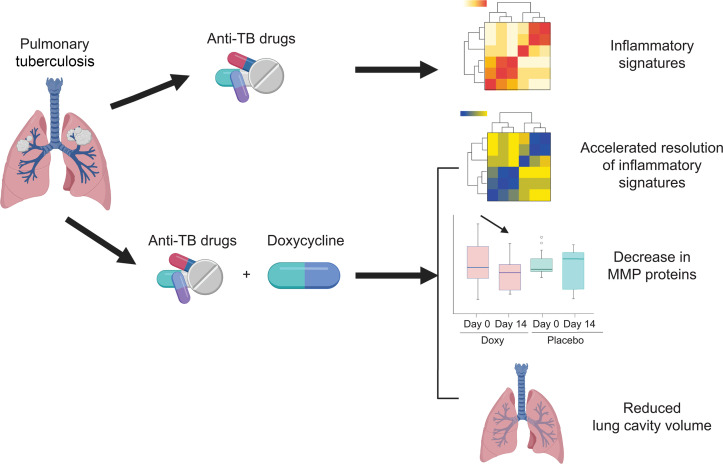
Abstract
BACKGROUND
Matrix metalloproteinases (MMPs) are key regulators of tissue destruction in tuberculosis (TB) and may be targets for host-directed therapy. We conducted a phase II double-blind, randomized, controlled trial investigating doxycycline, a licensed broad-spectrum MMP inhibitor, in patients with pulmonary TB.
METHODS
Thirty patients with pulmonary TB were enrolled within 7 days of initiating anti-TB treatment and randomly assigned to receive either 100 mg doxycycline or placebo twice a day for 14 days, in addition to standard care.
RESULTS
Whole blood RNA-sequencing demonstrated that doxycycline accelerated restoration of dysregulated gene expression in TB towards normality, rapidly down-regulating type I and II interferon and innate immune response genes, and up-regulating B-cell modules relative to placebo. The effects persisted for 6 weeks after doxycycline discontinuation, concurrent with suppressed plasma MMP-1. Doxycycline significantly reduced sputum MMP-1, -8, -9, -12 and -13, suppressed type I collagen and elastin destruction, reduced pulmonary cavity volume without altering sputum mycobacterial loads, and was safe.
CONCLUSION
Adjunctive doxycycline with standard anti-TB treatment suppressed pathological MMPs in PTB patients. Larger studies on adjunctive doxycycline to limit TB immunopathology are merited.
TRIAL REGISTRATION
ClinicalTrials.gov NCT02774993.
FUNDING
Singapore National Medical Research Council (NMRC/CNIG/1120/2014, NMRC/Seedfunding/0010/2014, NMRC/CISSP/2015/009a); the Singapore Infectious Diseases Initiative (SIDI/2013/013); National University Health System (PFFR-28 January 14, NUHSRO/2014/039/BSL3-SeedFunding/Jul/01); the Singapore Immunology Network Immunomonitoring platform (BMRC/IAF/311006, H16/99/b0/011, NRF2017_SISFP09); an ExxonMobil Research Fellowship, NUHS Clinician Scientist Program (NMRC/TA/0042/2015, CSAINV17nov014); the UK Medical Research Council (MR/P023754/1, MR/N006631/1); a NUS Postdoctoral Fellowship (NUHSRO/2017/073/PDF/03); The Royal Society Challenge Grant (CHG\R1\170084); the Sir Henry Dale Fellowship, Wellcome Trust (109377/Z/15/Z); and A*STAR.
Keywords: Clinical Trials, Infectious disease
Keywords: Tuberculosis
Introduction
Globally, an estimated 10 million people develop tuberculosis (TB) each year, and this disease remains a leading cause of death from a single infectious agent (1). Standard short-course anti-TB treatment still requires antimicrobial drugs of at least 6 months, and drug-resistant TB is an increasing public health threat. Even after microbiological cure of TB, patients often suffer from significant sequelae, such as residual or secondary lung diseases (2). A recent meta-analysis revealed that TB survivors have approximately 3 to 4 times greater mortality than their local population (3). Consequently, there is interest in adjunctive host-directed therapies that may modulate host immune responses to Mycobacterium tuberculosis (Mtb) to improve the efficacy of anti-TB drugs, shorten treatment duration, and limit associated tissue damage (4–12). These aims are unlikely to be met by antimicrobial treatment alone.
Mtb causes apical pulmonary disease in the immunocompetent host and drives destructive pathology, resulting in pulmonary cavity formation (13–15). Cavities are sites of high mycobacterial burden, and are poorly penetrated by anti-TB drugs, leading to the persistence of drug-tolerant bacilli and contributing to the transmission of infectious bacilli (16–18). After completion of anti-TB treatment, the sequelae of tissue damage include permanent respiratory dysfunction in the form of pulmonary fibrosis or post-TB bronchiectasis, which can lead to decreased effort tolerance, and infectious exacerbations resulting in repeated hospitalizations and reduced quality of life (6, 19–21). Furthermore, several population-based studies have demonstrated that a history of treated TB increases the risk of obstructive airway disease, independent of smoking and other clinical factors (22–24).
Pathological destruction of the highly stable network of collagen fibrils in the lung parenchyma is mainly mediated by proteases, in particular, matrix metalloproteinases (MMPs) (10, 15, 17, 18, 25, 26). MMPs are a family of zinc-dependent proteases that may collectively degrade all fibrillar components of the extracellular matrix at neutral pH and are involved in diverse physiological processes, including tissue modeling, organ development, and regulation of immune responses (27, 28). MMP concentrations (MMP-1, -2, -3, -7, -8, -9, and -10) and their matrix degradation products, procollagen III N-terminal propeptide (PIIINP) and desmosine, are consistently found to be elevated in respiratory samples from patients with pulmonary TB compared with patients with other respiratory diseases and healthy volunteers (29–32). Increased MMP concentrations associate with markers of pulmonary TB disease severity, such as sputum smear status, radiographic disease extent, and presence of cavitation (29, 33). In addition, Mtb infection and/or stimulation by conditioned media from Mtb-infected monocytes induced secretion of MMPs with proteolytic activity in cellular models of human bronchial epithelial cells, monocyte-derived macrophages, and neutrophils (14, 34–38), as well as in animal models (26, 39). These observations implicate MMPs as dominant effectors of lung matrix destruction in pulmonary TB and consequently, MMPs are attractive host targets for adjunctive host-directed therapies.
Currently, the only licensed MMP inhibitor is doxycycline, a tetracycline antibiotic with broad spectrum MMP inhibitory activity (40). Doxycycline suppresses Mtb-induced MMP secretion in cellular models (29) and limits collagen destruction by Mtb-induced MMPs (14, 31). Doxycycline treatment inhibits MMP activity in periodontal disease at 20 mg twice daily (41) and improves lung function in inflammatory lung disorders, such as chronic obstructive pulmonary disease (COPD) (42) and asthma (43). In a guinea pig model of TB, doxycycline monotherapy reduced the lung mycobacterial burden in a dose-dependent manner (29) and in the mouse model of TB, MMP inhibition improved drug efficacy (39). However, adjunctive doxycycline treatment in patients with TB has not been evaluated.
Here, we performed a first-in-human pilot phase II randomized, double-blind, placebo-controlled trial to investigate the effects of doxycycline on the host transcriptome, mycobacterial burden, and biological markers of tissue destruction, including pulmonary cavitation, as well as concentrations of MMPs, tissue inhibitor of metalloproteinases (TIMPs), and matrix degradation products in patients with drug-susceptible pulmonary TB. Concurrently, we assessed the safety of adjunctive doxycycline treatment with standard anti-TB therapy.
Results
Study profile and safety analyses.
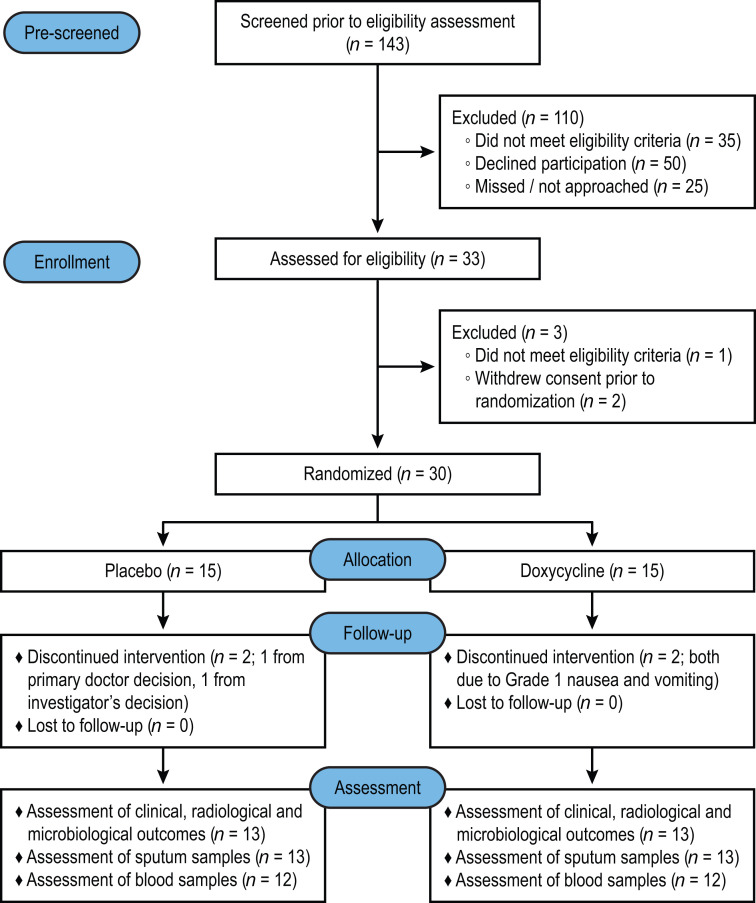
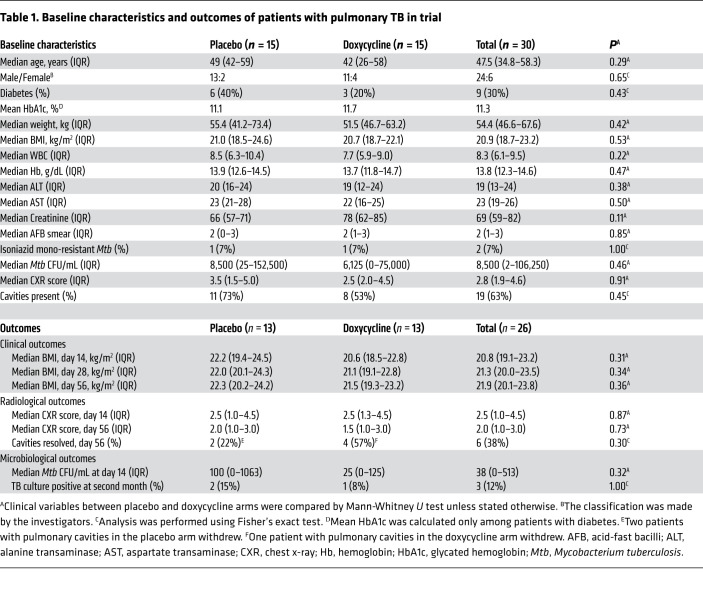
A total of 143 patients with pulmonary TB who were HIV negative were prescreened, of which 33 were assessed for eligibility. Thirty patients with pulmonary TB were enrolled within 7 days of initiating anti-TB treatment and randomly assigned to either doxycycline or placebo (Figure 1). The baseline clinical, laboratory, and radiologic characteristics at enrollment were similar between the study arms (Table 1). Six (20%) of the 30 patients with TB were female and 9 (30%) had diabetes mellitus with a mean HbA1c of 11.3%. The median chest x-ray (CXR) score was 2.8 (IQR 1.9–4.6) and 19 (63%) patients had pulmonary cavities. Two patients had isoniazid mono-resistant Mtb while the remaining 28 had fully drug-sensitive Mtb.
