Summary
Publicly available genetic summary data have high utility in research and the clinic, including prioritizing putative causal variants, polygenic scoring, and leveraging common controls. However, summarizing individual-level data can mask population structure, resulting in confounding, reduced power, and incorrect prioritization of putative causal variants. This limits the utility of publicly available data, especially for understudied or admixed populations where additional research and resources are most needed. Although several methods exist to estimate ancestry in individual-level data, methods to estimate ancestry proportions in summary data are lacking. Here, we present Summix, a method to efficiently deconvolute ancestry and provide ancestry-adjusted allele frequencies (AFs) from summary data. Using continental reference ancestry, African (AFR), non-Finnish European (EUR), East Asian (EAS), Indigenous American (IAM), South Asian (SAS), we obtain accurate and precise estimates (within 0.1%) for all simulation scenarios. We apply Summix to gnomAD v.2.1 exome and genome groups and subgroups, finding heterogeneous continental ancestry for several groups, including African/African American (∼84% AFR, ∼14% EUR) and American/Latinx (∼4% AFR, ∼5% EAS, ∼43% EUR, ∼46% IAM). Compared to the unadjusted gnomAD AFs, Summix’s ancestry-adjusted AFs more closely match respective African and Latinx reference samples. Even on modern, dense panels of summary statistics, Summix yields results in seconds, allowing for estimation of confidence intervals via block bootstrap. Given an accompanying R package, Summix increases the utility and equity of public genetic resources, empowering novel research opportunities.
Keywords: ancestry, population stratification, gnomAD, population structure, summary, allele frequency, common controls, external controls
Introduction
Genetic summary data are a cornerstone of modern analyses. Allele frequencies (AFs) from publicly available data such as the Genome Aggregation Database (gnomAD)1 and Allele Frequency Aggregator (ALFA) from dbSNP2 can be used to prioritize putative causal variants for rare diseases and as pseudo controls in case-control analysis.3, 4, 5, 6 Compared to individual-level data, genetic summary data often have fewer barriers to access, promoting open science and the broad use of valuable resources. However, summary-level genetic data frequently contain fine-scale and continental-level population structure. For instance, unquantified continental ancestry exists in gnomAD’s “African/African American,” “American/Latinx,” and “other” groups as well as in other publicly available data (e.g., the BRAVO server for TopMED).7 Using public data without accounting for the underlying population structure can lead to confounded associations and incorrect prioritization of putative rare causal variants.
The use of mixture models to estimate population structure has a history going back over two decades, beginning with IMMANC8 and later the commonly-used STRUCTURE,9 originally using Dirichlet priors for multinomial modeling. Inference was performed via Markov chain Monte Carlo, which limits tractability to datasets with thousands to tens of thousands of markers. As datasets grew with the advent of genome-wide arrays, and later with sequencing, new methods were designed with improved convergence characteristics, such as the maximum-likelihood methods FRAPPE10 and ADMIXTURE,11 as well as the variational method FastSTRUCTURE.12 Along the way, methods were developed to leverage pooled data,13 e.g., iAdmix,14 with improvements to enhance supervised analysis in the ADMIXTURE framework.15 However, no method was designed explicitly and efficiently to model mixtures with genome-wide, summary statistic data that are common in modern genomics.
Individuals and samples from understudied or admixed populations are most likely to lack large public resources with precisely matched ancestry data. As a result, researchers and clinicians working with these populations are often left with a suboptimal choice: use the closest but still poorly matched ancestral group or do not use the publicly available and highly useful resource.6,16 The former has the potential to produce biased results in the very populations where additional high-quality research is needed, while the latter is likely to suffer from smaller sample sizes and thus a loss of statistical power. This choice exacerbates inequities in research in understudied and admixed populations.17,18 These issues are magnified in the context of precision medicine where genetic summary data will most likely not be sufficiently matched for the majority of people who themselves are a mixture of continental or fine-scale ancestries. Thus, methods to estimate and adjust for population structure within publicly available genetic summary data are needed.
Here, we present Summix, an efficient method that identifies, estimates, and adjusts for the proportion of continental reference ancestry in publicly available summary genetic data. We demonstrate the effectiveness of Summix, including the ability to produce ancestry-adjusted AFs to tailor analyses to less-studied populations, in over 5,000 simulation scenarios and in gnomAD v.2.1. Ultimately, Summix and the accompanying R, Python, and Shiny app software help to increase the efficacy and, importantly, the equity of valuable publicly available resources, especially for understudied and admixed samples.
Material and methods
Summix
Estimating ancestry proportions
An observed single-nucleotide polymorphism (SNP) AF can be described as a mixture of AFs across unobserved subgroups (e.g., continental ancestral populations). We estimate the group-specific mixing proportions,, by minimizing the least-squares difference between vectors of N SNPs for the observed AF, , and the AF generated from a mixture of K reference ancestry groups, , as shown in Equation 1.
| (Equation 1) |
This objective function is quadratic and, as such, is continuous, convex, and easily differentiable; the inequality constraints are linear. Hence, a feasible minimizer will fulfill the Karush-Kuhn-Tucker (KKT) conditions for optimality. We use sequential quadratic programming (SQP),19,20 a gradient-based, iterative algorithm for constrained, nonlinear optimization, to efficiently estimate the proportion of each reference group. We obtain confidence intervals (CIs) for the continental ancestry proportions by using block bootstrapping as described below.
Ancestry-adjusted allele frequencies
Using estimated continental ancestry proportions, we update the AFs in the observed data matching the continental ancestry proportions of an individual or sample as follows in Equation 2. To estimate the ancestry-adjusted AF, K − 1 homogenous reference ancestries are used. The ancestry not used is indexed as . In theory, ancestry can be any of the non-zero reference ancestry groups. Here, we choose to be the most common ancestry present in the summary data.
| (Equation 2) |
where, is the ancestry-adjusted allele frequency, is the ancestry group for which the reference allele frequency data are not used, is ancestry group index, is population k ancestry proportion for target individual or sample, is estimated ancestry proportion of population k for observed publicly available summary data, is allele frequency for observed publicly available summary data (e.g., gnomAD), and is reference allele frequency for ancestry k; K − 1 homogenous reference ancestries are used.
Equation 2 can be used to estimate ancestry-adjusted AF for a homogenous or admixed sample or individual with given ancestry proportions. We evaluate both scenarios as described below.
Data
All data were on genome build GRCh37.
1000 Genomes and Indigenous American data
We used four continental ancestry groups from 1000 Genomes v.5 phase 3 20150502,20 African (AFR), East Asian (EAS), non-Finnish European (EUR), and South Asian (SAS), and an Indigenous American (IAM) sample for the reference data. The IAM Affymetrix 6.0 data had been previously harmonized with the 1000 Genomes data.21 1000 Genomes AFs were calculated as previously described.22 IAM AFs were calculated with PLINK 1.9. We excluded the admixed populations from the 1000 Genomes AFR continental ancestry: Americans of African ancestry in the southwestern USA (ASW) and African Caribbeans in Barbados (ACB). Related individuals were also removed, resulting in sample sizes of 504 AFR, 504 EAS, 404 EUR, 489 SAS, and 43 IAM. We merged the 1000 Genomes and IAM data and kept the subset of SNPs in both datasets. We limited further to bi-allelic non-palindromic SNPs with minor allele frequency (MAF) > 1% in at least one continental ancestral group, resulting in 613,298 SNPs.
gnomAD v.2.1
Variants from gnomAD v.2.1 (data accessed April 2019) were limited to bi-allelic and PASS, as defined by gnomAD,1 resulting in 13,742,683 and 196,606,976 SNPs in the exome and genome gnomAD samples, respectively. After we further limited to SNPs with MAF > 1% in at least one gnomAD group and merged these with the reference data, 9,763 and 582,156 SNPs remained in the exome and genome data, respectively. As described further below, ancestry proportions were estimated for African/African American (n = 8,128 exome; n = 4,359 genome), American/Latinx (n = 17,296 exome; n = 424 genome), other (n = 3,070 exome; n = 544 genome), non-Finnish European (n = 56,885 exome; n = 7,718 genome), East Asian (n = 9,197 exome; n = 780 genome), and South Asian (n = 15,308 exome) groups and all subsets (i.e., controls, non-cancer, non-neuro, and non-TopMED). Additionally, we used gnomAD v.2.1 Ashkenazi Jewish (n = 5,040 exome; n = 145 genome) and Finnish (n = 10,824 exome; n = 1,738 genome) to evaluate the performance of Summix for mismatched reference data.
