Abstract
Mutations are the raw material on which evolution acts, and knowledge of their frequency and genomic distribution is crucial for understanding how evolution operates at both long and short timescales. At present, the rate and spectrum of de novo mutations have been directly characterized in relatively few lineages. Our study provides the first direct mutation-rate estimate for a strepsirrhine (i.e., the lemurs and lorises), which comprises nearly half of the primate clade. Using high-coverage linked-read sequencing for a focal quartet of gray mouse lemurs (Microcebus murinus), we estimated the mutation rate to be among the highest calculated for a mammal at 1.52 × 10–8 (95% credible interval: 1.28 × 10−8–1.78 × 10−8) mutations/site/generation. Further, we found an unexpectedly low count of paternal mutations, and only a modest overrepresentation of mutations at CpG sites. Despite the surprising nature of these results, we found both the rate and spectrum to be robust to the manipulation of a wide range of computational filtering criteria. We also sequenced a technical replicate to estimate a false-negative and false-positive rate for our data and show that any point estimate of a de novo mutation rate should be considered with a large degree of uncertainty. For validation, we conducted an independent analysis of context-dependent substitution types for gray mouse lemur and five additional primate species for which de novo mutation rates have also been estimated. These comparisons revealed general consistency of the mutation spectrum between the pedigree-based and the substitution-rate analyses for all species compared.
Subject terms: Evolution, Molecular evolution
Introduction
Spontaneous germline mutations are errors that occur as DNA is transmitted from parent to offspring in sexually reproducing organisms. The accrual of these errors, often referred to as de novo mutations, provides not only the raw material for evolution but can also serve as a means for measuring evolutionary time along phylogenies (Kimura and Ohta 1971; Langley and Fitch 1974; Zuckerkandl and Pauling 1965). The rate at which these mutations are introduced into genomes is thus a crucial metric of evolution at the genomic level, as well as a measure of fundamental biological processes (Kondrashov and Kondrashov 2010). By characterizing mutation-rate variation across the genome and between generations, we may be able to shed light on the impacts of biological processes such as sex and parental age biases. Ultimately, by quantifying the variation in de novo mutation rates across the tree of life, we can refine hypotheses regarding the relationship between mutation rates and life-history characteristics (Agarwal and Przeworski 2019; Fazalova and Nevado 2020; Garimella et al. 2020; Wang et al. 2020; Wu et al. 2020).
Approaches for estimating rates of genomic change in vertebrates generally fall into one of two categories: phylogenetic (indirect) versus pedigree-based (direct) estimation. While phylogenetic methods have been the standard for many years, recent developments in sequencing technology have made whole-genome sequencing widely accessible and pedigree-based approaches are now increasingly being used to estimate de novo rates for nonmodel species. By comparing the genomes of individuals with known genealogical relationships—typically, parent to offspring—investigators can count mutations as they appear in single-generation transmissions (Feng et al. 2017; Koch et al. 2019; Pfeifer 2017; Scally and Durbin 2012; Smeds et al. 2016; Thomas et al. 2018). Phylogenetic approaches, on the other hand, use external calibrations such as fossils or geological events to obtain substitution rates in units of absolute time (Drummond et al. 2006; Sanderson 2002; Thorne and Kishino 2002; Thorne et al. 1998). Phylogenetic studies work from the fundamental assumption that the rate at which substitutions accumulate between species at putatively neutral sites is equal to the de novo mutation rate (Kimura 1983). If this assumption holds, pedigree-based and phylogenetic methods should in principle produce equivalent estimates of the rate of evolution.
Phylogenetic methods for estimating rates of evolution are known to suffer from various sources of uncertainty, however, including violation of the molecular clock (Thorne et al. 1998), inaccuracies in external calibration points (Benton and Donoghue 2007), incomplete lineage sorting (Angelis and dos Reis 2015), and the difficulties of recovering multiple overlapping changes (i.e., “multiple hits”) at any given site (Felsenstein 1981). Although a number of solutions to these problems have been proposed (Heath et al. 2014; Ogilvie et al. 2017), some limitations such as sampling biases or an absence of fossils are difficult to overcome (Herrera and Davalos 2016; Magallon and Sanderson 2005; Near et al. 2005). Pedigree-based mutation-rate estimates are not affected by the same complications and can help characterize variation among different types of mutations (Harris and Pritchard 2017) or among different regions of the genome (Segurel et al. 2014). Previously, these estimates have relied on well-assembled genomes available only in model organisms (Jonsson et al. 2017; Scally and Durbin 2012; Uchimura et al. 2015; Venn et al. 2014), and have therefore been limited in taxonomic scope. For example, mutation-rate estimates within mammals are dominated by primates (Table 1). Fortunately, recent genome assembly strategies (Rhie et al. 2020) have enabled chromosome-level assemblies of nonmodel organisms, including mouse lemurs (Larsen et al. 2017), and pedigree-based mutation-rate estimation is now feasible for virtually any species, as long as related individuals with known pedigrees are available (Feng et al. 2017; Harland et al. 2017; Koch et al. 2019; Martin et al. 2018; Pfeifer 2017; Smeds et al. 2016).
Table 1.
Directly estimated mammalian de novo mutation rates.
| Species | Common name | Citation | Ratea |
|---|---|---|---|
| Homo sapiensb | Human | Jonsson et al. (2017) | 1.29 |
| Pan troglodytes | Chimpanzee | Venn et al. (2014) | 1.2 |
| Tatsumoto et al. (2017) | 1.48 | ||
| Besenbacher et al. (2019) | 1.26 | ||
| Gorilla gorilla | Gorilla | Besenbacher et al. (2019) | 1.13 |
| Pongo abelii | Orangutan | Besenbacher et al. (2019) | 1.66 |
| Chlorocebus sabaeus | Green Monkey | Pfeifer (2017) | 0.94 |
| Papio anubis | Baboon | Wu et al. (2020) | 0.57 |
| Macaca mulatta | Rhesus Macaque | Wang et al. (2020) | 0.58 |
| Bergeron et al (2020) | 0.77 | ||
| Aotus nancymaae | Owl Monkey | Thomas et al. (2018) | 0.81 |
| Mus musculus | House Mouse | Lindsay et al. (2019) | 0.54 |
| Canis lupus | Wolf | Koch et al. (2019) | 0.45 |
| Felis catus | Domestic Cat | Wang et al. (2021) | 0.86 |
| Bos taurus | Cow | Harland et al. (2017) | 1.17 |
| Ornithorhynchus anatinus | Platypus | Martin et al. (2018) | 0.7 |
a×10−8 mutations per site per generation.
bMany more studies exist than can be listed here.
These advantages notwithstanding, pedigree-based studies also face substantial challenges. Perhaps foremost among them is the fact that mutation rates are orders of magnitude lower than the sequencing error rate, even for the most accurate sequencing methods. Furthermore, while de novo mutations are biologically distinct from somatic mutations, it can be hard to differentiate the two because new mutations can occur at any stage of embryonic development post fertilization (especially during the earliest cell divisions when mutagenesis is highly likely), and thus can affect both somatic and germline cells in the developing embryo. The mistaken identity of somatic mutations for de novo germline mutations (Li 2014), which can occur at a non-negligible rate (Muryas et al. 2020), can also be the consequence of the tissues sampled for genomic comparisons. Because the number of de novo mutations produced in a single generation can be difficult to differentiate from erroneous variant calls, stringent variant filtering is applied. While necessary, true mutations can be missed (i.e., false negatives can also be common), and the mutation rate can be under- rather than overestimated (Scally 2016). Thus, studies that attempt to accurately estimate de novo rates must deal with a high probability of detecting false positives as well as false negatives (Segurel et al. 2014).
In this study, we utilize two strategies for minimizing both false-negative and false-positive rates. First, linked short reads from 10x Genomics (Weisenfeld et al. 2017) provide improved mapping and increased accuracy of individual variant calls (Long et al. 2016; Winter et al. 2018), especially in repeat-rich mammalian genomes (Chaisson et al. 2015). In addition, the phasing information provided by linked reads can determine the parent-of-origin with just two generations of sequencing. Phased haplotypes with known parental origin then allow individual mutations to be assigned to either the maternal or paternal germline. We estimated the callable fraction of our genome using two approaches. The first was based on variant filtering criteria, while the second introduced synthetic mutations to the sequencing data for one individual and evaluated the accuracy of our bioinformatic pipeline in recovering these mutations (Keightley et al. 2015; Xie et al. 2016). Although the use of synthetic mutations recovered by mutation-calling pipelines has typically been applied to estimating false-negative rates (Bergeron et al. 2020; Keightley et al. 2015; Koch et al. 2019; Pfeifer 2017; Wu et al. 2020; Xie et al. 2016), callable sites and false-negative rates are not independent of each other (i.e., determining low-coverage sites as not callable will also remove a majority of false negatives). Here, we show that the two callable site estimators yield similar mutation rates. To estimate a false-negative and false-positive rate for our data, we sequenced a technical replicate of the father in our pedigree and show that any point estimate of a de novo mutation rate should be considered with a large degree of uncertainty. Last, we illustrate how the adjustment of key variant filtering steps, such as the number of callable sites and allelic balance, can affect the final rate estimate, whereas many features of the mutation spectrum are robust to likely variant calling errors.
We applied these sequencing and computational methods to produce the first pedigree-based mutation-rate estimate for a strepsirrhine primate, the gray mouse lemur (Microcebus murinus). Mouse lemurs comprise a radiation of morphologically cryptic primates distributed throughout Madagascar (Setash et al. 2017). Numerous studies have suggested that their rapid speciation dynamics may reflect climatic change through time in Madagascar (Andriatsitohaina et al. 2019; Poelstra et al. 2021; Setash et al. 2017) and that their unique life-history characteristics make them an ideal genetic model organism (Ezran et al. 2017; Hozer et al. 2019). Thus, an accurate mutation-rate estimate for these organisms can potentially yield valuable insight into both geological and biological phenomena. Even though previous divergence time studies exist, they have had to rely on either phylogenetic methods, wherein only distantly related external fossil calibrations are available (dos Reis et al. 2018; Yang and Yoder 2003), or on pedigree-based mutation-rate estimates from distant relatives (Yoder et al. 2016). Notably, fossil-calibrated phylogenetic and pedigree-based approaches have yielded highly divergent age estimates further emphasizing the need for accurate estimates of de novo rates in mouse lemurs, and more generally, in other recently radiated groups wherein divergence time estimation may be problematic (Tiley et al. 2020).
