Abstract
Background
Pullulanases are the significant industrial group in the 13 glycosyl hydrolases category, known as the α-amylases family. There are very few reports on pullulanase from fungal sources. Based on the above research gap, the present study was undertaken to explore the endophytic fungi for their pullulanase-producing capabilities.
Results
A total of 126 endophytes were isolated from Tradescantia pallida, Zea mays, and Trifolium alexandrinum. Aspergillus, Penicillium, and Ganoderma species recovered highest from the stem of Tradescantia palida. Fusarium was dominant in the stem and leaf of Zea mays. Penicillium, Aspergillus, Ganoderma, Cladosporium, Fusarium, and Alternaria were recovered from the Trifolium alexandrium. The Shannon index in Tradescantia pallida was highest in leaves while in Zea mays and Trifolium alexandrinum, it is highest in the stem. The Simpson’s index is highest in the case of Zea mays stem and root. Species richness was indicated by Menhinick’s index, and it was found that this value was highest in the roots of Trifolium alexandrinum. As per our knowledge, no comparative data is available on the endophytic diversity of the above plants taken for the study. Out of 126 endophytes, only 2.38% produced pullulanase while 7.94% produced amylase. The recovery of pullulanase-producing endophytic fungi was very less. But the importance of pullulanase is high as compared to amylase because it has both α-1,6 and α-1,4 hydrolyzing ability. Therefore, the most promising isolates were identified by ITS sequence analysis. Based on spore chain morphology, isolates BHU-25 and BHU-30 were identified as Penicillium sp. and Aspergillus species, respectively. This is the first report of pullulanase from endophytic Aspergillus and Penicillium.
Conclusion
Endophytes Aspergillus sp. and Penicillium sp. produce pullulanase enzyme. This is the first report of pullulanase from endophytic Aspergillus and Penicillium.
Supplementary Information
The online version contains supplementary material available at 10.1186/s43141-021-00208-0.
Keywords: Pullulanase, Endophytes, Fungi, Aspergillus, Penicillium, Bindu Naik is the first author of the manuscript.
Background
Endophytes are microorganisms that are found inside the tissues of the host and perform ecological relationships without causing any harm to the host. They have been distributed throughout nature and are the source of various novel biomolecules such as enzymes, antibiotics, antioxidants, and anticancer compounds [1]. The endophytes also provide resistance to the plants by secreting secondary metabolites [2]. The fungal diversity is associated with different tissues of the same host plant and is also dependent on its geographical distribution and climatic conditions [3]. Nowadays, it is an emerging challenge to analyze the diversity of conglomerated fungal endophytes for the discovery of novel biomolecules producing species and their role in the ecosystem. Hence, the diversity of various endophytic microbes have been explored for their metabolic potential [4–7]. Pullulanase constitutes an important group of industrial enzyme which belongs to a family of 13 glycosyl hydrolases, also called as the α-amylase family [8, 9]. They hydrolyze the glycosidic bonds in the starch during the saccharification process which leads to the production of glucose, maltose, and maltotriose syrups. Pullulanase is produced by animals, plants, fungi, and bacteria. Among bacteria, many mesophilic, thermophilic, and hyperthermophilic bacteria and archaea have been reported to produce pullulanase. This enzyme is distributed mostly among bacteria like Clostridium spp., Bacillus spp., certain species of Bacillus, and Geobacillus [10–12]. Bacterial pullulanase has a high production cost and low yield which are major limitations in the industrial production of this enzyme. The production cost of pullulanase can be minimized by selecting agro-industrial waste as the substrate for enzyme production under solid-state fermentation (SSF) processes, which can mainly be achieved by fungi. Various agricultural wastes such as wheat bran, rice bran, corn cobs, soy hull, and sugarcane bagasse have been successfully used for the production of various metabolites especially enzymes in solid-state fermentation [13, 14]. However, very little or limited information is available on fungal pullulanase. In this context, the present study was undertaken to explore the fungal diversity of endophytes associated with Tradescantia pallida, Trifolium alexandrinum, and Zea mays having pullulanase-producing capabilities.
Methods
The plants were collected from Varanasi, Uttar Pradesh, India, in January 2014. The samples which were used in the experiment and their voucher numbers are given in Table 1. The samples were identified from the Botanical Survey of India, Dehradun, Uttarakhand, India.
Table 1.
