Abstract
Coronavirus 3C-like protease (3CLpro) is a crucial target for treating coronavirus diseases including COVID-19. Our preliminary screening showed that Ampelopsis grossedentata extract (AGE) displayed potent SARS-CoV-2-3CLpro inhibitory activity, but the key constituents with SARS-CoV-2-3CLpro inhibitory effect and their mechanisms were unrevealed. Herein, a practical strategy via integrating bioactivity-guided fractionation and purification, mass spectrometry-based peptide profiling and time-dependent biochemical assay, was applied to identify the crucial constituents in AGE and to uncover their inhibitory mechanisms. The results demonstrated that the flavonoid-rich fractions (10-17.5 min) displayed strong SARS-CoV-2-3CLpro inhibitory activities, while the constituents in these fractions were isolated and their SARS-CoV-2-3CLpro inhibitory activities were investigated. Among all isolated flavonoids, dihydromyricetin, isodihydromyricetin and myricetin strongly inhibited SARS-CoV-2 3CLpro in a time-dependent manner. Further investigations demonstrated that myricetin could covalently bind on SARS-CoV-2 3CLpro at Cys300 and Cys44, while dihydromyricetin and isodihydromyricetin covalently bound at Cys300. Covalent docking coupling with molecular dynamics simulations showed the detailed interactions between the orthoquinone form of myricetin and two covalent binding sites (surrounding Cys300 and Cys44) of SARS-CoV-2 3CLpro. Collectively, the flavonoids in AGE strongly and time-dependently inhibit SARS-CoV-2 3CLpro, while the newly identified SARS-CoV-2 3CLpro inhibitors in AGE offer promising lead compounds for developing novel antiviral agents.
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID-19, coronavirus disease 2019; 3CLpro, Chymotrypsin-like protease; CoVs, coronaviruses; AGE, Ampelopsis grossedentata extract; IC50, half maximal inhibition concentration; EDTA, ethylene diamine tetraacetic acid; DTT, dithiothreitol; IAA, iodoacetamide; PMSF, phenylmethylsulfonyl fluoride; HEPES, 2-[4-(2-hydroxyethyl) piperazin-1-yl] ethane sulfonic acid; HCl, hydrochloric acid; NaCl, sodium chloride; DMSO, dimethyl sulfoxide; PBS, potassium phosphate buffer; FRET, fluorescence resonance energy transfer; PDA, photo diode array; TOF-MS/MS, time of flight tandem mass spectrometry; nanoLC-MS/MS, nano liquid chromatography-tandem mass spectrometer; HCD, higher energy collision dissociation; MD, molecular dynamics; QFM, the orthoquinone form of myricetin; ACE2, angiotensin-converting enzyme 2; UGTs, UDP-glucuronosyltransferases
Keywords: SARS-CoV-2 3CLpro, Ampelopsis grossedentata extract, Covalent inhibitors
1. Introduction
Over the last two decades, the growing occurrences of coronaviruses-related diseases with high mortality have been one of the long-standing and life-threatening issues to the global population [1]. Currently, the newly emerging coronavirus disease 2019 (COVID-19), a globally infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has brought a colossal threat to public health, economic development and society safety [2], [3]. In the past one-year, exhaustive efforts have been made by the scientists to discover efficacious therapeutics for treating COVID-19, via targeting on several validated therapeutic targets. Among all identified therapeutic targets for combating COVID-19, the chymotrypsin-like protease (3CLpro) has drawn great concerns and has been recognized as a pivotal therapeutic target for fighting this pandemic, due to its high conservative and indispensable role in viral replication [4], [5], [6]. It has been validated that strong inhibition or dysfunction of 3CLpro can successfully block SARS-CoV-2 replication, and further generate benefits in the treatment of COVID-19 [7].
Although a variety of SARS-CoV-2 3CLpro inhibitors have been identified recently, the majority of them were restricted to the reversible interactions with the target protease [8], [9], [10]. By contrast, the covalent inhibitors could significantly attenuate the proteolytic activity of SARS-CoV-2 3CLpro via forming a stable chemical bond, which then inactivated this key protease and blocked coronavirus replication. Generally, the covalent inhibitors of target hydrolases bear at least one electrophilic group (such as quinones [11], Michael receptors [12], or some metal elements [13]) that could covalently bind to the nucleophilic residues (such as cysteine). The covalent inhibitors possessed several inherent advantages (such as high specificity, good inhibition potency, and durable interactions) [14], which could bring benefits to both anti-COVID-19 and other CoVs-related diseases. Unfortunately, the promising warheads and lead compounds for the development of efficacious SARS-CoV-2 3CLpro covalent inhibitors for treating COVID-19 are rarely reported. Recently, to find more efficacious SARS-CoV-2 3CLpro inhibitors with good safety profile, a high-throughput screening campaign was implemented to screen the herbal products with strong SARS-CoV-2 3CLpro inhibition activities, by using a fluorescence-based biochemical assay [15], [16]. After a large-scale screening of herbal products, we noticed that Ampelopsis grossedentata extract (AGE) strongly inhibited SARS-CoV-2 3CLpro in both time- and dose-dependently inhibition manners, with the apparent IC50 value of 3.44 μg/mL after 60-min preincubation. This finding suggests that AGE should contain the naturally occurring covalent inhibitors of SARS-CoV-2 3CLpro.
Herein, a practical strategy via integrating bioactivity-guided fractionation and purification, mass spectrometry-based peptide identification and time-dependent inhibition assays, was utilized to recognize and characterize the key constituents in AGE with SARS-CoV-2-3CLpro inhibitory activities. The results clearly showed that the flavonoid-rich fractions (10–17.5 min on reverse phase liquid chromatography) displayed strong inhibitory activities against SARS-CoV-2-3CLpro. After then, the major constituents in these bioactive factions were isolated and their structures as well as inhibitory effects on SARS-CoV-2-3CLpro were carefully identified. Among all isolated constituents, dihydromyricetin, isodihydromyricetin and myricetin strongly inhibited SARS-CoV-2-3CLpro in dose- and time- dependent manners. On the basis of the chemical structures of these newly identified SARS-CoV-2-3CLpro inhibitors, we postulated their catecholic groups in B-ring were easily oxidated into orthoquinones, which in turn, covalently modifying SARS-CoV-2-3CLpro (Fig. S4). In these cases, further mass spectrometry-based peptide assays coupling with molecular dynamics simulations were carried out to reveal the covalent binding sites and to explore the inhibitory mechanisms of these naturally occurring flavonoids.
2. Materials and methods
2.1. Chemicals and reagents
The Smt3-SARS-CoV-2 3CLpro codon was cloned into the pET29a (+) vectors by GENEWIZ, Inc. (Beijing, China). Escherichia coli (E. coli) BL21 (DE3) was gained from Shanghai Weidi Biotechnology Co., Ltd. (Shanghai, China). Tris-Base was obtained from Amresco (USA). Ethylene Diamine Tetraacetic Acid (EDTA) was gained from Dalian Meilun Biotechnology Co. Ltd. (Dalian, China). Lysozyme, sodium chloride (NaCl), imidazole, dithiothreitol (DTT), phenylmethylsulfonyl fluoride (PMSF), and hydrochloric acid (HCl), were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). 2-[4-(2-hydroxyethyl) piperazin-1-yl] ethane sulfonic acid (HEPES), NH4HCO3, urea, iodoacetamide (IAA), chymotrypsin and trypsin were provided by Sigma-Aldrich (St. Louis, MO, USA). Super Nuclease was purchased from Sino Biological Inc. (Beijing, China). One hundred and two herbal products were provided by Tianjiang Pharmaceutical Co., Ltd. (Jiangsu, China), two standard extracts (St. John’s Wort and Ginkgo Folium) were bought from Baoji Guokang Bio-Technology Co.,Ltd and Hubei Nokete Pharmaceutical Co.,Ltd,. Ampelopsis grossedentata extract was attained from Eastsign Foods Co., Ltd. (Quzhou, China). The reported covalent inhibitor of SARS-CoV-2 3CLpro (ebselen) was provided by TCI (Shanghai, China) [17]. Fluorescent substrate (Dabcyl-KNSTLQSGLRKE-Edans) was purchased from Shanghai Sangon Biological Engineering & Technology and Service Co. Ltd. (Shanghai, China), with the purity of 99%. The stock solution of this fluorescent substrate was prepared by Millipore water and stored at 4 °C. HPLC grade methanol, acetonitrile, dimethyl sulfoxide (DMSO) and formic acid were all ordered from Tedia (Fairfield, USA). DMSO‑d 6 and MeOD were provided by Adamas-beta (Shanghai, China) for 1H and 13C NMR analyses. Millipore water (Millipore, Bedford, USA) was used for preparing PBS buffer (pH 7.4, 100 mM) that was stored at 4 °C until use.
