Abstract
Although glucose, through pentose phosphate pathway (PPP), is the main source to generate NADPH, solid tumors are often deprived of glucose, hence alternative metabolic pathways to maintain NADPH homeostasis in cancer cells are required. Here, we report that lactate and glutamine support NADPH production via isocitrate dehydrogenase 1 (IDH1) and malic enzyme 1 (ME1), respectively, under glucose-deprived conditions. Isotopic tracing demonstrates that lactate participates in the formation of isocitrate. Malate derived from glutamine in mitochondria shuttles to cytosol to produce NADPH. In cells cultured in the absence of glucose, knockout of IDH1 and ME1 decreases NADPH/NADP+ and GSH/GSSG, increases ROS level and facilitates cell necrosis. In 4T1 murine breast tumors, knockout of ME1 retards tumor growth in vivo, with combined ME1/IDH1 knockout more strongly suppressing tumor growth. Our findings reveal two alternative NADPH-producing pathways that cancer cells use to resist glucose starvation, reflecting the metabolic plasticity and flexibility of cancer cells adapting to nutrition stress.
Keywords: NADPH, Lactate, Glutamine, IDH1, ME1, Glucose deprivation
Abbreviations: PPP, pentose phosphate pathway; IDH1, isocitrate dehydrogenase 1; ME1, malic enzyme 1; ROS, reactive oxygen species; TME, tumor microenvironment; MTHFD1, methylene tetrahydrofolate dehydrogenase 1; ALDH1L1, 10-formyltetrahydrofolate dehydrogenase; G6PD, glucose-6-phosphate dehydrogenase; TCA, tricarboxylic acid; MCT1, monocarboxylate transporter; α-KG, α-ketoglutarate; MDH1, malate dehydrogenase 1; BPTES, bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide; LDH, lactate dehydrogenase; Nampt, Nicotinamide phosphoribosyltransferase; PDH, pyruvate dehydrogenase; HIF-1, hypoxia-inducible factor 1; PDK1, pyruvate dehydrogenase kinase 1; Ac-CoA, acetyl coenzyme A; OAA, oxaloacetate; TACE, transarterial chemoembolization; THF, tetrahydrofolate
Graphical abstract
Highlights
-
•
Lactate and glutamine are the sources of NADPH in tumors under glucose deprivation.
-
•
Lactate supports NADPH generation and maintains redox balance in tumors by IDH1.
-
•
Glutamine supports NADPH generation and maintains redox balance in tumors by ME1.
-
•
Lactate-dependent NADPH generation is conditional on thermodynamics.
-
•
Combination of IDH1 and ME1 is crucial to the growth and metastasis of solid tumors.
1. Introduction
Malignant tumors display reprogrammed metabolic phenotypes, which are dictated by both intrinsic and extrinsic factors [1]. While cell lineage and genetic alteration confer tumors with inherent metabolic characteristics [2], a variety of metabolic pathways are imposed by tumor microenvironment (TME) [1,3]. Due to the poor vascularization of solid tumors and rapid nutrient consumption by tumor cells, oxygen and nutrients such as glucose in TME are often limited [4,5]. Cancer cells develop metabolic adaptation to TME, exhibiting metabolic flexibility (alternative material and energy sources) and metabolic plasticity (rewired metabolic pathways) [6].
The deprivation of glucose in TME raises the question how cancer cells in solid tumors maintain NADPH/NADP+ homeostasis, because glucose is the main hydride ion source to reduce NADP+ through oxidative branch of pentose phosphate pathway (oxPPP) [7,8]. Cellular NADPH is essential for maintaining redox balance [9] and sustaining reductive biosynthesis [10], which are indispensable for tumor growth. Therefore, other NADPH-generation pathways must take over. Apart from oxPPP, other cytosolic generation pathways of NADPH include reactions catalyzed by isocitrate dehydrogenase 1 (IDH1), malic enzyme 1 (ME1), methylene tetrahydrofolate dehydrogenase 1 (MTHFD1) and 10-formyltetrahydrofolate dehydrogenase (ALDH1L1) [11]. It is reported that wild-type IDH1 promotes growth, migration and resistance to targeted therapies by regulating lipid biosynthesis and redox balance in glioblastoma [[12], [13], [14]]. ME1 is reported to regulate NADPH homeostasis to promote cancer growth and metastasis in gastric cancer [15] and nasopharyngeal carcinoma [16]. ME1 is found to be co-expressed with IDH1 and G6PD in diverse cancer cell lines [17], and ME1 can collaborate with mitochondrial IDH2 to maintain antioxidant systems to support tumor growth and metastasis [18]. Folate-dependent NADPH production by MTHFD and ALDH1L also contributes a large proportion to total NADPH pool in proliferating cells [10]. Nevertheless, these studies were not performed under glucose deprivation and it is not clear which of the above pathways are crucial for maintaining adequate NADPH levels in cancer cells when glucose is deprived.
Another prominent feature in TME is lactic acidosis (high lactate concentration with acidic pH), which is caused by Warburg effect and poor tumor vascularization [19]. Researchers begin to appreciate that lactate is not just a waste product, but also a universal fuel [20] or a signaling molecule [21,22]. Lactate is reported to contribute to the TCA cycle in lung cancer [23] and be a primary circulating TCA substrate in tumors [24]. Uptake of lactate through MCT1 in vivo supports the oxPPP and decreases ROS level, leading to an efficient metastasis [25]. We previously reported that lactic acidosis, but not lactosis nor acidosis, confers cancer cells with resistance to glucose-deprivation induced death via inhibiting apoptosis and activating autophagy [26]. Under lactic acidosis, cancer cells also maintain an adequate NADPH level to support cancer cell survival after glucose was exhausted, yet the underlying mechanism was unknown [26].
In this study, we sought to investigate how cancer cells maintain adequate NADPH level under glucose deprivation with or without lactic acidosis. Using Crispr/Cas9 knockout technology and isotopic tracing, we reveal that under glucose deprivation, lactate is used to synthesize isocitrate, which donates hydride ion to NADP+ to produce NADPH by the catalysis of IDH1. On the other hand, glutamine is metabolized to malate in mitochondria, which shuttles to cytosol to generate NADPH by ME1 when glucose and lactate are both depleted. Knockout of IDH1 and ME1 decreases NADPH production, exacerbates oxidative stress, enhances cell necrosis, and retards tumor growth.
2. Materials and methods
2.1. Cell lines and cell culture
Mouse breast cancer cell 4T1 (female), human cervical cancer cell HeLa (female) and human large cell lung cancer cell NCI–H460 (male) were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cell lines were all authenticated by DNA fingerprinting (SNP for 4T1, and STR for HeLa and NCI–H460), and tested for Mycoplasma-free. For regular culture, cells were all maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (Sijiqing, cat. no. 11011–8611), 100 μg/ml penicillin/streptomycin in a humidified incubator at 37 °C in a 5% CO2 atmosphere.
2.2. Specific culture conditions
For nutrition deprivation cultures, cells were washed with PBS (supplemented with 0.4 mM Ca2+ and Mg2+) three times and changed with glucose-free RPMI 1640 medium (Gibco, cat. no. 11879020) with 10% ultra-filtrated FBS (glucose deprivation), or with glucose & glutamine-free RPMI 1640 medium (Caisson Labs, cat. no. RPL10) with 10% ultra-filtrated FBS (glucose and glutamine deprivation). Lactic acidosis condition was created by adding lactic acid into glucose-free RPMI 1640 medium with 10% ultra-filtrated FBS to the final concentration of 20 mM lactate (for 4T1 and HeLa, pH 6.7). In experiments using H460 cells, a final concentration of 10 mM NaOH was also added to achieve an optimized lactic acidosis condition (pH 7.1) for H460. For hypoxia culture, except that the cells were incubated in a tri-gas incubator with 1% oxygen concentration, other conditions were the same as that under normoxia culture. Except survival curves and cell death test experiments, the incubation times under above conditions are determined to ensure the cells were all alive, e.g., as the IDH1 and ME1 knockout 4T1 cells under glucose deprivation started to die after 7 h, the incubation time is within 7 h under this condition.
2.3. Establishment of knockout cells
The knockout cells were established by Crispr-Cas9 system and conducted according to Feng Zhang's protocol [27]. The sequences of designed sgRNA were listed in Supplementary Table 1. We used pSpCas9(BB)-2A-Puro (PX459) V2.0 plasmid (Addgene, cat. no. 62988) as sgRNA expression vector, and the constructed plasmids were transiently transfected into cells using Lipofectamine 3000 (Invitrogen, cat. no. L3000015). The plasmid without insert was also transfected as a knockout control. After selection with 4.5 (for 4T1) or 2 (HeLa and H460) μg/ml puromycin for 48–72 h, cells were allowed to grow to confluence before plating in 96 wells for monoclonal selection. Using limited dilution, we obtained monoclonal cell lines. The microdeletion was confirmed by targeted genomic sequencing. Double knockouts of IDH1 and ME1 were generated from knocking out ME1 in IDH1 knockout cells. The gene knockouts were further confirmed by western blots and enzyme activity determinations.
2.4. Metabolite extraction
Cellular metabolites were extracted using Yuan et al.'s protocol [28] with some modifications. Briefly, after incubation, cultured cells were quickly washed with ice-cold PBS (supplemented with 0.4 mM Ca2+ and Mg2+) twice to remove residual medium. 80% methanol (precooled in −80 °C) was added to the plate wells or dishes (0.6 ml for a well of 6-well plate and 1.5 ml for a 6 cm dish), then the plates or dishes were placed at −80 °C for 20 min. The cells were scraped and the mixture was vortexed for 1 min. The debris was removed by centrifuging at 25,000×g for 5 min at 4 °C, and the supernatant was lyophilized by a freeze dryer. For tumor tissues, 0.6 ml 80% methanol (precooled in −80 °C) was added to tissue piece (~20 mg) and the tissue was grinded for 1–2 min with tissue grinder on dry ice. The tissue sample was vortexed for 1 min and placed at −80 °C for 4 h. The mixture was then centrifuged at 25,000×g for 5 min at 4 °C. The supernatant was collected and the precipitate was extracted again using the same procedure. The supernatants from both extractions were combined, and lyophilized by a freeze dryer. The dried samples were stored at −80 °C before determination (including determining NADPH/NADP+, NAD+/NADH, isotopic labeling, metabolites concentrations and GSH/GSSG).
2.5. NADPH/NADP+and NAD+/NADH determination
NADP and NAD determination were measured according to previous methods [29] with some modifications. During extraction, the centrifuged supernatant was equally divided into two tubes before freeze drying. One tube of the lyophilized sample was dissolved in 0.01 M NaOH (80 μl for 2.5 × 106 4T1 cells) to determine the NADPH and NADH concentration, and the other tube was dissolved in equal volume of 0.01 M HCl to determine NADP+ and NAD+. The tubes were incubated at 60 °C for 15 min and then cooled on ice before determination. For NADPH and NADP+, the assay buffer was 100 mM Tris-HCl (pH 8.1, with 5 mM EDTA) containing 2 mM G6P (Sigma, cat. no. V900924), 1 U/ml G6PDH (Sigma, cat. no. G7877), 10 μl MTS/PES (Promega, cat. no. G3581). For NADH and NAD+, the assay buffer was 100 mM Tris-HCl (pH 8.1, with 5 mM EDTA) containing 5% (v/v) ethanol, 30 U/ml ADH (Sigma, cat. no. A7011), 10 μl MTS/PES. 10 μl sample was added to a well of 96-well plate, and 190 μl assay buffer was added per well. The plate was incubated at 37 °C for 30 min, then the absorbances were read at 490 nm by a microplate reader. Standard curves of a gradient concentrations of NADP and NAD were simultaneously determined, and the concentrations of the samples were calculated according to the standard curves.
2.6. Isotopic tracing
Lactate labeling: cells were cultured in glucose-free medium with 10% ultra-filtrated FBS supplemented with 20 mM [U–13C3]-sodium lactate (Sigma, cat. no. 485926) and 20 mM HCl, or 20 mM [2-2H]-sodium lactate (Sigma, cat. no. 693987) and 20 mM HCl for 6 h (4T1) or 4 h (HeLa).
Glutamine labeling: cells were cultured in glucose & glutamine-free medium with 10% ultra-filtrated FBS supplemented with 2 mM [U–13C5]-glutamine (Sigma, cat. no. 605166) for 6 h (4T1) or 3 h (HeLa).
