Abstract
Background and aim
Gastric Cancer (GC) is a leading cause of morbidity and mortality worldwide, particularly in developing nations, only a few suitable gastric cancer serum biomarkers with acceptable sensitivity and specificity exist. This work aims to highlight and uncover miR-30a-5p and miR-182–5p′s diagnostic roles regarding gastric cancer and their roles in predicting prognosis.
Methods
148 patients participated in this study. Groups I, II, and III had 47 patients with GC, 54 patients with benign gastric lesions, and 47 apparently healthy subjects of coincided age and gender as controls, respectively. All participants were clinically evaluated and subjected to CBC, serum CEA, and CA19-9 by ELISA, and real-time PCR tests of miR-30a-5p and miR-182–5p.
Results
MiR30a-5p and miR-182–5p were down regulated in gastric cancer patients in Group I more than Groups II and III (P < 0.001). ROC curve analysis revealed that miR30a-5p had better AUC, sensitivity, and specificity (0.961%, 93.62%, and 90.74%respectively). When miR-182–5p was gathered with CEA and CA19-9, specificity raised to 98.15% and PPV to 97.6%. Lower miR-30a-5p levels are linked with the presence of distant metastases, advanced TNM stage, and degree of pathological differentiation of tumors in GC patients (p = 0.034, 0.019, 0.049) respectively. According to the multivariate analysis, miR30a-5p expression level could be an independent predictor of GC.
Conclusion
Our results exhibited that miRNAs, miR-30a-5p and miR182–5p, gene expression have a diagnostic power and can identify patients with GC. MiR-30a-5p displayed the highest diagnostic specificity and sensitivity. Besides other known tumor markers, they could offer simple noninvasive biomarkers that predict gastric cancer.
Keywords: Gastric cancer, miR-30a-5p, miR-182–5p, RT-PCR
Highlights
-
•
Gastric Cancer (GC) proceeds to be a leading cause of morbidity and mortality worldwide, espcially in developing nations.
-
•
We observed that miR-30a-5P, miR-182–5 P showed considerably lower values in gastric cancer patients.
-
•
Lower miR-30a-5P level was significantly linked with distant metastases, advanced TNM stage and pathological differentiation of tumor.
-
•
Lower miR-30a-5P, and miR182–5 P gene expressions in gastric cancer patients were significantly associated with decreased PFS.
-
•
Lower miR-30a-5P expression level could be independent predictor for Gastric Cancer.
1. Introduction
Gastric cancer (GC) is in the six place among the malignancies worldwide, and it is the third leading cause of cancer-related death [1]. In 2018, The World Health Organization estimates that gastric cancer accounted for 783,000 deaths worldwide [2].
The causes of gastric cancer are complex and multivariate, including dietary imbalance, alcohol consumption, smoking, and helicobacter pylori infection. Gastric cancer pathogenesis is linked with genetic variables such as methylation of DNA, gene amplifications and deletions, epigenetic inactivation of many genes, and erratic somatic mutations [3].
The lack of vivid clinical symptoms inhibits early gastric cancer diagnosis [4]. That is the reason why patients with gastric cancer are almost often diagnosed at late stages resulting in critical metastasis and a bad prognosis. As a result, the five-year survival rate is lower than thirty percent [5].
Despite the high diagnostic precision of endoscopy, it is inconvenient and could lead to more complications. Hence, extra markers to recognize early gastric cancer are vital [6].
MicroRNAs are about 22 nucleotides in length that bind to complementary sequences of mRNA at the 3′-untranslated portion (3′UTR), resulting in the down regulation of protein-coding target genes within the nucleus and the cytoplasm [7]. However, interaction of miRNAs with other regions, such as 5′UTR, has also been reported, and can enhance expression of target gene [8].
MicroRNAs were found to contribute to diverse biological procedures, such as cell growth, development, and differentiation, additionally; they play substantial roles in cell proliferation, angiogenesis, apoptosis, and metastasis [7]. It has been revealed that miRNAs can alter gastric cancer metastasis and growth by targeting the STAT3 signaling pathway [9].
The miR-30 family is a complex family with crucial roles in miRNAs functions in mammalian and human beings. It consists of five members [10]. Of which, MiR-30a as a tumor suppressor could organize cellular proliferation, apoptosis, migration, and invasion of diverse tumor cells [11].
MiR-182, a member of the miR-183 family, is located at 7q31-34 [12]. Multiple researchers illustrated that miR-182 is abnormally expressed in numerous cancer types [13,14].
Attempts to overcome diagnostic issues and restrictions should be the primary objective of modern medicine. In other cancers, a simple noninvasive blood-based test would be perfect, enabling gastric cancer to be detected at a time where curative action is still feasible. So, in this study, we aimed to highlight the diagnostic roles of miR-30a-5p and miR-182–5p in gastric cancer and uncover their roles in predicting prognosis in gastric patients.
2. Subjects and methods
SUBJECTS: 148 participants were included in this case–control study. It was carried out in the Medical Biochemistry and Molecular Biology department in partnership with the Department of Tropical Medicine, Faculty of Medicine. The participants were chosen from the Department of Tropical Medicine between September 2017 and December 2018 and categorized into three groups: Group I composed of 47 patients with gastric cancer. Group II included 54 patients with benign lesions in the stomach. Group III included 47 sound subjects of coincided age and gender as controls. The sample size was determined utilizing Epi Info (2000) program according to a previous study [15], where 36 cases were studied per group and the sample size has been calculated within a power of 95% and alpha error 0.05.
For each participant, history and clinical evaluation, laboratory investigations (including CBC, liver function tests, ESR, CEA, and CA19-9, miR-30a-5p and miR-182–5p, and H. pylori antigen in stool), and abdominal pelvic ultrasonography were done. Patients with malignancies in other places of their bodies were excluded. Control participants were healthy subjects with no history of weight loss, smoking, or NSAID consumption and had no GIT symptoms as hematemesis or melena, pain related to meal, nausea, vomiting, or postprandial fullness and negative H. pylori stool antigen test. Diagnosis of benign gastric lesions and malignancy was conducted by clinical assessment, and endoscopic evaluation with tissue biopsies was taken for histopathological examination. Confirmed gastric cancer patients underwent radiological assessment in the form of baseline-computed tomography of chest, abdomen, and pelvis besides a bone scan to detect distant metastases. TNM staging was done for all patients with gastric cancer [16]. Gastric cancer patients were followed up upon in the clinical oncology department for 24 months (until the end of November 2020). The Progression-Free Survival (PFS) rate has been realized as the period besides the date of disease diagnosis and the progression date in the form of local recurrence recently developed metastases for patients with localized cancer or excess in number and/or size of metastases in stage IV disease patients or final visit. The PFS rate was evaluated among patients according to various prognostic factors.
Ethical Approval: This study was performed per the Declaration of Helsinki. All participants gave informed consents assured by the ethical committee, Faculty of Medicine, Menoufia University.
Methods: All participants were subjected to history taking, clinical evaluation, and laboratory investigations, involving a complete blood count, liver and kidney function tests, H. pylori stool antigen, serum CEA and CA19-9 b y ELISA, and real-time PCR of miR-30a-5p and miR-182–5p.
