|---|---|---|---|
| VGG16 | without GAN | 0.8670 | 0.7447 | 0.8364 | 0.8799 | 0.7879 |
| with GAN | 0.9023 | 0.8024 | 0.8877 | 0.9083 | 0.8429 | |
| VGG19 | without GAN | 0.8836 | 0.7646 | 0.8758 | 0.8868 | 0.8194 |
| with GAN | 0.9086 | 0.8086 | 0.9048 | 0.9102 | 0.8540 | |
| Xception | without GAN | 0.9145 | 0.8039 | 0.9399 | 0.9039 | 0.8666 |
| with GAN | 0.9486 | 0.8779 | 0.9593 | 0.9440 | 0.9168 | |
| ResNet50 | without GAN | 0.8978 | 0.7900 | 0.8906 | 0.9007 | 0.8373 |
| with GAN | 0.9303 | 0.8644 | 0.9061 | 0.9404 | 0.8847 | |
| ResNet50v2 | without GAN | 0.8914 | 0.7755 | 0.8902 | 0.8919 | 0.8289 |
| with GAN | 0.9167 | 0.8272 | 0.9077 | 0.9205 | 0.8656 | |
| InceptionV3 | without GAN | 0.9061 | 0.8075 | 0.8955 | 0.9105 | 0.8492 |
| with GAN | 0.9498 | 0.8936 | 0.9423 | 0.9530 | 0.9173 | |
| InceptionResNetV2 | without GAN | 0.8983 | 0.7928 | 0.8875 | 0.9028 | 0.8374 |
| with GAN | 0.9373 | 0.8682 | 0.9287 | 0.9409 | 0.8974 | |
| DenseNet121 | without GAN | 0.9000 | 0.7953 | 0.8907 | 0.9039 | 0.8403 |
| with GAN | 0.9346 | 0.8673 | 0.9203 | 0.9407 | 0.8930 | |
| DenseNet169 | withoutGAN | 0.9032 | 0.7982 | 0.8997 | 0.9046 | 0.8459 |
| with GAN | 0.9319 | 0.8624 | 0.9156 | 0.9388 | 0.8882 |
Discussion
In this paper, two experiments with and without GAN were tested on two different datasets (655 internal and 14545 external) with different deep learning methods.
Table 4 analyzes the performance results of models with and without GAN on internal dataset. We clearly see that in all deep learning models with GAN, a slightly better performance ratio is achieved in all of the accuracy, precision, recall, specificity and F1-score values compared to the models without GAN. The best detection accuracy rate obtained in the InceptionV3 model with GAN on the internal dataset is and the F1-score rate is .
Table 5 analyzes the performance results of models with and without GAN on different external dataset that the system has never seen before. We clearly see that in all deep learning models with GAN, a better performance rate of to is achieved in all of the accuracy, precision, recall, specificity and F1-score values compared to the models without GAN. The best detection accuracy rate obtained in the InceptionV3 model with GAN on the external dataset is and the F1-score rate is .
The results of this study reveal that synthetic augments produced with GAN make an important contribution to the improvement of generalization performance, especially in external dataset.
Conclusion
In this research, we developed a GAN-based model to overcome the overfitting problem in CNNs and improve their generalization performance and proposed a model in which the dataset can be artificially amplified with synthetic CT images.
An internal dataset containing 3274 CT images was used for training two different experiments (with and without GAN). The improvement in the generalization ability of CNN models was investigated using synthetic data augmentation technique with GAN on this limited dataset. Both experiments were tested on an external dataset.
This study uses two classes (COVID-19 and normal) for classification. Synthetic data augmentation with GAN expands the dataset, providing more variability. When the CNN is trained on real images and synthetic images, a slight increase in accuracy and the other results is observed in the internal dataset, but between and in the external dataset. Performance results show that synthetic images produced with GAN make a significant contribution to the detection of COVID-19, especially in external dataset that the system has not seen before.
In conclusion, we proposed a method to increase the accuracy of COVID-19 detection with minimal dataset by producing synthetic images of CT images. The proposed method will improve the generalization performance of CNNs and lead to more robust systems.
Acknowledgements
We would like to thank referees for valuable suggestions.
Confusion values and performance results for each model
Confusion values and performance results for each model are given in Tables 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40 and 41.
Table 6.
Confusion values of VGG16 on internal dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 311 | 10 | 331 | 3 |
| Fold2 | 314 | 7 | 332 | 2 | |
| Fold3 | 313 | 8 | 330 | 4 | |
| Fold4 | 310 | 11 | 331 | 3 | |
| Fold5 | 312 | 9 | 331 | 3 | |
| With GAN | Fold1 | 315 | 6 | 331 | 3 |
| Fold2 | 314 | 7 | 332 | 2 | |
| Fold3 | 313 | 7 | 332 | 1 | |
| Fold4 | 312 | 7 | 332 | 1 | |
| Fold5 | 312 | 7 | 331 | 1 |
Table 7.
Performance results of VGG16 on internal dataset
| KFold | Accuracy | Precision | Recall | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.9787 | 0.9873 | 0.9688 | 0.9881 | 0.9780 |
| Fold2 | 0.9863 | 0.9937 | 0.9782 | 0.9940 | 0.9859 | |
| Fold3 | 0.9817 | 0.9874 | 0.9751 | 0.9880 | 0.9812 | |
| Fold4 | 0.9789 | 0.9904 | 0.9657 | 0.9910 | 0.9779 | |
| Fold5 | 0.9817 | 0.9905 | 0.9720 | 0.9910 | 0.9811 | |
| Overall | 0.9844 | 0.9898 | 0.9720 | 0.9904 | 0.9808 | |
| With GAN | Fold1 | 0.9863 | 0.9906 | 0.9813 | 0.9910 | 0.9859 |
| Fold2 | 0.9863 | 0.9937 | 0.9782 | 0.9940 | 0.9859 | |
| Fold3 | 0.9878 | 0.9968 | 0.9782 | 0.9970 | 0.9874 | |
| Fold4 | 0.9878 | 0.9968 | 0.9782 | 0.9970 | 0.9874 | |
| Fold5 | 0.9878 | 0.9968 | 0.9782 | 0.9970 | 0.9874 | |
| Overall | 0.9872 | 0.9949 | 0.9788 | 0.9952 | 0.9868 |
Table 8.
