Abstract
Resolving the genomic basis underlying phenotypic variations is a question of great importance in evolutionary biology. However, understanding how genotypes determine the phenotypes is still challenging. Centuries of artificial selective breeding for beauty and aggression resulted in a plethora of colors, long-fin varieties, and hyper-aggressive behavior in the air-breathing Siamese fighting fish (Betta splendens), supplying an excellent system for studying the genomic basis of phenotypic variations. Combining whole-genome sequencing, quantitative trait loci mapping, genome-wide association studies, and genome editing, we investigated the genomic basis of huge morphological variation in fins and striking differences in coloration in the fighting fish. Results revealed that the double tail, elephant ear, albino, and fin spot mutants each were determined by single major-effect loci. The elephant ear phenotype was likely related to differential expression of a potassium ion channel gene, kcnh8. The albinotic phenotype was likely linked to a cis-regulatory element acting on the mitfa gene and the double-tail mutant was suggested to be caused by a deletion in a zic1/zic4 coenhancer. Our data highlight that major loci and cis-regulatory elements play important roles in bringing about phenotypic innovations and establish Bettas as new powerful model to study the genomic basis of evolved changes.
Keywords: domestication, evolution, major-effect loci, cis-regulation, mitfa, zic1/zic4
Introduction
Already in the “On the Origin of Species” (1859) and later in the “The Variation of Animals and Plants under Domestication” (1868), Charles Darwin recognized that the same processes of selection that act in nature also apply to selective breeding where they are sped up orders of magnitude by breeders’ goals to obtain particular traits. Obviously, Darwin could not know the genomic basis that underlies the selected traits. This began to change, particularly in the last decade, as genome sequences could be obtained for more species. Yet, we are still only at the beginning of understanding how the genotype controls and determines the phenotype (Frazer et al. 2009). Both, mutations in coding sequences and polymorphisms in noncoding sequences are now known to play important roles in generating phenotypic variation (Wittkopp and Kalay 2012; Andersson et al. 2015; Petit et al. 2017; Kemble et al. 2019). Beyond a handful of genetic or developmental model systems (Lehner 2013), organisms under artificial selection not only provide outstandingly useful phenotypes but also permit studying the connection between evolutionary changes and their genomic bases. Knowledge about the bridge between genotypes and phenotypes poses a question, whether the same genomic mechanisms, as Darwin suggested, both in evolution as well as in animal breeding, are at work bringing about innovations (Shapiro et al. 2004; Karlsson et al. 2007; Rubin et al. 2010). Importantly, assays including CRISPR-Cas mediated genome modification that allow establishing functional associations between genotype and phenotype only became recently available.
The Siamese fighting fish (Betta splendens), one of the most popular ornamental fishes worldwide, is well-known for its aggressive behavior (Simpson 1968), extremely diverse color patterns, and huge variation in fin shapes (Lucas 1968). It belongs to the anabantoid fishes, characterized by a modified gill skeleton that forms the labyrinth organ, which permits air breathing as these fish tend to live in oxygen-deprived waters. Males build bubble nests and perform complex courting and parental care behaviors (Lucas 1968; Rüber et al. 2004; Monvises et al. 2009). It is a short-lived species and its generation interval is only 5–6 months, with each spawning producing up to several hundred eggs (Monvises et al. 2009). The initiation of domestication of fighting fish has been documented to have occurred as early as 600 years ago, with the purpose of using these fish in staged fighting contests by the Siamese in the current Thailand, leading to the “Plakat” betta (Smith 1945). Selection on other display traits, mainly including coloration and fin shapes, has a more recent origin traced back to the middle of the nineteenth century and was prompted by the use of these fish in exhibition contests (Lucas 1968). In the past decades since these fish became the object of a worldwide aquarium hobby, most artificial selection in fighting fish has focused on modifying the spectacular body colorations and the overgrowth of fins (supplementary fig. S1, Supplementary Material online). Therefore, Siamese fighting fish constitute an unparalleled system for identifying genetic variants underlying both simple and highly complex morphological traits to increase the understanding of the genetic basis of phenotypic variation, between “natural mutants” and domesticated “sports.”
Results and Discussion
Genome Assembly and Annotation
We sequenced a homozygous yellow single-tail female and a homozygous transparent double-tail male fighting fish (supplementary fig. S2 and table S1, Supplementary Material online), each with over 120-fold genome coverage. Genome assembly sizes for the female and male were 424.9 and 411.1 Mb, respectively (supplementary note 1, Supplementary Material online). The contig and scaffold N50 sizes for the female were 21.3 kb and 2.1 Mb, respectively, and for the male, 17.3 kb and 1.9 Mb, respectively (supplementary table S2, Supplementary Material online). Both the female and male assemblies showed complete and single-copy BUSCO scores of >95%, indicative of high quality (supplementary table S3 and figs. S3 and S4, Supplementary Material online). A total of 22,977 protein-coding genes were predicted. Transposable elements and other types of repetitive elements together accounted for only 10.7% of the genome sequences (supplementary table S4, Supplementary Material online), a relatively small fraction of the genome compared with other teleosts (Malmstrøm et al. 2017). We also annotated 160,595 conserved noncoding elements (CNEs) and 1,059 lncRNAs in the genome, with a total length of 34.1 Mb (∼8.0%) and 1.47 Mb (∼0.35%), respectively. In comparison to nine other fish genomes, the fighting fish therefore has, besides the pufferfishes, the shortest mean intergenic regions and the lowest overall proportion of noncoding elements within genes, indicating that it has a very compact genome (supplementary fig. S5, Supplementary Material online). Using two high-density linkage maps, 94.9% and 95.3% of scaffolds were anchored on 21 linkage groups corresponding to the chromosomes of the female and male karyotypes, respectively (supplementary tables S5–S7 and fig. S6, Supplementary Material online). Our genome assembly is an important addition to those published recently for the fighting fish (Fan et al. 2018; Prost et al. 2020). Therefore, they supply a useful tool for downstream genetic and genomic studies.
Genetic Diversity and Population Structure of the Fighting Fish
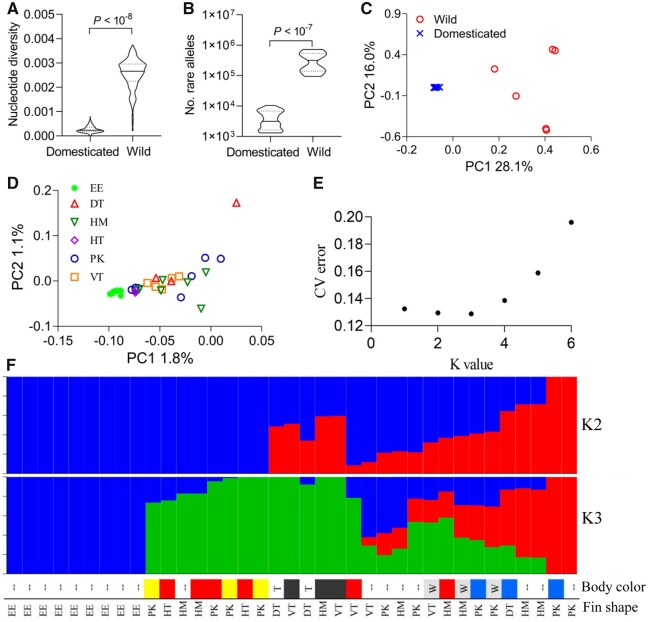
We sequenced the whole genomes of domesticated fish of diverse coloration and fin traits as well as several wild fish exhibiting ancestral phenotypes (supplementary table S8, Supplementary Material online). Domesticated species tend to lose genetic diversity compared with the wild type. Here, based on whole-genome resequencing data, we observed that the overall genetic diversity in domesticated fighting fish was reduced approximately 10-fold when compared with the fish from the wild (nucleotide diversity: 0.0003 vs. 0.0025, P < 10−8 for t-test, estimated using VCFtools [Danecek et al. 2011]; unbiased nucleotide diversity: 0.0004 vs. 0.0033, P < 10−8 for t-test, estimated using pixy [Korunes and Samuk 2021]). Remarkably, the average number of rare single nucleotide polymorphisms (SNPs) with a cutoff value of minor allele frequency of 0.01 was even more decreased, nearly 80 times (4,290 vs. 349,824, P < 10−7 for t-test; fig. 1A and B). Such rapid loss of rare alleles during domestication is likely due to genetic bottlenecks during establishment of strains and random genetic drift during domestication, rather than resulting from intense artificial selection imposed by selective breeding (Hyten et al. 2006).
Fig. 1.
