Abstract
Currently, no approved vaccine is available against the Middle East respiratory syndrome coronavirus (MERS-CoV), which causes severe respiratory disease. The spike glycoprotein is typically considered a suitable target for MERS-CoV vaccine candidates. A computational strategy can be used to design an antigenic vaccine against a pathogen. Therefore, we used immunoinformatics and computational approaches to design a multi-epitope vaccine that targets the spike glycoprotein of MERS-CoV. After using numerous immunoinformatics tools and applying several immune filters, a poly-epitope vaccine was constructed comprising cytotoxic T-cell lymphocyte (CTL)-, helper T-cell lymphocyte (HTL)-, and interferon-gamma (IFN-γ)-inducing epitopes. In addition, various physicochemical, allergenic, and antigenic profiles were evaluated to confirm the immunogenicity and safety of the vaccine. Molecular interactions, binding affinities, and the thermodynamic stability of the vaccine were examined through molecular docking and dynamic simulation approaches, during which we identified a stable and strong interaction with Toll-like receptors (TLRs). In silico immune simulations were performed to assess the immune-response triggering capabilities of the vaccine. This computational analysis suggested that the proposed vaccine candidate would be structurally stable and capable of generating an effective immune response to combat viral infections; however, experimental evaluations remain necessary to verify the exact safety and immunogenicity profile of this vaccine.
Subject terms: Computational biology and bioinformatics, Drug discovery
Introduction
Middle East respiratory syndrome coronavirus (MERS-CoV) originated in bats and causes an acute, infectious, viral respiratory disease characterized by symptoms including cough, fever, diarrhea, and occasional multi-organ failure1,2. MERS-CoV is covered by a single-stranded RNA and belongs to the Beta-coronavirus (β-CoV) genus in the family of Coronaviridae, which is distinct from the coronaviruses that cause the common cold coronavirus and severe acute respiratory syndrome (SARS)3–5. MERS-CoV was first identified in Saudi Arabia in 2012 and later spread to European countries. MERS-CoV is associated with an unusually high mortality rate of approximately 35%6–8. According to the WHO report released in May 2018, a total of 2,220 people contracted MERS-CoV, including 790 deaths, resulting in a mortality rate of 35.6%. In Saudi Arabia alone, 1,844 cases were identified, resulting in 716 deaths. In 2015, the South Korean population also became infected with this acute virus, resulting in 186 cases and 36 deaths5,9.
The genome for MERS-CoV encodes envelope (E), membrane (M), nucleocapsid (N), and spike (S) structural proteins, which are necessary to complete the structure of this viral particle10,11. The E protein is typically expressed when cells become infected and is distributed within the intracellular membranes of the endoplasmic reticulum (ER) and Golgi compartments12–14. The N protein binds to the genome of the CoV-RNA, forming the nucleocapsid15, whereas the M protein defines the shape of the viral envelope16. The spike (S) glycoprotein is another important protein because it is involved in host recognition and facilitates host cell entry17,18. The spike protein consists of S1 and S2 subunits19,20. To initiate infection, the S1 subunit binds to the dipeptidyl peptidase 4 (DPP4) receptor on the host cell surface19,20,21, and the S2 subunit mediates the fusion of the viral and host cell membranes20. As of 2020, no specific vaccines against MERS-CoV are available. The S glycoprotein has been considered to be a potential vaccine candidate because neutralizing antibodies against this protein would block viral entry and prevent viral infection23–25.
Using conventional methods, vaccines are designed using large proteins, and the use of inappropriate antigens can increase the potential for allergic reactions; however, a peptide-based multi-epitope vaccine that contains short antigenic peptide fragments, referred to as epitopes, might be able to overcome these limitations26. Epitopes, the antigenic portion of the pathogen that is recognized by the host immune system, and innate immunity are elicited against it27. The cell-mediated immune response is mainly dependent on the pattern recognition receptors recognizing the pathogen-associated molecular patterns of the pathogen28. Toll-like receptors are considered pathogen recognition receptors (PRRs), TLRs family consists of eleven proteins, and each of them uniquely interacts with diverse PAMPs, they are expressed on the surface of the cells29. Toll-like receptor-2 can recognize viral structural glycoproteins. The TLR4 has an essential role during host pathogenesis that can trigger the anti-viral host defense mechanisms against coronavirus30. After infection, cytotoxic T-cell lymphocytes (CTLs) become activated and kill infected cells31. Antigens bound to the major histocompatibility complex (MHC) are presented on the infected cell surface, allowing them to be recognized by CTLs32. MHC class I molecules exhibit cytosolic peptides antigens of the infected cells and phagocytosed antigens are presented on MHC class II molecules33–36. CD4 + T cells recognize the antigenic peptides that are displayed by class II MHC molecules whereas cytotoxic T-cell lymphocyte (CTL)-interact with class I MHC-peptide complexes37. Activated CD4 + T cells secrete cytokines and are responsible for the further activation of B cells required for producing proper antibodies38.
In this study, an immunoinformatics strategy was applied in a sequential manner to design a multi-epitope subunit vaccine to target the MERS-CoV S glycoprotein. This epitope-based vaccine prediction was designed to be antigenic and non-allergenic in nature. The final vaccine contained cytotoxic T-cell lymphocyte (CTL)-, helper T-cell lymphocyte (HTL)-, and interferon-γ (IFN-γ)-inducing and B-cell epitope sequences. Additional physicochemical, molecular docking and thermodynamic stability profiling were assessed to evaluate the safety and efficiency of the vaccine (Fig. 1).
Figure 1.
Graphical representation of the overall systematic study which shown: selection sequence (antigenic proteins) that suitable for phylogenetic analysis, epitope prediction from the goal protein, construction of vaccine, prediction of B cell epitope, feature assessment of target vaccine. Molecular docking of the vaccine with TLR-2 and TLR-4 immune receptor. Molecular dynamic simulations to estimate the stability of docked complexes. Finally, immune stimulation was employed to understand the immune efficiency of the target vaccine.
Results
Sequences retrieval and analysis of phylogenetic tree
A phylogenetic tree was built using all of the spike glycoprotein sequences obtained from various MERS-CoV isolates identified in different countries (Fig. 2). The results showed that all of the S proteins assembled together into a single clade and were closely related to one another. Therefore, a MERS-CoV vaccine that was designed against one single strain could potentially be effective against all of the strains.
Figure 2.
MERS-CoV spike glycoprotein that was taken from 11 different countries (Saudi Arabia, Lebanon, Yemen, Qatar, Egypt, Oman, Iran, United Arab Emirates, Jordan, England, and South Korea) and their phylogenetic analysis. The phylogenetic tree was constructed using the Maximum likelihood method with 1000 bootstrap replicates.
Antigenic and physicochemical evaluations of the target protein
The VaxiJen v2.0 predicted score was 0.4919, which signified that the sequence was potently antigenic. The ExPASy tool in ProtParam indicated that the virulent target protein being used in this study has an instability index score of 36.53, an aliphatic index score of 83.00, and a negative grand average of hydropathicity (GRAVY) value of − 0.064 (Supplementary Table 1). The targeted spike glycoprotein contains 1353 amino acids with the molecular weight of 149,505.26 kDa which specifies the sequences to be antigenic. The estimated half-life was computed to be 30 h for mammalian-reticulocytes, > 20 h for yeast, > 10 h for Escherichia coli.
The prediction of T-cell epitopes and assessment
The CTL epitopes can be used to eliminate virus-infected cells, generate cellular immunity, and reduce the level of circulating virus. In contrast, HTL epitopes can develop both cellular and humoral immune responses and have the ability to activate B-cells to produce antibodies39. Therefore, an effective vaccine should contain receptor-specific cytotoxic T-cell lymphocyte, helper T-cell lymphocyte epitopes. In this study, the NetCTL-1.2 server and the Infectious Disease Epidemiology Bureau (IDEB) stabilized matrix method (SMM) were employed for the prediction of the CTL epitope, whereas the HTL epitope was predicted using the Net MHCII pan 3.2 server (Supplementary Tables 2 and 3). In addition, the predicted sequences were subjected to numerous immune filters, including antigenicity, high-binding affinity, non-allergenicity, and promiscuously towards MHC (MHC-I and MHC-II) alleles. Finally, the best candidate was selected, as shown in Tables 1, 2, and Supplementary Table 4.
Table 1.
Selected promiscuous cytotoxic T-lymphocyte epitope.
| Epitopes | Position | HLA class 1 supertypes (Combined score) | MHC Class 1 alleles | IC50 | Antigenicity |
|---|---|---|---|---|---|
| ATDCSDGNY | 211 | A1 (3.5657) | HLA-A*01:01 | 102.93 | 0.7838 |
| HLA-C*12:03 | 6.55 | ||||
| HLA-C*05:01 | 31.89 | ||||
| HLA-C*14:02 | 205.8 | ||||
| HLA-B*15:02 | 82.46 | ||||
| HLA-A*30:02 | 83.84 | ||||
| KLQPLTFLL | 317 |
A2 (1.4454) A3 (0.7613) A24 (0.7629) B8 (0.8034) |
HLA-A*02:01 | 18.79 | 1.14 |
| HLA-A*32:01 | 50.22 | ||||
| HLA-A*02:06 | 70.07 | ||||
| HLA-C*12:03 | 94.02 | ||||
| HLA-C*07:02 | 123.8 | ||||
| LVRSESAAL | 1086 |
B7 (1.5250) B8 (1.2136) B62 (0.9772) |
HLA-C*12:03 | 84.41 | 0.41 |
| HLA-B*07:02 | 134.24 | ||||
| HLA-C*14:02 | 119.57 | ||||
| HLA-A*30:01 | 214.77 | ||||
| HLA-C*03:03 | 241.33 | ||||
| MLKRRDSTY | 696 |
A1 (0.8621) A3 (0.9329) B8 (1.7688) B62 (1.4024) |
HLA-A*30:02 | 117.88 | 0.90 |
| HLA-B*08:01 | 55.78 | ||||
| HLA-B*15:01 | 63.24 | ||||
| HLA-C*12:03 | 6.67 | ||||
| HLA-C*14:02 | 153.33 | ||||
| RRDSTYGPL | 699 |
B27 (1.5815) B39 (1.1004) |
HLA-C*07:02 | 41.60 | 1.42 |
| HLA-C*12:03 | 17.19 | ||||
| HLA-C*14:02 | 66.93 | ||||
| HLA-B*39:01 | 80.43 | ||||
| HLA-B*27:05 | 173.61 | ||||
| LSIPTNFSF | 780 |
B58 (2.112) B62 (1.39) |
HLA-B*58:01 | 19.24 | 1.127 |
| HLA-B*15:01 | 44.15 | ||||
| HLA-A*23:01 | 194.72 | ||||
| HLA-C*03:03 | 49.73 | ||||
| HLA-C*12:03 | 56.52 | ||||
| HLA-B*57:01 | 236.49 | ||||
| HLA-B*35:01 | 156.16 | ||||
| FSFGVTHEY | 786 |
A1 (2.44) A3 (0.8719) A26 (1.4849) B62 (1.5080) B58 (1.71) |
HLA-B*35:01 | 11.92 | 1.79 |
| HLA-B*58:01 | 94.25 | ||||
| HLA-A*30:02 | 131.05 | ||||
| HLA-A*01:01 | 230.96 | ||||
| HLA-C*07:01 | 21.94 | ||||
| HLA-C*12:03 | 9.27 | ||||
| HLA-A*29:02 | 25.22 | ||||
| HLA-B*46:01 | 47.2 | ||||
| HLA-A*68:01 | 93.86 | ||||
| HLA-B*15:01 | 159.59 |
The epitopes listed in the table are non-allergic, non-toxic, and displayed 100% conservancy among target protein sequence (as predicted using the IDEB conservancy tool). Epitopes with half-maximal inhibitory concentration (IC50) values below 250 were considered to bind suitably with respective human leukocyte antigen (HLA) alleles. The VaxiJen v2.0 server was utilized at 0.4 thresholds to predict the antigenicity of the epitopes.
