Abstract
Purpose
The purpose of this study was to develop an Asian polygenic risk score (PRS) to predict high myopia (HM) in Chinese children in the Singapore Cohort of Risk factors for Myopia (SCORM) cohort.
Methods
We included children followed from 6 to 11 years old until teenage years (12–18 years old). Cycloplegic autorefraction, ultrasound biometry, Illumina HumanHap 550, or 550 Duo Beadarrays, demographics, and environmental factors data were obtained. The PRS was generated from the Consortium for Refractive Error and Myopia genomewide association study (n = 542,934) and the Strabismus, Amblyopia, and Refractive Error in Singapore children Study (n = 500). The Growing Up in Singapore Towards healthy Outcomes Cohort study (n = 339) was the replication cohort. The outcome was teenage HM (≤ −5.00 D) with predictive performance assessed using the area under the curve (AUC).
Results
Mean baseline age ± SD was 7.85 ± 0.84 (n = 1004) and 571 attended the teenage visit; 23.3% had HM. In multivariate analysis, the PRS was associated with a myopic spherical equivalent with an incremental R2 of 0.041 (95% confidence interval [CI] = 0.010, 0.073; P < 0.001). AUC for HM (0.77 [95% CI = 0.71–0.83]) performed better (P = 0.02) with the PRS compared with a model without (0.72 [95% CI = 0.65, 0.78]). Children at the top 25% PRS risk had a 2.34-fold-greater risk of HM (95% CI = 1.53, 3.55; P < 0.001).
Conclusions
The new Asian PRS improved the predictive performance to detect children at risk of HM.
Translational Relevance
Clinicians may use the PRS with other predictive factors to identify high risk children and guide interventions to reduce the risk of HM later in life.
Keywords: high myopia, polygenic risk score, teenagers, East Asian
Introduction
The prevalence of myopia is increasing globally and is particularly high in urbanized East Asian countries where up to 80 to 97% of young adults have myopia.1–3 High myopia (HM) is associated with potentially blinding ocular complications, including myopic macular degeneration and retinal detachment.4
Myopia is a complex trait, arising from environmental factors, which include educational attainment and intensity, that are likely mediated by increased near work, and lack of outdoor time.5–9 Both genetic variation and gene-environment interactions are also important risk factors for myopia.10–13
Several large-scale genomewide association studies (GWAS) in Europeans and Asians have identified hundreds of loci associated with refractive error and myopia.14–16 These loci have enabled calculation of polygenic risk scores (PRS) that provide overall risk of individual genetic susceptibility to myopia. The PRS aggregates the effect of several genetic influences using single nucleotide polymorphisms (SNPs) allowing the estimation of specific individual risk at birth.17 The PRS in adult populations of European ancestry was able to explain 7.9% and 12.1% of the interindividual variation in spherical equivalent (SE) refractive error and self-reported age of myopia onset (area under the receiver-operating characteristic curve [AUC] of 0.75 to predict moderate myopia [MM],15,16 but it remains unclear if these findings are generalizable to populations of other ancestry and demographics. The Avon Longitudinal Study of Parents and Children (ALSPAC; n = 1516) found that individuals in the top 10% of the PRS distribution (HM ≤−5 D) had a 6.1-fold (95% CI = 3.4–10.9) higher risk than the remaining individuals.17 In a retrospective analysis in the ALSPAC (n = 2048), the combination of parental myopia and a genetic risk score gave the best performance to predict SE in children aged 7 and 15 years (P < 0.001).18
Although these studies demonstrate the utility of PRS to distinguish myopia risk in large-scale studies of mainly European ancestry, few studies have examined the generalizability of a PRS in East Asian children with the trans-ancestry portability of PRS remaining poor. In this study, we leverage data from Chinese Singaporean children (n = 1004) and summary statistics from the largest GWAS of myopia in Europeans16 to date, to generate a new PRS in East Asians.
We aim to develop a novel Asian PRS and use this PRS to predict HM in Singapore Chinese children in the Singapore Cohort of Risk Factors for Myopia (SCORM) cohort.
Methods
Singapore Cohort of Risk Factors for Myopia
SCORM is a prospective cohort whereby children from grades 1 to 3 were recruited from 3 Singapore schools in 1999 and 2001 (n = 1979), and has been described previously.19–21 Briefly, children were excluded if they had serious medical or eye disorders, such as congenital cataract. Institutional review board (IRB)/ethics committee approval was obtained. All human research was conducted according to the Declaration of Helsinki. Written informed consent was obtained after the nature of the study was explained.
Seven annual follow-up visits were conducted in the schools and children aged 12 to 18 years were seen at the last teenage follow-up (2007). Cycloplegic refraction was measured at every visit. After the instillation of 1 drop of 0.5% proparacaine, cycloplegia was achieved with 3 drops of 1% cyclopentolate instilled at 5-minute intervals. Subsequently, cycloplegic autorefraction was performed with a table-mounted autorefractor (model RK5; Canon, Japan) at least 30 minutes after the last eye drop. SE was calculated as sphere +1/2 cylinder. Individuals were further grouped into HM (≤ −5.00 D) and MM (−3.00 D to > −5.00 D). Axial length (AL) measurements were obtained after instillation of 1 drop of 0.5% proparacaine; contact ultrasound biometry was performed (Echoscan model US-800, probe frequency 10 mHz: Nidek Co., Ltd., Tokyo Japan).
Questionnaires were administered to ascertain information, such as number of books read per week and number of parents with myopia.20 Time outdoors (hours per week) in teenagers was recorded separately for school weekdays and school weekends using the Sydney Myopia Study questionnaire, and was defined as the sum of outdoor leisure and outdoor sporting activities.22
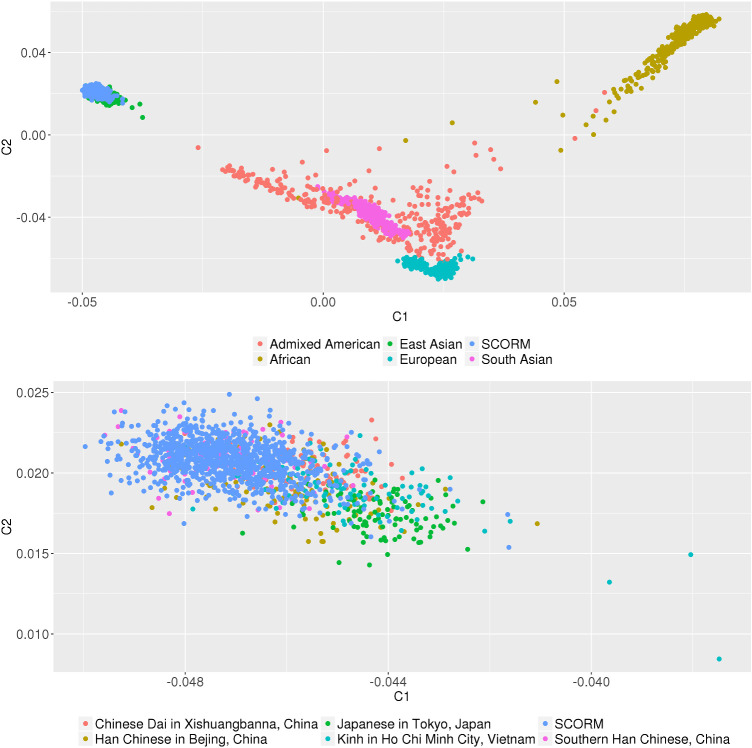
Genotyping was performed using the Illumina HumanHap 550 or 550 Duo Beadarrays23 data array for Chinese children (n = 1004) in 2006, with 571 children (57.4%) continuing through the last teenage follow-up. Quality control of the genotype data was achieved by excluding SNPs with call rates <95%, excluding minor allele frequencies (MAF) <0.05 and using the Hardy-Weinberg equilibrium (HWE) test P < 10−6. The East Asia (EAS) reference population in the 1000 Genomes reference panel was used to identify and exclude variants with differences in allele coding and SNPs with allele frequency differences >0.20. To ascertain and confirm genetic ancestry, the genotype data was combined with data from the 1000 Genomes comprised of 2504 individuals from 26 populations. Multidimensional scaling (MDS) analysis was performed on the combined set of 3508 individuals and 568,974 HapMap3 SNPs that were filtered at MAF <0.05, HWE test P < 10−6 and a genotype call rate of < 0.01. Figure 1 illustrates the first two components from the MDS analysis. The SNP data was then imputed to the 1000 Genomes reference panel using the Sanger Imputation Service for imputation with the “PBWT, no pre-phasing” pipeline. Quality control of the imputed data retained data on the basis of non-monomorphic (i.e. MAF > 0), biallelic SNPs with HWE test P > 10−6, MAF > 0.05, and an INFO score > 0.50.
