Abstract
Oogenesis is a complex process resulting in the production of a truly remarkable cell—the oocyte. Oocytes execute many unique processes and functions such as meiotic segregation of maternal genetic material, and essential life-generating functions after fertilization including posttranscriptional support of essential homeostatic and metabolic processes, and activation and reprogramming of the embryonic genome. An essential goal for understanding female fertility and infertility in mammals is to discover critical features driving the production of quality oocytes, particularly the complex regulation of oocyte maternal mRNAs. We report here the first in-depth meta-analysis of oocyte maturation-associated transcriptome changes, using eight datasets encompassing 94 RNAseq libraries for human, rhesus monkey, mouse, and cow. A majority of maternal mRNAs are regulated in a species-restricted manner, highlighting considerable divergence in oocyte transcriptome handling during maturation. We identified 121 mRNAs changing in relative abundance similarly across all four species (92 of high homology), and 993 (670 high homology) mRNAs regulated similarly in at least three of the four species, corresponding to just 0.84% and 6.9% of mRNAs analyzed. Ingenuity Pathway Analysis (IPA) revealed an association of these shared mRNAs with many shared pathways and functions, most prominently oxidative phosphorylation and mitochondrial function. These shared functions were reinforced further by primate-specific and species-specific differentially expressed genes (DEGs). Thus, correct downregulation of mRNAs related to oxidative phosphorylation and mitochondrial function is a major shared feature of mammalian oocyte maturation.
Keywords: maternal mRNA, meiosis, meta-analysis, mitochondria, oxidative phosphorylation
INTRODUCTION
Rodent models, particularly mice, comprise the predominant animal models in biomedical research, owing to small size, ease of manipulation and husbandry, available tools for genetic manipulation, and an ever-increasing legacy of genomics, genetic, and other data to enable rapid hypothesis testing. However, although rodent models are highly valuable for some basic studies of mammalian biology, significant differences across species limit the value of rodent models, particularly in reproductive biology, where litter-bearing rodents have clearly different modes of regulation compared with mono-ovular species. In addition, it is well-established that even some of the most fundamental developmental events in the life of every mammal, such as early cell lineage commitment of cells to inner cell mass or trophectoderm, can differ across species in key mechanistic respects (1–4). This suggests that a substantial amount of variation may exist in controlling mechanisms relevant to reproductive biology, and understanding that variation is important for understanding the limits to which any given model organism informs us about human reproductive biology.
That mice are useful models for some aspects of human reproduction, whereas other species (e.g., cow) are more useful for other aspects was noted nearly two decades ago (5). The implicit lessons are that there is much to be learned by taking advantage of multiple mammalian model species to better understand the human embryo or embryos of any given species. In addition, it is important to understand which aspects of each species are shared, which are species-specific, and which are relevant to understanding human biology. Despite these obvious conclusions, relatively little headway has been made to date on the incorporation of different mammalian models into our quest to understand human reproduction.
There are likely a variety of reasons underlying the limited use of diverse mammalian species to understand human reproduction, such as feasibility, cost, type of study (in vivo vs. in vitro), and biased perceptions of relevance. Despite such limitations, these other species have been extensively employed, including in recent studies using more current technologies such as transcriptome analysis (e.g., see Refs. 6–17). But efficient use of data from diverse species has been limited. Most data have been used for addressing immediate and narrowly focused questions of interest. Differences in developmental timing, assay platforms, and interlaboratory variations in methodology have presented barriers to the broader use of published data, and as a result, very few meta-analyses have been attempted for mammalian oocytes or preimplantation stage embryos.
Our goal here was to gain deeper insight into the fundamental mechanisms, pathways, and processes that contribute to mammalian oocyte maturation. Our strategy was to apply a novel combination of methods to complete a meta-analysis of transcriptome changes during oocyte maturation and compare these changes across multiple species.
Such an analysis must take into account the relationships between maternal mRNA storage, translation, and degradation. The controlling mechanisms of these processes are complex (18, 19). Within the cytoplasmic compartment, mRNAs can variably be translated, stored, or degraded, depending upon the actions of diverse RNA binding proteins, micro-RNAs, nonsense-mediated decay factors, and RNA degradation complexes (18–20). mRNA decay can occur without translation, during translation, or after translation. Deposition in storage granules or other depots can greatly extend mRNA half-life, particularly in oocytes (21), and exit from storage can lead to faster degradation. mRNA degradation is achieved by both 5′- and 3′-directed exonucleases. Inhibiting translation initiation can enhance the rates of 5′ degradation by exposing the mRNA to de-capping. Conversely, stress-mediated inhibition of translation initiation or elongation can inhibit decapping and stabilize mRNAs. Poly(A) tail lengthening can enhance translation, whereas poly(A) tail shortening can enhance 3′ degradation. Degradation can also be coupled to translation or to translational stalling. Because maternal mRNA degradation can be coupled to translational recruitment (i.e., removal from storage) and translation, one can infer that for many mRNAs, a high rate of degradation during maturation indicates production of protein within the cell. Increased degradation may also reflect a shift to translation inhibition and less protein production during maturation. To distinguish between such possibilities and capture information about mRNA translation, we coupled whole oocyte transcriptome meta-analysis with data from a previous analysis of changes in mRNA translation during the first 8 h of in vitro oocyte maturation (22). The combination of these datasets allows maternal mRNAs to be characterized according to both pattern of stability/degradation and translational regulation.
This analysis revealed mRNAs that are regulated similarly, mRNAs that are regulated species-specifically, and the pathways and cell physiological functions that are associated with these classes of mRNAs. The results reveal for the first time that only a limited number of mRNAs are regulated similarly across all four species examined, but certain pathways and functions nevertheless emerge that are regulated in all species, marking these pathways and functions as fundamental to the overall process of oocyte maturation. In addition, the data reveal cell physiological functions associated with cohorts of mRNAs that are stable, moderately degraded, or highly degraded during maturation, indicating apparent roles before and after fertilization.
MATERIALS AND METHODS
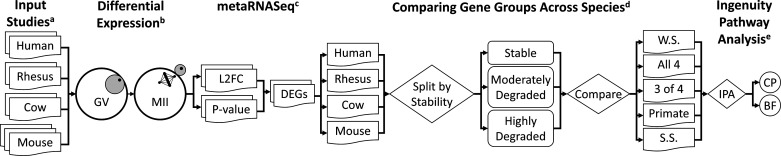
A summary of the process flow of the computational analysis for this study is depicted in Fig. 1. This included sample set processing for different species, identification of mRNAs of different stability classes, processing gene lists through QIAGEN Ingenuity Pathway Analysis (IPA; QIAGEN, Hilden, Germany), and interspecies comparisons of mRNA expression classes and associated IPA results. Differentially expressed gene (DEG) lists and IPA results are described in Supplemental Information (all Supplemental material is available at https://doi.org/10.6084/m9.figshare.14226368.v1).
Figure 1.
Flowchart of analysis. Input read counts for each study (a) were imported into DESeq2 and significant differences are calculated between germinal vesicle, immature oocyte (GV) and metaphase II, mature oocyte (MII) (b). Within each species, the P values for all included studies were input into metaRNASeq to calculate differentially expressed genes (DEGs) between GV and MII, resulting in whole species (WS) DEGs (c). Comparison of WS DEGs and derivation of gene groups (d). Each gene list was trifurcated based on direction (stable, moderately degrade, and highly degraded): 1) individual species results, 2) mRNAs regulated in the same direction from “All-4” species, 3) mRNAs regulated in the same direction by “3 of 4” species, 4) mRNAs regulated in the same direction by only human and rhesus, 5) mRNAs regulated in a species-specific (SS) manner, and each gene list was submitted to IPA (e) for Canonical Pathways (CP) and Biological Functions (BF) analysis.
Dataset Selection and Data Processing
We identified four mammalian species (human, mouse, cow, and rhesus monkey) for which RNAseq datasets could be identified that contained both germinal vesicle, immature oocyte (GV) and metaphase II, mature oocyte (MII) stage oocytes, at least three biological replicates at each stage, and meeting other quality parameters. To access these datasets, we used The European Nucleotide Archive. Study parameters are listed in Supplemental Table S1, including sequencing platform, sequencing read format/length, and RNA sequencing preparation kit. Unless otherwise noted, each study was processed with the following methods. Raw sequencing data in FASTQ format were downloaded for processing. Initial quality metrics were conducted using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Trimming was conducted with Fastp (v0.20.0) (23): minimum quality threshold of 20, minimum length of 20, and removal of low complexity/mononucleotide reads. Genomes index and abundance quantified with Kallisto (v0.44.0) (24), using standard settings.
