Abstract
Interindividual immune variability is driven predominantly by environmental factors, including exposure to chronic infectious agents such as cytomegalovirus (CMV). We investigated the effects of rhesus CMV (RhCMV) on composition and function of the immune system in young macaques. Within months of infection, RhCMV was associated with impressive changes in antigen presenting cells, T cells, and NK cells—and marked expansion of innate-memory CD8+ T cells. These cells express high levels of NKG2A/C and the IL-2 and IL-15 receptor beta chain, CD122. IL-15 was sufficient to drive differentiation of the cells in vitro and in vivo. Expanded NKG2A/C+CD122+CD8+ T cells in RhCMV-infected macaques, but not their NKG2-negative counterparts, were endowed with cytotoxicity against class I–deficient K562 targets and prompt IFN-γ production in response to stimulation with IL-12 and IL-18. Because RhCMV clone 68-1 forms the viral backbone of RhCMV-vectored SIV vaccines, we also investigated immune changes following administration of RhCMV 68-1–vectored SIV vaccines. These vaccines led to impressive expansion of NKG2A/C+CD8+ T cells with capacity to inhibit SIV replication ex vivo. Thus, CMV infection and CMV-vectored vaccination drive expansion of functional innate-like CD8 cells via host IL-15 production, suggesting that innate-memory expansion could be achieved by other vaccine platforms expressing IL-15.
Keywords: Immunology, Infectious disease
Keywords: Cellular immune response, Innate immunity, T cells
Introduction
The human and nonhuman primate immune systems vary markedly within outbred populations, both phenotypically and functionally. Of significant medical importance, people mount variable immune responses to vaccines and infectious challenges (1, 2). It is perhaps surprising that immune variation in humans is primarily driven by environmental factors (3). For example, we and others have shown that gut microbiota composition has a substantial and long-lasting impact on immune development (4–6). Chronic viral infections such as those caused by cytomegalovirus (CMV) (3) form another class of environmental exposure that alters the host immune system. Interestingly, the activity of subclinical infection with some viruses varies with host immunosuppression, resulting in a complex relationship between these infections and other factors affecting host immunity (7).
CMV is a β-herpesvirus that infects a large proportion of the adult human population, having an average global seroprevalence of 83% (8). The virus has a large genome (~236 kbp) with multiple immunomodulatory gene functions affecting host intrinsic, innate, and adaptive functions (>33% of the genomic coding capacity; ref. 9). Collectively, these viral gene products enable the virus to establish and maintain a lifelong persistence within immune-competent hosts (10). Human CMV (HCMV) infection influences nearly 60% of all immune phenotypes and functional responses, including a major impact on the memory T cell pool, with about 10% of the memory T cell repertoire (both CD4+ and CD8+) being CMV specific (11). This massive overall impact is perhaps most easily understood as the cumulative effect of lifelong expression of CMV-encoded immunomodulatory gene functions; therefore, the immunologic impact of CMV is considered by many to be categorically distinct from that of other viruses. However, another nonexclusive possibility is that the host response to CMV exaggerates mechanisms that are also at play in other viral infections.
HCMV infection is associated with expansion of and adaptive changes in subsets of NK cells, e.g., g-negative NK cells or g-NK cells, a class of cells with memory function that do not rely on germline-encoded, antigen-specific receptors (12). Some investigators have argued that these cells are essentially specific to CMV infection, since CMV-seronegative individuals with prevalent g-NK cells can frequently be shown to possess CMV-specific T cells. HCMV infection is also associated with expansion of adaptive immune cells with innate features, e.g., NK-CTLs, which are CD8+ cytotoxic T lymphocytes (CTLs) with NK-like surface markers and functional activity. NK-CTLs express HLA class I–specific inhibitory receptors most often associated with NK cells (e.g., NKG2A) and have the capacity to kill many allogeneic tumor cells, apparently due to recognition of HLA-E–restricted allogeneic peptides (13–16). HCMV-infected cells markedly downregulate the surface expression of HLA class Ia molecules (ligands for HLA class I–specific inhibitory NK receptors; ref. 17). HLA-E–restricted NK-CTLs may thus provide an additional source of effector cells capable of lysing such class Ia–deficient targets (17).
Other CD8+ T cells with an NK-like surface phenotype (KIR, NKG2A) have been identified in human PBMCs (18–20). Jacomet et al. (20) argued that those cells may represent the equivalent of murine innate/memory CD8+ T cells, also known as virtual memory CD8+ T cells (TVMs) (reviewed in ref. 21). In a recent study, Jin et al. (22) demonstrated a critical role for TVMs in controlling the HIV reservoir through KIR- but not TCR-mediated recognition. Importantly, generation of TVMs is dependent on IL-15 signaling. TVMs express IL-15Rβ (CD122 or IL2Rβ) (23) and eomesodermin (Eomes), a T-box transcription factor that promotes IL-15Rβ expression (24). TVMs also exhibit increased responsiveness to IL-12 and IL-18, which together promote cytokine production and even bystander killing from TVMs in an antigen-independent manner (25, 26). While TVMs lack access to the gut, they preferentially migrate to the liver to gain access to IL-15 (26). Recently, a third subset of innate-like human CD8+ T cells was identified, which expresses an array of NK-associated proteins and has antimicrobial activity against intracellular pathogens (amCTLs; ref. 27). amCTLs additionally express granzyme B (GZMB), perforin (PRF), and granulysin (GNLY), as well as IL-2Rβ. Interestingly, the authors found that epidermal Langerhans cells (LCs) can induce naive CD8+ T cells to differentiate into amCTLs (27).
CMV-vectored HIV vaccines were developed to exploit the unique and inimitable features of CMV-encoded immunomodulation. Salient characteristics of CMV include (a) the ability to reinfect hosts with prior CMV immunity, (b) viral-mediated persistence in hosts with vigorous anti-CMV immune responses, and (c) a large and manipulatable viral genome capable of expressing ectopic antigen cassettes (28). In the rhesus macaque model, rhesus CMV-vectored SIV (RhCMV/SIV) vaccines were shown to provide unprecedented protection against SIV disease in approximately 50% of recipient macaques (29–33). Remarkably, protection is associated in many cases with complete clearance of the pathogenic challenge virus, a feat that in humans has only rarely been achieved after bone marrow transplantation (34). RhCMV/SIV vaccines are based on a specific strain of RhCMV, 68-1, carrying several mutations compared with the WT virus that confer altered cell tropism (9). RhCMV-vectored vaccines provoke robust adaptive immune response to vaccine antigens driven by both MHC class II and especially HLA-E–restricted CD8+ T cell responses (35, 36). Notably, RhCMV-vectored vaccines retain much of the immunomodulatory character associated with WT RhCMV infection, and thus, act broadly on the host immune system.
At present, it remains unclear if such a broad impact on the host immune system could be an important part of the mechanism of protection mediated by RhCMV-vectored vaccines. To investigate how RhCMV infection affects the rhesus immune system, we explored RhCMV-mediated immune modulation at the protein and transcript levels in immune cells of RhCMV-seropositive versus -seronegative macaques. Dramatic expansion of NKG2A/C+CD8+ T cells was observed in animals infected by both WT and 68-1 vaccine strains of RhCMV. The transcriptomic profile of the expanded cells was more similar to that of NK cells than of NKG2A/C– CTLs, including high expression of NK-associated surface markers, granzymes, granulysin, inflammatory cytokines, and cytokine receptors. In fact, the transcriptional profile extensively overlapped that described for TVMs (21) or amCTLs (27). Furthermore, NKG2A/C+ CD8+ T cells exhibited functional capacities that appeared identical to those of TVMs. Thus, WT and vaccine-strain RhCMV causes innate-memory CD8+ T cell expansion that may contribute to RhCMV/SIV-mediated protection against SIV.
Results
Extensive effects of RhCMV infection on the macaque immune system.
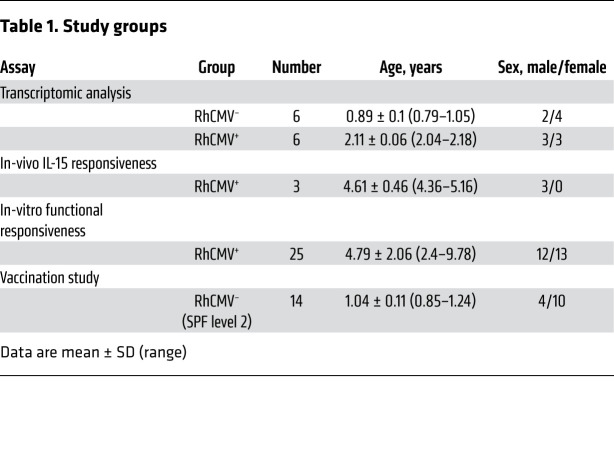
To investigate changes in cells of the circulating immune system associated with RhCMV infection, we characterized peripheral blood immunophenotypes in 42 RhCMV-seronegative (RhCMV–) and 29 RhCMV-seropositive (RhCMV+) juvenile rhesus macaques ranging in age from 5 to 10 months (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI148542DS1). The study animals were housed in outdoor corrals within their natal groups encompassing a broad range of ages (neonate to >20 years) in which RhCMV is endemic. Viral spread is mediated by horizontal transfer of virus in the bodily fluids of RhCMV-seropositive animals (saliva, urine) across mucosal surfaces of cohoused cohorts. Rhesus macaques rapidly seroconvert during the first year of life due to high levels of RhCMV shedding in bodily fluids of RhCMV-infected animals (37, 38). For the age range of animals in this study (5–10 months), prior reported seroprevalence data indicated that the frequency of seroprevalence would have been approximately 25% (5 months) to approximately 90% (10 months; ref. 37). Accordingly, the RhCMV+ macaques in this study would have been infected relatively recently (weeks to months) prior to sample collection. Since both seronegative and seropositive animals were housed together in outdoor corrals, all animals would have been comparably exposed to most other endemic infectious agents.
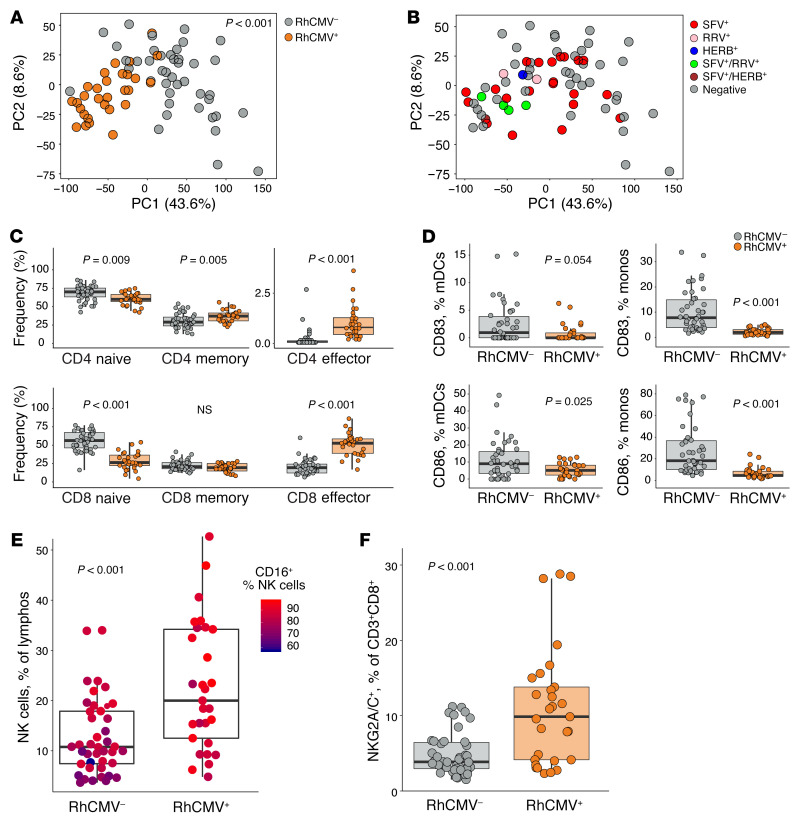
We assessed blood for a comprehensive set of immunophenotypes defined by 5 cytometry panels including T, Treg, NK, and antigen presenting cell (APC) markers and also tested the capacity for intracellular cytokine expression (gating strategies shown in Supplemental Figures 1–5). To summarize the variability in the data set, we first performed principal component analysis (PCA). RhCMV+ and RhCMV– animals were shown to differ substantially in several key components of the immune system, as demonstrated by almost complete separation of the 2 animal groups in a plot of the first 2 principal components (Figure 1A, Adonis P < 0.001). Some rhesus macaques in our study were also infected with additional endemic viral pathogens, including simian foamy virus (SFV), rhesus rhadinovirus (RRV), and/or Macacine alphaherpesvirus 1 (i.e., simian herpes B virus [HERB]). However, clear clusters were not observed when plotting according to infections with these viruses (Figure 1B).