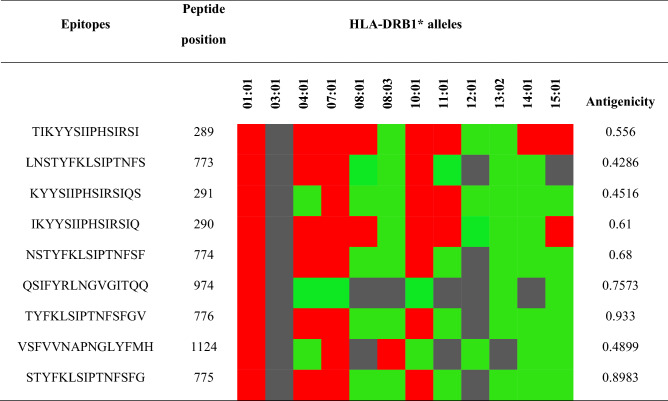
Table 2.
Selected helper T-lymphocyte epitopes, as predicted by the NetMHC II pan 3.2 server. The strong, intermediate, and non-binding affinities are represented by red, green, and black boxes, respectively. The epitope antigenicity was predicted by the VaxiJen v2.0 server at the 0.4-threshold level. The epitopes are promiscuous, non-allergic, non-toxic, and displayed 100% conservancy with the target sequence.
Vaccine construction, structure modeling, refinement, and validation
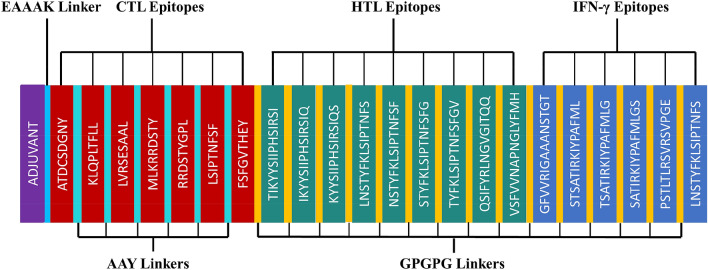
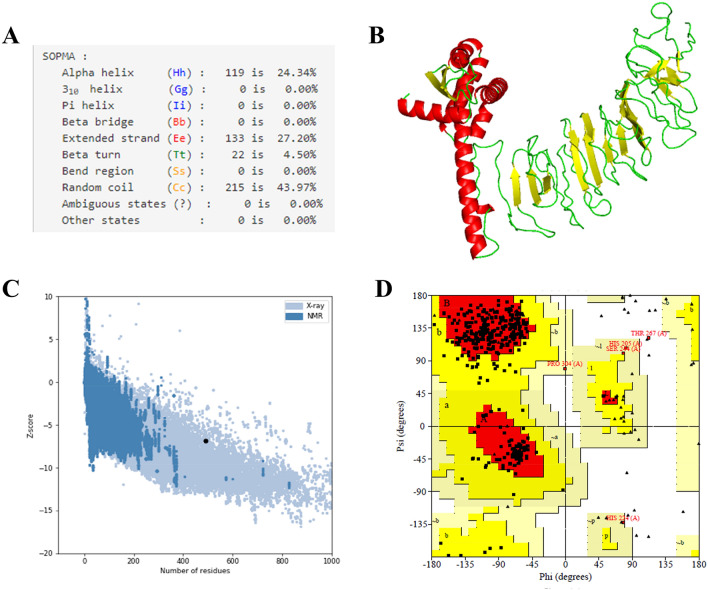
The criteria used for epitope selection during the construction of the linear vaccine included: (a) antigenic and non-allergic; (b) high-binding affinity with MHC alleles; (c) promiscuous; (d) containing overlapping CTL and HTL epitopes; (e) 100% conserved across S proteins; and (f) no overlapping capacity with any component of the human proteome. Epitopes that featured these properties were used to construct a linear vaccine sequence, comprising 7 CTL, 9 HTL, and 6 IFN-γ epitopes (Tables 1, 2, and Supplementary Tables 4–7). In addition, AYY and GPGPG linkers were added to the construction to prevent junctional epitope formation, facilitate the immune processing of the antigens and allow effective separation of each epitope within the human body40. To improve immunogenicity, cholera toxin subunit B (CTB) was added as an adjuvant attached to the N-terminal end (Fig. 3). The finalized vaccine sequence featured a molecular weight of 51.8 kDa and consisted of 489 amino acids (Supplementary Material SM1). The selected epitopes included in the vaccine were visualized on the 3D model of the S glycoprotein, which displayed the epitopes retaining their original positions (Fig. 4). The secondary structure prediction for the vaccine suggested that it would contain 24.34% alpha-helical regions, 27.20% extended strands, 4.50% beta turns, and 43.97% random coils (Fig. 5A). The 3D model of the finalized vaccine was prepared by the trRosetta web server which displayed five different models as per their Z-score ranging from 1.74 to 3.03 where the highest TM-score containing model (0.61 ± 0.05) was subjected to refinement. TM-score measures the similarity between two structures. A TM-score is higher than 0.5 is implying that a model of correct topology and less than 0.17 indicates random similarity41. Among all of the refined 3D structures, model 1 was selected which was predicted to be the best based on different parameters including MolProbity (2.727), GDT-HA (0.9059), and RMSD (0.519) (Fig. 5B and Supplementary Table 8). The poor rotamers and clash score of the finalized model were predicted to be 2.1 and 34.1, respectively. The global distance test-high accuracy (GDT-HA) is a global measure agreements between the experimental model and a predicted protein structure42. The Z-score of the finalized vaccine structure was − 6.85, as predicted by Pro-SA web (Fig. 5C), and the score was within the range for comparably-sized proteins, which indicated that the predicted model was reliable43. The results of the Ramachandran plot analysis showed that 87%, 12%, 0.8%, and 0.3% residues were in favoured, additionally allowed, allowed, and disallowed regions, respectively (Fig. 5D and Supplementary Materials SM2). The ERRAT value of the structure was predicted to be 56.017 (Supplementary Fig. S1). The ERRAT score higher than 50 indicating the good quality of a protein model44.
Figure 3.
Schematic representation of the finalized vaccine construct that shows 489-amino acid long and containing CTL, HTL, and IFN-γ inducing epitopes, an adjuvant (violet) linked in the constructs via EAAAK linker (blue) at the end of N-terminal. CTL epitopes are linked with AAY linkers (green). HTL and IFN- γ epitopes are linked through GPGPG linkers (yellow).
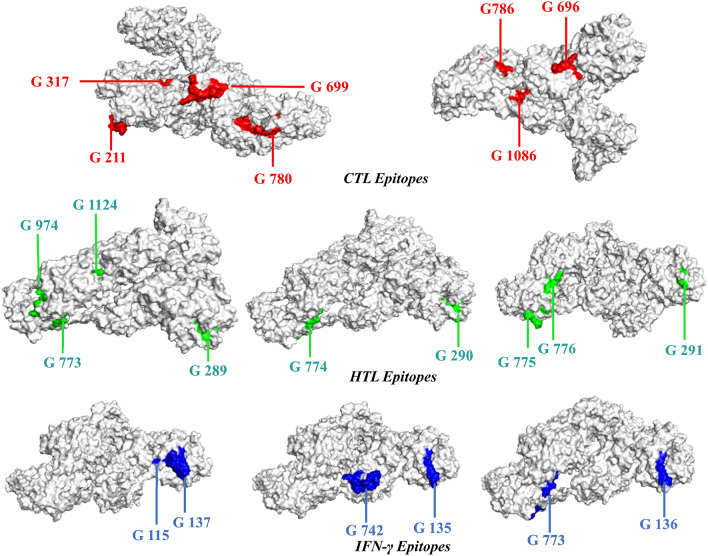
Figure 4.
The surface view of the tertiary structure of spike glycoprotein with epitopes that are involved in the vaccine construct. The selected CTL, HTL, and IFN-γ epitope denoted in the structure by red, green, and blue colors, respectively.
Figure 5.
(A) The secondary structural properties of the vaccine. (B) 3-D model of the final vaccine. In this structure, red, yellow, and green color represents the helical, sheet, and loop regions, respectively. (C) 3-D structure validation with a Z-score of -6.85 followed by Pro-SA. (D) Analysis of Ramachandran plot utilizing PROCHECK server showed 87%, 12%, 0.8%, and 0.3% residues laying in favoured, additional allowed, allowed, and disallowed regions, respectively.
Assessment of the antigenic, physicochemical, and allergenic profiles of the vaccine construct
The VaxiJen v2.0 server predicted a score of 0.5134, which confirms that the candidate vaccine is antigenic. Allergenicity was analyzed by AllerTOP v.2.0 and the AllergenFP v.1.0 server and was determined to be non-allergenic (Supplementary Fig. S2). The physicochemical properties of a vaccine must be determined to assess efficiency and safety45; therefore, several physical and chemical parameters of the vaccine construct were evaluated by ExPASy (Supplementary Materials SM3). The aliphatic index and theoretical pI values were found to be 71.10 and 9.74, respectively, which suggested that the candidate is thermostable. The estimated half-life was evaluated to be 30 h in mammalian reticulocytes, > 20 h in yeast, > 10 h in E. coli, The Grand average hydrophathicity (GRAVY) score is − 0.130 which specifies the candidate is hydrophilic in nature and has the ability to interact with the aqueous environment. The instability index was calculated as 26.58, which classified the protein as being stable46. In addition, our designed vaccine does not contain any signal peptides that would either specify or inhibit protein localization (Supplementary Fig. S3). The TMHMM server predicted that no production difficulties would be associated with expression (Supplementary Fig. S4).
Post-translational modification analysis
A wide range of post-translational modifications was predicted in both targeted spike glycoprotein and the finalized vaccine construct. Analyzation of glycosylation site within the target protein indicated that there are two N-glycosylation sites were existing (position 6 and 244) and one O-GlcNAc site was found in 878 positions (Supplementary Figs. S5 and S6) which specifies the epitopes which are used in vaccine designing are not overlapping in those regions (PTM sites). Additionally, there was no lipid PTMs as N-terminal glycines myristoyl and GPI-modification was predicted in the finalized vaccine construct. Prediction of phosphorylation modification displayed 45 sites (Ser: 24, Thr: 16, Tyr: 5) in the construct. The YinOYang server predicted one O_GlcNAc attachment site to exist in the vaccine construct and there is no N-glycosylation and N-acetylation was found by NetNGlyc 1.0 server and NetAct 1.0 server, respectively (Supplementary Table 9).