Figure 1. Trial profile.
Safety analysis included all participants who underwent randomization and received an initial dose of placebo or doxycycline. The downstream assessment included all participants who underwent randomization, took at least 24 of the 28 doses of placebo or doxycycline, and returned for follow-up on days 14 and 56 (94).
Table 1. Baseline characteristics and outcomes of patients with pulmonary TB in trial.
In the doxycycline arm, 2 patients withdrew due to adverse events (both grade 1 nausea and vomiting), while in the placebo arm, one patient developed neutropenia and was withdrawn by the managing physician, and one by the investigator’s decision as the patient was found to have cognitive impairment after randomization. Thus, there were 13 patients in each arm who completed the allocated intervention (took at least 24 of the 28 doses of the study drug) and were followed up for 2 months (Figure 1). Subsequently, 1 patient in the placebo arm declined phlebotomy at day 14, and another patient’s neutrophil and monocyte samples in the doxycycline arm were not processed due to electrical failure at the laboratory, which led to analysis of 12 blood samples in each arm.
Among all the 30 enrolled patients, there was no difference between the study arms due to any adverse events (relative risk [RR] 1.1, 95% CI 0.69–1.76, Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI141895DS1), or presumed toxic effects (any grade 3 or 4 events, or serious adverse events). Patients in the doxycycline arm tended to experience more nausea and vomiting (RR 3.0, 95% CI 0.72–12.56) and rash (RR 3.0, 95% CI 0.35–25.69), but there was no statistical significance between the arms (P = 0.21 and P = 0.60 by Fisher’s exact test, respectively, Supplemental Table 1). One patient in the doxycycline arm developed grade 4 neutropenia attributed to anti-TB treatment, which persisted after completion of the intervention. Two patients in the placebo arm developed serious adverse events: one had prolonged hospitalization due to paradoxical reaction and another developed dyspnea requiring a visit to the emergency department that resolved.
Ten healthy volunteers (5 males and 5 females) were also enrolled and received 100 mg doxycycline twice a day for 14 days. All 10 volunteers completed at least 24 of the 28 doses of doxycycline and were followed for 2 months. No subject discontinued participation in the study due to a doxycycline-related adverse event and no serious adverse events were observed (Supplemental Table 2). Two of 10 volunteers developed grade 1 nausea and vomiting, which resolved on completion of the drug course (Supplemental Table 2).
Clinical, radiological, and microbiological outcomes were similar between study arms.
There was no difference in median BMI and CXR score between doxycycline and placebo-treated patients at any time point (Table 1). Although doxycycline is bacteriostatic to Mtb in vitro (29, 44, 45), sputum Mtb burden was unchanged between the study arms. Two patients in the placebo arm and one in the doxycycline arm had positive sputum Mtb cultures at 2 months into anti-TB treatment (Table 1, P = 1.00). Pulmonary cavities resolved in 4 of 7 patients (57%) in the doxycycline arm compared with 2 of 9 (22%) in the placebo arm by the end of 2 months, but this finding was not statistically significant.
Gene expression of MMPs and immunoregulatory genes are dysregulated in patients with TB.
To analyze the effects of Mtb infection and doxycycline in an unbiased manner, we first studied baseline differences between blood transcriptomes of patients with TB and healthy volunteers. After normalization and filtering, principal component analysis (PCA) showed differentiation between healthy volunteers and patients with TB, although with significant inter-individual variation, as expected from human disease with diverse severity (Supplemental Figure 1A). Differentially expressed gene (DEG) analysis using generalized linear models (EdgeR) (46) identified 1657 DEGs, including 853 genes upregulated in patients with TB. As evaluated by gene ontology analyses, these genes mostly encoded for immune response and detection of bacterial molecules (Supplemental Figure 1B), while pathway enrichment identified immune, infectious, and inflammatory pathways (Supplemental Figure 1C). To visualize the extent of changes induced by Mtb infection in the blood cell transcriptome, significantly regulated genes were plotted onto the diagrams of individual KEGG pathways (47–49), including the TB pathway (Supplemental Figure 2) and the TNF signaling pathway (Supplemental Figure 3), which demonstrated extensive changes in gene expression in each pathway.
Coexpression module analysis using CEMITool (50) identified 3 modules (modules 1, 2, and 4) to be differentially enriched in patients with TB compared with healthy volunteers (Supplemental Figure 4A). Modules 2 and 4 were specifically overexpressed in TB, with normalized enrichment scores of 1.96 and 1.45, respectively. Module 2 was predominantly enriched for immune activation, neutrophils, extracellular matrix, and dendritic cells, all of which have been described to be important pathways in TB (Supplemental Figure 4B). Module 4 was dominated by B cell signatures, which are being recognized as being of increasing importance in the host immune response to TB (Supplemental Figure 4C). Of note, expression of MMPs was increased in module 2, including MMP8 and MMP9, as well as PLAUR, encoding a receptor for urokinase plasminogen activator involved in extracellular matrix degradation (Supplemental Figure 4D). In module 4, genes were involved in immuno-regulation, including IFIT3, MX2, and TNFAIP6 (Supplemental Figure 4E). Quantitative PCR (qPCR) validated upregulation of these genes in TB (Supplemental Figure 4, F and G).
Doxycycline modulates diverse host immune pathways, with effects persisting 6 weeks after doxycycline discontinuation.
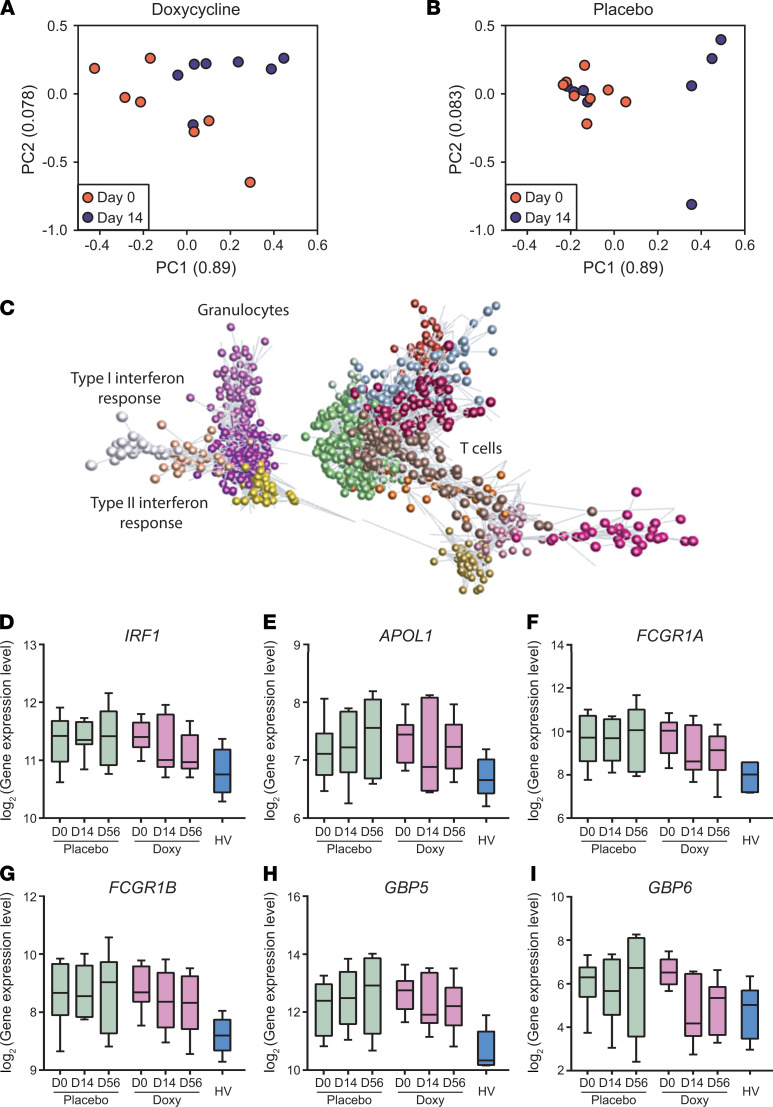
Next, we investigated the effect of doxycycline therapy over time on the blood transcriptome. There was significant donor-to-donor variability, consistent with studying human disease. In PCA analysis, the interindividual differences were reduced between day 0 and day 14 as analyzed in the doxycycline arm (Figure 2A), but in the placebo arm, greater dispersal between patients occurred between day 0 and day 14 (Figure 2B).
