Abstract
The relationships among the four major embryophyte lineages (mosses, liverworts, hornworts, vascular plants) and the timing of the origin of land plants are enigmatic problems in plant evolution. Here, we resolve the monophyly of bryophytes by improving taxon sampling of hornworts and eliminating the effect of synonymous substitutions. We then estimate the divergence time of crown embryophytes based on three fossil calibration strategies, and reveal that maximum calibration constraints have a major effect on estimating the time of origin of land plants. Moreover, comparison of priors and posteriors provides a guide for evaluating the optimal calibration strategy. By considering the reliability of fossil calibrations and the influences of molecular data, we estimate that land plants originated in the Precambrian (980–682 Ma), much older than widely recognized. Our study highlights the important contribution of molecular data when faced with contentious fossil evidence, and that fossil calibrations used in estimating the timescale of plant evolution require critical scrutiny.
Keywords: land plants, bryophytes, phylogeny, timescale, maximum bounds
Introduction
Streptophyta comprises all land plants (embryophytes) and six monophyletic groups of streptophyte green algae, and they have significantly contributed to global environmental change in geological history (Kenrick et al. 2012; Lenton et al. 2012, 2016). The colonization of the terrestrial realms by early land plants is one of most important events in the evolution of life on earth, causing soil formation, increasing primary productivity, impacting weathering and global climates, and establishing new habitats for animals that increased their diversity (Heckman et al. 2001; Parnell and Foster 2012). The origin and evolution of the various groups of land plants largely initiated our modern terrestrial ecosystems.
It is widely accepted that land plants evolved from streptophyte green algae which adapted to freshwater conditions early in their history (Becker and Marin 2009). During the invasion of plants onto land, they had to overcome enhanced ultraviolet (UV) radiation, water deficit, salinity, and other environmental stresses (Fang et al. 2017). Deciding which plant innovations were fundamental in the transformation from freshwater algae to land-dwelling forms has been long-debated (Ligrone et al. 2012; Hori et al. 2014; Buschmann and Zachgo 2016; Chater et al. 2017; Jill 2017; Reski 2018; Szovenyi et al. 2019). However, studies of trait evolution are hindered by the lack of resolution of early-diverging land plant phylogeny (Niklas and Kutschera 2010; Rensing 2018). Support for Zygnematophyceae as sister to land plants is strengthened by recent phylogenomic analyses (Zhong et al. 2013, 2015; Wickett et al. 2014; Cheng et al. 2019; One Thousand Plant Transcriptomes Initiative 2019; Jiao et al. 2020), but the relationships among the four major groups of land plants—mosses, liverworts, hornworts, vascular plants—are still unsettled (Cox 2018; Puttick et al. 2018; de Sousa et al. 2019; One Thousand Plant Transcriptomes Initiative 2019; Bell et al. 2020).
Bryophytes—that is, liverworts, mosses, and hornworts—are the second-most diverse group of land plants (Laenen et al. 2014; Tomescu et al. 2018). They play key roles in tracing the evolution of important characters associated with the terrestrialization process. However, the relationships of bryophytes (hornworts, liverworts, and mosses) and tracheophytes have long been controversial, hindering our ability to understand early land plant evolution. Among three bryophyte lineages, recent molecular studies have reached a consensus supporting mosses and liverworts as forming a natural group (Wickett et al. 2014; Cox 2018; Puttick et al. 2018; de Sousa et al. 2019; One Thousand Plant Transcriptomes Initiative 2019). Morphological similarities between the moss and liverwort locomotory apparatus (e.g., centrioles, flagella, unique microtubule, and lamellar arrays) also have supported the same conclusion (Renzaglia et al. 2018). In previous studies, hornworts were placed as sister to either the clade of ((mosses+liverworts)+tracheophytes) (Renzaglia et al. 2000; Wickett et al. 2014) or tracheophytes (Qiu et al. 2006; Puttick et al. 2018), or the clade of mosses and liverworts (Puttick et al. 2018; de Sousa et al. 2019; Zhang et al. 2020). The position of hornworts is the only uncertainty and the key point for resolving early land plant phylogeny. Puttick et al. (2018) and de Sousa et al. (2019) revisited the phylogenetic relationships among the four major lineages of land plants by considering compositional heterogeneity and substitutional saturation using the large-scale data set from Wickett et al. (2014). Although both studies recovered the monophyly of bryophytes, Puttick et al. (2018) could not reject two hypotheses where bryophytes were not monophyletic: liverworts-mosses or liverworts as sister group to the remaining land plants. Similarly, de Sousa et al. (2019) increased branch support for a monophyletic bryophyte group, but this hypothesis was supported using a small number of genes and taxa. Thus, a well-supported phylogenetic relationship among bryophytes (hornworts, liverworts, and mosses) and tracheophytes is crucial to better understand the evolutionary novelties for plant colonization of land.
Establishing the timescale of Streptophyta is essential for testing the hypotheses of codiversification between plants and animals. Although the fossil record offers windows into the history of plants, it is well known to be incomplete and insufficient to provide a coherent picture of land plant history. Inferring ancient evolutionary timescales has often suffered from limited taxon sampling (Foster et al. 2017), biases in methodologies (dos Reis et al. 2015), and especially the rarity of unambiguous fossils (Massoni et al. 2015; dos Reis et al. 2016). Acritarchs are taxonomically ambiguous microfossils from the Palaeoproterozoic to Early Ordovician. Many have been interpreted as phytoplankton, but their exact biological affinities are difficult to confirm for establishing a well-resolved timetable of early-diverging streptophyte algae (Moczydłowska et al. 2011). Cryptospores are critical for understanding the nature of the earliest land floras. Nevertheless, the fossil record of cryptospores is scattered and incomplete, and their affinities are contentious (Edwards et al. 2014). The lack of unambiguous bryophyte fossils has been a problem for calibrating molecular clock studies of early land plants (Tomescu et al. 2018). The poor fossil records of these key lineages make it difficult to provide reliable fossil constraints for divergence time estimation.
There have been a handful of molecular studies performed to estimate divergence times for early land plants (Heckman et al. 2001; Clarke et al. 2011; Magallon et al. 2013; Morris et al. 2018). Clarke et al. (2011) tested the impact of changing the maximum constraints on the three basal nodes (liverwort, moss, and hornwort) of early land plants. When the maximum constraints were changed from 1,042 to 509 Ma, the mean age estimate for the origin of embryophytes was younger and differed by 165 My. Compared with previous studies, Morris et al. (2018) estimated a much younger origin time of land plants (Middle Cambrian—Early Ordovician: 515–470 Ma), which was limited by a narrow temporal calibration (516–469 Ma). Hedges et al. (2018) questioned the validity of the maximum constraints of some nodes in Morris et al. (2018) because calibrations based on fossil absence are less reliable. They re-estimated the divergence times after removing some maximum bounds, and inferred an older origin time for embryophytes (793–560 Ma) than that obtained in Morris et al. (2018). In agreement with the findings from Battistuzzi et al. (2015), the maximum constraints of calibrated nodes have a pervasive and significant impact upon molecular clock estimates. Considering the rare reliable fossils and the difficulties of determining appropriate interpretations of the fossil record, there are great challenges for establishing the timescale of land plants.
In this study, we inferred the phylogeny and divergence times of all major lineages of Streptophyta, especially focusing on relationships among the three bryophyte groups and the time of origin of land plants. We increased taxon sampling among bryophytes (especially hornworts) and explored the effect of analytical errors, stemming from sequence biases, among the 1,440 genes in the data set. Species tree inferences under the multispecies coalescent model and concatenation approach supported the monophyly of bryophytes. To test the impact of fossil calibrations, we implemented three different maximum bounds in divergence time estimation using 100 clock-like genes and 22 fossil calibrations. Our new well-resolved phylogeny and timescale of land plants provide the foundation for accurate interpretation of the development of plant traits during the water-to-land transition.
