Abstract
We present the complete genome sequences of Caribbean watersnake bornavirus (CWBV) and Mexican black-tailed rattlesnake bornavirus (MRBV), which we identified in archived raw transcriptomic read data of a Caribbean watersnake (Tretanorhinus variabilis) and a Mexican black-tailed rattlesnake (Crotalus molossus nigrescens), respectively. The genomes of CWBV and MRBV have a length of about 8,900 nucleotides and comprise the complete coding regions and the untranslated regions. The overall genomic makeup and predicted gene content is typical for members of the genus Orthobornavirus within the family Bornaviridae. Alternative splicing was detected for the L and M genes. Based on a phylogenetic analysis of all viral proteins, we consider both viruses to be members of a single novel species within the genus Orthobornavirus. Both viruses form a distinct outgroup to all currently known orthobornaviruses. Based on the novel virus genomes, we furthermore identified closely related endogenous bornavirus-like nucleoprotein sequences in transcriptomic data of veiled chameleons (Chamaeleo calyptratus) and a common lancehead (Bothrops atrox).
The family Bornaviridae belongs to the order Mononegavirales [1] and comprises viruses with a monopartite single-stranded RNA(-) genome that form enveloped and spherical virions with a diameter of 70-130 nm. Their genomes are about 8.9 kb in length and encode the nucleoprotein (N), the accessory protein X, the phosphoprotein (P), the matrix protein (M), the glycoprotein (G), and the large protein (L), which contains the RNA-directed RNA polymerase. The organization of the respective open reading frames varies among the different bornaviruses, and additional protein isotypes produced by alternative splicing [2] or start codon skipping [3] have been reported. Bornaviruses are divided into the three taxonomic genera: Orthobornavirus, Carbovirus, and Cultervirus [1, 4].
Currently, orthobornaviruses have the widest known host spectrum within the family Bornaviridae, ranging from mammals and birds to reptiles, whereas carbo- and culterviruses have so far only been identified in reptiles [5, 6] and fish [7], respectively. Mammalian orthobornaviruses are known to be zoonotic agents and may be transmitted from reservoirs, such as shrews or squirrels, to humans, sheep and horses [8–10]. Reptilian carbo- and orthobornaviruses have so far been identified in Australian carpet pythons (Morelia spilota (Lacépède, 1804)) with neurological disease [5] and in a wild-caught Loveridge's garter snake (Elapsoidea loveridgei (Parker, 1949)) [6], respectively. Furthermore, partial sequences of exogenous orthobornavirus-like N, X, and P genes identified in a Gaboon viper (Bitis gabonica (Duméril, Bibron & Duméril, 1854)) [11] suggested a wider distribution of orthobornaviruses among snakes.
In this study, we used data mining of transcriptomic and metagenomic raw RNA read archives in order to identify hitherto undetected bornaviruses of reptiles. As a result, we determined the full genome sequence of two bornaviruses in datasets from colubrid and viperid snakes.
In detail, we initially employed the Serratus website [12] in order to identify datasets within the Sequence Read Archive (SRA) that potentially contain bornavirus-like sequences. We then downloaded promising SRA datasets (SRR5440420 and SRR9693197), trimmed them with respect to quality and adapter contamination using Trim Galore (v0.6.6), and subsequently used rnaSPAdes (v3.13.0) for de novo assembly. The resulting contigs were then screened for bornavirus-like sequences using DIAMOND BLASTX (v0.9.21.122) with a representative database of bornavirus proteins. For both SRA datasets, the initial assembly yielded a single contig representing the complete viral genome. These initial contigs were then further polished using an iterative mapping and assembly strategy [13]. The full genomes were annotated with respect to known bornaviruses using Geneious Prime (v2021.0.1). Furthermore, we predicted introns and splice sites using STAR (v2.7.7a) running in basic two-pass mode. The splice sites deduced from raw reads were further evaluated by in silico prediction using NNSPLICE (v0.9). Transcription termination sites were predicted using sequence similarity [14] and manual inspection of raw reads showing transition to polyA at the respective termination position.