Figure 1. Extensive effects of RhCMV on the macaque immune system.
(A) PCA summarizing variability in all immune parameters measured by flow cytometry. Dot plots represent values along the first and second principal component axes for RhCMV– macaques (gray dots) and RhCMV+ macaques (orange dots). (B) PCA plot with dots colored according to presence of other common viral infections: SFV (red dots), RRV (pink dots), HERB (blue dots), SFV+ and RRV+ (green dots), SFV+ and HERB+ (brown dots), and seronegative (gray dots). (C) Percentage of naive (CD28+CD95–), memory (CD28+CD95+), and effector (CD28–CD95+) cells among CD4+ and CD8+ T cells in RhCMV– and RhCMV+ groups. (D) Percentage of CD83+ and CD86+ among mDCs (CD3–CD20–CD16–CD14–HLADR+CD11c+CD123–) and monocytes (CD3–CD20–HLADR+CD14+) in RhCMV– and RhCMV+ groups. (E) Percentage of NK cells (CD3–CD20–CD8+NKG2A+) in RhCMV– and RhCMV+ groups, colored according to the fraction of NK cells that were CD16+. (F) Percentage of NKG2A/C+CD8+ CTLs (CD3+CD8+NKG2A+) in RhCMV– and RhCMV+ groups. Peripheral blood immunophenotypes were characterized in 42 RhCMV– and 29 RhCMV+ rhesus macaques. Box plots show the median value, 25th and 75th quartiles, and the range of values. Permutational MANOVA (function vegan:adonis) was used to assess correlations between dissimilarity of samples and RhCMV serostatus in A. Wilcoxon rank-sum tests were used to calculate P values in C–G.
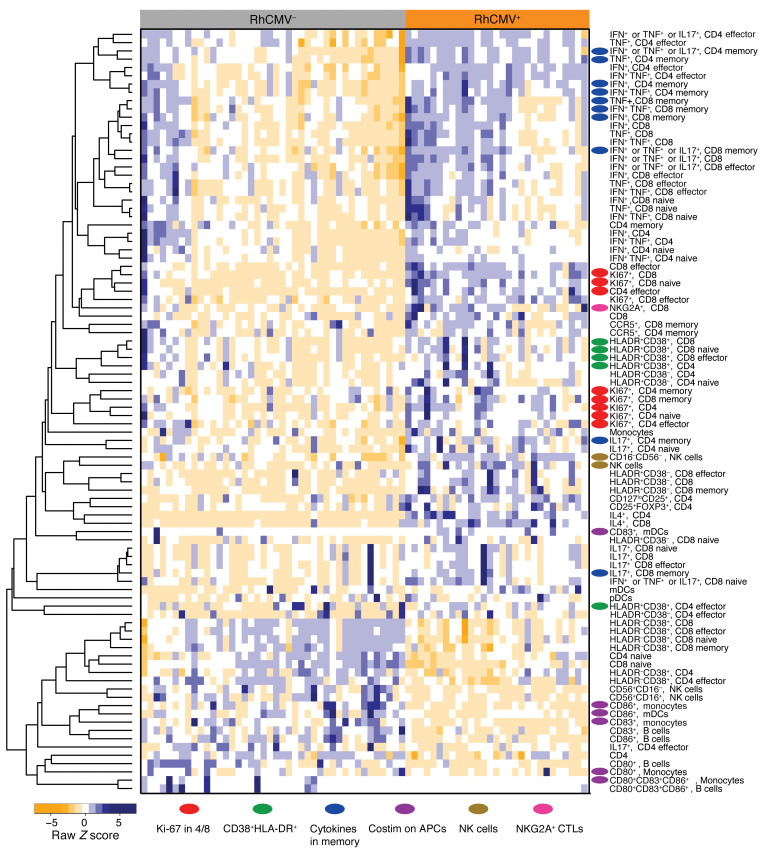
Significant reduction of naive T cells and accumulation of memory and, especially, effector cells were observed among RhCMV+ animals in both the CD4+ and CD8+ subsets (Figure 1C). This impact on immune homeostasis was accompanied by increased T cell proliferation (Ki-67+) and activation (CD38+HLADR+), as well as increased capacity for cytokine production by T cells stimulated with PMA and ionomycin (Figure 2, red or blue oval markers). As we have shown previously (1), only RhCMV+ animals developed circulating T cells capable of IL-4 expression, while simultaneously demonstrating higher frequencies of Th1 and Th17 T cells. Cellular activation and proliferation might be expected in all immune subsets as a natural inflammatory response to infection. Nonetheless, and although APCs (monocytes, myeloid, and plasmacytoid dendritic cells) were more frequently represented among the PBMCs of RhCMV+ animals (Figure 2), expression of costimulatory molecules, especially CD83 and CD86, were substantially lower on these cells (Figure 2, purple oval markers, and Figure 1D) in the infected rhesus macaques. RhCMV+ animals also had more circulating NK cells (Figure 1E) and a significant increase in the abundance of CD16+ compared with CD56+ NK cells (Figure 1E). Finally, we observed an expansion of CD3+CD8+ T cells expressing NKG2A/C (Figure 1F), a cell population that has been reported to be expanded in certain HCMV-seropositive individuals (14, 17). Note that the inhibitory NKG2A and activating NKG2C receptors of nonhuman primates are not distinguished by commercially available antibodies due to similarity of their extracellular domains (39). Approximately 60% of the CD3+CD8+NKG2A/C+ expressed TCR-αβ (average 60% ± 6.1%). Thus, recent RhCMV infection (< ~5 months) was associated with profound changes to a broad spectrum of the host immune systems of rhesus macaques.
Figure 2. Heatmap of immune phenotypes differently expressed by RhCMV groups.
Wilcoxon rank sum tests were used to identify immune subsets found to be significantly different between RhCMV+ and RhCMV- groups. Immune cell subsets that were found significantly different between groups (P < 0.05) were included to generate the heatmap.
Transcriptomes of APC CD11c+ dendritic cells, NK cells, and CD8+ T cells in RhCMV-seropositive versus -seronegative animals.
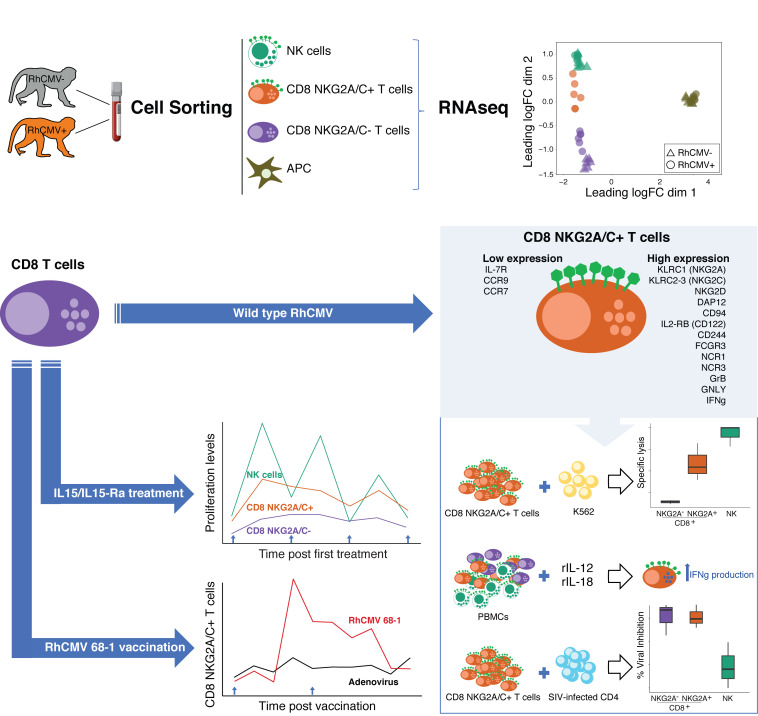
To better understand the immunologic changes imposed by RhCMV infection, we performed transcriptomic analysis by RNA 3′ tag sequencing of fresh, flow-sorted, CD11c+ dendritic cells (CD3–CD20– HLA-DR+CD11c+), NKG2A/C–CD8+ T cells (CD3+CD20–CD8+NKG2A/C–), or NK cells (CD3–CD20–CD8+NKG2A/C+) from 6 RhCMV+ and 6 RhCMV– rhesus macaques (Table 1 and Supplemental Figure 6, A–C for sort strategy and purity checks). The average postsort purity was 96.8% ± 1.9% for APC CD11c+ dendritic cells, 99.1% ± 2.1% for NKG2A/C–CD8+ T cells, and 99.9% ± 0.1% for NK cells. Differential expression analyses were conducted using the limma-voom Bioconductor pipeline, using a statistical model incorporating the flow-sorted cell population, RhCMV serostatus, and fragmentation time. Standard errors of estimates were adjusted for within-animal correlation. Differentially expressed genes for a given contrast were defined to have an adjusted P value of < 0.05.
Table 1. Study groups.