Results and Discussion
The Well-Supported Monophyletic Bryophytes
Transcriptomes provide a large amount of nuclear data for phylogenomic reconstruction of plants, whereas there exists a serious deficiency of available molecular data from hornworts. Only one study that used the transcriptomes of 1,124 species (One Thousand Plant Transcriptomes Initiative 2019) made efforts in increasing taxon sampling of hornworts so far. We expanded taxon sampling within bryophytes (72 species, including nine additional hornwort species) compared with Puttick et al. (2018) and de Sousa et al. (2019). Our molecular data included 1,440 nuclear genes from 120 streptophyte species with three Chlorophyta as outgroups. The multispecies coalescent model and concatenation approach were both used for reconstructing species trees.
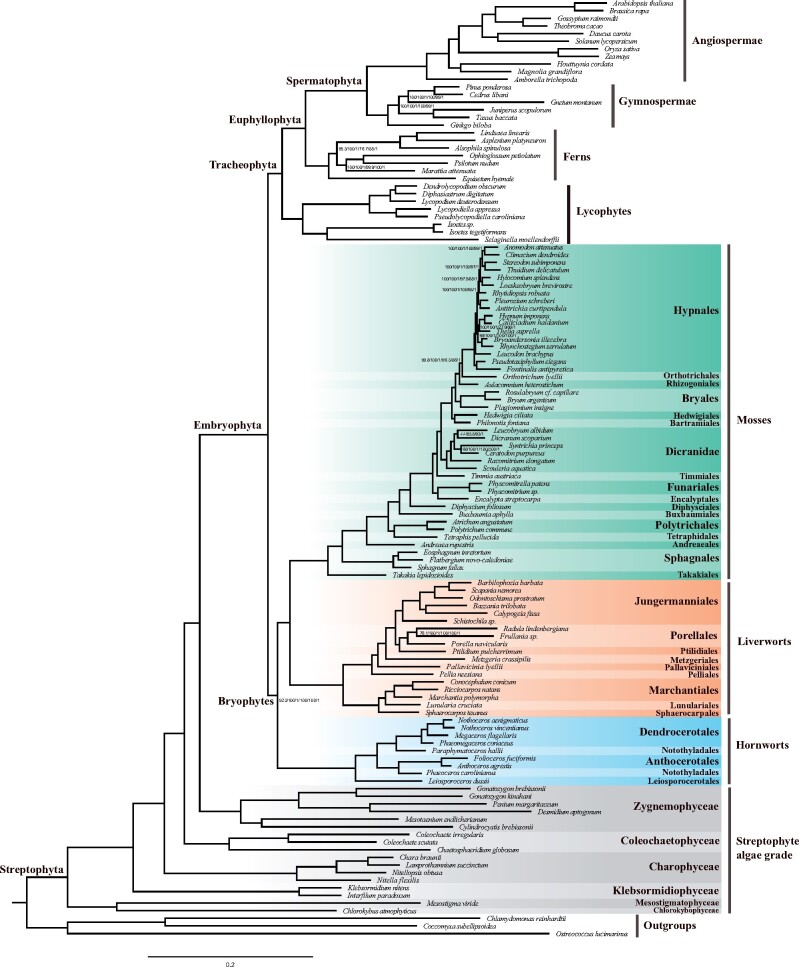
The concatenation analyses of nucleotide data using maximum likelihood and Bayesian methods supported hornworts as sister to all other land plants (supplementary figs. S1 and S2, Supplementary Material online). It has been widely reported that substitutional saturation can affect branch support and induce a phylogenetic artifact (Jeffroy et al. 2006; Liu et al. 2014). Recent molecular studies have indicated that fast-evolving synonymous substitutions may lead to the nonmonophyly of bryophytes using nucleotide data (e.g., Cox et al. 2014; Li et al. 2014; Liu et al. 2014; de Sousa et al. 2019). To mitigate this error stemming from fast-evolving synonymous substitutions, we reanalyzed our nucleotide data in which synonymous nucleotides at codon sites were recoded using nucleotide ambiguity codes and synonymous substitutions were eliminated. The three bryophyte lineages were recovered as a monophyletic group with strong support using codon-degenerated data (fig. 1 and supplementary figs. S3 and S4, Supplementary Material online). In addition, maximum likelihood and Bayesian analyses based on amino acid data also strongly recovered the monophyly of bryophytes and achieved well-supported resolution of relationships among all major lineages of bryophytes (fig. 1 and supplementary figs. S5 and S6, Supplementary Material online). These results indicated that reducing fast-evolving synonymous substitutions by codon-degeneracy recovered monophyly of bryophytes in agreement with the results using an amino acid matrix. To assess the effect of compositional heterogeneity, maximum likelihood analyses were employed on the concatenated amino acid data using a site-heterogeneous mixture model (LG+C20+F+R5) and Dayhoff recoding strategy, respectively. These analyses resulted in bryophyte monophyly with high support (supplementary figs. S7 and S8, Supplementary Material online), indicating that the compositional heterogeneity of 1,440 concatenated genes does not impose a significant bias in reconstructing the relationships among major lineages of land plants.
Fig. 1.
The concatenation species tree of land plants and their algal relatives. The cladogram is reconstructed using an amino acid supermatrix of 1,440 genes by IQ-TREE. Nodal support values are estimated by SH-aLRT test (SH) and ultrafast bootstrap (UFBS) in IQ-TREE, and Bayesian posterior probabilities (BPP) in ExaBayes. The first three are SH, UFBS, and BPP values based on codon-degenerate nucleotide data, and the last three are SH, UFBS, and BPP values based on amino acid data. The nodes without values indicate full support.
Considering the substitutional saturation in nucleotide data, only the amino acid data were used for coalescent analysis. The coalescent-based species tree resolved bryophytes as a robust monophyletic group (fig. 2 and supplementary figs. S9 and S10, Supplementary Material online). Mosses and liverworts constitute a clade with full support, which is consistent with previous analyses based on chloroplast genes (Nishiyama et al. 2004; Goremykin and Hellwig 2005; Karol et al. 2010; Ruhfel et al. 2014) and nuclear genes (Wickett et al. 2014; Puttick et al. 2018; de Sousa et al. 2019). The hornwort clade is identified as the sister-group to the clade of mosses plus liverworts (fig. 2). In mosses, the relationships among the 17 orders are similar to those obtained by Liu et al. (2019). In addition, our phylogenies provided strong support for a sister relationship between Zygnematophyceae and land plants that was strengthened by recent studies (Zhong et al. 2013, 2014; Wickett et al. 2014; Puttick et al. 2018; Cheng et al. 2019; Jiao et al. 2020).
Fig. 2.
The coalescent species tree of land plants and their algal relatives. The cladogram is reconstructed using 1,440 genes by ASTRAL. Nodal support values are estimated by local posterior probability and multilocus bootstrapping (gene+site resampling) (PP/MLBS), and the nodes without values indicate full support.
New Timescale for Early Land Plant Evolution
The Bayesian methods of clock dating can incorporate complex parameters (e.g., birth rate, death rate, sample fraction, and rate drift) to establish various relaxed clock models for describing the uncertainty in the fossil record and variation in evolutionary rate (Ho 2014). Many studies have found that fossil calibrations are the most important variables in molecular clock dating (Magallon et al. 2013; Barba-Montoya et al. 2018; Nie et al. 2020). Fossil evidence is the most common type of information for calibration, converting molecular sequence change into estimates of absolute times and rates (Barba-Montoya et al. 2017). We applied fossil calibrations following recommendations (Parham et al. 2012), emphasizing fewer but more reliable calibration points. However, well-dated fossils only provide reliable minimum constraints for divergence times, and a simple hard minimum bound is insufficient information for establishing a robust timescale.
Two approaches are typically used for maximum constraints. One is to assign a parametric distribution on the age of the fossil (minimum) calibration, such as the gamma, lognormal, or the truncated Gaussian distribution, whereas the shapes of these prior distributions are often established without basic biological justification (Chazot et al. 2019). The second approach assumes that a clade has not yet evolved in a geologic period if there is an absence of available fossils in that period. In this case, the upper bound of the period is the maximum constraint for the clade. It is challenging to prove that the absence of a taxon is true rather than due to the incompleteness of the fossil and rock records (Marshall 2019). Despite these two approaches, researchers have not reached a consensus on how to establish a suitable maximum age for a lineage divergence (Donoghue and Yang 2016). Because the choice of a maximum constraint for crown embryophytes has been controversial (e.g., Clarke et al. 2011: 1,042 Ma; Morris et al. 2018: 515.5 Ma), we focused on exploring the influence of different maximum bounds for the origin of land plants.
