Abstract
Cysteine sulfenic acids (Cys-SOH) are pivotal modifications in thiol-based redox signaling and central intermediates en route to disulfide and sulfinic acid states. A core mission in our lab is to develop bioorthogonal chemical tools with the potential to answer mechanistic questions involving cysteine oxidation. Our group, among others, has contributed to the development of nucleophilic chemical probes for detecting sulfenic acids in living cells. Recently, another class of Cys-SOH probes based on strained alkene and alkyne electrophiles has emerged. However, the use of different models of sulfenic acid and methodologies, has confounded clear comparison of these probes with respect to chemical reactivity, kinetics, and selectivity. Here, we perform a parallel evaluation of nucleophilic and electrophilic chemical probes for Cys-SOH. Among the key findings, we demonstrate that a probe for Cys-SOH based on the norbornene scaffold does not react with any of the validated sulfenic acid models in this study. Furthermore, we show that purported cross-reactivity of dimedone-like probes with electrophiles, like aldehydes and cyclic sulfenamides, is a not meaningful in a biological setting. In summary, nucleophilic probes remain the most viable tools for bioorthogonal detection of Cys-SOH.
Keywords: Cysteine, Sulfenic acid, Posttranslational modification, Chemical probes, Bioorthogonal probes
1. Introduction
The cysteine residue undergoes a broad variety of oxidative post-translational modifications (OxiPTMs), which highlights its diverse functions in redox regulation and signaling, and warrant delicate methods for detection and characterization [[1], [2], [3]]. Among these cysteine OxiPTMs, the sulfenic acid modification is the initial product of thiol oxidation by a common reactive oxygen species, H2O2, and is pivotal to its transformation to other OxiPTMs, including the sulfinic acid, sulfonic acid, disulfide, persulfide, and thiosulfinate modifications [4]. When stabilized by local environmental factors [5], such as hydrogen bond acceptors, limited solvent access, and lack of adjacent nucleophiles (e.g., free thiols), cysteine sulfenic acid modification can modulate protein functions by altering their activity, stability, localization and ability to bind metal ions or other proteins [6]. Most sulfenic acids are transient intermediates to other OxiPTMs, primarily disulfide bridges that confer protein structural stability, or in some cases redox-active disulfides with regulatory functions [7]. Overall, profiling protein sulfenic acid modification has paved the way for elucidation of redox-based mechanisms in various organisms [[8], [9], [10], [11], [12], [13], [14], [15], [16]].
The short lifetime of protein sulfenic acids limits the use of antibody, crystallography, or mass spectrometry (MS)-based approaches for direct detection [5]. Indirect approaches monitoring the changes in thiol population upon reduction also struggle to recognize sulfenic acids among other OxiPTMs due to the lack of specificity of the reducing agents. To date, analyses of the cysteine “sulfenylome” are enabled almost exclusively by chemical probes that target this modification [3]. An ideal probe should react rapidly with sulfenic acids before they are sequestered by cellular nucleophiles (mainly thiols), and should exhibit strong chemo-selectivity, especially against thiols, which constitute >90% of global cellular cysteines [17]. Therefore, it is critical to develop chemical reactions with excellent bioorthogonality, a term comprising virtues of reactivity (fast reaction kinetics), selectivity (no off-target reaction) and biocompatibility (stability, permeability, toxicity, etc. suitable for applications in complex biological systems) [18].
Cysteine sulfenic acids possess both nucleophilic and electrophilic properties [5], which lead to several strategies for chemical ligations. Analogous to sulfenyl halides, the electrophilic sulfur atom in sulfenic acid is subject to nucleophilic attack by various nucleophiles, including carbanions, amines, phosphines, and thiols [5]. However, heteroatom-based nucleophiles tend to give reversible adducts with sulfenicacids, because of their weak bonds (N–S bond D°298 = 467 kJ/mol, S–S bond D°298 = 425 kJ/mol; compare to C–S bond D°298 = 712 kJ/mol) [19]. Consequently, carbon nucleophiles have long been used for covalent labeling of sulfenic acids, with dimedone, a 1,3-diketone compound, being the most prominent example [5]. Several groups, including ours, have developed carbon-based nucleophile (C-nucleophile) probes with various reporter tags and a wide range of kinetics [[20], [21], [22]] (Fig. 1A and B, left). On the other hand, deprotonated sulfenic acids (sulfenate anions) display moderate to weak nucleophilicity, and react with strong electrophiles such as alkyl iodides or 4-chloro-7-nitrobenzofurazan (NBD-Cl) [23], albeit at low reaction rates [24]. Such reactions have found applications mostly in simple systems (small-molecules or purified proteins) due to significant cross-reactivity with abundant biological nucleophiles, such as protein cysteines and glutathione (GSH). Sulfenic acids are also known to react with alkenes and alkynes under heat [5], via a concerted cycloaddition mechanism (Fig. 1A and B, right). Ring strain-activated alkenes/alkynes have been used to promote reactivity for bioconjugation to sulfenic acids. Notable examples include a cyclooctyne reagent [25] typically used in strain-promoted azide-alkyne cycloaddition (SPAAC) reactions, and a trans-cycloctene derivative [26] originally used for tetrazine ligation. Norbornadiene and less strained norbornene derivatives are also purported to react with sulfenic acid [27,28].