Genetic diversity and population structure in fighting fish. (A and B) Differences of genetic diversity between domesticated and wild fish measured in nucleotide diversity and number of rare alleles, respectively. P values for t-test are shown above. (C and D) Population structure among domesticated and wild fish, and within domesticated fish, respectively, revealed by principal component analysis. EE, elephant ear; DT, double tail; HM, halfmoon tail; HT, horse tail; PK, Plakat tail; VT, veil tail. (E) The most likely number of genetic clusters (K) is inferred as 3, where shows the lowest cross-validation errors. (F) Population structure at individual level revealed by admixture analysis, at K = 2 and 3, in domesticated fish. Major traits including body color (– indicates too complicated color pattern to phenotype, whereas T and W indicate transparent and white coat color, respectively) and fin shape (codes are corresponding to those in D), for each individual, are also shown below.
Population structure analyses based on both principal component and admixture analyses consistently showed that the domesticated fish have significantly diverged from the fish from the wild, after several hundred years of selective breeding (fig. 1C and supplementary fig. S7, Supplementary Material online). Principal component analysis in the different breeding lines of domesticated fish showed that, except for the elephant ear phenotypes, there was no clear differentiation among the studied traits (fig. 1D). In admixture analysis, the most likely number of genetic clusters for hypothesized ancestral groups within domesticated fish was inferred to be three (fig. 1E). We observed that fish exhibiting the same particular trait, for example, the halfmoon tail, the double tail, and the Plakat fin shape, are not always assigned to the same genetic clusters (fig. 1F). The results imply that these traits are more independent of their genetic background than those determined by a number of minor-effect loci, as in some domesticated animal breeds (Do et al. 2013; Al-Mamun et al. 2015; Wei et al. 2015) and is likely determined by a single or a few loci with major effects.
Genetic Control of Diverse Pigment Patterns
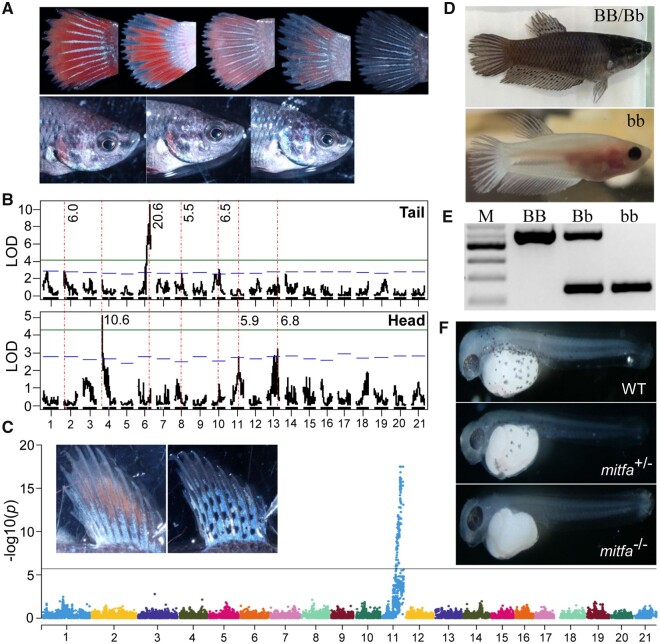
The fighting fish is famous for its diversity of striking pigment patterns generated by artificial selection (supplementary fig. S1, Supplementary Material online). First, we examined if pigment-related traits are monogenic or polygenic in several test crosses focusing first on red pigments. We noted that xanthophore density differed markedly between body segments (fig. 2A and supplementary fig. S8, Supplementary Material online). Haley–Knott regression quantitative trait loci (QTL) mapping revealed a major locus at LG6 for red pigment distribution in the caudal fin, with 20.6% of its phenotypic variation explained (PVE) by this QTL. Aside from this major QTL, three additional QTLs with significant but smaller effects were identified at LG2, LG8, and LG10, with PVE of 6.0%, 5.5%, and 6.5%, respectively (fig. 2B). For pigmentation in the head, we identified one significant and two suggestive QTLs at LG4, LG11, and LG13, with PVE of 10.6%, 5.9%, and 6.8%, respectively (fig. 2B). These data show that the distribution of red pigments is a polygenic trait. Interestingly, the QTLs found for tail had no overlap with those for head, implying the distribution of red pigments in different body sections is determined by different genetic loci. Although we did not identify the genes underlying red pigment distribution, our study provides first insights for a better understanding of various pigmentation patterns from a polygenic perspective.
Fig. 2.
Genetic mapping of distribution of red pigments (xanthophores) and fin-spotting pattern, albino mutant, and validation of mitfa gene as the candidate causal gene for albino mutant using CRISPR/Cas9 knockout. (A) Variation of the distribution of red pigments in caudal fin and head sections. (B) QTL mapping and comparison for distribution of red pigments in caudal fin and head sections, where blue and green horizontal lines indicate LOD cutoff values of chromosome- and genome-wide significance, respectively. PVE (%) by each QTL is shown at the top of each QTL region. Comparisons of QTL distributions between the two traits are indicated with vertical dashed lines. (C) Spotted versus nonspotted fin pigmentation patterns in fighting fish and association study using mixed linear modeling, which identified only one locus at LG11 for this trait. (D and E) The melanin (wild-type pigmented) and albino mutant and their corresponding genotypes based on a deletion flanking mitfa. (F) The wild-type pigmented fighting fish with regular pattern of melanized cells at 48 hpf (WT), mosaic mitfa-knockout fish showing less melanized cells at 48 hpf (mitfa+/−) and mitfa-knockout fish showing no melanized cells throughout the whole embryo at 48 hpf (mitfa-/-), where no wild-type haplotypes are detected.
The second pigmentation trait we investigated was dorsal fin spotting (fig. 2C). We phenotyped 156 fish from the F2 family, RM2, and found that this conforms to a pattern of Mendelian inheritance (supplementary fig. S9a, Supplementary Material online). Mixed linear modeling with a genetic relatedness matrix (GRM) based on ∼25 K SNPs revealed only one major locus on LG 11 responsible for this trait (fig. 2C and supplementary fig. S9b, Supplementary Material online). All fish with dorsal fin spots were homozygous at the most differentiating SNPs, suggesting that this trait is recessive. Using this data set, the locus was restricted to a region of ∼800 kb harboring ∼100 genes (supplementary fig. S9c, Supplementary Material online). This genomic region will be the focus for further investigation.
Finally, we studied the albino phenotype, which is characterized by a total lack of black pigments in the fins and body, except for the eyes, regardless of presence of the other colors. This recessive trait follows a monogenic Mendelian inheritance pattern (supplementary fig. S10a, Supplementary Material online). We mapped this trait to a locus on LG4 by using RAD-tag markers on our test crosses (supplementary fig. S10b and c, Supplementary Material online). Recombination analysis based on 293 fish revealed that this locus spans a genomic region of ∼438 kb (supplementary fig. S10d, Supplementary Material online), with 18 predicted genes (supplementary S11a, Supplementary Material online). Because the albino fish lacked melanin expression in the skin (fig. 2D and E), we compared the expression pattern of these genes in albino and wild-type pigmented fish and found that only microphthalmia-associated transcription factor a (mitfa) within this region was differentially expressed (supplementary fig. S11b, Supplementary Material online). However, the expression of mitfa was also decreased in the eye of the albino fish, which typically shows black pigmentation (supplementary fig. S11d, Supplementary Material online). Studying the expression of the mitfa gene, we found a paralog of mitfa, in the eye. The expression of mitfb was higher than that of mitfa in the eye of both albino and wild-type fish, and, interestingly, mitfb was more highly expressed in eyes of albino than wild-type fish (supplementary fig. S11d, Supplementary Material online). It is likely that mitfb has critical functions for retinal pigment formation and shows compensatory effects on mitfa in albino fish phenotype. Interestingly, this mechanism is consistent with the nacre mutant of zebrafish (Danio rerio), which is also a mutation of mitfa and also has black eyes (Lister et al. 1999, 2001).
To verify whether mitfa is associated with the albino phenotype, we knocked out this gene using CRISPR/Cas9 system by targeting the coding sequences (supplementary fig. S12, Supplementary Material online). Two G0 CRISPants without detectable wild-type alleles completely lost melanin pigmentation, whereas the mosaic fish with both wild-type and mutated haplotypes were markedly reduced in the density of melanin-containing cells, compared with wild-type controls, which consistently presented normal melanin pigmentation 48 hpf (fig. 2F and supplementary fig. S13, Supplementary Material online). This CRISPRant phenotype matches the phenotype of the albino mutant, suggesting that mitfa is the altered gene in this fighting fish mutant. However, we did not find any mutation in introns and exons of mitfa, implying that the mutant phenotype is associated with variation in cis-regulatory element acting on this gene. Comparison between homozygous albino and wild-type pigmented fish revealed a cluster of indels and SNPs about 25 kb upstream of mitfa, including a 366-bp deletion in the albino mutant. Genotyping this deletion in ∼1,000 fish revealed that this deletion was strictly correlated with the albino phenotype (supplementary fig. S14 and table S9, Supplementary Material online). These data suggest that the 366-bp deletion is a distant cis-regulatory element and could underlie the albino phenotype.
