Abstract
The use of enzyme-mediated reactions has transcended ancient food production to the laboratory synthesis of complex molecules. This evolution has been accelerated by developments in sequencing and DNA synthesis technology, bioinformatic and protein engineering tools, and the increasingly interdisciplinary nature of scientific research. Biocatalysis has become an indispensable tool applied in academic and industrial spheres, enabling synthetic strategies that leverage the exquisite selectivity of enzymes to access target molecules. In this Outlook, we outline the technological advances that have led to the field’s current state. Integration of biocatalysis into mainstream synthetic chemistry hinges on increased access to well-characterized enzymes and the permeation of biocatalysis into retrosynthetic logic. Ultimately, we anticipate that biocatalysis is poised to enable the synthesis of increasingly complex molecules at new levels of efficiency and throughput.
Short abstract
With advances in enabling technologies, biocatalysis has transcended early applications in fermentation to have an expanding footprint as an essential tool in the production of high value compounds.
Introduction
The utility of naturally occurring enzymes has been harnessed for thousands of years through fermentation and food preservation processes.1 Fascination with the chemistry of microbes originated at the dawn of the Neolithic era, nearly 12 000 years ago, when humans began domesticating grains and consuming alcohol, the evidence of which can be found in archeological records.2 In fact, arguments have been made that our use of alcohol produced enzymatically even predates archeological records, with evidence for ethanol-degrading enzymes present in primate species that lived before Homo sapiens.2 Driven by curiosity, people sought to understand how and why leaving cereals or grapes3 alone for some time caused them to adopt new properties, seeding fields like enzymology, molecular biology, and biocatalysis as an extension of this fascination.4
Within the past few decades, biocatalysis in fine chemical and pharmaceutical production has surged.5−7 This trend is driven in part by advances in DNA sequencing, bioinformatics, and protein engineering that allow for the identification of enzymes that meet the reactivity and selectivity needs of a given synthetic route.8 Biocatalytic reactions are now routinely used in scalable processes ranging from simple chemical manipulations such as chiral resolutions,9−11 reductive aminations,9,12 and alcohol oxidations,13 to complex, multistep chemoenzymatic cascades that enable access to high-value drug molecules on an industrial scale.14 The rapid developments in biocatalysis are also enabling a re-emergence of natural products in the current era of genomics.15 Natural products have a long history in drug discovery and development given their often potent biological activity. Still, the structural complexity of these compounds challenges chemists and demands a substantial time and resource investment to synthesize these compounds and analogs thereof.16 It seems undeniable that the next logical step in synthetic chemistry is to leverage the machinery that Nature has developed to access a similar breadth of complexity exhibited by these compounds.
Biocatalytic methods also offer several key advantages over traditional chemical processes. These advantages include increased safety and sustainability, procedural simplicity, and the tunability of an enzyme’s reactivity or selectivity through protein engineering.7 In particular, the sustainability profile of enzymatic transformations has motivated their adoption.17 In contrast to the organometallic catalysts developed using precious metals, which are being rapidly depleted from the Earth’s crust,18 enzymatic catalysts can be produced without the worry of exhausting limited resources. These advantages poise biocatalysis for adoption into the mainstream synthetic repertoire.19 It is easier now than ever before for anyone, from enzyme novices to global biotech companies, to tap into the powerful transformations biocatalysis can offer.
In this Outlook, we explore the diverse field of biocatalysis and highlight recent advances in technology that have dramatically improved the accessibility of enzymes and driven the transition from simple biocatalytic systems to sophisticated complexity-generating biocatalytic platforms. Additionally, we highlight the advantages that biocatalysts offer in organic synthesis, the current state-of-the-art in this field, and how advancing technologies will provide new opportunities for incorporating biocataytic strategies in the synthesis of target molecules.
Early Applications of Biocatalysis in Synthesis
Despite their use in the fermentation process for millennia, enzymatic methods were first appreciated on the molecular level beginning in the early 1800s20 when researchers began investigating yeasts for their fermentation abilities.21 Though it was first believed that the entire microorganism itself was functioning as the catalyst, the discovery of the first enzyme mixture, called “diastase”, fundamentally changed the field by demonstrating that observed reactions were mediated by only specific parts of the organism.22 This sparked increased interest in the then-new field of enzymology, and crucial milestones in understanding enzymes followed. These include the development of the lock and key model,23 cell-free fermentation,24 the realization that enzymes were in fact proteins,25 and the elucidation of the DNA structure;26 all of which have paved the way for modern biocatalysis (Figure 1).21 Finally, with the invention and adoption of X-ray crystallography, researchers were finally able to view the three-dimensional structure of these miraculous macromolecules in detail and gain insight into their functions and mechanisms.27 A more detailed account of the rich history of enzymology has been reviewed previously.21
Figure 1.
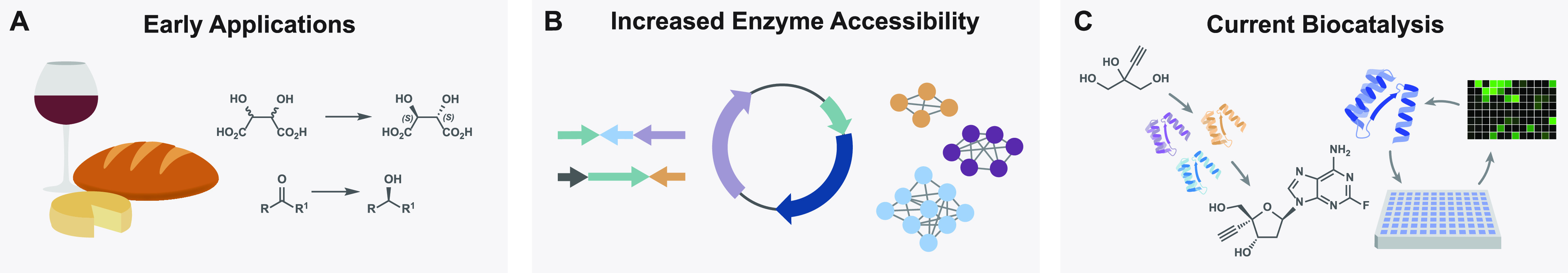
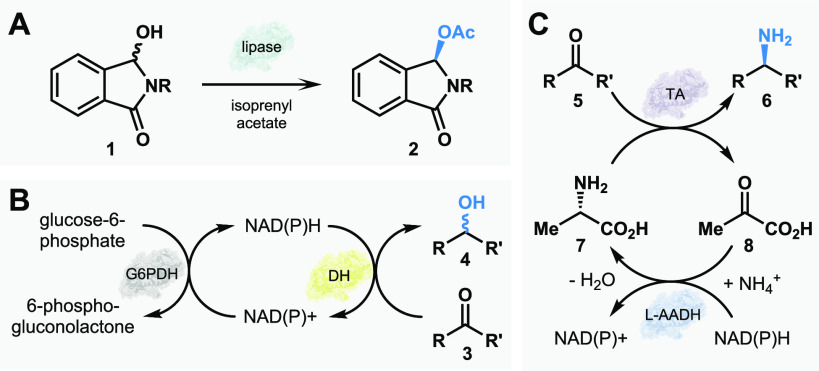
(A) Early uses of enzyme-mediated transformations, such as fermentation, chiral resolutions, and functional group interconversions. (B) Recent advances in genome sequencing, gene synthesis, and bioinformatics increase the accessibility of obtaining enzymes. (C) Select strategies in modern biocatalysis include cascades, chemoenzymatic synthesis, and enzyme evolution.
Over much of the 20th century and into the early 2000s, the use of enzymes to perform useful chemistry truly gained popularity.21 Enzyme-mediated kinetic resolutions were one of the most common initial uses of biocatalysis in synthesis. Though several different classes of enzymes have been applied to conduct these enantiomeric enrichments,9 lipases are commonly employed to affect this transformation based on their commercial availability, large substrate scope, high levels of selectivity, and cofactor-free catalysis.11 Also commonly used in the production of cheese products and laundry detergents,28 the first member of this enzyme class was discovered in 1848 by Claude Bernard in his investigation of pancreatic secretions.29 Initial experimentation with lipases in the 1930s30 and 1940s31 laid the groundwork for their use in kinetic resolutions and other biocatalytic transformations for the rest of the 1900s.11,28 A relatively recent example published by Kaga and co-workers in 2003 demonstrates the simplicity of using lipases in a more modern biocatalytic setting: to construct a small library of chiral hemiaminals through the dynamic kinetic resolution of racemic starting materials.10 The group first screened a set of commercially available lipase enzymes for acylation activity against their library of racemic N-acylhemiaminals and determined that lipase QL gave short reaction times and operated with high levels of enantioselectivity. With this enzyme, they constructed several O-acylated hemiaminals in quantitative yields and in high/exquisite enantioselectivities (Figure 2A). Lipase QL is just a select example among several others that demonstrates the early and widespread use of lipases and other kinetic resolution enzymes in academia and industry.28
Figure 2.
(A) Dynamic kinetic resolution of racemic N-acylhemiaminals by a lipase. (B) NAD(P)H recycling system developed by Wong and Whitesides. (C) Cascade system for construction of chiral amines using an ω-transaminase. Abbreviations: G6PDH glucose-6-phosphate dehydrogenase, DH dehydrogenase, TA transaminase, L-AADH l-α-amino acid dehydrogenase.