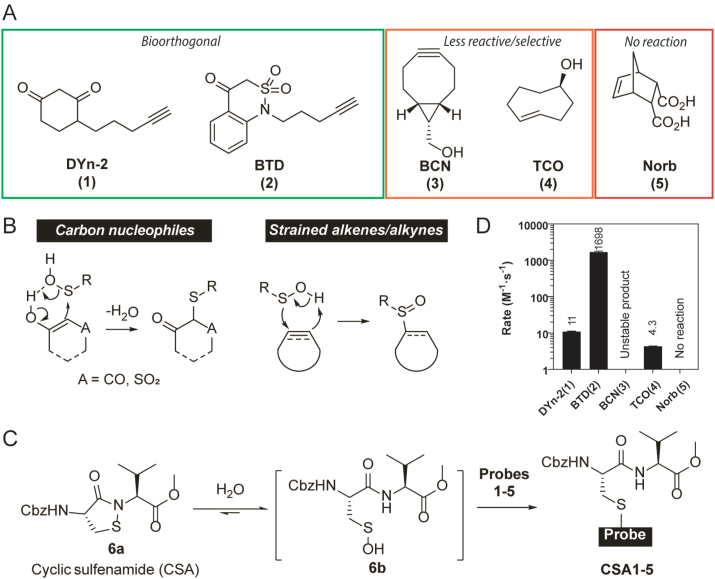
Fig. 1.
Kinetic rate studies of sulfenic acid probes with a cyclic sulfenamide (CSA) model. A) Structures of chemical probes involved in this study. B) Two classes of sulfenic acid probes and their mechanisms of action. C) CSA sulfenic acid model. D) Kinetic rate of probes 1–5 reacting with CSA.
The structure-activity relationship of C-nucleophile probes has been well-studied [21]. Generally speaking, destabilization of the carbanion at C-2 is linked to increased reactivity. The strength of the electron-withdrawing groups (inductive effect) is the primary factor, while resonance and steric effects also play a part. In contrast, kinetic profiles of strained alkene/alkyne-based sulfenic acid probes are inconclusive, because different small-molecule or protein models were used to evaluate reaction rates. In addition, questions and conjectures with respect to the cross-reactivity of C-nucleophile probes remain unanswered within a physiological context. Here, we implement multiple models to benchmark the reactivity of various types of sulfenic acid probes, and address the selectivity of C-nucleophiles with both the rate and occurrence of potential off-targets in mind.
2. Results and discussion
2.1. Comparison of probe reactivity using small-molecule sulfenic acid models
Several recently reported probes with applications in proteome-wide detection of protein S-sulfenylation, including two C-nucleophile probes DYn-2 (1) [8], BTD (2) [22], and three strained alkyne/alkene probes BCN (3) [25], TCO (4) [26], Norb (5) [28] were selected for our study (Fig. 1A). First, we used a dipeptide cyclic sulfenamide (CSA) 6 as a small-molecule model for sulfenic acid (Fig. 1C and D). Previously we reported that CSA readily hydrolyzes in aqueous solution to generate a transient sulfenic acid, which eventually becomes thiosulfenate and sulfinic acid in absence of chemical probes [21]. CSA has the advantages of representing a chemically unperturbed cysteine sulfenic acid in aqueous solutions, and easy handling due to its stability in organic solvents. It has been successfully used for the benchmark of a library of >100 C-nucleophiles, whose reaction kinetics with CSA correlate well with the labeling efficiency in sulfenylated proteins, cell lysates and living cells [22,29]. Furthermore, CSA kinetics are consistent with the number of MS-identified S-sulfenylation sites from the human proteome. For example, DYn-2 (1), which is an alkyne-functionalized nucleophilic probe based on dimedone, posed moderate reactivity with CSA at a rate of 10 M−1 s−1 and labeled 183 sites in RKO cell lysates, while BTD (2) is a recent probe with a vastly superior kinetic rate at 1700 M−1 s−1 and labeled 1186 sites in the same study [22]. The cyclooctyne derivative BCN (3) also reacted in the CSA model, but the product found by LC-MS, presumably from the alkyne attacking the electrophilic sulfur, was not stable and eventually converted to the disulfide of the dipeptide; the desired labeling product was only observed in trace quantity. In a two-step process, the trans-cyclooctene TCO (4) attacks the electrophilic sulfur with its alkene moiety, followed by participation of a neighboring axial hydroxyl group. It registered a slightly slower rate than DYn-2 at 4.3 M−1 s−1. It is worth noting that unlike the C-nucleophile probes, TCO functions at acidic conditions (e.g., 25% formic acid) because the alkene nucleophilicity is less affected by pH changes. On the other hand, Norb (5) apparently did not react in the CSA model as no adduct was observed. The results above indicate that the alkene nucleophile TCO is less reactive than C-nucleophile probes, while the cyclooctyne and norbornene derivatives are not compatible with the CSA model, likely due to their intrinsic reactivity towards the sulfenamide moiety.
