Abstract
APOBEC3s are innate single-stranded DNA cytidine-to-uridine deaminases that catalyze mutations in both pathogen and human genomes with significant roles in human disease. However, how APOBEC3s mutate a single-stranded DNA that is available momentarily during DNA transcription or replication in vivo remains relatively unknown. In this study, utilizing hepatitis B virus (HBV) viral mutations, we evaluated the mutational characteristics of individual APOBEC3s with reference to the HBV replication process through HBV whole single-strand (−)-DNA genome mutation analyses. We found that APOBEC3s induced C-to-T mutations from the HBV reverse transcription start site continuing through the whole (−)-DNA transcript to the termination site with variable efficiency, in an order of A3B >> A3G > A3H-II or A3C. A3B had a 3-fold higher mutation efficiency than A3H-II or A3C with up to 65% of all HBV genomic cytidines being converted into uridines in a single mutation event, consistent with the A3B localized hypermutation signature in cancer, namely, kataegis. On the other hand, A3C expression led to a 3-fold higher number of mutation-positive HBV genome clones, although each individual clone had a lower number of C-to-T mutations. Like A3B, A3C preferred both 5′-TC and 5′-CC sequences, but to a lesser degree. The APOBEC3-induced HBV mutations were predominantly detected in the HBV rcDNA but were not detectable in other intermediates including HBV cccDNA and pgRNA by primer extension of their PCR amplification products. These data demonstrate that APOBEC3-induced HBV genome mutations occur predominantly when the HBV RNA genome was reversely transcribed into (−)-DNA in the viral capsid.
Keywords: APOBEC3, HBV, cytidine deaminase, viral replication, mutagenesis, enzyme mutation, cancer, genome, nucleotide
Abbreviations: APOBEC3 or A3, apolipoprotein B mRNA editing catalytic polypeptide-like 3; cccDNA, covalently closed circular DNA; HBV, hepatitis B virus; pe, primer extension; pgRNA, pre-genome RNA; rcDNA, relaxed circular DNA
Human APOBEC3 (apolipoprotein B mRNA editing catalytic polypeptide-like 3) proteins are a family of seven cytidine deaminases (A3A/B/C/D/F/G/H) capable of causing cytidine-to-uridine (C-to-T as sequencing readout) mutations on single-stranded DNA (ssDNA) molecules (1). The seven APOBEC3s constitute vital defense enzymes of the innate immune system that respond to infection from exogenous DNA-based viruses and endogenous transposons by catalyzing C-to-T deamination of foreign ssDNA (1). Human immunodeficiency virus 1 (HIV-1) is the prototypical retrovirus that has been studied extensively for A3G-induced viral restriction and mutation activity. A3G has been defined as the major defense factor against HIV-1 through hypermutation (1). In addition to A3G, A3D/F/H are often expressed together with A3G in lymphoid cells that serve as targets for HIV-1 infection. The current model is that A3D/F/G/H incorporate into budding HIV-1 particles through a binding event between their N-terminal domain and HIV-1 RNA. Upon HIV-1 fusion with a target cell, A3D/F/G/H deaminate viral cDNA cytidines to uridines during the reverse transcription of viral replication (1, 2, 3). Up to 10% of the cytidines in the HIV-1 genome can be mutated into uridines by a single round of viral infection (1). The resulting cDNA mutant is either degraded or immortalized as G-to-A hypermutations in the viral genome (1, 2). In addition to the hypermutations, A3G could also inhibit HIV-1 through direct RNA binding (1). To counteract the deleterious effects of A3G, HIV-1 acquired the ability to degrade A3G and thus prevent it from packaging in virions through the HIV-1 accessory protein Vif (3, 4). The Vif protein binds to A3G, resulting in rapid degradation by the proteasome (4). By Vif countering activity, partially inactivated A3G could also lead to sublethal levels of HIV-1 genome mutation, resulting in better viral fitness (3).
In addition to restricting HIV-1, APOBEC3s also restrict many other viruses, such as hepatitis B, human T cell leukemia virus type 1, and human papilloma virus (1). Hepatitis B virus (HBV) is one of the smallest enveloped DNA viruses that can cause acute and chronic liver diseases that can progress to liver fibrosis, cirrhosis, and hepatocellular carcinoma. Similar to retroviral replication, HBV viruses replicate through RNA reverse transcription in a capsid intermediate that contains viral RNA, core protein, HBV polymerase, and various host proteins (5, 6). HBV DNA mutations induced by APOBEC3s occur when the viral RNA is converted by HBV polymerase into partially double-stranded relaxed circular DNA (rcDNA) through cDNA in the capsid (7). The mature HBV rcDNA nucleocapsid is then enveloped with surface proteins for secretion as an infectious virion (5). A3B/C/F/G/H-mediated HBV mutations have been described (1, 8, 9, 10). A3B is a unique nucleocytoplasmic shuttling protein with a predominant nuclear location, whereas A3C/F/G/H mainly or exclusively localize in the cytoplasm where HBV assembly occurs. Nevertheless, A3B can effectively incorporate into the HBV virial capsid and inhibit viral replication by inducing mutations (8). A3B expression knockdown by siRNA in HBV virus producing HepG2.2.15 cells increased HBV replication (8). The endogenous deaminases edited 10% to 25% of HBV rcDNA genomes in HBV-producing HepG2.2.15 cells (10). In contrast to A3G, HIV-1 Vif has no degradation effect on A3B (11).
Although described as restriction factors that impede replication of many viruses such as HIV-1 and HBV, this family of enzymes has also been associated with a distinct mutational signature in the genomes of many cancers, particularly those localized to the breast, lung, bladder, cervix, and head and neck (12, 13, 14).
APOBEC3 mutagenesis has been described as one of the most common mutational processes second only to “aging” (12). APOBEC3's signature mutations have been thought to contribute to subclonal diversity in tumors (13, 15), drug resistance (16), tumor metastases (17), or improved survival in patients with cancer (18, 19). Recent genome-wide sequencing data showed that A3B is a key molecular driver inducing mutations and is overexpressed in a wide spectrum of human cancers (20). Consistent with its unique nuclear location, A3B overexpression and aberrant activation lead to unexpected clusters of mutations in most cancers (14, 20). APOBEC3s have been considered as targets for viral and cancer therapies (3). However, how APOBEC3s, A3B in particular, access and mutate single-stranded DNA, which is only momentarily available during DNA replication or transcription in vivo, remains to be elucidated.
APOBEC3s are innate ssDNA cytidine-to-uridine deaminases that catalyze single-base substitution mutations in both pathogen and human genomes (3). Although each member of the family is different in protein size, cellular location, and preference of the nucleotide immediately adjoining the mutated cytidines, they share single-stranded DNA substrate specificity (3). Their ssDNA substrate specificity suggests that APOBEC3s must have access to ssDNA to be functional in either pathogen or human genomes. Although their mutations in cancer are complicated due to accumulated mutations over a long period of time, APOBEC3 mutations in viral genomes are relatively simple because viruses like HIV-1 and HBV replicate rapidly and most of the biological processes have been identified (1, 5). Compared with HBV, APOBEC3-induced mutations in HIV-1 are more complicated due to the recombination that occurs when the HIV-1 viral reverse transcriptase switches between two copackaged RNA templates during minus-strand DNA synthesis, resulting in a hybrid DNA copy that contains a portion of information from each RNA genome (21). Although A3D/F/G/H-mediated mutations in HIV-1 or A3B/C/G/H-induced mutations in HBV have been extensively studied, most viral genome mutations have been investigated only using representative short fragments of the viral DNA genome (8, 9, 10, 22, 23, 24). There have been several viral genome-wide mutational investigations in patients, but no conclusions were made regarding a mechanism relating APOBEC3-induced mutation and ssDNA owing to the complexity of viral mutation accumulation in vivo (10, 25, 26, 27, 28). The interrelationship between APOBEC3-induced mutation and viral reverse transcription remains to be clarified (10). In addition, the mutational characteristics of different APOBEC3s, especially A3B, have not been well established and compared with each other under the same conditions. In this study, we used the HBV viral model to investigate the distribution of A3B/C/G/H-induced C-to-T mutations over the entire HBV genome through genome-wide mutational landscape analyses. We found that mutation of the HBV viral genome occurred at the same time as when the HBV viral RNA genome was reversely transcribed into (−)-DNA. The HBV mutation model captures mutational reactions induced by APOBEC-3s and allows comparing mutational characteristics of multiple APOBEC3s.
Results
The HBV viral genome requires extensive denaturing for efficient and accurate PCR amplification
The HBV viral replication processes in vivo can be mimicked by transfecting an HBV genome encoding plasmid in a human liver cell line (5, 6, 29, 30, 31). HBV genome variants are generated in HBV-transfected HepG2 cells due to alternative RNA splicing, which is identical to what occurs in liver tissues or sera of HBV-infected patients (29, 30, 31). Cotransfection of APOBEC3s with an HBV genome encoding plasmid in hepatoma cells has been used in multiple publications of APOBEC3-induced mutations on HBV viral DNA (7, 32, 33). In this study, we used this same cellular system to investigate APOBEC3 mutational activities through HBV genome mutational landscape analyses. Briefly, the HBV genome encoding plasmid was transfected into HepG2 cells together with individual APOBEC3-encoding plasmids. HBV genomic DNAs were isolated from the capsids in the cytoplasm for HBV mutational analyses.
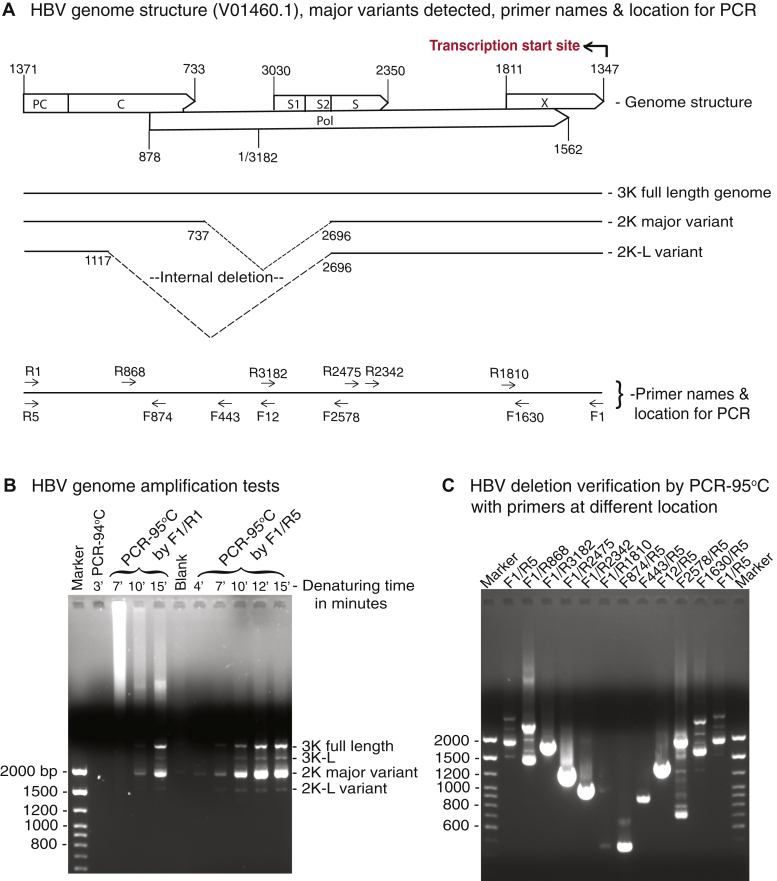
An ayw strain HBV viral genome was used in this study with a sequence identical to HBV V01460.1 in the GenBank (33). The HBV viral genome structure and genome variants are linearly presented in Figure 1A with a nucleotide numbering start of 1 and end of 3182 in accordance with HBV V01460.1. The full-length genome and the major genome variants due to alternative RNA splicing were named 3K, 2K, and 2K-L for the sake of convenience in this study. HBV reverse transcription is initiated from the X-gene 3′ DR1 (1347 nt) by the HBV polymerase, moves to 3182/1 nt, and then continues up to the PC gene 5′ DR1∗ (5) (see Fig. 1A, top panel). HBV genomic DNAs isolated from viral capsids are partially double-stranded rcDNAs that have a complete genomic (−)-DNA and a cohort of less than full-length (+)-DNA with an overlap bridging over the (−)-DNA two ends (5, 6). For HBV genome cloning, primers annealable to both ends of the (−)-DNA, i.e., F1/R1 or F1/R5 as shown in the lower panel of Figure 1A, were used to amplify the HBV genomes as previously described (34).
Figure 1.
HBV can only be accurately amplified by an extensive denaturing PCR at 95 °C.A, diagram of the ayw strain HBV genome structure with reverse transcription starting site, genome variants with internal deletion locations, and primers used at various locations. The nucleotide positions in the genome are numbered by reference to HBV V01460.1 in the GenBank. B, one percent agarose analyses of HBV genome PCR amplifications. The HBV-encoding plasmid was transfected into HepG2 cells. HBV genomic DNA was isolated from viral capsids after a 48-h transfection and was PCR amplified after removal of potential contamination of HBV plasmid from the initial transfection by micrococcal nuclease pretreatment. The HBV genomic DNA amplifications were tested with primers F1/R1 by regular ampliTaq with initial denaturing at 94 °C for 3 min or Gold Taq with initial denaturing at 95 °C for 7, 10, or 15 min. The HBV DNA amplifications were further tested with an improved primer pair F1/R5 by Gold Taq with denaturing at 95 °C for 4 to 15 min. The resultant HBV amplicons were analyzed by 1% agarose electrophoresis. C, one percent agarose analyses of HBV genome internal deletions. The internal deletions of HBV genomes were verified by PCR-95 °C amplifications with reverse and forward primer walks with or without inclusion of deleted regions. HBV, hepatitis B virus.
We found that, although the HBV viral genome was small (only 3.2 kb), it was difficult to amplify it by normal PCR-94 °C conditions without bias amplification. As shown in Figure 1B, the HBV genome could not be amplified by regular ampliTaq DNA polymerase from our HBV rcDNA preparations with DNA denaturing at 94 °C. HBV genomic DNA could only be efficiently amplified by Gold Taq DNA polymerase with an initial DNA denaturation at 95 °C for at least 10 min. The amplification was further improved by reverse primer alternatives, F1/R5 instead of F1/R1, as shown in Figure 1B, right side of the panel. In addition, we also found that primers designed without considering the HBV (−)-DNA gap in its native condition resulted in absent or compromised amplifications (data not shown).
As shown in Figure 1B, HBV genomic DNA amplification resulted in multiple bands. In addition to the full-length HBV genome 3.2 kb, there were multiple smaller-sized amplifications with 2 kb as the major one. These HBV genomes were cloned into a pCR4 vector through the TA cloning method and were identified by sequencing analyses as the 3K full-length HBV genome, 2K major variant with internal deletion of 1223 nt, and 2K-L variant with internal deletions of 1603 nt as shown in Figure 1A, middle panel. These data are consistent with previous reports that HBV viral genomes isolated from the blood of patients with chronic hepatitis B coexist in multiple alternative RNA splicing forms with internal deletions of 1223 nt (sp1) and 1603 nt (sp3) as the major ones (29, 30, 31, 35). The presence of HBV genome variants with the internal deletions were further verified by PCR amplifications using primer pairs at various locations. As shown in the Figure 1A, bottom panel and C, multiple PCR amplification bands were observed when primer pairs of F1/R5 or F1/R868 across the deletion region were used. Alternatively, PCR with the primer pairs of F1630/R5 or F2578/R5 also resulted in multiple bands due to the internal deletions. In contrast, the primer pairs of F1/R3182, F12/R5, or F443/R5 selectively amplified fragments from the 3K full-length HBV genome alone, whereas F1/R2475 or F874/R5 primer pairs amplified fragments from all HBV genomic variants including 3K, 2K, and 2K-L. Of note, there were extra bands detected by the F2578/R5 primer pair. The major top and bottom two bands corresponded to amplifications from 3K and 2K genomes. These minor bands between them were other HBV genome variants generated through a variety of viral RNA alternative splicing events as identified by sequencing with some of them (data not shown).