Figure 1.
Multidimensional scaling analysis of SCORM. The genotype data from the SCORM cohort (n = 1004) was combined with data from the 1000 Genomes (phase 1, version 3) comprised of 2504 individuals from 26 populations. Multidimensional scaling (MDS) analysis was performed on the combined set of 3508 individuals and 568,974 HapMap3 SNPs that were filtered on minor allele frequency <0.05, Hardy-Weinberg equilibrium test P < 10−6 and genotype call rate < 0.01. Shown are the first two components from the MDS analysis.
Strabismus, Amblyopia, and Refractive Error Study
Strabismus, Amblyopia, and Refractive Error Study (STARS) is a population-based survey of Chinese families with children aged 6 to 72 months residing in the southwestern and western region of Singapore. Details of the study design and methodology have been previously described.24 Data on 550 children were included in this study.
Growing Up in Singapore Towards Healthy Outcomes Cohort Study
Growing Up in Singapore Towards Healthy Outcomes (GUSTO) cohort study consists of offspring of ethnic Chinese (60%), Malay and Indian pregnant women, aged ≥18 years who attended the first trimester antenatal clinic at the National University Hospital (NUH) and the KK Women's and Children's Hospital (KKH) in 2009 to 2010.25 In this study, we only included data for Chinese children from the 9-year visit. Myopia was measured with cycloplegic refraction. Risk factors for myopia, including near work, time outdoors, mother's education, and parental myopia, were ascertained using questionnaires. The study was approved by the SingHealth Centralized Institutional Review Board and National Health Group's Domain Specific Review Board. Written informed consent was obtained from the parents after the nature of the study was explained.
Genotyping was performed using Infinium OmniExpressExome array. Quality control included excluding SNPs with call rates < 95%, MAF < 0.05, and HWE test P < 10−6. GUSTO Chinese participants were compared with the EAS reference population in the 1000 Genome reference panel, where variants with different allele codings than 1000 Genome as well as SNPs with frequencies that differ more than 0.20 were excluded. Following quality control, a total of 529,083 SNPs were available for analysis, and were pre-phased using SHAPEIT version 2.837 with family trio information. The SNP data was then imputed to the 1000 Genome reference panel using the Sanger Imputation Service for imputation with “PBWT, no pre-phasing” pipeline. Quality control of the imputed data retained non-monomorphic (MAF > 0), biallelic SNPs with HWE test P > 10−6, MAF > 0.05, and INFO score > 0.50.
Polygenic Risk Score
We used the input GWAS summary statistics from Hysi et al., a large GWAS study on refractive error conducted in individuals of European ancestry.16 The SNPs were selected using the MAF (equal to or higher than 0.01) in Chinese children and retained if they were directly associated, or in high linkage disequilibrium (LD) with SNPs significantly associated (P < 5e-10−8) with refractive error in European adults.26 SNPs with associations that were less statistically significant in the European analysis were not considered. Presence of multiple redundant signals in our subjects of Chinese descent was subsequently verified and eventually rectified with LD pruning as implemented in the PLINK software.27 In all cases, an independent (i.e. not used for the analyses whose results are described here) population of ethnic Chinese ancestry, the STARS (n = 500), was used to calculate MAF and LD between pairs of SNP markers. SNPs were selected if they had a probability of conditional association P < 10−8.
Because the original meta-analysis association was z-score based, linear logistic regression coefficients (in standardized units) were calculated using effective population sizes using the formula:
where Zscore is the reported meta-analysis Z-score, N.eff is the effective population number, and p is the MAF at the locus.
A PRS was calculated using the 655 SNPs for each individual in SCORM as the sum of risk alleles weighted by the effect sizes described above using the PRSice version 2.3.1.e (https://github.com/choishingwan/PRSice) software.28 To determine if the inclusion of additional SNPs in the PRS calculation improved the prediction accuracy in SCORM, we applied three recently developed methods (SbayesR,29 SbayesS, and SBayesRS30), which have been shown the perform better than LDpred and clumping and thresholding (P + T) approaches. These three approaches are genomewide Bayesian methods that take as input GWAS summary statistics from Hysi et al.16 and an LD reference panel. Each method effectively shrinks SNPs effect sizes while maximizing the variance explained by binning SNPs into a mixture of normally distributed priors while accounting for LD. Shrunk sparse LD matrices generated by Lloyd-Jones et al.29 (downloaded from: https://cnsgenomics.com/software/gctb/#Download) were used as the LD reference panel, which were built using 1.09 million HapMap3 SNPs from a subset of 50,000 unrelated Europeans from the UKBiobank. Each method was run with the default parameters: –pi 0.95, 0.02, 0.02, and 0.01; gamma 0, 0.01, and 0.1, 1; chain-length 50,000; burn-in 20,000; out-freq 10, and using the –exclude-mhc flag. The PRS was calculated for each individual in SCORM using a total of 683,970 HapMap3 SNPs and multiplying the best guess genotypes by the effect sizes reweighted by SBayesR, SBayesS, and SBayesRS using the PLINK –score function. The PRSs were subsequently standardized to have a mean of zero and variance of one to aid in the interpretation of results.