Plant taken during the study for the isolation of endophytic fungi
| V.No. | Plant | Plant parts | Place |
|---|---|---|---|
| BHUIAS-2 | Tradescantia pallida | Root, leaf, stem | Varanasi |
| BHUIAS-3 | Zea mays | Root, leaf, stem | Varanasi |
| BHUIAS-4 | Trifolium alexandrinum | Root, leaf, stem | Varanasi |
Isolation and screening of fungi-producing pullulan hydrolyzing enzyme
The different plant parts were used for the isolation of endophytic fungi. The parts of the plants were washed with running tap water. The surface sterilization was done according to the method described previously [15]. The plant parts were cut with a sterilized sharp blade into small pieces and plated on potato dextrose agar medium and incubated at 25°C for 4–5 days. The isolates were screened for their ability to hydrolyze the pullulan by the agar plate method. The fungal strains were inoculated on pullulan agar medium and incubated at 25°C for 72 h. The plates were flooded with iodine and observed for the clear zone around the fungal colonies. The solid-state fermentation was carried in 250 ml flask containing 05g (dry weight) powdered solid substrate supplemented with nutrient salts—1% (NH4)2SO4, 1% KH2PO4, 0.2% NaCl, and 0.2% MgSO4. The flasks containing the above media were sterilized (121°C for 20 min.) and cooled, then added 1 mL of spore suspension and incubated at 27°C for 5 days. Overall 60% initial moisture content was maintained in the flask. The agro-based wastes such as wheat bran were obtained from the local market of Varanasi have been used as a solid substrate. After fermentation, the flasks were flooded with phosphate buffer (pH 6.5) and the enzyme was harvested by centrifuging at 10,000g for 10 min. The extract was used as the crude enzyme. Pullulanase activity was assayed by the DNS method [16]. The same extract was used for well diffusion assay on pullulan agar plates (1% pullulan and 1.5% agar). 0.1 mL of the enzyme sample and 0.4 mL phosphate buffer (pH 6.5) were added to 0.5 mL of 1% (w/v) solution of pullulan. The reaction mixture (substrate plus enzyme) was incubated for 30 min at 40°C. Added 1 ml DNS reagent and incubation of test tubes were performed 5–10 min in a boiling water bath. Then, after cooling down, 0.5ml of 1% (w/v) sodium-potassium tartarate solution was also added. The final volume was adjusted to 5ml by adding 2.5ml of sterile distilled water. The absorption was measured at 570 nm afterward. The protein has been estimated by Lowry et al. [17] method.
Diversity index
The Shannon diversity index (H), Simpson’s Diversity Index, and Menhinick’s index were determined according to the method described by Chowdhary and Kaushik [18].
Identification of the endophytic fungi
The promising isolates were initially identified through colony and spore chain morphology according to the method described previously [19]. For genotypic identification, genomic DNA was isolated from the endophytic fungi by using a genomic DNA isolation kit (Chromous Biotech, Bangalore, India). The endophytic fungi were grown in potato dextrose broth (PDB) and after 48 h centrifuged at 10000 rpm for 10 min to obtain mycelium. This mycelium is used for the preparation of genomic DNA by using DNA extraction kit as per kit instructions (Chromous biotech, Bangalore, India). PCR amplification of the ITS region (650–700bp) of the fungal endophytes was performed using two primers ITS1: TCCGTRSGNGAACYTGHGG and ITS4: TCCTCCGCTTATTKATDTGC. The final reaction mixture of volume 100μL contained PCR buffer F, 1.5mM MgCl2, 200μM of each dNTP, 400ng of each primer, 2.5 U Taq DNA polymerase (Genei, Bangalore, India), and 100 ng template. The amplification was carried out in an Eppendorf Thermo-cycler 96 with the following protocol: a 5-min denaturation stage at 94°C, followed by 35 amplification cycles at 94°C for 30 s, 52°C for 30 s, and 72°C for 45 s, and a 5-min extension step at 72°C. Agarose gel electrophoresis was used for the detection of PCR products and was visualized by ultraviolet (UV) fluorescence after ethidium bromide staining. PCR products were purified by HiPurATM PCR product purification spin kit (HiMedia Laboratories, Mumbai, India) according to the manufacturer’s instructions. The ABI PRISM® Big Dye® Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) was used to sequence PCR products according to the manufacturer’s instructions using ITS1 and ITS4 primers. The sequences of selected isolates were also analyzed using the BLAST (Blastn) search engine (http://www.ncbi.nlm.nih.gov) and submitted to the GenBank database (NCBI).
Molecular characterization of most promising isolates
The software included in the MEGA X package was used for phylogenetic and molecular evaluation [20]. The ITS sequences of the fungal type strains were aligned with the respective type fungal nucleotide sequences obtained from GenBank using the CLUSTAL W [21] program. The maximum likelihood method was used to study evolutionary history by using the Kimura 2-parameter model [22]. The consensus bootstrap tree derived from 1000 replicates represents the evolutionary history of the analyzed species. Branches that match partitions in bootstrap replicates reproduced in 50% collapse [23].
Results
Biodiversity of endophytic fungi
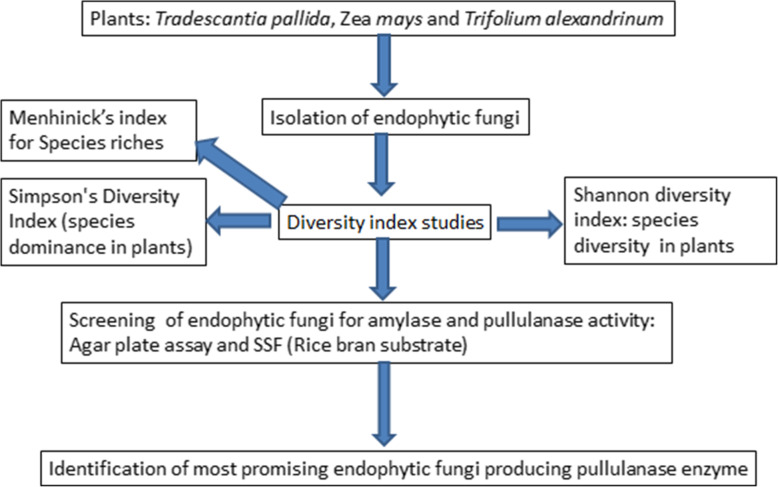
The schematic representation of the present study has been shown in Fig. 1. A total of 126 endophytes were isolated from plants under study and were identified based on cultural characteristics and spore chain morphology. The distribution of endophytic fungi in different parts (stem, root, and leaves) of Tradescantia pallida, Zea mays, and Trifolium alexandrinum has been given in Fig. 2. Aspergillus, Penicillium, and Ganoderma species recovered highest from the stem of Tradescantia palida. Fusarium was dominant in the stem and leaf of Zea mays. Penicillium, Aspergillus, Ganoderma, Cladosporium, Fusarium, and Alternaria were recovered from the Trifolium alexandrium. This finding suggests that Aspergillus is most commonly associated with plant endophytes.