The design of systems for in situ cofactor regeneration is a significant milestone in biocatalytic method development.32 Early studies of cofactor-dependent enzymes in synthesis relied on the addition of stoichiometric quantities of these cofactors, which limited the utility of the enzymatic reaction. Thus, the ability to continuously recycle these essential components in the reaction mixture was critical to certain biocatalysts’ practical use.33 The early work of Wong and Whitesides on the regeneration of NAD(P)H in situ to enable reductions by dehydrogenase enzymes demonstrates this method’s capability and has since made an enormous impact on the field.34−36 To apply these dehydrogenases toward the construction of chiral alcohols, they developed the use of glucose-6-phosphate dehydrogenase (G6PDH) from L. mesenteroides to reduce the NAD(P)+ cofactor in situ following its oxidation by the dehydrogenase. G6PDH relies on glucose-6-phosphate, which is inexpensive and easy to synthesize, to provide the equivalent of hydride needed to reduce NAD(P)+ to NAD(P)H (Figure 2B). With this recycling system in place, Wong and Whitesides completed the biocatalytic generation of optically pure D-lactic acid, threo-Ds(+)-isocritic acid, and (S)-benzyl-α-d1 alcohol.34 This early example of a biocatalytic cascade has since enabled the use of many enzyme-catalyzed reductions and has paved the way for application on an industrial scale.37
Since this preliminary work, methods relying on electrochemistry, photochemistry, and other hydride donor/acceptor systems have been developed. For example, readily available reagents like isopropanol have been used in cofactor regeneration systems, providing an alternative to the more expensive sugars used previously.32 A variety of more economical and industrially feasible sacrificial functional group donors have also been applied to improve efficiency, scalability, and ease of use of cofactor regeneration.38,39 Several reports describe the use of isopropyl amine as an amino donor for transamination reactions, providing a substitute for the cost prohibitive amino acids used conventionally.40,41 There is also a focus on the construction and regeneration of synthetic, biomimetic cofactors, which holds promise for increasing the effectiveness of these systems further.42
The rapid adoption of recycling system methods allowed for the broad application of biocatalytic reduction reactions.37 The use of transaminases for constructing chiral amines is a prime example, and their utility in synthesis has been showcased in the synthesis of drug molecules such as sitagliptin.43,44 Transaminases offer many advantages over their chemical counterparts, including improved stereoselectivity, mild reaction conditions, and reducing the reliance on harmful solvents and transition metals.43,45 A notable example of the application of ω-transaminases, a subgroup of transaminases that has drawn particular attention in the pharmaceutical industry,46 was developed by Koszelewski and co-workers. Ultimately relying on a system similar to that produced by Wong and Whitesides to power catalysis, the Koszelewski group constructed nine chiral amines with excellent enantiomeric excess through a reductive amination with the commercial ω-transaminase ATA-113.12 They also employed a second enzyme, l-α-amino acid dehydrogenase (L-AADH), to further streamline this reaction by regenerating the amino acid alanine in situ, which is the amine source for the transamination reaction. L-AADH, enabled by the NAD(P)H recycling system, utilizes an equivalent of ammonium as the ultimate nitrogen source to reduce pyruvate to the desired alanine (Figure 2C). Albeit a relatively simple transformation by today’s standards, this early work serves as a quintessential example of synthetic utility of transaminases.
The seminal work highlighted here, alongside other early examples of simple biocatalytic reactions such as isomerizations, redox manipulations, and ligations,47 brought to light the power of enzymes as catalysts in synthesis. In the more modern history of biocatalysis, there has been a paradigm shift from using enzymes to construct relatively simple building blocks or provide chiral intermediates for traditional syntheses,48 to relying on them for late-stage synthetic modifications,49 combining molecule fragments toward value-added compounds, and conducting multistep, biocatalytically mediated total syntheses.14,37 Additionally, the tools for investigating and leveraging biocatalysts for synthetic uses have reached a stage where they are widely accessible to the chemistry community: obtaining the knowledge and equipment needed for biocatalysis can be accomplished with just a few clicks.
Accessibility of Biocatalysis to Synthetic Chemists
Once relegated to the fields of biochemistry and molecular biology, recent advances in bioinformatics,50 DNA sequencing,51 protein engineering,52 and DNA synthesis have made it possible for virtually anyone to take advantage of enzymatic catalysts and tailor them to their own needs. The process of identifying, producing, isolating, and tuning the reactivity of biocatalysts for desired transformations is as accessible to synthetic chemists as obtaining and using small molecule catalysts. In particular, the recent exponential growth in annotated protein sequences available in online databases has created an enormous catalog of potential enzymes to serve many synthetic needs. Two of the most popular databases, UnitProt53 and Genbank,54 now house information on more than 420 000 individual species, representing over one billion total sequence records. Instead of taking to the field and collecting specimens by hand to examine their genes, these databases store a wealth of information on protein sequence and origin and are a valuable starting point for anyone looking to identify enzymes for a given synthetic purpose.55
Combining the vast amount of data stored in these online libraries with bioinformatic tools allows one to begin making predictions about the function of uncharacterized or “hypothetical” proteins,56 and to search for previously identified proteins that may also demonstrate activity in a noncanonical transformation.57 For example, the basic local alignment search tool (BLAST) is one of the most popular and easy to use for this type of analysis.58,59 Gaining popularity in the early 1990s and now available to use for free on the National Center for Biotechnology Information (NCBI) Web site,59 this tool relies on algorithms to search available online databases for protein sequences that resemble a given input sequence. By feeding the BLAST search engine a known nucleotide or amino acid sequence, or a protein identifier such as an accession number, the tool can align all known protein sequences that share similarity with the input sequence and rank them in a list. As minute changes in the order or position of amino acid residues can drastically alter function between homologous proteins with highly similar sequences, this type of search can be advantageous when trying to identify enzymes with improved stability and activity, complementary substrate scopes, or proteins that can perform desired transformations with the alternative site- and/or stereoselectivity to the one used to build the query.36,60,61 This tool also provides known information about each sequence, such as the originating organism and any characterized metabolic function of the protein within said organism. By displaying data on the degree of similarity between proteins based on how well their sequences align, a user can quickly identify any known proteins that may share functional characteristics with the input protein sequence.62
Albeit a useful starting point, this list format provided by BLAST can become cumbersome when the search yields thousands of potentially related protein sequences. To obtain a more comprehensive view of entire protein families, some of which can contain hundreds of thousands of proteins,63 tools have been developed that provide greater context for viewing connections within these groups. Phylogenetic trees are commonly used to examine relationships between homologous proteins and study changes in protein families over their evolution.64 This bioinformatic analysis technique relies on the alignment of homologous protein sequences to construct a visual representation of the evolutionary history of the related sequences in a phylogenetic tree (Figure 3A).64 Building and visualizing these trees has also been simplified by programs like Molecular Evolutionary Genetics Analysis (MEGA)65,66 and Ensembl67 that provide straightforward user interfaces. Once various algorithms and search tools are applied to analyze all available data and establish the most likely configuration, the trees can be examined to draw conclusions about relatedness among protein families and test hypotheses about their evolutionary origins.68,69
Figure 3.
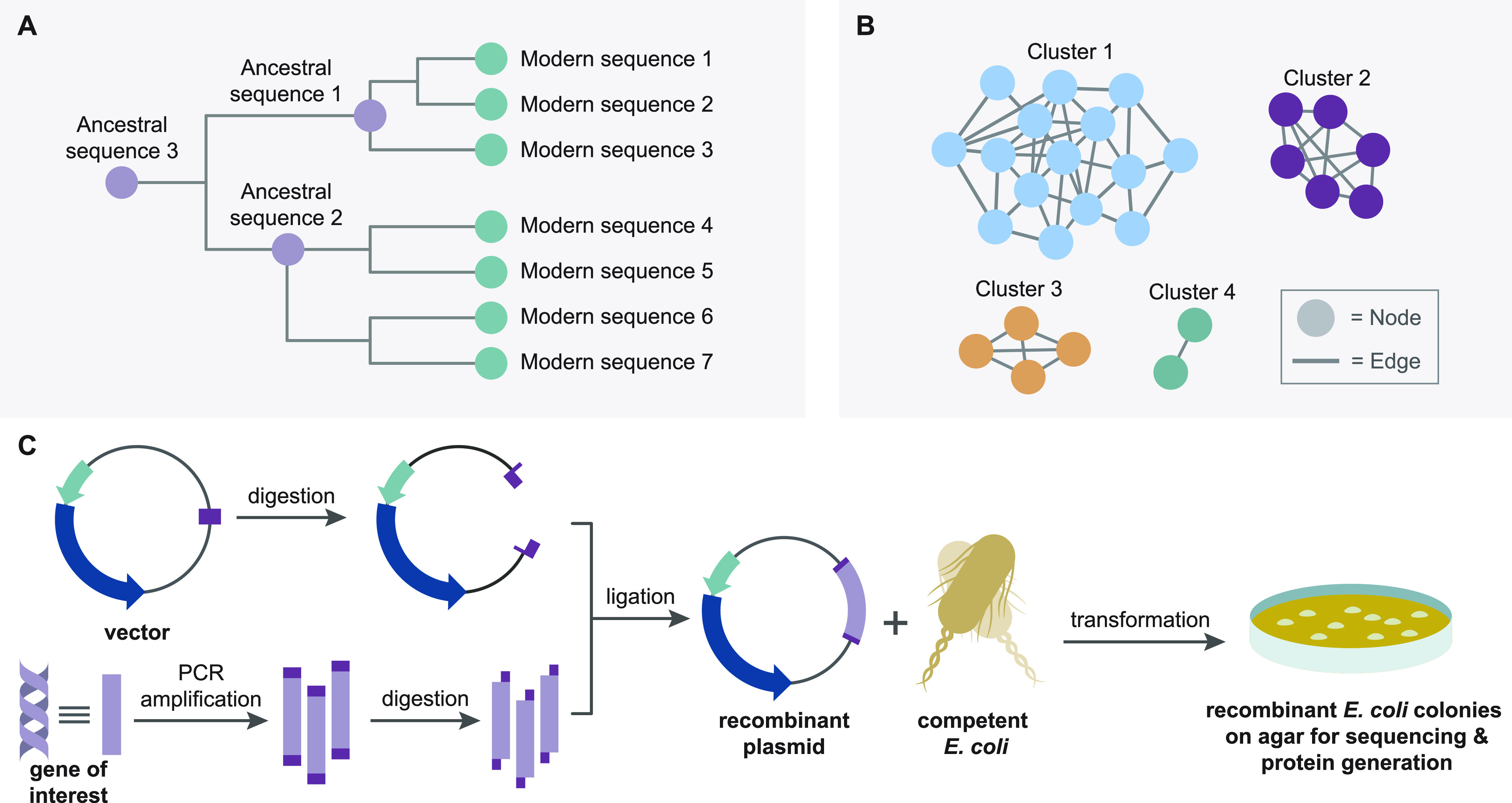
(A) Conceptual phylogenetic tree depicting locations of calculated ancestral sequences. (B) Conceptual SSN demonstrating nodes, edges, and clusters. (C) Workflow for a traditional cloning procedure.
