Abstract
Introduction
The mounting incidence of multidrug-resistant bacterial strains and the dearth of novel antibiotics demand alternate therapies to manage the infections caused by resistant superbugs. Bacteriophages and phage=derived proteins are considered as potential alternates to treat such infections, and have several applications in health care systems. The aim of this review is to explore the hidden potential of bacteriophage proteins which may be a practical alternative approach to manage the threat of antibiotic resistance.
Results
Clinical trials are in progress for the use of phage therapy as a tool for routine medical use; however, the existing regulations may hamper their development of routine antimicrobial agents. The advancement of molecular techniques and the advent of sequencing have opened new potentials for the design of engineered bacteriophages as well as recombinant bacteriophage proteins. The phage enzymes and proteins encoded by the lysis cassette genes, especially endolysins, holins, and spanins, have shown plausible potentials as therapeutic candidates.
Conclusion
This review offers an integrated viewpoint that aims to decipher the insights and abilities of bacteriophages and their derived proteins as potential alternatives to antibiotics.
Keywords: Bacteriophage proteins, Antibiotic resistance, Potential antibacterials, Alternative therapy
Key Summary Points
| The soaring crisis of antibiotic resistance has directed research towards the development of alternatives to antibiotics. |
| The use of engineered phages as therapy that is helpful in killing resistant bacteria is a promising tool to confront antibiotic resistance. |
| Pre-screening of phage-encoded proteins may be performed before in vitro or in vivo analysis of the candidate protein. |
| Bacteriophage lysins target only specific bacteria and do not damage the microflora as do antibiotics, thus phage therapy has an advantage over antibiotics. |
| Bacteriophages and associated proteins have been suggested as promising antibacterials, and may have a wide range of applications in humans, animals, and agriculture. |
| In future, further experimental and clinical investigations are essential regarding the safety profile, half-life, dose, and site specificity of the bacteriophages or their proteome to be used as novel therapeutic candidates. |
Digital Features
This article is published with digital features, including a summary slide, to facilitate understanding of the article. To view digital features for this article go to https://doi.org/10.6084/m9.figshare.14316113.
Introduction
In the current era, antibiotic resistance has become an emerging and important threat, globally jeopardizing healthy living. The non-judicious use of antibiotics is leading to the emergence of multidrug-resistant superbugs. Antibiotic resistance is posing huge economic and health losses in both the developing and the developed world. According to WHO estimates, multidrug-resistant infections account for approximately 25,000 deaths every year, which will increase to 10 million by 2050. Overall, it costs about €55 billion/year in the US, which could rise to US$100 trillion by 2050. The stay of patients suffering from such resistant infections is approximately 13 days, which costs about $29,000/patient treated with such resistant bacterial infections. In this regard, a total annual economic loss (productivity loss) of about $35 billion has been reported [1–4]. The global nature of this issue emphasizes the need to control the antibiotic resistance crisis [1, 5].
At present, there is increasing interest in the use of bacteriophages as well as their lytic proteins to treat resistant bacterial infections. Bacteriophages are known as natural enemies of bacteria, their self-replicating particles having stringent host specificity to recognize and kill bacteria by hijacking the cell machinery [6]. Phages destroy the pathogenic bacteria without destroying the microflora, hence prevent the side effects that are usually associated with antibiotic treatment, for example, antibiotic-associated diarrhea [7].
Phage-associated peptidoglycan hydrolasinges are potential antibacterials. During the last decade, bacteriophage research has evolved from basic to applied and up to clinical trials, e.g., use of endolysins as antibacterials. These hydrolases destroy the peptidoglycan layer of a host bacterial cell, which results in rapid killing of the host and the exit of phage progeny [8]. This useful killing ability of endolysins may be utilized by employing these hydrolases as chimeric proteins to destroy the resistant and pathogenic bacterial strains [9, 10]. This plan proved to work well against Gram-positive bacterial pathogens, which have no external protection for their cell walls, permitting direct access of recombinant endolysins to the peptidoglycan layer of the cells. Additionally, many studies to make lytic enzymes effective against Gram-negative bacteria have been reported in recent times [11].
The World Health Organization (WHO) has proclaimed the urgent need for antibiotics against ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) pathogens [12]. These pathogens are associated with severe nosocomial infections, especially in critical patients [13]. Several studies have reported the effectiveness of phage therapy in humans. Previously, a patient with P. aeruginosa-associated septicemia was treated with phage therapy [14], and another patient with P. aeruginosa-associated aortic graft infection has also been managed successfully by employing phage therapy [15].
Additionally, the role of bacteriophages in the food industry and as potential sanitizing agents against multidrug-resistant pathogens has also been recognized [16]. Antibiotics are unable to penetrate the matrix of biofilms to kill or destroy the bacterial species; however, the phages have been found to be very effective in killing bacterial biofilms, making them a viable alternative to antibiotics [17]. The prevalence rate of antimicrobial resistance has increased remarkably in food-borne pathogens, and the emergence of resistance in the food chain is an increasing public health concern across the globe. Phage therapy has profound beneficial effects on human and animal health, and can lead to a greater productivity rate in the livestock industry. Meeting the increasing demand for meat can be possible only by adopting alternative approaches to antibiotics, like phage therapeutics, to control infections in intensified production systems [18].
The increase of multidrug-resistant bacterial strains and the lack of new antibiotics calls for the development of new antibacterial drugs or alternative strategies, like phage therapy and the use of phage lytic proteins [19]. Moreover, a few pathogens can evade the action of antibiotics by hiding themselves in safe niches within the host, e.g., S. aureus, a recognized pathogen that may survive in the intracellular environment, biofilms, or abscesses, resulting in persistent infections. Thus, the development of novel, target-specific, safe, and effective antibacterials is urgently required [20–22].
The use of engineered phages as a therapy that helps to kill bacteria could be the next step to confront antibiotic resistance. Engineered phages precisely detect and control bacteria, serving as a new source of antimicrobials as well as a vehicle for the delivery of vaccine antigens and drugs. For example, endolysins have a modular structure that may be exploited for phage engineering by substituting and recombining the functional domain of diverse species, which could lead to the development of chimeric proteins with the desired features. This practice may also be a good option for other phage lytic proteins like virion-associated peptidoglycan hydrolases (VAPGHs). Additionally, it could be a promising tool for proteins with non-phage origins, like bacteriocins and autolysins [23]. The specificity of such engineered proteins would be a key advantage when used as antibacterials, for further bactericidal activity, with a negligible risk of resistance development, and good efficacy against persister cells, multidrug-resistant strains, and biofilms [9]. In this review, an attempt is made to explore the hidden potential of bacteriophage proteins, which may be a practical alternative approach to manage the threat of antibiotic resistance.
This article is based on previously conducted studies, and does not contain any new studies with human participants or animals performed by any of the authors.
Bacteriophage Proteins
Bacteria are all around the world, good ones and bad ones. The dominance of bad bacteria leads to infections and disease in the susceptible host. Antibiotics not only kill the bad bacteria but are also harmful to the good ones, and overuse of antibiotics introduces resistance in exposed bacterial populations. The development of bacterial resistance to many antibiotics has become a crucial therapeutic challenge across the globe. Often, these multidrug-resistant bacteria are associated with the community as well as with hospital-acquired infections. Because of the alarming situation of antibiotic resistance among bacterial species and complications in treatment, possible support or alternatives are required to reduce the infection rate [24].
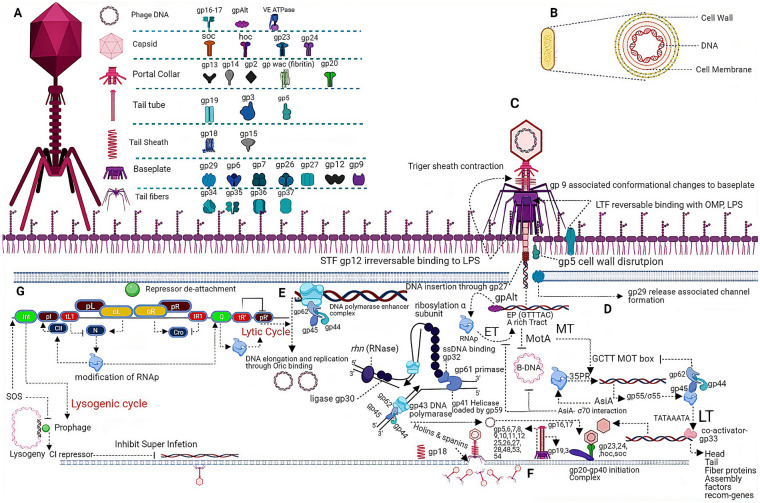
Bacteriophages have been suggested as antibacterial agents to combat bacterial infections in humans, animals, and crops. Since bacteriophages encode a variety of proteins that play a vital role in the bacteriophage replication cycle, from the recognition of the host to the lysis of the cell (Fig. 1), phage-encoded proteins like receptor binding proteins, VAPGHs, endolysins, and holins are being considered as a possible means to fight antibiotic-resistant bacteria [25].
Fig. 1.