Confusion values of VGG19 on internal dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 312 | 9 | 333 | 1 |
| Fold2 | 313 | 8 | 332 | 2 | |
| Fold3 | 314 | 7 | 332 | 2 | |
| Fold4 | 316 | 5 | 331 | 3 | |
| Fold5 | 315 | 6 | 332 | 2 | |
| With GAN | Fold1 | 315 | 6 | 330 | 4 |
| Fold2 | 315 | 6 | 330 | 4 | |
| Fold3 | 317 | 4 | 331 | 3 | |
| Fold4 | 317 | 4 | 331 | 3 | |
| Fold5 | 317 | 4 | 331 | 3 |
Table 9.
Performance results of VGG19 on internal dataset
| KFold | Accuracy | Precision | Recall | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.9847 | 0.9968 | 0.9720 | 0.9970 | 0.9842 |
| Fold2 | 0.9847 | 0.9937 | 0.9751 | 0.9940 | 0.9843 | |
| Fold3 | 0.9863 | 0.9937 | 0.9782 | 0.9940 | 0.9859 | |
| Fold4 | 0.9878 | 0.9906 | 0.9844 | 0.9910 | 0.9875 | |
| Fold5 | 0.9878 | 0.9937 | 0.9813 | 0.9940 | 0.9875 | |
| Overall | 0.9863 | 0.9937 | 0.9782 | 0.9940 | 0.9859 | |
| With GAN | Fold1 | 0.9847 | 0.9875 | 0.9813 | 0.9880 | 0.9844 |
| Fold2 | 0.9847 | 0.9875 | 0.9813 | 0.9880 | 0.9844 | |
| Fold3 | 0.9893 | 0.9906 | 0.9875 | 0.9910 | 0.9891 | |
| Fold4 | 0.9893 | 0.9906 | 0.9875 | 0.9910 | 0.9891 | |
| Fold5 | 0.9893 | 0.9906 | 0.9875 | 0.9910 | 0.9891 | |
| Overall | 0.9875 | 0.9894 | 0.9850 | 0.9898 | 0.9872 |
Table 10.
Confusion values of Xception on internal dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 317 | 4 | 332 | 2 |
| Fold2 | 318 | 3 | 331 | 3 | |
| Fold3 | 318 | 3 | 330 | 4 | |
| Fold4 | 318 | 3 | 330 | 4 | |
| Fold5 | 318 | 3 | 331 | 3 | |
| With GAN | Fold1 | 319 | 2 | 332 | 2 |
| Fold2 | 319 | 2 | 331 | 3 | |
| Fold3 | 319 | 2 | 331 | 3 | |
| Fold4 | 319 | 2 | 331 | 3 | |
| Fold5 | 319 | 2 | 331 | 3 |
Table 11.
Performance results of Xception without GAN on internal dataset
| KFold | Accuracy | Precision | Recall | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.9908 | 0.9937 | 0.9875 | 0.9940 | 0.9906 |
| Fold2 | 0.9908 | 0.9907 | 0.9907 | 0.9910 | 0.9907 | |
| Fold3 | 0.9893 | 0.9876 | 0.9907 | 0.9880 | 0.9891 | |
| Fold4 | 0.9893 | 0.9876 | 0.9907 | 0.9880 | 0.9891 | |
| Fold5 | 0.9908 | 0.9907 | 0.9907 | 0.9910 | 0.9907 | |
| Overall | 0.9902 | 0.9900 | 0.9900 | 0.9904 | 0.9900 | |
| With GAN | Fold1 | 0.9939 | 0.9938 | 0.9938 | 0.9940 | 0.9938 |
| Fold2 | 0.9924 | 0.9907 | 0.9938 | 0.9910 | 0.9922 | |
| Fold3 | 0.9924 | 0.9907 | 0.9938 | 0.9910 | 0.9922 | |
| Fold4 | 0.9924 | 0.9907 | 0.9938 | 0.9910 | 0.9922 | |
| Fold5 | 0.9924 | 0.9907 | 0.9938 | 0.9910 | 0.9922 | |
| Overall | 0.9927 | 0.9913 | 0.9938 | 0.9916 | 0.9925 |
Table 12.
Confusion values of ResNet50 on internal dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 311 | 10 | 325 | 9 |
| Fold2 | 314 | 7 | 326 | 8 | |
| Fold3 | 315 | 6 | 329 | 5 | |
| Fold4 | 313 | 8 | 330 | 4 | |
| Fold5 | 315 | 6 | 331 | 3 | |
| With GAN | Fold1 | 317 | 4 | 332 | 2 |
| Fold2 | 317 | 4 | 333 | 1 | |
| Fold3 | 317 | 4 | 334 | 0 | |
| Fold4 | 315 | 6 | 333 | 1 | |
| Fold5 | 315 | 6 | 333 | 1 |
Table 13.
Performance results of ResNet50 on internal dataset
| KFold | Accuracy | Precision | Recall | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.9710 | 0.9719 | 0.9688 | 0.9731 | 0.9704 |
| Fold2 | 0.9771 | 0.9752 | 0.9782 | 0.9760 | 0.9767 | |
| Fold3 | 0.9832 | 0.9844 | 0.9813 | 0.9850 | 0.9828 | |
| Fold4 | 0.9817 | 0.9874 | 0.9751 | 0.9880 | 0.9812 | |
| Fold5 | 0.9863 | 0.9906 | 0.9813 | 0.9910 | 0.9859 | |
| Overall | 0.9798 | 0.9819 | 0.9769 | 0.9826 | 0.9794 | |
| With GAN | Fold1 | 0.9908 | 0.9937 | 0.9875 | 0.9940 | 0.9906 |
| Fold2 | 0.9924 | 0.9969 | 0.9875 | 0.9970 | 0.9922 | |
| Fold3 | 0.9939 | 1.0000 | 0.9875 | 1.0000 | 0.9937 | |
| Fold4 | 0.9893 | 0.9968 | 0.9813 | 0.9970 | 0.9890 | |
| Fold5 | 0.9893 | 0.9968 | 0.9813 | 0.9970 | 0.9890 | |
| Overall | 0.9911 | 0.9969 | 0.9850 | 0.9970 | 0.9909 |
Table 14.