Statistical Analysis
Last teenage follow-up visit (children aged 12 to 18 years) SE and AL measurements (dependent variables) were tested for association with the standardized PRS by multivariable linear regression. First, we tested a model without the PRS (basic model), including age, sex, mother's education, school, and 10 genotyping principal components. The 10 principal components are the principal components derived from the genetic relationship matrix. Specifically, we calculated the GRM using 5,285,015 imputed SNPs in SCORM using the –make-grm command in GCTA.31 Principal components were then calculated using the –pca command in GCTA. The inclusion of 10 genotypic principal components in the analysis accounts for the subtle correlation between individuals due to population stratification and cryptic relatedness.32,33 Baseline height was also included in the analysis for AL. Second, we added the PRS to the basic model (basic model + PRS) and determined the incremental R2, defined as the gain in adjusted R2 when the PRS was added to the model. Third, we added parental myopia (basic model + parental myopia), time outdoors (basic model + time outdoors), and books read per week (basic model + books read per week) to the basic model and determined the incremental R2 when parental myopia, time outdoors, or books read per week were added to the model. We also analyzed a full multivariable model, adjusting for the basic model, time outdoors, and number of books read per week. AL at the last follow-up visit was additionally adjusted for baseline height. ANOVA was performed on the basic model versus a model with the inclusion of the PRS to determine the significance of the inclusion of the PRS in the model. For the replication in the GUSTO cohort, the basic model included sex and height (for analysis of AL) as covariates. The full multivariate model included the covariates from the basic model, time outdoors, and near-work. The effect size (in standard deviation units), standard error, 95% confidence interval (CI), P value, and the adjusted R2 were used to assess the strength of associations.
Logistic regression was performed with HM (≤ −5.00 D) and MM (−5.00 D < SE ≤ −3.00 D) at the last follow-up visit as a dichotomous outcome and PRS as the independent variable in multivariable models. Individuals with >−0.5 D at the last follow-up visit were placed in the “no myopia” group.
Time-to-HM and MM analysis considered all eight visits, where all subjects were measured consecutively every year. We determined the visit number (number between 1 and 8) at which each individual developed HM and MM. For example, an individual with SE of −3.00 D at baseline (visit 1) that progressed to −5.00 D by visit 3 had time-to-HM equal to 3 and the time-to-MM equal to 1. Individuals that did not develop HM or MM by visit eight were censored. Time-dependent Cox proportional hazard models were then used to test for association between the PRS and progression to HM and MM in a multivariate model.
Classification accuracy was assessed by AUC, which relates the false-positive rate (specificity) with the true-positive rate (sensitivity). Risks factors were progressively added comparing the discriminative ability using AUC to obtain the best clinical model. The results of 4 models were analyzed: model 1 with age, time outdoors, and parental myopia; model 2, with age, time outdoors, parental myopia, and PRS; model 3 only the PRS; and model 4 only parental myopia. To compare the AUC between two receiver operating characteristic (ROC) curves, we used DeLong's test implemented in the roc.test command from the pROC library in R version 3.6.0.
Finally, for each of the HM and MM groups, odds ratios were calculated for individuals in the top 25th percentile of the PRS versus the bottom 75th percentile, and the top 50th percentile versus the bottom. The P values were calculated with a χ2 test from the 2 × 2 table of myopia level versus the PRS-risk group. All analyses were performed using R version 3.6.0.
Results
Polygenic Risk Score
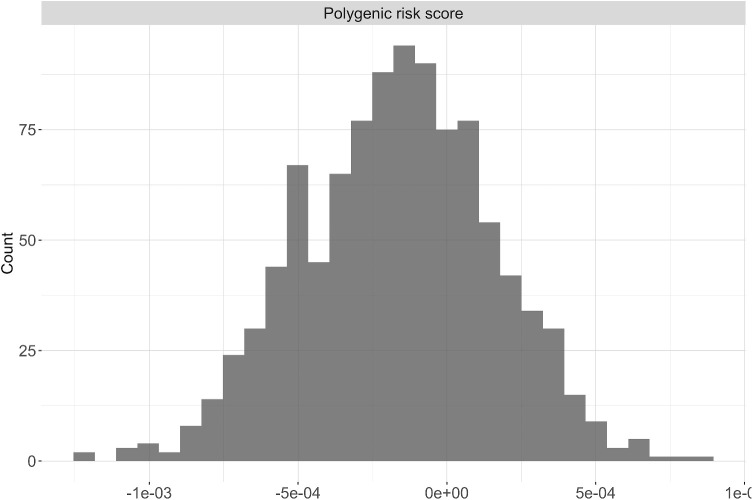
The PRS and cycloplegic auto-refraction data were available for a total of 1004 Chinese children in SCORM (mean age at baseline ± standard deviation [SD] = 7.85 ± 0.84). Of these, 571 attended the last follow-up visit with 22.8% having MM and 23.3% having HM. The mean SE at the last visit was −2.99 ± 2.57 D and mean AL at last visit was 24.75 ± 1.24 mm. The distribution of the PRS generated from the 655 SNPs in SCORM is illustrated in Figure 2.
Figure 2.
Distribution of the raw polygenic risk score in SCORM (n = 1 004).
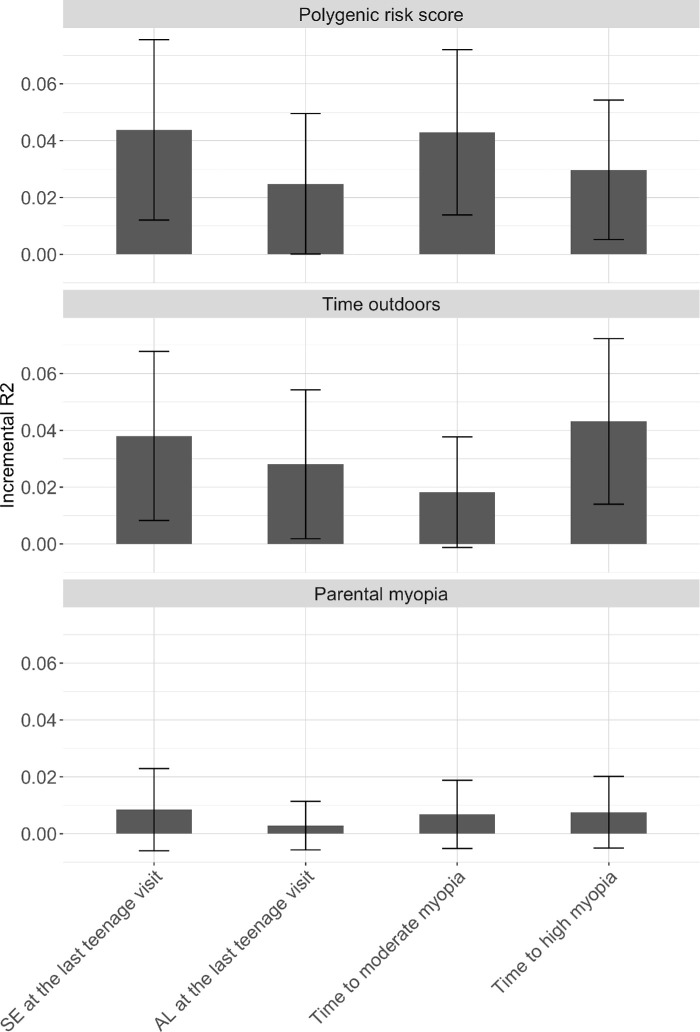
A model without the PRS (basic model), including age, sex, mother's education, school, and 10 genotyping principal components explained 4.0% and 10.8% of SE and AL variance, respectively (Table 1). Adding the PRS to the basic model showed an incremental R2 of 0.041 (95% CI = 0.010, 0.073) for SE (Fig. 3); that is, inclusion of the PRS in the basic model showed statistically significant improvement (ANOVA P < 0.001) in the prediction (i.e. increase in adjusted R2 of 4.1%) of SE. The PRS had an incremental R2 of 0.022 for AL (95% CI = −0.001, 0.046; ANOVA P < 0.001; see Fig. 3). The incremental R2 values for parental myopia and number of books read per week were lower (R2 ≤ 1%). The inclusion of parental myopia and time outdoors to the basic model showed statistically significant improvement over the basic model, although, in both cases, the incremental R2 was less than that observed with the inclusion of the PRS in the model (see Table 1). We found a small increase in the prediction accuracy of SE (i.e. incremental R2 of 4.1% vs. 6.7% with SbayesRS) and AL (i.e. incremental R2 of 2.2% vs. 3.7% with SbayesRS and SbayesS) when 683,970 HapMap3 SNPs were used in the calculation of the PRS versus our main approach that uses 655 SNPs from a GWAS of refractive error in Europeans.