2.1. Sampling and assay method
Through venipuncture, 6 ml of venous blood were withdrawn, of which 3 ml were left to clot into a plain tube and centrifuged at 4000 rpm for around 10 min. At −80 °C, the serum obtained was stored for subsequent use. The serum to estimate its carbohydrate antigen 19–9 (CA 19–9) and CEA by Enzyme-Linked Immunosorbent Assay kit (ELISA) provided by Chemux Bioscience, Inc. (USA). Utilizing the DIAMOND diagnostic kit (Germany), the liver functions as Alanine Aminotransferase (ALT) and Aspartate Aminotransferase (AST) (by LTEC kit, England) and serum albumin were estimated by enhanced specificity of bromocresol green colorimetric assay. Serum bilirubin (total and direct) was assessed by the quantitative assurance of bilirubin IVD utilizing the DIAMOND diagnostics kit. 2 ml of blood were put in an EDTA tube for Complete Blood Count (CBC) was obtained by KX21 N Hematology analyzer (Sysmex, Kobe, Japan). An automated hematology cell counter, besides the remaining blood, was collected in an EDTA-coated tube and then centrifuged for about 5 min at 4000 rpm. The separated plasma was kept frozen at −20 °C for microRNA extraction and subsequent quantitation of miR-30a-5P and 182–5 P expression by RT-PCR.
Helicobacter pylori Ag in stool was done by an ELISA Assay kit (Eagle Biosciences, Amherst, New Hampshire, USA).
2.2. Detection and quantification of MiR-30a-5p and MiR-182–5p gene expression
MicroRNA was extracted from the frozen plasma samples by utilizing a MiRNeasy extraction kit (QIAGEN, USA) as directed by the manufacturer's instructions. The isolated RNA was stored at −80 °C. Complementary DNA (cDNA) was synthesized by utilizing a reverse transcriptase kit (MiScript II RT kit, QIAGEN, USA). An Applied Biosystems 2720 thermal cycler (Singapore) was used to process the reaction mixture of 20 μl that included the following: 4 μl of miScript HI Spec RT buffer, 2 μl of miScript Nucleic Mix, 2 μl of miScript™ reverse transcriptase, and 2 μl of nuclease-free water that were pipetted into each well. Then, 10 μl of extracted microRNA was added. The reaction was at a temperature of 37 °C for 60 min and 95 °C for 5 min to inactivate the reverse transcriptase. The resulting cDNA was stored at −20 °C until the next amplification step. Then, the next step of conducting a real-time PCR was done via a miScript SYBR Green PCR kit (QIAGEN, USA). cDNA samples were diluted with nuclease-free water at a ratio of 1:5 before amplification, and a reaction volume of 25 μl was created (12.5 μl of SYBR Green Master Mix, 3.5 μl of nuclease-free water, 4 μl of diluted cDNA, 2.5 μl of miScript primer assay, and 2.5 μl of miScript universal primer) microRNA SNORD 68 was utilized for normalization. The following primers were used: mature miR-30a-5p (cat.no.MS00007350) UGUAAACAUCCUCGACUGGAAG, mature miR-182–5p (cat.no.MS00008855) UUUGGCAAUGGUAG AACUCACACU, and mature SNORD68 as a reference gene (cat.no.MS00033712) through miScript primer assay kit, QIAGEN, USA. The thermal cycler, Applied Biosystems® 7500 with the software version 2.0.1(Foster City, CA, USA), was used for the real-time PCR as shown in Fig. 1 (1 A, 1 B, 1C, and 1D), where a program of 95 °C for 15 min was used. Then, it was followed by three steps of 40 cycles (15 s at 94 °C, 30 s at 55 °C, and 30 s at 70 °C). The results were interpreted using the comparative Ct method (2−ΔΔCt), and the relative quantification (RQ) of miR-30–5p and miR-182–5p were normalized to those of SNORD68 and that of a control.
Fig. 1.
A: Amplification plot of miR-30a-5p expression (normalized fluorescence signal (ΔRn) plotted versus cycle number) B: Amplification plot of miR-182–5p expression (normalized fluorescence signal (ΔRn) plotted versus cycle number) C: Melting curve of miR-30a-5p expression D: Melting curve of miR-182–5p expression.
2.3. Statistical analysis of the data
Data were analyzed utilizing the IBM SPSS software package version 20.0. (Armonk, NY: IBM Corp). The Chi-square test (Monte Carlo) was utilized for the comparison between groups for categorical variables. The three studied groups were assessed and compared via an ANOVA test followed by a post hoc test (Tukey) for pairwise comparison. To compare different groups of abnormally distributed quantitative variables, a Kruskal Wallis test was used, in the meantime comparing between two groups for not normally distributed quantitative variables, a Mann-Whitney test was used. Spearman coefficient test was utilized for correlating between quantitative data. Receiver Operating Characteristic Curve (ROC) was used. Kaplan-Meier survival curve and Cox regression were illustrated for the significant relationship with progression-free survival. The significance of the obtained results was judged at a 5% level.
3. Results
This case–control study was conducted on 148 participants grouped into three groups: Group I included 47 gastric cancer patients of which 31 (66%) were males and 16 (34%) were females with a mean age of 60.9 ± 7.9. Group II included 54 patients (32 males and 22 females with a mean age of 60.5 ± 6.3) with benign lesions in the stomach (benign peptic ulcer, benign gastric polyps, and atrophic gastritis). And Group III included 47 (32 males and 15 females with a mean age of 57.6 ± 8.3) healthy individuals. No difference was observed between the three studied groups regarding age, sex, Total Leukocyte Count (TLC), liver function tests (ALT, AST, albumin, and total bilirubin). Hemoglobin Concentration (Hb) or Platelet Count (PLT), H. pylori stool antigen, and ESR were significantly different amongst the three examined groups (p < 0.001). Regarding tumor markers (CEA and CA19-9) statistically significant fluctuations (P < 0.001) amongst the three studied groups were detected, with considerably higher levels in gastric cancer patients compared with those in benign gastric lesions and controls. MiR-30-a-5p and miR-182–5p showed considerably lower values in Group I compared with those detected in Groups II and III (P < 0.001). Moreover, a non-significant difference was found between patients with benign gastric lesions and controls (p = 0.157 and 0.368), as shown in Table 1, Fig. 2A and B. Clinically, we noticed that the present symptoms did not differ between the two patient groups. Moreover, meals-related epigastric pain was the most frequent presenting symptom in Groups I and II, representing 36.2% and 53.7%. Manifestations of anemia represented 25.5% in the gastric cancer group. History of smoking and weight loss was significantly more prevalent among gastric cancer patients (p = 0.029 and < 0.001), while the history of NSAIDs consumption was not different (p = 0.215). For examination, pallor was more widespread in group I (p = 0.002) and Virchow lymph nodes were detected in five cases of gastric cancer group as displayed in Table 2.
Table 1.
Comparison between the three studied groups according to demographic and laboratory data.
