Summary
Human C2orf69 is an evolutionarily conserved gene whose function is unknown. Here, we report eight unrelated families from which 20 children presented with a fatal syndrome consisting of severe autoinflammation and progredient leukoencephalopathy with recurrent seizures; 12 of these subjects, whose DNA was available, segregated homozygous loss-of-function C2orf69 variants. C2ORF69 bears homology to esterase enzymes, and orthologs can be found in most eukaryotic genomes, including that of unicellular phytoplankton. We found that endogenous C2ORF69 (1) is loosely bound to mitochondria, (2) affects mitochondrial membrane potential and oxidative respiration in cultured neurons, and (3) controls the levels of the glycogen branching enzyme 1 (GBE1) consistent with a glycogen-storage-associated mitochondriopathy. We show that CRISPR-Cas9-mediated inactivation of zebrafish C2orf69 results in lethality by 8 months of age due to spontaneous epileptic seizures, which is preceded by persistent brain inflammation. Collectively, our results delineate an autoinflammatory Mendelian disorder of C2orf69 deficiency that disrupts the development/homeostasis of the immune and central nervous systems.
Keywords: C2ORF69, mitochondriopathy, inflammation, GBE1, encephalopathy, zebrafish, Elbracht-Işikay syndrome, lipase, glycogen, Mendelian genetics
Introduction
The mitochondrial proteome is composed of about ∼1,100 proteins, and fewer than a thousand are shared with yeast.1 With the exception of 13 proteins encoded by the mitochondrial genome (mtDNA), all other proteins of the mitochondrial proteome are encoded by the nuclear genome. As such, the vast majority of human diseases with mitochondrial defects, or mitochondriopathies, result from mutations in the nuclear rather than the mtDNA genome.2 In addition to their canonical role of ATP generation through oxidative phosphorylation (OXPHOS), mitochondria play a host of varied functions, ranging from regulation of apoptosis and metabolism of amino acids and lipids to calcium handling and reactive oxygen species (ROS) regulation. Primary mitochondrial disorders, such as Leigh syndrome (MIM: 256000), are often caused by pathogenic variants in mitochondrial proteins that result in bioenergetic defects.3 Other inherited mitochondrial defects arise from impairments in lipid metabolism,4 control of cell death,5 organellar and protein quality control,6 fission and fusion, metabolite biogenesis7 that impacts OXPHOS, and other cellular processes dependent on mitochondrial integrity. Despite extensive efforts to catalog the mitochondrial proteome,8 many proteins remain uncharacterized.
While mitochondrial disorders are clinically heterogeneous and can affect any organ system with varying degrees of severity and age of onset,9 mitochondriopathies often present as central nervous system diseases, consistent with their intense energy demand and high mitochondria content. Similarly, there is a preponderance of common neurological disorders with a mitochondrial basis.10 Even within the brain, systemic proteomic analysis has revealed differences in mitochondrial composition across neuronal subtypes,11 reflecting exquisite functional specificity. Brain mitochondria are required for axonal differentiation, synaptic branching, synaptic transmission, and metabolite production.12 In addition, mitochondria are intimately involved in regulating innate immunity in the brain.13 For these reasons, many mitochondriopathies are characterized by epilepsy,14 microcephaly, and neuroinflammation.15
Here, we identify and report on the clinical, genetic, and cellular characterization of a mitochondriopathy with autoinflammatory aspects that is driven by bi-allelic pathogenic variation in C2orf69 (MIM: 619219), a protein-coding gene with no known function.
Methods
Human study participants
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and proper informed consent was obtained. We withdrew peripheral blood samples from all available family members to extract genomic DNA (gDNA) by using DNeasy Blood and Tissue Kits (QIAGEN). All bio-specimens were obtained after written informed consents were signed from participants or their legal guardians. All human studies were reviewed and approved by the Rockefeller University Hospital (New York, US), Hacettepe University Ethics Committee (GO15/721, GO19/604), or the institutional review boards of A∗STAR (2019-087).
Exome and Sanger sequencing
Parents and their children were genotyped with Illumina Humancore-12v1 BeadChips following the manufacturer’s instructions. Call rates were above 99%. Gender and relationships were verified with Illumina BeadStudio. Exome sequencing was performed on gDNA from libraries prepared on an Ion OneTouch System and sequenced on an Ion Proton instrument (Life Technologies). Sequence reads were aligned to the human GRCh37/hg19 assembly (UCSC Genome browser). Each variant was annotated with the associated gene, location, protein position, amino acid change, quality score, and coverage. Variants were filtered for common SNPs with the NCBI’s “common and no known medical impacts” database, the Genome Aggregation Consortium, and the Exome Sequencing Project as well as in-house databases of sequenced individuals, mainly of Middle East origin. Homozygous variants were further filtered on the basis of functional prediction scores, including SIFT, PolyPhen-2, and M-CAP.16
CRISPR-Cas9 editing
Zebrafish were maintained and used according to the Singapore National Advisory Committee on Laboratory Animal Research Guidelines. Two guide RNAs against exon 1 of c2orf69 were used with the following targeting sequences: 5′-AGAGCAGAACATCATTGACG-3′ and 5′-GGAAAATGACCGCTGCAACG-3′. gRNAs were synthesized with a MEGAshortscript Kit (Thermo Fisher) according to the manufacturer’s protocol and purified with an RNeasy Mini Kit (QIAGEN). 1 nL of a mixture containing 500 ng/μL gRNA and 0.1 mg/mL Cas9 protein was injected into the yolk of 1-cell AB zebrafish embryos. We selected, raised, and outbred two independent lines to eliminate potential off-target mutations.
Pentylenetetrazole-treated convulsion test assay
Zebrafish behavioral experiments were conducted according to approved protocol (A∗STAR IACUC #: 191501). As described previously,17 videos of larval zebrafish at 50 fps (frames per second) were acquired and analyzed for locomotion changes in response to pentylenetetrazole (PTZ) 5 mM (final concentration in the well; Pentylenetetrazole Sigma, CAS Number 54-95-5) treatment. Briefly, 2-min videos of 7–11 days post fertilization (dpf) larvae in 24-well flat bottom plates were acquired on custom designed hardware via a Basler Ace (acA1300-200um; 1,280 × 1,024) camera with a 25 mm lens attachment placed 65 cm above the plate. The 24-well plate was prefilled with 1 mL 2% agarose and backlit by a uniform white light LED lightbox. Larvae were gently delivered singly into each well via a dropper and acclimated for 5 min. Five videos of 2 min each were acquired. PTZ or system water (control) was pipetted into each well after the first video. Larvae were tracked online during video recording on a custom written software in LabView. Low-, medium-, and high-speed swim bouts were defined as 0 to 8 mm/sec, 8 to 16 mm/sec, and >16 mm/sec, respectively, as suggested.18 Tracks, distance traveled, velocity, and immobility frequency were calculated automatically with custom written scripts in Python.
Adult fish study was performed in a similar manner. Briefly, fish were netted from the home tank in pairs and transferred to the behavior examination room. They were then transferred into two observation chambers with 150 mL of 5 mM PTZ placed against a black background and were uniformly illuminated by a white light LED lightbox. A Basler Ace (acA1300-200um; 1,280 × 1,024) camera placed in front of the tanks at ∼40 cm distance recorded videos at 50 fps for 15 min. Latency to tonic and clonic seizure-like behavior was scored according to the protocol chapter19 described for adult responses to PTZ treatment.
Quantitative real time PCR
Total RNA was isolated from fibroblast cultures with an RNeasy kit (QIAGEN, #74106) according to the manufacturer’s protocol. 1 μg of RNA was reverse transcribed with the Superscript III First-Strand Synthesis System (Invitrogen, #18080-051). Products were amplified with SYBR Green PCR Master Mix (Applied Biosystems, #4309155) on an Applied Biosystems 7500 Real-Time PCR System. We performed each reaction in triplicate and averaged and normalized data to mean β-actin RNA levels to obtain the respective ΔCT (cycle threshold) value.
Seahorse analysis
10,000 BJ-TERT fibroblast cells were plated on Seahorse XF96 Cell Culture Microplates (Agilent). For MitoStress assay, 2 μM of oligomycin (Sigma), 1 μM of FCCP (Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone) (Sigma), and 1 μM of Rotenone/Antimycin (Sigma/Sigma) were injected according to Seahorse MitoStress assay protocol (Agilent). MitoStress test data were obtained via XF96 Seahorse Wave software (Agilent). We performed citrate synthase normalization assay on the cultured cell plate to normalize the measured oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). 2,000 ReNcell VM neurons were plated on Seahorse XF96 Cell Culture Microplates and differentiated for 14 days prior to siRNA-mediated knockdown of C2orf69. Cells were subjected to MitoStress assay 3 days post transfection as described above.
