Summary
Despite release of the GRCh38 human reference genome more than seven years ago, GRCh37 remains more widely used by most research and clinical laboratories. To date, no study has quantified the impact of utilizing different reference assemblies for the identification of variants associated with rare and common diseases from large-scale exome-sequencing data. By calling variants on both the GRCh37 and GRCh38 references, we identified single-nucleotide variants (SNVs) and insertion-deletions (indels) in 1,572 exomes from participants with Mendelian diseases and their family members. We found that a total of 1.5% of SNVs and 2.0% of indels were discordant when different references were used. Notably, 76.6% of the discordant variants were clustered within discrete discordant reference patches (DISCREPs) comprising only 0.9% of loci targeted by exome sequencing. These DISCREPs were enriched for genomic elements including segmental duplications, fix patch sequences, and loci known to contain alternate haplotypes. We identified 206 genes significantly enriched for discordant variants, most of which were in DISCREPs and caused by multi-mapped reads on the reference assembly that lacked the variant call. Among these 206 genes, eight are implicated in known Mendelian diseases and 53 are associated with common phenotypes from genome-wide association studies. In addition, variant interpretations could also be influenced by the reference after lifting-over variant loci to another assembly. Overall, we identified genes and genomic loci affected by reference assembly choice, including genes associated with Mendelian disorders and complex human diseases that require careful evaluation in both research and clinical applications.
Keywords: Human Genome Reference, GRCh38, GRCh37, hg19, exome sequencing, clinical genome sequencing
Introduction
Decreasing costs and dramatic improvements in next-generation sequencing (NGS) have allowed research and clinical diagnostic laboratories to establish analytical pipelines centered on NGS genomic technologies such as exome sequencing (ES).1, 2, 3, 4 In particular, clinical laboratories often use ES as a first-tier testing tool to diagnose rare genetic disorders through clinical interpretation of detected variants.5, 6, 7 One of the first steps in this process involves the alignment of short reads generated from ES to a haploid human reference genome sequence.
A complete human reference genome is a prerequisite for an accurate, precise, and replicable alignment for genetic variant calling and subsequent variant interpretation. Despite standardized best practices and guidelines for short read variant detection,8, 9, 10 variant calling discrepancies still exist and hinder comparative and aggregated analyses between different laboratories.11, 12, 13 These variant identification discrepancies subsequently result in conflicting variant interpretations and prevent precise translation of clinical sequencing data into diagnostic targets for precision medicine.8 The variant calling discrepancies are in part due to inconsistent variant calling workflows and in part due to the use of different reference assemblies.14,15
The current “gold standard” human genome reference assemblies curated by The Genome Reference Consortium (GRC) are GRCh37 (also known as hg19) originally released in 200916 and periodically updated until 2013 when its successor GRCh38 (also known as hg38) was published.17 The GRC has improved reference assembly reconciliation by actively providing patches, fixes, and alternate scaffolds to the human reference genome. Fix patches provide changes to existing assembly sequences to correct errors on the human reference genome, while alternate loci and novel sequence patches enable the reference assembly to represent allelic diversity.16,17 The updates to GRCh38 from GRCh37 included alternate scaffolds for highly variable genomic regions, synthetic centromere sequences to fill large mega-base gaps, and corrections and gap filling of thousands of misassembled regions and artifacts.18 These updates not only resolved locus-specific issues, but also holistically improved genome-wide alignment and variant calling by correcting false alignment across the genome.17 For example, in an analysis of 121 whole genomes using the GRCh38 genome assembly, on average 52 of 178 known structurally variable regions corresponded better to an alternate locus rather than the primary assembly.19 In addition, a comparative study of 30 exomes demonstrated the superiority of variant calling on GRCh38 due to an improved alignment rate of short reads.15 Even with improved alignment, however, the GRCh38 reference includes an expanded repertoire of alternative contigs which if mishandled can lead to erroneous variant calls. This was recently demonstrated by a reanalysis of 50,000 exomes from the UK Biobank, which originally showed erroneous, zero-variation in 641 genes.20
To date, the GRCh37 reference has been the foundation for cataloging human genetic variation and mapping functional variants in many breakthrough projects.21, 22, 23 It is only recently that the human genetic variation catalogs and annotations have been directly mapped to GRCh38.18,24,25 The GRCh37 reference and associated genomic resources are, however, deeply embedded in many current workflows and as a consequence, even though GRCh38 was released more than seven years ago, GRCh37 remains the primary reference utilized in most human genetic tools, annotation resources, and NGS workflows. Systematic updates to GRCh38 have therefore lagged behind, particularly in clinical diagnostic applications, where alterations of informatic pipelines require substantial validation in order to comply with clinical standards. Universally updating from GRCh37 to GRCh38 requires a field-wide paradigm shift in the current curation of variant annotation resources.26
An alternative approach to complete adoption of GRCh38 in clinical settings is to first align sequencing data to GRCh38 and then “lift-over” the data to the GRCh37 reference in order to utilize existing GRCh37 variant annotations and pipelines.27 The difference between the reference sequences can complicate lift-overs, and careful curation of corresponding regions on both GRCh37 and GRCh38 is required in order to ensure that variants that map to the amended sequences or filled gaps (e.g., centromere regions) on GRCh38 will be properly identified in GRCh37. Notably, a study using whole-genome sequencing from the Genome in a Bottle consortia showed about 5% of detected variants could not be converted between GRCh38 and GRCh37.14 Further, lift-overs are unable to leverage contextual advantages such as increased sequence representation and assembly corrections in the GRCh38 reference.17
Thus, the full consequences of switching to GRCh38 remain unclear, especially when applied to the clinical diagnosis of rare genetic diseases. In the current study, the impact of reference assembly changes in exome variant identification and interpretation was evaluated. Exome data from 1,572 individuals with Mendelian disease and their family members were analyzed, revealing the list of genes and genomic loci most likely to be affected by reference assembly choice during variant identification. Using these data, the impact and mechanisms of reference differences on identification of variants associated with both rare genetic diseases and complex human disorders were assessed.
Subjects and methods
Study cohort
The ES data from 1,572 individuals were collected from the Baylor-Hopkins Center for Mendelian Genomics (CMG;28 ES from individuals with clinically suspected Mendelian disease and their family members). Of the 1,572 individuals, 55% were phenotypically unaffected and 45% were phenotypically affected (Figure S1). Further, 286 singletons as well as families of varying size (depending on consent status) including 233 trios were sequenced. In addition, while more than 80% of the individuals were of European descent, individuals of Hispanic, African American, East Asian, and South Asian genetic ancestries were represented in the cohort (Figure S1). Written informed consent for all individuals in this study was obtained during recruitment under a research protocol (H-29697) approved by the Institutional Review Board of Baylor College of Medicine.
Exome sequencing
Exome capture and sequencing were carried out at the Human Genome Sequencing Center at Baylor College of Medicine as part of the CMG project and have been described previously.29 Briefly, genomic DNA samples underwent exome capture using the HGSC VCRome2.1 (covering ∼24K genes; Roche) and were then sequenced on the Illumina NovaSeq platform to an average of 94% of targeted bases with >20× coverage.