Prediction of the B-cell epitope
B-cells have significant roles in humoral immunity through the secretion of antibodies. A B-cell epitope can be recognized by the B-cell receptor, providing long-term immunity. The Ellipro server result included 10 linear and four conformational B-cell epitopes (Tables 3 and 4). PyMOL was used to visualize the mapped epitopes in the vaccine construct (Supplementary Figs. S7 and S8).
Table 3.
Linear B-cell epitopes in the vaccine construct.
| No | Start | End | Peptide | No. of residues | Score |
|---|---|---|---|---|---|
| 1 | 427 | 489 | MLGGPGPGSATIRKIYPAFMLGSGPGPGPSTLTLRSVRSVPGEGPGPGLNSTYFKLSIPTNFS | 63 | 0.825 |
| 2 | 90 | 127 | NNKTPHAIAAISMANEAAAKATDCSDGNYAAYKLQPLT | 38 | 0.757 |
| 3 | 18 | 59 | IHTLNDKIFSYTESLAGKREMAIITFKNGATFQVEVPGSQHI | 42 | 0.753 |
| 4 | 145 | 170 | YMLKRRDSTYAAYRRDSTYGPLAAYL | 26 | 0.744 |
| 5 | 184 | 198 | FGVTHEYGPGPGTIK | 15 | 0.718 |
| 6 | 80 | 84 | EAKVE | 5 | 0.602 |
| 7 | 248 | 257 | IQSGPGPGLN | 10 | 0.597 |
| 8 | 385 | 398 | ANSTTGPGPGSTSA | 14 | 0.59 |
| 9 | 269 | 277 | FSGPGPGNS | 9 | 0.558 |
| 10 | 411 | 421 | PGPGTSATIRK | 11 | 0.544 |
Table 4.
ElliPro server predicted a total of 259 residues, located in four discontinuous B-cell epitopes.
| No | Residues | No. of residues | Score |
|---|---|---|---|
| 1 | A:M1, A:T2, A:P3, A:Q4, A:N5, A:I6, A:T7, A:D8, A:L9, A:C10, A:A11, A:H14, A:Q17, A:H19, A:T20, A:L21, A:N22, A:D23, A:K24, A:I25, A:F26, A:S27, A:Y28, A:T29, A:E30, A:S31, A:L32, A:A33, A:G34, A:K35, A:R36, A:E37, A:M38, A:A39, A:I40, A:I41, A:T42, A:F43, A:K44, A:N45, A:G46, A:A47, A:T48, A:F49, A:Q50, A:V51, A:E52, A:V53, A:P54, A:G55, A:S56, A:Q57, A:H58, A:I59, A:Q62, A:Y77, A:E80, A:A81, A:K82, A:V83, A:E84, A:C87, A:N90, A:N91, A:K92, A:T93, A:P94, A:I97, A:A98, A:A99, A:I100, A:S101, A:M102, A:A103, A:N104, A:E105, A:A106, A:A107, A:A108, A:K109, A:A110, A:T111, A:D112, A:C113, A:S114, A:D115, A:G116, A:N117, A:Y118, A:A119, A:A120, A:Y121, A:K122, A:L123, A:Q124, A:P125, A:L126, A:T127, A:T174 | 99 | 0.74 |
| 2 | A:A385, A:N386, A:S387, A:T388, A:T389, A:G390, A:P391, A:G392, A:P393, A:G394, A:S395, A:T396, A:S397, A:A398, A:T399, A:I400, A:R401, A:K402, A:P411, A:G412, A:P413, A:G414, A:T415, A:S416, A:A417, A:T418, A:I419, A:R420, A:I422, A:Y423, A:P424, A:M427, A:L428, A:G429, A:G430, A:P431, A:G432, A:P433, A:G434, A:S435, A:A436, A:T437, A:I438, A:R439, A:K440, A:I441, A:Y442, A:P443, A:A444, A:F445, A:M446, A:L447, A:G448, A:S449, A:G450, A:P451, A:G452, A:P453, A:G454, A:P455, A:S456, A:T457, A:L458, A:T459, A:L460, A:R461, A:S462, A:V463, A:R464, A:S465, A:V466, A:P467, A:G468, A:E469, A:G470, A:P471, A:G472, A:P473, A:G474, A:L475, A:N476, A:S477, A:T478, A:Y479, A:F480, A:K481, A:L482, A:S483, A:I484, A:P485, A:T486, A:N487, A:F488, A:S489 | 94 | 0.734 |
| 3 | A:Y145, A:M146, A:R149, A:R150, A:D151, A:S152, A:T153, A:Y154, A:A155, A:A156, A:Y157, A:R158, A:R159, A:D160, A:S161, A:T162, A:Y163, A:G164, A:P165, A:L166, A:A167, A:A168, A:Y169, A:L170, A:F184, A:G185, A:V186, A:T187, A:H188, A:E189, A:Y190, A:G191, A:P192, A:G193, A:P194, A:G195, A:T196, A:I197, A:K198, A:G213, A:P214, A:G215, A:I216, A:K217, A:P234, A:G235, A:K236 | 47 | 0.696 |
| 4 | A:I248, A:Q249, A:S250, A:G251, A:P252, A:G253, A:P254, A:G255, A:L256, A:N257, A:F269, A:S270, A:G271, A:P272, A:G273, A:P274, A:G275, A:N276, A:S277 | 19 | 0.578 |
Population coverage
Successful HLA allele calculations are necessary to develop a vaccine that will be effective for the entire global population. The IDEB Population Coverage tool indicated that the selected epitope included in our study would cover 100% of the worldwide population (Table 5 and Supplementary Table 10), with 100% coverage in East Asia, South Asia, Southwest Asia, North America, South America, and Europe. This result indicated that our designed vaccine could be used worldwide.
Table 5.
Population coverage of the selected epitope included in the vaccine construct.
| Population/area | Class combined | ||
|---|---|---|---|
| Coveragea | Average_hitb | pc90c | |
| Central Africa | 99.34% | 26.31 | 14.8 |
| East Africa | 99.25% | 25.78 | 14.64 |
| East Asia | 100.0% | 39.72 | 28.42 |
| Europe | 100.0% | 47.54 | 35.91 |
| North Africa | 99.8% | 30.56 | 17.21 |
| North America | 100.0% | 46.02 | 31.76 |
| Northeast Asia | 99.1% | 24.76 | 14.4 |
| Oceania | 99.35% | 24.71 | 14.5 |
| South Africa | 99.82% | 28.55 | 15.78 |
| South America | 100.0% | 31.57 | 21.27 |
| South Asia | 100.0% | 40.36 | 28.65 |
| Southeast Asia | 100.0% | 35.37 | 22.34 |
| Southwest Asia | 98.64% | 23.09 | 14.02 |
| West Africa | 99.55% | 27.53 | 15.21 |
| West Indies | 99.46% | 24.44 | 14.66 |
| World | 100.0% | 44.2 | 30.49 |
| Average | 95.83 | 30.82 | 19.71 |
| Standard deviation | 15.27 | 10.49 | 8.55 |
aCoverage of population on projected.
bPopulation recognized by HLA combinations/epitope hits on the average number.
c90% of the population recognized by HLA combinations/epitope hits on the minimum number.
Analysis of molecular docking
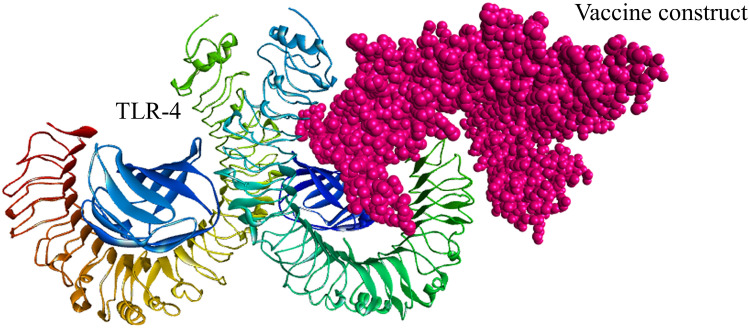
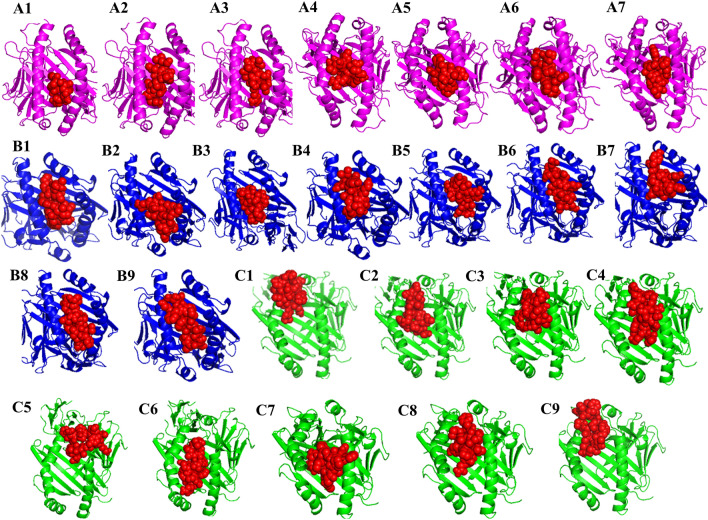
The interaction between immune cells and the vaccine is necessary for the development of stable immune response. Toll-like receptors (TLRs) serve as pathogen detectors and play crucial roles in innate immunity47. TLR2 and TLR4 can recognize viral structural glycoproteins, resulting in the production of inflammatory cytokines48. The molecular interactions pattern and binding affinities between the designed multi-epitope vaccine and the TLR-4 and TLR-2 immune receptors were analyzed via a protein–protein docking approach. Based on the electrostatic complementarity and geometry of the protein surface, the PatchDock server generates the interaction model of a receptor-ligand complex. The refinement of the best complexes was performed using the FireDock tool and found ten complexes for each TLR2-vaccine and TLR4-vaccine. Among the top-ten complexes generated when examining the interaction between the vaccine and TLR-4, solution 10 was characterized as having the best global energy (− 21.16), attractive van der Waals (VdW) energy (− 42.23), repulsive VdW energy (38.54), hydrogen bond (HB) energy (− 6.82), and atomic contact energy (ACE; 9.29) (Fig. 6 and Supplementary Table 11). Similarly, in the docking study between TLR-2 and the vaccine, the global energy (− 22.13), attractive VdW (− 34.45), repulsive VdW (26.29), HB (− 6.78), and ACE (23.41) values for solution 10 were better than those of the other models (Fig. 7 and Supplementary Table 12). In contrast, the selected CTL and HTL epitopes included in the finalized vaccine construct docked individually with three commonly occurring HLA alleles in the human population HLA-A*02:01, HLA-DRB1*01:01 and HLA-DRB1*15:01 (Fig. 8) and also compared the binding affinity with positive and negative control. We found that 7 CTL epitopes efficiently bind with HLA-A*02:01, the best interacting CTL model was KLQPLTFLL (lowest energy -795.6) as compared to the CTL positive (lowest energy -562.6) and negative (lowest energy 756.5) control (Supplementary Table 13). On the other hand, molecular interaction studies between HTL epitopes and HLA-DRB1 alleles have shown a strong binding affinity. The nine HTL epitopes were docked individually with HLA-DRB1*01:01 and HLA-DRB1*15:0, the best model was NSTYFKLSIPTNFSF, which shows the lowest energy (-987.0 with HLA-DRB1*01:01 and -1125.6 with HLA-DRB1*15:0) compared to the positive and negative control (Supplementary Table 14). Thus, these T-cell epitopes ensured their suitable binding affinity to be used in multi-epitope vaccine design against MERS-CoV.