A Various phage-associated proteins located at different structural components of a bacteriophage. B Key structural components of the bacterial host related to phage infection. C Attachment of bacteriophage to the host (bacterium); attachment initiates after long-term facilitation binds (reversibly) to host receptors, leads to conformational changes to the base plates and allows rotation, resulting in short-term facilitation binding (irreversibly) to the host receptors. Sheath contraction allows gp27-associated DNA translocation, while gp5 and gp29 play roles in channel formation. D–F Various steps along with different phage-encoded proteins in the lytic phase of the infection. D Ribosylation of host RNAp by phage-encoded gp Alt recruits RNAp to early gene transcription, while the MotA transcriptional regulator inhibits transcription from the host (bacterial) DNA and initiates middle transcription by binding to the MotA box. Middle phase transcription product, gp55, along with proteins of DNA polymerase enhancer complex and gp33 initiates late transcription, while subsequent products play an important role in phage assembly. E Phage DNA replication through phage-encoded DNA polymerase (gp43) depends on the DNA polymerase enhancer complex, while replisome-associated proteins include a DNA polymerase enhancer complex (clamp loader), Primase gp 61 (produces primers for DNA elongation), gp41 (helicase activity), gp30 (ligation), Rnh gene (RNase H), and gp32 (ssDNA binding activity). F Assembly of phage components takes place right after the formation of the gp20–gp40 initiation complex followed by head recruitment, while the DNA is transferred to head domain via ATPase-dependent gp16–gp17 DNA translocation activity, wile phage-encoded holins and spanins assist the phage existing from the host cell. G The CI repressor binds with the oL and oR operators and represses the transcription from pL and pR, retaining the virus in a prophage state, while SOS leads to CI de-attaching from the operators. Modification of RNAp by the N and Q proteins allows it to pass through the terminators tR1, tL1, and tR’, leading to lytic genes transcription, while the N protein-associated CII expression leads to the expression of Int and the integration of phage DNA into the host DNA
Receptor Binding Proteins (RBPs)
The role of RBPs as detecting tools for bacterial pathogens has been described, and they have also been suggested as probes for bacterial O-serotyping [26–28]. RBs-encoded depolymerases have been proposed to be applied in pathogen-specific targeting. Thus, a depolymerase has been used as a host immune system adjuvant against bacterial infection by inhibiting the bacterial capsule [29, 30]. On the other hand, the amazing capsule specificity of these depolymerases may be employed for diagnostics and capsular typing, particularly for K. pneumoniae [31, 32]. Thorough knowledge regarding the structural and molecular phenomena controlling the RBPs and phage–host interaction would substantially improve the applicability of phage-derived tools in the medical and biotechnological fields.
These proteins are particularly valuable for identifying particular receptors, like the membrane surface-anchored protein gamma phage receptor, GamR, on the surface of the cell wall of Bacillus anthracis, thus making it possible to detect particular bacteria [33]. RBPs can identify glycosylation patterns of O signature antigens (determinants of the immunogenicity of bacteria and disease spread). The binding affinity and specificity of these proteins have a major role in the detection of bacterial endotoxin contaminants [28]. The binding specificity of phages is mediated by RBPs, which form spikes or tail fibers at the caudal end. A monomeric RBP located at the T5 tail tip, called Pb5, showed irreversible binding with FhuA receptors of the bacterial host during the infection, i.e., E. coli [34, 35]. The outer membrane protein, FhuA has a key role in iron acquisition by E. coli from the environment as it transports the siderophore–ferrichrome very actively.
Recombinant phage-derived proteins have also been designed which introduced specific tags (Cys-tag) to the receptor binding proteins, which significantly improved their capturing performance and their efficiency as detection probes [36]. It has been identified that the C-terminal domain of these receptor binding proteins is responsible for the binding and recognition to the host receptors, such as cell wall teichoic acids [37]. Due to the higher affinity and specificity of receptor binding proteins, many of their applications are directed towards pathogen detection rather than pathogen control [38]. Therefore, the use of such proteins to detect bacterial pathogens can help to manage bacterial infections, including antibiotic-resistant bacteria.
Long Tail Fiber (gp37-gp38)
To infect the bacterial host cell by targeting specific receptors, tailed bacteriophages use their molecular machinery, having a wide range of RBPs, like tail spikes, central tail spikes, and tail fibers. After the recognition of the host, these tail fibers penetrate the cell and deliver the phage DNA into the host cytoplasm. Different receptors for these fibers include porins, lipopolysaccharides (LPS), outer membrane protein C (OmpC), and teichoic acids. The first contact of a long-tail fiber with any of the mentioned receptors is followed by a second contact of another long-tail before the dissociation of the first one. It is a repetitive process until the phage finds the proper site for permanent adsorption followed by DNA ejection [35, 39] (Fig. 2).
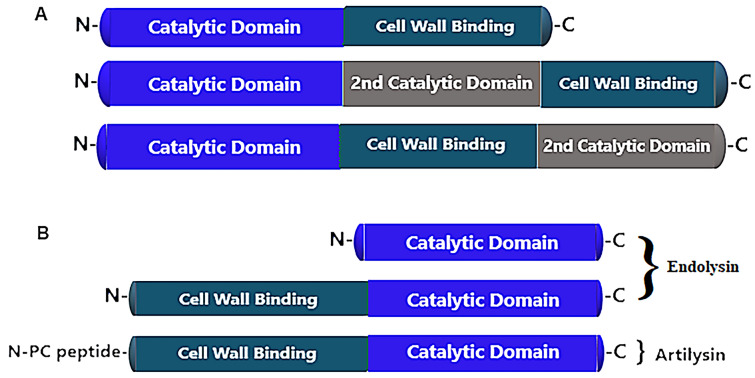
Fig. 2.
Schematic of the endolysin structure of Gram-positive and Gram-negative bacteria. A The endolysin of Gram-positive bacteria possesses two domains, namely the N-terminus catalytic domain and the C-terminus cell wall binding domain. Some endolysins also have a 2nd catalytic domain which is different from the 1st one. The cell wall binding domain contains binding motifs which may be single or multiple. B Endolysin of Gram-negative bacteria exhibit a catalytic domain. In those which contain a cell wall binding domain, this domain is situated on the N-terminus rather than the C-terminus of the protein. Artilysin, an engineered lysin, can be made by adding hydrophobic or amphipathic sequences at either the N-terminus or the C-terminus, and are effective only when applied externally
The molecular mechanisms used by myophages for the adsorption and infection of Gram-positive bacteria have been described previously. Phage phi812 uses its tail fibers to contact the host bacteria (S. aureus), which alters the shape of the base plate [40]. However, phi812 has a distinct base plate conformation as compared to T4; it possesses heavy ‘legs’, a trimer of RBPs and tripod proteins. These all lead to base plate reorganization by binding with specific receptors (receptor–tripod complex). The phi812 myophage with double-layered base plate machinery can infect both Gram-negative and Gram-positive bacteria [41, 42].
The long tail fiber proteins have been demonstrated as high-affinity molecules for the detection of Salmonella. The long tail fiber proteins have a great capability for diagnosing a broad spectrum of bacteria. These proteins are made particularly useful by coating them with paramagnetic particles for capturing S. typhimurium from food samples. A new method, enzyme-linked long tail fiber assay (EELTA), has now been suggested that detects as low as 102 CFU/ml of S. typhimurium. It has been observed that phage S16 long tail fiber paramagnetic beads have a great capability to bind and immobilize all strains of Salmonella. A 98% detection rate of S. typhimurium cells from tested suspensions has been demonstrated. It is suggested that the microflora does not affect these proteins, as these are also able to tolerate variable concentrations of salt and pH. These factors promote in situ detection of Salmonella by these proteins. Tail fibers may be used as a promising diagnostic tool for pathogen detection in health care settings [43]. The LPS of P. aeruginosa are of two types: the A-band (common polysaccharide antigen) and the B-band (O-specific antigen) [44]. The LKA1 phage of Pseudomonas primarily utilizes the LPS B-band O-specific antigen as a receptor. The tail spike protein LKAI gp49 has three basic domains: β-helix, β-barrel, and β-sandwich. This protein has a strong enzymatic activity and can degrade the LPS binding protein and ultimately destroy the biofilm matrix, allowing small molecules to rapidly diffuse across the membrane. Owing to its high specificity and enzymatic activity, LKA1gp49 can be used for various microbiological as well as biotechnological applications. The tail spike protein LKA1gp49 can act as a powerful antimicrobial agent [45]. The depolymerase activity of LKA1gp49 is considered a potential antibacterial. The LKA1 is a lyase and specifically degrades O5 serotype LPS molecules, thus disrupting the biofilm matrix [45].
Long-Tail Fiber Protein gp37
Phage T4 of E. coli has long tail fibers through which they interact with the LPS of their host, and thus recognizes and binds to it. Long-tail fibers consist of four different proteins: gp34, gp35, gp36, and gp37 [46]. The tip of the tail fiber protein gp37 comprises a length of 1600 Angstroms and divides into two portions (proximal and distal) of about 70 nm [47]. The distal portion of the long tail fiber interacts with the LPS of bacteria, thus determining the phage’s host range against Gram-negative bacteria [47]. Overall, gp37 is one of the key proteins in the distal segment of the phage tail, forming a needle-like foot containing the receptor-binding site at the C terminus. It is interlaced with a globular collar, an elongated needle, and head. It is the iron ions that hold the interlaced strand and residues in the head domains, which facilitate the binding to LPS, OmpC, or porins of the host [47, 48].
Tail Spikes
P22 typically infects serotype S. typhimurium, has icosahedral symmetry, and has a short tail. It is almost 60 nm in diameter, and belongs to the Podoviridae family. P22 has a tail spike gp9 protein which forms a homotrimer, and each unit contains three domains. The N terminus binding domain has two central β-sheets used to attach the tail of the phage [49]. The C terminus receptor binding domain facilitates the attachment with the O antigen and shows endorhamnosidase hydrolyzing activity which cuts the O polysaccharide chain [28, 50]. Bacteriophage infection occurs as the gp9 tail spike protein of bacteriophage P22 binds to the O antigen of S. typhimurium. This spike protein hydrolyzes the alpha-1, 3-O-glycosidic linkage of the O antigen polysaccharide. It then enters either in the lytic phase (in which the virus replicates itself and ultimately releases its progeny cells through cell lysis) or in the lysogenic phase (in which the cell divides by integrating the phage’s DNA into the chromosomes of the host) [50]. Also, the Det7 (myophage) and SP6 (podophage) display the same tail spikes (gp49 and gp207 respectively) and host specificity, as they both can infect Salmonella species [51].
Although tail spikes may vary in their host specificity or receptor binding domains, they generally contain conserved structures. Moreover, these proteins frequently carry different enzymes which destroy the host cell wall and expedite the ejection of phage DNA. Thus, these proteins are useful for serotyping purposes. and also helpful in visualizing phase variations during vaccine production [25].
