Abstract
Genome-wide association studies (GWASs) have identified regions associated with chronic obstructive pulmonary disease (COPD). GWASs of other diseases have shown an approximately 10-fold overrepresentation of nonsynonymous variants, despite limited exonic coverage on genotyping arrays. We hypothesized that a large-scale analysis of coding variants could discover novel genetic associations with COPD, including rare variants with large effect sizes. We performed a meta-analysis of exome arrays from 218,399 controls and 33,851 moderate-to-severe COPD cases. All exome-wide significant associations were present in regions previously identified by GWAS. We did not identify any novel rare coding variants with large effect sizes. Within GWAS regions on chromosomes 5q, 6p, and 15q, four coding variants were conditionally significant (P < 0.00015) when adjusting for lead GWAS single-nucleotide polymorphisms A common gasdermin B (GSDMB) splice variant (rs11078928) previously associated with a decreased risk for asthma was nominally associated with a decreased risk for COPD [minor allele frequency (MAF) = 0.46, P = 1.8e-4]. Two stop variants in coiled-coil α-helical rod protein 1 (CCHCR1), a gene involved in regulating cell proliferation, were associated with COPD (both P < 0.0001). The SERPINA1 Z allele was associated with a random-effects odds ratio of 1.43 for COPD (95% confidence interval = 1.17–1.74), though with marked heterogeneity across studies. Overall, COPD-associated exonic variants were identified in genes involved in DNA methylation, cell-matrix interactions, cell proliferation, and cell death. In conclusion, we performed the largest exome array meta-analysis of COPD to date and identified potential functional coding variants. Future studies are needed to identify rarer variants and further define the role of coding variants in COPD pathogenesis.
Keywords: chronic obstructive pulmonary disease, exome, exon, functional, genomics
INTRODUCTION
Chronic obstructive pulmonary disease (COPD) is characterized by irreversible airflow limitation (1) and is one of the leading causes of morbidity and mortality worldwide (2). Genome-wide association studies (GWASs) have lent considerable insight into the genetic risk to COPD (3–7). Most GWAS variants are noncoding and are thought to affect COPD susceptibility through gene regulation (8). As such, identifying disease-causing variants in COPD GWAS regions remains challenging. Whereas coding regions make up only ∼1% of the genome, ∼10% of GWAS signals in complex diseases are attributable to nonsynonymous variants (8). Specific rare coding variants may confer a particularly high risk for complex diseases such as COPD. For example, α-1 antitrypsin deficiency (AATD) is associated with an ∼15-fold increased odds for emphysema (9) and is most commonly caused by homozygosity for the Serpin Family A Member 1 (SERPINA1) Z allele (rs28929474), which is a missense variant found in ∼2%–3% of the United States population (10). Exome sequencing of a French-Canadian family with early-onset emphysema identified a rare nonsynonymous causal variant in protein tyrosine phosphatase nonreceptor type 6 (PTPN6) (11). Germline mutations in telomerase genes have been observed in severe COPD cases (12). Thus, examining coding variants across the genome may identify important functional (protein-altering) variants associated with COPD.
Exome arrays were designed to allow genotyping of a large fraction of functional (nonsynonymous, splice, stop-gain) variants across the genome (13), and association analyses have been reported for COPD and lung function (14–16). However, several important questions regarding the utility of exome array studies in COPD remain unanswered. It is not known whether increasing sample size and power will identify novel rare coding variants that markedly increase COPD risk. There have since been several large-scale GWASs for lung function and COPD, yet GWASs have poor coverage of exonic variants and are not intended to identify rare coding variants; exome array results have not been directly compared with GWAS results, which may elucidate the functional variants being tagged by GWAS-identified single-nucleotide polymorphisms (SNPs). Although the SERPINA1 Z allele (rs28929474) is a known risk factor for COPD—even in heterozygous individuals (17)—the largest GWASs to date (3, 4, 6, 7) did not identify an association of the SERPINA1 Z allele with COPD or lung function; one reason for this result may be the smoking-dependent effects of the Z allele and/or imputation inaccuracies, as the Z allele is not present on most genotyping arrays. However, the Z allele is present on exome arrays, allowing for direct assessment of the association of the Z allele with COPD risk. Further, many COPD case-control studies intentionally exclude ZZ individuals, which could introduce selection bias. A gene-by-environment interaction of cigarette smoking may be important for SERPINA1 variants to contribute to COPD (18), which may affect the association of the Z allele on COPD in population-based cohorts (as opposed to COPD cohorts enriched for cigarette smoking).
We hypothesized that a larger exome array meta-analysis would provide increased power to detect rare and poorly imputed functional exonic variants associated with COPD and to identify the most likely causal variants in previously defined COPD GWAS regions. We also leveraged the exome array data to assess effect size heterogeneity of the Z allele across studies.
METHODS
Study Cohorts
We included 12 cohorts in our analysis: ARIC (Atherosclerosis Risk in Communities) study with African ancestry (Aa) and European ancestry (Ea) participants (19); CHS (Cardiovascular Health Study) including Ea and Aa participants (20); COPDGene (Genetic Epidemiology of COPD) with non-Hispanic White (NHW) and African American (AA) participants (21); EOCOPD/ICGN [Boston Early-Onset COPD (22, 23) and International COPD Genetics Network (24)] studies; the FHS (Framingham Heart Study) (25–27); HABC (Health, Aging, and Body Composition) study with Ea and Aa participants (28); KARE (Korean Association Resource) study (29); MESA (Multi-Ethnic Study of Atherosclerosis) including non-Hispanic African American, Chinese American, Hispanic, and non-Hispanic White subpopulations (30, 31); RS (Rotterdam Study) (32, 33); TCGS (Transcontinental COPD Genetics Study) from Poland and South Korea (34); UK COPD Exome Chip Consortium (UKECC) (16); and UK Biobank (35). Moderate-to-severe COPD was primarily defined by prebronchodilator forced expiratory volume in 1 s (FEV1)/forced vital capacity (FVC) ratio < 0.7 and FEV1 < 80% predicted; postbronchodilator measures were only performed in a minority of studies and were used when available. Individual study details, including genotyping methods, are available in the Supplemental Materials.
Statistical Analyses
Genetic association analysis was performed for case-control moderate-to-severe COPD status using an additive genetic model adjusted for age, sex, cigarette smoking pack-years, and principal components of genetic ancestry. In the family-based studies, including FHS and EOCOPD/ICGN, we utilized logistic regression with generalized estimating equations to adjust for familial clustering. Quality control on summary statistics from all cohorts was performed with EasyQC (36) to ensure common variant names and reference strand across cohorts and minor allele count (MAC) > 10 within each cohort. In addition to these exome array results, we also included the subset of matching variants (MAC > 4) in a case-control association analysis of UK Biobank (4); models were adjusted for age, sex, pack-years of smoking, ever smoking (when available), and principal components of genetic ancestry.
Power calculations were performed using the genome association study (GAS) calculator available at http://csg.sph.umich.edu/abecasis/cats/gas_power_calculator/index.html (37), based on a COPD prevalence of 0.10 (38, 39). To account for relatedness among individuals, total effective sample size was calculated as previously described (40) and used in power calculations to approximate the number of independent individuals represented by our sample. The total effective sample size was 53,117.
We performed an inverse-variance fixed-effects meta-analysis of exome array results with METAL (41) and limited our analysis to putative functional (nonsynonymous, stop, and splice) variants, which were annotated using wANNOVAR (42). Variants were considered for analysis if they were present in the UK Biobank and at least half of the other cohorts. Exome-wide significance was determined using Bonferroni adjustment (P < 0.05/20,536 variants < 2.4e-6). Replication of signals from previously reported exome array studies (14–16) was defined as a consistent direction of effect and exome-wide statistical significance. Plink v1.9 (43) clump was used to choose a single “index” variant from all variants with R2 ≥ 0.2 in each significantly associated genetic locus. We also performed a targeted analysis of splice and stop variants, considering P values below a Bonferroni-adjusted threshold (P < 0.05/number-of-stop/splice-variants < 0.05/257 < 0.00019) to be nominally significant. We examined the relative association of exome-wide significant COPD variants with the spirometric parameters FEV1 and FEV1/FVC in the GWAS results from Shrine et al. (3). As genetic effects may vary with age, we examined whether age modifies the effect of exome-wide significant variants in the UK Biobank. In addition, we evaluated the association of alleles with age of COPD diagnosis in the COPDGene study. To examine the differential effects of associated variants based on smoking exposure, we performed stratified analyses in ever versus never smokers and heavy (>20 pack-years)versus light (≤ 20 pack-years) smokers in UK Biobank and compared effect sizes between strata.
To determine whether the exome signals were novel, or accounted for by previously described associations, index exonic variants from each locus were compared with prior COPD and lung function GWAS results (3, 4, 6, 7, 26) and were considered distinctly associated if outside of a 2-Mb window. For SNPs within this 2-Mb window, we assessed linkage disequilibrium (LD) between exonic variants and prior GWAS variants by calculating an R2 value using a reference panel of 10,000 randomly selected UK Biobank participants (4). To determine if exonic SNPs were distinct from previously described lead GWAS variants, we used results from GCTA-conditional and joint (COJO) analyses (44) from a prior GWAS, as exome arrays do not assay genome-wide variants (4). Being previously performed, this conditional and joint (COJO) analysis was necessarily limited to variants and cohorts present in the prior GWAS (i.e., all cohorts in the current analysis except HABC and UK COPD Exome Chip Consortium). We calculated a conditionally significant P value threshold by performing a Bonferroni correction for the total number of functional exonic variants genotyped within 2 Mb of the index GWAS variants in which exome-wide significant variants were found [0.05/340 variants (across all regions) = 0.000147].
To examine whether exonic SNPs explained the lead signal at previously reported GWAS loci, we examined whether the exonic variant was present within the 99% credible sets from a recent COPD GWAS (4), obtained using the method of Wakefield et al. (45). We also evaluated predicted functional consequences of amino acid mutations using PolyPhen 2.0 and scaled CADD (Combined Annotation-Dependent Depletion) scores (46, 47). Briefly, CADD scores are based on a support vector machine model predicting the relative deleteriousness of a mutation within a data set; scaling these scores on a rank order magnitude scale allows for external comparisons. For example, a scaled CADD score of 10 means the mutation is in the top 10% of deleterious mutations, a scaled CADD score of 20 means the mutation is in the top 1% of deleterious mutation, and so forth (46, 47). To gain insight into potential biological pathways affected by exonic variants, we also queried gene names at genetics.opentargets.org, which reports relevant biological pathways based on the Reactome Database of Pathways (48, 49).