After incubation, the metabolites were extracted as described above. The extract was dissolved in 80 μl 30% acetonitrile followed by LC-MS/MS analysis. Waters ACQUITY BEH Amide column (2.1 × 100 mm, 1.7 μm) was used on Waters ACQUITY UPLC system to perform liquid chromatography. Mobile phase A: 10 mM ammonium acetate in 85% acetonitrile, 15% water, pH 9.6; Mobile phase B: 10 mM ammonium acetate in 50% acetonitrile, 50% water, pH 9.6. Gradient program: 0–0.4 min, 100% A; 0.4–2 min, 100–30% A; 2–2.5 min, 30–15% A; 2.5–3 min, 15% A; 3–3.1 min, 15–100% A; 3.1–7.5 min, 100% A. Column was kept at 50 °C, injection volume was 7.5 μl, and the flow rate was 0.6 ml/min. The mass spectrometer was an AB SCIEX 4000 QTRAP equipped with an electrospray ionization ion source in multiple reaction monitoring (MRM) mode. All sample analyses except glutamine were performed in negative ion mode. Glutamine was performed in positive ion mode. The m/z, declustering potential (DP), collision energy (CE), entrance potential (EP), and collision cell exit potential (CXP) are listed in Supplementary Table 2. The MRM data were acquired and processed using Analyst 1.5.2 software equipped in the mass spectrometer. The observed mass isotopomer distribution (MID) of target intermediates was calculated by the peak areas of the ion fragmentations. The observed MID data were all corrected for nature isotopic abundance if not specifically indicated.
2.7. Metabolite assays
For cultured cells, cell numbers (N) and the average cell diameter (D) were measured by a flow cell counter before extraction. Then the total cell volume (V) was estimated by 4/3 × π × (D/2)3 × N. For tumor tissue, the tissue weight M (mg) was recorded and the total liquid volume in tumor tissue is M × 0.83 (μl) (The water content in solid tumor is about 0.83 (w/w) [30]). The volume was used to calculate the cellular concentrations of metabolites. After extraction, the lyophilized extract was dissolved in 50–100 μl water. The metabolites concentrations were measured according to methods described previously [31] with some modifications. For each assay, a parallel group that contained the same ingredients but a key enzyme was included to correct the background that caused by non-enzymatic reactions.
Assay of isocitrate: 10 μl sample was added to a 1.5 ml tube containing 90 μl reaction buffer (50 mM Tris-HCl, pH 8.1, with 100 μM MnCl2, 0.01% BSA, 300 μM NADP+ (Sigma, cat. no. N5755) and 0.4 U/ml isocitrate dehydrogenase (Sigma, cat. no. I2002)). The tube was incubated at room temperature for 15 min, then 10 μl 2 mol/L NaOH was added and the tube was incubated at 60 °C for 15 min to degrade NADP+. The concentration of formed NADPH (equal to isocitrate concentration) in the reaction was measured by the methods described above. Then the intracellular concentration of isocitrate was calculated. The percentage of m+2 isotopologue was determined in isotopic labeling experiment, thus the concentration of isocitrate (m+2) was calculated.
Assay of α-ketoglutarate: 10 μl sample was added to a 1.5 ml tube containing 90 μl reaction buffer (20 mM imidazole base, 20 mM imidazole acetate, 25 mM ammonium acetate, with 100 μM ADP (Sigma, cat. no. A5285), 100 μM NADH (Sigma, cat. no. N8129) and 0.15 U/ml glutamate dehydrogenase (Sigma, cat. no. G2501)). The tube was incubated at room temperature for 15 min, then 10 μl 2 mol/L HCl was added and the tube was incubated at 60 °C for 15 min to degrade NADH. The concentration of formed NAD+ (equal to α-KG concentration) in the reaction was measured by the methods described above. Then the intracellular concentration of α-KG was calculated. The percentage of m+2 isotopologue was determined in isotopic labeling experiment, thus the concentration of α-KG (m+2) was calculated.
Assay of malate: 10 μl sample was added to 90 μl reaction buffer (100 mM 2-Amino-2-methyl-1-propanol (AMP) buffer, pH 9.9, with 40 mM glutamate (Sigma, cat. no. G8415), 2 mM NAD+ (Sigma, cat. no. N0632), 3.5 U/ml malate dehydrogenase (Sigma, cat. no. M2634) and 0.9 U/ml aspartate transaminase (Sigma, cat. no. G2751)). The tube was incubated at room temperature for 15 min, then 10 μl 2 mol/L NaOH was added and the tube was incubated at 60 °C for 15 min to degrade NAD+. The concentration of formed NADH (equal to malate concentration) in the reaction was measured by the methods described above. Then the intracellular concentration of malate was calculated. The percentage of m+4 isotopologue was determined in isotopic labeling experiment, thus the concentration of malate (m+4) was calculated.
Assay of glucose: 30 μl sample was added to 600 μl reaction buffer (100 mM Tris-HCl, pH 8.1, with 1 mM MgCl2, 500 μM NAD+, and 500 μM ATP (Sigma, cat. no. A3377)). 0.5 U/ml G6PDH (Sigma, cat. no. G5760) was added to start the reaction and OD340 was read by a spectrophotometer to measure G6P concentration. Afterwards 0.5 U/ml hexokinase (Sigma, cat. no. H4502) was added, and the mixture was incubated for 10 min, then OD340 was read by a spectrophotometer to measure glucose concentration.
Assay of lactate: 30 μl sample was added to 600 μl reaction buffer (200 mM glycine, 170 mM hydrazine (Sigma, cat. no. H4766), pH 9.2, with 2 mM NAD+ and 10 U/ml LDH (Sigma, cat. no. L2500)). The mixture was incubated at room temperature for 1 h, and OD340 was read by a spectrophotometer to measure lactate concentration.
Assay of glutamine plus glutamate: 10 μl sample was added to 50 μl glutamine reaction buffer (50 mM acetate buffer, pH 4.9, with 0.2 mM EDTA and 1 U/ml glutaminase (Sigma, cat. no. G8880)). The mixture was incubated for 1 h at room temperature. Then 540 μl glutamate reaction buffer (50 mM Tris-HCl buffer, pH 8.4, with 400 μM NAD+, 100 μM ADP and 2 U/ml glutamate dehydrogenase (Sigma, cat. no. G2501)) was added and OD340 was read by a spectrophotometer after incubation of 30 min at room temperature. The results measured glutamine plus glutamate concentrations.
2.8. GSH/GSSG measurement
GSH/GSSG was measured according to the method described by Rahman et al. [32] with some modifications. After extraction, the lyophilized sample was dissolved in 400 μl 0.1 M KPE buffer, then equally divided into two tubes. 4 μl 2-vinylpyridine (Sigma, cat. no. 132292) (1:10 diluted) was added in one tube (tube A) to derivatize GSH and the reaction was taken place for 1 h at room temperature in a fume hood. Then 12 μl triethanolamine (Sigma, cat. no. T1377) (1:6 diluted) was added to tube A to neutralization for 10 min. Tube B was kept on ice before determination. 2 mg DTNB (Sigma, cat. no. D8130) in 3 ml KPE, 2 mg NADPH (Sigma, cat. no. N7505) in 3 ml KPE, and 40 μl GR (Sigma, cat. no. G3664) (250 U/ml) in 3 ml KPE were prepared fresh. 20 μl sample/blank/standard were added to wells of 96-well plate, then equal volumes of freshly prepared DTNB and GR solutions were mixed together and 120 μl mixture were added to each well. After 30 s, 60 μl NADPH was added and well mixed, then OD412 were immediately read in a microplate reader and measurements were taken every 30 s for 2 min. The change rate of the absorbance indicated the concentration of GSH or GSSG. Thus, the concentrations of the samples were calculated by standard curves. Tube A represented GSSG concentration while tube B represented total [GSH] ([GSH] + 2 × [GSSG]) concentration. Then GSH/GSSG was calculated.
2.9. Enzymatic activity assays
Cells (at least 106) were harvested by treating with trypsin-EDTA at 37 °C for 2–5 min. The digestion was stopped by adding complete culture medium and the medium was removed by centrifuging at 200×g for 5 min. The cell pellet was washed with PBS, then lysed with mammalian protein extraction reagent (Thermo fisher, cat. no. 78501) (100 μl/106 cells) containing protease inhibitor cocktail (Thermo fisher, cat. no. 78437) on ice for 20 min. The debris was removed by centrifuging at 25,000×g for 5 min at 4 °C, and the supernatant was kept on ice before determination. The protein concentration of the cell lysate was measured by BCA Protein Assay Kit (Thermo fisher, cat. no. 23225). The enzyme activity was measured by a spectrophotometer and the experiments were all performed at 37 °C according to methods described previously [31]:
For IDH activity determination: 40 μl lysate was added to 600 μl reaction buffer (50 mM Tris-HCl, pH 8.1, with 100 μM MnCl2, 1 mM isocitrate (Sigma, cat. no. I1252) and 1 mM NADP+). The absorbance at 340 nm was read every 10 s for 2 min. The activity was calculated from the linear part of the curve.
For ME activity determination: 40 μl lysate was added to 1 ml reaction buffer (50 mM Tris-HCl, pH 7.8, with 50 μM MnCl2, 5 mM malate (Sigma, cat. no. 02288) and 100 μM NADP+). The absorbance at 340 nm was read every 5 min for 30 min. The activity was calculated from the linear part of the curve.
For MDH activity determination: 40 μl lysate was added to 1 ml reaction buffer (50 mM Tris-HCl, pH 8.1, with 200 μM oxaloacetate (Sigma, cat. no. O7753) and 200 μM NADH). The absorbance at 340 nm was read every 10 s for 1 min. The activity was calculated from the linear part of the curve.
For LDH activity determination: 10 μl lysate was added to 1 ml reaction buffer (1 × PBS, with 1 mM pyruvate (Sigma, cat. no. 107360) and 200 μM NADH). The absorbance at 340 nm was read every 10 s for 1 min. The activity was calculated from the linear part of the curve.
2.10. Immunoblotting
The method to prepare cell lysate was the same as in 2.9. The lysates were diluted by 5 × loading buffer (Thermo fisher, cat. no. 39001) followed by boiling for 10 min. 50 μg protein was loaded onto the 10% SDS-PAGE gels and was separated. The proteins were transferred to polyvinylidene difluoride (PVDF) membranes and the membrane was blocked with 5% non-fat milk (w/v) at room temperature for 1 h followed with primary antibody incubation overnight at 4 °C. The primary antibodies used here include: anti-IDH1 (1:1000, Abcam, cat. no. ab172964), anti-ME1 (1:2000, Abcam, cat. no. ab97445), anti-MDH1 (1:10000, Abcam, cat. no. ab180152), anti-Visfatin (1:1000, Abcam, cat. no. ab236874), anti-LDHA (1:1000, Cell Signal Technology, cat. no. 2012), anti-LDHB (1:200, Santa Cruz Biotechnology, cat. no. sc-100775), anti-beta actin (1:10000, Abcam, cat. no. ab8226), anti-GAPDH (1:10000, Proteintech, cat. no. 60004-1-Ig). The membrane was washed with TBST for three times, and then incubated with HRP-conjugated secondary antibody (1:10000, BBI, cat. no. D110058 and D110087) at room temperature for 1 h. The membrane was washed with TBST for three times and protein bands were detected by adding ECL (PerkinElmer, cat. no. NEL103001EA) and captured on a Chemiluminescent Imaging System (Tanon, Shanghai, China).
2.11. Redox state determination of thioredoxin 1 and peroxiredoxin 1
For redox state determination assay, control and knockout cells were incubated under glucose deprivation with or without lactic acidosis. Control cells cultured in medium containing DTT (500 μM) or H2O2 (200 μM) were also included in the experiments as negative and positive controls.
The redox state of Trx1 was measured according to the method described by Y.M. Go et al. [33] with some modification. After treatment, the medium was aspirated and cells were washed by PBS three times. 200 μl lysis buffer (20 mM Tris, 150 mM NaCl, 1% Triton X-100, 50 mM iodoacetic acid, 1 × proteases inhibitor cocktail, pH 7.5) was added to a well of a 6-well plate. The cells were scraped, collected into a 1.5 ml tube, and were lysed by pipetting up and down until the suspension was clear. The lysate was incubated at 37 °C for 30 min in the dark. The cell debris was removed by centrifuging at 25,000×g for 5 min at 4 °C. The protein concentrations of the derivatized samples were determined using BCA assay, and the samples were mixed with 5 × native and non-reducing loading buffer. 25 μg proteins were loaded onto a 15% native PAGE gel and were electrophoresed in 1 × native running buffer at 150 V for 80 min. The proteins were transferred to PVDF membrane in 1 × native transfer buffer at 125 W for 2 h. The blocking and antibody incubation procedures were the same with that in 2.10. The primary antibody was anti-Trx1 (1:10000, Abcam, cat. no. ab109385). GAPDH is difficult to be transferred from the gel to the membrane as it charged little without SDS. Therefore, GAPDH was analyzed by regular SDS-PAGE instead.
The redox state of Prx1 was measured according to the method described by A.G. Cox et al. [34] with some modification. After treatment, the medium was aspirated and cells were washed by PBS three times. 200 μl alkylation buffer (40 mM HEPES, 50 mM NaCl, 1 mM EGTA, 100 mM N-Ethylmaleimide, 1 × proteases inhibitor cocktail, pH 7.4) was added to a well of a 6-well plate. The plate was incubated at room temperature for 10 min. Then 15 μl 15% CHAPS was added to a well and the cells were lysed at room temperature for 10 min. The cell debris were removed by centrifuging at 25,000×g for 5 min at 4 °C. The protein concentrations of the samples were determined using BCA assay, and the samples were mixed with 5 × non-reducing loading buffer. 25 μg proteins were loaded onto a 15% SDS-PAGE gel. The SDS-PAGE and immunoblotting procedures were the same with that in 2.10. The primary antibody was anti-Prx1 (1:10000, Abcam, cat. no. ab109498).