Confusion values of ResNet50v2 on internal dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 312 | 9 | 333 | 1 |
| Fold2 | 312 | 9 | 333 | 1 | |
| Fold3 | 315 | 6 | 331 | 3 | |
| Fold4 | 314 | 7 | 331 | 3 | |
| Fold5 | 317 | 4 | 331 | 3 | |
| With GAN | Fold1 | 317 | 4 | 332 | 2 |
| Fold2 | 317 | 4 | 333 | 1 | |
| Fold3 | 319 | 2 | 333 | 1 | |
| Fold4 | 318 | 3 | 334 | 0 | |
| Fold5 | 319 | 2 | 334 | 0 |
Table 15.
Performance results of ResNet50v2 on internal dataset
| KFold | Accuracy | Precision | Recall (Sensitivity) | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.9847 | 0.9968 | 0.9720 | 0.9970 | 0.9842 |
| Fold2 | 0.9847 | 0.9968 | 0.9720 | 0.9970 | 0.9842 | |
| Fold3 | 0.9863 | 0.9906 | 0.9813 | 0.9910 | 0.9859 | |
| Fold4 | 0.9847 | 0.9905 | 0.9782 | 0.9910 | 0.9843 | |
| Fold5 | 0.9893 | 0.9906 | 0.9875 | 0.9910 | 0.9891 | |
| Overall | 0.9860 | 0.9931 | 0.9782 | 0.9934 | 0.9856 | |
| With GAN | Fold1 | 0.9908 | 0.9937 | 0.9875 | 0.9940 | 0.9906 |
| Fold2 | 0.9924 | 0.9969 | 0.9875 | 0.9970 | 0.9922 | |
| Fold3 | 0.9954 | 0.9969 | 0.9938 | 0.9970 | 0.9953 | |
| Fold4 | 0.9954 | 1.0000 | 0.9907 | 1.0000 | 0.9953 | |
| Fold5 | 0.9969 | 1.0000 | 0.9938 | 1.0000 | 0.9969 | |
| Overall | 0.9942 | 0.9975 | 0.9907 | 0.9976 | 0.9941 |
Table 16.
Confusion values of InceptionV3 on internal dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 315 | 6 | 334 | 0 |
| Fold2 | 315 | 6 | 332 | 2 | |
| Fold3 | 315 | 6 | 332 | 2 | |
| Fold4 | 315 | 6 | 332 | 2 | |
| Fold5 | 314 | 7 | 332 | 2 | |
| With GAN | Fold1 | 321 | 0 | 333 | 1 |
| Fold2 | 318 | 3 | 333 | 1 | |
| Fold3 | 318 | 3 | 334 | 0 | |
| Fold4 | 316 | 5 | 334 | 0 | |
| Fold5 | 318 | 3 | 334 | 0 |
Table 17.
Performance results of InceptionV3 on internal dataset
| KFold | Accuracy | Precision | Recall (Sensitivity) | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.9908 | 1.0000 | 0.9813 | 1.0000 | 0.9906 |
| Fold2 | 0.9878 | 0.9937 | 0.9813 | 0.9940 | 0.9875 | |
| Fold3 | 0.9878 | 0.9937 | 0.9813 | 0.9940 | 0.9875 | |
| Fold4 | 0.9878 | 0.9937 | 0.9813 | 0.9940 | 0.9875 | |
| Fold5 | 0.9863 | 0.9937 | 0.9782 | 0.9940 | 0.9859 | |
| Overall | 0.9881 | 0.9949 | 0.9807 | 0.9952 | 0.9878 | |
| With GAN | Fold1 | 0.9985 | 0.9969 | 1.0000 | 0.9970 | 0.9984 |
| Fold2 | 0.9939 | 0.9969 | 0.9907 | 0.9970 | 0.9938 | |
| Fold3 | 0.9954 | 1.0000 | 0.9907 | 1.0000 | 0.9953 | |
| Fold4 | 0.9924 | 1.0000 | 0.9844 | 1.0000 | 0.9922 | |
| Fold5 | 0.9954 | 1.0000 | 0.9907 | 1.0000 | 0.9953 | |
| Overall | 0.9951 | 0.9949 | 0.9913 | 0.9952 | 0.9950 |
Table 18.
Confusion values of InceptionResNetV2 on internal dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 313 | 8 | 334 | 0 |
| Fold2 | 318 | 3 | 331 | 3 | |
| Fold3 | 317 | 4 | 332 | 2 | |
| Fold4 | 316 | 5 | 331 | 3 | |
| Fold5 | 317 | 4 | 331 | 3 | |
| With GAN | Fold1 | 319 | 2 | 332 | 2 |
| Fold2 | 318 | 3 | 333 | 1 | |
| Fold3 | 318 | 3 | 333 | 1 | |
| Fold4 | 318 | 3 | 333 | 1 | |
| Fold5 | 318 | 3 | 333 | 1 |
Table 19.
Performance results of InceptionResNetV2 on internal dataset
| KFold | Accuracy | Precision | Recall (Sensitivity) | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.9878 | 1.0000 | 0.9751 | 1.0000 | 0.9874 |
| Fold2 | 0.9908 | 0.9907 | 0.9907 | 0.9910 | 0.9907 | |
| Fold3 | 0.9908 | 0.9937 | 0.9875 | 0.9940 | 0.9906 | |
| Fold4 | 0.9878 | 0.9906 | 0.9844 | 0.9910 | 0.9875 | |
| Fold5 | 0.9893 | 0.9906 | 0.9875 | 0.9910 | 0.9891 | |
| Overall | 0.9893 | 0.9931 | 0.9850 | 0.9934 | 0.9890 | |
| With GAN | Fold1 | 0.9939 | 0.9938 | 0.9938 | 0.9940 | 0.9938 |
| Fold2 | 0.9939 | 0.9969 | 0.9907 | 0.9970 | 0.9938 | |
| Fold3 | 0.9939 | 0.9969 | 0.9907 | 0.9970 | 0.9938 | |
| Fold4 | 0.9939 | 0.9969 | 0.9907 | 0.9970 | 0.9938 | |
| Fold5 | 0.9939 | 0.9969 | 0.9907 | 0.9970 | 0.9938 | |
| Overall | 0.9893 | 0.9962 | 0.9913 | 0.9964 | 0.9938 |
Table 20.