Penetrating Proteins
There are some proteins, which are different both structurally and functionally from tail fibers and spikes, known as membrane-penetrating proteins. The exact mechanism of penetration into the inner membrane is not known, but it has been suggested that these proteins have the ability to breach the boundary of the host bacterium. In addition, a membrane-penetrating protein of λ phage80, i.e., gpJ, may have a role in adsorption or in determining the host range of the phage. The T4 myophage central spike protein, i.e., gp5, is a lysozyme; it hydrolyzes the glycosidic bonds in peptidoglycans. This gp5 forms a complex with gp27 and looks like a long torch. The cylindrical portion of the structure is made up of the gp27 trimer and contains three N terminus domains of gp5 trimers. The C terminus domain of gp5 is attached to these which then is folded into a β-helix [34, 52]. The neighboring three gp5 lysozyme domains are attached to the N-terminus and C-terminus domains via two linkers (long peptide) [34]. The binding of the substrate to the lysozyme is prevented by these linkers, while the polysaccharide binding site is blocked by the β-helix. Consequently, β-helix dissociation restores the activity of the lysozyme which causes the digestion of peptidoglycan, hence assisting in the tail tube penetration [34]. Although the puncturing mechanism of the inner membrane is vague, it is assumed that gp27 or gp29 are involved in the DNA ejection [34, 53].
Endolysins
Endolysins are bacteriophage lysins or peptidoglycan hydrolases that degrade the peptidoglycan layer of the bacterial host during the lytic cycle. Since the outer membrane is absent in the cell walls of Gram-positive bacteria, purified endolysins when applied from outside directly gain access or reach the peptidoglycan layer, suggesting them as potent antimicrobials [54].
The endolysin structure of Gram-positive bacteria differs slightly from those of Gram-negative bacteria (Fig. 1). The endolysin of Gram-positive bacteria have two domains: the N-terminal enzymatically active domains, and the C-terminal cell wall binding domain (see Fig. 2A). An enzymatically active domain contains certain enzymes that are endopeptidases, L-amidases, glucosaminidase, muramidase, or transglycosylase. Cell wall binding domains contain binding motifs which may be single or multiple [55]. Enzymatically active domains cleave the particular bonds in bacterial peptidoglycan, while the cell wall binding domain specifically targets the protein to its substrate. After lysis of the bacterial cell, it binds tightly to the cell wall debris, thereby preventing diffusion and destruction of those intact bacterial cells that have not been infected by phage cell [54]. Table 1 presents an overview of the endolysins which have been purified and tested against Gram-positive bacteria.
Table 1.
Some important endolysins against Gram-positive bacteria
| Endolysins | Phage source | Target pathogen | Main outcome | References |
|---|---|---|---|---|
| LysK | Lambda phage K | S. aureus | Has lysing activity against live and killed staphylococcal cells | [102] |
| SAL-1 | Phage SAP-1 | MRSA strain of S. aureus | Had increased lytic activity in synergism with Lysk, helpful in designing a recombinant-based drug SAL 200, helpful in treating MRSA staphylococcal infections | [103] |
| PlyGRCS | Phage GRCS | S. aureus | Has lysing activity against biofilms produced by S. aureus | [104] |
| MV-L | Phage MR11 | S. aureus and S. simulans | Helpful in treating staphylococcal infection, but has no lysing activity against S. epidermidis and E. coli | [105] |
| CHAPSH3b | Phage vB-SauS-philPLA88 | S. aureus | Reduces biofilm-forming activity of staphylococci | [106] |
| MR-10 | S. phage MR-10 | MRSA S. aureus | Given synergically with minocycline, effective against MRSA staphylococcal and burn wound infection | [107] |
| Stephefelet | Staphylococcal phage | MRSA S. aureus | A recombinant engineered phage-based lysin, helpful in treating skin infections (without disturbing normal flora) | [108] |
| PlyV12 | E. faecalis phage lysin PlyV12 | Vancomycin-resistant Enterococcus | Show broad lytic spectrum including VRE, Staphylococcus, group A and B streptococci | [109] |
| ABgp46 | Phage vB-AbaP-CEB1 | A. baumannii strains | Has lytic activity against A. baumannii, P. aeruginosa, S. typhimurium | [110] |
| PlyAB1 | Phage Abp1 | A. baumannii strains | Display highly effective lytic activity against hospital acquired pan-drug A. baumannii strains | [111] |
| Cpl-1 | Pneumococcal phage | S. pneumoniae | Effective against pneumococcal pneumoniae and endocarditis | [112] |
| Art-240 | S. agalactiae lambda phage Sa2 | S. pyogenes, S. uberis, S. equi, Group E and G Streptococcus | A derivative of endolysin λSa2lys, lethal to Streptococcus isolates, exhibits enhanced bactericidal activity | [106] |
| Art-175 | Pseudomonas phage varphiKZ | Targets A. baumannii and P. aeruginosa | Reduces the bacterial count, and displays high lytic activity against A. baumannii | [113] |
| Ply511 | L. lactis | L. monocytogenes | Has bio-preservative property against Listeria monocytogenes | [114] |
| Ply700 | Streptococcus spp. | S. uberis, S. pyogenes, and S. dysgalactiae | In milk, show antibacterial activity against S. uberis | [115] |
Endolysins of Gram-negative bacteria have a single domain of globular proteins, particularly without a cell wall binding domain. This means that phage endolysins against Gram-negative bacteria comprise only the catalytic domain (Fig. 2B). Since the outer membrane is present in Gram-negative bacteria, so phage endolysins against Gram-negative bacteria cannot access peptidoglycan, and are not active when applied externally against Gram-negative bacteria. To overcome this issue, endolysins have been engineered. One of the most important engineered endolysin is Artilysin, which is developed by adding hydrophobic/polycationic peptide sequences to the N-terminal portion or C-terminal portion of endolysins, which enables penetration to the outer membrane of Gram-negative bacteria [56]. Innolysins is a T5 endolysin combined with RBP Pb5, and it showed significant lytic activity against E. coli when Pb5 specific receptors FhuA are present. Additionally, innolysins displayed activity against other bacterial hosts carrying FhuA receptors, such as P. aeruginosa and Shigella sonnei [57]. Endolysins have a three-dimensional crystalline structure, and two endolysins have been successfully crystallized, namely pneumococcal phage lysin Cp1-1 and Listeria phage PSA [58, 59]. Various purified endolysins active against Gram-negative bacteria are listed in Table 2.
Table 2.
Some important endolysins against gram negative bacteria
| Endolysins | Source | Type of phage protein | Targeted bond | Host organisms | References |
|---|---|---|---|---|---|
| BcepC6gp22 | Phage BcepC6B | Globular structure with single catalytic domain | Glycoside bond between the MurNac and the GluNac components of the glycan in the peptidoglycan | Burkholderia cepacia | [116] |
| P2gp09 | Phage P2 | Globular structure with single catalytic domain | Hydrolyze the glycoside bond | E. coli | [116] |
| PsP3gp10 | Phage PsP3 | Globular structure with single catalytic domain | Hydrolyze glycoside bond | S. enterica | [116] |
| K11gp3.5 | Phage K11 | Globular or modular | Hydrolyze the amide bond | K. pneumoniae | [116] |
| KP32gp15 | Phage KP32, | Globular or modular type | Hydrolyze the amide bond | K. pneumoniae | [116] |
| Lys AB2 | Phage AB2 | Modular | Hydrolyze the amide bond | A. baumannii | [117] |
| KZ144 | KZ phage | Modular | Hydrolyze the glycoside bond | P. aeruginosa | [116] |
There is growing interest in the field of engineered phage-encoded lytic proteins, called enzybiotics, since their discovery. Lytic enzymes make a significant impact in therapeutics, such as endocarditis and bloodstream infection. Exebacase (CF-301) which is a prophage endolysin has good lytic activity against S. aureus [9, 60]. Recently, it has been observed that human serum albumin and serum lysozyme have a synergistic effect with CF-301 [61]. A hydrolase called SAL200 is currently under investigation for the treatment of bacterial-associated pneumoniae caused by P. aeruginosa, A. baumannii, S. pneumoniae, B. anthracis, and S. aureus. Likewise, a selected endolysin for Gram-negative bacteria, i.e., PlyPa91, showed 70% challenge protection in animal models against P. aeruginosa-induced pneumonia [62]. One commercially available pharmaceutical gel product, Staphefekt, is an engineered endolysin that is used to treat skin infections [63]. CF-301 was used systemically to treat experimental osteomyelitis, and showed a significant reduction in bone localization of S. aureus [64]. A phage-encoded protein, PlyCD, has been used to develop a chimeric protein, LHD, which showed strong lytic activity against C. difficile, mitigated C. difficile-associated diarrhea, and reduced the toxin concentration in patient feces [65].
Endolysins are considered as novel antibacterials; therefore, in vivo safety of endolysins is critically important. It has been observed that the phage-encoded endolysin, PlyC, has lytic activity [66] against S. uberis, showing no cytotoxicity, and did not affect the production of reactive oxygen species in leukocytes of lactating cows [67]. The advantage of endolysins over antibiotics is that they are highly specific for peptidoglycan and bind to the most conserved regions in the cell walls of bacteria which are immobile. This high specificity limits the risk of developing antimicrobial resistance. Bacteriophage lysins target only specific bacteria and do not damage the microflora as do antibiotics, thus phage therapy has an advantage over antibiotics [54].
An experimental study has reported that antibodies conjugated with bacteriophage-derived lysins or bacterial autolysins to form a functional homodimer called a lysibody have a protective effect against methicillin-resistant S. aureus (MRSA). Although the Gram-positive bacterial cell wall contains highly conserved carbohydrate molecules, these are considered poor immunogens. Autolysins or phage lysins have distinct high-affinity binding domains for carbohydrates present in the cell wall. Lysibodies may be produced by combining lysin binding domains with the Fc portion of human IgG, and it has ben observed that lysibodies have fixed the complement on the bacterial cell surface and augmented the phagocytic activity of macrophages and neutrophils. Based on these results, it is suggested that lysibodies could be potential antibacterials [62].