2.12. ROS level measurement
Cellular ROS level was measured by DCFH-DA (Sigma, cat. no. D6883) according to manufactures’ instruction. In brief, after a period of time of nutrition deprivation, cells were loaded with 10 μM DCFH-DA for 30 min (the deprivation condition was not changed at the time). Cells were washed with ice-cold PBS (supplemented with 0.4 mM Ca2+ and Mg2+) three times and were immediately observed under a Zeiss LSM710 laser confocal microscope. The images were photographed by the equipped Zen software.
2.13. Flow cytometric analysis of cell necrosis and apoptosis
Cells were harvested by accutase digestion, and centrifuged at 200×g for 5 min. The cells were washed with PBS and stained using Annexin V-FITC and Propidium iodide (PI) solution (Beyotime, cat. no. C1062L) for 10 min at room temperature in the dark. The percentage of necrotic and apoptotic cells for each sample was immediately measured by Beckman Coulter DxFLEX flow cytometer.
2.14. Cell survival curves and doubling times determination
In survival curves experiments, cells were seeded into 96-well plates to allow attachment overnight (70–80% confluence the next day). The medium was replaced with corresponding nutrition-deprived medium. Cell numbers were counted by CCK-8 assay (Boster, cat. no. AR1160) at indicated times. To determine doubling times, 2 × 103 cells were seeded into 96-well plates to allow attachment overnight. Cell numbers were counted every day (including day 0) by CCK-8 assay until 72 h. CCK-8 was used according to manufactures’ instruction. Briefly, the medium was changed with 100 μl fresh medium containing 10% CCK-8, and the plate was incubated at 37 °C for 30 min. OD450 were read in a microplate reader. Standard curves were created by plotting cell numbers versus OD450. The exact cell numbers were calculated by standard curves.
2.15. In vivo study
Balb/c mice (female, 6 weeks old) were obtained from SLAC Laboratory animal Co., Ltd, Shanghai, China, and maintained at the Laboratory Animal Research Center of Zhejiang Chinese Medical University. Mice were housed in a standard polypropylene cage containing sterile bedding with free access to standard rodent chow and water under a controlled condition of temperature (25 °C), humidity (50%), and light (10 and 14 h of light and dark). All the animal study were performed in accordance with the institutional guidelines and regulations, and were approved by the Animal Ethical Committee of Zhejiang Chinese Medical University (ethical agreement number: ZSLL-2017-125).
For syngeneic xenograft tumor model, 4T1 control, IDH1 knockout, ME1 knockout and IDH1&ME1 knockout cells were harvested by Trypsin-EDTA, washed by PBS twice, and temporarily (less than 0.5 h) resuspend in RPMI-1640 without FBS before inoculation. The final cell concentration was 106/ml. 100 μl cell suspension was inoculated in the right second mammary pad area of each Balb/c mouse (6 mice were randomly assigned to the same group). The tumor sizes were measured by vernier calipers every few days, and tumor volumes were estimated by the formula: V = (L × W2) × 0.5, where L is length and W is width. On day 28, mice were sacrificed and tumors were dissected, weighted and photographed. Halves of the tumors were stored in liquid nitrogen for metabolites determination, and the other halves were stored in formalin. The lung metastasis nodules were assessed by counting macroscopic metastatic nodules based on previous report [35].
For survival analysis study, 5 × 104 (suspended in 100 μl RPMI-1640 without FBS) 4T1 control, IDH1 knockout, ME1 knockout, and IDH1&ME1 knockout cells were inoculated in the right second mammary pad area of each Balb/c mouse, respectively (10 mice were randomly assigned to the same group). The mice were observed every day after 28 days and the death events were recorded until 90 days.
2.16. Quantification and statistical analysis
All statistical analyses were performed using GraphPad Prism 8.0. The statistical parameters (i.e., exact value of n, what n represents, representation of error bars, p values) were all indicated in each Fig. legend. The statistical significance of the difference between two groups was analyzed using unpaired two-tailed Student's t-test, and Kaplan Meier curves for survival were analyzed using log-rank test. p < 0.05 was considered as statistically significant.
3. Results
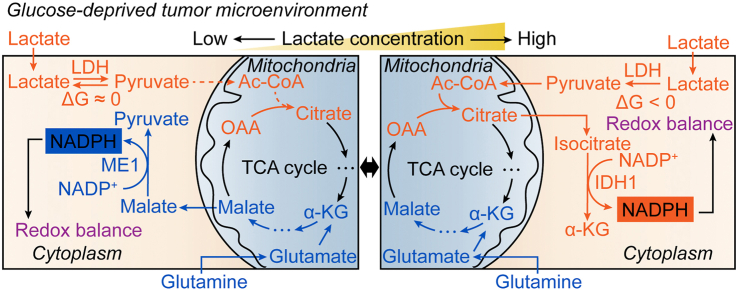
3.1. Lactate supports NADPH generation by IDH1 rather than ME1 under glucose-deprived condition with lactic acidosis
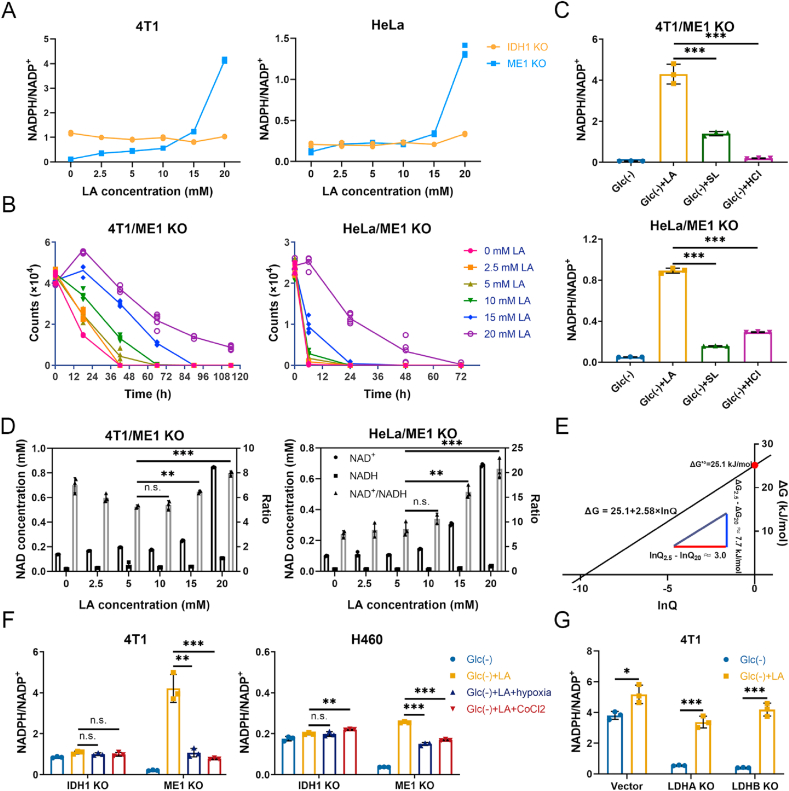
To investigate the glucose-independent NADPH producing pathways, we used glucose-free RPMI-1640 medium supplemented with 10% ultra-filtrated FBS to build a glucose-deprived environment. We found that the tolerances of cancer cell lines to glucose deprivation are different. To be more representative, we chose three cancer cell lines whose tolerances were different from each other (Supplementary Fig. 1A). The cellular NADPH/NADP+ ratios of the three cells were all declined over time under glucose deprivation (Supplementary Fig. 1B), and the decreases showed statistical significance in hours (Fig. 1A), indicating a major contribution of glucose to NADPH production. Lactic acidosis partially recovered the NADPH/NADP+ ratios of the three cancer cells (Fig. 1A), suggesting that lactate is involved in maintaining NADPH homeostasis when glucose is absent.
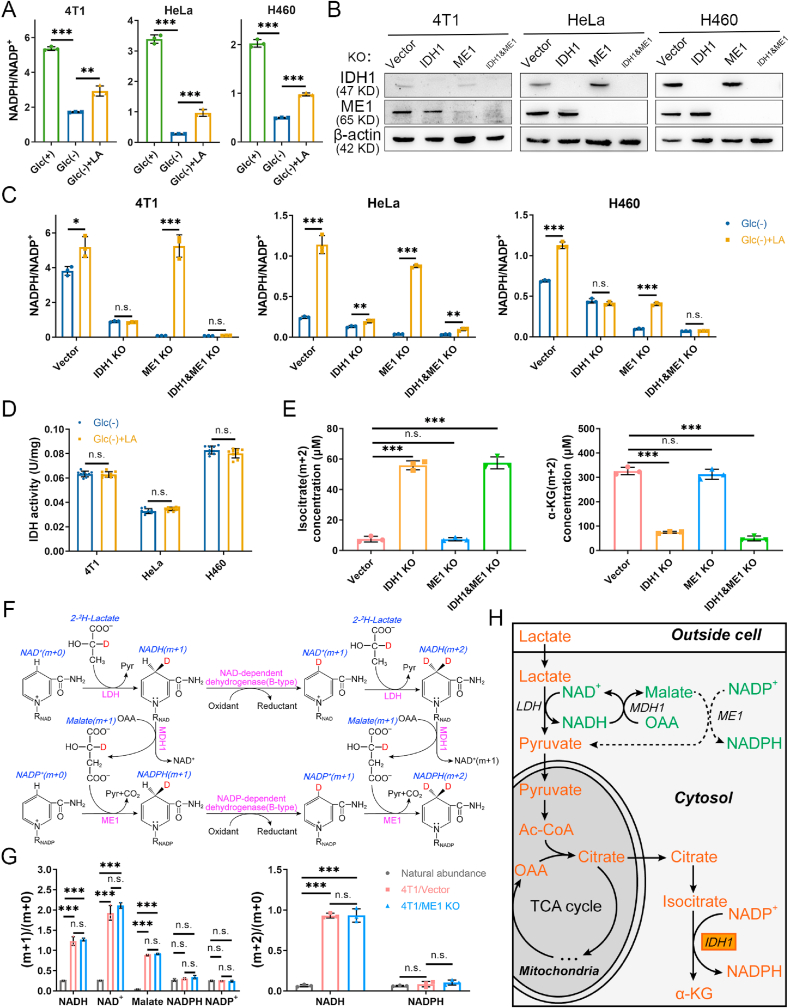
Fig. 1.
Lactate supports NADPH generation by IDH1 rather than ME1 under glucose-deprived condition with lactic acidosis. (A) Cellular NADPH/NADP+ ratios of three cancer cell lines under three different conditions. Incubation times were 9 h (4T1), 3 h (HeLa) and 6 h (H460), respectively. (B) Confirmation of IDH1 knockout, ME1 knockout, and double gene knockout in three cancer cell lines by immunoblot. The cells transfected with empty px459 vector were used as knockout controls. (C) The effect of lactic acidosis on NADPH/NADP+ ratios of IDH1 or/and ME1 knockout cancer cells under glucose-deprived condition. Incubation times were 6 h (4T1), 3 h (HeLa) and 4 h (H460), respectively. (D) The effect of lactic acidosis on total IDH activities of the three cancer cells. (E) Concentrations of isocitrate (m+2) and α-KG (m+2) after labeling with 20 mM [U–13C3]-lactate for 6 h in 4T1 knockout cells. (F) Theoretical pathways to transfer electron pairs from C-2 of lactate to the nicotinamide ring of NADPH. Except the nicotinamide ring, the remaining moiety of NAD(P) is denoted by RNAD(P). (G) The m+1 (2)/m+0 ratios of the intermediates in the electron transfer pathways after labeling with 20 mM [2-2H]-lactate for 6 h in 4T1 cells. The labeling data were not corrected for natural isotopic abundance. (H) Schematic diagram to illustrate NADPH maintenance by lactate under glucose-deprived conditions. The fluxes indicated by dash lines are marginal. Glc (+): Complete RPMI-1640 medium; Glc (−): Glucose-free RPMI-1640 medium; Glc (+) + LA: Glucose-free RPMI-1640 medium supplemented with 20 mM lactic acid (4T1 and HeLa, pH 6.7) or with 20 mM lactic acid and 10 mM NaOH (H460, pH 7.1). Data are shown as means ± SD, with n = 3 biological replicates in (A), (C), (E) and (G), and the results were all confirmed by three independent experiments; Data are means ± SD, from three independent experiments performed in triplicates in (D). n. s. not significant, *p < 0.05, **p < 0.01, ***p < 0.001 (Student's test). See also Supplementary Figs. 1–3.