Alignment, variant calling, and filtering
We generated variant calls for each of the 1,572 samples using both the GRCh37 and GRCh38 human reference assemblies. Specifically, for the GRCh37 variant calls, we selected the hs37d5 reference genome that includes the revised Cambridge reference sequence of the human mitochondrial DNA, human herpesvirus 4 type 1 sequences, and the concatenated decoy sequences;30 and for GRCh38 we selected the full analysis set (GenBank assembly accession: GCA_000001405.15) with decoy sequences, alternate loci scaffolds, and HLA sequences. Both of these references have been used by Phase 3 of the 1000 Genomes Project.30 Through a routine, functional equivalent genome sequencing analysis pipeline implemented at the Human Genome Sequencing Center,31 alignment was performed using BWA-MEM32 against the two reference assemblies for each sample followed by insertion/deletion (indel) Realignment and Base Quality Score Recalibration using GATK.9 Variant calling was then performed using xAtlas10 for single-nucleotide variants (SNVs) and indels independently to generate gvcf files, which then underwent joint-calling via GLnexus33 (default parameters; autosomal variants only). Notably, we used the same parameters to generate variant call sets on GRCh37 and GRCh38.
We then performed a series of quality controls (QC) following previously published procedures and criteria34,35 to ensure the quality of the aggregated variant calls for SNVs and indels. First, for each variant in each sample, we assigned a missing genotype (./.) to the variant if the variant fulfilled one of the following criteria: read depth less than 15; allelic ratio less than 0.25 or greater than 0.75; or non-PASS variant from xAtlas variant call results. We then filtered out variants with a missing genotyping rate greater than 15% to ensure variant quality. This aggressive filtering strategy prevents the identification of stochastic differential variant calls between the two assemblies due to poor variant quality. We kept variants within 100 bp to the boundaries of the exome capture targets. In addition, we performed sample-wise QC by ensuring that all samples had a missing genotyping rate < 10% and a heterozygosity rate < 3 standard deviations. We also calculated pairwise identity by descent across all individuals using PLINK36 to ensure the sample relatedness matches the reported pedigree structures.
Variant lift-over and comparisons
Variant loci were lifted-over from one reference assembly (source) to the other (target) using the LiftOver tool and the corresponding chain files obtained from the UCSC genome browser.37 We first removed monomorphic variants with alternative allele frequency (AF) of one (AF = 1) from both call sets. This removal prevents the comparison of AF = 1 variants in the source assembly to AF = 0 variants in the target assembly (which did not exist in the filtered vcf file) in cases where the reference and the alternative alleles swap between GRCh37 and GRCh38. During lift-over from GRCh37 to GRCh38, and from GRCh38 to GRCh37, a set of “unliftable” variants was identified on GRCh37 and GRCh38, respectively, due to unmatched loci on the target assembly and were considered as one source of discordant variants. Next, in each sample we determined the genotypic concordance of the “liftable” variants between GRCh37 and GRCh38. Specifically, variants called on GRCh38 were lifted-over to the GRCh37 coordinate and were compared to the variants called on the GRCh37 assembly to identify concordant and discordant variant calls in each sample using vcfeval from the RealTimeGenomics toolbox.38 We also lifted the concordant variants from GRCh37 to GRCh38 to ensure the liftover was bi-directional. Altogether, these analyses resulted in variants that were (1) concordant between GRCh37 and GRCh38, (2) variants found only on GRCh37 (unique to GRCh37), and (3) variants found only on GRCh38 (unique to GRCh38).
Identification of discordant reference patches (DISCREPs)
The analyses were performed on GRCh37 and GRCh38 separately. To identify DISCREPs regions within each assembly, we divided the genome into 10 kb windows, counted the number of total distinct variants across all samples in each window, and filtered to keep windows with greater than 10 distinct variants for analysis. Then, in each genomic window, we counted distinct variants uniquely detected by either the GRCh37 or GRCh38 assembly and compared them with the number of distinct concordant variants against the baseline level summed across all windows using one-sided Fisher’s exact test, followed by false-discovery rate (FDR; Benjamini-Hochberg procedure) adjustment. We considered q < 0.01 as a statistically-significant threshold. Illustrations of DISCREPs were conducted using R packages KaryoploteR39 and Circlize.40
Enrichment analyses for genomic features in DISCREPs
The locations of the following genomic features available for both GRCh37 and GRCh38 were downloaded from the UCSC genome browser:37 simple tandem repeats, microsatellite, segmental duplications, interrupted repeats, known assembly problems, loci with fix patches, loci with alternate haplotypes, loci with known genome assembly differences, and gaps in the assembly. Enrichment analyses for each of the above genomic features within the DISCREPs were performed using R package LOLA41 based on two-sided Fisher’s exact tests (compared to non- DISCREP genomic windows). The test statistics were adjusted by FDR with q < 0.01 considered to be statistically significant. For DISCREPs regions that overlapped between GRCh37 and GRCh38, we combined the counts of the genomic windows from both GRCh37 and GRCh38 for the Fisher’s exact tests.
Identification of genes influenced by the reference assembly
Gene annotation was performed with ANNOVAR42 (v2019.10.24) using the GENCODE database (v34). We considered only genes with GENCODE annotations on both the GRCh37 and GRCh38 references. Variants (including SNVs and indels) were found in a total of 19,003 genes, and we further analyzed 790 genes with at least one discordant variant call found in our cohort. For each gene, we counted the number of distinct concordant and discordant variants on GRCh37 or GRCh38 across all individuals and compared against the aggregated counts of distinct concordant and discordant variants from all the 790 genes using one-sided Fisher’s exact tests. The test statistic was adjusted by FDR; genes with greater than five discordant variants and q < 0.05 were considered to be statistically significant.
To assess whether genes enriched for discordant variants between GRCh37 and GRCh38 in our study (n = 206) significantly overlap with genes carrying erroneous variant calls found in the UK Biobank study (n = 641),20 a permutation test was performed (ten million permutations) to derive a null distribution of the number of overlapping genes by randomly sampling our gene sets from a total of 19,003 genes (number of genes assessed in our study). The empirical p value was calculated as the proportion of times that the number of overlapping genes from the permutation equaled or exceeded the observed number of overlapping genes (n = 28).
Gene and variant annotation
Variant effects (e.g., frameshift deletion, missense, stopgain, etc.) were annotated using ANNOVAR42 (v2019.10.24). Genes implicated in known Mendelian diseases were identified from the Online Mendelian Inheritance in Man (OMIM) database. Known pathogenic variants were identified from ClinVar43 (version 20190305) with one or more “stars” describing the level of support of clinical significance. To identify variants with a potential deleterious effect, we focused on rare variants with minor allele frequency (MAF) < 0.01 in all of the following population databases: 1000 Genomes Phase 3,30 gnomAD (v2.1.1 for GRCh37 and v3.0 for GRCh38),24 and ExAC (v0.3).44 We defined potential deleterious variants that fulfilled one of the following criteria: (1) rare potential loss-of-function variants (frameshift insertions/deletions, nonsense single-nucleotide variants, stoploss variants, or splicing variants) or (2) rare missense variants with CADD45 score > 20 and REVEL46 score > 0.8. Genes from previous genome-wide association studies (GWASs) were obtained from the GWAS Catalog v1.0.2.
