Abstract
Circadian rhythms evolved through adaptation to daily light/dark changes in the environment; they are believed to be regulated by the core circadian clock interlocking feedback loop. Recent studies indicate that each core component executes general and specific functions in metabolism. Here, we review the current understanding of the role of these core circadian clock genes in the regulation of metabolism using various genetically modified animal models. Additionally, emerging evidence shows that exposure to environmental stimuli, such as artificial light, unbalanced diet, mistimed eating, and exercise, remodels the circadian physiological processes and causes metabolic disorders. This Review summarizes the reciprocal regulation between the circadian clock and metabolism, highlights remaining gaps in knowledge about the regulation of circadian rhythms and metabolism, and examines potential applications to human health and disease.
To adapt to daily environmental changes caused by our Earth’s rotation, most organisms on the planet evolved near-24-hour cycles of behavioral, physiological, and metabolic rhythms (1). In addition to the entrained environmental stimuli, the internal timekeeping system of the circadian clock has evolved to anticipate external changes (2, 3). These conserved rhythms synchronize internal biological and behavioral processes to the external temporal environment, presumably providing organisms with selective advantages for survival. However, over the past century, modern industrialized society has profoundly changed our external environment (4). For example, the boundaries between day and night have been blurred by electric light and travel across different time zones. Disrupted circadian rhythms are highly associated with metabolic disorders (5). Conversely, obesity induced by overeating or overnutritional environment leads to circadian remodeling (6, 7). Understanding the reciprocal regulation of circadian rhythm and metabolism may provide mechanistic insights into circadian physiology and advance new chronotherapy approaches and therapeutic targets for metabolic disorders.
Intrinsic circadian clock machinery and metabolism
“Circadian rhythm” refers to an innate, endogenous, and entrainable rhythm whose cycle is approximately 24 hours. This rhythm is retained under constant conditions, in the absence of any external or environmental cue that entrains the circadian clock (referred to as zeitgebers), including light, temperature, eating patterns, exercise, and social interactions. More generally, a biological rhythm synchronized by light/dark cycles is known as a diurnal rhythm, which may or may not be a circadian rhythm (depending on whether it is independent of light/dark cycles). Organisms most active in the daytime (such as humans) are referred to as diurnal, whereas nocturnal organisms (such as mice and rats) sleep primarily during the light phase. Owing to the power of genetic manipulation and the ability to carefully and (relatively) inexpensively control housing conditions, mechanistic studies of circadian rhythms in mammals have largely used mice as models, which presents translational challenges to human chronomedicine since people are diurnal. Moreover, metabolic differences between rodents and humans need to be taken into account. For example, rodents feed frequently and have fast metabolic rates, such that 24 hours of fasting could result in a profound loss of body weight (up to 20%) and an even greater percentage loss of body fat (8).
Molecular circadian clock machinery.
The notion of intrinsic clock machinery was first posited in 1729 by the discovery that, in constant darkness, the leaves of the mimosa plant retained their daily pattern of opening and closing, indicating that this rhythm was programmed and not just a response to natural light (9). Two centuries later, in 1971, the genetic foundation of circadian rhythms was pioneered by Ron Konopka and Seymour Benzer (10). They reported that mutations at a single genetic locus produced abnormal circadian rhythms in the behavior of Drosophila melanogaster (10). In 1984, the laboratories of Hall, Rosbash, and Young cloned the first circadian rhythm gene, period (Per), in the fruit fly (11–15), and 10 years later Takahashi and colleagues discovered circadian locomotor output cycles kaput (Clock), the first mammalian circadian clock gene, in mice (16).
Interestingly, Per encodes a protein that represses its own transcription, resulting in daily Per rhythm, while CLOCK activates the transcription of Per, forming a negative-feedback loop that is highly conserved from flies to humans (17). In mammals, two basic helix-loop-helix (bHLH) transcriptional activators, CLOCK and brain and muscle ARNT-like 1 (ARNTL1; also known as BMAL1 and MOP3), form a heterodimer and bind to E-box element to activate the expression of the transcriptional repressors cryptochromes 1 and 2 (CRY1/2), periods 1, 2, and 3 (PER1/2/3), and REV-ERBα/β (also known as nuclear receptor subfamily 1 group D members 1 and 2 [NR1D1/2]) (17–19). CRY and PER repress CLOCK-BMAL1–dependent transcription (20-22). REV-ERBα and REV-ERBβ repress Bmal1 transcription via recruitment of nuclear receptor corepressor (NCoR) and histone deacetylase 3 (HDAC3) (23–25). Additionally, retinoic acid receptor–related orphan receptors α and γ (RORα/γ) are induced by CLOCK-BMAL1 and function as transcriptional activators to positively on Bmal1 expression in competition with REV-ERBα/β on ROR response elements (ROREs) (1, 19). Overall, the circadian clock oscillator uses multiple mechanisms to drive the temporally restricted gene expression pattern.
Core circadian clock genes and metabolism.
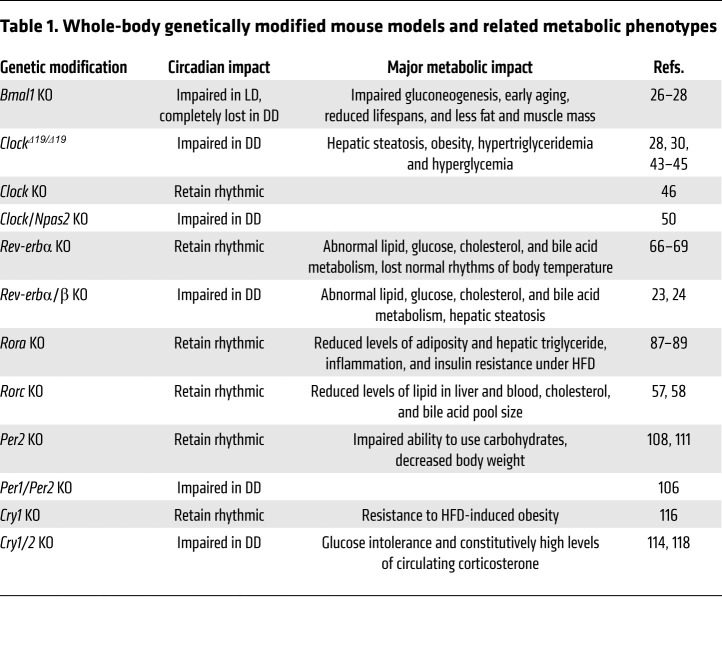
To understand the connection between circadian rhythm and metabolism, genetically modified mouse strains targeting circadian clock genes have been generated and phenotypically characterized. It is important to recognize that these targeted genes are transcription factors, and as such, their genetic loss affects both their role in the circadian clock and their more general function as direct regulators of gene expression. Figure 1 and Table 1 summarize the effects, described below, of knockout of these clock components on biological rhythms and metabolic function.
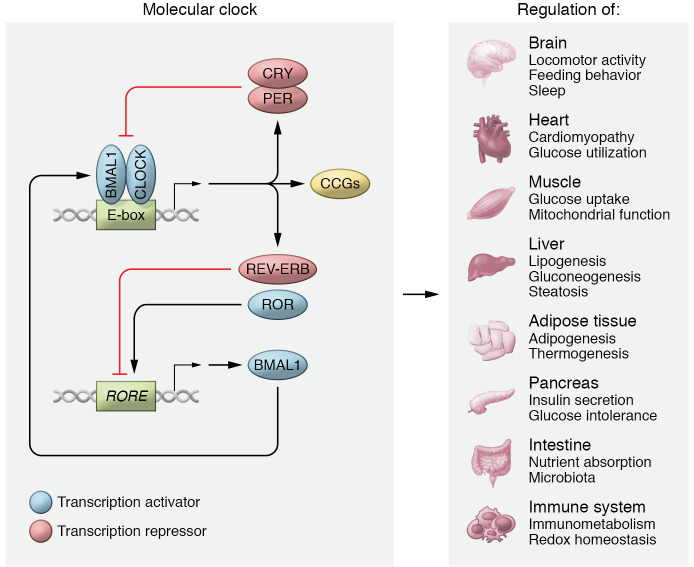
Figure 1. Intrinsic circadian clock machinery and metabolism.
Core circadian clock genes, including transcription activators and repressors, form an autoregulatory feedback loop present in most cells to regulate their circadian physiology. The figure summarizes the major metabolic functions regulated by core circadian clock genes in indicated tissues based on loss- and gain-of-function studies from genetically modified animal models.
Table 1. Whole-body genetically modified mouse models and related metabolic phenotypes.
Bmal1.
BMAL1 is the core clock component whose single knockout in a mouse model results in complete loss of rhythmicity (26). Bmal1-null mice have altered activity, reduced total activity levels on light/dark cycles, and complete abolition of behavioral circadian rhythms in constant darkness (26). Bmal1-knockout mice also show impaired gluconeogenesis, display various symptoms of early aging, and have reduced lifespan and body weight (27, 28). Intriguingly, constitutive expression of Bmal1 in brains of whole-body Bmal1-knockout mice restored the circadian rhythms of wheel-running activity, but activity levels and body weight were still lower than in WT mice (29). By contrast, re-expression of Bmal1 in the skeletal muscle of Bmal1-null mice normalized activity levels and body weight but not the circadian rhythm of behavior (29), suggesting distinct tissue-specific functions of BMAL1.
Tissue-specific knockout of Bmal1 has provided insight into its specific functions. Pancreatic β cell–specific Bmal1-knockout mice have impaired glucose tolerance and decreased insulin secretion, resulting in hyperglycemia without affecting the circadian rhythms of activity and feeding/fasting cycles (30). Liver-specific Bmal1-knockout mice have higher triglyceride and cholesterol accumulation in the liver and plasma, which is rescued by liver-specific Bmal1 re-expression (31, 32). Adipocyte-specific deletion of Bmal1 increases adipose tissue mass and body weight gain as a result of reduced energy expenditure and increased daytime food intake (33). In cardiomyocyte-specific Bmal1-knockout mice, hearts decrease glucose utilization, and the mice exhibit early-onset development of dilated cardiomyopathy and early mortality (34, 35). Skeletal muscle–specific Bmal1-knockout mice show impaired insulin-stimulated glucose uptake and decreased glucose oxidation, but circadian rhythms of locomotor activity are normal (36, 37). Bmal1 also modulates immunometabolism, as macrophage-specific Bmal1 knockout impairs mitochondrial function and exacerbates succinate dehydrogenase–mediated mitochondrial production of reactive oxygen species (38). In summary, these studies indicate that BMAL1 is not only a key regulatory signal in the suprachiasmatic nucleus (SCN) regulating overall rhythmic behavior, but also drives local tissue-specific metabolic functions.