| Group I (n = 47) | Group II (n = 54) | Group III (n = 47) | Test of Sig. | p | Sig. Bet. Grps. I vs II I vs III II vs III |
|||
|---|---|---|---|---|---|---|---|---|
| Sex | ||||||||
| Male | 31 (66%) | 32 (59.3%) | 32 (68.1%) | χ2= 0.945 |
0.623 | – | – | – |
| Female | 16 (34%) | 22 (40.7%) | 15 (31.9%) | |||||
| Age (years) | ||||||||
| Mean ± SD. | 60.9 ± 7.9 | 60.5 ± 6.3 | 57.6 ± 8.3 | F= 2.822 |
0.063 | – | – | – |
| Median (Min. – Max.) | 60 (44–79) | 60.5 (49–73) | 58 (36–75) | |||||
| BMI (kg/m2) | ||||||||
| Mean ± SD. | 23.9 ± 4.9 | 26.5 ± 5.1 | 21.5 ± 1.9 | F= 17.550* |
<0.001* | 0.007* | 0.019* | <0.001* |
| Median (Min. – Max) | 23.2 (17.7–31) | 27.23 (18.6–38) | 21.6 (18.6–24.6) | |||||
| TLC (103/mm3) | ||||||||
| Mean ± SD | 7.9 ± 1.3 | 7.7 ± 1.2 | 7.3 ± 1.3 | F = 2.963 | 0.055 | – | – | – |
| Median (Min. – Max.) | 8 (4.9–10.1) | 8.1 (4.9–9.7) | 7.6 (4.9–9.8) | |||||
| Hb(gm/dl) | ||||||||
| Mean ± SD | 10.3 ± 2.1 | 10 ± 1.7 | 13.9 ± 1.2 | F= 77.734* |
<0.001* | 0.478 | <0.001* | <0.001* |
| Median (Min. – Max) | 10.4 (5.5–15) | 9.7 (4.5–14.2) | 13.8 (11.8–16.1) | |||||
| Platelets (103/mm3) | ||||||||
| Mean ± SD | 294.2 ± 75.1 | 258.4 ± 79.1 | 255.5 ± 71.2 | F= 3.924* |
0.022* | 0.048* | 0.036* | 0.979 |
| Median (Min. – Max.) | 289 (165–420) | 276 (155–421) | 258 (164–421) | |||||
| Albumin (g/dl) | ||||||||
| Mean ± SD. | 3.9 ± 0.3 | 4 ± 0.3 | 4 ± 0.5 | F = 1.841 | 0.162 | – | – | – |
| Median (Min. – Max). | 3.8 (3.5–4.3) | 3.9 (3.7–4.9) | 3.9 (3.5–5.7) | |||||
| Total bilirubin (mg/dl) | ||||||||
| Mean ± SD | 1 ± 0.2 | 1 ± 0.2 | 1 ± 0.2 | F = 0.107 | 0.898 | – | – | – |
| Median (Min. – Max.) | 0.9 (0.6–1.4) | 1 (0.6–1.3) | 1 (0.6–1.3) | |||||
| ALT (U/L) | ||||||||
| Mean ± SD | 29.2 ± 9 | 28.4 ± 6.9 | 25.8 ± 6.1 | F = 2.804 | 0.064 | – | – | – |
| Median (Min. – Max) | 31 (17–56) | 29 (18–50) | 27 (17–38) | |||||
| AST (U/L) | ||||||||
| Mean ± SD. | 27.6 ± 5.5 | 26.9 ± 5.8 | 256 ± 5.9 | F = 2.494 | 0.086 | – | – | – |
| Median (Min. – Max.) | 28 (18–37) | 28 (18–35) | 27 (14–35) | |||||
| H. pylori | ||||||||
| No | 37 (78.7%) | 20 (37%) | 47 (100%) | χ2= 50.040* |
<0.001* | <0.001* | 0.001* | <0.001* |
| Yes | 10 (21.3%) | 34 (63%) | 0 (0%) | |||||
| CEA (U/ml) | ||||||||
| Mean ± SD | 56.91 ± 97.8 | 9.34 ± 7.40 | 6.38 ± 5.4 | H= 30.439* |
<0.001* | <0.001* | <0.001* | 0.055 |
| Median (Min. – Max) | 14.90 (3–575) | 5.10 (1.10–22) | 3.40 (0.80–13) | |||||
| CA19–9(U/ml) | ||||||||
| Mean ± SD | 54.91 ± 31.9 | 15.43 ± 8.67 | 13.36 ± 7.4 | H= 48.108* |
<0.001* | <0.001* | <0.001* | 0.353 |
| Median (Min. – Max.) | 54 (7.5–112) | 16 (1–37) | 12 (2–29) | |||||
| ESR | ||||||||
| Mean ± SD | 709 ± 32.17 | 10.81 ± 2.67 | 10.30 ± 2.96 | H= 87.755* |
<0.001* | <0.001* | <0.001* | 0.452 |
| Median (Min. – Max.) | 78 (10–110) | 11 (6–15) | 10 (5–17) | |||||
| miR-30a-5p | ||||||||
| Mean ± SD | 2.21 ± 0.83 | 7.96 ± 4.91 | 9.50 ± 4.45 | H= 84.210* |
<0.001* | <0.001* | <0.001* | 0.157 |
| Median (Min. – Max.) | 2.37 (0.05–3.50) | 6.17 (2.5–20) | 9.50 (2.7–25.7) | |||||
| miR-182-5p | ||||||||
| Mean ± SD. | 2.67 ± 1.26 | 7.98 ± 3.51 | 9.36 ± 4.90 | H= 65.801* |
<0.001* | <0.001* | <0.001* | 0.368 |
| Median (Min. – Max.) | 2.91 (0.1–4) | 8.13 (2–17) | 8.10 (2.4–20.9) | |||||
BMI: Body Mass Index, Hb: Hemoglobin concentration, TLC; Total Leukocyte Count, ALT: Alanine Aminotransferase, AST: Aspartate Aminotransferase, CEA; Carcinoembryonic Antigen, CA19-9; Carbohydrate Antigen 19–9, ESR: Erythrocyte Sedimentation Rate, χ2: Chi-square test, F: F for ANOVA test, Pairwise comparison bet. The two groups were done using a post hoc test (Tukey), H: H for the Kruskal Wallis test, pairwise comparison bet. The two groups were done using a post hoc test (Dunn's for multiple comparisons test), P: P-value for comparing between the studied groups.
*: Statistically significant at p < 0.05, IQR: Inter Quartile Range.
Group I: Gastric cancer Group II: Benign gastric lesion Group III: Control.
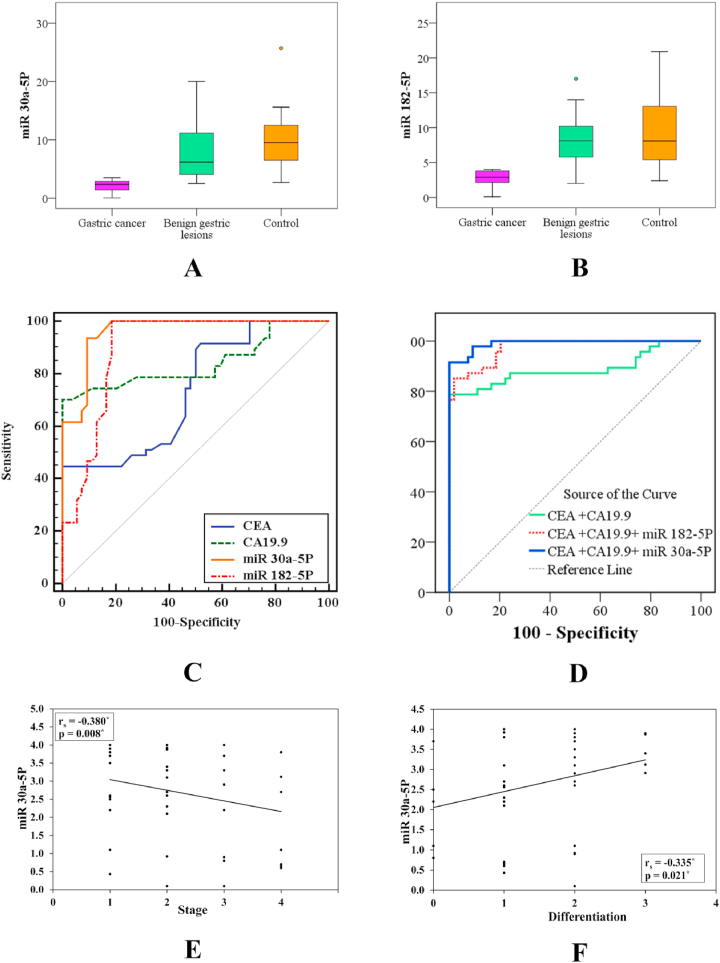
Fig. 2.
A: Comparison between the three studied groups according to miR-30a-5p B: Comparison between the three studied groups according to miR-182–5p C: ROC curve for miR-30a-5p and miR-182–5p to predict gastric cancer patients (n = 47) from benign gastric lesions (n = 54) D: ROC curve for a combination of different markers to predict gastric cancer patients (n = 47) from benign gastric lesions (n = 54) E: Correlation between miR-30a-5p and TNM stage of gastric cancer patients F: Correlation between MiR-30a-5p and degree of pathological differentiation in gastric cancer patients.
