Summary
EDEM3 encodes a protein that converts Man8GlcNAc2 isomer B to Man7-5GlcNAc2. It is involved in the endoplasmic reticulum-associated degradation pathway, responsible for the recognition of misfolded proteins that will be targeted and translocated to the cytosol and degraded by the proteasome. In this study, through a combination of exome sequencing and gene matching, we have identified seven independent families with 11 individuals with bi-allelic protein-truncating variants and one individual with a compound heterozygous missense variant in EDEM3. The affected individuals present with an inherited congenital disorder of glycosylation (CDG) consisting of neurodevelopmental delay and variable facial dysmorphisms. Experiments in human fibroblast cell lines, human plasma, and mouse plasma and brain tissue demonstrated decreased trimming of Man8GlcNAc2 isomer B to Man7GlcNAc2, consistent with loss of EDEM3 enzymatic activity. In human cells, Man5GlcNAc2 to Man4GlcNAc2 conversion is also diminished with an increase of Glc1Man5GlcNAc2. Furthermore, analysis of the unfolded protein response showed a reduced increase in EIF2AK3 (PERK) expression upon stimulation with tunicamycin as compared to controls, suggesting an impaired unfolded protein response. The aberrant plasma N-glycan profile provides a quick, clinically available test for validating variants of uncertain significance that may be identified by molecular genetic testing. We propose to call this deficiency EDEM3-CDG.
Keywords: CDG, EDEM3, N-glycan, Mannosidase, mouse, UPR, high-mannose, dysmorphism
Main text
The endoplasmic reticulum-associated degradation (ERAD) pathway recognizes misfolded proteins that will be targeted and translocated to the cytosol and degraded by the proteasome.1, 2, 3, 4 The best characterized system is dedicated to N-linked glycoproteins: the glycoprotein ERAD (gpERAD) pathway. Proteins that fail to fold or assemble properly are subjected to degradation via specific trimming of high-mannose-type glycans in the lumen of the ER.5,6 In mammals, EDEM2 and ERmanI are known to primarily catalyze the first step, conversion of Man9GlcNAc2 (M9) to Man8GlcNAc2 isomer B (M8B).7, 8, 9 EDEM3 and EDEM1 are further involved in catalyzing the second step, the conversion of M8B to Man7GlcNAc2 (M7).7, 8, 9, 10, 11, 12 These products are then recognized by lectin OS-9 in the ER lumen and targeted for degradation in the cytosol.13
Cells can degrade misfolded proteins from the secretory pathway by using the glycan-dependent and glycan-independent ERAD pathways.14, 15, 16 The presence of misfolded ER proteins is harmful to cells and is involved in several congenital disorders of glycosylation (CDGs),17, 18, 19, 20 such as PMM2-CDG (MIM: 212065), MPI-CDG (MIM: 602579), ALG6-CDG (MIM: 603147), DPM1-CDG (MIM: 608799), ALG12-CDG (MIM: 607143), and DPAGT1-CDG (MIM: 608093). If this degradative process fails to effectively remove misfolded and abnormal proteins from the ER, then the unfolded protein response (UPR) is initiated.17, 18, 19, 20 The UPR is critical to maintain cellular homeostasis. In mammals, three classical ER stress markers are known to reflect induction of the UPR: protein kinase RNA-like endoplasmic reticulum kinase (PERK), activating transcription factor 6 (ATF6), and inositol requiring enzyme 1 (IRE1).21,22 Upon protein overload in the ER, UPR signaling via the PERK, ATF6, and IRE1 pathways is activated to increase ER capacity, inhibit translation, and stimulate the removal of excess and misfolded proteins.23, 24, 25, 26 Persistent activation of UPR can lead to apoptosis.27, 28, 29
