Abstract
Diverging from traditional target inhibition, proteasomal protein degradation approaches have emerged as novel therapeutic modalities that embody distinct pharmacological profiles and can access previously undrugged targets. Small molecule degraders have the potential to catalytically destroy target proteins at substoichiometric concentrations, thus lowering administered doses and extending pharmacological effects. With this mechanistic premise, research efforts have advanced the development of small molecule degraders that benefit from stable and increased affinity ternary complexes. However, a holistic framework that evaluates different degradation modes from a catalytic perspective, including focusing on kinetically favored degradation mechanisms, is lacking. In this Outlook, we introduce the concept of an induced cooperativity spectrum as a unifying framework to mechanistically understand catalytic degradation profiles. This framework is bolstered by key examples of published molecular degraders extending from molecular glues to bivalent degraders. Critically, we discuss remaining challenges and future opportunities in drug discovery to rationally design and phenotypically screen for efficient degraders.
Short abstract
Applying a catalysis framework to proteasomal protein degradation paradigms is critical to advancing mechanistic understanding of protein degraders as well as identifying future opportunities.
I. Introduction
Cellular processes are governed by both subcellular compartmentalization and molecular recognition. These molecular interactions in turn provide a template to convey information as cues that can impact signaling, biosynthesis, and degradation pathways in live cells. Quantifying kinetic and thermodynamic contributions inherent to these cooperative interactions is essential to gain a deeper biological understanding and unlock novel biology.1 Despite remarkable biological complexity, hijacking and reprogramming these molecular recognition patterns with chemically induced proximity (CIP) approaches has been exploited to understand biological mechanisms and leverage this knowledge to identify novel therapeutics.2−4 In particular, proteasomal protein degradation has recently emerged as a privileged therapeutic strategy that enables the selective destruction of a protein of interest (POI) by reprogramming proteostasis machinery.5,6
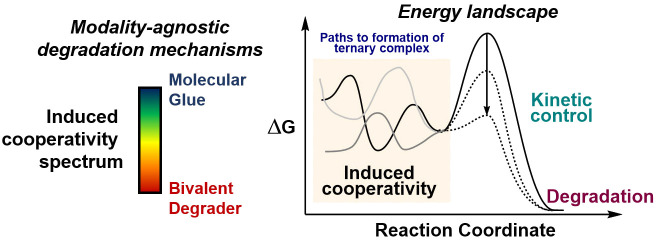
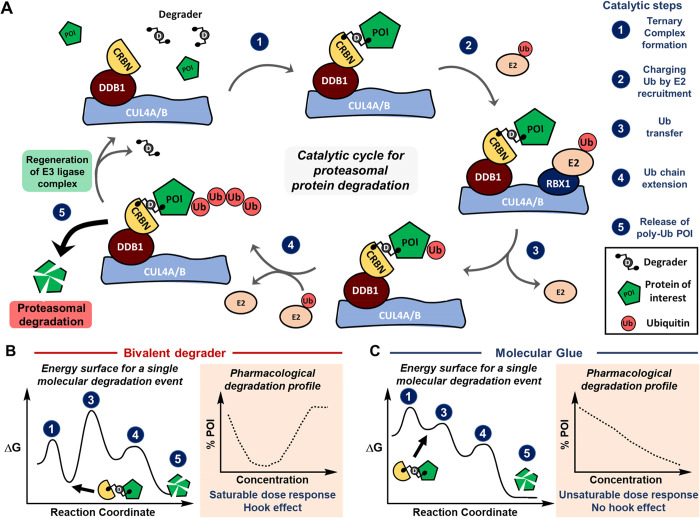
Productive proteasomal degradation relies on coordinated steps that include (1) the recognition of POI and formation of a ternary complex; (2, 3) charging and modification of POI with ubiquitin (Ub); (4) polyubiquitination of POI; and (5) release of poly-Ub POI followed by proteasomal destruction (Figure 1a). This defined catalytic cycle has been efficient and selectively commandeered by different modalities. Many recent reviews provide detailed accounts on the identification and development of proteolysis targeting chimeras, known as PROTACs, as well as molecular glues.7−9 At first glance, both degrader entities can showcase different energy landscapes and degradation profiles (Figure 1b,c). However, the distinction between bivalent degraders and molecular glues has blurred over the last several years into a conceptual continuum,10,11 as researchers build up molecular glues to affect the degradation profile while conversely trimming down PROTACs to optimize degradation and improve pharmacokinetic (PK) properties. As new degradation mechanisms are discovered, the detailed characterization of highly dynamic ternary complexes and corresponding catalytic implications will enable an improved mechanistic understanding and selection of opportunities for future investment.
Figure 1.
(A) The catalytic cycle for proteasomal protein degradation can be hijacked by unnatural degrader entities. This catalytic cycle is orchestrated by multiple enzymes, where the CUL4A-DDB1-CRBN complex is illustrated here. Sequence of events including (1) ternary complex formation, (2) charging Ub by E2 recruitment, (3) Ub transfer, (4) Ub chain extension, and (5) release of poly-Ub from the ternary complex are required for proteasomal degradation. (B, C) Molecular degraders exhibit different energy landscapes, which ultimately impact the degradation profile and saturability for degrader dose–response curves. Energy diagrams represent reaction coordinates for a single molecular degradation event, whereby the formation of binary complexes is omitted for clarity. The pharmacological degradation profile describes the average population of molecular degraders with corresponding equilibria that can be empirically measured.
With exponential growth in targeted protein degradation reports across academic and industry research,12 we believe that this area presents a novel opportunity to employ concepts from catalysis (chemical catalysis, enzymology, and pharmacology) that enable the development of innovative chemical probes and medicines. Here, we introduce the concept of induced cooperativity to provide a framework by which molecular glues and monovalent and bivalent degraders exemplify a spectrum of degradation mechanisms. In Section II, we highlight key examples from the literature with a focus on leveraging the importance of kinetics to pursue privileged ternary complexes and inform future degrader design. Section III weaves these catalytic considerations into cellular contexts to capitalize on cellular screening technologies and identify improved starting points to develop efficient degraders. In this Outlook, we use a catalysis lens to help contextualize opportunities and tackle challenges in order to enable the future discovery and development of efficient degraders.