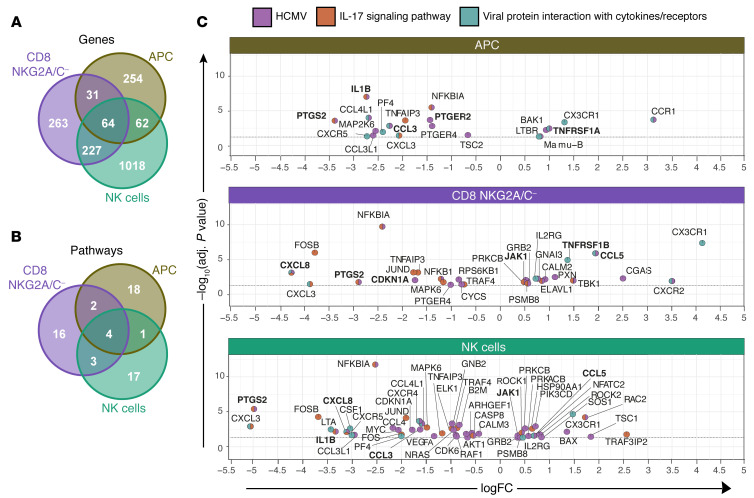
The markers used for sorting were differentially expressed in the expected populations, confirming successful sorting. Other subset-specific markers were seen in the expected patterns, e.g., Lck and ZAP70 in T cells but not dendritic cells. Subsequently focusing on comparable cell subsets from RhCMV+ versus RhCMV– animals, our analysis identified 411, 585, and 1371 protein-coding transcripts differentially expressed by sorted dendritic cells, NKG2A/C–CD8+ T cells, and NK cells, respectively, between RhCMV-infected and -uninfected animals (adjusted P < 0.05; Figure 3A). Sixty-four genes were differentially expressed in all subsets, and in 63 cases, these differences were of the same direction. This shared gene expression profile in different cell types demonstrated shared effects of RhCMV across cellular subsets. KEGG enrichment analyses were performed to determine the biological functions impacted by the identified differentially expressed transcripts in each subset. Interestingly, 4 pathways among the top 25 significant pathways (P < 0.05) were impacted in all 3 cell types: human cytomegalovirus infection (mcc05163), IL-17 signaling (mcc04657), viral protein interaction with cytokines and cytokine receptors (mcc04061), and parathyroid hormone synthesis, secretion, and action (mcc04928; Figure 3, B and C). Identification of common affected pathways in all cell subsets demonstrated the substantial global impact of RhCMV on intrinsic, innate, and adaptive immunity (40). Upregulated and downregulated genes in the first 3 more closely immune-related pathways are shown in volcano plots with pie charts used as markers for genes to indicate pathway membership (Figure 3C). Although the KEGG pathway database does not include IL-10 signaling, many of the coordinately regulated genes in different pathways are members of the Reactome IL-10 signaling pathway (shown in bold in Figure 3C). Indeed, analysis of differentially expressed genes in the Reactome pipeline demonstrates significantly different regulation of IL-10 signaling in the APC subset (P = 0.004). Higher levels of the IL10RA transcript were also found in APCs from RhCMV+ macaques (adjusted P = 0.106).
Figure 3. Gene expression profiles of dendritic cells, NK cells, and T cells in RhCMV– and RhCMV+ rhesus macaques.
(A) Venn diagram showing differently expressed genes (adjusted P < 0.05) in 3 sorted cell types examined for RhCMV– and RhCMV+ macaques. (B) Venn diagram showing the top 25 significant (P < 0.05) KEGG pathways. (C) Volcano-plot representations of differentially expressed genes in the indicated KEGG pathways among CD11c+ DCs (top), NKG2A/C–CD8+ T cells (middle), or NK cells (bottom) in RhCMV+ and RhCMV– macaques. Each colored pie slice indicates membership of the corresponding gene in the KEGG pathway; colors defined in the key. Transcriptomic analysis was performed of flow-sorted cell types from 6 RhCMV+ and 6 RhCMV– rhesus macaques. Differential expression analyses were conducted with the limma-voom Bioconductor pipeline, using a statistical model incorporating the flow-sorted cell population, RhCMV serostatus, and fragmentation time.
Innate transcriptional profile of flow-sorted NKG2A/C+ CD8+ T cells.
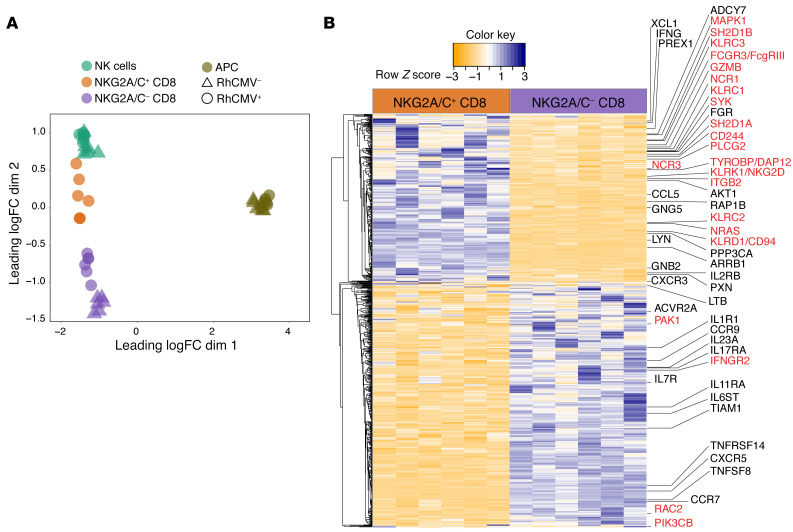
We hypothesized that expression of NKG2A/C by CD8+ T cells (Figure 1F) might reflect broader upregulation of many genes related to NK or other innate cells. Therefore, the transcriptional landscape and function of NKG2A/C+CD8+ T cells was analyzed (Supplemental Figure 6, A–C for sort strategy and purity check, averaging 96.8% ± 1.4%). Multidimensional scaling (MDS) was first used to place the cells within the relative transcriptional landscape of sorted subsets from RhCMV+ or RhCMV– animals (Table 1 and Figure 4A). An MDS plot based on leading log2 fold changes between samples (edgeR:plotMDS function) demonstrated substantial overall similarity between NKG2A/C+CD8+ T cells and NK cells. In fact, NKG2A/C+CD8+ T cells clustered more closely to NK cells than to NKG2A/C–CD8+ T cells, suggesting the existence of an overall NK-like transcriptional program associated with NKG2A/C expression.
Figure 4. Gene expression of flow-sorted NKG2A/C+CD8+ T cells.
(A) MDS plot of transcript abundances using distances based on log2-fold differences. (B) Heatmap showing scaled values for transcripts that were found to be statistically different (adjusted P < 0.05) between NKG2A/C+ and NKG2A/C– CD8+ T cells in 6 RhCMV+ macaques.
Differential gene expression analysis was then performed to identify protein-coding transcripts differentially expressed between NKG2A/C+CD8+ and NKG2A/C–CD8+ T cells from RhCMV+ animals, yielding a total of 862 differentially expressed transcripts (adjusted P < 0.05; Figure 4B). Among the significant 862 differentially expressed transcripts, as expected, KLRC1 (NKG2A) transcripts were significantly more abundant in NKG2A/C+CD8+ T cells than on NKG2A/C–CD8+ T cells. Of note, both KLRC2 and KLRC3 expression (NKG2C-1 and NKG2C-2; ref. 41) were also abundant among NKG2A/C+CD8+ T cells. Increased expression of DAP-12 (TYROBP), a transmembrane adapter that pairs with activating NK receptors, was also detected, as was increased expression of DAP-10 (HOPX). Additionally, we found that numerous genes that likely enhance NK cell responses were upregulated, including KLRD1 (CD94), the natural cytotoxicity triggering receptors 3 and 1 (NCR3 and NCR1, respectively), and cytokine receptors (e.g., IL2RB). Increased expression of genes related to cytolytic effector functions, including granulysin (GNLY) and granzymes A and B (GZMA and GZMB), was also seen. Gene set enrichment analysis for members of all KEGG pathways revealed that the single most enriched pathway was NK cell–mediated cytotoxicity (adjusted P < 0.01). Among the individual genes identified as differentially expressed in NKG2A/C+CD8+ T cells (adjusted P < 0.05), 24 were associated with this KEGG pathway (labeled in red in Figure 4B).
Innate-memory transcriptional profile and IL-15 and IL-12/IL-18 responsiveness of NKG2A/C+CD8+ T cells.
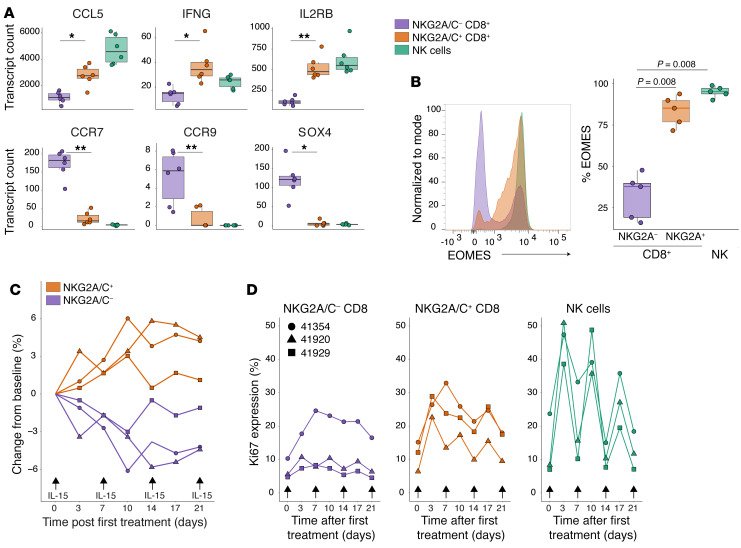
Recently, a population of human T cells with features similar to murine CD8+ TVMs was reported (20, 26). TVMs arise in the periphery, and their generation is dependent on IL-15 signaling. Thus, TVMs express IL-15Rβ (CD122 or IL-2Rβ; ref. 23). The transcriptional profile of TVMs has been reported several times and is well defined (reviewed in ref. 21). The transcriptome of NKG2A/C+CD8+ T cells overlaps in many respects with the TVM transcriptome, including high expression of GNLY as well as certain granzymes (GZMA and GZMB; Figure 4B), inflammatory cytokines (IFNG, CCL5), and cytokine receptors (IL-2Rβ; Figure 5A); lower expression of CCR7 and CCR9 (Figure 5A), which may limit the cells’ retention in lymph nodes and access to the small intestine respectively; decreased expression of SOX4, a transcription factor for memory cell differentiation (Figure 5A); and higher expression of Eomes (Figure 5B).
Figure 5. NKG2A/C+CD8+ T cells exhibit transcriptional and functional features of TVMs.
(A) Abundance of transcripts commonly expressed by TVMs in sorted cell populations from 6 RhCMV+ and 6 RhCMV– rhesus macaques. (B) Left: Representative histogram showing expression of EOMES among gated NKG2A/C+CD8+ T cells, NKG2A/C–CD8+ T cells or NK cells. Right: Percentage of EOMES+ in the 3 gated populations from 5 different macaques. Dynamics (C) and Ki-67 expression (D) of NKG2A/C+CD8+, NKG2A/C–CD8+ T cells and NK cells following treatment of 3 RhCMV+ rhesus macaques with recombinant IL-15 (10 μg/kg) and IL-15Rα (40 μg/kg). Box plots show the median value, 25th and 75th quartiles, and the range of values. Wilcoxon signed-rank tests were used to compare values. *Adjusted P < 0.05, **Adjusted P < 0.001.
