Abstract
Understanding how genes interact is a central challenge in biology. Experimental evolution provides a useful, but underutilized, tool for identifying genetic interactions, particularly those that involve non-loss-of-function mutations or mutations in essential genes. We previously identified a strong positive genetic interaction between specific mutations in KEL1 (P344T) and HSL7 (A695fs) that arose in an experimentally evolved Saccharomyces cerevisiae population. Because this genetic interaction is not phenocopied by gene deletion, it was previously unknown. Using “evolutionary replay” experiments, we identified additional mutations that have positive genetic interactions with the kel1-P344T mutation. We replayed the evolution of this population 672 times from six timepoints. We identified 30 populations where the kel1-P344T mutation reached high frequency. We performed whole-genome sequencing on these populations to identify genes in which mutations arose specifically in the kel1-P344T background. We reconstructed mutations in the ancestral and kel1-P344T backgrounds to validate positive genetic interactions. We identify several genetic interactors with KEL1, we validate these interactions by reconstruction experiments, and we show these interactions are not recapitulated by loss-of-function mutations. Our results demonstrate the power of experimental evolution to identify genetic interactions that are positive, allele specific, and not readily detected by other methods, shedding light on an underexplored region of the yeast genetic interaction network.
Keywords: experimental evolution, genetic interactions, yeast
Introduction
Genetic interactions drive evolutionary change (Breen et al. 2012), define functional relationships between genes (Costanzo et al. 2010; Costanzo et al. 2016), and contribute to disease, behaviors, and other complex traits (Manolio et al. 2009; Zuk et al. 2012). Despite the importance of genetic interactions, we lack a broad understanding of their prevalence and their properties. Most of our understanding of genetic interactions comes from systematic screens of double deletion mutants (Tong et al. 2004; Baryshnikova et al. 2010; Babu et al. 2014; Costanzo et al. 2016; Kuzmin et al. 2018). Double deletion analyses and alternative approaches, such as RNAi (Bassik et al. 2013) or CRISPR/Cas9 library screens (Han et al. 2017; Shen et al. 2017) and transposon mutagenesis (Horton and Kumar 2015) focus almost exclusively on loss-of-function mutations, which are rarely beneficial and account for only a small fraction of standing natural variation (Bergström et al. 2014; Saleheen et al. 2017). Conversely, methods that assess the phenotypic effects of natural variants, such as genome-wide association studies and quantitative trait locus (QTL) mapping experiments, are underpowered to detect genetic interactions (Marchini et al. 2005; Jiang et al. 2010; Zuk et al. 2012; Fang et al. 2019). Experimental evolution can bridge the gap between systematic deletion analyses and QTL mapping by identifying genetic interactions between mutations that are representative of natural genetic variation including non-loss-of-function mutations and mutations in essential genes.
Genetic interactions are thought to be prevalent among fixed mutations in experimental evolution (Chou et al. 2011; Khan et al. 2011; Buskirk et al. 2017). Previous experiments have identified genetic interactions statistically, most often as pairs of genes in which mutations co-occur more (or less) often than expected by chance across replicate populations (Kvitek and Sherlock 2011; Tenaillon et al. 2012; Good et al. 2017; Fisher et al. 2019). Most interactions identified in this way are negative genetic interactions, where the combined effect of two mutations is less beneficial than expected given the effects of the individual mutations. In contrast, relatively few positive genetic interactions, in which the combined effect of two mutations is more beneficial than expected, have been identified and characterized (Blount et al. 2008; Quan et al. 2012; Buskirk et al. 2017; Fisher et al. 2019). It is possible that beneficial positive genetic interactions are less common or, instead, they may be more difficult to detect by existing methods (Kryazhimskiy et al. 2009, 2014; Khan et al. 2011).
Previously, we identified a positive genetic interaction between two mutations that arose several hundred generations apart during the course of a yeast laboratory evolution experiment (Buskirk et al. 2017). One of the first mutations to arise was a missense mutation in KEL1 (P344T) that had little effect on fitness by itself. Later, a frameshift mutation in HSL7 (A695fs) arose that had a much larger fitness effect on the kel1-P344T background than it would have had on the ancestral background. These two genes, KEL1 and HSL7 were not previously known to interact genetically and this interaction may have been inaccessible to methods that systematically survey pairwise loss-of-function mutations.
Here, we use evolutionary-replay experiments to test the hypothesis that the kel1-P344T mutation “opened the door” to mutations that would otherwise be less favored. By replaying the evolution of this population hundreds of times we allow selection to enrich for mutations that, like the hsl7-A695fs mutation, interact positively with the kel1-P344T mutation. By initiating replays from timepoints before and after the kel1-P344T mutation arose we identify genes that fix preferentially in the kel1-P344T background. We identify both known and previously unknown KEL1 genetic interactors and validate putative genetic interactions by genetic reconstruction. We show that interactions identified in this way are positive, beneficial, and unlikely to be uncovered by systematic analysis of double deletions or knockdowns.
Results
Evolutionary Replay Experiments Reveal the Range and Likelihood of Alternative Outcomes
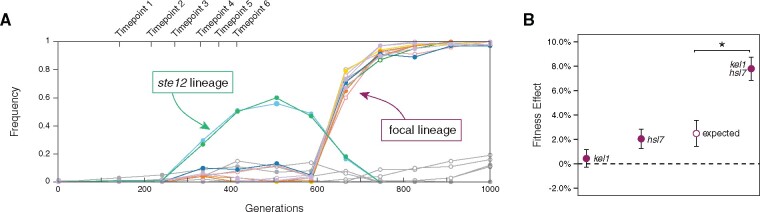
Previously, we identified a strong positive genetic interaction that arose in a 1,000-generation laboratory-evolved yeast population (Lang et al. 2013). In this population (BYS2-E01 from Lang et al. 2011) a low frequency lineage persisted for hundreds of generations and acquired a KEL1 mutation (P344T) by Generation 335 (fig. 1A). We showed that the kel1-P344T mutation is nearly neutral in the ancestral background and the success of this low-frequency lineage (referred to as our focal lineage) was due to a mutation in HSL7 (A695fs) that arose before Generation 600 and had a strong positive genetic interaction with kel1-P344T (fig. 1B) (Buskirk et al. 2017). This positive genetic interaction allowed our focal lineage to outcompete an ascendant ste12 lineage present in the population.