Clock and Npas2.
The Clock gene was the first gene identified to have a critical role in mammalian circadian rhythms (16, 39). Positional cloning and transgenic rescue studies identified a Clock point mutation (ClockΔ19) leading to altered circadian behavior (39, 40). This single point mutation within the intron between exons 18 and 19 caused aberrant mRNA splicing with resultant loss of 51 amino acids that comprise a putative transcriptional regulatory domain (41). This mutant CLOCKΔ19 protein could form heterodimers with BMAL1 but failed to activate transcription (42). Compared with WT mice, ClockΔ19/Δ19 double-mutant mice display metabolic disorders, including hepatic steatosis, obesity, hypertriglyceridemia and hyperglycemia, and increased absorption of lipids throughout the day (28, 30, 43–45).
Interestingly, although ClockΔ19/Δ19 double-mutant mice display abnormal circadian behavior in constant darkness (16, 39), mice with whole-body knockout of Clock (Clock−/−) display robust behavioral rhythmicity, although they do have altered responses to light (46). Neuronal PAS domain–containing protein 2 (NPAS2) is a paralog of CLOCK that can form a heterodimer with BMAL1 (47, 48). Like Clock-null mice, Npas2-null mice retain robust circadian rhythms, suggesting a potential complementary effect between CLOCK and NPAS2 (49). Indeed, mice with double knockout of CLOCK and NPAS2 (Clock–/– Npas2–/–) exhibit arrhythmic locomotor behavior in constant darkness (50). However, Npas2 and Clock do not equally contribute to circadian regulation. Npas2 is mainly expressed in the brain, and Clock is highly expressed in the peripheral tissues (51). Npas2-null mice adapt slowly to restricted feeding (52), while peripheral oscillators are arrhythmic without Clock (53).
Rev-erbs.
REV-ERBα (54) and REV-ERBβ (55) are closely related members of the nuclear receptor superfamily. In most nuclear receptors, activation function 2 (AF2) is required to interact with coactivators and activate gene transcription (56), but REV-ERBs lack AF2 at the C-terminal end of the ligand-binding domain. As a result, REV-ERBs are thought to be unable to activate transcription and act as constitutive repressors of transcription (57). Recent studies have indicated that REV-ERBs repress gene expression in two ways: First, they compete with the transcriptional activators RORs at sites known as ROREs (58, 59), passively blocking activation. Second, REV-ERBs actively repress gene expression from these sites by recruiting the NCoR-HDAC3 corepressor complex (25, 60, 61). This active repression function requires two REV-ERB moieties, binding as closely spaced monomers or cooperatively as a homodimer (62). In addition to direct DNA binding, some REV-ERBα binding activity occurs via tethering. One example of such indirect binding was revealed through the function of a REV-ERBα mutant that lacks the DNA-binding domain (60). In this case, REV-ERBα was tethered to DNA through hepatocyte nuclear factors (HNFs). Since HNFs, particularly HNF4 and HNF6 (63–65), are specific to the liver, this example demonstrates how lineage-specific factors can facilitate tissue-specific genomic binding of transcriptional modifiers like REV-ERBα.
REV-ERBα is an important metabolic regulator and modulates lipid, glucose, cholesterol, and bile acid metabolism, as well as adipogenesis and inflammation (66–69). Heme, a substance precursive to hemoglobin, has been identified as a direct ligand for REV-ERBα and REV-ERBβ (70). Although the physiological role of this interaction remains to be fully understood, heme suppresses hepatic gluconeogenic gene expression and glucose output through REV-ERBα–mediated gene expression (70, 71). Deficiency of both Rev-erb genes led marked hepatic steatosis and profoundly altered circadian wheel-running behavior and lipid homeostasis (23, 24). Although REV-ERBα is required for adipocyte differentiation in cell culture (72–74), adipose tissue mass is increased via fibroblast growth factor 21 (FGF-21) signaling in mice lacking REV-ERBα (75, 76). Rev-erbα–null mice were also shown to lose normal rhythms of body temperature, brown adipose tissue (BAT) activity, and improved cold tolerance at 5 pm, suggesting that REV-ERBα contributes to thermogenic oscillation (77). In skeletal muscle, Rev-erbα deficiency led to reduced mitochondrial content and oxidative function, upregulated autophagy, and compromised exercise capacity (78). Rev-erbα mutation (DBD domain deletion) and Rev-erbβ deletion in SCN GABAergic neurons disrupted the rhythms of hepatic glucose production in mice (79). Collectively, these studies using tissue-specific genetic models suggest tissue-specific functions of REV-ERBs in mice. Emerging studies are actively dissecting whether the metabolic effects of tissue-specific functions of REV-ERBs are mediated cell-autonomously or by modulation of systemic signals (80–82).
Rors.
The three RORs (RORα, RORβ, and RORγ) are members of the nuclear receptor superfamily. They were named based on sequence similarities to the retinoic acid receptor (83–85), but they share DNA-binding specificity with REV-ERBs (58, 59). Their endogenous ligands remain controversial, but they may be activated by oxysterols as well as being constitutively activated through the ligand-independent recruitment of transcriptional coactivators (86). Both RORα and RORγ have been implicated in the control of energy homeostasis and regulation of lipid and glucose metabolism.
Deficiency of Rora, but not Rorc, in mice fed an obesogenic high-fat diet (HFD) led to reduced levels of adiposity and hepatic triglyceride levels, inflammation, and insulin resistance in comparison with WT mice (87–89). Multiple organs contributed to this phenotype. In skeletal muscle, compared with WT mice, increased levels of AKT and phosphorylated AKT and enhanced glucose uptake were observed in Rora-deficient mice (89). Upon Rora knockout, genes related to lipid synthesis were downregulated in the liver, and inflammatory genes were also downregulated in white adipocyte tissue (87, 88). However, a previous study reported that RORα-null mice had increased triglyceride accumulation and lipogenic gene expression in the liver (90), leading researchers to revisit the function of RORs in liver. Liver-specific double knockout of Rora/Rorc shows the overactivation of the INSIG2/SREBP lipogenic response at the peak expression of RORs in mice (5 am) and exacerbation of diet-induced hepatic steatosis, demonstrating an important circadian rhythmic consideration for metabolic studies (91). In BAT, Ucp1 and other thermogenic genes were upregulated upon Rora knockout. Consistently, primary brown adipocytes from Rora-deficient mice displayed a higher metabolic rate (87).
Like RORα, RORγ plays important roles in the metabolic regulation of multiple organs. Rorc-deficient mice show decreased adipocyte sizes and high insulin sensitivity with improved control of circulating free fatty acids compared with WT controls. HFD-fed Rorc-deficient mice are also protected from hyperglycemia and insulin resistance (92). Consistently, Rorc expression in the adipose stromal vascular fraction from obese human subjects is positively correlated with adipocyte size and negatively correlated with adipogenesis and insulin sensitivity (92). In the liver, both whole-body Rorc-knockout and liver-specific Rorc-knockout mice display reduced levels of lipid in liver and blood, reduced cholesterol, and reduced bile acid pool size (93, 94). RORγ is highly expressed in skeletal muscle and controls the expression of genes that regulate muscle and fat mass, and modulates the production of reactive oxygen species (95). An isoform of RORγ, called RORγt, is unique to inflammatory Th17 lymphocytes (96), although its metabolic function has not been characterized.
Pers and Crys.
Period, the first clock gene to be identified, has three homolog genes (Per1, Per2, and Per3) in mammals (97–100). Cryptochrome (Cry) has two homologs, Cry1 and Cry2 (101). Although PERs and CRYs lack a DNA-binding domain and therefore are very unlikely to directly bind to DNA, they form a heterodimer that moves into the nucleus upon phosphorylation by casein kinase 1 (CK1), and inhibit the transcriptional activity of BMAL1-CLOCK heterodimer (102–105).
Per1- or Per2-deficient mice, but not Per3-deficient mice, display disrupted locomotor activity rhythms in extended exposure to constant darkness. Interestingly, the circadian disruption observed in Per1/Per3 and Per2/Per3 double-knockout mice resembles rhythms of circadian disruption of Per1- or Per2-deficient mice, while Per1/Per2 double-knockout mice are immediately arrhythmic in constant darkness (106). PERs have different functions in the regulation of metabolism (106). Per1-deficient mice display elevated blood pressure involving a mechanism of renal sodium reabsorption (107). Rhythms of glucocorticoid secretion and diurnal feeding rhythms are disrupted in normal chow– and HFD-fed Per2-deficient mice, resulting in decreased body weight gain in mice (108). Moreover, Per2-deficient mice exhibit increased oxidative capacity in white adipocyte tissue and enhanced adipocyte differentiation of cultured fibroblasts (109), and Per1/Per2–deficient mice have different circadian accumulation patterns of triacylglycerol compared with WT mice (110). Per2 deficiency in ischemic hearts impairs carbohydrate utilization for oxygen-efficient glycolysis (111). Per3- or Per1/Per2/Per3–deficient mice also gained more diet-induced body weight, suggesting an opposing function to Per2 (112), while another study indicated that Per2 knockout alters the body composition, with both increased adipose and decreased muscle tissue (113).
Mice lacking Cry1 or Cry2 alone display a phase-accelerated or a phase-delayed free-running period of locomotor activities, respectively, but the circadian rhythms are still robust. Cry1/Cry2 double-knockout mice show arrhythmic behavior in constant darkness (114). Mice deficient in Cry1, but not mice deficient in Cry2, are resistant to HFD-induced obesity (115). HFD-fed Cry1/Cry2 double-knockout mice rapidly gain weight and display hyperinsulinemia (116). CRY regulates glucose homeostasis through several mechanisms. CRY represses gluconeogenesis by inhibiting protein kinase A–mediated phosphorylation of cAMP response element–binding protein (CREB) during fasting via blocking of glucagon-mediated increases in intracellular cAMP concentration (117). CRY1 and CRY2 interact with glucocorticoid receptor (GR) in a ligand-dependent manner. Knockout of Cry1/Cry2 results in glucose intolerance and constitutively high levels of circulating corticosterone (118). Moreover, SREBP1c downregulates gluconeogenesis via CRY1-mediated FOXO1 degradation to regulate hyperglycemia in db/db mice (119).