Table 1.
Multivariable Linear Regression Models of Polygenic Risk Score, Parental Myopia, Time Outdoors, and Association With Teenage Spherical Equivalent and Axial Length, to Determine the Degree of Improvement in Prediction Accuracy in SCORM (n = 1004)
| Spherical Equivalent (D)a | Axial Length (mm)b | |||||||
|---|---|---|---|---|---|---|---|---|
| Adjusted R2, Full Model | Incremental Adjusted R2 | Incremental R2 95% CI | ANOVA P (vs. Basic) | Adjusted R2, Full Model | Incremental Adjusted R2 | Incremental R2 95% CI | ANOVA P (vs. Basic) | |
| Basic | 0.040 | – | – | – | 0.108 | – | – | – |
| Basic + PRS | 0.082 | 0.041 | 0.010, 0.073 | 4.85 × 10−7 | 0.130 | 0.022 | −0.001, 0.046 | 1.49 × 10−4 |
| Basic + Parental myopia | 0.051 | 0.011 | −0.006, 0.027 | 7.16 × 10−3 | 0.112 | 0.004 | −0.006, 0.014 | 0.065 |
| Basic + Time outdoors | 0.076 | 0.035 | 0.006, 0.064 | 3.36 × 10−6 | 0.133 | 0.026 | 0.0004, 0.051 | 5.06 × 10−5 |
| Basic + books read per week | 0.040 | 0 | 0, 0 | 0.466 | 0.106 | 0 | 0, 0 | 0.849 |
Basic model for spherical equivalent included age, sex, mother's education, school and 10 genotyping principal components.
Basic model for axial length included age, sex, height, mother's education, school, and 10 genotyping principal components.
Figure 3.
Prediction accuracy (incremental R2) of polygenic risk scores, time outdoors , and parental myopia in SCORM (n = 1004). Error bars represent 95% confidence intervals. Incremental R2 values represent the increase in R2 obtained by adding the PRS as predictor to a model with covariates. Multivariate models were adjusted for baseline age, sex, mother's education, school, time outdoors, number of books read per week, and 10 genotyping principal components. AL at last visit was additionally adjusted for baseline height.
The prediction accuracy of the full multivariable model was better, explaining between 11.9% and 15.7% of the variance for SE and AL, respectively (Table 2), as compared to lower prediction for parental myopia (R2 = 8.3% for SE; R2 = 13.5% for AL). Time outdoors provided similar accuracy to the PRS in relation to SE (R2 = 11.9%) and AL (R2 = 15.7%). Number of books read per week, an indicator of near work, was not significant associated with SE or AL (P > 0.05; data not shown).
Table 2.
Multivariable Linear Regression Models of Polygenic Risk Score and Teenage Spherical Equivalent and Axial Length in SCORM (n = 1004)
| Spherical Equivalent (D) | Axial Length (mm) | |||||
|---|---|---|---|---|---|---|
| Univariate | Multivariable | n | Univariate | Multivariable | n | |
| Polygenic risk score a | 568 | 548 | ||||
| β | −0.21 | −0.22 | 0.18 | 0.17 | ||
| Standard error | 0.04 | 0.04 | 0.04 | 0.04 | ||
| 95% CI | −0.30, −0.13 | −0.30, −0.14 | 0.09, 0.26 | 0.09, 0.25 | ||
| P | 6.21 × 10−7 | 1.49 × 10−7 | 4.70 × 10−5 | 5.39 × 10−5 | ||
| R2 | 0.041 | 0.119 | 0.038 | 0.157 | ||
| Parental myopia a | 568 | 548 | ||||
| β | −0.32 | −0.23 | 0.30 | 0.15 | ||
| Standard error | 0.09 | 0.09 | 0.09 | 0.09 | ||
| 95% CI | −0.49, −0.15 | −0.41, −0.05 | 0.13, 0.47 | −0.03, 0.33 | ||
| P | 2.10 × 10−4 | 0.01 | 6.05 × 10−4 | 0.10 | ||
| R2 | 0.022 | 0.083 | 0.029 | 0.135 | ||
| Time outdoors b | 568 | 548 | ||||
| β | 0.16 | 0.21 | −0.13 | −0.18 | ||
| Standard error | 0.04 | 0.04 | 0.04 | 0.04 | ||
| 95% CI | 0.08, 0.24 | 0.13, 0.29 | −0.21, −0.05 | −0.26, −0.10 | ||
| P | 1.08 × 10−4 | 8.77 × 10−7 | 2.09 × 10−3 | 1.94 × 10−5 | ||
| R2 | 0.024 | 0.119 | 0.025 | 0.157 | ||
Caption: Association effect sizes (β); 95% confidence interval (95% CI); P value (P); Adjusted R2.
Multivariate models were adjusted for baseline age, sex, mother's education, school, time outdoors, number of books read per week and 10 genotyping principal components. Axial length at last visit was additionally adjusted for baseline height.
Multivariate models were adjusted for baseline age, sex, mother's education, school, polygenic risk score, number of books read per week and 10 genotyping principal components. Axial length at last visit was additionally adjusted for baseline height.
The PRS was associated with both an altered risk to time to HM (HR = 1.49; R2 = 12.8%) and time to MM (HR = 1.39; R2 = 12.5%) with good R2 (full multivariable model; Table 3). An increase in the amount of time outdoors corresponded to a decreased risk in time to HM (HR = 0.58) and time to MM (HR = 0.79). Parental myopia had similar risk and only slightly lower R2. Books read per week, an indicator of near work, was not significant associated with time to MM (P = 0.17), but was nominally significantly associated with time to HM (P = 0.03; data not shown).
Table 3.
Cox Proportional Hazard Regression Models of Time to Moderate Myopia and Time to High Myopia in SCORM (n = 1004)
| Time to High Myopia (−5.00 D) | Time to Moderate Myopia (−3.00 D) | |||||
|---|---|---|---|---|---|---|
| Univariate | Multivariable | n | Univariate | Multivariable | n | |
| Polygenic risk score a | 672 | 672 | ||||
| HR | 1.42 | 1.49 | 1.33 | 1.39 | ||
| Standard error | 0.08 | 0.08 | 0.06 | 0.06 | ||
| 95% CI | 1.21, 1.67 | 1.26, 1.75 | 1.18, 1.49 | 1.24, 1.56 | ||
| P | 1.97 × 10−5 | 1.84 × 10−6 | 1.19 × 10−6 | 1.25 × 10−8 | ||
| R2 | 0.026 | 0.128 | 0.034 | 0.125 | ||
| Parental myopia a | 672 | 672 | ||||
| HR | 1.80 | 1.61 | 1.52 | 1.34 | ||
| Standard error | 0.19 | 0.21 | 0.12 | 0.13 | ||
| 95% CI | 1.25, 2.61 | 1.07, 2.41 | 1.19, 1.93 | 1.03, 1.74 | ||
| P | 1.71 × 10−3 | 0.02 | 7.60 × 10−4 | 0.03 | ||
| R2 | 0.016 | 0.106 | 0.018 | 0.089 | ||
| Time outdoors b | 672 | 672 | ||||
| HR | 0.71 | 0.58 | 0.89 | 0.79 | ||
| Standard error | 0.09 | 0.10 | 0.06 | 0.06 | ||
| 95% CI | 0.59, 0.85 | 0.47, 0.70 | 0.79, 1.00 | 0.70, 0.90 | ||
| P | 1.41 × 10−4 | 8.30 × 10−8 | 0.04 | 2.80 × 10−4 | ||
| R2 | 0.023 | 0.128 | 0.006 | 0.125 | ||
Caption: Hazard ratios (HR); 95% Confidence interval (95% CI); P value (P); Adjusted R2.