Cell lines, cell culture media, and reagents
The parental BJ-TERT fibroblasts, human newborn foreskin fibroblasts immortalized with the telomerase reverse transcriptase, were provided by Procter and Gamble (Cincinnati, USA). The cell line is maintained in Fibroblast Basal Medium (#PCS-201-030) and was supplemented with Fibroblast Growth Kit-Low serum (#PCS-201-041) with a final concentration of 5 ng/mL recombinant human fibroblast growth factor (FGF) basic, 7.5 mM L-glutamine, 50 μg/mL ascorbic acid, 1 μg/mL hydrocortisone hemisuccinate, 5 μg/mL recombinant human insulin, and 2% fetal bovine serum, purchased from the American Type Culture Collection (ATCC, Manassas, VA). HEK293T cells were maintained with DMEM containing 10% fetal bovine serum (FBS). ReNcell VM human neural progenitor cell lines (Sigma Aldrich) were maintained in ReNcell NSC Maintenance medium (Sigma Aldrich) supplemented with 20 ng/mL of epidermal growth factor (EGF) (Thermo Fisher) and basic fibroblast growth factor (bFGF) (Thermo Fisher). Cells were differentiated in the same medium, but EGF and bFGF were replaced with 10 ng/mL of glial cell line-derived neurotrophic factor (GDNF) (Thermo Fisher) and brain-derived neurotrophic factor (BDNF) (Thermo Fisher) for 2 weeks with a change of media every 3 days. Cells were seeded on culture flasks or plates pre-coated overnight at 4°C with 20 μg/mL laminin (Thermo Fisher) in DPBS. The cells were subcultured approximately every 5 days at 90% confluence by detaching them with Accutase (Millipore). All cells were grown at 37°C in a 5% CO2 incubator. MG132 was purchased from Sigma Aldrich. Cells were treated with 20 nM of MG132 for 20 h prior to lysis for immunoblot analysis.
The haploid HAP1 wild-type (WT) and knockout (KO) cells (Horizon Discovery: WT, C631; C2orf69 KO1, HZGHC005743c010; C2orf69 KO2, HZGHC005744c004; GBE1 KO, HZGHC006051c010) were cultured in Iscove's Modified Dulbecco's Media (Thermo Fisher, 12440053) with 10% FBS (Sigma, F2442) and 1× penicillin-streptomycin (Thermo Fisher, 15140122) at 37°C and 5% CO2.
Immunoblotting
We prepared protein lysates from cultured cells by scraping in lysis buffer composed of 150 mM NaCl, 1% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris (pH 8.0), 0.4 mM EDTA (pH 8.0), 10% glycerol, and protease inhibitors (Sigma, 11836170001) while on ice and transferred them into microcentrifuge tubes. The extracts were clarified by centrifugation (16,000 g, 15 min, 4°C), and the supernatants were transferred to new tubes. Then 20 μg of cleared whole-cell lysate, as determined by BCA assay (Thermo Fisher, 23225), was separated on a NuPAGE 4%–12% Bis-Tris gel (Thermo Fisher, NP0323BOX) with a protein standard (Thermo Fisher, LC5800), transferred to PVDF membrane (Sigma, IPFL00010), probed with primary antibodies (GBE1, Proteintech, 20313-1-AP; GBE1, Abcam, ab180596; VDAC1, Abcam, ab18988; ACTB, Abcam, ab8224), and analyzed with a LI-COR Odyssey CLx Imaging System and LI-COR Image Studio Software (v.5.2.5) with secondary antibodies (LI-COR: 926-32211, 926-68070).
Muscle histopathology
Skeletal muscle biopsy was fresh frozen in isopentane at liquid nitrogen temperature and 8 μm-thick cryosections were used for routine diagnostic batch of histochemical stains, including hematoxylin-eosin, modified Gomori trichrome, periodic acid-Schiff (PAS), PAS-diastase, Oil Red O, nicotinamide adenine dinucleotide tetrazolium reductase (NADH), succinate dehydrogenase (SDH), cytochrome c-oxidase (COX), COX-SDH (COX-succinate dehydrogenase), and adenosine triphosphatase (ATPase).20
Cellular fractionation and mitochondrial extraction
Cytosolic, membrane, and nuclear protein extracts were obtained via the Cell Fractionation Kit Standard (Abcam #ab109719) according to the manufacturer’s instructions. Proteinase K protection and extraction assays on isolated mitochondria were performed as previously described.21 We gently resuspended mitochondria pellet in the isotonic buffer to get protein concentration at 1 mg/mL. For extraction assays, aliquots of mitochondria were solubilized by increasing concentrations of digitonin (0% to 0.22%) for 1 h at 4 °C. Soluble and insoluble fractions were separated by centrifugation at 20,000 × g for 20 min. Insoluble fractions were fully resuspended in an equal amount of isotonic buffer to match the volume of supernatant. Both fractions were lysed in 1× Laemmli sample buffer and analyzed by SDS-PAGE and immunoblotting. For proteinase K (PK) protection assays, we then added PK to 100 μg/mL and incubated it for 30 min on ice to allow the complete digestion of accessible proteins. To terminate the protease digestion, we freshly prepared PMSF and added it to a concentration of 8 mM. Samples were analyzed by western.
Protein quantification was performed with the PierceTM BCA Protein Assay Kit (Thermo Fisher #23225). For immunoblotting, samples were reduced in Laemmli loading buffer containing dithiothreitol and denatured at 95°C for 5 min. Protein samples were loaded into 4%–20% Criterion TGX Precast Midi Protein Gels (Bio-Rad #5671093) in 1× running buffer (25 mM Tris, 200 mM Glycine, 0.1% sodium dodecyl sulfate) and electrophoresed at 120 V until desired separation. Proteins were transferred from the polyacrylamide gel onto a 0.2 μm Immun-Blot Low Fluorescence PVDF Membrane (Bio-Rad #1620261) via the Trans-Blot Turbo Transfer System (Bio-Rad) for 7 min. Membranes were blocked for 1 h at room temperature with 5% milk or 5% BSA in Tris-buffered saline, Tween and then incubated with the primary antibody overnight at 4°C. Antibodies used are listed in the supplemental information.
Respiratory enzymatic assays (RCAs)
We isolated zebrafish muscle homogenates from adult zebrafish skeletal muscle and brain by disrupting dissected tissues in mitochondria isolation buffer (67 mM sucrose, 50 mM KCl, 1 mM EDTA, 0.2% fatty acid free BSA, 50 mM Tri-HCl, pH 7.4) with a Dounce homogenizer tight pestle operated at 1,300 rpm. Homogenate were subjected to RCA according to Spinazzi et al.22 with the following input quantities: 8 μg for CI, 4 μg for CII, 1.5 μg for CIII, and 1 μg for CIV. We normalized all kinetic activity measurements with citrate synthase enzyme activity of the same sample to account for differences in mitochondrial content.
Transfection by electroporation
Electroporation of C2ORF69 constructs was performed with the Neon Transfection System (Invitrogen). Briefly, cells were trypsinized, pelleted, washed, and resuspended in Resuspension Buffer R at cell density of 7 × 105 cells. The cell suspensions were mixed with 30 μg of plasmid in sterile 1.5 mL microcentrifuge tube brought to a final volume of 120 μL cell suspension with Buffer R. Electroporation was then carried out at two pulses at 1,400 V and 10 ms according to the manufacturer’s instructions.
MitoTracker Red CMXRos and immunofluorescence
Transfected and control cells on coverslips were incubated with 200 nM of MitoTracker Red CMXRos (Invitrogen) for 15 min at 37°C prior to fixation with 4% paraformaldehyde in PBS. Cells were permeabilized with 0.2% Triton-X and incubated with a monoclonal mouse antibody raised against FLAG (Sigma Aldrich) at a 1:250 dilution. Mouse anti-FLAG staining was detected with an Alexa488 conjugated Donkey anti-mouse secondary antibody (Molecular Probes). Stained cells were mounted in FluoSave Mounting Medium (Millipore). Images were acquired with 40× oil immersion objective on a Zeiss LSM700 confocal microscope. Image analysis was performed with Fiji software.23
siRNA transfection
BJ-TERT fibroblasts were reverse transfected with final concentration of 25 nM of non-targeting control siRNA (#D-001210-03-50, Dharmacon) or C2ORF69 siRNA (#M-018633-01-0005, Dharmacon) complexed with Lipofectamine RNAiMAX (Invitrogen) in Opti-MEM (Invitrogen) according to operating instructions. The siRNA and transfection reagent were mixed for 20 min prior to transfection. siRNA silencing in differentiated ReNcell VM (10 days) was performed with similar preparation but via forward transfection with Lipofectamine RNAiMAX Reagent in 96-well format.
Blue and clear native PAGE
We further isolated mitochondria by centrifuging the homogenates described above at 600 × g for 5 min to clear intact myofibrils and heavy cell debris and then spun them at 7,000 × g for 15 min to isolate the desired mitochondrial fraction. 50 μg of purified mitochondria were extracted with 8 g/g digitonin and resolved on 3%–12% native PAGE gels (Thermo Fisher) for immunoblotting (blue native) or complex I in-gel assay (clear native) according to manufacturer's instructions.24
Mitochondrial membrane potential and ROS measurements
The concentrations used for TMRE (tetramethylrhodamine, ethyl ester) (Thermo Fisher T669), JC-1 (Life Technologies 65-0851-38), and mitoSOX (Thermo Fisher M36008) were 100 nM, 1 μM, and 5 μM, respectively. Cells were incubated in culture media (BJ fibroblasts) or HBSS (5.6 mM glucose + 1% BSA) with the dyes for 30 min at 37°C. The cells were then washed with warm PBS, trypsinized and pelleted before resuspending in warm media for acquisition by flow cytometry or imaged immediately on the Operetta system (Perkin Elmer). For JC-1 measurements, cells were imaged in both Y3 and GFP channels under non-saturating parameters. The ratio between Y3 and GFP (red/green) was obtained by segmenting on the Y3 channel at a threshold that accurately selected all JC-1-positive mitochondria.