Human Data Processing
Two human studies were identified meeting criteria: PRJNA377237 and PRJNA293908 (12, 25). FastQC identified aberrant nucleotide distribution in the first 13 bp in both studies. Therefore, the Fastp settings were set to include a hard trim of 13 bp from start of reads. The human cDNA genome (GRCh38, build 100) was downloaded from Ensembl. Quantification and differential expression were conducted as detailed under Dataset Selection and Data Processing. MII stage oocytes were matured in vivo.
Rhesus Data Processing
Two rhesus studies were identified meeting criteria: PRJNA343030 and PRJNA448148/PRJNA448150 (13, 17). FastQC identified aberrant nucleotide distribution in the first 13 bp in PRJNA343030 and 6 bp in PRJNA448148/PRJNA448150, which were hard trimmed accordingly. The rhesus cDNA genome (Mmul_10, build 100) was downloaded from Ensembl. Quantification and differential expression were conducted as detailed under Dataset Selection and Data Processing. The two studies used two distinct rhesus monkey populations (Indian-origin and Chinese-origin), providing for inclusion of genetic diversity in this analysis. MII stage oocytes were matured in vivo.
Cow Data Processing
Two cow studies were identified meeting criteria: PRJNA261946 and PRJNA228235 (26, 27). FastQC identified aberrant nucleotide distribution in the first 6 bp for PRJNA228235, which were hard trimmed. The cow cDNA genome (ARS-UCD1.20, build 100) was downloaded from Ensembl. Quantification and differential expression were conducted as detailed under Dataset Selection and Data Processing. MII stage oocytes were matured in vitro.
Mouse Data Processing
Two mouse studies were identified meeting criteria: PRJNA342001 and PRJNA464431 (15, 28). FastQC identified aberrant nucleotide distribution in the first 13 bp in PRJNA342001 and 6 bp in PRJNA464431, which were hard trimmed accordingly. The mouse cDNA genome (GRCm38, build 100) was downloaded from Ensembl. The study PRJNA464431 consisted of three different mouse strains (B6, D2, and BDF1), which were processed independently, therefore resulting in effectively four mouse datasets. Quantification and differential expression were conducted as detailed under Dataset Selection and Data Processing. MII stage oocytes were matured in vivo.
Differential Expression Calculation and Gene Homology
Kallisto output was imported into R (v4.0) and processed with DESeq2 (v1.30.0) (29), and transcript abundance was collapsed to gene using Ensembl identifiers converted with biomartR (v2.45.8), summing transcript isoform abundances for each gene. Two different lists of genes were processed through DESeq2 for each study: unfiltered gene lists and only genes with high level of homology across species. Because normalization and expression threshold selection were performed independently on each gene list, the homology lists were not simple subsets of the full gene lists. As the IPA gene set enrichment software is primarily based on human and mouse data, there is some concern in mapping genes incorrectly for other species. To address this potential concern, the metaPhOrs database (30) was used to identify genes with a high degree of homology, and IPA was repeated on this set of homologous genes. All pairwise species comparisons were retrieved from the database. Genes with high homology scores across all species were retained, numbering 11,272. Differential expression calculations were then repeated on this list of homologous genes. Both methods of filtering were conducted in parallel to ascertain the impact on mRNA overlap and gene set enrichment within IPA. For both methods, genes with an FPKM (fragments per kilobase of transcript per million mapped read) above 1 in at least one sample were included for differential expression calculation. For each study, DESeq2 (29) was used to calculate differentially expressed genes (DEGs) between GV and MII, where a positive log2(fold-change) indicates a higher expression in MII as compared with GV; the level of significance for genes was set at an adjusted P value (false discovery rate: FDR) below 0.05.
Differential Meta-Analysis
Within each species, the R package metaRNASeq (v1.0.3) (31) was used to calculate a meta P value between studies via the Fisher’s combination method. In short, the Fisher’s combination method assumes that gene counts follow a negative binomial distribution within each included study. For each gene in each study, the null hypothesis was tested; that each gene is not differentially expressed. Whereupon the Fisher’s exact test is applied to calculate gene- and study-wise P values.
In a review of different library preparation kits used in RNA-seq (32), there are inherent differences and biases. This was confirmed in this analysis and presented in Supplemental Table S1, such that within species, there is a level of variability in the number of captured genes with expression and number of mRNAs by stability classification. By leveraging the metaRNAseq method, these differences become a strength by allowing the integration of different preparation and sequencing methods for the derivation of a cohesive transcriptome. Volcano plots, plotting pre- and post-metaRNAseq log2(fold-change) versus FDR, for all species and datasets, can be found in Supplemental Figs. S1, S2, S3, and S4.
Gene Group Classification
During oocyte maturation, transcription is inactive and there are no new mRNAs being produced. Therefore, it is incorrect to state that a gene is “upregulated” in MII compared with GV. What is occurring is that many mRNAs are degraded during maturation, thereby drastically changing the “background” mRNA population being used to identify DEGs. Thus, ”upregulated” mRNAs are preferentially stabilized (i.e., display longer half-lives), degrading at a lesser rate compared with the background population. mRNAs calculated to be “downregulated” are those that undergo an elevated rate of degradation (i.e., become destabilized or have shorter half-lives). In addition, mRNAs exhibiting no significant change in expression undergo a moderate amount of degradation, which is less than that impacting the “degraded” class of mRNAs. This situation requires the reframing of directional classification of gene classes post-metaRNASeq. Genes were thus classified by actual change during maturation: upregulated (MII > GV, termed “stable”), no significant change (termed “moderately degraded”), and downregulated (MII < GV, termed “highly degraded”). These trifurcated lists were then compared across species, based on gene symbol, deriving all possible distinct gene group overlaps. There were a number that were found to have discordant directionality: genes regulated in opposite directions across species. As the underpinning goal of this study was to identify core shared features, these genes were not included in the analysis nor appear in the counts of overlaps.
Correlating Stability and Translational Changes during Early Oocyte Maturation
Raw sequencing data in FASTQ format were downloaded for processing for polysome-associated mRNAs at germinal vesicle stage and at metaphase I of meiosis for mouse oocytes (22). Processing of samples was conducted as described under Dataset Selection and Data Processing, with the inclusion of the Fastp parameter of a minimum length of 36 bp for reads, matching the original publication. Differential expression was calculated between 0 h and 8 h postmaturation induction, corresponding to GV and metaphase I stages. Although not equivalent to GV versus MII comparisons, this analysis nevertheless provides some insight into whether mRNAs are recruited to or depleted from polyribosomes in response to maturation induction. Three classifications of temporal translation pattern were derived: activated (higher in 8 vs. 0 h), repressed (lower in 8 vs. 0 h), and constitutive (no significant difference). These classifications were intersected with the three stability classifications, resulting in nine gene groups.
IPA Core Analysis
Gene lists were analyzed through the use of Ingenuity Pathway Analysis (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis), focusing on Canonical Pathway (CP) and Diseases and Functions (DF) analysis tools (IPA database v. 11/2020) (33). IPA is a software suite that allows for the enrichment of Canonical Pathways (CP) and Biological Functions (BF, a manually selected subset of Diseases and Functions) and the development of novel networks, based on submitted gene lists. IPA was selected due to the robustness of their database (>7M interactions, >700 pathways, >800,000 expression datasets, and >30 integrated third-party databases), and because it is manually curated and has the ability to compare multiple datasets. As a typical gene set enrichment methodology, submitted gene lists are compared with the genes in each CP/BF to calculate a level of significant overlap (P value; significance set at 0.05). In addition, with the known impact of up- or downregulating a gene on CP or BF, the IPA software can calculate a direction (activated or inhibited) as indicated by a positive or negative z-score (significance set at z > |1.96|). It should be noted that the magnitudes of gene expression changes do not factor into the calculations; only the direction of change is used. Each derived gene group was submitted to IPA and the CP/BF results were retrieved.
RESULTS
We first identified shared and species-specific DEGs (mRNAs that change during maturation from GV to MII stage) and nonchanging mRNAs, as well as shared and species-specific Ingenuity Pathway Analysis (IPA) CPs and BFs associated with gene sets. Through the analysis of these mRNAs and associated IPA results, we then assessed shared and species-specific aspects of maternal mRNA regulation during oocyte maturation.