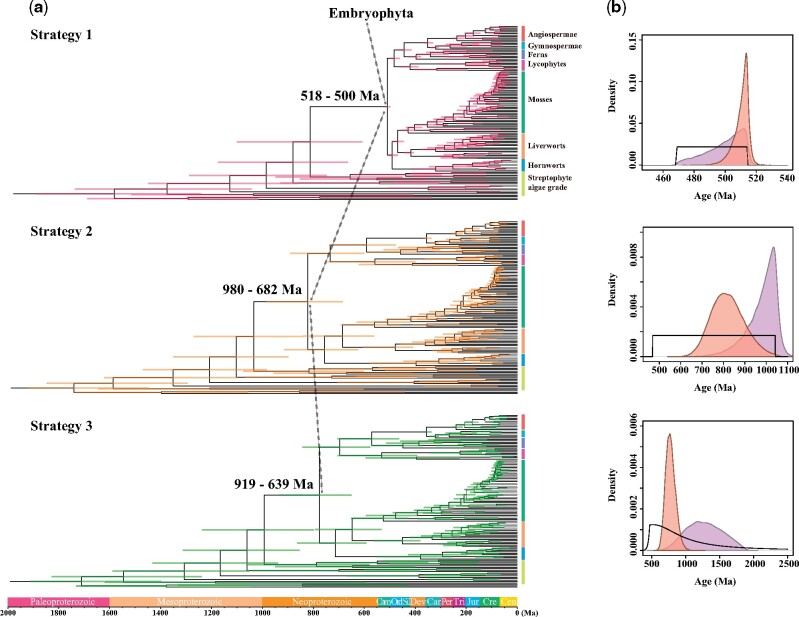
The extensive sampling of early embryophyte lineages in our study provides substantial transcriptomic data to estimate the divergence times for early land plants. Molecular clock analyses were performed using a Bayesian relaxed clock method (MCMCTree) (Yang 2007) based on 100 clock-like genes and the coalescent-based species tree (fig. 2). We employed three fossil calibration strategies to accommodate different maximum bounds of crown embryophytes and the internal calibration nodes among monophyletic bryophytes. In the first calibration strategy (hereinafter referred to as “Strategy 1”), the soft maximum constraints (515.5 Ma) were based on the first appearance of cryptospores following Morris et al. (2018). For the second calibration strategy (hereinafter referred to as “Strategy 2”), we applied a more conservative maximum bound (1,042 Ma) used by Clarke et al. (2011). In the third calibration strategy (hereinafter referred to as “Strategy 3”), we specified a truncated Cauchy distribution on the nodes.
Our results indicate that the divergence times of early embryophyte lineages are highly sensitive to the maximum limits of calibration nodes (fig. 3a and table 1). Strategy 1 estimated that crown embryophytes originated at 518–500 Ma (Cambrian) (fig. 3a and supplementary fig. S11, Supplementary Material online), which is consistent with the estimated time obtained from Morris et al. (2018) that used a similar maximum calibration. However, Strategies 2 and 3 produced much older estimates, inferring a Neoproterozoic origin of embryophytes (Strategy 2: 980–682 Ma; Strategy 3: 919–639 Ma) (figs. 3a and 4 and supplementary fig. S12, Supplementary Material online).
Fig. 3.
Comparison of results from three different strategies. (a) Comparison of divergence time estimates from three fossil strategies. Node ages are plotted at the posterior means, with horizontal bars representing 95% credibility intervals. (b) Comparison of the age distributions on the crown node of land plants for the specified priors (black line), effective priors (purple), and posteriors (orange red).
Table 1.
The 95% HPD Age Estimates of Major Lineages Using Three Strategies.
| Clade | Strategy 1 (Ma) | Strategy 2 (Ma) | Strategy 3 (Ma) |
|---|---|---|---|
| Embryophyta | 518–500 | 980–682 | 919–639 |
| Bryophytes | 507–472 | 902–614 | 855–584 |
| Mosses | 460–397 | 683–450 | 643–419 |
| Liverworts | 439–405 | 580–405 | 550–373 |
| Hornworts | 418–225 | 569–264 | 538–246 |
| Marchantiopsida | 303–227 | 311–227 | 295–191 |
| Jungermanniopsida | 386–241 | 454–280 | 418–251 |
| Tracheophyta | 503–462 | 880–593 | 830–566 |
| Lycopodiophyta | 459–386 | 722–388 | 688–382 |
| Euphyllophyta | 473–416 | 726–467 | 693–455 |
| Monilophyta | 425–376 | 568–372 | 557–361 |
| Spermatophyta | 368–332 | 369–338 | 369–338 |
| Gymnospermae | 338–308 | 339–308 | 340–308 |
| Angiospermae | 256–209 | 258–217 | 257–216 |
We further compared the user-specified priors, effective priors, and posteriors (supplementary figs. S13–S15, Supplementary Material online). The user-specified prior (also called “user prior”) is the temporal fossil calibration prior on individual node, and it is truncated in the construction of the effective prior (also called “marginal prior” or “joint prior”) on times by ensuring that ancestral nodes are older than descendant nodes (dos Reis et al. 2015; Warnock et al. 2015; Brown and Smith 2018). Effective priors are dependent on the interaction among user priors, topology, and the birth–death process that can specify the distribution of the ages of the noncalibration nodes. Although the effective and user-specified priors should be the same ideally, the effective priors are usually different from the user-specified calibration densities due to the effects of truncation (Barba-Montoya et al. 2017), and sometimes it implies the interaction of “pseudodata” in the user priors (Brown and Smith 2018). Posterior distributions are generated when using the data in divergence time estimation. A noticeable discordance between effective priors and posteriors indicates that posterior time estimates are not simply dependent on priors.
In calibration Strategy 1, there is a nearly half-overlap of the effective prior and posterior distribution, especially in their estimated peak values (fig. 3b). We did not expect such overfitting of joint priors and posteriors in view of uncertainties in the fossil record. Instead, the ideal situation is one in which the posteriors are different from the effective priors. Our results indicate that Strategy 1 is being largely constrained by the fossil calibrations instead of supported by the molecular data. If there are potential problems with the calibration information of Strategy 1, these comparisons imply that the sequence data lack sufficient information to overrule the narrow temporal range (515.5–469 Ma). Similar to Morris et al. (2018), we employed the maximum age of the oldest-possible nonmarine palynomorphs as the maximum bound for constraining the crown embryophytes in Strategy 1. Cryptospores are nonmarine sporomorphs, and there is little knowledge about their producers and affinity (Edwards et al. 2014). Yin et al. (2013) presented new cryptospore-like microfossils during the Cambrian period, which may have originated from land plants or primitive plant sporoderm types. These microfossils suggest that land plants possibly occurred earlier than the Cambrian. Using this age as maximum bound of crown embryophytes may mistakenly estimate a younger origin time if they actually belong to the crown group rather than the stem lineage of land plants.
In contrast, there is a considerable distinction between effective prior and posterior densities in Strategies 2 and 3 (fig. 3b), indicating that the molecular data contributed the information relevant to the age of embryophytes and the age estimates were not entirely driven by priors. The difference between the effective prior and user-specified calibration density in Strategy 3 implied the interaction of “pseudodata” (fig. 2b). The truncated Cauchy distributions (Strategy 3) employed at nodes 4–12 might be unwarranted. Although the Cauchy distribution attempts to assume a genuine condition rather than a diffuse uniform fossil calibration prior, it greatly constrained the age bounds. Compared with Strategies 1 and 3, employing broad uniform priors (i.e., Strategy 2) is a more fruitful approach for relaxing the excessive influence of fossil calibration distributions (Brown and Smith 2018).