Table 2.
Comparison between the two studied groups according to clinical data (history and examination).
| Group I (n = 47) | Group II (n = 54) | χ2 | P | |
| Presenting symptom | ||||
| Pain | 17 (36.2%) | 29 (53.7%) | 9.704 |
MCp= 0.075 |
| Dyspepsia | 4 (8.5%) | 4 (7.4%) | ||
| Hematemesis & melena | 9 (19.1%) | 6 (11.1%) | ||
| Anemia | 12 (25.5%) | 4 (7.4%) | ||
| Nausea & vomiting | 2 (4.3%) | 3 (5.6%) | ||
| Post-prandial fullness | 3 (6.4%) | 8 (14.8%) | ||
| History and General Examination | ||||
| History of smoking | 22 (46.8%) | 14 (25.9%) | 4.777* | 0.029* |
| History of weight loss | 23 (48.9%) | 0 (0%) | 34.218* | <0.001* |
| History of NSAIDs | 12 (25.5%) | 20 (37%) | 1.537 | 0.215 |
| Pallor | 20 (42.6%) | 8 (14.8%) | 9.649* | 0.002* |
NSAIDs: Nonsteroidal Anti-Inflammatory Drugs χ2: Chi-square test, MC: Monte Carlo.
P: p-value for comparing between the studied groups.
*: Statistically significant at p < 0.05.
Group I: Gastric cancer.
Group II: Benign gastric lesions.
ROC curve analysis was used to demonstrate the diagnostic value of the investigated biomarkers (CEA, CA19-9, miR-30a-5p, and miR-182–5p) to predict gastric cancer revealed that miR30a-5p had better AUC, sensitivity, and specificity (0.961%, 93.62%, and 90.74% respectively), and P < 0.001, at a cutoff ≤ 3.27. Combined diagnostic analysis of miR-30a-5p with CEA and CA19-9 showed that the sensitivity was 91.49% and specificity was 92.59%. Additionally, the combination of miR-182–5p with CEA and CA19-9 showed that specificity increased to 98.15% and PPV to 97.6% (shown in Table 3 and Fig. 2C and D).
Table 3.
Agreement (sensitivity, specificity) for different markers and combinations of markers to predict gastric cancer patients (n = 47) from benign gastric lesions (n = 54).
| AUC | p | 95% C·I | Cutoff | Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|---|---|---|---|
| CEA | 0.736 | <0.001* | 0.638–0.833 | >7.1 | 74.47 | 53.70 | 58.3 | 70.7 |
| CA19-9 | 0.840 | <0.001* | 0.755–0.925 | >21 | 78.72 | 72.22 | 71.2 | 79.6 |
| miR-30a-5p | 0.961 | <0.001* | 0.903–0.990 | ≤3.27 | 93.62 | 90.74 | 89.8 | 94.2 |
| miR-182–5p | 0.898 | <0.001* | 0.833–0.963 | ≤3.9 | 87.23 | 81.48 | 80.4 | 88.0 |
|
CEA + CA19-9 |
0.888 | <0.001* | 0.814–0.963 | 78.72 | 88.89 | 86.0 | 82.8 | |
| CEA + CA19-9 + miR–30a-5p | 0.991 | <0.001* | 0.979–1.0 | 91.49 | 92.59 | 91.5 | 92.6 | |
| CEA + CA19-9 + miR-182–5p | 0.974 | <0.001* | 0.950–0.997 | 85.11 | 98.15 | 97.6 | 88.3 |
CEA; Carcinoembryonic antigen CA19-9; Carbohydrate antigen 19-9.
AUC: Area under a curve p value: Probability value.
CI: Confidence intervals.
NPV: Negative predictive value PPV: Positive predictive value.
*: Statistically significant at p < 0.05.
Lower miR-30a-5P level was significantly linked with the existence of distant metastases, advanced TNM stage (stage III and IV than stage I and II) of disease, and a degree of pathological differentiation of tumor (no and low differentiation than moderate and well-differentiated cases) in gastric cancer patients (p = 0.034, 0.019, and 0.049), with no significant relations related to sex, presence of H. pylori stool antigen, performance status, history of smoking, weight loss and NSAIDs, T stage, nor nodal metastases. Furthermore, miR-182–5 P had no significant association with the previously mentioned parameters (Table 4). MiR-30a-5P showed a significant negative correlation with the stage of the tumor (rs = - 0.380 and p = 0.008) and a degree of pathological differentiation (rs = −0.335 and p = 0.021) as presented in Fig. 2E and F.
Table 4.
Relation between MiR-30a-5p and MiR-182–5p with different parameters in group I (n = 47).
| N | miR–30a-5P |
miR-182–5 P |
|||||
|---|---|---|---|---|---|---|---|
| Min. – Max. | Mean ± SD. | Median | Min. – Max. | Mean ± SD. | Median | ||
| Sex | |||||||
| Male | 31 | 0.05–3.50 | 2.14 ± 0.85 | 2.37 | 0.10–4.0 | 2.54 ± 1.23 | 2.90 |
| Female | 16 | 1.25–3.50 | 2.35 ± 0.78 | 2.40 | 0.10–4.0 | 2.92 ± 1.31 | 3.65 |
| U(p) | 213.50 (0.438) | 186.50 (0.167) | |||||
| H. pylori | |||||||
| No | 37 | 0.05–3.50 | 2.23 ± 0.83 | 2.37 | 0.43–4.0 | 2.88 ± 1.10 | 3.12 |
| Yes | 10 | 1.25–3.50 | 2.14 ± 0.86 | 1.93 | 0.10–3.90 | 1.87 ± 1.51 | 1.56 |
| U(p) | 176.0 (0.828) | 111.0 (0.055) | |||||
| PS | |||||||
| 0 | 22 | 1.25–3.50 | 2.22 ± 0.71 | 2.37 | 0.10–3.92 | 2.57 ± 1.23 | 2.80 |
| 1 | 23 | 1.25–3.50 | 2.34 ± 0.83 | 2.37 | 0.43–4.0 | 2.79 ± 1.28 | 3.30 |
| 2 | 2 | 0.05–1.27 | 0.66 ± 0.86 | 0.66 | 0.90–3.70 | 2.30 ± 1.98 | 2.30 |
| U(p) | 4.269 (0.118) | 1.103 (0.576) | |||||
| History & general examination | |||||||
| Smoking | |||||||