II. Induced Cooperativity and Pursuit of Privileged Ternary Complexes for Proteasomal Degradation
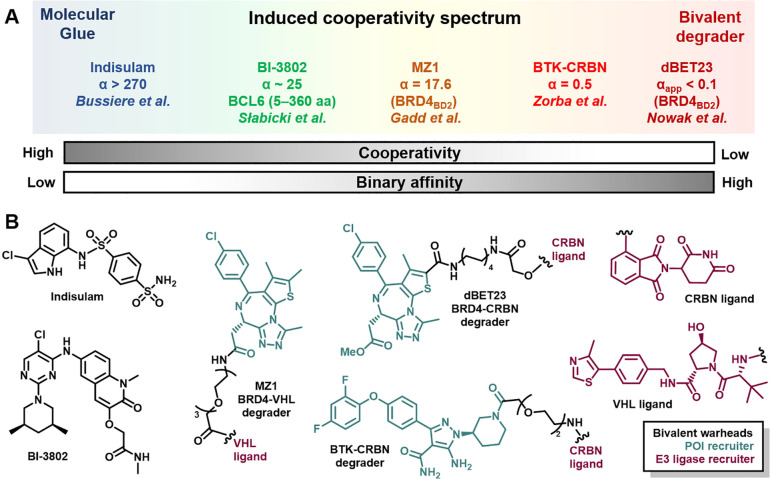
Induced cooperativity through proximal recruitment of artificial protein–protein interactions (PPIs) is essential for effective catalysis to take place in a protein degradation paradigm. A fundamental understanding of the kinetic and thermodynamic parameters that contribute to cooperativity can inform on the degrader design and optimization process. As a result, this understanding could drive efficient protein degradation and expedite the identification of key chemical matter through rational drug design and tailored screening approaches. To conceptualize the induced cooperativity spectrum, we can think of how small molecule degraders can toggle through both cooperativity and binary affinity axes (Figure 2a). In the leftmost side of the spectrum, a molecular glue theoretically may not have any detectable binary affinity toward either protein surface by itself but forms a highly cooperative ternary complex. Conversely, at the rightmost end of the spectrum, a bivalent degrader can exhibit a maximum binary affinity toward either POI or E3 ligase, with negative cooperativity arising from steric clashes within the ternary complex. To support these notions, we discuss selected examples of empirically discovered protein degraders and key considerations for ternary complexes across the induced cooperativity spectrum.
Figure 2.
The induced cooperativity spectrum encompasses multiple degrader entities. (A) Theoretical ends of the induced cooperativity spectrum are flanked by molecular glues (left) and bivalent degraders (right). This spectrum can be further dissected into contributions arising from cooperativity (α value) and binary affinity for either POI or E3 Ub ligase recruiter, or a combination thereof. Selected degrader examples are described based on cooperativity values reported or estimated from experimental data from indicated references. (B) Chemical structures for small molecule degraders from part A, with bivalent warheads corresponding to the POI recruiter in teal and E3 ligase in purple.
In less than two decades, researchers have advanced PROTACs from basic science to the clinic, where a handful of molecules have entered phase I trials.13 PROTACs typically feature an E3 ubiquitin ligase recruiting end, a linker, and a ligand for the POI (Figure 2b). Colocalization of the E3 ligase complex with a protein of interest can lead to productive polyubiquitination and proteasomal degradation. This approach, however, is not without several challenges including extensive medicinal chemistry to identify suitable E3 ligase recruiters and optimized linkers that afford bifunctional small molecules with improved cell permeability and efficient degradation. To access robust protein degradation, these challenges can be addressed by a careful balance of target binding potency, cellular permeability, and judicious choice of E3 ligase for a successful ubiquitination outcome. Beyond cellular degradation, developing efficient degraders also requires tackling unique hurdles to achieve the in vivo pharmacological phenotype with improved pharmacodynamics (PD) and PK.14 However, this drug discovery phase can be prohibitively long, and guidelines for accessing the desired degradation profile remain largely empirical.5
To highlight a single target class, kinase inhibitors have provided a ripe entry point to develop bivalent degraders against a highly characterized kinome with corresponding chemical ligandability.15,16 Several reports have demonstrated that degraders based on pan-kinase inhibitors could exhibit an exquisite degradation selectivity of a single kinase, which was unprecedented for the parent kinase ligand.17−19 Furthermore, Crews and co-workers developed a selective degrader for the p38-gamma isoform of the MAPK family by linking a foretinib warhead to a VHL recruiter.19 Collectively, these studies demonstrated that key design elements such as the E3 ligase recruiter and linker composition were required to achieve the desired target selectivity and degradation. However, these efforts also revealed that target engagement alone was insufficient to result in protein degradation. For this reason, significant work in the protein degradation field has gravitated toward identifying key determinants required for the productive formation of a ternary complex that lead to efficient degradation.
The recombinant expression of ternary complex components has facilitated the evaluation of the binding affinity of degraders to both POI and E3 ligase, as well as the determination of cooperativity (α value). Isothermal calorimetry (ITC), fluorescence polarization (FP), size exclusion chromatography (SEC), and luminescent proximity assay (i.e., aLISA) are among popular techniques that allow the measurement of these thermodynamic parameters under steady state conditions.20 In addition to the recent understanding emerging from crystal structures,21 these biophysical methods have helped define interactions at the protein–protein interface that can display both cooperative behavior and recently appreciated plasticity. Notably, bivalent degrader design can capitalize on protein–protein interactions (i.e., BRD4-VHL,22 BRD4-CRBN,23 BTK-CRBN24) to develop more efficient degraders that can also benefit from cooperative behavior (Figure 2b). This work was followed up by the cyclization of the MZ1 degrader to produce macroPROTAC-1, where Ciulli and co-workers demonstrated that reducing entropic cost can increase cooperativity and maintain cellular degradation efficiency.25 Harnessing cooperativity within ternary complexes can also have a positive impact in minimizing the hook effect from catalysis and safety perspectives.26,27 In addition to routine ternary complex characterization, prospective mathematical models such as three-body equilibria can also inform iterative degrader design.28−30 Furthermore, more recent in silico methods can also guide the prioritization of degraders as well as linker design that benefit from increased protein surface complementarity to reduce the time for exploring structure–activity relationships (SARs).24,31−33 Combining experimental and modeling approaches to prioritize stable ternary complexes has been productive11 but may also bias discovery of degraders toward molecules that primarily access static and long-lived populations. Thus, opportunities where sufficiently fast protein degradation occurs, such that a small fraction of ternary complex is present, could go unrecognized.