By estimating the mutation rate in mouse lemurs with a pedigree-based approach, we aim to simultaneously expand our knowledge of mutation-rate variation across lineages and to facilitate the estimation of divergence times within the mouse lemur radiation specifically. To do so, we deeply sequenced a pedigree of gray mouse lemurs, including a focal quartet of mother, father, and two offspring, to accurately identify de novo mutations and to assign mutations to their parent-of-origin. We found a relatively high mutation rate, an unexpectedly low rate of transitions at CpG sites, and a weak paternal sex bias compared with other primates. Given the surprising nature of these results, we take care to discuss mutation-rate estimates in the context of their uncertainty and with the caution they deserve. We also show, however, that some patterns observed in the de novo mutation spectrum are likely robust to mutation-calling errors and are validated by substitution-rate patterns derived from a statistically rigorous phylogenetic relaxed-clock model. We conclude that though unexpected, the results of our pedigree analysis offer reliable estimates of the de novo mutation rate and spectrum in mouse lemurs.
Materials and methods
Samples
Four individuals were selected from the Duke Lemur Center’s mouse lemur colony consisting a focal family of two parents with two offspring from separate litters, for de novo mutation identification. In addition, a half-sibling to the offspring, and three other individuals in the maternal lineage were sequenced to help correct for standing variation (Fig. S1). The sire in our focal quartet had an age of 4.1 and 5 years at the time of conception for the male offspring (Texas Pete) and female offspring (Floretta), respectively. The dam was 1 and 1.9 years old at the time of conception for these offspring. Four of the eight selected samples were colony founders, which were transferred in 2003 from the CNRS mouse lemur colony in Brunoy, Paris, France. Blood and tissue samples were collected from all individuals during annual veterinary checkups. High-molecular-weight DNA was extracted with the Qiagen MagAttract kit (Qiagen, Germantown, MD, USA) and 10x Genomics library preparation was performed at the Duke Molecular Genomics Core.
Sequencing
Nine sequencing libraries were produced from the eight individuals; every individual was sequenced once, except the focal paternal sample that was prepared twice and sequenced as two separate libraries to serve as a technical replicate. Libraries were sequenced at the Duke Center for Genomic Computational Biology (GCB) Sequencing and Genomic Technology Shared Resource across nine lanes of a HiSeq 4000. Paired-end sequencing of 150 basepair reads was performed with an average insert size of 554 bp (range: 527–574 bp). A single lane was run as a test of the 10x Genomics LongRanger analysis software and was analyzed to confirm successful indexing and preparation of the samples. Next, the remaining eight libraries were multiplexed across eight lanes of a single flowcell. Over 933 Gb were generated across nine libraries and nine lanes. Sequencing data are available through NCBI’s SRA database (SRR10130788-SRR10130796).
10x Genomics pipeline
Basecall files were demultiplexed and analyzed using 10x Genomic’s LongRanger v2.2.1 pipeline. Average genomic coverage after filtering was 34.5× across the nine samples. Sequences were aligned to the reference gray mouse lemur genome assembly (mmur3.0, GCF_000165445.3) and variant calling was performed using GATK v3.8 (McKenna et al. 2010; Van der Auwera et al. 2013), implemented within LongRanger v2.2.1 (Weisenfeld et al. 2017). The mean N50 scaffold length, across samples, generated by the 10x Genomics LongRanger alignment pipeline, was 1.18 Mb.
DeNovoGear
LongRanger alignments were used to find de novo mutations within the offspring in the focal family. Several methods were used to find mutations. First, DeNovoGear v1.1.1 (Ramu et al. 2013) was used to analyze the LongRanger variant call files with default settings. VarScan2 v2.4.3 was run with the LongRanger binary alignment files and the resulting variants were intersected with the de novo mutations found with DeNovoGear. Only mutations found by both approaches were retained.
De novo mutations were inferred separately with each replicate library from the sire, and mutations that differed by sire replicate were used to estimate both the false-positive and false-negative rates (see Supplementary Methods: Variant calling’s effect on de novo mutation rate estimation). Finally, we checked whether alleles produced by the inferred de novo mutations were absent in the nonquartet samples and in existing data from a sequenced diversity panel of gray mouse lemurs (NCBI SRA:SRP045300). The final list of mutations was filtered for de novo quality in the offspring (de novo quality of at least 100), offspring mapping quality (mapping quality of at least 50), for at least 10× depth of coverage in both parents, less than 85× depth (2.5-fold increase over average coverage) in the offspring, and allelic balance of >0.30 and <0.70 (e.g., Thomas et al. 2018; Wang et al. 2020). The total number of mutations in each offspring was used to estimate a credible interval for the per-generation mutation rate (see Supplementary Methods: De novo mutation rate credible intervals).
Estimating the number of callable sites
We estimated the proportion of the genome ultimately considered for variant calling using two approaches. First, we conducted an “allele drop” test (Keightley et al. 2015) by introducing synthetic mutations to the sequencing data for one individual and subsequently tested the accuracy of our bioinformatic pipeline for recovering these mutations to determine the number of sites at which we would expect to miss a true mutation. This test consisted of adding 1000 synthetic mutations into the pedigree with the software BAMsurgeon v1.0.0 (Ewing et al. 2015). These mutations were added as heterozygotes by changing half of the aligned bases in the bam file at a given site to the nonreference allele. Next, we again applied our pipeline to find de novo mutations and examined the results for the 1000 synthetic sites. By conducting this allele drop test, we were able to estimate the fraction of the genome for which de novo mutations should have been found. The proportion of detected synthetic mutations was multiplied by the genome size to approximate the callable sites in a way that jointly considers our data and the uncertainty introduced by bioinformatic pipelines. Second, we estimated the number of callable sites as the fraction of the genome that passed our minimum and maximum depth filters (e.g., Krasovec et al. 2019). We repeated the mutation-rate calculation for increasing depth of coverage to evaluate uncertainty in our mutation rate due to filtering criteria.
Mutation-rate calculation
To calculate the single-base mutation-rate estimate, we determined the weighted average of mutations across the genomes of two offspring. The weighted average is the number of mutations on the autosomes (m1a + m2a) and X chromosome (m1x + m2x) divided by the number of callable autosomal and X-chromosome sites (gca and gcx). The denominator of each weighted average was multiplied by the number of haplotype genomes tested; for autosomes, the number was four, but for X chromosomes, only three were tested as one offspring was male and the other female. After determining the weighted average, we made a direct adjustment for the estimated amount of false-positive (fp) and false-negative (fn) mutations. We subtracted the number of raw mutations by the estimated number of false positives and added the estimated number of false negatives (Eq. (1), see also Supplementary Methods: De novo mutation rate calculation). These corrections assumed that the variants not shared by our two technical replicates were equally contributed to by false positives and false negatives (see Supplementary Methods: Variant calling’s effect on de novo mutation rate estimation), although it is possible to weight the effects of false positives and negatives on erroneous variants differently.
| 1 |
Parent-of-origin
Phased variant call files produced by LongRanger were used to assign mutations to a maternal or paternal chromosome. In brief, these methods took input of the three family individuals and a mutation location. The surrounding haplotype that contained the mutation was directly compared with the parental haplotypes at the same location to determine a match. As these individuals are all genetically related members from a single colony, dam and sire often shared similar haplotypes. When the mutation-bearing haplotype was found in both parents, a parent-of-origin was not assigned, resulting in <100% parent-of-origin assignment of mutations.
CpG islands and mutation rates
CpG islands were identified by two independent methods and compared to measure the number of mutations within them. First, the EMBOSS cpgplot tool (Chojnacki et al. 2017) was run with the latest gray mouse lemur reference genome (mmur3.0, GCF_000165445.3) to identify regions that met the threshold of a CpG island (200 bp, over 50% CG content). Then, to confirm these annotations, a fasta file of CpG island annotations from the gray mouse lemur genome 2.0 (GCF_000165445.2) was downloaded from the UCSC genome browser. A blast (Alschul et al. 1990) database of the mmur3.0 genome was created and the mmur2.0 CpG islands were queried to determine their coordinates in the genome used for mapping and assembly. Only the CpG islands identified with both methods (a total of 67,673 annotations) were used to determine whether a mutation at a CpG site was contained in a CpG island.
Context-dependent substitution-rate estimation
Because the mutation spectrum determined in mouse lemur differed from those observed in other primates, and because our study is complicated by the challenges of robust mutation-rate estimation from a single pedigree, we performed additional analyses to estimate substitution rates across the primate phylogeny. To do so, we used molecular clock methods that allow rates to differ by substitution type, including C>T transitions at non-CpG and CpG sites. First, we downloaded high-coverage mammalian whole-genome alignments from Ensembl (ftp://ftp.ensembl.org/pub/current_emf/ensemblcompara/multiple_alignments/46_mammals.epo/; last accessed February 2020). Analyses used alignments that included seven taxa: Mus musculus, Microcebus murinus, Callithrix jacchus, Chlorocebus sabaeus, Pongo abelii, Pan troglodytes, and Homo sapiens. The M. murinus reference genome used in the whole-genome alignment was the same version used for calling mutations (Larsen et al. 2017). Sites that mapped to protein-coding genes and CpG islands based on human gene features were removed. Data processing was done with Perl scripts available through Dryad. We randomly sampled ten one-megabase lengths of concatenated alignment to keep analyses computationally tractable.