We analyzed the transcriptomes of the included mammalian species by two methods. The first method used the full gene list, mapping Ensembl gene identifiers to gene symbols, and the second method limited the analysis to those genes with a high level of homology across all four species. This second method addresses disparities in species and associated genome builds (gaps in sequencing, unannotated genes, evolutionary divergence, etc.) and possible impacts on results. Although the utilization of the full unfiltered gene lists may include genes not annotated in all species, the homology-based analysis can result in a decrease in power for the detection of DEGs. Presenting the outputs for both methods provides the most complete view of the analysis. Both methods are valuable and ultimately displayed highly similar results.
In addition to accounting for gene homologies, we leveraged the metaRNAseq method. This allows for the integration of multiple sequencing studies based on their respective generated P values based on a Fisher’s combination method. Because of different sequencing platforms, library preparation kits used, and breed/strain/ethnicity variation, and other methodological differences between datasets, this metaRNAseq method helps account for these variables. A total of 94 sequencing libraries were processed for this study (12 Cow, 38 Mouse, 18 Human, and 26 Rhesus). An average of 14,380 genes were captured per study (Supplemental Table S1).
Identification of mRNA Sets According to Cellular mRNA Stability during Maturation
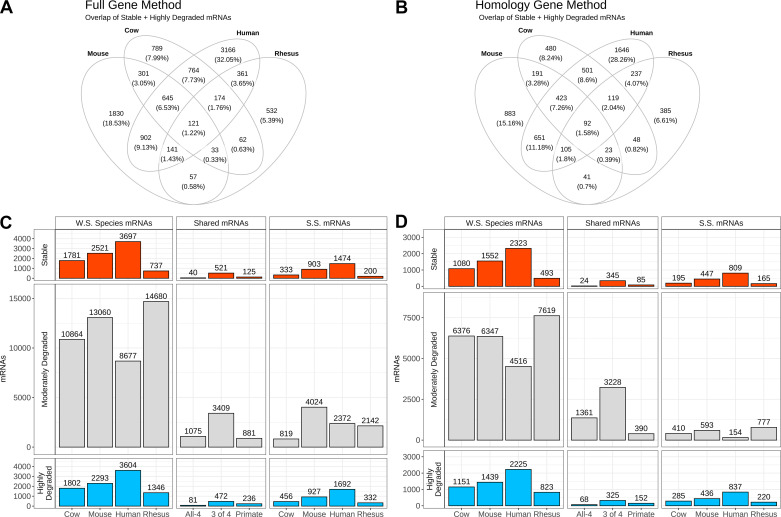
We categorized mRNAs for each species as highly degraded (MII < GV), moderately degraded (MII not significantly different from GV), and stable (MII > GV), and analyzed total (“full”) mRNAs detected and highly homologous mRNAs (Supplemental Tables S2–S5). Hereafter, numbers of mRNAs will be given in the format: “full mRNA number (highly homologous mRNA number).” Across the four species analyzed, we observed a median of 2,151 (1,316) stable, 11,962 (6,362) moderately degraded, and 2,048 (1,295) highly degraded mRNAs (Fig. 2 and Supplemental Tables S1, S2, S3, and –S5).
Figure 2.
Gene regulation groups during maturation. Visual representation of stability classes, numerological depiction of mRNAs, classified by regulation and species. A: Venn diagram overlap of stable + highly degraded mRNAs across the four species using the full gene method. B: Venn diagram overlap of stable + highly degraded mRNAs across the four species using the homology method. C and D: gene counts for full and homology method analyses, respectively, for mRNAs identified by stability classification. Column 1, WS mRNAs, denotes global classification of changes for each whole species gene lists. Column 2 (Shared mRNAs) shows selected mRNAs overlaps of: “All-4” species, “3 of 4” species, and primate-specific mRNAs. Column 3 (SS mRNAs) shows the number of mRNAs regulated in a species-specific manner. The associated mRNA numbers and gene lists are found in Supplemental Tables S2, S3, S4, S5, S6, and S7.
Shared and Species-Specific Members of Different mRNA Stability Classes
Our next step was to identify mRNAs that changed in abundance with a similar pattern across species. We examined transcriptomes for mRNAs regulated similarly across all four species (Fig. 2, designated hereafter as “All-4” mRNAs). We also identified mRNAs that were regulated across three of the four species (Fig. 2, designated as “3 of 4” mRNAs). The combined set of the All-4 plus the 3-species mRNAs is referred to as “4&3” mRNAs (Fig. 2). We identified 993 (670) “3 of 4” DEGs [include 645 (423) not in rhesus monkey, 33 (23) not in human, 174 (119) not in mouse, and 141 (105) not in cow for the full and homology methods]. Including the use of the 4&3 mRNA sets provided a less stringent look at shared mRNAs that considers possible impact of differences in genome annotation completeness across species. Such differences could artificially underestimate the degree of conservation of mRNA temporal expression profiles and impacts on cellular functions, pathways, and processes during oocyte maturation. We reasoned, therefore, that allowing a single species exception to a pattern reduced the risk of such an artifact impacting conclusions of the study. We therefore examined the mRNAs and associated IPA results for the 4&3, as well as the All-4 mRNAs alone.
A prominent result of our analysis was the limited number of All-4 mRNAs (Fig. 2, B and C, middle column “All-4” group). Although thousands of mRNAs changed in abundance during maturation within any one species, only 121 (92) were regulated similarly, comprised of 40 (24) stable and 81 (68) highly degraded, across all four species. These 121 (92) mRNAs accounted for an average of only 1.22% (full method) and 1.58% (Homology method) of the total number of 9,878 (5,825) DEGs identified across all four species combined. Even allowing for a single species exception, the number of 3-species mRNAs was still comparatively limited (Fig. 2, column “3 of 4” group in Shared mRNAs). These encompassed 993 (670), consisting of 521 (345) stable and 472 (325) highly degraded mRNAs (Supplemental Table S6). Further evidence of the dramatic interspecies differences in mRNA regulation was seen with respect to the moderately degraded mRNAs. Although a median number of 11,962 (6,362) mRNAs were categorized as moderately degraded (i.e., unchanged during maturation), the tremendous variation across species in mRNAs being stabilized or degraded resulted in a very limited number of mRNAs being classified as moderately degraded across all four species or even three of four species (Fig. 2).
Aspects of shared mRNA regulation specific to primate species were observed by considering mRNAs displaying changes in human and rhesus monkey only (Fig. 2 and Supplemental Table S6). Genes with shared changes in mRNA relative abundance specific to rhesus and human, exclusive of 3-species or All-4 mRNAs, included a total of 248 (237), consisting of 125 (85) stable and 236 (152) highly degraded mRNAs. There were an additional 881 (390) primate-specific mRNAs classified as moderately degraded.
We observed a range of species-specific changes for mRNAs across the species, 200–1,474 (165–809) stable and 332–1,692 (220–837) highly degraded (Fig. 2 and Supplemental Table S7). The rhesus monkey had the fewest species-specific changes, and the human had the most. A range of species-specific moderately degraded mRNAs was found for full and homology analyses (819–4,024 and 154–777, respectively). We also note that some mRNAs encoded by the mitochondrial genome appeared in the species-specific DEG lists but not in the shared All-4 and 4&3 DEG lists.
IPA Analysis of Shared and Overall Species Changes in mRNA Abundance
During maturation, some mRNAs are dramatically degraded in abundance, suggesting translation to produce cognate proteins contributing to the maturation process or terminating production of those proteins to downmodulate associated functions. Conversely, some mRNAs do not undergo degradation, and thus may be reserved to contribute to later functions or may become translationally silenced to downmodulate certain functions. The IPA terms “inhibition” and “activation” applied to the highly degraded and stable cohorts of mRNAs, respectively, thus could indicate which biological pathways and functions are used/terminated and which are reserved/sustained for later use. Defining the biological functions of the stable and highly degraded classes of mRNAs could thus provide insight into a core set of essential features of oocyte maturation that are shared across species, namely pathways and functions that are directly associated with the process of oocyte maturation and those that are associated with maternal mRNAs roles in the early embryo. To ascertain the overall functional systems enriched per species and the core shared features, IPA was applied to the stable and highly degraded whole species (WS) DEG lists for each species, the All-4 DEGs, the 4&3 DEGs, and the primate-specific DEGs. In addition, moderately degraded mRNAs seen specifically in primates were analyzed using IPA. The other moderately degraded gene lists were not processed through IPA due to the large number of gene members.