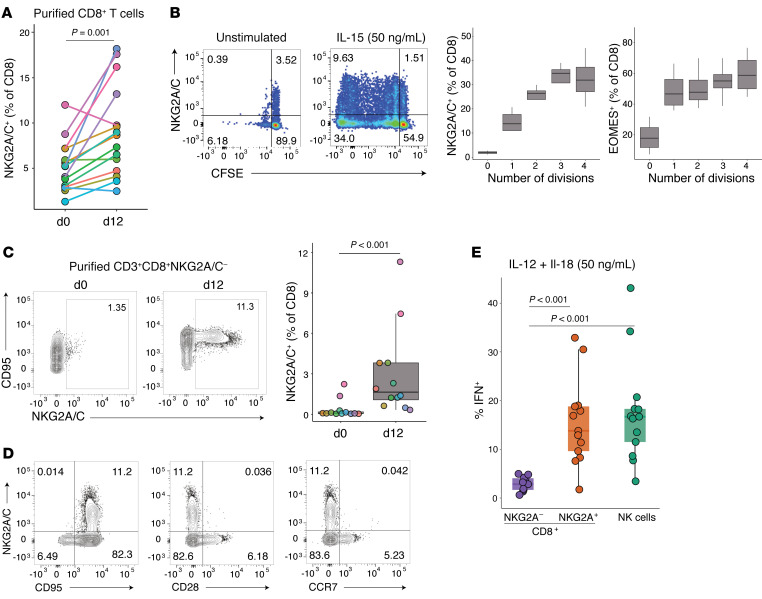
Given the importance of IL-15 in stimulating mouse TVM development (23), we next tested the IL-15 responsiveness of NKG2A/C+CD8+ T cells in vivo (Figure 5, C and D). Rhesus macaques were administered 4 doses of recombinant IL-15 (10 μg/kg) and IL-15Rα (40 μg/kg), and frequencies of T- and NK-cell subsets were followed. All treated macaques underwent expansion of NKG2A/C+CD8+ but not NKG2A/C–CD8+ T cells (Table 1 and Figure 5C), which was driven by increased proliferation as demonstrated by Ki-67 expression (Figure 5D). In vitro, similarly, after 12 days of culturing purified CD8+ T cells with recombinant IL-15 (50 ng/mL), a significant increase in the percentage of NKG2A/C+CD8+ T cells was observed (Table 1 and Figure 6A). NKG2A/C expression on CD8+ T cells increased during IL-15 culture with higher NKG2A and EOMES expression associating with increased proliferation (Figure 6B). IL-15 was able to stimulate de novo NKG2A/C expression after 12 days of culture of sorted NKG2A/C–CD8+ T cells (Figure 6C). IL-15–induced NKG2A/C+CD8+ T cells mainly exhibited a CD95+CCR7–CD28– phenotype (Figure 6D). We next tested if NKG2A/C+ CD8+ T cells share with TVM cells the ability to respond to IL-12 and IL-18 without simultaneous TCR stimulation (25, 26). Total PBMCs were cultured for 48 hours in the presence of IL-12 and IL-18. NKG2A/C+CD8+ T cells responded to IL-12 and IL-18 stimulation by generating IFN-γ, a capacity shared with NK cells but not found among NKG2A/C–CD8+ T cells (Table 1 and Figure 6E).
Figure 6. NKG2A/C+CD8+ T cells exhibit functional features of TVMs.
(A and B) Purified CD8+ T cells from 15 RhCMV+ rhesus macaques were cultured for 12 days with IL-15 (50 ng/mL). (A) Percentage of NKG2A-expressing CD8+ T cells on days 0 or 12. (B) Left: Representative plots showing NKG2A expression on dividing cells, which are marked by CFSE on day 12 in unstimulated and IL-15–stimulated cultures. Right: Graph showing the increase in NKG2A and EOMES expression on CD8+ T cells gated by division number (according to CFSE dilution), box plots show the median value, 25th and 75th quartiles, and the range of values, n = 5. (C and D) Purified NKG2A/C– CD8+ CTLs from 12 RhCMV+ rhesus macaques were cultured for 12 days with IL-15 (50 ng/mL). (C) Left: Representative plots showing NKG2A expression on days 0 and 12. Right: Percentage of NKG2A-expressing CD8+ T cells on days 0 or 12. (D) Flow cytometry plots showing NKG2A/C, CD95, CD28, and CCR7 on day 12. (E) PBMCs from 13 RhCMV+ rhesus macaques were cultured for 48 hours with IL-12 and IL-18. IFN-γ expression was analyzed after gating on NKG2A/C+CD8+ T cells, NKG2A/C–CD8+ T cells, or NK cells. The frequencies of IFN-γ–expressing cells among NKG2A/C+CD8+ T cells when cultured in medium alone was lower than 1.6%. Box plots show the median value, 25th and 75th quartiles, and the range of values. Wilcoxon signed-rank tests were used to compare values.
Cytotoxicity of NKG2A/C+CD8+ T cells elicited by RhCMV infection or RhCMV-vectored vaccine administration.
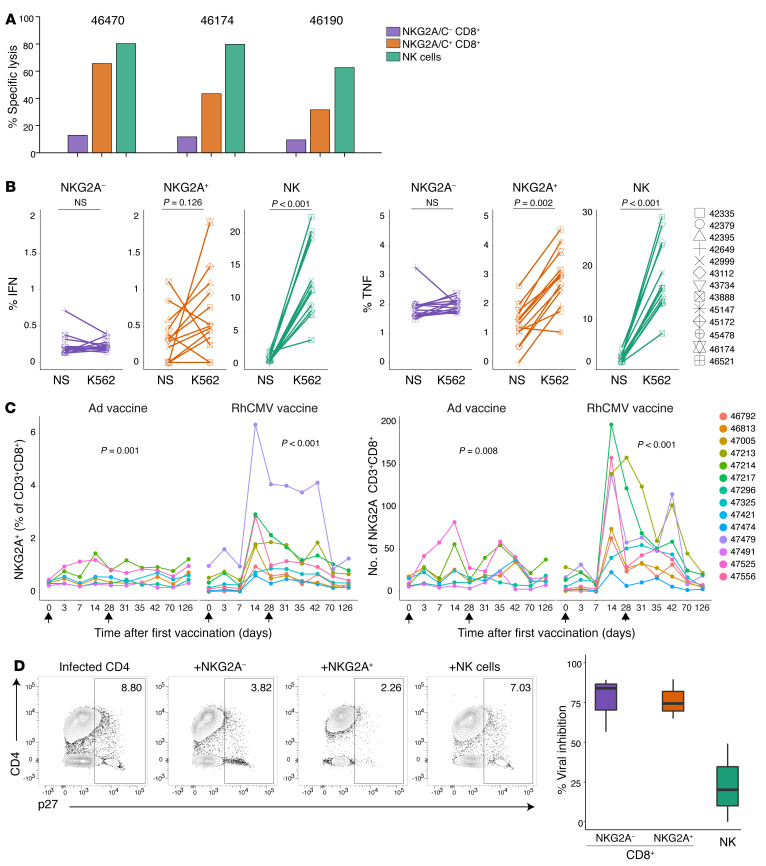
We tested if NKG2A/C+CD8+ T cells have NK-like killing function by analyzing their activity in the presence of MHC class I–deficient K562 target cells (Table 1 and Figure 7, A and B). A Calcein AM-based killing assay was used to assess cytolytic activity against K562 cells of sorted NKG2A/C–CD8+ T cells, NKG2A/C+CD8+ T cells, or NK cells (Figure 7A and Supplemental Figure 7, A and B for sort strategy and purity checks). After sorting, the average purity of NKG2A/C–CD8+ T cells was 99.3% ± 0.4%, for NKG2A/C+ CD8+ T cells was 96.3% ± 1.7% and for NK cells was 96.5% ± 0.8%. NK cells and NKG2A/C+CD8+ T cells, but not NKG2A/C–CD8+ T cells, efficiently killed the targets. Furthermore, both NK cells and NKG2A/C+ cells, but not NKG2A/C– cells, were stimulated to express cytokines by coincubation with K562s (Figure 7B).
Figure 7. NKG2A/C+CD8+ T cells have functional capacities similar to those of NK cells.
(A) Cytolytic activity of sorted NKG2A/C–CD8+ T cells, NKG2A/C+CD8+ T cells, or NK cells from 3 RhCMV+ rhesus macaques against Calcein AM-stained K562 at an E/T ratio of 5:1. (B) Resting PBMCs from 13 RhCMV+ rhesus macaques were incubated for 6 hours alone or with K562. IFN-γ and TNF-α expression were analyzed after gating on NKG2A/C–CD8+ T cells, NKG2A/C+CD8+ T cells, or NK cells. Wilcoxon signed-rank tests were used to compare IFN-γ and TNF-α values. (C) Longitudinal representation of the percentage and absolute numbers of NKG2A/C+CD8+ T cells among lymphocytes before and after vaccination of 14 RhCMV– rhesus macaques. Changes in numbers of NKG2A/C+CD8+ T cells with vaccination time were evaluated with Friedman tests. (D) Left: Representative FACS plots demonstrating viral outgrowth inhibition (assessed by intracellular Gag p27) observed when autologous, infected CD4+ T cells are cultured alone or in the presence of sorted NKG2A/C–CD8+ T cells, NKG2A/C+CD8+ T cells, or NK cells at an E/T ratio of 1:1. Right: Percentage of viral inhibition. The assay was performed with spleen cells and PBMCs from necropsies, box plots show the median value, 25th and 75th quartiles, and the range of values, n = 3.
We next tested if animals receiving a RhCMV-vectored vaccine would generate substantial populations of CD8+ T cells expressing NKG2 receptors, as observed in those infected by WT RhCMV (Figure 1F). These vaccines are based on the 68-1 strain of RhCMV, which lacks epithelial tropism due to the deletion of genes in the ULb′ region that code for members of the pentameric virion-surface complex essential for entry to epithelial cells (9). All RhCMV-vaccinated macaques manifested expansion of NKG2A/C+CD8+ T cells, which in most cases reached peak frequency in the blood 2 weeks after vaccination (Table 1 and Figure 7C). In contrast, only minor expansion of the cells was observed following administration of adenoviral vectors. Furthermore, although the vast majority of antigen-specific T cells elicited by RhCMV/SIV vaccination were NKG2A/C negative (35), viral inhibition assays showed that postvaccination NKG2A/C+ and NKG2A/C– T cells had equivalent capacity to inhibit SIV replication ex vivo (Figure 7D).
Discussion
RhCMV and HCMV infections have broad impact on the infected host’s immune system, which extends to phenotypic changes in nearly all subsets examined including APCs, T cells, and NK cells (3). The ubiquity of this impact is substantial, considering that active replication in immunocompetent hosts, as evidenced in plasma viremia, can be relatively brief after primary exposure (42). Another parameter of persistent infection, shedding of infectious virus in bodily fluids, can be relatively brief or intermittent when primary infection is after childhood, but shedding can persist for years when healthy immunocompetent children are infected during early childhood or in utero (43).
RhCMV-seropositive and -seronegative animals in this study were housed and fed together in outdoor corrals, minimizing the possible influence of environmental confounders on the results. Identical-twin studies have shown that the influences of nonheritable factors, such as HCMV infection, predominate over genetic influences in immunologic impact (3). Our results expand upon those previously obtained in studies of adolescent or adult, non-SPF or SPF rhesus macaques (1, 38). Previous authors demonstrated increased memory and effector CD4+ and CD8+ T cell populations in non-SPF (RhCMV seropositive) compared with SPF level 2 (RhCMV seronegative) animals, including higher IL-4 production among seropositive macaques. Our findings in recently infected infants demonstrate early effects of RhCMV infection on immune homeostasis, including reduction of naive T cells, accumulation of memory and effector cells, and capacity for production of Th2 cytokines. Thus, the effects of CMV seroconversion are established early and persist into adulthood.