Multivariate models were adjusted for baseline age, sex, mother's education, school, time outdoors, number of books read per week and 10 genotyping principal components.
Multivariate models were adjusted for baseline age, sex, mother's education, school, polygenic risk score, number of books read per week and 10 genotyping principal components.
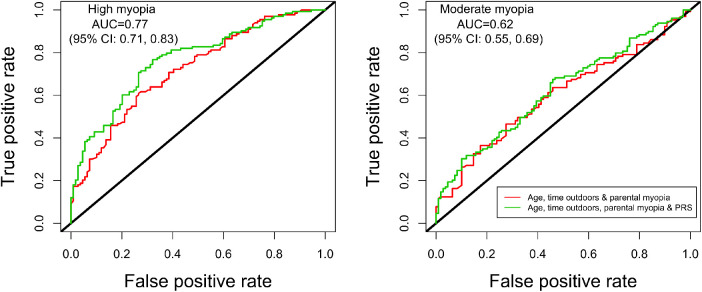
The AUC approach was used to assess the ability for the PRS to distinguish between HM (n = 133) and MM (n = 129, versus n = 109 no myopia controls), where an AUC of 1 and 0.5 represents a PRS with perfect and no discriminatory power, respectively. A model without the PRS (model 1: age, time outdoors, and parental myopia) showed an AUC of 0.72 (95% CI = 0.65, 0.78) for HM and 0.60 (95% CI = 0.53, 0.67) for MM. The AUC showed statistically significant performance improvement for HM but not MM when the PRS was added to the model (model 2: age, time outdoors, parental myopia, and PRS) with AUC for HM of 0.77 (95% CI = 0.71–0.83, DeLong's test P = 0.02; Fig. 4) and 0.62 (95% CI = 0.55–0.69; DeLong's test P = 0.36) for MM. A model with only the PRS (model 3: AUC of 0.64 [95% CI = 0.57–0.71] for HM and 0.57 [95% CI = 0.49–0.64] for MM) was more predictive than a model with only parental myopia (model 4: AUC of 0.61 [95% CI = 0.55–0.67] for HM and 0.56 [95% CI = 0.50–0.63] for MM), but the best model included both the PRS and parental myopia together with age and time outdoors (model 2). We observed a small improvement in the AUC for MM (e.g. 0.65 [95% CI = 0.58–0.72] with SbayesS) when the PRS was generated using 683,970 HapMap3 SNPs versus our main approach, but no significant improvement in the AUC was found for HM (Supplementary Fig. S1).
Figure 4.
Receiver Operating Characteristic (ROC) curve for detecting high myopia (≤ −5.00 D) and moderate myopia (≤ −3.00 D) versus no myopia controls using the polygenic risk score in SCORM. ROC curve for high myopia (≤ −5.00 D, n = 133) and moderate myopia (≤ −3.00 D, n = 129) versus no myopia controls (n = 109) with PRS, age, time outdoors, and parental myopia as predictors. The area under the curve (AUC) and 95% confidence interval corresponds to the PRS, age, time outdoors, and parental myopia model. The black line represents an AUC of 0.5.
We also analyzed the distribution of the PRS in our four myopia groups: HM (SE ≤ −5.00 D), MM (−5.00 D < SE ≤ −3.00 D), myopia (−3.00 D < SE ≤ −0.5 D), and no myopia (SE > −0.50 D). The PRS varied significantly across the four myopia groups (P < 0.001), where the average PRS increased with the severity of myopia. In particular, we found the PRS to be significantly higher in individuals with HM than children with no myopia (P < 0.001). Further, we found that individuals in the upper percentile of the PRS distribution had increased odds of HM and MM. Individuals with PRS in the top 25% had 2.34 (95% CI = 1.53, 3.55; P < 0.001) and 1.76 (95% CI = 1.20, 2.59; P < 0.001) times higher odds of HM and MM, respectively, as compared to individuals in the remaining 75% of the PRS distribution (Fig. 5). Similarly, individuals with time outdoors in the top 25% had 0.60 (95% CI = 0.37, 0.95; P = 0.032) times lower odds of HM, as compared to the remaining individuals. Books read per week could not distinguish between HM and MM (data not shown). The results were similar when comparing the top and bottom 50% of the PRS and time outdoors distribution. Individuals with PRS in the top 50% had 1.50 (95% CI = 1.07, 2.09; P = 0.02) and 1.77 (95% CI = 1.19, 2.64; P < 0.001) times higher odds of MM and HM, respectively, as compared to individuals in the bottom 50%. Further, individuals with time outdoors in the top 50% had 0.68 (95% CI = 0.49, 0.95; P = 0.02) and 0.49 (95% CI = 0.33, 0.73; P < 0.001) times lower odds of MM and HM, respectively, as compared to individuals in the bottom 50%. Again, the number of books read could not distinguish between HM and MM.
Figure 5.
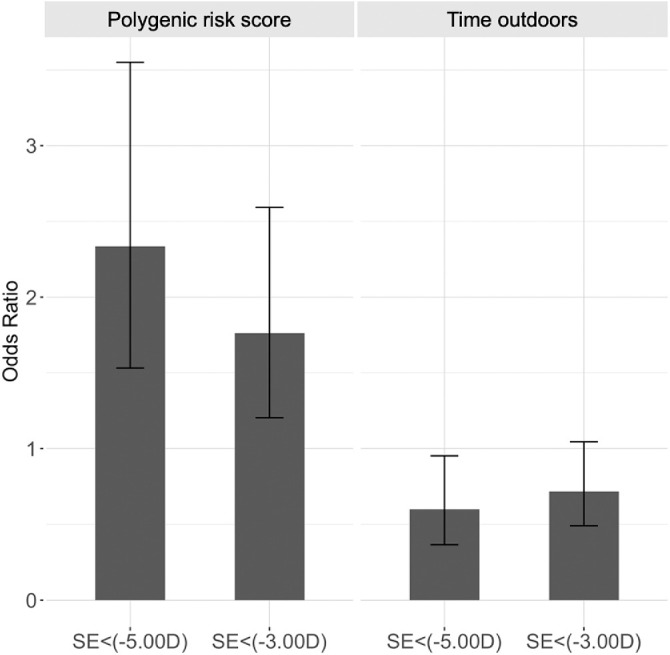
Odds ratios for teenage high myopia (≤ −5.00 D) and moderate myopia (≤ −3.00 D) for children classified as high risk using the polygenic risk score and time outdoors in SCORM (n = 1004). Odds ratios were calculated with a 2 × 2 table of myopia level (high myopia, ≤ −5.00 D and moderate myopia, ≤ −3.00 D) versus high-risk group where error bars represent 95% confidence intervals.