Endolysin activity can be measured both by quantitative and qualitative methods, namely turbidity reduction analysis, zymogram assays, viable count assays, minimal inhibitory concentration assays, or plate agar lysis assays [54]. The endolysins have a vast range of lytic activity displaying species specificity, thus avoiding selective pressure on commensals. Here are some possible reasons due to which resistance against endolysins is unlikely to develop. Endolysins bind to and cleave highly conserved bonds and linkages in the structures of the cell wall, and bacteria are unable to modify this property of phage endolysin, thus minimizing the chances of the development of resistance against endolysins. The majority of endolysins have two catalytic domains in their structure that cleave bonding within the peptidoglycan layer, which is unfavorable in the development of antibiotic resistance. Since most of the phage lysins are used externally and target the cell wall without entering the bacterial cells, the endolysins avoid possible mechanisms of antibacterial resistance, such as active efflux from the cell and reduced membrane permeability. It has also been suggested that, to increase their therapeutic property and to minimize the development of antimicrobial resistance, endolysins can be used in combination with antibiotics to treat infections. Thus, endolysins have been suggested as potential antibacterials to fight antibiotic-resistant bacteria [68].
Holins
The lysis mechanism of bacteriophages exhibits two different strategies: murein hydrolase activity, namely endolysin, and a membrane protein known as holin. Holins include a vast group of small proteins (Table 3). They form pores in the cell membrane of the host, through which lysins move to the murein layer and degrade the peptidoglycan layer which induces cell death and measures the timing of the infection cycle [69, 70]. Holins perform a distinct established function which includes (1) release of gene transfer agents, (2) facilitate various processes required for differentiation, like spore germination, (3) have a role in biofilm formation, (4) release toxins and related proteins, and (5) help in different responses to stress conditions [71]. In animals, programmed cell death is facilitated by Bax and Bak (holin-like proteins), which may have a bacterial origin. It has been suggested that holins are ubiquitous in the three domains of life as they have also been characterized from archaea.
Table 3.
Description of important holins proteins present in bacteriophage proteome
| Holins | Source | Belongs to | Expression medium | Main outcome | References |
|---|---|---|---|---|---|
| HolGH15 | S. aureus phage GH15 | Class 2 as well as phage holin 1 superfamily | E. coli BL21 cells |
(1) Showed vast range of antibacterial activity against S. aureus, B. subtilis, P. aeruginosa, K. pneumonia (2) Releases cellular contents, causing shrinkage and death of these organisms |
[76] |
| Hol 15 | S. aureus phage P68 | Class 1 | E. coli | (1) Complement phage lambdam mutation. (2) Retards growth of E. coli | [118] |
| Lambda holin S105 | Lambda phages | Holin superfamily 3 as well as belongs to class 1 holins | E. coli | By accumulating in membrane forms only one irregular pore releasing endolysin, causing lysis of cell within seconds | [119, 120] |
| Hol 3626 | C. perfringens phage 3626 | Class 2 holin | E. coli | (1) Has antibacterial activity against C. perfringens. (2) Helpful in control measures in food as well as microbial communities | [121] |
Structurally holins have been classified into three major classes. Holins of Class I have three hydrophobic transmembrane domains. The N-terminal portion is present inside the periplasm while the C-terminal portion is present inside the cytoplasm. Lambda phage holins and the S. aureus phage P68 Hol 15 protein have been categorized to belong to this group. Holins of Class II have two hydrophobic transmembrane domains, as well as – and C-terminal portions, but both terminals are found in cytoplasmic portions. The S protein of the lambda phage 21 and Hol 3626 holin protein from the Clostridium perfringens phage 3626 are examples of Class II holins. Holins of Class III also have – and C-terminal portions with the N-terminus inside the cytoplasm and the C inside the periplasm. Examples of this category include T4 holins and holins from phage CP 26F and phage CP390 [72, 73].
Mostly, genes encoded for holins are located adjacent to a gene encoding for endolysin. During the late phase of the phage infection, before activation holins accumulate in the plasma membrane, a gradual increase in holin concentration is required to achieve the threshold level, which, once it is achieved, leads to the formation of holes that permit the release of endolysins from the cytoplasm which destroys the peptidoglycan [74]. Canonical holins form large oligomer holes through which protein molecules of about 500 kDa can pass, thus allowing endolysins to escape and subsequently gain access to the peptidoglycan layer, and ultimately cleave various bonds in the peptidoglycan layer and digest it [75]. Pinholins form small pores, thus depolarizing the membrane. They are mostly associated with signal arrest release endolysins. In periplasm, they are present as an inactive-enzymatic form.
Due to the increased development of multidrug-resistant strains, holin proteins have been suggested as potent antibacterial agents for fighting against resistant strains of bacteria. The S. aureus GH15 phage-associated holin, i.e., HolGH15, has shown significant lytic activity against a wide host range [76]. Recently, the lytic activity of HolGH15 was observed against L. monocytogenes. HolGH15 showed significant lytic activity against the tested pathogen, and it was observed that HolGH15 showed a 240-µg/mL minimal bactericidal concentration against L. monocytogenes. It was concluded that HolGH15 displayed a similar sort of lytic activity against various serotypes of L. monocytogenes. Additionally, the antibacterial activity of HolGH15 has also been observed at refrigerator temperature against L. monocytogenes. Such abilities of HolGH15 make it a potential antibacterial agent against L. monocytogenes, and it may be used as a spray in the course of the handling and processing of various food items, like fruits, eggs, meat products, and vegetables [77]. Holins do not disturb the microflora and thus ultimately reduces the risk of the development of antimicrobial resistance [78].
Spanins
Since lysins and holins disrupt the cell wall and inner membrane, respectively, disruption of the Gram-negative bacterial outer membrane is also very necessary with the help of the third type of holin proteins, called spanins. Spanins disrupt the outer membrane by catalyzing both the outer and inner membranes. Without these spanin proteins, the lysis mechanism is supposed to be blocked, and viral particles would remain trapped inside the dead cells instead of being released [79]. There are two types of spanins used by phages during the last step in the course of host Gram-negative bacteria lysis, i.e., unimolecular spanin and bicomponent spanins [8, 80]. Structural details of the lambda phage Rz-Rz1 spanin (a bi-component spanin) revealed that it contains a basic internal membrane protein, i.e., I-spanin, and a lipoprotein of the external membrane, i.e., o-spanin, which produces a complex spanning the gel-like matrix called periplasm. Three different genetic configurations of bi-component spanins include separated units, embedded parts, or overlapped components. On the other hand, T1 gp11 (unimolecular spanin) has an N-terminal lipoylation sequence and a C-terminal domain, e.g., U-spanins have N-terminal signal protruding towards the outer membrane lipoproteins as well as the C-terminus transmembrane domain [81].
Holin proteins act as the clock in the phage lytic cycle. During phage assembly in the cytoplasm, these holin proteins disturb the integrity of the whole membrane with the help of spanins, create holes in the membrane, and allow phage lysins to get access to the cell wall which cleaves the bacterial peptidoglycan, hence allowing the release of phages from host bacterial cells. Holin also reduces the chances of biofilm development by incorporating an extracellular polymeric substances layer while antibiotics do not penetrate the biofilm layers. Thus, phage holins in conjunction with endolysins could be powerful alternative lytic proteins in confronting antibiotic-resistant bacteria [78].
Anti-CRISPR Proteins
From the origin of life, the fight between phages and bacteria never ends. The bacterial system uses an adaptive immune system, CRISPR-Cas, to destroy the phage’s attack and, in turn, the bacteriophages produce anti-CRISPR proteins to fight back. The CRISPR-Cas system is divided into two groups, the Class I and Class II CRISPR-Cas systems. The Class I system includes types I and III, as well as IV, with multiple subunits of ribonucleoproteins, while the Class II CRISPR-Cas system comprises the II, V, and VI types with a single ribonucleoprotein. Bacteriophages in turn develop anti-CRISPR protein systems to combat the CRISPR-Cas system of bacteria. These proteins also belong to two classes based on the classification of the target CRISPR-Cas system, namely Classes 1 and 2 anti-CRISPR proteins [82].
The most important and notable feature of anti-CRISPR proteins (Acr) is a lack of sequence similarity with any other protein. The most important anti-CRISPR proteins are AcrF1, AcrF2, and AcrF3. The first two proteins, AcrF1 and AcrF2, bind to the type I-F CRISPR-Cas system with the help of a binding complex formed by multiple subunits. AcrF1 binds to Csy3 subunits while AcrF2 binds to the Csy complex (Csy1 and Csy2). However, AcrF3 binds to the Cas-3 helicase-nuclease protein, preventing its recruitment to the DNA–Csy complex [83].
One of the possible approaches to manage antibiotic resistance is a genetic strategy that seems to include sensitization of bacteria to antibiotics and selective killing of antibiotic-resistant bacteria. Towards this approach, temperate phages have proved to be very helpful. By using temperate phages, the CRISPR-Cas system is inserted within the genome of the resistant bacteria and this engineered CRISPR-Cas system protects from lytic phages by degrading both lytic phages (genetically modified) as well as destroys plasmids having antibiotic-resistant genes. These lytic phages selectively destroy antibiotic-resistant bacteria and protect antibiotic-sensitive bacteria. A transferable CRISPR-Cas system has been designed to sensitize the bacteria having an antibiotic resistance gene. The CRISPR-Cas genes are amplified by PCR and introduced in lambda prophage via homologous recombination. The CRISPR array that specifically targets the conserved regions of resistant genes has also been introduced within the same lysogen. Then, induction of prophage and lysogenization of resistant bacteria with a lambda CRISPR-Cas and lambda-Cas is carried out to determine plasmid loss. Plasmid curing occurs with lambda CRISPR-Cas, but not with lambda-Cas (Fig. 3). It has also been seen that the transferable CRISPR-Cas system prevents horizontal gene transfer of antibiotic resistance elements, as well as protects bacteria from an engineered T7 phage, thus linking bacterial sensitization to antibiotics with resistance to lytic phages. This transferable system prevents the transformation of plasmids and protects lysogenized bacteria from the lytic phages. This lysogenization is then used to sensitize antibiotic-resistant bacteria. This genetic strategy may be used in hospital settings as well as in hand sanitizers to replace resistant bacteria with antibiotic-sensitive bacteria. The benefit of using this genetic strategy is that there is no need to administer the phages within the host tissue [84].