Taken together, in the fighting fish, a single or very few major loci can bring about phenotypic innovations for some colors. Thus, these traits are more easily affected by selection than polygenic traits. Certainly, many of the other color strains in the fighting fish are likely to be determined by major-effect loci. Further studies on these traits will provide valuable information to understand the mechanism of how selection affects phenotypic innovations.
Genetic Basis of Elephant Ear and Double-Tail Varieties
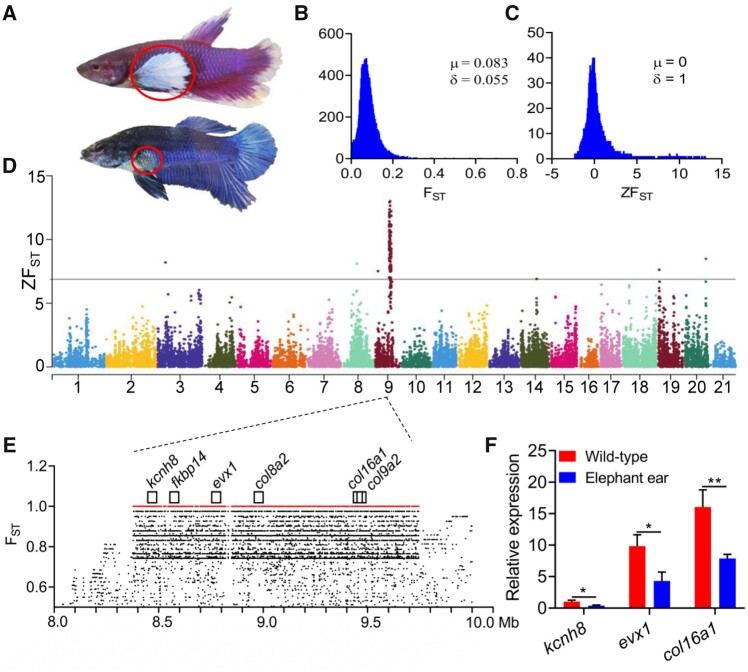
Another striking feature of domesticated fighting fish is the overgrowth of almost all types of fins. Using 47 sequenced fish including nine “elephant ear” phenotypes (supplementary table S8 and fig. S1f, Supplementary Material online), we firstly mapped the locus for the elephant ear mutation (fig. 3A), a recessive trait following Mendelian inheritance characterized by elongated pectoral fins (Lucas 1968). Using FST scans based on whole-genome resequencing data, this locus locates to a 1.3-Mb region on LG9 (fig. 3B–D). We further refined the haplotypes between elephant ear and wild-type fish and annotated 55 protein-coding genes in this region, of which six are known to play important roles in fin development and regeneration (fig. 3E). Examining their expression patterns in pectoral fins at 1 month posthatching when the elephant ear mutation becomes fully apparent, we identified three interesting candidate genes: potassium voltage-gated channel subfamily H member 8 (kcnh8), homeobox even-skipped homolog protein 1 (evx1), and collagen alpha-1(XVI) chain (col16a1) that were significantly downregulated in elephant ear phenotypes when compared with wild-type fish (fig. 3F), an observation that agrees with the recessive inheritance pattern of this trait (Lucas 1968). A previous study suggested that evx1 is required for joint formation in zebrafish fin dermoskeleton, but, apparently has no role in fin length (Schulte et al. 2011). Though important in fin regeneration and typically affected by domestication processes (Anastasiadi and Piferrer 2019), there was no obvious evidence that collagen genes are responsible for overgrowth of fins (Durán et al. 2011; Anastasiadi and Piferrer 2019). In particular, we found one paralog of kcnh8 at LG14, implying the functions of these two paralogs might have diverged with one fulfilling the general neural function and the other one regulating fin growth, a situation resembling what has been observed regarding the expression patterns of potassium channels in both zebrafish and goldfish (Carassius auratus) long-fin mutants (Perathoner et al. 2014; Lanni et al. 2019; Stewart et al. 2019; Kon et al. 2020). Interestingly, in one swordtail species, Xiphophorus hellerii, differential expression of kcnh8 was associated with development of a male ornamental trait, a ventral outgrowth of the caudal fin, called sword (Schartl et al. 2020). In zebrafish and goldfish long-fin mutants, mutations in paralogous potassium channel genes kcnh2a, kcnk5b, and kcc4a, cause overgrowth of different types of fins (Perathoner et al. 2014; Lanni et al. 2019; Stewart et al. 2019; Kon et al. 2020). Mutations disrupting ion channels and ion-dependent signaling often are related to abnormal organ development and regeneration via bioelectrical regulation (McLaughlin and Levin 2018). As discussed above, expression alteration and subfunctionalization of kcnh8 encoding a potassium voltage-gated channel are more likely related to the formation of the elongated pectoral fins (elephant ear) breed. We found a fixed missense mutation in the last exon (2912 A/G, exon16) of kcnh8, but not in the other candidate genes. This amino acid change (H/R) is neither evolutionary conserved across teleosts (supplementary fig. S15, Supplementary Material online) nor predicted to likely affect protein function, with a score of 1.00 as estimated by SIFT (Sim et al. 2012). However, an SNP in coding sequences might be less effective altering gene expression (Cowper-Sal et al. 2012). Except for this SNP, there are still some SNPs and short sequence variations in the noncoding sequences within and closely flanking these candidate genes that may affect expression. Therefore, the elephant ear breed is more likely caused by mutations that affect expression. In addition, it is also worth mentioning that the FKBP prolyl isomerase 14 (fkbp14), encoding a chaperone and calcium-binding protein, shows a similar expression pattern with kcnh8 and the statistical significance for differential expression is only slightly over 0.05 (fold change, 2.04 and P value for t-test, 0.07). In zebrafish, inhibition of fkbp14 function was shown to cause outgrowth of the caudal fin margin (Kujawski et al. 2014). In swordtails, the expression pattern of a paralogous gene fkbp9 was also observed to be associated with the development of sword of the tail fin in males (Schartl et al. 2020). These data imply that fkbp14 is another potential candidate gene for “elephant ear” phenotypes. Taken together, our results suggest that a variety of potassium channel and/or calcium-binding genes play critical roles to generate favored ornamental phenotypes of overgrowth of various fin types that are observed in artificial selective breeding in the fighting fish (Stewart et al. 2019; Kon et al. 2020; Schartl et al. 2020).
Fig. 3.
Mapping and identifying candidate genes for elephant ear mutant of fighting fish. (A) Elephant ear mutant showing overgrowth of pectoral fin (highlighted with circle), in contrast to wild-type fish. (B and C) Distribution of FST and Z-transformed FST of 30-kb window size for whole-genome-wide variants between elephant ear and wild-type samples, respectively. (D) Whole-genome scan identifies a major locus at LG9 for elephant ear using Z-transformed FST. Genome-wide significance cutoff value is denoted with horizontal line. (E) Six protein-coding genes associated with fin development and regeneration are predicted in the elephant ear haplotype with a length of ∼1.3 Mb. Fixed variants are denoted with red. (F) Three genes including kcnh8, exv1, and col16a1 are significantly downregulated in elephant ear mutants (*P < 0.05, **P < 0.01; n = 3, t-test).