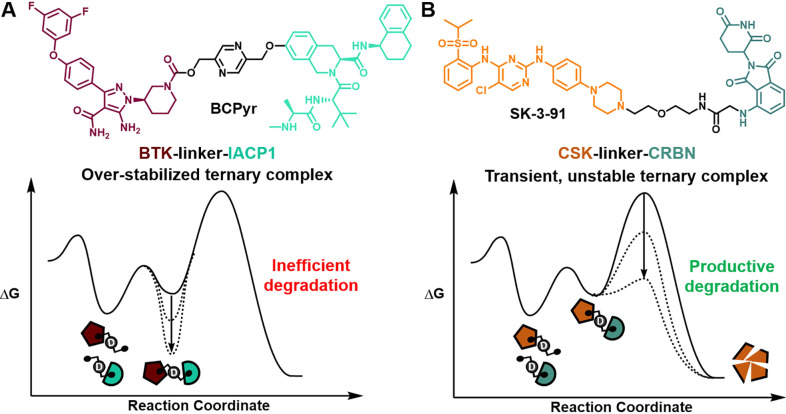
In contrast to the notion that stable ternary complexes can enhance degradation efficiency, Pfizer scientists found that increased rigidity and stability may negatively impact BTK degradation mediated by the recruitment of cIAP1.34 With a suite of biochemical, biophysical, and structural studies, they showed that in solution ensembles can lead to ternary complex conformations with different degradation profiles upon bivalent degrader recruitment, perhaps stemming from rigidification of the degrader linker or intrinsic rigidity of the ternary complex, or a combination of both.34 In this scenario, visualizing high-stability ternary complexes from an energy landscape perspective can illustrate the steep activation requirement for productive degradation (Figure 3a). In line with recognizing unique attributes of a given ternary complex, Donovan et al. conducted a tour de force effort to map the degradable kinome and answer fundamental questions regarding kinase tractability and degradability. Interestingly, this comprehensive study revealed (1) that high potency binders can be ineffective starting points for degraders, (2) that degradation efficiency is not predicted by the formation of stable ternary complexes, and (3) that catalytic degradation can result from transient and unstable ternary complexes (degrader example depicted as a reaction coordinate in Figure 3b).35 Both collective works highlighted above point out considerations for overstabilizing ternary complexes as well as opportunities to exploit degradation outcomes based on dynamic yet productive ternary complexes. Furthermore, plasticity in ternary complexes has been previously appreciated for bivalent degraders22,23 as well as next-generation immunophilins that engage a malleable FKBP12 surface.36 Therefore, a mechanistic understanding of dynamic protein complexes with refined methods as well as the role of privileged conformations and populations could provide untapped opportunities in protein degradation workflows.37,38
Figure 3.
Energy landscapes for specific examples where stable ternary complex formation leads to inefficient degradation (A), and unstable complexes can lead to productive degradation (B). Chemical structures of degraders BCPyr and SK-3-91 are color-coded by POI ligand, linker (black), and E3 ligase.
Beyond target selectivity and interprotein contacts, the premise of event-driven pharmacology, where a substoichiometric amount of a small molecule catalytically degrades POI, is a unique pharmacological attribute of degradation mechanisms. Indeed, Bondeson et al. demonstrated that bivalent degraders at substoichiometric concentrations can catalyze ubiquitination rates in vitro.39 The catalytic degradation of long-half-life proteins, such as AKT40 and RIPK2,41 has demonstrated profound and prolonged PD. Furthermore, electrophilic molecules can facilitate protein degradation at fractional E3 ligase (i.e., DCAF16) occupancy42 and with nanomolar doses,43 both viable strategies for the longer durability of POI removal. Reactivity profiling to engage nucleophilic residues has enabled the discovery and development of novel covalent warheads to pursue previously inaccessible E3 ligases.44−46 Using a covalent warhead to reprogram substrate recognition by a modified E3 ligase is an elegant strategy to enhance both selectivity and catalysis of degradation. Indeed, Nomura and co-workers achieved selective disruption of RNF114-substrate recognition with a nimbolide warhead thereby accessing the desired efficacy and on-target mechanism of action (MOA).43 Covalent degraders could also have the unique potential to address unresolved challenges of improved physicochemical properties and PD/PK relationships.46
Despite productive strides to harness thermodynamic cooperative interactions, fewer efforts have intentionally pursued kinetically driven ternary complexes that could address the hook effect, overly stable ternary complexes, and lack of catalysis. To do this successfully, a mechanistic understanding of kinetic parameters and enzymology of the protein degradation cycle is imperative.47 Biophysical methods, including surface plasma resonance (SPR) and biolayer interferometry (BLI), are better positioned to qualify kinetic parameters and have been utilized to inform bivalent degrader design.34,48 Complementary to measuring koff/kon rates and dissociation half-lives, native mass spectrometry can provide additional granularity of ternary complex formation and intermediate conformational states in a single label-free experiment.49 Future approaches to further explore degradation opportunities may require an in-depth kinetic analysis of protein complexes as those used for dynamic transcription factors,37 which microfluidics and single molecule studies may also be well positioned to address.50 Drawing from enzymology principles, a deeper appreciation and mechanistic understanding of energetics for protein ensembles could provide new avenues for protein degradation paradigms.51−54 Productive integration of these molecular contributions will continue to refine kinetic degradation models and translational frameworks to successfully develop bivalent degraders.30,55
Following the induced cooperativity spectrum, monovalent degraders offer a distinct degradation profile from bivalent degraders.56 A recent emergence in identifying monomeric degraders may be a result of both improved MOA deconvolution frameworks for molecules of a desired phenotype and more sensitive detection methods. These degraders can offer distinct physicochemical properties from bivalent counterparts with improved solubility and cell permeability, which are critical for selecting the dose and route of administration.56 Often serendipitously discovered, molecular perturbagens modulating B-cell lymphoma protein 6 (Bcl6), an oncogenic transcription factor, are great examples of how small changes in chemical structure can induce target inhibition or degradation.57,58 Recently, Ebert, Fischer, and co-workers unraveled the molecular MOA of BI-3802 (Figure 2b), a monomeric degrader that initially triggers the polymerization of Bcl6, followed by entrapment in cellular foci and, finally, destruction by the proteasome.59 Notably, a bivalent degrader derived from a structurally different Bcl6 inhibitor was not superior to the parent warhead and exhibited mild antiproliferative properties.60 This latter study raises important considerations when repurposing inhibitor warheads as degrader starting points, specifically, untangling mechanistic contributions of target inhibition from degradation to understand cellular phenotypes. Collectively, Bcl6 inhibition and degradation studies exemplify the need for rigorous MOA deconvolution for chemical matter of interest with an eye toward target dynamics that are governed by kinetic and thermodynamic processes.