Fig. 3.
CRISPR-CAS-based mechanism for re-sensitizing antibiotic-resistant bacteria. A vector phage is used to insert a CRISPR system into a targeted population of bacteria. The Cas9 RNA-guided nuclease is conveyed along with a guide RNA to cut a target sequence. The target carried on plasmids leads to a plasmid-curing phenomenon, i.e., re-sensitization to antibiotics, while the target carried on the chromosome leads to the death of the cell due to degradation of the chromosome
Virion-Associated Peptidoglycan Hydrolases (VAPGH’s)
Unlike endolysins, the VAPGHs (endopeptidases, glucosaminidase, etc.) facilitate the bacteriophage entry into the host cell by developing a hole in the bacterial cell wall. However, both these lytic proteins have a similar mode of action (the same catalytic domains), which degrades the bacterial cell wall, called the peptidoglycan layer [80, 85]. The exact location of these enzymes is not fixed, and they may be located at any place in the phage. They have been observed in the capsid, head, tail, and phage membrane [86, 87]. Hydrolases are appropriate to perform the activity against Gram-positive as well as Gram-negative bacteria [88]. VAPGHs have numerous benefits which make these proteins potential antibacterial agents against multidrug resistant bacteria. Additionally, they are very useful food decontaminants, particularly in processed foods and dairy products [87]. Recently, activity of VAPGHs against Agrobacterium tumefaciens-associated plant diseases has been observed [89]. The bactericidal activity of VAPGHs may be enhanced by creating chimeric proteins by exchanging the functional domains of various enzymes or via combining different proteins. Chimeric proteins have shown promising results against S. aureus infections [87].
It has been reported that VAPGHs have no certain cell wall binding domain as have endolysins. Also, it has been suggested that VAPGHs have highly conserved regions, but modification of these conserved regions is very rare. Thus, the development of bacterial resistance against these VAPGHs is a remote possibility, suggesting VAPGHs as novel and alternative antibacterials. VAPGHs derived from Gram-negative infecting phages have a broad host range compared to VAPGHs from Gram-positive infecting phages.
The VAPGHs proteins are highly thermostable, which could be exploited against extremophile bacteria. The VAPGH HydH5 is a novel VAPGH protein, produced from staphylococcal phage vB-SauS-PhilLA88. This protein, when combined with lysostaphin, exhibits augmented lytic activity against S. aureus in milk. HydH5 is a potential antibacterial that may be used in the dairy industry [87, 90]. VAPGH proteins act extracellularly, and can avoid the intracellular bacterial mechanism of resistance, i.e., as an efflux pump, and can also share a similar mode of action as for endolysins, this suggests them as promising antimicrobial agents to counter drug resistance. On the other hand, VAPGH-derived enzybiotics may be a significant tool that can be employed to cope with intracellular bacterial pathogens, as, due to the absence of receptors, the phage may encounter hurdles to obtain entry inside the eukaryotic cell.
Challenges to Face with Possible Solutions
In the past, enzymes used in experiments conducted for therapeutic evaluation of phage-encoded proteins were chosen randomly and are not suitable for particular applications. Additionally, in vivo experimental conditions are crucial to obtain good therapeutic efficacy [10]. To solve this problem, pre-screening of phage-encoded proteins may be performed before advance (in vitro or in vivo) analysis of suitable therapeutic candidate proteins. It has been successfully demonstrated in the case of engineered staphylococcal peptidoglycan hydrolases characterized from a collection that they showed significant efficacy in milk, human blood, and the inner environment of the cell [91–93].
The therapeutic efficacy of phage-encode proteins is dependent on the serum half-life, as most of the proteins feature a short half-life ranging from minutes to hours [94–96]. Generally, a long half-life is considered suitable because in this case boosting is not required, but the associated risk of anaphylaxis is also considerable in such cases. Recently, engineered PGH-ABD demonstrated a long half-life and augmented distribution, improved efficacy in human blood [93].
Some bacteria, like E. coli, S. aureus, and L. monocytogenes, can evade therapy and immunity by surviving inside the cells. Few phage proteins have the penetrating features to target these intracellular pathogens. Significant development has been made to solve this problem by combining lytic enzymes with cell-penetrating peptides, which facilitates the penetration of lytic enzymes into the host. For instance, short peptides which identify specific cells by surface markers and can deliver the biomolecules to a specific site are called homing peptides (HPs). These HPs have been used in cancer therapeutics and have been considered for clinical trials (e.g., NGR015) [97]. The same function of HPs may be exploited in phage-engineered therapeutics.
Preclinical development of phage-encoded proteins is dependent on different important concerns, like therapeutic safety, immunogenicity, and cell toxicity. A study conducted in the recent past described the safety of endolysins Pal and Cpl-1: experimental mice exposed to these endolysins showed an augmented IgG response and decreased level of IgE, which suggested that these enzymes have an adequate profile of safety and toxicity to be considered for therapeutic use [98]. Endolysins like lysostaphin may also be used to construct lysibodies by fusing them with the Fc portion of human IgG. It has been observed that lysibodies play a significant role in the opsonization of S. aureus, which facilitates the phagocytosis of this pathogen [99].
Industrial production is also a substantial contributing factor in the therapeutic application of phage-encoded proteins. Safety protocols and cost-effectiveness are most important concerns which should be kept in consideration during the production process. This should be related to the whole process, starting from the selection of the host, e.g., yeast bacteria or plants, to the removal of the endotoxins. In the end, regulatory approval for phage-encoded proteins to be used as therapeutics is mandatory [23, 95]. The approval process should be that followed in the case of therapeutic proteins. This authorization approach was successful in the past for the approval of recombinant therapeutic proteins as compared to the approval of new antibiotics, which is a lengthy process [100, 101].
Conclusions
The soaring crisis of antibiotic resistance has directed research towards the development of alternatives to antibiotics. In this context, bacteriophages and phage-encoded proteins are promising, effective, and novel classes of therapeutic agents to fight antibiotic resistance. Despite the novelty and aspiration to use bacteriophage proteome as an alternative to antibiotic therapy, it has also been proved to be worthy of further research for clinical applications of these therapeutic engineered proteins. Since the world is facing a serious threat of antimicrobial resistance, it is necessary and timely to look afresh at this approach with practical applications. In the future, further experimental and clinical investigations are essential regarding the safety profile, half-life, dose, and site-specificity of the bacteriophages or their proteome for them to be used as novel therapeutic candidates.
Acknowledgements
Higher Education Commission (HEC) Pakistan for Project No. 7166/Punjab/ NRPU/R&D/HEC/ 2017.
Funding
This study and the journal’s Rapid Service Fee was supported by grants from the major science and technology special project of Yunnan Province, No. 2019ZF004.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Author Contribution
Zulqarnain baloch, Bilal Aslam, Muhammad Imran Arshad, Muhammad Aamir Aslam, Saima Muzammil, and Abu Baker Siddique designed the concept. Bilal Aslam, Nafeesa Yasmeen, Moeed Ahmad, Mohsin Khurshid, Maria Rasool, and Muhammad Hidayat Rasool have drafted the manuscript. Xueshan Xia, Mohammad Fahim, and Riaz Hussain critically reviewed the manuscript. Zulqarnain Baloch and Bilal Aslam did the final drafting of the manuscript.
Disclosures
Bilal Aslam, Muhammad Imran Arshad, Muhammad Aamir Aslam, Saima Muzammil, Abu Baker Siddique, Nafeesa Yasmeen, Mohsin Khurshid, Maria Rasool, Moeed Ahmad, Muhammad Hidayat Rasool, Mohammad Fahim, Riaz Hussain, Xueshan Xia, Zulqarnain Baloch have nothing to disclose.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any new studies with human participants or animals performed by any of the authors.
Data Availability
The datasets generated and analyzed during the current study are not publicly available due to the risk of compromising the individual privacy of participants but are available from the corresponding author on reasonable request.
Contributor Information
Xueshan Xia, Email: oliverxia2000@aliyun.com.
Zulqarnain Baloch, Email: znbalooch@yahoo.com.