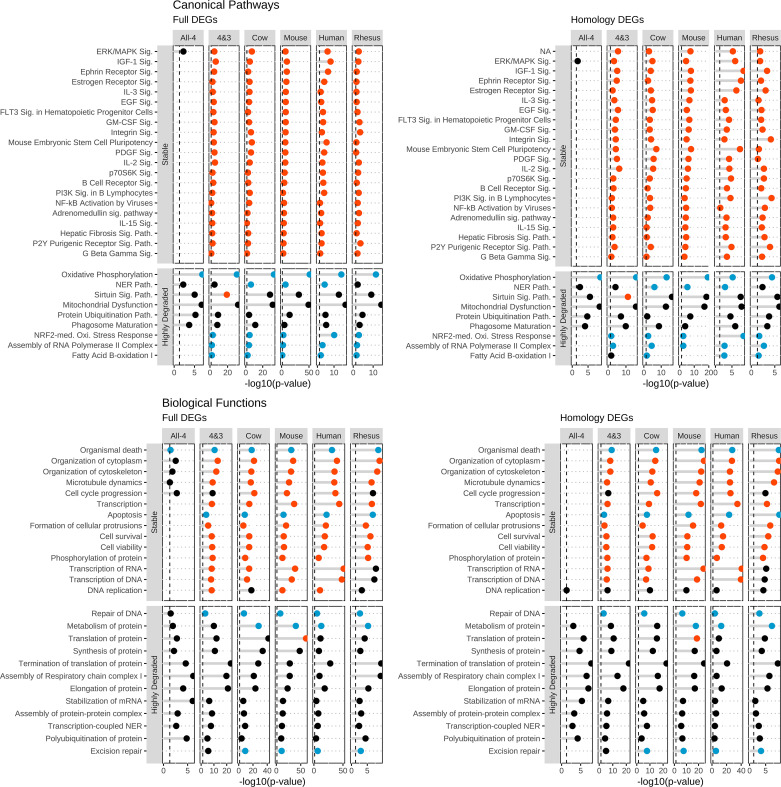
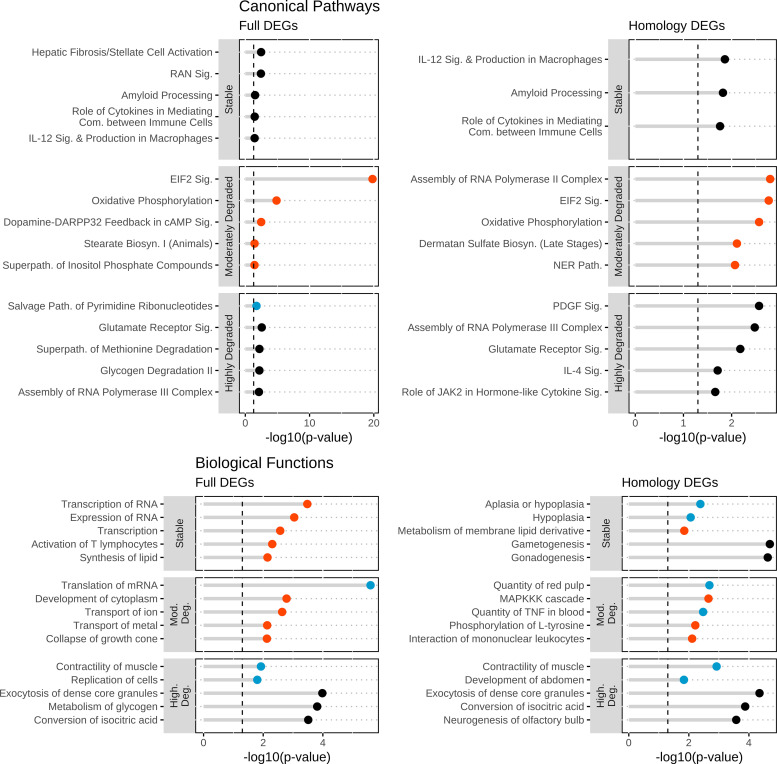
To identify the core shared processes of mammalian oocyte maturation, the IPA results of the All-4, 4&3, and the four whole species DEG lists were compared for stable and highly degraded mRNA classes (Fig. 3 and Supplemental Tables S8 and S9). The ERK/MAPK signaling pathway was significantly affected in IPA results for stable mRNAs identified in the All-4, 4&3, and individual whole species (WS)-DEG lists, with predicted activation in all of these except the All-4 set (Fig. 3 and Supplemental Table S8). Twenty additional pathways displayed significant activation (positive z-score) with stable mRNAs from the 4&3 and all individual WS-DEG sets, including IGF-1 signaling, ephrin receptor signaling, estrogen receptor signaling, IL-3 signaling, and Fms-related receptor tyrosine kinase 3 (FLT3) signaling in hematopoietic progenitor cells in all species (full and homologous DEGs; Fig. 3 and Supplemental Table S8).
Figure 3.
Ingenuity Pathway Analysis (IPA) features during maturation from shared differentially expressed genes (DEGs). The top selected IPA entries pertinent to oocyte maturation, derived from DEGs shared by all four (“All-4”) and at least three species (“4&3”). The four panels represent IPA CP and BF results obtained for DEGs identified by the full and homology methods. Each panel has two vertical facet plots for each mRNA stability classification: stable and highly degraded. For each plot, the x-axis denotes the −log10(P value) and the y-axis are the IPA entries. The color of each points represents the z-score: activated = red, inhibited = blue, no-significant = black. Vertical dashed lines at 1.3 equates to a P value of 0.05. Associated data and additional IPA entries are listed in Supplemental Tables S8 and S9.
When comparing the IPA CP results on the highly degraded genes, the most prominent result present across gene sets was for oxidative phosphorylation, which was inhibited for the All-4, 4&3, all individual WS-DEG lists, and for the primate-specific moderately degraded mRNAs, with additional effects observed among species-specific DEG sets (Fig. 3 and Supplemental Table S8). In addition, NRF2 (NFE2L2, nuclear factor erythroid 2-like 2)-mediated oxidative stress response, assembly of RNA polymerase II complex, and fatty acid beta-oxidation I had significantly inhibited z-scores for the 4&3 and all individual WS-DEG sets (Fig. 3 and Supplemental Table S8).
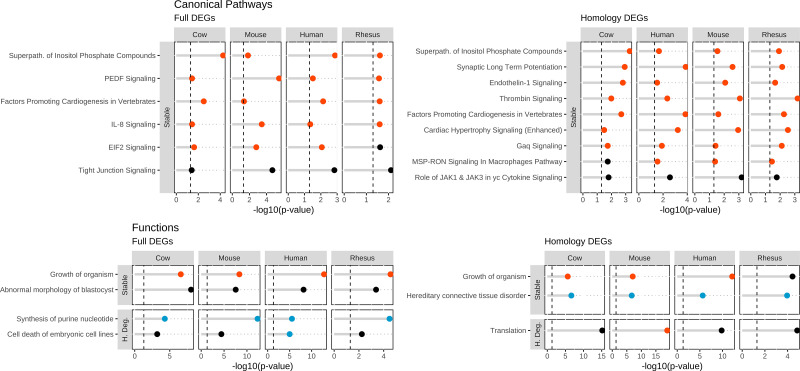
Additional similarities were seen comparing results obtained by IPA analysis of each individual WS-DEG list that were not present in the All-4 and 4&3 analyses. The shared CPs identified using the full method included ones with activation in all species, such as PEDF (SERPINF1, serpin peptidase inhibitor clade F member 1)-mediated and IL-8 signaling, and the superpathway of inositol phosphate compounds. The results obtained with the homology method included CPs with significant activation z-scores such as superpathway of inositol phosphate compounds, endotehlin-1, thrombin, and Gaq signaling (Fig. 4 and Supplemental Table S8).
Figure 4.
Additional Ingenuity Pathway Analysis (IPA) features during maturation shared by all species. IPA entries pertinent to oocyte maturation shared by all species while not present in All-4 and 4&3 analyses. The four panels show IPA CP and BF results obtained for DEGs identified by the full and homology methods. Each panel has two vertical facet plots for each mRNA stability classification: stable and highly degraded. For each plot, the x-axis denotes the −log10(P value) and the y-axis are the IPA entries. The color of each points represents the z-score: activated = red, inhibited = blue, no-significant = black. Vertical dashed lines at 1.3 equates to a P value of 0.05. Associated data and additional IPA entries are listed in Supplemental Tables S8 and S9.
The stable mRNA BF analysis revealed considerable similarities between using full or homology methods (Fig. 3 and Supplemental Table S9). For both methods, we identified an overall inhibition of functions relating to death and apoptosis, while finding activation for functions relating to survival and viability. For all datasets excluding All-4, we found activation for organization of cytoplasm/skeleton, microtubule dynamics, and transcription.