Many have hypothesized that development of effector-memory cells in CMV-infected individuals is due to the location, level, or long-term persistence of antigen expression (44). Our data show that RhCMV infection has a broad transcriptional effect on T and NK cells, without an exclusive requirement for antigen responsiveness. Collins et al. recently demonstrated that a transcription factor–based scheme was evolutionarily conserved between innate and adaptive lymphocytes (45), suggesting the possibility that alterations in adaptive cells in CMV infection are not necessarily related to antigen load. Marked changes in the activation phenotype of APCs from RhCMV-infected animals suggest that an altered costimulatory environment contributes to changes seen in the T cell compartment. Altered APC activation in young animals may also affect T cell development within the thymus, as it has been documented in mice that CD83 is essential for thymic maturation of CD4+ T cells (46–48).
Other transcriptomic associations of RhCMV infection were shared across innate and adaptive subsets. CX3CR1, a discriminative marker for HCMV-specific CD8+ T cells (49), was upregulated in sorted dendritic cells, CD8+NKG2A/C– T cells and NK cells from RhCMV-infected macaques. PTGS2, also known as cyclooxygenase-2 (COX-2), was downregulated in all sorted populations. COX-2 is a key rate-limiting enzyme in the eicosanoid synthetic pathway for prostaglandins from arachidonic acid. The presence of a virally encoded COX-2 enzyme is a unique characteristic of RhCMV (50). Human CMV, which does not encode a viral COX-2 isoform, has been shown to induce endogenous COX-2 (cellular, or cCOX-2) expression, and cCOX-2 activity has been shown to be essential for normal virus replication (51, 52). In rhesus macaques, expression of a virally encoded COX-2 may obviate the need for cCOX-2 induction during infection. Reduced PTGS2 expression was seen together with decreased IL1B, PTGER2, CCL3, CCL3L1, CXCL8, and CDKN1A as well as increased CCR1, TNFRSF1A, TNFRSF1B, JAK1, and CCL5—all associated with IL-10 signaling. Both HCMV and RhCMV encode cellular IL-10 (cIL-10) orthologs (cmvIL-10 and rhcmvIL-10, respectively) that bind to IL-10R1, in each case exhibiting comparable functionality to cIL-10 (53–55). Additionally, IL-4 and cIL-10 efficiently inhibit the production of proinflammatory cytokines, such as TNF-α, IL-1α, IL-1β, IL-6, and IL-8 by monocytes/macrophages (56–58) and, indeed, we observed statistically significantly lower expression of IL-1β within sorted dendritic cells (adjusted P = 9 × 10–8). cmvIL-10 also inhibits the expression of MHC class II and the costimulatory molecules CD80, CD86, and CD83 (59). Although only CD83 was shown significantly decreased at the transcript level, we observed clear downregulation of CD86 and CD83 at the protein level. IL-10RA expression was elevated in APCs from RhCMV+ macaques, which suggests that inhibitory signaling to APCs may be one mechanism by which cIL-10 regulates adaptive immunity (60).
RhCMV infection was also associated with expansion of NKG2A/C-expressing CD8+ T cells, which exhibited remarkable transcriptional similarity to NK and TVM cells (21). Effector functions of NKG2A/C+CD8+ T cells were identical to those described for TVMs, including prompt IFN-γ production in response to innate-like stimulation by IL-12 and IL-18 (20). NKG2A/C+CD8+ T cells acquired NK-like cytotoxic activity against MHC class I molecule–deficient K562 cells and secreted cytokines in response to the same targets. NKG2A/C+CD8+ T cells were expanded in RhCMV-seronegative animals vaccinated with RhCMV 68-1–based vaccines. Remarkably, NKG2A/C+CD8+ T cells from vaccinated animals demonstrated comparable capacity to control SIV replication ex vivo to that of NKG2A/C–CD8+ T cells, despite the fact that few NKG2A/C+CD8+ are specific for vaccine epitopes (35).
IL-15 is mainly produced by monocytes/macrophages and dendritic cells (61). Type-I IFNs, production of which is induced by infections such as RhCMV, can regulate IL-15 signaling by causing such APCs to increase the amount of IL-15 available (62). It has been also reported that type-I IFN signaling can increase sensitivity of CD8+ T cells to IL-15 by increasing expression of Eomes, a T-box transcription factor that promotes expression of IL-2Rβ (63).
IL-15 is known to be critical for TVM generation (23) and was shown in this work to be capable of inducing de novo NKG2A/C expression on purified NKG2A–CD8+ T cells (Figure 6C). In previous studies, Correia et al. (64, 65) showed that IL-15 could induce KIR, NKG2A, and NKp46 on peripheral blood CD8+ T cells. NKG2A/C+CD8+ T cells from RhCMV-infected macaques express high levels of the IL2Rβ transcript, whose protein product is required for IL-15 signaling. These cells also express elevated levels of the IL15RA transcript (P = 0.026) and similar levels of the IL2RG transcript compared with NKG2A/C–CD8+ T cells. Indeed, we observed a substantial expansion and proliferation of NKG2A/C+CD8+ T cells following recombinant IL-15/IL-15Rα administration, which was greater than observed for conventional, NKG2A/C–CD8+ T cells. Although the small sample size of our interventional experiment is a limitation, the expansion we observed fits neatly with similar expansion of NK cells, known to be IL-15 responsive, after HCMV infection.
Furthermore, Langerhans cells, which are the most abundant innate immune cells at oral and genital mucosae, natural routes of HCMV acquisition (66), can secrete more IL-15 than other conventional DC subtypes (67), and induce CD8+ naive T cells to express NK-associated markers and acquire antimicrobial activity. The resulting antimicrobial CTLs (amCTLs) are associated with protection against Mycobacterium tuberculosis (68) and Mycobacterium leprae (27). The possibility that CMV-induced NKG2A/C-expressing T cells are functionally similar to amCTLs suggests a mechanism for the promising efficacy of CMV-based vaccines against M. tuberculosis (36).
Thus, expansion of innate-like CD8+ T cells following RhCMV infection or vaccination is driven largely by host IL-15 expression and associated with invocation of an NK- and TVM-like transcriptional program, as well as acquisition of TVM-like effector functions. NKG2A/C+CD8+ T cells are able to kill NK-cell targets (K562), suggesting that the NK-associated receptors expressed on NKG2A/C+CD8+ T cells have similar functional implications for the expressing cell. Furthermore, NKG2A/C+CD8+ T cells were observed to expand after RhCMV 68-1/SIV vaccination and have the potential to control SIV replication ex vivo. A recent report supports the association of RhCMV/SIV vaccine-mediated protection with IL-15 signaling (69). We conclude that NKG2A/C+CD8+ T cells are innate-memory cells of macaques, whose expansion is driven by IL-15 and which may contribute to control over viral infections (26, 70, 71).
Methods
Study design.
The survey of infant macaques was an observational study aimed at identifying immunologic changes in cells of the circulating immune system associated with RhCMV infection. Specifically, we hypothesized between-group differences in the frequency of (a) innate immune cells with adaptive features, such as g-NK cells, and (b) adaptive immune cells with innate features, such as NK-CTLs. Healthy, genetically outbred rhesus macaques (Macaca mulatta) from the CNPRC, tested twice and confirmed to be either RhCMV– or RhCMV+, were used for immunophenotyping studies and transcriptomic analysis. Both RhCMV– and RhCMV+ animals were born, reared, and cohoused in outdoor corrals at the CNPRC and were exposed to the same diet and environment. The sample included all animals of the correct age and housing situation. The vaccine study used outbred rhesus macaques from the SPF level 2 colony at the CNPRC, which is maintained free of CMV, randomized to vaccination with adenovector or RhCMV. Conventional SPF level 1 rhesus macaques (RhCMV+) were used for IL-15 and IL-15Rα in vivo administration study and in vitro functional responsiveness studies. Sample sizes for the latter 2 studies were based upon likelihood of SIV control and of cytokine-driven T cell proliferation, respectively, which were most relevant to the prespecified hypotheses. Conventional RhCMV+ macaques were randomly selected and used for in vitro functional responsiveness studies. Blood samples were taken from each rhesus macaque for plasma and PBMCs. Plasma samples were stored at –70°C. PBMCs were isolated by gradient density purification using Lymphocyte Separation Medium (MP Biomedicals, LLC).
Immune cell phenotyping by flow cytometry.
Naive/memory/effector T cell composition, T cell activation, Tregs, NK cells, and APCs were evaluated using fresh unstimulated rhesus macaque PBMCs stained with predetermined optimal concentrations of the following antibodies: anti-CD3–Alexa 700 (clone SP34-2; catalog 557917, BD Biosciences), anti-CD3–Pacific Blue (clone SP34-2; catalog 558124, BD Biosciences), anti-CD95–APC (clone DX2; catalog 558814, BD Biosciences), anti-CD95–FITC (clone DX2; catalog 555673, BD Biosciences), anti-CD28–APC-H7 (clone CD28.2; catalog 561368, BD Biosciences), anti-CCR5–APC (clone 3A9; catalog 550856, BD Biosciences), anti-Ki67–Alexa 488 (clone B56; catalog 561165, BD Biosciences), anti-CD127–PE (clone HIL-7R-M21; catalog 557938, BD Biosciences), anti-CD25–PE-Cy7 (clone M-A251; catalog 557741, BD Biosciences), anti-CD123–peridinin chlorophyll protein–Cy5.5 (clone 7G3; catalog 558714, BD Biosciences), anti–HLA-DR–PE-Cy7 (clone L243; catalog 335795, BD Biosciences), anti-CD16–PacBlue (clone 3G8; catalog 558122, BD Biosciences), anti-CD83–PE (clone HB15e; catalog 550634, BD Biosciences), anti-CD80–FITC (clone L307.4; catalog 557226, BD Biosciences), anti-CD86–APC (clone 2331 [FUN-1]; catalog 555660, BD Biosciences), anti-CD56–AF700 (clone B159; catalog 557919, BD Biosciences), anti-CD8–PE-Cy5.5 (clone 3B5; catalog MHCD0818, Thermo Fisher Scientific), anti-CD14–Qdot 605 (clone TUK4; catalog Q10013, Thermo Fisher Scientific), anti–CD20 PE-Texas Red (clone HI47; catalog MHCD2017, Thermo Fisher Scientific), LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (catalog L34966, Thermo Fisher Scientific), anti-CD11c–Alexa 700 (clone 3.9; catalog 56-0116-42, Thermo Fisher Scientific), anti-FOXP3–PacBlue (clone 206D; catalog 320116, BioLegend), anti-CD28–Alexa 700 (clone CD28.2; catalog 302920, BioLegend), anti-CD4–Qdot 655 (19Thy5D7; catalog PR-4190, NIH Nonhuman Primate Reagent Resource), anti-CD38–PE (clone OKT10; catalog PR-3802, NIH Nonhuman Primate Reagent Resource), and anti–HLA-DR–ECD (clone Immu-357; catalog IM3636, Beckman Coulter), anti–CD45RA–ECD (clone 2H4; catalog IM2711U, Beckman Coulter), anti-CD159a–PECy7 (clone z199; catalog B10246, Beckman Coulter). FOXP3 Fix/Perm kit (catalog 421403, BioLegend) was used to intranuclearly stain with anti-FOXP3 and anti-Ki67, washed, fixed in PBS containing 1% paraformaldehyde, and acquired on a BD Fortessa cytometer. Analysis was performed using FlowJo version 10.3.
Cytokine flow cytometry assay.