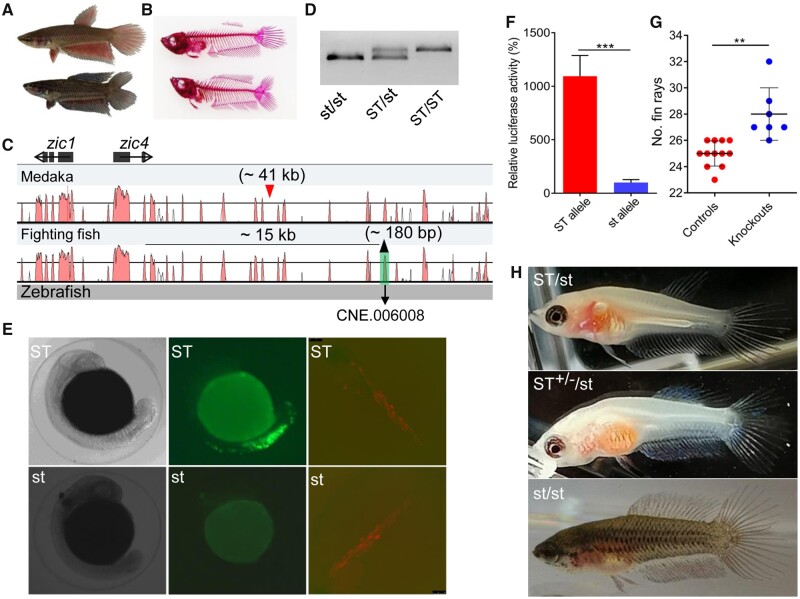
Double tail is one of the most well-known and most appreciated among various fin varieties of fighting fish. This mutant presents a unique ventralized pattern of dorsal trunk and tail, and features a doubling of the number of fin rays for both dorsal and caudal fins (fig. 4A and B and supplementary fig. S16, Supplementary Material online). Double-tail fighting fish was found to be a recessive homozygote (st) and we mapped the locus responsible for double tail to a ∼130-kb region on LG1 (st vs. ST) by RAD sequencing and fine mapping by examination of recombinants in 502 fish (supplementary fig. S17, Supplementary Material online). Sequence analysis revealed that this locus harbors three genes: zinc finger transcription factors Zic1 and Zic4 (zic1and zic4) and phospholipid scramblase 1 (plscr1), and overlaps with the Da locus of medaka (Oryzias latipes) for a double-tail mutant that contains only zic1 and zic4 (Moriyama et al. 2012). Consistent with medaka, expressions of both zic1 and zic4 were suppressed in double tail (supplementary fig. S18, Supplementary Material online) and no mutation was identified in the coding sequences (Kawanishi et al. 2013). We further sequenced the genomes of both homozygous single- and double-tail fish and found in double tail no large sequence variation except for a ∼180-bp deletion ∼15-kb downstream of zic4 (fig. 4C and supplementary fig. S19a, Supplementary Material online). This deletion was located in a cluster of CNEs and coincided with predicted CNE.006008 (supplementary fig. S19b, Supplementary Material online). Genotyping at this locus showed that the deletion was completely correlated with phenotypes in > 1000 examined fish (fig. 4D and supplementary table S10, Supplementary Material online). In medaka, both genes, zic1 and zic4, were verified to be responsible for double tail (Moriyama et al. 2012). However, the mechanism by which these genes induce this phenotype is still unclear. It was assumed that a transposon, Albatross (> 41 kb), inserted into the common regulatory region of both zic1 and zic4, ultimately leads to the double-tail mutation of medaka (Moriyama et al. 2012). Therefore, we hypothesized that the deletion in CNE.006008, an enhancer, is responsible for the double-tail phenotype in fighting fish. To test this hypothesis, first we inserted the CNE.006008 locus and its closely flanking sequences of ∼100 bp separately from single- and double-tail fish into Zebrafish Enhancer Detection (ZED) vectors (Bessa et al. 2009) and injected them into one-cell stage embryos. We observed that the wild-type ST allele significantly enhanced green fluorescent protein (GFP) expression in embryos at 24 hpf, when both zic1 and zic4 show differential expression between double-tail and wild-type fish (Moriyama et al. 2012), whereas no visible GFP expression was detected for the st allele (fig. 4E and supplementary table S11, Supplementary Material online). The efficiency of the two alleles as candidate enhancers was further examined using a Dual-Luciferase Reporter Assay, which showed that the ST allele enhanced luciferase expression by ∼10× relative to st allele in Singapore grouper embryonic cell line (fig. 4F).
Fig. 4.
Deletion in the putative enhancer of zic1 and zic4 is associated with double-tail mutant. (A) Overview of the wild-type (single-tail) and double-tail mutant fighting fish. (B) Skeleton staining shows the numbers of fin rays of both dorsal fin and caudal fin are significantly higher in double tail than in single tail. (C) Vista plotting of the genomic locus for double-tail mutation among zebrafish, fighting fish, and medaka. Zebrafish is used as reference. Approximately 180-bp deletion located at ∼15-kb downstream of zic4 is screened overlapping with predicted CNE.006008 of double-tail allele. The insert position of transposon Albatross (∼41 kb) in medaka Da locus is indicated with red triangle. (D) PCR screening of the deletion in single-tail and double-tail fish. (E) Representative fighting fish injected with enhancer detection vector ZED constructed with CNE.006008 from single-tail allele (ST) showing GFP expression predominantly in the dorsal fin and caudal fin positions, and those injected with double-tail allele (st) showing no GFP expression in the whole embryos at 24 hpf. RFP that is only detectable, particularly in muscles, since 72 hpf, is used as internal control. (F) Relative luciferase activity in Singapore grouper embryonic cell line transfected with pGL3-Promoter constructs including CNE.006008 region separately from the single-tail and double-tail alleles (Mann–Whitney U test, *** P < 0.001). (G) The total number of fin rays of dorsal and caudal fins between genetically modified fish (n = 7) and its corresponding controls (n = 12) in CNE.006008 (Mann–Whitney U test, ** P < 0.01). (H) The knockout fighting fish (ST+/−/st), with ∼60% of ST allele sequences deleted at CNE.006008, shows much more fin rays both in dorsal fin and caudal fin than the single-tail (ST/st) control, but less than double-tail control (st/st). Heterozygous ST/st fish were used as recipients for the CRISPR/Cas9 injections.
Finally, we deleted this enhancer using the CRISPR-Cas9 system in fighting fish. Considering the efficiency of tested gRNAs and the cluster of CNEs that could have unpredicted functions, we limited the modification to the CNE.006008 region and did not involve the other CNEs (supplementary fig. S20, Supplementary Material online). Genetic analysis revealed that none of these fish had completely deleted CNE.006008, suggesting nonsimultaneous cutting at multiple targeted gRNA positions. These mosaic fish (n = 7) had significantly more fin rays than the noninjected controls (P < 0.01; fig. 4G). We screened one modified fish, where >80% of sequenced clones were mutants with deleted sequences up to 56% of st allele (supplementary fig. S21, Supplementary Material online). Although this fish was not a pure knockout, we observed that the number of fin rays of both dorsal and caudal fins was significantly higher than in the single tail and approaching that of the double tail (fig. 4H). Taken together, deletion of the candidate coenhancer of zic1 and zic4, CNE.006008 was found to be the causative mutation for double-tail fighting fish.
To date, double-tail mutants are only reported in the fighting fish and medaka, and both are caused by mutations in the coregulatory regions of zic1 and zic4 (Moriyama et al. 2012). As shown in medaka loss of function of either gene is not able to cause a double-tail phenotype (Moriyama et al. 2012). Furthermore, loss of functions of both zic1 and zic4 causes fatal Dandy–Walker malformation-like disease in animals (Grinberg et al. 2004; Blank et al. 2011). Thus, selection on the coregulatory regions of multiple effector genes in a single locus becomes more efficient to bring about such phenotypic innovations. The occurrences of those kinds of mutations are scarcer than those determined by a single gene, which likely explains why only two double-tail cases in a number of domesticated teleosts have been observed so far. In comparison, traits that are determined by single genes are much more common. Microphthalmia-associated transcription factors have been extensively reported responsible for the albinism of a number of domesticated animals, such as dogs (Karlsson et al. 2007), pigs (Chen et al. 2016), ducks (Zhou et al. 2018), and quails (Minvielle et al. 2010). Interestingly, most of them are caused by mutations in the regulatory sequences (Karlsson et al. 2007; Hauswirth et al. 2012; Chen et al. 2016; Zhou et al. 2018; Hofstetter et al. 2019). Mutations in coding sequences are more likely to alter protein functions. In particular for pleiotropic factors, such mutations will be more harmful to the organisms than those occurring in regulatory regions, which only affect the expression level (Wittkopp and Kalay 2012; Petit et al. 2017). In this regard, mutations in the coding sequences of microphthalmia-associated transcription factors have been extensively reported to lead to various defects in animals (Tassabehji et al. 1994; Levy et al. 2006). This type of mutation is more prone to be eliminated by artificial selection. Above all, mutations in cis-regulatory elements provide valuable raw materials for selection during domestication and play critically important roles in phenotypic variation.
Conclusion
In this study, we sequenced the genomes of several fighting fish breeds and studied the genomic basis of most striking color and fin shape variants in this species. We found that phenotypes including some colorations and fin shapes were determined by major-effect loci, indicating that major loci can bring about phenotypic innovations rapidly. Using CRISPR/Cas-9 induced-mutations, we verified that both double-tail and albino phenotypes resulted from mutations in a regulatory element near zic1/zic4 and mutations in coding regions of mitfa, respectively. Our findings suggest that cis-regulatory elements play critically important roles in generating phenotypic variation during domestication by artificial selection as well as natural selection at evolutionary time scales. There are still other breeds varying in fin shapes and sizes as well as in pigment patterns and in aggression worthy of further investigation by the CRISPR/Cas9 methods we developed here. This will facilitate that this species will become a new model system since it is amenable for dissecting the genetic architecture underlying morphological and behavioral evolutionary innovations.