Figure 2. Doxycycline results in faster normalization of type II interferon and innate immune response genes relative to placebo.
(A and B) PCA analysis of doxycycline-treated (A) and placebo-treated (B) patients at day 0 (orange) and day 14 (blue). First 2 components of PCA are shown and their variances are shown in parenthesis. Doxycycline reduces variation between individuals over the first 2 weeks of treatment. Only patients with day 0 and day 14 samples are plotted. n = 8 placebo, n = 7 doxycycline. (C) Transcript to transcript clustering of 12,977 genes filtered and normalized using TMM identified 27 coexpressed clusters (Pearson r = 0.8; MCL = 1.7; n gene/cluster: 3/10) over the course of doxycycline treatment. The 15 largest clusters (Supplemental Table 3) are shown in different colors. Lines represent the similarity between transcripts, circles represent individual genes. Two major groupings identify genes preferentially expressed in innate immune response (granulocytes, interferon response) and adaptive immune response (T cells). (D–I) Longitudinal analysis of selected genes, IRF1 (D), APOL1 (E), FCGR1A (F), FCGR1B (G), GBP5 (H), and GBP6 (I) from a cluster encoding for type II interferon and innate immune responses. TMM normalized gene expression at days 0, 14, and 56 of patients with TB in placebo (n = 8, green) and doxycycline (Doxy, n = 7, purple) arms, and baseline expression of healthy volunteers (HV, n = 6, blue) are plotted. Box represents 25th and 75th percentile, line is median, with whiskers denoting extremes.
To investigate this phenomenon further, we performed paired analysis in each study arm to identify genes that were differentially expressed over time. Doxycycline led to a more rapid normalization of immune response genes toward expression levels in healthy volunteers, such as SLC26A8, IGSF6, PYGL, GBP6, and CLEC12B (Supplemental Figure 5, A–E). Similarly, in a parallel unbiased coexpression network analysis using GraphiaPro (51), 27 coexpressed clusters were identified (Figure 2C and Supplemental Table 3). These clusters comprised genes highly expressed in TB that were downregulated during the course of treatment with doxycycline but were upregulated or unaltered in the placebo arm (Figure 2, D–I). Of note, one cluster encoded type I interferon signaling pathway (adjusted P = 5.20 × 10–19), while another cluster encoded cellular response to interferon-γ (adjusted P = 5.05 × 10–14) and innate immune responses (adjusted P = 2.17 × 10–12). In addition, the latter cluster comprised IRF1, APOL1, FCGR1A, FCGR1B, GBP5, and GBP6, genes all related to the innate immune response, indicating doxycycline selectively modulates innate immunity (Figure 2, D–I). Furthermore, although doxycycline was only administered for the first 14 days of treatment, the effects on gene expression were still observed at day 56. Similar observations were found in the qPCR validation of these genes (Supplemental Figure 5, F–K).
To further assess the effect of doxycycline on specific signaling pathways in comparison with placebo, we performed Ensemble of Gene Set Enrichment Analyses (EGSEA) (52), combining results from 11 algorithms to calculate collective gene set scores for biological relevance of the highest ranked gene sets. Mapping of the divergently regulated genes onto the KEGG TB pathway (EGSEA adjusted P = 0.0047 for doxycycline treatment) visualized that doxycycline modulated numerous different stages of the pathway, with expression changes often in the opposite direction to placebo (Figure 3). A parallel analysis of biological processes and pathway enrichment in genes differentially regulated between day 14 and day 0 in doxycycline and placebo arms across all available Gene Ontology and Hallmark signatures demonstrated multiple pathways divergently regulated by doxycycline (Supplemental Figure 6, A and B).
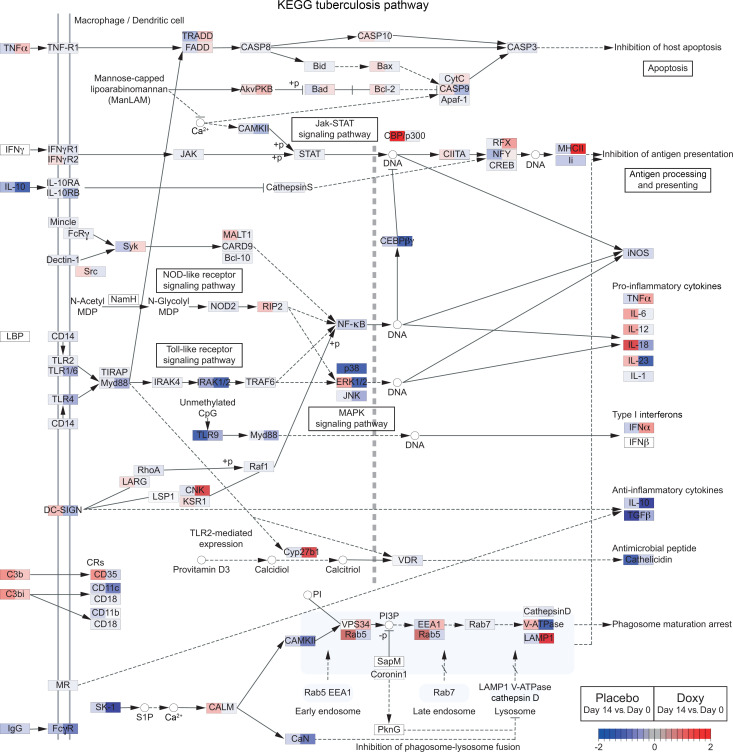
Figure 3. Doxycycline differentially regulates numerous genes in TB pathway relative to placebo.
Comparison of gene expression changes (day 14 vs. day 0) between placebo and doxycycline (Doxy) arms for genes in the KEGG TB pathway (map05152) (47–49). The pathway was identified by EGSEA (52) to be significantly regulated by doxycycline treatment (adjusted P = 0.0047). Gene expression changes (day 14 vs. day 0) in the placebo and doxycycline arms are shown on the left and right side, respectively, of each gene box. Red represents upregulation and blue represents downregulation of gene expression.
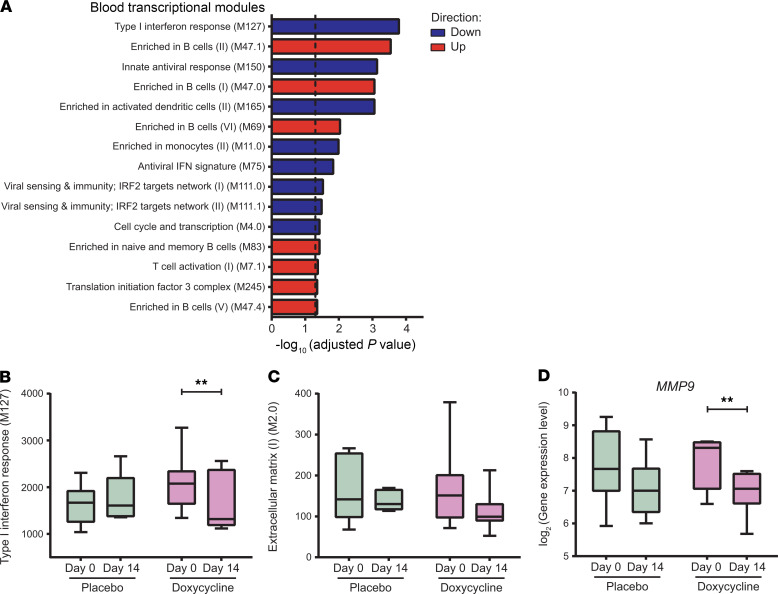
To further dissect which pathways were significantly changed in the doxycycline arm but not placebo, a comparison within linear model was used for identification of doxycycline-specific DEGs (EdgeR, contrasts: (Doxy_Day14-Doxy_Day0)-(Placebo_Day14-Placebo_Day0)). Gene Set Enrichment Analysis using Camera (53) with blood transcriptional modules (54) showed that several pathways were more significantly regulated in the doxycycline arm than the placebo arm (Figure 4A and Supplemental Figure 6C), including interferon responses (Figure 4B), T cell responses and innate responses. In addition, B cell responses increased with doxycycline treatment (Figure 4A). Finally, we studied gene expression of extracellular matrix related genes and showed a trend toward reduction with doxycycline (Figure 4C). Within this pathway, doxycycline led to a significant reduction of MMP9 gene expression compared with placebo (Figure 4D; adjusted P = 1.00 and P = 0.003 for placebo and doxycycline arms, respectively).
Figure 4. Doxycycline leads to greater downregulation of type I interferon responses and extracellular matrix genes, and upregulation of B cells markers relative to placebo.
(A) GSEA of DEGs specific to doxycycline treatment ((Doxy_Day14-Doxy_Day0)–(Placebo_Day14-Placebo_Day0)). Blood transcriptional modules (54) were used as gene sets. The top 15 enriched gene sets are shown. Dotted line marks adjusted P = 0.05. Red represents upregulation and blue represents downregulation of gene sets. (B–D) Longitudinal analysis of type I interferon response gene set (M127) (B), extracellular matrix gene set (M2.0) (C), and MMP9 (D) at day 0 and day 14 of placebo (n = 8, green) and doxycycline (n = 7, purple) arms. Median expression levels of genes in the gene sets are plotted, while TMM normalized gene expressions are shown for MMP9. Box represents 25th and 75th percentile, line is median, with whiskers denoting extremes. **Adjusted P < 0.01.