Levels of cytokine-producing cells were determined by cytokine flow cytometry after stimulation with phorbol 12-myristate 13-acetate (PMA) and ionomycin. Rhesus macaque PBMCs were resuspended to 106 cells/mL in RPMI 1640 supplemented with 10% FBS and penicillin/streptomycin (cRPMI). Cells were then incubated for 6 hours at 37°C in medium containing PMA (50 ng/mL) (Sigma-Aldrich), ionomycin (1 μg/mL) (Sigma-Aldrich), and Brefeldin A (5 μg/mL) (BioLegend). After incubation, cells were washed and stained with surface LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (catalog L34966, Thermo Fisher Scientific), anti-CD8–PE-Cy5.5 (clone 3B5; catalog MHCD0818, Thermo Fisher Scientific), anti-CD4–Qdot 655 (19Thy5D7; catalog PR-4190, NIH Nonhuman Primate Reagent Resource), anti-CD3–Pacific Blue (clone SP34-2; catalog 558124, BD Biosciences), anti-CD95–APC (clone DX2; catalog 558814, BD Biosciences), and anti-CD28–APC-H7 (clone CD28.2; catalog 561368, BD Biosciences). Cells were then washed, permeabilized using a Cytofix/Cytoperm kit (catalog 554714, BD Biosciences) according to the manufacturer’s instructions, intracellularly stained with anti-IL-17–PE (clone eBio64CAP17; catalog 12-7178-42, Thermo Fisher Scientific), anti-IL-4 FITC (clone MP4-25D2; catalog 500806, BioLegend), anti-IFN-γ–PE-Cy7 (clone B27; catalog 557643, BD Biosciences) and anti-TNF-α Alexa 700 (clone Mab11; catalog 557996, BD Biosciences), washed, fixed in PBS containing 1% paraformaldehyde, and acquired on a BD Fortessa cytometer. Analysis was performed using FlowJo version 10.3.
3′ RNA tag sequencing.
PBMCs from 6 RhCMV+ and 6 RhCMV– rhesus macaques were stained with surface markers anti-CD3–Pacific Blue (clone SP34-2; catalog 558124, BD Biosciences), anti-CD4–APC (clone L200; catalog 551980, BD Biosciences), anti-CD8–AF700 (clone RPA-T8; catalog 561453, BD Biosciences), anti-CD20–FITC (clone L27; catalog 347673, BD Biosciences), anti-CD11c–PE (clone 3.9; catalog 301606, BioLegend), and anti-CD159a–PECy7 (clone z199; catalog B10246, Beckman Coulter), sorted on FACS Aria (BD Biosciences) into 4 subpopulations with the following profiles: CD3–CD20–HLADR+CD11c+ (CD11c+ DC), CD3+CD20–CD8+NKG2A/C+ (NKG2A/C+CD8+ T cells), CD3+CD20–CD8+NKG2A/C– (NKG2A/C–CD8+ T cells) and CD3–CD20–CD8+NKG2A/C+ (NK cells), and used for RNA-seq analysis. Sorted cells with the profiles given above were resuspended in RNA later and stored at –70°C until RNA extraction. Total RNA was extracted from these sorted cells using the Total RNA Purification Micro Kit (Norgen Biotek) according to the manufacturer’s protocol. The quantity and quality of total RNA extracted was checked using the Nanodrop 2000 (Thermo Fisher Scientific) and the Agilent 2100 Bioanalyzer (Agilent Technologies). Libraries were prepared by the UC Davis DNA Technologies Core using the Quant Seq kit (Lexogen Inc.) following the manufacturer’s protocol for 3′ mRNA-sequencing. Libraries were pooled and sequenced using the Illumina HiSeq 4000 (Illumina) with 90 bp, single-ended stranded sequencing. Sequence quality was checked using fastqc (72) and aligned to Macaca mulatta genome assembly version Mmul_10 annotated with Macaca_mulatta.Mmul_10.99.gtf using STAR (73) to generate a matrix of feature counts. Differential gene expression was analyzed using the R package edgeR (74). Read counts were normalized using the TMM method. Prior to analysis, low-variability genes (defined here as having less than 10 normalized reads in all samples) were filtered, leaving 10,596 genes. Differential expression analyses were conducted with the limma-voom Bioconductor pipeline, using a model with factors for marker-RhCMV status interaction and fragmentation time. Standard errors of estimates were adjusted for within-animal correlation. Differentially expressed genes were defined to have an adjusted P value less than 0.05.
Recombinant IL-15 and IL-15Rα in vivo administration.
Starting from day 0, 3 animals were treated with 4 weekly doses (subcutaneous) of recombinant IL-15 (10 μg/kg) and IL-15Rα-Fc (40 μg/kg). Peripheral blood sample collections were performed on the day of administration of each dose and at 3 and 7 days after each dose, and PBMCs were cryopreserved. Frozen PBMCs were stained with surface markers LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (catalog L34966, Thermo Fisher Scientific), anti-CD3–APC-Cy7 (clone SP34-2; catalog 557757, BD Biosciences), anti-CD8–PerCP-Cy5.5 (clone SK1; catalog 341049, BD Biosciences) and anti-CD159a–PE (clone z199; catalog IM3291U, Beckman Coulter). Cells were then washed, permeabilized using a Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer’s instructions, intracellularly stained with anti-Ki67–Alexa 488 (clone B56; catalog 561165, BD Biosciences), washed, fixed in PBS containing 1% paraformaldehyde, and acquired on a BD Fortessa cytometer. Analysis was performed using FlowJo version 10.3.
Cytokine production and cytotoxicity assay.
For the cytokine production assay, PBMCs were stimulated with target tumor cells (K562, source CNPRC) at a ratio of 10:1 (E/T) in cRPMI for 6 hours at 37°C in the presence of Brefeldin A (BioLegend) or with rhesus rIL-12 and rIL18 (20 ng/mL or 50 ng/mL, each) from R&D Systems for 44 hours at 37°C followed by 4 hours of stimulation in the presence of Brefeldin A (BioLegend). After incubation, cells stimulated with K562 were washed and stained with surface markers LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (catalog L34966, Thermo Fisher Scientific), anti-CD3–Alexa 700 (clone SP34-2; catalog 557917, BD Biosciences), anti-CD20–APC-H7 (clone 2H7; catalog 560734, BD Biosciences), anti-CD159a–PE-Vio770 (clone REA110; catalog 130-105-694, Miltenyi Biotec), anti-CD8–Pacific Blue (clone SK1; catalog 740093, BD Biosciences). Cells were then washed, permeabilized using a Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer’s instructions, intracellularly stained with anti-IFN-γ–APC (clone B27; catalog 554702, BD Biosciences) and anti-TNFα–PE (clone Mab11; catalog 554513, BD Biosciences), washed, fixed in PBS containing 1% paraformaldehyde, and acquired on a BD Fortessa cytometer. Analysis was performed using FlowJo version 10.3.
After 48 hours incubation, cells stimulated with rhesus rIL-12 and rIL18 were washed and stained with surface markers LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (catalog L34966, Thermo Fisher Scientific), anti-CD8–PerCP-Cy5.5 (clone SK1; catalog 341049, BD Biosciences), anti-CD3–BUV737 (clone SP34-2; catalog 741872, BD Biosciences), anti-CD4–BV650 (clone L200; catalog 563737, BD Biosciences), and anti-CD159a (NKG2A)–VioBright FITC (clone REA110; catalog 130-113-568, Miltenyi Biotec). Cells were then washed, permeabilized using a Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer’s instructions, intracellularly stained with anti-IFN-γ–PE-Cy7 (clone B27; catalog 557643, BD Biosciences), washed, fixed in PBS containing 1% paraformaldehyde, and acquired on a BD FACSymphony Flow Cytometer. Analysis was performed using FlowJo version 10.3.
For the cytotoxicity assay, PBMCs were stained with surface markers anti-CD3–Pacific Blue (clone SP34-2; catalog 558124, BD Biosciences), anti-CD4–BV605 (clone L200; catalog 562843, BD Biosciences), anti-CD8–AF700 (clone RPA-T8; catalog 561453, BD Biosciences) and anti-CD159a–PECy7 (clone z199; catalog B10246, Beckman Coulter), and sorted on FACS Aria (BD Biosciences) into 3 subpopulations with the following profiles: CD3+CD8+NKG2A/C+ (NKG2A/C+CD8+ T cells), CD3+CD8+NKG2A/C– (NKG2A/C–CD8+ T cells), and CD3–CD8+NKG2A+ (NK cells). Sorted cells were incubated in cRMPI with K562 tumor cells stained with 2 μM calcein AM (BD Biosciences) at a ratio of 5:1 (E/T) in a 96-well U-bottom plate for 4 hours. Supernatant was collected from wells containing both effector and target cells (specific lysis), wells containing only target cells (spontaneous lysis), and wells containing target cells lysed with 0.1% Tween-20 (total lysis), and fluorescence was measured at 495/515 nm. The percentage of specific lysis was calculated using the measured fluorescence as follow: 100 × (specific lysis – spontaneous lysis) / (total lysis – spontaneous lysis).
Recombinant IL-15–induced NKG2A expression assay.
CD8+ T cells were purified from PBMCs using MACS nonhuman primate CD8 MicroBead kit (Miltenyi Biotec) according to the manufacturer’s instructions or sorted on FACS Aria (BD Biosciences) into CD3+CD8+NKG2A/C– (NKG2A/C–CD8+ T cells) and cultured for 12 days with recombinant IL-15 (50 ng/mL). After incubation, cells were washed and stained with surface markers for LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (catalog L34966, Invitrogen), anti-CD3–Pacific Blue (clone SP34-2; catalog 558124, BD Biosciences), anti-CD4–BV605 (clone L200; catalog 562843, BD Biosciences), anti-CD8–FITC (clone SK1; catalog 344704, BioLegend), anti-CD159a–PE-Cy7 (clone z199; catalog B10246, Beckman Coulter), anti-CD95–APC (clone DX2; catalog 558814, BD Biosciences), anti-CD28–APC-H7 (clone CD28.2; catalog 561368, BD Biosciences), and anti-CCR7–BV786 (clone 3D12; catalog 563710, BD Biosciences). For proliferation studies, purified CD8+ T cells were isolated using MACS nonhuman primate CD8 MicroBead kit (Miltenyi Biotec) according to the manufacturer’s instructions and labeled with 2.5 μM CFSE (Invitrogen). After incubation for 12 days in the presence of recombinant IL-15 (50 ng/mL), cells were washed and stained with surface markers for LIVE/DEAD Fixable Far Red Dead Cell Stain Kit (catalog L34973, Invitrogen), anti-CD3–Pacific Blue (clone SP34-2; catalog 558124, BD Biosciences), anti-CD4–BV605 (clone L200; catalog 562843, BD Biosciences), anti-CD8–BUV805 (clone SK1; catalog 612890, BD Biosciences), and anti-CD159a–PE-Cy7 (clone z199; catalog B10246, Beckman Coulter). Cells were then washed, permeabilized using BD Pharmingen Transcription Factor Buffer Set according to the manufacturer’s instructions, intranuclearly stained with anti-EOMES–efluor660 (catalog 50-4877-42, Thermo Fisher Scientific), washed, fixed in PBS containing 1% paraformaldehyde, and acquired on a BD FACSymphony Flow Cytometer. Analysis was performed using FlowJo version 10.3.
Vaccine administration.