Core clock component–specific regulation of metabolic cycles.
In addition to the common regulation of the core circadian clock interlocking feedback loop, groups of rhythmic genes are specifically regulated by certain core clock components. Consistently, knockouts of these components share some common phenotypes, including disrupted locomotor activities in constant dark and tumorigenesis in mice with chronic jet lag (120), but each knockout model also has component-specific metabolic outcomes as discussed above. To explore the underlying mechanisms of component-specific regulation, the genome occupancy of core clock components was determined, revealing that only a small proportion of binding sites are shared among all of the core components (25, 121). Further genome-wide rhythmic enhancer mapping using global run-on sequencing (GRO-Seq) identified that each phase of enhancers and downstream gene transcription is regulated by distinct core clock transcription factors (TFs) (122).
As summarized in Figure 2, the BMAL1-CLOCK complex can bind to E-box motif (123, 124), while REV-ERBs, competing with RORs, bind on RORE motifs to regulate the expression of target genes whose enhancers or promoters contain these motifs (58, 59, 122). In addition to the interaction with BMAL1-CLOCK, CRY1 broadly interacts with multiple nuclear receptors and modulates specific gene expression (125). In addition to competing with ROR on ROREs, REV-ERBs can be tethered by cell type–specific TFs and regulate the rhythmic expression of another specific group of genes involved in metabolism (60). The studies described above provide mechanisms of how core clock components bind in diverse ways on chromatin and directly regulate the oscillating expression of their target genes. Core clock components can also indirectly regulate their target genes via downstream TFs. For example, BMAL1 activates the oscillating expression of Hlf, Tef, and Dbp (which encode TFs in the PAR bZIP family) to indirectly regulate the expression of rhythmic genes whose regulatory elements contain D-box (123, 126, 127). REV-ERBs repress another D-box–binding transcription repressor, E4bp4, which subsequently regulates the rhythmic expression of E4BP4 target genes (122). These downstream TFs of each core clock component either independently or collaboratively regulate the rhythmic expression of circadian output genes.
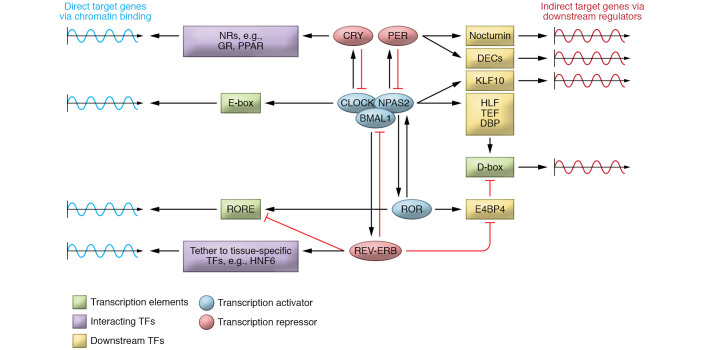
Figure 2. Mechanisms of core clock component–specific regulation of target genes.
Core clock components independently, or forming protein complexes, bind to specific chromatin regions to directly regulate circadian gene expression (left). Each core clock component indirectly mediates circadian gene expression via downstream TFs (right). GR, glucocorticoid receptor; NRs, nuclear receptors.
Environmental cues regulating biological rhythms and metabolism
Biological rhythms are the natural cycle of change in the body regarding biological processes, which are regulated by intrinsic circadian clock machinery and adjusted (entrained) by external environmental inputs and social cues, including light/dark cycles, nutrients, feeding schedule, and exercise time (128–130). Figure 3 and Table 2 summarize the effects of these zeitgebers on biological rhythms and metabolic function.
Figure 3. Interactions between circadian rhythms and human physiology.
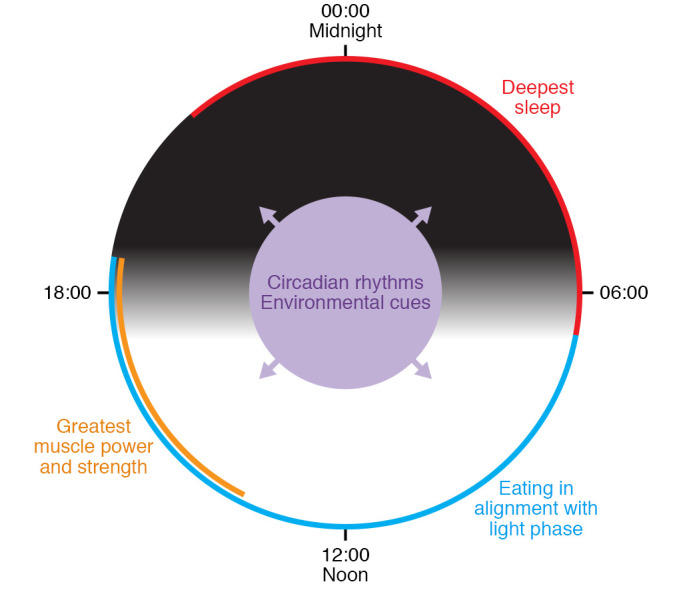
Light/dark cycles synchronize the intrinsic clock to a 24-hour cycle. In human physiology, alignment of wake/sleep cycles, eating patterns, and exercise schedule to certain circadian phases leads to beneficial metabolic outcomes, while misalignment exacerbates or causes metabolic disorders.
Table 2. Effects of environmental cues on biological rhythms and metabolism in humans.
Follow the light.
Photosensitive organisms are biologically programmed to follow light/dark cycles in order to entrain hormone production and bodily function. In mammals, the daily light/dark cycles entrain the circadian clock of the brain, specifically the SCN of the hypothalamus, to the 24-hour period in our environment. It has been suggested that disruption of circadian rhythms caused by abnormal light/dark cycles, including artificial light at night and the usage of screens with shorter-wavelength lights, results in metabolic disorders (131, 132). Night-shift workers display dyslipidemia, increased postprandial serum glucose and insulin (133), and increased circulating levels of several biomarkers of metabolic syndrome and inflammation (134). Moreover, night-shift workers and people with long working hours have a high risk of obesity and diabetes (135–142). Experimental animal models have been used to demonstrate that circadian misalignment causes metabolic disturbances. Here, we will discuss the effect of constant bright light (LL), alternating dim and bright light (dLL), and wavelengths of light on metabolism, including obesity, insulin resistance, and hepatic steatosis (143, 144).
Mice exposed to LL become behaviorally arrhythmic, and their SCNs become desynchronized (145). The disruption of the peripheral clock was also observed in LL-exposed mice. Mice exposed to LL developed obesity and hepatic steatosis, which was paralleled by an altered miRNA profile targeting the core clock gene Rev-erbα (143). When an obesogenic diet is superimposed on LL, mice display a reduced amplitude of rhythms in the SCN and a complete abolishment of circadian rhythms of feeding pattern, energy expenditure, and insulin sensitivity (146). During early development, the circadian system experiences a critical adjustment and is vulnerable to altered lighting conditions. During lactation, short-term LL in pups caused a loss of rhythmicity, a reduction in vasoactive intestinal polypeptide–positive (VIP-positive) and arginine vasopressin–positive (AVP-positive) cells in the SCN, a reduction of PER1 expression in the SCN, reduced body weight gain, and loss of daily rhythms in plasma glucose and triglycerides (147). These rhythmic metabolic disorders could not be restored in conditions of alternating light and dark (LD) after lactation (147). In adult rats, LL downregulated plasma melatonin (which is absent in most mouse models; refs. 148–150) and systematically induced complete circadian rhythmic suppression of body temperature and locomotor activity, but this circadian disruption could be restored within 1 week in constant darkness (DD) (151).
Like LL, dLL exposes mice to light over the course of the 24-hour day, but also provides a temporal cue for a 24-hour day via different light intensity between day and night. Compared with LL, dLL has lesser impacts on circadian rhythms. Interestingly, compared with LD controls, both LL- and dLL-exposed mice display increased body mass and reduced glucose tolerance, but caloric intake and total daily activity output are not affected (152). In the dLL-exposed mice, rhythms of Per1 and Per2 in the hypothalamus were attenuated, similarly to those of REV-ERB genes in the liver and adipose tissue (153).
In addition to LL, acute exposure to light in the night or a different wavelength also affects biological rhythms and metabolism. Very short exposures to nocturnal light inhibit melatonin release, alter clock gene expression, and increase c-Fos expression in the SCN, and this effect is wavelength dependent: blue light has the greatest effect, whereas red light has no effect (154–157). Therefore, light as a predominant zeitgeber entraining the clock in the SCN is a major contributor to maintenance of organismal metabolic homeostasis.
You are what you eat.
Diet composition is another important factor that affects the circadian clock. HFD disrupts circadian rhythms of locomotor and feeding activity in mice, with greater rhythmic expression of clock genes in fat than in liver (158). Rhythmic transcriptome profiling identified a genome-wide reprogramming of the clock in the liver (159). Using GRO-Seq to map HFD-specific circadian enhancers and quantify HFD-specific transcription rates, the DNA binding motifs for peroxisome proliferator–activated receptor (PPAR) and SREBP were shown to be enriched. Further functional studies revealed an unexpected synchronization of two opposing lipid processes, lipid synthesis and oxidation, at a similar time of the day. The synchronization could be a maladaptive response to the overnutrition environment (160).
Ketogenic diets (KDs) are high-fat, adequate-protein, very-low-carbohydrate diets that induce fatty acid oxidation as an energy source and lead to the synthesis of ketone bodies. This diet is used to treat epilepsy in children (161), to induce weight loss (162–164), and to decrease the risk of heart disease (165, 166). In mice, KD induced a profound circadian remodeling in the liver and gut in a tissue-specific manner. KD drastically alters BMAL1 target genes in the liver, but not the gut, while highly diurnal rhythms of PPARα are only observed in the gut (167). A low-calorie diet, which is known to boost fat metabolism and lifespan, enhances the magnitude of cyclic expression of circadian clock genes in Drosophila (168). These results highlight the intricate reciprocal relationship between metabolism and food content–regulated peripheral clocks.
Eat on time.