We performed expression quantitative trait locus (eQTL) lookups for COPD-associated exonic variants, extracting eQTL-regulated genes (eGenes) with PeQTL < 1e-8 from prior publications and publicly available data. We queried four previously published eQTL data sources, including GTEx (www.gtexportal.org) (50, 51) analysis release V6, cis- and trans-eQTLs from Westra et al. (52), lung tissue eQTLs from the Hao et al. (53) study of asthma, and cis- and trans-eQTLs from the Vosa et al. (54) study in the eQTLGen Consortium. For all eQTL sources, a false discovery rate (FDR) of < 0.05 was considered a statistically significant eQTL association. Due to sparsity inherent to exome array association analyses, colocalization with eQTLs could not be performed. Therefore, for each COPD-associated exonic variant that was an eQTL for an eGene, we calculated R2 values between the COPD-associated eQTL and the eGene’s sentinel eQTL SNP using the UK Biobank as an LD reference panel, considering an R2 > 0.2 to be indicative of a shared causal variant in the eQTL and exome array analyses. Sentinel eQTL SNPs for each eGene were defined as the eQTL SNP with the lowest P value. Protein QTL (pQTL) analyses were performed by querying SNP-regulated proteins from Sun et al. (56) and considering P values less than a Bonferroni-corrected threshold (0.05/128,037 SNP-protein pairs = 3.9e-7) to be statistically significant.
We also examined the association of the nonsynonymous SERPINA1 Z allele (rs28929474), the most common cause of α-1 antitrypsin deficiency, with COPD in our study. For the Z allele, we examined the impact of including Z allele homozygotes in a study. For COPD-associated functional exonic variants and the Z allele, we constructed forest plots using the meta R package (57). To examine heterogeneity across studies, we performed meta-regression (57) of COPD-associated variant effect sizes across studies, evaluating the contribution of age, FEV1% predicted, pack-years of smoking, and whether studies excluded ZZ homozygotes (for the Z allele) to individual variant effect sizes.
RESULTS
Characteristics of Cohorts
Characteristics of study participants in each cohort are shown in Table 1. In total, there were 218,399 controls and 33,851 moderate-to-severe COPD cases, which provided 99% power to detect variants with an MAF of 0.01 and an odds ratio of 1.3. (Supplemental Table S1; all Supplemental material is available at https://doi.org/10.6084/m9.figshare.14538222.v1). The cohorts were diverse with respect to case ascertainment, sex distribution, cigarette smoking history, and ancestry. For example, COPDGene, BEOCOPD, ICGN, TCGS (Korea and Poland), and the UK COPD Exome Chip Consortium were COPD case-control studies, and thus, the participants were enriched for COPD cases, ever-smoking status, pack-years of smoking, and lower FEV1% predicted compared with individuals in the population-based cohorts. TCGS-Korea had the highest percentage of males (>95%) in both case and control participants, whereas the lowest proportion of males was observed among cases in BEOCOPD (39.9%) and controls (27.6%) in CHS Aa. Among COPD cases, there was a predominance of males, with 14 cohorts reporting over 55% of cases to be male. With respect to genetic ancestry, there were 7,493 African (including African Americans), 236,312 European, 7,771 East Asian, and 674 Hispanic participants. Not surprisingly, with this sample size and the different cohort characteristics, all of the ANOVA or chi-squared P values across studies were significant (P < 1x10-3).
Table 1.
Characteristics of cohorts in meta-analysis
| Cohort |
n |
Age in Years, Mean ± SD |
Males, % |
Pack Years of Smoking, Mean ± SD |
FEV1 (% Predicted) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | |
| ARIC AA | 289 | 2,765 | 55.7 (5.84) | 53.06 (5.73) | 52.6 | 34.43 | 28.23 (29.4) | 9.18 (16) | 63.2 (13) | 100.3 (12.5) |
| ARIC EA | 1,284 | 7,405 | 56.33 (5.37) | 53.73 (5.64) | 56.15 | 44.09 | 37.73 (25.9) | 11.83 (17.9) | 65.5 (12.4) | 100.7 (11) |
| BEOCOPD | 366 | 560 | 50.8 (8.2) | 39.3 (12.2) | 39.9 | 41.6 | 37.5 (16.5) | 1.66 (14.2) | 29.2 (22.6) | 94.5 (8.5) |
| CHS AA | 107 | 232 | 72.25 (4.91) | 72.76 (5.25) | 49.5 | 27.6 | 29.36 (24.4) | 20.89 (20.4) | 61.38 (13.4) | 102.6 (15) |
| CHS EA | 914 | 1,690 | 73.51 (5.72) | 71.87 (5.11) | 60.2 | 32.3 | 44.59 (28.9) | 25.77 (23.7) | 59.97 (15.6) | 99.49 (13.6) |
| COPDGene AA | 796 | 1,715 | 58.2 (6.6) | 51.8 (3.8) | 55.5 | 58.1 | 37.8 (13.8) | 32.7 (11.1) | 54 (13.1) | 96.6 (9.4) |
| COPDGene NHW | 2,777 | 2,507 | 65.2 (5.8) | 59.3 (6.6) | 55.7 | 49.4 | 49.8 (20.7) | 35 (12) | 50 (15.5) | 95.5 (8.5) |
| FHS | 625 | 4,959 | 61.96 (12.1) | 51.63 (13.2) | 51.04 | 44.69 | 37.22 (24.2) | 15.47 (16.9) | 66.4 (11) | 102 (12) |
| HABC AA | 126 | 817 | 73.3 (2.84) | 73.43 (2.91) | 67.46 | 42.59 | 38.99 (24.8) | 27.34 (23.5) | 59.83 (13.6) | 101.6 (20.5) |
| HABC EA | 213 | 1,259 | 73.53 (2.74) | 73.75 (2.85) | 56.34 | 52.9 | 49.71 (34.7) | 33.36 (30.5) | 62.79 (12.7) | 97.7 (15.7) |
| ICGN | 1,769 | 696 | 59.4 (4.2) | 54.9 (5.8) | 58.6 | 48.3 | 45 (19.5) | 25.1 (13.7) | 39.2 (13.5) | 97.5 (10.5) |
| KARE | 106 | 6,862 | 57.39 (8.14) | 51.49 (8.64) | 78.3 | 45.07 | 23.67 (25) | 8.124 (14.7) | 70.5 (8.6) | 113.8 (16.3) |
| MESA Black | 95 | 551 | 67.5 (8.95) | 64.39 (9.39) | 67.4 | 41.4 | 35 (25.5) | 18.8 (17.8) | 64.1 (13.9) | 102.6 (14.4) |
| MESA Chinese | 32 | 403 | 69.56 (9.09) | 64.03 (9.34) | 53.1 | 47.9 | 31.88 (21) | 20.74 (18) | 65.7 (13.4) | 104.3 (14) |
| MESA Hispanic | 61 | 613 | 67.74 (9.48) | 62.82 (9.54) | 63.9 | 43.2 | 33.73 (31.5) | 15.82 (18) | 65.3 (12.9) | 100 (12.2) |
| MESA White | 180 | 824 | 68.42 (8.87) | 64.38 (9.61) | 51.1 | 47 | 42.42 (33.2) | 23.01 (22.7) | 66.2 (12.1) | 98.9 (11.7) |
| RS | 60 | 415 | 80.1 (5.3) | 79.6 (4.9) | 70 | 52 | 30.3 (23.8) | 14.3 (20.3) | 66.5 (11.1) | 111.6 (18.1) |
| TCGS Korea | 149 | 219 | 69 (5) | 53 (6) | 99.3 | 96.8 | 40 (12) | 25.5 (8.5) | 33.2 (6.9) | 93.6 (6.4) |
| TCGS Poland | 304 | 307 | 62.2 (5.5) | 58.3 (4.6) | 70.1 | 67.4 | 40.3 (12.6) | 32.3 (8.9) | 28.7 (6.7) | 102 (9) |
| UK Biobank | 21,081 | 179,711 | 59.4 (7.3) | 55.7 (8) | 52 | 58 | 19.7 (23.9) | 6.1 (12.5) | 65.1 (11.8) | 98.3 (11.4) |
| UK COPD Exome Chip Consortium |
2,517 | 3,889 | 66.1 (7.74) | 49 (6.08) | 55.24 | 56.08 | 41.2 (24.4) | 22.05 (15.6) | 51.93 (10) | 99.25 (10) |
In total, there were 218,399 controls and 33,851 moderate-to-severe COPD cases. Aa, African ancestry; ARIC, Atherosclerosis Risk in Communities; CHS, Cardiovascular Health Study; COPD, chronic obstructive pulmonary disease; EOCOPD/ICGN, Early-Onset COPD study and International COPD Genetics Network; Ea, European ancestry; FHS, Framingham Heart Study; HABC, Health, Aging, and Body Composition; KARE, Korean Association Resource; MESA, Multi-Ethnic Study of Atherosclerosis; NHW, non-Hispanic White; RS, Rotterdam Study; TCGS, Transcontinental COPD Genetics Study; UKECC, UK COPD Exome Chip Consortium.
Exome Chip Meta-Analysis
An overview of the study design is shown in Fig. 1. Exome arrays containing 109,036 nonsynonymous, stop, and splice variants from International COPD Genetics Consortium (ICGC) (n = 51,458) and UK Biobank (n = 200,792) were meta-analyzed; 20,536 variants were reported in UK Biobank and ≥ 50% of the other studies. The distribution of variant allele frequencies is shown in Supplemental Fig. S1. Of these, 80 variants reached exome-wide Bonferroni-adjusted level of significance (P < 2.4e-6) (Fig. 2 and Supplemental Table S2). After clumping, these 80 variants were represented by 35 lead variants. All 35 lead variants were within 2 Mb of previously reported GWAS SNPs (Table 2). Eight of these variants met the criteria (see methods) for replication of exonic signals from prior exome array and genome-wide studies (Supplemental Table S3). Twenty-one exonic variants were in low LD (R2 < 0.2) with nearby GWAS variants.
Figure 1.

Overview of study design. COPD, chronic obstructive pulmonary disease; GWAS, genome-wide association studies.
Figure 2.
Manhattan plot of exome array variants. The horizontal red line indicates an exome-wide significance level of 2.4e-6. The exonic variants reaching exome-wide significance are annotated.
Table 2.