Results
A recessive Mendelian disorder caused by C2orf69 deficiency
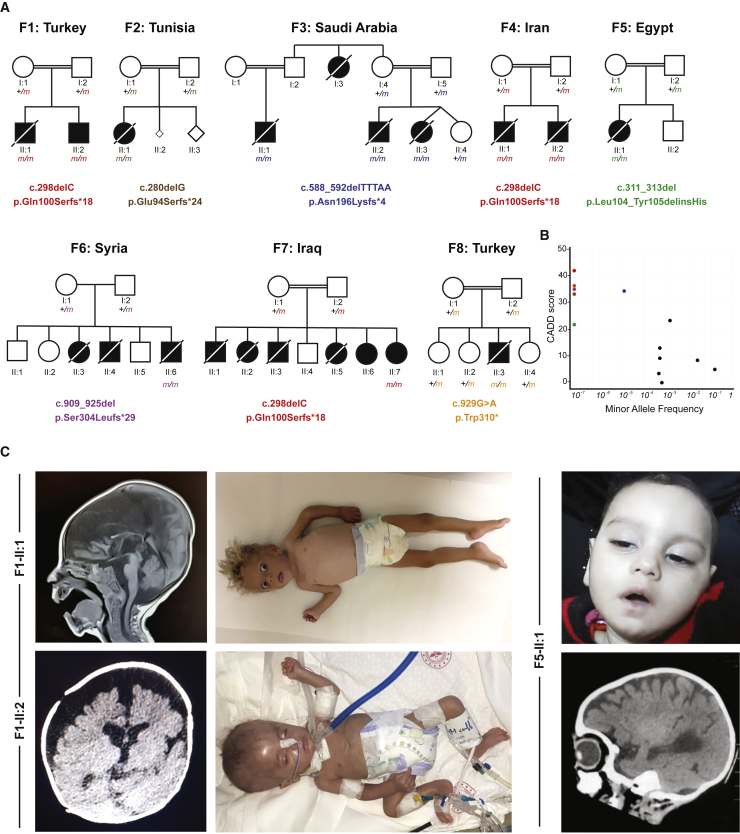
We initiated this study with the clinical investigation of two affected brothers born to consanguineous parents of Kurdish Turkish origin (Figures 1A and 1C). Both siblings presented with multisystem involvement in the first 3 months of the postnatal period (Table 1). The clinical findings were failure to thrive, global developmental delay, periodic fevers with elevated C-reactive protein, hypochromic microcytic anemia, and episodes of septic and aseptic osteomyelitis and/or arthritis. At 12 months of age, the index individual, I:2, showed progredient and severe microcephaly with a head circumference of 39 cm, exceeding −5 standard deviation (SD). Cranial MRI revealed prominent leukoencephalopathy with cerebellar atrophy/Dandy-Walker variant, corpus callosum dysgenesis, and diffuse hypomyelination (Figures 1C and 1D). Both brothers experienced recurrent seizures. Proband II:1 from family 1 (F1) died of pneumonia at 18 months of age. Exome sequencing (ES) revealed a potentially disease-causing germline homozygous frameshift variant (c.298del [p.Gln100Serfs∗18] [GenBank: NM_153689.5]) in chromosome 2 open-reading frame 69 (C2orf69, MIM: 619219). Sanger sequencing confirmed that both neurotypical parents are heterozygous for this variant, which was found to fulfill Mendelian expectations for an autosomal recessive trait in the younger affected brother II:2 (family 1).
Figure 1.
Eight families segregating recessive loss-of-function C2orf69 germline mutations
(A) Pedigrees of eight consanguineous families segregating homozygous C2orf69 loss-of-function variants. The identified germline homozygous mutations are shown for 12 affected individuals; eight additional children with similar symptoms died before they could be tested.
(B) C2orf69 is intolerant of genetic variation. Minor allele frequency (MAF) and combined annotation-dependent depletion (CADD) score of homozygous C2orf69 coding variants found in gnomAD v.2.1.1 (black dots) and those found in each family (color-coded dots).
(C) Photographs and brain MRIs taken at 10 months of age (F1-II:1), 5 months of age (F1-II:2), and 6 months of age (F5-II:1) showing cerebral atrophy with leukoencephalopathy.
Table 1.
Abridged clinical presentation of 12 children with homozygous loss-of-function C2orf69 variants causing Elbracht-Işikay syndrome
| Family 1 | Family 2 | Family 3 | Family 4 | Family 5 | Family 6 | Family 7 | Family 8 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individual | II:1 | II:2 | II:1 | II:1 | II:2 | II:3 | II:1 | II:2 | II:1 | II:6 | II:7 | II:3 |
| Origin | Turkey | Turkey | Tunisia | Saudi Arabia | Iran | Egypt | Syria | Iraq | Turkey | |||
| Sex | male | male | female | male | male | female | male | male | female | male | female | male |
| C2orf69 variant (GenBank: NM_153689.5) | c.298delC | c.280delG | c.588_592delTTTAA | c.298delC | c.311_313del | c.909_925del | c.298delC | c.929G>A | ||||
| Predicted/observed protein change (UniProt: Q8N8R5) | p.Gln100Serfs∗18/p.0 | p.Glu94Serfs∗24/NT | p.Asn196Lysfs∗4/NT | p.Gln100Serfs∗18/p.0 | p.Leu104_Tyr105delinsHis/NT | p.Ser304Leufs∗29/NT | p.Gln100Serfs∗18/p.0 | p.Trp310∗/NT | ||||
| Clinical synopsis | ||||||||||||
| Disease onset | 3 months | 3 months | congenital | NA | NA | congenital | 3 months | 4 months | 2 months | 4 months | neonatal | neonatal |
| Head circumference at birth (cm) | normal | NA | 35 | NA | NA | NA | normal | normal | 33 (−0.6 SD) | normal | NA | 35 (−0.4 SD) |
| Birth weight (kg)/birth length (cm) | 3.75/normal | 2.3/45 | 3.1/50 | NA | NA | 2.12/NA | 3.5/52 | 3.7/50 | 3.0/49 | 3.5/normal | 3.5/NA | 3.1/49 |
| Age at last exam (month)/head circumference (cm) | 12/39 (−5 SD) | 6/39 (−3.1 SD) | 18/39.5 (−6 SD) | NA | NA | 6/36 (−6 SD) | 24/NA | NA | 6/37 (−3.5 SD) | 6/NA | 6/NA | 7/37 (−5.5 SD) |
| Failure to thrive (weight at last exam [kg]) (HP: 0001508) | + (4.9, −5.3 SD) | + (4.3, −4.3 SD) | + (6.5, −3 SD) | NA | NA | + (4.6) | + | NA | + (4.7, −3.25 SD) | + (2.5, −5 SD) | + | + (5.8, −3 SD) |
| Post-natal short stature (length at last exam [cm]) (HP: 0004322) | + (68, −2.7 SD) | + (58, −3.7 SD) | + (85) | NA | NA | + (53) | NA | NA | + 59 (−3.4 SD) | NA | + | + (60, −3.5 SD) |
| Deceased (age of death) | + (18 months) | alive (in critical condition) | + (32 months) | + (NA) | + (18 months) | + (24 months) | + (29 months) | + (11 months) | + (9 months) | + (9 months) | alive (in critical condition) | + (12 months) |
| Cause of death | pneumonia | NA | status epilepticus | NA | NA | NA | pneumonia | pneumonia | pneumonia | cardiac arrest | NA | pneumonia |
| Brain anomalies | ||||||||||||
| Developmental delay (HP: 0001263) | + | + | + | + | + | + | + | + | + | + | + | + |
| Secondary microcephaly (HP: 0005484) | + | + | + | + | + | + | NA | NA | + | − | + | + |
| Dysgenesis of corpus callosum (HP: 0006989) | + | + | − | NA | NA | NA | + | + | + | + | + | − |
| CNS hypomyelination (HP: 0003429) | + | + | − | NA | NA | NA | NA | NA | + | + | + | + |
| Cerebral atrophy (HP: 0002059) | + | + | + | NA | NA | NA | NA | NA | + | + | + | + |
| Seizures (HP: 0001250) | + | + | + (focal, pharmacoresistant) | + (intractable) | + | + (focal) | + (absent seizures, several episodes) | NA | + (myoclonic, several times a day) | + (tonic) | + (focal and myoclonic) | + (intractable, focal) |
| Immune anomalies | ||||||||||||
| Recurrent fever (HP: 0001954) | + | + | + | NA | NA | NA | + | + | + | − | + | + |
| Inflammatory arthritis (HP: 0001369) | − | − | − | NA | NA | NA | + | + | − | − | − | + (knee, once) |
| Septic arthritis (HP: 0003095) | − | − | − | NA | NA | NA | + | + | − | − | − | − |
| Aseptic osteomyelitis (HP: 0002754) | + (tibia, elbow, hip) | − | − | NA | NA | NA | + (elbows, hip, clavicle) | NA | − | − | − | − |
| Elevated C-reactive protein level (HP: 0011227) | + | + | − | NA | NA | NA | + | NA | + | − | − | + |
| Hypochromic microcytic anemia (HP: 0004840) | + | + | NT | NA | NA | NA | + | NT | + | NA | NA | anemia due to congenital dyserythropoietic anemia |
| Other phenotypes | ||||||||||||
| Muscular hypotonia (HP: 0001252) | + | + | + | NA | + | + | − | − | + | + | + | + |
| General muscle wasting (HP: 0009055) | + | + | + | NA | NA | NA | + | NA | + | + | + | + |
| Abdominal distention (HP: 0003270) | + | + | − | NA | NA | NA | + | + | NA | − | − | − |
| Muscular spasticity (HP: 0001257) | − | − | + | NA | NA | NA | + | + | NA | + | NA | − |
| Hepatomegaly (HP: 0002240) | + | + | − | NA | NA | NA | − | − | − | − | NA | − |
Abbreviations are as follows: −, negative; +, affirmative; NT, not tested; NA, not available; CS, caesarean section; NVD, normal vaginal delivery. Additional information on each individual is presented in the supplemental information.