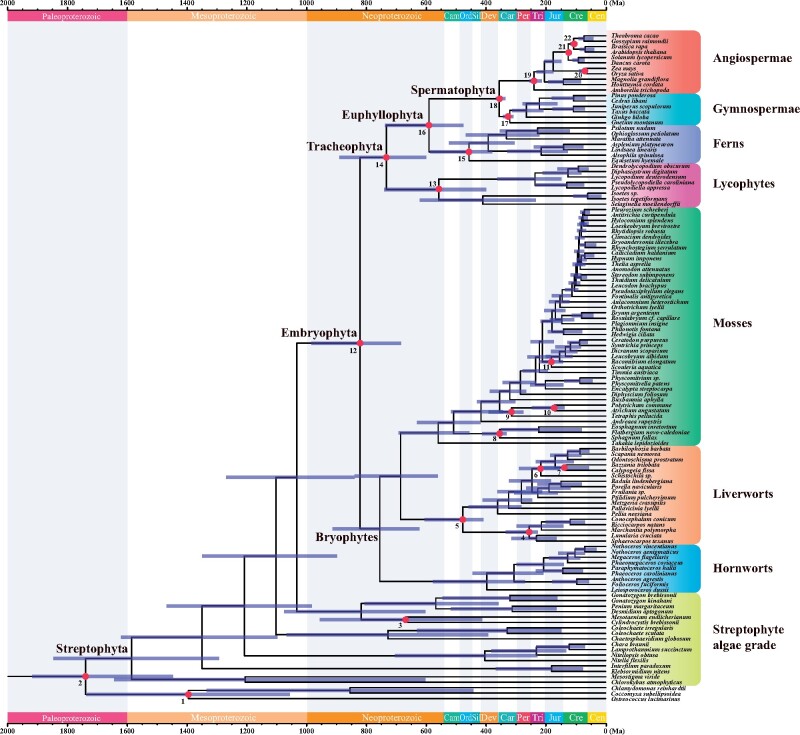
The fossil record provides important biological evidence to establish user-specified priors, and the user priors interact with the tree priors to establish effective priors. Although the data may correct prior assumptions if priors are unrealistic (Bromham 2019), the reliability and precision of fossil calibrations still have a great impact on the estimated divergence times even with an infinite amount of data (Rannala and Yang 2007). We should consider the reliability of fossil calibrations firstly, followed closely by the influences of molecular data. The maximum constraint of crown embryophytes in Strategy 1 is controversial, and the prior information of crown embryophytes from Strategy 1 affected posteriors most obviously among three strategies. Taken together, our analyses support that crown embryophytes originated in the Neoproterozoic (fig. 4, Strategy 2: 980–682 Ma).
Fig. 4.
Timetree of Streptophyta inferred from Strategy 2. Node ages are plotted at the posterior means and horizontal bars represent 95% credibility intervals. A total of 22 fossil calibration nodes are marked by red dots.
Bryophytes are the earliest-diverging embryophytes and the second-most diverse group of land plants, but they have few reliable fossils for deepening our understanding of the colonization of the land by plants. The important character of embryophytes, sporopollenin, is an extremely resistant polymer for the outer wall of all land-plant spores and pollen grains, improving the preservation potential of the plants themselves, and especially the spores (Li et al. 2019). The discovery of fossil spore assemblages offers new windows into the origin of early land plants (Edwards et al. 2014). Cryptospores with permanent tetrads are regarded as spores of land plants (Morris et al. 2018). The well-preserved cryptospores from the Dapingian Zanjon Formation in Argentina (Tetrahedraletes cf. Medinensis: 469 Ma) provide the earliest fossil record for land plants (Rubinstein et al. 2010; Morris et al. 2018). However, the accurate phylogenetic position of these cryptophytes remains unclear because of the highly fragmented fossils. Clarke et al. (2011) also indicated that crown land plants gradually enhanced fossilization potential, such as the development of a thickened cuticle and spore walls. The earliest land plants may have had low potential for fossil preservation. The source plants of many enigmatic cryptospores are not likely to belong to the first land plants.
Paleontologists have re-examined the fossil evidence of bryophytes, and suggested that the traditional view about the poor bryophyte fossil record needs to be revised (Tomescu et al. 2018). High potential and various modes for fossil preservation of bryophytic material, along with the extensive stratigraphic range of fossils, fully imply that many exquisite fossils of bryophytes will be discovered in the future (Tomescu et al. 2018). The similarities between the Dapingian cryptospore assemblage and younger cryptospore occurrences possibly indicate that phenotypic change in the early evolution of embryophytes was slow (Rubinstein et al. 2010). This evidence implies that a time gap may exist between the actual origin of land plants and the earliest fossil record based on cryptospores, which is consistent with our new timescale of land plant origin.
The recent discovery of a remarkably well-preserved green algal fossil from the Precambrian (1,000 Ma) (Tang et al. 2020) also offers circumstantial evidence. This fossil is a member of the Order Siphonocladales, Class Ulvophyceae, and is nested within the crown group Chlorophyta. The occurrence of this multicellular fossil confirms that Chlorophyta, at least, had originated before 1,000 Ma. Although fully consistent with our molecular timescale of plant evolution, it contradicts other studies including that of Morris et al. (2018), who obtained time estimates for the origin of Chlorophyta hundreds of millions of years younger than the new chlorophyte fossil.
Conclusions
Our phylogenomic analyses markedly improved the robustness of early land plant relationships, and strongly supported the monophyly of bryophytes using a large nuclear data set and dense taxon sampling. We further show that the controversy over land plant relationships has largely stemmed from nucleotide analyses that did not fully accommodate biases from fast-evolving synonymous substitutions. In evolutionary time estimation, fossil calibrations and molecular data are the crucial sources of actual biological information. Changing the calibrations will obviously influence time estimation, considering the importance of fossil calibrations in time priors. High consistency among different analyses will strengthen the confidence for a certain result, whereas inconsistency demands reasonable explanation. By comparing user priors, effective priors, and posteriors, we gained insight into why past studies have differed widely in their estimated time of the origin of land plants. We found that studies favoring a Neoproterozoic origin of land plants (980–682 Ma) are informed more by molecular data whereas those favoring a Phanerozoic origin (518–500 Ma) are informed more by fossil constraints. Our divergence time analyses highlighted the important contribution of the molecular data (time-dependent molecular change) when faced with contentious fossil evidence. Fossil calibrations used in estimating the timescale of plant evolution require greater scrutiny, and more efforts are needed to explain the results of disparate estimated times. A careful integration of fossil and molecular evidence will revolutionize our understanding of how land plants evolved.
Materials and Methods
Algal Strains and Culture Conditions
Lamprothamnium succinctum (NIES-1606), Nitella flexilis (NIES-1611), Nitellopsis obtusa (NIES-1638), and Gonatozygon brebissonii (NIES-138) strains were obtained from the Microbial Culture Collection at the National Institute for Environmental Studies, Tsukuba, Japan. All strains were grown in soil and cultured at 20 °C under alternating 12 h-light/12 h-dark periods. Lamprothamnium succinctum, N. flexilis, and Nitellopsis obtusa were grown in mSWC-2 medium (Okazaki et al. 1984; Sakayama et al. 2004), and G. brebissonii was grown in C medium (Ichimura 1971).
Data Processing, Assembly, and Annotation
We sequenced transcriptomes of four charophyte species using Illumina HiSeq technologies. Library construction and sequencing were performed at Novogene Bioinformatics Technology Co., Ltd (Beijing, China). Illumina raw data from Lamprothamnium succinctum, N. flexilis, Nitellopsis obtuse, and G. brebissonii were filtered by removing reads containing adapters, reads with more than 10% ambiguous bases (N), and low-quality reads (more than 50% bases with small Qphyred ≤20). We assembled four transcriptomes using Trinity with default settings (Grabherr et al. 2011), except that min_kmer_cov was set to 2. The transcripts were clustered into genes by using Corset (Davidson and Oshlack 2014) with default parameters. All of the assembled unigenes were BLAST against the NCBI nonredundant protein database (Nr), NCBI nonredundant nucleotide database (Nt) and Swiss-Prot to predict protein function with the E-value cutoff of 10−5, and Eukaryotic Ortholog Groups of protein database (KOG) with the E-value cutoff of 10−3 (Altschul et al. 1997). We identified for each gene the protein domains and unannotated regions using the Pfam database (Finn et al. 2014). All the unigenes were functionally annotated in the KEGG database (Moriya et al. 2007). The gene ontology (GO) classification of each gene model was carried out using Blast2GO v2.5 (Gotz et al. 2008).