Results
Discrepant variant calls between the GRCh37 and GRCh38 references
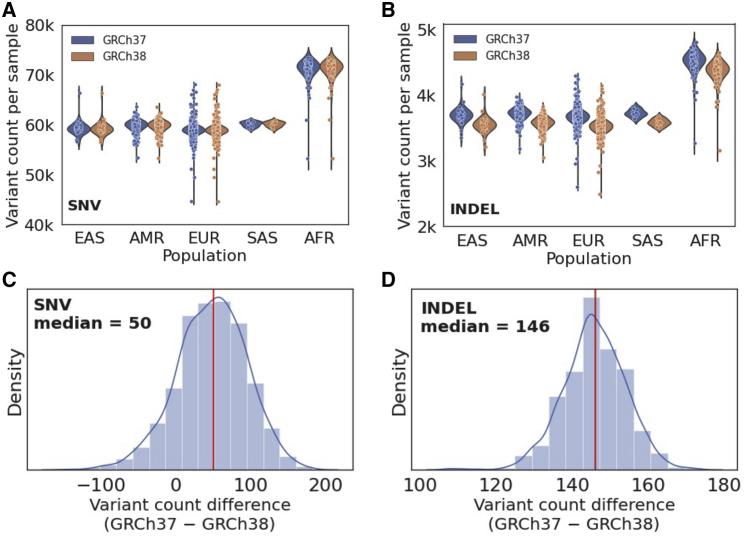
To determine the impact of reference genome choice on the identification of variants, we analyzed ES data from 1,572 individuals with Mendelian disease and their family members recruited through the Baylor-Hopkins CMG. Using both the GRCh37 and GRCh38 reference assemblies, we identified SNVs and indels on autosomes and found similar numbers of variants called by the two references for each sample (Figure 1; Table 1). Notably, on average, the GRCh37 reference generated 50 more SNVs and 146 more indels in each individual (Figure 1).
Figure 1.
Count of variants identified using the GRCh37 and GRCh38 references in each sample
(A and B) Distributions of variant count in each sample (A for SNV and B for indel), grouped by genetic ancestry of the individuals.
(C and D) Distribution of variant count differences between GRCh37 and GRCh38 in each sample (C for SNV; D for indel). The red line shows the median difference across all samples.
Note that the variant counts shown here include variants in exons and regions within 100 bp to exon boundaries (EAS, East Asian; AMR, admixed American; EUR, European; SAS, South Asian; AFR, African American).
Table 1.
Number of variants identified in the study cohort
|
SNV |
Indel |
|||
|---|---|---|---|---|
| GRCh37 | GRCh38 | GRCh37 | GRCh38 | |
| Variants per individual (median count with interquartile range) | ||||
| Heterozygous | 36,496 (1,498) | 36,470 (1,513) | 2,538 (127) | 2,537 (127) |
| Homozygous | 22,570 (803) | 22,537 (789) | 1,148 (50) | 1,002 (50) |
| Total | 59,110 (1,231) | 59,060 (1,242) | 3,687 (120) | 3,542 (121) |
| Total number of distinct variants across all samples | ||||
| 1,240,896 | 1,242,297 | 75,898 | 75,860 | |
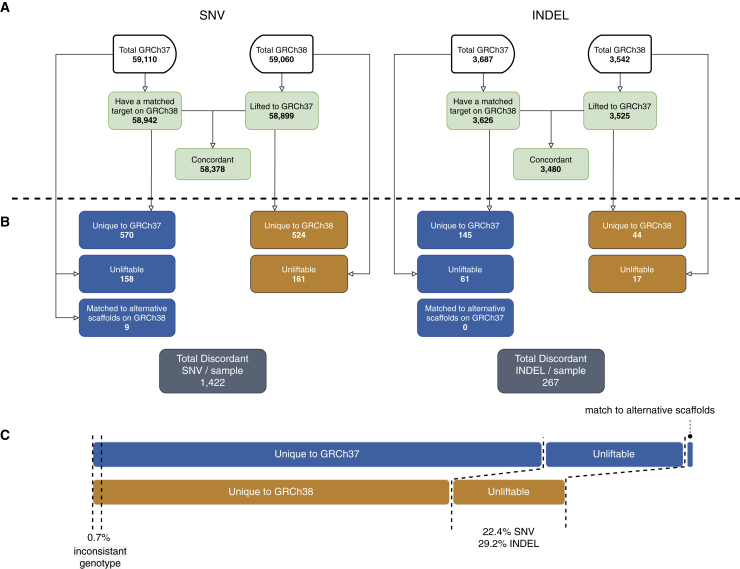
Despite the similar variant counts between GRCh37 and GRCh38 in each sample, we next sought to determine the systemic concordance of the variant calls between the two references. By lifting-over variants detected from one reference to the other and comparing variant calls in each sample, we found that the majority of variants had concordant genotypes between GRCh37 and GRCh38 (>98% SNVs and >93% indels across all samples; Figure 2). However, in each sample, an average of 1,422 SNVs and 267 indels were called discordantly by the two references (Figure 2). Among them, only 0.7% were due to inconsistent genotypes (i.e., the same variant locus with heterozygous versus homozygous alternative-allele calls); a majority of the discordant calls were caused by variants uniquely found on one reference only (Figure 2). Notably, among the discordant variants in each sample, an average of 22.4% SNVs and 29.2% indels did not have a matched target for lift-over (unliftable) and thus were not comparable to variants called by the other reference (Figure 2). Altogether, the discordant variant calls impacted 1.5% of total distinct SNVs (n = 18,477 / 1,248,403) and 2.0% of total distinct indels (n = 1,523 / 76,414) found in our cohort.
Figure 2.
Average number of concordant and discordant variants between GRCh37 and GRCh38 in each sample
(A) Total number of variants called on GRCh37 and GRCh38 in each sample (median number across all samples, same below) and the number of concordant variants between the two references.
(B) The number of discordant variants found on GRCh37 and GRCh38 in each sample and their sources.
(C) The relative proportion of the discordant variants (including both SNVs and indels). Blue boxes indicate discordant variants identified on GRCh37, whereas orange boxes indicate discordant variants identified on GRCh38.