2.2. SARS-CoV-2 3CLpro inhibition assay
A fluorescence-based enzyme inhibition assay in 96-well plate format was used to assess the SARS-CoV-2 3CLpro inhibition activity [15], [16]. The hydrolytic rates of 3CLpro-catalyzed Dabcyl-KNSTLQSGLRKE-Edans were monitored in a reaction mixture (100 μL, total volume) with or without each tested inhibitor. Briefly, the SARS-CoV-2 3CLpro (4 μg/mL, final concentration) was preincubated with analytes / DMSO (as a control group) in PBS (pH 7.4, 100 mM, 1 mM EDTA) at 37 °C for 60 min or 0.5 min. Then the reaction proceeded for 20 min after added the Dabcyl-KNSTLQSGLRKE-Edans (20 μM, final concentration). The generated fluorescent signals (excitation/emission, 340 nm/490 nm) were continuously monitored by the microplate reader (SpectraMax® iD5, Molecular Devices, Austria).
2.3. Bioactivity-guided fractionation of AGE
A bioactivity-guided fractionation strategy was applied to quickly find out the bioactive fractions in AGE, which was supported by one Shimadzu UFLC system (Kyoto, Japan) equipped with an SPD-M 30A PDA detector. Water-0.1% formic acid (A) and acetonitrile (B) were optimized as the mobile phases in gradient conditions: 0.01–2 min, 95% A; 15–20 min,70%–25% A; 22–25 min, 95% A. The AGE sample (5 μL, 10 mg/mL) was injected and separated on a Shim-pack VP-ODS C18 column (2.0 × 250 mm, 4.6 μm) with the flow rate of 0.4 mL/min at 40 °C, and the LC fractions were collected every 2.5 min. Ten collected LC fractions were dried under vacuum pressure, and then redissolved in DMSO for assessment of 3CLpro inhibition activity.
2.4. Isolation and identification of the major bioactive constituents in AGE
An LC-TOF-MS/MS system (Foster City, CA, USA) equipped with a Shimadzu UFLC system (Kyoto, Japan) was used to identify the major constituents in the bioactive fractions (10–17.5 min) of AGE in both positive and negative ion modes. The mass parameters were listed in Table S1. Meanwhile, five major constituents were isolated by using a preparative HPLC (Waters, USA). The AGE sample (10 mL, 10 mg/mL) was continuously injected and separated on a Acchrom-C18 column (50 × 450 mm, 7 μm), accompanied by 80% of water-0.1% formic acid and 20% of acetonitrile: methanol (4:1) with the flow rate of 65 mL/min. Five isolated constituents in AGE were enriched and dried in vacuo separately, the solid of each constituent was collected for structural characterization and SARS-CoV-2 3CLpro inhibition assay.
2.5. Identification of the covalent binding sites of three flavonoids on SARS-CoV-2 3CLpro
To identify the covalently modified sites for three flavonoids on SARS-CoV-2 3CLpro, the peptides of target enzyme co-incubated with or without each tested flavonoid were analyzed by using a nanoLC-MS/MS system [17], [18], [19], [20]. Firstly, the SARS-CoV-2 3CLpro (147 μg, total content) was co-incubated with inhibitors (400 μM, final concentration) at 37 °C overnight. After that, the urea (6 M, final concentration) was added into the mixtures at 75 °C to denature SARS-CoV-2-3CLpro. In order to complete the alkylation of protein, the modified-SARS-CoV-2-3CLpro were separately co-incubated with DTT (1 mM, final concentration) for 10 min at 95 °C, and then treated with IAA (3 mM, final concentration) at 30 °C for 30 min in the dark. The protein precipitations were collected after added ice-cold acetonitrile and centrifugated at 16000 g for 10 min, which further dried in a termovap sample concentrator (ATR, AutoVap S60, USA). After then, the enriched proteins were redissolved in NH4HCO3 solution (pH 8.0, 50 mM), digested by chymotrypsin and trypsin (the content of protein: SARS-CoV-2 3CLpro ratios were 1:40), respectively. The produced peptides were desalted on a MonoSpin C18 column (GL Sciences Inc.), the eluents were dried in vacuo and resolved in 0.1% formic acid for analyses.
The samples (contained 0.5–1 μg peptides) were injected into nanoLC system (EASY-nLC 1200, Thermo Fisher Scientific, USA), separated at a self-packed analytical C18 column (20 μm × 360 μm × 200 mm, 3 μm) with the flow-rate of 0.3 μL/min. Water containing 0.1% formic acid (A) and 80% acetonitrile-20% water (B) were used as the mobile phases, the elution conditions were as follow: 0–1 min, 1%–6% B; 1–47 min, 6%–35% B; 47–54 min, 35%–37% B; 54–56 min, 37%–95% B; 56–65 min, 95% B. The full MS data were recorded by using the data-dependent mode on the Hybrid Quadrupole-Orbitrap mass spectrometer (Q Exactive™ HF-X, Thermo Fisher Scientific, USA) from m/z 300–m/z 1800, with the resolution of 60,000 (AGC target 3e6, maximum IT 50 ms). The analytes were fragmented in the HCD (Higher Energy Collision Dissociation) mode with resolution of 15,000 at a 28% normalized collision energy (AGC target 1e5, maximum IT 30 ms).
2.6. Inactivation kinetic analysis for three flavonoids on SARS-CoV-2 3CLpro
The inactivation kinetics for three flavonoids were investigated as the reported procedure [21]. Firstly, two incubation mixture groups (group A and group B) were prepared for use. Group A (90 μL) were comprised of Dabcyl-KNSTLQSGLRKE-Edans (20 μM, final concentration) and PBS (pH 7.4, 100 mM, 1 mM EDTA). Group B (100 μL) contained flavonoid and SARS-CoV-2 3CLpro that were co-incubated at 37 °C in tubes filled with PBS (pH 7.4, 100 mM, 1 mM EDTA). Then the mixtures (10 μL) in group B were transferred to group A at different preincubation time to initiate the hydrolytic reaction. After incubation for another 20 min at 37 °C, the hydrolytic reaction was quenched by adding ice-cold acetonitrile (100 μL). The natural logarithm of the residual SARS-CoV-2 3CLpro activity was plotted against the preincubation time.
2.7. Covalent docking and molecular dynamics simulations
The covalent docking was carried out through the covalent docking module of MOE (Molecular Operating Environment 2019.01, Chemical Computing Group Inc., Montreal, Canada). Firstly, the crystal structure of SARS-CoV-2 3CLpro (PDB Code: 6XHU, [22]) was download for preliminary treatment by using the QuickPrep module, including adding hydrogens and partial charges, optimizing the hydrogen bond network and minimizing energies. Then, the orthoquinone form of myricetin (QFM) was constructed and minimized energy in the Builder panel. Next, the covalent reaction formula for QFM and cysteine residuals was generated by MarvinSketch and imported to MOE for covalent docking module. Finally, with the help of GBVI/WSA dG, these generated conformations of the rigid receptor were refined and estimated the binding scores [23]. The pose with the lowest S score was selected as the initial conformation of QFM-3CLpro complexes.
To perform the molecular dynamics (MD) simulations for QFM-3CLpro complexes, it was necessary to get the force field of non-standard amino acid (Cys-QFM), which was generated by AmberTools20 (AMBER 2020, University of California, San Francisco, USA). The detailed procedures were as follows. At first, AM1-BCC charges of the Cys-QFM complex were calculated [24]. Then, atoms, bonds, angles, and dihedral parameters of Cys-QFM complex were established via atom types of amber99sb-ildn force field and checked by parmchk2 [25]. Next, coordinate and topology files of Cys-QFM complex were created by leap program, and then translated to the GROMACS topology file via ACYPE [26], [27]. Finally, manually written residue topology parameter file (rtp file) as per the GROMACS topology file and manually written hydrogen database file (hdb file) were loaded into the libraries files of amber99sb-ildn force field.