Thirty-five lead exonic SNPs and nearby sentinel GWAS SNPs (within 2 Mb)
| Exon rs No. | Chr | Position (hg19) | Risk Allele | Other Allele | Gene Name | Risk Allele Frequency | OR (95% CI) | P | GWAS rs No. | GWAS Closest Gene | R2 | Source | Trait |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs11205303 | 1 | 149906413 | C | T | MTMR11 | 0.4 | 1.06 (1.03–1.08) | 7.41E-08 | rs11205354 | C1orf54 | 0.0055** | Shrine (3) | PEF |
| rs2571445 | 2 | 218683154 | A | G | TNS1 | 0.39 | 1.08 (1.05–1.1) | 4.17E-13 | rs2571445 | TNS1 | 1 | Shrine (3) | FEV1 |
| rs11938093 | 4 | 75675841 | A | T | BTC | 0.74 | 1.07 (1.04–1.09) | 4.44E-09 | rs4585380 | BTC | 1 | Sakornsakolpat (4) | Moderate COPD |
| rs2454206 | 4 | 106196951 | G | A | TET2 | 0.38 | 1.07 (1.05–1.09) | 1.67E-10 | rs10516526 | GSTCD | 0.011** | Wain (6) | FEV1 |
| rs10043775 | 5 | 147805120 | T | C | FBXO38 | 0.73 | 1.09 (1.06–1.11) | 1.86E-13 | rs10037493 | HTR4 | 0.31 | Sakornsakolpat (4) | Moderate COPD |
| rs1800888 | 5 | 148206885 | T | C | ADRB2 | 0.014 | 1.32 (1.22–1.42) | 9.39E-13 | rs1800888 | ADRB2 | 1 | Shrine (3) | FEV1 |
| rs2287749 | 5 | 156918850 | C | T | ADAM19 | 0.87 | 1.09 (1.06–1.12) | 3.61E-09 | rs72811310 | ADAM19 | 0.8 | Sakornsakolpat (4) | Moderate COPD |
| rs1422795 | 5 | 156936364 | C | T | ADAM19 | 0.35 | 1.09 (1.07–1.11) | 1.94E-17 | rs1990950 | ADAM19 | 0.31 | Wain (6) | FEV1/FVC |
| rs11740603 | 5 | 157098756 | T | G | C5orf52 | 0.14 | 1.08 (1.05–1.11) | 1.11E-07 | rs1990950 | ADAM19 | 0.029** | Wain (6) | FEV1/FVC |
| rs1334576 | 6 | 7211818 | A | G | RREB1 | 0.42 | 1.05 (1.03–1.07) | 5.11E-08 | rs1334576 | RREB1 | 1 | Sakornsakolpat (4) | Moderate COPD |
| rs13195509 | 6 | 26463660 | A | G | BTN2A1 | 0.12 | 1.11 (1.08–1.15) | 2.53E-12 | rs2070600 | AGER | 0.0023** | Shrine (3) | FEV1/FVC |
| rs61742093 | 6 | 27879982 | G | A | OR2B2 | 0.11 | 1.11 (1.08–1.14) | 5.98E-11 | rs2070600 | AGER | 0.0026** | Shrine (3) | FEV1/FVC |
| rs2232423 | 6 | 28366151 | G | A | ZSCAN12 | 0.11 | 1.11 (1.08–1.15) | 1.96E-11 | rs2070600 | AGER | 0.0023** | Shrine (3) | FEV1/FVC |
| rs3749971* | 6 | 29342775 | A | G | OR12D3 | 0.12 | 1.11 (1.08–1.14) | 2.60E-11 | rs2070600 | AGER | 0.003** | Shrine (3) | FEV1/FVC |
| rs2523989* | 6 | 30078275 | T | C | TRIM31 | 0.17 | 1.09 (1.06–1.12) | 1.40E-10 | rs2070600 | AGER | 0.0012** | Shrine (3) | FEV1/FVC |
| rs929156* | 6 | 30139699 | G | A | TRIM15 | 0.76 | 1.06 (1.04–1.09) | 8.17E-08 | rs2070600 | AGER | 0.044** | Shrine (3) | FEV1/FVC |
| rs9262143* | 6 | 30652781 | T | C | PPP1R18 | 0.14 | 1.11 (1.08–1.15) | 9.98E-14 | rs2070600 | AGER | 0.0044** | Shrine (3) | FEV1/FVC |
| rs2074506* | 6 | 30890483 | G | T | VARS2 | 0.65 | 1.06 (1.03–1.08) | 2.79E-07 | rs2070600 | AGER | 0.041** | Shrine (3) | FEV1/FVC |
| rs7750641* | 6 | 31129310 | T | C | TCF19 | 0.15 | 1.11 (1.08–1.14) | 1.11E-12 | rs2070600 | AGER | 0.0078** | Shrine (3) | FEV1/FVC |
| rs3101017* | 6 | 31733466 | C | T | VWA7 | 0.13 | 1.12 (1.09–1.15) | 1.92E-14 | rs2070600 | AGER | 0.0092** | Shrine (3) | FEV1/FVC |
| rs2070600* | 6 | 32151443 | C | T | AGER | 0.94 | 1.19 (1.15–1.25) | 1.31E-16 | rs2070600 | AGER | 1 | Shrine (3) | FEV1/FVC |
| rs7775397* | 6 | 32261252 | G | T | C6orf10 | 0.13 | 1.12 (1.08–1.15) | 3.27E-13 | rs2070600 | AGER | 0.011** | Shrine (3) | FEV1/FVC |
| rs1129740* | 6 | 32609105 | A | G | HLA-DQA1 | 0.59 | 1.07 (1.04–1.09) | 9.84E-10 | rs2070600 | AGER | 0.033** | Shrine (3) | FEV1/FVC |
| rs17280293 | 6 | 142688969 | A | G | ADGRG6 | 0.97 | 1.32 (1.24–1.41) | 8.13E-18 | rs17280293 | ADGRG6 | 1 | Shrine (3) | FEV1/FVC |
| rs11155242 | 6 | 142691549 | A | C | ADGRG6 | 0.81 | 1.14 (1.11–1.17) | 5.98E-26 | rs9399401 | ADGRG6 | 0.62 | Sakornsakolpat (4) | Moderate COPD |
| rs721917 | 10 | 81706324 | G | A | SFTPD | 0.42 | 1.05 (1.03–1.07) | 8.91E-08 | rs721917 | SFTPD | 1 | Shrine (3) | FEV1/FVC |
| rs11049488 | 12 | 28412372 | G | A | CCDC91 | 0.7 | 1.06 (1.04–1.08) | 4.67E-08 | rs11049386 | PTHLH; CCDC91 | 0.83 | Sakornsakolpat (4) | Moderate COPD |
| rs3829947 | 14 | 93118038 | A | G | RIN3 | 0.45 | 1.05 (1.03–1.07) | 5.35E-07 | rs117068593 | RIN3 | 0.18** | Wain (6) | FEV1 |
| rs3885951 | 15 | 78825917 | G | A | HYKK | 0.096 | 1.09 (1.06–1.13) | 4.71E-08 | rs28534575 | CHRNB4 | 0.026** | Sakornsakolpat (4) | Moderate COPD |
| rs16969968 | 15 | 78882925 | A | G | CHRNA5 | 0.33 | 1.11 (1.09–1.14) | 5.03E-25 | rs28534575 | CHRNB4 | 0.14** | Sakornsakolpat (4) | Moderate COPD |
| rs4842860 | 15 | 83680287 | A | G | C15orf40 | 0.38 | 1.05 (1.03–1.07) | 3.54E-07 | rs7181169 | BTBD1 | 0.44 | Sakornsakolpat (4) | Moderate COPD |
| rs17361375 | 15 | 83680329 | G | A | C15orf40 | 0.79 | 1.06 (1.04–1.09) | 3.72E-07 | rs7181169 | BTBD1 | 0.33 | Sakornsakolpat (4) | Moderate COPD |
| rs4842838 | 15 | 84582124 | G | T | ADAMTSL3 | 0.47 | 1.07 (1.05–1.09) | 2.92E-12 | rs1896797 | SH3GL3 | 0.28 | Shrine (3) | FEV1/FVC |
| rs9897794 | 17 | 28296327 | G | T | EFCAB5 | 0.48 | 1.05 (1.03–1.07) | 1.51E-07 | rs2244592 | SSH2 | 0.71 | Shrine (3) | FEV1/FVC |
| rs12373142 | 17 | 43924200 | G | C | SPPL2C | 0.22 | 1.08 (1.05–1.1) | 1.80E-10 | rs12373142 | SPPL2C | 1 | Sakornsakolpat (4) | Moderate COPD |
*Variant location within HLA region (hg19; chromosome 6: 28477797-33448354). **R2 < 0.2. Exonic variants were clumped prior to comparing with GWAS SNPs based on an R2 > 0.2. References to GWAS variants are for the most recent publication. COPD, chronic obstructive pulmonary disease; GWAS, genome-wide association study; HLA, human leukocyte antigen; PEF, peak expiratory flow; SNPs, single-nucleotide polymorphisms.
Of the 35 exome-wide significant lead variants, we identified four novel conditionally significant exonic SNPs (Table 3), meaning that these SNPs were within 2 Mb of COPD GWAS variants, though retained regional significance after conditioning on the lead COPD GWAS SNP using GCTA-COJO (Bonferroni P value = 0.05/340 variants = 0.000147; see materials and methods). Seven of the 35 exonic variants were index variants, so conditional and joint analyses were not performed for these variants (rs721917, rs28929474, rs12373142, rs11205303, rs2571445, rs1800888, and rs1334576) (4). The rs2454206 variant in tet methylcytosine dioxygenase 2 (TET2) was significant after conditioning on the rs34712979 index variant, though this variant exists at a locus with two additional independent variants (rs2047409 and rs10516528) (5). We observed that the exonic rs2454206 variant association was no longer significant after conditioning on rs2047409 (P = 0.24). In stepwise joint modeling considering these four TET2 locus variants, only rs2454206 and rs34712979 were selected for the final model.
Table 3.
Conditionally significant exome array SNPs (P < 0.000147) within 2 Mb of GWAS SNPs identified in UK Biobank (4)
| Chr | GWAS Index SNP | Exonic SNP | Gene Name | Exonic Variant Risk Allele Frequency | B | SE | P | b (Conditional) | SE (Conditional) | P (Conditional) |
|---|---|---|---|---|---|---|---|---|---|---|
| 5 | rs10866659 | rs2287749 | ADAM19 | 0.9 | −0.089 | 0.014 | 1.00E-09 | −0.057 | 0.014 | 4.30E-05 |
| 6 | rs2284174 | rs13195509 | BTN2A1 | 0.069 | 0.12 | 0.015 | 7.80E-15 | 0.056 | 0.014 | 3.40E-05 |
| 15 | rs10152300 | rs17361375 | C15orf40 | 0.81 | −0.058 | 0.012 | 1.60E-06 | −0.051 | 0.012 | 2.20E-05 |
| 15 | rs10152300 | rs4842860 | C15orf40 | 0.65 | 0.043 | 0.0099 | 1.60E-05 | 0.04 | 0.0099 | 5.20E-05 |
Exonic variant effects were adjusted for the index SNPs indicated in the table. Chromosome positions based on build hg19. Note that the rs2454206 variant in TET2 was significant after conditioning on the rs34712979 index variant, though this variant exists at a locus with two additional independent variants (rs2047409 and rs10516528) (5). We observed that the exonic rs2454206 variant was no longer significant after conditioning on rs2047409 (P = 0.24). Note that HLA imputation was not performed, so HLA region variants were not included in conditional and joint analyses. GWAS, genome-wide association study; HLA, human leukocyte antigen; SNPs, single-nucleotide polymorphisms.