Fig. 1.
Dynamics of adaptation of population BYS2-E01. (A) Frequency of mutations present in the original BYS2-E01 population as determined by whole genome sequencing (Lang et al. 2013). A lineage containing a beneficial ste12-Q151fs mutation was outcompeted when an hsl7-A695fs mutation arose on the low-frequency focal lineage. Replay experiments were initiated at the timepoints indicated above (Generations 140, 210, 270, 335, 375, and 415). The kel1-344T mutation was first detected by whole genome, whole population sequencing at Timepoint 4. (B) Average fitness effects and standard deviations of mutations that contribute to the fitness of the focal lineage (Buskirk et al. 2017). Expected double mutant fitness (open circle) was calculated by summing individual fitness effects and propagating uncertainty. Asterisk (*) indicates P < 0.001 (Welch’s modified t-test).
To investigate the role of the kel1-P344T mutation on the fate of our focal linage, we resurrected the population in which it arose from timepoints before and after the kel1-P344T mutation was detected: Generations 140, 210, and 270 and Generations 335, 375, and 415, respectively (fig. 1A). For each timepoint we restarted 48 replicate replay populations and evolved them for 630 generations under the same conditions as the original experiment, namely daily 1:210 dilutions into 128 µl of rich glucose medium (see Materials and Methods).
Each week (every 70 generations), we followed the fate of our focal lineage in all 288 replay populations using a two-step process: first by screening phenotypically for loss of the ste12 lineage using a fluorescent reporter of Ste12 activity, which is present in this strain background (Lang et al. 2011), then by genotyping for a mutation present only in our focal lineage. Since the ste12-Q151fs and kel1-P344T mutations arose on independent backgrounds, only in populations where the ste12 lineage goes extinct could our focal lineage have fixed. We find that the ste12 lineage “won” (either fixed or was at a frequency above 0.5 at Generation 630) in most populations from both the pre-kel1 (118 of 144) and post-kel1 (127 of 144) timepoints.
For those populations where the ste12 lineage “lost” (either went extinct or was at a frequency below 0.5 at Generation 630), we determined if our focal lineage had won. For this, we took advantage of a SspI restriction site that was introduced by the iqg1-S571N mutation that arose in our focal lineage around Generation 140, prior to the kel1-P344T mutation (note that, although the kel1-P344T mutation is only present in replay experiments initiated at Generation 335 and beyond, the iqg1-S571N mutation identifies our focal lineage in all six replay experiments and has no detectable effect on fitness [Buskirk et al. 2017]) (supplementary fig. S1, Supplementary Material online). In only one of the pre-kel1 replay populations did our focal lineage win. However, in the post-kel1 replay populations, our focal (iqg1-S571N, kel1-P344T) lineage won in 4 of the 17 populations where the ste12 lineage lost (fig. 2 and supplementary table S1, Supplementary Material online).
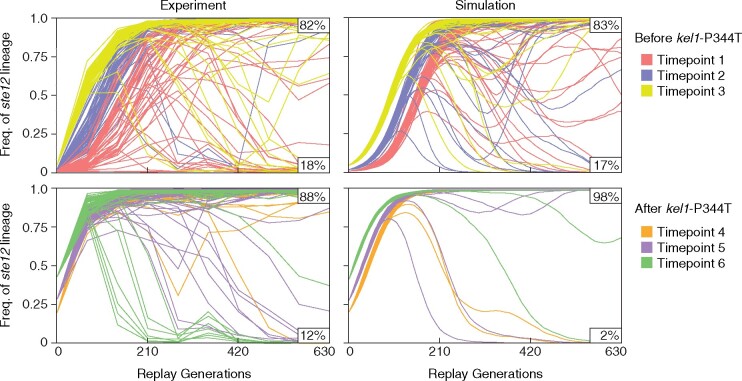
Fig. 2.
Fate of ste12 lineages in the replay populations and simulated populations. Frequency of sterile cells in the replay populations over the course of the replay experiments as measured by flow cytometry (left) and frequency of the ste12 lineage over time in simulated populations (right). Each line represents one population. Percentage of populations where the ste12 lineage wins (top value) or loses (bottom value) are shown for each graph. For the simulations we show the first 48 simulated trajectories as an unbiased representation. Initial frequencies of ste12 lineage in experimental populations were plotted from values reported in Lang et al. (2011).
Chance Alone Is Insufficient to Explain the Observed Lineage Dynamics
We sought to determine whether the appearance of the neutral kel1-P344T mutation in our low-frequency focal lineage affected the likelihood that the dominant ste12 lineage won or lost in the replay populations. Direct comparison between timepoints is not possible because each subsequent set of replay experiments starts with a higher initial frequency of the ste12 lineage (fig. 1A). To address this, we simulated the evolutionary replay experiments using a previously described computational model of our experiment (Frenkel et al. 2014).
For each of the initial frequencies of the ste12 lineage, we ran 10,000 forward evolution simulations using known values for the starting frequency of the ste12 lineage (Lang et al. 2013), the fitness of the ste12 lineage, and empirically derived values for the beneficial mutation rate and the distribution of fitness effects in our experiment (see Materials and Methods). In the simulations we find that the fraction of replay populations where the ste12 lineage wins is greater for each subsequent replay timepoint (supplementary table S1, Supplementary Material online). This is expected since the ste12 lineage starts at a higher frequency in each successive replay timepoint from <0.01 at Generation 140 to 0.42 at Generation 415.