Systemic MMPs are inhibited by doxycycline
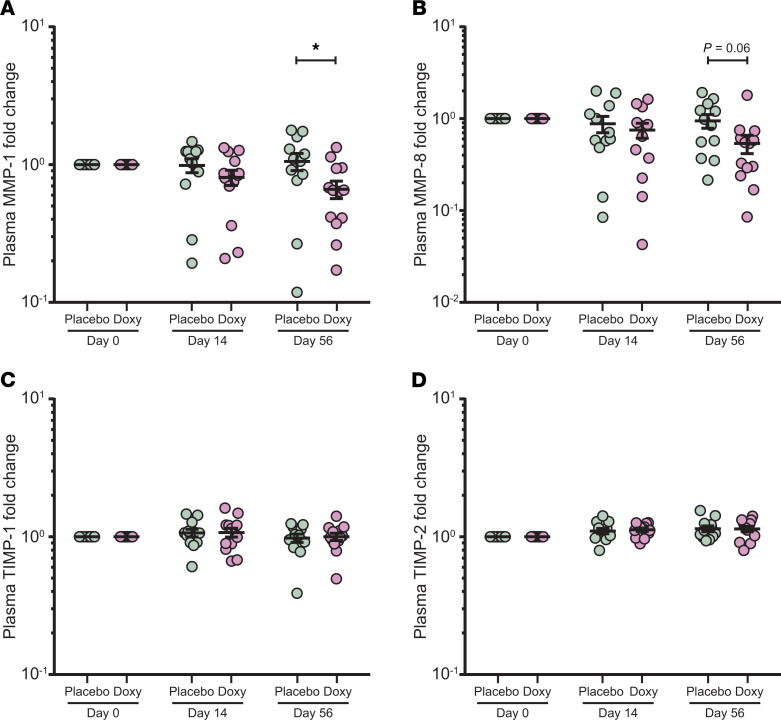
Since the unbiased blood transcriptomic analysis identified upregulated extracellular matrix-related genes in TB, and doxycycline suppressed MMP9 expression, we next investigated the effect of adjunctive doxycycline on plasma MMPs. Plasma MMP-1 was significantly suppressed at day 56 by doxycycline (Figure 5A, adjusted P < 0.05). A similar trend was observed for plasma MMP-8 (Figure 5B, adjusted P = 0.06 at day 56). These data show that systemic MMP-1 continues to be suppressed at a protein level at a late time point, even after doxycycline treatment was stopped after 14 days, consistent with the observations in the blood transcriptomic analysis (Figure 2, D–I). Fold change of the tissue inhibitor of MMPs TIMP-1 and -2 in the plasma were not different between both arms (Figure 5, C and D). Other plasma MMPs were unchanged between arms (Supplemental Figure 7, A–F).
Figure 5. Plasma MMP-1 is suppressed by doxycycline but TIMP-1 and -2 are not affected.
(A–D) Longitudinal analysis of plasma MMP-1 (A), MMP-8 (B), TIMP-1 (C), and TIMP-2 (D) concentrations at days 0, 14, and 56 of placebo (n = 12, green) and doxycycline (Doxy, n = 13, purple) arms. Protein concentrations of each subject were normalized to their day 0 values. Analysis by 2-way ANOVA with Sidak’s multiple comparisons. *Adjusted P < 0.05. Bars represent mean ± SEM.
Next, we analyzed the fold change of circulating matrix degradation products of type III collagen, PIIINP (32), and desmosine from elastin fibers (55). Both were unchanged in the plasma of patients with TB (Supplemental Figure 7, G and H). In addition, Mtb-induced MMP secretion from ex vivo culture of neutrophils (Supplemental Figure 8, A and B) and monocytes (Supplemental Figure 9, A–G) were not different between doxycycline and placebo-treated patients with TB.
Doxycycline suppresses MMPs in respiratory secretions and decreases extracellular matrix degradation with a concurrent decrease in pulmonary cavity volume.
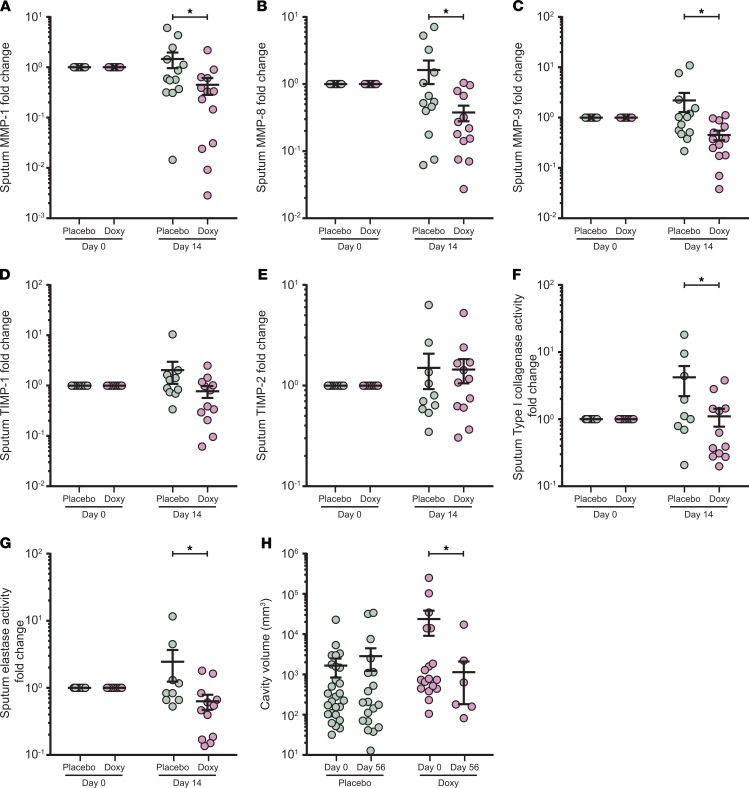
Since MMP activity is tightly regulated and may be compartmentalized (56), we next investigated MMPs in respiratory samples to determine the effect of doxycycline in the lung. Sputum MMP-1, -8, -9, -12, and -13 fold changes were significantly decreased by doxycycline relative to placebo (Figure 6, A–C and Supplemental Figure 10, A and B), while other MMPs were unchanged (Supplemental Figure 10, C–G). From day 0 to day 14, collagenases MMP-1, -8, and gelatinase MMP-9 were decreased in the doxycycline arm (mean fold change of day 14 vs. day 0 ± SEM: MMP-1, 0.45 ± 0.16; MMP-8, 0.38 ± 0.10; MMP-9, 0.45 ± 0.10), while in the placebo arm, they remained unchanged or increased (mean fold change of day 14 vs. day 0 ± SEM: MMP-1, 1.46 ± 0.50; MMP-8, 1.62 ± 0.62; MMP-9, 2.19 ± 0.90). Inhibitors TIMP-1 and -2 were not significantly different between the 2 arms (Figure 6, D and E), indicating an overall suppression of sputum MMP activity by doxycycline.
Figure 6. Sputum MMPs and extracellular matrix degradation activity in patients with TB treated with doxycycline are suppressed with a concurrent decrease in cavity volume.
(A–G) Longitudinal analysis of sputum MMP-1 (A), MMP-8 (B), MMP-9 (C), TIMP-1 (D), TIMP-2 (E), type I collagenase activity (F), and elastase activity (G) at days 0 and 14 of placebo (green) and doxycycline (Doxy, purple) arms. Protein concentrations, as well as functional activity, for each subject were normalized to their day 0 values. n = 13 placebo and n = 13 doxycycline for MMPs; n = 10 placebo and n = 12 doxycycline for TIMPs; n = 12 placebo and n = 9 doxycycline for functional activity. (H) Longitudinal analysis of cavity volume in patients with pulmonary cavities at days 0 and 56. n = 9 placebo (green) and n = 7 doxycycline (Doxy, purple). Analysis by 2-way ANOVA with Sidak’s multiple comparisons. *Adjusted P < 0.05. Bars represent mean ± SEM.
Next, we examined the effects of doxycycline on sputum enzymatic activity degrading type I collagen and elastin, which are key lung extracellular matrix proteins (57, 58). Type I collagen is the substrate of MMP-1 and -8, while elastin is a substrate of MMP-9 (59). In doxycycline-treated patients with TB, sputum type I collagenase and elastase activities were significantly decreased compared with placebo-treated patients (Figure 6, F and G), consistent with the observed decrease in sputum MMP-1, -8, and -9 concentrations (Figure 6, A–C). Furthermore, a decrease in pulmonary cavity volume was observed in the doxycycline arm at day 56, while no difference was observed in the placebo arm (Figure 6H; adjusted P = 0.045). Total sputum PIIINP fold change was unchanged between the 2 arms (Supplemental Figure 10H), while sputum desmosine showed a nonsignificant trend toward suppression in the doxycycline arm (Supplemental Figure 10I; adjusted P = 0.06).
Discussion
Our phase II randomized controlled trial was an exploratory study to investigate the effects of adjunctive doxycycline to standard anti-TB treatment in 30 patients with pulmonary TB. Doxycycline significantly regulated the host transcriptome and biological markers of tissue destruction. We demonstrate for the first time in patients with TB that adjunctive treatment with doxycycline regulated expression of immune and inflammatory response genes, and these effects persisted for 6 weeks after completion of 2 weeks doxycycline intervention. Pathway analyses revealed that doxycycline downregulated type I/II interferon and innate immune response genes and upregulated genes involved in B cell biology. Gene expression of the extracellular matrix–related gene set showed a trend toward reduction in the doxycycline arm, with significant MMP-9 gene suppression. At the protein level, doxycycline suppressed systemic MMP-1, while in the respiratory compartment, collagenases MMP-1 and -8 and gelatinase MMP-9 were reduced. Doxycycline reduced total functional type 1 collagenase and elastase activities in the sputum of patients with TB, with a concurrent significant (P = 0.045) decrease in pulmonary cavity volume. There was no change in BMI, which is reassuring as a low BMI is associated with mortality (60). Our findings in patients with TB extend previous in vitro observations that doxycycline inhibits MMP activity in cellular TB models (14, 29) and suggest that MMP inhibition may be associated with decreased tissue damage in TB. Finally, we show that adjunctive doxycycline with standard anti-TB treatment was safe.
Whole blood RNA-sequencing dissected the molecular pathways affected by adjunctive doxycycline in patients with pulmonary TB. Using coexpression network analysis, we found immune response genes, such as IRF1, APOL1, FCGR1A, FCGR1B, GBP5, and GBP6, to be specifically downregulated by doxycycline treatment toward the expression levels in healthy volunteers. Of note, these genes are often elevated in patients with TB and have been repeatedly identified in blood gene signatures proposed to diagnose TB with high sensitivity and specificity (61–64).