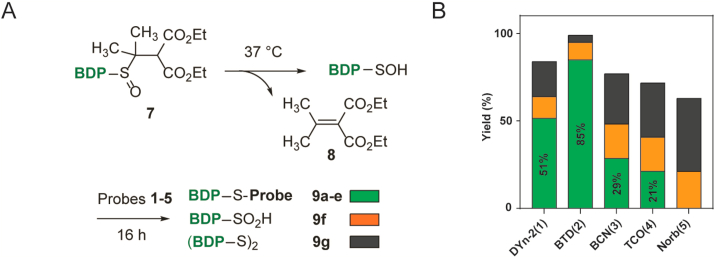
To develop a practical small-molecule sulfenic acid model with broad compatibility, we utilized a caged sulfenic acid precursor tagged with a fluorophore (Fig. 2A). Elimination of a sulfoxide produces a sulfenic acid and an alkene, and the rate of this reaction can be modulated by the strength of the electron-withdrawing groups on the β-carbon [30]. With this in mind, we synthesized DMDE (7), a BODIPY-tagged, cysteine-derived sulfoxide, which slowly releases a sulfenic acid and an alkene 8 over 16 h at 37 °C. DMDE was incubated with 10 equivalents of sulfenic acid probe 1–5, then the compositions of the reaction mixtures were analyzed by HPLC (493 nm absorption for the BODIPY tag) after completion. Each mixture was composed of probe-labeled product 9a-e, together with disulfide 9f and sulfinic acid 9g as byproducts. Similar to the prior findings with the CSA model, C-nucleophile probes captured more than half of the BODIPY-tagged cysteine sulfenic acids, with BTD achieving greater than 80% yield. In comparison, BCN and TCO labeled 30% and 20% of sulfenic acids, respectively, while the rest degraded to disulfides and sulfinic acids over time. In contrast, Norb did not give any detectable labeling product, casting doubts on its ability to capture transient sulfenic acids (Fig. 2B).
Fig. 2.
Yields of sulfenic acid capture from a fluorescent sulfoxide model. A) Elimination of a sulfoxide generated a transient sulfenic acid that was captured by probes. BDP, BODIPY-FL tag, see supplementary materials for full structures. B) Yield of captured sulfenic acids (probe adducts, green) and escaped side products (sulfinic acids in orange; disulfide in black). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.2. Comparison of probe reactivity using Gpx3 protein sulfenic acid
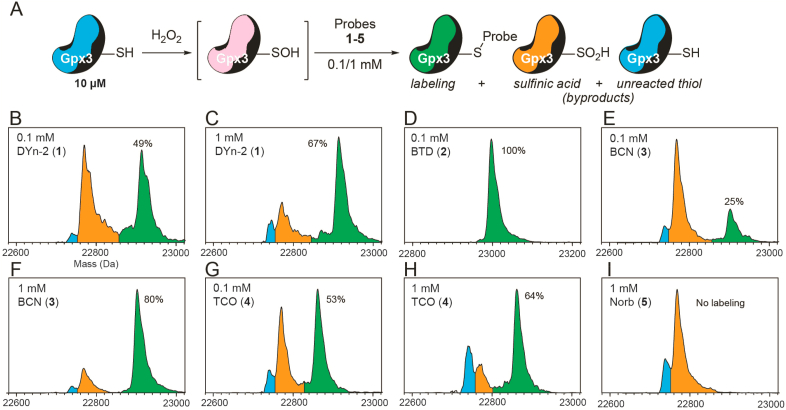
To field-test the aforementioned probes with their intended targets, a stabilized protein sulfenic acid in C64,82S glutathione peroxidase 3 (Gpx3) was employed (Fig. 3A). This Gpx3 mutant hosts one redox-sensitive cysteine at C36 that can be readily oxidized to a sulfenic acid by a stoichiometric amount of hydrogen peroxide (H2O2) and has been successfully validated in numerous protein sulfenic acid studies [20,21,24]. We observed a similar trend in protein sulfenic acid labeling: DYn-2 captured 49% and 67% of Gpx3 (10 μM) at 0.1 mM and 1 mM (10 and 100 equivalents compared to the protein), respectively (Fig. 3B and C). BTD was the most efficient probe that quantitatively captured Gpx3 when administrated at 0.1 mM (Fig. 3D). BCN labeled just 25% of Gpx3 at a lower concentration of 0.1 mM, but the yield increased to 80% at 1 mM (Fig. 3E and F). TCO labeled 53% and 64% of Gpx3 at 0.1 and 1 mM, respectively (Fig. 3G and H). Again, no labeling product was observed with up to 100 equivalents of Norb (Fig. 3I).
Fig. 3.
Reaction of protein Gpx3 sulfenic acid with chemical probes. A) In situ oxidation of C64,82S Gpx3 with quantitative H2O2 resulted in a sulfenic acid that was labeled by probes (green). Gpx3 sulfinic acid (orange) and the unreacted thiol form (blue) were also observed. B–I) Deconvoluted mass spectra of Gpx3 labeling by DYn-2 (B–C), BTD (D), BCN (E–F), TCO (G–H) and Norb (I). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In summary, the trend in reactivity of surveyed probes was BTD ≫ DYn-2>BCN ≈ TCO, whereas Norb was incapable of targeting sulfenic acids in all models tested. Generally speaking, C-nucleophiles were more potent than strained alkynes/alkenes, whose efficiency is mainly dictated by ring strain. This reinforces the idea that sulfenic acids reacts more efficiently with more localized electronegative charges, or “harder” nucleophiles, by the decreasing order of carbanions (e.g., BTD), enolates (e.g., DYn-2), strained alkynes (e.g., BCN) or alkenes (e.g., TCO). Norbornenes (e.g., Norb) simply cannot provide enough ring strain to facilitate the reaction with sulfenic acids at a useful rate.