ClinVar
gnomAD v.2.1 AF and ClinVar (GRCh37/hg19) variants were merged by chromosome, base pair, and alleles. For three variants, there were multiple ClinVar allele IDs in the same position with the same alleles. All duplicate positions were retained. After we merged and restricted to ClinVar classifications of “uncertain pathogenicity” and “conflicting reports of pathogenicity,” 42 and 122 variants were present in the exome and genome data, respectively. Ancestry-adjusted AFs were estimated for an African sample from the African/African-American gnomAD control AFs with gnomAD EUR as the reference sample.
The American College of Medical Genetics (ACMG) guidelines23 designate >5% MAF in a control population as stand-alone strong evidence of a variant’s having benign impact for a rare Mendelian disorder. As such, for the classifications of “uncertain Pathogenicity” and “conflicting reports of pathogenicity,” we identified variants where the ancestry-adjusted AF differed from the unadjusted AF with respect to the MAF > 5% threshold. Additionally, we identified variants with the classification “pathogenic,” “likely_pathogenic,” or “pathogenic/likely_pathogenic” with either adjusted or unadjusted AF above 5%.
Simulations
Using the 1000 Genomes as reference data, we simulated SNP genotypes for all combinations and subsets of the five continental populations. SNP genotypes were simulated with the rmultinom R function with probability defined from the AFs for the continental reference ancestral populations assuming Hardy-Weinberg Equilibrium. We chose ancestry proportions randomly within an assigned proportion bin to ensure coverage across the range of possible values, especially at the edges of the distribution: 0–0.015, 0.010–0.055, 0.05–0.105, 0.10–0.255, 0.25–0.505. The simulated proportion for the Kth ancestry group was chosen so that the ancestry proportions summed to one.
Simulating across all combinations of continental ancestry groups and the ancestry proportion bins resulted in 5,360 simulation scenarios. We used 1,000 replicates within each simulation scenario to assess accuracy and precision. For each simulation replicate, we randomly sampled 100,000 SNPs. We define accuracy as the difference between the mean estimated ancestry proportion and simulated ancestry proportion. We define precision as the standard deviation of the simulation replicates. The simulation code is provided on the manuscript’s GitHub site.
Real data application
Estimating ancestry proportions
Continental reference ancestry proportions were estimated with the summix R function for gnomAD v.2.1 African/African American, American/Latinx, East Asian, other, non-Finnish European, and South Asian exome and genome including all subgroups (e.g., controls, non-neuro). To estimate the ancestry group proportions, we used the filtered datasets with 9,763 and 582,156 SNPs in the exome and genome samples, respectively. To assess stability of the estimates over different numbers of SNPs, we estimated the ancestry proportions from 1,000 random samples of sets of N SNPs: 10, 50, 100, 500, 1,000, 2,500, 5,000, 10,000, 50,000, and 100,000 for genomes and 10, 50, 100, 500, 1,000, 2,500, 5,000, 7,500, and 9,000 for exomes.
Block bootstrapping
We used block bootstrapping to estimate uncertainty for the ancestry proportion estimates. We used the sex-averaged centimorgan (cM) map created from Bherer et al.24 to define 1 cM blocks throughout the genome. We used the na.approx function8,25 from the zoo R package25 to linearly interpolate cM for SNPs in our dataset that were not observed in Bherer et al. This resulted in 3,357 1 cM blocks across the genome. Five and 129 SNPs in the exome and genome data, respectively, that were outside the genetic regions contained in the Bherer et al. dataset were not linearly interpolated. This resulted in 9,763 and 582,156 SNPs in 2,206 and 3,353 1 cM blocks for the exome and genome gnomAD samples, respectively. This final sample was used for all real data analysis. We used 1,000 bootstrap replicates to estimate 95% block bootstrap CIs. The lower and upper CIs were estimated from the 2.5 and 97.5 percentiles of the block bootstrap distribution.
Estimating ancestry-adjusted allele frequencies
We estimated and assessed ancestry-adjusted AFs for two samples: (1) an African sample (100% African) estimated from the African/African American gnomAD AFs and (2) an admixed Peruvian sample with average ancestry proportions of 76.8% Indigenous American, 19.6% European, 2.7% African, and 0.9% East Asian ancestry estimated from the American/Latinx gnomAD AFs. The continental ancestry proportions for the admixed Peruvian population were estimated from a subset of unrelated individuals (n = 85) from the 1000 Genomes Peruvian sample via supervised ADMIXTURE.11 For both the African sample and the Peruvian sample, ancestry-adjusted AFs were estimated with reference AF from either 1000 Genomes or gnomAD. We used reference groups with ≥2% estimated ancestry proportion in the observed gnomAD v.2.1 group and normalized the estimated ancestry proportions to total 1. This resulted in K = 2 ancestry groups for the African/African American group and K = 4 for the American/Latinx group. For the African/African American group with K = 2 reference groups, non-Finnish European reference AFs were used (excluding African reference). For the Peruvian population where K = 4, non-Finnish European, African, and East Asian reference AFs were used (excluding Indigenous American reference). We estimated the Peruvian ancestry-adjusted AFs by using gnomAD non-Finnish European and East Asian reference populations and estimated ancestry-adjusted AFs for a 100% African ancestry from the gnomAD African/African American reference group.
To assess the accuracy of the ancestry-adjusted estimates, we compared the ancestry-adjusted and unadjusted gnomAD AFs to 1000 Genomes AFs for the target ancestral population (i.e., African or Peruvian Latinx). For these comparisons, we filtered out variants that were called in fewer than 25% of the gnomAD group. This removed 120 and 128 variants for African/African American and American/Latinx gnomAD exome groups, respectively, and 8 and 11 variants for African/African American and American/Latinx gnomAD genome groups, respectively. We calculated both the absolute and relative difference as shown in Equation 3.
| (Equation 3) |
where is the ancestry-adjusted or unadjusted AF.
To test whether differences varied by adjustment group, we use a linear mixed effects model with SNP and cM block as random effects with the lmer function from the lme4 package.26 We tested all SNPs as well as within AF bins defined by the target 1000 Genomes reference ancestry (AF bin: <0.01, 0.01–0.02, 0.02–0.05, 0.05–0.1, 0.1–0.3, 0.3–0.5, 0.5–0.7, 0.7–0.9, 0.9–0.95, 0.95–0.98, 0.98–0.99, ≥0.99). Pairwise comparisons were adjusted for post-hoc multiple testing with Tukey adjustment. We assessed agreement between 1000 Genomes and ancestry-adjusted AFs or unadjusted AFs by using Lin’s concordance correlation coefficient (Lin’s CCC)27,28 and 95% CIs estimated with the CCC function in the DescTools R package v.0.99.38 29.
Summix versus ADMIXTURE ancestry estimates
We compared continental ancestry proportions from Summix to estimates from ADMIXTURE11 by using individual-level data for a sample of n = 85 unrelated individuals from the Peruvian 1000 Genomes data. Estimates were obtained via supervised and unsupervised ADMIXTURE with default settings as well as projection with the learned AFs from unsupervised ADMIXTURE of the reference data. For both the unsupervised and supervised ADMIXTURE estimates, the AFR, EAS, EUR, and IAM reference individuals were included along with the Peruvian individuals. Bootstrap 95% CIs in ADMIXTURE were estimated with the -B command. Supervised estimates were used for all comparisons and further analyses. To compare computing time between ADMIXTURE and Summix, we ran the methods on a Dual Intel Xeon E5-2670v2 2.5 Ghz (10 core/20 thread) with 192GB DDR3-1600 ECC Registered Memory.
Reanalysis of PADI3
To demonstrate the utility of Summix’s ancestry-adjusted estimates in case-control analysis using external controls, we reanalyzed data from Malki et al.6 Malki et al. used gnomAD v.2.1 African/African American as controls in a case-control analysis to show that PADI3 (MIM: 606755) is associated with central centrifugal cicatricial alopecia (MIM: 618352) in women of African ancestry.6 We estimated the ancestry-adjusted AF for PADI3 variants identified by Malki et al. for a sample with 100% African ancestry. We estimated ancestry-adjusted allele counts (ACs) by multiplying the ancestry-adjusted AF by the variant-specific allele number. The number of individuals in gnomAD v.2.1 with at least one minor allele was calculated as the number of minor alleles minus the number of homozygotes. Using the case numbers reported from their manuscript, we repeated the case-control analysis by using the original unadjusted gnomAD v.2.1 data and the ancestry-adjusted gnomAD v.2.1 results for a homogeneous African sample. As in Malki et al., we provide p values for both a chi-square test of independence and Fisher’s exact test.