Coexpression network analysis revealed that doxycycline led to greater downregulation of type I/II interferon and innate immune response genes. A parallel blood transcriptional module analysis of the linear model showed type I interferon and innate response genes were more downregulated, while conversely genes involved in B cell biology were more upregulated after doxycycline treatment. In several other blood transcriptional studies, interferon-inducible genes (both type I and II interferon signaling) and innate immune-related genes were highly expressed in patients with TB (65–67) and their expression diminished to that of healthy individuals after successful standard anti-TB treatment (67–70). In addition, B cell markers increased in expression only at the later phases of treatment during disease resolution (69). Thus, the greater change of these pathways after 14 days of additional doxycycline treatment suggests that doxycycline hastens the normalization of gene expression relative to standard anti-TB treatment.
The doxycycline course lasted for 2 weeks, but the change in expression levels of TB-associated immune response genes persisted for 6 weeks after doxycycline discontinuation. Similar long-term suppressive effect was also observed on plasma MMP-1, which is often elevated in patients with active TB (31). These findings suggest that doxycycline host-directed therapy sustains modulations to the host immune responses.
Adjunctive doxycycline specifically suppresses MMPs and their functional activity in patients with pulmonary TB. In particular, MMP-1, -8, -9, -12, and -13, and cleavage of type I collagen and elastin in the respiratory compartment were downregulated by doxycycline, while the tissue inhibitor of MMPs TIMP-1 and -2 remained unchanged. This MMP suppression is likely due to the direct effect of doxycycline (71), since sputum Mtb loads were unchanged. Multiple studies have demonstrated collagenases MMP-1 and -8 and gelatinase MMP-9 to be elevated in respiratory samples of patients with TB, and are closely associated with parameters of immunopathology, such as cavitation and chest radiograph infiltration (14, 29, 30, 33). Although humans express more than 20 MMPs (72), the luminex bead array can only analyze for 10 MMPs, including EMMPRIN, and so our analysis does not cover all MMPs. Overall, our findings of MMP suppression by doxycycline suggest it may be useful in limiting TB immunopathology, and may also reduce the risk of pulmonary impairment after TB (20), as has been proposed for other inflammatory diseases (73, 74). Although PIIINP was listed as primary outcome measure at trial conception, no difference was observed between groups. These may be due to the interval before analysis, as PIIINP has been reported to be unstable in serum or EDTA plasma after 3 months of storage (75).
In serial 18-F fluorodeoxyglucose positron emission tomography–computed tomography (18-F FDG PET-CT) scans of patients with pulmonary TB, high cavity volume is strongly associated with poor treatment outcome (76). In murine studies, treatment with another broad spectrum MMP inhibitor, marimastat, increases the potency of frontline TB drugs isoniazid and rifampicin and enhances the delivery and/or retention of TB drugs in the infected tissue due to improved vascular integrity (39). Conversely, monotherapy of cipemastat, a selective inhibitor of MMP-1, -7, -8, and -13, paradoxically increased cavitation, immunopathology, and mortality in a murine model of cavitary TB (77). Similarly, in the rabbit model of cavitary TB, cipemastat monotherapy also did not prevent cavitation (78). In the current study, adjunctive doxycycline decreased the average volume of pulmonary cavities from 23,694 mm3 at day 0 to 1136 mm3 at day 56, suggesting an inhibitory effect on cavitation by MMP suppression when combined with standard anti-TB treatment. Further investigation of MMP inhibition by doxycycline on TB immunopathology and chronic lung tissue damage is merited.
Doxycycline at 100 mg twice daily is the maximum clinical dose and it is approved by the U.S. Food and Drug Administration (79), and so it was selected for our placebo-controlled trial. The dose required for MMP inhibition in periodontal disease is 20 mg twice daily (80). Murine studies have shown that doxycycline has high organ penetration and reached higher concentrations in both cellular and necrotic TB lesions compared with plasma (81). Combined treatment with rifampin can reduce the levels of doxycycline by approximately 67% in plasma, while clearance of doxycycline was increased approximately 2-fold in patients receiving concurrent rifampin (82). However, the interaction of doxycycline with standard anti-TB treatment was not evaluated in this study, and the pharmacokinetics of doxycycline should be considered in future trials. Regardless, adjunctive doxycycline with standard anti-TB treatment in patients with pulmonary TB is safe from our initial analyses and the safety profile is consistent with recent clinical trials examining doxycycline therapy in other diseases (83, 84).
Our study has limitations, including the sample size of 30 patients with pulmonary TB and the short doxycycline treatment regimen of 2 weeks. Sputum Mtb CFUs and CXR score remained the same in both arms, and although a higher proportion of patients in the doxycycline arm had resolved pulmonary cavities, this did not reach statistical significance, whereas analysis of cavity volume did differ. PET-CT would be more sensitive in detecting changes in cavity size and lung inflammation than CXR analysis used in this study. However, due to concerns of substantial radiation exposure by repeated scans and high operating costs, PET-CT was not used in our study. A larger sample size to sufficiently power future phase 3 clinical trials evaluating the effect of doxycycline on TB immunopathology is needed, which could incorporate weekly collection of sputum for TB cultures to document time to culture conversion. Lung function tests, pulmonary cavitation, time to conversion of sputum culture, pharmacokinetics studies, and adverse drug effects should be considered as outcome measures.
The duration of intervention was based on a prospective study showing the rapid decrease of levels of collagenases MMP-1, -3, and -8 after 2 weeks of anti-TB treatment (33). Given the current data that doxycycline is safe and suppressed biological markers of tissue destruction, a longer treatment duration in a larger cohort could be considered, similar to other clinical trials, which administered doxycycline for at least 4 weeks (41–43). Doxycycline, the only U.S. FDA–licensed drug for MMP inhibition, is routinely used as a broad-spectrum antibiotic and may have off-target effects other than MMPs. Indeed, doxycycline was shown to have comprehensive immunomodulation effects on cytokine and chemokine levels and production (85, 86), and may also influence the composition of the gastrointestinal microbiota (87).
In summary, adjunctive doxycycline with standard anti-TB treatment is associated with accelerated normalization of the increased type I/II interferon response and innate immune response genes in TB, and up-regulation of suppressed B cell markers. Doxycycline specifically suppresses systemic MMP-1 and respiratory MMP-1, -8, and -9, inhibits matrix destruction in respiratory samples of patients with pulmonary TB, and reduces pulmonary cavity volume. Adjunctive doxycycline is well-tolerated in patients with TB on standard anti-TB treatment. Overall, this study is a stepping stone for larger studies to further evaluate and validate the benefits of adjunctive doxycycline treatment for patients with TB. Doxycycline may prove to be a cheap and widely available host-directed therapy targeting TB-associated tissue destruction and the associated dysregulated inflammatory immune response.
Methods
Study design and participants.
This was a randomized, double-blind, placebo-controlled trial of doxycycline as an adjunct to standard anti-TB treatment in patients with pulmonary TB at the National University Hospital (NUH) and the Tuberculosis Control Unit (TBCU), Singapore. Patients were eligible if they were aged 21 to 70 years old, had confirmed pulmonary TB with positive acid-fast bacilli smear and/or positive GeneXpert and/or culture results, had a chest radiograph demonstrating pulmonary involvement, and were within 7 days of initiating anti-TB treatment. Patients with HIV coinfection, previous pulmonary TB, and severe preexisting lung diseases were excluded. Other exclusion criteria included pregnancy or breastfeeding; allergies to tetracyclines; on retinoic acid, neuromuscular blocking agents and pimozide treatment; autoimmune disease and/or on systemic immunosuppressants; hemoglobin lower than 8 g/dL; creatinine 2 times upper limit of normal (ULN); ALT above 3 times ULN; severe depression, schizophrenia, or mania. Healthy volunteers aged 21 to 70 years and in general good health were enrolled at the Investigational Medicine Unit, NUH. The complete list of eligibility criteria can be found in the study protocol (available at https://scholarbank.nus.edu.sg/handle/10635/171211). In total, 30 patients with pulmonary TB and 10 healthy volunteers were recruited from 2015 to 2017. Due to unintentional administrative oversight, the trial was registered in ClinicalTrials.gov (NCT02774993) after the full ethical approval, Singapore Health Science Authority clinical trial certificate (CTC1400221), and study start date. Monitoring of the clinical trial was performed by the Singapore Clinical Research Institute (SCRI).
Study procedures.
Patients with TB were randomly assigned (1:1) using www.randomisation.com to receive daily self-administered doxycycline 100 mg bd or matching placebo (Beacons Pharmaceuticals) for 14 days (Figure 1). Participants, clinicians, and the study team were masked to treatment allocations during the conduct of the trial. Patients with TB were prescribed standard anti-TB treatment in conjunction with the study drug. Healthy volunteers were given 14 days of doxycycline and were not randomized.
Participants were followed up at day 14 and 56 ± 2 days for safety monitoring and sample collection. Criteria for cessation of study intervention include serious adverse events related to the study drug, participant withdrawal of consent, and other reasons specified in the study protocol. A pill count was performed at the end of study intervention to determine adherence. Participants were excluded from downstream assessment if they missed 4 doses of the study drug. Induced sputum was collected from patients with TB at day 0 and 14, while chest radiograph and whole blood were taken at days 0, 14, and 56. For healthy volunteers, blood collection was performed at days 0, 14, and 56. All samples were sent for immediate processing in the BSL-3 laboratory at National University of Singapore (NUS).
Samples collection and processing.
Sputum induction was performed in designated collection rooms, where patients with TB were nebulized with 5% saline in 5-minute cycles, up to 20 minutes as tolerated. Sputum samples were liquefied as previously described (29) and sterile filtered through 0.22 μm Durapore PVDF membrane (Merck Millipore) to remove Mtb (88).