Systems of QFM-3CLpro complexes were established for simulations. Prior to MD simulations, an energy minimization of 50,000 steps steepest descent was performed. To neutralize the charges of the protein-solvent system, SARS-CoV-2 3CLpro and QFM-3CLpro complexes were separately solvated with the TIP3P water model including sodium ions. Then, the system was equilibrated, including 100 ps for NVT heating to 310 K, and 100 ps for NPT. Finally, the system was subjected to 50 ns MD at 310 K (V-rescale thermostat) under a pressure of 1 bar (Parrinello–Rahman barostat). To analyze the interactions of QFM-3CLpro complexes, we clustered the equilibrium conformations [28], and the largest center structure was selected to study the interactions by creating its stereoscopic picture via Discovery Studio Visualizer (BIOVIA Discovery Studio 2019, Dassault Systèmes, San Diego, USA).
2.8. Statistical analysis
IC50 and K I values were evaluated by nonlinear regression using Graph Pad Prism 7.0 software (GraphPad Software, Inc., La Jolla, USA). The nanoLC-MS/MS data was searched by Protein Discovery 2.4 (Thermo Scientific) by using the Sequest HT algorithm for peptide identification.
3. Results
3.1. Inhibition of SARS-CoV-2 3CLpro by herbal products
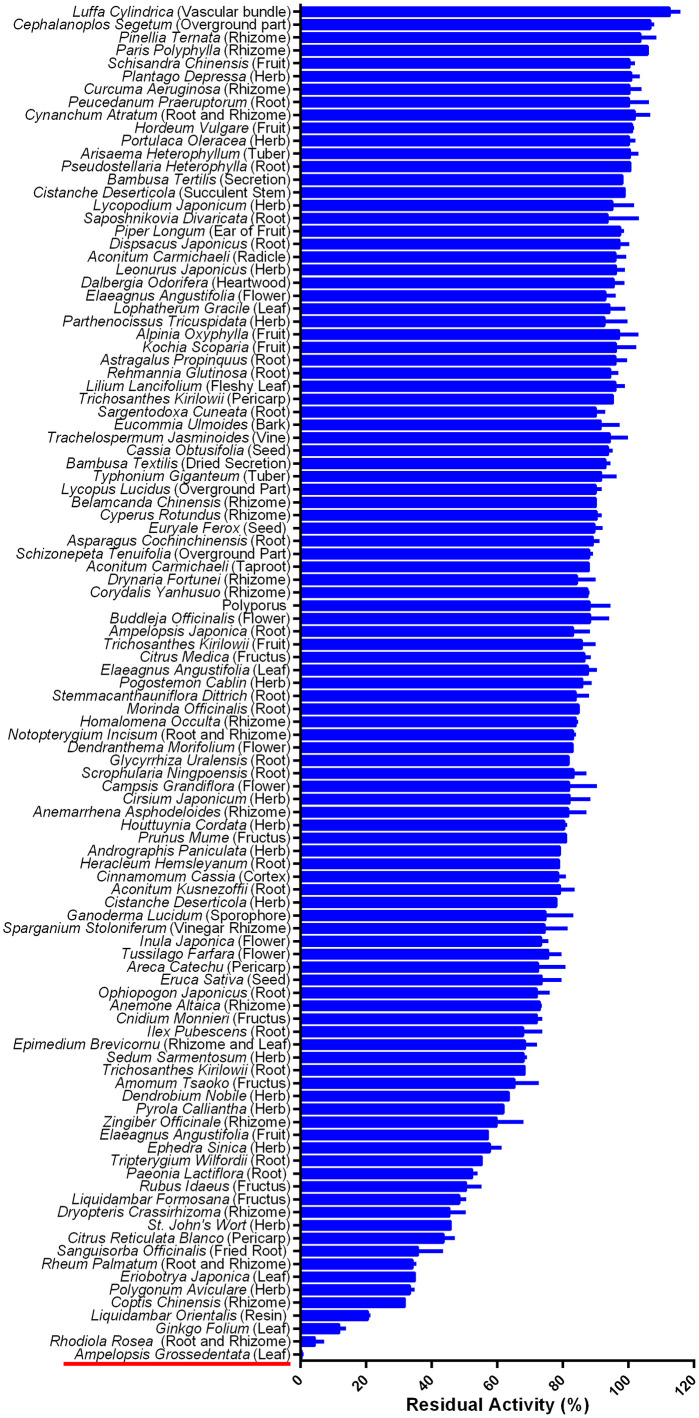
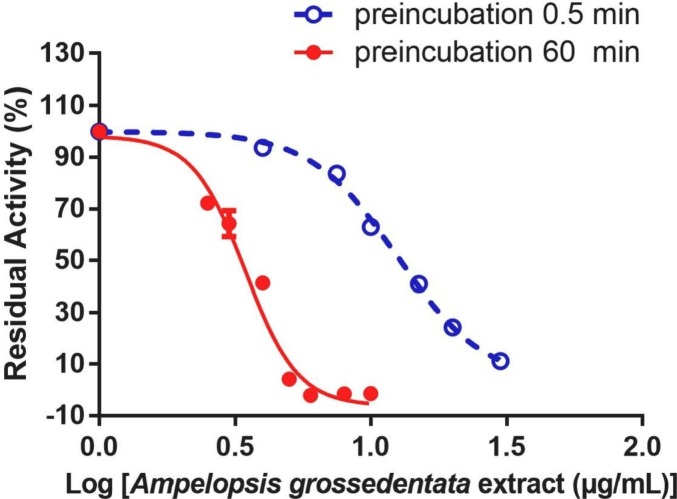
Firstly, the inhibitory potentials of 105 herbal extracts (100 μg/mL, final concentration) on SARS-CoV-2 3CLpro were assayed by using Dabcyl-KNSTLQSGLRKE-Edans as the fluorescent substrate. From the preliminary screening, AGE exhibited the most potent SARS-CoV-2 3CLpro inhibition activity (Fig. 1 ). The residual activity of SARS-CoV-2 3CLpro in the presence of AGE (100 μg/mL, final concentration) was 0.26%. As depicted in Fig. 2 , AGE could dose-dependently inhibit SARS-CoV-2 3CLpro-catalyzed Dabcyl-KNSTLQSGLRKE-Edans cleavage reaction, with IC50 value of 3.44 μg/mL. Meanwhile, time-dependent inhibition assays showed that AGE time-dependently inhibited SARS-CoV-2 3CLpro, with an obvious shift in apparent IC50 value (the ratio of IC50 = 3.63) when AGE was pre-incubated with SARS-CoV-2 3CLpro at various preincubation times. These findings demonstrate that AGE strongly inhibits SARS-CoV-2 3CLpro in dose- and time- dependent manners, implying that some natural constituents in AGE may covalently bind with SARS-CoV-2 3CLpro.
Fig. 1.
The inhibitory effects of 105 herbal products (100 μg/mL, final concentration) against SARS-CoV-2 3CLpro.
Fig. 2.
The dose-inhibition curves of 0.5 min and 60 min of AGE against SARS-CoV-2 3CLpro.
3.2. Identification of the naturally occurring SARS-CoV-2 3CLpro inhibitors in AGE
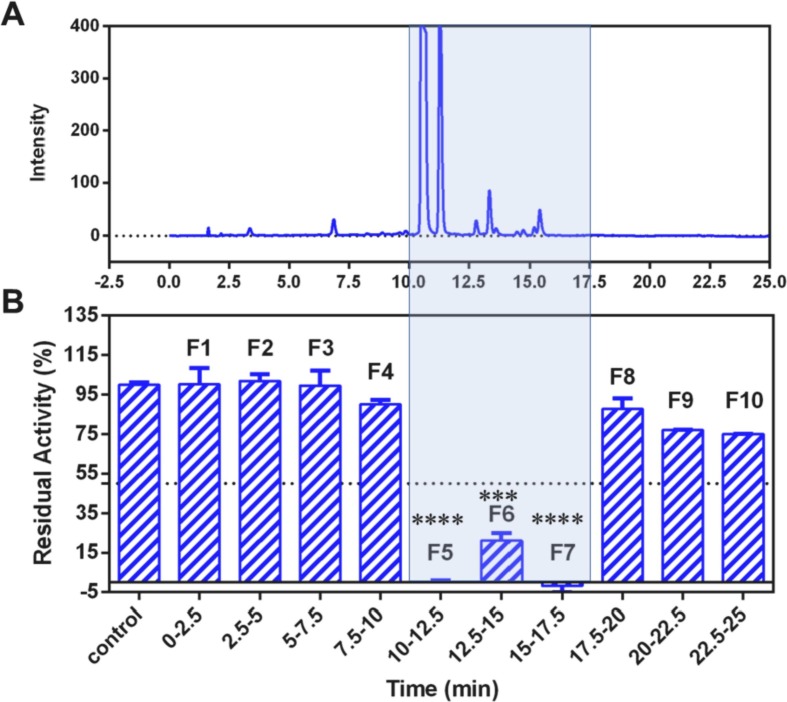
Then, a practical strategy via integrating bioactivity-guided fractionation and purification, as well as inhibition assay, was used to identify the key constituents in AGE. From the results exhibited in Fig. 3 , within ten fractions, F5, F6 and F7 possessed strong inhibitory properties against SARS-CoV-2 3CLpro. After then, five major peaks of the bioactive fractions (10–17.5 min) were isolated, while their structures and effects of inhibiting SARS-CoV-2-3CLpro were characterized. The MS1 and MS2 spectra of these natural compounds were shown in Table 1 , Figs. S6–S10, while their 1H and 13C NMR spectra were presented in Figs. S19–S23. These spectra data clearly suggested that five major constituents in the bioactive fractions of AGE (10–17.5 min) were five flavonoids, including dihydromyricetin, isodihydromyricetin, myricitrin, taxifolin and myricetin.