Growing Up in Singapore Towards Healthy Outcomes Cohort Study
The association between PRS and SE and AL were tested for replication in an independent dataset from the GUSTO cohort. Genetic and cycloplegic auto-refraction measurement information was available in a total of 339 Chinese children from GUSTO (9-year-olds). Among these children, 10.9% and 2.4% had MM and HM, respectively.
The prediction accuracy of the full multivariable model in the GUSTO cohort was similar to SCORM (Fig. 6), indicating good replicability across cohorts of similar Chinese ancestry. For example, in the full multivariable model (which included sex, time outdoors, and near-work), the association between the PRS and SE was of similar effect size in SCORM (β = −0.22; R2 = 11.9%) and in GUSTO (β = −0.24; R2 = 4.6%; P < 0.001), albeit with a much lower variation explained in GUSTO for SE at 9-year-olds. Time outdoors was associated with teenage SE in SCORM (β = 0.21; R2 = 11.9%) but was not significantly associated with SE at 9-year-olds in GUSTO (P = 0.18). The association between the PRS and AL in the full multivariable model (which included sex, height, time outdoors, and near-work) was of similar effect size in SCORM (β = 0.17; R2 = 15.7%) and in GUSTO (β = 0.19; R2 = 16.7%; P < 0.001). Adding the PRS to the basic model (which included sex as a covariate for SE, and sex and height as covariates for AL) in GUSTO showed an incremental R2 of 4.9% for SE (ANOVA P < 0.001) and 3.3% for AL (ANOVA P < 0.001); that is, inclusion of the PRS to the basic model showed statistically significant improvement in GUSTO.
Figure 6.
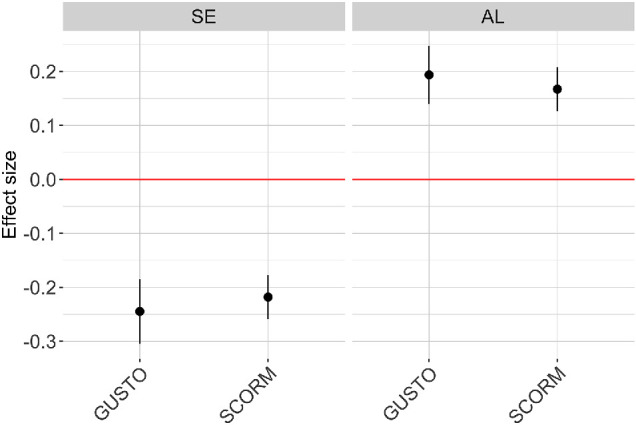
Association effects size (β) from multivariate linear regression of spherical equivalent (SE) and axial length (AL) on polygenic risk score (PRS) in SCORM (n = 1004) and GUSTO (n = 339).
Discussion
In our study, we found that our newly developed PRS based on the latest GWAS results was associated with HM. The model (model 2), including age, time outdoors, parental myopia, and the PRS, had the best predictive ability of HM. Clinicians may use the PRS with other predictive factors to identify high risk children and guide interventions to reduce the risk of HM later in life.
Our full multivariate model showed that the PRS explained 11.9% of the variance for SE, whereas parental myopia only explained 8.3%. We also found that the PRS explained about 13% of the phenotypic variance of time to HM. The PRS alone was able to incrementally explain 2.2% to 4.1% of the phenotypic variance for teenage AL and SE, respectively. These results were lower than previous studies in subjects of European ancestry that reported a genetic risk score explaining 7.9%15 (Rotterdam Study I–III; n = 10,792) and 12.1%16 of the interindividual variation in SE and self-reported age of myopia onset in a group of adults (AUC of 0.75 to predict MM). Thus, we hypothesize that the performance of the PRS may be better, as the SNPs selected on the basis of effects in adults are likely to perform better in predicting SE in adults. In a previous study from the ALSPAC mother's cohort (n = 1516 aged 24 to 51 years), the best PRS result was obtained for SE combined with age of onset of spectacle wear.17 The author's compared a genomewide approach using LDpred to a clumping and thresholding (P + T) approach applied to 1.1 million HapMap3 SNPs using a variety of P value thresholds to maximize the refractive error variance explained by the PRS. They found that a PRS generated from the top GWAS signals (R2 = 6.3% with 7372 top SNPs with P < 0.01 identified by the P + T approach) was less accurate than one generated from the 1.1 million SNPs (R2 = 11.2% by the LDpred approach). To our knowledge, this is one of the highest prediction R2 for refractive error to date.
Similar to our results, the ALSPAC birth cohort study (n = 2048; age 7–15 years) reported that the highest R2 value achieved using a genetic risk score and parental myopia was less than ∼ 7% for SE.18 The ALSPAC birth cohort study results were the first to demonstrate an independent effect of parental myopia and genetic risk on myopia in children. In our study, we found that the PRS, time outdoors, and parental myopia each have an independent effect on SE. This indicates that parental myopia may be capturing the risk of myopia from common environmental factors, and, in particular, is replicating the result from the ALSPAC birth cohort study18 in Singaporean Chinese children from SCORM. Thus, combining genetic testing with parental myopia may provide more accurate prediction of later HM and guide treatment decisions. For example, younger children with both parents having myopia may be targeted for more aggressive treatment to avoid progression to HM during their teenage years.
Our results showed acceptable accuracy of prediction of the PRS combined with parental myopia to predict HM, supporting the role of genetic factors in the progression of myopia. The transferability of the PRS across ancestry divergent populations has been shown to be influenced by differences in allele frequencies of casual variants, the magnitude and direction of effect sizes, and variation in the patterns of linkage disequilibrium between the training and target populations.34 Population-specific causal variants and gene-environment interactions may also contribute to differences in prediction performance between populations. Further, empirical and simulation studies have shown that the prediction accuracy of a genetic predictor decreases with greater genetic distance between the training and target samples.35 Therefore, there is some expectation that the prediction R2 generated in the present study will be lower than those observed in studies where the training and target samples are both of European ancestry; that is, this study does not expect to reach 11% to 12% prediction accuracy as demonstrated in other European studies. However, future large-scale GWAS studies in East Asians will likely close this gap.
We also found that the PRS alone (model 3) performed better in the prediction of HM than the conventional measurement of parental myopia (model 4). However, combining both the PRS and parental myopia with the other predictors (model 2) showed better predictive performance than using parental myopia alone to determine the risk of developing HM. Previously, genetic factors have mostly been measured by the number of myopic parents.10–12 Nevertheless, in our study, including both the PRS and number of myopic parents improved the AUC. The PRS explained higher phenotypic variance of the SE at adolescence, showing that parental myopia may be a less effective proxy for genetic factors then the PRS. For example, myopic parents may raise their children in a myopia-induced environment, with parental myopia being an imperfect proxy for genetic factors, reflecting gene-environmental interactions, such as with time outdoors, whereas the PRS allows a more accurate measurement of pure genetic risk.