Eighteen other similarly affected children, nine girls and nine boys, from seven additional families originating from Tunisia (family 2 [F2]), Saudi Arabia (family 3 [F3]), Iran (family 4 [F4]), Egypt (family 5 [F5]), Syria (family 6 [F6]), Iraq (family 7 [F7]), and Turkey (family 8 [F8]) were recruited through existing collaborations (Figure 1A). All children shared a triad of symptoms consisting of brain atrophy with progressive leukoencephalopathy and recurrent seizures, septic inflammation, and failure to thrive (Table 1). Of the 20 affected children, 12 could be shown to have inherited homozygous damaging C2orf69 variants, while eight others, who had died before this study began, were unavailable for testing (Figure 1A). In the affected individual from F8, developmental and epileptic encephalopathy is accompanied by congenital dyserythropoietic anemia (MIM: 615631). The detailed clinical features of each family can be found in the supplemental notes.
According to gnomAD (v.2.1.1 and v.3.1), no homozygous damaging variants have been reported for C2orf69. The three frameshift variants, c.298del (p.Gln100Serfs∗18) of families 1, 4, and 7, c.280delG (p.Glu94Serfs∗24) of family 2, and c.909_925del (p.Ser304Leufs∗29) of family 6, and stop-gained c.929G>A (p.Trp310∗) mutations of family 8 have not been reported in public databases (gnomAD, BRAVO/TOPmed) or in combined in-house databases consisting of >50,000 exomes/genomes. The c.588_592delTTTAA (p.Asn196Lysfs∗4) variant from family 3 is rare and was reported seven times before in the heterozygous state (rs775817125; MAF = 2 × 10−5). All five truncating variants were predicted to be deleterious and had combined annotation-dependent depletion (CADD) scores above 30 (Figure 1B). The in-frame deletion/insertion c.311_313del (p.Leu104_Tyr105delinsHis) variant identified in II:1 from family 5 is annotated as a possible non-damaging change by MutationTaster and has a computed CADD value of 22.4. C2orf69 has a residual variation intolerance score (RVIS) of 0.39 (placing it in the top 76% of human genes most intolerant to genetic variation) and a pLOF observed/expected score of 0.38 (gnomAD), suggesting that C2orf69 is a target of negative selection. Overall, these clinical and genetic findings suggest that homozygosity for pLOF variants is exceedingly rare in the general population and that the C2orf69 variants observed in these eight kindreds are probably deleterious, most likely revealing the genetic etiology for this heretofore unknown fatal autoinflammatory and neurodevelopmental disorder (NDD).
C2orf69 is an evolutionarily conserved protein in most eukaryotic species
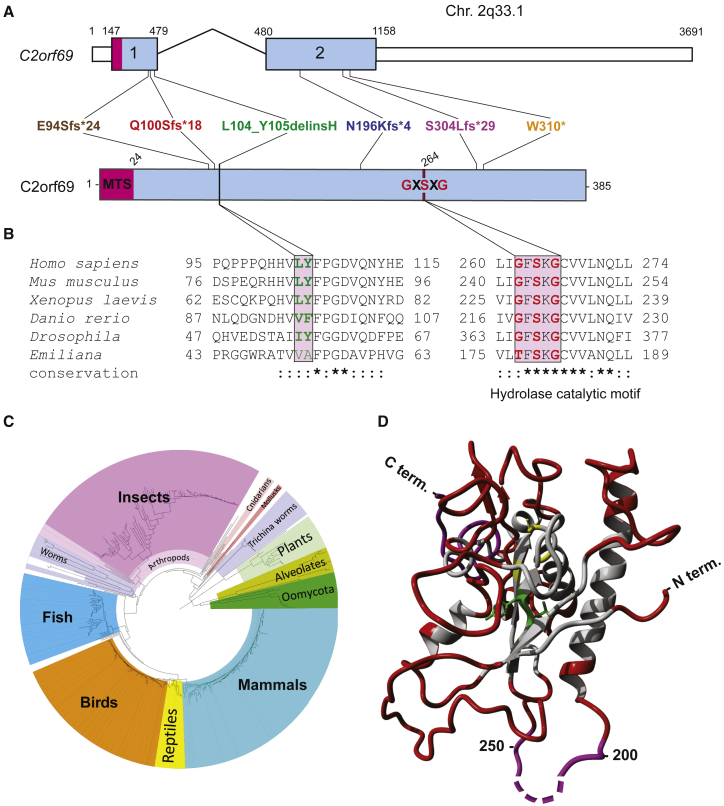
Genomic sequence analysis revealed that human C2orf69 is encoded by two exons on chromosome 2q33.1 (hg19) (Figure 2A). The four identified frameshift variants are expected to significantly alter the length of the encoded protein Q8N8R5, which in humans, is predicted to be 385 amino acids long and consist of an N-terminal mitochondrial targeting signal (MTS) rich in arginine followed by a UPF0565 domain of unknown function. C2orf69 orthologs, identified with reciprocal best BLAST hits search against GenBank,25 can be found in most eukaryotic genomes, in all metazoans, some plant genomes, and unicellular organisms, such as the phytoplankton Emilia huxleyi (Figure 2C). Surprisingly, no ortholog was detected in fungal genomes. Remote structure prediction and modeling with HHpred and Modeler26 identified multiple significant hits to proteins from the hydrolase fold family, suggesting C2ORF69 may encode an enzyme belonging to the class of esterases or lipases (Figure 2D). Consistently, the hydrophobic core and predicted catalytic site are invariant for the serine 264 in all sequences analyzed (Figure 2B). The low complexity portion between residues 200 and 250 is presumably not part of the globular fold. In insect sequences, such as Drosophila, this central loop is significantly extended by ∼100 residues. The p.Leu104_Tyr105delinsHis variant observed in subject II:1 of family 5 is part of a highly conserved portion of residues (Figure 2B) that are at the hydrophobic core in the center of the hydrolase fold (Figure 2D) and would most likely disrupt the 3D structure of this presumed enzyme.
Figure 2.
C2ORF69 is conserved in most eukaryotic species and possesses homology to esterase enzymes
(A) Exon-intron genomic organization of C2orf69 with positions of the six germline loss-of-function mutations identified.
(B) Protein organization of C2orf69 with positions of identified mutations.
(C) Amino acid sequence conservation of C2orf69 orthologs across all major eukaryotic phyla. With the exception of fungi, C2orf69 is recorded in all metazoans, plants, and phytoplankton.
(D) 3D structure prediction of human C2ORF69 with annotated residues L104_Y105 in green and predicted catalytic residue Ser264 in yellow.
C2ORF69 encodes an outer-membrane mitochondria-targeted protein
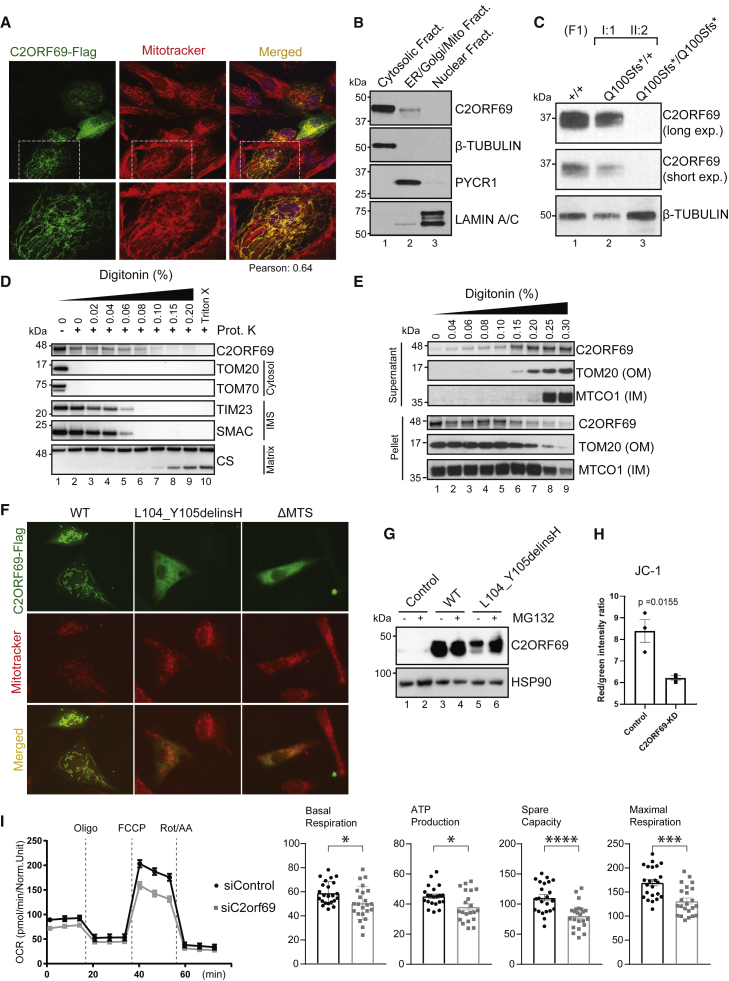
Automated sequence annotations by UniProt and the Human Protein Atlas indicate that C2ORF69 may contain a secretion signal peptide consistent with extracellular localization. However, the endogenous C2ORF69 in the supernatant of cultured fibroblasts was variable and sometimes undetectable. When overexpressed, a small fraction of the tagged protein could be detected in the cultured media (Figure S1A).