In our experiment, like in the simulations, the ste12 lineage wins the fewest times for replays initiated at the first timepoint when the ste12 lineage starts at a frequency of <0.01. Apart from the first timepoint, however, the ste12 lineage wins the fewest times in the final two replay timepoints (Timepoints 5 and 6) where the ste12 lineage started at its highest frequencies (0.28 and 0.42) and where in the simulations the ste12 lineage won most often. The key difference between the simulation and the experiment is that the simulation assumes that the distribution of fitness effects of new beneficial mutations is fixed. In reality, this distribution will be impacted by the presence of other mutations in the population, such as the low-frequency kel1-P344T mutation that arose around Generation 335. Grouping the first three (pre-kel1) timepoints we find the likelihood of the ste12 lineage winning in the replay experiments does not deviate from the null model given by the simulations (X2 = 0.05, P = 0.82). However, grouping the last three (post-kel1) timepoints we find that the ste12 lineage loses more often than expected by chance (X2 = 63, P < 0.001) (fig. 2 and supplementary table S1, Supplementary Material online).
Our hypothesis for why the ste12 lineage lost more often than expected in replay experiments initiated after Generation 335 is that the presence of the kel1-P344T mutation opens paths to higher-fitness mutations (e.g., the fitness effect of the hsl7-A695fs mutation is greater in the background of the kel1-P344T mutation). It would follow then that the deviations from the null expectation are due to populations where our focal lineage won. However, if we exclude from our analysis those populations where our focal lineage won, we find that the ste12 lineage still loses more often than expected by chance in the later three timepoints (X2 = 34, P < 0.001). Therefore, accounting for the kel1-P344T mutation can explain some, but not all, of the deviations between the simulated and actual replay experiments. This suggests that additional low-frequency lineages in the population influence the dynamics of adaptation.
Recurrent Targets of Selection Are Putative KEL1-Interacting Genes
Genes that acquire mutations more often than expected by chance only in populations where our focal lineage won are candidate genetic interactors with kel1-P344T. Because our focal lineage won in only 4 of the 144 populations in the initial replay experiments, we initiated an additional 384 replay populations from Timepoint 5 (in which our focal lineage won 3 out of 48 times) and evolved them for 350 generations. We used the same two-step method for identifying populations where our focal lineage won: screening for loss of the ste12 lineage followed by PCR/SspI digest to detect the presence of the iqg1-S571N mutation. The ste12 lineage lost in 45 of the 384 second-round replay populations and our focal lineage won in 26 of those 45.
We sequenced one clone from each population in which our focal lineage had won: five populations from the first round of replay experiments and 26 from the second round. Additionally, we sequenced 19 randomly selected control populations that saw either the ste12 lineage or another non-ste12, nonfocal lineage win (supplementary data set 1, Supplementary Material online). We identified 526 de novo mutations that arose during the replay experiments; an average of approximately 13 mutations per clone in the first-round replays and an average of approximately 9 per clone in the second-round replays. Autodiploidization events, a common outcome in our experimental system (Fisher et al. 2018), were detected in 17 out of the 50 sequenced clones (supplementary data set 1, Supplementary Material online).
We classified mutations as putative kel1-P344T interacting mutations if they occurred in genes previously known to interact with KEL1 and/or in genes with multiple unique mutations in independent replay populations. A total of 22 genes fulfill the first criterion, and four genes fulfill the second. One gene, HSL1, fulfills both criteria (supplementary data set 1, Supplementary Material online).
Reconstruction Experiments Validate Positive Genetic Interactions
We reconstructed putative kel1-interacting mutations in both the ancestral and kel1-P344T backgrounds. We included mutations in known KEL1 interacting genes, recurrent targets of selection, and/or essential genes, as well as a gene mutated in both kel1 and non-kel1 lineages as a negative control. Each mutation was assayed for fitness (alone and in combination with the kel1-P344T mutation) against a fluorescently labeled ancestral reference strain (supplementary data set 2, Supplementary Material online).
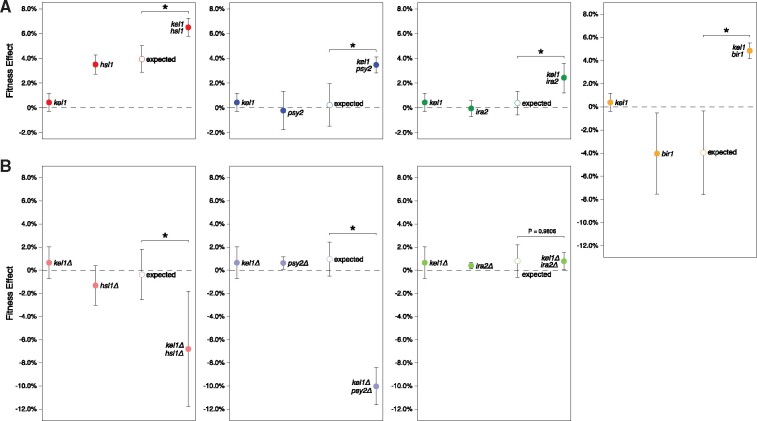
HSL1 is both a known KEL1 interactor (Sharifpoor et al. 2012; Kryazhimskiy et al. 2014; Kuzmin et al. 2018) and a recurrent target of selection, mutated in three independent replay populations. By reconstructing the hsl1-A262P mutation on the KEL1 background we find that it has a 3.50 ± 0.3% (α = 0.05) fitness effect. On the kel1-P344T background, however, it has a 6.53 ± 0.2% (α = 0.05) fitness effect, exceeding the additive expectation (P < 0.001 Welch’s modified t-test, fig. 3A). A mutation (D233fs) in another known KEL1 interactor, PSY2, is also more beneficial in the presence of kel1-P344T than in its absence (P < 0.001 Welch’s). Interestingly, in this second interaction, both the kel1-P344T and pys2-D233fs mutations are neutral in isolation yet produce a 3.48 ± 0.3% (α = 0.05) benefit when combined.