Reanalysis of p.Phe508del
Nappo et al. use gnomAD v.2.1 to estimate the prevalence of CFTR (MIM: 602421) variants defined as cystic fibrosis (CF [MIM: 219700])-causing or varying clinical consequence in non-European populations, including South Asian and African/African American.16 To highlight Summix’s utility in providing more precise AF adjusting for ancestry, we provide ancestry-adjusted AF for a 100% African target sample for p.Phe508del (c.1521_1523delCTT), the most common CF variant.
Sensitivity of ancestry estimates to reference data
We evaluated the sensitivity of Summix’s ancestry proportion estimates to mismatches in the reference data in simulations and gnomAD v.2.1. Specifically, we compared the least-squares loss function value in scenarios where the reference data were known to not represent the observed data well. For gnomAD v.2.1 exomes and genomes, we used the five reference ancestries (i.e., AFR, EAS, EUR, IAM, and SAS) to estimate the ancestry proportions for Ashkenazi Jewish and Finnish, which are known to diverge from European ancestry.
Additionally, we simulated 5-way admixed populations by using the five reference ancestries (i.e. AFR, EAS, EUR, IAM, and SAS) and the rmultinom function in R as described above for other simulations. For each simulation scenario, we held a specific reference ancestry at a fixed proportion (i.e., 0, 0.01, 0.05, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4, 0.45, 0.5, 0.55, 0.6, 0.65, 0.7, 0.75, 0.8, 0.85, 0.9, 0.95, 0.99) and allowed the other four ancestry proportions to be random with the constraint that the proportions sum to 1. We removed the fixed ancestry from the reference panel and used Summix to estimate the remaining four ancestry proportions. We recorded the least-squares loss of the solution. For each fixed ancestry and proportion combination, we performed 100 simulation replicates.
Software and code
Summix is available in R and Python. The Python (v.3.7) package, summixpy, contains two main Python scripts. The first function, summix.py, estimates the proportions of reference ancestry groups for an observed sample. The second function, adjAF.py, estimates the ancestry-adjusted AF. The package contains example genetic data and analysis presented in a Jupyter Notebook and is hosted on GitHub.
The Summix R package enables both estimation of reference ancestry groups via the summix function and ancestry-adjusted AFs via the adjAF function. More details, including example data and implementation, are available in our package, which is hosted on Bioconductor v.3.13 or later and our GitHub site.
Shiny app
Within the Shiny app, users can estimate and visualize ancestry proportions for three gnomAD ancestry groups (i.e., African/African American, American/Latinx, and other) for both the exome and genome data. The Shiny app is hosted on the University of Colorado Denver servers (see web resources).
Results
Simulations
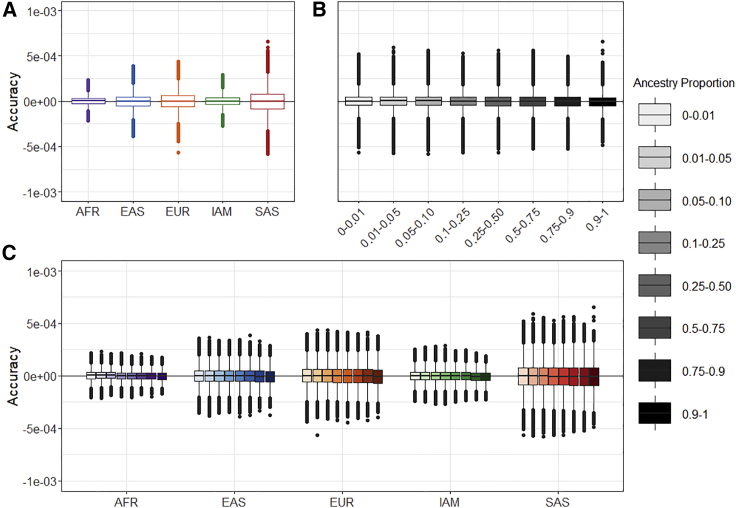
Summix achieved accuracy within 0.001% and precision within 0.1% across all simulation scenarios (Table S1, Table S10, Table S17, Figure 1, Figures S1–S4). Accuracy of the proportion estimates was consistent across the range of simulated mixture proportions with a slight increase in bias near 0 and 1. While bias and variability in the estimates was small for all ancestral groups, AFR had the lowest variability followed by IAM, EAS, EUR, and then SAS, which had the highest variability across simulation replicates (Table S1, Table S10, Table S17, Figure 1, Figures S1–S4). For simulation scenarios with non-zero proportions simulated for all five ancestry groups, the standard deviation was as follows: SDAFR = 4.31E−5, SDIAM = 5.53E−5, SDEAS = 7.58E−5, SDEUR = 9.18E−5, and SDSAS = 1.21E−4. This trend was consistent across all simulation scenarios.
Figure 1.
Simulation results for five ancestries
Accuracy is defined as the difference between the estimated ancestry proportions and given ancestry proportions within simulations. We used five reference ancestries to simulate genotypes of an admixed population.
(A) Accuracy separated by ancestry.
(B) Accuracy separated by ancestry proportion.
(C) Accuracy separated by both ancestry and ancestry proportion.
Application to gnomAD
Estimating ancestry proportions
We estimated the proportion of reference continental ancestry groups in gnomAD v.2.1 African/African American, American/Latinx, East Asian, other, non-Finnish European, and South Asian groups and all subgroups (e.g., controls, non-TopMED) for both the exome and genome data (Table 1, Tables S6 and S7). As expected, the African/African American groups have primarily AFR (>80%) and EUR ancestry (∼15%), most likely because of contribution from African American individuals. The exome and genome American/Latinx gnomAD groups had high proportions of both EUR and IAM ancestry (i.e., >35%) and ancestry proportion estimates between 1%–6% for the other reference groups. Interestingly, >1% SAS ancestry was estimated in both the exome and genome American/Latinx gnomAD groups, perhaps because of misspecification from a limited number of reference groups. The exome and genome “other” gnomAD groups were primarily EUR reference ancestry (>77%). The estimated reference proportions for non-Finnish Europeans and East Asian were very homogeneous and had >96% EUR- and 100% EAS-estimated reference ancestries, respectively. The South Asian gnomAD exome group had 85% estimated SAS ancestry and ∼15% estimated EUR reference ancestry as expected because of the known ancient admixture events in the region.30, 31, 32
Table 1.
Estimated ancestry proportions for gnomAD groups (95% block bootstrap CI)
| Ancestry group | AFR | EAS | EUR | IAM | SAS |
|---|---|---|---|---|---|
| Genome | |||||
| African/African American (n = 4,359) | 0.825 (0.824, 0.825) | 0.005 (0.004, 0.006) | 0.157 (0.156, 0.158) | 0.008 (0.008, 0.009) | 0.005 (0.004, 0.006) |
| American/Latinx (n = 424) | 0.058 (0.057, 0.059) | 0.038 (0.036, 0.041) | 0.505 (0.502, 0.507) | 0.380 (0.378, 0.382) | 0.0194 (0.016, 0.023) |
| Other (n = 544) | 0.047 (0.046, 0.048) | 0.034 (0.032, 0.036) | 0.793 (0.790, 0.796) | 0.043 (0.042, 0.045) | 0.084 (0.080, 0.087) |
| Non-Finnish European (n = 7,718) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) | 0.964 (0.962, 0.967) | 0.016 (0.015, 0.017) | 0.020 (0.017, 0.022) |
| East Asian (n = 780) | 0.000 (0.000, 0.000) | 1.000 (1.000, 1.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) |
| South Asian (n = 0) | – | – | – | – | – |
| Exome | |||||
| African/African American (n = 8,128) | 0.840 (0.838, 0.842) | 0.005 (0.001, 0.009) | 0.146 (0.142, 0.149) | 0.009 (0.006, 0.012) | 0.000 (0.000, 0.006) |
| American/Latinx (n = 17,296) | 0.0430 (0.039, 0.047) | 0.049 (0.040, 0.059) | 0.432 (0.423, 0.441) | 0.463 (0.455, 0.471) | 0.013 (0.000, 0.027) |
| Other (n = 3,070) | 0.034 (0.032, 0.037) | 0.046 (0.041, 0.051) | 0.780 (0.775, 0.787) | 0.051 (0.047, 0.054) | 0.089 (0.080, 0.098) |
| Non-Finnish European (n = 56,885) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) | 0.975 (0.969, 0.982) | 0.008 (0.005, 0.012) | 0.017 (0.009, 0.023) |
| East Asian (n = 9,197) | 0.000 (0.000, 0.000) | 1.000 (1.000, 1.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) |
| South Asian (n = 15,308) | 0.002 (0.000, 0.004) | 0.000 (0.000, 0.000) | 0.150 (0.144, 0157) | 0.002 (0.000, 0.005) | 0.845 (0.838, 0.852) |
Abbreviations: AFR, African continental ancestry group; EAS, East Asian continental ancestry group; EUR, European continental ancestry group; IAM, Indigenous American continental ancestry group; SAS, South Asian continental ancestry group.