Materials and Methods
Genome Sequencing and Assembly
Genomic DNA from one highly inbred yellow single-tail female and one transparent double-tail male were used for construction of both short-insert (∼270, 350, and 550 bp) and jumping libraries (3, 5, 10, 15, and 20 kb) (supplementary table S1 and fig. S2, Supplementary Material online). In addition, one female (albino and double-tail) and one male (also homozygous for melanin pigmentation and single-tail) were sequenced with 550-bp insert libraries. Genomic DNA was isolated with MagAttract HMW DNA Kit (Qiagen). Heterozygosity versus homozygosity of these fish was assessed at ten microsatellite loci development in a previous study (Chailertrit et al. 2014). The yellow female showed a low genetic heterozygosity of 0.1. Genotypes of these fish were determined by both test crosses and genetic markers developed as described below. All sequencing was carried out on Illumina Nextseq 500. Raw reads were cleaned using the program process_shortreads (-r -c -q -t 150) in the Stacks software package (Catchen et al. 2013). Genomes of the highly inbred yellow single-tail female and the transparent double-tail male of varying libraries were assembled using ALLPATHS-LG (Gnerre et al. 2011) with default parameters, whereas the other two unique libraries were assembled using ABYSS2.0 (Jackman et al. 2017) with default parameters. Gaps were filled with paired reads using GapFiller (Nadalin et al. 2012). The genome size of the fighting fish was estimated based on k-mer frequencies, de novo RADtags mapping and Q-PCR method (supplementary note 1, Supplementary Material online). Completeness of the genome assembly was evaluated using BUSCO (Simão et al. 2015) and by the mapping rate of transcripts and de novo RADtags.
RNA Sequencing and Analysis
Three mRNA libraries were separately constructed for one male and two females of 3 months’ age. Total RNA was isolated from brain, eye, skin, gill, muscle, intestine, spleen, liver, heart, kidney, and gonad, and then equal amounts from each tissue were pooled for library construction using Illumina TruSeq RNA sample preparation kit (Illumina). Moreover, RNA samples of another mature male and female, derived from the above pooling strategy, were used for total RNA library construction with rRNA depletion, using NEBNext Ultra RNA Library Prep Kit (NEB). Raw sequences were cleaned with process_shortreads (-r -c -q -t 150) in Stacks package (Catchen et al. 2013). Transcripts of individuals were assembled using Trinity (Haas et al. 2013) with default parameters and then used for genome annotation.
RADseq and SNP Genotyping
Samples from both mapping families and cultured strains with specific traits (see below), were genotyped using RADseq (Baird et al. 2008) with some modifications as described in our previous study (Bai et al. 2018). High-quality genomic DNA of 500 ng was digested with restriction enzyme PstI-HF (NEB) and ligated to barcoded adaptors with T4 DNA Ligase (NEB). DNA was then sheared with a peak of 500 bp for library construction. All libraries were sent to NextSeq500 (Illumina) for 150-bp single-end sequencing. Parental and offspring samples were sequenced with an average of 16.9 and 6.1 M reads, respectively, for accurate SNP calling (supplementary table S1, Supplementary Material online). Raw reads were filtered using process_radtags (-r -c -q -t 150) in Stacks package (Catchen et al. 2013). BWA-MEM (Bessa et al. 2009) was used for reference-based mapping with default parameters and only reads with unique targets were retained. SNPs were discovered and genotyped using Stacks package (Catchen et al. 2013) with parameters as described in our previous study (Bai et al. 2018).
Linkage Mapping and Chromosomal-Level Genome Assembly
Two F2 families: BM1 and RM2 were used for linkage mapping. These two F2 families: BM1 (92 fish) and RM2 (274 fish) were generated with two pairs of F1 parents (i.e., BM1female × BM1male and RM2female × RM2male), respectively, which were the offspring of P parents: DtY2female and F0B1male (see details about their phenotypes in the subsection “Genetic mapping for traits of interest”). Genotyping of the two F2 mapping families was conducted using RAD sequencing as described above. SNPs were firstly filtered for Mendelian segregation distortion using χ2 tests (P < 0.05). The cutoff value of missing genotypes across families was <15%, which left 80 and 213 samples for BM1 and RM2, respectively, for linkage mapping. Linkage group assignment and marker ordering were carried out using Lep-MAP3 (Rastas 2017) with logarithm of the odds (LOD) cutoff of 10. Both sex-averaged and sex-specific maps were constructed. The constructed linkage maps (Supplementary linkage maps, Supplementary Material online) were used to build a chromosome-level genome assembly. RAD sequences of mapped markers were aligned to scaffolds to examine the occurrence of chimeric assemblies using ALLMAPS (Tang et al. 2015), as linkage maps are not likely to generate among-chromosome grouping errors (Small et al. 2016). If there were more than three markers from the same scaffolds mapped to different linkage groups, the scaffolds were split at the longest gaps between mismatched fragments. The new scaffolds were then anchored onto genetic maps to generate chromosome-level assemblies using ALLMAPS (Tang et al. 2015) with default parameters.
Genome Annotation
Annotation of the highly inbreed female fighting fish genome was conducted using MAKER (Cantarel et al. 2008). The sequences were softmasked using RepeatMasker (Chen 2004) based on the repeat libraries obtained from RepeatModeler (http://www.repeatmasker.org), Repbase (Jurka et al. 2005), and MAKER (Cantarel et al. 2008) sequence repeat databases. Both evidence-based and ab initio gene models were used for annotation. Transcriptomes of fighting fish and protein sequences of zebrafish, medaka, stickleback, fugu, and Nile tilapia from Ensembl database (release 86) were used for evidence. SNAP (Korf 2004) and Augustus (Stanke and Waack 2003) were iteratively used for ab initio gene models training. Predicted protein sequences were annotated by blast to nr and RefSeq databases (Pruitt et al. 2005) with BLASTP (E-value < 1E−10).
Prediction of CNEs
Identification of CNEs was according to a previous method (Brawand et al. 2014). In brief, the fighting fish genome was used as reference for pairwise whole-genome alignment with zebrafish (D. rerio), medaka (O. latipes), stickleback (G. aculeatus), fugu (T. rubripes), and Nile tilapia (O. niloticus) (downloaded from Ensembl database, release 86) using LASTZ (Harris 2007). Multiple alignments were generated with MULTIZ (Blanchette et al. 2004) using the tree topology among the six species based on the phylogenetic study. Conserved sequences at least in one pair of alignments were predicted using PhastCons (Siepel et al. 2005) under both conserved and nonconserved models (coverage = 0.3 and length = 45 bp). The predicted CNEs were then filtered by comparison to the coding sequences, noncoding RNAs, pseudogenes, and transposable elements of the six studied species and also the transcripts of fighting fish (E-value < 1E−10). Only the elements of > 30 bp and with repetitive content <50% were retained. For studies on CNEs related to candidate genes, we manually aligned the genomic loci with more reference fish including nonmodel species to refine the CNEs and identify candidate regulatory elements that might be lineage- or species-specific, using the above standards.
Whole-Genome Resequencing and Genetic Diversity Analysis
Six wild (four from Thailand and two from Cambodia, respectively) and 28 randomly selected domesticated fish were sequenced with 500-bp insert libraries (supplementary table S8, Supplementary Material online). Nine “elephant ear” mutants, that is, with phenotype of overgrowth of pectoral fin, were further sequenced to identify the genetic locus (supplementary table S8, Supplementary Material online). Raw sequencing reads were filtered using the above method. Sequence mapping and variant calling were carried out using BWA-mem (Bessa et al. 2009) and Picard/GATK v4.0 best practices workflows (DePristo et al. 2011). SNPs were filtered with the following parameters: “QD < 2.0 ‖ FS > 60.0 ‖ MQ < 40.0 ‖ MQRankSum < −12.5 ‖ ReadPosRankSum < −8.0 ‖ SOR > 4.0,” and indels with “QD < 2.0 ‖ FS > 200.0 ‖ ReadPosRankSum < −20.0 ‖ SOR > 10.0.” We further filtered the variants with “minDP 7, –max-missing 0.925” using VCFtools (Danecek et al. 2011). A total of 6,735,573 genotypes were obtained for further analysis. Genetic diversity was estimated with a 100-kb window size using VCFtools (Danecek et al. 2011). Unbiased estimation of nucleotide diversity taking into consideration both segregating and nonsegregating sites were computed using the program pixy (Korunes and Samuk 2021). Population structure was firstly analyzed with principal component analysis (PCA) using Plink2.0 (Purcell et al. 2007). The program Admixture (Alexander et al. 2009) was then used to infer the genetic clusters at individual level.
Genetic Mapping for Traits of Interest
Three pigmentation traits, including distribution of red pigments in different body compartments (Fig. 2A), dorsal fin spotting (fig. 2C), and albino phenotype (fig. 2D), and two fin morphology traits (i.e., elephant ear and double tail, figs. 3Aand 4A) were studied. Before setting up mapping families by crossing parents with different phenotypes, test crosses were generated to examine phenotypic segregation of double tail, albino, and dorsal fin spotting. Parents that were homozygous for the two studied traits (i.e., double-tail vs. single-tail and melanin vs. albino) were selected as P (parental) generation (DtY2female and F0B1male) to set up mapping families (supplementary fig. S2, Supplementary Material online). Two F2 families: BM1 and RM2 were produced by crossing F1 parents (BM1female and BM1male) and (RM2female and RM2male), respectively, which were the offspring of P generation: DtY2female and F0B1male. The two F2 mapping families were genotyped using RAD sequencing as described above.