Fig. 3.
Positive genetic interactions between evolved mutations. Average fitness effects and standard deviations of (A) evolved mutations when reconstructed in the ancestral background and (B) gene deletions in the ancestral background. Observed fitness effects were determined by competitive fitness assays against a fluorescently labeled version of the ancestor. Expected double mutant fitness (open circles) was calculated by summing individual fitness effects and propagating uncertainty. Asterisk (*) indicates P < 0.001 (Welch’s modified t-test).
In addition to validating two known KEL1 interactors, we identify two genes that interact genetically with kel1-P344T but were not previously recognized as KEL1 interactors (fig. 3A). A nonsense mutation in IRA2 (R1575*) has a fitness cost of –0.47 ± 0.2% (α = 0.05) in the ancestral background but has a 2.44 ± 0.4% (α = 0.05) fitness advantage when combined with the kel1-P344T mutation (P < 0.001 Welch’s). A BIR1 mutation (P685T) has a substantial fitness cost of −4.51 ± 0.7% (α = 0.05) in the ancestral background but confers a fitness benefit of 4.26 ± 0.3% (α = 0.05) when combined with the kel1-P344T mutation (P < 0.001 Welch’s). Neither the fitness effect of a control mutation in ECM21 (P799fs) nor a mutation in YLR001C (R427fs), a recurrently mutated gene in the replay experiments, are influenced by the kel1-P344T mutation (P = 0.2060 and P = 0.0347, respectively, Welch’s, supplementary fig. S2, Supplementary Material online). Taken together, these results highlight the ability of our experimental system to identify known and previously unknown genetic interactions.
Genetic Interactions Revealed through Experimental Evolution Are Positive and Allele Specific
Experimental evolution samples all types of spontaneous mutations including loss, gain, or attenuation of function. As such, interactions identified in this way may be allele specific. We tested for allele specificity by determining whether the interactions we identified could be phenocopied by gene deletion. For each KEL1 interactor, we generated full-gene deletions in the ancestral background as well as in an isogenic KEL1 gene deletion (kel1Δ) mutant background (fig. 3B). Like the kel1-P344T mutation, the kel1Δ mutation by itself is neutral.
HSL1 and PSY2 have previously been identified as KEL1 genetic interactors using double-deletion mutants in yeast (Costanzo et al. 2016; Kuzmin et al. 2018). We also observe genetic interactions between kel1Δ and hsl1Δ as well as between kel1Δ and psy2Δ (both P < 0.001, Welch’s, fig. 3B). Importantly, however, the direction of the interactions differs between the gene deletions (which decrease fitness) and the evolved mutations (which increase fitness). Therefore, although KEL1/HSL1 and KEL1/PSY2 genetic interactions can be identified by gene deletion, the effects of these interactions on fitness are allele specific.
Although we observe a positive genetic interaction between evolved mutations in IRA2 and KEL1 we do not detect a genetic interaction between kel1Δ and ira2Δ (P = 0.9806, Welch’s, fig. 3B). A bir1Δ mutation was not constructed because it is an essential gene (Li et al. 2000; Widlund et al. 2006). Together, these results demonstrate the power of experimental evolution to identify genetic interactions that are not easily detectable by current methods.
Discussion
Genetic interactions impose constraints on the paths that evolution can take, and thus they determine the extent to which the evolutionary future of a population is dependent on its past. Retrospective analysis of the mutations that arose during laboratory experimental evolution has found evidence for both negative (Chou et al. 2011; Khan et al. 2011; Tenaillon et al. 2012; Kryazhimskiy et al. 2014) and positive genetic interactions (Blount et al. 2008; Quandt et al. 2015; Buskirk et al. 2017; Fisher et al. 2019). Here, we reverse the standard paradigm. Rather than identifying genetic interactions among mutations that arose during experimental evolution, we use experimental evolution as a tool to recover mutations that interact with a specific mutation of interest.
Experimental evolution provides an opportunity to identify genetic interactions that are potentially overlooked by current methods. Unlike suppressor screens, which require a selectable phenotype, continuous selective pressure exerted over hundreds of generations efficiently enriches for genotypes that contain positive genetic interactions whose effects may be subtle. Mutations arise spontaneously and cover all types of mutations including non-loss-of-function mutations and mutations in essential genes, making for a much larger and diverse search space, and one that is likely more reflective of natural genetic variation.
We previously detected a positive genetic interaction between mutations in KEL1 and HSL7 that arose during a laboratory evolution experiment (Lang et al. 2013; Buskirk et al. 2017). Though their protein products both localize to sites of polarized growth, these two genes were not previously known to interact physically or genetically. Two high-throughput screens have identified possible interactions between KEL1 and other cell cycle genes, yet the local genetic interaction network linking the cell polarity gene KEL1 and the cell cycle has not been fully explored (Ho et al. 2002; Ubersax et al. 2003).
Here, we use evolutionary replay experiments to identify other genetic interactors that, like hsl7-A695fs, have a significantly higher fitness effect on the kel1-P344T background. The population used here, BYS2-E01 from Lang et al. (2011), is ideal for this purpose for two reasons: 1) the nearly neutral kel1-P344T mutation arose and persisted at low frequency for several hundred generations prior to the hsl7-A695fs mutation, and 2) the presence of the beneficial ste12-Q151fs mutation and the restriction site created by the iqg1-S571N mutation allows individual lineages of interest to be tracked quickly and easily without sequencing.