We first estimated context-independent substitution rates. Branch lengths were optimized by maximum likelihood with the baseml program in PAML v4.8j (Yang 2007) using the HKY + gamma model. The approximate likelihood method (dos Reis and Yang 2011) was used to estimate absolute rates of evolution with fossil calibrations on all nodes (Table S1) that follow “calibration strategy A” from dos Reis et al. (2018). For each subsample, we ran four MCMC chains that discarded the first 50 million generations as burn-in and kept 10000 posterior samples for every 50,000 generations. Input alignments, control files, and the species tree are available through Dryad. Posteriors were analyzed in R v3.6.3 with the package CODA (Plummer et al. 2006).
The same subsampled alignments were used to estimate substitution rates for nine context-dependent substitution types (Table S2) following the method in (Lee et al. 2015). This method characterizes dinucleotide sites by integrating over uncertainty in substitution history for each site based on a sample of stochastic character maps. Substitution histories for each site were generated with PhyloBayes MPI v1.8 (Lartillot et al. 2013) under the CAT–GTR model (Lartillot and Philippe 2004). In total, 5000 samples were collected for two chains for each subsampled alignment while sampling every five generations. The first 1000 samples were discarded as burn-in. A total of 15 stochastic mappings were collected for each site. These were used to compute the variance–covariance matrices for the nine substitution types and approximate the likelihood surface of Bayesian relaxed-clock model. MULTIDIVTIME (Thorne et al. 1998) was then used to estimate absolute rates of evolution for each substitution type under an autocorrelated model (Thorne and Kishino 2002) with calibrations in Table S1. MULTIDIVTIME analyses collected 10,000 posterior samples for two chains, sampling every 10,000 generations after a 10-million-generation burn-in. Rate posteriors were evaluated for convergence and combined.
Comparing mutation and substitution rates across species
Because the de novo mutation rate should, in theory, be equivalent to the neutral substitution rate, we compared the mouse lemur mutation rate along with previously published third-codon position substitution-rate estimates from a recent study of primate divergence times (dos Reis et al. 2018). For species with a published per-generation de novo mutation rate, we took their terminal branch-specific substitution rate from an autocorrelated relaxed-clock model using “calibration strategy A.” However, substitution rates are measured per-year, as fossil calibrations are given in absolute time. To make per-year substitution rates comparable to the per-generation mutation rates, we scaled substitution rates by the averaged generation times from each pedigree-based study (Table S3). For example, to calculate the mouse lemur per-generation substitution rate, we multiplied its phylogenetic substitution rate (1.72 × 10−9 substitutions/site/year) by the average parent age at the time of conception averaged across offspring (3 years/generation) to get a mean substitution rate of 5.16 × 10−9 substitutions/site/generation. The same was done to the Bayesian credible intervals from substitution rates. Because males are expected to contribute more mutations over time (Thomas et al. 2018; Wang et al. 2020; Wu et al. 2020), we also rescaled substitution rates by the average paternal age at the time of reproduction.
Divergence time estimation
Using BPP v4.0 (Yang 2015), we re-evaluated divergence time estimates from a previous study (Yoder et al. 2016) using the pedigree-based mutation rate recovered by this study. We have written an R package, bppr (available at https://github.com/dosreislab/bppr), for calibrating node heights estimated by BPP to geological time using estimates of the mutation rate. Using bppr, we estimated mouse lemur divergence times twice (1) using the mutation rate prior of Yoder et al. (2016), which was based on estimates of mouse (genus, Mus) and human mutation rates, and (2) using the new estimates of the de novo rate generated by this study.
Results
Estimating the gray mouse lemur mutation rate
We assessed 4,542,770 potential variants across eight related individuals to discover 107 de novo mutations in two focal offspring (Fig. S2), which was reduced to 92 after filtering for allele balance (Fig. 1). Among these 92 mutations, 87 (46 in Floretta and 41 in Texas Pete) were located on autosomes and five (four in Floretta and one in Texas Pete) were located on the X chromosome. The average depth of coverage in the quartet for the 92 mutations was 59 reads (SD = 14.61). Our estimation of callable sites with synthetic mutations, similar to previous efforts to account for false-negative results (Keightley et al. 2015; Xie et al. 2016), detected 798 of 952 mutations on autosomes and 38 of 48 mutations on the X chromosome. Therefore, we estimate our detection rate to be 83.8% on autosomes and 79.2% on the X chromosome, which yields a total of 2.075 billion callable sites (out of a total genome size of 2.487 billion). When using depth-based criteria for determining callable sites, we estimated that between 88.9 and 62.2% of sites were callable for our quartet at 10× and 25× depth, respectively (Fig. 2). Thus, the number of callable sites was sensitive to depth criteria, although the number of de novo mutations was not. The number of de novo mutations was sensitive to filtering on allelic balance (Figs. 2 and S2). Most mutations that passed our filters appeared to be free of technical artifacts such as poor alignment of repeat-rich regions upon visual inspection (Figs. S3–S5). Although some mutations at higher depths appear suspect as potential paralogous alignments (Fig. S6), only ten mutations are between 2 and 2.5 times the average sequencing depth and there are no apparent systemic biases in mutation type among them (additional data available on Dryad).
Fig. 1. Focal family quartet.
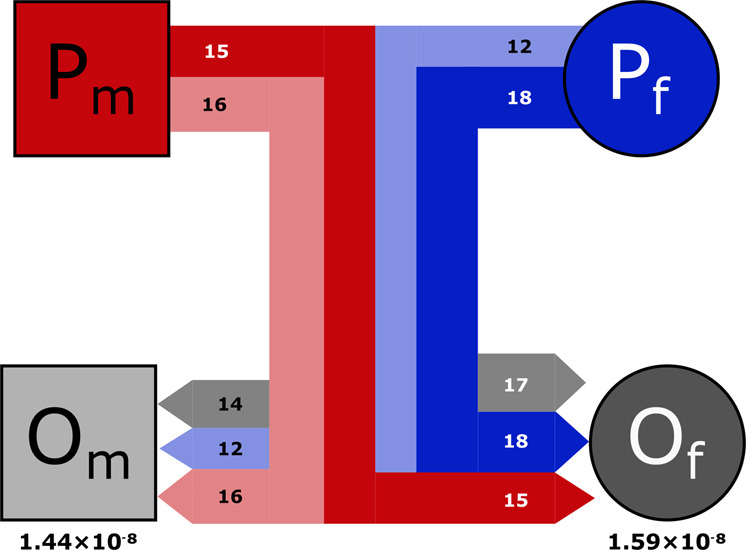
Parents (P) and offspring (O) are subscripted as male (m) or female (f). Lines represent familial relationships as in a traditional pedigree, with thickness and color reflecting the number and source of de novo mutations passed down (red is from male parent, blue from female parent, and gray is undetermined origin). Color of line represents source and shading represents destination (lighter shading to Om, darker to Of). Numbers within bars show mutation counts and the rate of each individual offspring is listed below.
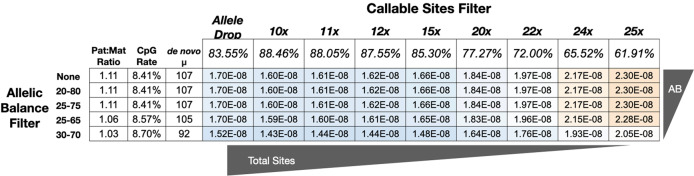
Fig. 2. Effect of filtering thresholds on mutation-rate estimation.
The mutation rate and spectrum of the gray mouse lemur, as a product of two main filtering decisions: (1) an allelic balance filter (along the rows) and (2) a callable site filter (along the columns). The first three columns display how the parent-of-origin, the mutation rate at CpG sites, and total number of mutations vary. The remainder of the table shows the combined effect of these filters on the calculated rate. Cells for lower rates are shaded blue and higher rates are shaded red. All mutation rates have been corrected for the estimated number of false positives and false negatives with their respective number of mutations and callable sites.
Based on an error rate of 0.021 from the number of variants unique to the two technical replicates, and assuming errors are caused equally by false positives and false negatives, we calculated 3.42 false positives and 34.46 false negatives from the total of 92 de novo mutations and 2.088 billion callable mutation sites. In an attempt to generate a more accurate estimate of the de novo mutation rate, we adjusted our raw rate (1.14 × 10−8) by accounting for the estimated false positives and false negatives, to arrive at a final rate estimate of 1.52 × 10−8 mutations per-site per-generation (95% credible interval: 1.28 × 10−8–1.78 × 10−8). This estimate is sensitive to assumptions about the proportion of unique variants between technical replicates that are due to false negatives and could be close to 1.28 × 10−8 if the contributions of false negatives are actually small (Fig. S7). This rate is also a median when considering depth filters on callable sites between 10× and 25× depth (Fig. 2).
The mouse lemur mutation spectrum
From the pedigree-based estimate of the mutation spectrum, a ratio of transitions to transversions (Ti:Tv) was estimated to be 0.96 (45 transitions and 47 transversions). The ratio of strong-to-weak mutations (SW; C/G>A/T) to weak-to-strong mutations (WS; A/T>C/G), SW:WS, was estimated to be 1.24 (41 SW and 33 WS mutations). The most common two categories of de novo mutation type were A>G and C>T (Fig. 3A). Eight mutations were detected at parental CpG sites, constituting 8.7% of all de novo mutations. This represents a roughly fourfold enrichment given that 1.9% of the genome consists of CpG sites. Because the elevated mutation rate at CpG sites is linked to methylation (Bird 1980), mutations are typically not expected in regions of the genome with high GC content (CpG islands), where CpG sites are much less likely to be methylated (Bird 1986; Molaro et al. 2011). As anticipated, none of the 92 de novo mutations were found within CpG islands, which constitute roughly 4% of the M. murinus genome.
Fig. 3. Mutation spectrum of the gray mouse lemur.