2.3. Chemical selectivity of C-nucleophilic probes
The aforementioned data demonstrate that exploiting the electrophilic character of the sulfur atom in sulfenic acid through reaction with nucleophilic probes is the preferred strategy for labeling. Although the cellular environment is also nucleophilic as a whole, it is critical to carefully evaluate the potential cross-reactivity with other biologically relevant electrophiles, such as cyclic sulfenamides, aldehydes, disulfides and hydrogen peroxide. Below, we delineate the reactivity of C-nucleophile probes with protein models and kinetic studies.
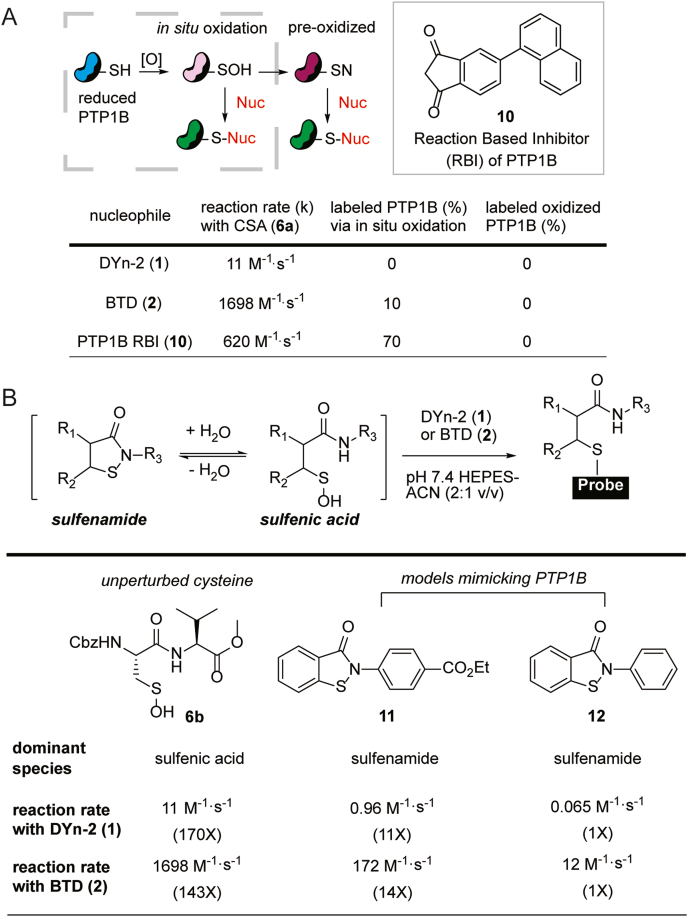
Cysteine sulfenamides are formed via an attack of an amide nitrogen on a sulfenic acid. Favored by conformation, the amide is from the backbone of the following amino acid in the sequence, yielding a five-membered cyclic structure. Like sulfenic acids, some cyclic sulfenamides can be good electrophiles and produce the same product when reacting with C-nucleophiles. For example, the dipeptide sulfenamide CSA can react with several thiol, phosphinate, and carbon nucleophiles in an anhydrous environment [31]. However, the formation of a (cyclic) sulfenamide will face a huge hurdle, because under normal circumstances, an amide (pKa ~17) is a very poor nucleophile and a sulfenic acid has a very short lifetime. Although the sulfenamide modification is rarely observed in proteins, a cyclic sulfenamide structure in protein tyrosine phosphatase 1B (PTP1B) was characterized crystallographically [32,33], which draws questions as to whether the target of C-nucleophile probes is actually the sulfenamide [34,35]. In fact, the formation of PTP1B sulfenamide is facilitated by two key factors, including a decreased pKa (increased nucleophilicity) of the Ser216 amide nitrogen due to hydrogen bonding, and steric shielding of the Cys215 sulfenic acid which becomes inaccessible by small-molecule nucleophiles, e.g., dimedone [36]. To analyze the reactivity of PTP1B, we added DYn-2 or BTD to PTP1B, which was either oxidized in situ, or pre-oxidized prior to nucleophile treatment, representing the sulfenic acid (-SOH) or sulfenamide (-SN) form of PTP1B, respectively (Fig. 4A). Neither nucleophile reacted with PTP1B-SN, while only a small percentage (10%) of PTP1B–SOH reacted with BTD. A PTP1B reaction-based inhibitor (RBI) 10 [37] which labeled the majority (70%) of PTP1B–SOH also failed to react with the PTP1B-SN form. These findings further indicate that the pocket harboring PTP1B-SN is not exposed for nucleophilic attack, regardless of the probe's reactivity towards sulfenic acids. Apart from PTP1B, the cyclic sulfenamide modification has proved to be exceedingly rare [38], having only been identified previously in recombinant OhrR [39] or an AhpC mutant when analyzed in the gas phase by MS [25].
Fig. 4.