Fig. 1.
The schematic representation of the present study
Fig. 2.
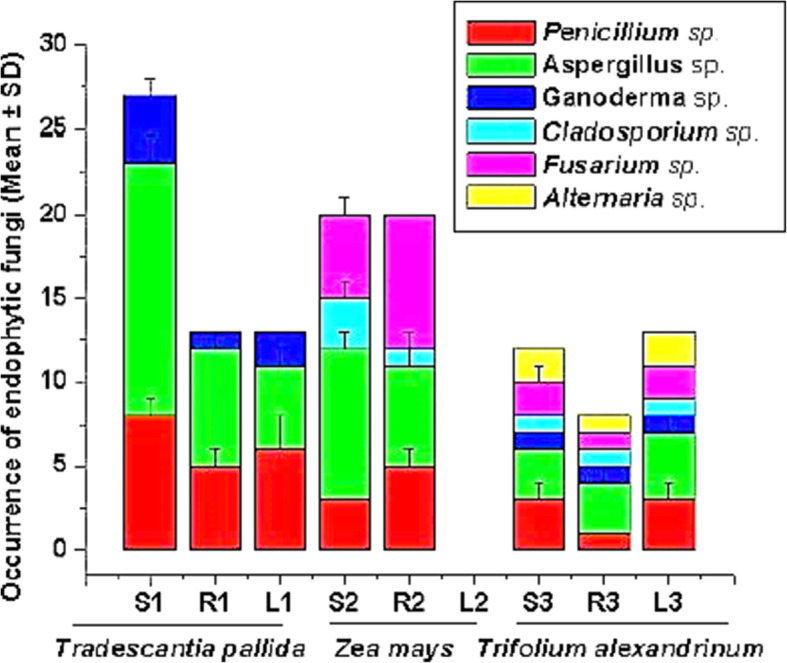
Distribution of endophytic fungi in different parts (stem, root, and leaves) of Tradescantia pallida (S1- the stem, R1-root, L1-leaf) Zea mays (S2- the stem, R2- root, L2-leaf) and Trifolium alexandrinum (S3-stem, R1-root, L3-leaf; bar indicates the ±standard deviation
The diversity index is a quantitative indicator that measures the number of different species and the degree of distribution of individuals among those species. The Shannon index in Tradescantia pallida is highest in leaves, While in Zea mays and Trifolium alexandrinum, it is highest in the stem. The Simpson’s index is highest in the case of Zea mays stem and root as shown in Table 2. Species riches are indicated by Menhinick’s index, and it is found that this value is highest in the roots of Trifolium alexandrinum. As per our knowledge, no comparative data is available on the endophytic diversity of the above plants taken for the study.
Table 2.
Alpha diversity of endophytic fungi
| Diversity index | Tradescantia pallida | Zea mays | Trifolium alexandrinum | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Stem | Root | Leaf | Stem | Root | Leaf | Stem | Root | Leaf | |
| Total species | 03 | 03 | 03 | 04 | 04 | 00 | 06 | 06 | 06 |
| Total isolates | 27 | 13 | 13 | 20 | 20 | 00 | 12 | 08 | 13 |
| Shannon index | 0.970 | 0.898 | 1.012 | 0.928 | 0.858 | 00 | 0.900 | 0.888 | .898 |
| Simpson index | 0.582 | 0.556 | 0.615 | 0.977 | 0.937 | 00 | 0.806 | 0.781 | 0.793 |
| Menhinick’s index (Dmn) | 0.577 | 0.832 | 0.832 | 0.894 | 0.894 | 00 | 1.732 | 2.121 | 1.664 |
Occurrence of endophytic fungi-producing amylase and pullulanase
The plates showing the clear zone around the colonies and wells indicate the presence of pullulanase enzyme (Fig. 3c, d and Table S1). Out of 126 endophytes, only 2.38% produced pullulanase while 7.94% produced amylase (Fig. 2). The recovery of pullulanase-producing endophytic fungi was very less. But the importance of pullulanase is high as compared to amylase because it has both α-1,6 and α-1,4 hydrolyzing ability. Therefore, the most promising isolates producing pullulanase were taken for secondary screening (in solid-state fermentation using rice bran as a substrate). In solid-state fermentation using rice bran (agro-waste) as substrate, an enzyme activity of 8.24±1.38 U/gds (protein, 2.1±0.36mg/mL) and 6.14 ±1.03 U/gds (protein 1.8±0.26 mg/mL) has been obtained by the isolates BHU-25 and BHU-46, respectively.
Fig. 3.