In the current study, the best predictive performance was found when the PRS was combined with age, time outdoors, and parental myopia (AUC = 0.77). Although there are differences in the HM rates in Asians compared with Europeans, the predictive performance of the AUC was considered acceptable, although not excellent. It is also important to note that the PRS alone had an AUC of 0.64 for HM and 0.57 for MM, showing that accuracy of prediction for the clinical setting must improve further. Thus, further studies using large-scale GWAS studies of myopia in East Asians are necessary to ascertain the predictive performance of the PRS before translation to clinical practice.
As our PRS may predict children who develop HM later, future gene testing may be implemented using the PRS with early identification of myopic children at risk of developing HM before the irreversible elongation of the eye sets in. These children may benefit from personalized counselling as well as treatment regimens to slow the progression of myopia to HM. Genetic predictions may be combined with information on child's age, parental myopia, and other lifestyle predictors, such as time outdoors, to prevent the progression of myopia to HM and later visually disabling complications. Clinically significant results should be communicated to families to aid in their decision making and taken into consideration by the eyecare professionals in patient management and evaluation. For instance, children at higher risk may be targeted for earlier and more aggressive pharmacology and optical interventions, such as atropine, novel myopia-control contact lenses, or spectacles lenses.36,37 Other predictors, such as time outdoors, may also have an impact on the development of teenage HM. Thus, clinicians may also advise parents that time outdoors in childhood can influence the onset of myopia. The onset of myopia usually occurs in childhood and thus early childhood outdoor patterns may influence the development of myopia later. A parent or clinician can advise their children to spend more time outdoors in a feasible and consistent sustained manner with the assistance of available structured outdoor facilities and programs. Nevertheless, it is important to note that the high predictive value of time outdoors in the present study may have resulted from the delayed onset of myopia. Compared to age at myopia onset, time outdoors is a much more changeable variable over time and harder to collect. As we used a one-time measurement, time outdoors may not be the most reliable measure in the prediction model. Quantifying time outdoors through verbal questioning is also more subjective compared to other ocular predictors.
Strengths of our study include the assessment of refractive error longitudinally with cycloplegic auto-refraction in developing a novel Asian PRS from a GWAS for refractive error with a large sample size, including 542,934 participants.16 However, there were several limitations to this work. In our cohort, there was a high rate of HM (23.3% in children aged 12 to 18 years) even using a lower cut off of ≤ −5.00 D. However, the prevalence of HM in teenagers and young adults from urban Asian regions has increased. The prevalence of HM (≤ −6.00 D) was 16.6% in students from grade 12 (n = 43,858) in Fenghua (Eastern China),38 19.5% in university students in Shanghai (n = 5083),39 and 21.60% in 19-year-old men (n = 23,616) in Seoul.40 We acknowledge the biases of this high rate, namely the participants who dropped out that may have a different severity of myopia compared with participants who were followed up (lost to follow up bias). For example, children who continued in the cohort are more likely to have high myopia compared with children who drop out. The PRS-high myopia relationship for children who were followed up may also be different from children who drop out before their teenage years. Nevertheless, having a relatively high proportion of cases is not a factor that inflates the predictive value and we have replicated our results using an independent cohort (GUSTO). Additionally, we also conducted time to event analyses that allowed the analyses of children who may have been lost to follow up, considering the time they remained in the study (Cox models).
The approach used to generate the PRS in our study included 655 SNPs in the PRS calculation. This approach may be limited due to the exclusion of potentially informative SNPs. Our results were consistent with those observed by Mojarrad et al.17 where a PRS generated from the top GWAS signals (R2 = 6.3% with 7372 top SNPs with P < 0.01 identified by the P + T approach) was less accurate than one generated from the 1.1 million SNPs (R2 = 11.2% by the LDpred approach). Nevertheless, the improvement observed in the incremental R2 with the genomewide Bayesian methods in our study was relatively small. This was confirmed in the AUC/ROC analysis, which did not show a notable improvement for classification of individuals with MM or HM.
Another limitation was that SNPs for the PRS in the current study were obtained from a GWAS conducted in Europeans. The genetic structure between European and Asian subjects has different haplotypic structure arising from LD and different frequency of genetic risk factors between these populations. Empirical and theoretical studies have shown that there is an expected decrease in performance (i.e. lower incremental R2) of the PRS when transferred across ancestries.41–45 The difference in genetic structure of the European and Asian population may have contributed to lower predictive ability of the PRS in our Asian cohort (SCORM and GUSTO). Further, there have also been an increasing number of mixed marriages, so the PRS based only on the highly significant SNPs may not perform as well in other ethnic groups or admixed ethnicity groups. There is therefore a strong need for future large-scale GWAS studies of myopia in East Asians and other non-European ancestries.
Conclusion
We found that adding the PRS to other clinical information, such as child's age, time outdoors, and parental myopia, improves the prediction of HM risk (AUC = 0.77) in teenagers. The PRS alone performed well in the prediction of HM, with children in the highest PRS risk percentile having increased odds of developing HM. Our findings suggest the potential clinical value of utilizing information on this new Asian PRS together with parental myopia to improve the predictive performance to detect children at risk of HM. Clinicians may use the PRS with other predictive factors to identify high risk children and guide interventions to reduce the risk of HM later in life. Further predictive studies with genetic loci from GWAS studies of myopia in East Asians, larger sample sizes, and detailed analyses of ocular and lifestyle factors may be important to increase the predictive performance to a level acceptable for use in clinical application.
Supplementary Material
Acknowledgments
Supported by the National Medical Research Council Individual Research Grant (NMRC/0975/2005) in Singapore. The sponsor or funding organization had no role in the design or conduct of this research.