Confusion values of DenseNet121 on internal dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 316 | 5 | 331 | 3 |
| Fold2 | 317 | 4 | 332 | 2 | |
| Fold3 | 317 | 4 | 332 | 2 | |
| Fold4 | 317 | 4 | 331 | 3 | |
| Fold5 | 316 | 5 | 331 | 3 | |
| With GAN | Fold1 | 318 | 3 | 331 | 3 |
| Fold2 | 317 | 4 | 331 | 3 | |
| Fold3 | 319 | 2 | 332 | 2 | |
| Fold4 | 319 | 2 | 331 | 3 | |
| Fold5 | 319 | 2 | 332 | 2 |
Table 21.
Performance results of DenseNet121 on internal dataset
| KFold | Accuracy | Precision | Recall (Sensitivity) | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.9878 | 0.9906 | 0.9844 | 0.9910 | 0.9875 |
| Fold2 | 0.9908 | 0.9937 | 0.9875 | 0.9940 | 0.9906 | |
| Fold3 | 0.9908 | 0.9937 | 0.9875 | 0.9940 | 0.9906 | |
| Fold4 | 0.9893 | 0.9906 | 0.9875 | 0.9910 | 0.9891 | |
| Fold5 | 0.9878 | 0.9906 | 0.9844 | 0.9910 | 0.9875 | |
| Overall | 0.9893 | 0.9919 | 0.9863 | 0.9922 | 0.9891 | |
| With GAN | Fold1 | 0.9908 | 0.9907 | 0.9907 | 0.9910 | 0.9907 |
| Fold2 | 0.9893 | 0.9906 | 0.9875 | 0.9910 | 0.9891 | |
| Fold3 | 0.9939 | 0.9938 | 0.9938 | 0.9940 | 0.9938 | |
| Fold4 | 0.9924 | 0.9907 | 0.9938 | 0.9910 | 0.9922 | |
| Fold5 | 0.9939 | 0.9938 | 0.9938 | 0.9940 | 0.9938 | |
| Overall | 0.9921 | 0.9919 | 0.9919 | 0.9922 | 0.9919 |
Table 22.
Confusion values of DenseNet169 on internal dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 317 | 4 | 334 | 0 |
| Fold2 | 318 | 3 | 332 | 2 | |
| Fold3 | 317 | 4 | 333 | 1 | |
| Fold4 | 317 | 4 | 331 | 3 | |
| Fold5 | 316 | 5 | 331 | 3 | |
| With GAN | Fold1 | 317 | 4 | 334 | 0 |
| Fold2 | 319 | 2 | 332 | 2 | |
| Fold3 | 318 | 3 | 333 | 1 | |
| Fold4 | 318 | 3 | 333 | 1 | |
| Fold5 | 317 | 4 | 332 | 2 |
Table 23.
Performance results of DenseNet169 on internal dataset
| KFold | Accuracy | Precision | Recall (Sensitivity) | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.9939 | 1.0000 | 0.9875 | 1.0000 | 0.9937 |
| Fold2 | 0.9924 | 0.9938 | 0.9907 | 0.9940 | 0.9922 | |
| Fold3 | 0.9924 | 0.9969 | 0.9875 | 0.9970 | 0.9922 | |
| Fold4 | 0.9893 | 0.9906 | 0.9875 | 0.9910 | 0.9891 | |
| Fold5 | 0.9878 | 0.9906 | 0.9844 | 0.9910 | 0.9875 | |
| Overall | 0.9911 | 0.9944 | 0.9875 | 0.9946 | 0.9909 | |
| With GAN | Fold1 | 0.9939 | 1.000 | 0.9875 | 1.000 | 0.9937 |
| Fold2 | 0.9939 | 0.9938 | 0.9938 | 0.9940 | 0.9938 | |
| Fold3 | 0.9939 | 0.9969 | 0.9907 | 0.9970 | 0.9938 | |
| Fold4 | 0.9939 | 0.9969 | 0.9907 | 0.9970 | 0.9938 | |
| Fold5 | 0.9908 | 0.9937 | 0.9875 | 0.9940 | 0.9906 | |
| Overall | 0.9933 | 0.9962 | 0.9900 | 0.9964 | 0.9931 |
Table 24.
Confusion values of VGG16 on external dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 3396 | 605 | 8467 | 1108 |
| Fold2 | 3323 | 678 | 8366 | 1179 | |
| Fold3 | 3296 | 705 | 8443 | 1102 | |
| Fold4 | 3412 | 589 | 8316 | 1229 | |
| Fold5 | 3305 | 696 | 8427 | 1118 | |
| With GAN | Fold1 | 3555 | 446 | 8703 | 842 |
| Fold2 | 3581 | 420 | 8640 | 905 | |
| Fold3 | 3499 | 502 | 8650 | 895 | |
| Fold4 | 3588 | 413 | 8629 | 916 | |
| Fold5 | 3536 | 465 | 8729 | 916 |
Table 25.
Performance results of VGG16 on external dataset
| KFold | Accuracy | Precision | Recall | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Withput GAN | Fold1 | 0.8738 | 0.7540 | 0.8488 | 0.8843 | 0.7986 |
| Fold2 | 0.8629 | 0.7381 | 0.8305 | 0.8765 | 0.7816 | |
| Fold3 | 0.8666 | 0.7494 | 0.8238 | 0.8845 | 0.7849 | |
| Fold4 | 0.8658 | 0.7352 | 0.8528 | 0.8712 | 0.7896 | |
| Fold5 | 0.8661 | 0.7472 | 0.8260 | 0. | 0.7847 | |
| Overall | 0.8670 | 0.7448 | 0.8364 | 0.8799 | 0.7879 | |
| With GAN | Fold1 | 0.9049 | 0.8085 | 0.8885 | 0.9118 | 0.8466 |
| Fold2 | 0.9022 | 0.7983 | 0.8950 | 0.9052 | 0.8439 | |
| Fold3 | 0.8969 | 0.7963 | 0.8745 | 0.9062 | 0.8336 | |
| Fold4 | 0.9019 | 0.7966 | 0.8968 | 0.9040 | 0.8437 | |
| Fold5 | 0.9054 | 0.8125 | 0.8838 | 0.9145 | 0.8466 | |
| Overall | 0.9023 | 0.8024 | 0.8877 | 0.9083 | 0.8429 |
Table 26.