BF analysis results for the highly degraded mRNAs yielded similar results for full and homology DEGs. Effects included numerous observations for protein translation-related functions and RNA processing/stability. All species analyzed by either method indicated inhibition for repair of DNA and excision repair (Fig. 3 and Supplemental Table S9).
IPA Analysis of Primate-Specific Stable and Highly Degraded mRNAs
The comparison of rhesus monkey and human oocyte maturation is of interest for better understanding processes that may be unique to primates, and which may cooperate with those features identified from examining the shared mRNA lists. In addition, common features of these two closely related species would provide corroboration of results obtained for each species. Primate-specific stable CPs included: role of cytokines in mediating communication between immune cells and IL-12 signaling and production of macrophages. Primate-specific moderately degraded mRNAs were significantly enriched in oxidative phosphorylation and eukaryotic translation initiation factor 2 (EIF2) pathway, both activated. The assembly of RNA POL-II complex, glutamate receptor signaling, and the superpathway of methionine degradation were found in the highly degraded DEG results (Fig. 5). The BF analysis (Fig. 5) showed no overlap between methods; the full method consisted of RNA processing and lipid synthesis, whereas the homology method identified hypoplasia and gamet/gonadogensis. A similar result was found for the moderately degraded mRNAs, with the full gene list method yielding effects on translation of RNA and transport of ion/metals. The homology method results included MAPKKK cascade and phosphorylation of l-tyrosine. Analysis of the highly degraded DEGs yielded effects associated with contractility of muscle, exocytosis of dense core granules, and conversion of isocitric acid (Fig. 5).
Figure 5.
Ingenuity Pathway Analysis (IPA) features during maturation from primate-specific regulated mRNAs. The top selected IPA entries pertinent to oocyte maturation from primate-specific regulated mRNAs. The four panels represent IPA CP and BF results obtained for DEGs identified by the full and homology methods. Each panel has three facet plots for each mRNA stability classification: stable, moderately degraded, and highly degraded. For each plot, the x-axis denotes the −log10(P value) and the y-axis are the IPA entries. The black color of the points denotes no significant z-score. Vertical dashed lines at 1.3 equates to a P value of 0.05.
IPA Analysis of Species-Specific Changes in mRNA Abundance
The aforementioned analysis focused on features of maternal mRNA regulation that were shared across species or that were primate specific. Because shared mRNAs accounted for a small fraction of the total number of mRNAs that were analyzed, and because each individual species displayed dynamic regulation of thousands of mRNAs, oocyte maturation is accompanied by species-specific modulations of maternal mRNAs. These species-specific modulations may signify species-specific requirements for oocyte function or early embryo development, which in turn may signify species-specific sensitivities to exogenous influences such as maternal health or environmental factors. In addition, species-specific mRNA modulations may act cooperatively with the shared changes. We therefore applied IPA analysis to the species-specific stable and highly degraded DEG sets to gain insight into pathways and functions associated with mRNAs that are regulated in a species-specific manner (Supplemental Tables S8 and S9).
Of note were those CPs/BFs overlapping from the All-4, 4&3, and species-specific datasets. Using both the full and homology DEGs, mouse-specific highly degraded DEGs were enriched for oxidative phosphorylation, mitochondrial dysfunction, and sirtuin signaling. These same pathways were also found for cow-specific highly degraded DEGs from the full method. In addition, human-specific highly degraded DEGs were enriched for the NRF2-mediated oxidative stress response. When comparing the BF results from the species-specific stable DEGs, the function organismal death was reinforced for all species, and cell cycle progression for human, rhesus, and mouse. There were no overlapping functions found from the highly degraded DEG functions.
Regulation of mRNAs Related to Oxidative Phosphorylation
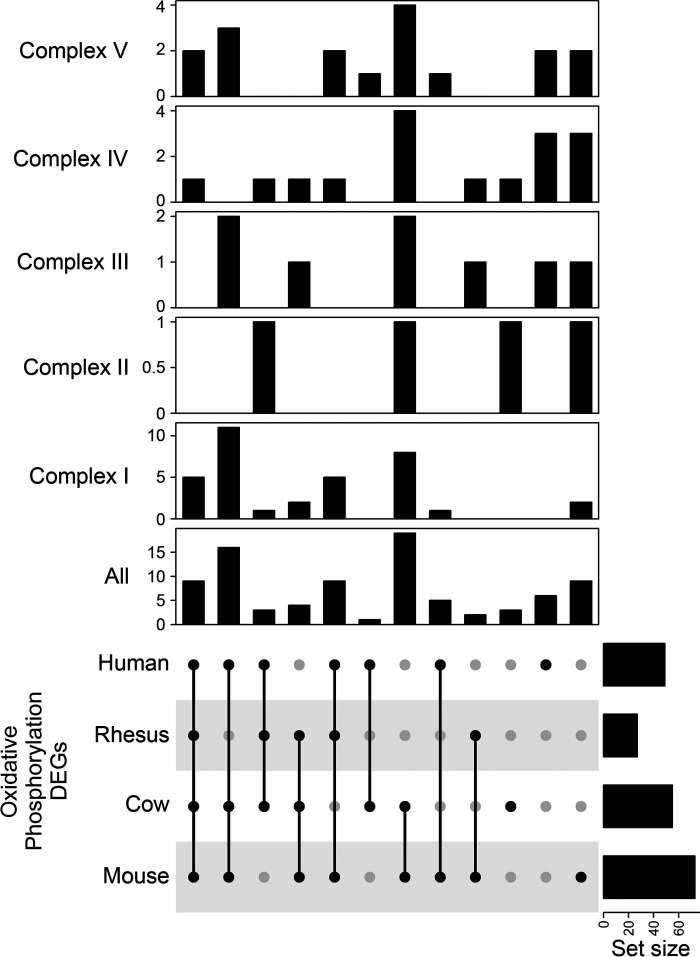
One of the most prominent results identified was the shared inhibition of the oxidative phosphorylation pathway across all species and datasets from the highly degraded DEGs. From the IPA database, the oxidative phosphorylation pathway has 109 member molecules. Interestingly, while an average of 47 of those 109 genes were among the DEGs for any given species (Human = 49, Rhesus = 27, Cow = 49, and Mouse = 73), 31 were shared by three of the four species and eight were shared by All-4 species. There were 24 DEGs shared between any two species and 17 species-specific DEGs. When splitting the DEGs by mitochondrial complexes (I–V), the majority of shared DEGs (n = 5) were from complex I and most of the species-specific DEGs were in complex IV (Fig. 6). In addition, when cross-referencing the mouse DEGs with the translational state, we found 68 entries with 19 constitutively translated and 49 repressed.
Figure 6.
Overlap of species regulation of differentially expressed genes (DEGs) in the oxidative phosphorylation pathway. UpSet plot depicting the overlap of highly degraded DEGs from each species that were found in the oxidative phosphorylation pathway. Figure depicts the overlaps for the entire pathway and DEG membership by mitochondrial complexes.
Relationship between Stability Classes and Early Maturational Changes in mRNA Translation
As previously stated, during oocyte maturation, transcription is inactive and stored mRNAs are either degraded, translated, or stabilized for use later. Luong et al. (22) employed the RiboTag method to explore the early maturation-related changes in mRNA translation during the first half of in vitro maturation period for mouse oocytes, from the GV to first meiotic metaphase stages. They defined three distinct groups of mRNAs based on changes in polyribosome abundances during this interval: activated (increased representation in polyribosomes), constitutive (constant representation in polyribosomes), and repressed (reduced representation in polyribosomes). The availability of this analysis in the mouse provides an opportunity for better understanding how initial changes in translation status relate to the stability classes identified here. This allowed comparison of the shared 4&3 class of mRNAs to the different translation classes defined on events during early oocyte maturation (Fig. 7).
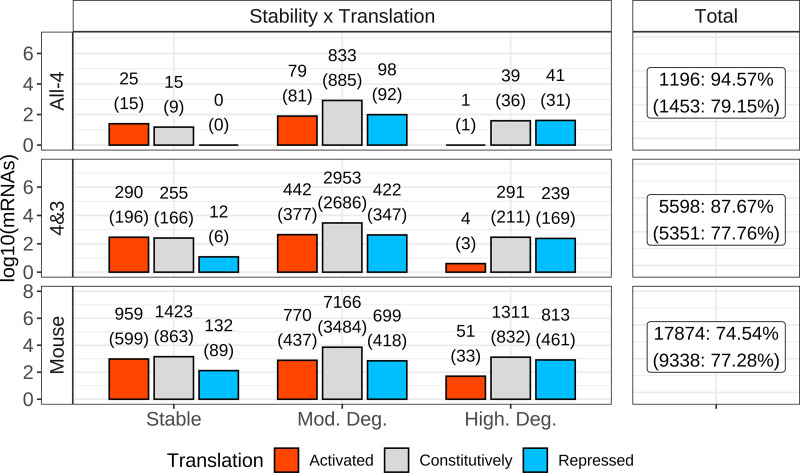
Figure 7.