Disclosure: C. Lanca, None; I. Kassam, None; K. Patasova, None; L.-L. Foo, None; J. Li, None; M. Ang, None; Q.V. Hoang, None; Y.-Y. Teo, None; P.G. Hysi, None; S.-M. Saw, None
References
- 1. Holden BA, Fricke TR, Wilson DA, et al.. Global Prevalence of myopia and high myopia and temporal trends from 2000 through 2050. Ophthalmology. 2016; 123(5): 1036–1042. [DOI] [PubMed] [Google Scholar]
- 2. Wu HM, Seet B, Yap EPH, et al.. Does education explain ethnic differences in myopia prevalence? A population-based study of young adult males in Singapore. Optom Vis Sci. 2001; 78(4): 234–239. [DOI] [PubMed] [Google Scholar]
- 3. Pan CW, Dirani M, Cheng CY, Wong TY, Saw SM.. The age-specific prevalence of myopia in Asia: a meta-analysis. Optom Vis Sci. 2015; 92(3): 258–266. [DOI] [PubMed] [Google Scholar]
- 4. Saw SM, Matsumura S, Hoang Q V. Prevention and management of myopia and myopic pathology. Investig Opthalmology Vis Sci. 2019; 60(2): 488. [DOI] [PubMed] [Google Scholar]
- 5. Mutti DO, Mitchell GL, Moeschberger ML, Jones LA, Zadnik K.. Parental myopia, near work, school achievement, and children's refractive error. Invest Ophthalmol Vis Sci. 2002; 43(12): 3633–3640. [PubMed] [Google Scholar]
- 6. Huang HM, Chang DST, Wu PC.. The association between near work activities and myopia in children—a systematic review and meta-analysis. Jhanji V, ed. PLoS One. 2015; 10(10): e0140419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jin J-X, Hua W-J, Jiang X, et al.. Effect of outdoor activity on myopia onset and progression in school-aged children in northeast China: the Sujiatun Eye Care Study. BMC Ophthalmol. 2015; 15(1): 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He M, Xiang F, Zeng Y, et al.. Effect of time spent outdoors at school on the development of myopia among children in China. JAMA. 2015; 314(11): 1142. [DOI] [PubMed] [Google Scholar]
- 9. Wu PC, Chen CT, Lin KK, et al.. Myopia prevention and outdoor light intensity in a school-based cluster randomized trial. Ophthalmology. 2018; 125(8): 1239–1250. [DOI] [PubMed] [Google Scholar]
- 10. Low W, Dirani M, Gazzard G, et al.. Family history, near work, outdoor activity, and myopia in Singapore Chinese preschool children. Br J Ophthalmol. 2010; 94(8): 1012–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lim DH, Han J, Chung T-Y, Kang S, Yim HW.. The high prevalence of myopia in Korean children with influence of parental refractive errors: The 2008-2012 Korean National Health and Nutrition Examination Survey. Tsai D-C, ed. PLoS One. 2018; 13(11): e0207690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yam JC, Tang SM, Kam KW, et al.. High prevalence of myopia in children and their parents in Hong Kong Chinese Population: the Hong Kong Children Eye Study. Acta Ophthalmol. 2020; 98(5): aos.14350. [DOI] [PubMed] [Google Scholar]
- 13. Chen Y, Wang W, Han X, Yan W, He M.. What twin studies have taught us about myopia. Asia-Pacific J Ophthalmol. 2016; 5(6): 411–414. [DOI] [PubMed] [Google Scholar]
- 14. Verhoeven VJM, Hysi PG, Wojciechowski R, et al.. Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat Genet. 2013; 45(3): 314–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tedja MS, Wojciechowski R, Hysi PG, et al.. Genome-wide association meta-analysis highlights light-induced signaling as a driver for refractive error. Nat Genet. 2018; 50(6): 834–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hysi PG, Choquet H, Khawaja AP, et al.. Meta-analysis of 542,934 subjects of European ancestry identifies new genes and mechanisms predisposing to refractive error and myopia. Nat Genet. 2020; 52(4): 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ghorbani Mojarrad N, Plotnikov D, Williams C, Guggenheim JA. Association between polygenic risk score and risk of myopia. JAMA Ophthalmol. 2020; 138(1): 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ghorbani Mojarrad N, Williams C, Guggenheim JA. A genetic risk score and number of myopic parents independently predict myopia. Ophthalmic Physiol Opt. 2018; 38(5): 492–502. [DOI] [PubMed] [Google Scholar]
- 19. Saw SM, Tong L, Chua W-H, et al.. Incidence and progression of myopia in Singaporean school children. Investig Opthalmology Vis Sci. 2005; 46(1): 51. [DOI] [PubMed] [Google Scholar]
- 20. Saw S-M, Chua WH, Hong CY, et al.. Nearwork in early-onset myopia. Invest Ophthalmol Vis Sci. 2002; 43(2): 332–339. [PubMed] [Google Scholar]
- 21. Saw SM, Shankar A, Tan S-B, et al.. A cohort study of incident myopia in Singaporean children. Invest Ophthalmol Vis Sci. 2006; 47(5): 1839–1844. [DOI] [PubMed] [Google Scholar]
- 22. Dirani M, Tong L, Gazzard G, et al.. Outdoor activity and myopia in Singapore teenage children. Br J Ophthalmol. 2009; 93(8): 997–1000. [DOI] [PubMed] [Google Scholar]
- 23. Li YJ, Goh L, Khor CC, et al.. Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmology. 2011; 118(2): 368–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dirani M, Chan YH, Gazzard G, et al.. Prevalence of refractive error in Singaporean Chinese children: the Strabismus, Amblyopia, and Refractive Error in Young Singaporean Children (STARS) Study. Invest Ophthalmol Vis Sci. 2010; 51(3): 1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soh SE, Tint MT, Gluckman PD, et al.. Cohort Profile: Growing Up in Singapore Towards healthy Outcomes (GUSTO) birth cohort study. Int J Epidemiol. 2014; 43(5): 1401–1409. [DOI] [PubMed] [Google Scholar]
- 26. Yang J, Ferreira T, Morris AP, et al.. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012; 44(4): 369–375, S1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Purcell S, Neale B, Todd-Brown K, et al.. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81(3): 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choi SW, O'Reilly PF. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience. 2019; 8(7): giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lloyd-Jones LR, Zeng J, Sidorenko J, et al.. Improved polygenic prediction by Bayesian multiple regression on summary statistics. Nat Commun. 2019; 10(1): 5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zeng J, Xue A, Jiang L, et al.. Widespread signatures of natural selection across human complex traits and functional genomic categories. Nat Commun. 2021; 12(1): 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang J, Lee SH, Goddard ME, Visscher PM.. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011; 88(1): 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patterson N, Price AL, Reich D.. Population Structure and Eigenanalysis. PLoS Genet. 2006; 2(12): e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D.. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006; 38(8): 904–909. [DOI] [PubMed] [Google Scholar]
- 34. Wang Y, Guo J, Ni G, Yang J, Visscher PM, Yengo L.. Theoretical and empirical quantification of the accuracy of polygenic scores in ancestry divergent populations. Nat Commun. 2020; 11(1): 3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Scutari M, Mackay I, Balding D. Using genetic distance to infer the accuracy of genomic prediction. Hickey JM, ed. PLOS Genet. 2016; 12(9): e1006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cooper J, Tkatchenko A V. A. Review of current concepts of the etiology and treatment of myopia. Eye Contact Lens. 2018; 44(4): 231–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Walline JJ, Lindsley KB, Vedula SS, et al.. Interventions to slow progression of myopia in children. Cochrane Database Syst Rev. 2020; 12: CD004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen M, Wu A, Zhang L, et al.. The increasing prevalence of myopia and high myopia among high school students in Fenghua City, Eastern China: a 15-year population-based survey. BMC Ophthalmol. 2018; 18(1): 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun J, Zhou J, Zhao P, et al.. High prevalence of myopia and high myopia in 5060 Chinese university students in Shanghai. Investig Opthalmology Vis Sci. 2012; 53(12): 7504. [DOI] [PubMed] [Google Scholar]
- 40. Jung S-K, Lee JH, Kakizaki H, Jee D.. Prevalence of myopia and its association with body stature and educational level in 19-year-old male conscripts in Seoul, South Korea. Investig Opthalmology Vis Sci. 2012; 53(9): 5579. [DOI] [PubMed] [Google Scholar]
- 41. Márquez-Luna C, Loh P-R, Price AL.. Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genet Epidemiol. 2017; 41(8): 811–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Veturi Y, de los Campos G, Yi N, Huang W, Vazquez AI, Kühnel B. Modeling heterogeneity in the genetic architecture of ethnically diverse groups using random effect interaction models. Genetics. 2019; 211(4): 1395–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ.. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019; 51(4): 584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Duncan L, Shen H, Gelaye B, et al.. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun. 2019; 10(1): 3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marnetto D, Pärna K, Läll K, et al.. Ancestry deconvolution and partial polygenic score can improve susceptibility predictions in recently admixed individuals. Nat Commun. 2020; 11(1): 1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.