Genomic loci enriched for discordant variant calls
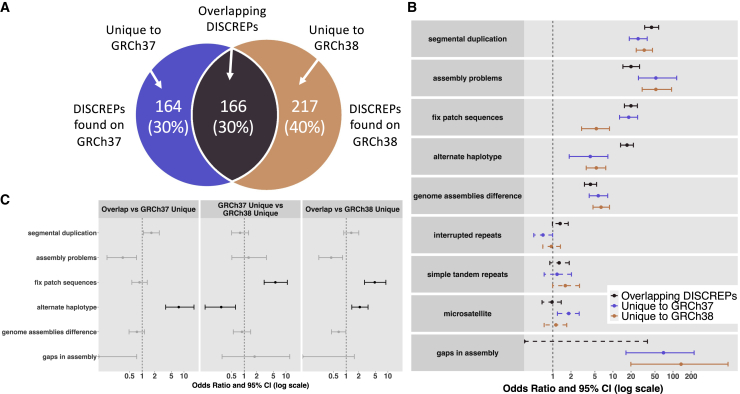
While the discordant variants were found across all chromosomes, we observed that they tended to cluster within certain genomic loci (Figures S2 and S3). To identify discordant reference patches (DISCREPs) with loci enriched for discrepant variant calls, we evaluated chromosomal windows (10 kb) on GRCh37 and GRCh38 separately to identify regions where the discordant variants were significantly enriched compared to the whole-exome baseline level. We found a total of 330 of 39,092 (0.8%) tested genomic windows as DISCREPs that were significantly enriched for variants only detected by the GRCh37 reference (false discovery rate q < 0.01; Table S1). These DISCREPs accounted for 72.9% of variants called only by GRCh37. On GRCh38, we found 383 DISCREPs out of 39,188 genomic windows (1.0%) that contained 79.9% of variants called only by the GRCh38 reference (Table S2). Therefore, a majority of discordant variants called on either GRCh37 or GRCh38 (76.6% altogether) were clustered within discrete genomic intervals.
We observed that the DISCREPs between GRCh37 and GRCh38 overlapped. Specifically, 1.38 Mb of the DISCREPs (138 10-kb genomic windows) overlapped between the two references and were enriched for discordant variants found on both GRCh37 and GRCh38 (Figure 3). To identify shared and distinct mechanisms for the regions enriched for discordant variant calls, we characterized these DISCREPs via enrichment analyses focusing on a series of genomic features. We found DISCREPs were significantly enriched in genomic loci containing segmental duplications, known assembly problems, fix patches, alternate haplotypes, and known differences between the two references (q < 0.01; Table S3), regardless of whether the DISCREPs were overlapping between GRCh37 and GRCh38 or unique to one of the references (Figure 3). However, we also found that genomic regions containing alternate haplotypes and fix patch sequences were more enriched in overlapping DISCREPs between GRCh37 and GRCh38 compared to the DISCREPs that were unique to GRCh37 or GRCh38 (Figure 3). Further, GRCh37 unique variants were more enriched for fix patch sequences while less enriched for alternative haplotypes compared to unique variants on GRCh38 (Figure 3). Therefore, the DISCREPs had shared mechanisms underlying certain genomic features such as segmental duplications, although the extent to which differ depending on the reference assembly.
Figure 3.
Genomic features enriched in the DISCREPs regions identified on GRCh37 and GRCh38
(A) Number of DISCREPs found on GRCh37 and GRCh38 (overlapping DISCREPs between the two references as well as unique DISCREPs on each assembly).
(B) Odds ratio of Fisher’s exact tests for the enrichment of various genomic features within different types of DISCREPs. Solid lines indicate statistically significant enrichment (q < 0.01), and dashed lines indicate non-significant results.
(C) Pairwise comparisons between different groups of DISCREPs on the enrichment of various genomic features. Black lines indicate statistically significant results (q < 0.01), whereas gray lines indicate non-significant results.
Genes influenced by the choice of reference assembly
As discordant variants were clustered within certain genomic regions, we next determined whether any genes were significantly impacted by the choice of reference genome in variant calling. Across the 19,003 genes that we evaluated, 790 genes (4.16%) had at least one variant that was only called on the GRCh37 or GRCh38 reference. Comparing variants called by only one reference to the concordant variants in each gene yielded a total of 206 genes significantly enriched for variants only detected by one reference genome (q < 0.05, Table S4): 120 enriched for unique GRCh37 variants, 144 enriched for unique GRCh38 variants, and 58 enriched for both GRCh37 and GRCh38 unique variants. Notably, 83.0% of these genes overlapped with DISCREPs found on GRCh37 and GRCh38.
Among the 206 genes significantly influenced by the reference assembly, a total of 34 and 26 genes had >90% of the variants called only by the GRCh37 and GRCh38 reference, respectively (Table S4). The discrepancy of the variant calls in these genes were all due to reads that were aligned to multiple loci (multi-mapped reads) on the reference assembly that lacked the variant call, which resulted in mapping scores of zero and failed variant calling.32 For instance, a total of 97.1% of variants (n = 136/140) from the CBS (MIM: 613381) gene were found only on the GRCh37 reference. The discrepancy of these unique variants on CBS were due to the split of two genes, CBS and CBSL, from the same locus on the GRCh37 reference genome (chr21:44,473,301–44,496,472) into two distinct loci on the GRCh38 reference (CBS: chr21:43,053,191–43,076,378; CBSL: chr21:6,444,869–6,468,040). This split led to multi-mapped reads aligned to two distinct regions of GRCh38 resulting in failed variant calls on the GRCh38 reference genome (Figure S4).
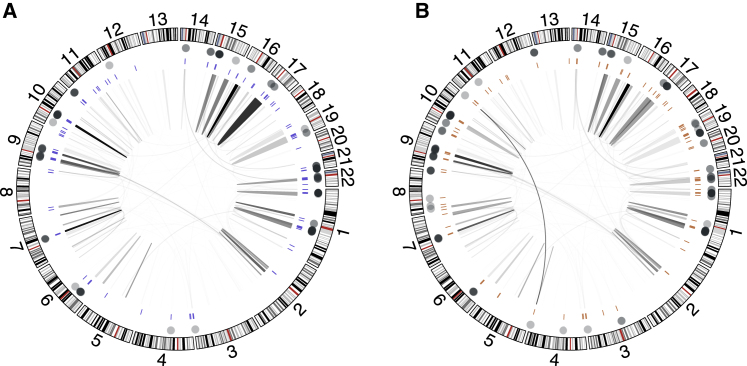
We found that the multi-mapped reads explained 64.1% of the 206 genes that were significantly influenced by the reference assembly. Throughout the whole exome, a majority of the multi-mapped reads were aligned to multiple loci within the same chromosome (>90% across all samples) with a few exceptions where multi-mapped reads aligned to different chromosomes (Figure 4). In fact, a total of 57 of the 206 genes (27.7%) were annotated as human paralogous genes based on genome-wide BLAST analyses,47 such as the PRAMEF gene cluster; and 28 genes (13.6%) are pseudogenes (Table S4). These results indicate that although the majority of the variants had concordant variant calls between the two references, certain regions and genes on the genome are vulnerable to the reference assembly changes due to similar mechanisms.
Figure 4.
Multi-mapped reads on GRCh37 and GRCh38
Results from GRCh37 (A) and GRCh38 (B). Dots on the outer track indicate genes with a significant enrichment of discordant variant calls. The bars on the intermediate track show the DISCREPs enriched for the discordant variants. The links within the inner track indicate the pairwise loci of multi-mapped reads that aligned to multiple loci on the genome, and the shade of the links are proportional to the density of such reads.