Despite large differences in sample sizes, the estimated proportion of reference ancestry groups was similar (i.e., within 2%) between exome and genome samples for all groups except American/Latinx where the exome and genome estimates differed by >5% for the EUR and IAM reference proportions. The reference ancestry proportion estimates for the gnomAD v.2.1 subgroups (i.e., controls, non-cancer, non-neuro, and non-TopMED) were mostly similar; they were within ∼2% of each other and of the overall gnomAD v.2.1 group estimates. The exception was for the American/Latinx and “other” genome groups, which sometimes varied by 5%–10%. These groups had sample sizes (N < 600) and, thus, were most likely more susceptible to the inclusion or exclusion of subsamples of individuals. Complete results are shown in Tables S6 and S7.
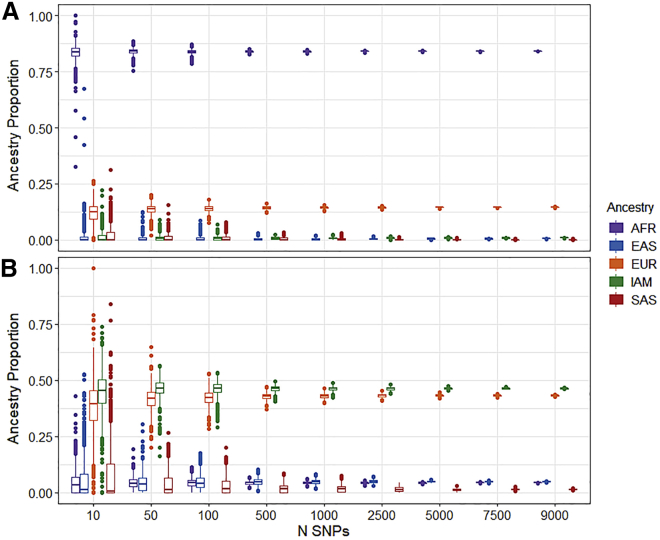
We evaluated Summix’s ability to estimate reference ancestry proportions by using smaller numbers of randomly chosen SNPs. We find that the ancestry proportion estimates stay unbiased regardless of the number of SNPs used to estimate ancestry, while the precision decreases as the number of SNPs decreases. The precision is still fairly tight down to ∼500 SNPs, especially when estimating the African/African American gnomAD samples (exomes: SDAFR = 0.0039, SDIAM = 0.0052, SDEAS = 0.0054, SDEUR = 0.0065, and SDSAS = 0.0067). This suggests that far fewer SNPs are most likely needed to arrive at sample estimates of ancestry proportions (Figure 2, Figure S5).
Figure 2.
Precision in ancestry estimates for African/African American and American/Latinx gnomAD groups by number of SNPs
Number of SNPs (x axis) and estimated ancestry proportion (y axis) for 1,000 replicates.
(A) African/African American exome.
(B) American/Latinx exome.
Ancestry-adjusted allele frequencies
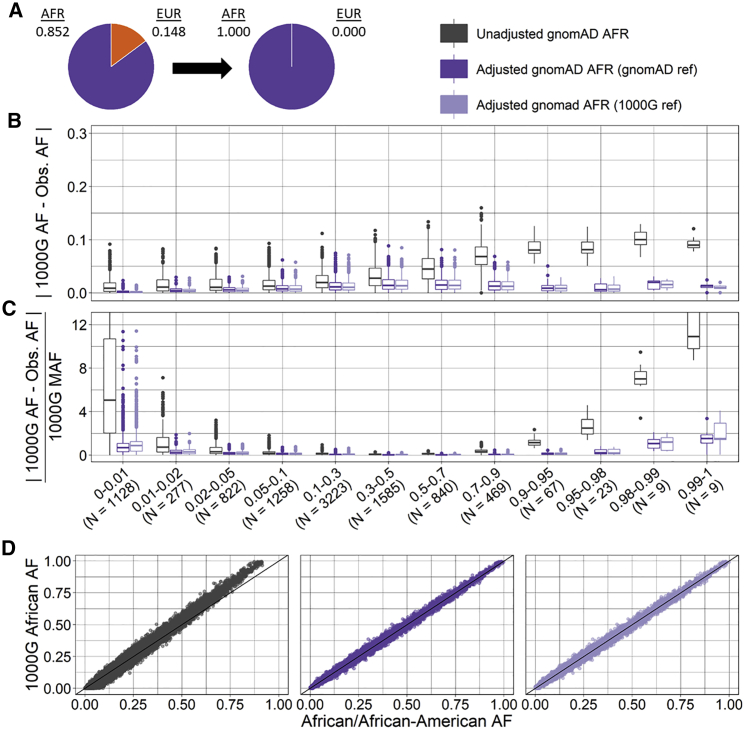
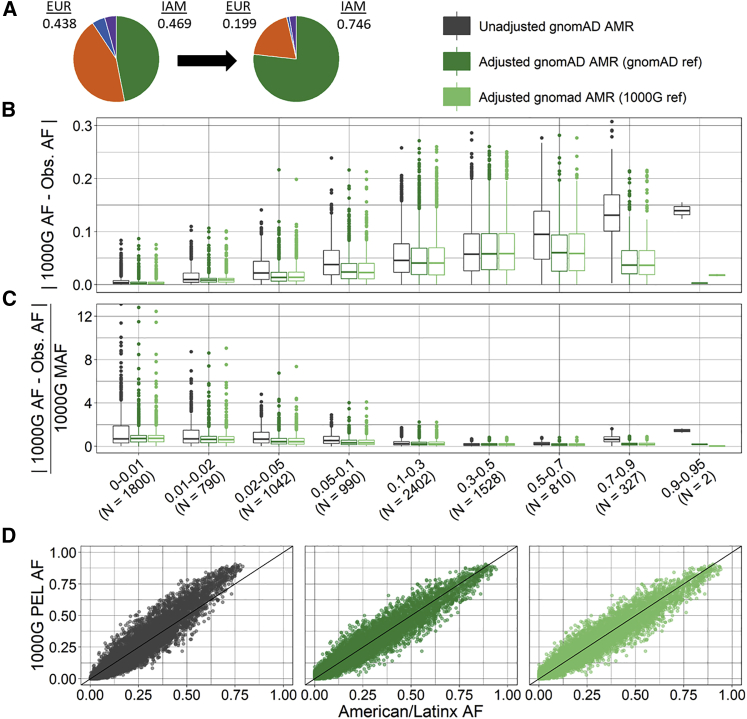
For both exome and genome data, we estimated the ancestry-adjusted AF for gnomAD African/African American for a target sample with 100% continental African ancestry and for gnomAD American/Latinx for a target Peruvian sample with 76.8% Indigenous American, 19.6% European, 2.7% African, and 0.9% East Asian ancestry proportions. Compared to the unadjusted AFs, the ancestry-adjusted AFs had significantly smaller absolute and relative differences with the target group AF (Table 2, Tables S8 and S9, p < 1E−16) regardless of reference data used (i.e., 1000 Genomes or gnomAD). The relative difference was greatest at small MAFs, while the absolute difference increased as AF increased, consistent with expectations. For SNPs with an alternative AF > 0.9 in 1000 Genomes, the absolute and relative difference between the unadjusted gnomAD and target 1000 Genomes AFs was especially large. This is most likely due to both a relatively small number of SNPs in these groups and differences between 1000 Genomes samples and the reference genome. The absolute and relative differences in SNPs with AF > 0.9 is considerably reduced for the ancestry-adjusted AFs (Figures 3 and 4, Table 2, Tables S8 and S9).
Table 2.