In the two F2 families (i.e., BM1 containing 92 fish and RM2 containing 274 fish), four traits: distribution of red pigment, dorsal fin spotting, albino, and double tail were recorded for each individual. In detail, F0B1male was a homozygous single-tail and melanin pigmented fish (wild type at both loci), whereas DtY2female was a homozygous double-tail and albino (loss of melanin) fish. Both traits (i.e., double-tail and albino) showed a recessive Mendelian inheritance pattern with a segregation ratio of 3:1 in F2 populations. Mixed linear model association analyses were separately conducted based on the two F2 families to map the two traits. To narrow down the genomic regions responsible for the two traits, 136 additional fish of confirmed phenotypes, from China, Thailand, Malaysia, Singapore, and Indonesia were genotyped using both RAD sequencing and SNP/indel markers developed from resequencing data, as described above. Genotypes were used to identify recombinants in the major loci determining double-tail and albino. For dorsal fin spotting, parents, DtY2female and F0B1male showed spotted and nonspotted dorsal fin, respectively, whereas F1 parents, RM2female and RM2male showed spotted and nonspotted dorsal fin, respectively. Due to a phenotypic interaction of iridescent pigmentation patterns and the albino condition, which also segregated in the F2 mapping crosses, only 156 individuals from the RM2 family could be phenotyped. This trait presented a segregation ratio of 1:1 for spotted versus nonspotted dorsal fin. We carried out both quantitative trait loci (QTL) mapping and mixed linear modeling for this trait using these 156 phenotyped fish. As the results in QTL mapping (peak LOD score and PVE were 282.79% and 100%, respectively) and mixed linear modeling were consistent, we only present the results of the mixed linear modeling. Finally, for distribution of red pigment, we observed red pigments in DtY2female, but not in F0B1male, whereas all F1 fish presented red pigments. In both F2 families, we observed the distribution pattern of red pigments varied evidently not only across different body compartments of each individual but also among individuals throughout the whole family. We developed a method to quantify and record red pigments in different body areas (supplementary note 2, Supplementary Material online). QTL mapping was conducted in the large RM2 family with 211 phenotyped offspring, rather than BM1 with only 77 phenotyped siblings, to map and estimate the effects of the loci.
Mixed linear modeling was performed with compressed mixed linear model implemented in the GAPIT R package, with the GRM and sex as covariates (Lipka et al. 2012). This mixed model incorporates and estimates component variance of both kinship relatedness matrix and sex, using VanRaden algorithm. P value was calculated for each marker and the statistical significance threshold was determined at 0.05 level with Bonferroni corrections (P = 0.05/N, where N is the number of total markers used for association test). QTL mapping was conducted using the Haley–Knott regression method (Haley and Knott 1992) implemented in the R package qtl (Broman et al. 2003). The linkage map of RM2 family was used for QTL mapping, with interval mapping algorithm. LOD thresholds for both chromosome- and genome-wide significance were estimated with permutation tests for 1,000 times. To screen the “elephant ear” locus, a genome-wide FST scan was performed between elephant ear mutants and the remaining resequenced samples, 9 and 28 fish respectively, using 30-kb window size with a step of 15 kb. Window-size FST values were then Z transformed (ZFST = (FST − µFST)/σFST) to compare among chromosomes. Variants within and flanking this locus were retrieved and analyzed to refine the haplotypes. Fixed or nearly fixed variants were annotated and protein-coding genes within this locus were individually analyzed by literature mining. Genes associated with fin development and regeneration were kept for expression analysis using real-time reverse transcription PCR (RT-PCR) to identify candidate genes.
Developing and Validating Trait-Associated DNA Markers
In order to quickly differentiate among genotypes, we developed indel markers for fast PCR assays for double-tail and albino traits. The shortest genomic region resulting from association mapping was used for marker screening. Homozygous individuals with regard to the above traits were resequenced for marker discovery using GATK pipeline (DePristo et al. 2011) according to our previous study (Wang et al. 2015). Indels were firstly validated by manual alignment of genome sequences between homozygous mutant and wild type. Primers of these indels of suitable length were then designed for PCR assays. The associations between phenotypes and these discovered markers were further tested in domesticated fish from different strains (from ∼500 to ∼1000 individuals for different traits) to examine recombination between markers for fine mapping.
Gene Expression Analysis Using RT-PCR and qRT-PCR
Gene expression was studied by RT-PCR. Total RNA from independent tissues or embryos was isolated using TRIzol reagent (Invitrogen). Two micrograms of total RNA were treated with DNase I (Roche) and then used for cDNA synthesis using Reverse Transcriptase M-MLV (Promega). Expression of genes of interest in different tissues and in different developmental stages was firstly examined using RT-PCR with gene-specific primers. The relative expression of candidate genes was then studied using real-time RT-PCR (qRT-PCR) using KAPA SYBR FAST qPCR Kits (KapaBiosystems) with CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Three replicates were performed for each sample and cDNA from 50 ng of total RNA was used for each reaction. Beta actin or EF1A was used as endogenous reference according to their expression stability. The 2−ΔΔCT method (Livak and Schmittgen 2001) was used to quantify relative gene expression.
Luciferase Reporter Assay
Candidate enhancer regions with closely flanking sequences from different alleles were cloned and constructed into the enhancer region of pGL3-Promoter vector that contains a basal SV40 promoter sequence (Promega). The reporter gene constructs together with pRL Renilla Luciferase Control Reporter Vector (Promega) were cotransfected into a Singapore grouper embryonic cell line (Chew-Lim et al. 1994) using TurboFect or Lipofectamine 3000 (Thermo Fisher). Luciferase activity was measured at 48 h post-transfection using Dual-Luciferase Reporter Assay System (Promega). Three independent transfections were carried out in six-well plates with each measured in triplicates.
Enhancer Reporter Assay
Candidate enhancer sequences of different alleles with closely flanking sequences were constructed into ZED Vector (Bessa et al. 2009) using Gateway Recombination Cloning Technology (Thermo Fisher Scientific). T7-Transposase (Khattak et al. 2014) (Addgene) was transcribed using mMESSAGE mMACHINE T7 kit (Life Technologies), according to the manufacturer’s instructions. A final concentration of 40 ng/µl ZED constructs, 50 ng/µl transposase mRNA, and 0.05% phenol red were coinjected into one-cell stage embryos. The embryos were imaged for GFP and internal control red fluorescent protein (RFP) expression at different time points using a Leica MZFLIII microscope. The elements were considered as candidate enhancers if there were more than 20% of injected embryos showing consistent expression pattern of GFP at the presence of RFP (Bessa et al. 2009; Sharma et al. 2015).
Knockout Using CRISPR/Cas9
CRISPR/Cas9 was used to introduce mutations into the genes or elements of interest. Guide RNA (gRNA) was designed using E-CRISP (Heigwer et al. 2014). gRNA sequences were blasted against the reference genome to avoid off-targets. Template of gRNA was assembled using PCR according to a previous method (Vejnar et al. 2016). In brief, gRNA was designed with common flanking adaptors as follows: 5′-TAATACGACTCACTATA[GGN(18)]GTTTTAGAGCTAGAA-3′. A universal primer was used to assemble gRNA temple with direct PCR, with the following sequences: 5′-AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3′. gRNA was transcribed using HiScribe T7 High Yield RNA Synthesis Kit (NEB) with 150 ng of purified DNA template and gRNA was subsequently purified using miRNeasy Mini Kit (Qiagen), according to the manufacturer’s instructions. Cas9 Nuclease NLS (NEB) and gRNA with a final concentration of 100 and 200 ng/µl, respectively, were coinjected into one-cell stage embryos. Both phenotypes and genotypes were screened for candidate mutants. DNA fragments spanning the targeted sequences of gRNAs were amplified using fragment-specific primers. PCR products were purified using QIAquick PCR Purification Kit (Qiagen) for mutant screening using T7 endonuclease assay (NEB). PCR products that showed cleavage in T7 endonuclease assay were then validated by TA cloning and Sanger sequencing. We developed a whole protocol for transgenic and CRISPR knockout technology for the fighting fish, a species of particular mating and brooding behaviors (supplementary note 3, Supplementary Material online).
Ethics Declarations
All procedures for handling of fish were according to the instructions of the Institutional Animal Care and Use Committee (IACUC) of Temasek Life Sciences Laboratory, Singapore (Approval no. TLL (F)-16-003).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Data Availability
The genome sequences and related annotations of fighting fish are hosted by the web server of Temasek Lifesciences Laboratory (https://genhua.tll.org.sg/) and archived in China National GeneBank (CNGB) and DDBJ with BioProject accession nos. CNP0001745 and PRJDB7253, respectively. Sequences used for whole-genome sequencing, RNA sequencing, and RAD sequencing are available with the DDBJ Sequencing Read Archive (SRA) through BioProject ID PRJDB7253–PRJDB7255.