Figure 6.
The docked complex of the designed multi-epitope vaccine protein and TLR-4 immune receptor. The vaccine construct has been shown in CPK molecular model (pink) in the figure and the global energy value of the finalized docked complex was − 21.16 kcal/mol.
Figure 7.
The docked complex of the designed multi-epitope vaccine protein and TLR-2 immune receptor. The vaccine construct has been shown in CPK molecular model (pink) in the figure. The global energy of the finalized docked complex was − 22.13 kcal/mol as predicted by the PatchDock server.
Figure 8.
The interaction patterns of selected epitopes included in the vaccine constructs with HLA alleles. A1–A7 are the binding pattern of 7 CTL epitopes (red) with HLA-A*02:01 allele (magentas). The interaction pattern of 9 HTL epitopes (red) with the HLA-DRB1*01:01 (blue) allele has been shown in figure B1–B9. Figures C1–C9 represents the binding pattern of 9 HTL epitopes with HLA-DRB1*15:01 allele (green).
Analysis of molecular dynamics and simulation
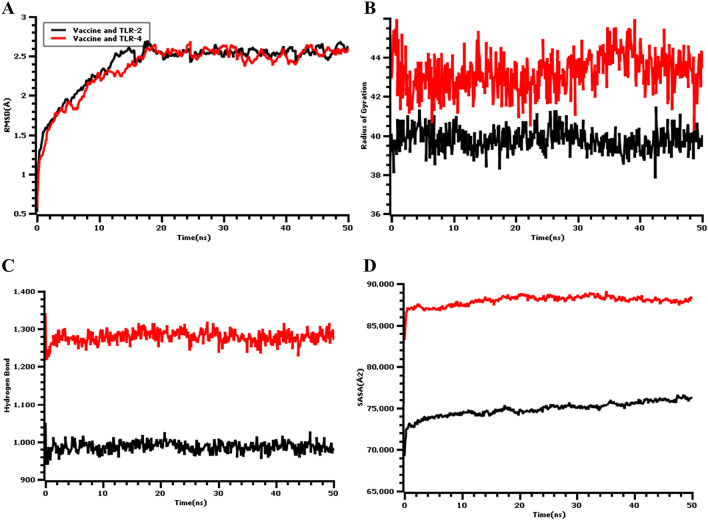
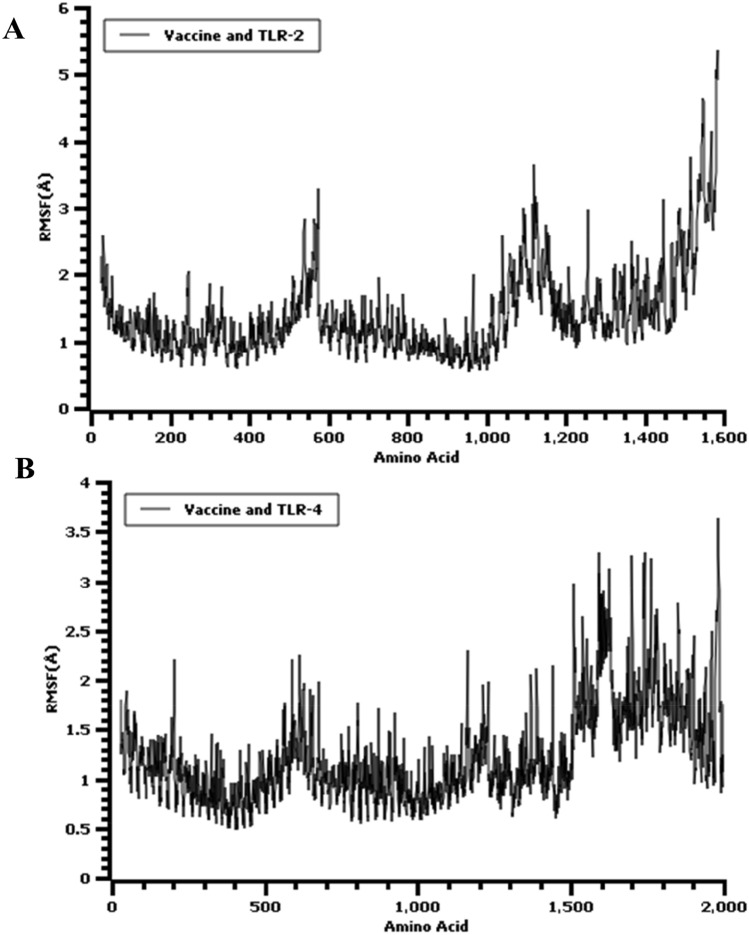
The root means square deviation (RMSD) of the c-alpha atoms of the vaccine complex was calculated for two vaccine complexes. The average RMSD values for the vaccine and TLR-2 complex and for the vaccine and TLR-4 complex were 2.043 Å and 2.357 Å, respectively, which demonstrates the stable nature of both complexes. As shown in Fig. 9A, both the vaccine and TLR-2 and vaccine and TLR-4 complexes experience an initial increase in RMSD descriptors until the 15 ns time point, after which the upward trend ceases. A low degree of fluctuation was observed for both two complexes, which may be responsible for structural integrity. Therefore, the solvent-accessible surface area (SASA) values for both complexes were analyzed to understand changes in protein volume, based on the simulation trajectories. The average SASA value of the vaccine and TLR-2 complex was found to be 74,896.758Å2, which is similar to the entire SASA profile (Fig. 9D) because no deviations were found. In contrast, the vaccine and TLR-4 complex had an average SASA value of 87,958.090Å2. Both simulated complexes displayed an initial rise in the SASA values, which indicated the expansion of the protein volume during the initial phase. The radius of gyration (Rg) for the simulation trajectory provides information regarding the compressed nature of the protein, in which a higher Rg profile denotes less rigidity in the biological system. As shown in Fig. 9B, the Rg profile of the two vaccine complexes could be demonstrated, and an initial increase in the Rg profile for the vaccine and TLR-2 complex was observed. Therefore, the Rg descriptor trend for the vaccine and TLR-2 complex was similar from 0 to 50 ns, although a few fluctuations were observed. In contrast, the Rg value of the vaccine and TLR-4 complex was similar until 35 ns, followed by a larger deviation observed from 35–38 ns, which might be responsible for the loose packaging of the system. The vaccine molecules and their degrees of stability were further assessed by examining the total hydrogen bond number during the entire simulation period. The formation of hydrogen bonds and the number of substantial changes in the simulated complexes can be used to define the rigidity of the complexes. The vaccine and TLR-2 and the vaccine and TLR-4 complexes both featured stable numbers of hydrogen bonds, as shown in Fig. 9C, indicating relative complex stability. Moreover, the protein flexibility across the amino acid residues was evaluated by assessing the root mean square fluctuation (RMSF) profile. The RMSF profiles of the TLR-2 and TLR-4 complexes indicated that most of the amino acid residues from both complexes had RMSF profiles below 2.5 Å, and larger changes were only observed for a few residues. This result, as shown in Fig. 10, defines the vaccine complex stability and stiffness. Furthermore, secondary structure content from TLR-4-vaccine, TLR-2, and vaccine complexes was analyzed to understand their structural integrity (Supplementary Fig. S9). The secondary structure content; alpha helix, beta-sheet, coil and turn the region from both biological complexes were steady state in whole molecular dynamics simulation.
Figure 9.
Molecular dynamic simulation result of the multi-epitope vaccine, and TLR-4 (red), TLR-2 (black) complexes at 50 ns. (A) The RMSD plot of the c-alpha atoms of the complexes. The mild fluctuations represent the stability of the complexes. (B) The Rg (Radius of Gyration) plot. (C) The Hydrogen bond (HB) plot of vaccine and TLRs docked complexes. (D) The SASA (Solvent-Accessible Surface Area) profile of the docked complexes.
Figure 10.
The RMSF plot of the multi-epitopic docked vaccine candidate and (A) TLR-2, (B) TLR-4 complexes at 50 ns.
In silico immune simulation
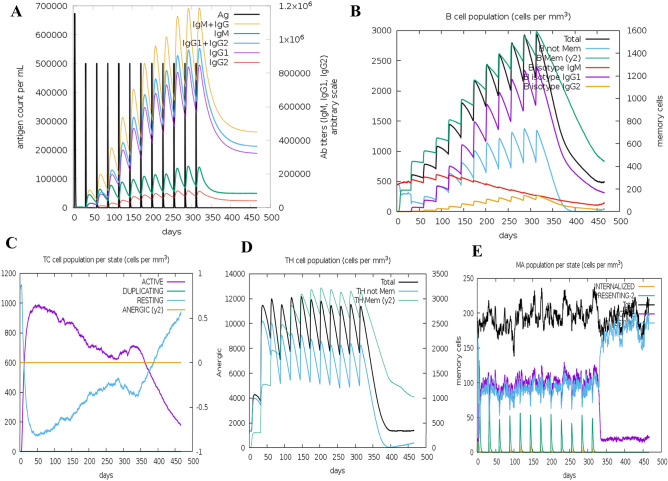
The immunogenic profile of the chimeric peptide vaccine is shown in Fig. 11 and Supplementary Fig. S10. The immune simulation results showed that the secondary and tertiary responses were considerably more immunogenic than the primary response. The antigenic concentration decreased, and the immunoglobulin activity became significantly increased during the secondary and tertiary responses (Fig. 11A). In addition, the formation of multiple B-cell isotypes was predicted (Fig. 11B). A similarly increased response was indicated by HTL and CTL populations during vaccination (Fig. 11C and D). Increased macrophage activity was detected in the form of natural killer (NK) and dendritic cells (Supplementary Fig. S10 and Fig. 11E). High levels of IFN-γ supported the activation of an adequate immune response, and IL-2 secretion was also elicited during the simulation (Supplementary Fig. S10E). This profile demonstrates the development of immune memory subsequently increased.
Figure 11.
The in silico immune simulation profile of vaccine which injected 12 doses over a period of 12 months. (A) shows both antigen concentration and relative antibody responses. The efficacy of the vaccination showed by the presence of protective IgGs. (B) shows the corresponding count of antibody generating plasma cells while (C−E) show the activity of macrophages cytotoxic T, and helper T cells.