For example, one intriguing use of phylogenetic analyses in the context of biocatalysis is the identification and reconstruction of ancestral protein sequences (Figure 3A) that can offer benefits in stability and biocatalytic activity over their modern “offspring”.70 This technique relies on software to compare related protein sequences that most likely evolved from a common ancestor to calculate or “infer” the exact sequence of that ancestral protein.71 The ability to now obtain any DNA sequence quickly and easily makes reconstructing ancestral proteins a potentially powerful tool in identifying novel enzymes with desirable functions. To this effect, Furukawa et al. have identified an ancestor of 3-isopropylmalate dehydrogenase (IPMDH), a key enzyme in the biosynthesis of leucine, which offers improvements in its stability and activity over extant IPMDH enzymes from present-day organisms through construction and analysis of a phylogenetic tree.72 Following inference and identification of two ancestral protein sequences, dubbed ancIPMDH-IQ and ancIPMDH-ML, the group successfully expressed each protein in E. coli and, after isolating the enzymes for further investigation, discovered they provided increased thermal stability and improved catalytic activity at low temperatures compared to their modern homologues.72 This work demonstrates just one of many potential uses for ancestral protein reconstruction, as other reports describe how ancestral proteins might possess higher degrees of substrate promiscuity compared to their modern offspring, thus offering potentially valuable characteristics to organic chemists seeking diverse and novel bond-forming activity.73
Despite their utility and newfound ease-of-use, phylogenetic trees can still prove overwhelming when examining extensive protein families or groups of sequences.74,75 Tools like sequence similarity networks (SSNs) have emerged to help overcome these challenges. SSNs have gained much attention since its introduction to the bioinformatics community in 2003.76 It provides a way to visualize family wide relationships and patterns in large groups of protein sequences by ranking sequences in “clusters” based on their alignment scores.74−77 These networks comprise groups of “nodes,” representing a protein sequence or group of sequences. These nodes are then connected by lines called “edges”, representing a threshold for sequence similarity that can be set by the user (Figure 3B). Changing this score controls which nodes group together, allowing for inferences to be made about protein structure and functions by examining and comparing the location of nodes within the clusters.77 These networks can be constructed and analyzed quickly and easily through a web-based tool called EFI-EST75 and the free-to-download software Cytoscape.76 Helpful tutorials and videos on how to construct, use, and manipulate SSNs with these programs are also available for free online.75,76
These networks can be beneficial for chemists looking to identify new enzymes for catalysis from families with a limited number of previously characterized proteins. Lewis and co-workers have recently applied SSNs to identify and profile novel flavin-dependent halogenase (FDH) enzymes.78 Using these networks to guide their search, the group elected 128 initial halogenase sequences to sample for useful halogenation activity. Following expression of the genes, they obtained 87 soluble proteins for preliminary activity screens with 12 initial substrates containing a mixture of phenols, indoles, and anilines. Overall, the group identified 39 previously uncharacterized halogenases that demonstrated unique bromination and/or chlorination activity against the substrate panel. After examining an additional 50 complex and bulky substrates, they discovered at least one member of their halogenase library that demonstrated activity with around 48% of the substrates tested. Ultimately, Lewis and co-workers examined and characterized the preference for these FDHs toward bromination and chlorination, their site-selectivity, and thermostability and could draw further conclusions about trends in their SSNs through this family wide profiling.78 This cutting-edge application of SSNs demonstrates how free and straightforward Internet-based software can be used to identify synthetically tractable biocatalysts without the need to perform more complex mutagenesis and directed evolution experiments.
Our group has also demonstrated the applicability of SSNs to examine previously uncharacterized enzymes with useful chemical functions.36,74 We sought to identify homologous flavin-dependent monooxygenase (FDMO) proteins to investigate the factors that control their site and facial selectivity in an oxidative dearomatization reaction and to identify enzymes suitable to enable a stereodivergent chemoenzymatic natural product synthesis campaign.36 Analysis of an SSN comprised of over 45 000 sequences from the flavin adenine dinucleotide (FAD) binding domain protein family (pfam01494) identified several FDMOs that are highly similar to those our group had investigated previously.35 Combining the experimental data gained from reactions of these enzymes in a model system with comparisons of their sequence information and location in the SSN allowed us to identify trends in the SSN that predict the site-selectivity of a putative FDMO based on which cluster it is located in. We envisioned this technique may also help predict the stereoselectivity of the dearomatization mediated by a given FDMO, but further studies suggest that this is much more finely controlled than what can be predicted by a precursory SSN. Additional studies suggested two key active site residues are crucial in controlling the stereochemical outcome of the dearomatization reaction known to these proteins.36,60 Though this does highlight a potential drawback of using SSNs in this way, the tool did ultimately demonstrate its utility in identifying other catalytically active proteins with desired activity. Work is currently underway to further characterize these enzymes in hopes of expanding our library of biocatalysts.
Before developing these tools for identifying and characterizing enzymes in silico, obtaining biocatalysts for chemical experimentation was a significant challenge. To investigate a wild-type or naturally occurring catalytic protein, a molecular biologist would first need to get the source DNA or RNA encoding the gene of interest from the native organism. Following isolation, it is necessary to amplify the DNA fragment through a polymerase chain reaction (PCR).79 These amplified fragments must then be digested with restriction enzymes and ligated into a circular piece of DNA called a plasmid that has also been prepared with the same enzymes to ensure the ends of these sequences are compatible.80 Inserting DNA into a vector such as this not only allows for the host organism to uptake the gene of interest but can also be used to impart properties like antibiotic resistance to the transfected cells to allow for the selection of individual cells that have successfully incorporated the plasmid. Following digestion, the prepared DNA fragment and cut vector are then combined in the presence of a DNA ligase enzyme, which efficiently joins the compatible ends of the fragment and vector, resulting in the production of a so-called “recombinant plasmid”.81 In the case of transforming E. coli, one of the most popular and easy-to-use host organisms for recombinant protein production, the recombinant plasmid is then added to competent bacterial cells (cells that are primed to uptake foreign DNA from their surroundings). The cells can then be grown on agar media possessing an antibiotic to prevent cells that do not contain the plasmid from growing. After allowing the cells to grow on the agar, a colony can be harvested and analyzed to ensure that it possesses the desired gene. Finally, after ensuring the gene is present and contains the correct sequence, the colony can be used to seed a larger culture to harvest usable amounts of the desired protein, as well as to produce more of the plasmid for additional studies or to transfect new cells with the desired gene without having to undergo the entire process from scratch (Figure 3C).81
In contrast to these traditional cloning techniques, technological breakthroughs in modern gene synthesis provide a highly streamlined process for chemists seeking DNA sequences and plasmids. Instead of using isolated DNA from native organisms as a template to amplify, solid-phase oligonucleotide synthesis allows for the de novo construction of any nucleotide sequence found online from individual nucleotide bases.82 Companies now offer customized DNA constructs for purchase on-demand: input your insert sequence of interest and choose the desired vector in their online interface, and the company will ship you a ready-to-use recombinant plasmid possessing your exact gene, or even a sample of host organisms containing the plasmid, in a matter of weeks. The cost is dependent on the number of base pairs in the DNA sequence and the particular plasmid desired, but these DNA constructs can typically be purchased for under 200 USD. Not only does this save time and effort in obtaining the recombinant vector, it also allows for nearly anyone to take advantage of this technology without the need for specialized equipment, reagents, and knowledge required for traditional cloning. Inexpensive and straightforward methods, reagents, and equipment for transforming, growing, and isolating recombinant protein from cells containing a mail order plasmid also lower the barrier for individuals and laboratories looking to enter the field of biocatalysis.83,84
These tools and techniques described above barely scratch the surface of what is available for anyone interested in using and tuning biocatalysts for a particular synthetic application.75,85,86 Advances in the fields of directed evolution52 and computer-guided enzyme engineering87 promise to construct enzymes with ever-greater efficiency, selectivity, stability, and reusability than those known today. Leveraging combinations of these strategies have already begun to provide highly applicable and useful biocatalysts to the synthetic community at large and will continue to improve biocatalytic methods as they are developed further.
State-of-the-Art Biocatalysis
Following this explosion of interest in enzyme-mediated catalysis, biocatalytic reactions are now increasingly employed in complex molecule synthesis. Biocatalytic methods that affect late-stage site- and stereoselective C–H functionalization constitute one of the best state-of-the-art transformations available today that maximize step efficiency and enable diversification of complex scaffolds. Select examples of biocatalytic C–H functionalization in complex molecule synthesis are shown in Figure 4A. Sherman and co-workers have carried out a late-stage hydroxylation of the macrolide natural product M-4365 G1 (9) to form antibiotic juvenimicin B1 (10) with P450 monooxygenase TylI.88 Late-stage biocatalytic C–H hydroxylation has also been explored in the pursuit of steroid-based drugs.89,90 Zhou and co-workers developed a biocatalytic C19 hydroxylation of cortexolone (11) to form 19-hydroxycortexolone (12) using TcP450-1, a cytochrome P450 enzyme.90 This strategy enables direct access to bioactive C19-hydroxylated steroids.90 It is worth mentioning that direct hydroxylation at the C19 position of steroids is extremely challenging using traditional chemical methods.91−93 Our research group’s long-standing interest in using enzymes to carry out C–H hydroxylation reactions has been channeled for the late-stage diversification of paralytic shellfish toxins.94−97 We have employed the Rieske oxygenase SxtT to carry out the site- and stereoselective hydroxylation of β-saxitoxinol (13), directly generating saxitoxin (14).95 The Renata group recently disclosed a nonheme iron (NHI) dependent enzymatic platform to enable late-stage biocatalytic hydroxylation of complex terpene scaffolds.98 The enzyme P450BM3 MERO1M177A was employed in carrying out selective C–H hydroxylation to form the oxidized terpene product 15.98 Direct C–H hydroxylation has also been developed for amino acid scaffolds. For example, Zaparucha discovered the NHI enzyme KDO1-3 that carried out selective hydroxylation of l-lysine.99,100 The enzymes KDO1 and KDO2/3 selectively hydroxylated the C3 and C4 positions of l-lysine, and the enzyme KDO3 carried out C4 hydroxylation of a pre-C3-hydroxylated l-lysine.99 Renata and co-workers employed the KDO1 mediated C3-selective hydroxylation of l-lysine in their total synthesis of tambromycin.101
Figure 4.