Absolute and relative difference between unadjusted and adjusted gnomAD AF and target sample in the exome data
|
Means (95% CI) |
p value |
|||||
|---|---|---|---|---|---|---|
| unadj (1) | anc-adj 1000G ref (2) | anc-adj gnomAD ref (3) | 1 versus 2 | 1 versus 3 | 2 versus 3 | |
| AFR absolute | 0.027 (0.0264, 0.0271) | 0.012 (0.011, 0.012) | 0.011 (0.011, 0.012) | <1E−16 | <1E−16 | 0.123 |
| AFR relative | 0.559 (0.536, 0.583) | 0.154 (0.130, 0.178) | 0.139 (0.115, 0.162) | <1E−16 | <1E−16 | 0.545 |
| AMR absolute | 0.046 (0.045, 0.047) | 0.0345 (0.034, 0.036) | 0.035 (0.034, 0.036) | <1E−16 | <1E−16 | 0.989 |
| AMR relative | 0.532 (0.517, 0.546) | 0.361 (0.347, 0.375) | 0.363 (0.349, 0.378) | <1E−16 | <1E−16 | 0.926 |
Abbreviations: AFR, African/African American gnomAD group; AMR, American/Latinx gnomAD group; unadj, unadjusted allele frequencies; anc-adj 1000G ref, ancestry-adjusted allele frequencies using 1000 Genomes reference data; anc-adj gnomAD ref, ancestry-adjusted allele frequencies using gnomAD reference data.
Figure 3.
Ancestry-adjusted versus unadjusted allele frequency for gnomAD African/African American exomes for a target sample with African ancestry
Ancestry-adjusted AF was estimated for a target sample with 100% African ancestry via gnomAD (dark purple) or 1000 Genomes (light purple) non-Finnish European as reference and compared to unadjusted AF (grey) for 9,710 SNPs.
(A) Ancestry proportions for gnomAD African/African American exomes (AFR = 0.852, EUR = 0.148) and target sample (AFR = 1).
(B) Absolute difference between target sample AF (1000 Genomes African ancestry) and unadjusted or ancestry-adjusted gnomAD AF by 1000 Genomes AF category.
(C) Relative difference between target 1000 Genomes African ancestry AF and unadjusted or ancestry-adjusted gnomAD AF by 1000 Genomes AF category; unzoomed versions of (B) and (C) are available in the supplemental information (Figure S10).
(D) Scatter plot of target sample 1000 Genomes AF (y axis) and unadjusted (left), ancestry-adjusted with gnomAD reference (center), and ancestry-adjusted with 1000 Genomes reference (right) gnomAD AF (x axis).
Figure 4.
Ancestry-adjusted versus unadjusted AF for gnomAD American/Latinx exomes for a target sample of Peruvian ancestry
Ancestry-adjusted AF was estimated for a target Peruvian sample via gnomAD (dark green) or 1000 Genomes (light green) East Asian, European, and African as reference ancestral populations and compared to unadjusted AF (grey) for 8,633 SNPs.
(A) Normalized ancestry proportions estimated for gnomAD American/Latinx exomes (purple, AFR = 0.044; blue, EAS = 0.049; orange, EUR = 0.438; green, IAM = 0.469) and target Peruvian ancestry proportions (purple, AFR = 0.028; blue, EAS = 0.027; orange, EUR = 0.199; green, IAM = 0.746).
(B) Absolute difference between target 1000 Genomes Peruvian ancestry AF and unadjusted or ancestry-adjusted gnomAD AF by 1000 Genomes AF category.
(C) Relative difference between target 1000 Genomes Peruvian ancestry AF and unadjusted or ancestry-adjusted gnomAD AF by 1000 Genomes AF category; unzoomed versions of (B) and (C) are available in the supplemental information (Figure S11).
(D) Scatter plot of target 1000 Genomes AF (y axis) and unadjusted (left), ancestry-adjusted with gnomAD reference (center), and ancestry-adjusted with 1000 Genomes reference (right) gnomAD AF (x axis).
The ancestry-adjusted AFs using 1000 Genomes or gnomAD as reference data were very similar. In the exome data, we observed no systematic significant absolute or relative differences between reference datasets (i.e., gnomAD reference versus 1000 Genomes reference) with the exception of relative difference for the African 0–0.01 bin. Most likely because of larger numbers of SNPs in the genome data, we do see significant, albeit very small, differences by reference ancestry between the ancestry-adjusted AF and the AFs in the target 1000 Genomes sample (Tables S8 and S9). These differences, while statistically significant, were 10 to 100 times smaller compared to the unadjusted AF.
We used Lin’s CCC to estimate agreement between the target sample and the gnomAD AF. In gnomAD exomes, Lin’s CCC estimates were higher for the ancestry-adjusted compared to the unadjusted AFs regardless of reference data for both the African group (estimate [95% CI]: adj. gnomAD_ref = 0.9977 [0.9976, 0.9978]; adj. 1000G_ref = 0.9976 [0.9975, 0.9977]; unadj. = 0.9866 [0.9862, 0.9870]) and the Peruvian group (adj. gnomAD_ref = 0.9689 [0.9676, 0.9702]; adj. 1000G_ref = 0.9691 [0.9678, 0.9704]; unadj. = 0.9438 [0.9417, 0.9458]). We found similar results for the gnomAD genome data. Both the ancestry-adjusted and unadjusted AFs differ more for the gnomAD American/Latinx group compared to the target 1000 Genomes Peruvian AFs than for the African comparison. This is perhaps due to a larger number of reference ancestry groups, more heterogeneity in Latinx compared to African American samples, or better representation in the reference data for African/African American than American/Latinx.
Within the gnomAD genome data, we identified some SNPs with a large mismatch between 1000 Genomes and gnomAD AF (Figures S6–S9). These SNPs appear to be mostly multi-allelic with one or more indels as the additional alleles (Tables S10 and S11). Additionally, the ancestry-adjusted AF is sometimes below 0 or above 1 (Table S12); for these variants, we rounded to 0 and 1, respectively. We recommend caution when using ancestry-adjusted AF estimates for multi-allelic variants or variants with an ancestry-adjusted AF close to or equal to 0 or 1. More research and potentially external validation are most likely needed to estimate the true AF of all alleles present.
Summix versus ADMIXTURE ancestry estimates
We estimated ancestry proportions for the 1000 Genomes Peruvian sample of 85 unrelated individuals by using AFs with Summix and individual-level data with ADMIXTURE. Assuming four reference ancestry groups, we estimate 0.033 AFR (0.032, 0.035), 0.035 EAS (0.031, 0.039), 0.209 EUR (0.206, 0.212), and 0.723 IAM (0.719, 0.726) for Summix compared to 0.027 AFR (0.018, 0.035), 0.010 EAS (0.003, 0.017), 0.196 EUR (0.166, 0.226), and 0.768 IAM (0.736, 0.799) for supervised ADMIXTURE. The ADMIXTURE ancestry estimates for unsupervised and supervised were nearly identical (Table S13). Overall, the estimates from Summix and ADMXITURE are similar and the 95% CIs overlap for AFR and EUR and are within 2% for EAS and IAM. The slight difference between Summix and ADMIXTURE supervised estimates may be due to reduced ability to distinguish between EAS and IAM. Indeed, the estimates between Summix and ADMIXTURE projection are nearly identical (Table S13), indicating that the minor differences between supervised ADMIXTURE and Summix are most likely due to using AFs versus individual-level data. The similarity between Summix estimates and ADMIXTURE estimates with individual-level data further supports Summix’s ability to estimate ancestry proportions from summary data without the need for individual-level data. Further, without the need to use individual reference data or estimate individual level ancestry, Summix is much more efficient solving in seconds, whereas ADMIXTURE takes a minimum of 24 minutes (Table S13).
Reanalysis of PADI3
Summix can be used to estimate and adjust for ancestry in analyses that use gnomAD and other summary data as external controls. As an exemplar, we repeated the case-control analysis of PADI3 from Malki et al.6 We found the p values were higher for the ancestry-adjusted allele counts (ACs) (chi-squared p value = 0.114, Fisher’s exact test p value = 0.101) compared to the unadjusted gnomAD v.2.1 African/African American ACs (chi-squared p value = 0.029, Fisher’s exact test p value = 0.031) (Tables S14 and S15). As expected, this shows that association results are not robust to differences in ancestry. It is likely that the cases used by Malki et al.6 were not 100% African ancestry. Summix could be used to estimate ancestry-adjusted AC in gnomAD given the exact ancestry proportions in the cases. We expect the p values of association would most likely lie between the unadjusted and adjusted p values provided given that the proportion of African ancestry is most likely between gnomAD unadjusted (0.852) and adjusted (1).
Reanalysis of p.Phe508del
To evaluate the prevalence of CF variants in non-European ancestral populations, Nappo et al. report the AF for CFTR variants with causal or varying clinical consequence in non-European ancestral groups from gnomAD v.2.1, such as African/African American and South Asian.16 As another exemplar of the utility for ancestry-adjusted AF, we estimated the adjusted AF for the most common CF variant, p.Phe508del, for the African/African American group assuming 100% African ancestry. As expected, given the higher AF in the non-Finnish European ancestry, the ancestry-adjusted AF is smaller than the unadjusted AF for both exome and genome African/African American groups (Table S16). This indicates that the prevalence of p.Phe508del may be lower in homogeneous African ancestry groups than the admixed African American group in gnomAD.