APOBEC3s induce variable HBV genome variant mutations as determined by primer extension analyses
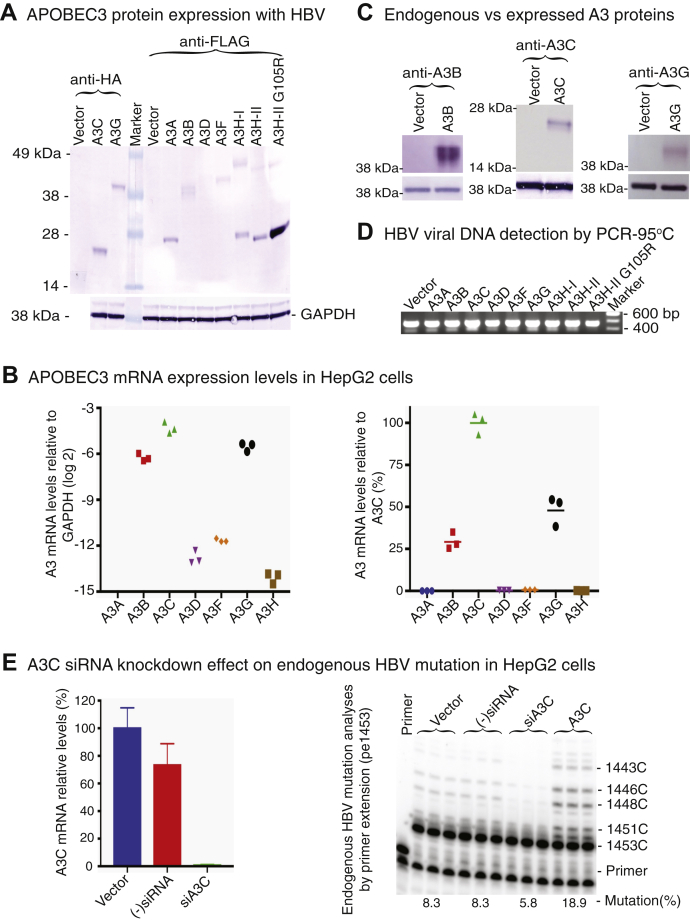
To investigate how APOBEC3s mutate HBV viral DNA during the viral life cycle, HBV and individual APOBEC3-encoding plasmids were cotransfected into HepG2 cells and the activity of APOBEC3-induced mutations was initially analyzed by primer extension. As shown in Figure 2A, all appropriate-sized APOBEC3 proteins were detected by the Western blotting in the cell lysates. All APOBEC3s had a comparable protein expression level except A3D, which was barely detectable, and the A3H-II G105R mutant (36), which was unusually high. The mock vector was used as the control. To investigate potential endogenous APOBEC3 expression in HepG2 cells, the total RNAs in the mock vector control cells were extracted and the APOBEC3 levels were analyzed by quantitative RT-PCR. As shown in Figure 2B, all APOBEC3s except A3A were detectable with relative expression levels of A3C > A3G > A3B >> A3F > A3D > A3H. A3C was the major form followed by A3G and A3B with about 50% and 25% of A3C mRNA levels, respectively. However, the endogenous protein expression of A3B, A3C, and A3G in the vector control were undetectable by Western blotting using antibodies against native A3B, A3C, or A3G, respectively, whereas the exogenous overexpressions were readily detectable as shown in Figure 2C. To further investigate the effect of the endogenous APOBEC3s, HBV viral DNAs were isolated from HepG2 cells with or without treatment of (−)siRNA control or A3C siRNA (siA3C) and HBV mutations were analyzed by primer extension (pe1453) as described in the next section. As shown in Figure 2E, A3C mRNA was knocked down to <5% of vector control by the siA3C treatment. The HBV mutation represented by ladder-like bands for C-to-T mutations at sites of 1453, 1452, 1451, 1448, 1446, and 1443 in the sequencing gel electrophoresis was significantly diminished by the siA3C treatment. These data suggest that the endogenous APOBEC3 protein levels in HepG2 cells are much lower than the overexpressed APOBEC3s and their effect should be negligible, but that endogenous A3C plays the major role on HBV mutation in the mock vector control.
Figure 2.
Coexpression of APOBEC3 proteins with HBV. The HBV-encoding plasmid was cotransfected with individual APOBEC3 into HepG2 cells. A, coexpressed APOBEC3 protein analyses. After a 48-h transfection, the cells were lysed and the expressed proteins in the cell lysates were analyzed by 10% polyacrylamide gel electrophoresis followed by Western blotting. The Western blot was cut into two parts along the protein marker lane for two different staining. The expressions of A3C and A3G were detected with an anti-HA antibody against their HA tag in the C terminus, and the expressions of A3A, A3B, A3D, A3F, and A3H were detected with anti-Flag antibody against their Flag tag in the C terminus. The GAPDH expression was also analyzed as an internal reference for each protein loading. Mock vector was used as control. Of note, the horizonal cut in the lower vector control lane by anti-HA staining was caused by a scotch tape. B, endogenous APOBEC3 mRNA expression analyses in the mock vector control. Total RNAs were extracted from the vector control HepG2 cells. APOBEC3 mRNA expression levels were analyzed by quantitative RT-PCR. Each APOBEC3 mRNA expression is presented in the left panel by their levels relative to house-keeping gene GAPDH. In addition, each APOBEC3 mRNA expression was also converted to the percentages relative to A3C and presented in the right panel for an easy comparison. C, endogenous APOBEC3 protein level analyses in the mock vector control. Additional endogenous protein expression of A3B, A3C, and A3G in the vector control was tested and compared with exogenous APOBEC3 expression by Western blotting using antibodies against native A3B, A3C, or A3G in upper panel. The GAPDH expression was also analyzed as an internal reference for each protein loading in the lower panel. D, coexpressed HBV viral DNA analyses. After removal of potential contamination of HBV-encoding plasmid from the initial transfection by micrococcal nuclease pretreatment, HBV genomic DNA was isolated from viral capsids in the cell lysate. The expression of HBV viral DNAs under each APOBEC3 treatment was verified by PCR-95 °C amplification using primer pair F874/R5 followed by 1% agarose gel electrophoresis. E, A3C siRNA knockdown effect on endogenous HBV mutation in HepG2 cells. HepG2 cells were reversely transfected by A3C siRNA or siRNA negative control overnight and then were further transfected by the HBV-encoding plasmid with mock vector or A3C. Total RNAs or HBV viral DNAs were isolated from duplicate experiments for analyses of A3C knockdown effect by quantitative RT-PCR or primer extension (pe1453). A3C mRNA was knocked down to <5% of vector control by siA3C in the left panel. The HBV mutation represented by ladder-like bands for C-to-T mutations at sites of 1453, 1452, 1451, 1448, 1446, and 1443 in the sequencing gel electrophoresis was significantly diminished by siA3C treatment in the right panel. HBV, hepatitis B virus.
To analyze APOBEC3 mutational activity on HBV, HBV DNAs were isolated from viral capsids in the cell lysates after removal of potential contamination of HBV-encoding plasmid from the initial transfection by a treatment of micrococcal nuclease. HBV viral DNAs were then PCR amplified using primer pairs of F874/R5 or F1/R2475. As shown in Figure 2D, HBV viral DNAs were detected in all APOBEC3 and HBV coexpression experiments. To evaluate APOBEC3s-induced mutation activities by the primer extension analyses, APOBEC3 mutation hot spots or regions were identified through sequencing studies described in the following sections and primers against specific HBV cytidine sites were designed as previously described (7, 32, 37). Briefly, primers against cytidines at sites 1453 and 1169, corresponding to the HBV viral reverse transcription initiation site and termination site, respectively, were representatively selected for primer extension analyses. During primer extension reactions, pe1453 or pe1169 primer annealed to the specific region of HBV PCR-95 °C amplifications and then the annealed primers were extended using DNA polymerase from a Sanger sequencing kit against the template in the presence of ddGTP. The mutation frequency of cytidine at the site 1453 or 1169 was determined by quantitating primer extension products due to ddGTP incorporation stop or not as described in Experimental procedures.
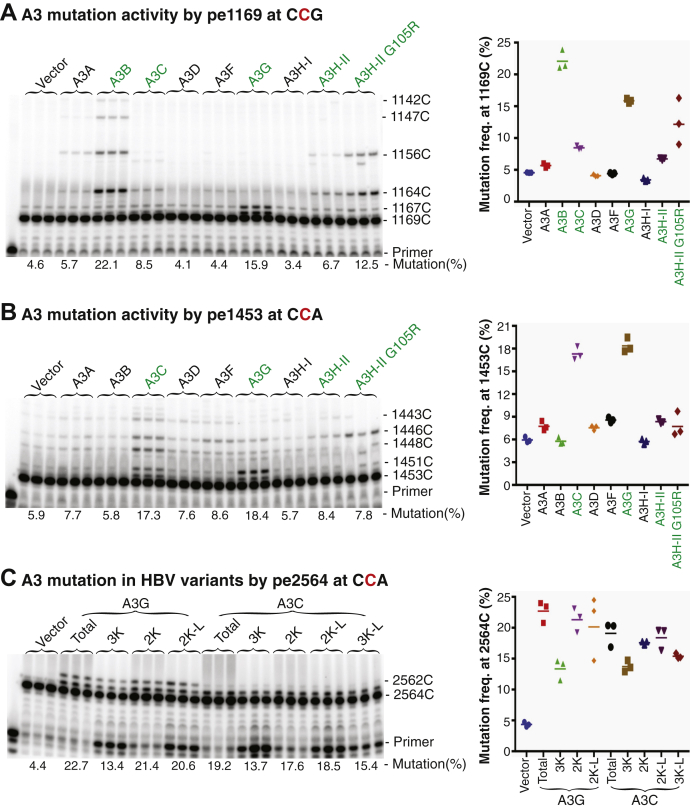
As shown in Figure 3, the primer extension products were clearly detected in a ladder-like form for A3B, A3C, A3G, and A3H-II. A3B had a 22.1% C-to-T mutational frequency for the cytidine at site 1169 by pe1169, whereas 5.8% was detected by pe1453. This likely indicates that A3B mutates 22.1% of HBV genomes without consideration of the potential run-through background contribution due to the sequencing gel separation limitation. The lower A3B mutational frequency for the cytidine at site 1453 by pe1453 likely reflects A3B-induced mutations at the pe1453 primer binding site that abolished primer extension and led to a lower mutation result (as shown in sequencing section below). A3G had comparable 15.9% and 18.4% mutation frequencies by pe1169 and pe1453, respectively. A3C had a 2-fold higher mutation frequency with up to 17.3% by pe1453 compared with its 8.5% mutational frequency by pe1169. A3H-II and its activity enhanced mutant, A3H-II G105R (36) had 6.7% and 12.5% mutational frequencies by pe1169, whereas 8.4% and 7.8% were detected by pe1453, respectively. These data indicate that A3B/C/G/H-II have strong mutational activities on HBV viral DNA and their mutation frequencies could be variable for cytidines at different genome location. On the other hand, weak activities were observed for A3A/D/F with mutational frequencies slightly above vector control by pe1169 or pe1453. Nevertheless, the primer extension products in a ladder-like form by pe1169 or pe1453 were clearly observable above that of the vector control, indicating that A3A/D/F were functional, although their mutational activities were weak. In contrast, A3H-I was indistinguishable compared with the vector control. These data indicate that all APOBEC3s except A3H-I can induce C-to-T mutations on HBV viral DNA, but their mutational activities vary significantly.
Figure 3.
APOBEC3-induced HBV DNA mutation analyses by primer extension. An APOBEC3-encoding plasmid was cotransfected with HBV into HepG2 cells as in Figure 2. HBV DNAs from the viral capsids were PCR amplified with primers of F874/R5 for pe1169 or F1/R2475 for pe1453 as in Figure 1.A and B, HBV mutational frequency analyses by primer extension with PCR-95 °C amplicons. After removal of dNTP, the PCR-95 °C amplicons were used to determine HBV C-to-T mutation frequencies through primer extension analyses for cytidine at site 1169 (A) by pe1169 and 1453 (B) by pe1453, respectively. The primer extension products were separated by an 8% polyacrylamide sequencing denaturing gel followed by PhosphorImager quantitation. The data are presented with the primer extension product gel analyses in the left panel and mutation frequency graph in the right panel. The “pe” stands for primer extension. Triplicate samples were used for each determination. C, HBV mutational frequency analyses of different HBV genome variants. HBV genome variants from PCR-95 °C amplifications using primers F1/R5 as in Figure 1 were separated by 1% agarose gel electrophoresis and the corresponding bands including full-length 3K, 3K-L, 2K, and 2K-L were gel isolated for primer extension analyses by pe2564. HBV, hepatitis B virus.
The strong mutational activities of A3B/C/G/H-II on HBV DNA made them good candidates for further mutation landscape analyses. Since the HBV genome had multiple variants due to alternative RNA splicing deletions, APOBEC3 mutational activity on different HBV genome variants were further investigated. The genomic PCR-95 °C amplifications from A3C/G-treated HBV were separated by 1% agarose electrophoresis and HBV genome variants were gel purified for mutational frequency analyses by the primer extension method. As shown in Figure 3C, A3G had 13.4%, 21.4%, and 20.6% mutational frequencies by pe2564 for 3K, 2K, and 2K-L isolations, respectively. A3C had 13.7%, 17.6%, 18.5%, and 15.4% mutational frequencies for 3K, 2K, 2K-L, and 3K-L isolations, respectively. Both A3G and A3C had the lowest mutational frequencies with 3K and increased mutational frequencies with 2K or 2K-L. The lower mutational activity of APOBEC3s with full-length 3K HBV genome indicates that the limited space in capsids probably restricts the access of APOBEC3s to the ssDNA substrate.
HBV mutations are only detected in rcDNA, not cccDNA or RNA by primer extension analyses
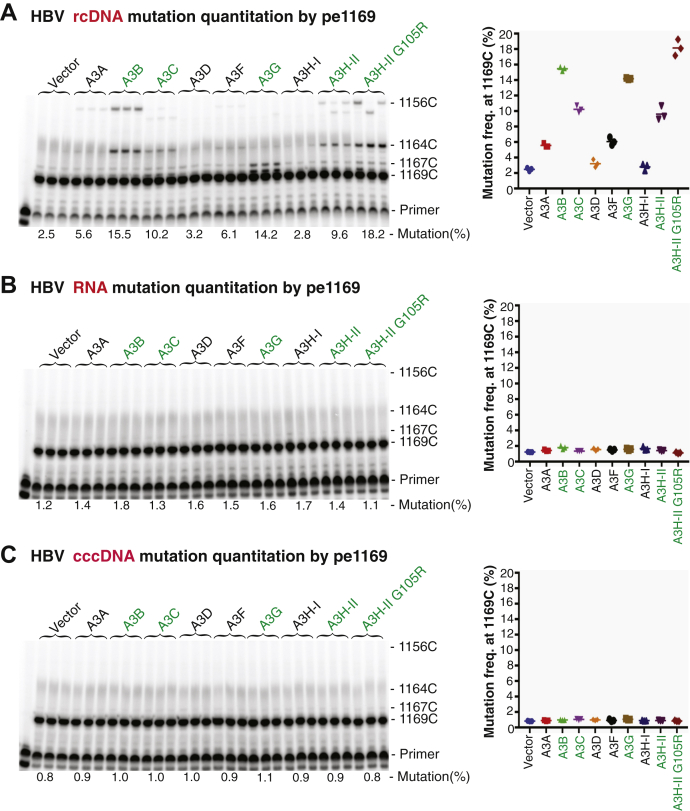
HBV viral genome mutations occur in the viral capsids when the viral pgRNA is reverse transcribed into (−)-DNA followed by conversion into rcDNA. The rcDNA capsid intermediate formed in the cytoplasm can potentially re-enter nuclei for another intracellular replication cycle or form mature virus through envelope formation and subsequent release from cells (5, 6). If the intracellular cycle occurs, HBV mutations should be detectable in covalently closed circular DNA (cccDNA) in the nuclei or viral RNA in the cytoplasm. In addition, viral mutations detected in rcDNA should represent cumulative effects of multiple HBV viral replication cycles. To investigate if cccDNA or viral RNA had mutations, total cellular RNAs or HBV cccDNAs were extracted from HepG2 cells after cotransfection of APOBEC3s with HBV. After removal of potential contamination by DNase for RNA or Dpn I for cccDNA, the corresponding HBV DNAs were amplified by either RT-PCR from the viral RNA or direct PCR from the cccDNA extractions using a 95 °C denaturing temperature. These HBV amplifications were then analyzed by the primer extension through pe1169 and pe2333 for the presence of C-to-T mutations. As shown in Figure 4, the C-to-T mutations in the HBV rcDNAs were readily detectable by pe1169 with mutation frequencies up to 15.5%, 10.2%, 14.2%, 9.6%, and 18.2% for A3B, A3C, A3G, A3H-II, and A3H-II G105R, respectively. However, parallel preparations from HBV RNA or cccDNA had no detectable C-to-T mutations except the background by both pe1169 and pe2333 analyses (data for pe2333 not shown). These data clearly demonstrate that there is no apparent intracellular replication cycle for the HBV viral capsids and that HBV mutations in rcDNA from the capsids reflect a one-time event of APOBEC3-induced HBV mutation.