We also evaluated the 35 exome-wide significant lead variants in the 99% credible sets for the COPD GWAS loci from Sakornsakolpat et al. (4) (Table 4); 18 lead exonic variants were present in the 99% credible sets. Three variants had a posterior probability of association (PPA) > 10%, and 10 variants were in the top 20% of their respective credible sets (Supplemental Fig. S2), suggesting these are more likely to be causal variants. Only rs1334576 in ras responsive element binding protein 1 (RREB1) had a PPA > 10%, ranked within the top 20% of its credible set, and was predicted to be damaging by PolyPhen and CADD. The remaining 17 top exonic variants (Supplemental Table S4) were not present in their respective 99% credible set.
Table 4.
Exome array variants identified in 99% credible sets derived from UK Biobank (4) using the method by Wakefield et al. (45) and wANNOVAR functional annotations
| Chr | Exonic SNP | Gene Name | GWAS Index SNP for Fine-Mapping* | PPA | Rank | Percentile | Polyphen Prediction | Polyphen Rank Score | Amino Acid Change | Scaled CADD |
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | rs2571445 | TNS1 | 218683154:A:G | 0.58 | 1 | 100 | Benign | 0.013 | W1197R | 5.551 |
| 4 | rs11938093 | BTC | 75673363:G:A | 0.032 | 4 | 93 | Probably damaging | 0.739 | L124M | 24.4 |
| 5 | rs10043775 | FBXO38 | 148059519:G:A | 0.00031 | 356 | 91 | Benign | 0.013 | S592P | 12.25 |
| 5 | rs10043775 | FBXO38 | 148203236:T:C | 4.70E-05 | 471 | 84 | Benign | 0.013 | S592P | 12.25 |
| 5 | rs10043775 | FBXO38 | 148611623:C:A | 7.60E-05 | 368 | 85 | Benign | 0.013 | S592P | 12.25 |
| 5 | rs2287749 | ADAM19 | 156948318:T:G | 0.023 | 6 | 100 | Possibly damaging | 0.494 | G660D | 24.3 |
| 5 | rs1422795 | ADAM19 | 156937043:A:G | 0.049 | 9 | 60 | Benign | 0.391 | S284G | 14.33 |
| 5 | rs11740603 | C5orf52 | 156948318:T:G | 1.00E-05 | 3708 | 6.2 | Possibly damaging | 0.451 | R45L | 24.2 |
| 5 | rs11740603 | C5orf52 | 157002695:C:T | 1.20E-05 | 1110 | 59 | Possibly damaging | 0.451 | R45L | 24.2 |
| 6 | rs1334576 | RREB1 | 7211818:G:A | 0.14 | 1 | 100 | Possibly damaging | 0.594 | G195R | 12.63 |
| 6 | rs17280293 | ADGRG6 | 142814991:C:T | 0.024 | 5 | 0 | Possibly damaging | 0.493 | S123G | 24.7 |
| 10 | rs721917 | SFTPD | 81706324:A:G | 0.23 | 1 | 100 | Benign | 0.013 | M31T | 0.003 |
| 12 | rs11049488 | CCDC91 | 28320536:T:A | 0.0022 | 93 | 81 | Benign | 0.08 | A36T | 11.08 |
| 14 | rs3829947 | RIN3 | 92600798:G:T | 5.20E-05 | 421 | 86 | Benign | 0.04 | H215R | 3.995 |
| 15 | rs3885951 | HYKK | 78388464:C:T | 6.60E-05 | 1189 | 67 | Benign | 0.104 | K343E | 18.98 |
| 15 | rs3885951 | HYKK | 78923845:T:G | 1.80E-05 | 944 | 66 | Benign | 0.104 | K343E | 18.98 |
| 15 | rs16969968 | CHRNA5 | 78898932:C:G | 0.016 | 10 | 83 | Benign | 0.145 | D398N | 15.07 |
| 15 | rs4842860 | C15orf40 | 83693513:T:C | 0.00088 | 70 | 78 | Benign | 0.013 | C25R | 0.029 |
| 15 | rs17361375 | C15orf40 | 83693513:T:C | 0.00021 | 149 | 53 | Benign | 0.093 | L11F | 6.178 |
| 15 | rs4842838 | ADAMTSL3 | 84515943:C:G | 3.30E-05 | 277 | 26 | Benign | 0.013 | V661L | 12.87 |
| 17 | rs9897794 | EFCAB5 | 28413129:T:C | 0.0039 | 73 | 79 | Benign | 0.013 | L237V | 12.25 |
| 17 | rs12373142 | SPPL2C | 43924200:C:G | 0.03 | 1 | 100 | Benign | 0.139 | P643R | 0.117 |
PolyPhen and CADD were used to predict consequences of mutation. CADD scores are based on a support vector machine model predicting the relative deleteriousness of a mutation within a dataset; scaling these scores on a rank order magnitude scale allows for external comparisons. For example, a scaled CADD score of 10 means the mutation is in the top 10% of deleterious mutations, a scaled CADD score of 20 means the mutation is in the top 1% of deleterious mutation, and so forth (46, 47). Chromosome positions based on hg19. Percentile indicates the ranking of the exonic variant within the credible set of the GWAS index SNP. CADD, combined annotation dependent depletion; GWAS, genome-wide association study; PPA, posterior probability of association.
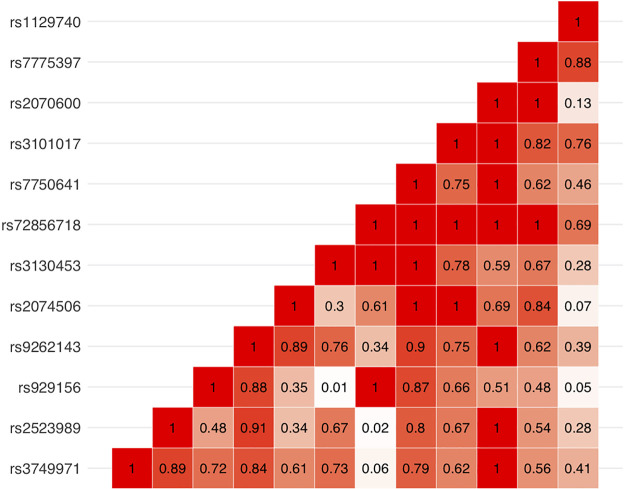
We also analyzed 257 stop or splice variants, of which two stop variants (rs3130453 and rs72856718) and one splice variant (rs11078928) reached Bonferroni-adjusted significance (P < 0.00019; see methods) (Supplemental Table S5). The stop variant rs3130453 (MAF = 0.49) in the coiled-coil α-helical rod protein 1 (CCHCR1) gene was associated with an odds ratio of 1.04 [95% confidence interval (CI) = 1.02–1.06, P = 1.3e-5] for COPD. The rs72856718 stop variant (MAF = 0.09), also in the CCHCR1 gene, was associated with an odds ratio of 1.08 (95% CI = 1.04–1.13, P = 8.8e-5) for COPD. The splice variant, leading to an exon 6 deletion in gasdermin B (GSDMB), had an odds ratio of 1.04 (95% CI = 1.02–1.06, P = 1.8e-4) in association with COPD. Forest plots for all exome-wide significant, stop, and splice variants are shown in Supplemental Fig. S3. Reactome pathways for the genes associated with conditionally significant, stop, and splice variants are shown in Supplemental Table S6. Twelve exonic variants (including stop/splice variants), many of which were highly correlated with each other (Fig. 3), are located within the complex human leukocyte antigen (HLA) region (hg19; chromosome 6:28477797–33448354).
Figure 3.
LD matrix (R2) of exome array variants within the HLA region (hg19; 6:28477797-33448354). HLA, human leukocyte antigen; LD, linkage disequilibrium.
Similar associations of variants in Table 2 were observed in a prior GWAS of FEV1 and FEV1/FVC, except that three variants (rs3885951, rs11078928, and rs28929474) did not reach our level of exome-wide significance for either GWAS phenotype (Supplemental Table S7). We also assessed for interactions of exome-wide significant variants in Table 2 with age in UK Biobank and found no significant interactions (all P > 0.05). Furthermore, we evaluated whether each of these variants was associated with earlier age of COPD diagnosis in the COPDGene cohort and observed that the smoking behavior-associated rs16969968 SNP in CHRNA5 was associated with earlier age of COPD diagnosis (P = 0.001; Supplemental Fig. S4). Comparing ever with never smokers and heavy with light smokers, the effect sizes are generally similar between strata (Supplemental Fig. S5). The exceptions include rs1422795, rs2571446, and rs3829947 in ever versus never smokers and rs1422795 in heavy versus light smokers.
eQTL and pQTL analyses.
The correlations between the 35 lead exonic variants and sentinel eQTL SNPs (i.e., the eQTL SNP with the lowest P value of all eQTL SNPs assigned to an eGene) in which the LD R2 is > 0.2 are shown in Table 5, and the full set of eQTL-regulated SNPs are shown in Supplemental Table S8. Several exonic variants are associated with eQTL SNPs that regulate the same gene within lung tissue, including FBXO38, ADGRG6, RREB1, C15orf40, and EFCAB5. In pQTL analyses (56), 12 exonic variants were significantly associated with protein expression (Table 6).
Table 5.