ClinVar
As an exemplar for the potential utility of ancestry-adjusted AF in clinical settings, we compare the ancestry-adjusted AF for 100% African ancestry to the unadjusted AF for the gnomAD African/African American sample for a subset of ClinVar variants labeled as pathogenic, uncertain pathogenicity, or conflicting reports (material and methods).
Based on previous ACMG guidelines,23 we focused on variants labeled as either “uncertain pathogenicity” or “conflicting reports of pathogenicity.” We identified 68 unique ClinVar variants at 67 positions on the exome and genome (including 11 variants that were identified on both the exome and genome) where the ancestry-adjusted AF was above 5% and unadjusted AF was below 5% and 42 variants at 41 positions where the ancestry-adjusted AF was below 5% and the unadjusted AF was above (five of these variants were identified on both the exome and genome). Overall, we find minor differences of less than 0.05 between ancestry-adjusted and unadjusted AF. Eleven variants had differences greater than 0.05 in AF (Table S17). Some of these variants most likely warrant further follow up.
We identified 29 variants with unique ClinVar IDs (18 in the genome data, two in the exome data, and nine in both the exome and genome data) listed as pathogenic or likely-pathogenic in ClinVAR with AF > 5% for either the unadjusted or adjusted gnomAD African/African American samples. All but three of these variants had adjusted and unadjusted AF > 5%, and most variants were very common (e.g., MAF > 20%) (Table S18). Further inspection of these variants indicated little to no support for pathogenicity. Most of these (n = 24) do not have assertion criteria provided and 13 were submitted to the ClinVar repository well before the 2015 update to the ACMG guidelines prompted by increased use of high-throughput sequencing.33 These variants may warrant further review. One of the variants identified as pathogenic and having a high AF is rs429358, one of two variants that defines the APOE-e4 (MIM: 107741) allele that incurs an increased risk of Alzheimer disease (MIM: 607822). The high AF for rs429358 observed in the gnomAD African/African American group is expected because AF > 0.1 has been observed in samples of African and African American individuals2,34. The APOE-e4 allele confers increased risk of Alzheimer disease in various ancestries, including European35 and African,36,37 although heterogeneity is observed in effect size and AF across ancestries.38
Sensitivity of ancestry estimates to reference data
In simulations, we find that the least-squares loss increases as the proportion of fixed continental ancestry not in the reference data increases. This increase in least-squares loss is seen for all continental reference ancestry groups removed from the reference data. However, continental reference ancestry groups that are known to be more distinct from other ancestries (e.g., AFR) result in larger loss, indicating a poorer fit of the model when these ancestries are not in the reference data (Figure S12). Based on these simulations, we find a loss above 0.5/1,000 SNPs indicates a moderate amount of missing ancestry and a loss above 1.5/1,000 SNPs most likely indicates a substantial amount of missing ancestry. Similarly, in gnomAD v.2.1 data, we find larger least-squares loss when estimating the continental ancestry proportions for Ashkenazi Jewish (loss/1,000 SNPs = 2.05 exome and 2.46 genome) and Finnish (loss/1,000 SNPs = 2.62 exome and 2.62 genome) groups, which are known to not be represented well by the five continental ancestry groups used here (i.e., AFR, EAS, EUR, IAM, and SAS) (Table S19).
Given these results, we recommend using the following loss thresholds to identify goodness of fit: <0.5 per 1,000 SNPs indicates a good fit, between 0.5 and 1.5 indicates a moderate fit, and >1.5 indicates the reference data are a poor fit. We do not recommend estimating ancestry-adjusted AF when the final loss/1,000 SNPs > 1.5. By these metrics, we achieve good fit for seven out of the eleven gnomAD v.2.1 groups for which we estimate ancestry proportions in Table 1 and moderate fit for four (American/Latinx exome = 1.080 and genome = 0.824, other genome = 0.680, and non-Finnish European genome = 0.500) (Table S19). Although we believe these thresholds will be good rules of thumb, loss per 1,000 SNPs is a continuum where higher values indicate poorer fit. Thus, even though both non-Finnish European genome and American/Latinx exome data are within the moderate fit range, the non-Finnish European genome is better represented by the reference data than the American/Latinx exome (loss/1,000 SNPs = 0.5 and 1.080, respectively). While similar loss values between the same exome and genome gnomAD groups suggest these thresholds should be reasonable for sequencing studies with an MAF > 1% filter such as that used here (material and methods), the thresholds may need to be reevaluated for fine-scale or array data.
Discussion
Here, we describe Summix, a fast, accurate, and precise method to estimate and adjust for population structure within publicly available genetic summary data. We evaluate Summix in over 5,000 simulation scenarios, showing the accuracy and precision are within 0.001% and 0.1%, respectively. In gnomAD, we find heterogeneous ancestry similar to what is expected in African/African American, American/Latinx, other, and South Asian groups. We provide ancestry proportion estimates for all gnomAD groups and subgroups as well as ancestry-adjusted AFs for an African sample and a Peruvian sample for others to use.
Using the estimated proportion of continental ancestry groups, we produce ancestry-adjusted AFs for target samples with either continental African ancestry or Peruvian ancestry. When comparing to a sample with matching ancestry, we find that the unadjusted AFs differ significantly more than the ancestry-adjusted AFs regardless of reference ancestry data used (i.e., gnomAD versus 1000 Genomes). The African ancestry-adjusted AFs are more similar to the target 1000 Genomes African AFs than the Peruvian ancestry-adjusted AFs are to the target 1000 Genomes Peruvian sample. The increased dissimilarity for the Peruvian sample may be due to more than two predominant reference ancestry groups or ancestral differences (including admixture) between gnomAD American/Latinx, the reference data, and 1000 Genomes Peruvian.
Although the AFs of putative causal variants from a breadth of ancestral populations in public databases are useful for assessing evidence for clinical pathogenicity of a genetic variant, checking the AF in an ancestral sample that matches the ancestry of the person with the disease is most useful. Summix can be used to provide ancestry-adjusted AFs to precisely match ancestry, increasing clinical utility of public datasets that may not have an ancestry that matches the patient. Additionally, Summix can produce ancestry-adjusted AFs matching the ancestry proportions for an external control sample. The use of gnomAD as a comparison dataset is increasingly common. Studies have used gnomAD as either the primary or secondary external control sample. While many of these studies are in European39, 40, 41 or East Asian42,43 ancestral groups, which we find to contain little to no other continental reference ancestry, some studies use gnomAD groups that contain admixture (e.g., African/African American, American/Latinx, and South Asian) for comparison without adjusting for population structure.6,16,44
We evaluate the potential utility of Summix’s ancestry-adjusted AF by producing ancestry-adjusted AFs for ClinVar variants, for the CF variant p.Phe508del in CFTR, and for a case-control analysis of PADI3, a gene where gnomAD was used as an external control sample to identify association with central centrifugal cicatricial alopecia in women with African ancestry. Although we find mostly minor discrepancies in the unadjusted and ancestry-adjusted AFs, we note that these differences can affect evidence of association for case-control analysis, as we demonstrate in PADI3, and prioritization of putative causal variants for follow up. For instance, under- or over-estimation of the frequency of clinically relevant or potentially relevant variants, e.g., p.Phe508del in CFTR or ClinVar variants, in certain ancestral populations could have clinical implications, such as when prioritizing variants to use for a screening tool. This emphasizes the importance of matching by or adjusting for ancestry differences whenever possible.
Here, we estimate genome-wide continental ancestry proportions. Although the mean local ancestry proportions for a sample often approach genome-wide ancestry proportions,45 there may exist regions of the genome, e.g., regions of selection, where the local ancestry for the sample differs substantially from genome-wide ancestry proportions.46,47 We expect that the ancestry-adjusted AF estimates may be less accurate in regions where the average local ancestry proportions differ from the genome-wide estimates. Our results suggest that Summix can estimate ancestry proportions accurately, although much less precisely, by using a relatively small number of randomly chosen SNPs (e.g., ∼100). This suggests that Summix may be able to estimate the proportion of local continental ancestry. Here, we evaluate subsets of randomly chosen SNPs, albeit reflecting genome-wide coverage. It could be that using ancestry informative markers (AIMs)48 or removing uninformative markers could increase the precision of Summix further enabling the estimation of local ancestry proportions.