Harnessing an occupancy-driven mechanism, PPI stabilizers, such as cyclosporin A, FK-506, and rapamycin, benefit from stable ternary complexes for driving immunosuppressive pharmacology.61 Conversely, event-driven pharmacology could advantageously exploit conditions where ternary complex formation is transient. Thus, kinetically processing this intermediate could lead to rapid and selective degradation of the protein substrate. The immunomodulatory (IMiD) drugs demonstrate this principle and will also be considered through the lens of the induced cooperativity spectrum. Often identified by empirical methods, molecular glues can create a neomorphic surface that can, in turn, selectively engage neosubstrates and funnel them for proteasomal degradation.8 This surface programmability has enabled drug discovery efforts to hone selectivity further for bespoke neosubstrates by diversifying chemical scaffolds of IMiDs.10 Remarkably, CRBN has been exploited as a privileged E3 ligase that can recognize structural degrons from over 100 Zn finger substrates.62−64 Moreover, this programmed complementarity has recently been exploited to modulate CAR-T cell activity.65,66 On the other hand, anticancer drugs with an arylsulfonamide scaffold (e.g., indisulam, Figure 2b) engaging DCAF15 also serve as molecular glues but only engage a handful of targets, as crystal structures have revealed a highly conserved peptide sequence in degradable neosubstrates.7,67−69 In a prospective effort to leverage structure-based drug design, scientists at Nurix Therapeutics were able to mimic a native phosphoepitope in β-catenin as a molecular glue that enhances association with SCFβ-TrCP, its cognate E3 ligase, to afford successful proteasomal degradation.70 Similarly to bivalent degraders, the development of molecular glues requires a long discovery phase with extensive medicinal chemistry campaigns and remains empirical.
As our structural understanding of matching the POI and E3 ligase continues to improve, the SAR cycle time to develop degraders that stabilize ternary complex formation and reduce entropic cost will also shorten. However, with this approach, biased attention is concentrated on an early step of the catalytic degradation cycle, which may not be the rate-limiting step for all degradation mechanisms across the induced cooperativity spectrum. Furthermore, we may not uncover novel starting points to kinetically alter the degradome of a ligand. Going forward, ligand screening platforms that could be adapted to select for catalytic degraders and provide new footholds for challenging targets will be important. In the next section, we focus on intrinsic attributes from the cellular milieu that can affect catalytic degradation and how those can be leveraged to identify efficient degraders.
III. Cellular Context Is Critical to Finding Efficient Degraders
When considering degrader catalytic efficiency in live cells, any biological factor that decreases saturation could diminish efficient degradation. Of potential factors, global protein abundance levels of POI or E3 ligase may not correlate with efficient degradation, perhaps in part due to a smaller labile or degradable pool.35 In addition to expression levels, interrogating relevant cellular contexts that recapitulate key biology is far more important. From a historical research perspective spanning the past decade, dissecting thalidomide’s toxicity mechanism epitomizes the necessity to thoroughly examine the molecular MOA of degraders in different cellular contexts. After the initial discovery of CRBN engagement in zebrafish,71 multiple neosubstrates have been identified,10 of which SALL4 degradation phenocopies have observed teratogenicity.72,73 Furthermore, our understanding of ligandable and recruitable E3 ubiquitin ligases has significantly expanded along with degradable neosubstrates.74,75 Leading this front, carefully defined phenotypic screens present unique advantages to discover and develop E3 ligase modulators.76 With growing multiomic data sets generated from cellular, preclinical, and clinical studies, future targeted protein degradation efforts could leverage systems biology approaches to nominate degradation hypotheses with greater confidence.77,78 In practice, the success of such strategies will be highly dependent on the available cellular and chemical tools to validate them.
Within recent years, several cellular technologies have emerged as valuable tools to interrogate protein degradation in live cells. Chemical genetic approaches such as the HaloPROTACs79 and dTAG platform80 have democratized access to visualize the degradation of your favorite POI in lieu of available small molecule ligands, including orphan cell surface transporters.81 Genetic tools such as haloTag and GFP fusions have enabled the early assessment of Ub ligase compatibility,82 while biodegraders have enabled swift scanning of POI degradation fitness for a number of E3 ligases83 and targeting specific conformational states of KRas.84 Collectively, these technologies offer a diverse menu to select tractable degrader starting points in live cells.
In order to drive SAR optimization of degraders, low- to high-throughput quantitative methods are routinely used.85 Of these methods, luminescence-based workflows developed by Promega surfaced as very powerful tools to kinetically characterize protein degradation events.86 In these experiments, POIs can be labeled with HiBiT tags at endogenous levels to interrogate competing biosynthesis and degradation rates.87 Furthermore, the portability of HiBiT constructs helps paint a crisp picture by reporting on specific events of the catalytic degradation cycle: cytosolic access, target engagement, ternary complex formation, Ub transfer, and proteasome processing.86 Of note, deconvolution of TL12-186, a pan-kinase PROTAC, illuminated the mechanistic profile with the required selectivity and temporal resolution to identify degradation of POI subpopulations.88 Collectively, these reports showcase that a detailed mechanistic understanding of degradation profiles for bivalent degraders can be achieved in relevant cellular contexts. Given the breadth and depth afforded by HiBiT tagging, this approach is also well positioned to characterize and exploit kinetically favored degradation mechanisms across the induced cooperativity spectrum.
In contrast to bivalent degraders, the prospective identification of privileged small molecule degraders remains challenging. In this context, functional genomic screens have unveiled degradation dependencies that accelerate the identification of novel molecular glues. For example, three independent studies identified distinct chemical scaffolds that stabilize DDB1-CDK12 interaction and thus lead to the enhanced degradation of Cyclin K, a CDK12 interactor.89−91 These findings represent a novel mechanism by which a molecular glue can induce POI degradation by a distant PPI rather than directly reprogramming the POI-E3 interface. Phenotypic screens that capitalize on degradation nodes, such as hyponeddylated cells,89 or exploit cancer vulnerabilities92−94 are elegant strategies to discover and enhance the druggability of novel degrader biology in relevant cellular contexts. Notably, Koduri et al. devised a creative screening strategy for novel IKZF1 degraders, which led to the identification of Spautin-1, a novel molecular glue that does not require CRBN, and subsequently deployed this strategy to uncover CDK2’s role in regulating the abundance of the oncogenic transcription factor ASCL1 pertinent to small cell lung cancers.93 Additionally, genetic screens have also provided early insights into potential resistance mechanisms emerging from multiple proteasomal degrader modalities.92,94,95 This collection of studies has embraced the tractability of genetic screens and translatability of phenotypic approaches to intentionally identify novel molecular glues and deconvolute underlying mechanisms of proteasomal degradation. Therefore, functional genomics will continue to play an important role in deconstructing desired phenotypes in relevant cellular contexts.