Taxon Sampling and Data Collection
The 124 Streptophyta taxa were sampled from 63 orders including 68% bryophyte orders (supplementary table S1, Supplementary Material online). Three Chlorophyta taxa were designated as outgroups. The public genome data were downloaded from Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html, last accessed January 1, 2019), GenBank (GCA_000708835.1), GigaDB (http://gigadb.org/dataset/view/id/100209, last accessed January 1, 2019), Dryad Digital Repository (https://datadryad.org/resource/doi:10.5061/dryad.0vm37.2, last accessed January 1, 2019), and Orcae (https://bioinformatics.psb.ugent.be/orcae/overview/Chbra, last accessed January 1, 2019). The 90 transcriptomes were obtained from the 1KP project. To address the limitation of sparse taxon sampling of bryophytes and charophytes, we newly sequenced four transcriptomes from previously unsampled charophyte species, and assembled 13 transcriptomes of bryophytes that were downloaded from NCBI SRA database (supplementary table S1, Supplementary Material online) by using Trinity with default settings (min_kmer_cov=2).
Ortholog Identification
We firstly used protein sequences predicted from the complete genomes of 15 selected species (Arabidopsis thaliana, Daucus carota, Oryza sativa, Amborella trichopoda, Gnetum montanum, Ginkgo biloba, Selaginella moellendorffii, Physcomitrella patens, Sphagnum fallax, Marchantia polymorpha, Chara braunii, Klebsormidium nitens, Chlamydomonas reinhardtii, Coccomyxa subellipsoidea, Ostreococcus lucimarinus) to generate putative ortholog groups by utilizing the tree-based approach (Yang and Smith 2014). The reduction of sequence redundancy was carried out using CD-HIT (-c 0.995 -n 5) (Fu et al. 2012). Homology searches were conducted using all-by-all BlastP from peptides of 15 complete genomes with an E value cutoff of 10 and -max_target_seqs 1,000. We used a hit_fraction cutoff of 0.5 to filter BlastP hits and set IGNORE_INTRASPECIFIC_HITS to be True. Markov clustering (MCL v14) (van Dongen 2000) was performed on filtered data with the E value cutoff of 10−5 and an inflation value of 2. We excluded small clusters that contained less than 13 species and employed the “fasta_to_tree_pxclsq.py” (Yang and Smith 2014) for building homolog tree of each remaining cluster with default parameters. Based on the resulting trees, we trimmed long tips longer than a relative length cutoff 0.5 and more than ten times longer than its sister or exceeded an absolute value set to 2. The monophyletic tips were masked by the “mask_tips_by_taxonID_genomes.py” (Yang and Smith 2014). We cut long internal branches that were longer than 0.8 for reducing deep paralogs, and only retained the clusters including no less than 13 species. We repeated the above processes including building homolog trees, trimming (a relative length cutoff: 0.5; an absolute value: 1.7), masking, and cutting deep paralogs (long internal branches >0.7). We further pruned final homologous gene trees using the “prune_paralogs_MO.py” (minimal_taxa: 13) (Yang and Smith 2014) and resulted in a set of 2,148 clusters of putative orthologs. The transcriptomic and genomic data of other 112 species were then incorporated into the 2,148 core orthologous clusters using Orthograph v0.6.3 (Petersen et al. 2017) by default value.
Phylogenetic Analyses
Amino acid sequences of each orthologous group (OG) were aligned using MAFFT v7.310 (Katoh and Standley 2013), with the option “-localpair -maxiterate 1,000.” We excluded poorly aligned regions using Gblocks 0.91b (Castresana 2000) with “Allowed Gap Positions” set to “half” and other default parameters. OGs were removed when their longest sequence was shorter than 100 amino acids. We eliminated short sequences of each OG using trimAl v1.4 (Capella-Gutierrez et al. 2009) with the option -resoverlap 0.5 -seqoverlap 50. All alignments that did not contain ≥80% species were discarded, leaving 1,440 OGs. To reduce the potential effects of missing data, we further removed four species, each of which covered less than 70% OGs (<1,008 OGs), thereby reducing the number of species to 123. The corresponding nucleotide alignments of 1,440 OGs were generated using PAL2NAL (Suyama et al. 2006), and poorly aligned positions were excluded using Gblocks 0.91b with the “codon” model, half gaps allowed, and otherwise default settings.
A concatenation-based method was used to infer phylogenetic trees for both nucleotide and amino acid data sets. Maximum likelihood (ML) analysis was performed using IQ-TREE v2.0.5 (Minh et al. 2020). Nodal support values were estimated using SH-aLRT test (Guindon et al. 2010) and ultrafast bootstrap (Minh et al. 2013) with 1,000 replicates. We applied ModelFinder (Kalyaanamoorthy et al. 2017) to select optimal partitioning schemes and appropriate amino acid or nucleotide substitution models, using -TESTMERGE and -rcluster 10 options (Lanfear et al. 2014). The best-fitting models of substitution for each matrix are presented in supplementary table S2, Supplementary Material online. Bayesian inferences were conducted by the MPI version of ExaBayes v1.5.1 (Aberer et al. 2014) under the GTR (nucleotide) and LG (amino acid) models with the same partitioning schemes as in ML analyses. Two independent MCMC runs with two chains were conducted from parsimony starting topologies sampling every 500 generations. ExaBayes runs continued until the termination condition of mean topological differences was less than 5% with at least 500,000 and 200,000 generations for nucleotide and amino acid matrix, respectively. Posterior distributions of trees were summarized using the “consense” script with 25% burn-in. Convergence was assumed when all parameters had effective sampling sizes (ESS) greater than 100 estimated with Tracer v1.6 (Rambaut et al. 2018), and potential scale reduction factors (PSRF) close to 1 using the “postProcParam” program in ExaBayes.
We used three strategies to alleviate the effect of systematic errors: 1) analyzing the concatenated amino acid data under a site-heterogeneous mixture model (LG+C20+F+R5) that accounts for compositional heterogeneity across sites, 2) recoding the 20 amino acids of the original supermatrix into six-state groups under the Dayhoff recoding scheme, which were assigned numbers 0–5: C, FWY, HKR, ILMV, EDNQ, and AGPST (Dayhoff et al. 1978), and 3) recoding the nucleotide matrix with codon-degenerate characters (de Sousa et al. 2019), which use nucleotide ambiguity codes to eliminate all possible synonymous substitutions among codon variants of amino acids. The ML analysis of amino acid supermatrix under LG+C20+F+R5 model was performed using IQ-TREE with same parameter settings as above. The Dayhoff recoding strategy recodes the 20 amino acids into six groups on the basis of their chemical and physical properties. This strategy is commonly used in phylogenomic studies to reduce the effects of lineage-specific compositional heterogeneity (Hrdy et al. 2004;Susko and Roger 2007; Rota-Stabelli et al. 2013; Puttick et al. 2018). The phylogenetic reconstruction using the Dayhoff-recoded data set was executed under a 6-state GTR model using IQ-TREE, and all the other aspects (e.g., tree search strategy) remain the same as original amino acid matrix. In addition, the codon-degenerate nucleotide data were analyzed both using the ML and Bayesian methods under the optimal partitioning schemes and appropriate substitution models. For the coalescent method, only amino acid alignments of 1,440 OGs were used. Individual gene trees were reconstructed by using RAxML v8.2.12 (Stamatakis 2014) with the best fitting model (-m PROTGAMMAAUTO –auto-prot=bic) and 200 rapid bootstrap replicates. The species tree was inferred using ASTRAL-III v5.6.3 (Zhang et al. 2018) with nodal support values estimated by local posterior probability and multilocus bootstrapping (gene+site resampling). To reduce gene tree estimation error, we further considered contracting low support branches (below 20% bootstrap support) from individual gene trees.