Confusion values VGG19 on external dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 3489 | 512 | 8348 | 1197 |
| Fold2 | 3506 | 495 | 8502 | 1042 | |
| Fold3 | 3501 | 500 | 8581 | 964 | |
| Fold4 | 3505 | 496 | 8392 | 1153 | |
| Fold5 | 3520 | 481 | 8498 | 1047 | |
| With GAN | Fold1 | 3599 | 402 | 8658 | 887 |
| Fold2 | 3612 | 389 | 8669 | 876 | |
| Fold3 | 3627 | 374 | 8721 | 824 | |
| Fold4 | 3653 | 348 | 8749 | 796 | |
| Fold5 | 3609 | 392 | 8642 | 903 |
Table 27.
Performance results of VGG19 on external dataset
| KFold | Accuracy | Precision | Recall | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.8738 | 0.7446 | 0.8720 | 0.8746 | 0.8033 |
| Fold2 | 0.8865 | 0.7709 | 0.8763 | 0.8908 | 0.8202 | |
| Fold3 | 0.8919 | 0.7841 | 0.8750 | 0.8990 | 0.8271 | |
| Fold4 | 0.8783 | 0.7525 | 0.8760 | 0.8792 | 0.8096 | |
| Fold5 | 0.8872 | 0.7707 | 0.8798 | 0.8903 | 0.8217 | |
| Overall | 0.8836 | 0.7646 | 0.8758 | 0.8868 | 0.8164 | |
| With GAN | Fold1 | 0.9048 | 0.8023 | 0.8995 | 0.9071 | 0.8401 |
| Fold2 | 0.9066 | 0.8048 | 0.9028 | 0.9082 | 0.8510 | |
| Fold3 | 0.9116 | 0.8149 | 0.9065 | 0.9137 | 0.8583 | |
| Fold4 | 0.9155 | 0.8211 | 0.9130 | 0.9166 | 0.8646 | |
| Fold5 | 0.9044 | 0.7999 | 0.9020 | 0.9054 | 0.8479 | |
| Overall | 0.9086 | 0.8086 | 0.9048 | 0.9102 | 0.8540 |
Table 28.
Confusion values of Xception on external dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 3757 | 244 | 8575 | 970 |
| Fold2 | 3786 | 215 | 8522 | 912 | |
| Fold3 | 3773 | 228 | 8684 | 861 | |
| Fold4 | 3725 | 276 | 8616 | 929 | |
| Fold5 | 3761 | 240 | 8631 | 914 | |
| With GAN | Fold1 | 3828 | 173 | 8989 | 556 |
| Fold2 | 3836 | 165 | 9033 | 512 | |
| Fold3 | 3829 | 172 | 9008 | 537 | |
| Fold4 | 3853 | 148 | 9026 | 513 | |
| Fold5 | 3845 | 156 | 8993 | 552 |
Table 29.
Performance results of Xception on external dataset
| KFold | Accuracy | Precision | Recall | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.9104 | 0.7948 | 0.9390 | 0.8984 | 0.8609 |
| Fold2 | 0.9168 | 0.8059 | 0.9463 | 0.9045 | 0.8704 | |
| Fold3 | 0.9196 | 0.8142 | 0.9430 | 0.9094 | 0.8739 | |
| Fold4 | 0.9110 | 0.8004 | 0.9310 | 0.9027 | 0.8608 | |
| Fold5 | 0.9148 | 0.8045 | 0.9400 | 0.9042 | 0.8670 | |
| Overall | 0.9145 | 0.8039 | 0.9399 | 0.9039 | 0.8666 | |
| With GAN | Fold1 | 0.9462 | 0.8732 | 0.9568 | 0.9417 | 0.9131 |
| Fold2 | 0.9500 | 0.8822 | 0.9588 | 0.9464 | 0.9189 | |
| Fold3 | 0.9477 | 0.8770 | 0.9570 | 0.9437 | 0.9153 | |
| Fold4 | 0.9512 | 0.8825 | 0.9630 | 0.9462 | 0.9210 | |
| Fold5 | 0.9477 | 0.8745 | 0.9610 | 0.9422 | 0.9157 | |
| Overall | 0.9486 | 0.8779 | 0.9593 | 0.9440 | 0.9168 |
Table 30.
Confusion values of ResNet50 on external dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 3557 | 444 | 8639 | 906 |
| Fold2 | 3578 | 423 | 8600 | 945 | |
| Fold3 | 3611 | 390 | 8570 | 975 | |
| Fold4 | 3507 | 494 | 8578 | 967 | |
| Fold5 | 3564 | 437 | 8601 | 944 | |
| With GAN | Fold1 | 3651 | 350 | 9026 | 519 |
| Fold2 | 3612 | 389 | 9009 | 536 | |
| Fold3 | 3623 | 378 | 8925 | 620 | |
| Fold4 | 3602 | 399 | 8953 | 592 | |
| Fold5 | 3639 | 362 | 8966 | 579 |
Table 31.
Performance results of ResNet50 on external dataset
| KFold | Accuracy | Precision | Recall | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.9003 | 0.7970 | 0.8890 | 0.9051 | 0.9404 |
| Fold2 | 0.8990 | 0.7911 | 0.8943 | 0.9010 | 0.8395 | |
| Fold3 | 0.8992 | 0.7874 | 0.9025 | 0.8979 | 0.8410 | |
| Fold4 | 0.8921 | 0.7839 | 0.8765 | 0.8987 | 0.8276 | |
| Fold5 | 0.8981 | 0.7906 | 0.8908 | 0.9011 | 0.8377 | |
| Overall | 0.8978 | 0.7900 | 0.8906 | 0.9007 | 0.8373 | |
| With GAN | Fold1 | 0.9358 | 0.8755 | 0.9125 | 0.9456 | 0.8936 |
| Fold2 | 0.9317 | 0.8708 | 0.9028 | 0.9438 | 0.8865 | |
| Fold3 | 0.9263 | 0.8539 | 0.9055 | 0.9350 | 0.8789 | |
| Fold4 | 0.9268 | 0.8588 | 0.9003 | 0.9380 | 0.8791 | |
| Fold5 | 0.9305 | 0.8627 | 0.9095 | 0.9393 | 0.8855 | |
| Overall | 0.9303 | 0.8644 | 0.9061 | 0.9404 | 0.8847 |
Table 32.