We followed the fate of ste12-Q151fs across 48 replicate replay experiments initiated at Generations 140, 210, 270, 335, 375, and 415. All else being equal, the ste12 lineage should win more often in populations initiated at later timepoints since this lineage starts at higher frequencies in each subsequent timepoint. We verified this null result by computational simulations that capture the details of our experiment (fig. 2). We predicted that after Timepoint 4 the evolutionary potential of our focal lineage would increase due to new beneficial mutations made accessible by the kel1-P344T mutation. Therefore, we expected to see a reduction in the fraction of replay populations in which the ste12 lineage won in our experiment relative to the simulations. Comparing our replay data to the simulations, we find that the ste12 lineage loses more often than expected in post-kel1 timepoints of the replay experiments. Though this difference is significant, the magnitude of the effect is not strong. This could indicate that the additional beneficial mutations available to the kel1-P344T-containing cells do not appreciably alter the distribution of fitness effects. Alternatively, the kel1-P344T mutation may “close” as many paths to higher fitness as it “opens,” resulting in a shift in the identity of beneficial mutations but not the distribution of fitness effects.
The deviations between experiment and simulation cannot be due only to the appearance of the kel1-P344T mutation. If we exclude populations where our focal kel1-P344T lineage won from our analysis, we still see a lower-than-expected number of ste12 lineages winning in replays from post-kel1 timepoints. This suggests that either the simulation parameters are not reflective of the experiment at these later time points or that there exists other low-frequency lineages containing beneficial mutations. Existence of other low-frequency lineages could also explain why we also see a lower-than-expected number of ste12 lineages winning in Timepoint 3 populations, despite being a pre-kel1 timepoint (supplementary table S1, Supplementary Material online).
Based on sequencing of clones from the replay experiments we do see several lineages that fixed in independent populations, suggesting that there were non-ste12, nonfocal lineages present in the original BYS2-E01 population (supplementary fig. S3, Supplementary Material online). Furthermore, we expected to find the most kel1-P344T-containing lineages winning in replays initiated from Timepoint 4, right after the kel1-P344T mutation was observed and before the ste12 lineage reached high frequency; however, no kel1-P344T lineages won in replays from Timepoint 4. Therefore, other mutations in the population must also be influencing the dynamics of the replay experiments. Additionally, mutations in the ste12 lineage (ste12-Q151fs or thi3-F404V) could be closing access to certain evolutionary paths. For example, additional mating pathway mutations would be less beneficial in the ste12-Q151fs background relative to the ancestral background.
We sequenced 50 populations from the first and second set of replay experiments and identified putative kel1-P344T genetic interactors as genes mutated multiple times in the kel1-P344T lineages. Only one of the four recurrently mutated genes, HSL1, was previously identified as a KEL1 interactor using methods that assay pairwise gene deletion mutants. Interestingly, a genetic interaction between kel1Δ and hsl1Δ is absent from the Cell Map data set (Costanzo et al. 2016), however, genetic interactions between kel1Δ and hsl1Δ have been reported elsewhere (Sharifpoor et al. 2012; Kuzmin et al. 2018). In addition, HSL1 was identified as a recurrent target of selection in another evolution experiment initiated with a strain containing a KEL1 mutation (Kryazhimskiy et al. 2014).
HSL7, the gene that we found to interact with the kel1-P344T mutation in Lang et al. (2013) was also identified as a recurrently mutated gene in the replay experiments. HSL7 is a known substrate of phosphorylation by HSL1 and association of Hsl1 and Hsl7 at the bud neck is required to advance through Swe1-mediated G2-M delay (Ma et al. 1996; Cid et al. 2001). The nonsynonymous mutations in HSL7 that arose in the original population (A695fs) and the replay experiments (L423*) both result in loss of residues phosphorylated by Hsl1, while maintaining residues important for Hsl1 association (Cid et al. 2001). Two of the three mutations in HSL1 that arose in our replay experiments (Q213K and A262P) fall within the protein kinase domain (residues 85–369), which could affect phosphorylation of HSL7 (Finnigan et al. 2016). This may suggest the positive fitness effects of the kel1-P344T/hsl7-A695fs interaction and kel1-P344T/hsl1-A262P interaction result from similar molecular mechanisms.
Here, we demonstrate the potential of experimental evolution to fish out genetic interactions to a query mutation. As a proof of principle, we use evolutionary replay experiments to identify genetic interactions with a mutant allele of KEL1 that arose in a previous evolution experiment (Lang et al. 2013). Because of the circumstances of population BYS2-E01 we were able to use evolutionary replay experiments to uncover KEL1 interactors. An alternative approach would be to use strains of known provenance either by evolving them independently and comparing the mutations that arise or as a marker displacement experiment (Hegreness et al. 2006).
We show here that experimental evolution effectively pulls out both known and previously unknown genetic interactors, showing that additional complexity exists in genetic interaction networks that is inaccessible to knockdown or gene deletion screens. Even for the known interactors, the effect of interactions between evolved mutations is beneficial, in contrast to genetic interactions between gene deletions which typically reduce fitness (Costanzo et al. 2016), which suggests different mechanisms of action. We show that experimental evolution, though underutilized for this purpose, is capable of identifying growth-promoting and allele specific genetic interactions that are not easily detectable by existing methods.