Reaction of cyclic sulfenamides with C-nucleophiles. A) In situ oxidation and “pre-oxidation” of PTP1B furnishes the sulfenic acid (-SOH) and sulfenamide form (-SN), respectively. The sulfenamide form did not react with nucleophiles, including the reaction-based inhibitor (RBI) of PTP1B. B) Equilibrium between sulfenamide and sulfenic acid with chemical trapping by C-nucleophiles. Unperturbed cysteines and synthetic models of PTP1B exhibit distinct behaviors.
In addition to its scarcity in the proteome, nucleophilic attack on sulfenamide is kinetically unfavorable and easily outcompeted by sulfenic acid [3]. We employed two small-molecule sulfenamide models, benzisothiazolinone derivatives 11 and 12, which include a low pKa thiol (~5.6, similar to the Cys215 of PTP1B [40]) and an activated amide nitrogen mimicking PTP1B. Their sulfenamide forms are the dominant species in aqueous-organic buffers, and they showed a significant decrease in reaction rates toward nucleophiles such as DYn-2 or BTD, compared to the unperturbed cysteine dipeptide 6b (Fig. 4B). The less electron-withdrawing sulfenamide 12 suffered a greater rate decrease (>100-fold) than sulfenamide 11 (>10 fold). Thus, current evidence does not support that cyclic sulfenamide is a universal protein cysteine modification, nor a meaningful target of C-nucleophile probes.
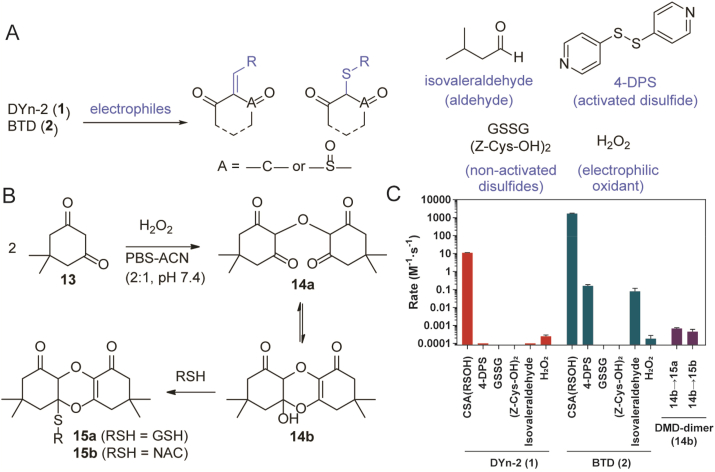
Next, we investigated the C-nucleophile probes' reactivity toward disulfides, which has redox potentials (E°) from −95 to −470 mV [41] that translate to a wide range of electrophilicity. We selected glutathione disulfide (GSSG) and a protected cystine (Z-Cys-OH)2 to represent redox-active disulfides which have similar redox potentials to classic oxidoreductases like thioredoxin-1 (E° = −270 mV) [42]. We also included a highly polarized disulfide known as 4,4′-dipyridyl disulfide (4-DPS). This electrophilic disulfide is not present in cells but is useful to model the upper limit of nucleophilic probe reactivity. Consistent with our earlier findings [21], both DYn-2 and BTD remain inactive toward GSSG or (Z-Cys-OH)2 (10 equivalents). Meanwhile, a rather sluggish but measurable reaction of BTD with 4-DPS (10 equivalents) was obtained (0.159 M−1 s−1), which is about 10,000 times slower than BTD reacting with the CSA sulfenic acid model (1700 M−1 s−1). A small portion of DYn-2 also reacted with 4-DPS, but the reaction remained incomplete after 24 h. These data indicate that C-nucleophiles and protein disulfides are highly unlikely to be biologically relevant reaction partners (Fig. 5A and C).
Fig. 5.
Reaction of nucleophilic probes for sulfenic acid with various electrophiles. A) Ostensible reaction between C-nucleophiles and electrophiles. B) Dimerization of dimedone and subsequent reaction with thiols. C) Rate constants of probes reacting with electrophiles, including sulfenic acids (intended targets), aldehydes, disulfides and hydrogen peroxide (cross-reactions). Note that the y-axis is plotted on logarithmic scale.
Another noteworthy class of biological electrophiles are aldehydes. They are mainly present at low levels as small-molecule byproducts of cellular metabolism [43], but some exist as modifications to biomolecules, such as the N-formylation of proteins [44]. We chose isovaleraldehyde, which possesses an alkyl side chain, to represent biologically relevant aldehydes. Similar to the reaction with 4-DPS, BTD presented poor reactivity with isovaleraldehyde at a rate of 0.079 M−1 s−1, about 21,000-fold less than that of CSA sulfenic acid model. The rate of DYn-2 and isovaleraldehyde was also too slow to be measured before aerobic degradation took place. With the kinetic data and concentration in mind, aldehydes also pose limited impact on the labeling of protein sulfenic acids by C-nucleophiles (Fig. 5A and C).