We sought to clarify the subcellular localization of C2ORF69, but available antibodies could not detect the endogenous protein by immunofluorescence. Therefore, we expressed the open-reading frame with a FLAG tag at its C terminus in human fibroblasts. The tagged protein was found to be in close proximity to mitochondria, as revealed by co-localization with MitoTracker Red CMXRos (Figure 3A). Next, we performed cell fractionation by using primary human fibroblasts and obtained a major cytosolic localization and a minor fraction of endogenous C2ORF69 with the mitochondrial and ER/Golgi membranes (Figure 3B, lanes 1–3). This result was not changed in fibroblasts that are deficient for MTX1 and MTX2,5 two proteins involved in the translocation of nuclear-encoded proteins into the outer mitochondrial membrane (OMM) (Figure S1B, lanes 4–6). The specificity of the commercial antibody was validated with cellular extracts of primary dermal fibroblasts obtained from the heterozygous mother (F1-I:1) and her affected son (F1-II:2), which showed that the p.Gln100Serfs∗18 variant results in a protein null allele (Figure 3C). To determine the topology and localization of mitochondrial-associated C2ORF69, we treated HEK293T mitochondrial extracts with proteinase K in the presence of increasing concentrations of digitonin. This revealed that the majority of C2ORF69 is associated with the OMM, facing the cytosolic side, as evidenced by the susceptibility of C2ORF69 to proteinase K in the absence of digitonin (Figure 3D, lane 1 versus lane 2). However, a minority of C2ORF69 displays evidence of import into the intermembrane space (IMS) (Figure 3D, lanes 3–5) with traces detectable even in the matrix (Figure 3D, lanes 6–9). Compared to OMM integral membrane proteins such as TOM20, C2ORF69 can be extracted by the lowest digitonin concentrations (Figure 3E), suggesting that it is not inserted into the OMM and remains primarily associated with the OMM on the cytosolic face. The low cytosolic signal observed by immunofluorescence suggests that most of the protein is loosely associated with mitochondrial membranes and may dissociate during cell fractionation (Figure 3B). Alternatively, the protein may primarily reside in the cytosol and become selectively trafficked to the mitochondrion.
Figure 3.
C2ORF69 associates with mitochondria and affects oxidative respiration of neurons
(A) Immunostaining of BJ-TERT fibroblasts overexpressed with C2ORF69-FLAG with FLAG antibodies (green) revealed co-localization with mitochondria marker, MitoTracker CXMRos (red). Pearson’s correlation coefficient was analyzed with Fiji software.
(B) Cellular fractionation of primary fibroblasts indicates that endogenous C2ORF69 is mostly cytoplasmic, but a small fraction is associated with membranous fractions including mitochondria.
(C) C2ORF69 protein is absent in primary dermal fibroblast with the homozygous p.Gln100Serfs∗18 variant.
(D) Proteinase K protection assay of HEK293T mitochondria to reveal topology and submitochondrial location of endogenous C2ORF69. The majority of the protein resides in the outer membrane vulnerable to proteinase K, and a small fraction displays evidence of translocation into the mitochondria. IMS, intermembrane space.
(E) Differential membrane extraction assays in HEK293T mitochondria confirm that endogenous C2ORF69 is both cytosolic and mitochondrial membrane associated.
(F) Immunostaining of BJ-TERT fibroblasts overexpressed FLAG epitope-tagged wild-type (WT) C2ORF69, L104_Y105delinsH, and C2ORF69 without mitochondria-targeting signal (ΔMTS). We immunostained cells with FLAG antibodies (green) and MitoTracker CXMRos (red) to determine co-localization.
(G) Immunoblot of fibroblasts overexpressing wild-type C2ORF69 or L104_Y105delinsH treated with proteasome inhibitor, MG132 (20 μM). Treatment with MG132 rescued expression level of p.Leu104_Tyr105delinsHis to near WT levels.
(H) Ratiometric JC-1 (1 μM) membrane potential measurement in ReN VM neurons. Each dot represents the average of all ratiometric measurements across three separate 40× fields of one well. Data are mean ± SEM, and p value is derived from unpaired t test.
(I) Agilent Seahorse Mito Stress Test on siControl- and siC2orf69-differentiated ReNcell VM neurons with basal respiration, ATP production, maximal respiration rate, and spare values indicated.
Importantly, overexpression of the proband-derived p.Leu104_Tyr105delinsHis variant resulted in the loss of mitochondrial targeting, as did the deletion of the predicted mitochondrial targeting or association signal (MTS) (Figure 3F). By immunoblot, we noted that the p.Leu104_Tyr105delinsHis mutant protein was present at a much lower level than its WT counterpart (Figure 3G, lane 3 versus lane 5). This most likely reflects decreased protein stability because addition of MG132, a proteasomal inhibitor, could readily rescue its half-life to WT levels (Figure 3G, lane 5 versus lane 6). These results indicate that this indel mutation is most likely a bona fide loss-of-function variant that disrupts the structure of C2ORF69, rendering the protein unstable and targeting it for proteasome-mediated degradation. These cellular and molecular results, which are congruent with those reported in children with highly deleterious truncating variants, are consistent with the phenotype observed in subject II:1 of family 5.
C2ORF69 depletion affects mitochondrial respiration
Epileptic seizures are a cardinal feature of mitochondrial diseases caused by mutations in mtDNA and nuclear-encoded mitochondrial genes.14 We next investigated the effects of C2ORF69 depletion on mitochondrial activity in primary fibroblasts by using siRNA-mediated knockdown of endogenous C2ORF69 (Figure S1C). C2orf69 knockdown (KD) did not overtly impair respiration (Figure S1D) but led to slightly reduced membrane potential (Figure S1E). Concurrently, the production of mitochondrial reactive oxygen species (mitoROS), as assayed by mitochondrial-targeted redox-sensitive dye mitoSOX, was significantly elevated (Figure S1F). Since the probands’ phenotypes largely manifest in the CNS, we reasoned that neurons are a more relevant cell lineage to address the etiology of this disease. We differentiated neuronal progenitors ReNcell VM into neurons and again depleted C2ORF69 by using siRNA (Figure S1G). On the other hand, mitochondrial membrane potential was significantly affected as measured by the ratiometric potential-sensitive JC-1 dye27 (Figure 3H, Figure S1H). A potential caveat is that JC-1 is sensitive to H2O2 levels, such that the observed results could be partially due to increased ROS production in siC2ORF69 neurons. The observed attenuation in mitochondrial membrane potential resulted in significantly reduced oxidative respiration in siC2ORF69 neurons (Figure 3I). Overall, these data suggest a mitochondrial defect that is cell type specific and more pronounced in neuronal lineages.