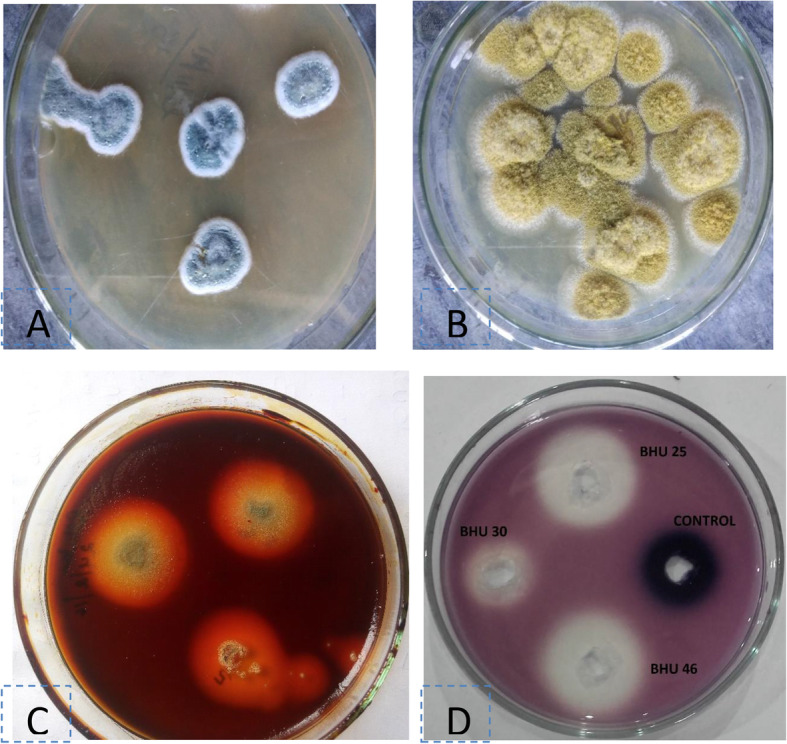
Colony morphology of isolates a BHU-25-1; b BHU-46, d hydrolysis of pullulan by the isolates BHU-25, and BHU-46 plate flooded with iodine
Characterization of promising isolates
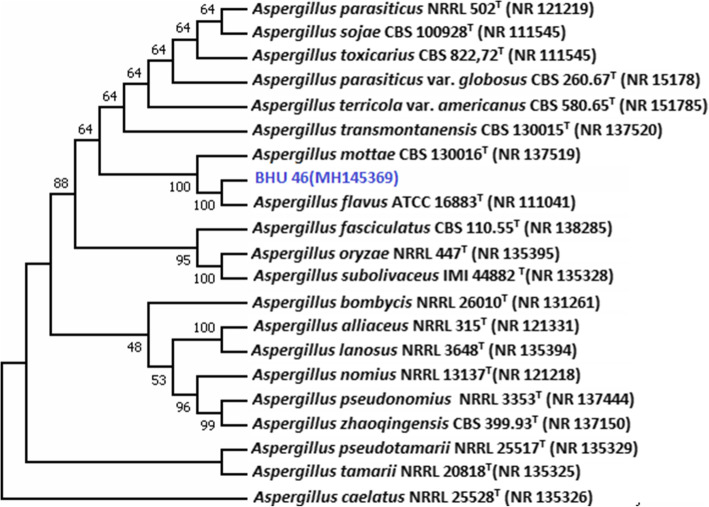
Based on colony morphology and spore chain morphology isolates, BHU-25 and BHU-46 were identified as Penicillium sp. and Aspergillus species, respectively (Figs. 3 and 4). ESEM study showed that spore chains of Penicillium were composed of spores, all of which had smooth surfaces (Fig. 4) while in Aspergillus species, the chains were composed entirely of rugose surfaces (Fig. 5). ITS sequences were submitted to the NCBI Genbank, the USA with the accession number given in Table 3. The pairwise sequence similarity index with type strains of most promising isolates has been given in Table 3. The pairwise sequence analysis showed that the isolates BHU-25 and BHU-46 are most closely related with Penicillium viridicatum FRR963T and Aspergillus flavus ATCC16883T with a sequence similarity of 99.1% and 99.4%, respectively. The evolutionary history was inferred by using the maximum likelihood method based on the Kimura 2-parameter model revealed the position of BHU-46 with Aspergillus flavus ATCC16883T with a confidence level of 100% while that of BHU-25 with Penicillium viridicatum FRR963Twith confidence of 99% (Figs. 6 and 7).
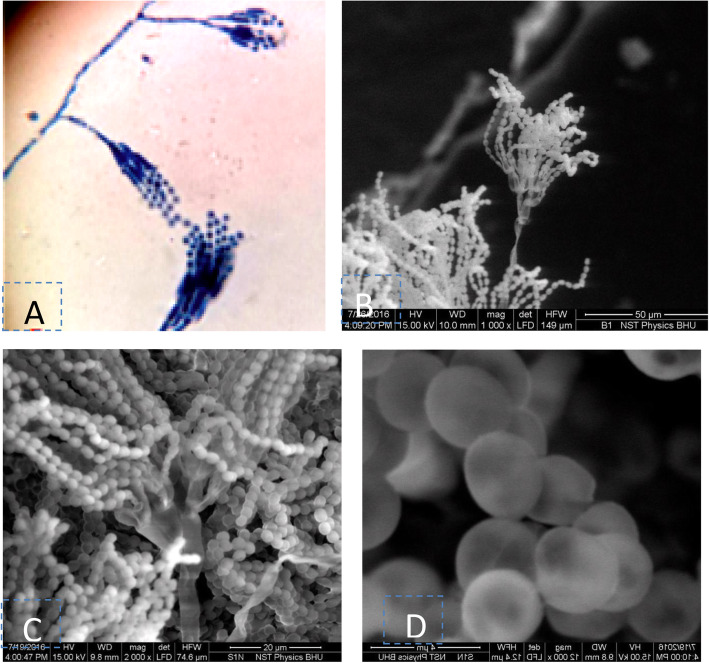
Fig. 4.