We next explored the mechanism by which lactate raises NADPH/NADP+ ratios under glucose deprivation. Lactate could convert to glucose via gluconeogenesis in some cancer cells [36], and the lactate-derived glucose may shunt to oxPPP and generate NADPH. As such, we first checked whether gluconeogenesis is active in the three cell lines or not. We used [U–13C3]-lactate as labeling substrate, however, [13C3] or [13C6]-glucose was not detected (Supplementary Fig. 1C). Hence, gluconeogenesis is excluded. Apart from oxPPP, isocitrate dehydrogenase 1 (IDH1) and malic enzyme 1 (ME1) are the main cytosolic NADPH producing enzymes. We then established IDH1 or ME1 or double gene knockout cell lines. The gene knockout cell lines were confirmed by immunoblot (Fig. 1B) and enzyme activity determination (Supplementary Fig. 1D and E). The gene knockout cell lines showed similar doubling times to control cells under regular culture conditions (Supplementary Fig. 2A). The effects of IDH1 or ME1 knockout on the NADPH/NADP+ ratios under regular culture showed difference in three cell lines (changes were statistically significant in 4T1 and HeLa cells but not in H460 cells), yet the three double gene knockout cells all exhibited a significant decrease of the NADPH/NADP+ ratios (Supplementary Fig. 2B), indicating a combinational contribution of IDH1 and ME1 to cellular NADPH pool. However, the decline of NADPH/NADP+ ratio caused by IDH1 and ME1 knockouts was much smaller than that caused by glucose deprivation (Supplementary Fig. 2C), indicating that the contribution of IDH1 and ME1 to NADPH pool in the presence of glucose is much smaller than the glucose-derived NADPH. Therefore, in the presence of glucose, though IDH1 and ME1 make an evident contribution to total NADPH pool (statistical significance in NADPH/NADP+ between control and double knockout cells), their contribution is minor (the decline in NADPH/NADP+ was relatively small compared to the total NADPH pool). Furthermore, the unabated growth rate of double gene knockout cells (Supplementary Fig. 2A) suggested that the loss of IDH1&ME1-dependent NADPH formation do not affect cell growth under regular culture. Thus, IDH1 and ME1 are not indispensable when glucose is present.
We next tested the effect of IDH1 or ME1 knockout on lactate-related NADPH production in cells under glucose deprivation. The NADPH/NADP+ in knockout cells were significantly decreased compared to the control cells under glucose starvation (Fig. 1C). When lactate was added, in 4T1 and H460 cells, the NADPH/NADP+ in control and ME1 knockout cells but not IDH1 or double gene knockout cells were markedly increased (Fig. 1C). In HeLa cells, NADPH/NADP+ in IDH1 or double gene knockout cells was also increased, yet the increments were much smaller than those in control and ME1 knockout cells. The results suggested that IDH1 but not ME1 is essential in the lactate-related NADPH production.
To investigate the relationship between lactate and IDH1 and its relevance with NADPH, we did the following experiments. First, lactate may upregulate IDH1, because lactate could be a signaling molecule [21]. However, we found that IDH activity and IDH1 protein expression did not change significantly under lactic acidosis (Fig. 1D and Supplementary Fig. 3A). Second, as lactate is a carbon source for the TCA cycle under glucose deprivation [37], it could be used to synthesize isocitrate. To verify this, we used [U–13C3]-lactate to trace the metabolic fate of lactate. Theoretically, m + 2 isotopologue of the intermediates is the product generated in the first TCA cycle (Supplementary Fig. 3B), and m+2 isocitrate was the most abundant in a short time of labeling (Supplementary Fig. 3C, upper panel). The concentrations of isocitrate (m+2) in IDH1 knockout and double gene knockout cells were significantly increased, while the concentrations of α-KG (m+2) were significantly reduced (Fig. 1E and Supplementary Fig. 3D), indicating an accumulation of lactate-carbon-incorporated isocitrate and blockage of oxidative decarboxylation of isocitrate with concomitant NADPH generation. The results confirmed that cancer cells use lactate carbon to synthesize isocitrate, which was used to generate NADPH by IDH1 under glucose-deprived condition with lactic acidosis.
On the other hand, why ME1 is independent of the lactate-involved NADPH production can be explained by [2-2H]-lactate labeling experiment (Fig. 1F). Theoretically, the deuterium at C-2 of lactate can be transferred to the nicotinamide ring of NAD+ to form NADH (m + 1) in the LDH-catalyzed reaction. Then the deuterium can be transferred to oxaloacetate to form malate (m + 1) via MDH1 or retained in NAD+ (m + 1) in the B-type dehydrogenase-catalyzed reactions. The deuterium leaves malate (m + 1) and is received by NADP+ to form NADPH (m +1) via ME1. NAD+ (m + 1) can receive another deuterium to form NADH (m + 2). NADPH (m + 2) can be formed in a similar way. The results showed that NADH, NAD+ and malate were all sufficiently labeled (Fig. 1G). However, the ratios of labeled NADPH to unlabeled ones ((m + 1)/(m + 0) and (m + 2)/(m + 0)) showed no significant increase compared to that of natural abundance (Fig. 1G). For NADP+, the value of (m + 1)/(m + 0) was almost equal to that of natural abundance. ME1 knockout had no significant effect on the labeling result (Fig. 1G). The rapid decrease of deuterium labeling from malate to NADPH implied that most deuterium from malate did not transfer directly to NADP+ or the transfer was diluted by other NADPH-producing fluxes (e.g., IDH1), hence the flux of ME1-catalyzed reaction was negligible compared to the total NADPH-producing flux under lactic acidosis and ME1 knockout had little effect to the rescue of NADPH by lactate. Taken together, we revealed the biochemical pathway of NADPH generation from lactate in cancer cells under glucose deprivation (Fig. 1H).
3.2. Glutamine supports NADPH generation by ME1 via reverse malate-shuttle under glucose-deprived condition without lactic acidosis
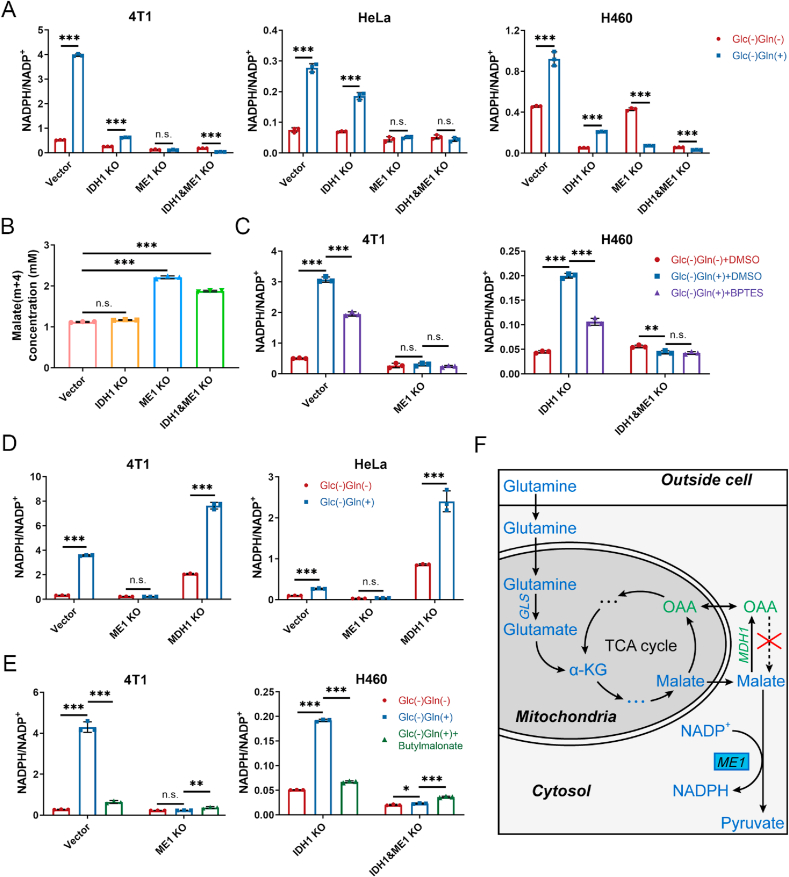
Although ME1 did not play a role in lactate-involved NADPH pathway, it was important for cancer cells when glucose and lactate were both depleted. Under this condition, the NADPH/NADP+ ratio of ME1 knockout cells decreased by 6–40 folds as compared with control cells (Fig. 1C). IDH1 knockout cells also showed a similar tendency, but not as marked as ME1 (Fig. 1C). Double gene knockout exerted an additive effect on NADPH/NADP+ decrease (Fig. 1C). The results indicated that ME1 plays a crucial role in maintaining cellular NADPH when cancer cells are deprived of glucose.
It has been reported that ME1-dependent NADPH production is associated with glutamine in pancreatic cancer [38]. To check whether it is the case here, we cultured the gene knockout cells in glucose and glutamine double free RPMI-1640 medium. When glutamine was replenished, control cells showed a marked increase of NADPH/NADP+ (Fig. 2A), IDH1 knockout cells showed a relatively moderate increase of NADPH/NADP+ (Fig. 2A), but the NADPH/NADP+ ratios of ME1 knockout cells were not changed (4T1 and HeLa) or even decreased (H460) (Fig. 2A). Therefore, ME1 is tightly associated with glutamine-related NADPH generation. To check whether glutamine directly participates in the formation of malate, we performed [U–13C5]-glutamine tracing experiment. M + 4 isotopologue of malate is produced from glutamine in the first cycle of TCA (Supplementary Fig. 3B), and it was the most abundant isotopologue (Supplementary Fig. 3C, lower panel). The concentrations of malate (m + 4) were significantly increased in the ME1 knockout cells (Fig. 2B and Supplementary Fig. 3E), indicating that malate converting to pyruvate (NADP+ reducing to NADPH) via ME1 was blocked. To further confirm this, we used glutaminase inhibitor BPTES (bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide) to test its effect on glutamine-involved NADPH recovery. BPTES significantly inhibited NADPH recovery in control and IDH1 knockout cells, but had no effect on ME1 knockout and double gene knockout cells (Fig. 2C).
Fig. 2.
Glutamine supports NADPH generation by ME1 via reverse malate-shuttle pathway under glucose-deprived condition without lactic acidosis. (A) Effect of supplementing glutamine (2 mM) on cellular NADPH/NADP+ ratios under glucose & glutamine-deprived condition in control, IDH1 knockout, ME1 knockout, and IDH1&ME1 knockout cancer cells. (B) Concentrations of malate (m + 4) after labeling with 2 mM [U–13C5]-glutamine for 6 h in 4T1 control, IDH1 knockout, ME1 knockout, and IDH1&ME1 knockout cells. (C) Effect of glutaminase inhibitor BPTES (100 μM) on cellular NADPH/NADP+ ratios under glucose-deprived condition in control or ME1 knockout cells (4T1) and IDH1 or IDH1&ME1 knockout cells (H460). As BPTES is dissolved in DMSO, DMSO is also set as a control. (D) Effect of supplementing glutamine (2 mM) on cellular NADPH/NADP+ ratios under glucose & glutamine-deprived condition in control, ME1 knockout and MDH1 knockout cancer cells. (E) Effect of mitochondrial malate transporter inhibitor butylmalonate (5 mM for 4T1 and 2.5 mM for H460) on cellular NADPH/NADP+ ratios under glucose-deprived condition in control or ME1 knockout cells (4T1) and IDH1 or IDH1&ME1 knockout cells (H460). (F) Schematic diagram to illustrate NADPH maintenance by glutamine under glucose-deprived condition without lactic acidosis. The red “ × ” refers the net flux of the MDH1-catalyzed reactions is from malate to oxaloacetate. Glc (−) Gln (−): Glucose and glutamine-free RPMI-1640 medium; Glc (−) Gln (+): Glucose and glutamine-free RPMI-1640 medium supplemented with 2 mM glutamine. Incubation times were 6 h (4T1), 3 h (HeLa) and 4 h (H460), respectively. Data are shown as means ± SD, with n = 3 biological replicates, and the results were all confirmed by three independent experiments. n. s. not significant, *p < 0.05, **p < 0.01, ***p < 0.001 (Student's test). See also Supplementary Fig. 3. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Previous studies demonstrated that glutamine-derived oxaloacetate is reduced to malate by MDH1 in cytoplasm and then produces NADPH by ME1 [[38], [39], [40]]. However, these experiments were not performed under glucose-deprived condition. We doubt whether there is enough NADH to reduce oxaloacetate in cytosol when glucose is deprived and glycolysis is abrogated. To test our assumption, we established MDH1 knockout cell lines. The knockout was confirmed by immunoblot (Supplementary Fig. 3F) and MDH activity determination (Supplementary Fig. 3G). If the ME1-dependent NADPH production is coupled with MDH1 reaction, MDH1 knockout should exert a similar effect to ME1 knockout on NADPH/NADP+ when glutamine is replenished. However, unlike ME1 knockout, MDH1 knockout significantly increased NADPH/NADP+ ratio (Fig. 2D). To our surprise, the increase of NADPH/NADP+ in MDH1 knockout cells are even larger than that of control cells (Fig. 2D), suggesting that the direction of MDH1 reaction is from malate to oxaloacetate under this condition. The results hence indicted that malate, the substrate of ME1, is not from the reduction of oxaloacetate by MDH1 in cytosol. We speculated that malate is directly transported from mitochondria to cytoplasm. Butylmalonate is used as a mitochondrial malate transporter inhibitor [41]. When butylmalonate was added, the glutamine-involved NADPH/NADP+ increments were impaired in control and IDH1 knockout cells (Fig. 2E). Taken together, when glucose is deprived, glutamine is converted to malate in a chain of reactions involving glutaminase, transaminase or glutamate dehydrogenase and the TCA cycle, then malate is shuttled from mitochondria to cytoplasm, where it is catalyzed by ME1 to produce NADPH (Fig. 2F).