Confusion values of ResNet50v2 on external dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 3548 | 453 | 8508 | 1037 |
| Fold2 | 3584 | 417 | 8523 | 1022 | |
| Fold3 | 3559 | 442 | 8581 | 964 | |
| Fold4 | 3540 | 461 | 8459 | 1086 | |
| Fold5 | 3578 | 423 | 8497 | 1048 | |
| With GAN | Fold1 | 3626 | 375 | 8822 | 723 |
| Fold2 | 3656 | 342 | 8797 | 748 | |
| Fold3 | 3612 | 389 | 8756 | 789 | |
| Fold4 | 3642 | 359 | 8787 | 758 | |
| Fold5 | 3619 | 382 | 8771 | 774 |
Table 33.
Performance results of ResNet50v2 on external dataset
| KFold | Accuracy | Precision | Recall | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.8900 | 0.7738 | 0.8868 | 0.8914 | 0.8265 |
| Fold2 | 0.8938 | 0.7781 | 0.8958 | 0.8929 | 0.8328 | |
| Fold3 | 0.8962 | 0.7869 | 0.8895 | 0.8990 | 0.8351 | |
| Fold4 | 0.8858 | 0.7652 | 0.8848 | 0.8862 | 0.8207 | |
| Fold5 | 0.8914 | 0.7735 | 0.8943 | 0.8902 | 0.8295 | |
| Overall | 0.8914 | 0.7735 | 0.8902 | 0.8919 | 0.8289 | |
| With GAN | Fold1 | 0.9189 | 0.8338 | 0.9063 | 0.9243 | 0.8685 |
| Fold2 | 0.9195 | 0.8302 | 0.9145 | 0.9216 | 0.8703 | |
| Fold3 | 0.9130 | 0.8207 | 0.9028 | 0.9173 | 0.8598 | |
| Fold4 | 0.9175 | 0.8277 | 0.9103 | 0.9206 | 0.8670 | |
| Fold5 | 0.9147 | 0.8238 | 0.9045 | 0.9189 | 0.8623 | |
| Overall | 0.9167 | 0.8272 | 0.9077 | 0.9205 | 0.8656 |
Table 34.
Confusion values of Inceptionv3 on external dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 3601 | 400 | 8691 | 854 |
| Fold2 | 3533 | 468 | 8699 | 846 | |
| Fold3 | 3570 | 431 | 8697 | 848 | |
| Fold4 | 3598 | 403 | 8651 | 894 | |
| Fold5 | 3613 | 388 | 8716 | 829 | |
| With GAN | Fold1 | 3800 | 201 | 9093 | 452 |
| Fold2 | 3769 | 232 | 9102 | 443 | |
| Fold3 | 3793 | 208 | 9096 | 449 | |
| Fold4 | 3776 | 225 | 9083 | 462 | |
| Fold5 | 3712 | 285 | 9106 | 439 |
Table 35.
Performance results of Inceptionv3 on external dataset
| KFold | Accuracy | Precision | Recall | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.9074 | 0.8083 | 0.9000 | 0.9105 | 0.8517 |
| Fold2 | 0.9030 | 0.8068 | 0.8830 | 0.9114 | 0.8432 | |
| Fold3 | 0.9056 | 0.8061 | 0.8923 | 0.9112 | 0.8481 | |
| Fold4 | 0.9043 | 0.8010 | 0.8993 | 0.9063 | 0.8473 | |
| Fold5 | 0.9102 | 0.8134 | 0.9030 | 0.9131 | 0.8559 | |
| Overall | 0.9061 | 0.8075 | 0.8955 | 0.9105 | 0.8492 | |
| With GAN | Fold1 | 0.9518 | 0.8937 | 0.9498 | 0.9526 | 0.9209 |
| Fold2 | 0.9502 | 0.8948 | 0.9420 | 0.9536 | 0.9178 | |
| Fold3 | 0.9515 | 0.8942 | 0.9480 | 0.9530 | 0.9203 | |
| Fold4 | 0.9493 | 0.8910 | 0.9438 | 0.9516 | 0.9166 | |
| Fold5 | 0.9493 | 0.8942 | 0.9278 | 0.9540 | 0.9107 | |
| Overall | 0.9494 | 0.8936 | 0.9423 | 0.9530 | 0.9173 |
Table 36.
Confusion values of InceptionResNetv2 on external dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 3414 | 587 | 8589 | 956 |
| Fold2 | 3505 | 496 | 8572 | 973 | |
| Fold3 | 3669 | 332 | 8656 | 889 | |
| Fold4 | 3545 | 456 | 8596 | 949 | |
| Fold5 | 3621 | 380 | 8674 | 871 | |
| With GAN | Fold1 | 3713 | 288 | 9017 | 528 |
| Fold2 | 3709 | 292 | 8935 | 610 | |
| Fold3 | 3725 | 276 | 8952 | 593 | |
| Fold4 | 3732 | 269 | 9046 | 599 | |
| Fold5 | 3700 | 301 | 8953 | 592 |
Table 37.
Performance results of InceptionResNetv2 on external dataset
| KFold | Accuracy | Precision | Recall | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.8861 | 0.7812 | 0.8533 | 0.8998 | 0.8157 |
| Fold2 | 0.8916 | 0.7827 | 0.8760 | 0.8981 | 0.8267 | |
| Fold3 | 0.9099 | 0.8050 | 0.9170 | 0.9069 | 0.8573 | |
| Fold4 | 0.8963 | 0.7888 | 0.8860 | 0.9006 | 0.8346 | |
| Fold5 | 0.9076 | 0.8061 | 0.9050 | 0.9087 | 0.8527 | |
| Overall | 0.8983 | 0.7928 | 0.8875 | 0.9028 | 0.8374 | |
| With GAN | Fold1 | 0.9398 | 0.8755 | 0.9280 | 0.9447 | 0.9010 |
| Fold2 | 0.9334 | 0.8588 | 0.9270 | 0.9361 | 0.8916 | |
| Fold3 | 0.9358 | 0.8627 | 0.9310 | 0.9379 | 0.8955 | |
| Fold4 | 0.9433 | 0.8821 | 0.9328 | 0.9477 | 0.9067 | |
| Fold5 | 0.9341 | 0.8621 | 0.9248 | 0.9380 | 0.8923 | |
| Overall | 0.9373 | 0.8682 | 0.9287 | 0.9409 | 0.8974 |
Table 38.