Materials and Methods
Evolutionary Replay Experiments
Replay experiments were initiated by resurrecting the BYS2-E01 population (described in Lang et al. 2013) at six timepoints corresponding to generations 140, 210, 270, 335, 375, and 415. These specific generations were chosen to encompass a range of timepoints before and after the kel1-P344T mutation was detectable. Our focal lineage was first detected by whole-genome, whole-population sequencing at Generation 140, via the initial mutation in that lineage, iqg1-S571N. For each timepoint, the population was resurrected by thawing the frozen stock and diluting 1:25 in Yeast extract Peptone Dextrose (YPD) broth. After incubation at 30 °C for 24 h, each culture was diluted 1:210 and distributed into 48 wells of a 96-well plate. The populations were then propagated in the same conditions as in the original experiment (Lang et al. 2011). Briefly, each day the populations were diluted 1:210 using a BiomekFX liquid handler into 128 µl of YPD plus 100 µg/ml ampicillin and 25 µg/ml tetracycline to prevent bacterial contamination. The dilution scheme equates to 10 generations of growth per day at an effective population size of approximately 105.
Phenotypic and Genotypic Tracking of Replay Populations
To track the dynamics of the ste12 lineage, we measured the frequency of sterile cells in the replay experiments every 70 generations. The saturated culture was diluted 1:25 into YPD plus α-factor (10 µg/ml) and incubated 6 h at 30 °C as was done previously (Lang et al. 2011). In the mating-competent ancestor, mating pheromone activates the mating pathway and induces expression of a downstream fluorescent reporter, yEVenus (Lang et al. 2009). However, cells that possess a sterile mutation do not respond to mating pheromone and do not fluoresce. Ratios of fluorescent to nonfluorescent cells were determined via flow-cytometry. Flow-cytometry data were analyzed with FlowJo version 10.5.3.
For populations where sterile frequency dropped below 0.5, two clones were isolated by streaking the population to singles on YPD agar. To identify nonsterile clones, cells were incubated with α-factor for 3 h and visually examined for shmoos. Nonsterile clones were then grown in 5 ml YPD and a sample was stocked for further analysis. Genomic DNA was harvested using phenol-chloroform extraction and precipitated in ethanol. To identify clones derived from our focal lineage we relied on a novel SspI cut site (AATATT) created by a G to A transition in IQG1 (iqg1-S571N). The IGQ1 locus was amplified via PCR and incubated with SspI (New England Biolabs) overnight. Digested products were visualized on an agarose gel. Correct genotyping of wildtype IQG1 and mutant iqg1 samples was verified by submitting 10% of samples for Sanger sequencing.
Quantifying Relative Fitness of the ste12 Lineage
The relative fitness of the ste12 lineage was determined using competitive fitness assays on clones isolated from the original BYS2-E01 population. A small sample of the population was struck to single colonies on YPD + α-factor to select for sterile cells. A ste12 lineage clone isolated this way was competed against a fluorescently labeled version of the ancestral strain after verifying the presence of the ste12-Q151fs mutation via Sanger sequencing. The fitness advantage of this lineage was determined to be 3.11 ± 0.2% (α = 0.05, supplementary data set 2, Supplementary Material online).
Computational Simulations
Simulations of ste12 lineage trajectories were performed using a forward-time algorithm designed to match the conditions (including population size, dilution bottleneck, and dilution frequency) in the evolution experiment (Frenkel et al. 2014). MATLAB scripts were downloaded from https://github.com/genya/asexual-lineage-adaptation, last accessed March 2021. Estimates for the distribution of beneficial fitness effects (an exponential distribution with mean = 0.85%) and beneficial mutation rate (Ub = 10−4) were used as described previously (Frenkel et al. 2014).
Simulations were performed with constant inputs for the distribution of fitness effects, beneficial mutation rate, and fitness effect of the ste12-Q151fs mutation (s0 = 3.11%). The initial frequency of the ste12 lineage varied (f0 = 0.0084, 0.0186, 0.0472, 0.2029, 0.2794, 0.4230) for each set of simulations based on estimated initial ste12-Q151fs frequency at each timepoint as previously reported (Lang et al. 2011). The inoculation time of the ste12 lineage in the simulations varied (t = 0.5), as some plates were frozen five generations after sterile frequencies were measured in Lang et al. (2011). For each starting frequency of the ste12 lineage, ten thousand simulations were performed. If the frequency of a simulated ste12 lineage was >0.5 at the end of the simulation, we considered it had “won.”
Whole-Genome Sequencing
Clones were grown to saturation in 5 ml YPD and spun down to pellets and frozen at −20 °C. Genomic DNA was harvested from frozen cell pellets using modified phenol-chloroform extraction and precipitated in ethanol. Total genomic DNA was used in a Nextera library preparation. The protocol used was described previously (Buskirk et al. 2017). Pooled clones were sequenced using an Illumina HiSeq 2500 sequencer by the Sequencing Core Facility at the Lewis-Sigler Institute for Integrative Genomics at Princeton. Average sequencing read depth was approximately 45.
Sequencing Analysis
Raw sequencing data were concatenated and then demultiplexed using a custom python script (barcodesplitter.py) from L. Parsons (Princeton University). Adapter sequences were trimmed using fastx_clipper from the FASTX Toolkit. Trimmed reads were aligned to a W303 reference genome (Matheson et al. 2017) using BWA v0.7.12 (Li and Durbin 2009) and mutations were called using FreeBayes v0.9.21-24-381 g840b412 (Garrison and Marth 2012). All calls were confirmed manually by viewing BAM files in IGV (Thorvaldsdóttir et al. 2013). Clones were predicted to be autodiploids if two or more mutations were called at an allele frequency of 0.5 and then manually confirmed by viewing BAM files in IGV.
Strain Construction
We selected several putative KEL1-interacting mutations to reconstruct. Recurrently mutated genes unique to our focal lineage (HSL1, IRA2, and YLR001C) were chosen. The other mutations chosen were done so based on several criteria including ease of construction (specifically, if the mutation fell within a Cas9 gRNA target sequence to avoid continual cutting by constitutively active Cas9), if there were no other candidate mutations present in that clone, and if the mutation was nonsynonymous. We also attempted to reconstruct mutations in LTE1 (S222*), BOI1 (G781D), and CLB2 (V172fs) but were unsuccessful.