3.3. IDH1 and ME1 play a key role in maintaining redox balance and resisting cell death under glucose-deprivation with or without lactic acidosis
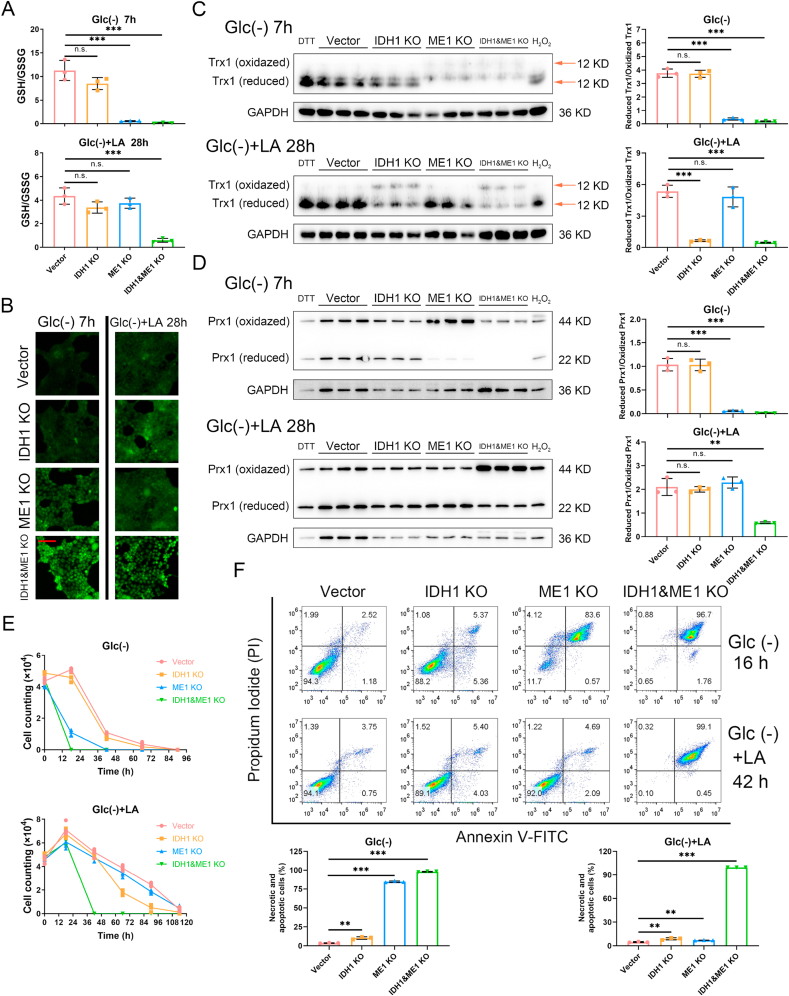
NADPH is essential to keep cellular redox balance. To check the importance of IDH1 and ME1 in redox homeostasis, we determined the GSH/GSSG ratio, ROS level and the redox states of antioxidant proteins (thioredoxin 1 and peroxiredoxin 1) in control and the gene knockout cells under glucose-deprivation with or without lactic acidosis. The GSH/GSSG ratio decreased significantly in 4T1 ME1 knockout and double gene knockout cells 7 h after glucose deprivation without lactic acidosis (Fig. 3A, upper panel). Under glucose deprivation with lactic acidosis, the GSH/GSSG ratio was significantly decreased in the double gene knockout cells after 28 h, while the ratio showed a minor and not statistically significant decrease in IDH1 knockout cells (Fig. 3A, lower panel). Similar results were obtained in HeLa (Supplementary Fig. 4A) and H460 (Supplementary Fig. 5A) cells.
Fig. 3.
IDH1 and ME1 play a key role in maintaining redox balance and resisting cell death in 4T1 cells under glucose-deprivation with or without lactic acidosis
(A) Cellular GSH/GSSG ratios of 4T1 control, IDH1 knockout, ME1 knockout and IDH1&ME1 knockout cells under glucose deprivation with or without lactic acidosis. (B) Cellular ROS levels of 4T1 control, IDH1 knockout, ME1 knockout and IDH1&ME1 knockout cells under glucose deprivation with or without lactic acidosis. ROS was probed by DCFH-DA. Bar = 25 μm. (C) The redox state of thioredoxin 1 (Trx1) in 4T1 control, IDH1 knockout, ME1 knockout and IDH1&ME1 knockout cells under glucose deprivation with or without lactic acidosis. The reduced form of Trx1 was derivatized with IAA, hence it had more negative charge than oxidized form and moved faster in the native PAGE. The reduced form/oxidized form ratios were calculated based on the gray values of the bands. (D) The redox state of peroxiredoxin 1 (Prx1) in 4T1 control, IDH1 knockout, ME1 knockout and IDH1&ME1 knockout cells under glucose deprivation with or without lactic acidosis. The reduced form of Prx1 is monomer and the oxidized form is dimer, hence they can be separated by non-reducing SDS-PAGE. The reduced form/oxidized form ratios were calculated based on the gray values of the bands.(E) Survival curves of 4T1 control, IDH1 knockout, ME1 knockout and IDH1&ME1 knockout cells under glucose deprivation with or without lactic acidosis. (F) The necrosis and apoptosis rates of 4T1 control, IDH1 knockout, ME1 knockout and IDH1&ME1 knockout cells under glucose deprivation with or without lactic acidosis. PI (+), Annexin V-FITC(+): necrosis or late apoptosis; PI (−), Annexin V-FITC(+): early apoptosis. Glc (−): Glucose-free RPMI-1640 medium; Glc (+) + LA: Glucose-free RPMI-1640 medium supplemented with 20 mM lactic acid, pH 6.7. Data are shown as means ± SD, with n = 3 biological replicates in (A) (C) (D) (F), n = 5 biological replicates in (E), and the results were all confirmed by three independent experiments. n. s. not significant, **p < 0.01, ***p < 0.001 (Student's test). See also Supplementary Figs. 4 and 5.
The ROS level of control, IDH1 knockout, ME1 knockout, and double gene knockout cells exhibited a stepwise increment under glucose deprivation without lactic acidosis (Fig. 3B, Supplementary Figs. 4B and 5B, left panel), indicating that the antioxidative activity of ME1 is larger than that of IDH1 and the effect of ME1 and IDH1 on ROS is additive. Under glucose deprivation with lactic acidosis, ROS was markedly and moderately increased in the double gene knockout and IDH1 knockout cells, respectively (Fig. 3B, Supplementary Figs. 4B and 5B, right panel).
The reduced form of thioredoxin 1 (Trx1) were decreased, while the oxidized form of Trx1 were increased in ME1 knockout and double gene knockout 4T1 cells under glucose deprivation without lactic acidosis (Fig. 3C, upper panel). Under glucose deprivation with lactic acidosis, Trx1 in IDH1 knockout and double gene knockout 4T1 cells showed an increase in oxidized form but a decrease in reduced form (Fig. 3C, lower panel). The variation tendencies of Prx1 in the knockout 4T1 cells were virtually the same with that of Trx1, except that IDH1 knockout did not affect the redox state of Prx1 under glucose deprivation with lactic acidosis (Fig. 3D). Similar results were also obtained in HeLa (Supplementary Fig. 4C and D) and H460 cells (Supplementary Fig. 5C and D). Altogether, IDH1 and ME1 play an important role in maintaining redox balance under glucose deprivation with and without lactic acidosis, respectively, and the combination of IDH1 and ME1 is more crucial for redox balance under both conditions.
Disrupted redox balance would threaten the survival of cancer cells. When we kept the cells under glucose deprivation with or without lactic acidosis, death occurred and the survival curves differed among the gene knockout cells (Fig. 3E, Supplementary Figs. 4E and 5E). Cultured in glucose-free medium without lactic acidosis, the IDH1 knockout cells showed a similar curve to control 4T1 cells (Fig. 3E), while ME1 knockout cells and the double gene knockout cells died much faster (Fig. 3E), suggesting that ME1 is more important than IDH1 for survival under glucose-deprived condition without lactic acidosis. In contrast, under lactic acidosis, IDH1 knockout cells died faster than ME1 knockout cells, and the double gene knockout cells still died fastest (Fig. 3E). Therefore, IDH1 contributes more than ME1 to survival under glucose-deprivation with lactic acidosis. The above results were recapitulated in HeLa and H460 cells (Supplementary Figs. 4E and 5E), with slight difference in death time caused by different sensitivities to glucose deprivation.
At last, we performed FACs experiments involving propidium iodide (PI) and Annexin V-FITC to measure cell necrosis and apoptosis rate under glucose deprivation with or without lactic acidosis. Pronounced necrosis or late apoptosis was observed in ME1 and double gene knockout 4T1 cells under glucose deprivation without lactic acidosis (Fig. 3F). Under glucose starvation with lactic acidosis, only the double gene knockout 4T1 cells showed evident necrosis or late apoptosis (Fig. 3F). HeLa knockout cells showed higher early apoptosis rates under glucose deprivation with or without lactic acidosis (Supplementary Fig. 4F). The results of H460 cells were similar with that of 4T1 cells (Supplementary Fig. 5F).
Taken together, the survival and death mode experiments were in agreement with the results regarding NADPH/NADP+ and redox state: when NADPH level falls (e.g., IDH1 + ME1 knockout cells under glucose deprivation), GSH/GSSG declines, ROS level rises and antioxidant proteins (Trx1 and Prx1) become more oxidized, then cells become vulnerable and cell deaths (necrosis and apoptosis) are triggered.
3.4. Lactate-involved NADPH production is constrained by the thermodynamic barrier of LDH-catalyzed reaction
To participate in the pathway to generate NADPH, lactate must first convert to pyruvate. This reaction is catalyzed by lactate dehydrogenase (LDH). The chemical equation of the reaction is as follows:
| Lactate + NAD+ ⇌ Pyruvate + NADH + H+ |
The standard change of Gibbs free energy (ΔG’°) of the reaction is 25.1 kJ/mol [42], hence unfavorable for lactate converting to pyruvate under standard condition. To convert lactate to pyruvate, the actual change of the Gibbs free energy (ΔG) must be negative. According to the Gibbs free energy equation, ΔG is dictated by reaction quotient (Q). Our previous studies showed that at steady state, intracellular pyruvate concentration is nearly constant [43], hence lactate concentration is the main determinant to change the Q value. We correlated lactate concentrations with NADPH/NADP+ in ME1 knockout cells (Fig. 4A). NADPH/NADP+ ratios increased moderately at lactate concentrations between 0 and 15 mM and then increased sharply by further increasing lactate concentration (Fig. 4A). Survival curves of ME1 knockout cells also showed a similar trend to that of NADPH (Fig. 4B). The survival curves were close (4T1) or nearly superposed (HeLa) at lactate concentrations between 0 and 10 mM. Rescue of cells by lactate was evident at lactate concentration higher than 15 mM (Fig. 4B). Changing lactate concentrations did not affect NADPH/NADP+ in IDH1 knockout cells (Fig. 4A) and only moderately affected the survival (Supplementary Fig. 6A), consistent with the conclusion regarding the relationship between lactate and IDH1.
Fig. 4.
Lactate-involved NADPH production is constrained by the thermodynamic barrier of LDH-catalyzed reaction. (A) The effect of lactate concentrations on cellular NADPH/NADP+ ratios of IDH1 or ME1 knockout cells under glucose-deprived condition. (B) The effect of lactate concentrations on the survival of ME1 knockout cells under glucose-deprived condition. (C) Cellular NADPH/NADP+ ratios of ME1 knockout cells under four different conditions: glucose-free, glucose free with lactic acidosis, glucose free with lactosis, and glucose free with acidosis. (D) The effect of lactate concentrations on cellular NAD+, NADH concentrations and NAD+/NADH ratios of ME1 knockout cells under glucose-deprived condition. (E) The linear relationship between ΔG and ln Q of the LDH-catalyzed reaction. The red line means the difference of ln Q between 2.5 mM and 20 mM lactate, the blue line means the corresponding difference of ΔG. (F) Effect of hypoxia (1% O2) or CoCl2 (200 μM) on cellular NADPH/NADP+ ratios of IDH1 or ME1 knockout cells under glucose deprivation with lactic acidosis. (G) The effect of lactic acidosis on cellular NADPH/NADP+ ratios of LDHA or LDHB knockout cancer cells under glucose-deprived condition. The ratios of 4T1 control here are the same with that of Fig. 1C, because the two experiments were in fact performed simultaneously. Glc (−): glucose-free RPMI-1640 medium; Glc (−) + LA: glucose-free RPMI-1640 medium supplemented with 20 mM lactic acid, pH 6.7. Glc (−) + SL: glucose-free RPMI-1640 medium supplemented with 20 mM sodium lactate, pH 7.4. Glc (−) + HCl: glucose-free RPMI-1640 medium supplemented with 20 mM hydrochloric acid, pH 6.7. Glc (−) + LA + hypoxia: glucose-free RPMI-1640 medium supplemented with 20 mM lactic acid under hypoxia (1% O2) condition. Glc (−) + LA + CoCl2: glucose-free RPMI-1640 medium supplemented with 20 mM lactic acid and 200 μM CoCl2. Incubation times were 6 h (4T1), 3 h (HeLa) and 4 h (H460), respectively. Data are shown as means ± SD, with n = 3 biological replicates in (A), (C), (D), (F) and (G), n = 5 biological replicates in (B), and the results were all confirmed by three independent experiments. n. s. not significant, *p < 0.05, **p < 0.01, ***p < 0.001 (Student's test). See also Supplementary Fig. 6. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Besides exogenous lactate, another factor that influences the intracellular lactate concentration is pH value [44]. Because the transporter of lactate, MCT, is a symporter for lactate and H+, low pH (high proton concentration) facilitates lactate influx. We showed that NADPH recovery by lactic acidosis (added with 20 mM lactic acid, pH 6.7) was significantly higher than by lactosis (added with 20 mM sodium lactate, pH 7.4) or by acidosis (added with 20 mM hydrochloric acid, pH 6.7) (Fig. 4C), confirmed the essentiality of both low pH and high lactate concentration.