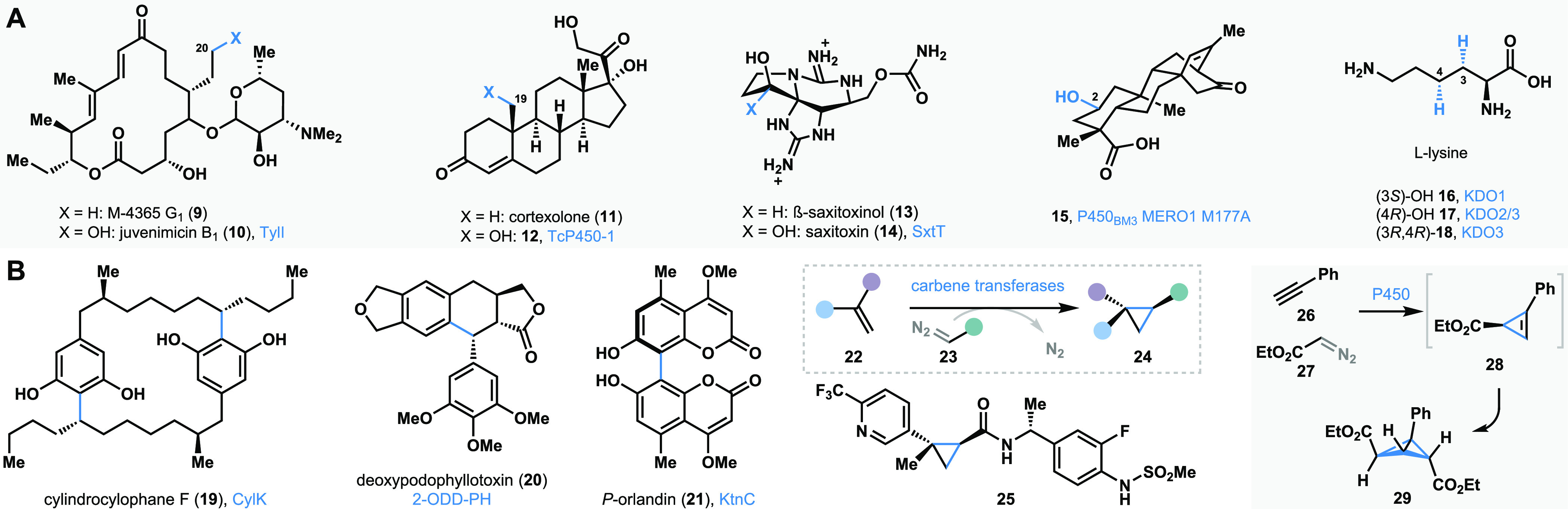
Biocatalysis in complex molecule synthesis: (A) selected C–H functionalization reactions. (B) Selected C–C bond forming reactions.
Rapid advances in biocatalysis have resulted in the identification of enzymes that can carry out carbon–carbon (C–C) bond-forming reactions (select examples in Figure 4B).102 Balskus and co-workers reported the enzyme CylK that carries out biocatalytic intermolecular Friedel–Crafts alkylation of two halogenated resorcinol derivatives to construct the cylindrocyclophane 19.103 The enzyme CylK has also been shown to be highly promiscuous, carrying out alkylation of a variety of resorcinol derivatives with secondary alkyl halides.104 Biocatalytic Friedel–Crafts alkylation has also been carried out to synthesize podophyllotoxin lignans.105,106 For example, the NHI enzyme 2-ODD-PH has been utilized to carry out the biocatalytic synthesis of deoxypodophyllotoxin (20) and related analogs.106−108 Biocatalytic oxidative phenolic coupling reactions are emerging as powerful tools to construct complex molecules.109−111 The Müller group recently reported fungal P450 enzymes capable of carrying out oxidative coupling of coumarin derivatives in a site- and stereoselective manner.109 For example, the enzyme KtnC catalyzes the synthesis of the bicoumarin P-orlandin (21).109 Biocatalytic C–C bond formation has been explored in carbene transfers to generate chiral cyclopropanes.112−114 Arnold and co-workers first reported an engineered P450BM3 that carried out carbene transfer reactions. Diazoacetate reagents were used as the carbene sources to carry out alkene cyclopropanation.112 Several other groups have contributed to the development of biocatalytic carbene transfer reactions, and these have been applied toward the synthesis of pharmacologically relevant compounds such as the TRPV1 inhibitor 25.115,116 Biocatalytic carbene transfer reactions can be extended to alkynes as well, where the first carbene transfer generates a cyclopropene product which is primed for a second carbene transfer reaction to generate stereopure bicyclobutane products.117 This transformation rivals the best of what synthetic chemistry has to offer in terms of building complexity through C–C bond formation.
In the case of selective C–H functionalization and C–C bond-forming reactions, biocatalysis is often employed at an advanced stage or in the final step of a synthetic campaign. Alternatively, biocatalysis can be engaged at an early stage in chemoenzymatic synthesis planning (Figure 5). In such cases, the product of a biocatalytic reaction is transformed into a target molecule of interest using modern synthetic organic chemistry tools. This strategic merge of biocatalysis and small molecule-based synthetic methods enables access to chemical scaffolds previously unattainable using traditional chemical methods alone. For example, Renata and co-workers developed a chemoenzymatic total synthesis of the natural product manzacidin C (32).118,119 The NHI-dependent enzyme GriE was employed to carry out selective hydroxylation of an l-leucine derivative 30 to form 31.118 The product 31 was taken through established synthetic steps to formally assemble manzacidin C (32).118 Our group has been interested in the hydroxylative dearomatization of resorcinol compounds using flavin-dependent monooxygenases (FDMOs).35,36 We have employed the site- and stereoselectivity of FDMOs in conjunction with small-molecule-based methods to enable the total synthesis of azaphilone natural products.36 For example, the enzyme AzaH was used to carry out the dearomatization of resorcinol 33 to form 34. The quinol product 34 was subsequently transformed to (S)-trichoflectin (35) using chemical methods.36 Our group has also focused on developing benzylic hydroxylation of o-cresol compounds using NHI-dependent monooxygenases.120 For example, we have employed the enzyme ClaD to carry out benzylic hydroxylation of resorcinol derivative 36, the product of which (37) undergoes spontaneous loss of water resulting in a biocatalytically generated o-quinone methide, which was trapped using a chiral dienophile to construct the bioactive natural product xyloketal D (38).120 α-Deuterated amino acids are important building blocks toward the synthesis of labeled pharmaceuticals and biological probes; however, traditional methods to access these compounds often require protecting group manipulations121 and can be difficult to perform in a stereoselective manner.122 We discovered that SxtA AONS, α-oxoamine synthase evolved for saxitoxin biosynthesis, is capable of deuterating a range of unprotected amino acids and their methyl esters using D2O as the deuteron source. For example, deuteration of alanine methyl ester (39) resulted in 40, which was subsequently transformed using chemical methods to access the deuterium-labeled Parkinson’s pharmaceutical safinamide (41).
Figure 5.
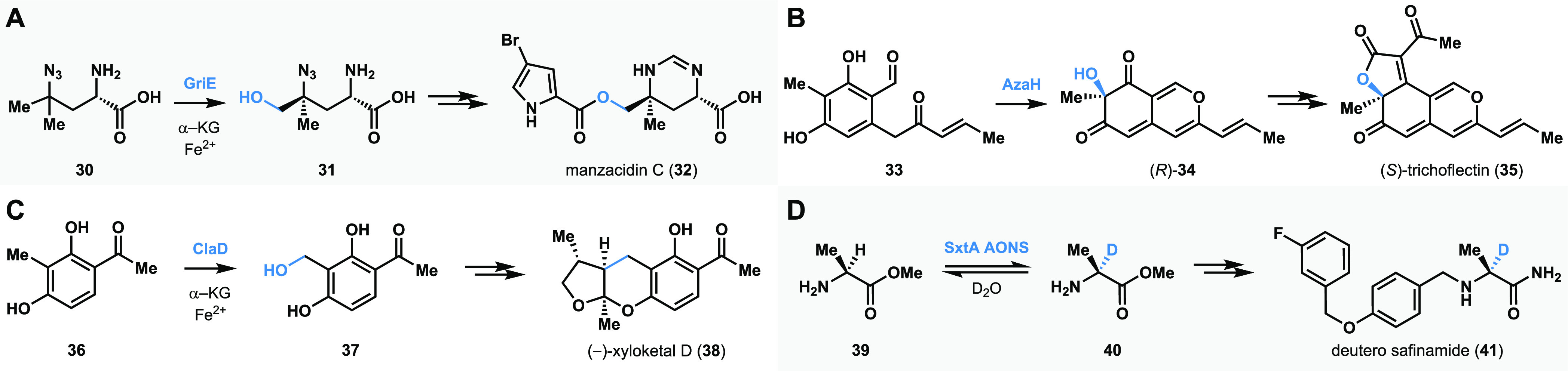
Chemoenzymatic sequences to complex molecules. (A) Amino-acid C–H hydroxylation in the synthesis of manzacidin C. (B) Hydroxylative dearomatization in the synthesis of azaphilone natural products. (C) Benzylic hydroxylation en route to xyloketal D synthesis. (D) Alpha deuteration of amino acids in the formation of deutero safinamide.
Multienzyme cascade reactions have been developed in industrial and academic laboratories to enable complex molecule synthesis (select examples in Figure 6). The process toward HIV treatment drug islatravir (48) developed by Merck and Codexis is a representative example of a multienzyme cascade employed on an industrial scale.14 The artificial nucleoside islatravir (48) was constructed using a combination of five enzymes from the nucleoside salvage pathway in bacteria, which were each engineered for a distinct purpose.14 This protecting group-free cascade yielded the product islatravir in markedly higher yields than previous chemical syntheses.14,123 Moore and co-workers developed a multienzyme synthesis of complex halogenated bacterial meroterpenoids napyradiomycins A1 and B1 (54 and 55) in a single pot.124 Starting with three organic substrates (tetrahydroxynaphthalene 49, dimethylallylpyrophosphate, and geranyl pyrophosphate), the team developed a catalytic sequence involving five enzymes: two aromatic prenyltransferases (NapT8 and T9) and three vanadium dependent haloperoxidase (VHPO) homologues (NapH1, H3, and H4) to assemble the complex halogenated metabolites in milligram quantities.124 Our group has leveraged the exquisite reactivity of FDMOs and NHI-dependent monooxygenases to construct tropolone natural products.35,125 Tropolones are a structurally diverse class of bioactive molecules that are characterized by a cycloheptatriene core bearing an α-hydroxyketone functional group. We developed a two-step, biocatalytic cascade to the tropolone natural product stipitatic aldehyde starting with the resorcinol 56. Hydroxylative dearomatization of 56 using TropB affords the quinol intermediate 57. The quinol intermediate undergoes oxidation by an α-KG dependent NHI enzyme TropC to form a radical intermediate which undergoes a net ring rearrangement to form stipitatic aldehyde 59.
Figure 6.