SRR5440420 contains raw reads from the Harderian gland transcriptome of a wild-caught adult Caribbean watersnake (Tretanorhinus variabilis (Duméril, Bibron & Duméril, 1854); family Colubridae) from Santa Fe, La Habana, Cuba [15]. SRR9693197 contains raw reads from the venom gland transcriptome of a wild-caught juvenile Mexican black-tailed rattlesnake (Crotalus molossus nigrescens (Gloyd, 1936); family Viperidae) from Nuevo León, Mexico. The bornaviruses identified and characterised in these datasets were named Caribbean watersnake bornavirus (CWBV) and Mexican black-tailed rattlesnake bornavirus (MRBV), respectively. The genomes of CWBV and MRBV are of similar length, with MRBV (8907 nt) being three nucleotides longer at the 5’ end than CWBV (8904 nt). The overall genomic makeup of the two viruses is very similar.
In detail, we predicted six protein coding open reading frames, encoding N, X, P, M, G, and L (Fig. 1). The gene order N-X/P-M-G-L is consistent with the genome organization of members of the genus Orthobornavirus but different from that of members of the genera Carbovirus and Cultervirus, which share the order N-X/P-G-M-L [5, 7]. Furthermore, we identified three conserved transcription initiation sites and four transcription termination sites, as well as alternative splicing for the M and L genes (Fig. 1). A third potential intron, located in the L gene, was identified only for MRBV. All predicted splicing events were supported by several reads missing the intron sequence in both datasets. All predicted transcription start sites (S1-3) correlated well with a steep increase in read coverage, while all predicted transcription termination sites (T1-4) correlated with an steep drop in read coverage and reads showing a transition to polyA at the respective termination site. When compared to representative orthobornaviruses, the 5’ and 3’ untranslated region of CWBV and MRBV can be considered complete, although further experiments need to be performed using molecular methods such as rapid amplification of cDNA ends (RACE) PCR.
Fig. 1.
The overall genome organization of Caribbean watersnake bornavirus (CWBV) and Mexican black-tailed rattlesnake bornavirus (MRBV) is highly conserved and comprises the canonical bornaviral gene order N-X/P-M-G-L (pink arrows) flanked by untranslated regions (UTR – blue arrows). Introns were detected for the M and L genes (grey arrows). Three transcription initiation sites (S1-3) and four transcription termination sites (T1-4) were predicted.
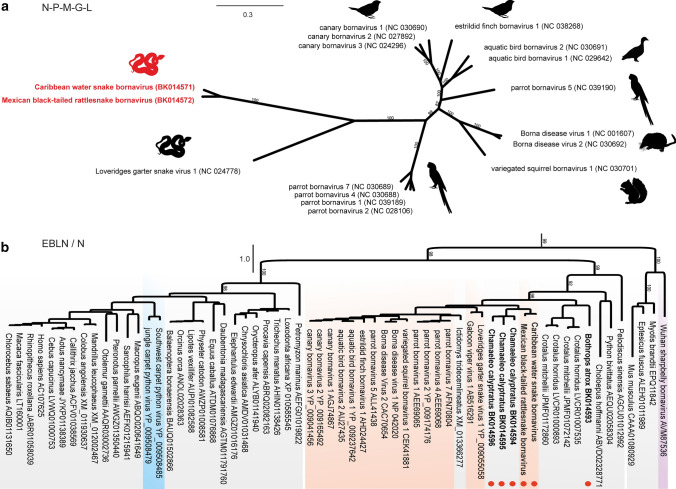
A phylogenetic analysis of a concatenated alignment of N-P-M-G-L amino acid sequences from both viruses along with representative bornavirus sequences showed that both viruses were distantly related to other members of the genus Orthobornavirus (Fig. 2a). Pairwise sequence comparison (PASC [16]) of complete genome sequences revealed 76.6% pairwise nucleotide sequence identity between CWBV and MRBV and 56.6-57.3% identity to the most closely related orthobornaviruses. Based on phylogenetic analysis and the conserved genome organisation, and in line with the species demarcation cutoff of 72 to 75% pairwise nucleotide sequence identity [1], we suggest that both viruses be assigned to a single new species within the genus Orthobornavirus.