Another substrate of LDH-catalyzed reaction is NAD+. We noticed that intracellular NAD+ concentration and NAD+/NADH ratio of ME1 knockout cells were increased with lactate concentration under lactic acidosis (Fig. 4D). The phenomenon was more obvious in HeLa cells, and it did not occur in IDH1 knockout cells (Supplementary Fig. 6B). As Nampt is the rate-limiting enzyme for NAD+ salvage synthesis [45], we tested its expression under lactic acidosis. The result showed that the expressions of Nampt in ME1 knockout HeLa cells were not changed under lactic acidosis (Supplementary Fig. 6C). Nevertheless, the raised NAD+/NADH ratio increased the Q value of LDH-catalyzed reaction, hence makes lactate converting to pyruvate more favorable. We can estimate the additive effect of the two substrates (Fig. 4E): take HeLa/ME1 knockout cells for example, the NAD+/NADH ratios at 20 mM and 2.5 mM lactate were about 20 and 8, respectively (Fig. 4D). Suppose the pyruvate concentrations are equal in two conditions, the Q values at 20 mM and 2.5 mM should be [Pyr] × (1/8)/2.5, and [Pyr] × (1/20)/20, respectively. Thus, Q2.5 is 20 times of Q20. Then, ln Q2.5 – ln Q20 ≈ 3. Then we could calculate (ΔG2.5 - ΔG20) according to the mathematic relationship between ΔG and ln Q (Fig. 4E), which is 7.7 kJ/mol, indicating the reaction is thermodynamically much more favorable to proceed from lactate to pyruvate at high lactate concentration (20 mM lactate, pH 6.7).
Besides Q values, the rate to remove pyruvate is also crucial for lactate converting to pyruvate. The major pathway to remove pyruvate is the TCA cycle and the rate-limiting enzyme pyruvate dehydrogenase (PDH) determines the rate of pyruvate into the TCA cycle. Because hypoxia-inducible factor-1 (HIF-1) is reported to inhibit the activity of PDH by activating PDK1 (Pyruvate Dehydrogenase Kinase 1) [46], we performed the lactic acidosis experiments under hypoxia or in CoCl2-containg medium. CoCl2 is reported to artificially induce hypoxia by blocking the degradation of HIF-1α [47]. As anticipated, hypoxia or CoCl2 inhibited the NADPH/NADP+ recovery in ME1 knockout cells, but not in IDH1 knockout cells (Fig. 4F).
Lactate use by cancer cells is constrained by thermodynamic but not kinetic barrier of the reaction catalyzed by LDH, because LDH activity in cancer cells is often excessively high (e.g., in 4T1 cells, the specific activity is about 3 U/mg (Supplementary Fig. 6E)). To test the hypothesis, we established LDHA or LDHB knockout 4T1 cell line. The knockout was confirmed by immunoblot and enzyme activity determination (Supplementary Fig. 6D and E). The decreased LDH activity did not affect lactate-involved generation of NADPH (Fig. 4G). In contrast, use of glutamine is constrained by kinetic but not thermodynamic barrier, because reaction from glutamine to glutamate is thermodynamically favorable (ΔG’° = −14.3 kJ/mol) [48], and inhibition of glutaminase suppressed glutamine-derived NADPH generation (Fig. 2C).
3.5. Combination of IDH1 and ME1 is crucial for solid tumors growth
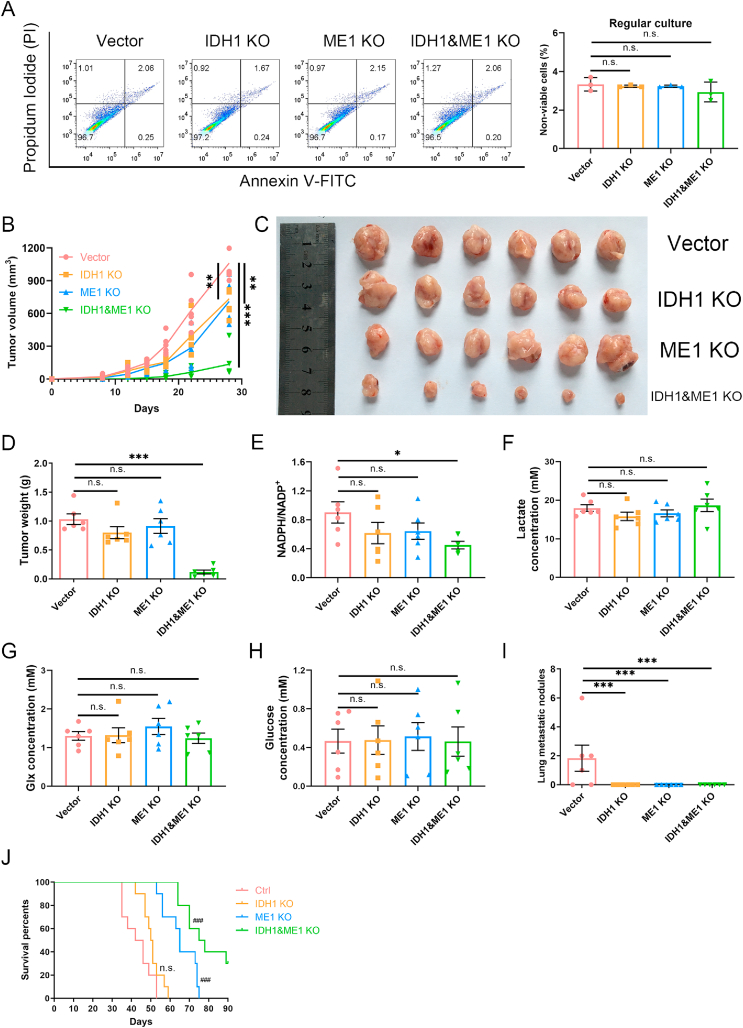
Tumor microenvironment is different from glucose-deprived conditions in vitro, hence we need to check the effect of IDH1 and/or ME1 knockout on solid tumor growth. Before in vivo studies, we used FACs method to confirm that the viabilities of the knockout cells were not affected (Fig. 5A). Doubling time experiment also showed that the growth rate under regular culture in vitro was not affected by knockout of IDH1 or (and) ME1 (Supplementary Fig. 2A). Then we used a 4T1 syngeneic xenograft model. In comparison to control, IDH1 or ME1 knockout moderately reduced tumor growth (Fig. 5B), yet there was no significant difference in the tumor weight on the day sacrificed (Fig. 5C and D). In contrast, double gene knockout remarkably inhibited tumor growth (Fig. 5B), and the final tumor weight was obviously lighter than control group (Fig. 5C and D).
Fig. 5.
Combination of IDH1 and ME1 are crucial for solid tumor growth. For solid tumor growth experiments, 105 4T1 control, IDH1 knockout, ME1 knockout, or IDH1&ME1 knockout cells (suspended in 100 μl RPMI-1640 without FBS) were inoculated in the right second mammary pad area of each Balb/c mouse. Twenty-four mice were randomly assigned into 4 groups with each group 6 mice. For survival analysis experiments, the inoculated cell number is 5 × 104, and 40 mice were randomly assigned into 4 groups with each group 10 mice. (A) The ratios of non-viable cells of 4T1 control, IDH1 knockout, ME1 knockout and IDH1&ME1 knockout cells before inoculation to mammary pad of the mice. (B) The growth curves of the primary tumors. (C) The photograph of the tumors on the day sacrificed. (D) The tumor weights on the day sacrificed. (E) NADPH/NADP+ ratios of samples from the primary tumors. (F) Lactate concentrations of samples from the primary tumors. (G) Glx (glutamine plus glutamate) concentrations of samples from the primary tumors. (H) Glucose concentrations of samples from the primary tumors. (I) The numbers of lung metastasis quantified by counting macroscopic metastatic nodules on the day sacrificed. (J) The survival curves of the tumor-bearing mice.
Data are shown as means ± SD, with n = 3 biological replicates in (A). Error bars represent the SEM of results from 6 mice for (B)–(H) or 10 mice for (J). n. s. not significant. *p < 0.05, **p < 0.01, ***p < 0.001 (Student's test). ###p < 0.001 vs control (Log-rank test).
To check whether the retarded tumor growth is associated with NADPH deficiency, we determined the NADPH/NADP+ ratios of the tumor samples. The NADPH/NADP+ ratios of gene knockout tumors all decreased compared to control group, yet only the double gene knockout tumors showed statistical significance (Fig. 5E). The drop extent is not as large as in culture cells, which may be caused by difference in nutrient supply. Thus, we determined the concentration of glucose, lactate and glx (glutamine + glutamate) in the tumor samples. The concentrations of these nutrients were not affected by the gene knockout (Fig. 5F–H). Lactate concentrations were all around 20 mM (Fig. 5F), which were nearly the same as the concentration used in vitro. Glx concentrations were mostly between 1 and 2 mM (Fig. 5G), also close to that in vitro (2.2 mM). Glucose concentrations were mostly lower than 1 mM (Fig. 5H), but not completely deprived like in vitro experiments, hence pentose phosphate pathway also contributed to NADPH generation in solid tumors.
We noticed that, except control group, there was no observable lung metastatic nodules in the ME1 or/and IDH1 knockout group (Fig. 5I), suggesting that IDH1 or ME1 knockout could repress tumor metastasis. It is reasonable because loss of matrix attachment during metastasis would inhibit glucose uptake and reduce oxPPP flux [49,50], resulting in a dependence on IDH1 and ME1 for maintaining redox balance. Therefore, metastasis may depend more on IDH1 and ME1 for maintaining redox balance. As lung metastasis is one of the main causes of death of tumor-bearing mice, we performed additional experiment to observe the survival (Fig. 5J). The average survival time of IDH1 knockout group was longer than that of control group, but the curves were not far apart (Fig. 5J). The curve of ME1 knockout group was separated from control and IDH1 knockout groups (Fig. 5J), suggesting that ME1 is more important than IDH1 for solid tumors in long-term. This could be explained by the unstable use of lactate, as lactate must kept in high concentration (Fig. 4A), and hypoxia, a common feature in solid tumor would impair the association between lactate and IDH1 (Fig. 4F). The survival time of double gene knockout group was significantly longer than other groups (Fig. 5J), further underlines the significance of combination of IDH1 and ME1 to solid tumors.
4. Discussion
Conclusion: In this study, we reveal that when glucose is deprived and oxPPP is abrogated, lactate and glutamine are the alternative hydride ion sources to reduce NADP+, and that IDH1 and ME1 are the corresponding enzymes to catalyze the final step to transfer hydride ion to NADP+. Deletion of both genes significantly reduces NADPH/NADP+ ratios, disrupts redox homeostasis and suppresses solid tumor growth. The finding is potentially applicable for interpreting how cancer cells in vivo maintain NADPH/NADP+ balance when glucose supply is insufficient.
TME is highly dynamic both temporally and spatially [1]. In the regions where vascular supply is sufficient, glucose could be used to generate NADPH by oxPPP. Unlike glucose, glutamine is not depleted in TME [4], thus it could provide NADPH by ME1 when glucose is deprived (Fig. 2). When glucose is deprived and lactate is accumulated to a high concentration, lactate makes a major contribution to NADPH production by IDH1 (Fig. 1). Therefore, tumor cells have at least 3 sets of programs to meet the demand of reducing power for proliferation in the context of dynamic TME, showing the metabolic plasticity and flexibility of cancer cells.
Noticeably, the two pathways that IDH1 and ME1 contribute to NADPH pool are both by virtue of partial reactions in the TCA cycle. Thus, TCA cycle becomes the central hub of the NADPH production under glucose-deprived conditions. This biochemical principle conveys that, apart from glutamine and lactate, nutrients or metabolic intermediates that replenish the TCA cycle can all contribute to NADPH generation, which include fatty acids, acetate, amino acids and etc. For instance, fatty acid oxidation (FAO) is reported to generate NADPH by providing Ac-CoA for the TCA cycle in cancer cells during energy stress [51,52], and the process is regulated by AMPK [52] or Nur77 [53]. The mitochondrial TCA cycle reactions are crucial to cytosolic redox homeostasis, reflecting the importance of mitochondrial metabolism to cancer cells.