Fig. 3.
Fingerprinting analysis of AGE by LC-UV in 290 nm (A), and the SARS-CoV-2 3CLpro inhibition profiles of the LC fractions collected at 2.5 min intervals (B). (***p < 0.001, ****p < 0.0001, compared with the control group).
Table 1.
Identification and characterization of five major constitutes in the bioactive fractions of AGE by LC-PDA-TOF-MS/MS.
| No. | tR (min) | λmax (nm) | Ionization | m/z | Formula | Fragment ions | Identification |
|---|---|---|---|---|---|---|---|
| 1 | 10.611 | 290 | [M-H]- | 319.0469 | C15H12O8 | 319.0469,301.0354, 283.0262, 257.0461,215.0352,193.0143,175.0035, 125.0245 | Dihydromyricetin |
| 2 | 11.413 | 293 | [M-H]- | 319.0462 | C15H12O8 | 319.0462,301.0370, 257.0464, 215.0358, 193.0149, 175.0047, 125.0252 | Isodihydromyricetin |
| 3 | 12.894 | 352 | [M-H]- | 463.0902 | C21H20O12 | 463.0902, 317.0330, 316.0245, 217.0263, 270.0187 | Myricitrin |
| 4 | 13.464 | 289 | [M-H]- | 303.0515 | C15H12O7 | 303.05-15, 275.0565, 241.0517, 217.0520, 125.0251 | Taxifolin |
| 5 | 15.521 | 370 | [M-H]- | 317.0310 | C15H10O8 | 317.0310, 289.0360, 271.0260, 178.9990 | Myricetin |
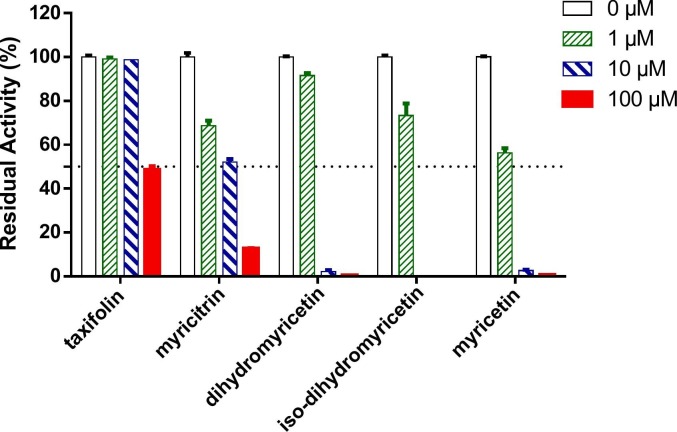
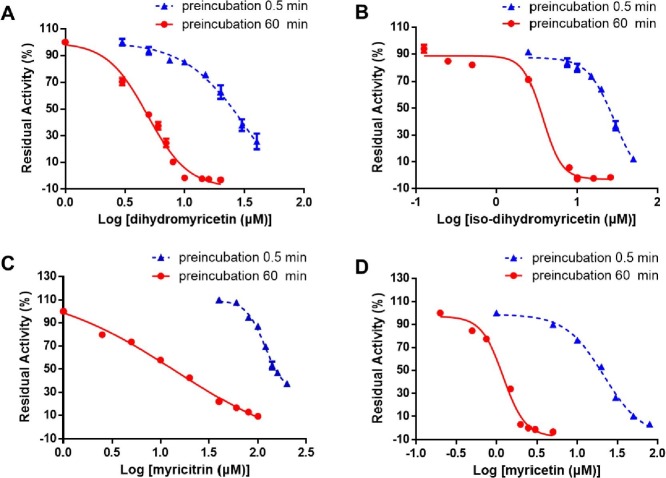
After then, the inhibition potentials of five isolated flavonoids against SARS-CoV-2 3CLpro were assayed by using three different doses. As depicted in Fig. 4 , the results presented that dihydromyricetin, isodihydromyricetin, as well as myricetin showed strong inhibition effects against SARS-CoV-2-3CLpro (IC50 < 5 μM). Meanwhile, taxifolin and myricitrin moderately inhibited SARS-CoV-2 3CLpro, with the IC50 values ranging from 10 μM to 100 μM. The dose-response curves of five flavonoids in AGE bioactive fractions (10–17.5 min) against SARS-CoV-2 3CLpro were also plotted by using increasing concentrations of each flavonoid (Fig. 5 , Fig. S12). As listed in Table 2 , dihydromyricetin, isodihydromyricetin and myricetin could strongly inhibit SARS-CoV-2 3CLpro, with IC50 values of 4.91 μM, 3.73 μM and 1.21 μM after 60-min preincubation, respectively. Notably, the inhibitory activity of myricetin was superior to the reported positive inhibitor ebselen (Fig. S11, a newly reported covalent inhibitor of SARS-CoV-2 3CLpro, IC50 = 2.62 μM).
Fig. 4.
Inhibitory effects of five major constituents in the bioactive fractions from AGE against SARS-CoV-2 3CLpro.
Fig. 5.
Dose- and time- dependent inhibition curves of dihydromyricetin (A), isodihydromyricetin (B), myricitrin (C) and myricetin (D) against SARS-CoV-2 3CLpro.
Table 2.
The inhibition parameters of the bioactive constituents in AGE against SARS-CoV-2 3CLpro.
| No. | Compound | Structure | IC50 (μM) |
Ratio | KI (μM) | Kinact (min−1) | |
|---|---|---|---|---|---|---|---|
| 0.5 (min) | 60 (min) | ||||||
| 1 | Dihydromyricetin | 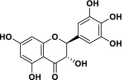 |
34.61 | 4.91 | 7.05 | 67.35 | 0.064 |
| 2 | Isodihydromyricetin | 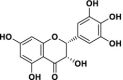 |
29.04 | 3.73 | 7.78 | 62.43 | 0.058 |
| 3 | Myricitrin | 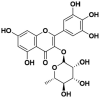 |
118.10 | 14.22 | 8.32 | – | – |
| 4 | Taxifolin | 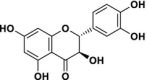 |
>200 | 72.72 | >2.75 | – | – |
| 5 | Myricetin | 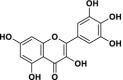 |
21.44 | 1.21 | 17.72 | 6.33 | 0.013 |
| 6 | Ebselena | 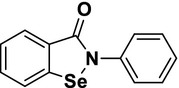 |
3.11 | 2.62 | 1.18 | – | – |
A known positive covalent inhibitor for SARS-CoV-2 3CLpro.
Meanwhile, time-dependent inhibition assessments for these five flavonoids were also carried out. The results clearly demonstrated that the inhibition potency of dihydromyricetin, isodihydromyricetin, myricitrin and myricetin against SARS-CoV-2 3CLpro would be enhanced with the pre-incubation time, with the IC50 ratios to be 7.05-fold, 7.78-fold, 8.32-fold, and 17.72-fold, respectively (Table 2, Fig. 5). These findings suggest that dihydromyricetin, isodihydromyricetin, myricitrin and myricetin are the key bioactive constituents in AGE that can dose- and time- dependently inhibit SARS-CoV-2 3CLpro.