Last but not least, we address the potential cross-reactivity with a ubiquitous reactive oxygen species, hydrogen peroxide (H2O2), which is also an electrophile. It has been reported that dimedone underwent oxidative dimerization when exposed to excessive (high millimolar) concentrations of H2O2 [45]. However, the impact of this reaction is unclear under practical labeling conditions. When 5 mM of dimedone-based probe DYn-2 (typical concentration used in lysate or cell labeling experiments) was treated with 10 mM of H2O2, less than 5% of oxidative dimer was found after 2 h at 37 °C (typical labeling condition). Even after overnight incubation, DYn-2 dimerization was only about 50% complete. When H2O2 concentration was further decreased to 1 mM, which is still a few orders of magnitude higher than the estimated cellular level (low micromolar range), almost no dimer was observed after 2 h, and about 12% of DYn-2 dimerized overnight. This indicates cellular H2O2 does not degrade DYn-2 at an observable level. The more reactive nucleophilic probe BTD was also surveyed, and found to react extremely slowly with H2O2 at a rate of 1.83 × 10−4 M−1 s−1, similar to that of DYn-2 (2.48 × 10−4 M−1 s−1) (Fig. 5B and C).
The oxidative dimerization of dimedone (13) was further investigated by NMR. Data showed the dimer existed as a mixture of enol-keto isomers in chloroform. As expected, the equilibrium shifted to the keto form in polar solvents like DMSO, yielding two singlets with a 2:3 ratio in 1H NMR, which suggested a symmetric structure 14a. We also observed that 14a reacted with thiols like GSH and NAC, likely through a hemiketal intermediate 14b and a thiol-acetal exchange reaction to furnish 15a and 15b. Nevertheless, these reactions occurred at decreased rates (6.81 × 10−4 M−1 s−1 and 4.42 × 10−4 M−1 s−1, respectively) (Fig. 5B and C). Taken together with the already sluggish dimerization reaction, cellular levels of H2O2 will not interfere with the specificity and efficacy of C-nucleophile probes for sulfenic acid, and dimedone is not expected to form covalent bonds with thiols via the oxidized dimer under physiological settings.
3. Conclusion
Reactivity and selectivity are the most critical features of chemical probes for bioorthogonal applications. With the recent emergence of new classes of Cys-SOH probes, we have evaluated these in parallel using two small-molecule models with focus on their kinetic rates or reaction yields, and further validated the trend in reactivity using a protein model. In contrast to C-nucleophiles (e.g., DYn-2 (1), BTD (2)), some strained alkene/alkyne derivatives (e.g., BCN–OH (3), TCO-OH (4)) target sulfenic acids via different mechanisms, but at a lower efficiency even compared to first-generation dimedone-based probes. We found no evidence of the norbornene derivative Norb (5) reacting with any validated sulfenic acid model in this study. This striking difference from original reports [28] is attributable to a flawed “model” where millimolar concentrations of N-acetylcysteine and H2O2 are used, which generates thiyl radicals that react with norbornene and related analogs. Aerobic oxidation of thiols can also lead to thiyl radicals and undergo thiol-ene reactions [46] with strained alkene/alkyne probes. On the other hand, although there are concerns that C-nucleophile probes might cross-react with cellular electrophiles, their population is limited by the overall nucleophilic environment of the cell. Our results indicated that biological electrophiles, including aldehydes, activated disulfides (those most prone to nucleophilic attack), and an electrophilic oxidant (H2O2), reacted with C-nucleophile probes at rates several orders of magnitude slower than the intended sulfenic acid target. Additionally, we have analyzed the cyclic sulfenamide modification, which is extremely rare in proteins. Benzisothiazolinone derivatives that mimic the Cys215-Ser216 motif of PTP1B spontaneously formed cyclic sulfenamides via sulfenic acid intermediates, but showed markedly reduced reactivity toward nucleophiles compared to an unperturbed cysteine. This finding, coupled with steric factors, illustrates why most nucleophiles, including inhibitors designed to fit the active site, fail to target the PTP1B sulfenamide.
Although small-molecule models are versatile tools for the generation of kinetic data, they can create discrepancies when comparing probes with separate models, because solvents, environments and surrounding groups can significantly impact their reactivity. For example, solvents dictate the dominating species of synthetic sulfenamides like CSA, which exists as stable cyclized form (6a) in organic solvents, but readily hydrolyzes to the sulfenic acid form (6b) in aqueous solutions. As another example, anthraquinone-1-sulfenic acid (Fries' Acid) is stabilized via an intramolecular hydrogen bond. It exhibits a vastly different reactivity profile than a protein sulfenic acid, and is hence not an ideal model for the latter [25]. Similarly, protein models can provide different rates depending upon accessibility and intrinsic reactivity. This limitation is showcased by the labeling of pre-oxidized protein PTP1B whose buried sulfenamide moiety is not targeted by most nucleophiles, despite the positive reactivity in small-molecule “models” of the PTP1B sulfenamide. Therefore, it is critical to establish standardized (preferably, protein-based) models for testing new probes. Assessing from one protein and two small-molecule models, we conclude that C-nucleophile probes DYn-2 and BTD remain the most viable bioorthogonal approach for trapping and tagging protein sulfenic acids.
4. Methods
All associated methods are included in the supplementary materials.
Declaration of competing interest
No authors have conflicts or competing interests.