With fatal seizures, C2orf69 knockout zebrafish phenocopy human syndrome
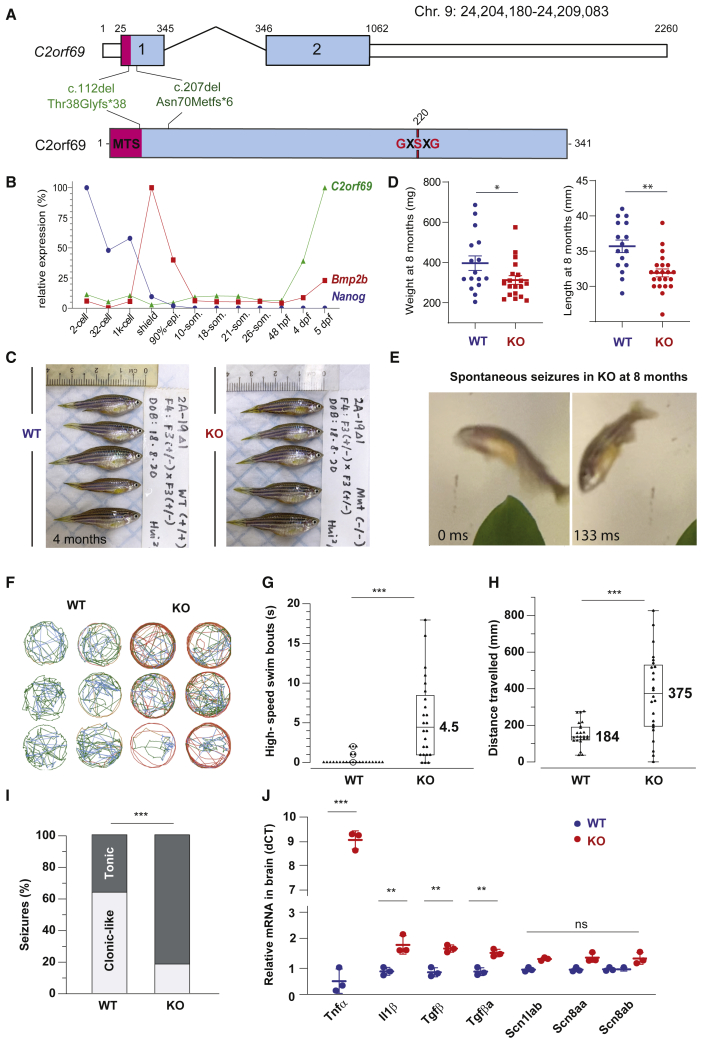
To better understand the physiological role of C2orf69, we set out to engineer knockout zebrafish by using CRISPR-Cas9 technology. Through the use of two gRNAs targeting exon 1 of the zebrafish C2orf69 ortholog (GenBank: NM_001077728.1, zgc153521), we identified, selected, and outbred two independent germline frameshift alleles: c.112del (p.Thr38Profs∗38) and c.207del (p.Asn70Metfs∗6) (Figure 4A). qPCR analysis indicated that C2orf69 mRNA is not maternally deposited in the egg (but its protein could be present by virtue of being attached to mitochondria), and a zygotic transcription began by 2 dpf (Figure 4B). Inbreeding of heterozygous mutant fish results in the expected 25% ratio of homozygous knockout animals, which by the age of 4 months, were indistinguishable in size from WT siblings (Figure 4C, Figure S2A). Zygotic null fish could be inbred allowing to produce 100% maternal zygotic (MZ) knockout fish. Whole larval respiration at 36 hours post fertilization (hpf) of MZ knockout embryos was not significantly altered (Figure S2B), possibly reflecting the lineage-specific effects of C2orf69 knockdown observed in human neurons (Figures 3H and 3I). While skeletal muscle mitochondria from adult MZ knockouts did not have gross electron transport chain (ETC) assembly defects as ascertained by BN-PAGE (Figure S2C) and respiratory chain enzymatic assays (Figure S2D), respiratory supercomplexes were mildly attenuated as revealed by in-gel complex I enzymology (Figure S2E). These results indicate that C2ORF69 does not directly impact the integrity of the ETC but nonetheless is required for proper oxidative respiration.
Figure 4.
KO C2orf69 zebrafish phenocopy the human syndrome with spontaneous fatal seizures
(A) Exon-intron structure of the C2orf69 ortholog in zebrafish. Annotations of the two distinct germline frameshift mutations generated by CRISPR-Cas9 editing at the genome and protein levels.
(B) Developmental expression of C2orf69 during early zebrafish embryogenesis. The transcription of C2orf69 begins at 48 hpf without any detectable maternal contribution.
(C) C2orf69 KO fish are indistinguishable from WT siblings at 4 months.
(D) C2orf69 KO fish show statistically significant reduced body mass and length at 8 months.
(E) Spontaneous seizures in adult 8-month-old KO fish lead to fully penetrant lethality.
(F) Six representative swimming tracks extracted from 2 min videos of 11 dpf KO and WT larvae each show high-speed swimming bouts (red) within 5 min of exposure to 5 mM PTZ.
(G) Quantification of high-speed swim bouts in 2 min (n = 24 each). The p value of the two-sided permutation t test is <0.0001
(H) Quantification of distance swam in 2 min (n = 24 each). The p value of the two-sided permutation t test is 0.0008
(I) The percentage of KO adult fish that show tonic seizures upon exposure to 5 mM PTZ (80%) within 12 min is 2.5-fold higher compared to WT (27%). The p value of the two-sided t test is 0.03.
(J) Molecular markers from 4-month-old whole brain extracts measured by qPCR reveal constitutive CNS inflammation in KO adult fish compared to WT siblings.
By 8 months of age, MZ knockout fish were significantly smaller both in terms of weight and length (Figure 4D), suggesting a possible growth retardation compared to WT fish. Under normal conditions, we observed that adult C2orf69Asn70Metfs∗3 knockout zebrafish died between 8 to 10 months of spontaneous epileptic seizures (Figure 4E and Video S1). This striking phenotype is reminiscent of the documented seizures seen in children lacking C2orf69. To further verify that mutant fish had increased susceptibility to seizures, we subjected 7- to 11-day-old larva to PTZ, a convulsant shown to induce seizure-like activity in zebrafish.28 Mutants showed a lower latency to reach stage 2 seizure-like activity (Figure 4F), characterized by frequent high-speed swimming bouts (Figure 4G) and traveling greater distances (Figure 4H) compared to WT larvae. The reduction in latency for induced seizure was also seen in 2- to 3-month-old adults with the same treatment. Mutant adults exhibit tonic seizure-like activity (Video S2) compared to WT siblings, which usually show clonic seizure-like activity (Figure 4I).
To examine whether mutant fish might exhibit tissue damage in the CNS, we collected whole brains from healthy 4-month-old WT and MZ null fish and measured a series of molecular markers by qPCR (Figure 4J). Consistent with sterile CNS inflammation, knockout fish revealed significantly increased expression of il1β, tgfβ, and tnfα (9-fold over WT). Levels of the Nav1.1 voltage-gated sodium channels, scn1ab, scn8aa, and scn8ab were unchanged, suggesting that the disease etiology is distinct from Dravet syndrome (MIM: 607208), another severe pediatric epilepsy syndrome that is successfully modeled in zebrafish.29 These results obtained in a surrogate animal model reveal that the in vivo function of C2orf69 is conserved between mammals and fish and that the product of this gene is essential for brain development and homeostasis in two distantly related vertebrate species.
C2ORF69 inactivation leads to glycogen metabolic defects in humans and zebrafish
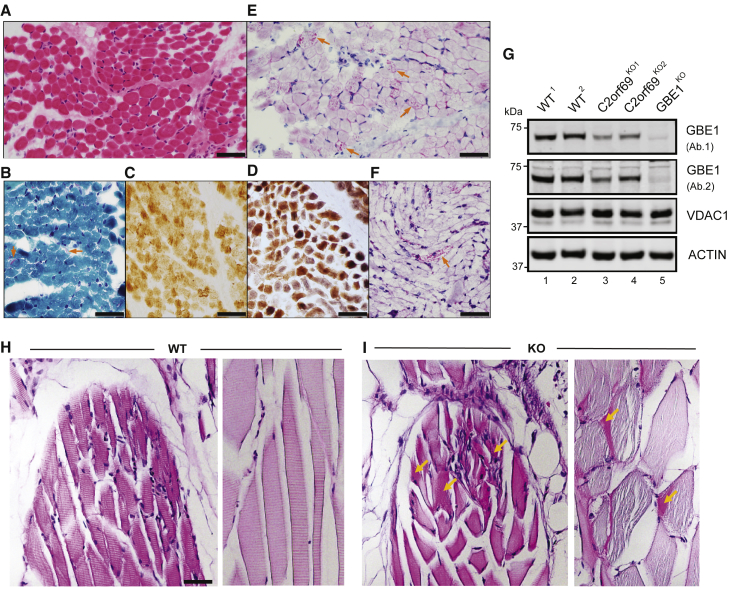
A muscle biopsy obtained during the diagnostic work-up of the affected individual (II:3) from family 8 at the age of 5 months revealed some changes suggestive of mitochondrial myopathy, such as subsarcolemmal accumulation of mitochondria and mild cytochrome c-oxidase (COX) deficiency (Figures 5A–5D). Although typical ragged-red fibers were not seen in sections stained by modified Gomori trichrome stain, few fibers showed abnormal mitochondrial aggregates (Figure 5B). COX staining was faint in some fibers, better detected by COX-SDH staining (Figure 5D). An intriguing finding was the presence of abnormal granular staining by PAS, which was partially resistant to diastase (amylase), in many fibers (Figures 5E and 5F). Diastase-resistant PAS staining indicates accumulation of polyglucosan bodies, which is pathognomonic for glycogen storage disease type IV (MIM: 232500), caused by defects in glycogen branching enzyme encoded by GBE1 (MIM: 607839). A possible relationship between GBE1 and C2ORF69 was also suggested by Kurth and colleagues.21 We thus generated two independent CRISPR-mediated C2orf69 KO lines and a GBE1 KO line in human haploid HAP1 cells. By immunoblot, compared to parental WT cells, endogenous GBE1 levels were decreased by approximately 50% in both C2orf69 KO lines (Figure 5G, lanes 1 and 2 versus lanes 3 and 4).
Figure 5.
C2orf69 deficiency leads to the accumulation of glycogen in skeletal muscles
(A) Skeletal muscle biopsy of proband II:3 from family 8 shows mild variation of fiber size in H&E sections.
(B) Subsarcolemmal accumulation of mitochondria is apparent in some fibers (arrows) by modified Gomori-trichrome stain.
(C) Several fibers show faint staining by cytochrome-c-oxidase (COX) stain.
(D) These COX-deficient fibers are more easily identified as bluish fibers by COX-SDH stain.
(E) Periodic acid Schiff (PAS) stain shows unusual granular staining in many fibers (arrows), which are partially resistant to diastase, as seen by PAS-diastase stain (F).
(F) PAS-positive material is seen in a few fibers after diastase treatment. Scale bars represent 50 μm.
(G) Approximately 50% decrease in endogenous GBE1 levels is seen in two independent C2orf69 KO HAP1 cell lines with two different antibodies (Ab1, Proteintech; Ab2, Abcam). Two WT and GBE1 KO HAP1 cell lines act as positive and negative controls. Antibodies against VDAC1 and ACTIN are used as loading controls.