3.3. Identification of the covalent binding sites of three flavonoids on SARS-CoV-2 3CLpro
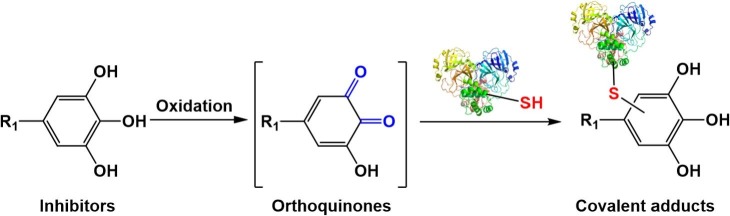
Next, the covalent binding sites of dihydromyricetin, isodihydromyricetin and myricetin on SARS-CoV-2 3CLpro were identified by using mass spectrometry. From the view of chemical structures of these naturally occurring flavonoids, all these compounds bear a catecholic group at the B-ring, which can be easily oxidized to form orthoquinones that can covalently bind on the biothiols or the cysteines in target proteins (Fig. 6 ) [28], [29]. In this case, the generated MS/MS spectra were analyzed by searching covalent modifications on cysteines of SARS-CoV-2 3CLpro, with the molecular mass increments of 316.24 Da (myricetin) and 318.25 Da (dihydromyricetin or isodihydromyricetin).
Fig. 6.
The proposed scheme of the newly identified flavonoid-type inhibitors covalently bind on the biothiols of SARS-CoV-2 3CLpro.
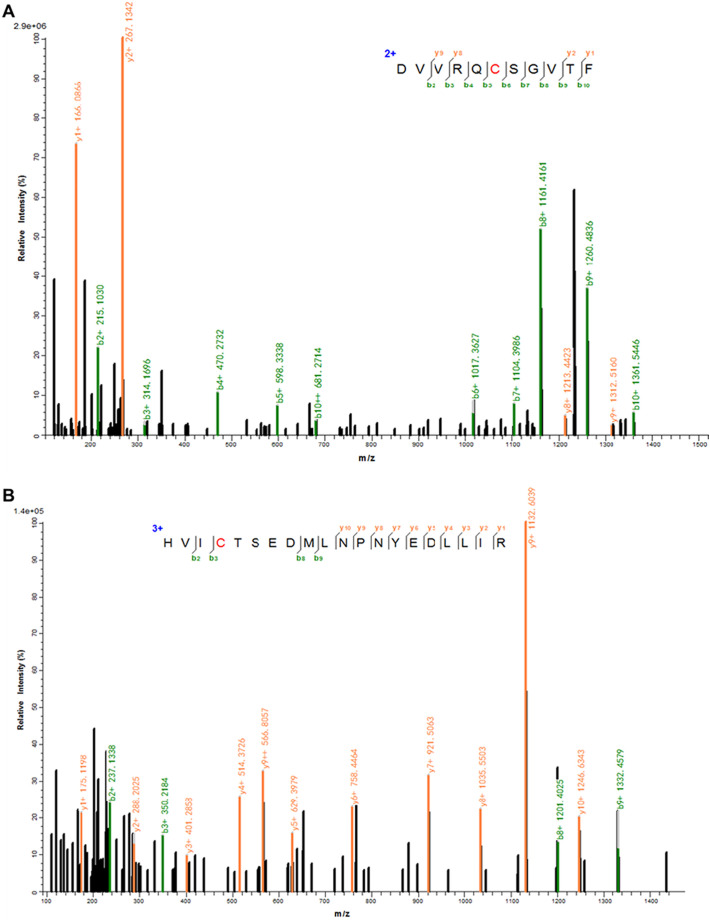
As shown in Table 3 , following co-incubation of SARS-CoV-2 3CLpro with each tested flavonoid, several cysteine residues in the peptides of SARS-CoV-2 3CLpro could be modified by the orthoquinone forms of these three flavonoids. From Fig. 7 , Fig. S14, Fig. S15, all tested flavonoids (myricetin, dihydromyricetin and isodihydromyricetin) could covalently modify Cys300, a key residue located at domain III (residuals 198-303) of SARS-CoV-2 3CLpro (Fig. S13). Several reports state that domain III (especially 290E-V303) functions as a crucial part to maintain the dimer conformation of active 3CLpro, mutation or modification of the key residuals (such as Gln290, Arg298 and Gln299) would result in the instability or inactivation of this key enzyme [[30], [31], [32], [33]]. Thus, it was easily conceivable that the surrounding micro-environment or the pivotal interactions for the formation of the dimer of active SARS-CoV-2 3CLpro might be changed or destroyed via modification of Cys300 by these naturally occurring flavonoids.
Table 3.
Identification of the covalent binding sites for three flavonoids on SARS-CoV-2 3CLpro by nanoLC-MS/MS.
| Inhibitor | Peptide | Modifications | Charge | m/z (Da) | MH+ (Da) | MH+ (Da) (Theoretical) |
Mass accuracy ΔMr (ppm) |
tR(min) |
|---|---|---|---|---|---|---|---|---|
| Dihydromyricetin | 295DVVRQC*SGVTF305 | Cys300 | 2 | 764.8166 | 1528.6260 | 1528.6260 | −0.04 | 41.157 |
| Isodihydromyricetin | 295DVVRQC*SGVTF305 | Cys300 | 2 | 764.8170 | 1528.6267 | 1528.6260 | 0.43 | 41.743 |
| Myricetin | 41HVIC*TSEDMLNPNYEDLLIR60 | Cys44 | 3 | 897.7310 | 2691.1785 | 2691.1587 | 7.36 | 48.006 |
| 295DVVRQC*SGVTF305 | Cys300 | 2 | 763.8091 | 1526.6108 | 1526.6102 | 0.46 | 42.820 |
*The amino acids modified by inhibitors.
Fig. 7.
The MS2 spectra of the peptide DVVRQCSGVTF (A) and HVICTSEDMLNPNYEDLLIR (B) covalently modified by myricetin.
In addition to Cys300, myricetin can also covalently bind on Cys44, which is near the catalytic site of SARS-CoV-2 3CLpro (Fig. 7, Fig. S13). Recently study has found that Cys44 is a hyper-reactive cysteine with higher nucleophilicity than Cys145, which is recognized as a promising binding site for designing and developing covalent inhibitors of this key enzyme [34]. The covalent binding of myricetin on Cys44 might block 3CLpro-catalyzed peptide-cleavage reactions. These findings suggest that myricetin and its analogous in AGE can covalently bind on some key cysteines of SARS-CoV-2 3CLpro, while myricetin can concurrently modify both Cys300 and Cys44. Meanwhile, these findings can partially explain the potent inhibition efficacy of myricetin, in comparison with its analogous (such as dihydromyricetin or isodihydromyricetin).
3.4. Inactivation kinetics of three flavonoids on SARS-CoV-2 3CLpro
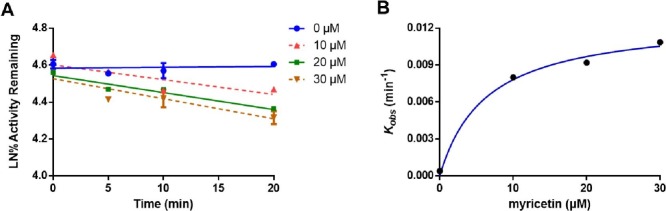
The inactivation kinetics for three flavonoids were further investigated to evaluate the inactivation potency of these naturally occurring SARS-CoV-2 3CLpro inhibitors. To this end, the inactivation kinetic curves were plotted by using various inhibitor concentrations with increasing pre-incubation times. As shown in Fig. S16 and Fig. 8 , dihydromyricetin, isodihydromyricetin and myricetin could inactivate SARS-CoV-2 3CLpro activity via dose- and time- dependent manners, their K I values were determined as 67.35 μM, 62.43 μM and 6.33 μM, respectively, while K inact values were calculated as 0.064 min−1, 0.058 min−1 and 0.013 min−1, respectively. These results suggest that three flavonoids are naturally occurring time-dependent SARS-CoV-2 3CLpro inhibitors, while myricetin exhibited the strongest inactivation potency, which encourages us to further investigate the interactional modes of this agent with SARS-CoV-2 3CLpro.
Fig. 8.
Time- and concentration-dependent inhibition of myricetin (A) on SARS-CoV-2 3CLpro. The hyperbolic plot of kobs of SARS-CoV-2 3CLprovs. myricetin (B) concentrations.