Starting from day 0, 6 and 8 SPF level 2 (RhCMV seronegative) animals received either an Adenoviral-vectored or a RhCMV-vectored vaccine prime and a booster on day 28 after priming. Whole blood samples were collected on the day of vaccination, and 3, 7, 14, and 28 days after vaccine prime and vaccine boost. Surface staining with markers anti-CD3–BUV737 (clone SP34-2; catalog 741872, BD Biosciences), anti-CD4–BUV395 (clone L200; catalog 564107, BD Biosciences), anti-CD8–BUV805 (clone SK1; catalog 612890, BD Biosciences), and anti-CD159a (NKG2A)–VioBright FITC (clone REA110; catalog 130-113-568, Miltenyi Biotec) was performed for 50 μL whole blood samples. Q-Prep was used according to the manufacturer’s recommended protocol that was provided in the product insert (Beckman Coulter, Inc.) and acquired on a BD FACSymphony Flow Cytometer.
Virus inhibition assay.
Virus-inhibitory activity (VIA) was measured as follows. PBMC targets were depleted of CD8+ cells using a MACS nonhuman primate CD8 MicroBead kit (Miltenyi Biotec) according to the manufacturer’s instructions. Targets were then resuspended in cRPMI and stimulated with PHA (5 μg/mL) in the presence of recombinant human interleukin-2 at a concentration of 20 U/mL (Miltenyi Biotec). After stimulation for 2 days, the medium was replenished and target PBMCs were infected at 1 to 3 × 106 cells/mL using a SIVmac251 stock at a MOI of 0.01 for one day at 37°C and 5% CO2. Following infection, cells were resuspended and plated at 5 × 104 cells per well in a U-bottom, 96-well tissue culture plate. The same day, PBMCs stained with surface markers anti-CD3–Pacific Blue (clone SP34-2; catalog 558124, BD Biosciences), anti-CD4–BV605 (clone L200; catalog 562843, BD Biosciences), anti-CD8–FITC (clone SK1; catalog 344704, BioLegend), and anti-CD159a–PECy7 (clone z199; catalog B10246, Beckman Coulter) were sorted into 3 subpopulations with the following profiles: CD3+CD8+NKG2A/C+ (NKG2A/C+CD8+ T cells), CD3+CD8+NKG2A/C– (NKG2A/C–CD8+ T cells), and CD3–CD8+NKG2A+ (NK cells). Effector cells, sorted as described above, were diluted and added to autologous infected targets at an E/T ratio of 1:1. Infectivity controls consisted of infected targets without added effectors. Duplicate cocultures were incubated at 37°C and 5% CO2 for 6 days. After incubation, cells were washed and stained with surface markers for LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (catalog L34966, Invitrogen), anti-CD3–Pacific Blue (clone SP34-2; catalog 558124, BD Biosciences), anti-CD4–BV605 (clone L200; catalog 562843, BD Biosciences), and anti-CD8–FITC (clone SK1; catalog 344704, BioLegend). Cells were then washed, permeabilized using a Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer’s instructions, intracellularly stained with anti-SIV Gag p27 (clone 55-2F12; catalog 1610, NIH AIDS Research Reagent Program), biotin-conjugated using the One-Step Antibody Biotinylation Kit (Miltenyi Biotec), washed and stained with APC-streptavidin (catalog 554067, BD Biosciences), washed, fixed in PBS containing 1% paraformaldehyde, and acquired on a BD FACSymphony Flow Cytometer. The percentage of viral inhibition was calculated using the following formula: 100 × (fraction of p27+ cells among CD4+ T cells cultured alone) — (fraction of p27+ cells among CD4+ T cells cultured with effector cells) / (fraction of p27+ cells among CD4+ T cells cultured alone).
Data availability.
The expression data generated by RNA-seq analysis are available in the GEO repository under accession number GSE171978.
Statistics.
R was used for all statistical analysis, and all plots were created using the ggplot2 package (75). Wilcoxon rank-sum tests were used for comparisons of immune phenotypes by RhCMV status. Wilcoxon signed-rank test was used for comparisons in paired observations as indicated in figure legends. Friedman tests were used for comparisons of repeated measured observations. For all statistical tests, differences between groups were considered significant when the P value was less than 0.05. PCA was performed using the ade4 package (76). The function adonis from the R package, Vegan (77), was used to assess correlations between dissimilarity of samples and factors such as CMV infection. Heatmaps were generated in R using the heatmap2 package.
Study approval.
The UC Davis animal care program is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care, International (AAALAC). All animal procedures were approved prior to implementation by the UC Davis Institutional Animal Care and Use Committee and were consistent with the requirements of the Animal Welfare Act. Activities related to animal care were performed as per standard operating procedures at the California National Primate Research Center.
Author contributions
GML, SK, CB, PAL, PAB, and DJHO designed research studies. GML, WLWC, JL, HTK, WL, and LA conducted experiments and performed data acquisition. GML, WLWC, NC, DM, and JL analyzed the data and prepared figures and publication materials. MRU provided advice for VIA measurement and analysis. GML wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Lewis Lanier for insightful comments on the manuscript. We thank the veterinary, Colony Research Clinical Laboratories, and pathology faculty and staff at CNPRC for expert technical assistance. The authors also thank Francois Villinger for provision of recombinant Mamu IL-15 and Mamu IL-15-Rα-Fc. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: anti-SIVmac p27 monoclonal (55-2F12), mouse anti-human CD4 Qdot 655 (19Thy5D7), and mouse anti-human CD38 PE (clone OKT10). The sequencing was carried by the DNA Technologies and Expression Analysis Core at the UC Davis Genome Center, supported by NIH Shared Instrumentation Grant 1S10OD010786-01. This work was supported by NIH grants AI 118451 (to DJHO), AI 143554 (to DJHO and PAB), AI 131568 (to DJHO, PAB, and CB), AI 150554 (to DJHO), OD P51 OD011107 (to CNPRC), by the California HIV/AIDS Research Foundation under award F13-D-312 (to GML), and by the UCSF-Gladstone Institute for Virology and Immunology, Center for AIDS Research (CFAR) under award 4P30AI027763-25 (to GML).
Version 1. 06/21/2021
In-Press Preview
Version 2. 08/02/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(14):e148542.https://doi.org/10.1172/JCI148542.
Contributor Information
Gema Méndez-Lagares, Email: gemendez@ucdavis.edu.
Ning Chin, Email: nchin@ucdavis.edu.
W.L. William Chang, Email: wlchang@ucdavis.edu.
Jaewon Lee, Email: jwwlee@ucdavis.edu.
Míriam Rosás-Umbert, Email: mru@clin.au.dk.
Hung T. Kieu, Email: htkieu@ucdavis.edu.
David Merriam, Email: david.merriam@ucdenver.edu.
Wenze Lu, Email: wzelu@ucdavis.edu.
Sungjin Kim, Email: sjikim@ucdavis.edu.
Lourdes Adamson, Email: laadamson@ucdavis.edu.
Christian Brander, Email: cbrander@irsicaixa.es.
Paul A. Luciw, Email: paluciw@ucdavis.edu.
Peter A. Barry, Email: pabarry@ucdavis.edu.
Dennis J. Hartigan-O’Connor, Email: dhartigan@ucdavis.edu.
References
- 1.Santos Rocha C, et al. Subclinical cytomegalovirus infection is associated with altered host immunity, gut microbiota, and vaccine responses. J Virol. 2018;92(13):e00167-18. doi: 10.1128/JVI.00167-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakaya HI, et al. Systems analysis of immunity to influenza vaccination across multiple years and in diverse populations reveals shared molecular signatures. Immunity. 2015;43(6):1186–1198. doi: 10.1016/j.immuni.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodin P, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160(1-2):37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ardeshir A, et al. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci Transl Med. 2014;6(252):252ra120. doi: 10.1126/scitranslmed.3008791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narayan NR, et al. Persistent effects of early infant diet and associated microbiota on the juvenile immune system. Gut Microbes. 2015;6(4):284–289. doi: 10.1080/19490976.2015.1067743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Vlaminck I, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. 2013;155(5):1178–1187. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuhair M, et al. Estimation of the worldwide seroprevalence of cytomegalovirus: a systematic review and meta-analysis. Rev Med Virol. 2019;29(3):e2034. doi: 10.1002/rmv.2034. [DOI] [PubMed] [Google Scholar]
- 9.Barry PA, et al. Cytomegalovirus-vectored vaccines for HIV and other pathogens. AIDS. 2020;34(3):335–349. doi: 10.1097/QAD.0000000000002396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang A, et al. Genome sequence divergences and functional variations in human cytomegalovirus strains. FEMS Immunol Med Microbiol. 2009;55(1):23–33. doi: 10.1111/j.1574-695X.2008.00489.x. [DOI] [PubMed] [Google Scholar]
- 11.Sylwester AW, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005;202(5):673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang T, et al. Cutting edge: antibody-dependent memory-like NK cells distinguished by FcRγ deficiency. J Immunol. 2013;190(4):1402–1406. doi: 10.4049/jimmunol.1203034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mingari MC, et al. Cytolytic T lymphocytes displaying natural killer (NK)-like activity: Expression of NK-related functional receptors for HLA class I molecules (p58 and CD94) and inhibitory effect on the TCR-mediated target cell lysis or lymphokine production. Int Immunol. 1995;7(4):697–703. doi: 10.1093/intimm/7.4.697. [DOI] [PubMed] [Google Scholar]
- 14.Pietra G, et al. The analysis of the natural killer-like activity of human cytolytic T lymphocytes revealed HLA-E as a novel target for TCR alpha/beta-mediated recognition. Eur J Immunol. 2001;31(12):3687–3693. doi: 10.1002/1521-4141(200112)31:12<3687::aid-immu3687>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 15.Romagnani C, et al. Identification of HLA-E-specific alloreactive T lymphocytes: a cell subset that undergoes preferential expansion in mixed lymphocyte culture and displays a broad cytolytic activity against allogeneic cells. Proc Natl Acad Sci U S A. 2002;99(17):11328–11333. doi: 10.1073/pnas.172369799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moretta L, et al. NK-CTLs, a novel HLA-E-restricted T-cell subset. Trends Immunol. 2003;24(3):136–143. doi: 10.1016/s1471-4906(03)00031-0. [DOI] [PubMed] [Google Scholar]
- 17.Mazzarino P, et al. Identification of effector-memory CMV-specific T lymphocytes that kill CMV-infected target cells in an HLA-E-restricted fashion. Eur J Immunol. 2005;35(11):3240–3247. doi: 10.1002/eji.200535343. [DOI] [PubMed] [Google Scholar]
- 18.Björkström NK, et al. CD8 T cells express randomly selected KIRs with distinct specificities compared with NK cells. Blood. 2012;120(17):3455–3465. doi: 10.1182/blood-2012-03-416867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Veken LT, et al. Functional analysis of killer Ig-like receptor-expressing cytomegalovirus-specific CD8+ T cells. J Immunol. 2009;182(1):92–101. doi: 10.4049/jimmunol.182.1.92. [DOI] [PubMed] [Google Scholar]
- 20.Jacomet F, et al. Evidence for eomesodermin-expressing innate-like CD8(+) KIR/NKG2A(+) T cells in human adults and cord blood samples. Eur J Immunol. 2015;45(7):1926–1933. doi: 10.1002/eji.201545539. [DOI] [PubMed] [Google Scholar]
- 21.Hussain T, Quinn KM. Similar but different: virtual memory CD8 T cells as a memory-like cell population. Immunol Cell Biol. 2019;97(7):675–684. doi: 10.1111/imcb.12277. [DOI] [PubMed] [Google Scholar]