Exon SNPs and sentinel eQTL SNPs with R2 > 0.2 and P value (eQTL) < 1e-8
| Chr | Exonic SNP | Gene Name | eQTL rs No. | eQTL-Regulated Gene | Direction of Effect | P Value | R2 | Source |
|---|---|---|---|---|---|---|---|---|
| 5 | rs10043775 | FBXO38 | rs6876982 | FBXO38 | + | 2.00E-20 | 0.7 | Hao et al. (53); lung cis-eQTL |
| 6 | rs11155242 | ADGRG6 | rs11155242 | ADGRG6 | − | 5.20E-136 | 1 | Vosa et al. (54); cis-eQTL |
| 6 | rs13195509 | BTN2A1 | rs35304979 | BTN3A2 | + | 3.10E-82 | 0.84 | GTeX lung |
| rs3117425 | BTN3A2 | − | 3.80E-45 | 0.59 | Hao et al. (53); lung trans-eQTL | |||
| rs149959 | ZNF165 | + | 2.00E-09 | 0.27 | Hao et al. (53); lung cis-eQTL | |||
| 6 | rs61742093 | OR2B2 | rs3117425 | BTN3A2 | − | 3.80E-45 | 0.79 | Hao et al. (53); lung trans-eQTL |
| rs35304979 | BTN3A2 | + | 3.10E-82 | 0.62 | GTeX lung | |||
| rs149959 | ZNF165 | + | 2.00E-09 | 0.32 | Hao et al. (53); lung cis-eQTL | |||
| 6 | rs2232423 | ZSCAN12 | rs3117425 | BTN3A2 | − | 3.80E-45 | 0.85 | Hao et al. (53); lung trans-eQTL |
| rs35304979 | BTN3A2 | + | 3.10E-82 | 0.58 | GTeX lung | |||
| rs149959 | ZNF165 | + | 2.00E-09 | 0.31 | Hao et al. (53); lung cis-eQTL | |||
| 6 | rs3749971* | OR12D3 | rs3117425 | BTN3A2 | − | 3.80E-45 | 0.92 | Hao et al. (53); lung trans-eQTL |
| rs35304979 | BTN3A2 | + | 3.10E-82 | 0.5 | GTeX lung | |||
| rs149959 | ZNF165 | + | 2.00E-09 | 0.26 | Hao et al. (53); lung cis-eQTL | |||
| 6 | rs2523989* | TRIM31 | rs3117425 | BTN3A2 | − | 3.80E-45 | 0.54 | Hao et al. (53); lung trans-eQTL |
| rs35304979 | BTN3A2 | + | 3.10E-82 | 0.28 | GTeX lung | |||
| 6 | rs929156* | TRIM15 | rs9261468 | TRIM10 | − | 4.60E-11 | 0.89 | Hao et al. (53); lung cis-eQTL |
| 6 | rs9262143* | PPP1R18 | rs3117425 | BTN3A2 | − | 3.80E-45 | 0.58 | Hao et al. (53); lung trans-eQTL |
| rs35304979 | BTN3A2 | + | 3.10E-82 | 0.32 | GTeX lung | |||
| 6 | rs7750641* | TCF19 | rs114810457 | AGER | + | 4.20E-82 | 0.29 | Vosa et al. (54); cis-eQTL |
| rs3117425 | BTN3A2 | − | 3.80E-45 | 0.41 | Hao et al. (53); lung trans-eQTL | |||
| rs35304979 | BTN3A2 | + | 3.10E-82 | 0.22 | GTeX lung | |||
| 6 | rs3101017* | VWA7 | rs114810457 | AGER | + | 4.20E-82 | 0.37 | Vosa et al. (54); cis-eQTL |
| rs3117425 | BTN3A2 | − | 3.80E-45 | 0.34 | Hao et al. (53); lung trans-eQTL | |||
| 6 | rs7775397* | C6orf10 | rs114810457 | AGER | + | 4.20E-82 | 0.39 | Vosa et al. (54); cis-eQTL |
| rs3117425 | BTN3A2 | − | 3.80E-45 | 0.29 | Hao et al. (53); lung trans-eQTL | |||
| 6 | rs1129740* | HLA-DQA1 | rs9273500 | HLA-DRA | − | 2.60E-29 | 0.36 | Vosa et al. (54); cis-eQTL |
| 6 | rs1334576 | RREB1 | rs2714341 | RREB1 | − | 1.20E-86 | 0.34 | Vosa et al. (54); cis-eQTL |
| 15 | rs16969968 | CHRNA5 | rs931794 | PSMA4 | + | 5.60E-107 | 0.91 | Vosa et al. (54); cis-eQTL |
| rs12591557 | PSMA4 | − | 3.30E-20 | 0.37 | Hao et al. (53); lung cis-eQTL | |||
| 15 | rs4842860 | C15orf40 | rs6603041 | C15orf40 | + | 3.10E-110 | 0.64 | Vosa et al. (53); cis-eQTL |
| rs6603041 | C15orf40 | + | 9.70E-11 | 0.64 | GTeX lung | |||
| 17 | rs9897794 | EFCAB5 | rs7501472 | CORO6 | + | 3.00E-61 | 0.78 | Vosa et al. (54); cis-eQTL |
| rs4567782 | EFCAB5 | + | 2.30E-09 | 0.97 | Hao et al. (53); lung cis-eQTL | |||
| rs3936006 | EFCAB5 | − | 5.10E-74 | 0.93 | Vosa et al. (54); cis-eQTL |
*Indicates HLA region. eQTL, expression quantitative trait locus; HLA, human leukocyte antigen; SNPs, single-nucleotide polymorphisms.
Table 6.
Exonic genes associated with pQTL-regulated proteins at Bonferroni-corrected significance level (P < 3.9e-7)
| Marker | Exon rs No. | Exon HGNC Symbol | Effect | P Value | pQTL-Regulated Protein |
|---|---|---|---|---|---|
| 6:26463660 | rs13195509 | BTN2A1 | 0.3635 | 1.90E-10 | MICB |
| 6:27879982 | rs61742093 | OR2B2 | −0.4214 | 1.40E-13 | MICB |
| −0.3019 | 2.20E-07 | PDE4D | |||
| 6:28366151 | rs2232423 | ZSCAN12 | −0.4332 | 2.50E-14 | MICB |
| −0.333 | 8.40E-09 | PDE4D | |||
| 6:29342775 | rs3749971 | OR12D3 | 0.4299 | 1.20E-14 | MICB |
| 0.3077 | 6.60E-08 | PDE4D | |||
| 6:30078275 | rs2523989 | TRIM31 | 0.2574 | 3.80E-07 | GRIA4 |
| 0.4292 | 1.80E-18 | MICB | |||
| 0.2725 | 6.80E-08 | PDE4D | |||
| 6:31124849 | rs3130453 | CCHCR1 | −0.2171 | 1.60E-08 | PRSS3 |
| 6:31125257 | rs72856718 | CCHCR1 | −0.7092 | 3.10E-22 | CREB3L4 |
| 6:31129310 | rs7750641 | TCF19 | 0.3488 | 4.30E-11 | C4A |
| 0.3216 | 1.50E-09 | CD96 | |||
| 0.4254 | 3.60E-16 | GRIA4 | |||
| 0.5975 | 3.30E-32 | MICB | |||
| 0.4388 | 3.50E-17 | PDE4D | |||
| 6:31733466 | rs3101017 | VWA7 | −0.4409 | 2.10E-15 | C4A |
| −0.4089 | 2.40E-13 | CD96 | |||
| −0.305 | 8.50E-08 | DEFB119 | |||
| −0.5098 | 1.70E-20 | GRIA4 | |||
| −0.3301 | 5.70E-09 | HLA-DQA2 | |||
| −0.3009 | 1.30E-07 | IL21 | |||
| −0.6693 | 5.00E-36 | MICB | |||
| −0.5366 | 9.90E-23 | PDE4D | |||
| 0.3086 | 5.50E-08 | PRSS3 | |||
| 6:32151443 | rs2070600 | AGER | 0.5749 | 8.40E-15 | AGER |
| 0.5391 | 2.90E-13 | PRSS3 | |||
| 0.5014 | 1.90E-11 | RACGAP1 | |||
| 6:32261252 | rs7775397 | C6orf10 | −0.4155 | 1.70E-13 | C4A |
| −0.3764 | 3.20E-11 | CD96 | |||
| −0.3222 | 1.90E-08 | DEFB119 | |||
| −0.4735 | 2.20E-17 | GRIA4 | |||
| −0.3798 | 2.10E-11 | HLA-DQA2 | |||
| −0.5861 | 1.00E-26 | MICB | |||
| −0.5085 | 5.10E-20 | PDE4D | |||
| 0.315 | 3.90E-08 | PRSS3 | |||
| 14:94844947 | rs28929474 | SERPINA1 | −1.1964 | 1.20E-21 | ACP2 |
| −0.7288 | 1.90E-08 | DNAJB9 | |||
| −1.7387 | 8.60E-48 | MRPL33 | |||
| −1.0455 | 1.40E-16 | NCF2 | |||
| −1.0295 | 4.40E-16 | PIM1 | |||
| −0.9825 | 1.10E-14 | SNAP25 | |||
| −0.7328 | 1.60E-08 | TXNDC5 | |||
| −0.7449 | 8.80E-09 | WISP3 | |||
| −1.3836 | 3.20E-29 | ZNF175 |
*Indicates variant is in HLA region. pQTL, protein quantitative trait locus; HGNC, HUGO Gene Nomenclature Committee; HLA, human leukocyte antigen; SNPs, single-nucleotide polymorphisms.
SERPINA1 Z allele effects.
The SERPINA1 Z allele rs28929474 was associated with a 1.18 odds ratio for COPD in the fixed-effects analysis (95% CI = 1.10–1.26, P = 1.74e-6) (Supplemental Table S5). In the Z allele meta-analysis, there is evidence of heterogeneity (I2 = 0.6), and the African Americans from the ARIC cohort exhibited an opposite direction of effect, which was not statistically significant (Fig. 4). Given the observed Z allele effect size heterogeneity, we performed a random-effects meta-analysis, and the rs28929474 variant demonstrated association with a 1.43 odds ratio for COPD (95% CI = 1.17–1.74, P = 0.0043).
Figure 4.
Forest plot of the SERPINA1 Z allele (rs28929474). The Z allele was associated with a 1.18 odds ratio for COPD in fixed-effects analysis (95% CI = 1.10–1.26, P = 1.74e-6) and 1.43 odds ratio for COPD in random-effects analysis (95% CI = 1.17–1.74, P = 0.0043) (I2 = 0.60). See methods for cohort abbreviations. Aa, African ancestry; ARIC, Atherosclerosis Risk in Communities; CHS, Cardiovascular Health Study; CI = confidence interval; COPD, chronic obstructive pulmonary disease; EOCOPD/ICGN, Early-Onset COPD study and International COPD Genetics Network; Ea, European ancestry; FHS, Framingham Heart Study; HABC, Health, Aging, and Body Composition; MESA, Multi-Ethnic Study of Atherosclerosis; NHW, non-Hispanic White; TCGS, Transcontinental COPD Genetics Study; UKECC, UK COPD Exome Chip Consortium.
Meta-regression.
For each exome-wide significant variant, we performed meta-regression to examine the cohort-specific effects of FEV1% predicted and pack-years of smoking; for the Z allele (rs28929474), we also examined the effects of inclusion of Z allele homozygotes on the reported variant effect sizes (Supplemental Table S9). Mean differences in FEV1% predicted from individual cohorts did not account for the observed heterogeneity, nor did whether a study excluded Z allele homozygotes. Heterogeneity of effect sizes was at least partially attributable to mean differences in pack-years of smoking for several variants (rs12373142, rs1334576, rs16969968, rs2523989, rs3130453, and rs7750641).
DISCUSSION
In this study, we meta-analyzed exome array data from 33,851 moderate-to-severe COPD cases and 218,399 controls. We report four exonic variants on chromosomes 5q, 6p, and 15q, as well as two stop variants and one splice variant associated with COPD. We also examined the association of the SERPINA1 Z allele (rs28929474) with COPD and heterogeneity of effect sizes across cohorts. These results lend further insight into the potential pathogenesis of this disease and identify potential loci for laboratory-based validation.