3.5. Covalent docking and molecular dynamics simulations
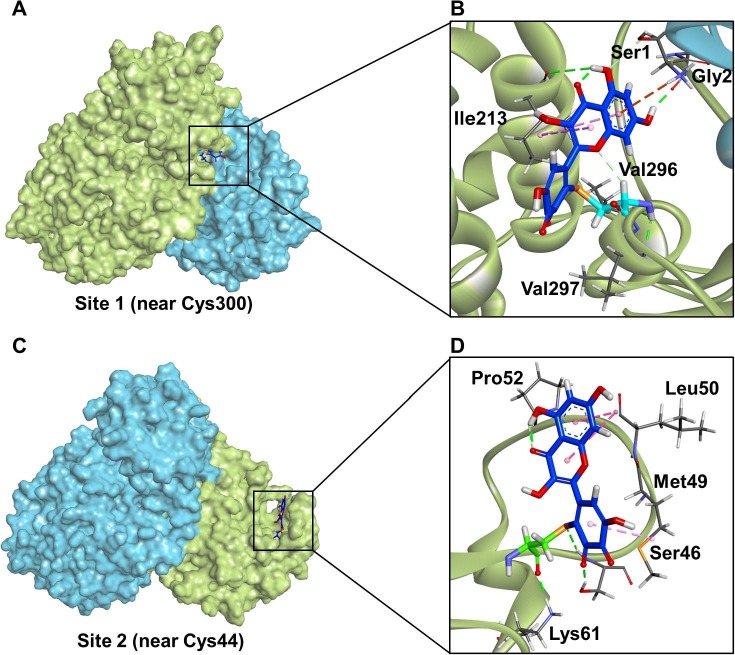
Finally, covalent docking simulations and molecular dynamics simulations were carefully conducted for QFM that covalently bound in site 1 (near Cys300) and site 2 (near Cys44) of SARS-CoV-2 3CLpro to explore the key interactions between this agent and the target enzyme. As shown in Fig. 9B and Fig. S17, when QFM covalently bound to the sulfur atom of Cys300, this agent mainly interacted with the surrounding amino acid residuals (including Val296, Val297, Gly2 and Ile213) by hydrogen bonding. As for site 2 (near Cys44), it was observed from Fig. 9D and Fig. S18 that QFM could form an adduct via covalently bound on Cys44, while this agent interacted with some residuals in site 2 through hydrogen bonding (via forming a carbon hydrogen bond and conventional hydrogen bonds) and hydrophobic bonding (amide-Pi stacked & Pi-alkyl). These results manifest that the orthoquinone form of myricetin can covalently bind in both site 1 (near Cys300) and site 2 (near Cys44) on SARS-CoV-2 3CLpro, while the covalent modifications at these two sites may lead SARS-CoV-2 3CLpro to its inactive forms.
Fig. 9.
The stereo view of the crystal structure SARS-CoV-2 3CLpro (PDB Code: 6XHU) that was covalently bound on the orthoquinone form of myricetin at Cys300 (A) or Cys44 (C). The detailed interactions between SARS-CoV-2 3CLpro and the orthoquinone form at Cys300 (B) or Cys44 (D).
4. Discussion
Currently, the COVID-19 global pandemic has already brought strong impact on human health, economic growth and social stability. To efficiently fight against COVID-19, the scientists have made great endeavors to develop novel therapeutics via targeting the validated therapeutic targets. Among all validated therapeutic targets, 3CLpro has been validated as a key target for treating SARS-CoV-2 and other coronavirus, owing to the exceptionally important role of 3CLpro during the viral life cycle [35]. Over the past one year, a variety of SARS-CoV-2 3CLpro inhibitors have been found, but only several compounds are identified as the covalent inhibitors of this vital enzyme via forming chemically stable and irreversible bonds [13]. Given that the covalent inhibitors always display good inhibition potency in living systems, the covalent inhibitors of 3CLpro are considered as a good choice to block the SARS-CoV-2 multiplication via inactivating the proteolytic activity of 3CLpro. Thus, it is urgent and highly desirable to find more efficacious SARS-CoV-2 3CLpro covalent inhibitors with improved safety profiles, which may offer the promising lead compounds for developing novel anti-COVID-19 agents.
In these cases, a high-throughput screening campaign was conducted for discovering effective SARS-CoV-2 3CLpro covalent inhibitors from herbal products. Among all tested herbal products, AGE demonstrated the most potent SARS-CoV-2 3CLpro inhibition activity, while this herbal extract inhibited this key enzyme in time- and dose- dependent manners. This finding intrigued us to reveal the key bioactive constituents in AGE that could covalently bind to SARS-CoV-2 3CLpro. In Southeast China, Ampelopsis grossedentata is a flavonoid-rich (w/w > 40%) medicinal herb, whose dried leaves and stems are popularly used as healthy tea to prevent chronic disorders by reducing hypertension, regulating plasma lipids and blood glucose [36], [37], [38], [39]. Herein, we identified that three abundant flavonoids (dihydromyricetin, isodihydromyricetin and myricetin) in AGE could strongly inhibit SARS-CoV-2 3CLpro by covalently binding with two key cysteines on this target enzyme. Previous reports have reported that the flavonoids (myricetin and dihydromyricetin) in AGE exhibit multiple beneficial effects including anti-inflammatory, anti-coagulative, as well as pulmonary fibrosis inhibition activities [34], [40], [41], [42]. Thus, Ampelopsis grossedentata could be used as a healthy plant-based supplement for treating COVID-19, which might relieve the COVID-19-related symptoms in both the respiratory tract system and alimentary system. Moreover, this edible herb can be co-administrated with other antiviral herbs (such as Scutellaria baicalensis [43], Glycyrrhiza uralensis [44] and Ephedra sinica [45]) that contain other anti-SARS-CoV-2 phytochemicals with diverse inhibition mechanisms or various binding targets, in which the combination use may bring additive or synergistic antiviral effects for the treatment of COVID-19.
Notably, three newly identified SARS-CoV-2 3CLpro covalent inhibitors (dihydromyricetin, isodihydromyricetin and myricetin) in AGE were also found with the inhibition against SARS-CoV helicase [46] and SARS-CoV3CLpro [47], as well as high affinities with the ACE2 receptor [40], suggesting that these agents hold sufficient potentials to develop as the broad-spectrum anti-coronavirus agents via targeting multiple key druggable targets. However, the poor cell-permeability and poor metabolic stability of these natural flavonoids strongly hampered the wide applications of these flavonoids in clinical settings [42]. Therefore, it is necessary to used more practical approaches (such as structural optimizations or drug delivery technologies) to develop more efficacious agents for combating COVID-19 pandemic. Considering that the naturally occurring flavonoids could be extensively metabolized by UDP-glucuronosyltransferases (UGTs) or other conjugative enzymes in humans [48], AGE could be co-administrated with other herbal medicines containing strong inhibitors against human UGTs, such as Fructus Psoraleae [49], or UGTs inhibitors like amentoflavone [50] and licochalcone A [51], which might improve the in vivo therapeutic effects of AGE against COVID-19. Furthermore, this study also offered several leading compounds and a key warhead for designing and developing more efficacious SARS-CoV-2 3CLpro covalent inhibitors. Among all tested flavonoids isolated from AGE, myricetin was identified as the most potent SARS-CoV-2 3CLpro covalent inhibitor, owing to this agent could concurrently label both Cys300 and Cys44 of this key enzyme. Our findings suggested that the ortho-trihydroxyl group in the B ring of these flavonoids was a crucial pharmacophore for covalently binding on 3CLpro, as well as SARS-CoV-2-3CLpro inhibitory activities. Meanwhile, the plane structures without glycosides were propitious to the process of covalent reactions. In future, this key warhead will be conducive to design a new generation of efficacious anti-COVID-19 medications, while these pyrogallol-containing compounds can be used as practical probes or tools to identify the covalent inhibitors for cysteine proteases. More importantly, compared with the cysteine residuals in SARS-CoV-2 3CLpro (such as Cys44 and Cys145), Cys300 is suggested as a more desired ligand-binding site for developing covalent inhibitors of SARS-CoV-2 3CLpro, owing to its unique location at the dimeric surface of SARS-CoV-2 3CLpro that is more liable to be covalently modified by small molecules. The mutation or molecule-modifications of the conserved Cys300 may destroy the dimerization of 3CLpro and further cause the active form of this enzyme into an inactive monomer, which provide new insight to design novel agents for combating fatal β-coronavirus, including SARS-CoV and SARS-CoV-2.