Cytosolic NADPH production by IDH1 requires replenishment of citrate in mitochondria. Formation of citrate needs condensation of Ac-CoA and OAA. Given that glutamine is one of the major sources of OAA, we speculated that glutamine was necessary to lactate-involved NADPH generation. We did the lactic acidosis experiments in ME1 knockout cells under glucose and glutamine double deprived condition. However, instead of impairing the rescue of NADPH level, deprivation of glutamine did not change or even enhanced the rescue effect (data not shown). Similar results are reported: Goji et al. found that removing amino acids prolongs the survival of glioblastoma under glucose deprivation, and it is caused by blocking cystine uptake [54]. Cystine is absorbed through the cystine/glutamine antiporter xCT, then it is quickly reduced to cysteine in cytosol by NADPH [55,56]. Thus, glutamine facilitate the uptake of cystine at the cost of NADPH. Consequently, glutamine has dual effect on cytosolic NADPH pool, when ME1 is knockout, the negative effect of glutamine predominates. It could explain why NADPH/NADP+ of H460 ME1 knockout is decreased when glutamine is replenished (Fig. 2A). Nevertheless, in solid tumors, the negative effect of glutamine on NADPH pool may be weaker, as cystine level in TME is relatively low [4].
That IDH1-mediated NADPH generation is more favorable under lactic acidosis than under lactosis (Fig. 4C) may provide a rationale for cancer treatment. We previously developed a protocol named TILA-TACE (targeting-intratumoral-lactic-acidosis-transarterial chemoembolization) [57]. In this protocol, bicarbonate was infused alternatively with anticancer drugs via tumor feeding artery into hepatocellular carcinoma, where bicarbonate is to neutralize lactic acidosis to lactosis. Bicarbonate markedly enhances the therapeutic efficacy of transarterial chemoembolization (TACE) in local control of hepatocellular carcinoma [57]. After glucose supply is blocked by embolizing tumor feeding arteries, cancer cells would depend on IDH1- and ME1-mediated NADPH production. Inhibition of IDH1-mediated NADPH by bicarbonate via converting lactic acidosis to lactosis may contribute a part to the therapeutic efficacy of TILA-TACE.
IDH1 and ME1 double knockouts under glucose deprivation almost completely abolish NADPH production (Fig. 1C), suggesting that IDH1- and ME1-mediated NADPH generations are more important than other NADPH generation pathways (e.g., folate-dependent NADPH generation) in cancer cells when glucose is deprived. This is consistent with the previous report that oxPPP, IDH1 and ME1 are the major sources of cytosolic NADPH [58]. Folate-dependent NADPH generation requires oxidation of tetrahydrofolate (THF), which derives from the reduction of folate with 2 molecules of NADPH [59]. G6PD knockout abrogates the supplement of NADPH that is needed to reduce folate to THF, result in a lack of THF [58]. In line with this, glucose starvation should have similar effect like G6PD knockout, hence folate-dependent NADPH formation is suppressed when glucose is absent. Kinds of mitochondrial NADPH-producing enzymes exist [60], however, mitochondrial NADPH cannot directly shuttle to cytosol [61]. Some researchers reported that mitochondrial NADPH can be shuttled indirectly to cytosol by coupling with IDH1 or ME1-catalyzed reaction [18,62]. Therefore, knockouts of IDH1 and ME1 abolish the indirect shuttles. Taken together, knockouts of IDH1 and ME1 are sufficient to block most cytosolic NADPH generations under glucose deprivation.
In solid tumors, the glucose supply is more complex, as it depends on the blood glucose supply. It is known that glucose concentration is inversely correlated to the distance to blood vessels, hence glucose concentrations vary markedly in different regions in a tumor [63]. It is also known that glucose could be intermittently, temporarily, and persistently deprived in tumors [64]. In our xenograft model, the average glucose concentrations in tumors were mostly lower than 1 mM, and the glucose concentrations between 4 groups (control, IDH1 knockout, ME1 knockout, and IDH1+ME1 knockout) were not significantly different from each, but only IDH1+ME1 knockout showed a markedly reduced NADPH/NADP+, accompanied with retarded growth of tumor (Fig. 5). Based on the above lines of evidence, in solid tumors where glucose is not completely deprived but deficient, the contribution of IDH1 and ME1 to NADPH is significant, and knockout of them would impair the growth and metastasis of the solid tumors, which is close to the experiment results performed under glucose deprivation in vitro. On the other hand, it is reported that G6PD is not essential for K-Ras-driven tumors growth or metastasis [65], suggesting a less dependence on oxPPP of solid tumors in the context of glucose-deficient TME. Furthermore, the expression of IDH1 in prostate tumors and the expression of ME1 in gastric/colon/rectal/cancers were reported to be higher than adjacent non-tumor tissues [17,66]. Consequently, tumors possibly depend on IDH1 and ME1 more than adjacent non-tumor tissues, resulting in a therapeutic liability that can be exploited by targeting both IDH1 and ME1. However, there is a paucity of specific inhibitors of the two enzymes to date. GSK-864, which is designed to inhibit R132H point-mutant IDH1 (IC50: 15.2 nM), is reported to also inhibit wild-type IDH1 at higher doses (IC50: 466.5 nM) [67]. Zhang et al. designed a apiperazine-1-pyrrolidine-2,5-dione scaffold-based malic enzyme inhibitor [68], and it suppressed growth of human CRC cells in vitro [69]. However, the inhibitor did not distinguish cytosolic or mitochondrial malic enzyme, and the further utilization of them is lacking. Lanthanide (e.g. Lu2+), which competitively binding to ME with Mn2+ [70], is also reported to show inhibitory effect [71]. However, as Mn2+ is involved in a variety of biochemical reactions, the off-target effects of lanthanide should be carefully considered. Another optional strategy is targeting the post-translational regulations of ME1, as acetylation of ME1 enhances its activity while phosphorylation of ME1 has opposite effect [72]. Overall, the studies of inhibitors of wild-type IDH1 and ME1 with high specificity remain inadequate, and relevant clinical trials remain a blank [60]. More drug development effort should be made to exploit the therapeutic benefits.
Author contributions
Conceptualization, H.X. and Y.M.; Methodology, Y.M. and Y.D.; Investigation, Y.M., Y.D., Z.X., C.L. and Z.S.; Writing – Original Draft, Y.M.; Writing – Review & Editing, H.X. and Y.M.; Funding Acquisition, H.X.; Resources, H.X.; Supervision, H.X.
Declaration of interests
None.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
This work has been supported in part by a key project (2018C03009) funded by Zhejiang Provincial Department of Sciences and Technologies (to XH & MC), China Natural Sciences Foundation project (81470126 to XH, 82073038 to XH), and the Fundamental Research Funds for the Central Universities (2017XZZX001-01, 2019FZJD009, to XH).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102065.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Kim J., DeBerardinis R.J. Mechanisms and implications of metabolic heterogeneity in cancer. Cell Metabol. 2019;30:434–446. doi: 10.1016/j.cmet.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vander Heiden M.G., DeBerardinis R.J. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metabol. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sullivan M.R., V Danai L., Lewis C.A., Chan S.H., Gui D.Y., Kunchok T., Dennstedt E.A., Vander Heiden M.G., Muir A. Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. Elife. 2019;8 doi: 10.7554/eLife.44235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaupel P., Kallinowski F., Okunieff P. Blood-flow, oxygen and nutrient supply, and metabolic microenvironment of human-tumors - a Review. Canc. Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- 6.Fendt S.M., Frezza C., Erez A. Cancer Discov; 2020. Targeting Metabolic Plasticity and Flexibility Dynamics for Cancer Therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patra K.C., Hay N. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 2014;39:347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z., Chen L., Liu L., Su X., Rabinowitz J.D. Chemical basis for deuterium labeling of fat and NADPH. J. Am. Chem. Soc. 2017;139:14368–14371. doi: 10.1021/jacs.7b08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tao R., Zhao Y., Chu H., Wang A., Zhu J., Chen X., Zou Y., Shi M., Liu R., Su N., Du J., Zhou H.M., Zhu L., Qian X., Liu H., Loscalzo J., Yang Y. Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nat. Methods. 2017;14:720–728. doi: 10.1038/nmeth.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan J., Ye J., Kamphorst J.J., Shlomi T., Thompson C.B., Rabinowitz J.D. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis C.A., Parker S.J., Fiske B.P., McCloskey D., Gui D.Y., Green C.R., Vokes N.I., Feist A.M., Vander Heiden M.G., Metallo C.M. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calvert A.E., Chalastanis A., Wu Y., Hurley L.A., Kouri F.M., Bi Y., Kachman M., May J.L., Bartom E., Hua Y., Mishra R.K., Schiltz G.E., Dubrovskyi O., Mazar A.P., Peter M.E., Zheng H., James C.D., Burant C.F., Chandel N.S., V Davuluri R., Horbinski C., Stegh A.H. Cancer-associated IDH1 promotes growth and resistance to targeted therapies in the absence of mutation. Cell Rep. 2017;19:1858–1873. doi: 10.1016/j.celrep.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen X., Wu S., Zhang J., Li M., Xu F., Wang A., Lei Y., Zhu G. Wildtype IDH1 affects cell migration by modulating the PI3K/AKT/mTOR pathway in primary glioblastoma cells. Mol. Med. Rep. 2020;22 doi: 10.3892/mmr.2020.11250. 1949–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahl D.R., Dresser J., Wilder-Romans K., Parsels J.D., Zhao S.G., Davis M., Zhao L., Kachman M., Wernisch S., Burant C.F., Morgan M.A., Feng F.Y., Speers C., Lyssiotis C.A., Lawrence T.S. Glioblastoma therapy can Be augmented by targeting IDH1-mediated NADPH biosynthesis. Canc. Res. 2017;77:960–970. doi: 10.1158/0008-5472.CAN-16-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Y.X., Ju H.Q., Liu Z.X., Chen D.L., Wang Y., Zhao Q., Wu Q.N., Zeng Z.L., Qiu H.B., Hu P.S., Wang Z.Q., Zhang D.S., Wang F., Xu R.H. ME1 regulates NADPH homeostasis to promote gastric cancer growth and metastasis. Canc. Res. 2018;78 doi: 10.1158/0008-5472.CAN-17-3155. 1972–1985. [DOI] [PubMed] [Google Scholar]
- 16.Zheng F.J., Ye H.B., Wu M.S., Lian Y.F., Qian C.N., Zheng Y.X. Repressing malic enzyme 1 redirects glucose metabolism, unbalances the redox state, and attenuates migratory and invasive abilities in nasopharyngeal carcinoma cell lines. Chin. J. Canc. 2012;31:519–531. doi: 10.5732/cjc.012.10088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmen F.A., Alhallak I., Simmen R.C.M. Malic enzyme 1 (ME1) in the biology of cancer: it is not just intermediary metabolism. J. Mol. Endocrinol. 2020;65:R77–R90. doi: 10.1530/JME-20-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao C., Lu W., Du Y., Yan W., Bao Q., Tian Y., Wang G., Ye H., Hao H. Cytosolic ME1 integrated with mitochondrial IDH2 supports tumor growth and metastasis. Redox Biol. 2020;36:101685. doi: 10.1016/j.redox.2020.101685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de la Cruz-Lopez K.G., Castro-Munoz L.J., Reyes-Hernandez D.O., Garcia-Carranca A., Manzo-Merino J. Lactate in the regulation of tumor microenvironment and therapeutic approaches. Front Oncol. 2019;9:1143. doi: 10.3389/fonc.2019.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabinowitz J.D., Enerback S. Lactate: the ugly duckling of energy metabolism. Nat Metab. 2020;2:566–571. doi: 10.1038/s42255-020-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ippolito L., Morandi A., Giannoni E., Chiarugi P. Lactate: a metabolic driver in the tumour landscape. Trends Biochem. Sci. 2019;44:153–166. doi: 10.1016/j.tibs.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Mendes C., Serpa J. Revisiting lactate dynamics in cancer-a metabolic expertise or an alternative attempt to survive? J. Mol. Med. 2020;98:1397–1414. doi: 10.1007/s00109-020-01965-0. [DOI] [PubMed] [Google Scholar]