There are several drawbacks to our current method and implementation. First, our method is currently only able to estimate the proportion of provided reference ancestry groups. We recommend the user include all expected ancestral populations in the reference dataset. Using least-squares loss per 1,000 SNPs as a measure of fit, we provide guidance as to how well the reference data matches the observed summary data. A value below 0.5 loss/1,000 SNPs indicates good fit, between 0.5 to 1.5 indicates moderate fit, and above 1.5 indicates poor fit. We do not recommend estimating ancestry-adjusted AF when the reference data are a poor fit. These thresholds are for sequencing studies using continental reference ancestry data. The thresholds will most likely need to be re-evaluated for other applications, such as estimation of fine-scale ancestry proportions and genome-wide association study (GWAS) arrays. Second, here we only evaluate the ability of Summix to estimate five broad continental ancestries. We are actively working on an extension to identify and estimate the proportion of ancestry not in the reference data and are evaluating the performance of Summix on a broader reference panel, including fine-scale ancestry. Lastly, differences in ancestry is not the only aspect of public databases that can cause confounding in analyses using external public controls. Differences in sequencing technology and computational variant calling pipelines can also cause biases in AFs due to non-exchangeability of individuals. Many methods have been and are being developed to adjust for this bias when using external controls.3, 4, 5,49,50
There are many extensions and applications for Summix beyond those evaluated here. First, Summix can be used with any reference ancestry data, needing only AFs. While we provide ancestry-adjusted AFs for a sample, the ancestry-adjusted AFs could be used for individuals, providing potential utility in the clinic. Summix has the potential to be applied to other summary datasets where AFs are provided or can be derived, such as from GWAS summary statistics, which are widely available online.51 While we found least-squares loss to perform well in our simulations and application to real data, other objective functions, such as the log likelihood, may be optimal, especially in the context of fine-scale ancestry. Lastly, Summix has similarities to deconvolution methods used in single-cell and other omics data types,52, 53, 54 suggesting paths of future development and application.
We provide an R package, Python function, Shiny app, and GitHub site to encourage reproducibility, broad use, and further development of our method. We hope that the methods presented here will be used and extended to improve the utility of valuable publicly available resources, especially for individuals and studies with admixed or understudied ancestry.
Declaration of interests
C.R.G. owns stock in 23and Me, Inc.
Acknowledgments
We thank the Education Through Undergraduate Research and Creative Activities (EURēCA!) Program and Undergraduate Research Opportunities Program (UROP) through the University of Colorado Denver and a gift from Dr. Ferdinand Baer for supporting many of the undergraduate researchers on this project. This work was supported by the National Human Genome Research Institute (R35HG011293 and U01HG009080 to A.E.H. and C.G.R.; U01HG009080-05S1 to C.G.R.).
Published: June 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.05.016.
Data and code availability
Code, final merged gnomAD and 1000 Genomes data, and ancestry-adjusted AF results are available at https://github.com/hendriau/Summix_manuscript. Public data used: gnomAD v.2 data were downloaded from https://gnomad.broadinstitute.org/downloads on October 11, 2018; 1000 Genomes data were downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/ on May 31, 2018; IAM Affymetrix 6.0 data were downloaded from ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/working/20130711_native_american_admix_train on October 11, 2018; and ClinVar data were downloaded from https://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh37/ on September 25, 2020.
Web resources
OMIM, https://omim.org/
Shiny app, https://shiny.clas.ucdenver.edu/Summix/
Summix Python package, https://github.com/jordanrhall/summix_py
Summix R package, https://github.com/hendriau/Summix
Summix R package, http://www.bioconductor.org/packages/release/bioc/html/Summix.html
Supplemental information
Figures S1–S12 and Tables S12–S16 and S19
Tables S1–S5
gnomAD group exome ancestry estimation
gnomAD group genome ancestry estimations
Tables S8 and S9
Tables S10 and S11
Tables S17 and S18
Article plus supplemental information
References
- 1.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phan L., Jin Y., Zhang H., Qiang W., Shekhtman E., Shao D., Revoe D., Villamarin R., Ivanchenko E., Kimura M. US National Library of Medicine; 2020. ALFA: Allele Frequency Aggregator. National Center for Biotechnology Information.https://www.ncbi.nlm.nih.gov/snp/docs/gsr/alfa/ [Google Scholar]
- 3.Guo M.H., Plummer L., Chan Y.-M., Hirschhorn J.N., Lippincott M.F. Burden Testing of Rare Variants Identified through Exome Sequencing via Publicly Available Control Data. Am. J. Hum. Genet. 2018;103:522–534. doi: 10.1016/j.ajhg.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendricks A.E., Billups S.C., Pike H.N.C., Farooqi I.S., Zeggini E., Santorico S.A., Barroso I., Dupuis J. ProxECAT: Proxy External Controls Association Test. A new case-control gene region association test using allele frequencies from public controls. PLoS Genet. 2018;14:e1007591. doi: 10.1371/journal.pgen.1007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S., Kim S., Fuchsberger C. Improving power for rare-variant tests by integrating external controls. Genet. Epidemiol. 2017;41:610–619. doi: 10.1002/gepi.22057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malki L., Sarig O., Romano M.-T., Méchin M.-C., Peled A., Pavlovsky M., Warshauer E., Samuelov L., Uwakwe L., Briskin V. Variant PADI3 in Central Centrifugal Cicatricial Alopecia. N. Engl. J. Med. 2019;380:833–841. doi: 10.1056/NEJMoa1816614. [DOI] [PubMed] [Google Scholar]
- 7.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Gagliano Taliun S.A., Corvelo A., Gogarten S.M., Kang H.M. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. bioRxiv. 2019 doi: 10.1101/563866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rannala B., Mountain J.L. Detecting immigration by using multilocus genotypes. Proc. Natl. Acad. Sci. USA. 1997;94:9197–9201. doi: 10.1073/pnas.94.17.9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang H., Peng J., Wang P., Risch N.J. Estimation of individual admixture: analytical and study design considerations. Genet. Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 11.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raj A., Stephens M., Pritchard J.K. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics. 2014;197:573–589. doi: 10.1534/genetics.114.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiang C.W.K., Gajdos Z.K.Z., Korn J.M., Kuruvilla F.G., Butler J.L., Hackett R., Guiducci C., Nguyen T.T., Wilks R., Forrester T. Rapid assessment of genetic ancestry in populations of unknown origin by genome-wide genotyping of pooled samples. PLoS Genet. 2010;6:e1000866. doi: 10.1371/journal.pgen.1000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bansal V., Libiger O. Fast individual ancestry inference from DNA sequence data leveraging allele frequencies for multiple populations. BMC Bioinformatics. 2015;16:4. doi: 10.1186/s12859-014-0418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shringarpure S.S., Bustamante C.D., Lange K., Alexander D.H. Efficient analysis of large datasets and sex bias with ADMIXTURE. BMC Bioinformatics. 2016;17:218. doi: 10.1186/s12859-016-1082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nappo S., Mannucci L., Novelli G., Sangiuolo F., D’Apice M.R., Botta A. Carrier frequency of CFTR variants in the non-Caucasian populations by genome aggregation database (gnomAD)-based analysis. Ann. Hum. Genet. 2020;84:463–468. doi: 10.1111/ahg.12396. [DOI] [PubMed] [Google Scholar]
- 17.Sirugo G., Williams S.M., Tishkoff S.A. The Missing Diversity in Human Genetic Studies. Cell. 2019;177:26–31. doi: 10.1016/j.cell.2019.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin A.R., Gignoux C.R., Walters R.K., Wojcik G.L., Neale B.M., Gravel S., Daly M.J., Bustamante C.D., Kenny E.E. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am. J. Hum. Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonnans J.-F., Gilbert J.C., Lemarechal C., Sagastizábal C.A. Springer Science & Business Media; 2006. Numerical Optimization: Theoretical and Practical Aspects. [Google Scholar]