Connecting stability and translational classification. For the three stability classifications (stable, moderately degraded, and highly degraded), mRNAs were intersected with translational classifications (activated, constitutively, and repressed). The x-axis represents the three stability classes, and the y-axis the number of mRNAs identified. The left facet of the figure is split into three rows, representing gene list origin: All-4, 4&3, and whole species (WS) mouse. Labels above bars denote number of genes from the full method on top, with the number generated from the homology method within parenthesis. The summation of the total number of identified genes, per gene list, are shown in the right facet. Coloring denotes translational classifications: red = activated, gray = constitutively, and blue = repressed.
Comparing the three translational groups with the three stability groups identified here for the mouse revealed that a large fraction of the stable DEGs (n = 959; 38.15%)) were classified as translationally activated. Most (n = 1,311, 60%) highly degraded mRNAs were in the constitutively translated class with another 813 (37.38%) of the highly degraded mRNAs in the translationally repressed class, and very few in the activated class. Most (n = 7,166, 82.99%) of moderately degraded mRNAs were constitutively translated (Fig. 7). Similar patterns were seen when comparing the All-4 and the 4&3 DEG sets to the mouse results, although the proportion of activated highly degraded mRNAs was much lower, as was the proportion of stable-repressed mRNAs (Fig. 7).
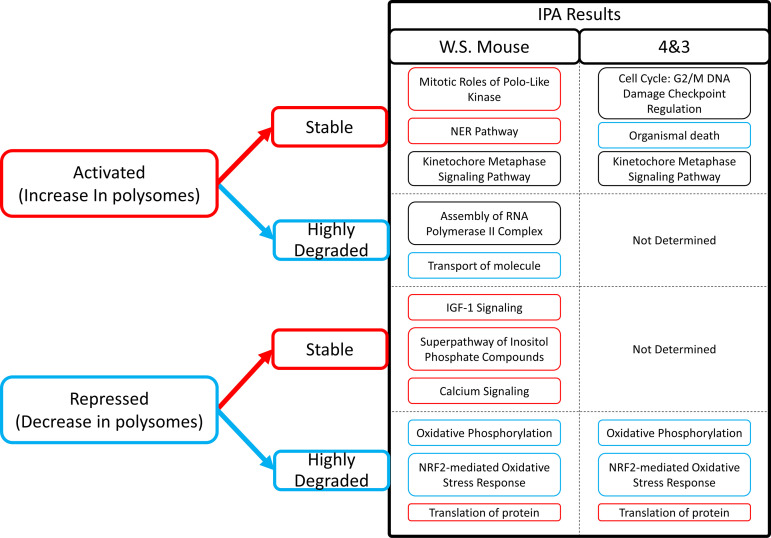
Subjecting the different translation-stability groups to IPA revealed prominent pathways and functions associated with particular combinations (Fig. 8 and Supplemental Tables S10 and S11). The stable-repressed category could only be examined for the mouse DEGs due to the small number of genes for the 4&3 group. The mouse stable-repressed group yielded significant associations with many signaling pathways (vascular endothelial growth factor VEGF, ciliary neurotrophic factor CNTF, insulin-like growth factor 1 IGF1, and Ephrin) and a prominent association with superpathway of inositol phosphate compounds, along with numerous entries for myoinositol, inositol, and phosphoinositide signaling and a relevant significant effect on calcium signaling. The stable-activated category for the 4&3 shared DEGs yielded significant effects for many CPs and BFs related to cell cycle and cell division such as G2/M DNA damage checkpoint, mitotic roles of polo-like kinase, kinetochore metaphase signaling, and cyclins and cell cycle regulation, as well as associations with cell viability, organismal death (inhibited), and mRNA degradation. These results were also seen for the mouse DEGs, along with effects on many other CPs and BFs. One striking result of this analysis was that the top results obtained for highly degraded-repressed category for both 4&3 and mouse DEGs were for strong inhibition of oxidative phosphorylation, and an effect on mitochondrial function, both with very strong P values [−log10(P) > 20] and associated with effects on numerous mRNAs encoding mitochondrial proteins. Fatty acid beta-oxidation was also inhibited. Affected BFs included inhibition of ATP synthesis and oxidative phosphorylation and activation of oxidative stress. This result was accompanied by predicted activation of sirtuin signaling, a result that may emerge from IPA due to the role for sirtuin signaling in regulating mitochondrial functions (34). The highly degraded-repressed DEGs were also associated with inhibition of NRF2-mediated oxidative stress response and effects on multiple BF entries related to protein synthesis.
Figure 8.
Key Ingenuity Pathway Analysis (IPA) features of translation-stability classified groups. Key IPA features for the stable and highly degraded mRNAs subdivided into activated or repressed translational categories. IPA results are presented for two submitted gene lists: whole species (WS) mouse and the 4&3 gene group. Exterior textbox coloring denotes the predicted z-score for each listed Canonical Pathways (CP)/Biological Functions (BF). Red = activated, Blue = inhibited. IPA entries are grouped by classified translation-stability groups.
DISCUSSION
An essential question in reproductive biology is what constitutes a high-quality oocyte in mammals. To answer this question, it is instructive to consider data obtained from different species to identify fundamental characteristics of normal oocyte maturation and strategies for managing the rich oocyte endowment to ensure not only oocyte maturation but also preservation of the requisite endowment of mRNAs to support early embryogenesis. We provide here the first meta-analysis to convey a comprehensive cross-species comparison of oocyte transcriptome changes associated with mammalian oocyte maturation. We applied a methodology that was designed to account for differences in library quality, molecular reagents, genome annotations, and sequencing platforms.
One main conclusion from this analysis is that, although each individual species displays many thousands of mRNAs that change in abundance, there emerged a small set of just 121 (92) mRNAs regulated in common (i.e., highly degraded, or stable) across all four species, and just 993 (670) additional mRNAs that changed in at least three of the four species analyzed, which averaged to just over one quarter of DEGs observed between GV and MII stage oocytes for each species. Thus, the degree of species conservation for transcriptome change during oocyte maturation is limited. This discovery highlights a surprising degree of divergence, given the presumed central importance of maternal mRNA regulation in oocyte function and early embryo development, and suggests that essential functions may not be strictly enforced at the individual gene level but rather at the level of overall pathway and function.
Indeed, despite the limited number of shared DEGs, our analysis was successful in highlighting many pathways and functions that are either used/terminated (highly degraded mRNAs) or reserved/sustained (stable mRNAs) in common across four mammalian species by maternal mRNA regulation during oocyte maturation, denoted by the IPA terms inhibition and activation, respectively. Shared aspects of transcriptome regulation during early maturation were observed both for shared DEG lists and attendant IPA results, and by comparing the IPA analysis obtained for WS-DEG lists for each individual species. This suggests species divergence in the regulation of stability of specific mRNAs, but with an underlying adherence to an essential set of functional outcomes; i.e., different mRNAs may be regulated to achieve effects on shared pathways or biological functions.
The most prominent shared functional outcomes to emerge from examining pathways and functions associated with the shared DEGs was the downregulation of mitochondrial function, reflected in inhibition of oxidative phosphorylation. Inhibition of NRF2-mediated oxidative stress response was also prominent. A majority of the shared DEGs related to oxidative phosphorylation encode components of complex I, whereas many of the species-specific DEGs related to oxidative phosphorylation encode proteins in complex IV. The highly degraded, translationally repressed (reduced in polyribosomes) subset of 4&3 DEG mRNAs were strongly associated with inhibition of oxidative phosphorylation and ATP synthesis, involving many mRNAs encoding mitochondrial proteins (Fig. 8). Such early maturational mRNA degradation coupled with reduced polyribosomal abundance during the first 8 h of maturation suggests that this downmodulation of oxidative phosphorylation is an early, shared, regulated process that begins with translational repression followed by transcript degradation. This result indicates that across all four species there is a dramatic exit of mRNAs associated with mitochondrial function and ATP synthesis from the polyribosomes followed by degradation. We note that downmodulation of oxidative phosphorylation and mitochondrial function may be protective by reducing mitochondrial activity and limiting reactive oxygen species production (35). The simultaneous shared degradation-translational repression of the NRF2-mediated oxidative stress response pathway further suggests the importance of downregulating oxidative phosphorylation. Indeed, a deficiency in the degradation of mRNAs related to oxidative phosphorylation is a key feature of human and rhesus monkey oocytes that fail to mature (13, 36). mRNAs related to nucleotide excision repair, fatty acid beta oxidation, and assembly or RNA polymerase II complex were also seen for degraded mRNAs. This suggests that these functions are also used/terminated across species.