Impact of the reference assembly on variants associated with rare genetic diseases
We next focused on the impact of discrepant variant calls on possible molecular diagnoses of Mendelian disorders. From the 206 genes with significant enrichment of discordant variants between GRCh37 and GRCh38, we identified a total of eight genes associated with known Mendelian phenotypes based on OMIM (Table 2). Among these genes, all or most variants in and near PRODH (MIM: 606810), SIK1 (MIM: 605705), CBS, H19 (MIM: 103280), CRYAA (MIM: 123580), and KCNE1 (MIM: 176261) were detectable only by the GRCh37 reference, whereas RPS17 (MIM: 180472) and ADAMTSL2 (MIM: 612277) were enriched for variants that could only be detected using the GRCh38 reference. Discordant variants in each of these eight genes were due to multi-mapped reads on the reference without the variant calls. Therefore, molecular diagnoses of Mendelian disorders associated with these genes were influenced by the choice of reference assembly.
Table 2.
Genes implicated in Mendelian disease enriched for unique GRCh37 or GRCh38 variants
| Gene |
Distinct variant count |
p value for the enrichment of unique variants |
OMIM phenotype | |||
|---|---|---|---|---|---|---|
| Concordant across all samples | Unique variants on GRCh37 | Unique variants on GRCh38 | GRCh37 | GRCh38 | ||
| PRODH | 0 | 155 | 0 | 1.28E−164 | 1.00 | hyperprolinemia |
| SIK1 | 0 | 133 | 0 | 1.92E−141 | 1.00 | epileptic encephalopathy |
| CBS | 4 | 139 | 0 | 1.06E−140 | 1.00 | homocystinuria; thrombosis |
| H19 | 0 | 75 | 0 | 3.32E−80 | 1.00 | Wilms tumor; Silver-Russell syndrome; Beckwith-Wiedemann syndrome |
| CRYAA | 6 | 75 | 1 | 6.26E−72 | 0.53 | cataract |
| KCNE1 | 4 | 13 | 0 | 2.69E−11 | 1.00 | long QT syndrome; Jervell and Lange-Nielsen syndrome |
| ADAMTSL2 | 98 | 1 | 99 | 1.00 | 5.00E−45 | geleophysic dysplasia |
| RPS17 | 0 | 0 | 24 | 1.00 | 1.81E−24 | Diamond-Blackfan anemia |
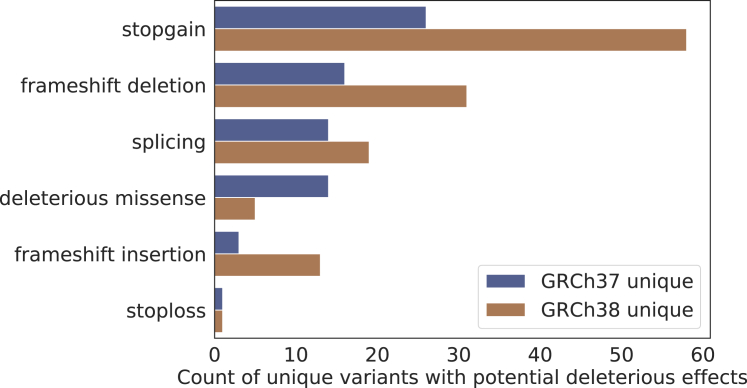
Within our cohort, we identified three known pathogenic or likely pathogenic (P/LP; from ClinVar) variants that were called differently by the two reference assemblies. All three P/LP variants were only called by the GRCh37 reference and were found in CBS, a gene associated with homocystinuria and thrombosis (MIM: 236200). We did not find any known ClinVar P/LP variants called only by the GRCh38 reference assembly within our cohort. Nonetheless, across all the discordant variants identified in our cohort, we found a total of 201 rare (MAF < 0.01) variants with potential deleterious effects in 128 genes (Figure 5; Table S5): 74 identified only by GRCh37 and 127 identified only by GRCh38; and 15 of these variants (7.5%) belong to genes implicated in known Mendelian disease.
Figure 5.
Categories of the unique variants with potential deleterious effect found on GRCh37 and GRCh38
We also found that the interpretation of the variant pathogenicity also differs due to the usage of different annotation resources derived from GRCh37 or GRCh38. For instance, out of the 201 rare, potentially deleterious variants that were called by only one reference assembly, 15 of them (7.5%) switched to a non-deleterious variant type when lifted-over and annotated by databases derived from the other reference assembly (Table S5). The majority of these discrepancies were due to different gene annotations between the two references. Therefore, the discordant variant calls due to the choice of reference assembly can influence both the identification and interpretations of potential pathogenic variants associated with rare genetic diseases.
Impact of the reference assembly on common variants
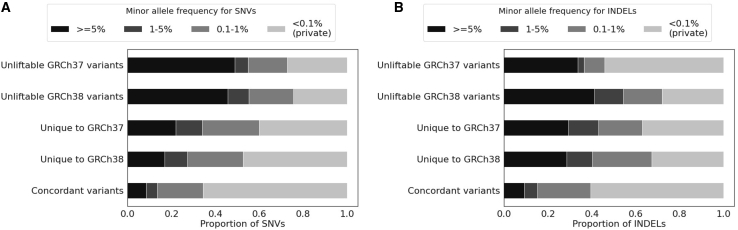
We next evaluated whether the choice of reference assembly had the same impact for both common and rare variants detected by ES. We found that discordant variants between GRCh37 and GRCh38 tended to have higher MAF than concordant variants. Specifically, within our cohort, 30.4% of the SNVs that were unique to GRCh37 or GRCh38 were common (MAF > 0.01), compared to that 13.4% of the concordant SNVs were common (p < 0.001; OR = 2.81; 95% CI = 2.72–2.90; Figures 6 and S5). Further, a larger proportion of the unique indels on GRCh37 and GRCh38 were common (MAF > 0.01) compared to the concordant indels (41.6% versus 15.1%; p < 0.001; OR = 3.99; 95% CI = 3.59–4.45; Figures 6 and S5). Similar trends were also observed for unliftable variants on GRCh37 and GRCh38 where unliftable variants tended to be more common than the concordant variants (Figures 6 and S5). Therefore, the discordant variant calls had relatively more impact on common variants than rare variants.
Figure 6.
Proportion of variants from concordant and discordant variant sets with different minor allele frequencies (MAFs)
Results for SNVs (A) and for indels (B).
Although common, non-coding variants associated with complex human diseases were not designed to be captured by ES, common functional variants within genic regions (e.g., promoter, 3′ untranslated region) are equally impacted by reference assembly, as discordant variant calls were clustered on the genome. Therefore, we evaluated the impact of reference assembly on genes associated with complex human disease. Across the 206 genes that showed significantly enriched discordant variant calls, we found 53 were associated with one or more of the 278 distinct phenotypes or traits from previous GWASs (Table S6). In particular, many were immune-related diseases associated with HLA genes. In summary, discordant variants due to reference assembly differences are present in genes associated with complex human disorders.