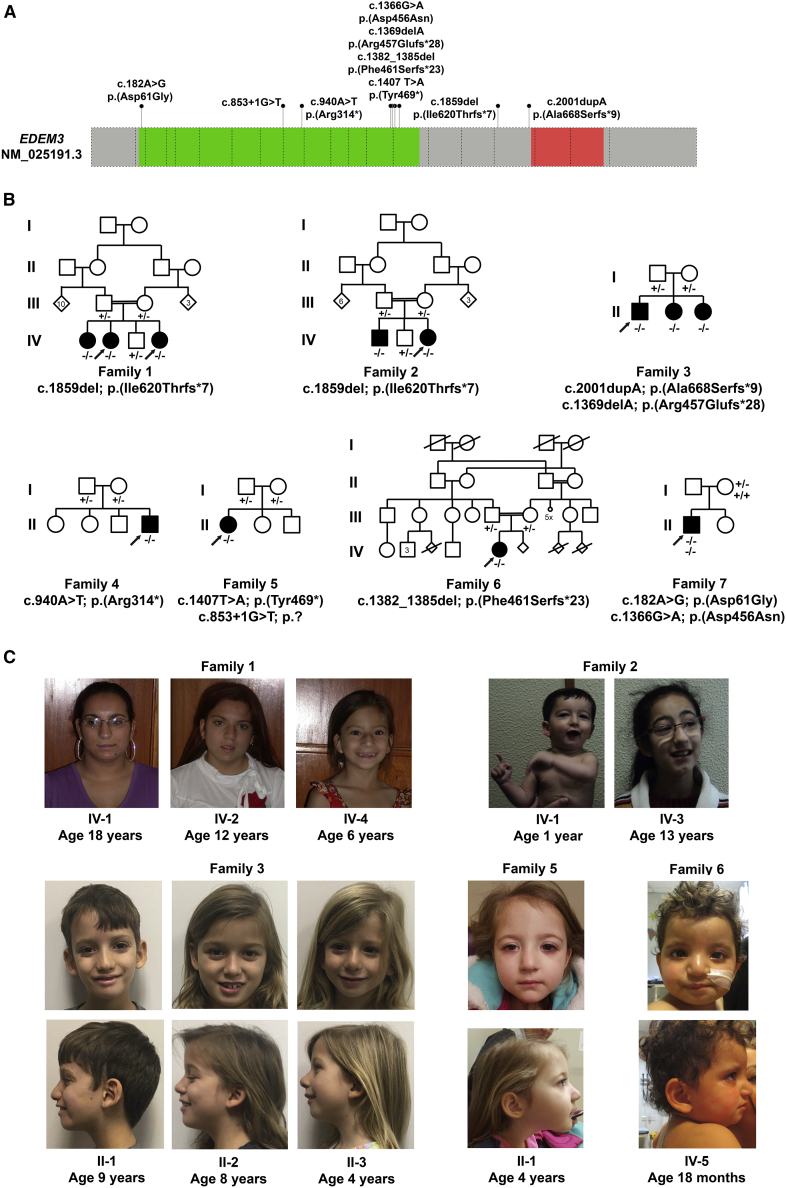
Here, we describe seven independent families with 11 individuals with bi-allelic protein-truncating variants and one additional individual with a compound heterozygous missense variant in EDEM3 (MIM: 610214) identified by exome sequencing (Figure 1) and collected through GeneMatcher.30 The parents or legal guardians provided written informed consent. All relevant approvals from the institutional review boards and ethics committees of the participating institutions were obtained. In affected individuals from families 1 and 2, a bi-allelic frameshift deletion, c.1859del (p.Ile620Thrfs∗7), was identified in EDEM3 (GenBank: NM_025191.3; Figure 1). Additionally, two frameshift variants, c.2001dup (p.Ala668Serfs∗9) and c.1369del (p.Arg457Glufs∗28), were identified in family 3. In family 4, a bi-allelic nonsense variant, c.940A>T (p.Arg314∗), was found, resulting from maternal uniparental isodisomy of chromosome 1. In family 5, a canonical splice site donor variant, c.853+1G>T, and a nonsense variant, c.1407T>A (p.Tyr469∗), were identified, and in family 6, a bi-allelic frameshift deletion, c.1382_1385del (p.Phe461Serfs∗23) was identified. Finally, in family 7, the compound heterozygous changes c.182A>G (p.Asp61Gly) and c.1366G>A (p.Asp456Asn) were identified. Sanger sequencing showed that the affected individuals were either homozygous or compound heterozygous for the identified EDEM3 variants. The unaffected parents were all heterozygous carriers. The p.Asp61Gly missense variant was found in six heterozygous individuals, out of 218,508 alleles, in gnomAD (rs777353823) and was predicted to be “disease causing” by MutationTaster31 but “tolerated’” by SIFT.32 The p.Asp456Asn missense variant was not present in gnomAD and was predicted to be “disease causing” by both MutationTaster and SIFT. Both are present in the so-called “Glyco_Hydro_47 domain,” which is essential for the α-mannosidase activity of the protein.
Figure 1.
Bi-allelic variants in EDEM3 lead to developmental delay or intellectual disabilities and dysmorphic features
(A) Schematic representation of human EDEM3 including the positions and the predicted effect on protein level of the seven identified variants in this study. The green box represents the Glyco_Hydro_47 domain, and the red box represents the protease-associated (PA) domain. Dashed boxes represent exons.
(B) Pedigrees of EDEM3-CDG families including the presence of the mutation. −, mutation present; +, wild-type.
(C) Craniofacial features of affected individuals, including narrow palpebral fissures, epicanthal folds, increased nasal height, bulbous nasal tip, hypoplastic alae nasi, short philtrum, thin upper lip, and retrognathia.