Compared with prior studies, this exome array meta-analysis includes significantly more participants and extends prior findings by providing an in-depth characterization of exonic variants. Using the criteria that exome-wide significance is reached and the direction of effect and risk allele are the same in both the current and prior studies, eight exonic variants from prior genome-wide and exome array studies were replicated (3, 4, 6, 7, 14–16, 58). Of these, multiple lines of evidence suggest that rs1334576 in RREB1 is likely a functional variant; RREB1 is a zinc-finger transcription factor that binds to Ras-related elements in gene promoters and has been implicated in cell differentiation (59–61). Smoking history was associated with the CHRNA5 variant in meta-regression, which is not surprising given the well-established role of this locus in smoking behavior (62). This variant was also associated with early age of COPD diagnosis. Although we had adequate power to detect variants with an MAF of 0.01 with an effect size of 1.3 or greater, we did not identify any novel rare variants with large effect sizes. These data suggest that low-frequency protein-coding variants (down to 1%) with large effect sizes do not play a substantial role in COPD pathogenesis. However, this study was not powered to assess the impact of very rare variants (MAF < 0.01), nor the effects of low frequency and common protein-coding variants on COPD subtypes.
Although all variants were near previously identified COPD GWAS loci, applying a more relaxed multiple testing threshold (63, 64) led to the identification of four independently associated exonic variants that remained significant after conditioning on the lead nearby GWAS variant. The rs2287749 variant may be a causal variant based on conditional, credible set, and PolyPhen analyses and is located within ADAM19, a metalloproteinase (65) involved in cell-matrix interactions and invadopodia formation in cancer cells (49) and previously implicated in COPD risk (4, 66). Three independent variants have been previously reported at the TET2 4q24 locus (5). Conditional and joint analyses suggest that rs2047409 and rs34712979 account for the signal observed at this locus. TET2 is involved in DNA demethylation and regulation of gene expression, and variants have been associated with extremes of FEV1 (5) and linked to age-associated clonal hematopoiesis and self-reported COPD and/or asthma (67). Our results further highlight the importance of TET2 in COPD risk. Two novel variants, rs4842860 and rs17361375, in C15orf40 were identified. In eQTL analyses, the former SNP was correlated with eQTL SNP rs6603041 in both lung and blood. Neither variant was found in their respective 99% credible sets from a COPD GWAS. This finding might indicate that these variants are not causal or could indicate that this locus was not adequately characterized by GWAS. A deeper characterization of this locus could help clarify its role in COPD pathogenesis.
Examining only the most deleterious (stop and splice) variants, we identified three nominally significant associations. We found two stop variants in the CCHCR1 gene. CCHCR1 has a role in regulating cell proliferation and differentiation, both of which are important in the pathogenesis of emphysema. The splice variant rs11078928 encodes a polymorphism at a splice acceptor site in gasdermin B (GSDMB) on chromosome 17q21 (67). GSDMB is important in pyroptosis, a type of programmed cell death that releases inflammatory mediators. Pyroptosis is activated by caspase-mediated cleavage of the inhibitory C-terminus of gasdermin B, releasing the functional N-terminus (68–70). The rs11078928 variant leads to a deletion of exon 6 in the N-terminus, rendering gasdermin B unable to activate pyroptosis (71). The minor allele (C) has been associated with lower asthma risk (71) and was associated with lower COPD risk in our study. This finding is consistent with the notion that subpopulations of individuals have features of both asthma and COPD; indeed, childhood asthma is associated with lower lung function and increased risk for COPD in adulthood (72, 73). Thus, GSDMB may contribute to the pathobiology of asthma-COPD overlap.
Prior investigations into the Z allele association with COPD have been conflicting. Many prior lung function and COPD genetic association studies (GWASs and exome-wide) have not reported associations with the SERPINA1 Z allele (3–6, 14–16). Yet, the Z allele has also been associated with severe COPD (74) and lung function (75) at genome-wide significance in cohorts enriched for COPD and heavy smoking, potentially with a gene-by-smoking interaction (76). We evaluated the effect of a directly genotyped (rather than imputed) Z allele and applied a random-effects meta-analysis. The Z allele was associated with a 1.17–1.74 odds ratio for COPD.
We observed an asymmetric distribution of effect sizes and standard errors for the Z allele across cohorts, suggesting that there may be cohort-specific selection bias with regard to the inclusion of individuals with the Z allele. The combination of the exclusion of homozygous Z allele individuals (PiZZ) from many COPD case-control studies and the inclusion of a large number of individuals with little to no smoking history in population-based cohorts likely diminished the power to detect an overall effect for the Z allele. To explore this issue, we used meta-regression to assess the impact of intentional exclusion of ZZ individuals, lung function severity, and cigarette smoke exposure on Z allele effect size heterogeneity. None of these factors clearly explained the observed Z allele effect size heterogeneity across studies.
This study has several strengths and limitations. The primary strengths of this study are the large sample size and the direct assessment of protein-coding variant associations with COPD in the context of GWAS findings. We were not well powered to detect variants with an MAF < 0.01, so we are unable to assess the impact of very rare protein-coding variants on COPD risk. Larger studies are needed to replicate our findings and assess the impact of very rare variants. Exome array data provide sparse coverage of variants across the genome, making colocalization analyses between COPD exonic variant associations and eQTLs or pQTLs impossible. However, we attempted to address this limitation by identifying eQTLs and pQTLs in the highest LD with COPD exonic variants. Finally, we used a limited set of bioinformatic prediction tools to identify functional variants, but the accuracy of such tools for predicting biologically important changes in protein structure and function is not clear (77). Laboratory-based validation is critical to understanding the causal influence of the exome array variants reported here.
In conclusion, we performed the largest exome array meta-analysis of moderate-to-severe COPD to date. We were unable to identify any protein-altering coding variants at exome-wide significance in regions not previously identified by GWAS. However, at previously described GWAS loci, we report multiple coding variants associated with COPD, including four conditionally significant nonsynonymous variants, two stop variants, and a splice variant. These variants exist in genes important in cell-matrix interactions, cell proliferation, DNA demethylation, regulation of proteases, and regulation of cell death. We further identify the heterogeneity of effects of the SERPINA1 Z allele across cohorts. Future studies will be needed to replicate and validate these identified exonic variants, identify rarer variants, and further describe the role of coding variants in COPD pathogenesis.
GRANTS
This work was supported by Grant T32HL007427 (to M.M.); NIH Grants K08HL136928 and R01 HL089856 (to B.D.H.); NIH Grants R01HL137927 and R01HL135142 (to M.H.C.); NIH Grants R01 HL137927, R01 HL147148, R01 HL133135, and P01 HL114501 (to E.K.S.); Wellcome Trust Investigator Award WT202849/Z/16/Z (to M.D.T); and the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences Grant ZO1 ES43012 to (S.J.L.). The research was partially supported by the National Institute for Health Research (NIHR) Leicester Biomedical Research Centre. The views expressed are those of the authors and not necessarily those of the National Health Service (NHS), the NIHR, or the Department of Health. L.V.W. holds a GSK/British Lung Foundation Chair in Respiratory Research (C17-1). I.P.H. holds an NIHR Senior Investigator Award.
DISCLOSURES
E.K.S. received grant support from GlaxoSmithKline and Bayer. M.H.C. has received grant support from GlaxoSmithKline and Bayer, consulting fees from Genentech and AstraZeneca, and speaking fees from Illumina. M.D.T. receives grant support from GlaxoSmithKline and Orion. L.V.W. received grant support from GlaxoSmithKline and Orion. I.P.H. has received grant support from GlaxoSmithKline and Orion. None of the other authors has any conflicts of interest, financial or otherwise, to disclose.
AUTHOR CONTRIBUTIONS
M.M., A.M., V.E.J., L.V.W., E.K.S., M.H.C., B.D.H., S.J.L., S.A.G., and J.D. conceived and designed research; M.M., H.X., J.P., K.H.K., B.H., V.E.J., J.N.N., N.T., P.S., J.L., C.J.B., B.Y., M.H.C., B.D.H., M.L.G., and T.M.B. analyzed data; M.M., V.E.J., M.D.T., L.V.W., E.K.S., M.H.C., B.D.H., and J.D. interpreted results of experiments; M.M. prepared figures; M.M., M.H.C., and B.D.H. drafted manuscript; M.M., H.X., P.A.C., B.K.P., W.K., J.P., K.H.K., B.H., A.M., V.E.J., J.N.N., S.S.R., L.L., N.T., G.B., P.S., C.J.B., I.P.H., M.D.T., B.Y., L.V.W., E.K.S., M.H.C., B.D.H., M.L.G., S.J.L., S.A.G., T.M.B., C.M.S., and J.D. edited and revised manuscript; M.M., G.T.O., H.X., P.A.C., B.K.P., W.K., J.P., K.H.K., B.H., R.B., A.M., V.E.J., J.N.N., S.S.R., L.L., N.T., G.B., P.S., J.L., C.J.B., I.P.H., M.D.T., B.Y., L.V.W., E.K.S., M.H.C., B.D.H., M.L.G., S.J.L., S.A.G., T.M.B., C.M.S., and J.D. approved final version of manuscript.