Discussion
In this largest study of its kind for evaluating the impact of the reference genome using ES data on the identification of both rare and common variants, we found that although the majority of the variants were not influenced by the reference change (and the results did not depend on the QC protocol we used [Figure S6]), certain regions of the genome were significantly impacted. Specifically, 206 genes were enriched for variants uniquely called by one reference. Among these genes, 8 have been associated with known Mendelian phenotypes and 53 have been associated with common traits from previous GWASs. We suggest that researchers and clinicians should pay increased attention to these genes and regions when interpreting variant calls from one reference assembly with an annotation database generated using another reference assembly.
The GRCh38 assembly is an improved version of the human genome reference.17 However, we found that certain disease-associated variants were missed by the GRCh38 reference and were only detectable from the GRCh37 assembly. In particular, we found six genes associated with known Mendelian phenotypes carried variants that were entirely or mostly detected by the GRCh37 reference (Table 2). Therefore, in our study although relatively few variants detected from ES data were discordantly detected by the two references, these genes warrant further investigation when using the GRCh38 reference, especially during the molecular diagnoses for individuals with clinical manifestations consistent with these conditions. We only identified two genes implicated in known Mendelian disease enriched for unique GRCh38 variants; however, this could result from ascertainment bias due to most groups reporting to ClinVar using the GRCh37 reference. Notably, none of the 59 genes recommended by the American College of Medical Genetics and Genomics (ACMG) for incidental findings48 contained an exome variant that was discrepantly called in our cohort.
One other consideration for switching from an old reference assembly to a new assembly is whether the disease of interest tends to associate with genes impacted the most by the reference assembly. Although only 1.5% of SNVs and 2.0% of indels were discordantly detected when different haploid human genome references were used, re-evaluation of retrospective data focusing on genes resulting in Mendelian disease in loci known to be improved in GRCh38 may provide opportunity for new discovery. For example, one of the most impacted regions were the major histocompatibility complex (MHC) loci, carrying many genes associated with autoimmune diseases.49 The GRCh38 reference is known to result in an improved variant call quality by introducing additional alternate haplotypes for the MHC region,17,19 and thus researchers interested in genes within the MHC loci may choose to use GRCh38. On the other hand, laboratories that study genes like CBS or diseases like homocystinuria or thrombosis may not choose to switch to GRCh38, as the majority of the variants in these genes could only be called on GRCh37.
Given the practical challenges of a systemic switch from the existing automated annotation pipelines based on GRCh37, many clinical diagnostic laboratories have proposed to utilize the GRCh38 reference genome assembly to perform variant calling followed by the lift-over of the variants to the GRCh37 coordinates for annotation. However, we found that not all potential deleterious variants were liftable to the GRCh37 reference genome; and, for variants that were able to be lifted-over to GRCh37, variant interpretation following lift-over could be altered due to the usage of annotation databases generated from a different reference genome. For instance, we found that 7.5% of the discordant variants with a potential deleterious effect switched to non-deleterious types using gene annotations derived from the other assembly. Therefore, the lift-over approach would not be applicable to all variants, especially for those within genomic regions enriched for discordant variant calls between GRCh37 and GRCh38.
Further, as most discoveries recorded in ClinVar and OMIM to date have been made using the GRCh37 reference, the probability of finding a P/LP ClinVar variant that is only called by GRCh37 should be higher relative to finding a P/LP variant called only by GRCh38. Nonetheless, of the 99 solved exomes on GRCh37 in our cohort who have had a thorough molecular diagnostic evaluation, by far, all P/LP variants have been successfully lifted-over from GRCh37 to GRCh38; and when re-mapped to GRCh38, 100% of these P/LP variants were still discovered (Figure S1).
In addition, variant interpretation efforts using annotation resources derived from a different assembly do not take full advantage of the improved human genome references and gene annotations. For instance, Steinberg et al. reported a genome assembly built from a haploid hydatidiform mole and identified 549 new genes absent from the GRCh37 assembly.50 These new genes would not be included in a GRCh37 gene annotation for variant interpretation even after a successful liftover to GRCh37. In addition, discrepancies of pathogenic variant interpretations also lie in the usage of the different gene and transcript annotation resources generated using different approaches, even from the same reference assembly (e.g., RefSeq versus GENCODE51). The impact of such annotation differences in pathogenic variant interpretation warrants further investigations.
Sixty-four percent of genes enriched for discordant variants between the two reference assemblies were due to multi-mapped reads. Taking CBS as an example, the introduction of additional reference sequences in GRCh38 for CBSL which contains high percent identity with CBS resulted in multi-mapped reads when aligned against GRCh38 but not GRCh37 (Figures S4 and S7). Pathogenic mutations in CBS result in a known, autosomal-recessive Mendelian disease. Although all the participants who carried pathogenic or likely pathogenic variants in CBS in our cohort were heterozygous for the variants, this discrepancy at the CBS locus does impact the evaluation of carrier status for pathogenic variants. This finding that the majority of discordant variant calls due to reference difference can be attributed to multi-mapped reads even in GRCh38 was corroborated by a study reanalyzing nearly 50,000 exomes from the UK Biobank,20 which shows that zero-variation is erroneously noted on GRCh38 in 641 genes including MYH11 (MIM: 160745), one of the 59 genes in which incidental variants should be reported according to the ACMG.48 Further, a statistically significant overlap of 28 of these 641 genes from the UK biobank study were found to be enriched for discordant variants in our study (p < 1 × 10−7, permutation test), and 23 of the 28 genes (82.1%) were specifically enriched for discordant variants on the GRCh38 reference (Table S4).
Also in the UK biobank study, the authors found that by including alternative contigs during alignment using BWA-MEM (alternative contigs-aware by default given an .alt index file) will significantly improve variant calling in 95% of genes that had alternate haplotypes potentially accounting for the multi-mapped reads. Further, in our study we also found that by including the alternative contig index during variant calling, only 6% of genes enriched for discordant variants were due to multi-mapped reads aligned to an alternative contig. Therefore, we recommend the inclusion of the alternative contig index to improve variant calling results.
Although our ES data do not represent the entire human genome, we hypothesize that the alignment of whole-genome data from short read sequencing would be equally impacted by reference assembly changes and thus influence detection of structural and non-coding variants as well. As the discordant variants were enriched in regions with segmental duplications, future evaluation should focus on whether third-generation sequencing techniques, such as long-read sequencing, can mitigate the issues found in genomic regions enriched for discrepant variant calls between GRCh37 and GRCh38.52 Additionally, given the short-read nature of exome sequencing, certain genomic regions with high homology to other genomic regions will be prone to accruing multi-mapped reads regardless of the reference genome assembly, and therefore long-read-based whole-genome sequencing could be able to resolve discordant variants in regions enriched for multi-mapped reads since the length of long reads typically allows them to map uniquely to the genome.53 Also, given the foreseeable availability of telomere-to-telomere assemblies of the whole genome,54 this study can serve as a future framework for describing how discrepancies between different references affect downstream variant identification.