References
- 1.Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645–1658. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khurshid M, Rasool MH, Ashfaq UA, Aslam B, Waseem M, Ali MA, et al. Acinetobacter baumannii sequence types harboring genes encoding aminoglycoside modifying enzymes and 16SrRNA methylase; a multicenter study from Pakistan. Infect Drug Resist. 2020;13:2855–2862. doi: 10.2147/IDR.S260643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saeed M, Rasool MH, Rasheed F, Saqalein M, Nisar MA, Imran AA, et al. Extended-spectrum beta-lactamases producing extensively drug-resistant Salmonella Typhi in Punjab, Pakistan. J Infect Dev Countries. 2020;14(2):169–176. doi: 10.3855/jidc.12049. [DOI] [PubMed] [Google Scholar]
- 4.Khurshid M, Rasool MH, Ashfaq UA, Aslam B, Waseem M. Emergence of ISAba1 harboring carbapenem-resistant Acinetobacter baumannii isolates in Pakistan. Future Microbiol. 2017;12:1261–1269. doi: 10.2217/fmb-2017-0080. [DOI] [PubMed] [Google Scholar]
- 5.Baloch Z, Aslam B, Muzammil S, Khurshid M, Rasool MH, Ma K. Selection inversion: a probable tool against antibiotic resistance. Infect Drug Resist. 2018;11:1903–1905. doi: 10.2147/IDR.S176759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutiérrez D, Fernández L, Martínez Fernández B, Rodríguez González A, García Suárez MP. Bacteriophages: the enemies of bad bacteria are our friends! Front. Young Mind. 2016;4:30. [Google Scholar]
- 7.Pirisi A. Phage therapy—advantages over antibiotics? Lancet. 2000;356(9239):1418. doi: 10.1016/S0140-6736(05)74059-9. [DOI] [PubMed] [Google Scholar]
- 8.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56(3):430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dams D, Briers Y. Enzybiotics: enzyme-based antibacterials as therapeutics. Adv Exp Med Biol. 2019;1148:233–253. doi: 10.1007/978-981-13-7709-9_11. [DOI] [PubMed] [Google Scholar]
- 10.Oliveira H, São-José C, Azeredo J. Phage-derived peptidoglycan degrading enzymes: challenges and future prospects for in vivo therapy. Viruses. 2018;10(6):292. doi: 10.3390/v10060292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerstmans H, Criel B, Briers Y. Synthetic biology of modular endolysins. Biotechnol Adv. 2018;36(3):624–640. doi: 10.1016/j.biotechadv.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Kakasis A, Panitsa G. Bacteriophage therapy as an alternative treatment for human infections. A comprehensive review. Int J Antimicrob Agents. 2019;53(1):16–21. doi: 10.1016/j.ijantimicag.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Esposito S, De Simone G. Update on the main MDR pathogens: prevalence and treatment options. Infez Med. 2017;25(4):301–310. [PubMed] [Google Scholar]
- 14.Jennes S, Merabishvili M, Soentjens P, Pang KW, Rose T, Keersebilck E, et al. Use of bacteriophages in the treatment of colistin-only-sensitive Pseudomonas aeruginosa septicaemia in a patient with acute kidney injury-a case report. Crit care (London, England). 2017;21(1):129. doi: 10.1186/s13054-017-1709-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan BK, Turner PE, Kim S, Mojibian HR, Elefteriades JA, Narayan D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Public Health. 2018;2018(1):60–66. doi: 10.1093/emph/eoy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez L, Gutierrez D, Rodriguez A, Garcia P. Application of bacteriophages in the agro-food sector: a long way toward approval. Front Cell Infect Microbiol. 2018;8:296. doi: 10.3389/fcimb.2018.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 18.Gigante A, Atterbury RJ. Veterinary use of bacteriophage therapy in intensively-reared livestock. Virol J. 2019;16(1):1–9. doi: 10.1186/s12985-019-1260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmelcher M, Loessner MJ. Bacteriophage endolysins—extending their application to tissues and the bloodstream. Curr Opin Biotechnol. 2021;2021(68):51–59. doi: 10.1016/j.copbio.2020.09.012. [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharya M, Wozniak DJ, Stoodley P, Hall-Stoodley L. Prevention and treatment of Staphylococcus aureus biofilms. Expert Rev Anti-infect Therapy. 2015;13(12):1499–1516. doi: 10.1586/14787210.2015.1100533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraunholz M, Sinha B. Intracellular Staphylococcus aureus: live-in and let die. Front Cell Infect Microbiol. 2012;2:43. doi: 10.3389/fcimb.2012.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singer AJ, Talan DA. Management of skin abscesses in the era of methicillin-resistant Staphylococcus aureus. N Engl J Med. 2014;370(11):1039–1047. doi: 10.1056/NEJMra1212788. [DOI] [PubMed] [Google Scholar]
- 23.Gutiérrez D, Fernández L, Rodríguez A, García P. Are phage lytic proteins the secret weapon to kill Staphylococcus aureus? mBio. 2018;9(1):e01923–7. doi: 10.1128/mBio.01923-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B, Delattre A-S, Lavigne R, Science P. Learning from bacteriophages-advantages and limitations of phage and phage-encoded protein applications. Curr Protein. 2012;13(8):699–722. doi: 10.2174/138920312804871193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santos SB, Costa AR, Carvalho C, Nóbrega FL, Azeredo J. Exploiting bacteriophage proteomes: the hidden biotechnological potential. Trends Biotechnol. 2018;36(9):966–84. doi: 10.1016/j.tibtech.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 26.Evans TJ, Crow MA, Williamson NR, Orme W, Thomson NR, Komitopoulou E, et al. Characterization of a broad-host-range flagellum-dependent phage that mediates high-efficiency generalized transduction in, and between, Serratia and Pantoea. Microbiology (Reading, England). 2010;156(Pt 1):240–247. doi: 10.1099/mic.0.032797-0. [DOI] [PubMed] [Google Scholar]
- 27.Tay LL, Huang PJ, Tanha J, Ryan S, Wu X, Hulse J, et al. Silica encapsulated SERS nanoprobe conjugated to the bacteriophage tailspike protein for targeted detection of Salmonella. Chem Commun (Cambridge, England). 2012;48(7):1024–1026. doi: 10.1039/C1CC16325F. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt A, Rabsch W, Broeker NK, Barbirz S. Bacteriophage tailspike protein based assay to monitor phase variable glucosylations in Salmonella O-antigens. BMC Microbiol. 2016;16(1):207. doi: 10.1186/s12866-016-0826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan YJ, Lin TL, Lin YT, Su PA, Chen CT, Hsieh PF, et al. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrob Agents Chemother. 2015;59(2):1038–1047. doi: 10.1128/AAC.03560-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutiérrez D, Briers Y, Rodríguez-Rubio L, Martínez B, Rodríguez A, Lavigne R, et al. Role of the pre-neck appendage protein (Dpo7) from phage vB_SepiS-phiIPLA7 as an anti-biofilm agent in Staphylococcal species. Front Microbiol. 2015;6:1315. doi: 10.3389/fmicb.2015.01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin TL, Hsieh PF, Huang YT, Lee WC, Tsai YT, Su PA, et al. Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: implication in typing and treatment. J Infect Dis. 2014;210(11):1734–1744. doi: 10.1093/infdis/jiu332. [DOI] [PubMed] [Google Scholar]
- 32.Hsieh PF, Lin HH, Lin TL, Chen YY, Wang JT. Two T7-like bacteriophages, K5–2 and K5–4, each encodes two capsule depolymerases: isolation and functional characterization. Sci Rep. 2017;7(1):4624. doi: 10.1038/s41598-017-04644-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertozzi Silva J, Storms Z, Sauvageau D. Host receptors for bacteriophage adsorption. FEMS Microbiol Lett. 2016;363(4):fnw002. doi: 10.1093/femsle/fnw002. [DOI] [PubMed] [Google Scholar]
- 34.Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, et al. Morphogenesis of the T4 tail and tail fibers. Virol J. 2010;7:355. doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nobrega FL, Vlot M, de Jonge PA, Dreesens LL, Beaumont HJE, Lavigne R, et al. Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol. 2018;16(12):760–773. doi: 10.1038/s41579-018-0070-8. [DOI] [PubMed] [Google Scholar]
- 36.Singh A, Arya SK, Glass N, Hanifi-Moghaddam P, Naidoo R, Szymanski CM, et al. Bacteriophage tailspike proteins as molecular probes for sensitive and selective bacterial detection. Biosensors. 2010;26(1):131–138. doi: 10.1016/j.bios.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 37.Le S, He X, Tan Y, Huang G, Zhang L, Lux R, et al. Mapping the tail fiber as the receptor binding protein responsible for differential host specificity of Pseudomonas aeruginosa bacteriophages PaP1 and JG004. PLoS ONE. 2013;8(7):e68562. doi: 10.1371/journal.pone.0068562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waseh S, Hanifi-Moghaddam P, Coleman R, Masotti M, Ryan S, Foss M, et al. Orally administered P22 phage tailspike protein reduces Salmonella colonization in chickens: prospects of a novel therapy against bacterial infections. PLoS ONE. 2010;5(11):e13904. doi: 10.1371/journal.pone.0013904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu B, Margolin W, Molineux IJ, Liu J. Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc Natl Acad Sci USA. 2015;112(35):E4919–E4928. doi: 10.1073/pnas.1501064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nováček J, Šiborová M, Benešík M, Pantůček R, Doškař J, Plevka P. Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate. Proc Natl Acad Sci USA. 2016;113(33):9351–9356. doi: 10.1073/pnas.1605883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kutter EM, Skutt-Kakaria K, Blasdel B, El-Shibiny A, Castano A, Bryan D, et al. Characterization of a ViI-like phage specific to Escherichia coli O157:H7. Virol J. 2011;8:430. doi: 10.1186/1743-422X-8-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arachchi GJ, Mutukumira AN, Dias-Wanigasekera BM, Cruz CD, McIntyre L, Young J, et al. Characteristics of three listeriaphages isolated from New Zealand seafood environments. J Appl Microbiol. 2013;115(6):1427–1438. doi: 10.1111/jam.12332. [DOI] [PubMed] [Google Scholar]