Discussion
Although MERS is a highly infectious disease49 with a high mortality rate, no approved vaccines and or treatment drugs are currently available to prevent MERS-CoV infection. Vaccines are urgently necessary for the prevention and treatment of this disease. The application of an immunoinformatics strategy and the associated tools represents a rationally effective approach to the development of a peptide-based vaccine26. This approach is time-consuming, cost-effective, and can allow researchers to predict antigenic epitopes that may be potential candidates for use in an effective multi-epitope vaccine. In this investigation, our aim was to design a peptide-based poly-epitope vaccine against MERS-CoV. By applying a cutting-edge immunoinformatics strategy against the MERS-CoV proteome, the sequence of the most recognizable virulent factor, the S glycoprotein, was targeted. The antibody-mediated neutralization of the viral S protein is a primary goal in vaccine development because the spike glycoprotein is associated with viral attachment and entry to host cells. Here, we constructed a multi-epitope vaccine by using several computational tools. The vaccine was designed to provide immunity based on the use of several small antigenic peptide fragments; in contrast to vaccines that utilize the whole genome or large proteins, this approach does not produce any allergenic reactions in the host50,51. In addition, our designed vaccine has several advantages compared with conventional and single-epitope vaccines owing to the following distinctive characteristics: (a) the presence of multiple MHC epitopes, allowing the vaccine to be recognized by several T-cell receptors; (b) the use of overlapping CD4 + and CD8 + T-cell epitopes; (c) the presence of multiple epitopes from the targeted virulent protein; and (d) the inclusion of an immunostimulator (adjuvant) for producing long term immunity52–55. The design of a vaccine using a similar method has been demonstrated to result in the development of protective efficiency in vivo56–58, and some of these vaccines have entered the clinical trial phase53,59–61. The selection of antigenic epitopes is crucial for vaccine development62. The cytotoxic T-cell lymphocyte, helper T-cell lymphocyte, and interferon-gamma inducing epitopes of the S protein were identified based on the use of several immune filters. Epitopes were screened to identify antigenic but non-allergenic sequences that were capable of interacting with multiple HLA alleles were conserved 100% among the various S protein sequences and did not overlap with any components of the human proteome to reduce the possibility of autoimmunity. Through the application of various in silico analysis techniques, the designed poly-epitope (CTL, HTL, IFN-γ inducing)-containing vaccine was constructed with the addition of an N-terminal-linked Cholera toxin B (CTB) adjuvant (Fig. 3). In addition, GPGPG and AAY linkers were added to the sequences to prevent junctional epitope formation. Adjuvant and epitopes (CTL-HTL-IFN-γ) are ordered in such way have been shown to antigen-specific immune response stimulation reported in many studies63,64, the adjuvant is attached in the N-terminal of the construct because binding of it with TLR displays enhanced results in producing immune-response whereas TLRs associated to the activation of the humoral and cellular immunity65.
Cholera toxin B is potentially used as a viral adjuvant66–68. The use of specialized sequences including linkers has the ability to improve vaccine construct. Previously many studies demonstrated that GPGPG and AAY linkers66,69 were added between predicted HTL and CTL epitopes sequences respectively and produced junctional immunogenicity, consequently allowing the rational design construction of a potent poly-epitope vaccine70. Arai et al. reported that the EAAAK linker was incorporated between epitopes and adjuvant for improving bioactivity of fused protein and reaching a high level of expression and increasing the stability of the vaccine construct71. Similarly, Bazan and colleagues designed a T-cell-based multi-epitope subunit vaccine against the Ebola virus. They used the Immune Epitope Database (IDEB) to predict antigenic epitopes that were used to construct a vaccine candidate that was found to be immunogenic when expressed in mice63.
During vaccine development, allergenicity can be a major problem. In our final vaccine, allergenicity was not detected. Various physicochemical properties were determined using the ProtParam ExPASy tool, which indicated an instability index value of 26.58, which indicated that the vaccine would be stable. The theoretical PI value was calculated as 9.74. The aliphatic index of the vaccine was evaluated to be 71.10, which indicates that the protein would be thermostable. The GRAVY value was calculated to be − 0.130, which suggested that the vaccine can interact with water, has a polar nature, and a high degree of solubility. Foroutan and other researchers designed a vaccine and then validated their candidate through experimental evaluation, reporting that their vaccine was able to induce strong cellular and humoral immune responses in mice64. The aliphatic and instability index values for our designed vaccine were better than those reported for the vaccine candidate designed and tested by Foroutan et al. The Ramachandran plot analysis for the vaccine showed that 87%, 12.0%, 0.8%, and 0.3% residues were found in the favored, allowed, additionally allowed, and disallowed regions, respectively, with an ERRAT value and a Z-score of 56.017 and − 6.85, respectively, which indicates that the protein falls in the plot which consists of the Z-scores of the already determined structures solved by NMR and X-ray crystallographic experiments43. TLR-4 is expressed in monocytes, granulocytes, and immature dendritic and macrophage cells72. The direct interaction between CTB and TLR-4 facilitates the activation of TLR-4 by CTB73. An enzyme-linked immunosorbent assay (ELISA) indicated that CTB is capable of inducing the activation of NF-κB in TLR-4 receptor cells through direct binding73. The viral E glycoprotein can also be recognized by TLR-274. Molecular docking studies were used to analyze the molecular interactions and binding affinity patterns between the vaccine and both TLR-2 and TLR-4. The global energies of the best-docked complexes between the vaccine and TLR-4 and TLR-2 were − 21.16 kj/mol and − 22.13 kJ/mol, respectively, which denoted favorable binding affinities. The molecular dynamics simulation study of the vaccine candidate and TLR-4 and TLR-2 complexes were conducted to confirm their stable nature at atomistic conditions. The simulation data by combining RMSD, RMSF, SASA, Rg descriptors from trajectories correlates with the structural rigidity of the vaccine complexes. The RMSD and RMSF profile of the vaccine candidates were below 2.5 Å for most of the simulation time. These results define the vaccine complexes' integrity and less mobility at the simulation conditions. The immune simulation study indicated that our designed vaccine candidate could likely generate an appropriate immune response during secondary exposure after the final injection (Fig. 11). Many researchers have recently applied similar immunoinformatics approaches to the design of multi-epitopic vaccine candidates against Kaposi’s sarcoma26, Klebsiella pneumoniae75, Pseudomonas aeruginosa76, dengue69, Nipah virus77, SARS-CoV-278, Hendra virus79, and malaria80. A similar strategy has also been used to develop a vaccine against cancerous antigens81. Therefore, the construction of a vaccine using epitopes appears to be capable of inducing the activation of immune cells in the host, which may further trigger other immune cells via a complex signaling pathway.
Materials and methods
Retrieval of protein sequence and the analysis of the phylogenetic tree
The NCBI database was utilized to retrieve the S glycoprotein amino acid sequences for MERS-CoV (Accession no: ANI69889.1), which were stored in a FASTA format. The sequences were aligned using the MUSCLE tool82, and a phylogenetic tree was constructed by using MEGA-X83. The Jones-Taylor-Thornton (JTT) model was used to an estimated algorithm for a matrix of pairwise distances to select superior log-likelihood values.
Antigenic and physiochemical evaluation of the target protein
The prediction of antigenicity is an important step when attempting to isolate the most antigenic protein sequences. The stored protein sequence was submitted to the VaxiJen v2.0 Server84 in plain sequence format to determine antigenicity, using default parameters. The physicochemical properties of the protein were analyzed by the ExPASy ProtParam tool46.
T-cell epitope prediction and assessment
CTL epitope prediction
Cytotoxic (CD8+) T-cell epitopes were predicted by submitting FASTA sequences of the target protein to the NetCTL-1.285 server where thresholds were set for epitope identification, TAP transport efficiency, and proteasomal C-terminal cleavage were 0.75, 0.05, and 0.15, respectively. NetCTL 1.2 can predict epitope from the query sequence on the basis of the training dataset. The CTL epitopes were predicted by recognizing the commonly occurring HLA 12 Class I supertypes and further IEDB SMM method was used to evaluate the binding affinities of the epitopes with MHC class I alleles86. Each of the output epitope from the IDEB server is assigned as IC50 values which characterize the binding affinity of peptide molecules in the MHC allele. The IC50 values < 50 nM specifies high-binding affinity, lesser than 500 nM categorize intermediate affinity, whereas < 5000 nM indicate low affinity66. Epitopes with binding affinities below 250 nM (IC50) were chosen for further analyses.
HTL epitope prediction
The Net MHCII pan 3.287 server was utilized to predict 15-mer-long epitopes capable of recognizing human leukocyte antigen class II DRB1 alleles:01:01, 03:01, 04:01, 07:01, 08:01, 08:03, 10:01, 11:02, 12:01, 13:02, 14:01, and 15:01. The server uses artificial neuron networks to predicts the peptide that binding to HLA-DQ, HLA-DR, and HLA-DP alleles, and the prediction of epitopes was performed based on receptor affinity, which is assigned a percentile rank for each predicted output. The predicted epitopes were divided into strong, weak, and non-binder, based on percentile scores of less than 2%, 2%–10%, and greater than 10%, respectively. The VaxiJen v.2.0 server84 was used for assessing the antigenicity of each epitope at 0.4 thresholds. AllerTOP v.2.088, AllergenFP v.1.089, AllerCatPro v.1.7, and ToxinPred server were applied to screen out epitopes based on non-allergenicity and non-toxicity. The visualization of the epitope in the S glycoprotein was performed using PyMOL.
Prediction of B-cell epitopes
Linear and conformational B-cell epitopes were predicted by the ElliPro server90 of IDEB. This method is driven by three algorithms that perform the calculation of the protein shape as an ellipsoid, the residues protrusion index (PI) calculation, and neighboring residues clustering on the basis of PI values. For the output of each B-cell epitope, ElliPro gives a score described as PI values for each residues. The residue Protrusion Index (PI) was also calculated. ElliPro indicated that 90% of protein residues were associated with PI values of 0.9 and 10% of residues contain without ElliPsoid. Ellipsoid and ElliPro are considered the most significant prediction tools for all proteins90.
Prediction of interferon-gamma inducing epitopes
In both cell-mediated and adaptive immune systems, interferon-gamma (IFN-γ) cytokine plays a significant role to stimulate natural killer cells and macrophages for exerting immunity against viral and bacterial infections. For the prediction of IFN-γ in the target protein, we used the IFN epitope server (http://crdd.osdd.net/raghava/ifnepitope/)91, which is based on an IFN-γ dataset. The main aim of this server is to design and predict IFN-γ inducing capacity containing peptide sequences from the query protein and it works based on a training dataset of 10,433 experimentally validated helper T-cell epitopes from the IDEB database91. From the input protein, the server generates overlapping IFN-γ inducing epitopes as displayed by the numerical score. The support vector machine (SVM) and a motif hybrid method were used to perform the prediction.
Population coverage and epitope conservancy calculation
The expression and distribution of HLA alleles could vary throughout the world according to the difference in regions. Thus, it is necessary to assess the HLA allele distribution around the world population. population coverage of the selected epitope was analyzed by the IEDB population coverage tool92 and the conservancy of each epitope was evaluated by the IEDB analysis resource.
Molecular interaction pattern analysis of selected epitopes with HLA alleles
The three-dimensional structure of selected CTL and HTL epitopes was generated by an online server named PEPFOLD 3.593 and the 3D crystal structure of the three commonly occurring HLA alleles in the human population HLA-A*02:01, HLA-DRB1*01:01, and HLA-DRB1*15:01 was downloaded from the RCSB PDB database ID- 1QEW, 2G9H, and 1BX2, respectively. The HLA class I and class II alleles used in the docking study are expected to cover more than 95% of the worldwide population94. The PDB structure was prepared before running P-P docking by removing water and ligand and energy minimization of the structure was carried out. In order to analyze the interaction pattern of screened out best epitopes with HLA alleles, ClusPro 2.095 P-P docking server was employed. This web-based docking tool accomplishes rigid docking by sampling billions of conformations, energy minimization, and pairwise RMSD of the complexes and estimate binding energy score of the Protein–Protein docked complex on the basis of shaped complementarity, Deocys as references states, and desolvation contribution. Molecular docking was carried out under the hydrophobic environment and the best cluster (complex) of epitopes and alleles was selected based on the lowest docking energy score. Finally, PyMol was used to visualize the interaction between epitopes and alleles.