We also observed that the African American population had a relatively lower rate of discordant variants compared to other genetic ancestry groups (Figure S8). Specifically, an average of 2.37% of variants were discordantly detected in the African American population, compared to 2.65% within samples of European descent (p = 1.26 × 10−153). The African population is the most genetically diverse population yet is the least represented population in the current human genome reference,30 which could minimize the observed impact upon reference change. A recent report suggests that using a pan-genome reference assembled from 910 subjects of African descent55 and a graph-based genome alignment strategy can improve variant calling from the African population.56
In summary, our data show that the intrinsic differences between the GRCh37 and GRCh38 references significantly impact variant calling for certain genomic regions including 206 genes (57 paralogous genes, 28 pseudogenes, 8 genes implicated in Mendelian diseases on OMIM, no ACMG genes). Only 3 known P/LP variants and 15 rare, putatively deleterious variants were discordantly called due to reference differences. Fifteen more variants had altered pathogenicity when lifted-over and annotated on the other assembly. On average in each exome, 1,422 SNVs and 267 indels were discordantly called representing <3% of variant calls per exome. Discordant variants clustered in DISCREP genomic regions, the majority of which included genomic intervals that were already known to be explicitly affected by segmental duplications, assembly issues, or alternate haplotypes. We recommend that for variant interpretation related to Mendelian disease molecular diagnosis or analysis of complex disease in the 206 genes enriched for discordant variants, or in a DISCREP, reference assembly differences should be accounted in the analysis, especially for researchers and clinicians who are lifting over variant coordinates from one reference to the other or using annotation resources generated from a different reference.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Human Genome Research Institute (NHGRI)/National Heart, Lung, and Blood Institute (NHLBI) UM1 HG006542 to J.R.L. J.E.P. was supported by K08 HG008986. M.D. is in the Medical Scientist Training Program at the Baylor College of Medicine. Additional funding was provided in part by a NHGRI grant to Baylor College of Medicine Human Genome Sequencing Center (U54HG003273 to R.A.G.) and U.S. National Institute of Neurological Disorders and Stroke (NINDS) (R35NS105078 to J.R.L.). H.L. was supported by the 2020 Xia-Gibbs Society Research Grant.
Published: June 14, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.05.011.
Data and code availability
The exome sequencing data used in this study are available at the database of Genotypes and Phenotypes (dbGaP: phs000711.v7.p2). The code generated in this study is available at https://github.com/hurleyLi/discreps. Additionally, we have made public sessions on the UCSC genome browser containing tracks of the DISCREP regions: https://genome.ucsc.edu/s/mdawood/DISCREPS_GRCh37; https://genome.ucsc.edu/s/mdawood/DISCREPS_GRCh38.
Web resources
GENCODE, https://www.gencodegenes.org
GWAS Catalog, https://www.ebi.ac.uk/gwas
OMIM, https://www.omim.org/
The Genome Reference Consortium, https://www.ncbi.nlm.nih.gov/grc
Supplemental information
References
- 1.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biesecker L.G., Green R.C. Diagnostic clinical genome and exome sequencing. N. Engl. J. Med. 2014;371:1170. doi: 10.1056/NEJMc1408914. [DOI] [PubMed] [Google Scholar]
- 3.Smith H.S., Swint J.M., Lalani S.R., Yamal J.M., de Oliveira Otto M.C., Castellanos S., Taylor A., Lee B.H., Russell H.V. Clinical Application of Genome and Exome Sequencing as a Diagnostic Tool for Pediatric Patients: a Scoping Review of the Literature. Genet. Med. 2019;21:3–16. doi: 10.1038/s41436-018-0024-6. [DOI] [PubMed] [Google Scholar]
- 4.Hayeems R.Z., Dimmock D., Bick D., Belmont J.W., Green R.C., Lanpher B., Jobanputra V., Mendoza R., Kulkarni S., Grove M.E., Medical Genome Initiative Clinical utility of genomic sequencing: a measurement toolkit. NPJ Genom. Med. 2020;5:56. doi: 10.1038/s41525-020-00164-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee H., Deignan J.L., Dorrani N., Strom S.P., Kantarci S., Quintero-Rivera F., Das K., Toy T., Harry B., Yourshaw M. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monies D., Abouelhoda M., Assoum M., Moghrabi N., Rafiullah R., Almontashiri N., Alowain M., Alzaidan H., Alsayed M., Subhani S. Lessons Learned from Large-Scale, First-Tier Clinical Exome Sequencing in a Highly Consanguineous Population. Am. J. Hum. Genet. 2019;104:1182–1201. doi: 10.1016/j.ajhg.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koboldt D.C. Best practices for variant calling in clinical sequencing. Genome Med. 2020;12:91. doi: 10.1186/s13073-020-00791-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farek J., Hughes D., Mansfield A., Krasheninina O., Nasser W., Sedlazeck F.J., Khan Z., Venner E., Metcalf G., Boerwinkle E. xAtlas: Scalable small variant calling across heterogeneous next-generation sequencing experiments. bioRxiv. 2018 doi: 10.1101/295071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Supernat A., Vidarsson O.V., Steen V.M., Stokowy T. Comparison of three variant callers for human whole genome sequencing. Sci. Rep. 2018;8:17851. doi: 10.1038/s41598-018-36177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J., Li X., Zhong H., Meng Y., Du H. Systematic comparison of germline variant calling pipelines cross multiple next-generation sequencers. Sci. Rep. 2019;9:9345. doi: 10.1038/s41598-019-45835-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumaran M., Subramanian U., Devarajan B. Performance assessment of variant calling pipelines using human whole exome sequencing and simulated data. BMC Bioinformatics. 2019;20:342. doi: 10.1186/s12859-019-2928-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan B., Kusko R., Xiao W., Zheng Y., Liu Z., Xiao C., Sakkiah S., Guo W., Gong P., Zhang C. Similarities and differences between variants called with human reference genome HG19 or HG38. BMC Bioinformatics. 2019;20(Suppl 2):101. doi: 10.1186/s12859-019-2620-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y., Dai Y., Yu H., Zhao S., Samuels D.C., Shyr Y. Improvements and impacts of GRCh38 human reference on high throughput sequencing data analysis. Genomics. 2017;109:83–90. doi: 10.1016/j.ygeno.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 16.Church D.M., Schneider V.A., Graves T., Auger K., Cunningham F., Bouk N., Chen H.C., Agarwala R., McLaren W.M., Ritchie G.R. Modernizing reference genome assemblies. PLoS Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider V.A., Graves-Lindsay T., Howe K., Bouk N., Chen H.C., Kitts P.A., Murphy T.D., Pruitt K.D., Thibaud-Nissen F., Albracht D. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017;27:849–864. doi: 10.1101/gr.213611.