- 43.Denyes JM, Dunne M, Steiner S, Mittelviefhaus M, Weiss A, Schmidt H, et al. Modified bacteriophage S16 long tail fiber proteins for rapid and specific immobilization and detection of Salmonella cells. Appl Environ Microbiol. 2017;83(12):e00277-17. doi: 10.1128/AEM.00277-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burrows LL, Chow D, Lam JS. Pseudomonas aeruginosa B-band O-antigen chain length is modulated by Wzz (Ro1) J Bacteriol. 1997;179(5):1482–1489. doi: 10.1128/jb.179.5.1482-1489.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olszak T, Shneider MM, Latka A, Maciejewska B, Browning C, Sycheva LV, et al. The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence. Sci Rep. 2017;7(1):16302. doi: 10.1038/s41598-017-16411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yap ML, Rossmann MG. Structure and function of bacteriophage T4. Future Microbiol. 2014;9(12):1319–1327. doi: 10.2217/fmb.14.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartual SG, Otero JM, Garcia-Doval C, Llamas-Saiz AL, Kahn R, Fox GC, et al. Structure of the bacteriophage T4 long tail fiber receptor-binding tip. Proc Natl Acad Sci USA. 2010;107(47):20287–20292. doi: 10.1073/pnas.1011218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Washizaki A, Yonesaki T, Otsuka Y. Characterization of the interactions between Escherichia coli receptors, LPS and OmpC, and bacteriophage T4 long tail fibers. MicrobiologyOpen. 2016;5(6):1003–1015. doi: 10.1002/mbo3.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinbacher S, Miller S, Baxa U, Budisa N, Weintraub A, Seckler R, et al. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. J Mol Biol. 1997;267(4):865–880. doi: 10.1006/jmbi.1997.0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andres D, Hanke C, Baxa U, Seul A, Barbirz S, Seckler R. Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro. J Biol Chem. 2010;285(47):36768–36775. doi: 10.1074/jbc.M110.169003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walter M, Fiedler C, Grassl R, Biebl M, Rachel R, Hermo-Parrado XL, et al. Structure of the receptor-binding protein of bacteriophage det7: a podoviral tail spike in a myovirus. J Virol. 2008;82(5):2265–2273. doi: 10.1128/JVI.01641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, et al. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415(6871):553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 53.Nakagawa H, Arisaka F, Ishii S. Isolation and characterization of the bacteriophage T4 tail-associated lysozyme. J Virol. 1985;54(2):460–466. doi: 10.1128/jvi.54.2.460-466.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmelcher M, Donovan DM, Loessner MJ. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012;7(10):1147–1171. doi: 10.2217/fmb.12.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seal BS, Drider D, Oakley BB, Brüssow H, Bikard D, Rich JO, et al. Microbial-derived products as potential new antimicrobials. Vet Res. 2018;49(1):66. doi: 10.1186/s13567-018-0563-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Briers Y, Walmagh M, Grymonprez B, Biebl M, Pirnay J-P, Defraine V, et al. Art-175 is a highly efficient antibacterial against multidrug-resistant strains and persisters of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2014;58(7):3774–3784. doi: 10.1128/AAC.02668-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zampara A, Sørensen MCH, Grimon D, Antenucci F, Vitt AR, Bortolaia V, et al. Exploiting phage receptor binding proteins to enable endolysins to kill Gram-negative bacteria. Sci Rep. 2020;10(1):12087. doi: 10.1038/s41598-020-68983-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korndörfer IP, Danzer J, Schmelcher M, Zimmer M, Skerra A, Loessner MJ. The crystal structure of the bacteriophage PSA endolysin reveals a unique fold responsible for specific recognition of Listeria cell walls. J Mol Biol. 2006;364(4):678–689. doi: 10.1016/j.jmb.2006.08.069. [DOI] [PubMed] [Google Scholar]
- 59.Domingo-Calap P, Delgado-Martínez J. Bacteriophages: protagonists of a post-antibiotic era. Antibiotics. 2018;7(3):66. doi: 10.3390/antibiotics7030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gilmer DB, Schmitz JE, Euler CW, Fischetti VA. Novel bacteriophage lysin with broad lytic activity protects against mixed infection by Streptococcus pyogenes and methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2013;57(6):2743–2750. doi: 10.1128/AAC.02526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Indiani C, Sauve K, Raz A, Abdelhady W, Xiong YQ, Cassino C, et al. The antistaphylococcal lysin, CF-301, activates key host factors in human blood to potentiate methicillin-resistant Staphylococcus aureus bacteriolysis. Antimicrob Agents Chemother. 2019;63(4):e02291-18. doi: 10.1128/AAC.02291-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raz A, Serrano A, Hernandez A, Euler CW, Fischetti VA. Isolation of phage lysins that effectively kill Pseudomonas aeruginosa in mouse models of lung and skin infection. Antimicrob Agents Chemother. 2019;63(7):e00024–19. doi: 10.1128/AAC.00024-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gutiérrez D, Garrido V, Fernández L, Portilla S, Rodríguez A, Grilló MJ, et al. Phage Lytic Protein LysRODI Prevents Staphylococcal Mastitis in Mice. Front Microbiol. 2020;11:7. doi: 10.3389/fmicb.2020.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karau MJ, Schmidt-Malan SM, Yan Q, Greenwood-Quaintance KE, Mandrekar J, Lehoux D, et al. Exebacase in addition to daptomycin is more active than daptomycin or exebacase alone in methicillin-resistant Staphylococcus aureus osteomyelitis in rats. Antimicrob Agents Chemother. 2019;63(10):e01235-19. doi: 10.1128/AAC.01235-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peng Z, Wang S, Gide M, Zhu D, Patabendige LWHM, Li C, et al. A novel bacteriophage lysin-human defensin fusion protein is effective in treatment of Clostridioides difficile infection in mice. Front Microbiol. 2018;9:3234. doi: 10.3389/fmicb.2018.03234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nelson D, Schuch R, Chahales P, Zhu S, Fischetti VA. PlyC: a multimeric bacteriophage lysin. Proc Natl Acad Sci USA. 2006;103(28):10765–10770. doi: 10.1073/pnas.0604521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scholte CM, Nelson DC, Garcia M, Linden SB, Elsasser TH, Kahl S, et al. Short communication: recombinant bacteriophage endolysin PlyC is nontoxic and does not alter blood neutrophil oxidative response in lactating dairy cows. J Dairy Sci. 2018;101(7):6419. doi: 10.3168/jds.2017-13908. [DOI] [PubMed] [Google Scholar]
- 68.Kashani HH, Schmelcher M, Sabzalipoor H, Hosseini ES, Moniri R. Recombinant endolysins as potential therapeutics against antibiotic-resistant Staphylococcus aureus: current status of research and novel delivery strategies. Clin Microbiol Rev. 2018;31(1):e00071–e117. doi: 10.1128/CMR.00071-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dewey JS, Savva CG, White RL, Vitha S, Holzenburg A, Young R. Micron-scale holes terminate the phage infection cycle. Proc Natl Acad Sci. 2010;107(5):2219–2223. doi: 10.1073/pnas.0914030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang I-N, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54(1):799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 71.Saier MH, Reddy BL. Holins in bacteria, eukaryotes, and archaea: multifunctional xenologues with potential biotechnological and biomedical applications. J Bacteriol. 2015;197(1):7–17. doi: 10.1128/JB.02046-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi Y, Yan Y, Ji W, Du B, Meng X, Wang H, et al. Characterization and determination of holin protein of Streptococcus suis bacteriophage SMP in heterologous host. Virol J. 2012;9(1):70. doi: 10.1186/1743-422X-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramanculov E, Young R. Genetic analysis of the T4 holin: timing and topology. Gene. 2001;265(1):25–36. doi: 10.1016/S0378-1119(01)00365-1. [DOI] [PubMed] [Google Scholar]
- 74.Reddy BL, Saier MH., Jr Topological and phylogenetic analyses of bacterial holin families and superfamilies. Biochim Biophys acta. 2013;1828(11):2654–2671. doi: 10.1016/j.bbamem.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang N, Deaton J, Young R. Sizing the holin lesion with an endolysin-β-galactosidase fusion. J Bacteriol. 2003;185(3):779–787. doi: 10.1128/JB.185.3.779-787.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Song J, Xia F, Jiang H, Li X, Hu L, Gong P, et al. Identification and characterization of HolGH15: the holin of Staphylococcus aureus bacteriophage GH15. J Gen Virol. 2016;97(5):1272–1281. doi: 10.1099/jgv.0.000428. [DOI] [PubMed] [Google Scholar]
- 77.Song J, Niu W, Wu R, Wang J, Lei L, Han W, et al. The phage holin HolGH15 exhibits potential as an antibacterial agent to control Listeria monocytogenes. Foodborne Pathogens Dis. 2020 (PubMed PMID: 32955931. Epub 2020/09/22. eng). [DOI] [PubMed]
- 78.Lin DM, Koskella B, Lin HC. Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Therap. 2017;8(3):162. doi: 10.4292/wjgpt.v8.i3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Young R, Way J, Way S, Yin J, Syvanen M. Transposition mutagenesis of bacteriophage lambda: a new gene affecting cell lysis. J Mol Biol. 1979;132(3):307–322. doi: 10.1016/0022-2836(79)90262-6. [DOI] [PubMed] [Google Scholar]
- 80.Young R, Wang N, Roof WD. Phages will out: strategies of host cell lysis. Trends Microbiol. 2000;8(3):120–128. doi: 10.1016/S0966-842X(00)01705-4. [DOI] [PubMed] [Google Scholar]
- 81.Berry JD, Rajaure M, Young R. Spanin function requires subunit homodimerization through intermolecular disulfide bonds. Mol Microbiol. 2013;88(1):35–47. doi: 10.1111/mmi.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu Y, Zhang F, Huang Z. Structural insights into the inactivation of CRISPR-Cas systems by diverse anti-CRISPR proteins. BMC Biol. 2018;16(1):32. doi: 10.1186/s12915-018-0504-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Borges AL, Davidson AR, Bondy-Denomy J. The discovery, mechanisms, and evolutionary impact of anti-CRISPRs. Annu Rev Virol. 2017;4:37–59. doi: 10.1146/annurev-virology-101416-041616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yosef I, Manor M, Kiro R, Qimron U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci. 2015;112(23):7267–72. doi: 10.1073/pnas.1500107112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Domingo-Calap P, Delgado-Martínez J. Bacteriophages: protagonists of a post-antibiotic era. Antibiot. 2018;7(3):66. doi: 10.3390/antibiotics7030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin H, Paff ML, Molineux IJ, Bull JJ. Therapeutic application of phage capsule depolymerases against K1, K5, and K30 capsulated E. coli in mice. Front Microbiol. 2017;8:2257. doi: 10.3389/fmicb.2017.02257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodríguez-Rubio L, Martínez B, Donovan DM, Rodríguez A, García P. Bacteriophage virion-associated peptidoglycan hydrolases: potential new enzybiotics. Crit Rev Microbiol. 2013;39(4):427–434. doi: 10.3109/1040841X.2012.723675. [DOI] [PubMed] [Google Scholar]