- 22.Jin JH, et al. Virtual memory CD8+ T cells restrain the viral reservoir in HIV-1-infected patients with antiretroviral therapy through derepressing KIR-mediated inhibition. Cell Mol Immunol. 2020;17(12):1257–1265. doi: 10.1038/s41423-020-0408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sosinowski T, et al. CD8α+ dendritic cell trans presentation of IL-15 to naive CD8+ T cells produces antigen-inexperienced T cells in the periphery with memory phenotype and function. J Immunol. 2013;190(5):1936–1947. doi: 10.4049/jimmunol.1203149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Intlekofer AM, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6(12):1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 25.Lee JY, et al. Virtual memory CD8 T cells display unique functional properties. Proc Natl Acad Sci U S A. 2013;110(33):13498–13503. doi: 10.1073/pnas.1307572110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White JT, et al. Virtual memory T cells develop and mediate bystander protective immunity in an IL-15-dependent manner. Nat Commun. 2016;7:11291. doi: 10.1038/ncomms11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balin SJ, et al. Human antimicrobial cytotoxic T lymphocytes, defined by NK receptors and antimicrobial proteins, kill intracellular bacteria. Sci Immunol. 2018;3(26):eaat7668. doi: 10.1126/sciimmunol.aat7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burwitz BJ, et al. Cross-species rhesus cytomegalovirus infection of cynomolgus macaques. PLoS Pathog. 2016;12(11):e1006014. doi: 10.1371/journal.ppat.1006014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen SG, et al. Immune clearance of highly pathogenic SIV infection. Nature. 2013;502(7469):100–104. doi: 10.1038/nature12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen SG, et al. Effector-memory T cell responses are associated with protection of rhesus monkeys from mucosal SIV challenge. Nat Med. 2009;15(3):293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen SG, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473(7348):523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen SG, et al. A live-attenuated RhCMV/SIV vaccine shows long-term efficacy against heterologous SIV challenge. Sci Transl Med. 2019;11(501):eaaw2607. doi: 10.1126/scitranslmed.aaw2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall EE, et al. Enhancing safety of cytomegalovirus-based vaccine vectors by engaging host intrinsic immunity. Sci Transl Med. 2019;11(501):eaaw2603. doi: 10.1126/scitranslmed.aaw2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta RK, et al. HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature. 2019;568(7751):244–248. doi: 10.1038/s41586-019-1027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen SG, et al. Broadly targeted CD8+ T cell responses restricted by major histocompatibility complex E. Science. 2016;351(6274):714–720. doi: 10.1126/science.aac9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen SG, et al. Prevention of tuberculosis in rhesus macaques by a cytomegalovirus-based vaccine. Nat Med. 2018;24(2):130–143. doi: 10.1038/nm.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogel P, et al. Seroepidemiologic studies of cytomegalovirus infection in a breeding population of rhesus macaques. Lab Anim Sci. 1994;44(1):25–30. [PubMed] [Google Scholar]
- 38.Oxford KL, et al. The interplay between immune maturation, age, chronic viral infection and environment. Immun Ageing. 2015;12:3. doi: 10.1186/s12979-015-0030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ram DR, et al. Tracking KLRC2 (NKG2C)+ memory-like NK cells in SIV+ and rhCMV+ rhesus macaques. PLoS Pathog. 2018;14(5):e1007104. doi: 10.1371/journal.ppat.1007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wills MR, et al. The immunology of human cytomegalovirus latency: could latent infection be cleared by novel immunotherapeutic strategies? Cell Mol Immunol. 2015;12(2):128–138. doi: 10.1038/cmi.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walter L, Petersen B. Diversification of both KIR and NKG2 natural killer cell receptor genes in macaques - implications for highly complex MHC-dependent regulation of natural killer cells. Immunology. 2017;150(2):139–145. doi: 10.1111/imm.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cannon MJ, et al. Review of cytomegalovirus shedding in bodily fluids and relevance to congenital cytomegalovirus infection. Rev Med Virol. 2011;21(4):240–255. doi: 10.1002/rmv.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim EY, et al. The CD4+ T cell response to human cytomegalovirus in healthy and immunocompromised people. Front Cell Infect Microbiol. 2020;10:202. doi: 10.3389/fcimb.2020.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cicin-Sain L, Arens R. Exhaustion and inflation at antipodes of T cell responses to chronic virus infection. Trends Microbiol. 2018;26(6):498–509. doi: 10.1016/j.tim.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 45.Collins PL, et al. Gene regulatory programs conferring phenotypic identities to human NK cells. Cell. 2019;176(1):348–360. doi: 10.1016/j.cell.2018.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujimoto Y, et al. CD83 expression influences CD4+ T cell development in the thymus. Cell. 2002;108(6):755–767. doi: 10.1016/s0092-8674(02)00673-6. [DOI] [PubMed] [Google Scholar]
- 47.García-Martínez LF, et al. A novel mutation in CD83 results in the development of a unique population of CD4+ T Cells. J Immunol. 2004;173(5):2995–3001. doi: 10.4049/jimmunol.173.5.2995. [DOI] [PubMed] [Google Scholar]
- 48.Lüthje K, et al. Transgenic expression of a CD83-immunoglobulin fusion protein impairs the development of immune-competent CD4-positive T cells. Eur J Immunol. 2006;36(8):2035–2045. doi: 10.1002/eji.200636068. [DOI] [PubMed] [Google Scholar]
- 49.Hertoghs KM, et al. Molecular profiling of cytomegalovirus-induced human CD8+ T cell differentiation. J Clin Invest. 2010;120(11):4077–4090. doi: 10.1172/JCI42758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen SG, et al. Complete sequence and genomic analysis of rhesus cytomegalovirus. J Virol. 2003;77(12):6620–6636. doi: 10.1128/JVI.77.12.6620-6636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reynolds AE, Enquist LW. Biological interactions between herpesviruses and cyclooxygenase enzymes. Rev Med Virol. 2006;16(6):393–403. doi: 10.1002/rmv.519. [DOI] [PubMed] [Google Scholar]
- 52.Zhu H, et al. Inhibition of cyclooxygenase 2 blocks human cytomegalovirus replication. Proc Natl Acad Sci U S A. 2002;99(6):3932–3937. doi: 10.1073/pnas.052713799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slobedman B, et al. Virus-encoded homologs of cellular interleukin-10 and their control of host immune function. J Virol. 2009;83(19):9618–9629. doi: 10.1128/JVI.01098-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lockridge KM, et al. Primate cytomegaloviruses encode and express an IL-10-like protein. Virology. 2000;268(2):272–280. doi: 10.1006/viro.2000.0195. [DOI] [PubMed] [Google Scholar]
- 55.Kotenko SV, et al. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10) Proc Natl Acad Sci U S A. 2000;97(4):1695–1700. doi: 10.1073/pnas.97.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fiorentino DF, et al. IL-10 inhibits cytokine production by activated macrophages. J Immunol. 1991;147(11):3815–3822. [PubMed] [Google Scholar]
- 57.Hart PH, et al. Potential antiinflammatory effects of interleukin 4: suppression of human monocyte tumor necrosis factor alpha, interleukin 1, and prostaglandin E2. Proc Natl Acad Sci U S A. 1989;86(10):3803–3807. doi: 10.1073/pnas.86.10.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.te Velde A, et al. Interleukin-4 (IL-4) inhibits secretion of IL-1 beta, tumor necrosis factor alpha, and IL-6 by human monocytes. Blood. 1990;76(7):1392–1397. [PubMed] [Google Scholar]
- 59.Chan A, et al. Maturation and function of human dendritic cells are inhibited by orf virus-encoded interleukin-10. J Gen Virol. 2006;87(11):3177–3181. doi: 10.1099/vir.0.82238-0. [DOI] [PubMed] [Google Scholar]
- 60.Mocellin S, et al. The multifaceted relationship between IL-10 and adaptive immunity: putting together the pieces of a puzzle. Cytokine Growth Factor Rev. 2004;15(1):61–76. doi: 10.1016/j.cytogfr.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 61.Musso T, et al. Human monocytes constitutively express membrane-bound, biologically active, and interferon-gamma-upregulated interleukin-15. Blood. 1999;93(10):3531–3539. [PubMed] [Google Scholar]
- 62.Zhang X, et al. Potent and selective stimulation of memory-phenotype CD8+ T cells in vivo by IL-15. Immunity. 1998;8(5):591–599. doi: 10.1016/s1074-7613(00)80564-6. [DOI] [PubMed] [Google Scholar]
- 63.Martinet V, et al. Type I interferons regulate eomesodermin expression and the development of unconventional memory CD8(+) T cells. Nat Commun. 2015;6:7089. doi: 10.1038/ncomms8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Correia MP, et al. IL-15 induces CD8+ T cells to acquire functional NK receptors capable of modulating cytotoxicity and cytokine secretion. Immunobiology. 2011;216(5):604–612. doi: 10.1016/j.imbio.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 65.Correia MP, et al. Distinct human circulating NKp30+FcεRIγ+CD8+ T cell population exhibiting high natural killer-like antitumor potential. Proc Natl Acad Sci U S A. 2018;115(26):E5980–E5989. doi: 10.1073/pnas.1720564115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crough T, Khanna R. Immunobiology of human cytomegalovirus: from bench to bedside. Clin Microbiol Rev. 2009;22(1):76–98. doi: 10.1128/CMR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ratzinger G, et al. Mature human Langerhans cells derived from CD34+ hematopoietic progenitors stimulate greater cytolytic T lymphocyte activity in the absence of bioactive IL-12p70, by either single peptide presentation or cross-priming, than do dermal-interstitial or monocyte-derived dendritic cells. J Immunol. 2004;173(4):2780–2791. doi: 10.4049/jimmunol.173.4.2780. [DOI] [PubMed] [Google Scholar]
- 68.Busch M, et al. Lipoarabinomannan-responsive polycytotoxic T cells are associated with protection in human tuberculosis. Am J Respir Crit Care Med. 2016;194(3):345–355. doi: 10.1164/rccm.201509-1746OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. doi: 10.1101/2021.01.11.426199. Barrenäs F, et al. Sustained IL-15 response signature predicts RhCMV/SIV vaccine efficacy [preprint]. Posted on bioRxiv January 11, 2021. [DOI]
- 70.Rolot M, et al. Helminth-induced IL-4 expands bystander memory CD8+ T cells for early control of viral infection. Nat Commun. 2018;9(1):4516. doi: 10.1038/s41467-018-06978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barbarin A, et al. Phenotype of NK-like CD8(+) T cells with innate features in humans and their relevance in cancer diseases. Front Immunol. 2017;8:316. doi: 10.3389/fimmu.2017.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. fastqcr: Quality Control of Sequencing Data. Version 0.1.2. Kassambara A; 2019. Accessed June 7, 2021. https://cran.r-project.org/web/packages/fastqcr.
- 73. STAR: Spike Train Analysis with R. Version 0.3-7. Pouzat C; 2012. Accessed June 7, 2021. https://sites.google.com/site/spiketrainanalysiswithr/Home.
- 74.Robinson MD, et al. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wickham H, ed. ggplot2: Elegant Graphics for Data Analysis. Springer;2016. [Google Scholar]
- 76.Bougeard S, Dray S. Supervised Multiblock Analysis in R with the ade4 Package. J Stat Softw. 2018;86(1):1–17. [Google Scholar]
- 77. ESaHW. vegan: Community Ecology Package. Version 2.5-7. Jari Oksanen et al.; 2019. Accessed June 7, 2021. https://cran.r-project.org/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The expression data generated by RNA-seq analysis are available in the GEO repository under accession number GSE171978.