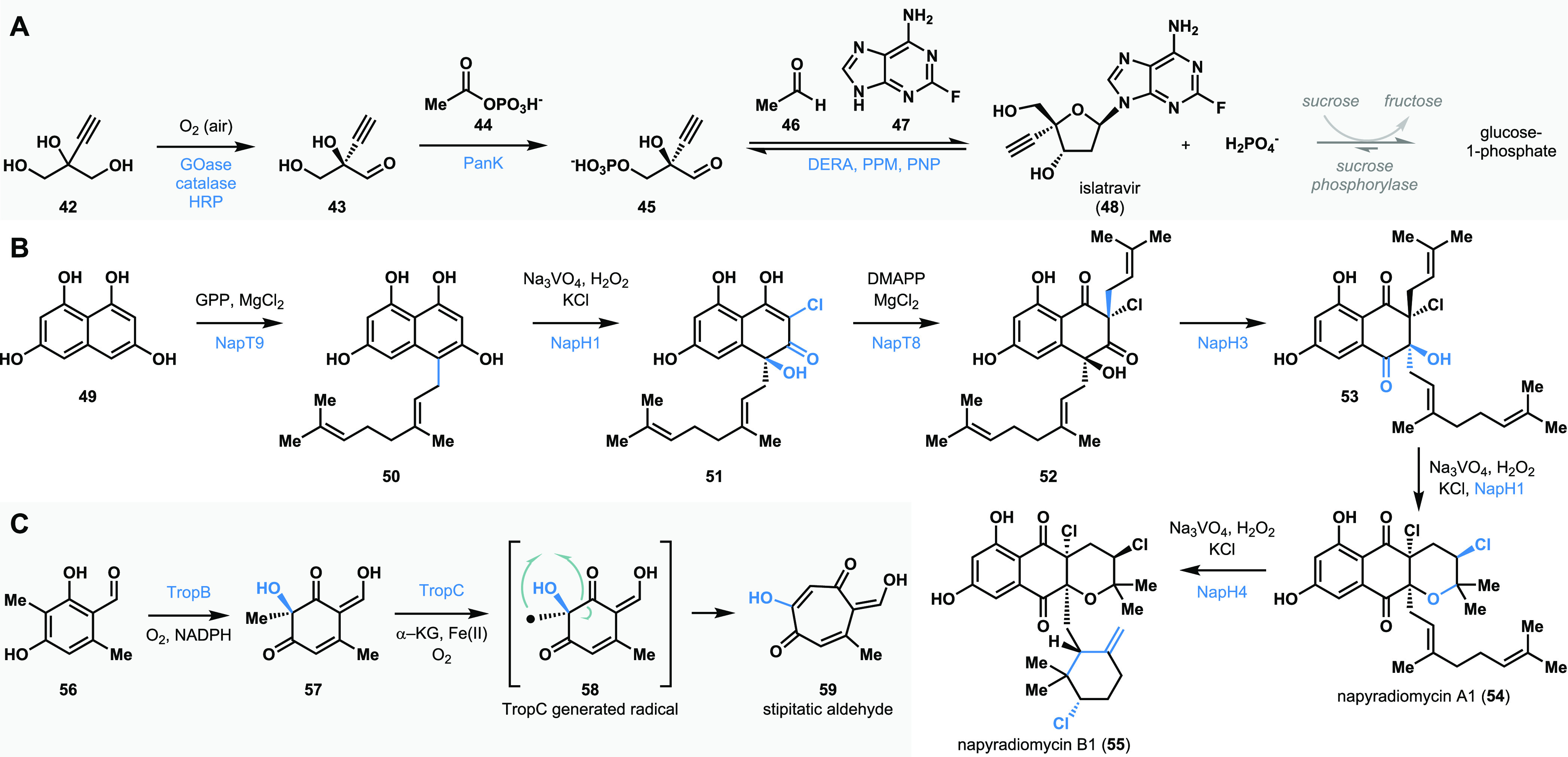
Multienzyme biocatalytic sequences: (A) Merck’s biocatalytic synthesis of islatravir. (B) Multienzyme synthesis of napyradiomycin A1 and B1. (C) Multienzyme sequence toward the synthesis of tropolone stipitatic aldehyde.
Biocatalytic methods are poised to significantly expand the repertoire of transformations possible in an organic chemist’s toolbox, allowing greater access to chemical space than previously possible. This creates an incentive for academic and industrial laboratories to embrace biocatalytic methods. As interest in this field continues to grow, it will most certainly inform the retrosynthetic logic of modern organic synthesis and shape the next generation of methods.
Outlook and Conclusion
New technology and approaches in biocatalysis continue to pave the way for innovation and paint a bright future for this field. Enzymatic catalysis has demonstrated utility in the construction of simple molecules and holds promise for expanding synthetic access to new corners of chemical space. The rapid technological advances surrounding biocatalyst discovery, characterization, and application naturally raises the question as to what comes next in the field. We anticipate that the amenability of biocatalysis to high-throughput experimentation will shape the application of enzymatic catalysis in synthesis. For example, we envision generation of compound libraries in plates will be possible through biocatalysis. Considering the benign nature of biocatalytic reactions, we anticipate biocatalytically generated compound libraries can be directly coupled with biological assays as well, matching the pace of compound generation with established high-throughput biological assays to ultimately accelerate drug discovery.126,127
Continued progress in biocatalysis would benefit combinatorial platforms for the synthesis of small-molecule-based compound libraries. The idea of combinatorial biocatalysis platforms for library synthesis has been around since the early 2000s; however, its widespread adoption has been hindered by the lack of resources to identify and develop promiscuous catalytic enzymes.128,129 Combinatorial biocatalytic syntheses are now taking shape with recent advances in contemporary organic chemistry, synthetic biology, and bioinformatics. In addition, studies of enzyme cocktails have shown that biocatalysts can operate synergistically to complement each other’s substrate scopes, creating useful catalyst mixtures to perform sequential chemical transformations.130,131 With this precedent, as well as equipment for high-throughput experimentation becoming more advanced and commonplace,126 it seems only a matter of time before the high-throughput synthesis of vast and diverse small molecule libraries mediated by combinatorial biocatalysis is realized.
Without question, biocatalysis has become a valued approach in modern organic synthesis126 and is a methodology we will rely heavily on as the need to develop green alternatives in chemistry grows.17,132 With the rapid advances in the field over the past few decades and the wealth of sequence data now widely available, biocatalytic methods are more accessible than ever before. As the global community adapts these techniques to their individual needs, new ideas and strategies will take hold and continue to push biocatalysis into the forefront of synthetic chemistry.
Acknowledgments
A.R.H.N. thanks the National Institutes of Health R35 GM124880 for support. E.O.R. acknowledges support from the NSF graduate research fellowship (DGE 1841052). J.B.P. acknowledges support from a Ruth L. Kirschstein National Research Service Award (1F31GM139387-01). The authors thank Dr. Kendrick Smith and Lara Zetzsche for helpful discussion and feedback on this manuscript.
The authors declare no competing financial interest.
References
- Bornscheuer U. T.; Buchholz K. Highlights in Biocatalysis - Historical Landmarks and Current Trends. Eng. Life Sci. 2005, 5, 309–323. 10.1002/elsc.200520089. [DOI] [Google Scholar]
- Dominy N. J. Ferment in the family tree. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 308–309. 10.1073/pnas.1421566112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers P. J.; Pretorius I. S. Fermenting knowledge: the history of winemaking, science and yeast research. EMBO Rep. 2010, 11, 914–920. 10.1038/embor.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland R. A. A Brief History of Enzymology. Enzymes: A practical introduction to structure, mechanism, and data analysis 2000, 1–10. 10.1002/0471220639.ch1. [DOI] [Google Scholar]
- Hughes G.; Lewis J. C. Introduction: Biocatalysis in Industry. Chem. Rev. 2018, 118, 1–3. 10.1021/acs.chemrev.7b00741. [DOI] [PubMed] [Google Scholar]
- Devine P. N.; Howard R. M.; Kumar R.; Thompson M. P.; Truppo M. D.; Turner N. J. Extending the application of biocatalysis to meet the challenges of drug development. Nat. Rev. Chem. 2018, 2, 409–421. 10.1038/s41570-018-0055-1. [DOI] [Google Scholar]
- Truppo M. D. Biocatalysis in the Pharmaceutical Industry: The Need for Speed. ACS Med. Chem. Lett. 2017, 8, 476–480. 10.1021/acsmedchemlett.7b00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelraheem E. M. M.; Busch H.; Hanefeld U.; Tonin F. Biocatalysis explained: from pharmaceutical to bulk chemical production. React. Chem. Eng. 2019, 4, 1878–1894. 10.1039/C9RE00301K. [DOI] [Google Scholar]
- Shin J. S.; Kim B. G.; Liese A.; Wandrey C. Kinetic resolution of chiral amines with omega-transaminase using an enzyme-membrane reactor. Biotechnol. Bioeng. 2001, 73, 179–87. 10.1002/bit.1050. [DOI] [PubMed] [Google Scholar]
- Sharfuddin M.; Narumi A.; Iwai Y.; Miyazawa K.; Yamada S.; Kakuchi T.; Kaga H. Lipase-catalyzed dynamic kinetic resolution of hemiaminals. Tetrahedron: Asymmetry 2003, 14, 1581–1585. 10.1016/S0957-4166(03)00313-6. [DOI] [Google Scholar]
- de Miranda A. S.; Miranda L. S. M.; de Souza R. O. M. A. Lipases: Valuable catalysts for dynamic kinetic resolutions. Biotechnol. Adv. 2015, 33, 372–393. 10.1016/j.biotechadv.2015.02.015. [DOI] [PubMed] [Google Scholar]
- Koszelewski D.; Lavandera I.; Clay D.; Guebitz G. M.; Rozzell D.; Kroutil W. Formal Asymmetric Biocatalytic Reductive Amination. Angew. Chem., Int. Ed. 2008, 47, 9337–9340. 10.1002/anie.200803763. [DOI] [PubMed] [Google Scholar]
- Akoh C. C.; Chang S.-W.; Lee G.-C.; Shaw J.-F. Biocatalysis for the Production of Industrial Products and Functional Foods from Rice and Other Agricultural Produce. J. Agric. Food Chem. 2008, 56, 10445–10451. 10.1021/jf801928e. [DOI] [PubMed] [Google Scholar]
- Huffman M. A.; Fryszkowska A.; Alvizo O.; Borra-Garske M.; Campos K. R.; Canada K. A.; Devine P. N.; Duan D.; Forstater J. H.; Grosser S. T.; et al. Design of an in vitro biocatalytic cascade for the manufacture of islatravir. Science 2019, 366, 1255–1259. 10.1126/science.aay8484. [DOI] [PubMed] [Google Scholar]
- Expanding biocatalysis for a sustainable future. Nat. Catal. 2020, 3, 179–180. 10.1038/s41929-020-0447-8 [DOI] [Google Scholar]
- Hayler J. D.; Leahy D. K.; Simmons E. M. A Pharmaceutical Industry Perspective on Sustainable Metal Catalysis. Organometallics 2019, 38, 36–46. 10.1021/acs.organomet.8b00566. [DOI] [Google Scholar]
- Chakrabarty S.; Wang Y.; Perkins J. C.; Narayan A. R. H. Scalable biocatalytic C−H oxyfunctionalization reactions. Chem. Soc. Rev. 2020, 49, 8137–8155. 10.1039/D0CS00440E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty S.; Romero E. O.; Pyser J. B.; Yazarians J. A.; Narayan A. R. H. Chemoenzymatic Total Synthesis of Natural Products. Acc. Chem. Res. 2021, 54, 1374–1384. 10.1021/acs.accounts.0c00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance H.; Wetzel S.; Kumar K.; Waldmann H. Charting, Navigating, and Populating Natural Product Chemical Space for Drug Discovery. J. Med. Chem. 2012, 55, 5989–6001. 10.1021/jm300288g. [DOI] [PubMed] [Google Scholar]
- Reetz M. T. Biocatalysis in Organic Chemistry and Biotechnology: Past, Present, and Future. J. Am. Chem. Soc. 2013, 135, 12480–12496. 10.1021/ja405051f. [DOI] [PubMed] [Google Scholar]
- Heckmann C. M.; Paradisi F. Looking Back: A Short History of the Discovery of Enzymes and How They Became Powerful Chemical Tools. ChemCatChem 2020, 12, 6082–6102. 10.1002/cctc.202001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong E. F. Enzymes: A Discovery and its Consequences. Nature 1933, 131, 535–537. 10.1038/131535a0. [DOI] [Google Scholar]