- 88.Criscuolo E, Spadini S, Lamanna J, Ferro M, Burioni R. Bacteriophages and their immunological applications against infectious threats. J Immunol Res. 2017;2017:3780697. doi: 10.1155/2017/3780697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Attai H, Rimbey J, Smith GP, Brown PJB. Expression of a peptidoglycan hydrolase from lytic bacteriophages Atu_ph02 and Atu_ph03 triggers lysis of Agrobacterium tumefaciens. Appl Environ Microbiol. 2017;83(23):e01498–17. doi: 10.1128/AEM.01498-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rodríguez-Rubio L, Martínez B, Donovan DM, García P, Rodríguez A. Potential of the virion-associated peptidoglycan hydrolase HydH5 and its derivative fusion proteins in milk biopreservation. PLoS ONE. 2013;8(1):e54828. doi: 10.1371/journal.pone.0054828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Verbree CT, Dätwyler SM, Meile S, Eichenseher F, Donovan DM, Loessner MJ, et al. Identification of peptidoglycan hydrolase constructs with synergistic staphylolytic activity in cow's milk. Appl Environ Microbiol. 2017;83(7):e03445-16. doi: 10.1128/AEM.03445-16. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 92.Röhrig C, Huemer M, Lorgé D, Luterbacher S, Phothaworn P, Schefer C, et al. Targeting hidden pathogens: cell-penetrating enzybiotics eradicate intracellular drug-resistant Staphylococcus aureus. mBio. 2020;11(2):e0020920. doi: 10.1128/mBio.00209-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sobieraj AM, Huemer M, Zinsli LV, Meile S, Keller AP, Röhrig C, et al. Engineering of long-circulating peptidoglycan hydrolases enables efficient treatment of systemic Staphylococcus aureus infection. mBio. 2020;11(5):e01781–20. doi: 10.1128/mBio.01781-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jun SY, Jang IJ, Yoon S, Jang K, Yu KS, Cho JY, et al. Pharmacokinetics and tolerance of the phage endolysin-based candidate drug SAL200 after a single intravenous administration among healthy volunteers. Antimicrob Agents Chemother. 2017;61(6):e02629-16. doi: 10.1128/AAC.02629-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Abdelkader K, Gerstmans H, Saafan A, Dishisha T, Briers Y. The preclinical and clinical progress of bacteriophages and their lytic enzymes: the parts are easier than the whole. Viruses. 2019;11(2):96. doi: 10.3390/v11020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Seijsing J, Sobieraj AM, Keller N, Shen Y, Zinkernagel AS, Loessner MJ, et al. Improved biodistribution and extended serum half-life of a bacteriophage endolysin by albumin binding domain fusion. Front Microbiol. 2018;9:2927. doi: 10.3389/fmicb.2018.02927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vrettos EI, Mező G, Tzakos AG. On the design principles of peptide-drug conjugates for targeted drug delivery to the malignant tumor site. Beilstein J Organic Chem. 2018;14:930–954. doi: 10.3762/bjoc.14.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harhala M, Nelson DC, Miernikiewicz P, Heselpoth RD, Brzezicka B, Majewska J, et al. Safety studies of pneumococcal endolysins Cpl-1 and Pal. Viruses. 2018;10(11). (PubMed PMID: 30445722. Pubmed Central PMCID: PMC6266847. Epub 2018/11/18. eng). [DOI] [PMC free article] [PubMed]
- 99.Raz A, Serrano A, Thaker M, Alston T, Fischetti VA. Lysostaphin lysibody leads to effective opsonization and killing of methicillin-resistant Staphylococcus aureus in a murine model. Antimicrob Agents Chemother. 2018;62(10):e01056-18. doi: 10.1128/AAC.01056-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cooper CJ, Khan Mirzaei M, Nilsson AS. adapting drug approval pathways for bacteriophage-based therapeutics. Front Microbiol. 2016;7:1209. doi: 10.3389/fmicb.2016.01209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kinch MS. An overview of FDA-approved biologics medicines. Drug Discov Today. 2015;20(4):393–398. doi: 10.1016/j.drudis.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 102.Haddad Kashani H, Fahimi H, Dasteh Goli Y, Moniri R. A novel chimeric endolysin with antibacterial activity against methicillin-resistant Staphylococcus aureus. Front Cell Infect Microbiology. 2017;7:290. doi: 10.3389/fcimb.2017.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jun SY, Jang IJ, Yoon S, Jang K, Yu K-S, Cho JY, et al. Pharmacokinetics and tolerance of the phage endolysin-based candidate drug SAL200 after a single intravenous administration among healthy volunteers. Antimicrob Agents Chemother. 2017;61(6):e02629-16. doi: 10.1128/AAC.02629-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Linden SB, Zhang H, Heselpoth RD, Shen Y, Schmelcher M, Eichenseher F, et al. Biochemical and biophysical characterization of PlyGRCS, a bacteriophage endolysin active against methicillin-resistant Staphylococcus aureus. Appl Microbiol Biotechnol. 2015;99(2):741–752. doi: 10.1007/s00253-014-5930-1. [DOI] [PubMed] [Google Scholar]
- 105.Rashel M, Uchiyama J, Ujihara T, Uehara Y, Kuramoto S, Sugihara S, et al. Efficient elimination of multidrug-resistant Staphylococcus aureus by cloned lysin derived from bacteriophage ϕMR11. J Infect Dis. 2007;196(8):1237–1247. doi: 10.1086/521305. [DOI] [PubMed] [Google Scholar]
- 106.Rodríguez-Rubio L, Chang W-L, Gutiérrez D, Lavigne R, Martínez B, Rodríguez A, et al. ‘Artilysation’of endolysin λSa2lys strongly improves its enzymatic and antibacterial activity against streptococci. Sci Rep. 2016;6:35382. doi: 10.1038/srep35382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chopra S, Harjai K, Chhibber S. Potential of combination therapy of endolysin MR-10 and minocycline in treating MRSA induced systemic and localized burn wound infections in mice. Int J Med Microbiol. 2016;306(8):707–716. doi: 10.1016/j.ijmm.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 108.Totté JE, van Doorn MB, Pasmans SG. Successful treatment of chronic Staphylococcus aureus-related dermatoses with the topical endolysin Staphefekt SA. 100: a report of 3 cases. Case Rep Dermatol. 2017;9(2):19–25. doi: 10.1159/000473872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fenton M, McAuliffe O, O’Mahony J, Coffey A. Recombinant bacteriophage lysins as antibacterials. Bioeng Bugs. 2010;1(1):9–16. doi: 10.4161/bbug.1.1.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Oliveira H, Vilas Boas D, Mesnage S, Kluskens LD, Lavigne R, Sillankorva S, et al. Structural and enzymatic characterization of ABgp46, a novel phage endolysin with broad anti-Gram-negative bacterial activity. Front Microbiol. 2016;7:208. doi: 10.3389/fmicb.2016.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang G, Shen X, Gong Y, Dong Z, Zhao X, Shen W, et al. Antibacterial properties of Acinetobacter baumannii phage Abp1 endolysin (PlyAB1) BMC Infect Dis. 2014;14(1):681. doi: 10.1186/s12879-014-0681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Entenza J, Loeffler J, Grandgirard D, Fischetti V, Moreillon P. Therapeutic effects of bacteriophage Cpl-1 lysin against Streptococcus pneumoniae endocarditis in rats. Antimicrob Agents Chemother. 2005;49(11):4789–4792. doi: 10.1128/AAC.49.11.4789-4792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Defraine V, Scheurmans J, Grymonprez B, Govers SK, Aertsen A, Fauvart M, et al. Efficacy of Artilysin® Art-175 against resistant and persistent Acinetobacter baumannii. Antimicrob Agents Chemother. 2016; 00285-16. [DOI] [PMC free article] [PubMed]
- 114.Gaeng S, Scherer S, Neve H, Loessner MJ. Gene cloning and expression and secretion of Listeria monocytogenes bacteriophage-lytic enzymes in Lactococcus lactis. Appl Environ Microbiol. 2000;66(7):2951–2958. doi: 10.1128/AEM.66.7.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Celia LK, Nelson D, Kerr DE. Characterization of a bacteriophage lysin (Ply700) from Streptococcus uberis. Vet Microbiol. 2008;130(1–2):107–117. doi: 10.1016/j.vetmic.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 116.Walmagh M, Boczkowska B, Grymonprez B, Briers Y, Drulis-Kawa Z, Lavigne R. Characterization of five novel endolysins from Gram-negative infecting bacteriophages. Appl Microbiol Biotechnol. 2013;97(10):4369–4375. doi: 10.1007/s00253-012-4294-7. [DOI] [PubMed] [Google Scholar]
- 117.Lin N-T, Chiou P-Y, Chang K-C, Chen L-K, Lai M-J. Isolation and characterization of ϕAB2: a novel bacteriophage of Acinetobacter baumannii. Res Microbiol. 2010;161(4):308–314. doi: 10.1016/j.resmic.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 118.Takáč M, Witte A, Bläsi U. Functional analysis of the lysis genes of Staphylococcus aureus phage P68 in Escherichia coli. Microbiology. 2005;151(7):2331–2342. doi: 10.1099/mic.0.27937-0. [DOI] [PubMed] [Google Scholar]
- 119.To KH, Young R. Probing the structure of the S105 hole. J Bacteriol. 2014;196(21):3683–9. doi: 10.1128/JB.01673-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.White R, Chiba S, Pang T, Dewey JS, Savva CG, Holzenburg A, et al. Holin triggering in real time. Proc Natl Acad Sci USA. 2011;108(2):798–803. doi: 10.1073/pnas.1011921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zimmer M, Vukov N, Scherer S, Loessner MJ. The murein hydrolase of the bacteriophage φ3626 dual lysis system is active against all tested Clostridium perfringens strains. Appl Environ Microbiol. 2002;68(11):5311–5317. doi: 10.1128/AEM.68.11.5311-5317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are not publicly available due to the risk of compromising the individual privacy of participants but are available from the corresponding author on reasonable request.