REFERENCES
- 1.Vogelmeier CF, Criner GJ, Martinez FJ, Anzueto A, Barnes PJ, Bourbeau J, Celli BR, Chen R, Decramer M, Fabbri LM, Frith P, Halpin DM, López Varela MV, Nishimura M, Roche N, Rodriguez-Roisin R, Sin DD, Singh D, Stockley R, Vestbo J, Wedzicha JA, Agustí A. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017. Am J Respir Crit Care Med 195: 557–582, 2017. doi: 10.1164/rccm.201701-0218PP. [DOI] [PubMed] [Google Scholar]
- 2.Soriano JB, Lamprecht B, Ramírez AS, Martinez-Camblor P, Kaiser B, Alfageme I, Almagro P, Casanova C, Esteban C, Soler-Cataluna JJ, de-Torres JP, Miravitlles M, Celli BR, Marin JM, Puhan MA, Sobradillo P, Lange P, Stenberg AL, Garcia-Aymerich J, Turner AM, Han MK, Langhammer A, Leivseth L, Bakke P, Johannessen A, Roche N, Sin DD. Mortality prediction in chronic obstructive pulmonary disease comparing the GOLD 2007 and 2011 staging systems: a pooled analysis of individual patient data. Lancet Respir Med 3: 443–450, 2015. doi: 10.1016/S2213-2600(15)00157-5. [DOI] [PubMed] [Google Scholar]
- 3.Shrine N, Guyatt AL, Erzurumluoglu AM, Jackson VE, Hobbs BD, Melbourne CA; Understanding Society Scientific Group, et al. New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat Genet 51: 481–493, 2019. doi: 10.1038/s41588-018-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakornsakolpat P, Prokopenko D, Lamontagne M, Reeve NF, Guyatt AL, Jackson VE, SpiroMeta Consortium, et al. Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nat Genet 51: 494–505, 2019. doi: 10.1038/s41588-018-0342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wain LV, Shrine N, Miller S, Jackson VE, Ntalla I, Soler Artigas M, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med 3: 769–781, 2015. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wain LV, Shrine N, Artigas MS, Erzurumluoglu AM, Noyvert B, Bossini-Castillo L; Understanding Society Scientific Group, et al. Genome-wide association analyses for lung function and chronic obstructive pulmonary disease identify new loci and potential druggable targets. Nat Genet 49: 416–425, 2017. doi: 10.1038/ng.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hobbs BD, de Jong K, Lamontagne M, Bossé Y, Shrine N, Artigas MS; COPDGene Investigators, et al. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet 49: 426–432, 2017. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Monolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci SA 106: 9362–9367, 2009. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eriksson S. Studies in alpha 1-antitrypsin deficiency. Acta Med Scand Suppl 432: 1–85, 1965. [PubMed] [Google Scholar]
- 10.Hobbs BD, Silverman EC. Genetics and epidemiology. In: α1-Antitrypsin Deficiency (ERS Monograph) , edited by Strnad P, Brantly M BR.. Sheffield: European Respiratory Society, 2019. p. 27–38. [Google Scholar]
- 11.Bossé Y, Lamontagne M, Gaudreault N, Racine C, Levesque M-H, Smith BM, Auger D, Clemenceau A, Pare ME, Laviolette L, Tremblay V, Maranda B, Morissette MC, Maltais F. Early-onset emphysema in a large French-Canadian family: a genetic investigation. Lancet Respir Med 7: 427–436, 2019. doi: 10.1016/S2213-2600(19)30056-6. [DOI] [PubMed] [Google Scholar]
- 12.Stanley SE, Chen JJL, Podlevsky JD, Alder JK, Hansel NN, Mathias RA, Qi X, Rafaels NM, Wise RA, Silverman EK, Barnes KC, Armanios M. Telomerase mutations in smokers with severe emphysema. J Clin Invest 125: 563–570, 2015. doi: 10.1172/JCI78554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abecasis Lab at the University of Michigan Center for Statistical Genetics. Exome chip design [Internet]. [cited 2019. August 8]. Available from: [http://genome.sph.umich.edu/wiki/https://genome.sph.umich.edu/wiki/Exome_Chip_Design].
- 14.Hobbs BD, Parker MM, Chen H, Lao T, Hardin M, Qiao D, et al. Exome array analysis identifies a common Variant in IL27 associated with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 194: 48–57, 2016. doi: 10.1164/rccm.201510-2053OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson VE, Latourelle JC, Wain LV, Smith AV, Grove ML, Bartz TM; Understanding Society Scientific Group, et al. Meta-analysis of exome array data identifies six novel genetic loci for lung function. Wellcome Open Res 3: 4, 2018. doi: 10.12688/wellcomeopenres.12583.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson VE, Ntalla I, Sayers I, Morris R, Whincup P, Casas JP, et al. Exome-wide analysis of rare coding variation identifies novel associations with COPD and airflow limitation in MOCS3, IFIT3 and SERPINA12. Thorax 71: 501–509, 2016. doi: 10.1136/thoraxjnl-2015-207876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foreman MG, Wilson C, DeMeo DL, Hersh CP, Beaty TH, Cho MH, Ziniti J, Curran-Everett D, Criner G, Hokanson JE, Brantly M, Rouhani FN, Sandhaus RA, Crapo JD, Silverman EK;Genetic Epidemiology of COPD (COPDGene) Investigators. Alpha-1 antitrypsin PiMZ genotype is associated with chronic obstructive pulmonary disease in two racial groups. Ann Am Thorac Soc 14: 1280–1287, 2017. doi: 10.1513/AnnalsATS.201611-838OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castaldi PJ, DeMeo DL, Kent DM, Campbell EJ, Barker AF, Brantly ML, Eden E, McElvaney NG, Rennard SI, Stoller JM, Strange C, Turino G, Sandhaus RA, Griffith JL, Silverman EK. Development of predictive models for airflow obstruction in alpha-1-antitrypsin deficiency. Am J Epidemiol 170: 1005–1013, 2009. doi: 10.1093/aje/kwp216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The atherosclerosis risk in communities (ARIC) study: design and objectives. The ARIC investigators. Am J Epidemiol 129: 687–702, 1989. [2646917] [PubMed] [Google Scholar]
- 20.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol 1: 263–276, 1991. doi: 10.1016/1047-2797(91)90005-W. [DOI] [PubMed] [Google Scholar]
- 21.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic Epidemiology of COPD (COPDGene) Study Design. COPD 7: 32–43, 2011. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silverman EK, Weiss ST, Drazen JM, Chapman HA, Carey V, Campbell EJ, Denish P, silverman RA, Celedon JC, Reilly JJ, Ginns LC, Speizer FE. Gender-related differences in severe, early-onset chronic obstructive pulmonary disease. Am J Respir Crit Care Med 162: 2152–2158, 2000. doi: 10.1164/ajrccm.162.6.2003112. [DOI] [PubMed] [Google Scholar]
- 23.Patel BD, Coxson HO, Pillai SG, Agustí AGN, Calverley PMA, Donner CF, Make BJ, Müller NL, Rennard SI, Vestbo J, Wouters EFM, Hiorns MP, Nakano Y, Camp PG, Fauerbach PVN, Screaton NJ, Campbell EJ, Anderson WH, Paré PD, Levy RD, Lake SL, Silverman EK, Lomas DA; International COPD Genetics Network. Airway wall thickening and emphysema show independent familial aggregation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 178: 500–505, 2008. doi: 10.1164/rccm.200801-059OC. [DOI] [PubMed] [Google Scholar]
- 24.Zhu G, Warren L, Aponte J, Gulsvik A, Bakke P, Anderson WH, Lomas DA, Silverman EK, Pillai SG; The International COPD Genetics Network (ICGN) Investigators. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med 176: 167–173, 2007. July doi: 10.1164/rccm.200611-1723OC. [DOI] [PubMed] [Google Scholar]
- 25.Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ, Agostino RB, Fox CS, Larson MG, Murabito JM, O’Donnell CJ, Vasan RS, Wolf PA, Levy D. The Third Generation Cohort of the National Heart, Lung, and Blood Institute’s Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol 165: 1328–1335, 2007. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 26.Wilk JB, Chen T-H, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, Myers RH, Borecki IB, Silverman EK, Weiss ST, O’Cornnor GT. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet 5: e1000429, 2009. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol 110: 281–290, 1979. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 28.Simonsick EM, Newman AB, Nevitt MC, Kritchevsky SB, Ferrucci L, Guralnik JM, Haris T; Health ABC Study Group. Measuring higher level physical function in well-functioning older adults: expanding familiar approaches in the Health ABC study. J Gerontol A Biol Sci Med Sci 56: M644–9, 2001. doi: 10.1093/gerona/56.10.M644. [DOI] [PubMed] [Google Scholar]
- 29.Hong CB, Kim YJ, Moon S, Shin Y-A, Cho YS, Lee J-Y. KAREBrowser: SNP database of Korea Association REsource Project. BMB Rep 45: 47–50, 2012. doi: 10.5483/BMBRep.2012.45.1.47. [DOI] [PubMed] [Google Scholar]
- 30.Hankinson JL, Kawut SM, Shahar E, Smith LJ, Stukovsky KH, Barr RG. Performance of American Thoracic Society-recommended spirometry reference values in a multiethnic sample of adults: the multi-ethnic study of atherosclerosis (MESA) lung study. Chest 137: 138–145, 2010. doi: 10.1378/chest.09-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez RAV, Folsom AR, Greenland P, Jacob DR Jr, Kronmal R, Liu K, Nelson JC, O'Leary D, Saad MF, Shea S, Szklo M, Tracy RP. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol 156: 871–881, 2002. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 32.Hofman A, Darwish Murad S, van Duijn CM, Franco OH, Goedegebure A, Ikram MA, Klaver CC, Nijsten TE, Peeters RP, Sticker BH, Uitterlinden AG, Vernooij MW. The Rotterdam Study: 2014 objectives and design update. Eur J Epidemiol 28: 889–926, 2013. doi: 10.1007/s10654-013-9866-z. [DOI] [PubMed] [Google Scholar]
- 33.Ikram MA, van der Lugt A, Niessen WJ, Koudstaal PJ, Krestin GP, Hofman A, Boss D, Vernooij MW. The Rotterdam Scan Study: design update 2016 and main findings. Eur J Epidemiol 30: 1299–1315, 2015. doi: 10.1007/s10654-015-0105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X, Baron RM, Hardin M, Cho MH, Zielinski J, Hawrylkiewicz I, Sliwinski P, Hersh Lugt CP, Mancini JD, Lu KE, Thibault D, Donahue AL, Klanderman BJ, Rosner B, Raby BA, Lu Q, Geldart AM, Layne MD, Perrella MA, Weiss ST, Choi AMK, Silverman EK. Identification of a chronic obstructive pulmonary disease genetic determinant that regulates HHIP. Hum Mol Genet [Internet] 21: 1325–1335, 2012. doi: 10.1093/hmg/ddr569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12: e1001779, 2015. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winkler TW, Day FR, Croteau-Chonka DC, Wood AR, Locke AE, Mägi R, Ferreira T, Fall T, Graff M, Justice AE, Luan J, Gustafsson S, Randall JC, Vedantam S, Workalemahu T, Kilpelainen TO, Scherag A, Esko T, Kutalik Z, Heid IM, Loos RJF;The Genetic Investigation of Anthropometric Traits (GIANT) Consortium. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc 9: 1192–1212, 2014. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 38: 209–213, 2006. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 38.Buist AS, McBurnie MA, Vollmer WM, Gillespie S, Burney P, Mannino DM, Menezes AMB, Sullivan SD, Lee TA, Weiss KB, Jensen RL, Marks GB, Gulsvik A, Nizankowska-Mogilnicka E; BOLD Collaborative Research Group. International variation in the prevalence of COPD (the BOLD Study): a population-based prevalence study. Lancet 370: 741–750, 2007. doi: 10.1016/s0140-6736(07)61377-4. [DOI] [PubMed] [Google Scholar]