Meal time is known to be a dominant zeitgeber for peripheral tissue clocks (such as the clock in the liver) (81, 169). Eating during the active phase has healthy consequences for metabolism, while mistimed eating leads to metabolic disorders (170, 171). Restricting feeding to the sleep phase (here referred to as reverse-phase feeding [RPF]) uncouples circadian oscillators in peripheral tissues (including liver, kidney, and heart) from the SCN (169) and desynchronizes peripheral clocks (172). Coupling RPF with HFD exacerbates increased adiposity, decreased glucose tolerance, and dyslipidemia, a metabolic profile often observed in subjects with night-eating syndrome (173). Human epidemiological studies suggest that skipping breakfast is associated with high risks of developing obesity and related metabolic disorders (174–177). Early nocturnal meal skipping in mice, equivalent to breakfast skipping in humans, disturbs the peripheral clock, increases lipid synthesis, and favors body mass gain (178).
Conversely, restricting mice to HFD feeding in the active phase without reducing caloric intake prevents weight gain and metabolic disturbances, including hyperglycemia, insulin resistance, hepatic steatosis, and hypercholesterolemia (179–181). Even in mice lacking core circadian clock genes, time-restricted feeding (TRF) during the active phase (from zeitgeber time points ZT 13 to ZT 22) can prevent HFD-induced metabolic disorders (182). In humans, because of the variation among time-restricted feeding protocols regarding eating time and period, it is unclear whether TRF contributes to weight loss, but TRF showed beneficial metabolic outcomes in several independent studies (183–188).
Exercise with a schedule.
Exercise is a crucial intervention in the prevention and treatment of metabolic disorders (189). Scheduled exercise has been shown to entrain circadian rhythms in skeletal muscle (190–192). However, the optimal timing of exercise for preventing the effects of disrupted circadian rhythm and maximizing the health benefits is still largely unknown. Several studies have indicated that exercise performance shows diurnal rhythmicity (193). A recent study in mice indicated that exercise causes circadian remodeling involving carbohydrate exhaustion, usage of alternative energy sources, and adaptation of systemic energy expenditure (194). More world records have been broken by athletes in early evening, as strength, power, and endurance are increased in the early evening compared with early morning (195–197). Interestingly, when the training period exceeded 12 weeks, individuals who exercise in the evening gained more muscle mass than individuals who exercise in the morning (198).
In addition to the above zeitgebers, other external environmental and internal physiological cues, including temperature (199, 200), alcohol (201), aging (202), sexual phenotype (203, 204), cancer (205), microbiota (206, 207), and oxygen levels (208–211), impose significant impacts on biological rhythms and chronometabolism. Moreover, the interactions among these timing cues can collaboratively entrain peripheral clocks. For example, nutrient catabolism, maintenance of body temperature, and exercise are tightly linked to oxygen consumption (208, 209). Heart attack and obstructive sleep apnea caused by metabolic disorders lead to hypoxia (210, 211). These changes in oxygenation affect the circadian clock in an HIF-1α–dependent manner (208–211).
Conclusions and perspective
Molecular circadian biology originated with a genetic screen in Drosophila (identifying the Per mutant), and then extended to mammals through genetic screens in mice. As core clock components have been uncovered, circadian whole-body gene-knockout mouse models have revealed the function of the molecular clock in regulating circadian behavior, including sleep/wake and feeding/fasting cycles, as well as maintenance of metabolic homeostasis. Emerging core circadian gene tissue-specific knockout mouse models and state-of-the-art “omics” analysis across different tissues under various environmental stimuli have shed light on tissue-specific circadian clocks. The recognition that multiple TFs function in the core clock, and that each of these has thousands of genomic DNA binding sites, suggests that the concept of “clock output genes” needs to be revised. Each of the core clock genes contributes directly to individual gene regulation in addition to its role in the reciprocal and homeostatic regulation of other clock genes by transcriptional-translational feedback loops that define the clock itself.
Interorgan rhythmic communications.
Future studies are expected to further determine the interorgan rhythmic communications and how they are integrated to perform physiological functions. Multidirectional interorgan interactions, including those between the nervous system and peripheral metabolic organs as well as between metabolic organs, are essential for adaption to external cues and maintain whole-body energy homeostasis. The nervous system coordinates whole-body metabolism not only by direct innervation of the target tissues but also by the production of neurohormones (212). Peripheral organs perform intercellular signaling in an autocrine, paracrine, or endocrine manner (213). Interorgan communications have been explored in feeding, fasting, cold exposure, and exercise conditions (214–220). However, how tissue metabolism is linked and gated to specific temporal windows, and how this coordinated communication and coherence among tissue clocks are remodeled in response to environmental stimuli, need further investigation.
Intraorgan rhythmic communications.
In addition to interorgan communication, intraorgan communication has attracted increasing attention due to the improved technique and computing methods of single-cell sequencing (221–225). We recently discovered that the disruption of clocks in hepatocytes via deletion of the core clock genes REV-ERBα and REV-ERBβ remodels the rhythmic enhancers, transcriptomes, and metabolomes of multiple cell types within the liver (81). These results suggest rhythmic communication of time signals between different cell types within an organ to coordinately perform a given physiological function. Interestingly, even within the same cell type, hepatocyte disruption of REV-ERBα/β regulates target gene expression in a liver zonation–dependent manner. Consistent with these findings, another independent study indicated that core circadian clock genes are expressed in a non-zonated pattern, but the rhythmicity of some oscillating genes is zonation dependent (226), suggesting that the microenvironment, including intraorgan communication, plays an important role in their rhythmic expression. Future studies would be important to determine the underlying mechanism and physiological consequences of intraorgan communication.
Circadian versus non-circadian functions of clock genes.
Tissue-specific knockout mouse models have been used to partially solve the above questions, but also raise another question: is it the rhythmicity or the expression level of these core clock genes that is important for circadian regulation? In Bmal1 whole-body knockout mice, constitutive re-expression of BMAL1 in brain and muscle tissues partially rescues the disrupted rhythmic behavior (29), suggesting that the rhythmicity of the expression level of Bmal1 mRNA is not essential for rhythmic behavior. This could be due to the post-transcriptional effects of an intact PER/CRY rhythm, but this needs to be determined. Yet in Bmal1-deficient mice, rhythmic re-expression of BMAL1 in the liver and skin cannot restore most of the rhythmic gene expression (227, 228), suggesting that additional signals are essential in these tissues. Comparison of rhythmic and constitutive re-expression of these core circadian genes with similar mean expressions across the day in tissue-specific knockout mouse models could be useful for dissecting the respective role of gene rhythmicity and gene expression level.
Non-canonical rhythmic regulators.
In addition to canonical or core clock regulators that are critical to rhythmic biology, there are also non-canonical regulators that either collaboratively or independently regulate various rhythmic processes. For example, diet-induced obesity leads to a profound circadian remodeling in the livers of mice despite only modest changes in expression of all core clock genes (158, 160). Similarly in human muscle, TRF induces a massive circadian remodeling of amino acids and other metabolites without perturbing core clock gene expression (193). These studies suggest there are additional non-canonical rhythmic regulators mediating the circadian remodeling. Interestingly, knockout of REV-ERBs in livers of adult mice disrupts rhythmicity of other core clock genes, yet the majority of circadian output genes retained their rhythms, indicating the role of non-canonical regulators of these genes (81). Moreover, TRF protects mice lacking circadian clocks from diet-induced obesity and metabolic syndrome, also suggesting functional roles for non-canonical regulators (182). Identification of the mechanism and functions of non-canonical rhythmic regulators could provide insights into the molecular hierarchy of circadian regulation and provide targets for chronotherapy.
Last but not least, expanding our knowledge of the reciprocal regulation of circadian rhythm and metabolism is valuable for boosting the circadian clocks to improve quality of life, including chrononutritional and chronopharmacological approaches. Determination of the optimal times to rest, eat, exercise, and administer drugs throughout the day to restore the perturbed circadian machinery in shift workers and individuals with sleep disorders will ultimately contribute to improving the outcome of metabolic disorders.
Acknowledgments
We thank members of the Lazar laboratory (P. Dierickx, M. Tackenberg, and A. Tom) for their constructive feedback and careful reading of the manuscript. Figures were created with BioRender. We also acknowledge the many investigators who have contributed to this area of research and whose work, in many cases, could not be cited owing to the limitation of references allowed in this Review. This work was supported by the JPB Foundation (to MAL) and the Cox Medical Research Institute (to MAL) as well as by NIH grants (R01-DK045586 to MAL and K01-DK125602 to DG).
Version 1. 08/02/2021
Electronic publication
Footnotes
Conflict of interest: MAL is a member of the advisory board of, and receives research support from, Pfizer Inc.; is a consultant for Novartis; and is a member of the advisory board of, and holds equity in, Flame Therapeutics.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(15):e148278.https://doi.org/10.1172/JCI148278.
Contributor Information
Dongyin Guan, Email: dongyin@pennmedicine.upenn.edu.
Mitchell A. Lazar, Email: lazar@pennmedicine.upenn.edu.