Spore chain morphology of Penicillium sp. BN-1, a-at 1000X (light microscope); b-ESEM at 1000X; c-2000X; d-Spore surface (12,000X)
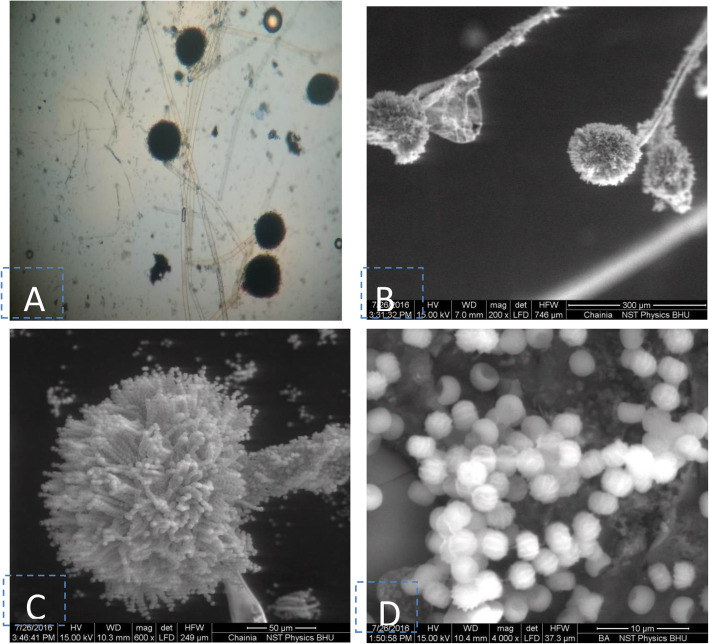
Fig. 5.
Spore chain morphology of Aspergillius sp.BN-2 , a-light microscope 10X;b- ESEM (200X); c-500X; d-spore surface (4000 X)
Table 3.
Genbank accession number of promising isolates
Fig. 6.
Phylogenetic tree of isolate BHU-46 obtained by maximum likelihood method based on the Kimura 2-parameter model. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The position of Isolate BHU 46 is shown with blue color. It is most closely related with Aspergillus flavus and supported by boot strap value of 100
Fig. 7.
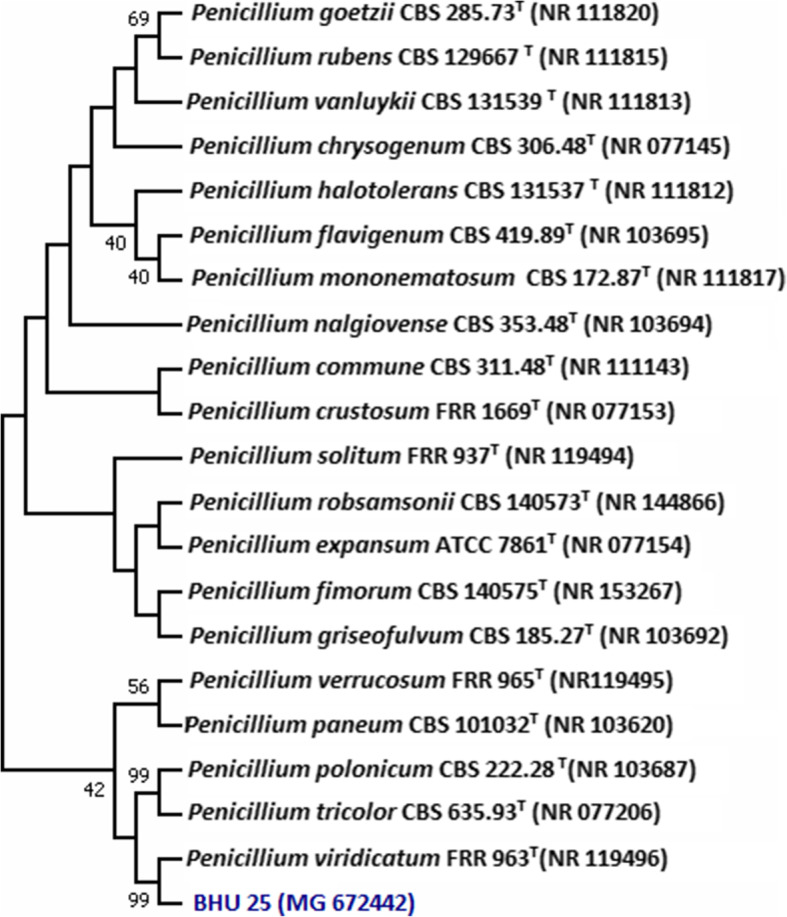
Phylogenetic tree of Isolate BHU-25 obtained by maximum likelihood method based on the Kimura 2-parameter model. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The position of Isolate BHU 46 is shown with blue color. It is most closely related with Penicillium viridicatum and supported by boot strap value of 99
Discussion
Endophytic fungi have been explored from various plant sources for their industrial potential. Therefore, researchers focused on terrestrial plants for endophytic fungi. In this context, the present study plants the diversity of endophytic fungi associated with Tradescantia pallida, Zea mays, and Trifolium alexandrinum has been explored. Aspergillus, Penicillium, and Ganoderma species recovered highest from the stem of Tradescantia palida. Fusarium was dominant in the stem and leaf of Zea mays, Penicillium, Aspergillus, Ganoderma, Cladosporium, Fusarium, and Alternaria were recovered from the Trifolium alexandrium. This finding suggests that Aspergillus is most commonly associated with plants endophytes. These findings are similar to previous reports by other authors [24]. However, most dominating fungal endophytes belonging to the genus Fusarium, Sarocladium, Aspergillus, and Penicillium are not tissue-specific [25].