Fig. 2.
Maximum-likelihood phylogenetic analysis of orthobornaviruses (a) and endogenous bornavirus-like nucleoproteins (b). (a) An unrooted phylogenetic tree based on the concatenated amino acid sequence alignments of N-P-M-G-L of the novel snake bornaviruses (highlighted in red) together with all available complete genome sequences of members of the genus Orthobornavirus. (b) Phylogenetic relationship between endogenous bornavirus-like nucleoprotein amino acid sequences (grey) and those of members of the bornavirus genera Carbovirus (blue), Orthobornavirus (orange) and Cultervirus (purple). Sequences determined in this study are indicated by red dots. Trees were calculated using IQ-TREE (version 2.1.2; 1 million ultrafast bootstraps; optimal substitution model for each alignment/alignment partition). The bars represent amino acid substitutions per site, and numbers in italics indicate bootstrap support for the major branches.
Finally, we used the CWBV and MRBV protein sequences to further search the SRA. For this purpose, we downloaded 2-8 million reads of all available transcriptomics or metagenomics datasets related to members of the taxonomic order Squamata and used DIAMOND BLASTX to match sequences. Datasets with promising hits were then assembled as described. As a result, we identified three endogenous bornavirus-like nucleoprotein (EBLN) sequence elements in veiled chameleons (Chamaeleo calyptratus (Duméril & Bibron, 1851); SRR6662597) [17] and one EBLN in a common lancehead (Bothrops atrox (Linnaeus, 1758); SRR1953004) [18] (Fig. 2b). The chameleon EBLNs exhibited 68.8-75.1% pairwise nucleotide sequence identity to each other and 65.2-66.3% to the CWBV and MRBV N genes, whereas the common lancehead EBLN was more distantly related. A phylogenetic comparison of EBLNs and circulating bornaviruses based on the frameshift-corrected protein alignment of Hyndman et al. [5] showed that a common ancestor of both novel bornaviruses left its genetic fingerprint in the genomes of non-avian reptiles. Currently, there is no evidence that these viruses cause any disease in the sampled snakes, and further screening is needed to evaluate their distribution and clinical relevance. However, these sequences will improve bornavirus diagnostic procedures and help researchers to understand the evolutionary origins of these viruses.
Acknowledgements
We want to thank the scientists worldwide who share their raw sequencing data with the scientific community. These data sets contain valuable information beyond their initial purpose.
Funding
Open Access funding enabled and organized by Projekt DEAL. This study was funded in part by the Federal Ministry of Education and Research within the Research Network Zoonotic Infections to the ‘Zoonotic Bornavirus Consortium’ (ZooBoCo; Grant no. 01KI2005A to Dennis Rubbenstroth).
Availability of data and material
The annotated genome sequences generated during and/or analysed during the current study are available in the DDBJ/EMBL/GenBank databases under the TPA accession numbers BK014571, BK014572, BK014593-BK014596. Additional metadata and raw sequencing data are available under the BioProject ID PRJNA382075 (SAMN06706898, SRR5440420) and PRJNA554814 (SAMN12284706, SRR9693197).