| No | 25 | 1.25–3.50 | 2.11 ± 0.73 | 2.27 | 0.10–4.0 | 2.57 ± 1.31 | 2.70 |
| Yes | 22 | 0.05–3.50 | 2.33 ± 0.93 | 2.55 | 0.43–4.0 | 2.78 ± 1.21 | 3.20 |
| U(p) | 229.50 (0.331) | 260.0 (0.749) | |||||
| Weight loss | |||||||
| No | 24 | 1.25–3.50 | 2.32 ± 0.76 | 2.43 | 0.10–4.0 | 2.48 ± 1.24 | 2.70 |
| Yes | 23 | 0.05–3.50 | 2.09 ± 0.89 | 2.32 | 0.43–4.0 | 2.87 ± 1.27 | 3.30 |
| U(p) | 234.0 (0.370) | 212.0 (0.173) | |||||
| Nsaid | |||||||
| No | 35 | 0.05–3.50 | 2.28 ± 0.86 | 2.37 | 0.10–4.0 | 2.68 ± 1.26 | 2.90 |
| Yes | 12 | 1.25–3.50 | 2.02 ± 0.74 | 2.01 | 0.10–4.0 | 2.65 ± 1.31 | 3.02 |
| U(p) | 156.50 (0.191) | 208.0 (0.961) | |||||
| T | |||||||
| 1 | 14 | 1.25–3.50 | 2.16 ± 0.73 | 2.27 | 0.70–4.0 | 2.78 ± 1.05 | 2.65 |
| 2 | 7 | 1.40–3.50 | 2.56 ± 0.77 | 2.79 | 0.10–4.0 | 2.28 ± 1.72 | 3.10 |
| 3 | 21 | 1.25–3.22 | 2.22 ± 0.77 | 2.37 | 0.10–4.0 | 2.67 ± 1.29 | 2.90 |
| 4 | 5 | 0.05–3.50 | 1.83 ± 1.38 | 1.42 | 0.90–3.80 | 2.89 ± 1.17 | 3.12 |
| H(p) | 1.789 (0.617) | 0.193 (0.979) | |||||
| N | |||||||
| 0 | 20 | 0.05–3.50 | 2.27 ± 0.86 | 2.40 | 0.10–4.0 | 2.79 ± 1.27 | 3.16 |
| 1 | 19 | 1.25–3.50 | 2.18 ± 0.87 | 2.37 | 0.65–4.0 | 2.66 ± 1.23 | 3.10 |
| 2 | 7 | 1.25–2.90 | 2.26 ± 0.70 | 2.37 | 0.10–4.0 | 2.64 ± 1.30 | 2.70 |
| 3 | 1 | 1.25 | 0.60 | ||||
| H(p) | 2.502 (0.475) | 2.338 (0.505) | |||||
| M | |||||||
| 0 | 41 | 0.05v3.50 | 2.29 ± 0.80 | 2.37 | 0.10–4.0 | 2.76 ± 1.23 | 3.10 |
| 1 | 6 | 1.25–3.50 | 1.66 ± 0.91 | 1.28 | 0.65–3.80 | 2.01 ± 1.36 | 1.90 |
| U(p) | 57.50* (0.034*) | 83.50 (0.214) | |||||
| Stage | |||||||
| I | 13 | 1.42–3.50 | 2.38 ± 0.78 | 2.32 | 0.43v4.0 | 2.94 ± 1.17 | 3.50 |
| II | 16 | 1.40–3.22 | 2.59 ± 0.55 | 2.72 | 0.10–4.0 | 2.87 ± 1.11 | 3.10 |
| III | 10 | 0.05–2.97 | 1.75 ± 0.93 | 1.42 | 0.10–4.0 | 2.48 ± 1.42 | 2.91 |
| IV | 8 | 1.25–3.50 | 1.76 ± 0.90 | 1.28 | 0.60–3.80 | 2.06 ± 1.44 | 1.90 |
| H(p) | 9.896*(0.019*) | 2.353 (0.502) | |||||
| Differentiation | |||||||
| No | 5 | 1.25–2.49 | 1.77 ± 0.61 | 1.49 | 0.80–3.70 | 2.06 ± 1.16 | 2.20 |
| Low | 18 | 1.25–3.25 | 2.18 ± 0.73 | 2.35 | 0.43–4.0 | 2.58 ± 1.28 | 2.65 |
| Moderate | 17 | 0.05–3.50 | 2.08 ± 0.90 | 2.77 | 0.10–4.0 | 2.57 ± 1.38 | 3.10 |
| Well | 7 | 1.42–3.50 | 2.92 ± 0.73 | 3.20 | 2.91–3.90 | 3.57 ± 0.42 | 3.87 |
| H(p) | 7.871 (0.049*) | 5.444 (0.142) | |||||
U: Mann Whitney test H: H for Kruskal Wallis test p: p-value for the association between different categories *: Statistically significant at p < 0.05.
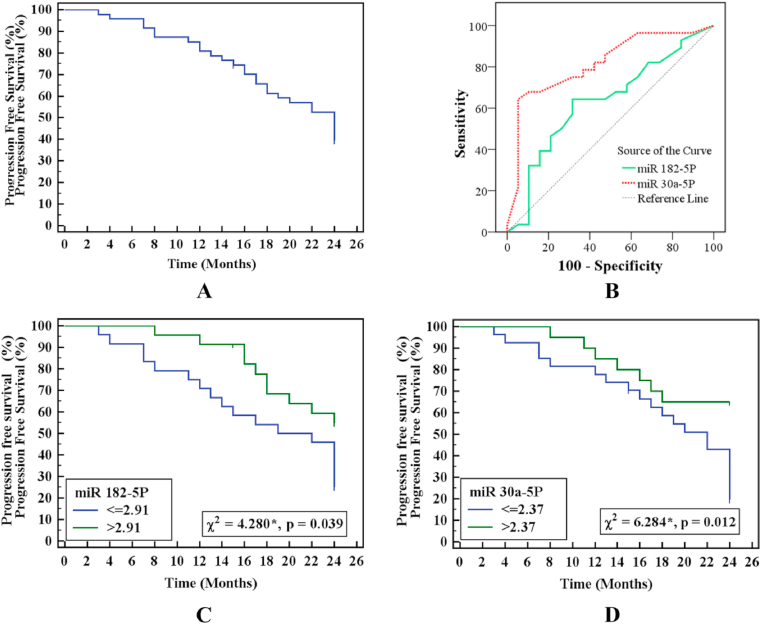
ROC curve analysis was used to explore the prognostic role of the investigated miR-30a-5p and miR-182–5p, which revealed that miR30a-5p level (≤2.37) was a considerable predictor of bad prognosis among patients with gastric cancer (p < 0.001) with sensitivity and specificity of 75% and 68.42%. On the other hand, miR182–5 P was not a significant predictor (p = 0.129) (Table 5 and Fig. 3A and B). Using the Kaplan–Meier survival curve, log-rank analyses showed that low miR-30a-5p was≤2.37 and miR-182–5p≤2.91. Gene expression in gastric cancer patients was significantly associated with decreased Progression-Free Overall Rates (PFS) (P < 0.012 and 0.039) (Fig. 3C and D).
Table 5.
Agreement (sensitivity and specificity) for miR-30a-5p and miR-182–5p to predict prognosis in gastric cancer (n = 47).
| AUC | p | 95% C·I | Cut off | Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|---|---|---|---|
| miR-30a-5p | 0.815 | <0.001* | 0.688–0.942 | ≤2.37 | 75.0 | 68.42 | 77.8 | 65.0 |
| miR–182–5p | 0.632 | 0.129 | 0.467–0.796 | ≤2.91 | 64.29 | 68.42 | 75.0 | 56.5 |
AUC: Area under a curve p value: Probability value.
CI: Confidence intervals.
NPV: Negative predictive value PPV: Positive predictive value.
*: Statistically significant at p ≤ 0.05.
Fig. 3.
A: Kaplan-Meier survival curve for progression-free survival of gastric cancer patients B: ROC curve for miR-182–5p and miR-30a-5p to predict prognosis in gastric cancer (n = 47) C: Kaplan-Meier survival curve for progression-free survival with miR-182–5p D: Kaplan-Meier survival curve for progression-free survival with miR-30a-5p.
For the parameters influencing gastric cancer patients, the univariate and multivariate logistic regression analyses revealed that the following: With the univariate test, miR-182–5p had p = 0.045 OR 2.164 (1.108–4.692) and miR-30a-5p p = 0.020 and 2.768 (1.175–6.523), which could be meaningful in disease prediction. On the other hand, in the multivariate analysis miR-30a-5p expression level, p = 0.041 OR 2.472 (1.037–5.893), which could be an independent predictor for gastric cancer; however, miR-182–5p expression level was not p = 0.121 OR 1.858 (0.848–4.071) (Table 6).
Table 6.