(H) PAS stain in skeletal muscles of WT zebrafish at 6 months of age.
(I) PAS-positive aggregates (marked by orange arrows) in skeletal muscles of C2orf69 KO zebrafish are suggestive of glycogen accumulation. Sarcomeres were partially disorganized in mutant striated muscle fibers. Scale bar represents 50 μm.
Finally, we sought to assess whether polyglucosan bodies could also be seen in KO C2orf69 zebrafish. At 6 months of age, compared to WT siblings, mutant fish displayed sporadic aggregates of PAS-positive staining in skeletal but not cardiac muscles (Figures 5H and 5I). Evident vacuolation was also seen in muscle fibers positive for PAS aggregates in mutant fish. These myocytes appeared disorganized and without a regular striated organization seen in control fish of the same age (Figure 5I). By recapitulating the accumulation of polyglucosan bodies observed in human probands, C2orf69 knockout zebrafish further substantiate a functional link between GBE1-driven glycogen metabolism and C2orf69 deficiency.
Discussion
In summary, we have identified eight unrelated families with autosomal recessive C2orf69 deficiency, which is characterized by autoimmune defects, failure to thrive, progressive neurodegeneration of the central nervous system, and early demise. A concurrent study has identified a similar condition in eight additional children,30 which independently confirms our clinical, genetic, and molecular findings.
Using sequence analysis and cellular assays, we demonstrated that the six germline C2orf69 variants are likely to have deleterious effects at the protein level. In particular, we confirm that the in-frame indel allele identified in the Egyptian family is more likely to compromise the stability and subcellular localization of C2ORF69. C2orf69 is highly conserved in eukaryotes but not indispensable for all species because fungi were found not to have a orthologous gene. Structural analysis suggests a single globular domain protein with distant homology with bacterial esterases. The protein contains an N-terminal sequence with similarity to a signal peptide and can be detected in supernatants of high-density cell cultures. However, in most cell culture conditions, the endogenous protein remains cytosolic and associates with the outer mitochondrial membrane. These data suggest that C2ORF69 could be acting on the lipid composition of membranes. The mitochondrial assays suggest a mild perturbation of the electron transport chain and the production of ROS. These anomalies may manifest as knockon effects driven by alterations of the outer mitochondrial membrane.
A combination of hepatic and neurological findings in the C2orf69 loss-of-function-associated phenotypes indicates a strong clinical suspicion for mitochondriopathy. However, microscopic identification of diastase-resistant PAS staining in the muscle biopsy from the proband of family 8, without any pathogenic variants in GBE1, indicated accumulation of polyglucan, which is uncommon for mitochondrial disorders. The clinical presentation of this newly recognized syndrome is indeed a singular combination of the clinically heterogeneous glycogen storage disease type IV and mitochondriopathy. Furthermore, the selective decrease in glycogen branching enzyme protein levels in C2orf69 knockout HAP1 cells supports a link between GBE1 and C2orf69. An independent study also supports this finding through a combination of biopsy findings from heart, muscle, and liver tissues and a decrease in GBE1 enzymatic activity.21 Whether C2orf69’s control over GBE1 is caused by a decrease in expression or increased degradation and how they link the two important energy metabolism-related processes in the cell, glycogen synthesis, and OXPHOS still needs to be addressed. Therefore, this intertwined phenotype is particularly challenging for pathological diagnosis of mitochondriopathies because muscle biopsy samples are commonly obtained for assessment of mitochondrial activity but findings about glycogen storage may be overlooked. In particular, a combination of intractable developmental and epileptic encephalopathy, autoinflammatory dysregulation, and hepatic involvement should urge the clinical teams to look for glycogen-storage-related mitochondriopathy.
Despite being ubiquitously expressed, the clinical presentation suggests that specific tissues such as neuronal lineages are particularly affected by C2ORF69 depletion. The protein localization at the mitochondria suggests that the children’s symptoms are at least in part due to defective mitochondrial function in neurons and/or glia of the CNS. This notion is supported by in vivo experiments where the lethality of C2orf69 mutant zebrafish was apparently linked to defects in the CNS. Lending credence to the notion that C2ORF69 plays an active role in the development/homeostasis of the immune and central nervous systems, genome-wide association studies (GWASs) have revealed genome-wide significant non-coding variants in the intron of C2orf69 to be positively associated with susceptibility to chickenpox infections (rs191220855, p = 3 × 10−7)31 and schizophrenia (rs1658810, p = 2 × 10−13).32 A subsequent GWAS has found that common C2orf69 variants are associated with eight other neuropsychiatric disorders, such as autism and Tourette syndrome.32 This was further validated in a transcriptome-wide association study where endogenous C2orf69 levels were significantly associated with schizophrenia and changes in chromatin architecture.33 Finally, a genome-wide methylation quantitative trait loci (meQTLs) study also corroborated this finding by demonstrating a cross-tissue genetic-epigenetic effect of C2orf69 in schizophrenia.34 Our results and these independent studies implicate that both common and rare C2orf69 variants contribute to brain and immune malfunction in humans.
The autoinflammation observed in children with Elbracht-Işikay syndrome is not commonly seen with mitochondriopathies. On the other hand, glycogen metabolism has recently been linked to acute inflammatory response in macrophages.35 Thus, the autoinflammation may be linked to metabolic perturbation in immune cells. The late onset lethality observed in the zebrafish model is a salient difference with the early demise seen in humans. The zebrafish genome does not seem to have other C2orf69 paralogs that could functionally compensate. As C2ORF69 appears involved in bioenergetics, a possible explanation is that tissues of warm-blooded animals, with higher metabolic needs, are more sensitive to the loss of the protein. In addition to these questions, future studies will need to address whether C2ORF69 serves as a bespoke lipase/esterase. If so, efforts will need to be invested to find substrate(s) and enzymatic product(s), the accumulation or absence of which, respectively, may be responsible for the observed autoinflammatory symptoms.
Declaration of interests
J.R.L. has stock ownership in 23andMe and is a paid consultant for the Regeneron Genetics Center. The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics (BG) Laboratories. J.R.L. serves on the Scientific Advisory Board of BG. A.B.A. and P.B. are employees of CENTOGENE GmbH. S.C. is a shareholder of Intergen Genetic Diagnosis Center. J.J.C. is a consultant for Thermo Fisher Scientific. All other authors declare no competing interests.
Acknowledgments
We thank all the families for partaking in this study and the referring clinicians for their generous help. We are grateful to all members of the Reversade, Ho, and Bard laboratories for swift support. We thank Z. Ekim Taskiran for facilitating WES studies in the Hacettepe University Exome Facility and Can Kosukcu for obtaining WES data. We thank Mohd Agus and the IMCB aquatics facility for zebrafish husbandry. H.H.W. and S.H.S. are supported by a grant from Procter and Gamble. J.L.C. is supported by the Howard Hughes Medical Institute (HHMI), the Rockefeller University, the St. Giles Foundation, Institut National de la Santé et de la Recherche Médicale (INSERM), and the “Université de Paris.” S.U. is supported by E-Rare grants for EuroDBA Project (TUBITAK, #315S192). S.Z. is supported by a Khoo Teck Puat postdoctoral fellowship. A.C. is supported by the EU’s Horizon 2020 research and innovation program under the EJP RD COFUND-EJP no. 825575 (TUBİTAK, #319S062). D.J.P. is supported by NIH awards R35 GM131795 and P41 GM108538, a UW2020 award, and funds from the BJC Investigator Program. J.J.C. is supported by NIH awards, P41 GM108538, and a UW2020 award. S.X. is supported by NMRC/OFYIRG/062/2017. A.S.M. and F.M.N. are supported by the Ministry of Education, Singapore and Yale-NUS College (IG19-BG106 and SUG). L.H. is supported by fellowships NRF-NRFF2017-05 and HHMI-IRSP55008732. T.M. is supported by The Uehara Memorial Foundation. D.P. is supported by International Rett Syndrome Foundation (#3701-1). J.R.L. is supported by the National Human Genome Research Institute, the National Heart, Lung, and Blood Institute, The Baylor-Hopkins Center for Mendelian Genomics (#HG006542), and the National Institutes of Neurological Disease and Stroke (R35NS105078) and Muscular Dystrophy Association (#512848). B.R. is an investigator of the National Research Foundation (Singapore) and is supported by a use-inspired basic research grant from the Agency for Science & Technology and Research (A∗STAR) in Singapore.
Published: May 25, 2021; Corrected online: June 16, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.05.003.
Contributor Information
Frederic A. Bard, Email: fbard@imcb.a-star.edu.sg.
Bruno Reversade, Email: bruno@reversade.com.
Data and code availability
The data that support the findings of this study are available within the paper or from the corresponding authors upon reasonable request.