Figure 4.
HBV rcDNA alone, not pgRNA or cccDNA, had C-to-T mutations as determined by primer extension analyses. APOBEC3-encoding plasmids were cotransfected with HBV into HepG2 cells as in Figure 2. After a 48-h transfection, HBV rcDNA viral capsids in the cytoplasm were obtained by lysing the cellular membrane with 0.5% NP-40 buffer. The remaining nuclei of the cells were further lysed with 1% SDS buffer for cccDNA extraction. In addition, HBV viral RNAs were obtained from total cellular RNA extraction in a duplicate experiment. After precleaning of potential contamination of HBV-encoding plasmid from the initial transfection, HBV rcDNA, cccDNA, or RNA were isolated and HBV mutations were analyzed by primer extensions using PCR-95 °C or RT-PCR-95 °C amplifications with the primer pair F874/R5. The representative pe1169 primer extension analyses were shown for HBV rcDNA (A), HBV pgRNA (B), and HBV cccDNA (C), respectively. The mutational analyses are presented with the primer extension product gel analyses in the left panel and mutation frequency graph in the right panel as in Figure 3. Triplicate samples were used for each determination. cccDNA, covalently closed circular DNA; HBV, hepatitis B virus; pgRNA, pre-genome RNA; rcDNA, relaxed circular DNA
Evaluating A3B/C/G/H-II mutational activities using short HBV fragment sequencing analyses
The primer extension analyses described above demonstrate that A3B/C/G/H-II have strong mutational activities on HBV viral DNA. Using primer extension, target cytidines at sites 1169 and 1453 were selected and primers for pe1169 and pe1453 were designed to anneal to the PCR template region immediately adjoining the target cytidines for primer extension reactions. A potential problem with primer extension is that the primer-binding site could be mutated, which would lead to lower estimates of mutational frequencies like the case of the A3B mutation analyses by pe1453 as shown in Figure 3B. On the other hand, the HBV mutational activities can alternatively be analyzed by cloning and sequencing of a short fragment PCR amplification (7, 32). To further evaluate A3B/C/G/H-II mutational activities, HBV 1360 to 2090 nt region was selected and amplified by PCR-95 °C for cloning and sequencing analyses.
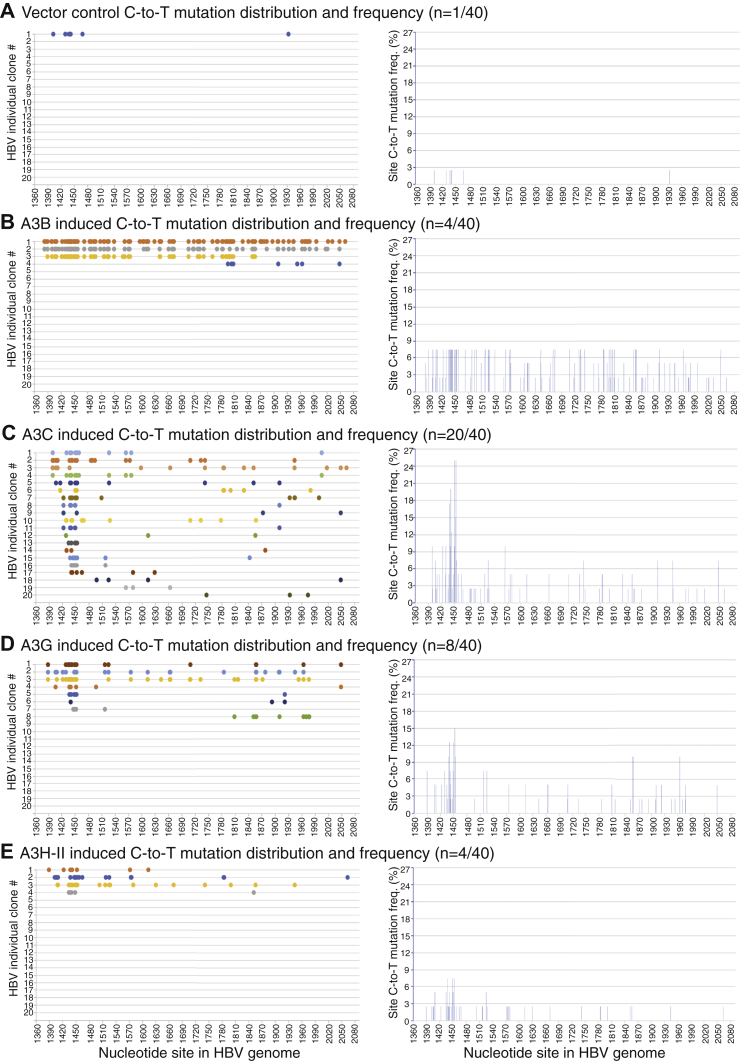
As shown in Figure 5 and Table 1, there was only one mutation-positive clone of 40 randomly selected clones in the mock vector control. A3B, A3C, A3G, and A3H-II coexpression resulted in 4, 20, 8, and 4 mutation-positive clones of 40 randomly selected clones, respectively. In addition, as shown in Table 1, the total detected number of C-to-T changes were seven for the vector control in contrast to 247, 162, 102, and 55 for A3B/C/G/H-II, respectively, demonstrating that the endogenous APOBEC3s have a negligible effect on overexpressed A3B/C/G/H-II-induced HBV mutations. Among the four A3B-induced mutation clones, three of them had extensive C-to-T mutations, and in one of these, A3B mutated 55% of 180 cytidines available in the fragment region, which was defined as mutational efficiency in a single mutation reaction as shown in Table 1. A3B had an average of 34.3% mutational efficiency that was much higher than the 3.9% in the vector control and average mutational efficiencies of 4.5%, 7.1%, and 7.7% for A3C, A3G, and A3H-II, respectively. As shown in Figure 5B, right panel, A3B induced an up to a 7.5% cytidine mutational frequency on most cytidine sites across the PCR analyzed region. Of note, the site C-to-T mutational frequencies represent the number of C-to-T mutations for a specific cytidine site in mutation-positive clones divided by the sequenced clone number. For example, the C-to-T mutation at site1453 appeared three times among the four mutation-positive clones in the total 40 sequenced clones by A3B. Therefore, the cytidine mutational frequency at site 1453 was 3/40 = 7.5%.
Figure 5.
Evaluation of A3B/C/G/H-II mutational activity using short HBV fragment sequencing analyses. APOBEC3-encoding plasmids were cotransfected with HBV into HepG2 cells as in Figure 2. After a 48-h transfection, HBV relaxed circular DNAs were isolated from viral capsids in the cytoplasm and amplified by PCR-95 °C using primers F1/R2065 targeting the HBV 1360 to 2090 nt region fragment. The resultant 730-bp HBV amplicons were TA-cloned into a pCR4 vector for sequencing analyses. Mutation-positive clones of 40 randomly selected clones for (A) vector control, (B) A3B, (C) A3C, (D) A3G, and (E) A3H-II are presented as the C-to-T mutation distribution in each clone against the corresponding cytidine sites in the HBV 1360 to 2090 nt region. The cytidine site locations are linearly presented with their site numbering reference to HBV V01460.1. The site mutational frequencies for each cytidine in the HBV 1360 to 2090 nt region represent the number of C-to-T mutations for a specific cytidine site in mutation-positive clones divided by the sequenced clone number and are presented as percentage bars in the right panel under each treatment. HBV, hepatitis B virus.
Table 1.
APOBEC3 mutational activities by sequencing analyses of HBV 1360-2090 nt PCR
| Treatment | Total sequenced clones (n) | Mutation-positive clones (x/n) | Total C-to-T # detected | Average C-to-T# per clone | Mutation efficiency (% of 180 cytidinesa) | Mutation efficiency average (%) |
|---|---|---|---|---|---|---|
| Vector | 40 | 1/40 | 7 | 7 | 3.9 | 3.9 |
| A3B | 40 | 4/40 | 247 | 61.8 | 3.9–55 | 34.3 |
| A3C | 40 | 20/40 | 162 | 8.1 | 1.7–11.7 | 4.5 |
| A3G | 40 | 8/40 | 102 | 12.8 | 1.7–17.2 | 7.1 |
| A3H-II | 40 | 4/40 | 55 | 13.8 | 2.8–12.8 | 7.7 |
The # stands for C-to-T mutation number.
There are 180 cytidines available to potential mutation in the HBV 1360 to 2090 nt.
As shown in Figure 5C, A3C coexpression resulted in 20 mutation-positive clones of 40 random selections. Most of the mutation-positive clones had C-to-T mutations for cytidines at sites 1449 and 1452 with an up to 25% mutational frequency, which was more than 3-fold higher compared with A3B's 7.5%. However, A3C mutational efficiency averaged 4.5% with the highest 11.7% by their single clonal mutation reaction over 180 cytidines available in the region as shown in the Table 1. As shown in Figure 5, D and E, A3G and A3H-II had eight and four mutation-positive clones of 40 random selections with up to 15% and 7.5% mutational frequencies for the cytidine at site 1452, respectively. As shown in Table 1, A3G and A3H-II had similar mutational efficiencies, averaging 7.1% and 7.7%, respectively. These data confirm the mutational activities of A3B/C/G/H-II on HBV viral DNA in contrast to the mock vector control. In addition, the data also demonstrate that A3B has a uniquely high mutational efficiency with an up to 5-fold greater C-to-T mutations per clone than A3G/H-II, indicating that A3B should be a better candidate for investigating HBV mutation genome-wide distribution. On the other hand, A3C has more than a 3-fold higher mutational frequency compared with A3B/G/H-II, indicating that A3C has a higher accessibility to its ssDNA substrate.
A3B/G-induced HBV C-to-T mutations occur during viral pregenomic RNA reverse transcription
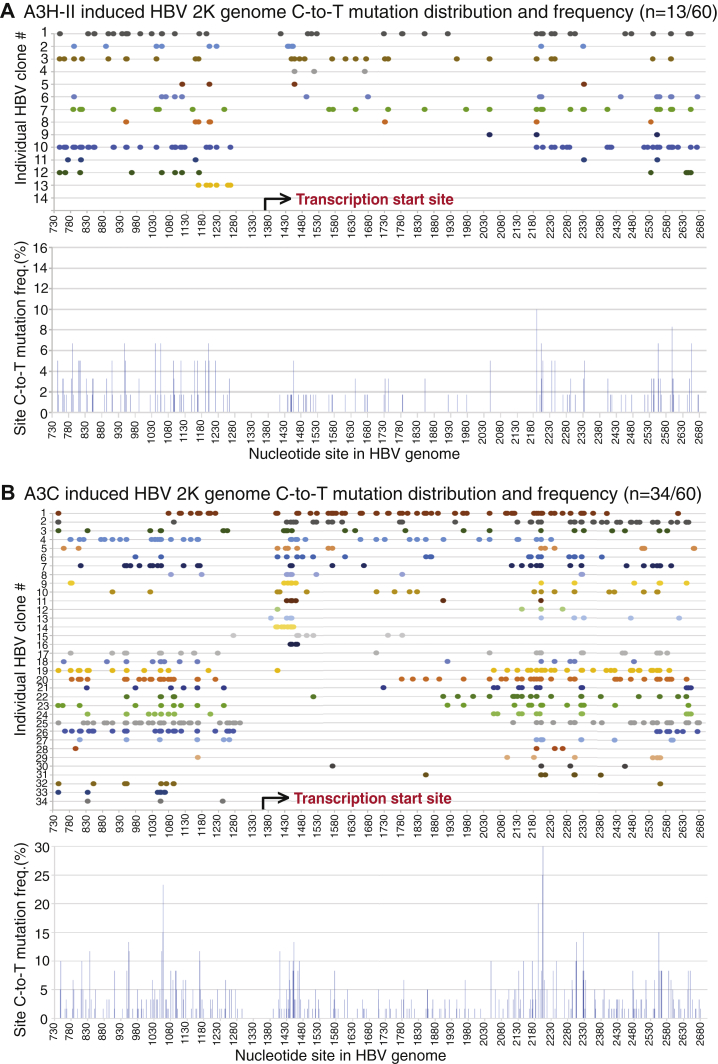
To investigate how APOBEC3s mutate HBV viral DNA during the viral life cycle, especially concerning availability of ssDNA substrate, HBV and individual A3B/C/G/H-II-encoding plasmids were cotransfected into HepG2 cells and resultant HBV rcDNA mutations were analyzed through genomic landscape. Briefly, HBV genome variants were amplified by PCR-95 °C using the primer pair F1/R5, which annealed to the terminal ends of HBV (−)-DNA as in Figure 1. The resultant genomic amplicons corresponding to HBV full-length 3K and the major internal deletions 2K and 2K-L were gel purified and cloned into pCR4 vector. A total of 60 to 120 clones of each HBV genome variant for each treatment including A3B, A3G, A3H-II, and A3C were randomly selected for Sanger sequencing analyses.