Supplementary Material
Acknowledgments
We thank Betta hobbyists from both Betta Club Singapore and Wild Betta Club Singapore for providing samples and photographs. We acknowledge the fish facility, biocomputing facility and IT department of Temasek Lifesciences Laboratory for help in fish culture, high-performance computing and data processing. We are grateful to Dr José Bessa and Dr Ying Yang for providing ZED and luciferase vectors, respectively. Our grateful thanks are also extended to Dr. Jinlu Wu for providing the Singapore grouper embryonic cell lines. This project was supported by the Temasek Lifesciences Laboratory, Singapore.
Author Contributions
G.H.Y. initiated the project “Genetics and genomics of the fighting fish.” G.H.Y., L.W., M.S., and A.M. conceived the study. G.H.Y., F.P., M.S., and A.M. supervised the whole study. L.W. and F.S. performed genome sequencing. L.W., Z.Y.W., and F.S. performed RNA sequencing. L.W., F.S., Z.Y.W., B.Y., and B.B. performed RAD sequencing. F.S., L.W., H.L., Y.W., and H.P. set up mapping families and cultured fish. L.W. assembled and annotated genomes and transcriptomes. L.W., B.Y., and Z.Y.W. analyzed sequences. L.W. and B.Y. constructed linkage maps. F.S., L.W., Y.W., and B.Y. conducted fine mapping of QTL. L.W., Z.Y., F.S., and Z.Y.W. constructed vectors. L.W., F.S., Z.Y.W., and Y.W. performed knockout and injections. F.S., L.W., and Z.Y.W. examined gene expression. F.S. and Y.W. contributed to genotyping and sequencing of phenotypes. Z.M., B.F., Y.A., Y.S., and M.L. collection and phenotyping of samples. L.W., B.Y., F.S., and G.H.Y. wrote the paper with inputs from the other authors. G.H.Y., L.W., F.P., M.S., and A.M. interpreted the findings in biological context and commented on the manuscript. All authors discussed the results and approved the final version of the paper.
References
- Al-Mamun HA, Kwan P, Clark SA, Ferdosi MH, Tellam R, Gondro C.. 2015. Genome-wide association study of body weight in Australian Merino sheep reveals an orthologous region on OAR6 to human and bovine genomic regions affecting height and weight. Genet Sel Evol. 47(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiadi D, Piferrer F.. 2019. Epimutations in developmental genes underlie the onset of domestication in farmed European sea bass. Mol Biol Evol. 36(10):2252–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L, Archibald AL, Bottema CD, Brauning R, Burgess SC, Burt DW, Casas E, Cheng HH, Clarke L, Couldrey C, et al. ; The FAANG Consortium. 2015. Coordinated international action to accelerate genome-to-phenome with FAANG, the Functional Annotation of Animal Genomes project. Genome Biol. 16(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B, Wang L, Zhang YJ, Lee M, Rahmadsyah R, Alfiko Y, Ye BQ, Purwantomo S, Suwanto A, Chua N-H, et al. 2018. Developing genome-wide SNPs and constructing an ultrahigh-density linkage map in oil palm. Sci Rep. 8(1):691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA.. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 3(10):e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa J, Tena JJ, de la Calle-Mustienes E, Fernández-Miñán A, Naranjo S, Fernández A, Montoliu L, Akalin A, Lenhard B, Casares F, et al. 2009. Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis‐regulatory regions in zebrafish. Dev Dyn. 238(9):2409–2417. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AFA, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14(4):708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank MC, Grinberg I, Aryee E, Laliberte C, Chizhikov VV, Henkelman RM, Millen KJ.. 2011. Multiple developmental programs are altered by loss of Zic1 and Zic4 to cause Dandy-Walker malformation cerebellar pathogenesis. Development. 138(6):1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Wagner CE, Li YI, Malinsky M, Keller I, Fan S, Simakov O, Ng AY, Lim ZW, Bezault E, et al. 2014. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 513(7518):375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen Ś, Churchill GA.. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 19(7):889–890. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, Holt C, Alvarado AS, Yandell M.. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18(1):188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA.. 2013. Stacks: an analysis tool set for population genomics. Mol Ecol. 22(11):3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chailertrit V, Swatdipong A, Peyachoknagul S, Salaenoi J, Srikulnath K.. 2014. Isolation and characterization of novel microsatellite markers from Siamese fighting fish (Betta splendens, Osphronemidae, Anabantoidei) and their transferability to related species, B. smaragdina and B. imbellis. Genet Mol Res. 13(3):7157–7162. [DOI] [PubMed] [Google Scholar]
- Chen L, Guo W, Ren L, Yang M, Zhao Y, Guo Z, Yi H, Li M, Hu Y, Long X, et al. 2016. A de novo silencer causes elimination of MITF-M expression and profound hearing loss in pigs. BMC Biol. 14(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N. 2004. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics 5(1):11–14. [DOI] [PubMed] [Google Scholar]
- Chew-Lim M, Ngoh G, Ng M, Lee J, Chew P, Li J, Chan Y, Howe J.. 1994. Grouper cell line for propagating grouper viruses. Singapore J Primary Ind. 22:113–116. [Google Scholar]
- Cowper-Sal R, Zhang X, Wright JB, Bailey SD, Cole MD, Eeckhoute J, Moore JH, Lupien M.. 2012. Breast cancer risk–associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet. 44(11):1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. ; 1000 Genomes Project Analysis Group. 2011. The variant call format and VCFtools. Bioinformatics. 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do DN, Strathe AB, Ostersen T, Jensen J, Mark T, Kadarmideen HN.. 2013. Genome-wide association study reveals genetic architecture of eating behavior in pigs and its implications for humans obesity by comparative mapping. PLoS One. 8(8):e71509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán I, Marí-Beffa M, Santamaría J, Becerra J, Santos-Ruiz L.. 2011. Actinotrichia collagens and their role in fin formation. Dev Biol. 354(1):160–172. [DOI] [PubMed] [Google Scholar]
- Fan G, Chan J, Ma K, Yang B, Zhang H, Yang X, Shi C, Chun-Hin LH, Ren Z, Xu Q.. 2018. Chromosome-level reference genome of the Siamese fighting fish Betta splendens, a model species for the study of aggression. GigaScience. 7:giy087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Murray SS, Schork NJ, Topol EJ.. 2009. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 10(4):241–251. [DOI] [PubMed] [Google Scholar]
- Gnerre S, MacCallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 108(4):1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg I, Northrup H, Ardinger H, Prasad C, Dobyns WB, Millen KJ.. 2004. Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy-Walker malformation. Nat Genet. 36(10):1053–1055. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley CS, Knott SA.. 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity (Edinb). 69(4):315–324. [DOI] [PubMed] [Google Scholar]
- Harris RS. 2007. Improved Pairwise Alignmnet of Genomic DNA. Pennsylvania: The Pennsylvania State University.
- Hauswirth R, Haase B, Blatter M, Brooks SA, Burger D, Drögemüller C, Gerber V, Henke D, Janda J, Jude R, et al. 2012. Mutations in MITF and PAX3 cause “splashed white” and other white spotting phenotypes in horses. PLoS Genet. 8(4):e1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F, Kerr G, Boutros M.. 2014. E-CRISP: fast CRISPR target site identification. Nat Methods. 11(2):122–123. [DOI] [PubMed] [Google Scholar]
- Hofstetter S, Seefried F, Häfliger IM, Jagannathan V, Leeb T, Drögemüller C.. 2019. A non‐coding regulatory variant in the 5′‐region of the MITF gene is associated with white‐spotted coat in Brown Swiss cattle. Anim Genet. 50(1):27–32. [DOI] [PubMed] [Google Scholar]
- Hyten DL, Song Q, Zhu Y, Choi I-Y, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB.. 2006. Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci U S A. 103(45):16666–16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman SD, Vandervalk BP, Mohamadi H, Chu J, Yeo S, Hammond SA, Jahesh G, Khan H, Coombe L, Warren RL, et al. 2017. ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 27(5):768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J.. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110(1–4):462–467. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, Salmon Hillbertz NHC, Zody MC, Anderson N, Biagi TM, Patterson N, Pielberg GR, Kulbokas EJ, et al. 2007. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 39(11):1321–1328. [DOI] [PubMed] [Google Scholar]
- Kawanishi T, Kaneko T, Moriyama Y, Kinoshita M, Yokoi H, Suzuki T, Shimada A, Takeda H.. 2013. Modular development of the teleost trunk along the dorsoventral axis and zic1/zic4 as selector genes in the dorsal module. Development. 140(7):1486–1496. [DOI] [PubMed] [Google Scholar]
- Kemble H, Nghe P, Tenaillon O.. 2019. Recent insights into the genotype–phenotype relationship from massively parallel genetic assays. Evol Appl. 12(9):1721–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattak S, Murawala P, Andreas H, Kappert V, Schuez M, Sandoval-Guzmán T, Crawford K, Tanaka EM.. 2014. Optimized axolotl (Ambystoma mexicanum) husbandry, breeding, metamorphosis, transgenesis and tamoxifen-mediated recombination. Nat Protoc. 9(3):529–540. [DOI] [PubMed] [Google Scholar]
- Kon T, Omori Y, Fukuta K, Wada H, Watanabe M, Chen Z, Iwasaki M, Mishina T, Matsuzaki S-IS, Yoshihara D, et al. 2020. The genetic basis of morphological diversity in domesticated goldfish. Curr Biol. 30(12):2260–2274. [DOI] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics. 5(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korunes KL, Samuk K.. 2021. pixy: Unbiased estimation of nucleotide diversity and divergence in the presence of missing data. Mol Ecol Resour. 21(4):1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujawski S, Lin W, Kitte F, Börmel M, Fuchs S, Arulmozhivarman G, Vogt S, Theil D, Zhang Y, Antos CL.. 2014. Calcineurin regulates coordinated outgrowth of zebrafish regenerating fins. Dev Cell. 28(5):573–587. [DOI] [PubMed] [Google Scholar]
- Lanni JS, Peal D, Ekstrom L, Chen H, Stanclift C, Bowen ME, Mercado A, Gamba G, Kahle KT, Harris MP.. 2019. Integrated K+ channel and K+ Cl-cotransporter functions are required for the coordination of size and proportion during development. Dev Biol. 456(2):164–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B. 2013. Genotype to phenotype: lessons from model organisms for human genetics. Nat Rev Genet. 14(3):168–178. [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE.. 2006. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 12(9):406–414. [DOI] [PubMed] [Google Scholar]
- Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z.. 2012. GAPIT: genome association and prediction integrated tool. Bioinformatics. 28(18):2397–2399. [DOI] [PubMed] [Google Scholar]
- Lister JA, Close J, Raible DW.. 2001. Duplicate mitf genes in zebrafish: complementary expression and conservation of melanogenic potential. Dev Biol. 237(2):333–344. [DOI] [PubMed] [Google Scholar]
- Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW.. 1999. Nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 126(17):3757–3767. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 25(4):402–408. [DOI] [PubMed] [Google Scholar]
- Lucas GA. 1968. A study of variation in the Siamese Fighting Fish, Betta splendens, with emphasis on color mutants and the problem of sex determination. Iowa: Iowa State University.