References
- 1.Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science. 2016;354(6315):994–999. doi: 10.1126/science.aah4965. [DOI] [PubMed] [Google Scholar]
- 2.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330(6009):1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerhart-Hines Z, Lazar MA. Circadian metabolism in the light of evolution. Endocr Rev. 2015;36(3):289–304. doi: 10.1210/er.2015-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim YH, Lazar MA. Transcriptional control of circadian rhythms and metabolism: a matter of time and space. Endocr Rev. 2020;41(5):707–732. doi: 10.1210/endrev/bnaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fonken LK, Nelson RJ. The effects of light at night on circadian clocks and metabolism. Endocr Rev. 2014;35(4):648–670. doi: 10.1210/er.2013-1051. [DOI] [PubMed] [Google Scholar]
- 6.Allada R, Bass J. Circadian mechanisms in medicine. N Engl J Med. 2021;384(6):550–561. doi: 10.1056/NEJMra1802337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asher G, Sassone-Corsi P. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161(1):84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 8.Dohm GL, et al. Influence of fasting on glycogen depletion in rats during exercise. J Appl Physiol Respir Environ Exerc Physiol. 1983;55(3):830–833. doi: 10.1152/jappl.1983.55.3.830. [DOI] [PubMed] [Google Scholar]
- 9.Gardner MJ, et al. How plants tell the time. Biochem J. 2006;397(1):15–24. doi: 10.1042/BJ20060484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68(9):2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sehgal A. Physiology flies with time. Cell. 2017;171(6):1232–1235. doi: 10.1016/j.cell.2017.11.028. [DOI] [PubMed] [Google Scholar]
- 12.Reddy P, et al. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell. 1984;38(3):701–710. doi: 10.1016/0092-8674(84)90265-4. [DOI] [PubMed] [Google Scholar]
- 13.Zehring WA, et al. P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell. 1984;39(2 pt 1):369–376. doi: 10.1016/0092-8674(84)90015-1. [DOI] [PubMed] [Google Scholar]
- 14.Bargiello TA, et al. Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature. 1984;312(5996):752–754. doi: 10.1038/312752a0. [DOI] [PubMed] [Google Scholar]
- 15.Bargiello TA, Young MW. Molecular genetics of a biological clock in Drosophila. Proc Natl Acad Sci U S A. 1984;81(7):2142–2146. doi: 10.1073/pnas.81.7.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitaterna MH, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264(5159):719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18(3):164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dierickx P, et al. Circadian clocks: from stem cells to tissue homeostasis and regeneration. EMBO Rep. 2018;19(1):18–28. doi: 10.15252/embr.201745130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papazyan R, et al. Genetic and epigenomic mechanisms of mammalian circadian transcription. Nat Struct Mol Biol. 2016;23(12):1045–1052. doi: 10.1038/nsmb.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kume K, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98(2):193–205. doi: 10.1016/S0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 21.Zheng B, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105(5):683–694. doi: 10.1016/S0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 22.Zheng B, et al. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400(6740):169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 23.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485(7396):123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bugge A, et al. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26(7):657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331(6022):1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunger MK, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103(7):1009–1017. doi: 10.1016/S0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondratov RV, et al. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20(14):1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudic RD, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2(11):e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDearmon EL, et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314(5803):1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcheva B, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466(7306):627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan X, et al. Global and hepatocyte-specific ablation of Bmal1 induces hyperlipidaemia and enhances atherosclerosis. Nat Commun. 2016;7:13011. doi: 10.1038/ncomms13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobi D, et al. Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metab. 2015;22(4):709–720. doi: 10.1016/j.cmet.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paschos GK, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012;18(12):1768–1777. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durgan DJ, et al. Evidence suggesting that the cardiomyocyte circadian clock modulates responsiveness of the heart to hypertrophic stimuli in mice. Chronobiol Int. 2011;28(3):187–203. doi: 10.3109/07420528.2010.550406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young ME, et al. Cardiomyocyte-specific BMAL1 plays critical roles in metabolism, signaling, and maintenance of contractile function of the heart. J Biol Rhythms. 2014;29(4):257–276. doi: 10.1177/0748730414543141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harfmann BD, et al. Muscle-specific loss of Bmal1 leads to disrupted tissue glucose metabolism and systemic glucose homeostasis. Skelet Muscle. 2016;6:12. doi: 10.1186/s13395-016-0082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dyar KA, et al. 2014. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol Metab. 2013;3(1):29–41. doi: 10.1016/j.molmet.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexander RK, et al. Bmal1 integrates mitochondrial metabolism and macrophage activation. Elife. 2020;9:e54090. doi: 10.7554/eLife.54090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.King DP, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89(4):641–653. doi: 10.1016/S0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antoch MP, et al. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89(4):655–667. doi: 10.1016/S0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King DP, et al. The mouse Clock mutation behaves as an antimorph and maps within the W19H deletion, distal of Kit. Genetics. 1997;146(3):1049–1060. doi: 10.1093/genetics/146.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gekakis N, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280(5369):1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 43.Pan X, et al. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab. 2010;12(2):174–186. doi: 10.1016/j.cmet.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308(5724):1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kudo T, et al. Attenuating effect of clock mutation on triglyceride contents in the ICR mouse liver under a high-fat diet. J Biol Rhythms. 2007;22(4):312–323. doi: 10.1177/0748730407302625. [DOI] [PubMed] [Google Scholar]
- 46.Debruyne JP, et al. A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50(3):465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 47.Zhou YD, et al. Molecular characterization of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc Natl Acad Sci U S A. 1997;94(2):713–718. doi: 10.1073/pnas.94.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hogenesch JB, et al. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J Biol Chem. 1997;272(13):8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 49.Garcia JA, et al. Impaired cued and contextual memory in NPAS2-deficient mice. Science. 2000;288(5474):2226–2230. doi: 10.1126/science.288.5474.2226. [DOI] [PubMed] [Google Scholar]
- 50.DeBruyne JP, et al. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10(5):543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reick M, et al. NPAS2: an analog of clock operative in the mammalian forebrain. Science. 2001;293(5529):506–509. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- 52.Wu X, et al. NPAS2 deletion impairs responses to restricted feeding but not to metabolic challenges. Physiol Behav. 2010;99(4):466–471. doi: 10.1016/j.physbeh.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeBruyne JP, et al. Peripheral circadian oscillators require CLOCK. Curr Biol. 2007;17(14):R538–R539. doi: 10.1016/j.cub.2007.05.067. [DOI] [PubMed] [Google Scholar]
- 54.Lazar MA, et al. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA alpha transcriptional unit. Mol Cell Biol. 1989;9(3):1128–1136. doi: 10.1128/mcb.9.3.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonnelye E, et al. Rev-erb beta, a new member of the nuclear receptor superfamily, is expressed in the nervous system during chicken development. Cell Growth Differ. 1994;5(12):1357–1365. [PubMed] [Google Scholar]
- 56.Burke L, et al. Transcriptional repression by the orphan steroid receptor RVR/Rev-erb beta is dependent on the signature motif and helix 5 in the E region: functional evidence for a biological role of RVR in myogenesis. Nucleic Acids Res. 1996;24(18):3481–3489. doi: 10.1093/nar/24.18.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harding HP, Lazar MA. The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Mol Cell Biol. 1995;15(9):4791–4802. doi: 10.1128/MCB.15.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/S0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 59.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19(6):1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, et al. Gene regulation. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science. 2015;348(6242):1488–1492. doi: 10.1126/science.aab3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Welch RD, et al. Rev-Erb co-regulates muscle regeneration via tethered interaction with the NF-Y cistrome. Mol Metab. 2017;6(7):703–714. doi: 10.1016/j.molmet.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zamir I, et al. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev. 1997;11(7):835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- 63.Beaudry JB, et al. Threshold levels of hepatocyte nuclear factor 6 (HNF-6) acting in synergy with HNF-4 and PGC-1alpha are required for time-specific gene expression during liver development. Mol Cell Biol. 2006;26(16):6037–6046. doi: 10.1128/MCB.02445-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Odom DT, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303(5662):1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rubins NE, et al. Transcriptional networks in the liver: hepatocyte nuclear factor 6 function is largely independent of Foxa2. Mol Cell Biol. 2005;25(16):7069–7077. doi: 10.1128/MCB.25.16.7069-7077.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang C, et al. The nuclear receptor REV-ERBα modulates Th17 cell-mediated autoimmune disease. Proc Natl Acad Sci U S A. 2019;116(37):18528–18536. doi: 10.1073/pnas.1907563116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Duez H, et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology. 2008;135(2):689–698. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 68.Raspe E, et al. Identification of Rev-erbalpha as a physiological repressor of apoC-III gene transcription. J Lipid Res. 2002;43(12):2172–2179. doi: 10.1194/jlr.M200386-JLR200. [DOI] [PubMed] [Google Scholar]
- 69.Lam MT, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498(7455):511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yin L, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318(5857):1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 71.Wu N, et al. Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbalpha. Genes Dev. 2009;23(18):2201–2209. doi: 10.1101/gad.1825809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J, Lazar MA. Bifunctional role of Rev-erbalpha in adipocyte differentiation. Mol Cell Biol. 2008;28(7):2213–2220. doi: 10.1128/MCB.01608-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chawla A, Lazar MA. Induction of Rev-ErbA alpha, an orphan receptor encoded on the opposite strand of the alpha-thyroid hormone receptor gene, during adipocyte differentiation. J Biol Chem. 1993;268(22):16265–16269. doi: 10.1016/S0021-9258(19)85415-7. [DOI] [PubMed] [Google Scholar]