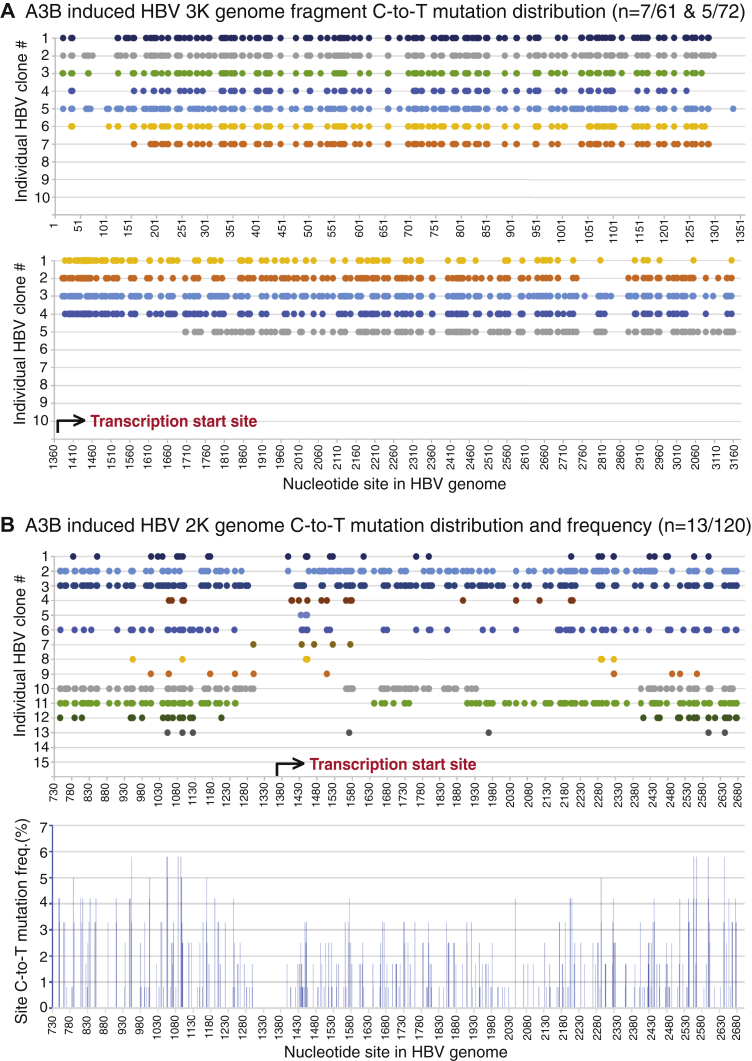
As shown in the Table 2, there was surprisingly no mutation-positive clones found of 120 randomly selected clones for A3B-treated HBV 3K full-length genomes. A similar situation was also observed with A3G-treated HBV 3K genomes where none was found of 60 randomly selected clones. However, 29 of 60 A3C-treated HBV 3K genome randomly selected clones were mutation positive. Since A3B had a >7-fold higher mutational efficiency than A3C as described above in Table 1, the data indicate that A3B-induced HBV 3K full-length genome C-to-T mutations could be too high for the full-length form to be cloned. Since the HBV 3K full-length genome can be selectively amplified with primers F1/R3182 and F12/R5 as described in Figure 1, we split the 3K HBV genome into two fragments (3K-I for 1–1360 nt and 3K-II for 1370–3182 nt) for further cloning and sequencing analyses with A3B. By this approach, A3B-induced extensive mutations in the 3K HBV full-length genomes were detected. As shown in Figure 6A and the Table 2, seven of 61 clones for 3K-I and five of 72 clones for 3K-II had extensive C-to-T mutations. On average 142.7 and 175 C-to-T mutations per clone were detected for 3K-I and 3K-II, respectively, indicating that on average 50.4% and 42.7% cytidines available in 3K-I (total 283 cytidines) and 3K-II (total 410 cytidines), respectively, were mutated. The A3B-induced C-to-T mutations occurred across all the HBV fragment regions in all clones except two as shown in Figure 6A. The reverse transcription of the viral RNA genome started from 1370 nt and moved toward 3182/1 nt and then continued from 1 nt to 1360 nt (see Fig. 1A) (5). As shown in the Figure 6A lower panel, four of the five mutation-positive clones for region 1370 to 3182 nt had C-to-T mutations starting from the reverse transcription site around 1380 nt and continuing along the (−)-DNA chain up to 3182 nt. The remaining single clone had C-to-T mutations starting from 1710 nt and continued sequentially to 3182 nt. On the other hand, all clones for region 1 to 1360 nt had sequential C-to-T mutations along the chain and all ended around 1300 nt. Although these mutation-positive clones of fragment 3K-I and 3K-II might not necessarily come from the same 3K genome molecule, the C-to-T mutation distribution pattern suggests that A3B-induced HBV 3K genome mutations probably occur during the viral reverse transcription process.
Table 2.
APOBEC3 mutational activities by sequencing analyses of HBV genome variants
| Treatment & genome variants | Total sequenced clones (n) | Mutation-positive clones (x/n) | Total detected C-to-T # |
Average C-to-T# per clone | Average C-to-T#/total Ca in a single clone (%) | C-to-T #/total Ca in a single clone (%) range | Preference for 5′ immediate nucleotide |
|---|---|---|---|---|---|---|---|
| A3B 3K | 120 | 0/120 | n/a | n/a | n/a | n/a | |
| A3B 3K-I | 61 | 7/61 | 999 | 142.7 | 50.4 | 34.6–64.7 | 5′TC & 5′CC |
| A3B 3K-II | 72 | 5/72 | 875 | 175 | 42.7 | 28.5–55.4 | (+++) |
| A3B 2K | 120 | 13/120 | 818 | 68.2 | 15.1 | 1.1–41.2 | |
| A3G 3K | 60 | 0/60 | n/a | n/a | n/a | n/a | |
| A3G 2K | 60 | 8/60 | 229 | 28.6 | 6.3 | 0.7–14.4 | 5′TC |
| A3G 2K-L | 60 | 10/60 | 447 | 44.7 | 12.2 | 1.4–21.9 | (+++) |
| 3H-II 3K | 60 | 11/60 | 305 | 27.7 | 4 | 0.4–11.4 | 5′TC |
| 3H-II 2K | 60 | 13/60 | 225 | 17.3 | 3.8 | 0.7–13.1 | (+++) |
| 3H-II 2K-L | 60 | 8/60 | 133 | 16.6 | 4.5 | 1.4–11.2 | |
| A3C 3K | 60 | 29/60 | 456 | 15.7 | 2.3 | 0.4–5.3 | 5′TC & 5′CC |
| A3C 2K | 60 | 34/60 | 728 | 21.4 | 4.7 | 0.7–16 | (+) |
| A3C 2K-L | 60 | 37/60 | 582 | 15.7 | 4.3 | 0.8–27.9 |
The underline indicates where the cytidine deamination occurs. The "+" and "+++" indicate weak and strong preference of APOBEC3s for 5' immediate nucleotide, respectively. The "#" stands for C-to-T mutation number.
There are 693, 451, 365, 283, and 410 cytidines in HBV 3K, 2K, 2K-L, 3K-I, and 3K-II, respectively, available for mutation.
Figure 6.
A3B-induced HBV 3K and 2K genomic mutational landscape analyses. A3B- and HBV-encoding plasmids were cotransfected into HepG2 cells as in Figure 2. HBV genomic DNA was isolated from viral capsids and amplified by PCR-95 °C using primers F12/R5 for 3K-I (1–1360 nt), F1/R3182 for 3K-II (1370–3182 nt), or F1/R5 for 2K genome as in Figure 1. The resultant genomic amplicons were separated by 1% agarose electrophoresis. and the corresponding bands were gel-purified and TA-cloned into a pCR4 vector for sequencing analyses. A, A3B-induced 3K genome mutation analyses through two split fragments, 3K-I and 3K-II. Mutation-positive clones obtained from 3K-I (n = 61) and 3K-II (n = 72) randomly selected clonal sequencing are collectively presented as the C-to-T mutation distribution in each clone against the corresponding cytidine site in the HBV genome with reference to HBV V01460.1. B, A3B-induced 2K genome mutation analyses. Mutation-positive clones from 2K genome (n = 120) clonal sequencing are presented as the C-to-T mutation distribution in each clone against the corresponding cytidine site in the HBV 2K genome. The cytidine site locations are linearly presented with their site numbering reference to HBV V01460.1, although the actual clonal sequences have an opening at 1340 to 1360 nt due to PCR primer design and the two ends of 737 nt and 2696 nt are covalently connected. The site mutation frequencies for each cytidine in A3B 2K genomes represent the number of C-to-T mutations for a specific cytidine site in mutation-positive clones divided by the sequenced clone number and are presented as percentage bars in the lower panel. HBV, hepatitis B virus.
Although no mutation-positive clones were obtained with HBV 3K full-length genome by A3B coexpression, 13 of 120 clones of the 2K genome variant were found to be C-to-T mutation positive. As shown in Figure 6B, A3B-induced HBV 2K genome mutations varied significantly. Of note, the HBV 2K genome clone had actual start and end sequences around 1360 nt due to the two cloning primer locations and the two ends of 2696 and 737 nt were covalently connected owing to an internal deletion (see Fig. 1A). However, a linear 730 to 2700 nt region was presented for an overview of C-to-T distributions by referencing to the HBV genome sequence V01460.1. As shown in Figure 6B upper panel, ten of 13 mutant clones had C-to-T mutations beginning from or near the viral reverse transcription starting site (1400–1450 nt) and continuing sequentially along the chain until the other transcription end (1240–1300 nt) in a continuous on or off manner with significantly varied efficiency from clone to clone. Nevertheless, clone No. 2 had 166 C-to-T mutations sequentially distributed over the entire genome from the reverse transcription start site to the termination end with 36.8% of 451 cytidines mutated in a single 2K molecular mutation, indicating that the A3B-induced mutation co-occurred along with the viral reverse transcription process. Besides these, one clone (No. 12) started C-to-T mutations around the 2400 nt location and then continued to the other end to around 1200 nt, indicating that A3B-induced HBV 2K genome mutations could also start at a downstream site. As shown in Table 2, the A3B HBV 2K mutation efficiency varied from 1.1% to 41.2% with an average of 15.1% when considering a total of 451 cytidines being available in the 2K genome, indicating that A3B was a potent mutational agent and could mutate up to 41.2% cytidines in a single HBV 2K genome. The mutational frequencies for each cytidine in the genome were collectively calculated using the number of C-to-T mutations at a specific cytidine site in all mutation-positive clones divided by the number of sequenced 120 HBV 2K clones. The resulting percentage is presented as a bar at each C-to-T mutation site in the HBV V01460.1 genome. As shown in Figure 6B lower panel, the overall cytidine mutation frequencies were about the same with gradual increases from the viral transcription start site toward the other end of the genome. These data clearly demonstrate that A3B mutates HBV viral DNA during the viral reverse transcription process when single-stranded (−)-DNA becomes available.
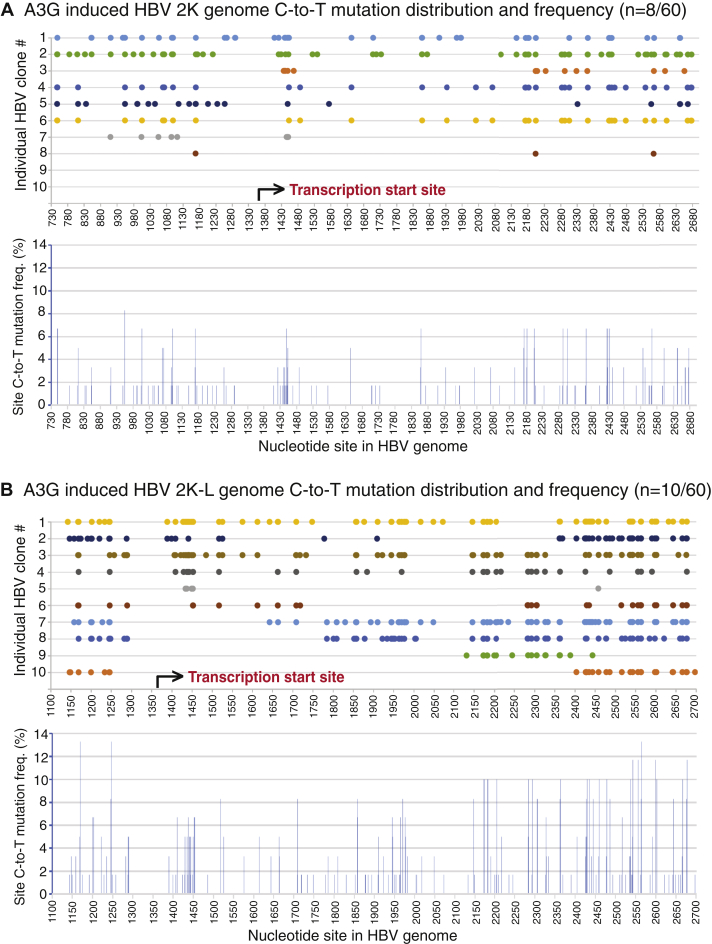
A3G-induced HBV genome mutations were investigated by sequencing 60 randomly selected clones for each HBV genome variant including 3K, 2K, and 2K-L. No mutation-positive HBV 3K full-length clone was found except for one clone with a minor mutation, which was not consistent with A3G activities observed by primer extension analyses above. This indicates that like that of A3B 3K above, C-to-T mutations in A3G 3K could be too high for cloning. As shown in Table 2 and Figure 7, eight mutation-positive clones from the 2K genome and ten from 2K-L of 60 randomly selected clones were found with average 28.6 and 44.7 C-to-T mutations per clone, or average 6.3% and 12.2% mutation efficiency, respectively. A3G HBV 2K-L had a 2-fold increased mutation efficiency compared with A3G HBV 2K, indicating that A3G mutated the smaller HBV genome more effectively within the limited space of the HBV viral capsids. As shown in Figure 7A, all 2K mutation-positive clones except one had C-to-T mutations from the viral reverse transcription starting region 1410 to 1460 nt through the other end in an on and off manner, indicating that like A3B, A3G HBV mutations also co-occurred along with HBV viral reverse transcription. As shown in Figure 7B, 2K-L had a distribution pattern similar to the 2K genome. Of ten clones, six had C-to-T mutations from the reverse transcription starting region 1390 to 1450 nt. The remaining clones had C-to-T mutations beginning downstream sites of the reverse transcription starting site. All clones except two (clone #5 and #9) had C-to-T mutations sequentially along the chain to 1250 to 1300 nt in an on and off manner. Compared with the 2K genome, 2K-L had C-to-T mutations closer to both ends by reference to the viral reverse transcription process. 2K-L had mutations as early as 1389 nt and as late as 1289 nt. These data suggest that A3G has a higher accessibility to the ssDNA substrate with the smaller HBV 2K-L genome and like A3B, A3G-induced mutation is strongly associated with the HBV viral reverse transcription process.
Figure 7.
A3G-induced HBV 2K and 2K-L genomic mutational landscape analyses. A3G- and HBV-encoding plasmids were cotransfected into HepG2 cells as in Figure 2. HBV genomic DNA was isolated from viral capsids and amplified by PCR-95 °C with primers F1/R5 as in Figure 1. The resultant 2K and 2K-L genomes were gel-purified and TA-cloned into a pCR4 vector for sequencing analyses. A, A3G-induced 2K genome (n = 60) mutation analyses. The clonal C-to-T mutation distribution against the corresponding cytidine site in the HBV genome are linearly presented with site numbering reference to HBV V01460.1 in the upper panel, although the actual clonal sequences have an opening at 1340 to 1360 nt and the two ends of 737 nt and 2696 nt are covalently connected. The collective site mutation frequencies for each cytidine in HBV 2K genome represent the number of C-to-T mutations for a specific cytidine site in mutation-positive clones divided by the sequenced clone number and are presented as percentage bars in the lower panel. B, A3G-induced 2K-L genome (n = 60) mutation analyses. Clonal C-to-T mutation distributions and frequencies for each cytidine mutation are collectively presented in upper and lower panels, respectively, as in Figure 7A.