Affected individuals from family 1 and 2 have the identical c.1859del (p.Ile620Thrfs∗7) EDEM3 variant, but there are no records of inter-familial consanguinity in these families. Comparison of the exome data from two affected individuals from family 1 and from one affected individual from family 2 indicated that these three individuals carried a common identical region of homozygosity of 3.16 Mb (chr1: 182,993,025–186,157,274, Hg19) surrounding EDEM3. In addition, 96 healthy control individuals in the Romani population in Portugal were screened for the presence of this variant, among whom one heterozygous allele was detected. Because families 1 and 2 are of Portuguese Romani origin, this suggests a possible founder effect.
All affected individuals presented with developmental delay and/or intellectual disability (ID) and speech delay (Table S1). Hypotonia was present in six out of 12 persons. Dysmorphic facial features, such as narrow palpebral fissures (6/12), epicanthal folds (6/12), increased nasal height (8/12), bulbous nasal tip (6/12), hypoplastic alae nasi (9/12), short philtrum (6/12), thin upper lip (9/12), and retrognathia (6/12) were also found in half or more of the affected individuals. Additionally, gastroesophageal reflux was observed in three persons, and two individuals had early feeding difficulties requiring a nasogastric tube; of these, one individual needed a percutaneous endoscopic gastrostomy placement. Brain magnetic resonance imaging (MRI) of affected individuals from families 2, 3, and 5 did not detect structural abnormalities or myelination defects.
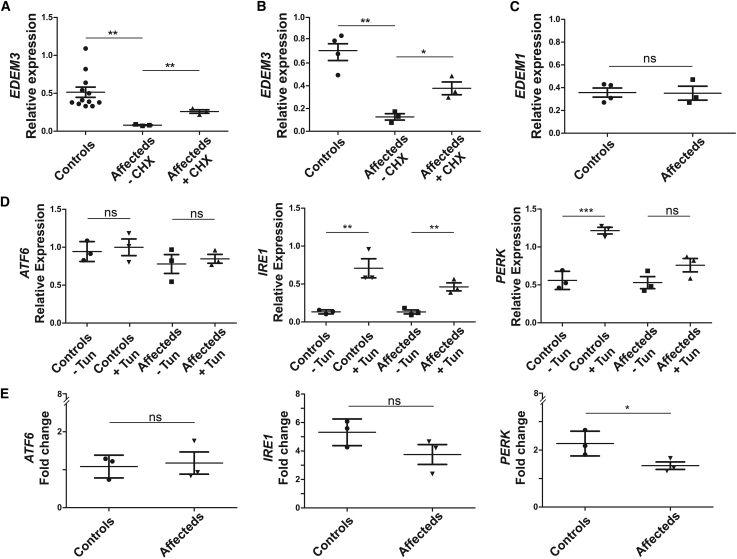
The protein-truncating variants identified in this study are expected to result in either reduced protein levels, due to degradation of mRNA via nonsense-mediated decay (NMD), or a truncated protein.33,34 Accordingly, quantitative real-time PCR analysis showed that EDEM3 mRNA levels in Epstein-Barr virus-transformed lymphoblastoid cell lines (EBV-LCLs) from affected individuals in family 1 were significantly decreased to 17% as compared to healthy controls (p = 0.0078; Figure 2A) and could be rescued with cycloheximide (CHX), an inhibitor of NMD, suggesting that the bi-allelic c.1859del frameshift variant triggers NMD. We repeated these experiments in fibroblasts from affected individuals in families 1 and 3 and confirmed that EDEM3 is degraded to 18% of normal levels by NMD (p = 0.0015; Figure 2B). Immunoblots of EDEM3 were performed in two individual fibroblast cell lines from these two families. These demonstrated the absence of EDEM3 in individual IV-4 (family 1) and individual II-1 (family 3) consistent with loss of function of EDEM3 (Figure 3C). Because the function of EDEM3 could theoretically be compensated by EDEM1,35 we quantified EDEM1 (MIM: 607673) RNA levels in affected and control fibroblast cell lines. EDEM1 levels were at 97% of normal levels (p = 0.9373) in fibroblast cell lines from affected individuals (Figure 2C). The comparable levels of EDEM1 in affected and control fibroblast cell lines seems to suggest that EDEM1 expression does not compensate for the reduction of EDEM3 in these cells.
Figure 2.
Relative expression of EDEM3 mRNA and expression analysis of UPR markers
(A) A significant decrease of mRNA levels is seen between relative expressions of EDEM3 in EBV-LCL from three affected individuals of family 1 with the bi-allelic c.1859del frameshift variant (n = 3) compared to healthy controls (n = 12; p = 0.0078) and samples treated with cycloheximide (CHX; n =3; p = 0.0016), normalized according to three housekeeping genes.