- ΡMalmstrøm M, Matschiner M, Tørresen OK, Jakobsen KS, Jentoft S.. 2017. Whole genome sequencing data and de novo draft assemblies for 66 teleost species. Sci Data. 4(1):160132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin KA, Levin M.. 2018. Bioelectric signaling in regeneration: mechanisms of ionic controls of growth and form. Dev Biol. 433(2):177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle F, Bed'Hom B, Coville J-L, Ito S, Inoue-Murayama M, Gourichon D.. 2010. The “silver“ Japanese quail and the MITF gene: causal mutation, associated traits and homology with the “blue” chicken plumage. BMC Genet. 11:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monvises A, Nuangsaeng B, Sriwattanarothai N, Panijpan B.. 2009. The Siamese fighting fish: well-known generally but little-known scientifically. ScienceAsia. 35(1):8–16. [Google Scholar]
- Moriyama Y, Kawanishi T, Nakamura R, Tsukahara T, Sumiyama K, Suster ML, Kawakami K, Toyoda A, Fujiyama A, Yasuoka Y, et al. 2012. The medaka zic1/zic4 mutant provides molecular insights into teleost caudal fin evolution. Curr Biol. 22(7):601–607. [DOI] [PubMed] [Google Scholar]
- Nadalin F, Vezzi F, Policriti A.. 2012. GapFiller: a de novo assembly approach to fill the gap within paired reads. BMC Bioinformatics. 13(S14):S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perathoner S, Daane JM, Henrion U, Seebohm G, Higdon CW, Johnson SL, Nüsslein-Volhard C, Harris MP.. 2014. Bioelectric signaling regulates size in zebrafish fins. PLoS Genet. 10(1):e1004080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit F, Sears KE, Ahituv N.. 2017. Limb development: a paradigm of gene regulation. Nat Rev Genet. 18(4):245–258. [DOI] [PubMed] [Google Scholar]
- Prost S, Petersen M, Grethlein M, Hahn SJ, Kuschik-Maczollek N, Olesiuk ME, Reschke J-O, Schmey TE, Zimmer C, Gupta DK.. 2020. Improving the chromosome-level genome assembly of the Siamese fighting fish (Betta splendens) in a university master's course. G3: Genes, Genomes, Genetics. 10:2179–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR.. 2005. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33(Database issue):D501–D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastas P. 2017. Lep-MAP3: robust linkage mapping even for low-coverage whole genome sequencing data. Bioinformatics. 33(23):3726–3732. [DOI] [PubMed] [Google Scholar]
- ΘRüber L, Britz R, Tan HH, Ng PK, Zardoya R.. 2004. Evolution of mouthbrooding and life‐history correlates in the fighting fish genus Betta. Evolution. 58(4):799–813. [DOI] [PubMed] [Google Scholar]
- Rubin C-J, Zody MC, Eriksson J, Meadows JRS, Sherwood E, Webster MT, Jiang L, Ingman M, Sharpe T, Ka S, et al. 2010. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 464(7288):587–591. [DOI] [PubMed] [Google Scholar]
- Schartl M, Kneitz S, Ormanns J, Schmidt C, Anderson JL, Amores A, Catchen J, Wilson C, Geiger D, Du K.. 2020. The developmental and genetic architecture of the sexually selected male ornament of swordtails. Curr Biol. 31:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte CJ, Allen C, England SJ, Juárez-Morales JL, Lewis KE.. 2011. Evx1 is required for joint formation in zebrafish fin dermoskeleton. Dev Dyn. 240(5):1240–1248. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jónsson B, Schluter D, Kingsley DM.. 2004. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 428(6984):717–723. [DOI] [PubMed] [Google Scholar]
- Sharma S, Londono D, Eckalbar WL, Gao X, Zhang D, Mauldin K, Kou I, Takahashi A, Matsumoto M, Kamiya N, et al. ; Japan Scoliosis Clinical Research Group. 2015. A PAX1 enhancer locus is associated with susceptibility to idiopathic scoliosis in females. Nat Commun. 6(1):6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15(8):1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim N-L, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC.. 2012. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40(Web Server issue):W452–W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Simpson M. 1968. The display of the Siamese fighting fish, Betta splendens. Anim Behav Monogr. 1:i–73. [Google Scholar]
- Small C, Bassham S, Catchen J, Amores A, Fuiten A, Brown R, Jones A, Cresko W.. 2016. The genome of the Gulf pipefish enables understanding of evolutionary innovations. Genome Biol. 17(1):258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HM. 1945. The freshwater fishes of Siam, or Thailand. Bull US Natl Mus. 188:1–633. [Google Scholar]
- Stanke M, Waack S.. 2003. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 19(Suppl 2):ii215–ii225. [DOI] [PubMed] [Google Scholar]
- Stewart S, Le Bleu HK, Yette GA, Henner AL, Braunstein JA, Stankunas K.. 2019. Longfin causes cis-ectopic expression of the kcnh2a ether-a-go-go K+ channel to autonomously prolong fin outgrowth. BioRxiv. doi:10.1101/790329. [DOI] [PMC free article] [PubMed]
- ΘTang H, Zhang X, Miao C, Zhang J, Ming R, Schnable JC, Schnable PS, Lyons E, Lu J.. 2015. ALLMAPS: robust scaffold ordering based on multiple maps. Genome Biol. 16(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassabehji M, Newton VE, Read AP.. 1994. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet. 8(3):251–255. [DOI] [PubMed] [Google Scholar]
- Vejnar CE, Moreno-Mateos MA, Cifuentes D, Bazzini AA, Giraldez AJ.. 2016. Optimized CRISPR–Cas9 system for genome editing in zebrafish. Cold Spring Harb Protoc. 2016(10):pdb.prot086850. [DOI] [PubMed] [Google Scholar]
- Wang L, Huang SQ, Xia JH, Liu P, Wan ZY, Yue GH.. 2015. Genome-wide discovery of gene-related SNPs in Barramundi Lates calcarifer. Conservation Genet Resour. 7(3):605–608. [Google Scholar]
- Wei C, Wang H, Liu G, Wu M, Cao J, Liu Z, Liu R, Zhao F, Zhang L, Lu J, et al. 2015. Genome-wide analysis reveals population structure and selection in Chinese indigenous sheep breeds. BMC Genomics. 16(1):194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G.. 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 13(1):59–69. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Li M, Cheng H, Fan W, Yuan Z, Gao Q, Xu Y, Guo Z, Zhang Y, Hu J, et al. 2018. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat Commun. 9(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences and related annotations of fighting fish are hosted by the web server of Temasek Lifesciences Laboratory (https://genhua.tll.org.sg/) and archived in China National GeneBank (CNGB) and DDBJ with BioProject accession nos. CNP0001745 and PRJDB7253, respectively. Sequences used for whole-genome sequencing, RNA sequencing, and RAD sequencing are available with the DDBJ Sequencing Read Archive (SRA) through BioProject ID PRJDB7253–PRJDB7255.