Evolved mutations were introduced into the ancestral background via CRISPR/Cas9 allele swaps as described previously (Fisher et al. 2019). Briefly, evolved mutations were reconstructed by amplifying 500 bp fragments centered around the mutation of interest. These linear PCR products were transformed into the ancestor, yGIL432 (MATa, ade2-1, CAN1, his3-11, leu2-3,112, trp1-1, URA3, bar1Δ::ADE2, hmlαΔ::LEU2, GPA1::NatMX, ura3Δ::pFUS1-yEVenus), along with a plasmid encoding Cas9 and gRNAs targeting near the mutation site (Addgene #67638) (Laughery et al. 2015). Successful genetic reconstructions were confirmed via Sanger sequencing.
Deletion mutants were generated by integrating KanMX markers, amplified from the yeast deletion collection, into the targeted loci of yGIL432. Double deletion mutants were generated by making the KanMX deletion of KEL1 in the MATα version of the ancestor and crossing it to the corresponding MATa deletion mutants, sporulating, and selecting MATa spores with both deletions. Integration of the KanMX cassettes at the correct loci was confirmed via Sanger sequencing.
Competitive Fitness Assays
We measured the effect of evolved mutations on fitness using competitive fitness assays described previously (Buskirk et al. 2017; Fisher et al. 2018). Briefly, query strains are mixed 1:1 with a fluorescently labeled version of the ancestral strain. Cocultures are propagated in 96-well plates in the same conditions in which they evolved for 50 generations. Saturated cultures were sampled for flow-cytometry every 10 generations. Each genotype assayed was done so with at least 24 technical replicates of 2–3 biological replicates, except kel1Δ/hsl1Δ, kel1Δ/psy2Δ, and kel1Δ/ira2Δ which were assayed using at least 24 technical replicates of a single biological replicate each.
Quantifying Genetic Interactions
Additive expectation of double mutant fitness effects was calculated by summing individual fitness effects of mutations and propagating uncertainty using the following formula: , where and are the uncertainty (standard deviation) of measured fitness effects and . If the measured fitness effect of a double mutant differed significantly from the expected value (using Welch’s modified t-test, cutoff of P < 0.01) we considered the two mutations to interact genetically.
Phylogenetic Analysis
The evolutionary history of our 50 sequenced clones was inferred using the Maximum Likelihood method and Tamura–Nei model (Tamura and Nei 1993). The tree with the highest log likelihood (–4,514.24) was used. The tree is drawn to scale with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA X (Kumar et al. 2018). Note there is a thi3-F404V mutation present in the ste12 lineage not previously identified in Lang et al. (2013).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank members of the Lang Lab for comments on the manuscript. This study was supported by the National Institutes of Health (R01GM127420 to G.I.L.).
Data Availability
The short-read sequencing data reported in this article have been deposited at the NCBI BioProject database (accession number: PRJNA693025).
References
- Babu M, Arnold R, Bundalovic-Torma C, Gagarinova A, Wong KS, Kumar A, Stewart G, Samanfar B, Aoki H, Wagih O, et al. 2014. Quantitative genome-wide genetic interaction screens reveal global epistatic relationships of protein complexes in Escherichia coli. PLOS Genet. 10(2):e1004120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryshnikova A, Costanzo M, Kim Y, Ding H, Koh J, Toufighi K, Youn J-Y, Ou J, San Luis B-J, Bandyopadhyay S, et al. 2010. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Methods. 7(12):1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, et al. 2013. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 152(4):909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström A, Simpson JT, Salinas F, Barré B, Parts L, Zia A, Nguyen Ba AN, Moses AM, Louis EJ, Mustonen V, et al. 2014. A high-definition view of functional genetic variation from natural yeast genomes. Mol Biol Evol. 31(4):872–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD, Borland CZ, Lenski RE.. 2008. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci USA. 105(23):7899–7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen MS, Kemena C, Vlasov PK, Notredame C, Kondrashov FA.. 2012. Epistasis as the primary factor in molecular evolution. Nature 490(7421):535–538. [DOI] [PubMed] [Google Scholar]
- Buskirk SW, Peace RE, Lang GI.. 2017. Hitchhiking and epistasis give rise to cohort dynamics in adapting populations. Proc Natl Acad Sci USA. 114(31):8330–8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H-H, Chiu H-C, Delaney NF, Segrè D, Marx CJ.. 2011. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332(6034):1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid VJ, Shulewitz MJ, McDonald KL, Thorner J.. 2001. Dynamic localization of the Swe1 regulator Hsl7 during the Saccharomyces cerevisiae cell cycle. Mol Biol Cell. 12(6):1645–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, Toufighi K, Mostafavi S, et al. 2010. The genetic landscape of a cell. Science 327(5964):425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, et al. 2016. A global genetic interaction network maps a wiring diagram of cellular function. Science 353(6306):aaf1420–aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Wang W, Paunic V, Heydari H, Costanzo M, Liu X, Liu X, VanderSluis B, Oately B, Steinbach M, et al. 2019. Discovering genetic interactions bridging pathways in genome-wide association studies. Nat Commun. 10(1):4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnigan GC, Sterling SM, Duvalyan A, Liao EN, Sargsyan A, Garcia G, Nogales E, Thorner J.. 2016. Coordinate action of distinct sequence elements localizes checkpoint kinase Hsl1 to the septin collar at the bud neck in Saccharomyces cerevisiae. Mol Biol Cell. 27(14):2213–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher KJ, Buskirk SW, Vignogna RC, Marad DA, Lang GI.. 2018. Adaptive genome duplication affects patterns of molecular evolution in Saccharomyces cerevisiae. PLoS Genet. 14(5):e1007396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher KJ, Kryazhimskiy S, Lang GI.. 2019. Detecting genetic interactions using parallel evolution in experimental populations. Philos Trans R Soc Lond B Biol Sci. 374(1777):20180237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel EM, Good BH, Desai MM.. 2014. The fates of mutant lineages and the distribution of fitness effects of beneficial mutations in laboratory budding yeast populations. Genetics 196(4):1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 [q-bio] [Internet]. Available from: http://arxiv.org/abs/1207.3907.