(B) EDEM3 expression in fibroblast cell lines from family 1 (individual IV-4) and family 3 (individuals II-1 and II-2) confirming these results (p = 0.0015 and p = 0.0161 for CHX). Four controls were used.
(C) EDEM1 expression in fibroblast cell lines from family 1 (individual IV-4) and family 3 (individuals II-1 and II-2) showing normal expression levels (p = 0.9373). Four controls were used.
(D) Analysis of total mRNA from EBV-LCL cell lines from controls revealed increased mRNA expression after treatment with tunicamycin (Tun) of IRE1 and EIF2AK3 (PERK) but not of ATF6.
(E) Affected individuals’ mRNA from family 1 showed significantly decreased induction of PERK expression when compared to controls (p = 0.020), whereas ATF6 and IRE1 did not change significantly (p = 0.423 and p = 0.091, respectively). Experiments in (D) and (E) were performed with three control cell lines and three cell lines from affected individuals. Bars indicate mean values. Error bars represent standard deviation. ns, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
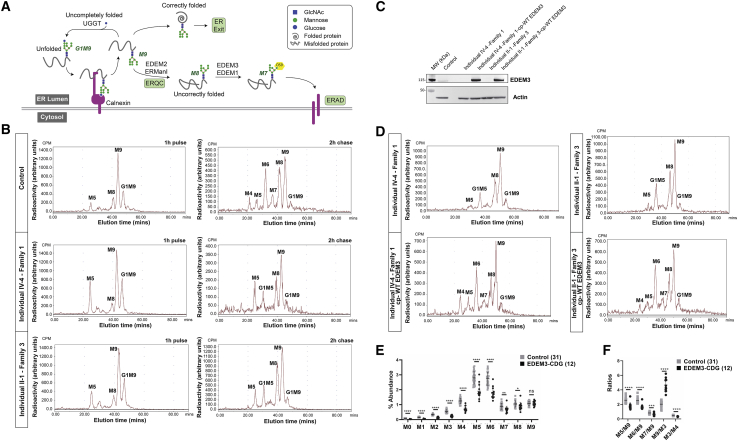
Figure 3.
N-glycosylation pathway and N-glycan analysis of fibroblasts and plasma of human subjects
(A) N-glycosylaton pathway and the role of EDEM proteins in the mannose trimming and targeting for protein degradation in the cytosol.
(B) Accumulation of G1M5 and M5 and a lack of M7, M6, and M4 on newly synthesized N-glycoproteins in two EDEM3-CDG fibroblast cell lines from affected individuals. Fibroblasts from family 1 (individual IV-4) and family 3 (individual II-1) and one control individual were incubated with [2-3H]mannose for 1 h and chased during 2 h.
(C) Immunoblot analysis of fibroblasts from individual IV-4 (family 1) and individual II-1 (family 3) before and after complementation with wild-type (WT) EDEM3. Total cell lysates were prepared and subjected to SDS-PAGE and immunoblot analysis with indicated antibodies.
(D) N-glycan analysis of fibroblasts from individual IV-4 and individual II-1 complemented with WT EDEM3 after incubation with [2-3H]mannose for 1 h and chasing for 2 h.
(E) Percent abundance of plasma N-glycan species obtained from EDEM3-CDG-affected individuals and control individuals.