- 74.Fontaine C, et al. The orphan nuclear receptor Rev-Erbalpha is a peroxisome proliferator-activated receptor (PPAR) gamma target gene and promotes PPARgamma-induced adipocyte differentiation. J Biol Chem. 2003;278(39):37672–37680. doi: 10.1074/jbc.M304664200. [DOI] [PubMed] [Google Scholar]
- 75.Jager J, et al. The nuclear receptor Rev-erbα regulates adipose tissue-specific FGF21 signaling. J Biol Chem. 2016;291(20):10867–10875. doi: 10.1074/jbc.M116.719120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Delezie J, et al. The nuclear receptor REV-ERBα is required for the daily balance of carbohydrate and lipid metabolism. FASEB J. 2012;26(8):3321–3335. doi: 10.1096/fj.12-208751. [DOI] [PubMed] [Google Scholar]
- 77.Gerhart-Hines Z, et al. The nuclear receptor Rev-erbα controls circadian thermogenic plasticity. Nature. 2013;503(7476):410–413. doi: 10.1038/nature12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woldt E, et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med. 2013;19(8):1039–1046. doi: 10.1038/nm.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ding G, et al. REV-ERB in GABAergic neurons controls diurnal hepatic insulin sensitivity. Nature. 2021;592(7856):763–767. doi: 10.1038/s41586-021-03358-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Adlanmerini M, et al. Hypothalamic REV-ERB nuclear receptors control diurnal food intake and leptin sensitivity in diet-induced obese mice. J Clin Invest. 2021;131(1):e140424. doi: 10.1172/JCI140424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guan D, et al. The hepatocyte clock and feeding control chronophysiology of multiple liver cell types. Science. 2020;369(6509):1388–1394. doi: 10.1126/science.aba8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dierickx P, et al. SR9009 has REV-ERB-independent effects on cell proliferation and metabolism. Proc Natl Acad Sci U S A. 2019;116(25):12147–12152. doi: 10.1073/pnas.1904226116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Giguere V, et al. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8(5):538–553. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- 84.Carlberg C, et al. RZRs, a new family of retinoid-related orphan receptors that function as both monomers and homodimers. Mol Endocrinol. 1994;8(6):757–770. doi: 10.1210/mend.8.6.7935491. [DOI] [PubMed] [Google Scholar]
- 85.Hirose T, et al. ROR gamma: the third member of ROR/RZR orphan receptor subfamily that is highly expressed in skeletal muscle. Biochem Biophys Res Commun. 1994;205(3):1976–1983. doi: 10.1006/bbrc.1994.2902. [DOI] [PubMed] [Google Scholar]
- 86.Solt LA, Burris TP. Action of RORs and their ligands in (patho)physiology. Trends Endocrinol Metab. 2012;23(12):619–627. doi: 10.1016/j.tem.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lau P, et al. Rorα deficiency and decreased adiposity are associated with induction of thermogenic gene expression in subcutaneous white adipose and brown adipose tissue. Am J Physiol Endocrinol Metab. 2015;308(2):E159–E171. doi: 10.1152/ajpendo.00056.2014. [DOI] [PubMed] [Google Scholar]
- 88.Kang HS, et al. Transcriptional profiling reveals a role for RORalpha in regulating gene expression in obesity-associated inflammation and hepatic steatosis. Physiol Genomics. 2011;43(13):818–828. doi: 10.1152/physiolgenomics.00206.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lau P, et al. Homozygous staggerer (sg/sg) mice display improved insulin sensitivity and enhanced glucose uptake in skeletal muscle. Diabetologia. 2011;54(5):1169–1180. doi: 10.1007/s00125-011-2046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wada T, et al. Identification of oxysterol 7alpha-hydroxylase (Cyp7b1) as a novel retinoid-related orphan receptor alpha (RORalpha) (NR1F1) target gene and a functional cross-talk between RORalpha and liver X receptor (NR1H3) Mol Pharmacol. 2008;73(3):891–899. doi: 10.1124/mol.107.040741. [DOI] [PubMed] [Google Scholar]
- 91.Zhang Y, et al. The hepatic circadian clock fine-tunes the lipogenic response to feeding through RORα/γ. Genes Dev. 2017;31(12):1202–1211. doi: 10.1101/gad.302323.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meissburger B, et al. Adipogenesis and insulin sensitivity in obesity are regulated by retinoid-related orphan receptor gamma. EMBO Mol Med. 2011;3(11):637–651. doi: 10.1002/emmm.201100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takeda Y, et al. Retinoid acid-related orphan receptor γ, RORγ, participates in diurnal transcriptional regulation of lipid metabolic genes. Nucleic Acids Res. 2014;42(16):10448–10459. doi: 10.1093/nar/gku766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takeda Y, et al. Retinoic acid-related orphan receptor γ (RORγ): a novel participant in the diurnal regulation of hepatic gluconeogenesis and insulin sensitivity. PLoS Genet. 2014;10(5):e1004331. doi: 10.1371/journal.pgen.1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Raichur S, et al. Retinoid-related orphan receptor gamma regulates several genes that control metabolism in skeletal muscle cells: links to modulation of reactive oxygen species production. J Mol Endocrinol. 2007;39(1):29–44. doi: 10.1677/jme.1.00010. [DOI] [PubMed] [Google Scholar]
- 96.Eberl G, et al. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5(1):64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- 97.Sun ZS, et al. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90(6):1003–1011. doi: 10.1016/S0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 98.Shearman LP, et al. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19(6):1261–1269. doi: 10.1016/S0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 99.Zylka MJ, et al. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20(6):1103–1110. doi: 10.1016/S0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 100.Takumi T, et al. A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J. 1998;17(16):4753–4759. doi: 10.1093/emboj/17.16.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van der Spek PJ, et al. Cloning, tissue expression, and mapping of a human photolyase homolog with similarity to plant blue-light receptors. Genomics. 1996;37(2):177–182. doi: 10.1006/geno.1996.0539. [DOI] [PubMed] [Google Scholar]
- 102.Ishida N, et al. Biological clocks. Proc Natl Acad Sci U S A. 1999;96(16):8819–8820. doi: 10.1073/pnas.96.16.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Akashi M, et al. Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells. Mol Cell Biol. 2002;22(6):1693–1703. doi: 10.1128/MCB.22.6.1693-1703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Eide EJ, et al. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon. J Biol Chem. 2002;277(19):17248–17254. doi: 10.1074/jbc.M111466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xu Y, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434(7033):640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 106.Bae K, et al. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30(2):525–536. doi: 10.1016/S0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 107.Stow LR, et al. The circadian protein period 1 contributes to blood pressure control and coordinately regulates renal sodium transport genes. Hypertension. 2012;59(6):1151–1156. doi: 10.1161/HYPERTENSIONAHA.112.190892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang S, et al. The role of mPer2 clock gene in glucocorticoid and feeding rhythms. Endocrinology. 2009;150(5):2153–2160. doi: 10.1210/en.2008-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Grimaldi B, et al. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 2010;12(5):509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Adamovich Y, et al. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 2014;19(2):319–330. doi: 10.1016/j.cmet.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Eckle T, et al. Adora2b-elicited Per2 stabilization promotes a HIF-dependent metabolic switch crucial for myocardial adaptation to ischemia. Nat Med. 2012;18(5):774–782. doi: 10.1038/nm.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dallmann R, Weaver DR. Altered body mass regulation in male mPeriod mutant mice on high-fat diet. Chronobiol Int. 2010;27(6):1317–1328. doi: 10.3109/07420528.2010.489166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Costa MJ, et al. Circadian rhythm gene period 3 is an inhibitor of the adipocyte cell fate. J Biol Chem. 2011;286(11):9063–9070. doi: 10.1074/jbc.M110.164558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.van der Horst GT, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398(6728):627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 115.Griebel G, et al. Mice deficient in cryptochrome 1 (cry1 (-/-)) exhibit resistance to obesity induced by a high-fat diet. Front Endocrinol (Lausanne) 2014;5:49. doi: 10.3389/fendo.2014.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barclay JL, et al. High-fat diet-induced hyperinsulinemia and tissue-specific insulin resistance in Cry-deficient mice. Am J Physiol Endocrinol Metab. 2013;304(10):E1053–E1063. doi: 10.1152/ajpendo.00512.2012. [DOI] [PubMed] [Google Scholar]
- 117.Zhang EE, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16(10):1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lamia KA, et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480(7378):552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jang H, et al. SREBP1c-CRY1 signalling represses hepatic glucose production by promoting FOXO1 degradation during refeeding. Nat Commun. 2016;7:12180. doi: 10.1038/ncomms12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kettner NM, et al. Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell. 2016;30(6):909–924. doi: 10.1016/j.ccell.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fang B, et al. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014;159(5):1140–1152. doi: 10.1016/j.cell.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38(3):369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 124.Munoz E, et al. Modulation of BMAL/CLOCK/E-Box complex activity by a CT-rich cis-acting element. Mol Cell Endocrinol. 2006;252(1–2):74–81. doi: 10.1016/j.mce.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 125.Kriebs A, et al. Circadian repressors CRY1 and CRY2 broadly interact with nuclear receptors and modulate transcriptional activity. Proc Natl Acad Sci U S A. 2017;114(33):8776–8781. doi: 10.1073/pnas.1704955114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ripperger JA, et al. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14(6):679–689. [PMC free article] [PubMed] [Google Scholar]
- 127.Rey G, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9(2):e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mistlberger RE, Skene DJ. Social influences on mammalian circadian rhythms: animal and human studies. Biol Rev Camb Philos Soc. 2004;79(3):533–556. doi: 10.1017/S1464793103006353. [DOI] [PubMed] [Google Scholar]
- 129.Eisenstein M. Chronobiology: stepping out of time. Nature. 2013;497(7450):S10–S12. doi: 10.1038/497S10a. [DOI] [PubMed] [Google Scholar]
- 130.Panda S. Circadian physiology of metabolism. Science. 2016;354(6315):1008–1015. doi: 10.1126/science.aah4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fukuda Y, Morita T. Effects of the light-dark cycle on diurnal rhythms of diet-induced thermogenesis in humans. Chronobiol Int. 2017;34(10):1465–1472. doi: 10.1080/07420528.2017.1362422. [DOI] [PubMed] [Google Scholar]