Acknowledgements
This work was supported by the US National Institutes of Health (R01 GM102187 and R01 CA174864 to K.S.C).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102072.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Paulsen C.E., Carroll K.S. Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem. Rev. 2013;113:4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcock L.J., Perkins M.V., Chalker J.M. Chemical methods for mapping cysteine oxidation. Chem. Soc. Rev. 2018;47:231–268. doi: 10.1039/c7cs00607a. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y., Carroll K.S. Activity-based sensing for site-specific proteomic analysis of cysteine oxidation. Acc. Chem. Res. 2020;53:20–31. doi: 10.1021/acs.accounts.9b00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Autréaux B., Toledano M.B. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 5.Gupta V., Carroll K.S. Sulfenic acid chemistry, detection and cellular lifetime. Biochim. Biophys. Acta Gen. Subj. 2014;1840:847–875. doi: 10.1016/j.bbagen.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmström K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 7.Bechtel T.J., Weerapana E. From structure to redox: the diverse functional roles of disulfides and implications in disease. Proteomics. 2017;17:1600391. doi: 10.1002/pmic.201600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paulsen C.E. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat. Chem. Biol. 2011;8:57. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Truong T.H. Molecular basis for redox activation of epidermal growth factor receptor kinase. Cell Chem. Biol. 2016;23:837–848. doi: 10.1016/j.chembiol.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pei J.F. Diurnal oscillations of endogenous H2O2 sustained by p66Shc regulate circadian clocks. Nat. Cell Biol. 2019;21:1553–1564. doi: 10.1038/s41556-019-0420-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J. Mining for protein S-sulfenylation in Arabidopsis uncovers redox-sensitive sites. Proc. Natl. Acad. Sci. U.S.A. 2019;116:20256–20261. doi: 10.1073/pnas.1906768116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng J., Fu L., Liu K., Tian C., Wu Z., Jung Y., Ferreira R.B., Carroll K.S., Blackwell T.K., Yang J. Global profiling of distinct cysteine redox forms reveals wide-ranging redox regulation in C. Elegans. Nat. Commun. 2021;12(1):1415. doi: 10.1038/s41467-021-21686-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y., Li Z., Zhang L., Tang H., Zhang H., Wang C., Chen S.Y., Bu D., Zhang Z., Zhu Z. Endogenous SO2-dependent Smad3 redox modification controls vascular remodeling. Redox Biol. 2021;41:101898. doi: 10.1016/j.redox.2021.101898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang M., Li W., Harberg C., Chen W., Yue H., Ferreira R.B., Wynia-Smith S.L., Carroll K.S., Zielonka J., Flaumenhaft R., Silverstein R.L., Smith B.C. Cysteine sulfenylation by CD36 signaling promotes arterial thrombosis in dyslipidemia. Blood Adv. 2020;4(18):4494–4507. doi: 10.1182/bloodadvances.2020001609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulsen C.E., Carroll K.S. Chemical dissection of an essential redox switch in yeast. Chem. Biol. 2009;16:217–225. doi: 10.1016/j.chembiol.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Seo Y.H., Carroll K.S. Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc. Natl. Acad. Sci. Unit. States Am. 2009;106:16163–16168. doi: 10.1073/pnas.0903015106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen R.E., Roth D., Winther J.R. Quantifying the global cellular thiol-disulfide status. Proc. Natl. Acad. Sci. Unit. States Am. 2009;106:422–427. doi: 10.1073/pnas.0812149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian Y., Lin Q. Fitness factors for bioorthogonal chemical probes. ACS Chem. Biol. 2019;14:2489–2496. doi: 10.1021/acschembio.9b00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo Y.R. CRC Press; Boca Raton: 2007. Comprehensive Handbook of Chemical Bond Energies. [Google Scholar]
- 20.Gupta V., Carroll K.S. Rational design of reversible and irreversible cysteine sulfenic acid-targeted linear C-nucleophiles. Chem. Commun. 2016;52(16):3414–3417. doi: 10.1039/c6cc00228e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta V., Carroll K.S. Profiling the reactivity of cyclic C-nucleophiles towards electrophilic sulfur in cysteine sulfenic acid. Chem. Sci. 2016;7(1):400–415. doi: 10.1039/c5sc02569a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta V., Yang J., Liebler D.C., Carroll K.S. Diverse redoxome reactivity profiles of carbon nucleophiles. J. Am. Chem. Soc. 2017;139(15):5588–5595. doi: 10.1021/jacs.7b01791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellis H.R., Poole L.B. Novel application of 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole to identify cysteine sulfenic acid in the AhpC component of alkyl hydroperoxide reductase. Biochemistry. 1997;36(48):15013–15018. doi: 10.1021/bi972191x. [DOI] [PubMed] [Google Scholar]