The diversity index is a quantitative indicator that measures the number of different species and the degree of distribution of individuals among those species. The Shannon diversity index (H) is commonly used to characterize species diversity in a community. The Shannon index in Tradescantia pallida is highest in leaves. These findings are similar to the results of Choudhary et al. [18], in which they also reported the highest Shannon index in leaves. While in Zea mays and Trifolium alexandrinum, it is highest in the stem which is comparable with previous studies [18, 26]. Simpson’s diversity index tells us about species dominance. It considers both the number of species present and their relative abundance. Simpson’s diversity index (species dominance) is a measure of diversity that takes into account the number of species present, as well as the relative abundance of each species. The diversity of species increases as the richness and evenness increase. Its value (D) ranges between 0 and 1, where 1 represents infinite diversity and 0 represents no diversity. The Simpson’s index was highest in the case of Zea mays while Menhinick’s index was highest in roots of Trifolium alexandrinum. Species riches are indicated by Menhinick’s index. As per our knowledge, no comparative data is available on the endophytic diversity of the above plants taken for the study. When we compared the diversity with other plants the results are comparable. Li et al. [27] reported that in the root, the fungal richness was significantly higher in Salsola nitraria other plants. Furthermore, the fungal richness was significantly higher in roots than in stems. Moreover, in recent times, the diversity analysis of fungal endophytes has been performed by several authors and revealed the discovery of new species producing novel metabolites. Diversity analysis is also helpful in understanding the role of endophytes in ecosystems [28, 29].
Among the various microbial enzymes available, starch processing enzymes are one of the prominent groups applied in processes like brewing, baking, and pharmaceuticals. Amylases are the group of enzymes that are generally used for the processing of starch [30, 31]. The starch processing enzymes are classified into four different categories which include exoamylases, endoamylase, transferases, and debranching enzymes. Among the different hydrolyzing enzymes, the α-Amylases and pullulanase are the more versatile enzymes used in the industrial sector and their contribution is about 25% of the whole enzyme market [32]. These enzymes act randomly on starches, glycogen, and oligosaccharides to yield-reducing sugar. Pullulanase is the significant industrial group in the 13 glycosyl hydrolases category, known as the α-amylases family [8, 9]. They hydrolyze the glycosidic bonds in the starch during the saccharification process and yield glucose, maltose, and maltotriose syrups. These products have found their significant applications in food and other related industries. Being a member of starch hydrolyzing enzymes, pullulanase hydrolyzes both α-1,6 and α-1,4 bond in pullulan and on other carbohydrates [33–35]. Therefore, the endophytes were screened for their pullulanase and amylase-producing capabilities. The recovery of pullulanase-producing endophytic fungi was very less. But the importance of pullulanase is high as compared to amylase because it has both α-1,6 and α-1,4 hydrolyzing ability. In SSF using rice bran (agro-waste) as substrate, a good yield was recovered and merit future interest for scale up the process.
Based on colony morphology, spore chain morphology, and ITS sequence analysis, the isolates BHU-25 and BHU-46 were identified as Penicillium sp. and Aspergillus species, respectively. Waqas et al. [36] reported Penicillium and Aspergillus species from tissues of sunflower (Helianthus annuus L.). Endophytic fungi like Synnematous sp., Nodilusporium sp., and Acremonium sp. have reported starch degrading enzymes [37, 38] reported forty-four endophytic fungal strains belonging to genus Penicillium, Cladosporium, Monodictys, Phoma, Tetraploa, and Acremonium, producing enzymes of industrial importance. However, pullulanase-producing endophytic fungi have not been reported so far.
Conclusions
Tradescantia pallida, Zea mays, and Trifolium alexandrinum are rich sources of endophytic fungi. The Shannon index in Tradescantia pallida was highest in leaves while in Zea mays and Trifolium alexandrinum, it is highest in the stem. The Simpson’s index is highest in the case of Zea mays stem and root. Species richness was indicated by Menhinick’s index, and it was found that this value was highest in the roots of Trifolium alexandrinum. As per our knowledge, no comparative data is available on the endophytic diversity of the above plants taken for the study. The endophytes from these plants can produce pullulanase (2.38%) and amylase (7.94%). The recovery of pullulanase-producing endophytic fungi was very less. But the importance of pullulanase is high as compared to amylase because it has both α-1,6 and α-1,4 hydrolyzing ability. This is the first report of pullulanase from endophytic Aspergillus and Penicillium producing pullulanase.
Supplementary Information
Additional file 1. The additional file contains Table-S1 showing preliminary screening for polysaccharides hydrolyzing activity. It also contains Fig. S1 which shows the occurrence of amylase and pullulanase-producing endophytes. Table S1 shows major isolates producing amylase and Pullulanase. Three isolates BHU-20, BHU25, and BHU46 showed both amylase and pullulanase activity. Isolates BHU-25 and BHU-46 showed higher activity in Preliminary screening hence taken for further studies. Fig. S1 shows that amylase-producing isolates were dominant as compared to pullulanase-producing fungi.