- 23.Faubert B., Li K.Y., Cai L., Hensley C.T., Kim J., Zacharias L.G., Yang C., Do Q.N., Doucette S., Burguete D., Li H., Huet G., Yuan Q., Wigal T., Butt Y., Ni M., Torrealba J., Oliver D., Lenkinski R.E., Malloy C.R., Wachsmann J.W., Young J.D., Kernstine K., DeBerardinis R.J. Lactate metabolism in human lung tumors. Cell. 2017;171:358–371. doi: 10.1016/j.cell.2017.09.019. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hui S., Ghergurovich J.M., Morscher R.J., Jang C., Teng X., Lu W., Esparza L.A., Reya T., Le Z., Yanxiang Guo J., White E., Rabinowitz J.D. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551:115–118. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tasdogan A., Faubert B., Ramesh V., Ubellacker J.M., Shen B., Solmonson A., Murphy M.M., Gu Z., Gu W., Martin M., Kasitinon S.Y., Vandergriff T., Mathews T.P., Zhao Z., Schadendorf D., DeBerardinis R.J., Morrison S.J. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature. 2020;577:115–120. doi: 10.1038/s41586-019-1847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H., Ding Z., Hu D., Sun F., Dai C., Xie J., Hu X. Central role of lactic acidosis in cancer cell resistance to glucose deprivation-induced cell death. J. Pathol. 2012;227:189–199. doi: 10.1002/path.3978. [DOI] [PubMed] [Google Scholar]
- 27.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan M., Breitkopf S.B., Yang X., Asara J.M. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibon Y., Larher F. Cycling assay for nicotinamide adenine dinucleotides: NaCl precipitation and ethanol solubilization of the reduced tetrazolium. Anal. Biochem. 1997;251:153–157. doi: 10.1006/abio.1997.2283. [DOI] [PubMed] [Google Scholar]
- 30.Simplăceanu V. Tissue water content and nuclear magnetic resonance in normal and tumor tissues. Canc. Res. 1975;35:1164–1167. [PubMed] [Google Scholar]
- 31.V Passonneau J., Lowry O.H. Humana Press; Totowa, NJ: 1993. Enzymatic Analysis: a Practical Guide. [Google Scholar]
- 32.Rahman I., Kode A., Biswas S.K. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 2006;1:3159–3165. doi: 10.1038/nprot.2006.378. [DOI] [PubMed] [Google Scholar]
- 33.Go Y.M., Jones D.P. Thioredoxin redox western analysis. Curr. Protoc. Toxicol. 2009 doi: 10.1002/0471140856.tx1712s41. [DOI] [PubMed] [Google Scholar]
- 34.Cox A.G., Winterbourn C.C., Hampton M.B. Methods Enzymol. 2010. Measuring the redox state of cellular peroxiredoxins by immunoblotting. [DOI] [PubMed] [Google Scholar]
- 35.Pulaski B.A., Ostrand‐Rosenberg S. Mouse 4T1 breast tumor model. Curr. Protoc. Im. 2000 doi: 10.1002/0471142735.im2002s39. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z., Dong C. Gluconeogenesis in cancer: function and regulation of PEPCK, FBPase, and G6Pase. Trends in Cancer. 2019;5:30–45. doi: 10.1016/j.trecan.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Ying M., Guo C., Hu X. The quantitative relationship between isotopic and net contributions of lactate and glucose to the tricarboxylic acid (TCA) cycle. J. Biol. Chem. 2019;294:9615–9630. doi: 10.1074/jbc.RA119.007841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Son J., Lyssiotis C.A., Ying H., Wang X., Hua S., Ligorio M., Perera R.M., Ferrone C.R., Mullarky E., Shyh-Chang N., Kang Y., Fleming J.B., Bardeesy N., Asara J.M., Haigis M.C., DePinho R.A., Cantley L.C., Kimmelman A.C. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y.P., Zhou W., Wang J., Huang X., Zuo Y., Wang T.S., Gao X., Xu Y.Y., Zou S.W., Liu Y.B., Cheng J.K., Lei Q.Y. Arginine methylation of MDH1 by CARM1 inhibits glutamine metabolism and suppresses pancreatic cancer. Mol. Cell. 2016;64:673–687. doi: 10.1016/j.molcel.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 40.Yang S., Hwang S., Kim M., Seo S.B., Lee J.H., Jeong S.M. Mitochondrial glutamine metabolism via GOT2 supports pancreatic cancer growth through senescence inhibition. Cell Death Dis. 2018;9:55. doi: 10.1038/s41419-017-0089-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huypens P., Pillai R., Sheinin T., Schaefer S., Huang M., Odegaard M.L., Ronnebaum S.M., Wettig S.D., Joseph J.W. The dicarboxylate carrier plays a role in mitochondrial malate transport and in the regulation of glucose-stimulated insulin secretion from rat pancreatic beta cells. Diabetologia. 2011;54:135–145. doi: 10.1007/s00125-010-1923-5. [DOI] [PubMed] [Google Scholar]
- 42.Nelson D.L., Cox M.M. W.H. Freeman and Company; New York: 2008. Lehninger Principles of Biochemistry. [Google Scholar]
- 43.Sun F., Dai C., Xie J., Hu X. Biochemical issues in estimation of cytosolic free NAD/NADH ratio. PloS One. 2012;7 doi: 10.1371/journal.pone.0034525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie J., Wu H., Dai C., Pan Q., Ding Z., Hu D., Ji B., Luo Y., Hu X. Beyond Warburg effect--dual metabolic nature of cancer cells. Sci. Rep. 2014;4:4927. doi: 10.1038/srep04927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gardell S.J., Hopf M., Khan A., Dispagna M., Hampton Sessions E., Falter R., Kapoor N., Brooks J., Culver J., Petucci C., Ma C.T., Cohen S.E., Tanaka J., Burgos E.S., Hirschi J.S., Smith S.R., Sergienko E., Pinkerton A.B. Boosting NAD(+) with a small molecule that activates NAMPT. Nat. Commun. 2019;10:3241. doi: 10.1038/s41467-019-11078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J.W., Tchernyshyov I., Semenza G.L., V Dang C. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metabol. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y.B., Wang X., Meister E.A., Gong K.R., Yan S.C., Lu G.W., Ji X.M., Shao G. The effects of CoCl2 on HIF-1α protein under experimental conditions of autoprogressive hypoxia using mouse models. Int. J. Mol. Sci. 2014 doi: 10.3390/ijms150610999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benzinger T., Kitzinger C., Hems R., Burton K. Free-energy changes of the glutaminase reaction and the hydrolysis of the terminal pyrophosphate bond of adenosine triphosphate. Biochem. J. 1959;71:400–407. doi: 10.1042/bj0710400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang L., Shestov A.A., Swain P., Yang C., Parker S.J., Wang Q.A., Terada L.S., Adams N.D., McCabe M.T., Pietrak B., Schmidt S., Metallo C.M., Dranka B.P., Schwartz B., DeBerardinis R.J. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532:255–258. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schafer Z.T., Grassian A.R., Song L., Jiang Z., Gerhart-Hines Z., Irie H.Y., Gao S., Puigserver P., Brugge J.S. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carracedo A., Cantley L.C., Pandolfi P.P. Cancer metabolism: fatty acid oxidation in the limelight. Nat. Rev. Canc. 2013;13:227–232. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeon S.M., Chandel N.S., Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X.X., Wang Z.J., Zheng Y., Guan Y.F., Yang P.B., Chen X., Peng C., He J.P., Ai Y.L., Wu S.F., Chien K.Y., Wu Q., Chen H.Z. Nuclear receptor Nur77 facilitates melanoma cell survival under metabolic stress by protecting fatty acid oxidation. Mol. Cell. 2018;69:480–492. doi: 10.1016/j.molcel.2018.01.001. e7. [DOI] [PubMed] [Google Scholar]
- 54.Goji T., Takahara K., Negishi M., Katoh H. Cystine uptake through the cystine/glutamate antiporter xCT triggers glioblastoma cell death under glucose deprivation. J. Biol. Chem. 2017;292:19721–19732. doi: 10.1074/jbc.M117.814392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joly J.H., Delfarah A., Phung P.S., Parrish S., Graham N.A. A synthetic lethal drug combination mimics glucose deprivation-induced cancer cell death in the presence of glucose. J. Biol. Chem. 2020;295:1350–1365. doi: 10.1074/jbc.RA119.011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X., Olszewski K., Zhang Y., Lim E.W., Shi J., Zhang X., Zhang J., Lee H., Koppula P., Lei G., Zhuang L., You M.J., Fang B., Li W., Metallo C.M., V Poyurovsky M., Gan B. Cystine transporter regulation of pentose phosphate pathway dependency and disulfide stress exposes a targetable metabolic vulnerability in cancer. Nat. Cell Biol. 2020;22:476–486. doi: 10.1038/s41556-020-0496-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chao M., Wu H., Jin K., Li B., Wu J., Zhang G., Yang G., Hu X. A nonrandomized cohort and a randomized study of local control of large hepatocarcinoma by targeting intratumoral lactic acidosis. Elife. 2016;5 doi: 10.7554/eLife.15691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen L., Zhang Z., Hoshino A., Zheng H.D., Morley M., Arany Z., Rabinowitz J.D. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat Metab. 2019;1:404–415. doi: 10.1038/s42255-019-0043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ducker G.S., Rabinowitz J.D. One-carbon metabolism in health and disease. Cell Metabol. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ju H.Q., Lin J.F., Tian T., Xie D., Xu R.H. NADPH homeostasis in cancer: functions, mechanisms and therapeutic implications. Signal Transduct Target Ther. 2020;5:231. doi: 10.1038/s41392-020-00326-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goodman R.P., Calvo S.E., Mootha V.K. Spatiotemporal compartmentalization of hepatic NADH and NADPH metabolism. J. Biol. Chem. 2018;293:7508–7516. doi: 10.1074/jbc.TM117.000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Balsa E., Perry E.A., Bennett C.F., Jedrychowski M., Gygi S.P., Doench J.G., Puigserver P. Defective NADPH production in mitochondrial disease complex I causes inflammation and cell death. Nat. Commun. 2020;11:2714. doi: 10.1038/s41467-020-16423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Konerding M.A., Fait E., Gaumann A. 3D microvascular architecture of pre-cancerous lesions and invasive carcinomas of the colon. Br. J. Canc. 2001 doi: 10.1054/bjoc.2001.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fukumura D., Jain R.K. Tumor microvasculature and microenvironment: targets for anti-angiogenesis and normalization. Microvasc. Res. 2007 doi: 10.1016/j.mvr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghergurovich J.M., Esposito M., Chen Z., Wang J.Z., Bhatt V., Lan T., White E., Kang Y., Guo J.Y., Rabinowitz J.D. Glucose-6-Phosphate dehydrogenase is not essential for K-Ras-Driven tumor growth or metastasis. Canc. Res. 2020;80:3820–3829. doi: 10.1158/0008-5472.CAN-19-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gonthier K., Poluri R.T.K., Weidmann C., Tadros M., Audet-Walsh E. Reprogramming of isocitrate dehydrogenases expression and activity by the androgen receptor in prostate cancer. Mol. Canc. Res. 2019 doi: 10.1158/1541-7786.MCR-19-0020. [DOI] [PubMed] [Google Scholar]
- 67.Okoye-Okafor U.C., Bartholdy B., Cartier J., Gao E.N., Pietrak B., Rendina A.R., Rominger C., Quinn C., Smallwood A., Wiggall K.J., Reif A.J., Schmidt S.J., Qi H., Zhao H., Joberty G., Faelth-Savitski M., Bantscheff M., Drewes G., Duraiswami C., Brady P., Groy A., Narayanagari S.R., Antony-Debre I., Mitchell K., Wang H.R., Kao Y.R., Christopeit M., Carvajal L., Barreyro L., Paietta E., Makishima H., Will B., Concha N., Adams N.D., Schwartz B., McCabe M.T., Maciejewski J., Verma A., Steidl U. New IDH1 mutant inhibitors for treatment of acute myeloid leukemia. Nat. Chem. Biol. 2015;11:878–886. doi: 10.1038/nchembio.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y.J., Wang Z., Sprous D., Nabioullin R. In silico design and synthesis of piperazine-1-pyrrolidine-2,5-dione scaffold-based novel malic enzyme inhibitors. Bioorg. Med. Chem. Lett. 2006;16:525–528. doi: 10.1016/j.bmcl.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 69.Fernandes L.M., Al-Dwairi A., Simmen R.C.M., Marji M., Brown D.M., Jewell S.W., Simmen F.A. Malic enzyme 1 (ME1) is pro-oncogenic in apc(min/+) mice. Sci. Rep. 2018;8:14268. doi: 10.1038/s41598-018-32532-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Z., Batra R., Floyd D.L., Hung H.C., Chang G.G., Tong L. Potent and competitive inhibition of malic enzymes by lanthanide ions. Biochem. Biophys. Res. Commun. 2000;274:440–444. doi: 10.1006/bbrc.2000.3163. [DOI] [PubMed] [Google Scholar]
- 71.Nakashima C., Yamamoto K., Fujiwara-Tani R., Luo Y., Matsushima S., Fujii K., Ohmori H., Sasahira T., Sasaki T., Kitadai Y., Kirita T., Kuniyasu H. Expression of cytosolic malic enzyme (ME1) is associated with disease progression in human oral squamous cell carcinoma. Canc. Sci. 2018;109:2036–2045. doi: 10.1111/cas.13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu Y., Gu L., Lin X., Liu C., Lu B., Cui K., Zhou F., Zhao Q., V Prochownik E., Fan C., Li Y. Dynamic regulation of ME1 phosphorylation and acetylation affects lipid metabolism and colorectal tumorigenesis. Mol. Cell. 2020;77:138–149. doi: 10.1016/j.molcel.2019.10.015. e5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.