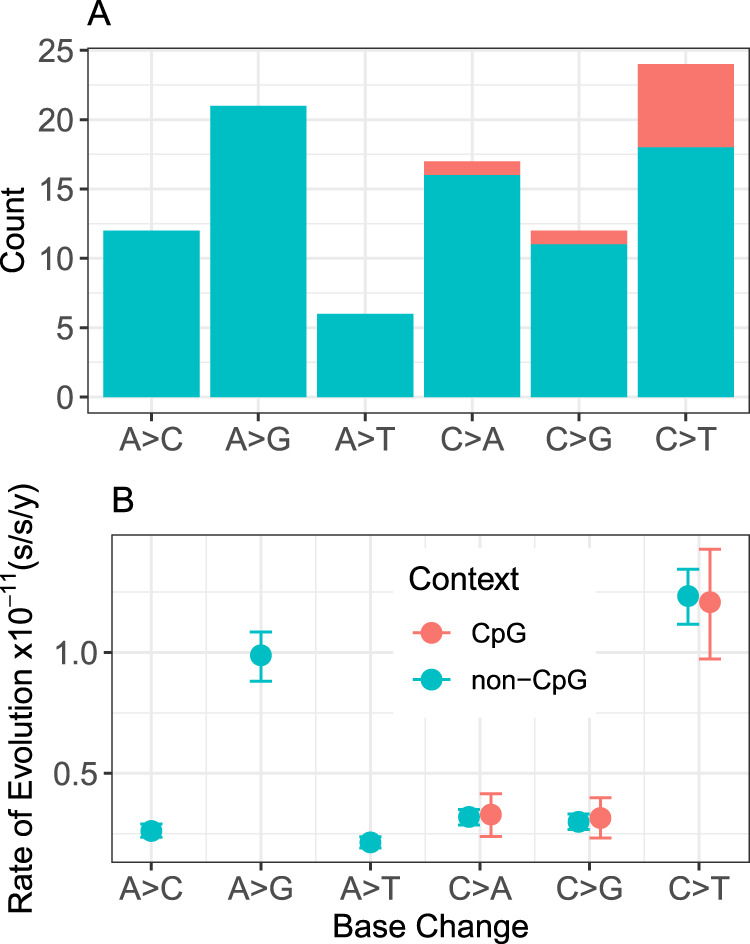
A Counts of de novo mutations from the pedigree analysis. Mutation types are broken down by weak-to-strong transversions (A>C and T>G), weak-to-strong transitions (A>G and T>C), weak-to-weak transversions (A>T and T>A), strong-to-weak transversions (C>A and G>T), strong-to-strong transversions (C>G and G>C), and strong-to-weak transitions (C>T and G>A). Complementary mutation types are shown together. B Context-dependent substitution-rate estimates. Nine possible substitution-type parameters are shown for the gray mouse lemur terminal branch, which are categorized similarly as the de novo mutation spectrum. Error bars on substitution rates represent 95% highest posterior densities.
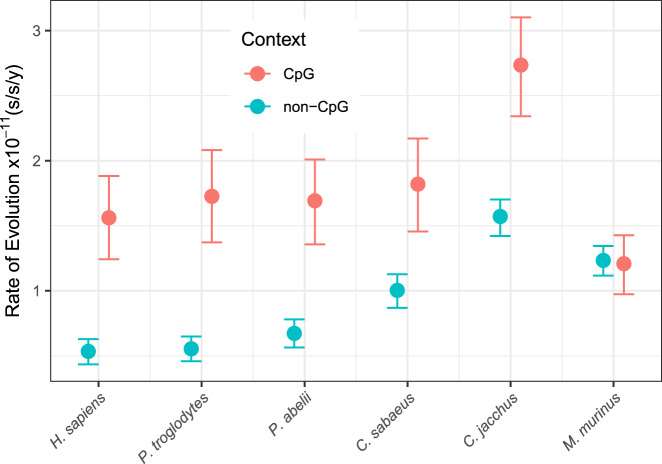
The mutation spectrum in mouse lemur was further investigated with an independent approach based on absolute substitution rates (substitutions/site/year; s/s/y) and fossil-calibrated relaxed-clock models. All clock model parameters (Fig. S8) converged across ten one-megabase replicates (Figs. S9–S19) and revealed a higher global substitution rate in mouse lemurs compared with apes and Old World monkeys (Fig. S20). We then estimated context-dependent substitution rates for the same alignments (Lee et al. 2015). All rate parameters converged (Figs. S21–S30) and transitions at CpG sites (Group 9) were the only substitution type to clearly break from the pattern expected by not partitioning across substitution types (Fig. S20). Mouse lemur had the lowest rate of C>T transitions at CpG sites of all primates (Figs. S31–S40), thereby supporting the results of the pedigree-based approach. Notably, in mouse lemur, the rate of C>T transitions at CpG sites is slightly lower than the rate of C>T transitions at non-CpG sites (Group 5), whereas the converse is true for all other primates across all ten subsampled alignments (Figs. S31–S40). Specifically, the mean rate estimate for C>T transitions at CpG sites is 98% of the rate of C>T transitions at non-CpG sites (1.210 × 10−11 s/s/y vs 1.234 × 10−11 s/s/y) in mouse lemur. The C>T transition rate is 2.92, 3.11, 2.51, 1.81, and 1.74 times higher for CpG versus non-CpG sites in human, chimp, orangutan, Old World monkey, and New World monkey, respectively. The pattern of rate variation across substitution types generally agrees with the observed mutation spectrum from our focal quartet (Fig. 3B) and corroborates the low rate of CpG mutations in the gray mouse lemur relative to other primates (Fig. 4).
Fig. 4. Context-dependent relaxed-clock analysis shows low rates of C>T substitution rates at CpG sites in the gray mouse lemur.
C>T substitution-rate estimates at non-CpG versus CpG sites are compared for six species of primate, including the gray mouse lemur (M. murinus). Note that with the exception of M. murinus, all primates examined show significantly higher CpG rates than non-CpG rates. The C>T substitution rates at non-CpG and CpG sites are nearly identical in M. murinus. Error bars represent 95% highest posterior densities.
Discrepancies of magnitude when comparing pedigree-based mutation rates and phylogenetic substitution rates
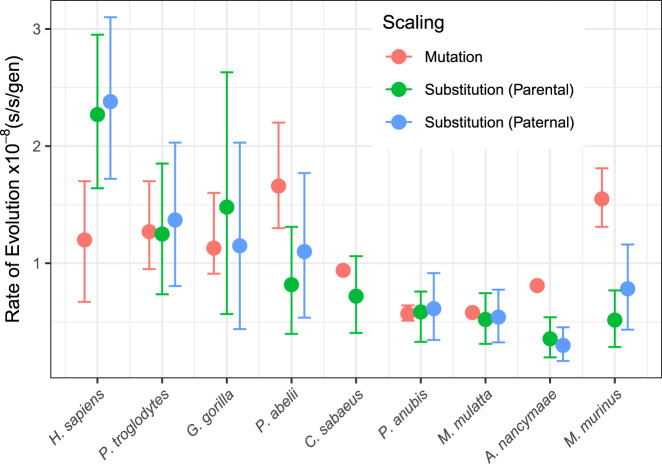
We compared pedigree-based estimates of the mutation rate for mouse lemurs together with published mutation-rate estimates from other primates (Table 1) with substitution rates estimated from a recent relaxed-clock analysis of the same species (dos Reis et al. 2018). Phylogenetic substitution rates are estimated per-year, so we rescaled them by generation time to represent them as substitutions per-site per-generation (s/s/gen), considering the average parent age as well as the average age of fathers (Table S3), for direct comparison with per-generation mutation rates from pedigrees. There are three notable observations: (1) the mean pedigree-based mutation-rate estimates are contained by the phylogenetic-based substitution-rate estimate highest posterior density intervals for all but three cases: human, owl monkey, and mouse lemur, (2) substitution rates are not consistently lower than mutation rates as demonstrated by humans, and (3) scaling phylogenetic substitution rates with the average age of the father closes the distance between mutation and substitution rates in cases where there are differences between the ages of fathers and mothers, as observed in orangutan and mouse lemur. For most great apes and Old World monkeys, their pedigree-based mutation-rate estimates are consistent with their third-codon substitution rates, especially when scaling by the average age of the father as opposed to average parent age for orangutan (P. abelii, Fig. 5).
Fig. 5. Difference between mutation and substitution rates among primates.
Error bars around substitution rates are 95% highest posterior density intervals from a Bayesian relaxed-clock analysis. Credible intervals are given for mutation rates where available from published data. Substitution rates are scaled from per-year to per-generation based on the average parent (parental) age at the time of conception, except for C. sabaeus where data were not available and the generation time assumed from external information. Where age information on parents was available, substitution rates were also scaled by the average father (paternal) age. Data are given in Table S3.
Sex bias
Using the long phasing blocks generated by the linked-read method, we were able to determine the parent-of-origin for 61 out of 92 (66%) de novo mutations. The number of mutations confidently assigned to a parent are notably higher in the analysis presented here compared with previous studies that used short-read sequencing alone, which found only 35% (Venn et al. 2014) or 38% (Thomas et al. 2018). Among the assigned mutations, 51% (n = 31) were found on the offsprings’ paternal haplotype, while the remaining 49% (n = 30) were found on the offsprings’ maternal haplotype; a ratio of male-to-female mutations of 1.03. This is considerably lower than the observation of approximately 4:1 typically observed in other primate studies (Wu et al. 2020).