Vaccine construction, structure modeling, and validation
The selected CTL, HTL, and IFN-γ epitopes were linked by using AYY and GPGPG linkers, and an adjuvant CTB was added to the N-terminal of the sequences via an EAAAK linker. Secondary structural features of the vaccine construct including extended strands, alpha-helical regions, random coils, and beta turns were predicted by the SOPMA server. The trRosetta (transform-restrained Restta) online tool96 (https://yanglab.nankai.edu.cn/trRosetta/) was applied to generate the three dimensional model of the linear vaccine constructs, and the GalaxyRefine web server97 was used to refine the model. The trRoseta server has been considered to be the accurate and fast algorithms for de novo protein structure prediction and the principle of this algorithm is to attempt to mimic the interplay of global and local interaction in defining protein model. On the basis of direct energy minimizations with a retrained Rosetta this computational tool generates three dimensional structure from the input protein sequence. The restrained comprises orientation distributions and inter-residues distance98. Galaxy Refine web server primarily reconstructs side chains and accomplishes repacking of the side chain and then utilizes molecular dynamic simulation to attain overall structure relaxation. This algorithm improves the quality of local structure according to CASP10 calculation techniques99. Validation of the vaccine structure was performed based on the ERRAT and Z-score43. ProSA-web determines and assigns Z-score for input protein structure which is shown on the plot in the context of all known protein models whereas structures have been evaluated by X-ray crystallography and NMR, this tool also displayed any problematic part of the structure as highlighted in three-dimensional molecule viewer. When the score was outside the range for comparably-sized proteins it specifies structure error43. The ERRAT web server evaluates non-bonded atom–atom interaction compared to reliable high-resolution crystallography structures. Finally, the overall structural quality was validated by a Ramachandran plot analysis, followed by the PROCHECK server100.
Assessment of the physicochemical and antigenic properties of the vaccine
Antigenicity was measured by the VaxiJen v2.0 web tool84, where allergenicity was checked by the AllerTOP v.2.088 and AllergenFP v.1.089 tools. VaxiJen v2.0 is a freely available server that can predict the antigenicity of a sequence on the basis of auto- and cross-covariance (ACC) transformation of protein sequences into uniform vectors of principal amino acid properties. AllerTOP v2.0 classifies the allergens of a protein using amino acid E-descriptors, k nearest neighbor’s machine learning methods, and the auto- and cross-covariance (ACC) transformation. Alternatively, AllergenFP is a descriptor-based and alignment-free strategy to recognize allergens and non-allergens. It is essential to determine physical and chemical parameters associated with the vaccine construct. Therefore,various physicochemical properties of the vaccine like instability index, aliphatic index, molecular weight, GRAVY values, isoelectric point, and half-life were evaluated by using the ExPASy ProtParam tool46. SignalP 4.1101 and TMHMM v2.0102 servers were used to check any transmembrane helices in the vaccine and the existence of any signal peptides.
Analysis of post-translational modification
In order to analyze post-translational modification in both the target spike glycoprotein and the designed vaccine construct including glycosylation, phosphorylation, and acetylation, we have used YinOYang 1.2, NetNGlyc 1.0, NetPhos 3.0, and NetAct 1.0 servers which are available at http://www.cbs.dtu.dk/services. The NetPhos 3.2 web tool can predict phosphorylation sites at threonine, tyrosine, and serine for amino acid sequences103. The NetNGlyc and YinOYang server use an artificial neural network to predict N-terminal glycosylation and O_GlcNAc attachment sites in mammalian protein104.
Molecular docking and refinement
To calculate the binding affinity and interaction patterns between the designed multi-epitope vaccine and Toll-like receptor 2 (PDB: 2Z7X) and Toll-like receptor 4 (PDB: 3FXI), the structures were retrieved from the RCSB PDB database in pdb format. The PatchDock server105 was used to perform molecular docking, and the refinement of the best complex was performed by the FireDock server106. PatchDock server calculates surface fix coordinating scores, separating scores, and portrayal of atomic shape for protein–protein molecular docking. This algorithm splits the TLRs receptor and the vaccine molecules into small patches in agreement with the surface. These small patches resemble distinctive shapes, which can separate puzzle pieces visually. The lowest docking energy score was used to select the top-ranked vaccine-TLRs complex structure. In addition, molecular interactions were visualized through PyMol and Discovery Studio 2020.
Molecular dynamic simulation
The YASARA software package107 was implemented to perform molecular dynamics simulations of the vaccine/TLR complexes. The AMBER14 force field108 was used for the system, and the vaccine complex was initially cleaned and optimized for hydrogen bond formation. The simulation system was established with the aid of a cubic simulation cell. For the initial energy minimization process, a simulated annealing method was applied with steep gradient approaches. The system was neutralized by the addition of water molecules and 0.9% NaCl salt. The physiological conditions of the system were set to pH 7.4 and a temperature of 310 K109. The system temperature was maintained with a Berendsen thermostat. The Part6icle Mesh Ewald (PME) method was applied for the calculation of long-range electrostatic interactions and short-range Columb and vdW interactions. A cut-off radius was established at 8 Å110. The simulation time step was set to 1.25 fs, and simulation trajectories were saved after every 100-ps interval107. Finally, the simulation was performed for 50 ns, and the RMSF, RMSD, Rg, and SASA values and the numbers of hydrogen bonds formed during the trajectories were analyzed to reveal the stabilities of the vaccine complexes111–115.
In silico immune simulation
The C-IMMSIM webserver was utilized to perform immune stimulations, assess immunogenicity, and determine the immune response profile for our vaccine116. This server is an agent-based model that uses position-specific scoring matrices to predict peptides derived from machine learning techniques for predicting immune interactions. It simulates three compartments which are three anatomical regions in mammals: (1) the bone marrow (the simulation region of hematopoietic stem cell for producing new myeloid and lymphoid cells); (2) the thymus (where native T-cells are selected to avoid autoimmunity); and (3) lymph node116. The minimum recommended time between dose 1 and dose 2 for most of the vaccines currently in use, is 4 weeks117. The entire simulation ran for 1,400-time steps which are about 15 months (a time step is about 8 h). Two peptide injections were given four weeks apart at time step 10, 94, 178, 262, 346, 430, 514, 598, 682, 766, 850, 934.
Conclusions
Multi-epitope vaccines have already gained importance and demonstrated protective efficiency, capable of generating immunity in vivo, with some entering clinical trials. The present study was based on an immunoinformatics-driven method that was used to identify potential antigenic epitopes for use in a vaccine candidate against MERS-CoV. Three antigen categories, including CTL, HTL, and IFN-γ epitopes, of the S glycoprotein, were used to construct a multi-epitope vaccine. The physicochemical and antigenic profiles of the vaccine were studied computationally. The stability profile and molecular interactions between the designed vaccine and immune receptors were assessed through molecular dynamics simulations and molecular docking studies. In silico immune simulation indicated the vaccine’s ability to trigger an immune response. A series of immunoinformatics strategies were applied sequentially to design and evaluation of a vaccine that may produce a protective immunity against viral infection, however, the experimental evaluation is required to assess the exact efficiency. The experimental assay may include the synthesis of the vaccine followed by the in vitro and in vivo assay. Additionally, we propose more studies that include synthesis and biological activities of the designed multi-epitope vaccine.
Supplementary Information
Author contributions
S.M., M.O.R., G.K.P. and M.A.S. together planned and designed the research. M.A.S. and M.A.M. arranged the whole facilities for the research and supervised the whole research. S.M. and M.O.R. imparted in study design and interpreted the results putting efforts on statistical analysis with M.M.P., M.S.S.S., S.B. and T.B.E. S.M., M.O.R., and G.K.P. participated in the manuscript draft and has thoroughly checked. T.B.E., K.D.,S.A.A., M.A.M., and M.A.S. revised the manuscript for necessary changes in format, grammar and English standard. All authors read and agreed on the final version of the manuscript.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shafi Mahmud and Md. Oliullah Rafi.
Contributor Information
Mohammad Ali Moni, Email: m.moni@unsw.edu.au.
Md. Abu Saleh, Email: saleh@ru.ac.bd.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-92176-1.