Whole blood was collected separately into Tempus Blood RNA tubes (Applied Biosystems) for RNA-sequencing, and 50 mL tube containing EDTA (1st BASE) for plasma processing, as well as neutrophils and monocytes isolation. Plasma obtained after blood centrifugation was sterile filtered through 0.22 μm Durapore PVDF membrane. Neutrophils and monocytes were respectively isolated from blood samples using CD15- and CD14-antibody conjugated pluriBeads (pluriSelect), following manufacturer’s instructions. Cell viability was greater than 99% by trypan blue assay, and neutrophil and monocyte purities were greater than 95% and greater than 80% by flow cytometry respectively.
Outcome measures.
The primary outcome measure was change of PIIINP in patients with TB. Other outcome measures include change of MMPs and TIMPs in plasma and sputum samples, change of collagenase and elastase activities in sputum samples, change of Mtb-induced MMP secretion from isolated neutrophils and monocytes, change of whole blood transcriptome, and change of Mtb CFU in sputum samples.
Chest radiograph analysis and Mtb CFU measurement.
Chest radiographs of patients with TB were scored for extent of pulmonary consolidation as previously described (29) by 2 specialists blinded to the intervention and the absence or presence of cavitation by 2 of 3 specialists. CXR was scored on a scale of 0 to 10 depending on the number of segments involved, on a modified scoring system developed by Lawson et al. (29, 89). Pulmonary cavity volume was estimated by measuring its radius in ImageJ (90), and calculated as v = 4/3πr3. Liquefied sputum was serially diluted and plated for CFU enumeration on Middlebrook 7H11 agar (BD Biosciences) supplemented with oleic albumin dextrose catalase growth enrichment, polymyxin B, carbenicillin, amphotericin B, and trimethoprim (Sigma-Aldrich).
Whole blood RNA-sequencing.
Blood samples collected in Tempus Blood RNA tubes were processed following the manufacturer’s protocol to obtain the RNA pellet in step 8 (Thermo Fisher Scientific). The pellet was resuspended in 1 mL TRIzol (Thermo Fisher Scientific), and total RNA was extracted by acid guanidinium thiocyanate-phenol-chloroform extraction followed by Qiagen RNeasy Micro clean-up procedure. All RNAs were analyzed on an Agilent Bioanalyzer for quality assessment. Only 15 patients with TB (8 placebo and 7 doxycycline) and 6 healthy volunteers had a complete set of samples (days 0, 14, and 56) that qualified for RNA-sequencing, with RNA Integrity Number (RIN) ranging from 6.4 to 9.2 and median RIN of 8. cDNA libraries were prepared as described previously (91) using SMARTSeq v2 protocol (54). The length distribution of the cDNA libraries was monitored using a DNA High Sensitivity Reagent Kit on the Perkin Elmer Labchip GX system. All samples were subjected to an indexed paired-end sequencing run of 2 × 151 cycles on an Illumina HiSeq 4000 (69 samples in 4 lanes).
Paired-end sequence reads were quantified to transcript abundance using Kallisto (92) with bias correction and 50 bootstrap samples. Reads were mapped to ENSEMBL release 95. On average, the percentage of aligned reads is 89.3%. The transcript abundance was then summarized to gene level using Sleuth (93).
DEG analysis.
Raw counts from RNA-sequencing were processed in Bioconductor package EdgeR (46), variance was estimated, and size factor normalized using TMM. Genes with minimum 2 reads at minimum 50% samples were included in the downstream analyses. All fit models included a term-to-model individual variation. For the identification of DEGs over time (day 14 vs. day 0), a paired model was used, whereas for the comparison of treatment effect between placebo and doxycycline, a nested design was applied ((Doxy_Day14-Doxy_Day0)-(Placebo_Day14-Placebo_Day0)). Genes with a FDR-corrected P value less than 0.05 were identified as differentially expressed, resulting from a likelihood ratio test using a negative binomial generalized linear model fit.
Transcript-to-transcript coexpression analysis
Transcript-to-transcript coexpression analysis was done on filtered, TMM-normalized genes using GraphiaPro (ref. 51; Pearson r = 0.8, Markov clustering algorithm [MCL] = 1.7) and CEMITool (ref. 50; min_ngen = 30, diss_thresh = 0.8, r2 = 0.8247, beta = 3).
Gene ontology and pathway enrichment analysis.
Gene ontology and pathway enrichment analysis were done using Camera (53) or EGSEA (52). Additionally, blood transcriptional modules (BTMs) (54) were used as gene sets. BTM activity was calculated using the BTM package (version 1.015) in Python using the normalized counts as input.
Ex vivo culture and stimulation.
Mtb H37Rv laboratory strain was first cultured in supplemented Middlebrook 7H9 medium (BD Biosciences) and used at mid-logarithmic growth (OD = 0.60) for infection of the isolated neutrophils and monocytes as described (14). Neutrophils were infected at a MOI of 10 for 4 hours, while monocytes were infected at MOI of 1 for 24 hours. Culture supernatants were sterile filtered through 0.22 μm Durapore PVDF membrane to remove Mtb.
ELISAs for TIMP-1/2, PIIINP, and desmosine.
TIMP-1 and -2 concentrations were measured using Duoset ELISA Development System (R&D Systems), with minimum detection limit of 31.2 pg/mL for both; PIIINP was measured using human PIIINP ELISA kit (Cloud-Clone Corporation), with 25.9 pg/mL detection limit; desmosine was measured using human desmosine ELISA kit (Cusabio), with 39.0 pg/mL detection limit. All ELISAs were performed according to the manufacturers’ instructions. Plasma samples were diluted to 1:200 for TIMP-1 and -2, 1:100 for PIIINP, and 1:300 for desmosine. Sputum samples were diluted to 1:500 for TIMP-1, 1:5 for TIMP-2, 1:150 for PIIINP, and 1:100 for desmosine. Results from sputum samples were normalized by their total protein concentrations, which were qualified by Bradford assay (Bio-Rad).
Luminex array for MMPs.
EMMPRIN, MMP-1, -2, -3, -7, -8, -9, -10, -12, and -13 concentrations were analyzed by the Magnetic Luminex Performance Assay (R&D Systems) on the Bio-Plex analyzer (Bio-Rad), according to the manufacturer’s protocol. The minimum detection limit for the 10 analytes were 5.6, 1.1, 12.6, 2.9, 6.6, 7.8, 5.7, 3.2, 0.7, and 63.5 pg/mL, respectively. Culture supernatants from ex vivo neutrophil cultures were diluted to 1:50 for both MMP-8 and -9. For all analytes, supernatants from monocyte cultures were assayed undiluted. Plasma and sputum samples were diluted to 1:5 for all analytes, except for plasma MMP-2 (1:50), and sputum MMP-8 and -9 (1:200 for both). Results from sputum samples were normalized by their total protein concentrations.
DQ collagen and elastin degradation assays.
Type I collagen and elastin degradation by sputum samples were respectively assessed at 1:8 dilution using EnzChek Gelatinase/Collagenase and EnzChek Elastase assay kits (Molecular Probes), as described previously (14). Samples were activated with 2 mM of p-aminophenylmercuric acetate (APMA) for 1 hour at 37°C, before mixing with the DQ collagen (10 μg) or elastin (5 μg). Matrix degradation activity was measured after 24 hours with a fluorescence plate reader (BioTek Instruments). Results were normalized by the total protein concentration.
Statistics.
Results of the trial were reported in accordance with the CONSORT guidelines. Data were analyzed using GraphPad Prism, version 7 (GraphPad Software). Multiple intervention experiments were compared using 2-way ANOVA with Sidak’s multiple comparisons, while continuous variables between 2 sets of data were assessed using 2-tailed Mann-Whitney U test. Categorical data were analyzed using Fisher’s exact test. Data are expressed as the mean ± SEM unless stated otherwise. An adjusted P value of less than 0.05 was considered statistically significant.
Data and materials availability.
All the notebooks used for the RNA-sequencing analysis are available at https://github.com/afvallejo/Doxycycline-as-host-directed-therapy-in-pulmonary-tuberculosis The raw data were deposited at the European Nucleotide Archive (ENA) under the accession number PRJEB38126.
Study approval.
The domain-specific review board from National Healthcare Group Singapore approved this study (reference: 2014/0222) and written informed consent or a thumbprint was obtained from all participants prior to inclusion in the study. Two people on a data safety and monitoring board provided oversight during the study.
Author contributions
CWMO, JSF, and PTE conceived the clinical trial. YD, FSWT, ADYW, SHG, HWS, CBEC, YTW, and CWMO carried out the trial. JSF, NIP, PAT, PTE, and CWMO analyzed the clinical trial. JL, AT, and AS performed the RNA-sequencing. QHM, YW, JMH, CB, and HRL performed the biological assays. QHM, PTE, TZT, AFV, MEP, AS, JSF, and CWMO analyzed the RNA-sequencing data and biological data. QHM, PTE, and CWMO wrote the first draft of the paper which was reviewed and revised by all authors.
Supplementary Material
Acknowledgments
We are grateful to the subjects who participated in this study. We thank the operations team of NUS BSL-3 core facility for logistical support of the study. We express gratitude to Sophia Archuleta and David Lye as members of the data safety and monitoring board and Singapore Clinical Research Institute (SCRI) for study monitoring. We thank Duan Kaibo for initial analysis of the RNA-sequencing data. We acknowledge the use of high-performance computing (HPC) platform and thank Elena Vataga (Computational Modelling Group, University of Southampton, United Kingdom) for assistance. This work was supported by the Singapore National Medical Research Council (NMRC/CNIG/1120/2014, NMRC/Seedfunding/0010/2014, NMRC/CISSP/2015/009a); the Singapore Infectious Diseases Initiative (SIDI/2013/013); National University Health System (PFFR-28 January 14, NUHSRO/2014/039/BSL3-SeedFunding/Jul/01); the Singapore Immunology Network Immunomonitoring platform (BMRC/IAF/311006, H16/99/b0/011, NRF2017_SISFP09); an ExxonMobil Research Fellowship, NUHS Clinician Scientist Program (NMRC/TA/0042/2015, CSAINV17nov014 to CWMO); the UK Medical Research Council (MR/P023754/1, MR/N006631/1 to PTE); a NUS Postdoctoral Fellowship (NUHSRO/2017/073/PDF/03 to QHM); The Royal Society Challenge Grant (CHG\R1\170084 to AFV); the Sir Henry Dale Fellowship, Wellcome Trust (109377/Z/15/Z to MEP); and A*STAR (to AS).