(F) Plot of ratio of plasma N-glycan species. Bars indicate mean values. Error bars represent standard deviation. Indicated p values from Student’s t test, ns, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Previous studies have shown that EDEM3 belongs to a group of proteins that accelerate the degradation of misfolded glycoproteins in the ER. It is one of the ER 1,2-alpha mannosidases and specifically trims M8B to M7 (Figure 3A), which is then recognized by lectin OS-9 and targeted for degradation.8, 9, 10, 11 We hypothesized that loss of M8B to M7 trimming activity would result in accumulation of abnormal N-glycans. Therefore, we determined the N-glycan profiles of total cellular glycoproteins prepared from a fibroblast cell line of a control individual and of an affected individual. Total N-glycan analysis via mass spectrometry in the affected individual’s fibroblast cell line showed accumulation of M9 and M8 and reduced M7 and Man6GlcNAc2 (M6) levels (Figure S1). To study more precisely the impact of EDEM3 deficiency, we also analyzed the N-glycan profile after a pulse-chase experiment by using [2-3H]mannose as precursor. We tested whether the variants in EDEM3 affect the level of glycoproteins in fibroblast cell lines of individuals from families 1 and 3. In control fibroblast cells, we observed the anticipated levels of M8, M9, and Glc1Man9GlcNAc2 (G1M9; Figure 3B). EDEM3 deficient cells from members of family 1 and 3 exhibited a similar pattern of N-glycan structures, although with increased accumulation of M5 and G1M5. The structure of these two species was confirmed with either saitoi or jack bean α-mannosidase treatment, as the G1M5 terminal glucose conferred substantial protection of the peak from the mannosidases (Figure S3). After 2 h of chase, the peaks for M7 and M4 did not appear in the cells of affected individuals (Figure 3B), indicating that the 1,2-alpha mannose residues were not removed from the M8B and M5 N-glycans during the chase, which is consistent with the absence of the biological function of EDEM3 (Figure 3A). We speculate that the M5 corresponds to the biosynthetic oligosaccharide precursor synthesized as a lipid-linked oligosaccharide and not generated through mannose trimming. The lipid-linked oligosaccharides profiles did not show M5 accumulation (data not shown). All together, these results may indicate that EDEM3 deficiency impairs lipid-linked oligosaccharide synthesis and that the M5 is transferred onto newly synthesized proteins and glucosylated to form G1M5 by the UDP-Glc:glycoprotein glucosyltransferase during calnexin cycle (Figure 3A).36
To evaluate the N-glycosylation of secreted glycoproteins, we studied the N-glycan profile of total plasma glycoproteins from all affected individuals, including the individual with compound heterozygous missense variants, by using our recently described and clinically validated semiquantitative N-glycan assay.37 The N-glycan profiles from affected individuals showed decreased levels of low mannose N-glycan species M3–M7 (Figure 3C) with preserved normal M8 and M9 or sometimes mildly increased abundance of M9 as compared to control subjects. Ratios of the N-glycan abundances were also explored, and affected individuals had prominently decreased ratios of M5:M9, M6:M9, and M7:M9 as well as decreased M3:M4 (Figure 3D; Table S2). M6:M9 ratio provided the highest discrimination between tested obligate heterozygotes (parents, n = 4) and affected individuals (n = 12; Table S3). In theory, M3 can be produced by multiple pathways with or without EDEM3; thus, we used M9:M3 ratio to normalize M9 abundance. M9:M3 was increased in all 12 affected individuals. Stepwise ratio analysis for plasma N-linked polymannose species showed a reduction of M7:M8 that is significant in affected individuals, consistent with EDEM3’s being the key enzyme in trimming M8B to M7 on secreted glycoproteins (Table S3).9,10 Interestingly, M3:M4 ratio was also reduced in all 12 affected individuals tested, consistent with a possible role of EDEM3 in trimming M5 to shorter polymannose glycans, as suggested by the pulse chase data. EDEM3’s involvement in trimming of both M8 and M5 emphasizes that it is an essential ER mannosidase in humans. Of note, human transferrin was normally glycosylated in the common clinical screening test for CDG in the three affected individuals from family 3.
N-glycan profiles were also obtained from Edem3 knockout (KO) mice (Figure S4). Although Edem3 KO mice did not present with any obvious phenotype, subtle changes have been noted, such as reduced weight of brains and body and largely skewed ratios of homozygous KO pups versus heterozygous and wild-type pups. The profiles were acquired via a previously described method.37 Similar to affected individuals, plasma from mice showed decreased ratios of M5:M9, M6:M9, and M7:M9 (p < 0.02; Figure S5B). In addition, mouse plasma proteins had significantly increased abundance of M8 and M9 (p < 0.01; Figure S5A). We also assayed mouse brain lysate for N-glycan profiles (Figure S5C) and ratios (Figure S5D). While plasma N-glycans reflect glycosylation of mature secreted glycoproteins and glycopeptides, N-glycan profiling of mouse tissue also includes cellular glycoproteins. Similar to mouse plasma N-glycan profiling, mouse brain showed increased high-mannose M8 and M9 N-glycan species (Figure S5C) and decreased ratios of M5:M9 and M6:M9 (Figure S5D).
N-glycan analysis of both Edem3 KO mouse brain and plasma showed significant increases of the abundance M8 and M9. Thus, the known function of EDEM3 in trimming M8B is most likely shared by both humans and mice. In humans, not all the EDEM3-deficient individuals have increased M8 or M9 in the plasma. Instead, M9:M3 ratio is increased in all 12 affected individuals, providing a more sensitive diagnostic biomarker than M8 or M9 abundance in plasma. N-linked M3 and M4 are newly discovered small high-mannose species with low abundance on normal plasma glycoproteins.37 Increases of M3 and M4 abundances have been reported as important diagnostic biomarkers for type I CDG subtypes, including PMM2-CDG, MPI-CDG, ALG3-CDG, and ALG9-CDG.37, 38, 39 Significantly reduced abundance of N-linked M3 with a decreased M3:M4 ratio (p < 0.0001), however, has not been reported before. The reduction of M3 in all of the affected individuals suggests that a large portion of N-linked M3 in the control population is probably derived from glycan trimming ultimately involving human EDEM3 (Figure 3A). In the plasma from Edem3 KO mice, M3:M4 ratio is not decreased (Figures S5B and S5D). Instead of increased M7:M8 ratio in humans, the increased M6:M7 ratio is the most significant, which may reflect a difference in overall substrate specificity between human and mouse EDEM3. The glycosylation abnormalities in both cellular or tissue proteins and secreted mature proteins indicate a global impact on protein N-glycosylation in EDEM3 deficiency. Consistent with our findings, it was previously demonstrated that EDEM3 participates in mannose trimming from total glycoproteins and not just misfolded proteins for ERAD.10 Therefore, we propose this deficiency to be EDEM3-CDG.