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate and publication
All authors confirm that they have read the manuscript and participated in the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kuhn JH, Dürrwald R, Bào Y, et al. Taxonomic reorganization of the family Bornaviridae. Arch Virol. 2015;160:621–632. doi: 10.1007/s00705-014-2276-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider PA, Schneemann A, Lipkin WI. RNA splicing in Borna disease virus, a nonsegmented, negative-strand RNA virus. J Virol. 1994;68:5007–5012. doi: 10.1128/jvi.68.8.5007-5012.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pyper JM, Gartner AE. Molecular basis for the differential subcellular localization of the 38- and 39-kilodalton structural proteins of Borna disease virus. J Virol. 1997;71:5133–5139. doi: 10.1128/JVI.71.7.5133-5139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker PJ, Siddell SG, Lefkowitz EJ, et al. Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2019) Arch Virol. 2019;164:2417–2429. doi: 10.1007/s00705-019-04306-w. [DOI] [PubMed] [Google Scholar]
- 5.Hyndman TH, Shilton CM, Stenglein MD, et al. Divergent bornaviruses from Australian carpet pythons with neurological disease date the origin of extant Bornaviridae prior to the end-Cretaceous extinction. PLoS Pathog. 2018;14:e1006881. doi: 10.1371/journal.ppat.1006881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stenglein MD, Leavitt EB, Abramovitch MA, et al. Genome Sequence of a Bornavirus Recovered from an African Garter Snake (Elapsoidea loveridgei) Genome Announc. 2014 doi: 10.1128/genomeA.00779-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi M, Lin X-D, Chen X, et al. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann B, Tappe D, Höper D, et al. A variegated squirrel bornavirus associated with fatal human encephalitis. N Engl J Med. 2015;373:154–162. doi: 10.1056/NEJMoa1415627. [DOI] [PubMed] [Google Scholar]
- 9.Niller HH, Angstwurm K, Rubbenstroth D, et al. Zoonotic spillover infections with Borna disease virus 1 leading to fatal human encephalitis, 1999–2019: an epidemiological investigation. Lancet Infect Dis. 2020;20:467–477. doi: 10.1016/S1473-3099(19)30546-8. [DOI] [PubMed] [Google Scholar]
- 10.Rubbenstroth D, Schlottau K, Schwemmle M, et al. Human bornavirus research: back on track! PLoS Pathog. 2019;15:e1007873. doi: 10.1371/journal.ppat.1007873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horie M, Honda T, Suzuki Y, et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature. 2010;463:84. doi: 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar RC, Taylor J, Altman T et al. (2020) Petabase-scale sequence alignment catalyses viral discovery. bioRxiv. 10.1101/2020.08.07.241729 [DOI] [PubMed]
- 13.Pfaff F, Schulze C, König P, et al. A novel alphaherpesvirus associated with fatal diseases in banded Penguins. J Gen Virol. 2017;98:89–95. doi: 10.1099/jgv.0.000698. [DOI] [PubMed] [Google Scholar]
- 14.Schneemann A, Schneider PA, Kim S, et al. Identification of signal sequences that control transcription of borna disease virus, a nonsegmented, negative-strand RNA virus. J Virol. 1994;68:6514–6522. doi: 10.1128/jvi.68.10.6514-6522.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domínguez-Pérez D, Durban J, Agüero-Chapin G, et al. The Harderian gland transcriptomes of Caraiba andreae, Cubophis cantherigerus and Tretanorhinus variabilis, three colubroid snakes from Cuba. Genomics. 2019;111:1720–1727. doi: 10.1016/j.ygeno.2018.11.026. [DOI] [PubMed] [Google Scholar]
- 16.Bao Y, Chetvernin V, Tatusova T. PAirwise sequence comparison (PASC) and its application in the classification of filoviruses. Viruses. 2012;4:1318–1327. doi: 10.3390/v4081318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinto BJ, Card DC, Castoe TA, et al. The transcriptome of the veiled chameleon (Chamaeleo calyptratus): a resource for studying the evolution and development of vertebrates. Dev Dyn. 2019;248:702–708. doi: 10.1002/dvdy.20. [DOI] [PubMed] [Google Scholar]
- 18.Freitas-de-Sousa LA, Amazonas DR, Sousa LF, et al. Comparison of venoms from wild and long-term captive Bothrops atrox snakes and characterization of Batroxrhagin, the predominant class PIII metalloproteinase from the venom of this species. Biochimie. 2015;118:60–70. doi: 10.1016/j.biochi.2015.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The annotated genome sequences generated during and/or analysed during the current study are available in the DDBJ/EMBL/GenBank databases under the TPA accession numbers BK014571, BK014572, BK014593-BK014596. Additional metadata and raw sequencing data are available under the BioProject ID PRJNA382075 (SAMN06706898, SRR5440420) and PRJNA554814 (SAMN12284706, SRR9693197).