Acknowledgements
Bindu Naik is highly thankful to the University Grants Commission, India, for providing national fellowship.
Abbreviations
- ITS
Internal transcribed spacer
- SSF
Solid-state fermentation
- DNA
Deoxyribonucleic acid
- PCR
Polymerase chain reaction
- MgCl2
Magnesium chloride
- dNTP
Deoxynucleotide triphosphates
- SSF
Solid-state fermentation
- UV
Ultraviolet
- ESEM
Environmental scanning electron microscope
- NCBI
National Centre for Biotechnology Information
Authors’ contributions
SKG, ADT, and VK contributed to the conceptualization, methodology, validation, and formal analysis of results. BN contributed to the conceptualization and methodology, performed the experiments, analyzed the data, wrote the original draft, and funded the acquisition. The authors read and approved the final manuscript.
Funding
BN is thankful to the University Grants Commission, India, for providing the national OBC fellowship (JRF, junior research fellowship)
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pinheiro EAA, Carvalho JM, dos Santos DCP, Feitosa ADO, Marinho PSB, Guilhon GMSP, de Souza ADL, da Silva FMA, Marinho AMDR. Antibacterial activity of alkaloids produced by endophytic fungus Aspergillus sp. EJC08 isolated from medical plant Bauhinia guianensis. Nat Prod Res. 2013;27(18):1633–1638. doi: 10.1080/14786419.2012.750316. [DOI] [PubMed] [Google Scholar]
- 2.Tan RX, Zou WX. Endophytes: a rich source of functional metabolites. Nat Prod Rep. 2001;18(4):448–459. doi: 10.1039/B100918O. [DOI] [PubMed] [Google Scholar]
- 3.Saikkonen K. Forest structure and fungal endophytes. Fungal Biol Rev. 2007;21(2-3):67–74. doi: 10.1016/j.fbr.2007.05.001. [DOI] [Google Scholar]
- 4.White JF, Jr, Cole GT, Morgan-Jones G. Endophyte-host associations in forage grasses. VI. A new species of Acremonium isolated from Festuca arizonica. Mycologia. 1987;79(1):148–152. doi: 10.1080/00275514.1987.12025383. [DOI] [Google Scholar]
- 5.Soares MA, Li H-Y, Kowalski KP, Bergen M, Torres MS, White JF. Functional role of bacteria from invasive Phragmites australis in promotion of host growth. Microb Ecol. 2016;72(2):407–417. doi: 10.1007/s00248-016-0793-x. [DOI] [PubMed] [Google Scholar]
- 6.Soares MA, Li H-Y, Kowalski KP, Bergen M, Torres MS, White J. Evaluation of the functional roles of fungal endophytes of Phragmites australis from high saline and low saline habitats. Biol Invasions. 2016;18(9):2689–2702. doi: 10.1007/s10530-016-1160-z. [DOI] [Google Scholar]
- 7.Dighton J, White J, Oudemans P. The fungal community. 4. Boca Raton: CRC Press; 2017. [Google Scholar]
- 8.Henrissat BA. classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280(2):309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matzke J, Herrmann A, Schneider E, Bakker EP. Gene cloning, nucleotide sequence and biochemical properties of a cytoplasmic cyclomaltodextrinase (neopullulanase) from Alicyclobacillus acidocaldarius, reclassification of a group of enzymes. FEMS Microbiol Lett. 2000;183(1):55–61. doi: 10.1111/j.1574-6968.2000.tb08933.x. [DOI] [PubMed] [Google Scholar]
- 10.Saha BC, Lamed R, Lee CY, Mathupala SP, Zeikus JG. Characterization of an endo-acting amylopullulanase from Thermoanaerobacter strain B6A. Appl Environ Microbiol. 1990;56(4):881–886. doi: 10.1128/aem.56.4.881-886.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melasniemi H. Genetics and Molecular Biology of Anaerobic Bacteria. New York: Springer; 1993. The α-Amylase-pullulanase (apu) gene from clostridium thermohydrosulfuricum: nucleotide sequence and expression in Escherichia coli; pp. 432–442. [Google Scholar]
- 12.Mathupala SP, Zeikus JG. Improved purification and biochemical characterization of extracellular amylopullulanase from Thermoanaerobacter ethanolicus 39E. Appl Microbiol Biotechnol. 1993;39(4-5):487–493. doi: 10.1007/BF00205038. [DOI] [Google Scholar]
- 13.Singhania RR, Patel AK, Soccol CR, Pandey A. Recent advances in solid-state fermentation. Biochem Eng J. 2009;44(1):13–18. doi: 10.1016/j.bej.2008.10.019. [DOI] [Google Scholar]
- 14.Sridevi V, Padmaja M, Sahitya A, Vardhan NH, Rao GH. Application of box-behnken design for the optimized production of lactic acid by newly isolated Lactobacillus plantarum jx183220 using cassava (Manihot esculenta crantz) flour. British Biotech J. 2015;9(2):1–9. doi: 10.9734/BBJ/2015/20236. [DOI] [Google Scholar]
- 15.Costa LE, Queiroz MV, Borges AC, Moraes CA, Araújo EF. Isolation and characterization of endophytic bacteria isolated from the leaves of the common bean (Phaseolus vulgaris) Braz J Microbiol. 2012;43(4):1562–1575. doi: 10.1590/S1517-838220120004000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31(3):426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 17.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–275. doi: 10.1016/S0021-9258(19)52451-6. [DOI] [PubMed] [Google Scholar]