The finding of abnormal N-glycan profiling pattern with reduced M3:M4, M5:M9, M6:M9, and M7:M9 ratios and increased M9:M3 among EDEM3-CDG-affected individuals provides additional diagnostic biomarkers for validating variants of uncertain significance, i.e., missense variants in EDEM3. One of the 12 EDEM3-CDG-affected individuals showed normal plasma of M5:M9 and M6:M9 ratios but also showed the lowest plasma N-linked M3:M4 ratio at 0.27 (normal 0.39–0.56) and a significantly increased M9:M3 ratio at 3.30 (normal 1.16–2.92) in this cohort. Therefore, the combination of high M9:M3 and low M3:M4 ratios might also provide diagnostic clues for EDEM3-CDG when M5:M9 and M6:M9 ratios are normal.
G1M5 accumulation is a marker for an impaired UPR.36 Given the role of EDEM3 in gpERAD and its association with the UPR, we tested whether the UPR, a key quality-control process in the cell that is activated in response to the accumulation of misfolded proteins,22 is affected in EDEM3-CDG. Quantitative real-time PCR analysis of total mRNA from EBV-LCLs from affected individuals showed decreased stimulation of ERN1 (IRE1 [MIM: 604033]) and EIF2AK3 (PERK [MIM: 604032]) when cells were treated with tunicamycin, a UPR-inducing compound, as compared to controls (Figure 2D). ATF6 (MIM: 605537) expression levels were similar. The difference in IRE1 expression levels was not significant (p = 0.091), whereas the decreased stimulation of PERK expression levels in cell lines from affected individuals was significant (p = 0.020; Figure 2E), suggesting that the UPR is impaired in EDEM3-CDG or that these cell lines have an increased capacity to eliminate misfolded proteins.
EDEM3 was recently implicated in triglyceride metabolism. A low-frequency EDEM3 missense variant in the protease-associated domain (rs78444298, p.Pro746Ser, minor allele frequency ∼1.5%) was associated with an approximately 5% decrease in triglyceride levels.40 In our subjects for whom fasting triglyceride levels are available (Table S1), all triglyceride measurements were within the normal range. This most likely reflects the multiple metabolic pathways influencing triglyceride levels and makes the use of triglycerides as a diagnostic marker for EDEM3-CDG challenging.
In conclusion, we show that bi-allelic EDEM3 variants cause EDEM3-CDG, a CDG with non-specific developmental delay and/or intellectual disability. Several affected individuals also have mild facial dysmorphisms. Given the increased accessibility of blood for clinical testing, semiquantitative N-glycan analysis provides additional diagnostic biomarkers for validating variants of uncertain significance that may be identified on molecular genetic testing. Further functional studies are necessary to determine the precise pathophysiological mechanism of EDEM3-CDG.
Declaration of interests
The authors declare no competing interests.
Consortia
The members of the CAUSES Study are Shelin Adam, Christele Du Souich, Alison Elliott, Anna Lehman, Jill Mwenifumbo, Tanya Nelson, Clara Van Karnebeek, and Jan Friedman.
Acknowledgments
We are grateful to the families that have participated to this study. This work was supported by the EU FP7 large-scale integrating project Genetic and Epigenetic Networks in Cognitive Dysfunction (241995) (to H.v.B.); National Institutes of Health (NIH) grants 5R01GM115730-03 (to M.H.), U54 NS115198 (to A.C.E. and M.H.), and T32GM008638 (to A.C.E.); and the Transatlantic Network of Excellence grant (10CVD03) from the Fondation Leducq and NIH NHGRI U01HG006398 (to D.J.R.). Family 4 was enrolled in the CAUSES Study; investigators include Shelin Adam, Christele Du Souich, Alison Elliott, Anna Lehman, Jill Mwenifumbo, Tanya Nelson, Clara Van Karnebeek, and Jan Friedman; it is funded by Mining for Miracles, British Columbia Children’s Hospital Foundation (grant number F15-01355) and Genome British Columbia (grant number F16-02276). D.L.P. is recipient of a CAPES Fellowship (99999.013311/2013-01). X.B. is supported by an AHA career development award (19CDA34630032).