Version 1. 06/15/2021
In-Press Preview
Version 2. 08/02/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(15):e141895.https://doi.org/10.1172/JCI141895.
See the related Commentary at Remodeling the matrix: doxycycline modulates tuberculosis immunopathology.
Contributor Information
Qing Hao Miow, Email: eddymiow@nus.edu.sg.
Andres F. Vallejo, Email: afvp1f17@soton.ac.uk.
Yu Wang, Email: koalawangyu@gmail.com.
Jia Mei Hong, Email: h.jiamei@gmail.com.
Chen Bai, Email: mdcbach@nus.edu.sg.
Felicia S.W. Teo, Email: felicia_sw_teo@nuhs.edu.sg.
Hong Rong Loh, Email: sparkloh@yahoo.com.sg.
Tuan Zea Tan, Email: csittz@nus.edu.sg.
Ying Ding, Email: Ying_Ding@ncid.sg.
Hoi Wah She, Email: Hoi_Wah_SHE@ncid.sg.
Suay Hong Gan, Email: suay_hong_gan@ttsh.com.sg.
Nicholas I. Paton, Email: mdcnijp@nus.edu.sg.
Josephine Lum, Email: Josephine_LUM@immunol.a-star.edu.sg.
Alicia Tay, Email: alicia_tay@immunol.a-star.edu.sg.
Cynthia B.E. Chee, Email: Cynthia_Chee@ncid.sg.
Paul A. Tambyah, Email: mdcpat@nus.edu.sg.
Marta E. Polak, Email: m.e.polak@soton.ac.uk.
Yee Tang Wang, Email: yee_tang_wang@ttsh.com.sg.
Amit Singhal, Email: Amit_Singhal@immunol.a-star.edu.sg.
Jon S. Friedland, Email: jonfriedland@sgul.ac.uk.
Catherine W.M. Ong, Email: Catherine.ong@nus.edu.sg.
References
- 1. World Health Organization. Global Tuberculosis Report 2019. World Health Organization; 2019. [Google Scholar]
- 2.van Kampen SC, et al. International research and guidelines on post-tuberculosis chronic lung disorders: a systematic scoping review. BMJ Glob Health. 2018;3(4):e000745. doi: 10.1136/bmjgh-2018-000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romanowski K, et al. Long-term all-cause mortality in people treated for tuberculosis: a systematic review and meta-analysis. Lancet Infect Dis. 2019;19(10):1129–1137. doi: 10.1016/S1473-3099(19)30309-3. [DOI] [PubMed] [Google Scholar]
- 4.Hawn TR, et al. Host-directed therapeutics for tuberculosis: can we harness the host? Microbiol Mol Biol Rev. 2013;77(4):608–627. doi: 10.1128/MMBR.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallis RS, et al. Tuberculosis — advances in development of new drugs, treatment regimens, host-directed therapies, and biomarkers. Lancet Infect Dis. 2016;16(4):34–46. doi: 10.1016/S1473-3099(16)00070-0. [DOI] [PubMed] [Google Scholar]
- 6.Wallis RS, Hafner R. Advancing host-directed therapy for tuberculosis. Nat Rev Immunol. 2015;15(4):255–263. doi: 10.1038/nri3813. [DOI] [PubMed] [Google Scholar]
- 7.Kaufmann SH, et al. Progress in tuberculosis vaccine development and host-directed therapies — a state of the art review. Lancet Respir Med. 2014;2(4):301–320. doi: 10.1016/S2213-2600(14)70033-5. [DOI] [PubMed] [Google Scholar]
- 8.Tiberi S, et al. Tuberculosis: progress and advances in development of new drugs, treatment regimens, and host-directed therapies. Lancet Infect Dis. 2018;18(7):e183–e198. doi: 10.1016/S1473-3099(18)30110-5. [DOI] [PubMed] [Google Scholar]
- 9.Phelan JJ, et al. Modulating iron for metabolic support of TB host defense. Front Immunol. 2018;9:2296. doi: 10.3389/fimmu.2018.02296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ong CW, et al. Tuberculosis, pulmonary cavitation, and matrix metalloproteinases. Am J Respir Crit Care Med. 2014;190(1):9–18. doi: 10.1164/rccm.201311-2106PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatima S, et al. Mycobacterium tuberculosis programs mesenchymal stem cells to establish dormancy and persistence. J Clin Invest. 2020;130(2):655–661. doi: 10.1172/JCI128043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khader SA, et al. Targeting innate immunity for tuberculosis vaccination. J Clin Invest. 2019;129(9):3482–3491. doi: 10.1172/JCI128877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkington PT, Friedland JS. Permutations of time and place in tuberculosis. Lancet Infect Dis. 2015;15(11):1357–1360. doi: 10.1016/S1473-3099(15)00135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ong CW, et al. Neutrophil-derived MMP-8 drives AMPK-dependent matrix destruction in human pulmonary tuberculosis. PLoS Pathog. 2015;11(5):e1004917. doi: 10.1371/journal.ppat.1004917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urbanowski ME, et al. Cavitary tuberculosis: the gateway of disease transmission. Lancet Infect Dis. 2020;20(6):e117–e128. doi: 10.1016/S1473-3099(20)30148-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan G, et al. Mycobacterium tuberculosis growth at the cavity surface: a microenvironment with failed immunity. Infect Immun. 2003;71(12):7099–7108. doi: 10.1128/IAI.71.12.7099-7108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elkington PT, et al. Tuberculosis immunopathology: the neglected role of extracellular matrix destruction. Sci Transl Med. 2011;3(71):71ps6. doi: 10.1126/scitranslmed.3001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohlwink UK, et al. Matrix metalloproteinases in pulmonary and central nervous system tuberculosis-a review. Int J Mol Sci. 2019;20(6):E1350. doi: 10.3390/ijms20061350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasipanodya JG, et al. Pulmonary impairment after tuberculosis and its contribution to TB burden. BMC Public Health. 2010;10:259. doi: 10.1186/1471-2458-10-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravimohan S, et al. Tuberculosis and lung damage: from epidemiology to pathophysiology. Eur Respir Rev. 2018;27(147):170077. doi: 10.1183/16000617.0077-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meghji J, et al. Patient outcomes associated with post-tuberculosis lung damage in Malawi: a prospective cohort study. Thorax. 2020;75(3):269–278. doi: 10.1136/thoraxjnl-2019-213808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amaral AF, et al. Tuberculosis associates with both airflow obstruction and low lung function: BOLD results. Eur Respir J. 2015;46(4):1104–1112. doi: 10.1183/13993003.02325-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byrne AL, et al. Tuberculosis and chronic respiratory disease: a systematic review. Int J Infect Dis. 2015;32:138–146. doi: 10.1016/j.ijid.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 24.Menezes AM, et al. Tuberculosis and airflow obstruction: evidence from the PLATINO study in Latin America. Eur Respir J. 2007;30(6):1180–1185. doi: 10.1183/09031936.00083507. [DOI] [PubMed] [Google Scholar]
- 25.Sabir N, et al. Matrix metalloproteinases: expression, regulation and role in the immunopathology of tuberculosis. Cell Prolif. 2019;52(4):e12649. doi: 10.1111/cpr.12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubler A, et al. Mycobacterium tuberculosis dysregulates MMP/TIMP balance to drive rapid cavitation and unrestrained bacterial proliferation. J Pathol. 2015;235(3):431–444. doi: 10.1002/path.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kessenbrock K, et al. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parks WC, et al. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol. 2004;4(8):617–629. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 29.Walker NF, et al. Doxycycline and HIV infection suppress tuberculosis-induced matrix metalloproteinases. Am J Respir Crit Care Med. 2012;185(9):989–997. doi: 10.1164/rccm.201110-1769OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elkington P, et al. MMP-1 drives immunopathology in human tuberculosis and transgenic mice. J Clin Invest. 2011;121(5):1827–1833. doi: 10.1172/JCI45666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker NF, et al. Matrix degradation in human immunodeficiency virus type 1-associated tuberculosis and tuberculosis immune reconstitution inflammatory syndrome: a prospective observational study. Clin Infect Dis. 2017;65(1):121–132. doi: 10.1093/cid/cix231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seddon J, et al. Procollagen III N-terminal propeptide and desmosine are released by matrix destruction in pulmonary tuberculosis. J Infect Dis. 2013;208(10):1571–1579. doi: 10.1093/infdis/jit343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ugarte-Gil CA, et al. Induced sputum MMP-1, -3 & -8 concentrations during treatment of tuberculosis. PLoS One. 2013;8(4):e61333. doi: 10.1371/journal.pone.0061333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elkington PT, et al. Mycobacterium tuberculosis, but not vaccine BCG, specifically upregulates matrix metalloproteinase-1. Am J Respir Crit Care Med. 2005;172(12):1596–1604. doi: 10.1164/rccm.200505-753OC. [DOI] [PubMed] [Google Scholar]
- 35.Elkington PT, et al. Synergistic up-regulation of epithelial cell matrix metalloproteinase-9 secretion in tuberculosis. Am J Respir Cell Mol Biol. 2007;37(4):431–437. doi: 10.1165/rcmb.2007-0011OC. [DOI] [PubMed] [Google Scholar]
- 36.Ong CWM, et al. Hypoxia increases neutrophil-driven matrix destruction after exposure to Mycobacterium tuberculosis. Sci Rep. 2018;8(1):11475. doi: 10.1038/s41598-018-29659-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ong CW, et al. Complex regulation of neutrophil-derived MMP-9 secretion in central nervous system tuberculosis. J Neuroinflammation. 2017;14(1):31. doi: 10.1186/s12974-017-0801-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parasa VR, et al. Inhibition of tissue matrix metalloproteinases interferes with Mycobacterium tuberculosis-induced granuloma formation and reduces bacterial load in a human lung tissue model. Front Microbiol. 2017;8:2370. doi: 10.3389/fmicb.2017.02370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y, et al. Matrix metalloproteinase inhibitors enhance the efficacy of frontline drugs against Mycobacterium tuberculosis. PLoS Pathog. 2018;14(4):e1006974. doi: 10.1371/journal.ppat.1006974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sang QX, et al. Matrix metalloproteinase inhibitors as prospective agents for the prevention and treatment of cardiovascular and neoplastic diseases. Curr Top Med Chem. 2006;6(4):289–316. doi: 10.2174/156802606776287045. [DOI] [PubMed] [Google Scholar]
- 41.Gapski R, et al. Systemic MMP inhibition for periodontal wound repair: results of a multi-centre randomized-controlled clinical trial. J Clin Periodontol. 2009;36(2):149–156. doi: 10.1111/j.1600-051X.2008.01351.x. [DOI] [PubMed] [Google Scholar]