- 18.Chowdhary K, Kaushik N. Fungal endophyte diversity and bioactivity in the Indian medicinal plant Ocimum sanctum Linn. PLoS ONE. 2015;10(11):e0141444. doi: 10.1371/journal.pone.0141444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naik B, Goyal SK, Tripathi AD, Kumar V. Use of environmental scanning electron microscope for taxonomy of fungi. J Adv Microsc Res. 2017;12(3):163–166. doi: 10.1166/jamr.2017.1337. [DOI] [Google Scholar]
- 20.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 23.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 24.Palencia ER (2012) Endophytic associations of species in the Aspergillus section Nigri with maize (Zea mays) and peanut (Arachis hypogea) hosts, and their mycotoxins (Doctoral dissertation, University of Georgia), Athens pp. 1–186
- 25.Potshangbam M, Devi SI, Sahoo D, Strobel GA. Functional characterization of endophytic fungal community associated with Oryza sativa L. and Zea mays L. Front Microbiol. 2017;8:325. doi: 10.3389/fmicb.2017.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandes EG, Pereira OL, da Silva CC, Bento CBP, de Queiroz MV (2015) Diversity of endophytic fungi in Glycine max. Microbiol Res 181:84–92. 10.1016/j.micres.2015.05.010 [DOI] [PubMed]
- 27.Li J-L, Sun X, Zheng Y, Lü P-P, Wang Y-L, Guo L-D. Diversity and community of culturable endophytic fungi from stems and roots of desert halophytes in northwest China. MycoKeys. 2020;62:75–95. doi: 10.3897/mycokeys.62.38923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez RJ, White JFJR, Arnold AE, Redman RS. Fungal endophytes: diversity and functional roles-Tansley review. New Phytol. 2009;182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- 29.Robl D, Delabona PdS, Mergel CM, Rojas JD, Costa PDS, Pimentel IC, Vicente VA, Pradella JGDC, Padilla G. The capability of endophytic fungi for production of hemicellulases and related enzymes. BMC Biotechnol. 2013;13:94. doi: 10.1186/1472-6750-13-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams RC. Use of polylysine for adsorption of nuclei acids and enzymes to electron microscope specimen films. Proc Natl Acad Sci. 1977;74(6):2311–2315. doi: 10.1073/pnas.74.6.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nigam P, Singh D. Enzyme and microbial systems involved in starch processing. Enzym Microb Technol. 1995;17(9):770–778. doi: 10.1016/0141-0229(94)00003-A. [DOI] [Google Scholar]
- 32.Sidhu JS, Caceres PG, Behbehani M. Measurement of starch properties during staling of Arabic bread. Starch. 1997;49(5):180–186. doi: 10.1002/star.19970490503. [DOI] [Google Scholar]
- 33.Domań-Pytka M, Bardowski J. Pullulan degrading enzymes of bacterial origin. Crit Rev Microbiol. 2004;30(2):107–121. doi: 10.1080/10408410490435115. [DOI] [PubMed] [Google Scholar]
- 34.Kuroiwa T, Shoda H, Ichikawa S, Sato S, Mukataka S. Immobilization and stabilization of pullulanase from Klebsiella pneumoniae by a multipoint attachment method using activated agar gel supports. Process Biochem. 2005;40:2637–2642. doi: 10.1016/j.procbio.2004.10.002. [DOI] [Google Scholar]
- 35.Hii LS, Rosfarizan M, Ling TC, Ariff AB. Statistical optimization of pullulanase production by Raoultella planticola DSMZ 4617 using sago starch as carbon and peptone as nitrogen sources. Food Bioprocess Technol. 2012;5(2):729–737. doi: 10.1007/s11947-010-0368-7. [DOI] [Google Scholar]
- 36.Waqas M, Khan AL, Hamayun M, Shahzad R, Kang SM, Jong-Guk Kim JG, Lee IJ. Endophytic fungi promote plant growth and mitigate the adverse effects of stem rot: an example of Penicillium citrinum and Aspergillus terreus. J Plant Interact. 2015;10(1):280–287. doi: 10.1080/17429145.2015.1079743. [DOI] [Google Scholar]
- 37.Marlida Y, Saari N, Hassan Z, Radu S. Raw Starch degrading enzyme from isolated strains of endophytic fungi. World J Microbiol Biotechnol. 2000;16:573–578. doi: 10.1023/A:1008935814516. [DOI] [Google Scholar]
- 38.Bezerra JDP, Santos MGS, Svedese VM, Lima DM M, Fernande MJS, Paiva LM, Souza-Motta CM (2012) Richness of endophytic fungi isolated from Opuntia ficus-indica Mill. (Cactaceae) and preliminary screening for enzyme production. World J Microbiol Biotechnol 28:1989–1995. 10.1007/s11274-011-1001-2 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The additional file contains Table-S1 showing preliminary screening for polysaccharides hydrolyzing activity. It also contains Fig. S1 which shows the occurrence of amylase and pullulanase-producing endophytes. Table S1 shows major isolates producing amylase and Pullulanase. Three isolates BHU-20, BHU25, and BHU46 showed both amylase and pullulanase activity. Isolates BHU-25 and BHU-46 showed higher activity in Preliminary screening hence taken for further studies. Fig. S1 shows that amylase-producing isolates were dominant as compared to pullulanase-producing fungi.
Data Availability Statement
All data generated or analyzed during this study are included in this article.