Univariate and multivariate COX regression analysis for the parameters affecting Group I (gastric cancer patients) (n = 47) vs. Group II benign gastric lesion patients (n = 54).
| Univariate |
Multivariate |
|||
|---|---|---|---|---|
| P | HR (95%C·I) | p | HR (95%C·I) | |
| Sex (female) | 0.957 | 0.978 (0.442–2.165) | ||
| Age (years) | 0.611 | 1.013 (0.963–1.065) | ||
| BMI (kg/m2) | 0.185 | 1.053 (0.976–1.136) | ||
| Dyspepsia | 0.701 | 0.755 (0.179–3.181) | ||
| Hematemesis and melena | 0.892 | 1.064 (0.431–2.627) | ||
| Hb | 0.986 | 0.999 (0.844–1.181) | ||
| Platelet count | 0.850 | 1.0 (0.996–1.0) | ||
| History of smoking | 0.222 | 0.622 (0.291–1.332) | ||
| History of weight loss | 0.116 | 0.543 (0.254–1.161) | ||
| Pallor | 0.400 | 1.378 (0.652–2.912) | ||
| CEA | 0.443 | 0.998 (0.994–1.0) | ||
| CA19-9 | 0.771 | 0.998 (0.987–1.0) | ||
| ESR | 0.114 | 1.010 (0.998–1.0) | ||
| Size (cm) | 0.407 | 0.895 (0.689–1.163) | ||
| Family H. of gastric cancer | 0.212 | 0.280 (0.038–2.066) | ||
| miR-30a-5p (≤2.37) | 0.020* | 2.768 (1.175–6.523) | 0.041* | 2.472 (1.037–5.893) |
| miR–182–5p (≤2.91) | 0.045* | 2.164 (1.108–4.692) | 0.121 | 1.858 (0.848–4.071) |
BMI: body mass index, Hb: Hemoglobin concentration.
CEA; Carcinoembryonic antigen, CA19-9; Carbohydrate antigen 19–9.
ESR: Erythrocyte sedimentation rate.
HR: Hazard ratio.
C·I: Confidence interval LL: Lower limit UL: Upper limit.
#: All variables with p < 0.05 were included in the multivariate.
*: Statistically significant at p < 0.05.
4. Discussion
Gastric cancer is a leading cause of morbidity and mortality worldwide, especially in developing nations. Despite the improved survival over the recent years due to improved endoscopic and imaging procedures and medical and surgical services, its prognosis continues to be inconvenient [17]. Despite scientific efforts, there are few suitable gastric cancer serum biomarkers with acceptable sensitivity and specificity for screening and monitoring [18].
We noticed that the platelet count was significantly higher in gastric cancer patients compared with other studied groups. These results agree with previous literature, where platelets were raised in gastric cancer patients [19]. In a gastric cancer scenario, the mechanism of platelet augmentation is still uncertain and not exceptionally clear. It is conceivably linked to bone marrow stimulation by a thrombopoietin-like hormone that is released from the inflammatory portion of the tumor [18]. Moreover, numerous studies have recorded the function of cytokines, especially IL-6, in thrombocytosis pathogenesis in cancer, especially in gastrointestinal cancers [20].
MiRNAs have been attracting growing attention among investigators in recent years, especially cancer investigators, as pivotal cellular molecules implicated within normal and pathological conditions.
Numerous pieces of research have illustrated the MiRNAs aberrant expression in diversified cancer groups and that they could be utilized as incoming biomarkers for tumor recognition and follow-up. It was elucidated that oncogenic MiRNAs were considerably up regulated in cancer scenarios, whilst tumor-suppressive MiRNAs were repeatedly down regulated. The effectiveness of using miRNAs as diagnostic or prognostic biomarkers began to gain fundamental consideration in cancer research [21,22].
In this study, we focused on investigating two oncogenic MiRNAs that are recently associated with cancers encompassing miR-30a-5p and miR-182–5p and displayed prominent results of down regulation of both miRNAs levels in gastric cancer patients than in patients with benign gastric lesions and controls.
These results agree with previous literature, where it was reported that MiR-30a-5p could function as a suppressor gene by inhibiting the invasion and migration in numerous cancers including breast, pancreatic ductal adenocarcinoma, and no small cell lung cancer [[23], [24], [25]]. Additionally, in former research, Xue and his colleagues noticed that the invasion and migration of hepatocellular carcinoma cells were reinforced by the overexpression loc339803 which was blocked by mimics of miR-30a-5p. Additionally, they found that loc339803 overexpression could suppress the expression of miR-30a-5p. Hence, they reported that miR-30a-5p could be a target gene of loc339803 [26].
MiR-30 family is an imperative and a complex family that includes five members and six separate mature MiRNAs (miR-30a, −30 b, −30c-1, -30c-2, -30 d, and −30e) that are encoded by six genes that exist on human chromosomes 1, 6, and 8 [10]. These MiRNAs possess the same seed sequence existing near the 5′ end with diverse compensating sequences located near the 3’ end that enhance the organization process of diverse genes and pathways. Occasionally, they maintain entirely inverse manners. An assortment of physiological conditions, as well as pathological disorders, in vivo, agrees with the differential expression of the members of the miR-30 family through modifying targeted gene expression [27]. MiR-30a-5p, an intragenic MiR (chromosome 6, 71, 403, 551–71,403,621 [- strand]), is thought to have a pivotal role in cellular development and differentiation [28].
We displayed remarkable associations between lower miR 30a-5p levels and the presence of distant metastases, advanced TNM stage of disease, and a degree of differentiation of tumor in gastric cancer patients. It was formerly stated that in Clear Cell Renal Cell Carcinoma (ccRCC), an onco-suppressor role was suggested for miR-30a-5p, as its down regulation was correlated with development metastasis [29,30]. Additionally, it was reported that miR-30a-5p inhibits autophagy through targeting BECN1, the gene encoding for beclin-1 and a key protein for forming autophagosome [31]. It was illustrated that miR-30a-5p could minimize tumor micro vessel density via targeting endothelial DLL4, which is enlisted in the angiogenesis of tumors [27].
Amongst known miRNAs, miR-182 (that has a place in miR-183-96-182 cluster) is deemed as a micro-oncogene. Recently, comprehensive profiling studies have related that miR-182 deregulated expression to various kinds of cancer, such as Colorectal Cancer (CRC), glioma, lung and bladder cancers [[32], [33], [34]].
In the present study, miR-182–5p was significantly down regulated in gastric cancer patients. Moreover, ROC curve analysis demonstrated its diagnostic role in predicting gastric cancer, revealing that miR30a-5p had a good diagnostic value with AUC 0.898.
Kong and his colleagues previously reported that miR‐182 was down regulated in tissue samples of human gastric adenocarcinoma, suggesting that miR‐182 might have a substantial role in the development of gastric adenocarcinoma in the form of a tumor suppressor gene [35]. Furthermore, MiR‐182 could deregulate RGS17 (Regulator of G-protein signaling 17) by targeting its 3′‐UTR, ultimately suppressing the development of lung cancer [36]. Similarly, MiR-182 was down regulated in tissues and cell lines of osteosarcoma; miR-182 restoration could minimize osteosarcoma cell invasion and proliferation [37].
In vitro studies have shown the negative effect of miR‐182 on cellular proliferation in BGC‐823, MGC‐803, and SGC‐7901 cells. Besides, bioinformatics analyses proposed a miR‐182–binding location on the cAMP‐responsive element-binding protein-1 gene (CREB1) transcript [35]. Additionally, it was reported that ANUBL1 is a target of miR-182 which is down regulated in human gastric cancer. It can suppress cell growth by down regulating ANUBL1 expression [38].
These results propose that miR-182 might function as a tumor suppressor gene with down regulation that gives a share in the progression and metastasis of cancers. However, former studies have documented that miR-182 expression is up regulated in some human cancers and acts as an oncogene like in melanoma, where it enhances tumor growth and metastasis and invasion. Besides, up regulation of miR-182 in Colorectal Cancer (CRC) was observed and found to be linked with adverse clinical characteristics and bad prognosis [39,40]. This difference can be attributed to the difference in the sample size, types of cancer and sample used (tissues or serum), or methods of detection and cutoff values of MiR-182.