Impacts for divergence time estimation
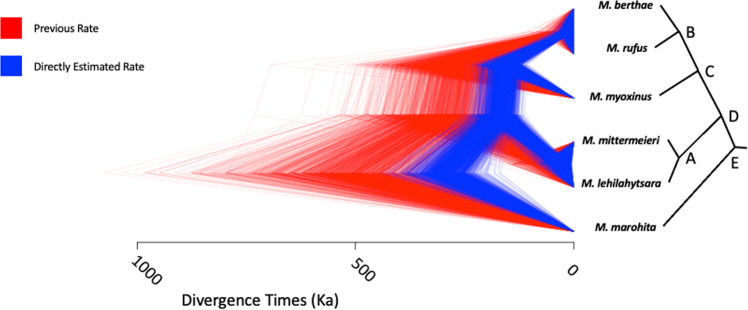
We recalibrated branch lengths in absolute time for a genus-level phylogeny of mouse lemurs (Yoder et al. 2016) based on the new mutation-rate estimate of 1.52 × 10−8 mutations/site/generation derived from this study. Previously, the mutation rate was modeled on a gamma distribution from mouse (Uchimura et al. 2015) and human (Scally and Durbin 2012) estimates, with a mean of 0.87 × 10−8 mutations/site/generation. The higher mutation rate calculated here yields considerably more recent divergence times (Fig. 6) with reduced uncertainty compared with sampling from the previously wide gamma distribution (Table S4).
Fig. 6. Estimated divergence times among mouse lemur species.
Trees are posterior samples from BPP based on a fixed previously published topology. The directly estimated mutation rate (blue) is nearly twice as high as the previously assumed rate (red). Divergence times estimated with the new mutation rate are nearly half of the previous estimates. Summary statistics are given in Table S4, matched by node labels (A–E).
Discussion
A high mutation rate in mouse lemurs
In this study, we provide the first pedigree-based estimate of the de novo mutation rate in a strepsirrhine primate. Our mean mutation-rate estimate was calculated to be 1.52 × 10−8 mutations/site/generation, which is high compared with previously characterized primates with the exception of orangutan that shows a similarly high rate (Besenbacher et al. 2019). We took several measures to ensure accurate mutation-rate estimation, including the use of simulations to determine the appropriate denominator for mutation-rate calculations and a technical replicate to estimate both false-positive and false-negative rates. Even so, any point estimate of the de novo mutation rate should be interpreted with caution as there are numerous variables that can impact rate estimates, including biological factors such as rate variation among the pedigrees themselves (Smith et al. 2018). Moreover, any pedigree-based estimate is the direct result of accumulated study-design decisions made regarding available animals, experimental planning, and data-quality thresholds. The rate we present is a product of these decisions and to change any of these inputs could potentially yield a change in the final estimate. For example, narrowing the allelic balance threshold would eliminate called mutations and thus lower the rate, while increasing the coverage requirement would decrease the number of callable sites and thus raise the rate (Fig. 2). We adjusted the allele balance and depth-based callable site filters to estimate a range of mutation rates, the majority of which are within the 95% CI of our allele drop-based estimate of 1.52 × 10−8 mutations/site/generation. Although the mutation-rate estimate was sensitive to various filters, the fraction of mutations found at CpG sites as well as the ratio of mutations from dam and sire was not.
We used linked-read sequencing technology that improves mapping accuracy to produce high-quality variants for the de novo mutations identified here. The linked reads also allowed us to recover parental haplotypes, and subsequently, the parent-of-origin for observed mutations in offspring (Fig. 1). The number of mutations with an assigned parent-of-origin is higher (66%) in the present study than in analyses that used short reads and three generations of sequencing (Thomas et al. 2018; Venn et al. 2014). Although a number of factors such as sequencing depth, heterozygosity, and recombination rate may vary across investigations and limit the value of cross-study comparisons, the prospect of successfully phasing more mutations while also eliminating the need to sequence across more than two generations with linked-read data is appealing.
Low numbers of mutations at CpG sites
CpG sites have generally been found to have higher mutation rates relative to other site classes, a pattern discovered several decades ago using DNA sequence comparisons (Bird 1980) and ascribed to the frequent deamination of methylated cytosines (Friedberg et al. 2005). Only a fourfold enrichment of mutations at CpG sites (8 mutations, 8.7% of all mutations) was found in mouse lemur, which is less than the at-least tenfold enrichment (12–25% of total mutations) found in other primate studies (Besenbacher et al. 2019; Gao et al. 2019; Thomas et al. 2018; Venn et al. 2014). Though surprising, we are confident that the result here reported has biological relevance. The findings from our relaxed-clock analyses of different substitution types are consistent with the observed de novo mutation spectrum (Fig. 3). Notably, the rate of C>T transitions at CpG sites breaks from the pattern expected without partitioning (Fig. S20), including C>T transitions at non-CpG sites (Figs. S31–S40) where mouse lemurs show a higher substitution rate than great apes and Old World monkeys but a lower rate than New World monkeys. Mouse lemurs have the lowest rate of C>T transitions at CpG sites of all primates analyzed here (Figs. S31–S40). This leads to the hypothesis that methylation of CpG sites in mouse lemur germ cell lines may actually be lower relative to that in other primates (Rahbari et al. 2016), thus ultimately contributing fewer hits to their mutation spectrum (Figs. 3 and 4).
A lowered rate of C>T transitions at CpG sites is surprising for primates. Because these mutations are caused by deamination of methylated cytosines, they are expected not to be affected by variation in generation times and are thus predicted to evolve more clock-like than other substitution types (Kim et al. 2006). Substitution rates are consistent with a molecular clock when there is no among-branch variation, such that the expected number of substitutions increases linearly over time. Previous studies of relative substitution rates using similar whole-genome alignments have found that transitions at CpG sites are much more clock-like than transitions at non-CpG sites when comparing great apes to Old World monkeys or New World monkeys (Moorjani et al. 2016a). These same analyses of context-dependent substitution rates also demonstrated clock-like behavior of C>T transitions at CpG sites across anthropoids (Lee et al. 2015). In both of these studies, a single stepsirrhine (Otolemur garnetii) was treated as an outgroup and rates within strepsirrhines were not estimated. However, earlier approaches for estimating context-dependent substitution rates on a 1.7-Mb region across mammals (Hwang and Green 2004) also discovered lowered relative C>T transition rates at CpG sites in lemurs and their common ancestor when compared with anthropoids, although we also found a notably elevated rate in New World monkeys (Callithrix jacchus; Fig. 4). New World monkeys have been shown to have rates of transitions at CpG sites approximately 20% higher than great apes (Moorjani et al. 2016a), but past analyses with context-dependent substitution rates on a 0.15-Mb alignment have suggested much more clock-like behavior (Lee et al. 2015). We anticipate that future analyses with denser sampling of New World monkeys and strepsirrhines will be necessary to rigorously test clock-like behavior of C>T transitions at CpG sites in primates.
The mouse lemur mutation spectrum
Our estimates of the Ti:Tv and SW:WS ratios at 0.96 and 1.24 each are also lower than values found in other animals. For instance, the Ti:Tv ratio found in previous pedigree-based studies in other species varied between 1.97 and 2.67 (Agier and Fischer 2012; Assaf et al. 2017; Besenbacher et al. 2019; Kong et al. 2012; Smeds et al. 2016; Thomas et al. 2018; Venn et al. 2014). Our finding of a lower Ti:Tv ratio is likely a consequence of the relatively low number of C>T transitions at CpG sites. For example, C>T transitions are twice as frequent as A>G transitions in human, chimp, and owl monkey (Thomas et al. 2018; Venn et al. 2014), but these two mutation classes occur in equal frequency in mouse lemur (Fig. 3A). These findings also explain the SW:WS ratio closer to 1 than previous studies, since C>T mutations are strong-to-weak transitions. For instance, without an elevation in the mutation rate at CpG sites, the Ti:Tv and SW:WS ratios would drop from 2.06 and 2.11 to 1.46 and 1.33, respectively, in a study of chimpanzees (Venn et al. 2014). Thus, reduced numbers of C>T transitions at CpG sites can simultaneously explain several aspects of the measured mouse lemur mutation spectrum that deviate from previous studies of other primate mutation rates.
The Ti:Tv and SW:WS ratios observed in the mouse lemur mutation spectrum are also supported by the context-dependent substitution-rate analysis. Taking the average of transition and transversion rate classes (Table S2) yields a Ti:Tv of 1.64. When considering substitution classes by strong-to-weak and weak-to-strong types, we find a SW:WS of 0.85. Although not equivalent to the spectrum-based estimates, these ratios are much lower than the branch rates observed in other species, for example, where Ti:Tv ranges from 2.05 to 2.58, while SW:WS ranges from 1.04 to 1.33 in C. jaccus and P. troglodytes, respectively. The lower ratios of Ti:Tv and SW:WS rates in mouse lemur are both explained by the lower-than-expected C>T transition rate at CpG sites. In total, the independent substitution-rate analysis of the primate reference genomes validates our findings that the C>T transition rate at CpG sites, Ti:Tv ratio, and SW:WS ratio of the de novo mutation spectrum in mouse lemurs deviates from those in other primates.
Reduced male mutational bias
A paternal mutational bias has long been hypothesized for diploid sexually reproducing organisms based on the idea that the increased number of cell divisions in sperm versus egg should lead to higher numbers of mutations in the male germline than the female germline (Haldane 1946; Kong et al. 2012; Lindsay et al. 2019). Indeed, a strong paternal mutation-rate bias has been observed in the vast majority of pedigree-based mutation-rate estimates to date (Gao et al. 2019; Lindsay et al. 2019; Rahbari et al. 2016; Thomas et al. 2018; Venn et al. 2014) and in many studies of phylogenetically based rates (Axelsson et al. 2004; Ellegren and Fridolfsson 1997; Goetting-Minesky and Makova 2006; Shimmin et al. 1993; Zhang 2004). The cell-division hypothesis has lately been challenged, however, with the suggestion made that observed paternal biases relate instead to more complicated relationships among DNA repair mechanisms and life-history traits (Wu et al. 2020).