A3C mutates HBV genomes with unique high mutation frequency
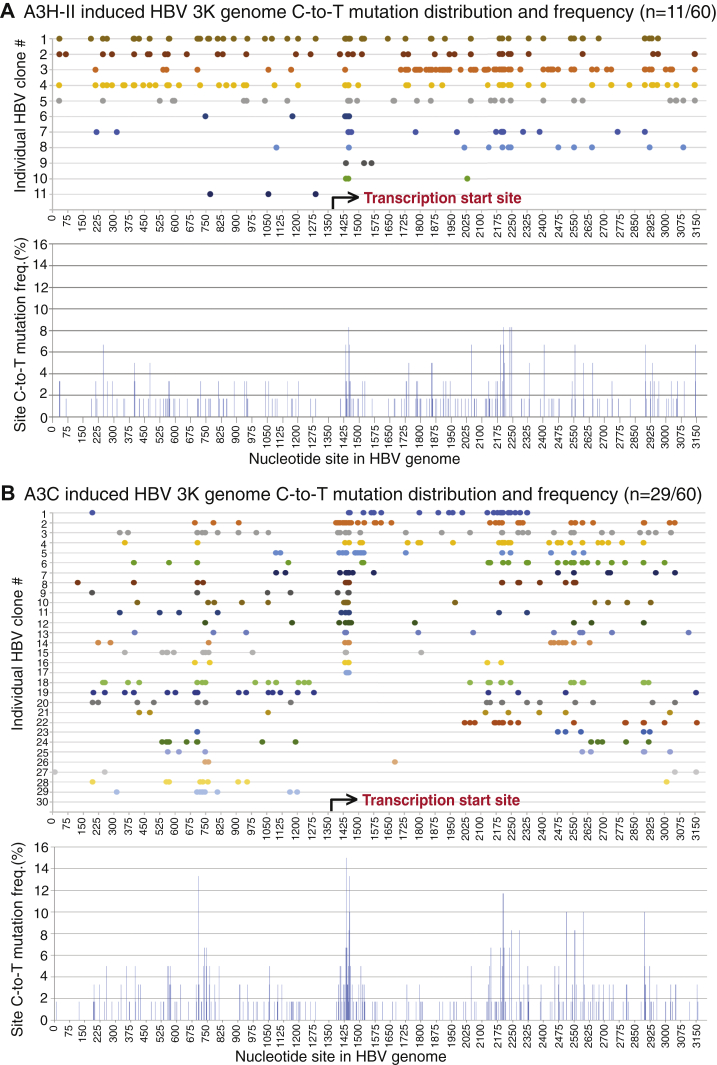
A3A and A3H have attracted attention in the cancer genome mutation field because they are potential mutation-inducing proteins, especially in the absence of A3B. However, A3A and A3H-I had no significant mutational effect on HBV in HepG2 cells by primer extension analyses as described in Figure 3. On the other hand, the other major A3H variant, A3H-II, and A3C were highly effective. Both A3H-II and A3C were cotransfected with HBV into HepG2 cells, and their mutation activities were analyzed through HBV genome variants including full-length 3K, 2K, and 2K-L. The mutation-positive clones of the 3K full-length genome were obtained for both A3H-II and A3C in contrast to A3B or A3G where no mutants were obtained even with screening up to 120 clones. As shown in Table 2, 60 randomly selected clones from A3H-II-treated HBV genomes resulted in 11, 13, and 8 mutation-positive clones with an average of 27.7, 17.3, and 16.6 C-to-T mutations per clone for 3K, 2K, and 2K-L, respectively. Based on the total number of cytidines available in each HBV genome variant, the average mutation efficiencies were about the same, 4%, 3.8%, and 4.5% for 3K, 2K, and 2K-L, respectively. On the other hand, significantly more mutation-positive clones were obtained for A3C-treated HBV genomes. As shown in Table 2, of 60 randomly selected clones, 29, 34, and 37 mutants were obtained for A3C-treated HBV 3K, 2K, and 2K-L with an average of 15.7, 21.4, and 15.7 C-to-T mutations per clone, respectively. The average cytidine mutation efficiencies per clone were 2.3%, 4.7%, and 4.3% with the highest mutation efficiencies of 5.3%, 16%, and 27.9% for 3K, 2K, and 2K-L, respectively. These data demonstrate that A3C is a unique highly effective mutation agent toward HBV with about a 3-fold increased mutant frequency compared with others, although the individual mutation efficiency in a single clone was lower for 3K or about the same for 2K and 2K-L genomes compared with A3H-II.
The 3K C-to-T mutation distributions for each individual clone were presented in Figure 8 with a linear reference for each cytidine site in the HBV V01460.1 genomic sequence, although the actual sequence beginning and ending was within 1340 to 1360 nt and the two ends of 1 nt and 3182 nt were covalently connected (see Fig. 1A). As shown in Figure 8A, all A3H-II 3K mutants except one (clone #11) had C-to-T mutations beginning from the viral reverse transcription starting region 1410 to 1440 nt and continuing along the chain to the other end at 1200 to 1290 nt with variable efficiency. In contrast, 17 of 29 A3C HBV 3K mutants had C-to-T mutations beginning from the transcription starting region 1389 to 1450 and then continuing variably along the chain, but fewer than five clones reach the other end at 1125 to 1200 nt as shown in Figure 8B. The other 12 clones had C-to-T mutations beginning later than 2000 nt site and only five of them reached the other end. The overall mutation frequency for individual cytidine across the whole 3K genome was about same for both A3H-II and A3C. However, there were four high mutation cluster regions located around 1450, 2200, 2550, and 700 nt for A3C with up to 15% mutation frequency for the cytidine at site 1437, whereas A3H-II had two highly mutated clusters around 1450 and 2200 nt with up to 8.3% cytidine mutation frequency at sites 1449 and 2235. These data suggest that the A3H-II-induced HBV 3K mutations are similar to those of A3B and A3G, having clear starting and ending regions co-occurring with the viral reverse transcription process. However, most of A3C-induced HBV 3K mutations co-occur with the starting point of viral reverse transcription, but the rest of the clones are much less restricted to the initial site. Overall, A3C has a much higher mutation frequency on HBV 3K genome population (29 versus 11 mutants of 60 3K clones for A3C and A3H-II, respectively) but less mutational efficiency considering all cytidines available in 3K genome (2.3% versus 4% for A3C and A3H-II, respectively, see Table 2).
Figure 8.
A3H-II- and A3C-induced HBV full-length 3K genomic mutational landscape analyses. An A3H-II- or A3C-encoding plasmid was cotransfected with HBV into HepG2 cells as in Figure 2. HBV genomic DNA was isolated from viral capsids and amplified by PCR-95 °C with primers F1/R5 as in Figure 1. The resultant full-length 3K genomes were gel-purified and TA-cloned into a pCR4 vector for sequencing analyses. A, A3H-II-induced 3K genome (n = 60) mutation analyses and (B) A3C-induced 3K genome (n = 60) mutation analyses. The clonal C-to-T mutation distributions against the corresponding cytidine site in HBV genome are linearly presented with site numbering reference to HBV V01460.1 in the upper panel. The collective site mutation frequencies for each cytidine in HBV 3K genome represent the number of C-to-T mutations for a specific cytidine site in mutation-positive clones divided by the sequenced clone number and are presented as percentage bars in the lower panel. HBV, hepatitis B virus.
The 2K genome mutation distributions for both A3H-II and A3C were presented in Figure 9. As shown in Figure 9A, seven of 13 clones for A3H-II had C-to-T mutations starting around the viral reverse transcription site and then continuing along the chain to the other end with variable mutation efficiency. The remaining five clones had C-to-T mutation starting at downstream sites and then continuing to the other end. The highly mutated cluster observed with A3H-II 3K around 1450 nt was decreased significantly in the 2K genome. The overall cytidine mutation frequency across the A3H-II 2K genome increased from the viral reverse transcription start site toward the other termination end. In contrast, 17 of 34 clones for A3C 2K had C-to-T mutations starting around the transcription start site in 1390 to 1450 nt and continuing variably along the chain with only seven of them reaching the other termination end in 1150 to 1300 nt as shown in Figure 9B. The remaining 17 clones had mutations starting later and most of them continuing to the other end. The overall individual cytidine mutation frequency across the 2K genome varied with four highly mutated clusters occurring around 1450, 2200, 2550, and 1167 nt. The mutation frequency around 2200 nt cluster in A3C 2K increased up to 30% for the cytidine at site 2205, which was about 2-fold higher than those around the starting site with 13.3% for cytidine at site 1452. In contrast, A3C 3K had similar mutation levels in the two regions (see Fig. 8B). The C-to-T mutation distribution pattern with both A3H-II and A3C 2K provide further evidence that A3H-II- and A3C-induced mutations start from the HBV viral reverse transcription start site or downstream and then move toward the other termination end with increased cytidine mutation frequencies. These data also indicate that both A3H-II- and A3C-induced mutations become less restricted for their mutation starting points with the smaller-sized HBV 2K genome and have a higher chance to start later along the reverse transcription process as more space is available in the size-limited viral capsids. Similar A3H-II and A3C mutational patterns were also observed on HBV 2K-L (data not shown).
Figure 9.
A3H-II- and A3C-induced HBV 2K genomic mutational landscape analyses. An A3H-II- or A3C-encoding plasmid was cotransfected with HBV into HepG2 cells as in Figure 2. HBV genomic DNA was isolated from viral capsids and amplified by PCR-95 °C with primers F1/R5 as in Figure 1. The resultant 2K genomes were gel-purified and TA-cloned into a pCR4 vector for sequencing analyses. A, A3H-II-induced 2K genome (n = 60) mutation analyses and (B) A3C-induced 2K genome (n = 60) mutation analyses. The clonal C-to-T mutation distributions against the corresponding cytidine site in HBV 2K genome are linearly presented with site numbering reference to HBV V01460.1 in the upper panel. The collective site mutation frequencies for each cytidine in HBV 2K genome represent the number of C-to-T mutations for a specific cytidine site in mutation-positive clones divided by the sequenced clone number and are presented as percentage bars in the lower panel. HBV, hepatitis B virus.
The comparison of A3C-induced mutations in vitro versus in vivo
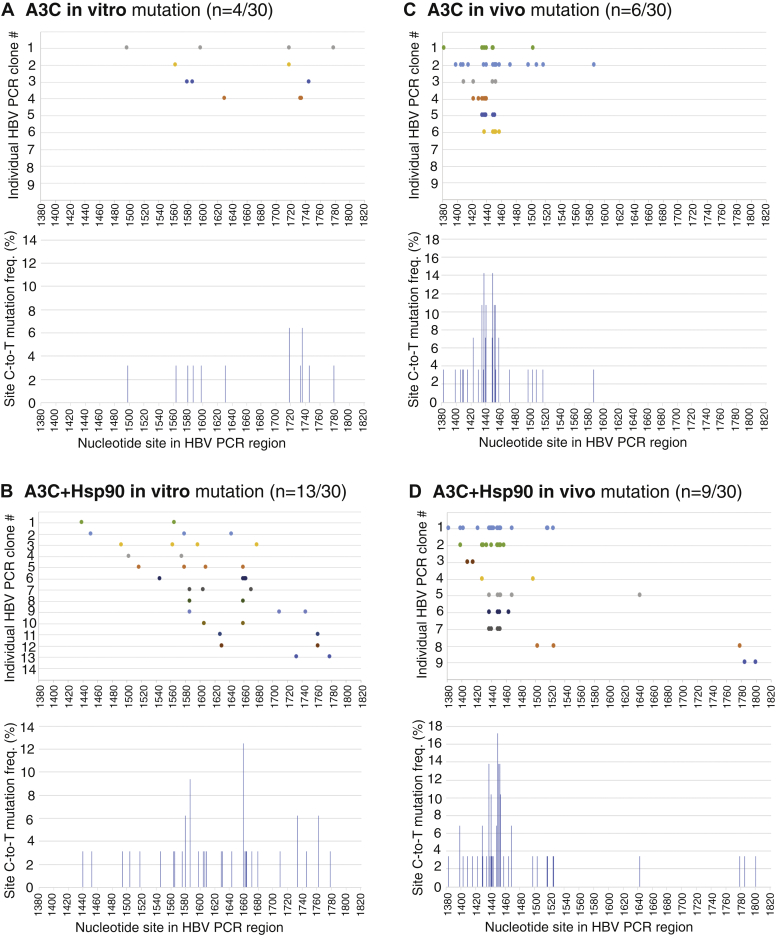
The APOBEC3 mutation distribution patterns as described above suggest that A3-induced C-to-T mutations occur during the momentary availability of single-stranded (−)-DNA (ssDNA), whereas HBV RNA is reversely transcribed and the mutations continue along with the reverse transcription process. It is reported that HBV ssDNA should be protected through binding to capsid interior (38). Mutations should be random if the ssDNA is available for a length of time without protection. A3G mutation in vitro analyses showed that A3G randomly scans ssDNA for cytidine targets by sliding, jumping, or intersegmental transfer motions (39, 40). To clarify if HBV ssDNA availability was restrictive or not, A3C-induced mutations were compared by in vitro versus in vivo assays. Since A3C in vitro mutation activity was very low, Hsp90 was included to increase A3C activity (7). In addition, 3D-PCR 88 °C amplification was used to enhance the detection sensitivity of A3C in vitro mutation. Briefly, A3C in vitro induced mutation was performed by an incubation of purified A3C with an HBV ssDNA fragment (1175–2089 nt) in the presence or absence of Hsp90. The HBV DNAs from the in vitro reaction were then amplified by PCR-94 °C and followed by a further 3D-PCR 88 °C using an inner primer pair to recover the region of 1380 to 1820 nt. The resultant 3D-PCR 88 °C amplicons were cloned for sequencing analyses. For the cellular in vivo mutation experiments, A3C with or without Hsp90 were cotransfected together with HBV encoding plasmid into HepG2 cells as previously reported (7). HBV rcDNAs were then isolated from viral capsids in the cytoplasm. HBV DNAs within the same region of 1380 to 1820 nt were amplified by PCR-94 °C and then followed by 3D-PCR 88 °C as in vitro for cloning and sequencing analyses.
As shown in Figure 10, A and B, A3C in vitro C-to-T mutations were distributed over an abroad region and the mutation-positive clones increased from 4 to 13 of 30 randomly selected clones in the presence of Hsp90, consistent with the Hsp90 stimulation effect as previously reported (7). However, C-to-T mutations per clone were about the same with average 3 and 3.5 for A3C and A3C+Hsp90, respectively. Overall, A3C in vitro mutations appear to be random with most of C-to-T mutations in the middle region of 1550 to 1740 nt. In contrast, as shown in Figure 10, C and D, A3C in vivo mutations showed a cluster around 1440 nt with little mutations in the region of 1550 to 1740 nt. Similarly, Hsp90 increased the A3C mutation-positive clones from six to nine of 30 randomly selected clones. The average C-to-T mutations per clone were 7.2 and 6.1 for A3C and A3C+Hsp90, respectively, a 2-fold increase compared with the in vitro results. These data demonstrate that A3C-induced mutation on HBV genome in vivo is a result of its dynamic interactions with components involved in the HBV reverse transcription process.
Figure 10.
In vitro and in vivo APOBEC3C mutation comparison. For in vitro mutation, purified A3C with or without Hsp90 was incubated with HBV cDNA fragment (nt 1175–2089) substrate at 30 °C for 3 h. The HBV DNAs were then extracted and amplified by PCR-94 °C followed by 3D-PCR at 88 °C using an inner primer pair to recover the region 1380 to 1820 nt. The resultant 88 °C 3D-PCR amplicons were TA-cloned into a pCR4 vector for sequencing analyses. For in vivo mutation, A3C with or without Hsp90 was cotransfected with an HBV-encoding plasmid into HepG2 cells. After 48-h transfection, HBV relaxed circular DNAs were isolated from HBV capsids in the cytoplasm. HBV DNAs in the region of 1380 to 1820 nt were amplified by PCR-94 °C followed by 3D-PCR at 88 °C. The resultant cellular 88 °C 3D-PCR amplicons were cloned for sequencing analyses in parallel with the in vitro samples. Thirty clones (n = 30) were randomly selected from each treatment for sequencing analyses. The C-to-T mutation distribution against the HBV corresponding cytidine site (reference to HBV V01460.1) and collective mutation frequencies for each treatment are graphically presented for (A) A3C in vitro; (B) A3C+Hsp90 in vitro; (C) A3C in vivo; (D) A3C+Hsp90 in vivo mutations. HBV, hepatitis B virus.