Finally, the complexity of the ubiquitin proteasome system (UPS) should not go unrecognized. As the understanding of proteostasis machinery accumulates across cellular environments, we will have a better appreciation for the intricacies of Ub code,74,96 redundancies of 600 Ub ligases,97 or lack thereof, regulation of CRL4 network dynamics,98 and protein turnover rates.99 Going forward, the identification of underlying mechanisms that amplify POI destruction upon degrader recruitment, such as potentiating proteasomal flux100 or enhancing Ub-chain elaboration,101 can unlock unique synergies to complement degrader approaches. Equally exciting, the degradation of disease-relevant protein aggregates102 and polymerized Bcl659 have already shifted naïve perceptions on processing and unfolding activities by the proteasome. Consequently, converging the elucidation of degradation mechanisms with compelling cellular contexts presents bright prospects to access therapeutically relevant human biology with catalytically efficient molecules.
IV. Conclusions and Outlook
Capitalizing on catalytic considerations for the induced cooperativity spectrum can advance future degrader discovery campaigns. Importantly, exploiting kinetically privileged protein degradation and leveraging phenotypically relevant cellular contexts present untapped opportunities. Of broader significance, the concept of using catalytic molecules to kinetically control cellular processes extends beyond the area of proteasomal degradation, including pioneering studies where novel modalities can hijack autophagy and lysosomal recycling mechanisms.103−105 Theoretically, any cellular process can be modulated with an approach that colocalizes cellular machinery to a target of interest, where applying induced cooperativity principles is critical. Though the vast majority of examples to date have focused in targeted protein degradation, early results for controlling both installation and removal of posttranslational modifications, like phosphorylation106,107 and glycosylation,108,109 have also been reported. In the pursuit of innovative chemical tools and medicines, the growth and success of this budding field could be significantly advanced by experimental workflows leveraged by catalysis researchers. We hope that this Outlook contextualizes the opportunities and challenges for this field under the lens of catalysis and brings forward new ideas that will ultimately benefit patients.
Acknowledgments
The authors would like to thank Raphaëlle Berger, Simon Bushell, John Caldwell, and Jason Imbriglio for helpful discussions and thoughtful comments on this paper. F.P.R.-R. and S.M.L. are employees of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA, and Pfizer, Inc., respectively.
The authors declare no competing financial interest.
References
- Whitty A. Cooperativity and biological complexity. Nat. Chem. Biol. 2008, 4, 435–439. 10.1038/nchembio0808-435. [DOI] [PubMed] [Google Scholar]
- Stanton B. Z.; Chory E. J.; Crabtree G. R. Chemically induced proximity in biology and medicine. Science 2018, 359, eaao5902. 10.1126/science.aao5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerry C. J.; Schreiber S. L. Unifying principles of bifunctional, proximity-inducing small molecules. Nat. Chem. Biol. 2020, 16, 369–378. 10.1038/s41589-020-0469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R. J. Multispecific drugs herald a new era of biopharmaceutical innovation. Nature 2020, 580, 329–338. 10.1038/s41586-020-2168-1. [DOI] [PubMed] [Google Scholar]
- Burslem G. M.; Crews C. M. Proteolysis-Targeting Chimeras as Therapeutics and Tools for Biological Discovery. Cell 2020, 181, 102–114. 10.1016/j.cell.2019.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R.; Mohl D.; Deshaies R. J. Harnessing the Power of Proteolysis for Targeted Protein Inactivation. Mol. Cell 2020, 77, 446–460. 10.1016/j.molcel.2020.01.010. [DOI] [PubMed] [Google Scholar]
- Wu T.; et al. Targeted protein degradation as a powerful research tool in basic biology and drug target discovery. Nat. Struct. Mol. Biol. 2020, 27, 605–614. 10.1038/s41594-020-0438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzl A.; Winter G. E. Targeted protein degradation: current and future challenges. Curr. Opin. Chem. Biol. 2020, 56, 35–41. 10.1016/j.cbpa.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S. L. The Rise of Molecular Glues. Cell 2021, 184, 3–9. 10.1016/j.cell.2020.12.020. [DOI] [PubMed] [Google Scholar]
- Chamberlain P. P.; et al. Evolution of Cereblon-Mediated Protein Degradation as a Therapeutic Modality. ACS Med. Chem. Lett. 2019, 10, 1592–1602. 10.1021/acsmedchemlett.9b00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust T. B.; Donovan K. A.; Yue H.; Chamberlain P. P.; Fischer E. S. Small-Molecule Approaches to Targeted Protein Degradation. Annu. Rev. Cancer Biol. 2021, 5, 181–201. 10.1146/annurev-cancerbio-051420-114114. [DOI] [Google Scholar]
- Kostic M.; Jones L. H. Critical Assessment of Targeted Protein Degradation as a Research Tool and Pharmacological Modality. Trends Pharmacol. Sci. 2020, 41, 305–317. 10.1016/j.tips.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. Targeted protein degraders crowd into the clinic. Nat. Rev. Drug Discovery 2021, 20, 247. 10.1038/d41573-021-00052-4. [DOI] [PubMed] [Google Scholar]
- Cantrill C.; et al. Fundamental aspects of DMPK optimization of targeted protein degraders. Drug Discovery Today 2020, 25, 969–982. 10.1016/j.drudis.2020.03.012. [DOI] [PubMed] [Google Scholar]
- Manning G.; Whyte D. B.; Martinez R.; Hunter T.; Sudarsanam S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Hu Y.; Furtmann N.; Bajorath J. Current compound coverage of the kinome. J. Med. Chem. 2015, 58, 30–40. 10.1021/jm5008159. [DOI] [PubMed] [Google Scholar]