Divergence Time Estimation
Data Set Assembly for Molecular Dating
We used MCMCTree v.4.9h from the PAML package (Yang 2007) to estimate divergence times, and conducted preliminary tests with the concatenated data set (1,440 OGs of 123 species) and fossil calibrations. Increasing the number of generations failed to improve convergence but raised computational burden for our large-scale data set (376,109 amino acid positions). Large amounts of data will generate an unpredictable computational burden under parameter-rich models, and different topologies and rate heterogeneity across genes may lead to mis-specified models (Smith et al. 2018). Foster et al. (2017) suggested selecting a subset of informative genes in molecular dating analyses, considering that time estimates are largely consistent in full compared with reduced data sets. “Clock-like” genes are defined as those that evolve in a clock-like manner (Jarvis et al. 2014), and they can minimize errors associated with model mis-specification (Smith et al. 2018). Therefore, we identified clock-like or nearly clock-like genes with low root-to-tip variance and minimal conflict using SortaDate (Smith et al. 2018). We calculated the variance of root-to-tip length for each rooted gene tree, and compared the individual gene trees with species trees by implementing bipartition-comparison analyses on each tree. The screening criteria were “1 = root-to-tip variance, 3 = bipartiton, 2 = tree-length.” Finally, 100 genes of 1,440 OGs were selected to be eligible as clock-like genes. These selected genes were likely to reduce the deviations caused by lineage-specific rate variation. The MCMC analyses were run for 20 million generations sampled every 500 generations after a burnin of 2,000,000 iterations. The chain convergence was assessed by running MCMC analyses twice simultaneously, and the effective sample size (ESS) of all parameters was confirmed to be >200 using Tracer v.1.6 (Rambaut et al. 2018).
Rate Priors
Two relaxed-clock models are often used for divergence time estimation, that is, the independent-rates (IR) model and the autocorrelated-rates (AR) model. Recent phylogenomic analyses have shown that the AR model is more suitable for the analysis of closely related species, and the IR model better fits distantly related taxa (e.g., Foster et al. 2017; Barba-Montoya et al. 2018; Nie et al. 2020). Our phylogenomic data set consists of many distantly related species, and the degree of autocorrelation would decrease as the rate differences among lineages amplify. Therefore, we used the IR model to estimate the divergence time of Streptophyta. The time unit was set to 100 My. Parameter σ2 was assigned a gamma prior G (1, 10) to determine the degree of rate variation across branches. In order to obtain a suitable prior on the mean of the rate μ (representing the overall rate), we compared the amino acid pairwise distance between Arabidopsis thaliana and Bryoandersonia illecebra (0.138 substitutions/site) using the LG + Γ4 + F model in CODEML (Yang 2007). The assumed divergence time between the two species is approximately 469 Ma (Edwards et al. 2014; Morris et al. 2018; Tomescu et al. 2018), and hence the mean rate was 0.138/4.69 = 0.0294 for 100 clock-like genes (meaning 2.94 × 10−10 amino acid substitutions per site per year). The shape parameter of the gamma distribution prior on rate was fixed to 2 following Morris et al. (2018), so that the scale parameter was set to 68.
Time Priors
Fossil calibrations and the birth–death process are the important sources of information for constructing the prior on times. The parameters for the birth–death process were set as λ = μ = 1 and ρ = 0.0003. The sampling proportion (ρ) of 0.03% was based on our sample size (120 taxa) compared with the number of extant Streptophyte species (∼386,969) (http://www.theplantlist.org, last accessed September 20, 2019; Shaw et al. 2011; Pteridophyte Phylogeny Group 2016). The ML estimates of the branch lengths were calculated based on the LG + Γ4 + F amino acid substitution model using CODEML program. The MCMCTree combined the calibration distributions and the birth–death process model to generate the joint priors. We ran the MCMC analyses without sequence data to obtain the effective priors.
Fossil Constraints
We applied 22 fossil calibrations in our analyses (fig. 3). The details of 22 selected fossil calibrations following contemporary standards (Parham et al. 2012) are available in the supplementary table S3, Supplementary Material online. Uniform distributions were employed at nodes 1–3 and 17–22 with a hard minimum age (pL = 1e-300) and a soft maximum age (pU = 0.025). We applied Cauchy distributions with 2.5% left tail probability at nodes 13–16. We tested the impact of different maximum bounds on nodes 4–12 owing to the controversy on the maximum constraint of land plants (Clarke et al. 2011; Hedges et al. 2018; Morris et al. 2018). In Strategy 1, we used uniform distributions with a hard minimum bound (pL = 1e-300) and a soft maximum bound (pU = 0.025) corresponding to 515.5 Ma for nodes 4–12. In Strategy 2, we changed the soft maximum ages from 515.5 to 1,042 Ma. In Strategy 3, no maximum bound was imposed on these nodes, and the minimum bound was represented using a truncated Cauchy distribution with 2.5% left tail probability. We constructed calibration densities for three calibration strategies by MCMCTreeR (Puttick 2019).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31970229 and 31570219), the State Key Laboratory of Paleobiology and Stratigraphy (Nanjing Institute of Geology and Paleontology, CAS), the Key Laboratory of Vertebrate Evolution and Human Origins of Chinese Academy of Sciences (IVPP, CAS), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the U.S. National Science Foundation (1932765). We thank Stephen A. Smith, Ya Yang, Joseph Walker, Linhua Sun, Yuan Nie, and Xi Li for helpful discussions, and Jiangsu Collaborative Innovation Center for Modern Crop Production for technical support. We also thank the editor and anonymous reviewers for their helpful suggestions.
Data Availability
The sequencing data that support the findings of this study are openly available in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA674414. The alignment data are available from the Figshare: https://figshare.com/s/29c09214f1dc9f72aeaf.
References
- Aberer AJ, Kobert K, Stamatakis A.. 2014. ExaBayes: massively parallel Bayesian tree inference for the whole-genome era. Mol Biol Evol. 31(10):2553–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba-Montoya J, dos Reis M, Yang Z.. 2017. Comparison of different strategies for using fossil calibrations to generate the time prior in Bayesian molecular clock dating. Mol Phylogenet Evol. 114:386–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba-Montoya J, Dos RM, Schneider H, Donoghue P, Yang Z.. 2018. Constraining uncertainty in the timescale of angiosperm evolution and the veracity of a Cretaceous terrestrial revolution. New Phytol. 218(2):819–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S.. 2015. A protocol for diagnosing the effect of calibration priors on posterior time estimates: a case study for the Cambrian explosion of animal phyla. Mol Biol Evol. 32(7):1907–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B, Marin B.. 2009. Streptophyte algae and the origin of embryophytes. Ann Bot. 103(7):999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell D, Lin Q, Gerelle WK, Joya S, Chang Y, Taylor ZN, Rothfels CJ, Larsson A, Villarreal JC, Li FW, et al. 2020. Organellomic data sets confirm a cryptic consensus on (unrooted) land-plant relationships and provide new insights into bryophyte molecular evolution. Am J Bot. 107(1):91–115. [DOI] [PubMed] [Google Scholar]
- Bromham L. 2019. Six impossible things before breakfast: assumptions, models, and belief in molecular dating. Trends Ecol Evol. 34(5):474–486. [DOI] [PubMed] [Google Scholar]
- Brown JW, Smith SA.. 2018. The past sure is tense: on interpreting phylogenetic divergence time estimates. Syst Biol. 67(2):340–353. [DOI] [PubMed] [Google Scholar]
- Buschmann H, Zachgo S.. 2016. The evolution of cell division: from streptophyte algae to land plants. Trends Plant Sci. 21(10):872–883. [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T.. 2009. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. [DOI] [PubMed] [Google Scholar]