Published: June 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.05.010.
Contributor Information
Miao He, Email: hem@email.chop.edu.
Arjan P.M. de Brouwer, Email: arjan.debrouwer@radboudumc.nl.
Data and code availability
All data that are relevant to this research are presented in this paper.
Web resources
OMIM, https://www.omim.org/
Supplemental information
References
- 1.Brodsky J.L. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151:1163–1167. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Needham P.G., Brodsky J.L. How early studies on secreted and membrane protein quality control gave rise to the ER associated degradation (ERAD) pathway: the early history of ERAD. Biochim. Biophys. Acta. 2013;1833:2447–2457. doi: 10.1016/j.bbamcr.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith M.H., Ploegh H.L., Weissman J.S. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thibault G., Ng D.T.W. The endoplasmic reticulum-associated degradation pathways of budding yeast. Cold Spring Harb. Perspect. Biol. 2012;4:a013193. doi: 10.1101/cshperspect.a013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aebi M., Bernasconi R., Clerc S., Molinari M. N-glycan structures: recognition and processing in the ER. Trends Biochem. Sci. 2010;35:74–82. doi: 10.1016/j.tibs.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Molinari M. N-glycan structure dictates extension of protein folding or onset of disposal. Nat. Chem. Biol. 2007;3:313–320. doi: 10.1038/nchembio880. [DOI] [PubMed] [Google Scholar]
- 7.Hosokawa N., Tremblay L.O., You Z., Herscovics A., Wada I., Nagata K. Enhancement of endoplasmic reticulum (ER) degradation of misfolded Null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J. Biol. Chem. 2003;278:26287–26294. doi: 10.1074/jbc.M303395200. [DOI] [PubMed] [Google Scholar]
- 8.Mast S.W., Diekman K., Karaveg K., Davis A., Sifers R.N., Moremen K.W. Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology. 2005;15:421–436. doi: 10.1093/glycob/cwi014. [DOI] [PubMed] [Google Scholar]
- 9.Ninagawa S., Okada T., Sumitomo Y., Kamiya Y., Kato K., Horimoto S., Ishikawa T., Takeda S., Sakuma T., Yamamoto T., Mori K. EDEM2 initiates mammalian glycoprotein ERAD by catalyzing the first mannose trimming step. J. Cell Biol. 2014;206:347–356. doi: 10.1083/jcb.201404075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirao K., Natsuka Y., Tamura T., Wada I., Morito D., Natsuka S., Romero P., Sleno B., Tremblay L.O., Herscovics A. EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J. Biol. Chem. 2006;281:9650–9658. doi: 10.1074/jbc.M512191200. [DOI] [PubMed] [Google Scholar]
- 11.Hosokawa N., Tremblay L.O., Sleno B., Kamiya Y., Wada I., Nagata K., Kato K., Herscovics A. EDEM1 accelerates the trimming of alpha1,2-linked mannose on the C branch of N-glycans. Glycobiology. 2010;20:567–575. doi: 10.1093/glycob/cwq001. [DOI] [PubMed] [Google Scholar]
- 12.Olivari S., Cali T., Salo K.E.H., Paganetti P., Ruddock L.W., Molinari M. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem. Biophys. Res. Commun. 2006;349:1278–1284. doi: 10.1016/j.bbrc.2006.08.186. [DOI] [PubMed] [Google Scholar]
- 13.Hosokawa N., Kamiya Y., Kamiya D., Kato K., Nagata K. Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J. Biol. Chem. 2009;284:17061–17068. doi: 10.1074/jbc.M809725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita E., Kouroku Y., Isoai A., Kumagai H., Misutani A., Matsuda C., Hayashi Y.K., Momoi T. Two endoplasmic reticulum-associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: ubiquitin/proteasome ERAD(I) and autophagy/lysosome ERAD(II) Hum. Mol. Genet. 2007;16:618–629. doi: 10.1093/hmg/ddm002. [DOI] [PubMed] [Google Scholar]
- 15.Hirsch C., Gauss R., Horn S.C., Neuber O., Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- 16.Rashid H.-O., Yadav R.K., Kim H.-R., Chae H.-J. ER stress: Autophagy induction, inhibition and selection. Autophagy. 2015;11:1956–1977. doi: 10.1080/15548627.2015.1091141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerriero C.J., Brodsky J.L. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol. Rev. 2012;92:537–576. doi: 10.1152/physrev.00027.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shang J., Körner C., Freeze H., Lehrman M.A. Extension of lipid-linked oligosaccharides is a high-priority aspect of the unfolded protein response: endoplasmic reticulum stress in Type I congenital disorder of glycosylation fibroblasts. Glycobiology. 2002;12:307–317. doi: 10.1093/glycob/12.5.307. [DOI] [PubMed] [Google Scholar]