- Huang H. T.; et al. A Chemoproteomic Approach to Query the Degradable Kinome Using a Multi-kinase Degrader. Cell Chem. Biol. 2018, 25, 88–99. 10.1016/j.chembiol.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeson D. P.; et al. Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell Chem. Biol. 2018, 25, 78–87. 10.1016/j.chembiol.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B. E.; et al. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat. Commun. 2019, 10, 1–13. 10.1038/s41467-019-12407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. J.; Ciulli A. Molecular recognition of ternary complexes: A new dimension in the structure-guided design of chemical degraders. Essays Biochem. 2017, 61, 505–516. 10.1042/EBC20170041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S.; Ciulli A. Building ubiquitination machineries: E3 ligase multi-subunit assembly and substrate targeting by PROTACs and molecular glues. Curr. Opin. Struct. Biol. 2021, 67, 110–119. 10.1016/j.sbi.2020.10.009. [DOI] [PubMed] [Google Scholar]
- Gadd M. S.; et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 2017, 13, 514–521. 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak R. P.; et al. Plasticity in binding confers selectivity in ligand-induced protein degradation article. Nat. Chem. Biol. 2018, 14, 706–714. 10.1038/s41589-018-0055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorba A.; et al. Delineating the role of cooperativity in the design of potent PROTACs for BTK. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E7285–E7292. 10.1073/pnas.1803662115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa A.; Hughes S. J.; Lucas X.; Wright J. E.; Ciulli A. Structure-Based Design of a Macrocyclic PROTAC. Angew. Chem. 2020, 132, 1744–1751. 10.1002/ange.201914396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R. D.; Rosenmund C.; Stefan M. I. Cooperative binding mitigates the high-dose hook effect. BMC Syst. Biol. 2017, 11, 74. 10.1186/s12918-017-0447-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K.; et al. Proteolysis-targeting chimeras in drug development: A safety perspective. Br. J. Pharmacol. 2020, 177, 1709–1718. 10.1111/bph.15014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass E. F.; Miller C. J.; Sparer G.; Shapiro H.; Spiegel D. A. A comprehensive mathematical model for three-body binding equilibria. J. Am. Chem. Soc. 2013, 135, 6092–6099. 10.1021/ja311795d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B. A suite of mathematical solutions to describe ternary complex formation and their application to targeted protein degradation by hetero-bifunctional ligands. J. Biol. Chem. 2020, 295, 15280. 10.1074/jbc.RA120.014715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett D. W.; Gilbert A. M. A kinetic proofreading model for bispecific protein degraders. J. Pharmacokinet. Pharmacodyn. 2021, 48, 149. 10.1007/s10928-020-09722-z. [DOI] [PubMed] [Google Scholar]
- Pérez-Benito L.; et al. The size matters? A computational tool to design bivalent ligands. Bioinformatics 2018, 34, 3857–3863. 10.1093/bioinformatics/bty422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond M. L.; Henry A.; Li H.; Williams C. I. Improved Accuracy for Modeling PROTAC-Mediated Ternary Complex Formation and Targeted Protein Degradation via New In Silico Methodologies. J. Chem. Inf. Model. 2020, 60, 5234–5254. 10.1021/acs.jcim.0c00897. [DOI] [PubMed] [Google Scholar]
- Bai N.; et al. Rationalizing PROTAC-Mediated Ternary Complex Formation Using Rosetta. J. Chem. Inf. Model. 2021, 61, 1368. 10.1021/acs.jcim.0c01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiemer J.; et al. Snapshots and ensembles of BTK and cIAP1 protein degrader ternary complexes. Nat. Chem. Biol. 2021, 17, 152–160. 10.1038/s41589-020-00686-2. [DOI] [PubMed] [Google Scholar]
- Donovan K. A.; et al. Mapping the Degradable Kinome Provides a Resource for Expedited Degrader Development. Cell 2020, 183, 1714–1731. 10.1016/j.cell.2020.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigdel U. K.; et al. Genomic discovery of an evolutionarily programmed modality for small-molecule targeting of an intractable protein surface. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 17195–17203. 10.1073/pnas.2006560117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley M. J.; et al. Unexpected specificity within dynamic transcriptional protein-protein complexes. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 27346–27353. 10.1073/pnas.2013244117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlick J. M.; Mapp A. K. Selective Modulation of Dynamic Protein Complexes. Cell Chemical Biology 2020, 27, 986–997. 10.1016/j.chembiol.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeson D. P.; et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 2015, 11, 611–617. 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You I.; et al. Discovery of an AKT Degrader with Prolonged Inhibition of Downstream Signaling. Cell Chem. Biol. 2020, 27, 66–73. 10.1016/j.chembiol.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mares A.; et al. Extended pharmacodynamic responses observed upon PROTAC-mediated degradation of RIPK2. Commun. Biol. 2020, 3, 1–13. 10.1038/s42003-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Crowley V. M.; Wucherpfennig T. G.; Dix M. M.; Cravatt B. F. Electrophilic PROTACs that degrade nuclear proteins by engaging DCAF16. Nat. Chem. Biol. 2019, 15, 737–746. 10.1038/s41589-019-0279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradlin J. N.; et al. Harnessing the anti-cancer natural product nimbolide for targeted protein degradation. Nat. Chem. Biol. 2019, 15, 747–755. 10.1038/s41589-019-0304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C. C.; et al. Covalent Ligand Screening Uncovers a RNF4 E3 Ligase Recruiter for Targeted Protein Degradation Applications. ACS Chem. Biol. 2019, 14, 2430–2440. 10.1021/acschembio.8b01083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.; et al. Chemoproteomics-enabled discovery of covalent RNF114-based degraders that mimic natural product function. Cell Chem. Biol. 2021, 28, 559. 10.1016/j.chembiol.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabizon R.; London N. The rise of covalent proteolysis targeting chimeras. Curr. Opin. Chem. Biol. 2021, 62, 24–33. 10.1016/j.cbpa.2020.12.003. [DOI] [PubMed] [Google Scholar]
- Fisher S. L.; Phillips A. J. Targeted protein degradation and the enzymology of degraders. Curr. Opin. Chem. Biol. 2018, 44, 47–55. 10.1016/j.cbpa.2018.05.004. [DOI] [PubMed] [Google Scholar]