References
- 1.Parry RL. Travel alert after eighth camel flu death. The Times. 2015;2:10. [Google Scholar]
- 2.O’Keefe LC. Middle east respiratory syndrome coronavirus. Work. Heal. Saf. 2016;64:184–186. doi: 10.1177/2165079915607497. [DOI] [PubMed] [Google Scholar]
- 3.Chan JFW, Lau SKP, To KKW, Cheng VCC, Woo PCY, Yue KY. Middle East Respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan JFW. Group, A. of the C.S. Middle east respiratory syn 1 drome coronavirus (MERS-CoV) J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durai P, Batool M, Shah M, Choi S. Middle East respiratory syndrome coronavirus: transmission, virology and therapeutic targeting to aid in outbreak control. Exp. Mol. Med. 2015;47:e181–e181. doi: 10.1038/emm.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alqahtani FY, Aleanizy FS, Mohamed RAEH, Alanazi MS, Mohamed N, Alrasheed MM, Abanmy N, Alhawassi T. Prevalence of comorbidities in cases of Middle East respiratory syndrome coronavirus: a retrospective study. Epidemiol. Infect. 2019;147:165923. doi: 10.1017/S0950268818002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaki AM, Van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 8.Bermingham A, Chand MA, Brown CS, Aarons E, Tong C, Langrish C, Hoschler K, Brown K, Galiano M, Myers R. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Eurosurveillance. 2012;17:20290. doi: 10.2807/ese.17.40.20290-en. [DOI] [PubMed] [Google Scholar]
- 9.Ki M. MERS outbreak in Korea: hospital-to-hospital transmission. Epidemiol. Health. 2015;2015:37. doi: 10.4178/epih/e2015033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotten M, Lam TT, Watson SJ, Palser AL, Petrova V, Grant P, Pybus OG, Rambaut A, Guan Y, Pillay D. Full-genome deep sequencing and phylogenetic analysis of novel human betacoronavirus. Emerg. Infect. Dis. 2013;19:736. doi: 10.3201/eid1905.130057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus ADME, Haagmans BL, Gorbalenya AE, Snijder EJ. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3:69. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raamsman MJB, Locker JK, De Hooge A, De Vries AAF, Griffiths G, Vennema H, Rottier PJM. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 2000;74:2333–2342. doi: 10.1128/JVI.74.5.2333-2342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nal B, Chan C, Kien F, Siu L, Tse J, Chu K, Kam J, Staropoli I, Crescenzo-Chaigne B, Escriou N. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S M and E. J. Gen. Virol. 2005;86:1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- 14.Lim KP, Liu DX. The missing link in coronavirus assembly retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 2001;276:17515–17523. doi: 10.1074/jbc.M009731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Haan CAM, Rottier PJM. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 2005;64:165–230. doi: 10.1016/S0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, Droese B, Klaus JP, Makino S, Sawicki SG. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirchdoerfer RN, Cottrell CA, Wang N, Pallesen J, Yassine HM, Turner HL, Corbett KS, Graham BS, McLellan JS, Ward AB. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siu YL, Teoh KT, Lo J, Chan CM, Kien F, Escriou N, Tsao SW, Nicholls JM, Altmeyer R, Peiris JSM, et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82:11318–11330. doi: 10.1128/jvi.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, Guo D, Fu L, Cui Y, Liu X. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Lu G, Qi J, Li Y, Wu Y, Deng Y, Geng H, Li H, Wang Q, Xiao H. Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of middle east respiratory syndrome coronavirus. J. Virol. 2013;87:13134–13140. doi: 10.1128/JVI.02433-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raj VS, Mou H, Smits SL, Dekkers DHW, Müller MA, Dijkman R, Muth D, Demmers JAA, Zaki A, Fouchier RAM. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y, Zhang Y, Zhang W, Yuan Y, Bao J. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du L, Yang Y, Zhou Y, Lu L, Li F, Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin. Ther. Targets. 2017;21:131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coleman CM, Liu YV, Mu H, Taylor JK, Massare M, Flyer DC, Glenn GM, Smith GE, Frieman MB. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine. 2014;32:3169–3174. doi: 10.1016/j.vaccine.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Wong G, Lu G, Yan J, Gao GF. MERS-CoV spike protein: targets for vaccines and therapeutics. Antiviral Res. 2016;133:165–177. doi: 10.1016/j.antiviral.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chauhan V, Rungta T, Goyal K, Singh MP. Designing a multi-epitope based vaccine to combat Kaposi Sarcoma utilizing immunoinformatics approach. Sci. Rep. 2019;9:1–15. doi: 10.1038/s41598-019-39299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lund JE, Larsen O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:146. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nürnberger T, Brunner F, Kemmerling B, Piater L. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 2004;198:249–266. doi: 10.1111/j.0105-2896.2004.0119.x. [DOI] [PubMed] [Google Scholar]
- 29.Netea MG, der Graaf C, der Meer JWM, Kullberg BJ. Toll-like receptors and the host defense against microbial pathogens: bringing specificity to the innate-immune system. J. Leukoc. Biol. 2004;75:749–755. doi: 10.1189/jlb.1103543. [DOI] [PubMed] [Google Scholar]
- 30.Astuti I. Others severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndr. Clin. Res. Rev. 2020;14:93337. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan A, Junaid M, Kaushik AC, Ali A, Ali SS, Mehmood A, Wei D-Q. Computational identification, characterization and validation of potential antigenic peptide vaccines from hrHPVs E6 proteins using immunoinformatics and computational systems biology approaches. PLoS ONE. 2018;13:e0196484. doi: 10.1371/journal.pone.0196484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ali A, Khan A, Kaushik AC, Wang Y, Ali SS, Junaid M, Saleem S, Cho WCS, Mao X, Wei D-Q. Immunoinformatic and systems biology approaches to predict and validate peptide vaccines against Epstein-Barr virus (EBV) Sci. Rep. 2019;9:720. doi: 10.1038/s41598-018-37070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shastri N, Cardinaud S, Schwab SR, Serwold T, Kunisawa J. All the peptides that fit: the beginning, the middle, and the end of the MHC class I antigen-processing pathway. Immunol. Rev. 2005;207:31–41. doi: 10.1111/j.0105-2896.2005.00321.x. [DOI] [PubMed] [Google Scholar]
- 34.Jutras I, Desjardins M. Phagocytosis: at the crossroads of innate and adaptive immunity. Annu. Rev. Cell Dev. Biol. 2005;21:511–527. doi: 10.1146/annurev.cellbio.20.010403.102755. [DOI] [PubMed] [Google Scholar]
- 35.Joffre OP, Segura E, Savina A, Amigorena S. Cross-presentation by dendritic cells. Nat. Rev. Immunol. 2012;12:557–569. doi: 10.1038/nri3254. [DOI] [PubMed] [Google Scholar]
- 36.Groothuis TAM, Griekspoor AC, Neijssen JJ, Herberts CA, Neefjes JJ. MHC class I alleles and their exploration of the antigen-processing machinery. Immunol. Rev. 2005;207:60–76. doi: 10.1111/j.0105-2896.2005.00305.x. [DOI] [PubMed] [Google Scholar]
- 37.Krogsgaard M, Davis MM. How T cells “see” antigen. Nat. Immunol. 2005;6:239–245. doi: 10.1038/ni1173. [DOI] [PubMed] [Google Scholar]
- 38.Ahlers JD, Belyakov IM. Molecular pathways regulating CD4(+) T cell differentiation, anergy and memory with implications for vaccines. Trends Mol. Med. 2010;16:478–491. doi: 10.1016/j.molmed.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Wilson CC, Palmer B, Southwood S, Sidney J, Higashimoto Y, Appella E, Chesnut R, Sette A, Livingston BD. Identification and antigenicity of broadly cross-reactive and conserved human immunodeficiency virus type 1-derived helper T-lymphocyte epitopes. J. Virol. 2001;75:4195–4207. doi: 10.1128/jvi.75.9.4195-4207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saadi M, Karkhah A, Nouri HR. Development of a multi-epitope peptide vaccine inducing robust T cell responses against brucellosis using immunoinformatics based approaches. Infect. Genet. Evol. 2017;51:227–234. doi: 10.1016/j.meegid.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 42.Huang YJ, Mao B, Aramini JM, Montelione GT. Assessment of template-based protein structure predictions in CASP10. Proteins. 2014;82(Suppl 2):43–56. doi: 10.1002/prot.24488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Messaoudi A, Belguith H, Ben Hamida J. Homology modeling and virtual screening approaches to identify potent inhibitors of VEB-1 β-lactamase. Theor. Biol. Med. Model. 2013;10:22. doi: 10.1186/1742-4682-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dey AK, Malyala P, Singh M. Physicochemical and functional characterization of vaccine antigens and adjuvants. Expert Rev. Vaccines. 2014;13:671–685. doi: 10.1586/14760584.2014.907528. [DOI] [PubMed] [Google Scholar]
- 46.Walker JM. The proteomics protocols handbook. Springer; 2005. [Google Scholar]
- 47.Carty M, Bowie AG. Recent insights into the role of Toll-like receptors in viral infection. Clin. Exp. Immunol. 2010;161:397–406. doi: 10.1111/j.1365-2249.2010.04196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lester SN, Li K. Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 2014;426:1246–1264. doi: 10.1016/j.jmb.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Memish ZA, Cotten M, Meyer B, Watson SJ, Alsahafi AJ, Al Rabeeah AA, Corman VM, Sieberg A, Makhdoom HQ, Assiri A, et al. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg. Infect. Dis. 2014;20:1012–1015. doi: 10.3201/eid2006.140402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sette A, Fikes J. Epitope-based vaccines: an update on epitope identification, vaccine design and delivery. Curr. Opin. Immunol. 2003;15:461–470. doi: 10.1016/s0952-7915(03)00083-9. [DOI] [PubMed] [Google Scholar]
- 51.Jabbar B, Rafique S, Salo-Ahen OMH, Ali A, Munir M, Idrees M, Mirza MU, Vanmeert M, Shah SZ, Jabbar I, et al. Antigenic peptide prediction from E6 and E7 oncoproteins of HPV types 16 and 18 for therapeutic vaccine design using immunoinformatics and MD simulation analysis. Front. Immunol. 2018;9:3000. doi: 10.3389/fimmu.2018.03000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu C, Meng S, Jin Y, Zhang W, Li Z, Wang F, Wang-Johanning F, Wei Y, Liu H, Tu H, et al. A novel multi-epitope vaccine from MMSA-1 and DKK1 for multiple myeloma immunotherapy. Br. J. Haematol. 2017;178:413–426. doi: 10.1111/bjh.14686. [DOI] [PubMed] [Google Scholar]
- 53.Jiang P, Cai Y, Chen J, Ye X, Mao S, Zhu S, Xue X, Chen S, Zhang L. Evaluation of tandem Chlamydia trachomatis MOMP multi-epitopes vaccine in BALB/c mice model. Vaccine. 2017;35:3096–3103. doi: 10.1016/j.vaccine.2017.04.031. [DOI] [PubMed] [Google Scholar]
- 54.Lu I-N, Farinelle S, Sausy A, Muller CP. Identification of a CD4 T-cell epitope in the hemagglutinin stalk domain of pandemic H1N1 influenza virus and its antigen-driven TCR usage signature in BALB/c mice. Cell. Mol. Immunol. 2017;14:511–520. doi: 10.1038/cmi.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He R, Yang X, Liu C, Chen X, Wang L, Xiao M, Ye J, Wu Y, Ye L. Efficient control of chronic LCMV infection by a CD4 T cell epitope-based heterologous prime-boost vaccination in a murine model. Cell. Mol. Immunol. 2018;15:815–826. doi: 10.1038/cmi.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo L, Yin R, Liu K, Lv X, Li Y, Duan X, Chu Y, Xi T, Xing Y. Immunological features and efficacy of a multi-epitope vaccine CTB-UE against H. pylori in BALB/c mice model. Appl. Microbiol. Biotechnol. 2014;98:3495–3507. doi: 10.1007/s00253-013-5408-6. [DOI] [PubMed] [Google Scholar]
- 57.Cao Y, Li D, Fu Y, Bai Q, Chen Y, Bai X, Jing Z, Sun P, Bao H, Li P, et al. Rational design and efficacy of a multi-epitope recombinant protein vaccine against foot-and-mouth disease virus serotype A in pigs. Antiviral Res. 2017;140:133–141. doi: 10.1016/j.antiviral.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 58.Zhou W-Y, Shi Y, Wu C, Zhang W-J, Mao X-H, Guo G, Li H-X, Zou Q-M. Therapeutic efficacy of a multi-epitope vaccine against Helicobacter pylori infection in BALB/c mice model. Vaccine. 2009;27:5013–5019. doi: 10.1016/j.vaccine.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 59.Lennerz V, Gross S, Gallerani E, Sessa C, Mach N, Boehm S, Hess D, von Boehmer L, Knuth A, Ochsenbein AF, et al. Immunologic response to the survivin-derived multi-epitope vaccine EMD640744 in patients with advanced solid tumors. Cancer Immunol. Immunother. 2014;63:381–394. doi: 10.1007/s00262-013-1516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slingluff CLJ, Lee S, Zhao F, Chianese-Bullock KA, Olson WC, Butterfield LH, Whiteside TL, Leming PD, Kirkwood JM. A randomized phase II trial of multiepitope vaccination with melanoma peptides for cytotoxic T cells and helper T cells for patients with metastatic melanoma (E1602) Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013;19:4228–4238. doi: 10.1158/1078-0432.CCR-13-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toledo H, Baly A, Castro O, Resik S, Laferté J, Rolo F, Navea L, Lobaina L, Cruz O, Míguez J, et al. A phase I clinical trial of a multi-epitope polypeptide TAB9 combined with Montanide ISA 720 adjuvant in non-HIV-1 infected human volunteers. Vaccine. 2001;19:4328–4336. doi: 10.1016/s0264-410x(01)00111-6. [DOI] [PubMed] [Google Scholar]