- 42.Dalvi PS, et al. Effect of doxycycline in patients of moderate to severe chronic obstructive pulmonary disease with stable symptoms. Ann Thorac Med. 2011;6(4):221–226. doi: 10.4103/1817-1737.84777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhattacharyya P, et al. Long-term use of doxycycline can improve chronic asthma and possibly remodeling: the result of a pilot observation. J Asthma Allergy. 2012;5:33–37. doi: 10.2147/JAA.S31402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lougheed KE, et al. New anti-tuberculosis agents amongst known drugs. Tuberculosis (Edinb) 2009;89(5):364–370. doi: 10.1016/j.tube.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collins L, Franzblau SG. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob Agents Chemother. 1997;41(5):1004–1009. doi: 10.1128/AAC.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson MD, et al. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanehisa M, et al. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47(d1):D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russo PST, et al. CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses. BMC Bioinformatics. 2018;19(1):56. doi: 10.1186/s12859-018-2053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Theocharidis A, et al. Network visualization and analysis of gene expression data using BioLayout Express(3D) Nat Protoc. 2009;4(10):1535–1550. doi: 10.1038/nprot.2009.177. [DOI] [PubMed] [Google Scholar]
- 52.Alhamdoosh M, et al. Combining multiple tools outperforms individual methods in gene set enrichment analyses. Bioinformatics. 2017;33(3):414–424. doi: 10.1093/bioinformatics/btw623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9(1):171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 55.Ma S, et al. The detection and quantitation of free desmosine and isodesmosine in human urine and their peptide-bound forms in sputum. Proc Natl Acad Sci U S A. 2003;100(22):12941–12943. doi: 10.1073/pnas.2235344100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murphy G, Nagase H. Localizing matrix metalloproteinase activities in the pericellular environment. FEBS J. 2011;278(1):2–15. doi: 10.1111/j.1742-4658.2010.07918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mithieux SM, Weiss AS. Elastin. Adv Protein Chem. 2005;70:437–461. doi: 10.1016/S0065-3233(05)70013-9. [DOI] [PubMed] [Google Scholar]
- 58.Seyer JM, et al. Collagen polymorphism in idiopathic chronic pulmonary fibrosis. J Clin Invest. 1976;57(6):1498–1507. doi: 10.1172/JCI108420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cathcart J, et al. Targeting matrix metalloproteinases in cancer: bringing new life to old ideas. Genes Dis. 2015;2(1):26–34. doi: 10.1016/j.gendis.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yen YF, et al. Association of body mass index with tuberculosis mortality: a population-based follow-up study. Medicine (Baltimore) 2016;95(1):e2300. doi: 10.1097/MD.0000000000002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singhania A, et al. The value of transcriptomics in advancing knowledge of the immune response and diagnosis in tuberculosis. Nat Immunol. 2018;19(11):1159–1168. doi: 10.1038/s41590-018-0225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blankley S, et al. A 380-gene meta-signature of active tuberculosis compared with healthy controls. Eur Respir J. 2016;47(6):1873–1876. doi: 10.1183/13993003.02121-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zak DE, et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. 2016;387(10035):2312–2322. doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sweeney TE, et al. Genome-wide expression for diagnosis of pulmonary tuberculosis: a multicohort analysis. Lancet Respir Med. 2016;4(3):213–224. doi: 10.1016/S2213-2600(16)00048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scriba TJ, et al. Sequential inflammatory processes define human progression from M. tuberculosis infection to tuberculosis disease. PLoS Pathog. 2017;13(11):e1006687. doi: 10.1371/journal.ppat.1006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bloom CI, et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One. 2013;8(8):e70630. doi: 10.1371/journal.pone.0070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berry MP, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466(7309):973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bloom CI, et al. Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PLoS One. 2012;7(10):e46191. doi: 10.1371/journal.pone.0046191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cliff JM, et al. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J Infect Dis. 2013;207(1):18–29. doi: 10.1093/infdis/jis499. [DOI] [PubMed] [Google Scholar]
- 70.Cliff JM, et al. The human immune response to tuberculosis and its treatment: a view from the blood. Immunol Rev. 2015;264(1):88–102. doi: 10.1111/imr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hanemaaijer R, et al. Matrix metalloproteinase-8 is expressed in rheumatoid synovial fibroblasts and endothelial cells. Regulation by tumor necrosis factor-alpha and doxycycline. J Biol Chem. 1997;272(50):31504–31509. doi: 10.1074/jbc.272.50.31504. [DOI] [PubMed] [Google Scholar]
- 72.Jackson BC, et al. Update of human and mouse matrix metalloproteinase families. Hum Genomics. 2010;4(3):194–201. doi: 10.1186/1479-7364-4-3-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Henehan M, et al. Doxycycline as an anti-inflammatory agent: updates in dermatology. J Eur Acad Dermatol Venereol. 2017;31(11):1800–1808. doi: 10.1111/jdv.14345. [DOI] [PubMed] [Google Scholar]
- 74.Garrido-Mesa J, et al. Immunomodulatory tetracyclines shape the intestinal inflammatory response inducing mucosal healing and resolution. Br J Pharmacol. 2018;175(23):4353–4370. doi: 10.1111/bph.14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cavalier E, et al. Critical analytical evaluation of promising markers for sarcopenia. Eur Geriatr Med. 2016;7(3):239–242. doi: 10.1016/j.eurger.2015.11.002. [DOI] [Google Scholar]
- 76.Malherbe ST, et al. Quantitative 18F-FDG PET-CT scan characteristics correlate with tuberculosis treatment response. EJNMMI Res. 2020;10(1):8. doi: 10.1186/s13550-020-0591-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ordonez AA, et al. Matrix metalloproteinase inhibition in a murine model of cavitary tuberculosis paradoxically worsens pathology. J Infect Dis. 2019;219(4):633–636. doi: 10.1093/infdis/jiy373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Urbanowski ME, et al. Repetitive aerosol exposure promotes cavitary tuberculosis and enables screening for targeted inhibitors of extensive lung destruction. J Infect Dis. 2018;218(1):53–63. doi: 10.1093/infdis/jiy127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. FDA. Drug Approval Package — Application No.: 050795. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2005/050795s000_DoryxTOC.cfm Updated April 21, 2006. Accessed April 23, 2021.
- 80.Gapski R, et al. Effect of systemic matrix metalloproteinase inhibition on periodontal wound repair: a proof of concept trial. J Periodontol. 2004;75(3):441–452. doi: 10.1902/jop.2004.75.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gelhaus HC, et al. Efficacy of post exposure administration of doxycycline in a murine model of inhalational melioidosis. Sci Rep. 2013;3:1146. doi: 10.1038/srep01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Colmenero JD, et al. Possible implications of doxycycline-rifampin interaction for treatment of brucellosis. Antimicrob Agents Chemother. 1994;38(12):2798–2802. doi: 10.1128/AAC.38.12.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eliassen KE, et al. Comparison of phenoxymethylpenicillin, amoxicillin, and doxycycline for erythema migrans in general practice. A randomized controlled trial with a 1-year follow-up. Clin Microbiol Infect. 2018;24(12):1290–1296. doi: 10.1016/j.cmi.2018.02.028. [DOI] [PubMed] [Google Scholar]
- 84.Baxter BT, et al. Effect of doxycycline on aneurysm growth among patients with small infrarenal abdominal aortic aneurysms: a randomized clinical trial. JAMA. 2020;323(20):2029–2038. doi: 10.1001/jama.2020.5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fredeking TM, et al. Dengue patients treated with doxycycline showed lower mortality associated to a reduction in IL-6 and TNF Levels. Recent Pat Antiinfect Drug Discov. 2015;10(1):51–58. doi: 10.2174/1574891X10666150410153839. [DOI] [PubMed] [Google Scholar]
- 86.Sun J, et al. Tetracyclines downregulate the production of LPS-induced cytokines and chemokines in THP-1 cells via ERK, p38, and nuclear factor-κB signaling pathways. Biochem Biophys Rep. 2015;4:397–404. doi: 10.1016/j.bbrep.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Elvers KT, et al. Antibiotic-induced changes in the human gut microbiota for the most commonly prescribed antibiotics in primary care in the UK: a systematic review. BMJ Open. 2020;10(9):e035677. doi: 10.1136/bmjopen-2019-035677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Elkington PT, et al. Filter sterilization of highly infectious samples to prevent false negative analysis of matrix metalloproteinase activity. J Immunol Methods. 2006;309(1-2):115–119. doi: 10.1016/j.jim.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 89.Lawson L, et al. Clinical presentation of adults with pulmonary tuberculosis with and without HIV infection in Nigeria. Scand J Infect Dis. 2008;40(1):30–35. doi: 10.1080/00365540701509899. [DOI] [PubMed] [Google Scholar]
- 90.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chakarov S, et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science. 2019;363(6432):eaau0964. doi: 10.1126/science.aau0964. [DOI] [PubMed] [Google Scholar]
- 92.Bray NL, et al. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34(5):525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 93.Pimentel H, et al. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. 2017;14(7):687–690. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- 94.Eldridge SM, et al. CONSORT 2010 statement: extension to randomised pilot and feasibility trials. BMJ. 2016;355:i5239. doi: 10.1136/bmj.i5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.