To our knowledge, this is the first study to examine the connection between maternal mRNA stability and translation status during early oocyte maturation. Conservation of these relationships is seen across species. Across stability categories, constitutively translated (mouse) mRNAs comprised the bulk of mRNAs detected. mRNAs that are moderately degraded across all four species are mostly in the mouse constitutively translated class. A large proportion of the shared (All-4 and 4&3) stable mRNAs are in the mouse translationally activated category. Conversely, a large proportion of the shared highly degraded mRNAs are in the mouse translationally repressed and constitutively translated categories. This indicates that the regulation of degradation at least during early oocyte maturation is connected to translation for large cohorts of mRNAs.
Stable-repressed mRNAs in the mouse are associated with multiple signaling pathways, particularly inositol and calcium signaling effects (Fig. 8). Early translational repression of this subset of stable mRNAs may sequester them to support later functions, such as oocyte activation. The stable-activated subset of mRNAs were associated with numerous G2/M and M-phase pathways and functions, checkpoint controls, transcription, as well as shared activation of pathways related to stress response, endocrine and cytokine signaling, pluripotency, and microtubule dynamics, but apparent shared inhibition of functions related to death and chromosomal instability (Fig. 8). We also found that stable mRNAs were strongly associated with inhibition of apoptosis and organismal death and activation of cell survival and viability functions. These were accompanied by predicted activation of other basic functions such as cytoskeletal and cytoplasmic organization and DNA replication. These observations indicate that mRNAs that are stable and translationally activated during early maturation may endow the oocyte with numerous essential proteins that support key signaling processes and cell survival and proteins that support early embryogenesis.
Beyond the early oocyte maturation period, other maternal mRNAs may be reserved/sustained for later use in the embryo. Stable mRNAs across species were associated with a variety of signaling pathways such as ERK/MAPK, IGF1, Ephrin, Estrogen, IL3, and EGF, as well as DNA damage checkpoint regulation, and several biological functions related to viability, cytoskeleton regulation, and transcription. Several of these functions were previously associated with mRNAs present on both MII stage oocyte and 1-cell stage polysomes (37). Earlier studies revealed maternal mRNAs encoding transcription regulators that act postfertilization (35, 38, 39) and both DNA repair and checkpoint control can be vital processes following fertilization (40–42). DNA repair and checkpoint control functions were also associated with polysomes in MII stage oocytes and fertilized embryos (37). Successful expression of proteins related to inhibiting apoptosis was previously proposed as an important embryo quality surveillance mechanism after fertilization (43).
Overall, this meta-analysis of oocyte maturation-associated transcriptome changes across four mammalian species used an improved approach that identified key shared functions that are driven by limited numbers of shared DEGs. Chief among these is the downmodulation of mRNAs related to mitochondrial activity, oxidative phosphorylation, and ATP synthesis. This is the first study to conduct such a meta-analysis in mammalian oocytes. Previous studies compared expressed mRNAs between oocytes of different species (16, 44), but a detailed look at maturation-related changes in the transcriptome conserved across species has not been reported. One previously published meta-analysis explored published microarray oocyte datasets for human, mouse, rhesus, and cow (44). Our study employed a more sensitive mRNA expression detection method (RNAseq vs. microarray), thereby quantifying a larger number of mRNAs, and employed a more rigorous approach to functional interpretation of the DEGs. These combined approaches extend insights gained previously. Indeed, we note that no DEG overlaps were observed across all species during oocyte maturation in the earlier study, whereas we identified 121 (92) shared DEGs. However, we also note that the previously reported Gene Ontology enrichment of DEGs associated with oocyte developmental competence shared by at least two of the included species were also revealed in our findings of effects on mitochondrial function. This further supports the conclusion that the regulation of the oocyte transcriptome related to mitochondrial function and oxidative phosphorylation is a key aspect of oocyte maturation.
The limited degree of conservation of transcriptome changes (i.e., mRNAs displaying significant change in relative abundance) across species echoes recent observations comparing different mouse inbred strains and their F1 hybrids (15). In that study, substantial variation was also observed for transcriptome changes during maturation. These observations indicate that the dynamic process of transcriptome change during oocyte maturation is subject to considerable genetic variability. Our data reveal a core set of features that are indeed shared across species. However, there is a vast amount of maturational change in the oocyte transcriptome that is highly specific to individual species or strains. Even looking at the level of IPA pathways and functions, there are many differences across species. This remarkable species divergence poses significant challenges for efforts to identify molecular markers of oocyte quality, as well as endeavors to optimize in vitro manipulation systems, because oocytes and early embryos of different species may accordingly have very different optima for in vitro culture and other procedures. In addition, the finding that such differences exist in maternal mRNA regulation during oocyte maturation indicates that there are likely multiple strategies for the generation of high-quality oocytes employed in different mammalian species or even different strains. This discovery sets the stage for many interesting future studies to understand why such divergent strategies exist and what exogenous factors and forces have driven the emergence of such diversity among mammals. In addition, the results provide an important baseline against which to judge the extent to which transcriptome alterations impacting essential features emerge under conditions that compromise oocyte quality. The understanding gained here of essential shared and species-specific aspects of oocyte maturation may thus be useful for designing novel methods for predicting oocyte quality.
SUPPLEMENTAL DATA
Supplemental Tables S1–S11: https://doi.org/10.6084/m9.figshare.14226368.v1.
Supplemental Figs. S1–S4: https://doi.org/10.6084/m9.figshare.14226368.v1.
GRANTS
This work was supported in part by National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development Grant T32HD087166, Michigan State University (MSU) AgBioResearch, and Michigan State University.
DISCLAIMERS
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
P.Z.S. and K.E.L. analyzed data; interpreted results of experiments; prepared figures; drafted manuscript; edited and revised manuscript; and approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Lane Christenson for constructive comments on the manuscript.