Confusion values of DenseNet121 on external dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 3544 | 457 | 8649 | 896 |
| Fold2 | 3533 | 468 | 8643 | 902 | |
| Fold3 | 3587 | 414 | 8588 | 957 | |
| Fold4 | 3581 | 420 | 8658 | 887 | |
| Fold5 | 3573 | 428 | 8600 | 945 | |
| With GAN | Fold1 | 3700 | 301 | 8767 | 579 |
| Fold2 | 3712 | 289 | 8983 | 562 | |
| Fold3 | 3677 | 324 | 9002 | 543 | |
| Fold4 | 3659 | 342 | 8958 | 587 | |
| Fold5 | 3662 | 339 | 8997 | 548 |
Table 39.
Performance results of DenseNet121 on external dataset
| KFold | Accuracy | Precision | Recall | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.9001 | 0.7982 | 0.8858 | 0.9061 | 0.8397 |
| Fold2 | 0.8989 | 0.7966 | 0.8830 | 0.9055 | 0.8376 | |
| Fold3 | 0.8988 | 0.7894 | 0.8965 | 0.8997 | 0.8396 | |
| Fold4 | 0.9035 | 0.8015 | 0.8950 | 0.9071 | 0.8457 | |
| Fold5 | 0.8986 | 0.7908 | 0.8930 | 0.9010 | 0.8388 | |
| Overall | 0.9000 | 0.7953 | 0.8907 | 0.9039 | 0.8403 | |
| With GAN | Fold1 | 0.9341 | 0.8649 | 0.9248 | 0.9381 | 0.8938 |
| Fold2 | 0.9372 | 0.8685 | 0.9278 | 0.9411 | 0.8972 | |
| Fold3 | 0.9360 | 0.8713 | 0.9190 | 0.9431 | 0.8945 | |
| Fold4 | 0.9314 | 0.8618 | 0.9145 | 0.9385 | 0.8874 | |
| Fold5 | 0.9345 | 0.8698 | 0.9153 | 0.9426 | 0.8920 | |
| Overall | 0.9346 | 0.8673 | 0.9203 | 0.9407 | 0.8930 |
Table 40.
Confusion values of DenseNet169 on external dataset
| KFold | TP | FN | TN | FP | |
|---|---|---|---|---|---|
| Without GAN | Fold1 | 3667 | 334 | 8621 | 924 |
| Fold2 | 3650 | 351 | 8656 | 886 | |
| Fold3 | 3585 | 416 | 8628 | 917 | |
| Fold4 | 3542 | 459 | 8603 | 942 | |
| Fold5 | 3555 | 446 | 8666 | 879 | |
| With GAN | Fold1 | 3704 | 297 | 8951 | 594 |
| Fold2 | 3710 | 291 | 8973 | 572 | |
| Fold3 | 3653 | 348 | 8953 | 592 | |
| Fold4 | 3644 | 357 | 8956 | 589 | |
| Fold5 | 3606 | 395 | 8969 | 576 |
Table 41.
Performance results of DenseNet169 without GAN on external dataset
| KFold | Accuracy | Precision | Recall | Specificity | F1-score | |
|---|---|---|---|---|---|---|
| Without GAN | Fold1 | 0.9071 | 0.7987 | 0.9165 | 0.9032 | 0.8536 |
| Fold2 | 0.9085 | 0.8041 | 0.9123 | 0.9069 | 0.8548 | |
| Fold3 | 0.9016 | 0.7963 | 0.8960 | 0.9039 | 0.8432 | |
| Fold4 | 0.8966 | 0.7899 | 0.8853 | 0.9013 | 0.8349 | |
| Fold5 | 0.9022 | 0.8018 | 0.8885 | 0.9079 | 0.8429 | |
| Overall | 0.9032 | 0.7982 | 0.8997 | 0.9046 | 0.8459 | |
| With GAN | Fold1 | 0.9342 | 0.8618 | 0.9258 | 0.9378 | 0.8926 |
| Fold2 | 0.9363 | 0.8664 | 0.9273 | 0.9401 | 0.8958 | |
| Fold3 | 0.9306 | 0.8605 | 0.9130 | 0.9380 | 0.8860 | |
| Fold4 | 0.9302 | 0.8609 | 0.9108 | 0.9383 | 0.8851 | |
| Fold5 | 0.9283 | 0.8623 | 0.9013 | 0.9397 | 0.8813 | |
| Overall | 0.9319 | 0.8624 | 0.9156 | 0.9388 | 0.8882 |
Author Contributions
All authors contributed to the study conception and design. Erdi Acar, Engin Şahin and İhsan Yılmaz contributed to conceptualization; Erdi Acar contributed to data curation; Erdi Acar and Engin Şahin contributed to formal analysis; Erdi Acar, Engin Şahin and İhsan Yılmaz contributed to investigation; Erdi Acar and İhsan Yılmaz contributed to methodology; Erdi Acar contributed to resources and software; İhsan Yılmaz supervised the study; Engin Şahin contributed to visualization, writing–original draft preparation and writing–review and editing; all authors read and approved the final manuscript.
Declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Funding
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Erdi Acar, Email: erdiacar@stu.comu.edu.tr.
Engin Şahin, Email: enginsahin@comu.edu.tr.
İhsan Yılmaz, Email: iyilmaz@comu.edu.tr.