The 1.03 ratio of paternal-to-maternal mutations in gray mouse lemur observed here, among the 66% of mutations that could be assigned with parent-of-origin, is considerably lower than the range observed in primates between 2.1 in owl monkey (Thomas et al. 2018) and 5.5 in chimpanzee (Venn et al. 2014), with most human studies falling around 3.6 (Gao et al. 2019; Rahbari et al. 2016) and 2.7 in mouse (Lindsay et al. 2019). It is similar, however, to the ratio of 1.2 found in collared flycatchers where the F1 male was only 1-year old (Smeds et al. 2016), which suggests that the low sex bias ratio observed in the gray mouse lemur is not unreasonable in the larger context of vertebrate diversity. Also, it is worth noting that one of the driving factors of the paternal mutational bias has been hypothesized to relate to the time of first reproduction after puberty, with rate increasing as time between puberty and first reproduction increases (Segurel et al. 2014). Here, mouse lemurs are exceptional in the primate clade given that puberty and time of first reproduction occur nearly simultaneously (Blanco et al. 2011; Blanco et al. 2015; Zohdy et al. 2014). Mouse lemurs are reproductively mature in the first year of life with females typically producing their first litter by the age of 10 months. It is less clear when males first become successful sires as they must compete with older more experienced males in their first year. The sire for our focal quartet was 4.1 and 5 years old at the time of conception of the male and female offspring, respectively (Figs. 1 and S1). Though mature regarding life-history stage, this timeframe may nonetheless be insufficient for producing a strong male mutational bias relative to longer-lived species where more mutations in the male germline would be anticipated (Kong et al. 2012; Thomas et al. 2018). In addition, there are differences in the methylation process within male and female germline cells, with male cells experiencing more methylation (Kobayashi et al. 2013; Reik and Dean 2001). This discrepancy yields more methylation-related mutations in males than females as mammals age (Gao et al. 2019; Jonsson et al. 2017). Thus, fewer methylation-related (i.e., CpG) mutations, and a short time to puberty in mouse lemurs may in combination lead to the observed, limited sex bias. As a potential caveat, mouse lemurs have both behavioral (Dammhahn and Kappeler 2005; Eberle and Kappeler 2004) and morphological signs (i.e., enlarged testes during the mating season) of sperm competition (Kappeler 1997) that in other primates may be correlated with high substitution rates (Wong 2014). though this appears not to be the case in mouse lemurs.
Mutation and substitution rates
Our analyses show that there is incongruence between the magnitude of mutation rates and of phylogenetic substitution rates when attempting to directly compare the two (Fig. 5). Several sources of uncertainty underlie both. Pedigree-based mutation rates offer only a sample of the present, and both mutation rate and generation time may have varied through time (Moorjani et al. 2016b). For example, one revelation in the rapidly developing literature on de novo mutation rates has been that the estimated rate in humans is less than half that predicted by phylogenetic studies, and is recapitulated here (Fig. 5), suggesting that the mutation rate has slowed down over time in humans and that rates can change rapidly within primates (Scally and Durbin 2012), and presumably other clades. Substitution and mutation rates from apes, aside from humans and Old World monkeys observed here, agree when considering the average paternal age for rescaling absolute substitution rates to per-generation. Mouse lemur, however, had a significantly elevated mutation rate to the point where credible intervals with their paternal age-rescaled substitution rates did not overlap, although they would if not correcting for the estimated number of false-positive and false-negative mutations.
Phylogenetically based estimates may be biased downward if substitutions are not fully neutral. Substitution rates used for comparison with generation times and mutation rates were based on third-codon positions from a supermatrix of different data types (dos Reis et al. 2018; Springer et al. 2012) and may be under weak purifying selection. Indeed, previous studies have found evidence for low phylogenetically based compared with pedigree-based estimates (Denver et al. 2000; Howell et al. 2003; Winter et al. 2018). For pedigree-based estimates, the degree to which somatic mutations and/or interindividual variation might impact these estimates is not clear (Segurel et al. 2014). Additional data and analyses will be needed to reconcile the differences between pedigree-based and phylogenetic estimates of the mutation rate.
Mutation rates and divergence time estimates
Application of the pedigree-based mutation-rate estimate observed in this study leads to more recent divergence times among mouse lemur species than previously inferred (Fig. 6 and Table S4). These divergence times are obtained by rescaling branch lengths in substitutions per-site to absolute time given a mutation rate and generation time (Burgess and Yang 2008) as opposed to relaxed-clock phylogenetic methods that estimated older species divergences within mouse lemurs (dos Reis et al. 2018; Yang and Yoder 2003). A previous analysis made assumptions regarding mutation rate in mouse lemurs (Yoder et al. 2016) that resulted in divergence times nearly twice as old as those presented here (Fig. 6 and Table S4). Although such assumptions regarding mutation rates are reasonable in the absence of data, direct mutation rates from pedigrees can arguably produce more accurate divergence time estimates, especially when no fossils are available for the target clade mandating that deeply diverged taxa must be included for external calibration in relaxed-clock studies (Tiley et al. 2020). Unfortunately, a complete lack of lemuriform fossils means that we cannot evaluate the accuracy of divergence time estimates for mouse lemurs in the context of the fossil record. Given the endangered status of many mouse lemur species, and virtually all other strepsirrhine species, an enhanced ability to provide a temporal context to speciation and to estimate demographic parameters such as effective population size may yield critical information for directing ongoing conservation policy and efforts. We caution though that pedigree-based mutation rates can also lead to poor estimation of divergence times when species are distantly related and de novo mutation rates vary significantly among lineages or have changed through time (Scally and Durbin 2012), as in the case of dating the common ancestor of apes and Old World monkeys (Wu et al. 2020).
Conclusions
Our study emphasizes the importance of increased sampling across the tree of life for gaining insight into the nature and causes of mutation-rate evolution. Critically, it also sheds light on the effect that data processing has on the final estimate of mutation rate. We emphasize that mutation-rate estimates are highly sensitive to variant filtering, and by using a technical replicate, identify assumptions about the sources of error for false-positive and false-negative rate estimation and their respective impacts on de novo rate estimation. Further, as this is the first pedigree-based mutation- rate estimate for a strepsirrhine primate, it is not clear whether the high mutation rate, low CpG mutation rate, and weak sex bias are specific to mouse lemurs or may be representative of strepsirrhines more generally. Although variation in the mutation rate and spectrum is anticipated among different pedigrees, and our study is largely based on a single quartet, the results of our context-dependent substitution-rate analysis validate the most surprising aspect of a low rate of C>T transitions in CpG sites. Reconciling the disparity in magnitude between mutation rates from pedigrees and substitution rates from phylogenetic methods will be a focus of future work as more pedigree-based mutation rates become available. As demonstrated by this study in mouse lemurs, de novo mutation-rate estimates stand to drastically revise divergence times, especially in recent evolutionary radiations.
Supplementary information
Acknowledgements
The authors thank our handling editor, as well as Matt Hahn and two anonymous reviewers, who provided critical feedback that improved the paper. We thank the Duke Lemur Center staff, especially Erin Ehmke, Bobby Schopler, and Cathy Williams, for providing tissue samples. Priya Moorjani, Susanne Pfeifer, Jonathan Pritchard, Molly Przeworski, and Meredith Yeager all provided helpful discussions in the development of this project. We especially thank Jonathan Pritchard for his suggestion that substitution-rate analysis could be useful for verifying the observed mutation-rate spectrum. We would like to acknowledge the assistance of the Duke Molecular Physiology Institute Molecular Genomics core for the generation of data for the paper. This study was funded by a National Science Foundation Grant DEB-1354610 and Duke University research funds to ADY and she gratefully acknowledges support from the John Simon Guggenheim Foundation and the Alexander von Humboldt Foundation during the writing phase of this project. JLT was supported by National Science Foundation Grant DEB-1754142. This is a Duke Lemur Center publication.
Data availability
Data are available from the Dryad Digital Repository 10.5061/dryad.8pk0p2njx. Raw sequence data are available through NCBI under BioProject PRJNA512515.
COMPETING INTERESTS
The authors declare no competing interests.
Footnotes
Associate editor Armando Caballero
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: C. Ryan Campbell, George P. Tiley
Supplementary information
The online version contains supplementary material available at 10.1038/s41437-021-00446-5.