REFERENCES
- 1.Berg DK, Smith CS, Pearton DJ, Wells DN, Broadhurst R, Donnison M, Pfeffer PL. Trophectoderm lineage determination in cattle. Dev Cell 20: 244–255, 2011. doi: 10.1016/j.devcel.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Daigneault BW, Rajput S, Smith GW, Ross PJ. Embryonic POU5F1 is required for expanded bovine blastocyst formation. Sci Rep 8: 7753, 2018. doi: 10.1038/s41598-018-25964-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozawa M, Sakatani M, Yao J, Shanker S, Yu F, Yamashita R, Wakabayashi S, Nakai K, Dobbs KB, Sudano MJ, Farmerie WG, Hansen PJ. Global gene expression of the inner cell mass and trophectoderm of the bovine blastocyst. BMC Dev Biol 12: 33, 2012. doi: 10.1186/1471-213X-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossant J. Developmental biology: a mouse is not a cow. Nature 471: 457–458, 2011. doi: 10.1038/471457a. [DOI] [PubMed] [Google Scholar]
- 5.Menezo YJ, Herubel F. Mouse and bovine models for human IVF. Reprod Biomed Online 4: 170–175, 2002. doi: 10.1016/s1472-6483(10)61936-0. [DOI] [PubMed] [Google Scholar]
- 6.Assidi M, Montag M, Van der Ven K, Sirard MA. Biomarkers of human oocyte developmental competence expressed in cumulus cells before ICSI: a preliminary study. J Assist Reprod Genet 28: 173–188, 2011. doi: 10.1007/s10815-010-9491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cagnone G, Sirard M-A. The impact of exposure to serum lipids during in vitro culture on the transcriptome of bovine blastocysts. Theriogenology 81: 712–722.e1-3, 2014. doi: 10.1016/j.theriogenology.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Chitwood JL, Burruel VR, Halstead MM, Meyers SA, Ross PJ. Transcriptome profiling of individual rhesus macaque oocytes and preimplantation embryos. Biol Reprod 97: 353–364, 2017. doi: 10.1093/biolre/iox114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu T, Dufort I, Sirard MA. Effect of ovarian stimulation on oocyte gene expression in cattle. Theriogenology 77: 1928–1938, 2012. doi: 10.1016/j.theriogenology.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 10.Khan DR, Landry DA, Fournier E, Vigneault C, Blondin P, Sirard MA. Transcriptome meta-analysis of three follicular compartments and its correlation with ovarian follicle maturity and oocyte developmental competence in cows. Physiol Genomics 48: 633–643, 2016. doi: 10.1152/physiolgenomics.00050.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labrecque R, Sirard MA. The study of mammalian oocyte competence by transcriptome analysis: progress and challenges. Mol Hum Reprod 20: 103–116, 2014. doi: 10.1093/molehr/gat082. [DOI] [PubMed] [Google Scholar]
- 12.Reyes JM, Silva E, Chitwood JL, Schoolcraft WB, Krisher RL, Ross PJ. Differing molecular response of young and advanced maternal age human oocytes to IVM. Hum Reprod 32: 2199–2208, 2017. doi: 10.1093/humrep/dex284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruebel ML, Schall PZ, Midic U, Vincent KA, Goheen B, VandeVoort CA, Latham KE. Transcriptome analysis of rhesus monkey failed-to-mature oocytes: deficiencies in transcriptional regulation and cytoplasmic maturation of the oocyte mRNA population. Mol Hum Reprod 24: 478–494, 2018. doi: 10.1093/molehr/gay032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schall PZ, Ruebel ML, Midic U, VandeVoort CA, Latham KE. Temporal patterns of gene regulation and upstream regulators contributing to major developmental transitions during Rhesus macaque preimplantation development. Mol Hum Reprod 25: 111–123, 2019. doi: 10.1093/molehr/gaz001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Severance AL, Midic U, Latham KE. Genotypic divergence in mouse oocyte transcriptomes: possible pathways to hybrid vigor impacting fertility and embryogenesis. Physiol Genomics 52: 96–99, 2020. doi: 10.1152/physiolgenomics.00078.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sylvestre EL, Robert C, Pennetier S, Labrecque R, Gilbert I, Dufort I, Leveille MC, Sirard MA. Evolutionary conservation of the oocyte transcriptome among vertebrates and its implications for understanding human reproductive function. Mol Hum Reprod 19: 369–379, 2013. doi: 10.1093/molehr/gat006. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Liu D, He D, Suo S, Xia X, He X, Han JJ, Zheng P. Transcriptome analyses of rhesus monkey preimplantation embryos reveal a reduced capacity for DNA double-strand break repair in primate oocytes and early embryos. Genome Res 27: 567–579, 2017. doi: 10.1101/gr.198044.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huch S, Nissan T. Interrelations between translation and general mRNA degradation in yeast. Wiley Interdiscip Rev RNA 5: 747–763, 2014. doi: 10.1002/wrna.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J 27: 471–481, 2008. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chalabi Hagkarim N, Grand RJ. The regulatory properties of the Ccr4-not complex. Cells 9: 2379, 2020. doi: 10.3390/cells9112379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarke HJ. Post-transcriptional control of gene expression during mouse oogenesis. Results Probl Cell Differ 55: 1–21, 2012. doi: 10.1007/978-3-642-30406-4_1. [DOI] [PubMed] [Google Scholar]
- 22.Luong XG, Daldello EM, Rajkovic G, Yang CR, Conti M. Genome-wide analysis reveals a switch in the translational program upon oocyte meiotic resumption. Nucleic Acids Res 48: 3257–3276, 2020. doi: 10.1093/nar/gkaa010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34: i884–i890, 2018. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527, 2016. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 25.Hendrickson PG, Dorais JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, Nix DA, Peterson CM, Tapscott SJ, Carrell DT, Cairns BR. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 49: 925–934, 2017. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graf A, Krebs S, Zakhartchenko V, Schwalb B, Blum H, Wolf E. Fine mapping of genome activation in bovine embryos by RNA sequencing. Proc Natl Acad Sci USA 111: 4139–4144, 2014. doi: 10.1073/pnas.1321569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reyes JM, Chitwood JL, Ross PJ. RNA-Seq profiling of single bovine oocyte transcript abundance and its modulation by cytoplasmic polyadenylation. Mol Reprod Dev 82: 103–114, 2015. doi: 10.1002/mrd.22445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franke V, Ganesh S, Karlic R, Malik R, Pasulka J, Horvat F, Kuzman M, Fulka H, Cernohorska M, Urbanova J, Svobodova E, Ma J, Suzuki Y, Aoki F, Schultz RM, Vlahovicek K, Svoboda P. Long terminal repeats power evolution of genes and gene expression programs in mammalian oocytes and zygotes. Genome Res 27: 1384–1394, 2017. doi: 10.1101/gr.216150.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550, 2014. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chorostecki U, Molina M, Pryszcz LP, Gabaldon T. MetaPhOrs 2.0: integrative, phylogeny-based inference of orthology and paralogy across the tree of life. Nucleic Acids Res 48: W553–W557, 2020. doi: 10.1093/nar/gkaa282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rau A, Marot G, Jaffrezic F. Differential meta-analysis of RNA-seq data from multiple studies. BMC Bioinformatics 15: 91, 2014. doi: 10.1186/1471-2105-15-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Y, Milon B, Ott S, Zhao X, Sadzewicz L, Shetty A, Boger ET, Tallon LJ, Morell RJ, Mahurkar A, Hertzano R. A comparative analysis of library prep approaches for sequencing low input translatome samples. BMC Genomics 19: 696, 2018. doi: 10.1186/s12864-018-5066-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kramer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30: 523–530, 2014. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci 35: 669–675, 2010. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruebel ML, Latham KE. Listening to mother: long-term maternal effects in mammalian development. Mol Reprod Dev 87: 399–408, 2020. doi: 10.1002/mrd.23336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruebel ML, Zambelli F, Schall PZ, Barragan M, VandeVoort CA, Vassena R, Latham KE. Shared aspects of mRNA expression associated with oocyte maturation failure in humans and rhesus monkeys indicating compromised oocyte quality. Physiol Genomics 53: 137–149, 2021. doi: 10.1152/physiolgenomics.00155.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potireddy S, Vassena R, Patel BG, Latham KE. Analysis of polysomal mRNA populations of mouse oocytes and zygotes: dynamic changes in maternal mRNA utilization and function. Dev Biol 298: 155–166, 2006. doi: 10.1016/j.ydbio.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 38.Midic U, Vincent KA, Wang K, Lokken A, Severance AL, Ralston A, Knott JG, Latham KE. Novel key roles for structural maintenance of chromosome flexible domain containing 1 (Smchd1) during preimplantation mouse development. Mol Reprod Dev 85: 635–648, 2018. doi: 10.1002/mrd.23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruebel ML, Vincent KA, Schall PZ, Wang K, Latham KE. SMCHD1 terminates the first embryonic genome activation event in mouse two-cell embryos and contributes to a transcriptionally repressive state. Am J Physiol Cell Physiol 317: C655–C664, 2019. doi: 10.1152/ajpcell.00116.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menezo Y, Dale B, Cohen M. DNA damage and repair in human oocytes and embryos: a review. Zygote 18: 357–365, 2010. doi: 10.1017/s0967199410000286. [DOI] [PubMed] [Google Scholar]
- 41.Adiga SK, Toyoshima M, Shiraishi K, Shimura T, Takeda J, Taga M, Nagai H, Kumar P, Niwa O. p21 provides stage specific DNA damage control to preimplantation embryos. Oncogene 26: 6141–6149, 2007. doi: 10.1038/sj.onc.1210444. [DOI] [PubMed] [Google Scholar]
- 42.Song Y, Li Z, Wang B, Xiao J, Wang X, Huang J. Phospho-Cdc25 correlates with activating G2/M checkpoint in mouse zygotes fertilized with hydrogen peroxide-treated mouse sperm. Mol Cell Biochem 396: 41–48, 2014. doi: 10.1007/s11010-014-2140-1. [DOI] [PubMed] [Google Scholar]
- 43.Jurisicova A, Latham KE, Casper RF, Casper RF, Varmuza SL. Expression and regulation of genes associated with cell death during murine preimplantation embryo development. Mol Reprod Dev 51: 243–253, 1998. doi:. [DOI] [PubMed] [Google Scholar]
- 44.Biase FH. Oocyte developmental competence: insights from cross-species differential gene expression and human oocyte-specific functional gene networks. OMICS 21: 156–168, 2017. doi: 10.1089/omi.2016.0177. [DOI] [PubMed] [Google Scholar]