- Chater C, Caine RS, Fleming AJ, Gray JE.. 2017. Origins and evolution of stomatal development. Plant Physiol. 174(2):624–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazot N, Wahlberg N, Freitas AVL, Mitter C, Labandeira C, Sohn J-C, Sahoo RK, Seraphim N, de Jong R, Heikkilä M.. 2019. Priors and posteriors in Bayesian timing of divergence analyses: the age of butterflies revisited. Syst Biol. 68(5):797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Xian W, Fu Y, Marin B, Keller J, Wu T, Sun W, Li X, Xu Y, Zhang Y, et al. 2019. Genomes of subaerial zygnematophyceae provide insights into land plant evolution. Cell 179(5):1057–1067. [DOI] [PubMed] [Google Scholar]
- Clarke JT, Warnock RC, Donoghue PC.. 2011. Establishing a time-scale for plant evolution. New Phytol. 192(1):266–301. [DOI] [PubMed] [Google Scholar]
- Cox CJ. 2018. Land plant molecular phylogenetics: a review with comments on evaluating incongruence among phylogenies. Crit Rev Plant Sci. 37(2–3):113–127. [Google Scholar]
- Cox CJ, Li B, Foster PG, Embley TM, Civán P.. 2014. Conflicting phylogenies for early land plants are caused by composition biases among synonymous substitutions. Syst Biol. 63(2):272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson NM, Oshlack A.. 2014. Corset: enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 15(7):410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff MO, Schwartz RM, Orcutt BC.. 1978. A model of evolutionary change in proteins. In: Dayhoff MO, editor. Atlas of protein sequence and structure. Vol. 5. Washington (DC: ): National Biomedical Research Foundation. p. 345–352. [Google Scholar]
- de Sousa F, Foster PG, Donoghue P, Schneider H, Cox CJ.. 2019. Nuclear protein phylogenies support the monophyly of the three bryophyte groups (Bryophyta Schimp.). New Phytol. 222(1):565–575. [DOI] [PubMed] [Google Scholar]
- Donoghue PC, Yang Z.. 2016. The evolution of methods for establishing evolutionary timescales. Philos Trans R Soc B. 371(1699):20160020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Donoghue PC, Yang Z.. 2016. Bayesian molecular clock dating of species divergences in the genomics era. Nat Rev Genet. 17(2):71–80. [DOI] [PubMed] [Google Scholar]
- dos Reis M, Thawornwattana Y, Angelis K, Telford MJ, Donoghue PC, Yang Z.. 2015. Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Curr Biol. 25(22):2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D, Morris JL, Richardson JB, Kenrick P.. 2014. Cryptospores and cryptophytes reveal hidden diversity in early land floras. New Phytol. 202(1):50–78. [DOI] [PubMed] [Google Scholar]
- Fang H, Huangfu L, Chen R, Li P, Xu S, Zhang E, Cao W, Liu L, Yao Y, Liang G, et al. 2017. Ancestor of land plants acquired the DNA-3-methyladenine glycosylase (MAG) gene from bacteria through horizontal gene transfer. Sci Rep. 7(1):9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42(Database issue):D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster C, Sauquet H, van der Merwe M, McPherson H, Rossetto M, Ho S.. 2017. Evaluating the impact of genomic data and priors on Bayesian estimates of the angiosperm evolutionary timescale. Syst Biol. 66(3):338–351. [DOI] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W.. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23):3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goremykin VV, Hellwig FH.. 2005. Evidence for the most basal split in land plants dividing bryophyte and tracheophyte lineages. Plant Syst Evol. 254(1–2):93–103. [Google Scholar]
- Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A.. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36(10):3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB.. 2001. Molecular evidence for the early colonization of land by fungi and plants. Science 293(5532):1129–1133. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Tao Q, Walker M, Kumar S.. 2018. Accurate timetrees require accurate calibrations. Proc Natl Acad Sci U S A. 115(41):E9510–E9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SY. 2014. The changing face of the molecular evolutionary clock. Trends Ecol Evol. 29(9):496–503. [DOI] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N, et al. 2014. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun. 5(1): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdy I, , HirtRP, , DolezalP, , BardonováL, , FosterPG, , TachezyJ, , Embley TM.. 2004. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature 432(7017):618–622. [DOI] [PubMed] [Google Scholar]
- Ichimura T. 1971. Sexual cell division and conjugation-papilla formation in sexual reproduction of Closterium strigosum. In: Nishizawa K, editor. Proceedings of the Seventh International Seaweed Symposium. Tokyo: University of Tokyo Press. p. 208–214. [Google Scholar]
- Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SY, Faircloth BC, Nabholz B, Howard JT, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346(6215):1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffroy O, , BrinkmannH, , DelsucF, , Philippe H.. 2006. Phylogenomics: the beginning of incongruence?. Trends Genet. 22(4):225–231. [DOI] [PubMed] [Google Scholar]
- Jiao C, Sorensen I, Sun X, Sun H, Behar H, Alseekh S, Philippe G, Palacio LK, Sun L, Reed R, et al. 2020. The Penium margaritaceum genome: hallmarks of the origins of land plants. Cell 181(5):1097–1111. [DOI] [PubMed] [Google Scholar]
- Jill HC. 2017. Development and genetics in the evolution of land plant body plans. Philos Trans R Soc Lond B Biol Sci. 372(1713):20150490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karol KG, Arumuganathan K, Boore JL, Duffy AM, Everett KD, Hall JD, Hansen SK, Kuehl JV, Mandoli DF, Mishler BD, et al. 2010. Complete plastome sequences of Equisetum arvense and Isoetes flaccida: implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evol Biol. 10(1):321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P, Wellman CH, Schneider H, Edgecombe GD.. 2012. A timeline for terrestrialization: consequences for the carbon cycle in the Palaeozoic. Philos Trans R Soc Lond B Biol Sci. 367(1588):519–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laenen B, Shaw B, Schneider H, Goffinet B, Paradis E, Desamore A, Heinrichs J, Villarreal JC, Gradstein SR, McDaniel SF, et al. 2014. Extant diversity of bryophytes emerged from successive post-Mesozoic diversification bursts. Nat Commun. 5:5134. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A.. 2014. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol Biol. 14:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenton T, Crouch M, Johnson M, Pires N, Dolan L.. 2012. First plants cooled the Ordovician. Nat Geosci. 5(2):86–89. [Google Scholar]
- Lenton TM, Dahl TW, Daines SJ, Mills BJ, Ozaki K, Saltzman MR, Porada P.. 2016. Earliest land plants created modern levels of atmospheric oxygen. Proc Natl Acad Sci U S A. 113(35):9704–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Lopes JS, Foster PG, Embley TM, Cox CJ.. 2014. Compositional biases among synonymous substitutions cause conflict between gene and protein trees for plastid origins. Mol Biol Evol. 31(7):1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FS, Phyo P, Jacobowitz J, Hong M, Weng JK.. 2019. The molecular structure of plant sporopollenin. Nat Plants. 5(1):41–46. [DOI] [PubMed] [Google Scholar]
- Ligrone R, Duckett JG, Renzaglia KS.. 2012. Major transitions in the evolution of early land plants: a bryological perspective. Ann Bot. 109(5):851–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Cox CJ, Wang W, Goffinet B.. 2014. Mitochondrial phylogenomics of early land plants: mitigating the effects of saturation, compositional heterogeneity, and codon-usage bias. Syst Biol. 63(6):862–878. [DOI] [PubMed] [Google Scholar]
- Liu Y, Johnson MG, Cox CJ, Medina R, Devos N, Vanderpoorten A, Hedenäs L, Bell NE, Shevock JR, Aguero B, et al. 2019. Resolution of the ordinal phylogeny of mosses using targeted exons from organellar and nuclear genomes. Nat Commun. 10(1):1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallon S, Hilu KW, Quandt D.. 2013. Land plant evolutionary timeline: gene effects are secondary to fossil constraints in relaxed clock estimation of age and substitution rates. Am J Bot. 100(3):556–573. [DOI] [PubMed] [Google Scholar]
- Marshall CR. 2019. Using the fossil record to evaluate timetree timescales. Front Genet. 10:1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoni J, Doyle J, Sauquet H.. 2015. Fossil calibration of Magnoliidae, an ancient lineage of angiosperms. Palaeontol Electron. 18:1–25. [Google Scholar]