Each APOBEC3 has a unique preference for the nucleotide immediately adjoining the mutated cytidines
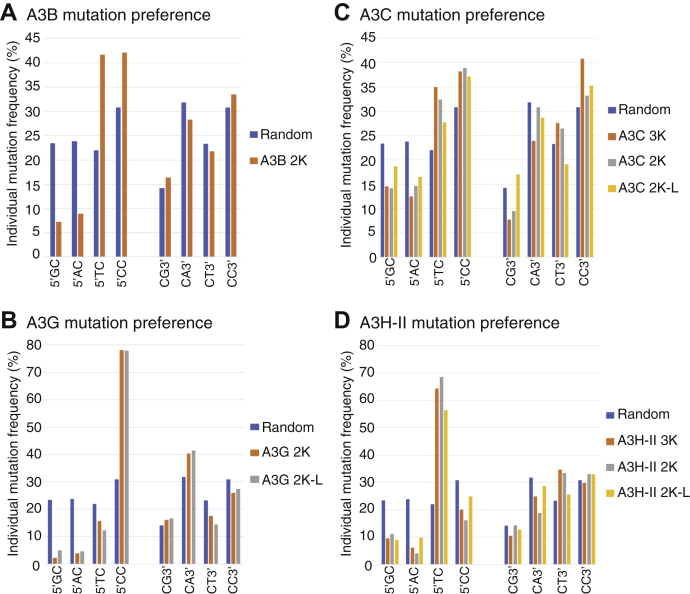
It is known that each APOBEC3 has a different preference for the immediate adjoining 5′ or 3′ nucleotide. All HBV C-to-T mutations in the HBV genome sequences above were collected and sorted according to their immediate 5′ or 3′ nucleotide associated with each corresponding mutated cytidine in HBV genomes. The occurring frequencies of adjoining 5′G, 5′A, 5′T, and 5′C or 3′G, 3′A, 3′T, and 3′C with individual mutated C were calculated by the mutated cytidine number in each sorted category divided by the total mutated cytidine number under each APOBEC3 treatment. As shown in Figure 11A, A3B had a strong preference for 5′T and 5′C but no preference for 3′ nucleotides. The occurring frequencies for 5′T and 5′C increased from random levels of 22% and 30.8% to 41.7% and 42.1%, respectively. The A3B preference for both 5′T and 5′C in HBV viral genome mutation clearly contrasts to its 5′T alone preference in cancer mutation signature analyses (41, 42, 43).
Figure 11.
APOBEC3 mutation preference analyses. HBV C-to-T mutations in all mutation-positive genome clones above were collected and sorted according to their immediate 5′ or 3′ nucleotide associated with each corresponding mutated cytidine in the HBV genomes. The frequencies of 5′G, 5′A, 5′T, and 5′C or 3′G, 3′A, 3′T, and 3′C for each mutated cytidine were calculated by the mutated cytidine number in each sorted category divided by the total mutated cytidine number and presented as percentage bars according to each APOBEC3 treatment including (A) A3B, (B) A3G, (C) A3C, (D) A3H-II. The expected random frequencies are used as reference. HBV, hepatitis B virus.
As shown in Figure 11B, A3G had a strong preference for 5′C with a frequency of 78.1% in contrast to the random value of 30.8%. In addition, A3G also had a weak preference for 3′A with a 40.4% frequency in contrast to the random value of 31.8%. As shown in Figure 11D, A3H-II had a strong preference for 5′T with variable observed frequencies of 64.3% for 3K, 68.6% for 2K, and 56.4% for 2K-L in contrast to the random value of 22%. In addition, A3H-II also had a weak preference for 3′T with observed frequencies of 34.8% for 3K, 33.6% for 2K, and 25.6% for 2K-L in contrast to the random value of 23.3%. The preference of A3G for 5′C and A3H-II for 5′T is consistent with previous reports (22, 44).
A3C had a large mutational effect on HBV genomes. As shown in Figure 11C, A3C had preferences for both 5′T and 5′C with variable frequencies depending upon the HBV genome variant. The 5′T frequencies varied from 27.7% to 34.9% in contrast to the random value of 22%. The 5′C varied from 37.1% to 38.9% in contrast to the random value of 30.8%. In addition, A3C also had a 3′C preference with a frequency varying from 33.2% to 40.8% in contrast to the random value of 30.8%. These data suggest that A3C may work like A3B with similar 5′T and 5′C preferences, although A3C has a lower mutation efficiency.
Discussion
APOBEC3s are ssDNA-specific deaminases that catalyze C-to-T mutations on both pathogen and human genomes. However, the ssDNA is usually available only momentarily during RNA reverse transcription or DNA replication. Here we used a cellular HBV replication model by cotransfecting HBV with APOBEC3 in HepG2 cells to investigate how APOBEC3s mutate HBV DNA during viral replication. A3B/C/G/H-II mutational activities were evaluated by genome-wide mutational analyses referencing to the HBV viral reverse transcription start and termination sites. We found that there was a close relationship between APOBEC3-induced mutation and the HBV viral reverse transcription processes. In addition, we also found that there were significantly different mutational characteristics between A3B/C/G/H-II under the same conditions.
To investigate how APOBEC3s mutate HBV DNA during viral replication, we first evaluated the mutational activities of all seven APOBEC3s on HBV viral DNA by primer extension analyses and found that A3B/C/G/H-II had sufficiently high mutational activities to warrant further HBV genome mutation landscape analyses (see Fig. 3). A3A/D/F also had mutational activity on HBV viral DNA, but their activities were very low. A3H-I showed little difference compared with the control mock vector. There is no apparent reason for the significant difference of the mutational activities between APOBEC3s. Although these APOBEC3s might have different abilities to enter the HBV viral capsid or accessibility to the HBV ssDNA substrate, we only focused on the genome-wide distribution of C-to-T mutations induced by the highly active A3B/C/G/H-II. A concern raised in this study was that the hepatoma cell line HepG2 used for the coexpression of HBV and APOBEC3s has endogenous APOBEC3 expression that could interfere with data interpretation. When evaluating this potential concern, we found that HepG2 cells had detectable A3B/C/G mRNA levels with A3C as the major one responsible for the endogenous HBV mutation, but their protein expression was not detectable by Western blotting using antibodies against the native A3B/C/G. In contrast, all exogenous APOBEC3 proteins in the coexpression experiment were readily detectable (see Fig. 2). In addition, a side-by-side mutational activity comparison by HBV short fragment PCR cloning and sequencing analyses demonstrated that A3B/C/G/H-II had mutational activities well above the vector control, especially with A3B and A3C (see Fig. 5). We conclude that the endogenous APOBEC3 contribution is minor compared with exogenous A3B/C/G/H-II overexpression. The estimate of A3B/C/G/H-II mutational activities by HBV short fragment PCR cloning and sequencing analyses also showed that A3B had very extensive mutational activity on individual clones, which made it the best candidate for evaluating ssDNA availability and A3B activity during HBV viral replication.
Of note, the HBV viral genome used in this study is an ayw strain with a sequence identical to HBV V01460.1 in the GenBank (33). The nucleotides in V01460.1 are numbered differently from HBV U95551.1, which has often been used as an HBV sequence reference in the HBV research field, including those of HBV in the HepG2.2.15 and HepAD38 cell lines (29, 34). Actually, V01460.1 referenced in this study is the (−) strand DNA and U95551.1 referenced in publications is the (+) strand DNA of HBV genome. HBV reverse transcription is initiated from the 3′ DR1 of HBV pgRNA by the HBV polymerase-linked DNA oligonucleotide that is generated from the 5′ ε bulge and then translocated to the RNA 3′ end (5). The HBV pgRNA is reversely transcribed into (−) strand DNA up to the 5′ DR1∗ (5). Based on our sequence reference of V01460.1, the HBV reverse transcription starts at 1360 nt, moves to 3182/1 nt, and then continues to the other end at 1340 to 1360 nt (see Fig. 1A, top panel). Owing to primer designs, the actual HBV sequencing covers 1380 nt to 3182 plus 1 to 1340 nt.
The A3B-induced C-to-T mutations in the HBV viral genome are distributed sequentially over the entire genome from 1382 to 3182 and then from 1 to 1337 nt, i.e., from the HBV reverse transcription start site to the termination site. Although no mutants were obtained directly for A3B 3K full-length genome cloning and sequencing, the half split 3K fragment cloning approach showed that A3B-induced clonal C-to-T mutations were sequential and very efficient with up to 65% of cytidines being mutated as shown in one genomic fragment clone (1–1360 nt) (see Fig. 6 and Table 2). In addition, A3B had the starting point of C-to-T mutations as early as 1382 and the ending point as late as 1337 nt. The C-to-T mutation sequential distribution along with the viral reverse transcription process was unambiguously further observed in A3B 2K genome clones, where multiple highly mutated A3B clones showed sequential C-to-T mutation distributions from the viral reverse transcription starting site to the termination site (see Fig. 6B). In addition, the overall site cytidine mutation frequencies across A3B 2K genome were about the same with gradual increases from the viral transcription starting site to the terminating end of the genome. These data indicate that A3B mutates HBV single-stranded (−)-DNA, parallel to the HBV reverse transcription process and A3B could enter the mutation process at the start site of the reverse transcription or late downstream and then continues to the termination site along with the reverse transcription process. Similar C-to-T mutation distribution patterns are also consistently observed with A3C/H-II-induced HBV 3K full-length genome mutations (see Fig. 8) and A3G/C/H-II-induced HBV 2K or 2K-L mutations (see Figs. 7 and 9). In addition, the C-to-T mutations at the starting site of reverse transcription are predominant for A3C/H-II-induced HBV 3K full-length genome, but it becomes less restrictive with smaller HBV 2K or 2K-L genome variants, indicating that the APOBEC3 access to the HBV ssDNA in 3K full-length genome is closely associated with the reverse transcription machinery. Of interest, the C-to-T mutation distribution pattern observed here is consistent with the clinical observation that HBV genome mutations are distributed over the entire HBV genome with about the same cytidine mutation frequency across the genome in HBV viruses obtained from sera of patients at different phases of HBV infection (25). The C-to-T mutation distribution pattern in A3B HBV genomes suggest that A3B-induced mutation is very closely associated with the process of viral reverse transcription and may occur spontaneously along with the reverse transcription process.
It has been reported that HBV genome-length ssRNA or ssDNA can be entirely bound to the capsid interior, forming a protein-nucleic acid complex that protects ssDNA from enzymatic modification (38). This indicates that HBV (−)-DNA is not freely available for APOBEC3 mutation owing to the protective role of the HBV core protein in the capsid. The HBV (−)-DNA could only be available momentarily, and A3B-induced mutations on the ssDNA must occur immediately when the (−)-DNA substrate becomes available before it is protected by the capsid protein. Consistent with this proposal, in vitro A3C-induced mutations randomly occurred over an abroad region with most of mutations in the middle region, and the mutation efficiency per single clone was low. In contrast, in vivo A3C-induced mutation was region specific and had a higher mutation efficiency (see Fig. 8), indicating that A3C-induced mutation in vivo involves a dynamic interaction with the HBV reverse transcription process. These data provide further evidence that APOBEC3 mutation on HBV single-stranded (−)-DNA occur along with the reverse transcription process while the substrate (−)-DNA is being generated.
Among the APOBEC3s, A3B is the most efficient mutation-inducing agent with an average of 15.1% mutation efficiency in the 2K HBV genome, whereas A3G, A3H-II, and A3C only have on average 6.3%, 3.8%, and 4.7%, respectively (see Table 2). Overall HBV mutation efficiency varies with different APOBEC3s with an order of A3B >> A3G > A3H-II or A3C. An A3B-induced 3K HBV mutation-positive clone could not be found when screening up to 120 clones. However, extensively mutated clones are found in HBV 3K half split fragment clones with an average of 50.4% and 42.7% for A3B 3K-I and 3K-II, respectively (see Fig. 6 and Table 2). A3B mutates up to 65% of cytidines in a single HBV 3K-I clone. These data demonstrate that A3B is a very strong mutational agent, consistent with the kataegis-like A3B mutation signature in cancer (45). A3B hypermutation results in extensive C-to-T mutations in the HBV 3K genome, which could lead to genome instability for cloning. This could be the reason for the inability to obtain A3B or A3G 3K mutant clones. Consistent with these observations, up to a 9% mutation frequency in specific A3B 3K PCR amplicons were detectable by primer extension analyses with pe1169 (data not shown). In contrast, A3C 3K mutant cloning was easily obtained because A3C only has a 2.3% average cytidine mutation efficiency in HBV 3K with the highest one up to 5.3% only (see Table 2).
In addition to highly mutated clones with C-to-T mutation distributions over the entire genome, there are also many other clones that showed variable patterns with different starting points and low mutation efficiencies for each APOBEC3. The cytidine mutation efficiency for any given APOBEC3 also varies significantly, even with A3B (see Fig. 6B). These data suggest that there are likely other factors within the viral capsids that could affect APOBEC3 mutation activity or substrate accessibility. For example, heat shock proteins were shown to enhance APOBEC3 mutation activity (7). In addition, the limited space within the HBV viral capsid could also be an important factor that affects APOBEC3-induced mutation on HBV genomes. In fact, the HBV 3K full-length genome mutation frequency was generally lower compared with the other HBV genome variants with reduced size (see Fig. 3 and Table 2). On the other hand, the proximity of C-to-T mutations at or near the viral reverse transcription initiation site was consistently kept for the full-length 3K genome mutations with A3H-II and A3C as well as 2K genome mutations with A3B and A3G in sequencing analyses (see Figure 6, Figure 7, Figure 8, Figure 9). However, the starting point of C-to-T mutations became more variable and more often downstream when the HBV genome size became smaller as shown in 2K genome mutations with A3H-II and A3C (see Fig. 9). The dominant mutation region also shifted to the lower genome region along the HBV reverse transcription process, especially for A3C 2K. These data suggest that APOBEC3-induced HBV genome mutations result from a dynamic interaction between APOBEC3 and multiple factors directly or indirectly associated with the viral reverse transcription process.
During A3C-induced HBV mutation analyses, we surprisingly found that more than half of HBV genomic clones including 3K, 2K, and 2K-L were mutation positive, which was about a 3-fold higher frequency than other APOBEC3s, including A3B, A3G, and A3H-II. However, the A3C clonal mutation efficiency was much lower than others (i.e., 2.3% with A3C 3K) (see Table 2). These data indicate that A3C has a higher accessibility to the HBV (−)-DNA substrate, even though each mutation activity on the affected HBV genome is relatively lower. In addition, the immediate nucleotide preference analyses show that A3C has a similar preference for 5′T and 5′C like A3B but with lower frequencies. It is reported that A3C is highly expressed in both normal human tissues and tumors, whereas other APOBEC3s' expressions are often low in normal tissues but increased in cancer (46). In the cancer genome mutation researches, A3B has been thought to be the potential mutation-inducing agent because A3B expression is often specifically increased in cancer. However, as A3C is generally highly expressed in both normal tissues and cancer (46), our data on A3C's unique high genome mutation frequency with a low mutation efficiency suggests that A3C could have an unexpectedly important role in the cancer genome mutation.
When evaluating HBV mutations, we isolated HBV rcDNA, cccDNA, and pgRNA (pre-genome RNA) separately from HepG2 cells cotransfected with HBV and APOBEC3s. APOBEC3-induced mutation effects on these different HBV intermediates during HBV viral replication were analyzed using the PCR-primer extension method. It is surprising that we found that HBV rcDNA alone had up to a 20% mutation frequency in HBV PCR-95 °C amplifications by A3B/C/G/H-II but none was detectable in HBV pgRNA or cccDNA under the same conditions (see Fig. 4). No mutations with HBV pgRNA have been reported previously when fewer than 20 clone sequencing analyses were performed on HBV pgRNA RT-PCR at 94 °C (8, 10). Using this few numbers of clones, however, does raise a question on the detection sensitivity. Here we used the much more sensitive PCR-primer extension method that analyzed a pool of HBV amplifications and found no sign of HBV pgRNA mutation. Regarding HBV cccDNA mutation, A3G-induced detectable mutations on duck HBV cccDNA were previously reported through a selective amplification by 3D-PCR when a base excision repair enzyme, uracil DNA glycosylase was inhibited (47). However, cccDNA mutation was undetectable here by a more sensitive PCR-primer extension method, whereas the rcDNA mutation was readily detectable. These data suggest that the level of mutations in HBV cccDNA or pgRNA should be very low or neglectable.