- Fischer E. The influence of configuration on enzyme activity (Translated from German). Ber. Dtsch. Chem. Ges. 1894, 27, 2985–2993. 10.1002/cber.18940270364. [DOI] [Google Scholar]
- Kohler R. The Background to Eduard Buchner’s Discovery of Cell-Free Fermentation. J. Hist. Bio. 1971, 4, 35–61. 10.1007/BF00356976. [DOI] [PubMed] [Google Scholar]
- Sumner J. B. The Isolation and Crystallization of the Enzyme Urease: Preliminary Paper. J. Biol. Chem. 1926, 69, 435–441. 10.1016/S0021-9258(18)84560-4. [DOI] [Google Scholar]
- Watson J. D.; Crick F. H. C. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature 1953, 171, 737–738. 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Feiten M. C.; Di Luccio M.; Santos K. F.; de Oliveira D.; Oliveira J. V. X-Ray Crystallography as a Tool to Determine Three-Dimensional Structures of Commercial Enzymes Subjected to Treatment in Pressurized Fluids. Appl. Biochem. Biotechnol. 2017, 182, 429–451. 10.1007/s12010-016-2336-9. [DOI] [PubMed] [Google Scholar]
- Kazlauskas R. J.; Bornscheuer U. T. Biotransformations with Lipases. Biotechnology 2008, 36–191. 10.1002/9783527620906.ch3.18384693 [DOI] [Google Scholar]
- de Romo A. C. Tallow and the Time Capsule: Claude Bernard’s Discovery of the Pancreatic Digestion of Fat. Hist. Philos. Life Sci. 1989, 11, 253–274. [PubMed] [Google Scholar]
- Sym E. A. Action of esterase in the presence of organic solvents. Biochem. J. 1936, 30, 609–617. 10.1042/bj0300609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry W. M.; Brand F. C. A Study of Cholesterol Esterase in Liver and Brain. J. Biol. Chem. 1941, 137, 377–387. 10.1016/S0021-9258(18)73009-3. [DOI] [Google Scholar]
- Liu W.; Wang P. Cofactor regeneration for sustainable enzymatic biosynthesis. Biotechnol. Adv. 2007, 25, 369–384. 10.1016/j.biotechadv.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Richter M. Functional diversity of organic molecule enzyme cofactors. Nat. Prod. Rep. 2013, 30, 1324–1345. 10.1039/c3np70045c. [DOI] [PubMed] [Google Scholar]
- Wong C.-H.; Whitesides G. M. Enzyme-catalyzed organic synthesis: NAD(P)H cofactor regeneration by using glucose-6-phosphate and the glucose-5-phosphate dehydrogenase from Leuconostoc mesenteroides. J. Am. Chem. Soc. 1981, 103, 4890–4899. 10.1021/ja00406a037. [DOI] [Google Scholar]
- Baker Dockrey S. A.; Lukowski A. L.; Becker M. R.; Narayan A. R. H. Biocatalytic site- and enantioselective oxidative dearomatization of phenols. Nat. Chem. 2018, 10, 119–125. 10.1038/nchem.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyser J. B.; Baker Dockrey S. A.; Benítez A. R.; Joyce L. A.; Wiscons R. A.; Smith J. L.; Narayan A. R. H. Stereodivergent, Chemoenzymatic Synthesis of Azaphilone Natural Products. J. Am. Chem. Soc. 2019, 141, 18551–18559. 10.1021/jacs.9b09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wildeman S. M. A.; Sonke T.; Schoemaker H. E.; May O. Biocatalytic Reductions: From Lab Curiosity to “First Choice”. Acc. Chem. Res. 2007, 40, 1260–1266. 10.1021/ar7001073. [DOI] [PubMed] [Google Scholar]
- Wichmann R.; Vasic-Racki D. Cofactor Regeneration at the Lab Scale. Adv. Biochem. Eng./Biotechnol. 2005, 92, 225–260. 10.1007/b98911. [DOI] [PubMed] [Google Scholar]
- Hughes D. L. Biocatalysis in Drug Development—Highlights of the Recent Patent Literature. Org. Process Res. Dev. 2018, 22, 1063–1080. 10.1021/acs.oprd.8b00232. [DOI] [Google Scholar]
- Cassimjee K. E.; Branneby C.; Abedi V.; Wells A.; Berglund P. Transaminations with isopropyl amine: equilibrium displacement with yeast alcohol dehydrogenase coupled to in situ cofactor regeneration. Chem. Commun. 2010, 46, 5569–5571. 10.1039/c0cc00050g. [DOI] [PubMed] [Google Scholar]
- Truppo M. D.; Rozzell J. D.; Moore J. C.; Turner N. J. Rapid screening and scale-up of transaminase catalysed reactions. Org. Biomol. Chem. 2009, 7, 395–398. 10.1039/B817730A. [DOI] [PubMed] [Google Scholar]
- Zachos I.; Nowak C.; Sieber V. Biomimetic cofactors and methods for their recycling. Curr. Opin. Chem. Biol. 2019, 49, 59–66. 10.1016/j.cbpa.2018.10.003. [DOI] [PubMed] [Google Scholar]
- Kelly S. A.; Mix S.; Moody T. S.; Gilmore B. F. Transaminases for industrial biocatalysis: novel enzyme discovery. Appl. Microbiol. Biotechnol. 2020, 104, 4781–4794. 10.1007/s00253-020-10585-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savile C. K.; Janey J. M.; Mundorff E. C.; Moore J. C.; Tam S.; Jarvis W. R.; Colbeck J. C.; Krebber A.; Fleitz F. J.; Brands J.; et al. Biocatalytic Asymmetric Synthesis of Chiral Amines from Ketones Applied to Sitagliptin Manufacture. Science 2010, 329, 305–309. 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- Slabu I.; Galman J. L.; Lloyd R. C.; Turner N. J. Discovery, Engineering, and Synthetic Application of Transaminase Biocatalysts. ACS Catal. 2017, 7, 8263–8284. 10.1021/acscatal.7b02686. [DOI] [Google Scholar]
- Kelly S. A.; Pohle S.; Wharry S.; Mix S.; Allen C. C. R.; Moody T. S.; Gilmore B. F. Application of ω-Transaminases in the Pharmaceutical Industry. Chem. Rev. 2018, 118, 349–367. 10.1021/acs.chemrev.7b00437. [DOI] [PubMed] [Google Scholar]
- Wandrey C.; Liese A.; Kihumbu D. Industrial Biocatalysis: Past, Present, and Future. Org. Process Res. Dev. 2000, 4, 286–290. 10.1021/op990101l. [DOI] [Google Scholar]
- Chênevert R.; Gagnon R.; Desjardins M.; Dickman M.; Bureau P.; Fortier G.; Martin R.; Létourneau M.; Thiboutot S.; Bel-Rhlid R. Chemoenzymatic Synthesis of Natural Products and Bioactive Compounds. Microbial Reagents in Organic Synthesis 1992, 135–147. 10.1007/978-94-011-2444-7_11. [DOI] [Google Scholar]
- McKean I. J. W.; Hoskisson P. A.; Burley G. A. Biocatalytic Alkylation Cascades: Recent Advances and Future Opportunities for Late-Stage Functionalization. ChemBioChem 2020, 21, 2890–2897. 10.1002/cbic.202000187. [DOI] [PubMed] [Google Scholar]
- Roumpeka D. D.; Wallace R. J.; Escalettes F.; Fotheringham I.; Watson M. A Review of Bioinformatics Tools for Bio-Prospecting from Metagenomic Sequence Data. Front. Genet. 2017, 8, 1–10. 10.3389/fgene.2017.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J.; Balasubramanian S.; Church G. M.; Gilbert W.; Rogers J.; Schloss J. A.; Waterston R. H. DNA sequencing at 40: past, present and future. Nature 2017, 550, 345–353. 10.1038/nature24286. [DOI] [PubMed] [Google Scholar]
- Packer M. S.; Liu D. R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015, 16, 379–394. 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E. W.; Cavanaugh M.; Clark K.; Ostell J.; Pruitt K. D.; Karsch-Mizrachi I. GenBank. Nucleic Acids Res. 2019, 47, D94–D99. 10.1093/nar/gky989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt J. A, Tools and strategies for discovering novel enzymes and metabolic pathways. Perspectives in Science 2016, 9, 24–32. 10.1016/j.pisc.2016.07.001. [DOI] [Google Scholar]
- Naveed M.; Chaudhry Z.; Ali Z.; Amjad M.; Zulfiqar F.; Numan A. W. Annotation and curation of hypothetical proteins: prioritizing targets for experimental study. Adv. Life Sci. 2018, 5, 73–87. [Google Scholar]
- Sandoval B. A.; Hyster T. K. Emerging strategies for expanding the toolbox of enzymes in biocatalysis. Curr. Opin. Chem. Biol. 2020, 55, 45–51. 10.1016/j.cbpa.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F.; Gish W.; Miller W.; Myers E. W.; Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Boratyn G. M.; Camacho C.; Cooper P. S.; Coulouris G.; Fong A.; Ma N.; Madden T. L.; Matten W. T.; McGinnis S. D.; Merezhuk Y.; Raytselis Y.; Sayers E. W.; Tao T.; Ye J.; Zaretskaya I. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013, 41, W29–W33. 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez Benítez A.; Tweedy S. E.; Baker Dockrey S. A.; Lukowski A. L.; Wymore T.; Khare D.; Brooks C. L.; Palfey B. A.; Smith J. L.; Narayan A. R. H. Structural Basis for Selectivity in Flavin-Dependent Monooxygenase-Catalyzed Oxidative Dearomatization. ACS Catal. 2019, 9, 3633–3640. 10.1021/acscatal.8b04575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R. An introduction to sequence similarity (“homology“) searching. Curr. Protoc. Bioinf. 2013, 42, 3.1.1–3.1.8. 10.1002/0471250953.bi0301s42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden T. The BLAST Sequence Analysis Tool. NCBI Handbook 2013, 2, 361–370. [Google Scholar]