In our study, we detected down regulation of miR-30a-5p and miR-182–5p gene expression in gastric cancer patients that were significantly linked to the reduction of progression-free survival rates. On the other hand, in multivariate regression analysis, it was shown that miR-30a-5P expression level was down regulated rather than miR-182–5 P, which could be an independent predictor for gastric cancer.
In conclusion, our results point out that MiRNAs, miR-30a-5p and miR-182–5p, gene expression has a diagnostic power and can identify patients with gastric cancer. MiR30a-5p displayed the highest diagnostic specificity and sensitivity. Together with other known tumor markers, it could offer simple noninvasive biomarkers that predict gastric cancer alerting physicians to perform the upper GIT endoscopy, the gold standard in gastric cancer screening and diagnosis to improve the early discovery of gastric cancer and can help with suspect prognosis.
Conflicting interests
The author(s) declared no potential conflicts of interest.
Ethical Approval
This work was performed per the Declaration of Helsinki and the principles of the “Ethical Committee of Medical Research”, Faculty of Medicine, and Menoufia University. Written consent was provided by all participants.
Funding
The author(s) received no financial support for this research.
Author contribution
SES performed the lab investigation and the molecular analysis and selected the study design. NSE was responsible for samples and data collections and evaluation of the involved patients. MFA shared an in-lab investigation, and SME participated in the selection of the study design. All authors participated in writing and revising the paper and approved the final manuscript.
Acknowledgment
The author(s) acknowledge Dr Suzy F. Gohar (assistant professor at Clinical Oncology department, Faculty of Medicine, Menoufia University) for her valuable input about the follow-up of patients with gastric cancer.
Contributor Information
Shimaa E. Soliman, Email: shaimaa_alshafaay@med.menofia.edu.eg, dr.shelshafey2010@yahoo.com.
Naglaa S. Elabd, Email: naglaa.alabd.12@med.menofia.edu.eg.
Salah M. EL-Kousy, Email: Salah.elqusy@science.menofia.edu.eg.
Mohamed F. Awad, Email: mohamedfathy1652@gmail.com.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018 Nov;68(6):394–424. doi: 10.3322/caac.21492. ([Medline]. [Full Text]) [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization Cancer. WHO. 2021. http://www.who.int/mediacentre/factsheets/fs297/en/ Available at: 12 September 2018; Accessed: February 22.
- 3.Guggenheim D.E., Shah M.A. Gastric cancer epidemiology and risk factors. J. Surg. Oncol. 2013 Mar;107(3):230–236. doi: 10.1002/jso.23262. Epub 2012 Nov 5. PMID: 23129495. [DOI] [PubMed] [Google Scholar]
- 4.Jiang Y., Ajani J.A. Multidisciplinary management of gastric cancer. Curr. Opin. Gastroenterol. 2010 Nov;26(6):640–646. doi: 10.1097/MOG.0b013e32833efd9b. PMID: 20827183. [DOI] [PubMed] [Google Scholar]
- 5.Terry M.B., Gaudet M.M., Gammon M.D. The epidemiology of gastric cancer. Semin. Radiat. Oncol. 2002 Apr;12(2):111–127. doi: 10.1053/srao.30814.PMID:11979413. [DOI] [PubMed] [Google Scholar]
- 6.Yun Z.Y., Li N., Zhang X., Zhang H., Bu Y., Sun Y., Liu T., Wang R.T., Yu K.J. Mean platelet volume, platelet distribution width and carcinoembryonic antigen to discriminate gastric cancer from gastric ulcer. Oncotarget. 2017 Mar 4;8(37):62600–62605. doi: 10.18632/oncotarget.15898. PMID: 28977972; PMCID: PMC5617532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling H., Fabbri M., Calin G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013 Nov;12(11):847–865. doi: 10.1038/nrd4140.PMID:24172333. PMCID: PMC4548803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashrafizadeh M., Zarrabi A., Hushmandi K., Hashemi F., Moghadam E.R., Owrang M., Hashemi F., Makvandi P., Goharrizi M.A.S.B., Najafi M., Khan H. Lung cancer cells and their sensitivity/resistance to cisplatin chemotherapy: role of microRNAs and upstream mediators. Cell. Signal. 2021 Feb;78:109871. doi: 10.1016/j.cellsig.2020.109871. Epub 2020 Dec 3. PMID: 33279671. [DOI] [PubMed] [Google Scholar]
- 9.Ashrafizadeh M., Zarrabi A., Orouei S., Zarrin V., Rahmani Moghadam E., Zabolian A., Mohammadi S., Hushmandi K., Gharehaghajlou Y., Makvandi P., Najafi M., Mohammadinejad R. STAT3 pathway in gastric cancer: signaling, therapeutic targeting and future prospects. Biology. 2020 Jun 12;9(6):126. doi: 10.3390/biology9060126. PMID: 32545648; PMCID: PMC7345582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croset M., Pantano F., Kan C.W.S., Bonnelye E., Descotes F., Alix-Panabières C. miRNA-30 family members inhibit breast cancer invasion, osteomimicry, and bone destruction by directly targeting multiple bone metastasis-associated genes. Canc. Res. 2018;78:5259–5273. doi: 10.1158/0008-5472.CAN-17-3058. [DOI] [PubMed] [Google Scholar]
- 11.Wang W., Lin H., Zhou L., Zhu Q., Gao S., Xie H., Liu Z., Xu Z., Wei J., Huang X., Zheng S. MicroRNA-30a-3p inhibits tumor proliferation, invasiveness and metastasis and is downregulated in hepatocellular carcinoma. Eur. J. Surg. Oncol. 2014 Nov;40(11):1586–1594. doi: 10.1016/j.ejso.2013.11.008. Epub 2013 Nov 19. PMID: 24290372. [DOI] [PubMed] [Google Scholar]
- 12.Wang M., Wang Y., Zang W., Wang H., Chu H., Li P., Li M., Zhang G., Zhao G. Downregulation of microRNA-182 inhibits cell growth and invasion by targeting programmed cell death 4 in human lung adenocarcinoma cells. Tumour Biol. 2014 Jan;35(1):39–46. doi: 10.1007/s13277-013-1004-8. Epub 2013 Jul 23. PMID: 23877371. [DOI] [PubMed] [Google Scholar]
- 13.Pignot G., Cizeron-Clairac G., Vacher S., Susini A., Tozlu S., Vieillefond A., Zerbib M., Lidereau R., Debre B., Amsellem-Ouazana D., Bieche I. microRNA expression profile in a large series of bladder tumors: identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int. J. Canc. 2013 Jun 1;132(11):2479–2491. doi: 10.1002/ijc.27949. Epub 2013 Feb 25. PMID: 23169479. [DOI] [PubMed] [Google Scholar]
- 14.Perilli L., Tessarollo S., Albertoni L., Curtarello M., Pastò A., Brunetti E. Silencing of miR-182 is associated with modulation of tumorigenesis through apoptosis induction in an experimental model of colorectal cancer. BMC Canc. 2019;19:821. doi: 10.1186/s12885-019-5982-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Chen S., Shan Z., Bi L., Yu S., Li Y., Xu S. miR-182-5p improves the viability, mitosis, migration, and invasion ability of human gastric cancer cells by down-regulating RAB27A. Biosci. Rep. 2017 Jun 27;37(3) doi: 10.1042/BSR20170136. PMID: 28546229; PMCID: PMC6434084. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Edge S.B., Compton C.C. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg Oncol. 2010 Jun;17(6):1471–1474. doi: 10.1245/s10434-010-0985-4.PMID:20180029. [DOI] [PubMed] [Google Scholar]