- 132.Plano SA, et al. Circadian and metabolic effects of light: implications in weight homeostasis and health. Front Neurol. 2017;8:558. doi: 10.3389/fneur.2017.00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lund J, et al. Postprandial hormone and metabolic responses amongst shift workers in Antarctica. J Endocrinol. 2001;171(3):557–564. doi: 10.1677/joe.0.1710557. [DOI] [PubMed] [Google Scholar]
- 134.Sookoian S, et al. Effects of rotating shift work on biomarkers of metabolic syndrome and inflammation. J Intern Med. 2007;261(3):285–292. doi: 10.1111/j.1365-2796.2007.01766.x. [DOI] [PubMed] [Google Scholar]
- 135.Lee JY, et al. Dose-response relationship between night work and the prevalence of impaired fasting glucose: the Korean worker’s special health examination for night workers cohort. Int J Environ Res Public Health. 2021;18(4):1854. doi: 10.3390/ijerph18041854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lalla-Edward ST, et al. Cross-sectional study of the health of southern African truck drivers. BMJ Open. 2019;9(10):e032025. doi: 10.1136/bmjopen-2019-032025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kozlowska L, et al. Health risk in transport workers. Part II. Dietary compounds as modulators of occupational exposure to chemicals. Int J Occup Med Environ Health. 2019;32(4):441–464. doi: 10.13075/ijomeh.1896.01434. [DOI] [PubMed] [Google Scholar]
- 138.Ritonja J, et al. The relationship between night work, chronotype, and cardiometabolic risk factors in female hospital employees. Chronobiol Int. 2019;36(5):616–628. doi: 10.1080/07420528.2019.1570247. [DOI] [PubMed] [Google Scholar]
- 139.Gatford KL, et al. Simulated shift work disrupts maternal circadian rhythms and metabolism, and increases gestation length in sheep. J Physiol. 2019;597(7):1889–1904. doi: 10.1113/JP277186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shan Z, et al. Rotating night shift work and adherence to unhealthy lifestyle in predicting risk of type 2 diabetes: results from two large US cohorts of female nurses. BMJ. 2018;363:k4641. doi: 10.1136/bmj.k4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Itani O, et al. Short sleep duration, shift work, and actual days taken off work are predictive life-style risk factors for new-onset metabolic syndrome: a seven-year cohort study of 40,000 male workers. Sleep Med. 2017;39:87–94. doi: 10.1016/j.sleep.2017.07.027. [DOI] [PubMed] [Google Scholar]
- 142.Kivimaki M, et al. Long working hours, socioeconomic status, and the risk of incident type 2 diabetes: a meta-analysis of published and unpublished data from 222 120 individuals. Lancet Diabetes Endocrinol. 2015;3(1):27–34. doi: 10.1016/S2213-8587(14)70178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Borck PC, et al. Nighttime light exposure enhances Rev-erbα-targeting microRNAs and contributes to hepatic steatosis. Metabolism. 2018;85:250–258. doi: 10.1016/j.metabol.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mukherji A, et al. The circadian clock and liver function in health and disease. J Hepatol. 2019;71(1):200–211. doi: 10.1016/j.jhep.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ohta H, et al. Constant light desynchronizes mammalian clock neurons. Nat Neurosci. 2005;8(3):267–269. doi: 10.1038/nn1395. [DOI] [PubMed] [Google Scholar]
- 146.Coomans CP, et al. Detrimental effects of constant light exposure and high-fat diet on circadian energy metabolism and insulin sensitivity. FASEB J. 2013;27(4):1721–1732. doi: 10.1096/fj.12-210898. [DOI] [PubMed] [Google Scholar]
- 147.Madahi PG, et al. Constant light during lactation programs circadian and metabolic systems. Chronobiol Int. 2018;35(8):1153–1167. doi: 10.1080/07420528.2018.1465070. [DOI] [PubMed] [Google Scholar]
- 148.Kasahara T, et al. Genetic variation of melatonin productivity in laboratory mice under domestication. Proc Natl Acad Sci U S A. 2010;107(14):6412–6417. doi: 10.1073/pnas.0914399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kennaway DJ, et al. Melatonin in mice: rhythms, response to light, adrenergic stimulation, and metabolism. Am J Physiol Regul Integr Comp Physiol. 2002;282(2):R358–R365. doi: 10.1152/ajpregu.00360.2001. [DOI] [PubMed] [Google Scholar]
- 150.Roseboom PH, et al. Natural melatonin ‘knockdown’ in C57BL/6J mice: rare mechanism truncates serotonin N-acetyltransferase. Brain Res Mol Brain Res. 1998;63(1):189–197. doi: 10.1016/S0169-328X(98)00273-3. [DOI] [PubMed] [Google Scholar]
- 151.Depres-Brummer P, et al. Light-induced suppression of the rat circadian system. Am J Physiol. 1995;268(5 pt 2):R1111–R1116. doi: 10.1152/ajpregu.1995.268.5.R1111. [DOI] [PubMed] [Google Scholar]
- 152.Fonken LK, et al. Light at night increases body mass by shifting the time of food intake. Proc Natl Acad Sci U S A. 2010;107(43):18664–18669. doi: 10.1073/pnas.1008734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fonken LK, et al. Dim light at night disrupts molecular circadian rhythms and increases body weight. J Biol Rhythms. 2013;28(4):262–271. doi: 10.1177/0748730413493862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bedrosian TA, et al. Nocturnal light exposure impairs affective responses in a wavelength-dependent manner. J Neurosci. 2013;33(32):13081–13087. doi: 10.1523/JNEUROSCI.5734-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nagai N, et al. Suppression of blue light at night ameliorates metabolic abnormalities by controlling circadian rhythms. Invest Ophthalmol Vis Sci. 2019;60(12):3786–3793. doi: 10.1167/iovs.19-27195. [DOI] [PubMed] [Google Scholar]
- 156.Masana MI, et al. Light-induced c-fos mRNA expression in the suprachiasmatic nucleus and the retina of C3H/HeN mice. Brain Res Mol Brain Res. 1996;42(2):193–201. doi: 10.1016/S0169-328X(96)00031-9. [DOI] [PubMed] [Google Scholar]
- 157.Aronin N, et al. Light regulates expression of a Fos-related protein in rat suprachiasmatic nuclei. Proc Natl Acad Sci U S A. 1990;87(15):5959–5962. doi: 10.1073/pnas.87.15.5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6(5):414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 159.Eckel-Mahan KL, et al. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155(7):1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Guan D, et al. Diet-induced circadian enhancer remodeling synchronizes opposing hepatic lipid metabolic processes. Cell. 2018;174(4):831–842. doi: 10.1016/j.cell.2018.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Freeman JM, et al. The ketogenic diet: one decade later. Pediatrics. 2007;119(3):535–543. doi: 10.1542/peds.2006-2447. [DOI] [PubMed] [Google Scholar]
- 162.Yang MU, Van Itallie TB. Composition of weight lost during short-term weight reduction. Metabolic responses of obese subjects to starvation and low-calorie ketogenic and nonketogenic diets. J Clin Invest. 1976;58(3):722–730. doi: 10.1172/JCI108519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gumbiner B, et al. Effects of diet composition and ketosis on glycemia during very-low-energy-diet therapy in obese patients with non-insulin-dependent diabetes mellitus. Am J Clin Nutr. 1996;63(1):110–115. doi: 10.1093/ajcn/63.1.110. [DOI] [PubMed] [Google Scholar]
- 164.Willi SM, et al. The effects of a high-protein, low-fat, ketogenic diet on adolescents with morbid obesity: body composition, blood chemistries, and sleep abnormalities. Pediatrics. 1998;101(1 pt 1):61–67. doi: 10.1542/peds.101.1.61. [DOI] [PubMed] [Google Scholar]
- 165.Dashti HM, et al. Ketogenic diet modifies the risk factors of heart disease in obese patients. Nutrition. 2003;19(10):901–902. doi: 10.1016/S0899-9007(03)00161-8. [DOI] [PubMed] [Google Scholar]
- 166.Aubert G, et al. The failing heart relies on ketone bodies as a fuel. Circulation. 2016;133(8):698–705. doi: 10.1161/CIRCULATIONAHA.115.017355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tognini P, et al. Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metab. 2017;26(3):523–538. doi: 10.1016/j.cmet.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 168.Katewa SD, et al. Peripheral circadian clocks mediate dietary restriction-dependent changes in lifespan and fat metabolism in Drosophila. Cell Metab. 2016;23(1):143–154. doi: 10.1016/j.cmet.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14(23):2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chaix A, et al. Time-restricted eating to prevent and manage chronic metabolic diseases. Annu Rev Nutr. 2019;39:291–315. doi: 10.1146/annurev-nutr-082018-124320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Depner CM, et al. Mistimed food intake and sleep alters 24-hour time-of-day patterns of the human plasma proteome. Proc Natl Acad Sci U S A. 2018;115(23):E5390–E5399. doi: 10.1073/pnas.1714813115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bray MS, et al. Quantitative analysis of light-phase restricted feeding reveals metabolic dyssynchrony in mice. Int J Obes (Lond) 2013;37(6):843–852. doi: 10.1038/ijo.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Arble DM, et al. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 2009;17(11):2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Alexander KE, et al. Association of breakfast skipping with visceral fat and insulin indices in overweight Latino youth. Obesity (Silver Spring) 2009;17(8):1528–1533. doi: 10.1038/oby.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ma Y, et al. Association between eating patterns and obesity in a free-living US adult population. Am J Epidemiol. 2003;158(1):85–92. doi: 10.1093/aje/kwg117. [DOI] [PubMed] [Google Scholar]
- 176.Resnicow K. The relationship between breakfast habits and plasma cholesterol levels in schoolchildren. J Sch Health. 1991;61(2):81–85. doi: 10.1111/j.1746-1561.1991.tb03242.x. [DOI] [PubMed] [Google Scholar]
- 177.Pereira MA, et al. Breakfast frequency and quality may affect glycemia and appetite in adults and children. J Nutr. 2011;141(1):163–168. doi: 10.3945/jn.109.114405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yoshida C, et al. Early nocturnal meal skipping alters the peripheral clock and increases lipogenesis in mice. Nutr Metab (Lond) 2012;9(1):78. doi: 10.1186/1743-7075-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hatori M, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15(6):848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tsai JY, et al. Influence of dark phase restricted high fat feeding on myocardial adaptation in mice. J Mol Cell Cardiol. 2013;55:147–155. doi: 10.1016/j.yjmcc.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chaix A, et al. Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 2014;20(6):991–1005. doi: 10.1016/j.cmet.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chaix A, et al. Time-restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metab. 2019;29(2):303–319. doi: 10.1016/j.cmet.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.de Cabo R, Mattson MP. Effects of intermittent fasting on health, aging, and disease [published correction appears in N Engl J Med. 2020;382(10):978] N Engl J Med. 2019;381:2541–2551. doi: 10.1056/NEJMra1905136. [DOI] [PubMed] [Google Scholar]
- 184.Chow LS, et al. Time-restricted eating effects on body composition and metabolic measures in humans who are overweight: a feasibility study. Obesity (Silver Spring) 2020;28(5):860–869. doi: 10.1002/oby.22756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hutchison AT, et al. Time-restricted feeding improves glucose tolerance in men at risk for type 2 diabetes: a randomized crossover trial. Obesity (Silver Spring) 2019;27(5):724–732. doi: 10.1002/oby.22449. [DOI] [PubMed] [Google Scholar]