- Cai X.-H.; Jaroszewski L.; Wooley J.; Godzik A. Internal organization of large protein families: relationship between the sequence, structure, and function-based clustering. Proteins: Struct., Funct., Genet. 2011, 79, 2389–2402. 10.1002/prot.23049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A. Phylogenetic Analysis of Protein Sequence Data Using the Randomized Axelerated Maximum Likelihood (RAXML) Program. Curr. Protoc. Mol. Biol. 2011, 96, 19.11.1–19.11.14. 10.1002/0471142727.mb1911s96. [DOI] [PubMed] [Google Scholar]
- Tamura K.; Peterson D.; Peterson N.; Stecher G.; Nei M.; Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G. Building Phylogenetic Trees from Molecular Data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. 10.1093/molbev/mst012. [DOI] [PubMed] [Google Scholar]
- Yates A. D.; Achuthan P.; Akanni W.; Allen J.; Alvarez-Jarreta J.; Amode M. R.; Armean I. M.; Azov A. G.; Bennett R.; Bhai J.; et al. Ensembl 2020. Nucleic Acids Res. 2019, 48, D682–D688. 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. M.; Stres B.; Rosenquist M.; Hallin S. Phylogenetic Analysis of Nitrite, Nitric Oxide, and Nitrous Oxide Respiratory Enzymes Reveal a Complex Evolutionary History for Denitrification. Mol. Biol. Evol. 2008, 25, 1955–1966. 10.1093/molbev/msn146. [DOI] [PubMed] [Google Scholar]
- Cavalcanti J. H. F.; Esteves-Ferreira A. A.; Quinhones C. G. S.; Pereira-Lima I. A.; Nunes-Nesi A.; Fernie A. R.; Araújo W. L. Evolution and Functional Implications of the Tricarboxylic Acid Cycle as Revealed by Phylogenetic Analysis. Genome Biol. Evol. 2014, 6, 2830–2848. 10.1093/gbe/evu221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiq M. A.; Hochberg G. K. A.; Thornton J. W. Evolution of protein specificity: insights from ancestral protein reconstruction. Curr. Opin. Struct. Biol. 2017, 47, 113–122. 10.1016/j.sbi.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton J. W. Resurrecting ancient genes: experimental analysis of extinct molecules. Nat. Rev. Genet. 2004, 5, 366–375. 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- Furukawa R.; Toma W.; Yamazaki K.; Akanuma S. Ancestral sequence reconstruction produces thermally stable enzymes with mesophilic enzyme-like catalytic properties. Sci. Rep. 2020, 10, 15493. 10.1038/s41598-020-72418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien P. J.; Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 1999, 6, R91–R105. 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- Rodríguez Benítez A.; Narayan A. R. H. Frontiers in Biocatalysis: Profiling Function across Sequence Space. ACS Cent. Sci. 2019, 5 (11), 1747–1749. 10.1021/acscentsci.9b01112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt J. A.; Bouvier J. T.; Davidson D. B.; Imker H. J.; Sadkhin B.; Slater D. R.; Whalen K. L. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta, Proteins Proteomics 2015, 1854, 1019–1037. 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; Baliga N. S.; Wang J. T.; Ramage D.; Amin N.; Schwikowski B.; Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson H. J.; Morris J. H.; Ferrin T. E.; Babbitt P. C. Using Sequence Similarity Networks for Visualization of Relationships Across Diverse Protein Superfamilies. PLoS One 2009, 4, e4345 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher B. F.; Snodgrass H. M.; Jones K. A.; Andorfer M. C.; Lewis J. C. Site-Selective C−H Halogenation Using Flavin-Dependent Halogenases Identified via Family-Wide Activity Profiling. ACS Cent. Sci. 2019, 5, 1844–1856. 10.1021/acscentsci.9b00835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wages J. M.Polymerase Chain Reaction. Encyclopedia of Analytical Science, Second ed.; Worsfold P., Townshend A., Poole C., Eds.; Elsevier: Oxford, 2005; pp 243–250. [Google Scholar]
- Smalla K.; Jechalke S.; Top E. M. Plasmid Detection, Characterization, and Ecology. Microbiol. Spectrum 2015, 3, 3. 10.1128/microbiolspec.PLAS-0038-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B.; Johnson A.; Lewis J.; Raff M.; Roberts K.; Walter P.. Molecular Biology of the Cell, 4th ed.; Taylor and Francis, 2002. [Google Scholar]
- Hughes R. A.; Ellington A. D. Synthetic DNA Synthesis and Assembly: Putting the Synthetic in Synthetic Biology. Cold Spring Harbor Perspect. Biol. 2017, 9, a023812. 10.1101/cshperspect.a023812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.; Rogers E. J. Transformation of chemically competent E. coli. Methods Enzymol. 2013, 529, 329–336. 10.1016/B978-0-12-418687-3.00028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield P. T. Overview of the purification of recombinant proteins. Curr. Protoc. Protein Sci. 2015, 80, 6.1.1–6.1.35. 10.1002/0471140864.ps0601s80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S. K.; Bandyopadhyay S.; Ray S. S. Evolutionary computation in bioinformatics: a review. IEEE Transactions on Systems, Man, and Cybernetics, Part C (Applications and Reviews) 2006, 36, 601–615. 10.1109/TSMCC.2005.855515. [DOI] [Google Scholar]
- Yang P.; Yang Y. H.; Zhou B.; Zomaya A. A Review of Ensemble Methods in Bioinformatics. Curr. Bioinf. 2010, 5, 296–308. 10.2174/157489310794072508. [DOI] [Google Scholar]
- Wilding M.; Scott C.; Warden A. C. Computer-Guided Surface Engineering for Enzyme Improvement. Sci. Rep. 2018, 8, 11998. 10.1038/s41598-018-30434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell A. N.; DeMars M. D.; Slocum S. T.; Yu F.; Anand K.; Chemler J. A.; Korakavi N.; Priessnitz J. K.; Park S. R.; Koch A. A.; et al. Chemoenzymatic Total Synthesis and Structural Diversification of Tylactone-Based Macrolide Antibiotics through Late-Stage Polyketide Assembly, Tailoring, and C—H Functionalization. J. Am. Chem. Soc. 2017, 139, 7913–7920. 10.1021/jacs.7b02875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-K.; Dai C.-F.; Duh C.-Y. Cytotoxic Pregnane Steroids from the Formosan Soft Coral Stereonephthya crystalliana. J. Nat. Prod. 2006, 69, 103–106. 10.1021/np050384c. [DOI] [PubMed] [Google Scholar]
- Wang J.; Zhang Y.; Liu H.; Shang Y.; Zhou L.; Wei P.; Yin W.-B.; Deng Z.; Qu X.; Zhou Q. A biocatalytic hydroxylation-enabled unified approach to C19-hydroxylated steroids. Nat. Commun. 2019, 10, 3378. 10.1038/s41467-019-11344-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renata H.; Zhou Q.; Baran P. S. Strategic Redox Relay Enables A Scalable Synthesis of Ouabagenin, A Bioactive Cardenolide. Science 2013, 339, 59–63. 10.1126/science.1230631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renata H.; Zhou Q.; Dünstl G.; Felding J.; Merchant R. R.; Yeh C.-H.; Baran P. S. Development of a Concise Synthesis of Ouabagenin and Hydroxylated Corticosteroid Analogues. J. Am. Chem. Soc. 2015, 137, 1330–1340. 10.1021/ja512022r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Ju W.; Tian H.; Tian W.; Gui J. Scalable Synthesis of Cyclocitrinol. J. Am. Chem. Soc. 2018, 140, 9413–9416. 10.1021/jacs.8b06444. [DOI] [PubMed] [Google Scholar]
- Lukowski A. L.; Denomme N.; Hinze M. E.; Hall S.; Isom L. L.; Narayan A. R. H. Biocatalytic Detoxification of Paralytic Shellfish Toxins. ACS Chem. Biol. 2019, 14, 941–948. 10.1021/acschembio.9b00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowski A. L.; Ellinwood D. C.; Hinze M. E.; DeLuca R. J.; Du Bois J.; Hall S.; Narayan A. R. H. C−H Hydroxylation in Paralytic Shellfish Toxin Biosynthesis. J. Am. Chem. Soc. 2018, 140, 11863–11869. 10.1021/jacs.8b08901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowski A. L.; Liu J.; Bridwell-Rabb J.; Narayan A. R. H. Structural basis for divergent C−H hydroxylation selectivity in two Rieske oxygenases. Nat. Commun. 2020, 11, 2991. 10.1038/s41467-020-16729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowski A. L.; Mallik L.; Hinze M. E.; Carlson B. M.; Ellinwood D. C.; Pyser J. B.; Koutmos M.; Narayan A. R. H. Substrate Promiscuity of a Paralytic Shellfish Toxin Amidinotransferase. ACS Chem. Biol. 2020, 15, 626–631. 10.1021/acschembio.9b00964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; King-Smith E.; Dong L.-B.; Yang L.-C.; Rudolf J. D.; Shen B.; Renata H. Divergent synthesis of complex diterpenes through a hybrid oxidative approach. Science 2020, 369, 799–806. 10.1126/science.abb8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud D.; Saaidi P.-L.; Monfleur A.; Harari M.; Cuccaro J.; Fossey A.; Besnard M.; Debard A.; Mariage A.; Pellouin V.; et al. Synthesis of Mono- and Dihydroxylated Amino Acids with New α-Ketoglutarate-Dependent Dioxygenases: Biocatalytic Oxidation of C-H Bonds. ChemCatChem 2014, 6, 3012–3017. 10.1002/cctc.201402498. [DOI] [Google Scholar]