During HBV replication, cccDNA works as the DNA template and is transcribed into pgRNA in the nucleus. After translation into viral proteins, pgRNA is assembled with HBV polymerase and core protein to form a nucleocapsid in the cytoplasm. The pgRNA is then converted into rcDNA by HBV polymerase within the nucleocapsid to become a mature capsid, which can either be enveloped with surface protein for secretion as a virion or recycled to the nucleus to enter the cccDNA pool (5, 6). However, the analyses of HBV replication intermediates including cccDNA, pgRNA, and rcDNA under our cellular replication model showed that only rcDNA had readily detectable mutations. These data suggest that the HBV viral capsids generated in the cytoplasm probably do not recycle to the nuclei for further generation of cccDNA or pgRNA as expected. The C-to-T mutations in the rcDNA from the capsids should be considered as a singular event, reflecting the mutation reactions of APOBEC3s.
We note that Nair and Zlotnick (10) similarly investigated HBV mutations using HBV viral genome sequencing analyses and their conclusion was different from ours. The authors isolated HBV viral DNA from virus released from Hep2.2.15 and HepAD38 cells with or without transient transfection of A3A or A3G for investigating HBV genome mutations. They split the HBV viral genome into two fragments, Region I: 1 to 1630 nt and Region II: 1631 to 3182 nt, for PCR amplifications. With the limited fragment PCR clone sequencing (n = 6–20), they found that the mutational frequency in the first half (Region I) of HBV (−)-DNA was much higher than in the last half (Region II) with up to a 5-fold difference. They also found that expressing A3A or A3G made no significant difference. They concluded that the newly synthesized minus-sense ssDNA is protected from deaminase by interaction with the virus capsid; during plus-strand synthesis, when enough dsDNA has been synthesized to displace the remaining minus strand DNA from the capsid surface, the ssDNA becomes deaminase sensitive.
Compared with our study, the authors split HBV full-length genome into two fragments without considering the native gap around 1820 nt in the complete genomic (−)-DNA within Region-II, 1631 to 3182 nt (which corresponds to HBV V01460.1 region 1–1552 nt with the gap around 1360 nt in this study). Based on our results and others, PCR amplifications across the native HBV genome gap would be significantly compromised and could result in recombinant products (48) (data not shown). Therefore, the Region-II results may not truly represent actual HBV mutation. When evaluating the effect of A3A or A3G, the authors transiently expressed A3A or A3G into tat-off controlled HBV persistent expression cells. This means that the HBV cell transfection efficiency is much higher than that of A3G or A3A and many cells produce HBV virus without the presence of A3G or A3A. Therefore, little difference was observed with A3A or A3G expression compared with control. In contrast, we cotransfected HBV and individual APOBEC3s in a ratio of 1:11 into HepG2 cells to ensure that most HBV-expressing cells coexpressed APOBEC3. Using cellular viral capsid as the source of HBV genomic DNA, we cloned full-length HBV genome variants for mutation sequencing analyses and obtained an unbiased evaluation of HBV mutation activity over the entire genome.
In summary, utilizing the HBV cellular replication system, we investigated the APOBEC3 HBV mutation landscape over the entire genome variants by a reference to HBV viral reverse transfection process. We found that (1) APOBEC3-induced HBV mutations occurred sequentially on HBV (−)-DNA from the reverse transcription start to termination site in parallel with the reverse transcription process; (2) APOBEC3 mutation efficiency varied significantly with an order of A3B >> A3G > A3H-II or A3C with variation observed even with the same APOBEC3, suggesting the presence of other factors affecting APOBEC3 mutation activity; (3) Among APOBEC3s, A3B had a 3-fold higher mutation efficiency with up to 65% of cytidines being mutated, consistent with the kataegis-like A3B mutation signature in cancer. On the other hand, A3C had a unique higher mutation frequency on HBV genome populations with a preference for both 5′TC and 5′CC; and (4) APOBEC3-induced HBV mutation was detected in HBV rcDNA, not in cccDNA and pgRNA, suggesting that APOBEC3-induced mutation on HBV genomes occurs essentially as a singular event, capturing the mutational reactions of APOBEC3s.
Experimental procedures
Plasmid DNA constructs
The plasmid encoding the HBV viral genome was a gift from Dr Josef Kock (33). It was generated from an HBV ayw strain having a sequence identical to HBV V01460.1 in the GenBank. APOBEC3C and A3G were RT-PCR amplified from the human acute monocytic leukemia cell line THP-1 and were cloned into a pcDNA3.2 vector with a nine-amino-acid HA tag at the C terminus as reported (7, 37). Other APOBEC3s including A3A, A3B, A3D, A3F, A3H-I (variant-1, V-200 with N15, L18, G105, K121, and E178), and A3H-II (variant-2, V183 with N15, L18, G105, K121, and E178) in the pCMV6-Entry vector with a FLAG tag at the C terminus were purchased from Origene company. The A3H-II G105R mutant was generated from A3H-II using the QuickChange II XL kit (Agilent) according to the manufacturer's instructions.
Cell culture and extractions of HBV viral rcDNA, pgRNA, and cccDNA
The human liver cell line HepG2 was purchased from ATCC and was maintained in eagle's minimum essential medium containing 10% fetal bovine serum. For the investigation of APOBEC3-induced mutations on HBV viral DNA, HepG2 cells were plated on 35-mm 6-well collagen-coated plates the day before transfection. Plasmids of APOBEC3s and HBV in a ratio of 11:1 were mixed and cotransfected into HepG2 cells by X-tremeGENE HP (Roche). A total of 2 μg of plasmid DNA and 6 μl of transfection reagents were added to HepG2 cells in each well. Three separate samples were prepared for each evaluation. An empty pcDNA3.1 vector was used as a control. Forty-eight hours post transfection, the intracellular HBV viral capsids were harvested by cell lysis in a 300 μl buffer containing 50 mM Tris-HCl, pH 8.0, 1.5 mM MgCl2, and 0.5% Nonidet P-40. After removal of potential contamination of HBV encoding plasmid from the initial transfection by a pretreatment with 10 units of micrococcal nuclease (New England Biolabs) in the presence of 2 mM CaCl2 at 37 °C for 1 h, the cell cytoplasm lysate was digested with 20 μl of 20 mg/ml proteinase K for 15 min at 55 °C, and the HBV rcDNA released from the viral capsids was isolated by a Pure Link genomic DNA purification kit (Thermo Fisher Scientific) according to the manufacturer's instructions.
After lysing cells with 0.5% NP-40 buffer for HBV rcDNA isolation as described above, the remaining cell nuclei on the HepG2 plates were further lysed for HBV cccDNA extraction as reported (49). Briefly, the cell nuclei were lysed in a 400 μl buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 1% SDS. To the nuclei lysates, 100 μl 2.5 M KCl solution was added and the mixtures were incubated at room temperature for 30 min. After removal of cell debris and genomic DNA through a 12,000-rpm centrifugation at 4 °C for 15 min, the cccDNA in the supernatants were twice extracted by one volume of phenol followed by one volume of phenol-chloroform (1:1) and the cccDNAs were precipitated by two volumes of ethanol with an incubation at −20 °C overnight. The resultant cccDNA extracts in the pellets were dissolved in 50 μl tris-EDTA buffer and further treated by digestion with 10 U Dpn I at 37 °C for 1 h to remove potential HBV plasmid remaining from the initial transfection. The precleaned HBV cccDNA extracts were used for further mutation analyses through PCR amplifications.
For extraction of HBV pgRNA, APOBEC3s and HBV were cotransfected into HepG2 cells under the same conditions as described above. The cells were lysed by 1 ml Trizol reagent (Thermo Fisher Scientific), and the total RNA was extracted according to the manufacturer's instructions. After precleaning of any potential DNA contamination, including HBV-encoding plasmid from the initial transfection through a digestion of 2 U DNase I at 37 °C for 1 h, the total RNAs were extracted by one volume of phenol-chloroform (1:1) and recovered by ethanol precipitation. After brief heat denaturing at 90 °C for 2 min, the precleaned RNAs were reverse transcribed into cDNAs by M-MLV RTase with random primers at 37 °C for 1 h. The resultant cDNA from the total RNA extract was used for further pgRNA analyses after PCR amplification.
For the protein expression analyses, APOBEC3s and HBV were cotransfected into HepG2 cells under the same conditions as described above. The cells were lysed by 0.5% NP-40 buffer in the presence of proteinase inhibitor mixtures (Roche) or directly by SDS sample buffer. The expressed protein levels were determined by electrophoresis in a 10% NuPAGE BisTris gel followed by Western blotting analyses using antibodies against the HA tag in A3C and A3G, or FLAG tag in A3A, A3B, A3D, A3F, and A3H (Sigma). Additional Western blot analyses were also performed using antibodies against native A3B, A3C, and A3G for the presence of endogenous A3/B/C/G protein expression in the empty vector control, but none of them were detectable (see Fig. 2). For A3C siRNA knockdown effect on the endogenous HBV mutation in HepG2 cells, HepG2 cells were reversely transfected by A3C siRNA or siRNA negative control (Dharmacon siGENOME SMARTpool) using Lipofectamine RNAiMax (Thermo Fisher Scientific). Twenty-four hours after siRNA transfection, HBV and A3C or vector were cotransfected into the HepG2 cells as described above. Total RNAs or HBV viral DNAs were isolated from the duplicate experiments for the analyses of A3C mRNA knockdown or HBV DNA mutation, respectively.
HBV genomic DNA cloning and sequencing analyses
HBV genomic DNAs were cloned for sequencing analyses by a modified method based upon previous reports (29, 34). Briefly, primers against the region flanking the HBV replication gap were designed for HBV viral genome cloning. Primers F1 and R5 were routinely used for HBV genome amplification through Gold AmpliTaq. The PCRs were performed by initially denaturing at 95 °C for 15 min and then 40 cycles of 95 °C × 1 min, 58 °C × 1 min, and 72 °C × 4 min followed by another hold at 72 °C × 7 min. The resultant PCR amplifications were separated by 1% agarose gel electrophoresis, and the bands corresponding to the full-length 3K HBV and major internal deletion genome variants 2K and 2K-L were cut out and purified by a QIAEX II gel extraction kit (Qiagen). 3′-dA was added to the gel-purified HBV genome amplicons by incubating with 1 U regular AmpliTaq at 72 °C for 20 min, and then the genomic amplicons were cloned into a pCR4 vector by using a TOPO TA-cloning kit (Thermo Fisher Scientific) following the manufacturer's instructions. A total of 60 clones were randomly selected for each HBV genome variant investigation except A3B HBV genomes where additional 60 were selected for data accuracy. Insert-containing plasmids in the clones were extracted for sequencing analyses by Sanger sequencing. Generally, the full HBV insert sequences could be obtained for HBV 2K and 2K-L genome by Sanger sequencing using the vector primers, T7 promoter and T3 primer extension. But for the HBV full-length 3K genome, one more primer, R317 (tggggcagaatctttccacc) should be added to get the full sequences. C-to-T mutations in each HBV genome clone were identified by comparing with the HBV genome V01460.1 using the online program BLAST from National Center for Biotechnology Information (NCBI), USA. The sequencing information was presented as a C-to-T mutation distribution for each HBV genome clone against the cytidine sites in the HBV V01460.1. In addition, the mutation frequencies for each cytidine in the HBV genome were calculated by the number of C-to-T mutations for a specific cytidine site in mutation-positive clones divided by the sequenced clone number under the same treatment. Mutation efficiency was also individually calculated by the number of C-to-T mutations in a single mutation-positive clone divided by the total cytidines available in the clone. Of note, there were other kinds of mutations in addition to the predominant C-to-T mutation, but they were very minor and were not shown in the data presentation. The summative data were presented in the figures and tables. The original raw sequence data can be provided if requested.
For A3B mutation analyses, additional fragment cloning was performed for further information with the full-length 3K genome through two split fragments, 3K-I (1360–3182 nt) and 3K-II (1–1360 nt). Briefly, the A3B 3K genome was amplified by PCR-95 °C as described above using primer pairs F1/R3182 or F12/R5 for 3K-I (1360–3182 nt) and 3K-II (1–1360 nt), respectively. The resultant 3K fragment amplicons were TA-cloned into the pCR4 vector for sequencing analyses as described above. For the side-by-side comparison sequencing analyses of A3/B/C/G/H-II mutational activities versus mock vector control, HBV region 1360 to 2090 nt was amplified by PCR-95 °C as described above using primer pairs F1/R2065. The 730-bp PCR amplicons were TA-cloned into the pCR4 vector, and 40 clones were randomly selected for sequencing analyses. For verification of the HBV genome internal deletions, a list of primers shown in Figure 1A and Table 3 were used to PCR amplify HBV genome fragments under the same PCR conditions as described above and the resultant amplicons were analyzed by 1% agarose gel electrophoresis.
Table 3.
Sequences for primers used in this study
| Primer name | Primer sequence (5′-->3′) |
|---|---|
| F1 | GAAAAAGTTGCATGGTGCTGGTG |
| F12 | GGAAGGTTGTGGAATTCCACTGCA |
| F443 | TCCATAGAGTGTGTAAATAGTG |
| F874 | CGGAAGTGTTGATAGGATAGGG |
| F1630 | ACGGGGAGTCCGCGTAAAGAGAGG |
| F2578 | GAAAGCCCAGGATGATGGGATG |
| R1 | TTTTTCACCTCTGCCTAATCA |
| R5 | CAAGCCTCCAAGCTGTGCCTTG |
| R317 | TGGGGCAGAATCTTTCCACC |
| R868 | AGACCACCAAATGCCCCTATC |
| R1810 | CGCAAATATACATCGTTTCCAT |
| R2065 | TTCTCGCCAACTTACAAGGCCTTTC |
| R2342 | TGGGTATACATTTAAACCCTAAC |
| R2475 | TGTTCAGTGGTTCGTAGGGC |
| R3182 | AGAGTTTGGTGGAAGGTTGTG |
| pe1169 | CTAGATACCGCCTCAGCTCTG |
| pe2564 | CCTGTATTCCCATCCCATC |
| pe2333 | ACATTTAAACCCTAACAAAAC |
| pe1453 | CAAAGACTGTTTGTTTAAAGA |
A3C in vitro mutation reactions
The single-stranded HBV DNA fragment (1175–2089 nt by reference to HBV V01460.1) was prepared as reported (7, 32). For the in vitro A3C mutation analyses, purified A3C (0.24 μg) with or without Hsp90 (0.7 μg) (purchased from Abnova) were incubated with the HBV cDNA substrate (20 ng) for 3 h at 30 °C in a buffer containing 20 mM Tris acetate, pH7.9, 50 mM potassium acetate, 10 mM magnesium acetate, and 100 μg/ml bovine serum albumin. After the reactions, the HBV DNAs were recovered by extraction with one volume of phenol/chloroform (1:1) twice and chloroform once, followed by ethanol precipitation. The HBV DNAs were amplified by PCR at 94 °C and followed by 3D-PCR at 88 °C using primer pairs for HBV region of 1380 to 1820 nt as described (7, 32). The 88 °C 3D-PCR amplicons were TA-cloned into a pCR4 vector for sequencing analyses.
HBV DNA mutation analyses by the PCR-primer extension method
HBV mutation frequencies were quantitatively determined by the primer extension method as described (7, 32). Briefly, HBV DNAs were amplified by PCR-95 °C from the precleaned HBV DNA extracts including rcDNA, cDNA from pgRNA, and cccDNA, using primer pair F1/R2475 for pe1453 and pe2333 or primer pair F874/R5 for pe1169. PCR was performed by initial denaturing at 95 °C for 15 min and then 38 cycles of 95 °C × 1 min, 55 °C × 1 min, and 72 °C × 2 min followed by another hold at 72 °C × 7 min.