- Roy M. J.; et al. SPR-Measured Dissociation Kinetics of PROTAC Ternary Complexes Influence Target Degradation Rate. ACS Chem. Biol. 2019, 14, 361–368. 10.1021/acschembio.9b00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge R.; et al. Native Mass Spectrometry Can Effectively Predict PROTAC Efficacy. ACS Cent. Sci. 2020, 6, 1223–1230. 10.1021/acscentsci.0c00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aditham A. K.; Markin C. J.; Mokhtari D. A.; DelRosso N.; Fordyce P. M. High-Throughput Affinity Measurements of Transcription Factor and DNA Mutations Reveal Affinity and Specificity Determinants. Cell Syst. 2021, 12, 112. 10.1016/j.cels.2020.11.012. [DOI] [PubMed] [Google Scholar]
- Benkovic S. J.; Hammes G. G.; Hammes-Schiffer S. Free-energy landscape of enzyme catalysis. Biochemistry 2008, 47, 3317–3321. 10.1021/bi800049z. [DOI] [PubMed] [Google Scholar]
- Hammes G. G.; Benkovic S. J.; Hammes-Schiffer S. Flexibility, diversity, and cooperativity: Pillars of enzyme catalysis. Biochemistry 2011, 50, 10422–10430. 10.1021/bi201486f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P. K. A Biophysical Perspective on Enzyme Catalysis. Biochemistry 2019, 58, 438–449. 10.1021/acs.biochem.8b01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P. K.; Bernard D. N.; Bafna K.; Doucet N. Enzyme Dynamics: Looking Beyond a Single Structure. ChemCatChem 2020, 12, 4704–4720. 10.1002/cctc.202000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak R. P.; Jones L. H. Target Validation Using PROTACs: Applying the Four Pillars Framework. SLAS Discovery Adv. Sci. Drug Discovery 2021, 26, 474. 10.1177/2472555220979584. [DOI] [PubMed] [Google Scholar]
- Hanan E. J.; et al. Monomeric Targeted Protein Degraders. J. Med. Chem. 2020, 63, 11330–11361. 10.1021/acs.jmedchem.0c00093. [DOI] [PubMed] [Google Scholar]
- Kerres N.; et al. Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6. Cell Rep. 2017, 20, 2860–2875. 10.1016/j.celrep.2017.08.081. [DOI] [PubMed] [Google Scholar]
- Bellenie B. R.; et al. Achieving in Vivo Target Depletion through the Discovery and Optimization of Benzimidazolone BCL6 Degraders. J. Med. Chem. 2020, 63, 4047–4068. 10.1021/acs.jmedchem.9b02076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Słabicki M.; et al. Small-molecule-induced polymerization triggers degradation of BCL6. Nature 2020, 588, 164–168. 10.1038/s41586-020-2925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoull W.; et al. Development of a Novel B-Cell Lymphoma 6 (BCL6) PROTAC to Provide Insight into Small Molecule Targeting of BCL6. ACS Chem. Biol. 2018, 13, 3131–3141. 10.1021/acschembio.8b00698. [DOI] [PubMed] [Google Scholar]
- Schreiber S. L.; Crabtree G. R. The mechanism of action of cyclosporin A and FK506. Immunology Today 1992, 13, 136–142. 10.1016/0167-5699(92)90111-J. [DOI] [PubMed] [Google Scholar]
- Lu G.; et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of ikaros proteins. Science 2014, 343, 305–309. 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krönke J.; et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 2014, 343, 301–305. 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers Q. L.; et al. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science 2018, 362, eaat0572. 10.1126/science.aat0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan M.; et al. Reversible ON- and OFF-switch chimeric antigen receptors controlled by lenalidomide. Sci. Transl. Med. 2021, 13, eabb6295 10.1126/scitranslmed.abb6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonneau S.et al. An IMiD-inducible degron provides reversible regulation for chimeric antigen receptor expression and activity. Cell Chem. Biol. 2020, in press. 10.1016/j.chembiol.2020.11.012. [DOI] [PubMed] [Google Scholar]
- Han T.; et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 2017, 356, eaal3755. 10.1126/science.aal3755. [DOI] [PubMed] [Google Scholar]
- Uehara T.; et al. Selective degradation of splicing factor CAPERα By anticancer sulfonamides. Nat. Chem. Biol. 2017, 13, 675–680. 10.1038/nchembio.2363. [DOI] [PubMed] [Google Scholar]
- Bussiere D. E.; et al. Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex. Nat. Chem. Biol. 2020, 16, 15–23. 10.1038/s41589-019-0411-6. [DOI] [PubMed] [Google Scholar]
- Simonetta K. R.; et al. Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction. Nat. Commun. 2019, 10, 1–12. 10.1038/s41467-019-09358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T.; et al. Identification of a primary target of thalidomide teratogenicity. Science 2010, 327, 1345–1350. 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- Matyskiela M. E.; et al. SALL4 mediates teratogenicity as a thalidomide-dependent cereblon substrate. Nat. Chem. Biol. 2018, 14, 981–987. 10.1038/s41589-018-0129-x. [DOI] [PubMed] [Google Scholar]
- Donovan K. A.; et al. Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane radial ray syndrome. eLife 2018, 7, 38430. 10.7554/eLife.38430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M. Post-Translational Modifications: Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 59–70. 10.1038/nrm.2017.83. [DOI] [PubMed] [Google Scholar]
- Schapira M.; Calabrese M. F.; Bullock A. N.; Crews C. M. Targeted protein degradation: expanding the toolbox. Nat. Rev. Drug Discovery 2019, 18, 949–963. 10.1038/s41573-019-0047-y. [DOI] [PubMed] [Google Scholar]
- Ege N.; Bouguenina H.; Tatari M.; Chopra R. Phenotypic screening with target identification and validation in the discovery and development of E3 ligase modulators. Cell Chemical Biology 2021, 28, 283–299. 10.1016/j.chembiol.2021.02.011. [DOI] [PubMed] [Google Scholar]
- Joshi A.; Rienks M.; Theofilatos K.; Mayr M. Systems biology in cardiovascular disease: a multiomics approach. Nat. Rev. Cardiol. 2021, 18, 313. 10.1038/s41569-020-00477-1. [DOI] [PubMed] [Google Scholar]