References
- 1.Verity R, Okell LC, Dorigatti I, Winskill P, Whittaker C, Imai N, et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis. 2020;20(6):669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He K, Zhao W, Xie X, Ji W, Liu M, Tang Z, et al. Synergistic learning of lung lobe segmentation and hierarchical multi-instance classification for automated severity assessment of COVID-19 in CT images. Pattern Recognit. 2021;113:107828. doi: 10.1016/j.patcog.2021.107828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esteva A, Chou K, Yeung S, Naik N, Madani A, Mottaghi A, et al. Deep learning-enabled medical computer vision. NPJ Digital Med. 2021;4(1):1–9. doi: 10.1038/s41746-020-00373-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakator M, Radosav D. Deep learning and medical diagnosis: a review of literature. Multimodal Technol Interact. 2018;2(3):47. doi: 10.3390/mti2030047. [DOI] [Google Scholar]
- 5.Liu Y, Liu G, Zhang Q. Deep learning and medical diagnosis. Lancet. 2019;394(10210):1709–1710. doi: 10.1016/S0140-6736(19)32501-2. [DOI] [PubMed] [Google Scholar]
- 6.Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J Big Data. 2019;6(1):1–48. doi: 10.1186/s40537-019-0197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang G, Fouladvand S, Zhang J, Brooks MA, Jacobs N, Chen J (2019) Ganai: Standardizing ct images using generative adversarial network with alternative improvement. In: 2019 IEEE International Conference on Healthcare Informatics (ICHI), IEEE, pp 1–11
- 8.Sandfort V, Yan K, Pickhardt PJ, Summers RM. Data augmentation using generative adversarial networks (CycleGAN) to improve generalizability in CT segmentation tasks. Sci Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-52737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao D, Zhu D, Lu J, Luo Y, Zhang G. Synthetic medical images using. Symmetry. 2018;10(10):519. doi: 10.3390/sym10100519. [DOI] [Google Scholar]
- 10.Chuquicusma MJ, Hussein S, Burt J, Bagci U (2018) How to fool radiologists with generative adversarial networks? a visual turing test for lung cancer diagnosis. In: 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), IEEE, pp 240–244
- 11.Frid-Adar M, Klang E, Amitai M, Goldberger J, Greenspan H (2018) Synthetic data augmentation using GAN for improved liver lesion classification. In: 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), IEEE, pp 289–293
- 12.Guibas JT, Virdi TS, Li PS (2017) Synthetic medical images from dual generative adversarial networks. arXiv preprint arXiv:1709.01872
- 13.Shin HC, Tenenholtz NA, Rogers JK, Schwarz CG, Senjem ML, Gunter JL et al (2018) Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In: International workshop on simulation and synthesis in medical imaging. Springer, Cham, pp 1–11
- 14.Wu X, Hui H, Niu M, Li L, Wang L, He B. Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: a multicentre study. Europ J Radiol. 2020;128:109041. doi: 10.1016/j.ejrad.2020.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu X, Jiang X, Ma C, Du P, Li X, Lv S, et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang B, Jin S, Yan Q, Xu H, Luo C, Wei L, et al. AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system. Appl Soft Comput. 2021;98:106897. doi: 10.1016/j.asoc.2020.106897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yousefzadeh M, Esfahanian P, Movahed SMS, Gorgin S, Rahmati D, Abedini A, et al. Ai-corona: radiologist-assistant deep learning framework for covid-19 diagnosis in chest ct scans. PloS one. 2021;16(5):e0250952. doi: 10.1371/journal.pone.0250952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput Biol Med. 2020;121:103795. doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Wu L, Zhang J, Zhang L, Gong D, Zhao Y, et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci Rep. 2020;10(1):1–11. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng C, Deng X, Fu Q, Zhou Q, Feng J, Ma H et al (2020) Deep learning-based detection for COVID- 19 from chest CT using weak label. medRxiv:2020.03.12.20027185
- 21.Shi F, Wang J, Shi J, Wu Z, Wang Q, Tang Z, Shen D (2020) Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for covid-19. IEEE Rev Biomed Eng [DOI] [PubMed]
- 22.Roberts M, Driggs D, Thorpe M, Gilbey J, Yeung M, Ursprung S, et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell. 2021;3(3):199–217. doi: 10.1038/s42256-021-00307-0. [DOI] [Google Scholar]
- 23.Tayarani-N MH. Applications of artificial intelligence in battling against Covid-19: a literature review. Solitons & Fractals: Chaos; 2020. p. 110338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalkreuth R, Kaufmann P (2020) COVID-19: a survey on public medical imaging data resources. arXiv preprint arXiv:2004.04569
- 25.Ning W, Lei S, Yang J, Cao Y, Jiang P, Yang Q, et al. Open resource of clinical data from patients with pneumonia for the prediction of COVID-19 outcomes via deep learning. Nat Biomed Eng. 2020;4(12):1197–1207. doi: 10.1038/s41551-020-00633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azad R, Asadi-Aghbolaghi M, Fathy M, Escalera S (2019) Bi-directional ConvLSTM U-Net with densley connected convolutions. arXiv:1909.00166
- 27.Rother C, Kolmogorov V, Blake A. GrabCut interactive foreground extraction using iterated graph cuts. ACM Trans Graph. 2004;23(3):309–314. doi: 10.1145/1015706.1015720. [DOI] [Google Scholar]
- 28.Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S et al (2014) Generative adversarial networks. arXiv preprint arXiv:1406.2661
- 29.Creswell A, White T, Dumoulin V, Arulkumaran K, Sengupta B, Bharath AA. Generative adversarial networks: an overview. IEEE Signal Process Magazine. 2018;35(1):53–65. doi: 10.1109/MSP.2017.2765202. [DOI] [Google Scholar]
- 30.Gui J, Sun Z, Wen Y, Tao D, Ye J (2020) A review on generative adversarial networks: Algorithms, theory, and applications. arXiv preprint arXiv:2001.06937
- 31.Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L (2009) Imagenet: A large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition, IEEE, pp 248–255
- 32.Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
- 33.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
- 34.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826
- 35.Szegedy C, Ioffe S, Vanhoucke V, Alemi A (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. In: Proceedings of the AAAI conference on artificial intelligence, Vol 31, No 1
- 36.Chollet F (2017) Xception: Deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1251–1258
- 37.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
- 38.Chollet F (2015) Keras: Deep Learning for Humans. [Online]. Available: https://keras.io/