- 24.Gupta V., Paritala H., Carroll K.S. Reactivity, selectivity, and stability in sulfenic acid detection: a comparative study of nucleophilic and electrophilic probes. Bioconjugate Chem. 2016;27(5):1411–1418. doi: 10.1021/acs.bioconjchem.6b00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poole T.H., Reisz J.A., Zhao W., Poole L.B., Furdui C.M., King S.B. Strained cycloalkynes as new protein sulfenic acid traps. J. Am. Chem. Soc. 2014;136(17):6167–6170. doi: 10.1021/ja500364r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scinto S.L., Ekanayake O., Seneviratne U., Pigga J.E., Boyd S.J., Taylor M.T., Liu J., am Ende C.W., Rozovsky S., Fox J.M. Dual-reactivity trans-cyclooctenol probes for sulfenylation in live cells enable temporal control via bioorthogonal quenching. J. Am. Chem. Soc. 2019;141(28):10932–10937. doi: 10.1021/jacs.9b01164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barton D.H.R., Greig D.G.T., Lucente G., Sammes P.G., Taylor M.V., Cooper C.M., Hewitt G., Underwood W.G.E. On the trapping of sulphenic acids from penicillin sulphoxides. J. Chem. Soc. D Chem. Commun. 1970;24:1683–1684. [Google Scholar]
- 28.Alcock L.J., Farrell K.D., Akol M.T., Jones G.H., Tierney M.M., Kramer H.B., Pukala T.L., Bernardes G.J.L., Perkins M.V., Chalker J.M. Norbornene probes for the study of cysteine oxidation. Tetrahedron. 2018;74(12):1220–1228. [Google Scholar]
- 29.Fu L., Liu K., Ferreira R.B., Carroll K.S., Yang J. Proteome-wide analysis of cysteine S-sulfenylation using a benzothiazine-based probe. Curr. Protein Pept. Sci. 2019;95(1):e76. doi: 10.1002/cpps.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barattucci A., Aversa M.C., Mancuso A., Salerno T.M., Bonaccorsi P. Transient sulfenic acids in the synthesis of biologically relevant products. Molecules. 2018;23(5):1030. doi: 10.3390/molecules23051030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiau T.P., Erlanson D.A., Gordon E.M. Selective reduction of peptide isothiazolidin-3-ones. Org. Lett. 2006;8(25):5697–5699. doi: 10.1021/ol062077j. [DOI] [PubMed] [Google Scholar]
- 32.van Montfort R.L.M., Congreve M., Tisi D., Carr R., Jhoti H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature. 2003;423(6941):773–777. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- 33.Salmeen A., Andersen J.N., Myers M.P., Meng T.-C., Hinks J.A., Tonks N.K., Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423(6941):769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 34.Forman H.J., Davies M.J., Krämer A.C., Miotto G., Zaccarin M., Zhang H., Ursini F. Protein cysteine oxidation in redox signaling: caveats on sulfenic acid detection and quantification. Arch. Biochem. Biophys. 2017;617:26–37. doi: 10.1016/j.abb.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pople J.M.M., Chalker J.M. A critical evaluation of probes for cysteine sulfenic acid. Curr. Opin. Chem. Biol. 2021;60:55–65. doi: 10.1016/j.cbpa.2020.07.011. [DOI] [PubMed] [Google Scholar]
- 36.Tsutsumi R., Harizanova J., Stockert R., Schröder K., Bastiaens P.I.H., Neel B.G. Assay to visualize specific protein oxidation reveals spatio-temporal regulation of SHP2. Nat. Commun. 2017;8(1):466. doi: 10.1038/s41467-017-00503-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carroll K.S. Targeted covalent probes and inhibitors of proteins containing redox-sensitive cysteines. WO2014089546A1. 2013;8 December. [Google Scholar]
- 38.Shi Y., Carroll K.S. Comments on ’A critical evaluation of probes for cysteine sulfenic acid. Curr. Opin.Chem. Biol. 2021;60:55–65. doi: 10.1016/j.cbpa.2021.01.004. [DOI] [PubMed] [Google Scholar]
- 39.Lee J.-W., Soonsanga S., Helmann J.D. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. Unit. States Am. 2007;104(21):8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohse D.L., Denu J.M., Santoro N., Dixon J.E. Roles of aspartic acid-181 and serine-222 in intermediate formation and hydrolysis of the mammalian protein-tyrosine-phosphatase PTP1. Biochemistry. 1997;36(15):4568–4575. doi: 10.1021/bi963094r. [DOI] [PubMed] [Google Scholar]
- 41.Wouters M.A., Fan S.W., Haworth N.L. Disulfides as redox switches: from molecular mechanisms to functional significance. Antioxidants Redox Signal. 2009;12(1):53–91. doi: 10.1089/ars.2009.2510. [DOI] [PubMed] [Google Scholar]
- 42.Krause G., Lundström J., Barea J.L., Pueyo de la Cuesta C., Holmgren A. Mimicking the active site of protein disulfide-isomerase by substitution of proline 34 in Escherichia coli thioredoxin. J. Biol. Chem. 1991;266(15):9494–9500. [PubMed] [Google Scholar]
- 43.Fritz K.S., Petersen D.R. An overview of the chemistry and biology of reactive aldehydes. Free Radic. Biol. Med. 2013;59:85–91. doi: 10.1016/j.freeradbiomed.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang T., Zhou X., Taghizadeh K., Dong M., Dedon P.C. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc. Natl. Acad. Sci. Unit. States Am. 2007;104(1):60–65. doi: 10.1073/pnas.0606775103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alcock L.J., Langini M., Stühler K., Remke M., Perkins M.V., Bernardes G.J.L., Chalker J.M. Proteome-wide survey of cysteine oxidation by using a norbornene probe. Chembiochem. 2020;21(9):1329–1334. doi: 10.1002/cbic.201900729. [DOI] [PubMed] [Google Scholar]
- 46.Hoyle C.E., Bowman C.N. Thiol–ene click chemistry. Angew. Chem. Int. Ed. 2010;49(9):1540–1573. doi: 10.1002/anie.200903924. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.