References
- Agarwal I, Przeworski M. Signatures of replication timing, recombination, and sex in the spectrum of rare variants on the human X chromosome and autosomes. Proc Natl Acad Sci USA. 2019;116:17916–17924. doi: 10.1073/pnas.1900714116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agier N, Fischer G. The mutational profile of the yeast genome is shaped by replication. Mol Biol Evol. 2012;29:905–913. doi: 10.1093/molbev/msr280. [DOI] [PubMed] [Google Scholar]
- Alschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andriatsitohaina B, Ramsay MS, Kiene F, Lehman SM, Rasoloharijaona S, Rakotondravony R et al. (2020) Ecological fragmentation effects in mouse lemurs and small mammals in northwestern Madagascar. Am J Primatol 82(4):e23059 [DOI] [PubMed]
- Angelis K, dos Reis M. The impact of ancestral population size and incomplete lineage sorting on Bayesian estimation of species divergence times. Curr Zool. 2015;61:874–885. doi: 10.1093/czoolo/61.5.874. [DOI] [Google Scholar]
- Assaf ZJ, Tilk S, Park J, Siegal ML, Petrov DA. Deep sequencing of natural and experimental populations of Drosophila melanogaster reveals biases in the spectrum of new mutations. Genome Res. 2017;27:1988–2000. doi: 10.1101/gr.219956.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E, Smith NG, Sundstrom H, Berlin S, Ellegren H. Male-biased mutation rate and divergence in autosomal, z-linked and w-linked introns of chicken and Turkey. Mol Biol Evol. 2004;21:1538–1547. doi: 10.1093/molbev/msh157. [DOI] [PubMed] [Google Scholar]
- Benton MJ, Donoghue PC. Paleontological evidence to date the tree of life. Mol Biol Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- Bergeron LA, Besenbacher S, Bakker J, Zheng J, Li P, Pacheco G, Sinding M-HS, Kamilari M, Gilbert MTP, Schierup MH, Zhang G (2020) The germline mutational process in rhesus macaque and its implications for phylogenetic dating. bioRxiv 10.1101/2020.06.22.164178 [DOI] [PMC free article] [PubMed]
- Besenbacher S, Hvilsom C, Marques-Bonet T, Mailund T, Schierup MH. Direct estimation of mutations in great apes reconciles phylogenetic dating. Nat Ecol Evol. 2019;3:286–292. doi: 10.1038/s41559-018-0778-x. [DOI] [PubMed] [Google Scholar]
- Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP (1986) CpG-rich islands and the function of DNA methylation. Nature 321:209-213 [DOI] [PubMed]
- Blanco MB, Rasoazanabary E, Godfrey LR. Reproductive opportunism in unpredictable environments: the comparison of two wild mouse lemur species (Microcebus rufus and M. griseorufus) from eastern and western Madagascar. Am J Phy Anthr. 2011;144:91–91. doi: 10.1002/ajp.22423. [DOI] [PubMed] [Google Scholar]
- Blanco MB, Rasoazanabary E, Godfrey LR. Unpredictable environments, opportunistic responses: Reproduction and population turnover in two wild mouse lemur species (Microcebus rufus and M. griseorufus) from eastern and western Madagascar. Am J Prima. 2015;77:936–947. doi: 10.1002/ajp.22423. [DOI] [PubMed] [Google Scholar]
- Burgess R, Yang Z. Estimation of hominoid ancestral population sizes under Bayesian coalescent models incorporating mutation rate variation and sequencing errors. Mol Biol Evol. 2008;25:1979–1994. doi: 10.1093/molbev/msn148. [DOI] [PubMed] [Google Scholar]
- Chaisson MJ, Wilson RK, Eichler EE. Genetic variation and the de novo assembly of human genomes. Nat Rev Genet. 2015;16:627–640. doi: 10.1038/nrg3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chojnacki S, Cowley A, Lee J, Foix A, Lopez R. Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res. 2017;45:W550–W553. doi: 10.1093/nar/gkx273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammhahn M, Kappeler PM. Social system of Microcebus berthae, the world’s smallest primate. Int J Primatol. 2005;26:407–435. doi: 10.1007/s10764-005-2931-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Vassilieva LL, Thomas WK. High direct estimate of the mutation rate in the mitochondrial genome of Caenorhabditis elegans. Science. 2000;289:2342–2344. doi: 10.1126/science.289.5488.2342. [DOI] [PubMed] [Google Scholar]
- dos Reis M, Gunnell GF, Barba-Montoya J, Wilkins A, Yang Z, Yoder AD. Using phylogenomic data to explore the effects of relaxed clocks and calibration strategies on divergence time estimation: primates as a test case. Syst Biol. 2018;67:594–615. doi: 10.1093/sysbio/syy001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Yang ZH. Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Mol Biol Evol. 2011;28:2161–2172. doi: 10.1093/molbev/msr045. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle M, Kappeler PM. Sex in the dark: determinants and consequences of mixed male mating tactics in Microcebus murinus, a small solitary nocturnal primate. BehavEcolSociobiol. 2004;57:77–90. [Google Scholar]
- Ellegren H, Fridolfsson A-K. Male-driven evolution of DNA sequences in birds. Nat Genet. 1997;17:182–184. doi: 10.1038/ng1097-182. [DOI] [PubMed] [Google Scholar]
- Ewing AD, Houlahan KE, Hu Y, Ellrott K, Caloian C, Yamaguchi TN, et al. Combining tumor genome simulation with crowdsourcing to benchmark somatic single-nucleotide-variant detection. Nat Methods. 2015;12:623–630. doi: 10.1038/nmeth.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezran C, Karanewsky CJ, Pendleton JL, Sholtz A, Krasnow MR, Willick J, et al. The mouse lemur, a genetic model organism for primate biology, behavior, and health. Genetics. 2017;206:651–664. doi: 10.1534/genetics.116.199448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazalova V, Nevado B. Low spontaneous mutation rate and pleistocene radiation of pea aphids. Mol Biol Evol. 2020;37:2045–2051. doi: 10.1093/molbev/msaa066. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Feng C, Pattersson M, Lamichhaney S, Rubin CJ, Rafati N, Casini M et al. (2017) Moderate nucleotide diversity in the Atlantic herring is associated with a low mutation rate. Elife 6:e23907 [DOI] [PMC free article] [PubMed]
- Friedberg EC, Walker GC, Siede W, Wood RD (2005) DNA repair and mutagenesis. American Society for Microbiology Press Washington DC
- Gao Z, Moorjani P, Sasani TA, Pedersen BS, Quinlan AR, Jorde LB, et al. Overlooked roles of DNA damage and maternal age in generating human germline mutations. Proc Natl Acad Sci U S A. 2019;116:9491–9500. doi: 10.1073/pnas.1901259116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garimella KV, Iqbal Z, Krause MA, Campino S, Kekre M, Drury E, et al. Detection of simple and complex de novo mutations with multiple reference sequences. Genome Res. 2020;30:1154–1169. doi: 10.1101/gr.255505.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetting-Minesky MP, Makova KD. Mammalian male mutation bias: impacts of generation time and regional variation in substitution rates. J Mol Evol. 2006;63:537–544. doi: 10.1007/s00239-005-0308-8. [DOI] [PubMed] [Google Scholar]
- Haldane HBS (1946) The mutation rate of the gene for haemophilia, and its segregation ratios in males and females. Ann Eugen 13(1):262-271 [DOI] [PubMed]
- Harland C, Charlier C, Karim L, Cambisano N, Deckers M, Mni M et al. (2017) Frequency of mosaicism points towards mutation-prone early cleavage cell divisions in cattle. bioRxiv
- Harris K, Pritchard JK (2017) Rapid evolution of the human mutation spectrum. Elife 6:e24284 [DOI] [PMC free article] [PubMed]
- Heath TA, Huelsenbeck JP, Stadler T. The fossilized birth-death process for coherent calibration of divergence-time estimates. Proc Natl Acad Sci U S A. 2014;111:E2957–2966. doi: 10.1073/pnas.1319091111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera JP, Davalos LM. Phylogeny and divergence times of lemurs inferred with recent and ancient fossils in the tree. Syst Biol. 2016;65:772–791. doi: 10.1093/sysbio/syw035. [DOI] [PubMed] [Google Scholar]
- Howell N, Smejkal CB, Mackey DA, Chinnery PF, Turnbull DM, Herrnstadt C. The pedigree rate of sequence divergence in the human mitochondrial genome: there is a difference between phylogenetic and pedigree rates. Am J Hum Genet. 2003;72:659–670. doi: 10.1086/368264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozer C, Pifferi F, Aujard F, Perret M. The biological clock in gray mouse lemur: adaptive, evolutionary and aging considerations in an emerging non-human primate model. Front Physiol. 2019;10:1033. doi: 10.3389/fphys.2019.01033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DG, Green P. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci U S A. 2004;101:13994–14001. doi: 10.1073/pnas.0404142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson H, Sulem P, Kehr B, Kristmundsdottir S, Zink F, Hjartarson E, et al. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature. 2017;549:519–522. doi: 10.1038/nature24018. [DOI] [PubMed] [Google Scholar]
- Kappeler PM. Intrasexual selection and testis size in strepsirhine primates. Behav Ecol. 1997;8:10–19. doi: 10.1093/beheco/8.1.10. [DOI] [Google Scholar]
- Keightley PD, Pinharanda A, Ness RW, Simpson F, Dasmahapatra KK, Mallet J, et al. Estimation of the spontaneous mutation rate in Heliconius melpomene. Mol Biol Evol. 2015;32:239–243. doi: 10.1093/molbev/msu302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Petko Z, Dzieciatkowski S, Lin L, Ghiassi M, Stain S, et al. CpG island methylation of genes accumulates during the adenoma progression step of the multistep pathogenesis of colorectal cancer. Genes Chromosomes Cancer. 2006;45:781–789. doi: 10.1002/gcc.20341. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Kimura M, Ohta T. On the rate of molecular evolution. J Mol Evol. 1971;1:1–17. doi: 10.1007/BF01659390. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Miura F, Imai M, Mochiduki K, Yanagisawa E, et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res. 2013;23:616–627. doi: 10.1101/gr.148023.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch E, Schweizer RM, Schweizer TM, Stahler DR, Smith DW, Wayne RK et al. (2019) De novo mutation rate estimation in wolves of known pedigree. Mol Biol Evol 36(11):2536-2547 [DOI] [PMC free article] [PubMed]
- Kondrashov FA, Kondrashov AS. Measurements of spontaneous rates of mutations in the recent past and the near future. Philos Trans R Soc Lond, B. 2010;365:1169–1176. doi: 10.1098/rstb.2009.0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasovec M, Sanchez-Brosseau S, Piganeau G. First estimation of the spontaneous mutation rate in diatoms. Genome Biol Evol. 2019;11:1829–1837. doi: 10.1093/gbe/evz130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley CH, Fitch WM. An examination of the constancy of the rate of molecular evolution. J Mol Evol. 1974;3:161–177. doi: 10.1007/BF01797451. [DOI] [PubMed] [Google Scholar]