- 17.Toyokawa T., Muguruma K., Tamura T., Sakurai K., Amano R., Kubo N. Comparison of the prognostic impact and combination of preoperative inflammation-based and/or nutritional markers in patients with stage II gastric cancer. Oncotarget. 2018;9(50):29351–29364. doi: 10.18632/oncotarget.25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng S., Han F., Wang Y., Xu Y., Qu T., Ju Y., Lu Z. The red distribution width and the platelet distribution width as prognostic predictors in gastric cancer. BMC Gastroenterol. 2017 Dec 20;17(1):163. doi: 10.1186/s12876-017-0685-7. PMID: 29262773; PMCID: PMC5738162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Lehleh A.M., El-Abd N.S., Gohar S.F., Zarad M.O. Diagnostic value of platelet indices, carbohydrate antigen 19-9 and carcinoembryonic antigen in differentiating malignant from benign gastric ulcers. Menoufia Med J. 2019;32:1452–1458. [Google Scholar]
- 20.Xinting L.V., Li Y., Chen T., Li N. Relationship between platelet count and gastric cancer stage and prognosis. Chin. Ger. J. Clin. Oncol. 2010;9(4):213–215. doi: 10.1007/s10330-010-0030-x. [DOI] [Google Scholar]
- 21.Hayes J., Peruzzi P.P., Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 2014 Aug;20(8):460–469. doi: 10.1016/j.molmed.2014.06.005. Epub 2014 Jul 12. PMID: 25027972. [DOI] [PubMed] [Google Scholar]
- 22.Saleh A.A., Soliman S.E., Habib M.S.E., Gohar S.F., Abo-Zeid G.S. Potential value of circulatory microRNA122 gene expression as a prognostic and metastatic prediction marker for breast cancer. Mol. Biol. Rep. 2019 Jun;46(3):2809–2818. doi: 10.1007/s11033-019-04727-5. Epub 2019 Mar 5. PMID: 30835039. [DOI] [PubMed] [Google Scholar]
- 23.Li L., Kang L., Zhao W., Feng Y., Liu W., Wang T. miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated Warburg effect. Canc. Lett. 2017;400:89–98. doi: 10.1016/j.canlet.2017.04.034. [DOI] [PubMed] [Google Scholar]
- 24.Zhou L., Jia S., Ding G., Zhang M., Yu W., Wu Z. Down-regulation of miR-30a-5p is associated with poor prognosis and promotes chemoresistance of gemcitabine in pancreatic ductal adenocarcinoma. J. Canc. 2019;10:5031–5040. doi: 10.7150/jca.31191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu J., Zeng Y., Li W., Qin H., Lei Z., Shen D. CD73/NT5E is a target of miR-30a-5p and plays an important role in the pathogenesis of non-small cell lung cancer. Mol. Canc. 2017;16:34. doi: 10.1186/s12943-017-0591-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue C., Zhang X., Gao P., Cui X., Zhu C., Qin X. LncRNA loc339803 acts as CeRNA of miR-30a-5p to promote the migration and invasion of hepatocellular carcinoma cells. J. Canc. January 2021;12(4):1061–1072. doi: 10.7150/jca.52413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X., Chen Y., Chen L. The versatile role of microRNA-30a in human Cancer. Cell. Physiol. Biochem. 2017;41(4):1616–1632. doi: 10.1159/000471111. [DOI] [PubMed] [Google Scholar]
- 29.Ran L., Liang J., Deng X., Wu J. miRNAs in prediction of prognosis in clear cell renal cell carcinoma. BioMed Res. Int. 2017;2017:4832931. doi: 10.1155/2017/4832931. Epub 2017 Dec 17. PMID: 29392135; PMCID: PMC5748131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Y.H., Chen C., Shi Z. The biological roles and clinical implications of microRNAs in clear cell renal cell carcinoma. J. Cell. Physiol. 2018;233(6):4458–4465. doi: 10.1002/jcp.26347. [DOI] [PubMed] [Google Scholar]
- 31.Aguiari G. MicroRNAs in clear cell renal cell carcinoma: biological functions and applications. J Kidney Cancer VHL. 2015;2(4):140–152. doi: 10.15586/jkcvhl.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S., Yang M.H., Wang X.Y., Lin J., Ding Y.Q. Increased expression of miRNA-182 in colorectal carcinoma: an independent and tissue-specific prognostic factor. Int. J. Clin. Exp. Pathol. 2014 May 15;7(6):3498–3503. PMID: 25031782; PMCID: PMC4097234. [PMC free article] [PubMed] [Google Scholar]
- 33.Casanova-Salas I., Rubio-Briones J., Calatrava A., Mancarella C., Masiá E., Casanova J., Fernández-Serra A., Rubio L., Ramírez-Backhaus M., Armiñán A., Domínguez-Escrig J., Martínez F., García-Casado Z., Scotlandi K., Vicent M.J., López-Guerrero J.A. Identification of miR-187 and miR-182 as biomarkers of early diagnosis and prognosis in patients with prostate cancer treated with radical prostatectomy. J. Urol. 2014 Jul;192(1):252–259. doi: 10.1016/j.juro.2014.01.107. Epub 2014 Feb 8. PMID: 24518785. [DOI] [PubMed] [Google Scholar]
- 34.Stenvold H., Donnem T., Andersen S., Al-Saad S., Busund L.T., Bremnes R.M. Stage and tissue-specific prognostic impact of miR-182 in NSCLC. BMC Canc. 2014;14(1) doi: 10.1186/1471-2407-14-138. article 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong W.Q., Bai R., Liu T., Cai C.L., Liu M., Li X., Tang H. MicroRNA-182 targets cAMP-responsive element-binding protein 1 and suppresses cell growth in human gastric adenocarcinoma. FEBS J. 2012 Apr;279(7):1252–1260. doi: 10.1111/j.1742-4658.2012.08519.x. Epub 2012 Mar 2. PMID: 22325466. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y., Fang R., Li C., Li L., Li F., Ye X., Chen H. Hsa-mir-182 suppresses lung tumorigenesis through down regulation of RGS17 expression in vitro. Biochem. Biophys. Res. Commun. 2010 May 28;396(2):501–507. doi: 10.1016/j.bbrc.2010.04.127. Epub 2010 Apr 24. PMID: 20420807. [DOI] [PubMed] [Google Scholar]
- 37.Hu J., Lv G., Zhou S., Zhou Y., Nie B., Duan H., Zhang Y., Yuan X. The downregulation of MiR-182 is associated with the growth and invasion of osteosarcoma cells through the regulation of TIAM1 expression. PloS One. 2015 May 14;10(5) doi: 10.1371/journal.pone.0121175. PMID: 25973950; PMCID: PMC4431740. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Tang L., Chen F., Pang E., Zhang Z., Jin B., Dong W. MicroRNA-182 inhibits proliferation through targeting oncogenic ANUBL1 in gastric cancer. Oncol. Rep. 2015;33:1707–1716. doi: 10.3892/or.2015.3798. [DOI] [PubMed] [Google Scholar]
- 39.Segura M.F., Hanniford D., Menendez S., Reavie L., Zou X., Alvarez-Diaz S. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc. Natl. Acad. Sci. U.S.A. 2009;106(6):1814–1819. doi: 10.1073/pnas.0808263106. [PMC free article] [PubMed] [CrossRef] [Google Scholar] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu H., Du L., Wen Z., Yang Y., Li J., Wang L. Up-regulation of miR-182 expression in colorectal cancer tissues and its prognostic value. Int. J. Colorectal Dis. 2013;28(5):697–703. doi: 10.1007/s00384-013-1674-0. [PubMed] [CrossRef] [Google Scholar] [DOI] [PubMed] [Google Scholar]