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowy-Gallego E., Fairley S., Zheng-Bradley X., Ruffier M., Clarke L., Flicek P., 1000 Genomes Project Consortium Variant calling on the GRCh38 assembly with the data from phase three of the 1000 Genomes Project. Wellcome Open Res. 2019;4:50. doi: 10.12688/wellcomeopenres.15126.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jäger M., Schubach M., Zemojtel T., Reinert K., Church D.M., Robinson P.N. Alternate-locus aware variant calling in whole genome sequencing. Genome Med. 2016;8:130. doi: 10.1186/s13073-016-0383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia T., Munson B., Lango Allen H., Ideker T., Majithia A.R. Thousands of missing variants in the UK Biobank are recoverable by genome realignment. Ann. Hum. Genet. 2020;84:214–220. doi: 10.1111/ahg.12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ballouz S., Dobin A., Gillis J.A. Is it time to change the reference genome? Genome Biol. 2019;20:159. doi: 10.1186/s13059-019-1774-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luu P.L., Ong P.T., Dinh T.P., Clark S.J. Benchmark study comparing liftover tools for genome conversion of epigenome sequencing data. NAR Genom Bioinform. 2020;2:a054. doi: 10.1093/nargab/lqaa054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Posey J.E., O’Donnell-Luria A.H., Chong J.X., Harel T., Jhangiani S.N., Coban Akdemir Z.H., Buyske S., Pehlivan D., Carvalho C.M.B., Baxter S., Centers for Mendelian Genomics Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet. Med. 2019;21:798–812. doi: 10.1038/s41436-018-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen A.W., Murugan M., Li H., Khayat M.M., Wang L., Rosenfeld J., Andrews B.K., Jhangiani S.N., Coban Akdemir Z.H., Sedlazeck F.J., Task Force for Neonatal Genomics A Genocentric Approach to Discovery of Mendelian Disorders. Am. J. Hum. Genet. 2019;105:974–986. doi: 10.1016/j.ajhg.2019.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regier A.A., Farjoun Y., Larson D.E., Krasheninina O., Kang H.M., Howrigan D.P., Chen B.J., Kher M., Banks E., Ames D.C. Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat. Commun. 2018;9:4038. doi: 10.1038/s41467-018-06159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 1303.3997v2. [Google Scholar]
- 33.Lin M.F., Rodeh O., Penn J., Bai X., Reid J.G., Krasheninina O., Salerno W.J. GLnexus: joint variant calling for large cohort sequencing. bioRxiv. 2018 doi: 10.1101/343970. [DOI] [Google Scholar]
- 34.Sabo A., Mishra P., Dugan-Perez S., Voruganti V.S., Kent J.W., Jr., Kalra D., Cole S.A., Comuzzie A.G., Muzny D.M., Gibbs R.A., Butte N.F. Exome sequencing reveals novel genetic loci influencing obesity-related traits in Hispanic children. Obesity (Silver Spring) 2017;25:1270–1276. doi: 10.1002/oby.21869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Sisoudiya S.D., Martin-Giacalone B.A., Khayat M.M., Dugan-Perez S., Marquez-Do D.A., Scheurer M.E., Muzny D., Boerwinkle E., Gibbs R.A. Germline Cancer-Predisposition Variants in Pediatric Rhabdomyosarcoma: A Report from the Children’s Oncology Group. J. Natl. Cancer Inst. 2020:djaa204. doi: 10.1093/jnci/djaa204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cleary J.G., Braithwaite R., Gaastra K., Hilbush B.S., Inglis S., Irvine S.A., Jackson A., Littin R., Rathod M., Ware D. Comparing Variant Call Files for Performance Benchmarking of Next-Generation Sequencing Variant Calling Pipelines. bioRxiv. 2015 doi: 10.1101/023754. [DOI] [Google Scholar]
- 39.Gel B., Serra E. karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics. 2017;33:3088–3090. doi: 10.1093/bioinformatics/btx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu Z., Gu L., Eils R., Schlesner M., Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 41.Sheffield N.C., Bock C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2016;32:587–589. doi: 10.1093/bioinformatics/btv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ouedraogo M., Bettembourg C., Bretaudeau A., Sallou O., Diot C., Demeure O., Lecerf F. The duplicated genes database: identification and functional annotation of co-localised duplicated genes across genomes. PLoS ONE. 2012;7:e50653. doi: 10.1371/journal.pone.0050653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalia S.S., Adelman K., Bale S.J., Chung W.K., Eng C., Evans J.P., Herman G.E., Hufnagel S.B., Klein T.E., Korf B.R. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 2017;19:249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 49.Dendrou C.A., Petersen J., Rossjohn J., Fugger L. HLA variation and disease. Nat. Rev. Immunol. 2018;18:325–339. doi: 10.1038/nri.2017.143. [DOI] [PubMed] [Google Scholar]
- 50.Steinberg K.M., Schneider V.A., Graves-Lindsay T.A., Fulton R.S., Agarwala R., Huddleston J., Shiryev S.A., Morgulis A., Surti U., Warren W.C. Single haplotype assembly of the human genome from a hydatidiform mole. Genome Res. 2014;24:2066–2076. doi: 10.1101/gr.180893.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frankish A., Uszczynska B., Ritchie G.R., Gonzalez J.M., Pervouchine D., Petryszak R., Mudge J.M., Fonseca N., Brazma A., Guigo R., Harrow J. Comparison of GENCODE and RefSeq gene annotation and the impact of reference geneset on variant effect prediction. BMC Genomics. 2015;16(Suppl 8):S2. doi: 10.1186/1471-2164-16-S8-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vollger M.R., Dishuck P.C., Sorensen M., Welch A.E., Dang V., Dougherty M.L., Graves-Lindsay T.A., Wilson R.K., Chaisson M.J.P., Eichler E.E. Long-read sequence and assembly of segmental duplications. Nat. Methods. 2019;16:88–94. doi: 10.1038/s41592-018-0236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Logsdon G.A., Vollger M.R., Eichler E.E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 2020;21:597–614. doi: 10.1038/s41576-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miga K.H., Koren S., Rhie A., Vollger M.R., Gershman A., Bzikadze A., Brooks S., Howe E., Porubsky D., Logsdon G.A. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 2020;585:79–84. doi: 10.1038/s41586-020-2547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherman R.M., Forman J., Antonescu V., Puiu D., Daya M., Rafaels N., Boorgula M.P., Chavan S., Vergara C., Ortega V.E. Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat. Genet. 2019;51:30–35. doi: 10.1038/s41588-018-0273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tetikol H.S., Narci K., Turgut D., Budak G., Kalay O., Arslan E., Demirkaya-Budak S., Dolgoborodov A., Jain A., Kabakci-Zorlu D. Population-specific genome graphs improve high-throughput sequencing data analysis: A case study on the Pan-African genome. bioRxiv. 2021 doi: 10.1101/2021.03.19.436173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The exome sequencing data used in this study are available at the database of Genotypes and Phenotypes (dbGaP: phs000711.v7.p2). The code generated in this study is available at https://github.com/hurleyLi/discreps. Additionally, we have made public sessions on the UCSC genome browser containing tracks of the DISCREP regions: https://genome.ucsc.edu/s/mdawood/DISCREPS_GRCh37; https://genome.ucsc.edu/s/mdawood/DISCREPS_GRCh38.