5. Conclusion
In summary, this study reported that both AGE and the major constituents or the flavonoid-rich fractions of this herbal extract could strongly inhibit SARS-CoV-2 3CLpro in dose- and time-dependent manner. Bioactivity-guided fractionation and purification revealed that three flavonoids in the bioactive fractions (10–17.5 min) of AGE, including dihydromyricetin, isodihydromyricetin and myricetin, were the key constituents in AGE contribute to SARS-CoV-2 3CLpro inactivation. Among the newly identified flavonoid-type 3CLpro inhibitors, myricetin displayed the most potent SARS-CoV-2 3CLpro inhibition activity, with the apparent IC50 value of 1.21 μM. Mass spectrometry-based peptide profiling demonstrated that dihydromyricetin and isodihydromyricetin could covalently bind on Cys300 of SARS-CoV-2 3CLpro, while myricetin acted as a dual-site (Cys300 and Cys44) covalent inhibitor against this key enzyme. Collectively, this study provides a framework example for deciphering and characterizing the key active ingredients in herbal extract responsible for SARS-CoV-2 3CLpro inactivation, while the newly identified SARS-CoV-2-3CLpro inhibitors in AGE and their key pharmacophores for covalently binding on 3CLpro offers new insights into the design and development of novel therapeutics for treating COVID-19 or other CoVs-related diseases.
CRediT authorship contribution statement
Xiong Yuan: Conceptualization, Methodology, Data Curation, Writing-Original Draft. Zhu Guang-Hao: Software, Methodology, Data curation. Zhang Ya-Ni: Methodology, Validation. Hu Qing: Methodology, Investigation. Wang Hao-Nan: Resources. Yu Hao-Nan: Methodology, Visualization, Data curation. Qin Xiao-Ya: Data curation. Guan Xiao-Qing: Software, Data curation. Xiang Yan-Wei: Supervision, Formal analysis. Tang Hui: Supervision, Conceptualization. Ge Guang-Bo: Supervision, Funding acquisition, Project administration, Writing- Reviewing and Editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank professor Guo-Qiang Lin and Dr. Ding-Ding Gao for the technical supports. This work was supported by the National Key Research and Development Program of China (2020YFC0845400), the NSF of China (81922070, 81973286, 81860614), Shanghai Science and Technology Innovation Action Plans (20S21901500 & 20S21900900) supported by Shanghai Science and Technology Committee, the Three-year Action Plan of Shanghai TCM Development (ZY-(2018-2020)-CCCX-5001), Shuguang Program (18SG40) & the Project on the Prevention and Treatment of COVID-19 with Chinese and Western Medicines supported by Shanghai Education Development Foundation and Shanghai Municipal Education Commission, Program of Shanghai Academic/Technology Research Leader (18XD1403600) and Program for Key Research and Development of Xinjiang Province (2017B03013).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2021.07.167.
Appendix A. Supplementary data
Supplementary material
References
- 1.Piret J., Boivin G. Pandemics throughout history. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.631736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma A., Ahmad Farouk I., Lal S.K. COVID-19: a review on the novel coronavirus disease evolution, transmission, detection, control and prevention. Viruses. 2021;13:202. doi: 10.3390/v13020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang P.H., Wang X.L. COVID-19: a new challenge for human beings. Cell. Mol. Immunol. 2020;17(5):555–557. doi: 10.1038/s41423-020-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian D.K., Liu Y.Z., Liang C.Y., Xin L., Xie X.L., Zhang D.Z., Wan M.G., Li H., Fu X.Q., Liu H., Cao W.Q. An update review of emerging small-molecule therapeutic options for COVID-19. Biomed. Pharmacother. 2021;137 doi: 10.1016/j.biopha.2021.111313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shamsi A., Mohammad T., Anwar S., Amani S., Khan M.S., Husain F.M., Rehman M.T., Islam A., Hassan M.I. Potential drug targets of SARS-CoV-2: from genomics to therapeutics. Int. J. Biol. Macromol. 2021;177:1–9. doi: 10.1016/j.ijbiomac.2021.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q.X., Kang C.B. Progress in developing inhibitors of SARS-CoV-2 3C-like protease. Microorganisms. 2020;8(8):1250. doi: 10.3390/microorganisms8081250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mengist H.M., Mekonnen D., Mohammed A., Shi R., Jin T. Potency, safety, and pharmacokinetic profiles of potential inhibitors targeting SARS-CoV-2 main protease. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.630500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abian O., Ortega-Alarcon D., Jimenez-Alesanco A., Ceballos-Laita L., Vega S., Reyburn H.T., Rizzuti B., Velazquez-Campoy A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int. J. Biol. Macromol. 2020;164:1693–1703. doi: 10.1016/j.ijbiomac.2020.07.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y., Liang C., Xin L., Ren X., Tian L., Ju X., Li H., Wang Y., Zhao Q., Liu H., Cao W., Xie X., Zhang D., Wang Y., Jian Y. The development of coronavirus 3C-like protease (3CL) inhibitors from 2010 to 2020. Eur. J. Med. Chem. 2020;206 doi: 10.1016/j.ejmech.2020.112711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jukic M., Janežic D., Bren U. Ensemble docking coupled to linear interaction energy calculations for identification of coronavirus main protease (3CL) non-covalent small-molecule inhibitors. Molecules. 2020;25(24):5808. doi: 10.3390/molecules25245808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caruso F., Singh M., Belli S., Berinato M., Rossi M. Interrelated mechanism by which the methide quinone celastrol, obtained from the roots of Tripterygium wilfordii, inhibits main protease 3CL pro of COVID-19 and acts as superoxide radical scavenger. Int. J. Mol. Sci. 2020;21(23):9266. doi: 10.3390/ijms21239266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y., Mandadapu S.R., Groutas W.C., Chang K.O. Potent inhibition of feline coronaviruses with peptidyl compounds targeting coronavirus 3C-like protease. Antivir. Res. 2013;97:161–168. doi: 10.1016/j.antiviral.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karges J., Kalaj M., Bembicky M., Cohen S.M. Re(I) tricarbonyl complexes as coordinate covalent inhibitors for the SARS-CoV-2 Main cysteine protease. Angew. Chem. Int. Ed. Engl. 2021 Feb 19 doi: 10.1002/anie.202016768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutanto F., Konstantinidou M., Dömling A. Covalent inhibitors: a rational approach to drug discovery. RSC. Med. Chem. 2020;11(8):876–884. doi: 10.1039/d0md00154f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L.L., Chen S., Gui C., Shen J., Shen X., Jiang H. Discovering severe acute respiratory syndrome coronavirus 3CL protease inhibitors: virtual screening, surface plasmon resonance, and fluorescence resonance energy transfer assays. J. Biomol. Screen. 2006;118(8):915–921. doi: 10.1177/1087057106293295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong Y., Zhu G.H., Wang H.N., Hu Q., Chen L.L., Guan X.Q., Li H.L., Chen H.Z., Tang H., Ge G.B. Discovery of naturally occurring inhibitors against SARS-CoV-2 3CL from Ginkgo biloba leaves via large-scale screening. Fitoterapia. 2021;152 doi: 10.1016/j.fitote.2021.104909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 18.Boberg M., Vrana M., Mehrotra A., Pearce R.E., Gaedigk A., Bhatt D.K., Leeder J.S., Prasad B. Age-dependent absolute abundance of hepatic carboxylesterases (CES1 and CES2) by LC-MS/MS proteomics: application to PBPK modeling of oseltamivir in vivo pharmacokinetics in infants. Drug Metab. Dispos. 2017;45(2):216–223. doi: 10.1124/dmd.116.072652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian Y., Weerapana E. A quantitative mass-spectrometry platform to monitor changes in cysteine reactivity. Methods Mol. Biol. 2017;1491:11–22. doi: 10.1007/978-1-4939-6439-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang C.M., Ku M.C., Chang C.K., Liang H.C., Wang T.F., Wu C.H., Chen S.H. Identification of endogenous site-specific covalent binding of catechol estrogens to serum proteins in human blood. Toxicol. Sci. 2015;148(2):433–442. doi: 10.1093/toxsci/kfv190. [DOI] [PubMed] [Google Scholar]