- Petralia F.; et al. Integrated Proteogenomic Characterization across Major Histological Types of Pediatric Brain Cancer. Cell 2020, 183, 1962–1985. 10.1016/j.cell.2020.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley D. L.; et al. HaloPROTACS: Use of Small Molecule PROTACs to Induce Degradation of HaloTag Fusion Proteins. ACS Chem. Biol. 2015, 10, 1831–1837. 10.1021/acschembio.5b00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabet B.; et al. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 2018, 14, 431–441. 10.1038/s41589-018-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensimon A.; et al. Targeted Degradation of SLC Transporters Reveals Amenability of Multi-Pass Transmembrane Proteins to Ligand-Induced Proteolysis. Cell Chem. Biol. 2020, 27, 728–739. 10.1016/j.chembiol.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottis P.; et al. Assessing Different E3 Ligases for Small Molecule Induced Protein Ubiquitination and Degradation. ACS Chem. Biol. 2017, 12, 2570–2578. 10.1021/acschembio.7b00485. [DOI] [PubMed] [Google Scholar]
- Lim S.; et al. BioPROTACs as versatile modulators of intracellular therapeutic targets including proliferating cell nuclear antigen (PCNA). Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 5791–5800. 10.1073/pnas.1920251117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.; et al. Exquisitely Specific anti-KRAS Biodegraders Inform on the Cellular Prevalence of Nucleotide-Loaded States. ACS Cent. Sci. 2021, 7, 274. 10.1021/acscentsci.0c01337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard J. R.; et al. High-Throughput Quantitative Assay Technologies for Accelerating the Discovery and Optimization of Targeted Protein Degradation Therapeutics. SLAS Discovery Adv. life Sci. R D 2021, 26, 503. 10.1177/2472555220985049. [DOI] [PubMed] [Google Scholar]
- Daniels D. L.; Riching K. M.; Urh M. Monitoring and deciphering protein degradation pathways inside cells. Drug Discovery Today: Technol. 2019, 31, 61–68. 10.1016/j.ddtec.2018.12.001. [DOI] [PubMed] [Google Scholar]
- Riching K. M.; et al. Quantitative Live-Cell Kinetic Degradation and Mechanistic Profiling of PROTAC Mode of Action. ACS Chem. Biol. 2018, 13, 2758–2770. 10.1021/acschembio.8b00692. [DOI] [PubMed] [Google Scholar]
- Riching K. M.; et al. CDK Family PROTAC Profiling Reveals Distinct Kinetic Responses and Cell Cycle-Dependent Degradation of CDK2. SLAS Discovery Adv. life Sci. R D 2021, 26, 560. 10.1177/2472555220973602. [DOI] [PubMed] [Google Scholar]
- Mayor-Ruiz C.; et al. Rational discovery of molecular glue degraders via scalable chemical profiling. Nat. Chem. Biol. 2020, 16, 1199–1207. 10.1038/s41589-020-0594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Słabicki M.; et al. The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K. Nature 2020, 585, 293–297. 10.1038/s41586-020-2374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L.; et al. Discovery of a molecular glue promoting cdk12-ddb1 interaction to trigger cyclin k degradation. eLife 2020, 9, 1–34. 10.7554/eLife.59994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers Q. L.; Gasser J. A.; Cowley G. S.; Fischer E. S.; Ebert B. L. Genome-wide screen identifies cullin-RING ligase machinery required for lenalidomide-dependent CRL4CRBN activity. Blood 2018, 132, 1293–1303. 10.1182/blood-2018-01-821769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koduri V.; et al. Targeting oncoproteins with a positive selection assay for protein degraders. Sci. Adv. 2021, 7, eabd6263 10.1126/sciadv.abd6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki R.; et al. Functional Genomics Identify Distinct and Overlapping Genes Mediating Resistance to Different Classes of Heterobifunctional Degraders of Oncoproteins. Cell Rep. 2021, 34, 108532. 10.1016/j.celrep.2020.108532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottis P.; et al. Cellular Resistance Mechanisms to Targeted Protein Degradation Converge Toward Impairment of the Engaged Ubiquitin Transfer Pathway. ACS Chem. Biol. 2019, 14, 2215–2223. 10.1021/acschembio.9b00525. [DOI] [PubMed] [Google Scholar]
- Clague M. J.; Heride C.; Urbé S. The demographics of the ubiquitin system. Trends Cell Biol. 2015, 25, 417–426. 10.1016/j.tcb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Morreale F. E.; Walden H. Types of Ubiquitin Ligases. Cell 2016, 165, 248–248. 10.1016/j.cell.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Reichermeier K. M.; et al. PIKES Analysis Reveals Response to Degraders and Key Regulatory Mechanisms of the CRL4 Network. Mol. Cell 2020, 77, 1092–1106. 10.1016/j.molcel.2019.12.013. [DOI] [PubMed] [Google Scholar]
- Mathieson T.; et al. Systematic analysis of protein turnover in primary cells. Nat. Commun. 2018, 9, 1–10. 10.1038/s41467-018-03106-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leestemaker Y.; et al. Proteasome Activation by Small Molecules. Cell Chem. Biol. 2017, 24, 725–736. 10.1016/j.chembiol.2017.05.010. [DOI] [PubMed] [Google Scholar]
- Kaiho-Soma A.; et al. TRIP12 promotes small-molecule-induced degradation through K29/K48-branched ubiquitin chains. Mol. Cell 2021, 81, 1411. 10.1016/j.molcel.2021.01.023. [DOI] [PubMed] [Google Scholar]
- Silva M. C.; et al. Targeted degradation of aberrant tau in frontotemporal dementia patient-derived neuronal cell models. eLife 2019, 8, 45457. 10.7554/eLife.45457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi D.; et al. AUTACs: Cargo-Specific Degraders Using Selective Autophagy. Mol. Cell 2019, 76, 797–810. 10.1016/j.molcel.2019.09.009. [DOI] [PubMed] [Google Scholar]
- Li Z.; et al. Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature 2019, 575, 203–209. 10.1038/s41586-019-1722-1. [DOI] [PubMed] [Google Scholar]
- Banik S. M.; et al. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 2020, 584, 291–297. 10.1038/s41586-020-2545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriwardena S. U.; et al. Phosphorylation-Inducing Chimeric Small Molecules. J. Am. Chem. Soc. 2020, 142, 14052–14057. 10.1021/jacs.0c05537. [DOI] [PubMed] [Google Scholar]
- Yamazoe S.; et al. Heterobifunctional Molecules Induce Dephosphorylation of Kinases-A Proof of Concept Study. J. Med. Chem. 2020, 63, 2807–2813. 10.1021/acs.jmedchem.9b01167. [DOI] [PubMed] [Google Scholar]
- Ramirez D. H.; et al. Engineering a Proximity-Directed O-GlcNAc Transferase for Selective Protein O-GlcNAcylation in Cells. ACS Chem. Biol. 2020, 15, 1059–1066. 10.1021/acschembio.0c00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y.; et al. Target protein deglycosylation in living cells by a nanobody-fused split O-GlcNAcase. Nat. Chem. Biol. 2021, 17, 593. 10.1038/s41589-021-00757-y. [DOI] [PMC free article] [PubMed] [Google Scholar]