Web resources
1000 Genomes Project Database, https://www.internationalgenome.org/
CENTOGENE, https://www.centogene.com/downloads
CRISPRScan, https://www.crisprscan.org/
Exome Aggregation Consortium (ExAC), http://exac.broadinstitute.org
Greater Middle East (GME) Variome, http://igm.ucsd.edu/gme/index.php
NCBI dbSNP, https://www.ncbi.nlm.nih.gov/SNP/
OMIM, https://omim.org
The Exome Variant Server (ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh37) from NHLBI Exome Sequencing Project (ESP), https://evs.gs.washington.edu/EVS/
Supplemental information
References
- 1.Song J., Herrmann J.M., Becker T. Quality control of the mitochondrial proteome. Nat. Rev. Mol. Cell Biol. 2021;22:54–70. doi: 10.1038/s41580-020-00300-2. [DOI] [PubMed] [Google Scholar]
- 2.Gorman G.S., Chinnery P.F., DiMauro S., Hirano M., Koga Y., McFarland R., Suomalainen A., Thorburn D.R., Zeviani M., Turnbull D.M. Mitochondrial diseases. Nat. Rev. Dis. Primers. 2016;2:16080. doi: 10.1038/nrdp.2016.80. [DOI] [PubMed] [Google Scholar]
- 3.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tein I., Demaugre F., Bonnefont J.P., Saudubray J.M. Normal muscle CPT1 and CPT2 activities in hepatic presentation patients with CPT1 deficiency in fibroblasts. Tissue specific isoforms of CPT1? J. Neurol. Sci. 1989;92:229–245. doi: 10.1016/0022-510x(89)90139-1. [DOI] [PubMed] [Google Scholar]
- 5.Elouej S., Harhouri K., Le Mao M., Baujat G., Nampoothiri S., Kayserili H., Menabawy N.A., Selim L., Paneque A.L., Kubisch C. Loss of MTX2 causes mandibuloacral dysplasia and links mitochondrial dysfunction to altered nuclear morphology. Nat. Commun. 2020;11:4589. doi: 10.1038/s41467-020-18146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narendra D., Tanaka A., Suen D.-F., Youle R.J. Parkin-induced mitophagy in the pathogenesis of Parkinson disease. Autophagy. 2009;5:706–708. doi: 10.4161/auto.5.5.8505. [DOI] [PubMed] [Google Scholar]
- 7.Escande-Beillard N., Loh A., Saleem S.N., Kanata K., Hashimoto Y., Altunoglu U., Metoska A., Grandjean J., Ng F.M., Pomp O. Loss of PYCR2 Causes Neurodegeneration by Increasing Cerebral Glycine Levels via SHMT2. Neuron. 2020;107:82–94.e6. doi: 10.1016/j.neuron.2020.03.028. [DOI] [PubMed] [Google Scholar]
- 8.Calvo S.E., Clauser K.R., Mootha V.K. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44(D1):D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calvo S.E., Mootha V.K. The mitochondrial proteome and human disease. Annu. Rev. Genomics Hum. Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macdonald R., Barnes K., Hastings C., Mortiboys H. Mitochondrial abnormalities in Parkinson’s disease and Alzheimer’s disease: can mitochondria be targeted therapeutically? Biochem. Soc. Trans. 2018;46:891–909. doi: 10.1042/BST20170501. [DOI] [PubMed] [Google Scholar]
- 11.Fecher C., Trovò L., Müller S.A., Snaidero N., Wettmarshausen J., Heink S., Ortiz O., Wagner I., Kühn R., Hartmann J. Cell-type-specific profiling of brain mitochondria reveals functional and molecular diversity. Nat. Neurosci. 2019;22:1731–1742. doi: 10.1038/s41593-019-0479-z. [DOI] [PubMed] [Google Scholar]
- 12.Rangaraju V., Lewis T.L., Jr., Hirabayashi Y., Bergami M., Motori E., Cartoni R., Kwon S.-K., Courchet J. Pleiotropic Mitochondria: The Influence of Mitochondria on Neuronal Development and Disease. J. Neurosci. 2019;39:8200–8208. doi: 10.1523/JNEUROSCI.1157-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joshi A.U., Minhas P.S., Liddelow S.A., Haileselassie B., Andreasson K.I., Dorn G.W., 2nd, Mochly-Rosen D. Fragmented mitochondria released from microglia trigger A1 astrocytic response and propagate inflammatory neurodegeneration. Nat. Neurosci. 2019;22:1635–1648. doi: 10.1038/s41593-019-0486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zsurka G., Kunz W.S. Mitochondrial dysfunction and seizures: the neuronal energy crisis. Lancet Neurol. 2015;14:956–966. doi: 10.1016/S1474-4422(15)00148-9. [DOI] [PubMed] [Google Scholar]
- 15.Moehlman A.T., Youle R.J. Mitochondrial Quality Control and Restraining Innate Immunity. Annu. Rev. Cell Dev. Biol. 2020;36:265–289. doi: 10.1146/annurev-cellbio-021820-101354. [DOI] [PubMed] [Google Scholar]
- 16.Jagadeesh K.A., Wenger A.M., Berger M.J., Guturu H., Stenson P.D., Cooper D.N., Bernstein J.A., Bejerano G. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 2016;48:1581–1586. doi: 10.1038/ng.3703. [DOI] [PubMed] [Google Scholar]
- 17.Hengel H., Bosso-Lefèvre C., Grady G., Szenker-Ravi E., Li H., Pierce S., Lebigot É., Tan T.-T., Eio M.Y., Narayanan G. Loss-of-function mutations in UDP-Glucose 6-Dehydrogenase cause recessive developmental epileptic encephalopathy. Nat. Commun. 2020;11:595. doi: 10.1038/s41467-020-14360-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pena I.A., Roussel Y., Daniel K., Mongeon K., Johnstone D., Weinschutz Mendes H., Bosma M., Saxena V., Lepage N., Chakraborty P. Pyridoxine-Dependent Epilepsy in Zebrafish Caused by Aldh7a1 Deficiency. Genetics. 2017;207:1501–1518. doi: 10.1534/genetics.117.300137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalueff A.V., Stewart A.M. Humana Press; 2012. Zebrafish Protocols for Neurobehavioral Research. [Google Scholar]
- 20.Gaspar B.L., Vasishta R.K., Radotra B.D. Springer; 2018. Myopathology: A Practical Clinico-pathological Approach to Skeletal Muscle Biopsies. [Google Scholar]
- 21.Zhang S., Reljić B., Liang C., Kerouanton B., Francisco J.C., Peh J.H., Mary C., Jagannathan N.S., Olexiouk V., Tang C. Mitochondrial peptide BRAWNIN is essential for vertebrate respiratory complex III assembly. Nat. Commun. 2020;11:1312. doi: 10.1038/s41467-020-14999-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spinazzi M., Casarin A., Pertegato V., Salviati L., Angelini C. Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat. Protoc. 2012;7:1235–1246. doi: 10.1038/nprot.2012.058. [DOI] [PubMed] [Google Scholar]
- 23.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jha P., Wang X., Auwerx J. Analysis of Mitochondrial Respiratory Chain Supercomplexes Using Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE) Curr. Protoc. Mouse Biol. 2016;6:1–14. doi: 10.1002/9780470942390.mo150182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurer-Stroh S., Koranda M., Benetka W., Schneider G., Sirota F.L., Eisenhaber F. Towards complete sets of farnesylated and geranylgeranylated proteins. PLoS Comput. Biol. 2007;3:e66. doi: 10.1371/journal.pcbi.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gabler F., Nam S.-Z., Till S., Mirdita M., Steinegger M., Söding J., Lupas A.N., Alva V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinformatics. 2020;72:e108. doi: 10.1002/cpbi.108. [DOI] [PubMed] [Google Scholar]
- 27.Garner D.L., Thomas C.A. Organelle-specific probe JC-1 identifies membrane potential differences in the mitochondrial function of bovine sperm. Mol. Reprod. Dev. 1999;53:222–229. doi: 10.1002/(SICI)1098-2795(199906)53:2<222::AID-MRD11>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 28.Baraban S.C., Taylor M.R., Castro P.A., Baier H. Pentylenetetrazole induced changes in zebrafish behavior, neural activity and c-fos expression. Neuroscience. 2005;131:759–768. doi: 10.1016/j.neuroscience.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 29.Baraban S.C., Dinday M.T., Hortopan G.A. Drug screening in Scn1a zebrafish mutant identifies clemizole as a potential Dravet syndrome treatment. Nat. Commun. 2013;4:2410. doi: 10.1038/ncomms3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lausberg E., Gießelmann S., Dewulf J.P., Wiame E., Holz A., Salvarinova R. A human multisystem disorder with autoinflammation, leukoencephalopathy and hepatopathy is caused by mutations in C2orf69. J. Clin. Invest. 2021 doi: 10.1172/JCI143078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian C., Hromatka B.S., Kiefer A.K., Eriksson N., Noble S.M., Tung J.Y., Hinds D.A. Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat. Commun. 2017;8:599. doi: 10.1038/s41467-017-00257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cross-Disorder Group of the Psychiatric Genomics Consortium Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell. 2019;179:1469–1482.e11. doi: 10.1016/j.cell.2019.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gusev A., Mancuso N., Won H., Kousi M., Finucane H.K., Reshef Y., Song L., Safi A., McCarroll S., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 2018;50:538–548. doi: 10.1038/s41588-018-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin D., Chen J., Perrone-Bizzozero N., Bustillo J.R., Du Y., Calhoun V.D., Liu J. Characterization of cross-tissue genetic-epigenetic effects and their patterns in schizophrenia. Genome Med. 2018;10:13. doi: 10.1186/s13073-018-0519-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J., Wei K., Liu J., Tang K., Zhang H., Zhu L., Chen J., Li F., Xu P., Chen J. Glycogen metabolism regulates macrophage-mediated acute inflammatory responses. Nat. Commun. 2020;11:1769. doi: 10.1038/s41467-020-15636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available within the paper or from the corresponding authors upon reasonable request.