- Baud D.; Peruch O.; Saaidi P.-L.; Fossey A.; Mariage A.; Petit J.-L.; Salanoubat M.; Vergne-Vaxelaire C.; de Berardinis V.; Zaparucha A. Biocatalytic Approaches towards the Synthesis of Chiral Amino Alcohols from Lysine: Cascade Reactions Combining alpha-Keto Acid Oxygenase Hydroxylation with Pyridoxal Phosphate-Dependent Decarboxylation. Adv. Synth. Catal. 2017, 359, 1563–1569. 10.1002/adsc.201600934. [DOI] [Google Scholar]
- Zhang X.; King-Smith E.; Renata H. Total Synthesis of Tambromycin by Combining Chemocatalytic and Biocatalytic C−H Functionalization. Angew. Chem., Int. Ed. 2018, 57, 5037–5041. 10.1002/anie.201801165. [DOI] [PubMed] [Google Scholar]
- Zetzsche L. E.; Narayan A. R. H. Broadening the scope of biocatalytic C-C bond formation. Nat. Rev. Chem. 2020, 4, 334–346. 10.1038/s41570-020-0191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H.; Schultz E. E.; Balskus E. P. A new strategy for aromatic ring alkylation in cylindrocyclophane biosynthesis. Nat. Chem. Biol. 2017, 13, 916–921. 10.1038/nchembio.2421. [DOI] [PubMed] [Google Scholar]
- Schultz E. E.; Braffman N. R.; Luescher M. U.; Hager H. H.; Balskus E. P. Biocatalytic Friedel-Crafts Alkylation Using a Promiscuous Biosynthetic Enzyme. Angew. Chem., Int. Ed. 2019, 58, 3151–3155. 10.1002/anie.201814016. [DOI] [PubMed] [Google Scholar]
- Lau W.; Sattely E. S. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science 2015, 349, 1224–1228. 10.1126/science.aac7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W.-C.; Yang Z.-J.; Tu Y.-H.; Chien T.-C. Reaction Mechanism of a Nonheme Iron Enzyme Catalyzed Oxidative Cyclization via C−C Bond Formation. Org. Lett. 2019, 21, 228–232. 10.1021/acs.orglett.8b03670. [DOI] [PubMed] [Google Scholar]
- Lazzarotto M.; Hammerer L.; Hetmann M.; Borg A.; Schmermund L.; Steiner L.; Hartmann P.; Belaj F.; Kroutil W.; Gruber K.; et al. Chemoenzymatic Total Synthesis of Deoxy-, epi-, and Podophyllotoxin and a Biocatalytic Kinetic Resolution of Dibenzylbutyrolactones. Angew. Chem., Int. Ed. 2019, 58, 8226–8230. 10.1002/anie.201900926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Zhang X.; Renata H. Asymmetric Chemoenzymatic Synthesis of (−)-Podophyllotoxin and Related Aryltetralin Lignans. Angew. Chem., Int. Ed. 2019, 58, 11657–11660. 10.1002/anie.201904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzaferro L. S.; Hüttel W.; Fries A.; Müller M. Cytochrome P450-Catalyzed Regio- and Stereoselective Phenol Coupling of Fungal Natural Products. J. Am. Chem. Soc. 2015, 137, 12289–12295. 10.1021/jacs.5b06776. [DOI] [PubMed] [Google Scholar]
- Präg A.; Grüning B. A.; Häckh M.; Lüdeke S.; Wilde M.; Luzhetskyy A.; Richter M.; Luzhetska M.; Günther S.; Müller M. Regio- and Stereoselective Intermolecular Oxidative Phenol Coupling in Streptomyces. J. Am. Chem. Soc. 2014, 136, 6195–6198. 10.1021/ja501630w. [DOI] [PubMed] [Google Scholar]
- Obermaier S.; Müller M. Biaryl-Forming Enzymes from Aspergilli Exhibit Substrate-Dependent Stereoselectivity. Biochemistry 2019, 58, 2589–2593. 10.1021/acs.biochem.9b00291. [DOI] [PubMed] [Google Scholar]
- Coelho P. S.; Brustad E. M.; Kannan A.; Arnold F. H. Olefin Cyclopropanation via Carbene Transfer Catalyzed by Engineered Cytochrome P450 Enzymes. Science 2013, 339, 307–310. 10.1126/science.1231434. [DOI] [PubMed] [Google Scholar]
- Coelho P. S.; Wang Z. J.; Ener M. E.; Baril S. A.; Kannan A.; Arnold F. H.; Brustad E. M. A serine-substituted P450 catalyzes highly efficient carbene transfer to olefins in vivo. Nat. Chem. Biol. 2013, 9, 485–487. 10.1038/nchembio.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeaux M.; Tyagi V.; Fasan R. Highly Diastereoselective and Enantioselective Olefin Cyclopropanation Using Engineered Myoglobin-Based Catalysts. Angew. Chem., Int. Ed. 2015, 54, 1744–1748. 10.1002/anie.201409928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj P.; Sreenilayam G.; Tyagi V.; Fasan R. Gram-Scale Synthesis of Chiral Cyclopropane-Containing Drugs and Drug Precursors with Engineered Myoglobin Catalysts Featuring Complementary Stereoselectivity. Angew. Chem., Int. Ed. 2016, 55, 16110–16114. 10.1002/anie.201608680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandgude A. L.; Ren X.; Fasan R. Stereodivergent Intramolecular Cyclopropanation Enabled by Engineered Carbene Transferases. J. Am. Chem. Soc. 2019, 141 (23), 9145–9150110. 10.1021/jacs.9b02700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.; Huang X.; Kan S. B. J.; Zhang R. K.; Arnold F. H. Enzymatic construction of highly strained carbocycles. Science 2018, 360, 71–75. 10.1126/science.aar4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick C. R.; Renata H. Remote C−H Hydroxylation by an α-Ketoglutarate-Dependent Dioxygenase Enables Efficient Chemoenzymatic Synthesis of Manzacidin C and Proline Analogs. J. Am. Chem. Soc. 2018, 140, 1165–1169. 10.1021/jacs.7b12918. [DOI] [PubMed] [Google Scholar]
- Zwick C. R.; Renata H. Evolution of Biocatalytic and Chemocatalytic C−H Functionalization Strategy in the Synthesis of Manzacidin C. J. Org. Chem. 2018, 83, 7407–7415. 10.1021/acs.joc.8b00248. [DOI] [PubMed] [Google Scholar]
- Doyon T. J.; Perkins J. C.; Baker Dockrey S. A.; Romero E. O.; Skinner K. C.; Zimmerman P. M.; Narayan A. R. H. Chemoenzymatic o-Quinone Methide Formation. J. Am. Chem. Soc. 2019, 141, 20269–20277. 10.1021/jacs.9b10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda R.; Abe H.; Shibata N.; Moriwaki H.; Izawa K.; Soloshonok V. A. Asymmetric synthesis of α-deuterated α-amino acids. Org. Biomol. Chem. 2017, 15, 6978–6983. 10.1039/C7OB01720K. [DOI] [PubMed] [Google Scholar]
- Chatterjee B.; Krishnakumar V.; Gunanathan C. Selective α-Deuteration of Amines and Amino Acids Using D2O. Org. Lett. 2016, 18, 5892–5895. 10.1021/acs.orglett.6b02978. [DOI] [PubMed] [Google Scholar]
- McLaughlin M.; Kong J.; Belyk K. M.; Chen B.; Gibson A. W.; Keen S. P.; Lieberman D. R.; Milczek E. M.; Moore J. C.; Murray D.; Peng F.; Qi J.; Reamer R. A.; Song Z. J.; Tan L.; Wang L.; Williams M. J. Enantioselective Synthesis of 4′-Ethynyl-2-fluoro-2′-deoxyadenosine (EFdA) via Enzymatic Desymmetrization. Org. Lett. 2017, 19, 926–929. 10.1021/acs.orglett.7b00091. [DOI] [PubMed] [Google Scholar]
- McKinnie S. M. K.; Miles Z. D.; Jordan P. A.; Awakawa T.; Pepper H. P.; Murray L. A. M.; George J. H.; Moore B. S. Total Enzyme Syntheses of Napyradiomycins A1 and B1. J. Am. Chem. Soc. 2018, 140, 17840–17845. 10.1021/jacs.8b10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler J. D.; Kevin S.; Di Y.; Leena M.; Troy W.; Markos K.; Paul M. Z.; Alison N. Radical Tropolone Biosynthesis. ChemRxiv 2020, 10.26434/chemrxiv.12780044.v1. [DOI] [Google Scholar]
- Mennen S. M.; Alhambra C.; Allen C. L.; Barberis M.; Berritt S.; Brandt T. A.; Campbell A. D.; Castañón J.; Cherney A. H.; Christensen M.; et al. The Evolution of High-Throughput Experimentation in Pharmaceutical Development and Perspectives on the Future. Org. Process Res. Dev. 2019, 23, 1213–1242. 10.1021/acs.oprd.9b00140. [DOI] [Google Scholar]
- Dandapani S.; Rosse G.; Southall N.; Salvino J. M.; Thomas C. J. Selecting, Acquiring, and Using Small Molecule Libraries for High-Throughput Screening. Curr. Protoc. Chem. Biol. 2012, 4, 177–191. 10.1002/9780470559277.ch110252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich J. O.; Michels P. C.; Khmelnitsky Y. L. Combinatorial biocatalysis. Curr. Opin. Chem. Biol. 2002, 6, 161–167. 10.1016/S1367-5931(02)00299-5. [DOI] [PubMed] [Google Scholar]
- Altreuter D. H.; Clark D. S. Combinatorial biocatalysis: taking the lead from Nature. Curr. Opin. Biotechnol. 1999, 10, 130–136. 10.1016/S0958-1669(99)80022-6. [DOI] [PubMed] [Google Scholar]
- Agrawal R.; Semwal S.; Kumar R.; Mathur A.; Gupta R. P.; Tuli D. K.; Satlewal A. Synergistic Enzyme Cocktail to Enhance Hydrolysis of Steam Exploded Wheat Straw at Pilot Scale. Front. Energy Res. 2018, 6, 122. 10.3389/fenrg.2018.00122. [DOI] [Google Scholar]
- Poppe J. K.; Matte C. R.; de Freitas V. O.; Fernandez-Lafuente R.; Rodrigues R. C.; Záchia Ayub M. A. Enzymatic synthesis of ethyl esters from waste oil using mixtures of lipases in a plug-flow packed-bed continuous reactor. Biotechnol. Prog. 2018, 34, 952–959. 10.1002/btpr.2650. [DOI] [PubMed] [Google Scholar]
- Sheldon R. A.; Woodley J. M. Role of Biocatalysis in Sustainable Chemistry. Chem. Rev. 2018, 118, 801–838. 10.1021/acs.chemrev.7b00203. [DOI] [PubMed] [Google Scholar]