- 39.Pauwels RA, Rabe KF. Burden and clinical features of chronic obstructive pulmonary disease (COPD. ) Lancet 364: 613–620, 2004. doi: 10.1016/s0140-6736(04)16855-4. [DOI] [PubMed] [Google Scholar]
- 40.van Belle G. Statistical Rules of Thumb (2nd Ed.). New York: John Wiley and Sons, 2008, p. 45–46. [Google Scholar]
- 41.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191, 2010. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet 49: 433–436, 2012. doi: 10.1136/jmedgenet-2012-100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaSci 4: 7, 2015. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, Ferreira T, Morris AP, Medland SE, Madden PAF, Heath AC, Martin NG, Montgomery GW, Weedon MN, Loos RJ, Frayling TM, McCarthy MI, Hirschhorn JN, Goddard ME, Visscher PM; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 44: 369–375, 2012. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wakefield J. Bayes factors for genome-wide association studies: comparison with P-values. Genet Epidemiol 33: 79–86, 2009. doi: 10.1002/gepi.20359. [DOI] [PubMed] [Google Scholar]
- 46.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46: 310–315, 2014. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 47: D886–D894, 2019. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu G, Haw R. Functional interaction network construction and analysis for disease discovery. Methods Mol Biol 1558: 235–253, 2017. doi: 10.1007/978-1-4939-6783-4_11. [DOI] [PubMed] [Google Scholar]
- 49.Fabregat A, Sidiropoulos K, Viteri G, Forner O, Marin-Garcia P, Arnau V, D’Eustachio P, Stein L, Hermjakob H. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics 18: 142, 2017. doi: 10.1186/s12859-017-1559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–585, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660, 2015. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westra H-J, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45: 1238–1243, 2013. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hao K, Bossé Y, Nickle DC, Paré PD, Postma DS, Laviolette M, Sandford A, Hackett TL, Daley D, Hogg JC, Elliott WM, Couture C, Lamontagne M, Brandsma C-A, van den Berge M, Koppelman G, Reicin AS, Nicholson DW, Malkov V, Derry JM, Suver C, Tsou JA, Kulkarni A, Zhang C, Vessey R, Opiteck GJ, Curtis SP, Timens W, Sin DD. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet 8: e1003029, 2012. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Võsa U, Claringbould A, Westra H-J, Bonder MJ, Deelen P, Zeng B, Kirsten H, Saha A, Kreuzhuber R, Kasela S, Pervjakova N, Alvaes I, Fave M-J, Agbessi M, Christiansen M, Jansen R, Seppälä I, Tong L, Teumer A, Schramm K, Hemani G, Verlouw J, Yaghootkar H, Sönmez R, Andrew AA, Kukushkina V, Kalnapenkis A, Rüeger S, Porcu E, Kronberg-Guzman J, Kettunen J, Powell J, Lee B, Zhang F, Arindrarto W, Beutner F, Brugge H, Dmitrieva J, Elansary M, Fairfax BP, Georges M, Heijmans BT, Kähönen M, Kim Y, Knight JC, Kovacs P, Krohn K, Li S, Loeffler M, Marigorta UM, Mei H, Momozawa Y, Müller-Nurasyid M, Nauck M, Nivard M, Penninx B, Pritchard J, Raitakari O, Rotzschke O, Slagboom EP, Stehouwer CDA, Stumvoll M, Sullivan P, t Hoen PAC, Thiery J, Tönjes A, van Dongen J, van Iterson M, Veldink J, Völker U, Wijmenga C, Swertz M, Andiappan A, Montgomery GW, Ripatti S, Perola M, Kutalik Z, Dermitzakis E, Bergmann S, Frayling T, van Meurs J, Prokisch H, Ahsan H, Pierce B, Lehtimäki T, Boomsma D, Psaty BM, Gharib SA, Awadalla P, Milani L, Ouwehand WH, Downes K, Stegle O, Battle A, Yang J, Visscher PM, Scholz M, Gibson G, Esko T, Franke L. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis (Preprint). bioRxiv 2018. doi: 10.1101/447367. [DOI]
- 56.Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran P, Oliver-Williams C, Kamat MA, Prins BP, Wilcox SK, Zimmerman ES, Chi A, Bansal N, Spain SL, Wood AM, Morrell NW, Bradley JR, Janjic N, Roberts DJ, Ouwehand WH, Todd JA, Soranzo N, Suhre K, Paul DS, Fox CS, Plenge RM, Danesh J, Runz H, Butterworth AS. Genomic atlas of the human plasma proteome. Nature 558: 73–79, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwarzer G. meta: An R package for meta-analysis. R News 7: 40–45, 2007. [Google Scholar]
- 58.Busch R, Hobbs BD, Zhou J, Castaldi PJ, McGeachie MJ, Hardin ME, et al. Genetic association and risk scores in a chronic obstructive pulmonary disease meta-analysis of 16,707 subjects. Am J Respir Cell Mol Biol 57: 35–46, 2017. doi: 10.1165/rcmb.2016-0331oc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang L, Zhao J, Edenberg HJ. A human Raf-responsive zinc-finger protein that binds to divergent sequences. Nucleic Acids Res 27: 2947–2956, 1999. doi: 10.1093/nar/27.14.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mukhopadhyay NK, Cinar B, Mukhopadhyay L, Lutchman M, Ferdinand AS, Kim J, Chung LWK, Adam RM, Ray SK, Leiter AB, Richie JP, Liu Brain C-S, Freeman MR. The zinc finger protein ras-responsive element binding protein-1 is a coregulator of the androgen receptor: implications for the role of the Ras pathway in enhancing androgenic signaling in prostate cancer. Mol Endocrinol 21: 2056–2070, 2007. doi: 10.1210/me.2006-0503. [DOI] [PubMed] [Google Scholar]
- 61.Thiagalingam A, De Bustros A, Borges M, Jasti R, Compton D, Diamond L, Mabry M, Ball DW, Baylin SB, Nelkin BD. RREB-1, a novel zinc finger protein, is involved in the differentiation response to Ras in human medullary thyroid carcinomas. Mol Cell Biol 16: 5335–5345, 1996. doi: 10.1128/MCB.16.10.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hartz SM, Short SE, Saccone NL, Culverhouse R, Chen L, Schwantes-An T-H, et al. Increased genetic vulnerability to smoking at CHRNA5 in early-onset smokers. Arch Gen Psychiatry 69: 854–860, 2012. doi: 10.1001/archgenpsychiatry.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Glubb DM, Maranian MJ, Michailidou K, Pooley KA, Meyer KB, Kar S, et al. Fine-scale mapping of the 5q11.2 breast cancer locus reveals at least three independent risk variants regulating MAP3K1. Am J Hum Genet 96: 5–20, 2015. doi: 10.1016/j.ajhg.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gallagher MD, Chen-Plotkin AS. The Post-GWAS Era: from association to function. Am J Hum Genet 102: 717–730, 2018. doi: 10.1016/j.ajhg.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei P, Zhao YG, Zhuang L, Ruben S, Sang QX. Expression and enzymatic activity of human disintegrin and metalloproteinase ADAM19/meltrin beta. Biochem Biophys Res Commun 280: 744–755, 2001. doi: 10.1006/bbrc.2000.4200. [DOI] [PubMed] [Google Scholar]
- 66.Pérez-Rubio G, Silva-Zolezzi I, Fernández-López JC, Camarena Á, Velázquez-Uncal M, Morales-Mandujano F, Hernandez-Zenteno RDJ, Flores-Trujillo F, Sanchez-Romero C, Velazque-Montero A, de Los Monteros CE, Sansores RH, Ramirez-Venegas A, Falfan-Valencia R. Genetic variants in IL6R and ADAM19 are associated with COPD severity in a Mexican Mestizo population. COPD 13: 610–615, 2016. doi: 10.3109/15412555.2016.1161017. [DOI] [PubMed] [Google Scholar]
- 67.Buscarlet M, Provost S, Zada YF, Barhdadi A, Bourgoin V, Lépine G, Mollica L, Szuber N, Dube MP, Busque L. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood 130: 753–762, 2017. doi: 10.1182/blood-2017-04-777029. [DOI] [PubMed] [Google Scholar]
- 68.Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, Dixit VM. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526: 666–671, 2015. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 69.Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang DC, Shao F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535: 111–116, 2016. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- 70.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F . Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526: 660–665, 2015. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 71.Panganiban RA, Sun M, Dahlin A, Park H-R, Kan M, Himes BE, Mitchel JA, Iribarren C, Jorgenson E, Randell SH, Israel E, Tantisira E, Shore S, Park JA, Weiss ST, Wu AC, Lu Q. A functional splice variant associated with decreased asthma risk abolishes the ability of gasdermin B to induce epithelial cell pyroptosis. J Allergy Clin Immunol 142: 1469–1478.e2, 2018. doi: 10.1016/j.jaci.2017.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hayden LP, Cho MH, Raby BA, Beaty TH, Silverman EK, Hersh CP; COPDGene Investigators. Childhood asthma is associated with COPD and known asthma variants in COPDGene: a genome-wide association study. Respir Res 19: 209, 2018. doi: 10.1186/s12931-018-0890-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McGeachie MJ, Yates KP, Zhou X, Guo F, Sternberg AL, Van Natta ML, et al. Patterns of Growth and decline in lung function in persistent childhood asthma. N Engl J Med 374: 1842–1852, 2016. doi: 10.1056/NEJMoa1513737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Busch R, Hobbs BD, Zhou J, Castaldi PJ, McGeachie MJ, Hardin ME, Hawrylkiewicz I, Sliwinski P, Yim JJ, Kim WJ, Kim DK, Agusti A, Make BJ, Crapo D, Calverley PM, Donner CF, Lomas DA, Wouters EF, Vestbo J, Tal-Singer R, Bakke P, Gulsvik A, Litonjua AA, Sparrow D, Paré PD, Levy RD, Rennard SI, Beaty TH, Hokanson J, Silverman EK, Cho MH; National Emphysema Treatment Trial Genetics; Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-Points; International COPD Genetics Network; COPDGene Investigators. Genetic association and risk scores in a chronic obstructive pulmonary disease meta-analysis of 16,707 subjects. Am J Respir Cell Mol Biol 57: 35–46, 2017. doi: 10.1165/rcmb.2016-0331OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li X, Ortega VE, Ampleford EJ, Graham Barr R, Christenson SA, Cooper CB, Dransfield MT, Han MLK, Hanse NN, Hoffman EA, Kanner RE, Kleerup EC, Martinez FJ, Paine R, Woodruff PG, Hawkins GA, Bleecker ER, Meyers DA; for the SPIROMICS Research Group. Genome-wide association study of lung function and clinical implication in heavy smokers. BMC Med Genet 19: 134, 2018. doi: 10.1186/s12881-018-0656-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Silverman EK, Province MA, Campbell EJ, Pierce JA, Rao DC, Boerwinkle E. Family study of alpha 1-antitrypsin deficiency: effects of cigarette smoking, measured genotype, and their interaction on pulmonary function and biochemical traits. Genet Epidemiol 9: 317–331, 1992. doi: 10.1002/gepi.1370090504. [DOI] [PubMed] [Google Scholar]
- 77.Miosge LA, Field MA, Sontani Y, Cho V, Johnson S, Palkova A, Balakishnan B, Liang R, Zhang Y, Lyon S, Beutler B, Whittle B, Bertram EM, Enders A, Goodnow CC, Andrews TD. Comparison of predicted and actual consequences of missense mutations. Proc Natl Acad Sci USA 112: E5189–E5198, 2015. doi: 10.1073/pnas.1511585112. [DOI] [PMC free article] [PubMed] [Google Scholar]