For HBV mutation frequency analyses by primer extension (pe), PCR amplifications were purified by a QIAEX II gel extraction kit (Qiagen). An aliquot of the purified DNA amplicons was subjected to a thermal cycle primer extension reaction in the presence of ddGTP with a 5′-32P-end-labeled primer for the specifically selected cytidine including pe1169, pe1453, pe2333, or pe2564. The primer extension reactions were performed by 5 min at 95 °C followed by 50 cycles of 45 s at 95 °C, 30 s at 56 °C, and 45 s at 72 °C, and a final 4 min at 72 °C using Sanger sequencing kit. The primer extension products were separated by electrophoresis in an 8% polyacrylamide-urea sequencing gel and quantitated by a PhosphorImager. Primer extension should be stopped at the target cytosine site due to ddGTP incorporation. When the target cytosine was mutated, the primer extension continued through the site until the next cytosine downstream and resulted in longer oligonucleotide products that appeared as ladder-like bands on the sequencing gel. The C-to-T mutational frequency at the target cytosine site was determined by the percentage of these bands above the target cytosine oligonucleotide divided by the total product counts in the lane.
Data availability
The summative data of HBV clonal sequencing analyses by Sanger sequencing methods were presented in Figure 5, Figure 6, Figure 7, Figure 8, Figure 9, Figure 10, Figure 11 and Tables 1 and 2. The original raw sequence data can be provided, if requested, by Zhigang Chen, Clinical Center, National Institute of Health, email: chenz2@cc.nih.gov. The rest of the data are presented within the article.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
Z. C., T. L. E., and A. P. P. conceptualization; Z. C. methodology; Z. C., T. L. E., A. V. B., I. N. B., T. G. V., and A. P. P. validation; Z. C. formal analysis; Z. C., T. L. E., and A. P. P. investigation; Z. C. resources; Z. C., T. L. E., A. V. B., I. N. B., T. G. V., and A. P. P. data curation; Z. C. writing-original draft; Z. C., T. L. E., and A. P. P. writing-review and editing; Z. C., T. L. E., A. V. B., I. N. B., T. G. V., and A. P. P. visualization; T. L. E. and A. P. P. supervision; Z. C., T. L. E., and A. P. P. project administration; T. L. E. and A. P. P. funding acquisition.
Funding and additional information
This research is supported by the intramural program of the National Institute of Health with funds from NIAID and NIDDK. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health.
Edited by Patrick Sung
References
- 1.Harris R.S., Dudley J.P. APOBECs and virus restriction. Virology. 2015;479-480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris R.S., Bishop K.N., Sheehy A.M., Craig H.M., Petersen-Mahrt S.K., Watt I.N., Neuberger M.S., Malims M.H. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 3.Olson M.E., Harris R.S., Harki D.A. APOBEC enzymes as targets for virus and cancer therapy. Cell Chem. Biol. 2018;25:36–49. doi: 10.1016/j.chembiol.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wissing S., Galloway N.L., Greene W.C. HIV-1 Vif versus the APOBEC3 cytidine deaminases: An intracellular duel between pathogen and host restriction factors. Mol. Aspects Med. 2010;31:383–397. doi: 10.1016/j.mam.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck J., Nassal M. Hepatitis B virus replication. World J. Gastroenterol. 2007;13:48–64. doi: 10.3748/wjg.v13.i1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glebe D., Bremer C.M. The molecular virology of hepatitis B virus. Semin. Liver Dis. 2013;33:103–112. doi: 10.1055/s-0033-1345717. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z., Eggerman T.L., Bocharov A.V., Baranova I.N., Vishnyakova T.G., Kurlander R., Patterson A.P. Heat shock proteins stimulate APOBEC-3-mediated cytidine deamination in the hepatitis B virus. J. Biol. Chem. 2017;292:13459–13479. doi: 10.1074/jbc.M116.760637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y., Hu J., Cai X., Huang Y., Zhou X., Tu Z., Hu J., Tavis J.E., Tang N., Huang A., Hu Y. APOBEC3B edits HBV DNA and inhibits HBV replication during reverse transcription. Antivir. Res. 2018;149:16–25. doi: 10.1016/j.antiviral.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Kanagaraj A., Sakamoto N., Que L., Li Y., Mohiuddin M., Koura M., Wakae K., Kurachi M., Muramatsu M., Kitamura K. Different antiviral activities of natural APOBEC3C, APOBEC3G, and APOBEC3H variants against hepatitis B virus. Biochem. Biophys. Res. Commun. 2019;518:26–31. doi: 10.1016/j.bbrc.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Nair S., Zlotnick A. Asymmetric modification of hepatitis B virus (HBV) genomes by an endogenous cytidine deaminase inside HBV cores informs a model of reverse transcription. J. Virol. 2018;92 doi: 10.1128/JVI.02190-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doehle B.P., Schafer A., Cullen B.R. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology. 2005;339:281–288. doi: 10.1016/j.virol.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Borresen-Dale A.L., Boyault S., Burkhardt B., Butler A.P., Caldas C., Davies H.R. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts S.A., Lawrence M.S., Klimczak L.J., Grimm S.A., Fargo D., Stojanov P., Kiezun A., Kryukov G.V., Carter S.L., Saksena G., Harris S., Shah R.R., Resnick M.A., Getz G., Gordenin D.A. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green A.M., Weitzman M.D. The spectrum of APOBEC3 activity: From anti-viral agents to anti-cancer opportunities. DNA Repair (Amst.) 2019;83:102700. doi: 10.1016/j.dnarep.2019.102700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jamal-Hanjani M., Wilson G.A., McGranahan N., Birkbak N.J., Watkins T.B.K., Veeriah S., Shafi S., Johnson D.H., Mitter R., Rosenthal R., Salm M., Horswell S., Escudero M., Matthews N., Rowan A. Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 16.Law E.K., Sieuwerts A.M., LaPara K., Leonard B., Starrett G.J., Molan A.M., Temiz N.A., Vogel R.I., Meijer-van Gelder M.E., Sweep F.C., Span P.N., Foekens J.A., Martens J.W., Yee D., Harris R.S. The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci. Adv. 2016;2:e1601737. doi: 10.1126/sciadv.1601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roper N., Gao S., Maity T.K., Banday A.R., Zhang X., Venugopalan A., Cultraro C.M., Patidar R., Sindiri S., Brown A.L., Goncearenco A., Panchenko A.R., Biswas R., Thomas A., Rajan A. APOBEC mutagenesis and copy-number alterations are drivers of proteogenomic tumor evolution and heterogeneity in metastatic thoracic tumors. Cell Rep. 2019;26:2651–2666.e2656. doi: 10.1016/j.celrep.2019.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris R.S., Serebrenik A.A., Argyris P., Jarvis M.C., Brown W.L., Bazzaro M., Vogel R.I., Erickson B.K., Lee S.H., Goergen K.M., Maurer M.J., Oberg A.L., Hou X., Weroha S.J., Kaufmann S.H. The DNA cytosine deaminase APOBEC3B is a molecular determinant of platinum responsiveness in clear cell ovarian cancer. Clin. Cancer Res. 2020;26:3397–3407. doi: 10.1158/1078-0432.CCR-19-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu B., Xiao Y., Yeager M., Clifford G., Wentzensen N., Cullen M., Boland J.F., Bass S., Steinberg M.K., Raine-Bennett T., Lee D., Burk R.D., Pinheiro M., Song L., Dean M. Mutations in the HPV16 genome induced by APOBEC3 are associated with viral clearance. Nat. Commun. 2020;11:886. doi: 10.1038/s41467-020-14730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou J., Wang C., Ma X., Wang E., Peng G. APOBEC3B, a molecular driver of mutagenesis in human cancers. Cell Biosci. 2017;7:29. doi: 10.1186/s13578-017-0156-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawson J.M.O., Nikolaitchik O.A., Keele B.F., Pathak V.K., Hu W.S. Recombination is required for efficient HIV-1 replication and the maintenance of viral genome integrity. Nucleic Acids Res. 2018;46:10535–10545. doi: 10.1093/nar/gky910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suspene R., Guetard D., Henry M., Sommer P., Wain-Hobson S., Vartanian J.P. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8321–8326. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vartanian J.P., Henry M., Marchio A., Suspene R., Aynaud M.M., Guetard D., Cervantes-Gonzalez M., Battiston C., Mazzaferro V., Pineau P., Dejean A., Wain-Hobson S. Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis. PLoS Pathog. 2010;6:e1000928. doi: 10.1371/journal.ppat.1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pak V., Heidecker G., Pathak V.K., Derse D. The role of amino-terminal sequences in cellular localization and antiviral activity of APOBEC3B. J. Virol. 2011;85:8538–8547. doi: 10.1128/JVI.02645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z.T., Huang S.Y., Chen L., Liu F., Cai X.H., Guo Y.F., Wang M.J., Han Y., Yu D.M., Jiang J.H., Zhang D.H., Gong Q.M., Zhang G.Q., Zang G.Q., Lu Z.H. Characterization of full-length genomes of hepatitis B virus quasispecies in sera of patients at different phases of infection. J. Clin. Microbiol. 2015;53:2203–2214. doi: 10.1128/JCM.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson M., Choga W.T., Moyo S., Bell T.G., Mbangiwa T., Phinius B.B., Bhebhe L., Sebunya T.K., Lockman S., Marlink R., Kramvis A., Essex M., Musonda R.M., Blackard J.T., Gaseitsiwe S. Molecular characterization of near full-length genomes of hepatitis B virus isolated from predominantly HIV infected individuals in Botswana. Genes (Basel) 2018;9 doi: 10.3390/genes9090453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou H., Gewaily D., Ahn S.H., Preskill C., Wang Y., Zong L., Zhang J., Han K.H., Wands J., Li J., Tong S. Sequence analysis and functional characterization of full-length hepatitis B virus genomes from Korean cirrhotic patients with or without liver cancer. Virus Res. 2017;235:86–95. doi: 10.1016/j.virusres.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alves B.M., Siqueira J.D., Garrido M.M., Botelho O.M., Prellwitz I.M., Ribeiro S.R., Soares E.A., Soares M.A. Characterization of HIV-1 near full-length proviral genome quasispecies from patients with undetectable viral load undergoing first-line HAART therapy. Viruses. 2017;9 doi: 10.3390/v9120392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J., Wu M., Wang F., Zhang W., Wang W., Zhang X., Zhang J., Liu Y., Liu Y., Feng Y., Zheng Y., Hu Y., Yuan Z. Hepatitis B virus spliced variants are associated with an impaired response to interferon therapy. Sci. Rep. 2015;5:16459. doi: 10.1038/srep16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam A.M., Ren S., Espiritu C., Kelly M., Lau V., Zheng L., Hartman G.D., Flores O.A., Klumpp K. Hepatitis B virus capsid assembly modulators, but not nucleoside analogs, inhibit the production of extracellular pregenomic RNA and spliced RNA variants. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.00680-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu H.L., Chen P.J., Tu S.J., Lin M.H., Lai M.Y., Chen D.S. Characterization and genetic analysis of alternatively spliced transcripts of hepatitis B virus in infected human liver tissues and transfected HepG2 cells. J. Virol. 1991;65:1680–1686. doi: 10.1128/jvi.65.4.1680-1686.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z., Eggerman T.L., Bocharov A.V., Baranova I.N., Vishnyakova T.G., Csako G., Patterson A.P. Hypermutation induced by APOBEC-1 overexpression can be eliminated. RNA. 2010;16:1040–1052. doi: 10.1261/rna.1863010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kock J., Blum H.E. Hypermutation of hepatitis B virus genomes by APOBEC3G, APOBEC3C and APOBEC3H. J. Gen. Virol. 2008;89:1184–1191. doi: 10.1099/vir.0.83507-0. [DOI] [PubMed] [Google Scholar]
- 34.Gunther S., Li B.C., Miska S., Kruger D.H., Meisel H., Will H. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J. Virol. 1995;69:5437–5444. doi: 10.1128/jvi.69.9.5437-5444.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunther S., Sommer G., Iwanska A., Will H. Heterogeneity and common features of defective hepatitis B virus genomes derived from spliced pregenomic RNA. Virology. 1997;238:363–371. doi: 10.1006/viro.1997.8863. [DOI] [PubMed] [Google Scholar]
- 36.Harari A., Ooms M., Mulder L.C., Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol. 2009;83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Z., Eggerman T.L., Bocharov A.V., Baranova I.N., Vishnyakova T.G., Kurlander R.J., Csako G., Patterson A.P. Hypermutation of ApoB mRNA by rat APOBEC-1 overexpression mimics APOBEC-3 hypermutation. J. Mol. Biol. 2012;418:65–81. doi: 10.1016/j.jmb.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J.C., Dhason M.S., Zlotnick A. Structural organization of pregenomic RNA and the carboxy-terminal domain of the capsid protein of hepatitis B virus. PLoS Pathog. 2012;8:e1002919. doi: 10.1371/journal.ppat.1002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chelico L., Pham P., Calabrese P., Goodman M.F. APOBEC3G DNA deaminase acts processively 3'--> 5' on single-stranded DNA. Nat. Struct. Mol. Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- 40.Nowarski R., Britan-Rosich E., Shiloach T., Kotler M. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat. Struct. Mol. Biol. 2008;15:1059–1066. doi: 10.1038/nsmb.1495. [DOI] [PubMed] [Google Scholar]
- 41.Burns M.B., Lackey L., Carpenter M.A., Rathore A., Land A.M., Leonard B., Refsland E.W., Kotandeniya D., Tretyakova N., Nikas J.B., Yee D., Temiz N.A., Donohue D.E., McDougle R.M., Brown W.L. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burns M.B., Temiz N.A., Harris R.S. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silvas T.V., Schiffer C.A. APOBEC3s: DNA-editing human cytidine deaminases. Protein Sci. 2019;28:1552–1566. doi: 10.1002/pro.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desimmie B.A., Burdick R.C., Izumi T., Doi H., Shao W., Alvord W.G., Sato K., Koyanagi Y., Jones S., Wilson E., Hill S., Maldarelli F., Hu W.S., Pathak V.K. APOBEC3 proteins can copackage and comutate HIV-1 genomes. Nucleic Acids Res. 2016;44:7848–7865. doi: 10.1093/nar/gkw653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikkila J., Kumar R., Campbell J., Brandsma I., Pemberton H.N., Wallberg F., Nagy K., Scheer I., Vertessy B.G., Serebrenik A.A., Monni V., Harris R.S., Pettitt S.J., Ashworth A., Lord C.J. Elevated APOBEC3B expression drives a kataegic-like mutation signature and replication stress-related therapeutic vulnerabilities in p53-defective cells. Br. J. Cancer. 2017;117:113–123. doi: 10.1038/bjc.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng J.C.F., Quist J., Grigoriadis A., Malim M.H., Fraternali F. Pan-cancer transcriptomic analysis dissects immune and proliferative functions of APOBEC3 cytidine deaminases. Nucleic Acids Res. 2019;47:1178–1194. doi: 10.1093/nar/gky1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitamura K., Wang Z., Chowdhury S., Simadu M., Koura M., Muramatsu M. Uracil DNA glycosylase counteracts APOBEC3G-induced hypermutation of hepatitis B viral genomes: Excision repair of covalently closed circular DNA. PLoS Pathog. 2013;9:e1003361. doi: 10.1371/journal.ppat.1003361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suspene R., Thiers V., Vartanian J.P., Wain-Hobson S. PCR mediated recombination impacts the analysis of hepatitis B Virus covalently closed circular DNA. Retrovirology. 2016;13:84. doi: 10.1186/s12977-016-0318-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X., Zhao J., Yuan Q., Xia N. Detection of HBV covalently closed circular DNA. Viruses. 2017;9 doi: 10.3390/v9060139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The summative data of HBV clonal sequencing analyses by Sanger sequencing methods were presented in Figure 5, Figure 6, Figure 7, Figure 8, Figure 9, Figure 10, Figure 11 and Tables 1 and 2. The original raw sequence data can be provided, if requested, by Zhigang Chen, Clinical Center, National Institute of Health, email: chenz2@cc.nih.gov. The rest of the data are presented within the article.