- Larsen PA, Harris RA, Liu Y, Murali SC, Campbell CR, Brown AD, et al. Hybrid de novo genome assembly and centromere characterization of the gray mouse lemur (Microcebus murinus) BMC Biol. 2017;15:110. doi: 10.1186/s12915-017-0439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 2013;62:611–615. doi: 10.1093/sysbio/syt022. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Rodrigue N, Thorne JL. Relaxing the molecular clock to different degrees for different substitution types. Mol Biol Evol. 2015;32:1948–1961. doi: 10.1093/molbev/msv099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay SJ, Rahbari R, Kaplanis J, Keane T, Hurles ME. Similarities and differences in patterns of germline mutation between mice and humans. Nat Commun. 2019;10:4053. doi: 10.1038/s41467-019-12023-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H, Winter DJ, Chang AY, Sung W, Wu SH, Balboa M, et al. Low base-substitution mutation rate in the germline genome of the ciliate Tetrahymena thermophil. Genome Biol Evol. 2016;8:3629–3639. doi: 10.1093/gbe/evw286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallon SA, Sanderson MJ. Angiosperm divergence times: the effect of genes, codon positions, and time constraints. Evolution. 2005;59:1653–1670. doi: 10.1554/04-565.1. [DOI] [PubMed] [Google Scholar]
- Martin HC, Batty EM, Hussin J, Westall P, Daish T, Kolomyjec S, et al. Insights into platypus population structure and history from whole-genome sequencing. Mol Biol Evol. 2018;35:1238–1252. doi: 10.1093/molbev/msy041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, et al. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146:1029–1041. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorjani P, Amorim CE, Arndt PF, Przeworski M. Variation in the molecular clock of primates. Proc Natl Acad Sci U S A. 2016;113:10607–10612. doi: 10.1073/pnas.1600374113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorjani P, Gao Z, Przeworski M. Human germline mutation and the erratic evolutionary clock. PLoS Biol. 2016;14:e2000744. doi: 10.1371/journal.pbio.2000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muryas F, Zapata L, Guigo R, Ossowski S. The rate and spectrum of mosaic mutations during embryogenesis revealed by RNA sequencing of 49 tissues. Genome Med. 2020;12:49. doi: 10.1186/s13073-020-00746-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, Bolnick DI, Wainwright PC. Fossil calibrations and molecular divergence time estimates in centrarchid fishes (Teleostei: Centrarchidae) Evolution. 2005;59:8. doi: 10.1111/j.0014-3820.2005.tb01825.x. [DOI] [PubMed] [Google Scholar]
- Ogilvie HA, Bouckaert RR, Drummond AJ. StarBEAST2 brings faster species tree inference and accurate estimates of substitution rates. Mol Biol Evol. 2017;34:2101–2114. doi: 10.1093/molbev/msx126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer SP. Direct estimate of the spontaneous germ line mutation rate in African green monkeys. Evolution. 2017;71:2858–2870. doi: 10.1111/evo.13383. [DOI] [PubMed] [Google Scholar]
- Plummer M, Best N, Cowles K, Vines K. CODA: convergence diagnosis and output analysis for MCMC. R News. 2006;6:7–11. [Google Scholar]
- Poelstra JW, Salmona J, Tiley GP, Schüßler D, Blanco MB, Andriambeloson JB, et al. Cryptic patterns of speciation in cryptic primates: microendemic mouse lemurs and the multispecies coalescent. Syst Biol. 2021;70:203–218. doi: 10.1093/sysbio/syaa053. [DOI] [PubMed] [Google Scholar]
- Rahbari R, Wuster A, Lindsay SJ, Hardwick RJ, Alexandrov LB, Turki SA, et al. Timing, rates and spectra of human germline mutation. Nat Genet. 2016;48:126–133. doi: 10.1038/ng.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu A, Noordam MJ, Schwartz RS, Wuster A, Hurles ME, Cartwright RA, et al. DeNovoGear: de novo indel and point mutation discovery and phasing. Nat Methods. 2013;10:985–987. doi: 10.1038/nmeth.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Dean W. DNA methylation and mammalian epigenetics. Electrophoresis. 2001;22:2838–2843. doi: 10.1002/1522-2683(200108)22:14<2838::AID-ELPS2838>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S et al. (2020) Towards complete and error-free genome assemblies of all vertebrate species. bioRxiv [DOI] [PMC free article] [PubMed]
- Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Scally A. The mutation rate in human evolution and demographic inference. Curr Opin Genet Dev. 2016;41:36–43. doi: 10.1016/j.gde.2016.07.008. [DOI] [PubMed] [Google Scholar]
- Scally A, Durbin R. Revising the human mutation rate: implications for understanding human evolution. Nat Rev Genet. 2012;13:745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- Segurel L, Wyman MJ, Przeworski M. Determinants of mutation rate variation in the human germline. Annu Rev Genom Hum Genet. 2014;15:47–70. doi: 10.1146/annurev-genom-031714-125740. [DOI] [PubMed] [Google Scholar]
- Setash CM, Zohdy S, Gerber BD, Karanewsky CJ. A biogeographical perspective on the variation in mouse lemur density throughout Madagascar. Mammal Rev. 2017;47:212–229. doi: 10.1111/mam.12093. [DOI] [Google Scholar]
- Shimmin LC, Chang BH-J, Li W-H. Male-driven evolution of DNA sequences. Nature. 1993;362:745–747. doi: 10.1038/362745a0. [DOI] [PubMed] [Google Scholar]
- Smeds L, Qvarnstrom A, Ellegren H. Direct estimate of the rate of germline mutation in a bird. Genome Res. 2016;26:1211–1218. doi: 10.1101/gr.204669.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TCA, Arndt PF, Eyre-Walker A. Large scale variation in the rate of germ-line de novo mutation, base composition, divergence and diversity in humans. PLoS Genet. 2018;14:e1007254. doi: 10.1371/journal.pgen.1007254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer MS, Meredith RW, Gatesy J, Emerling CA, Park J, Rabosky DL, et al. Macroevolutionary dynamics and historical biogeogrphy of primate diversification inferred from a species supermatrix. PLoS ONE. 2012;7:e49521. doi: 10.1371/journal.pone.0049521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumoto S, Go Y, Fukuta K, Noguchi H, Hayakawa T, Tomonaga M, Hirai H, Matsuzawa T, Agata K, Fujiyama A (2017) Direct estimation of de novo mutation rates in a chimpanzee parent- offspring trio by ultra- deep whole genome sequencing. Scientific Reports 7(1):13561 [DOI] [PMC free article] [PubMed]
- Thomas GWC, Wang RJ, Puri A, Harris RA, Raveendran M, Hughes DST, et al. Reproductive longevity predicts mutation rates in primates. Curr Biol. 2018;28:3193–3197.e5. doi: 10.1016/j.cub.2018.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- Thorne JL, Kishino H, Painter IS. Estimating the rate of evolution of the rate of evolution. Mol Biol Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- Tiley GP, Poelstra JW, dos Reis M, Yang Z, Yoder AD (2020) Molecular clocks without rocks: new solutions for old problems. Trends Genet 36(11):845-856 [DOI] [PubMed]
- Uchimura A, Higuchi M, Minakuchi Y, Ohno M, Toyoda A, Fujiyama A, et al. Germline mutation rates and the long-term phenotypic effects of mutation accumulation in wild-type laboratory mice and mutator mice. Genome Res. 2015;25:1125–1134. doi: 10.1101/gr.186148.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinform. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venn O, Turner I, Mathieson I, de Groot N, Bontrop R, McVean G. Strong male bias drives germline mutation in chimpanzees. Science. 2014;344:1272–1275. doi: 10.1126/science.344.6189.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RJ, Raveendran M, Harris RA, Murphy WJ, Lyons LA, Rogers J, Hahn MW (2021) De novo mutations in domestic cat are consistent with an effect of reproductive longevity on both the rate and spectrum of mutations. bioRxiv 10.1101/2021.04.06.438608 [DOI] [PMC free article] [PubMed]
- Wang RJ, Thomas GWC, Raveendran M, Harris RA, Doddapaneni H, Muzny DM, et al. Paternal age in rhesus macaques is positively associated with germline mutation accumulation but not with measures of offspring sociability. Genome Res. 2020;30:826–834. doi: 10.1101/gr.255174.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld NI, Kumar V, Shah P, Church DM, Jaffe DB. Direct determination of diploid genome sequences. Genome Res. 2017;27:757–767. doi: 10.1101/gr.214874.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter DJ, Wu SH, Howell AA, Azevedo RBR, Zufall RA, Cartwright RA. accuMUlate: a mutation caller designed for mutation accumulation experiments. Bioinformatics. 2018;34:2659–2660. doi: 10.1093/bioinformatics/bty165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. Covariance between testes size and substitution rates in primates. Mol Biol Evol. 2014;31:1432–1436. doi: 10.1093/molbev/msu091. [DOI] [PubMed] [Google Scholar]
- Wu FL, Strand AI, Cox LA, Ober C, Wall JD, Moorjani P, et al. A comparison of humans and baboons suggests germline mutation rates do not track cell divisions. PLoS Biol. 2020;18:e3000838. doi: 10.1371/journal.pbio.3000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Wang L, Wang L, Wang Z, Lu Z, Tian D, et al. Mutation rate analysis via parent-progeny sequencing of the perennial peach. I. A low rate in woody perennials and a higher mutagenicity in hybrids. Proc Biol Sci. 2016;283:1841. doi: 10.1098/rspb.2016.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang ZH. The BPP program for species tree estimation and species delimitation. Curr Zool. 2015;61:854–865. doi: 10.1093/czoolo/61.5.854. [DOI] [Google Scholar]
- Yang ZH, Yoder AD. Comparison of likelihood and Bayesian methods for estimating divergence times using multiple gene loci and calibration points, with application to a radiation of cute-looking mouse lemur species. Syst Biol. 2003;52:705–716. doi: 10.1080/10635150390235557. [DOI] [PubMed] [Google Scholar]
- Yoder AD, Campbell CR, Blanco MB, Dos Reis M, Ganzhorn JU, Goodman SM, et al. Geogenetic patterns in mouse lemurs (genus Microcebus) reveal the ghosts of Madagascar’s forests past. Proc Natl Acad Sci U S A. 2016;113:8049–8056. doi: 10.1073/pnas.1601081113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Evolution of DMY, a newly emergent male sex-determination gene of medaka fish. Genetics. 2004;166:1887–1895. doi: 10.1093/genetics/166.4.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohdy S, Gerber BD, Tecot S, Blanco MB, Winchester JM, Wright PC, et al. Teeth, sex, and testosterone: aging in the world’s smallest primate. PLoS ONE. 2014;9:e109528. doi: 10.1371/journal.pone.0109528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J Theor Biol. 1965;8:357–366. doi: 10.1016/0022-5193(65)90083-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from the Dryad Digital Repository 10.5061/dryad.8pk0p2njx. Raw sequence data are available through NCBI under BioProject PRJNA512515.