- 20.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao X., Bigham A.W., Mei R., Gutierrez G., Weiss K.M., Brutsaert T.D., Leon-Velarde F., Moore L.G., Vargas E., McKeigue P.M. A genomewide admixture mapping panel for Hispanic/Latino populations. Am. J. Hum. Genet. 2007;80:1171–1178. doi: 10.1086/518564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wojcik G.L., Fuchsberger C., Taliun D., Welch R., Martin A.R., Shringarpure S., Carlson C.S., Abecasis G., Kang H.M., Boehnke M. Imputation-Aware Tag SNP Selection To Improve Power for Large-Scale, Multi-ethnic Association Studies. G3 (Bethesda) 2018;8:3255–3267. doi: 10.1534/g3.118.200502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalia S.S., Adelman K., Bale S.J., Chung W.K., Eng C., Evans J.P., Herman G.E., Hufnagel S.B., Klein T.E., Korf B.R. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 2017;19:249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 24.Bhérer C., Campbell C.L., Auton A. Refined genetic maps reveal sexual dimorphism in human meiotic recombination at multiple scales. Nat. Commun. 2017;8:14994. doi: 10.1038/ncomms14994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeileis A., Grothendieck G. zoo: S3 Infrastructure for Regular and Irregular Time Series. J. Stat. Softw. 2005;14:1–27. [Google Scholar]
- 26.Bates D., Mächler M., Bolker B., Walker S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015;67:1–48. [Google Scholar]
- 27.Watson P.F., Petrie A. Method agreement analysis: a review of correct methodology. Theriogenology. 2010;73:1167–1179. doi: 10.1016/j.theriogenology.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Lin L.I. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255–268. [PubMed] [Google Scholar]
- 29.Signorell A., Aho K., Anderegg N., Aragon T., Arppe A., Baddeley A., Bolker B., Caeiro F., Champely S., Chessel D. R Package Version 0.99.24; 2018. DescTools: Tools for descriptive statistics. [Google Scholar]
- 30.Nakatsuka N., Moorjani P., Rai N., Sarkar B., Tandon A., Patterson N., Bhavani G.S., Girisha K.M., Mustak M.S., Srinivasan S. The promise of discovering population-specific disease-associated genes in South Asia. Nat. Genet. 2017;49:1403–1407. doi: 10.1038/ng.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narasimhan V.M., Patterson N., Moorjani P., Rohland N., Bernardos R., Mallick S., Lazaridis I., Nakatsuka N., Olalde I., Lipson M. The formation of human populations in South and Central Asia. Science. 2019;365:eaat7487. doi: 10.1126/science.aat7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reich D., Thangaraj K., Patterson N., Price A.L., Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klarin D., Damrauer S.M., Cho K., Sun Y.V., Teslovich T.M., Honerlaw J., Gagnon D.R., DuVall S.L., Li J., Peloso G.M. Genetics of blood lipids among ∼300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 2018;50:1514–1523. doi: 10.1038/s41588-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farrer L.A., Cupples L.A., Haines J.L., Hyman B., Kukull W.A., Mayeux R., Myers R.H., Pericak-Vance M.A., Risch N., van Duijn C.M., APOE and Alzheimer Disease Meta Analysis Consortium Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 36.Graff-Radford N.R., Green R.C., Go R.C.P., Hutton M.L., Edeki T., Bachman D., Adamson J.L., Griffith P., Willis F.B., Williams M. Association between apolipoprotein E genotype and Alzheimer disease in African American subjects. Arch. Neurol. 2002;59:594–600. doi: 10.1001/archneur.59.4.594. [DOI] [PubMed] [Google Scholar]
- 37.Logue M.W., Schu M., Vardarajan B.N., Buros J., Green R.C., Go R.C.P., Griffith P., Obisesan T.O., Shatz R., Borenstein A., Multi-Institutional Research on Alzheimer Genetic Epidemiology (MIRAGE) Study Group A comprehensive genetic association study of Alzheimer disease in African Americans. Arch. Neurol. 2011;68:1569–1579. doi: 10.1001/archneurol.2011.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blue E.E., Horimoto A.R.V.R., Mukherjee S., Wijsman E.M., Thornton T.A. Local ancestry at APOE modifies Alzheimer’s disease risk in Caribbean Hispanics. Alzheimers Dement. 2019;15:1524–1532. doi: 10.1016/j.jalz.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marenne G., Hendricks A.E., Perdikari A., Bounds R., Payne F., Keogh J.M., Lelliott C.J., Henning E., Pathan S., Ashford S., INTERVAL, UK10K Consortium Exome Sequencing Identifies Genes and Gene Sets Contributing to Severe Childhood Obesity, Linking PHIP Variants to Repressed POMC Transcription. Cell Metab. 2020;31:1107–1119.e12. doi: 10.1016/j.cmet.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diez-Fairen M., Makarious M.B., Bandres-Ciga S., Blauwendraat C., International Parkinson’s Disease Genomics Consortium (IPDGC) Assessment of LIN28A variants in Parkinson’s disease in large European cohorts. Neurobiol. Aging. 2021;100:118.e1–118.e3. doi: 10.1016/j.neurobiolaging.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan J.-H., Schulman B.R., Effraim P.R., Sulayman D.-H., Jacobs D.S., Waxman S.G. Genomic analysis of 21 patients with corneal neuralgia after refractive surgery. Pain Rep. 2020;5:e826. doi: 10.1097/PR9.0000000000000826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X., Chen W., Li W., Priest J.R., Fu Y., Pang K., Ma B., Han B., Liu X., Hu S., Zhou Z. Exome-Based Case-Control Analysis Highlights the Pathogenic Role of Ciliary Genes in Transposition of the Great Arteries. Circ. Res. 2020;126:811–821. doi: 10.1161/CIRCRESAHA.119.315821. [DOI] [PubMed] [Google Scholar]
- 43.Li C., Huang Q., Yang R., Guo X., Dai Y., Zeng J., Zeng Y., Tao L., Li X., Zhou H., Wang Q. Targeted next generation sequencing of nine osteoporosis-related genes in the Wnt signaling pathway among Chinese postmenopausal women. Endocrine. 2020;68:669–678. doi: 10.1007/s12020-020-02248-x. [DOI] [PubMed] [Google Scholar]
- 44.Lu H.-M., Li S., Black M.H., Lee S., Hoiness R., Wu S., Mu W., Huether R., Chen J., Sridhar S. Association of Breast and Ovarian Cancers With Predisposition Genes Identified by Large-Scale Sequencing. JAMA Oncol. 2019;5:51–57. doi: 10.1001/jamaoncol.2018.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montana G., Pritchard J.K. Statistical tests for admixture mapping with case-control and cases-only data. Am. J. Hum. Genet. 2004;75:771–789. doi: 10.1086/425281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Q., Zhao L., Guan Y. Strong Selection at MHC in Mexicans since Admixture. PLoS Genet. 2016;12:e1005847. doi: 10.1371/journal.pgen.1005847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hodgson J.A., Pickrell J.K., Pearson L.N., Quillen E.E., Prista A., Rocha J., Soodyall H., Shriver M.D., Perry G.H. Natural selection for the Duffy-null allele in the recently admixed people of Madagascar. Proc. Biol. Sci. 2014;281:20140930. doi: 10.1098/rspb.2014.0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown R., Pasaniuc B. Enhanced methods for local ancestry assignment in sequenced admixed individuals. PLoS Comput. Biol. 2014;10:e1003555. doi: 10.1371/journal.pcbi.1003555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang L., Jiang H., Dai S., Chen Y., Song Y., Tang C.S.-M., Wang B., Garcia-Barcelo M.-M., Tam P., Cherny S.S. Deviation from baseline mutation burden provides powerful and robust rare-variants association test for complex diseases. bioRxiv. 2020 doi: 10.1101/2020.07.04.186619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y., Lee S. Novel score test to increase power in association test by integrating external controls. Genet. Epidemiol. 2020;44:293–304. doi: 10.1002/gepi.22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gong T., Szustakowski J.D. DeconRNASeq: a statistical framework for deconvolution of heterogeneous tissue samples based on mRNA-Seq data. Bioinformatics. 2013;29:1083–1085. doi: 10.1093/bioinformatics/btt090. [DOI] [PubMed] [Google Scholar]
- 53.Hao Y., Yan M., Heath B.R., Lei Y.L., Xie Y. Fast and robust deconvolution of tumor infiltrating lymphocyte from expression profiles using least trimmed squares. PLoS Comput. Biol. 2019;15:e1006976. doi: 10.1371/journal.pcbi.1006976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Racle J., de Jonge K., Baumgaertner P., Speiser D.E., Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife. 2017;6:e26476. doi: 10.7554/eLife.26476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S12 and Tables S12–S16 and S19
Tables S1–S5
gnomAD group exome ancestry estimation
gnomAD group genome ancestry estimations
Tables S8 and S9
Tables S10 and S11
Tables S17 and S18
Article plus supplemental information
Data Availability Statement
Code, final merged gnomAD and 1000 Genomes data, and ancestry-adjusted AF results are available at https://github.com/hendriau/Summix_manuscript. Public data used: gnomAD v.2 data were downloaded from https://gnomad.broadinstitute.org/downloads on October 11, 2018; 1000 Genomes data were downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/ on May 31, 2018; IAM Affymetrix 6.0 data were downloaded from ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/working/20130711_native_american_admix_train on October 11, 2018; and ClinVar data were downloaded from https://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh37/ on September 25, 2020.