- 19.Lecca M.R., Wagner U., Patrignani A., Berger E.G., Hennet T. Genome-wide analysis of the unfolded protein response in fibroblasts from congenital disorders of glycosylation type-I patients. FASEB J. 2005;19:240–242. doi: 10.1096/fj.04-2397fje. [DOI] [PubMed] [Google Scholar]
- 20.Yuste-Checa P., Vega A.I., Martín-Higueras C., Medrano C., Gámez A., Desviat L.R., Ugarte M., Pérez-Cerdá C., Pérez B. DPAGT1-CDG: Functional analysis of disease-causing pathogenic mutations and role of endoplasmic reticulum stress. PLoS ONE. 2017;12:e0179456. doi: 10.1371/journal.pone.0179456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter P., Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 22.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 23.Kim I., Xu W., Reed J.C. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 24.Kimata Y., Kohno K. Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr. Opin. Cell Biol. 2011;23:135–142. doi: 10.1016/j.ceb.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Termine D.J., Moremen K.W., Sifers R.N. The mammalian UPR boosts glycoprotein ERAD by suppressing the proteolytic downregulation of ER mannosidase I. J. Cell Sci. 2009;122:976–984. doi: 10.1242/jcs.037291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang C., Wang Y., Zhang H., Han F. The role of endoplasmic reticulum stress in neurodegenerative disease. Apoptosis. 2017;22:1–26. doi: 10.1007/s10495-016-1296-4. [DOI] [PubMed] [Google Scholar]
- 27.Fribley A., Zhang K., Kaufman R.J. Regulation of apoptosis by the unfolded protein response. Methods Mol. Biol. 2009;559:191–204. doi: 10.1007/978-1-60327-017-5_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sano R., Reed J.C. ER stress-induced cell death mechanisms. Biochim. Biophys. Acta. 2013;1833:3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabas I., Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 32.Sim N.-L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452-7. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maquat L.E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 34.Baker K.E., Parker R. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr. Opin. Cell Biol. 2004;16:293–299. doi: 10.1016/j.ceb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Shenkman M., Ron E., Yehuda R., Benyair R., Khalaila I., Lederkremer G.Z. Mannosidase activity of EDEM1 and EDEM2 depends on an unfolded state of their glycoprotein substrates. Commun. Biol. 2018;1:172. doi: 10.1038/s42003-018-0174-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foulquier F., Duvet S., Klein A., Mir A.-M., Chirat F., Cacan R. Endoplasmic reticulum-associated degradation of glycoproteins bearing Man5GlcNAc2 and Man9GlcNAc2 species in the MI8-5 CHO cell line. Eur. J. Biochem. 2004;271:398–404. doi: 10.1046/j.1432-1033.2003.03938.x. [DOI] [PubMed] [Google Scholar]
- 37.Chen J., Li X., Edmondson A., Meyers G.D., Izumi K., Ackermann A.M., Morava E., Ficicioglu C., Bennett M.J., He M. Increased Clinical Sensitivity and Specificity of Plasma Protein N-Glycan Profiling for Diagnosing Congenital Disorders of Glycosylation by Use of Flow Injection-Electrospray Ionization-Quadrupole Time-of-Flight Mass Spectrometry. Clin. Chem. 2019;65:653–663. doi: 10.1373/clinchem.2018.296780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W., James P.M., Ng B.G., Li X., Xia B., Rong J., Asif G., Raymond K., Jones M.A., Hegde M. A Novel N-Tetrasaccharide in Patients with Congenital Disorders of Glycosylation, Including Asparagine-Linked Glycosylation Protein 1, Phosphomannomutase 2, and Mannose Phosphate Isomerase Deficiencies. Clin. Chem. 2016;62:208–217. doi: 10.1373/clinchem.2015.243279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis K., Webster D., Smith C., Jackson S., Sinasac D., Seargeant L., Wei X.-C., Ferreira P., Midgley J., Foster Y. ALG9-CDG: New clinical case and review of the literature. Mol. Genet. Metab. Rep. 2017;13:55–63. doi: 10.1016/j.ymgmr.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y.-X., Peloso G.M., Nagai T.H., Mizoguchi T., Deik A., Bullock K., Lin H., Musunuru K., Yang Q., Vasan R.S. EDEM3 Modulates Plasma Triglyceride Level through Its Regulation of LRP1 Expression. iScience. 2020;23:100973. doi: 10.1016/j.isci.2020.100973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that are relevant to this research are presented in this paper.