- 186.Jamshed H, et al. Early time-restricted feeding improves 24-hour glucose levels and affects markers of the circadian clock, aging, and autophagy in humans. Nutrients. 2019;11(6):E1234. doi: 10.3390/nu11061234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lowe DA, et al. Effects of time-restricted eating on weight loss and other metabolic parameters in women and men with overweight and obesity: the TREAT randomized clinical trial. JAMA Intern Med. 2020;180(11):1491–1499. doi: 10.1001/jamainternmed.2020.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Peeke PM, et al. Effect of time restricted eating on body weight and fasting glucose in participants with obesity: results of a randomized, controlled, virtual clinical trial. Nutr Diabetes. 2021;11(1):6. doi: 10.1038/s41387-021-00149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Egan B, Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013;17(2):162–184. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 190.Wolff G, Esser KA. Scheduled exercise phase shifts the circadian clock in skeletal muscle. Med Sci Sports Exerc. 2012;44(9):1663–1670. doi: 10.1249/MSS.0b013e318255cf4c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Marchant EG, Mistlberger RE. Entrainment and phase shifting of circadian rhythms in mice by forced treadmill running. Physiol Behav. 1996;60(2):657–663. doi: 10.1016/s0031-9384(96)80045-x. [DOI] [PubMed] [Google Scholar]
- 192.Hong S, et al. Dissociation of muscle insulin sensitivity from exercise endurance in mice by HDAC3 depletion. Nat Med. 2016;23(2):223–234. doi: 10.1038/nm.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lundell LS, et al. Time-restricted feeding alters lipid and amino acid metabolite rhythmicity without perturbing clock gene expression. Nat Commun. 2020;11(1):4643. doi: 10.1038/s41467-020-18412-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sato S, et al. Time of exercise specifies the impact on muscle metabolic pathways and systemic energy homeostasis. Cell Metab. 2019;30(1):92–110. doi: 10.1016/j.cmet.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 195.Atkinson G, Reilly T. Circadian variation in sports performance. Sports Med. 1996;21(4):292–312. doi: 10.2165/00007256-199621040-00005. [DOI] [PubMed] [Google Scholar]
- 196.Facer-Childs E, Brandstaetter R. The impact of circadian phenotype and time since awakening on diurnal performance in athletes. Curr Biol. 2015;25(4):518–522. doi: 10.1016/j.cub.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 197.Chtourou H, Souissi N. The effect of training at a specific time of day: a review. J Strength Cond Res. 2012;26(7):1984–2005. doi: 10.1519/JSC.0b013e31825770a7. [DOI] [PubMed] [Google Scholar]
- 198.Kuusmaa M, et al. Effects of morning versus evening combined strength and endurance training on physical performance, muscle hypertrophy, and serum hormone concentrations. Appl Physiol Nutr Metab. 2016;41(12):1285–1294. doi: 10.1139/apnm-2016-0271. [DOI] [PubMed] [Google Scholar]
- 199.Buhr ED, et al. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330(6002):379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Brown SA, et al. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol. 2002;12(18):1574–1583. doi: 10.1016/S0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- 201.Gaucher J, et al. Distinct metabolic adaptation of liver circadian pathways to acute and chronic patterns of alcohol intake. Proc Natl Acad Sci U S A. 2019;116(50):25250–25259. doi: 10.1073/pnas.1911189116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sato S, et al. Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell. 2017;170(4):664–677. doi: 10.1016/j.cell.2017.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bailey M, Silver R. Sex differences in circadian timing systems: implications for disease. Front Neuroendocrinol. 2014;35(1):111–139. doi: 10.1016/j.yfrne.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Boivin DB, et al. Diurnal and circadian variation of sleep and alertness in men vs. naturally cycling women. Proc Natl Acad Sci U S A. 2016;113(39):10980–10985. doi: 10.1073/pnas.1524484113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Masri S, et al. Lung adenocarcinoma distally rewires hepatic circadian homeostasis. Cell. 2016;165(4):896–909. doi: 10.1016/j.cell.2016.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Thaiss CA, et al. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell. 2016;167(6):1495–1510. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 207.Tuganbaev T, et al. Diet diurnally regulates small intestinal microbiome-epithelial-immune homeostasis and enteritis. Cell. 2020;182(6):1441–1459. doi: 10.1016/j.cell.2020.08.027. [DOI] [PubMed] [Google Scholar]
- 208.Adamovich Y, et al. Rhythmic oxygen levels reset circadian clocks through HIF1α. Cell Metab. 2017;25(1):93–101. doi: 10.1016/j.cmet.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 209.Peek CB, et al. Circadian clock interaction with HIF1α mediates oxygenic metabolism and anaerobic glycolysis in skeletal muscle. Cell Metab. 2017;25(1):86–92. doi: 10.1016/j.cmet.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wu Y, et al. Reciprocal regulation between the circadian clock and hypoxia signaling at the genome level in mammals. Cell Metab. 2017;25(1):73–85. doi: 10.1016/j.cmet.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 211.Manella G, et al. Hypoxia induces a time- and tissue-specific response that elicits intertissue circadian clock misalignment. Proc Natl Acad Sci U S A. 2020;117(1):779–786. doi: 10.1073/pnas.1914112117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Clasadonte J, Prevot V. The special relationship: glia-neuron interactions in the neuroendocrine hypothalamus. Nat Rev Endocrinol. 2018;14(1):25–44. doi: 10.1038/nrendo.2017.124. [DOI] [PubMed] [Google Scholar]
- 213.Richards JS, Ascoli M. Endocrine, paracrine, and autocrine signaling pathways that regulate ovulation. Trends Endocrinol Metab. 2018;29(5):313–325. doi: 10.1016/j.tem.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 214.Bruning JC, et al. Role of brain insulin receptor in control of body weight and reproduction. Science. 2000;289(5487):2122–2125. doi: 10.1126/science.289.5487.2122. [DOI] [PubMed] [Google Scholar]
- 215.Liu S, et al. A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature. 2013;502(7472):550–554. doi: 10.1038/nature12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Obici S, et al. Hypothalamic insulin signaling is required for inhibition of glucose production. Nat Med. 2002;8(12):1376–1382. doi: 10.1038/nm1202-798. [DOI] [PubMed] [Google Scholar]
- 217.Whittle AJ, et al. BMP8B increases brown adipose tissue thermogenesis through both central and peripheral actions. Cell. 2012;149(4):871–885. doi: 10.1016/j.cell.2012.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Simcox J, et al. Global analysis of plasma lipids identifies liver-derived acylcarnitines as a fuel source for brown fat thermogenesis. Cell Metab. 2017;26(3):509–522. doi: 10.1016/j.cmet.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Inagaki T, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5(6):415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 220.Dyar KA, et al. Atlas of circadian metabolism reveals system-wide coordination and communication between clocks. Cell. 2018;174(6):1571–1585. doi: 10.1016/j.cell.2018.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Xiong X, et al. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol Cell. 2019;75(3):644–660. doi: 10.1016/j.molcel.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hu P, et al. Single-nucleus transcriptomic survey of cell diversity and functional maturation in postnatal mammalian hearts. Genes Dev. 2018;32(19–20):1344–1357. doi: 10.1101/gad.316802.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Park J, et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360(6390):758–763. doi: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wang H, et al. Single-cell in vivo imaging of cellular circadian oscillators in zebrafish. PLoS Biol. 2020;18(3):e3000435. doi: 10.1371/journal.pbio.3000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ofengeim D, et al. Single-cell RNA sequencing: unraveling the brain one cell at a time. Trends Mol Med. 2017;23(6):563–576. doi: 10.1016/j.molmed.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Droin C, et al. Space-time logic of liver gene expression at sub-lobular scale. Nat Metab. 2021;3(1):43–58. doi: 10.1038/s42255-020-00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Koronowski KB, et al. Defining the independence of the liver circadian clock. Cell. 2019;177(6):1448–1462. doi: 10.1016/j.cell.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Welz PS, et al. BMAL1-driven tissue clocks respond independently to light to maintain homeostasis. Cell. 2019;177(6):1436–1447. doi: 10.1016/j.cell.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sharma A, et al. Glucose metabolism during rotational shift-work in healthcare workers. Diabetologia. 2017;60(8):1483–1490. doi: 10.1007/s00125-017-4317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wefers J, et al. Circadian misalignment induces fatty acid metabolism gene profiles and compromises insulin sensitivity in human skeletal muscle. Proc Natl Acad Sci U S A. 2018;115(30):7789–7794. doi: 10.1073/pnas.1722295115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Morris CJ, et al. Endogenous circadian system and circadian misalignment impact glucose tolerance via separate mechanisms in humans. Proc Natl Acad Sci U S A. 2015;112(17):E2225–E2234. doi: 10.1073/pnas.1418955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Leproult R, et al. Circadian misalignment augments markers of insulin resistance and inflammation, independently of sleep loss. Diabetes. 2014;63(6):1860–1869. doi: 10.2337/db13-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Scheer FA, et al. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 2009;106(11):4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Knutsson A, et al. Increased risk of ischaemic heart disease in shift workers. Lancet. 1986;2(8498):89–92. doi: 10.1016/s0140-6736(86)91619-3. [DOI] [PubMed] [Google Scholar]
- 235.Spiegel K, et al. Impact of sleep debt on metabolic and endocrine function. Lancet. 1999;354(9188):1435–1439. doi: 10.1016/S0140-6736(99)01376-8. [DOI] [PubMed] [Google Scholar]
- 236.Jakubowicz D, et al. High caloric intake at breakfast vs. dinner differentially influences weight loss of overweight and obese women. Obesity (Silver Spring) 2013;21(12):2504–2512. doi: 10.1002/oby.20460. [DOI] [PubMed] [Google Scholar]
- 237.Jakubowicz D, et al. High-energy breakfast with low-energy dinner decreases overall daily hyperglycaemia in type 2 diabetic patients: a randomised clinical trial. Diabetologia. 2015;58(5):912–919. doi: 10.1007/s00125-015-3524-9. [DOI] [PubMed] [Google Scholar]
- 238.Ravussin E, et al. Early time-restricted feeding reduces appetite and increases fat oxidation but does not affect energy expenditure in humans. Obesity (Silver Spring) 2019;27(8):1244–1254. doi: 10.1002/oby.22518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Gill S, Panda S. A smartphone app reveals erratic diurnal eating patterns in humans that can be modulated for health benefits. Cell Metab. 2015;22(5):789–798. doi: 10.1016/j.cmet.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Sutton EF, et al. Early time-restricted feeding improves insulin sensitivity, blood pressure, and oxidative stress even without weight loss in men with prediabetes. Cell Metab. 2018;27(6):1212–1221. doi: 10.1016/j.cmet.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Carter S, et al. Effect of intermittent compared with continuous energy restricted diet on glycemic control in patients with type 2 diabetes: a randomized noninferiority trial. JAMA Netw Open. 2018;1(3):e180756. doi: 10.1001/jamanetworkopen.2018.0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ezagouri S, et al. Physiological and molecular dissection of daily variance in exercise capacity. Cell Metab. 2019;30(1):78–91. doi: 10.1016/j.cmet.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 243.Savikj M, et al. Afternoon exercise is more efficacious than morning exercise at improving blood glucose levels in individuals with type 2 diabetes: a randomised crossover trial. Diabetologia. 2019;62(2):233–237. doi: 10.1007/s00125-018-4767-z. [DOI] [PMC free article] [PubMed] [Google Scholar]