- Good BH, McDonald MJ, Barrick JE, Lenski RE, Desai MM.. 2017. The dynamics of molecular evolution over 60,000 generations. Nature 551(7678):45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Jeng EE, Hess GT, Morgens DW, Li A, Bassik MC.. 2017. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol. 35(5):463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegreness M, Shoresh N, Hartl D, Kishony R.. 2006. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 311(5767):1615–1617. [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams S-L, Millar A, Taylor P, Bennett K, Boutilier K, et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415(6868):180–183. [DOI] [PubMed] [Google Scholar]
- Horton BN, Kumar A.. 2015. Genome-wide synthetic genetic screening by transposon mutagenesis in Candida albicans. Methods Mol Biol. 1279:125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Barmada MM, Visweswaran S.. 2010. Identifying genetic interactions in genome-wide data using Bayesian networks. Genet Epidemiol. 34(6):575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AI, Dinh DM, Schneider D, Lenski RE, Cooper TF.. 2011. Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332(6034):1193–1196. [DOI] [PubMed] [Google Scholar]
- Kryazhimskiy S, Rice DP, Jerison ER, Desai MM.. 2014. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 344(6191):1519–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S, Tkačik G, Plotkin JB.. 2009. The dynamics of adaptation on correlated fitness landscapes. Proc Natl Acad Sci USA 106(44):18638–18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin E, VanderSluis B, Wang W, Tan G, Deshpande R, Chen Y, Usaj M, Balint A, Usaj MM, Leeuwen JV, et al. 2018. Systematic analysis of complex genetic interactions. Science 360(6386):eaao1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitek DJ, Sherlock G.. 2011. Reciprocal sign epistasis between frequently experimentally evolved adaptive mutations causes a rugged fitness landscape. PLOS Genet. 7(4):e1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Botstein D, Desai MM.. 2011. Genetic variation and the fate of beneficial mutations in asexual populations. Genetics 188(3):647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Murray AW, Botstein D.. 2009. The cost of gene expression underlies a fitness trade-off in yeast. Proc Natl Acad Sci USA. 106(14):5755–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, Botstein D, Desai MM.. 2013. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500(7464):571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughery MF, Hunter T, Brown A, Hoopes J, Ostbye T, Shumaker T, Wyrick JJ.. 2015. New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae. Yeast 32(12):711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Flanary PL, Altieri DC, Dohlman HG.. 2000. Cell division regulation by BIR1, a member of the inhibitor of apoptosis family in yeast. J Biol Chem. 275(10):6707–6711. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XJ, Lu Q, Grunstein M.. 1996. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 10(11):1327–1340. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. 2009. Finding the missing heritability of complex diseases. Nature 461(7265):747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Donnelly P, Cardon LR.. 2005. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 37(4):413–417. [DOI] [PubMed] [Google Scholar]
- Matheson K, Parsons L, Gammie A.. 2017. Whole-genome sequence and variant analysis of W303, a widely-used strain of Saccharomyces cerevisiae. G3. G3 (Bethesda)) 7(7):2219–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan S, Ray JCJ, Kwota Z, Duong T, Balázsi G, Cooper TF, Monds RD.. 2012. Adaptive evolution of the lactose utilization network in experimentally evolved populations of Escherichia coli. PLOS Genet. 8(1):e1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt EM, Gollihar J, Blount ZD, Ellington AD, Georgiou G, Barrick JE.. 2015. Fine-tuning citrate synthase flux potentiates and refines metabolic innovation in the Lenski evolution experiment. eLife 4:e09696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleheen D, Natarajan P, Armean IM, Zhao W, Rasheed A, Khetarpal SA, Won H-H, Karczewski KJ, O’Donnell-Luria AH, Samocha KE, et al. 2017. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature 544(7649):235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifpoor S, van Dyk D, Costanzo M, Baryshnikova A, Friesen H, Douglas AC, Youn J-Y, VanderSluis B, Myers CL, Papp B, et al. 2012. Functional wiring of the yeast kinome revealed by global analysis of genetic network motifs. Genome Res. 22(4):791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, Licon K, Klepper K, Pekin D, Beckett AN, et al. 2017. Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nat Methods. 14(6):573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M.. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10(3):512–526. [DOI] [PubMed] [Google Scholar]
- Tenaillon O, Rodríguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS.. 2012. The molecular diversity of adaptive convergence. Science 335(6067):457–461. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP.. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14(2):178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AHY, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. 2004. Global mapping of the yeast genetic interaction network. Science 303(5659):808–813. [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO.. 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425(6960):859–864. [DOI] [PubMed] [Google Scholar]
- Widlund PO, Lyssand JS, Anderson S, Niessen S, Yates JR, Davis TN.. 2006. Phosphorylation of the chromosomal passenger protein Bir1 is required for localization of Ndc10 to the spindle during anaphase and full spindle elongation. Mol Biol Cell. 17(3):1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES.. 2012. The mystery of missing heritability: genetic interactions create phantom heritability. Proc Natl Acad Sci USA. 109(4):1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The short-read sequencing data reported in this article have been deposited at the NCBI BioProject database (accession number: PRJNA693025).