- 21.Harrelson J.P., Stamper B.D., Chapman J.D., Goodlett D.R., Nelson S.D. Covalent modification and time-dependent inhibition of human CYP2E1 by the meta-isomer of acetaminophen. Drug Metab. Dispos. 2012;408(8):1460–1465. doi: 10.1124/dmd.112.045492. [DOI] [PubMed] [Google Scholar]
- 22.Kneller D.W., Phillips G., Tan K., Joachimiak A., Coates L., Kovalevsky A., O'Neill H.M. Room temperature X-ray crystallography reveals oxidation and reactivity of cysteine residues in SARS-CoV-2 3CL Mpro: insights for enzyme mechanism and drug design. IUCrJ. 2020;7(6):1028–1035. doi: 10.1107/S2052252520012634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labute P. The generalized Born/Volume integral implicit solvent model: estimation of the free energy of hydration using London dispersion instead of atomic surface area. J. Comput. Chem. 2008;29(10):1693–1698. doi: 10.1002/jcc.20933. [DOI] [PubMed] [Google Scholar]
- 24.Jakalian A., Jack D.B., Bayly C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002;23(16):1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 25.Alviz-Amador A., Galindo-Murillo R., Pineda-Alemán R., Pérez-González H., Rodríguez-Cavallo E., Vivas-Reyes R., Méndez-Cuadro D. Development and benchmark to obtain AMBER parameters dataset for non-standard amino acids modified with 4-hydroxy-2-nonenal. Data. Brief. 2018;21:2581–2589. doi: 10.1016/j.dib.2018.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sousa da Silva A.W., Vranken W.F. ACPYPE - AnteChamber PYthon parser interfacE. BMC. Res. Notes. 2012;5:367. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 28.Pagoni A., Grabowiecka A., Tabor W., Mucha A., Vassiliou S., Berlicki L. Covalent inhibition of bacterial urease by bifunctional catechol-based phosphonates and phosphinates. J. Med. Chem. 2021;64(1) doi: 10.1021/acs.jmedchem.0c01143. [DOI] [PubMed] [Google Scholar]
- 29.Bittner S. When quinones meet amino acids: chemical, physical and biological consequences. Amino Acids. 2006;30(3):205–224. doi: 10.1007/s00726-005-0298-2. [DOI] [PubMed] [Google Scholar]
- 30.Li C.M., Teng X., Qi Y.F., Tang B., Shi H.L., Ma X.M., Lai L.H. Conformational flexibility of a short loop near the active site of the SARS-3CLpro is essential to maintain catalytic activity. Sci. Rep. 2016;6:20918. doi: 10.1038/srep20918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi J., Sivaraman J., Song J. Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease. J. Virol. 2008;829(9):4620–4629. doi: 10.1128/JVI.02680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goyal B., Goyal D. Targeting the dimerization of the Main protease of coronaviruses: a potential broad-Spectrum therapeutic strategy. ACS Comb. Sci. 2020;22(6):297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 33.Muramatsu T., Takemoto C., Kim Y.T., Wang H., Nishii W., Terada T., Shirouzu M., Yokoyama S. SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc. Natl. Acad. Sci. U. S. A. 2016;113(46):12997–13002. doi: 10.1073/pnas.1601327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verma N., Henderson J.A., Shen J. Proton-coupled conformational activation of SARS coronavirus Main proteases and opportunity for designing small-molecule broad-Spectrum targeted covalent inhibitors. J. Am. Chem. Soc. 2020;142(52):21883–21890. doi: 10.1021/jacs.0c10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carneiro R.C.V., Ye L.Y., Baek N., Teixeira G.H.A., O’Keefe S.F. Vine tea (Ampelopsis grossedentata): a review of chemical composition, functional properties, and potential food applications. J. Funct. Foods. 2021;76 doi: 10.1016/j.jff.2020.104317. [DOI] [Google Scholar]
- 37.Zhao J., Deng J.W., Chen Y.W., Li S.P. Advanced phytochemical analysis of herbal tea in China. J. Chromatography. A. 2013;1313:2–23. doi: 10.1016/j.chroma.2013.07.039. [DOI] [PubMed] [Google Scholar]
- 38.Xie K., He X., Chen K., Chen J., Sakao K., Hou D.X. Antioxidant properties of a traditional vine tea, Ampelopsis grossedentata. Antioxidants. 2019;8(8):295. doi: 10.3390/antiox8080295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang J., Lv Q., Yi F., Song Y., Le L., Jiang B., Xu L., Xiao P. Dietary supplementation of vine tea ameliorates glucose and lipid metabolic disorder via akt signaling pathway in diabetic rats. Molecules. 2019;24(10):1866. doi: 10.3390/molecules24101866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song X., Tan L., Wang M., Ren C., Guo C., Yang B., Ren Y., Cao Z., Li Y., Pei J. Myricetin: a review of the most recent research. Biomed. Pharmacother. 2021;134 doi: 10.1016/j.biopha.2020.111017. [DOI] [PubMed] [Google Scholar]
- 41.Chen S., Lv K., Sharda A., Deng J., Zeng W., Zhang C., Hu Q., Jin P., Yao G., Xu X., Ming Z., Fang C. Anti-thrombotic effects mediated by dihydromyricetin involve both platelet inhibition and endothelial protection. Pharmacol. Res. 2021;167 doi: 10.1016/j.phrs.2021.105540. [DOI] [PubMed] [Google Scholar]
- 42.Liskova A., Samec M., Koklesova L., Samuel S.M., Zhai K., Al-Ishaq R.K., Abotaleb M., Nosal V., Kajo K., Ashrafizadeh M., Zarrabi A., Brockmueller A., Shakibaei M., Sabaka P., Mozos I., Ullrich D., Prosecky R., La Rocca G., Caprnda M., Büsselberg D., Rodrigo L., Kruzliak P., Kubatka P. Flavonoids against the SARS-CoV-2 induced inflammatory storm. Biomed. Pharmacother. 2021;138 doi: 10.1016/j.biopha.2021.111430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su H.X., Yao S., Zhao W.F., Li M.J., Liu J., Shang W.J., Xie H., Ke C.Q., Hu H.C., Gao M.N., Yu K.Q., Liu H., Shen J.S., Tang W., Zhang L.K., Xiao G.F., Ni L., Wang D.W., Zuo J.P., Jiang H.L., Bai F., Wu Y., Ye Y., Xu Y.C. Anti-SARS-CoV-2 activities in vitro of shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020;41(9):1167–1177. doi: 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z., Yang L. Chinese herbal medicine: fighting SARS-CoV-2 infection on all fronts. J. Ethnopharmacol. 2021;270 doi: 10.1016/j.jep.2021.113869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lv Y., Wang S., Liang P., Wang Y., Zhang X., Jia Q., Fu J., Han S., He L. Screening and evaluation of anti-SARS-CoV-2 components from Ephedra sinica by ACE2/CMC-HPLC-IT-TOF-MS approach. Anal. Bioanal. Chem. 2021:1–10. doi: 10.1007/s00216-021-03233-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu M.S., Lee J., Lee J.M., Kim Y., Chin Y.W., Jee J.G., Keum Y.S., Jeong Y.J. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 2012;22:4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen T.T., Woo H.J., Kang H.K., Nguyen V.D., Kim Y.M., Kim D.W., Ahn S.A., Xia Y., Kim D. Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnol. Lett. 2012;34(5):831–838. doi: 10.1007/s10529-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fraga C.G., Croft K.D., Kennedy D.O., Tomás-Barberán F.A. The effects of polyphenols and other bioactives on human health. Food Funct. 2019, 2019;10(2):514–528. doi: 10.1039/c8fo01997e. [DOI] [PubMed] [Google Scholar]
- 49.Wang X.X., Lv X., Li S.Y., Hou J., Ning J., Wang J.Y., Cao Y.F., Ge G.B., Guo B., Yang L. Identification and characterization of naturally occurring inhibitors against UDP-glucuronosyltransferase 1A1 in fructus psoraleae (Bu-gu-zhi) Toxicol. Appl. Pharmacol. 2015;289(1):70–80. doi: 10.1016/j.taap.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Lv X., Zhang J.B., Wang X.X., Hu W.Z., Shi Y.S., Liu S.W., Hao D.C., Zhang W.D., Ge G.B., Hou J., Yang L. Amentoflavone is a potent broad-spectrum inhibitor of human UDP-glucuronosyltransferases. Chem. Biol. Interact. 2018;284:48–55. doi: 10.1016/j.cbi.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 51.Xin H., Qi X.Y., Wu J.J., Wang X.X., Li Y., Hong J.Y., He W., Xu W., Ge G.B., Yang L. Assessment of the inhibition potential of licochalcone a against human UDP-glucuronosyltransferases. Food Chem. Toxicol. 2016;90:112–122. doi: 10.1016/j.fct.2016.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material