- Minh BQ, Nguyen MA, von Haeseler A.. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R.. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczydłowska M, Landing ED, Zang W, Palacios T.. 2011. Proterozoic phytoplankton and timing of chlorophyte algae origins. Palaeontology 54(4):721–733. [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M.. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35(Web Server issue):W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JL, Puttick MN, Clark JW, Edwards D, Kenrick P, Pressel S, Wellman CH, Yang Z, Schneider H, Donoghue P.. 2018. The timescale of early land plant evolution. Proc Natl Acad Sci U S A. 115(10):E2274–E2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y, Foster C, Zhu T, Yao R, Duchene DA, Ho S, Zhong B.. 2020. Accounting for uncertainty in the evolutionary timescale of green plants through clock-partitioning and fossil calibration strategies. Syst Biol. 69(1):1–16. [DOI] [PubMed] [Google Scholar]
- Niklas KJ, Kutschera U.. 2010. The evolution of the land plant life cycle. New Phytol. 185(1):27–41. [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Wolf PG, Kugita M, Sinclair RB, Sugita M, Sugiura C, Wakasugi T, Yamada K, Yoshinaga K, Yamaguchi K, et al. 2004. Chloroplast phylogeny indicates that bryophytes are monophyletic. Mol Biol Evol. 21(10):1813–1819. [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Shimmen T, Tazawa M.. 1984. Turgor regulation in a brackish charophyte, Lamprothamnium succinctum I. Artificial modification of intracellular osmotic pressure. Plant Cell Physiol. 25:565–571. [Google Scholar]
- One Thousand Plant Transcriptomes Initiative. 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574:679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham JF, Donoghue PC, Bell CJ, Calway TD, Head JJ, Holroyd PA, Inoue JG, Irmis RB, Joyce WG, Ksepka DT, et al. 2012. Best practices for justifying fossil calibrations. Syst Biol. 61(2):346–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell J, Foster S.. 2012. Ordovician ash geochemistry and the establishment of land plants. Geochem Trans. 13(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Meusemann K, Donath A, Dowling D, Liu S, Peters RS, Podsiadlowski L, Vasilikopoulos A, Zhou X, Misof B, et al. 2017. Orthograph: a versatile tool for mapping coding nucleotide sequences to clusters of orthologous genes. BMC Bioinformatics 18(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pteridophyte Phylogeny Group. 2016. A community-derived classification for extant lycophytes and ferns: PPG I. J Syst Evol. 54:563–603. [Google Scholar]
- Puttick MN. 2019. MCMCtreeR: functions to prepare MCMCtree analyses and visualize posterior ages on trees. Bioinformatics 35(24):5321–5322. [DOI] [PubMed] [Google Scholar]
- Puttick MN, Morris JL, Williams TA, Cox CJ, Edwards D, Kenrick P, Pressel S, Wellman CH, Schneider H, Pisani D, et al. 2018. The interrelationships of land plants and the nature of the ancestral embryophyte. Curr Biol. 28(5):733–745. [DOI] [PubMed] [Google Scholar]
- Qiu YL, Li L, Wang B, Chen Z, Knoop V, Groth-Malonek M, Dombrovska O, Lee J, Kent L, Rest J, et al. 2006. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci U S A. 103(42):15511–15516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst Biol. 67(5):901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Yang Z.. 2007. Inferring speciation times under an episodic molecular clock. Syst Biol. 56(3):453–466. [DOI] [PubMed] [Google Scholar]
- Rensing SA. 2018. Plant evolution: phylogenetic relationships between the earliest land plants. Curr Biol. 28(5):R210–R213. [DOI] [PubMed] [Google Scholar]
- Renzaglia KS, Duff R, Nickrent DL, Garbary DJ.. 2000. Vegetative and reproductive innovations of early land plants: implications for a unified phylogeny. Philos Trans R Soc Lond B Biol Sci. 355(1398):769–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzaglia KS, Villarreal JC, Garbary DJ.. 2018. Morphology supports the setaphyte hypothesis: mosses plus liverworts form a natural group. Bry Div Evo. 40(2):11–017. [Google Scholar]
- Reski R. 2018. Enabling the water-to-land transition. Nat Plants. 4(2):67–68. [DOI] [PubMed] [Google Scholar]
- Rota-Stabelli O, , LartillotN, , PhilippeH, , Pisani D.. 2013. Serine codon-usage bias in deep phylogenomics: pancrustacean relationships as a case study. Syst Biol. 62(1):121–133. [DOI] [PubMed] [Google Scholar]
- Rubinstein CV, Gerrienne P, de la Puente GS, Astini RA, Steemans P.. 2010. Early Middle Ordovician evidence for land plants in Argentina (eastern Gondwana). New Phytol. 188(2):365–369. [DOI] [PubMed] [Google Scholar]
- Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG.. 2014. From algae to angiosperms-inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol Biol. 14:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakayama H, Hara Y, Nozaki H.. 2004. Taxonomic re-examination of six species of Nitella (Charales, Charophyceae) from Asia, and phylogenetic relationships within the genus based on rbcL and atpB gene sequences. Phycologia 43(1):91–104. [Google Scholar]
- Shaw AJ, Szovenyi P, Shaw B.. 2011. Bryophyte diversity and evolution: windows into the early evolution of land plants. Am J Bot. 98(3):352–369. [DOI] [PubMed] [Google Scholar]
- Smith SA, Brown JW, Walker JF.. 2018. So many genes, so little time: a practical approach to divergence-time estimation in the genomic era. PLoS One 13:e197433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susko E, , Roger AJ.. 2007. On reduced amino acid alphabets for phylogenetic inference. Mol Biol Evol. 24(9):2139–2150. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szovenyi P, Waller M, Kirbis A.. 2019. Evolution of the plant body plan. Curr Top Dev Biol. 131:1–34. [DOI] [PubMed] [Google Scholar]
- Tang Q, Pang K, Yuan X, Xiao S.. 2020. A one-billion-year-old multicellular chlorophyte. Nat Ecol Evol. 4(4):543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomescu AMF, Bomfleur B, Bippus AC, Savoretti A.. 2018. Chapter 16 – why are bryophytes so rare in the fossil record? A spotlight on taphonomy and fossil preservation. In: Krings M, Harper CJ, Cuneo NR, Rothwell GW, editors. Transformative plaeobotany: papers to commemorate the life and legacy of Thomas N. Taylor. San Francisco (CA): Academic Press. p. 375–416. [Google Scholar]
- van Dongen S. 2000. Graph clustering by flow simulation [PhD thesis]. Utrecht, the Netherlands: University of Utrecht. Center for Math and Computer Science (CWI).
- Warnock RC, Parham JF, Joyce WG, Lyson TR, Donoghue PC.. 2015. Calibration uncertainty in molecular dating analyses: there is no substitute for the prior evaluation of time priors. Proc Biol Sci. 282(1798):20141013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA, et al. 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci U S A. 111(45):E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Smith SA.. 2014. Orthology inference in nonmodel organisms using transcriptomes and low-coverage genomes: improving accuracy and matrix occupancy for phylogenomics. Mol Biol Evol. 31(11):3081–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yin L, Zhao Y, Bian L, Peng J.. 2013. Comparison between cryptospores from the Cambrian Log Cabin Member, Pioche Shale, Nevada, USA and similar specimens from the Cambrian Kaili Formation, Guizhou, China. Sci China Earth Sci. 56(5):703–709. [Google Scholar]
- Zhang C, Rabiee M, Sayyari E, Mirarab S.. 2018. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19(S6):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Fu XX, Li RQ, Zhao X, Liu Y, Li MH, Zwaenepoel A, Ma H, Goffinet B, Guan YL, et al. 2020. The hornwort genome and early land plant evolution. Nat Plants. 6(2):107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Liu L, Yan Z, Penny D.. 2013. Origin of land plants using the multispecies coalescent model. Trends Plant Sci. 18(9):492–495. [DOI] [PubMed] [Google Scholar]
- Zhong B, Sun L, Penny D.. 2015. The origin of land plants: a phylogenomic perspective. Evol Bioinform Online. 11:137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Xi Z, Goremykin VV, Fong R, McLenachan PA, Novis PM, Davis CC, Penny D.. 2014. Streptophyte algae and the origin of land plants revisited using heterogeneous models with three new algal chloroplast genomes. Mol Biol Evol. 31(1):177–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data that support the findings of this study are openly available in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA674414. The alignment data are available from the Figshare: https://figshare.com/s/29c09214f1dc9f72aeaf.