- 62.Yin D, Li L, Song X, Li H, Wang J, Ju W, Qu X, Song D, Liu Y, Meng X, et al. A novel multi-epitope recombined protein for diagnosis of human brucellosis. BMC Infect. Dis. 2016;16:219. doi: 10.1186/s12879-016-1552-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bazhan SI, Antonets DV, Karpenko LI, Oreshkova SF, Kaplina ON, Starostina EV, Dudko SG, Fedotova SA, Ilyichev AA. In silico designed ebola virus T-cell multi-epitope DNA vaccine constructions are immunogenic in mice. Vaccines. 2019;7:50. doi: 10.3390/vaccines7020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Foroutan M, Ghaffarifar F, Sharifi Z, Dalimi A. Vaccination with a novel multi-epitope ROP8 DNA vaccine against acute Toxoplasma gondii infection induces strong B and T cell responses in mice. Comput. Immunol. Microbiol. Infect. Dis. 2020;69:101413. doi: 10.1016/j.cimid.2020.101413. [DOI] [PubMed] [Google Scholar]
- 65.Mizel SB, Bates JT. Flagellin as an adjuvant: cellular mechanisms and potential. J. Immunol. 2010;185:5677–5682. doi: 10.4049/jimmunol.1002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khatoon N, Pandey RK, Prajapati VK. Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-08842-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hou J, Liu Y, Hsi J, Wang H, Tao R, Shao Y. Cholera toxin B subunit acts as a potent systemic adjuvant for HIV-1 DNA vaccination intramuscularly in mice. Hum. Vaccin. Immunother. 2014;10:1274–1283. doi: 10.4161/hv.28371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim HJ, Kim J-K, Seo SB, Lee HJ, Kim H-J. Intranasal vaccination with peptides and cholera toxin subunit B as adjuvant to enhance mucosal and systemic immunity to respiratory syncytial virus. Arch. Pharm. Res. 2007;30:366–371. doi: 10.1007/BF02977620. [DOI] [PubMed] [Google Scholar]
- 69.Ali M, Pandey RK, Khatoon N, Narula A, Mishra A, Prajapati VK. Exploring dengue genome to construct a multi-epitope based subunit vaccine by utilizing immunoinformatics approach to battle against dengue infection. Sci. Rep. 2017;7:9232. doi: 10.1038/s41598-017-09199-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meza B, Ascencio F, Sierra-Beltrán AP, Torres J, Angulo C. A novel design of a multi-antigenic, multistage and multi-epitope vaccine against Helicobacter pylori: an in silico approach. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2017;49:309–317. doi: 10.1016/j.meegid.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 71.Arai R, Ueda H, Kitayama A, Kamiya N, Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001;14:529–532. doi: 10.1093/protein/14.8.529. [DOI] [PubMed] [Google Scholar]
- 72.Vaure C, Liu Y. A comparative review of toll-like receptor 4 expression and functionality in different animal species. Front. Immunol. 2014;5:316. doi: 10.3389/fimmu.2014.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Phongsisay V, Iizasa E, Hara H, Yoshida H. Evidence for TLR4 and FcRγ-CARD9 activation by cholera toxin B subunit and its direct bindings to TREM2 and LMIR5 receptors. Mol. Immunol. 2015;66:463–471. doi: 10.1016/j.molimm.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 74.Boehme KW, Compton T. Innate sensing of viruses by toll-like receptors. J. Virol. 2004;78:7867–7873. doi: 10.1128/JVI.78.15.7867-7873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dar HA, Zaheer T, Shehroz M, Ullah N, Naz K, Muhammad SA, Zhang T, Ali A. Immunoinformatics-aided design and evaluation of a potential multi-epitope vaccine against klebsiella pneumoniae. Vaccines. 2019;7:10. doi: 10.3390/vaccines7030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Solanki V, Tiwari M, Tiwari V. Prioritization of potential vaccine targets using comparative proteomics and designing of the chimeric multi-epitope vaccine against Pseudomonas aeruginosa. Sci. Rep. 2019;9:1–19. doi: 10.1038/s41598-019-41496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ojha R, Pareek A, Pandey RK, Prusty D, Prajapati VK. Strategic development of a next-generation multi-epitope vaccine to prevent nipah virus zoonotic infection. ACS Omega. 2019;4:13069–13079. doi: 10.1021/acsomega.9b00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kar T, Narsaria U, Basak S, Deb D, Castiglione F, Mueller DM, Srivastava AP. A candidate multi-epitope vaccine against SARS-CoV-2. Sci. Rep. 2020;10:10895. doi: 10.1038/s41598-020-67749-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kamthania M, Srivastava S, Desai M, Jain A, Shrivastav A, Sharma DK. Immunoinformatics approach to design T-cell epitope-based vaccine against hendra virus. Int. J. Pept. Res. Ther. 2019;25:1627–1637. doi: 10.1007/s10989-018-09805-z. [DOI] [Google Scholar]
- 80.Pandey RK, Bhatt TK, Prajapati VK. Novel immunoinformatics approaches to design multi-epitope subunit vaccine for malaria by investigating anopheles salivary protein. Sci. Rep. 2018;8:1125. doi: 10.1038/s41598-018-19456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mishra S, Sinha S. Immunoinformatics and modeling perspective of T cell epitope-based cancer immunotherapy: a holistic picture. J. Biomol. Struct. Dyn. 2009;27:293–306. doi: 10.1080/07391102.2009.10507317. [DOI] [PubMed] [Google Scholar]
- 82.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Larsen MV, Lelic A, Parsons R, Nielsen M, Hoof I, Lamberth K, Loeb MB, Buus S, Bramson J, Lund O. Identification of CD8+ T cell epitopes in the West Nile virus polyprotein by reverse-immunology using NetCTL. PLoS One. 2010;5:e12697. doi: 10.1371/journal.pone.0012697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dhanda SK, Mahajan S, Paul S, Yan Z, Kim H, Jespersen MC, Jurtz V, Andreatta M, Greenbaum JA, Marcatili P. IEDB-AR: immune epitope database—analysis resource in 2019. Nucleic Acids Res. 2019;47:W502–W506. doi: 10.1093/nar/gkz452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jensen KK, Andreatta M, Marcatili P, Buus S, Greenbaum JA, Yan Z, Sette A, Peters B, Nielsen M. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology. 2018;154:394–406. doi: 10.1111/imm.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dimitrov I, Flower DR, Doytchinova I. AllerTOP–a server for in silico prediction of allergens. BMC Bioinf. 2013;6:S4. doi: 10.1186/1471-2105-14-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dimitrov I, Naneva L, Doytchinova I, Bangov I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014;30:846–851. doi: 10.1093/bioinformatics/btt619. [DOI] [PubMed] [Google Scholar]
- 90.Ponomarenko J, Bui H-H, Li W, Fusseder N, Bourne PE, Sette A, Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dhanda SK, Vir P, Raghava GPS. Designing of interferon-gamma inducing MHC class-II binders. Biol. Direct. 2013;8:30. doi: 10.1186/1745-6150-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Q, Wang P, Kim Y, Haste-Andersen P, Beaver J, Bourne PE, Bui H-H, Buus S, Frankild S, Greenbaum J. Immune epitope database analysis resource (IEDB-AR) Nucleic Acids Res. 2008;36:W513–W518. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016;44:W449–W454. doi: 10.1093/nar/gkw329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moise L, McMurry JA, Buus S, Frey S, Martin WD, De Groot AS. In silico-accelerated identification of conserved and immunogenic variola/vaccinia T-cell epitopes. Vaccine. 2009;27:6471–6479. doi: 10.1016/j.vaccine.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, Vajda S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang J, Anishchenko I, Park H, Peng Z, Ovchinnikov S, Baker D. Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. U. S. A. 2020;117:1496–1503. doi: 10.1073/pnas.1914677117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.HeeShin W. others Prediction of protein structure and interaction by GALAXY protein modeling programs. Biodesign. 2014;2:1–11. [Google Scholar]
- 98.Rohl CA, Strauss CEM, Misura KMS, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 99.Lee GR, Won J, Heo L, Seok C. GalaxyRefine2: simultaneous refinement of inaccurate local regions and overall protein structure. Nucleic Acids Res. 2019;47:W451–W455. doi: 10.1093/nar/gkz288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Laskowski RA, MacArthur MW, Thornton JM. PROCHECK: validation of protein-structure coordinates. Science. 2012;5:684–687. doi: 10.1107/97809553602060000882. [DOI] [Google Scholar]
- 101.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 102.Krogh A, Larsson B, Heijne G, Sonnhammer E, Bioinformatics S. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 2001;305:115. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 103.Cai CZ, Han LY, Ji ZL, Chen X, Chen YZ. SVM-Prot: Web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acids Res. 2003;31:3692–3697. doi: 10.1093/nar/gkg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT-BG, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, et al. Precision mapping of the human O-GalNAc glycoproteome through simplecell technology. EMBO J. 2013;32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Andrusier N, Nussinov R, Wolfson HJ. FireDock: fast interaction refinement in molecular docking. Proteins. 2007;69:139–159. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 107.Krieger E, Darden T, Nabuurs SB, Finkelstein A, Vriend G. Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins Struct. Funct. Bioinf. 2004;57:678–683. doi: 10.1002/prot.20251. [DOI] [PubMed] [Google Scholar]
- 108.Dickson CJ, Madej BD, Skjevik AA, Betz RM, Teigen K, Gould IR, Walker RC. Lipid14: the amber lipid force field. J. Chem. Theory Comput. 2014;10:865–879. doi: 10.1021/ct4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Krieger E, Vriend G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015;36:996–1007. doi: 10.1002/jcc.23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Krieger E, Nielsen JE, Spronk CAEM, Vriend G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model. 2006;25:481–486. doi: 10.1016/j.jmgm.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 111.Samiul Islam M, Mahmud S, Sultana R, Dong W. Identification and in silico molecular modelling study of newly isolated Bacillus subtilis SI-18 strain against S9 protein of Rhizoctonia solani. Arab. J. Chem. 2020 doi: 10.1016/j.arabjc.2020.09.044. [DOI] [Google Scholar]
- 112.Khan MA, Mahmud S, Alam ASMRU, Rahman ME, Ahmed F, Rahmatullah M. Comparative molecular investigation of the potential inhibitors against SARS-CoV-2 main protease: a molecular docking study. J. Biomol. Struct. Dyn. 2020;3:1–7. doi: 10.1080/07391102.2020.1796813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bappy SS, Sultana S, Adhikari J, Mahmud S, Khan MA, Kibria KMK, Rahman MM, Shibly AZ. Extensive immunoinformatics study for the prediction of novel peptide-based epitope vaccine with docking confirmation against envelope protein of Chikungunya virus: a computational biology approach. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1726815. [DOI] [PubMed] [Google Scholar]
- 114.Rafi MO, Bhattacharje G, Al-Khafaji K, Taskin-Tok T, Alfasane MA, Das AK, Parvez MAK, Rahman MS. Combination of QSAR, molecular docking, molecular dynamic simulation and MM-PBSA: analogues of lopinavir and favipiravir as potential drug candidates against COVID-19. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1850355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rafi MO, Al-Khafaji K, Tok TT, Rahman MS. Computer-based identification of potential compounds from Salviae miltiorrhizae against Neirisaral adhesion A regulatory protein. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1856189. [DOI] [PubMed] [Google Scholar]
- 116.Rapin N, Lund O, Bernaschi M, Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE. 2010;5:1–14. doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Castiglione F, Mantile F, De Berardinis P, Prisco A. How the interval between prime and boost injection affects the immune response in a computational model of the immune system. Comput. Math. Methods Med. 2012;2012:842329. doi: 10.1155/2012/842329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).