Abstract
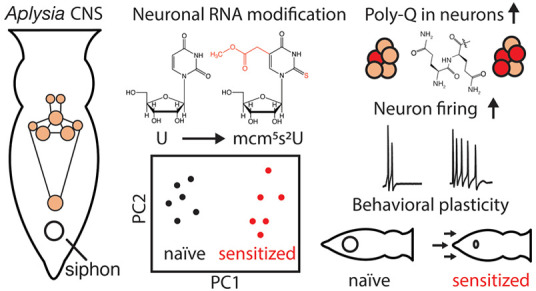
Subtle changes in the landscape of post-transcriptional modifications have emerged as putative regulators of central nervous system plasticity and activity-induced protein synthesis. However, simultaneous characterization of multiple RNA modifications and their covariation during learning and memory paradigms has been impeded by the complexity of animal models and lack of untargeted approaches for identifying pathway-relevant RNA modifications in small-volume samples. Here, we used mass spectrometry to profile spatiotemporal changes in dozens of neuronal RNA modifications in Aplysia californica during behavioral sensitization of a simple defensive reflex. Unique RNA modification patterns were observed in the major ganglia of trained and naı̇ve animals, with two tRNA modifications, namely, 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) and 1-methyladenosine (m1A), at significantly higher levels in trained subjects. We report that tRNAs, and their modifications, correlate with increased polyglutamine synthesis and excitability in neurons, characterizing the first link between noncoding RNA modifications and non-associative learning.
Short abstract
Neuronal RNA modifications contribute to poly-Q synthesis, neuron excitability, and behavioral change in Aplysia californica, comprising an additional layer of regulation in non-associative learning.
Introduction
Dynamic post-transcriptional modifications to RNA are increasingly recognized for their roles in tuning the translation of the cellular proteome. Over 100 structurally unique RNA modifications have been discovered that influence folding, stability, and translation efficiency in coding and noncoding RNAs alike.1−6 Some of these modifications are reversible,7,8 while others are deposited substoichiometrically on RNA molecules,9,10 resulting in a mosaic of differentially modified cellular RNAs (i.e., the epitranscriptome) that have been implicated in the regulation of cell stress response,11−13 development,14 cell differentiation,15 and cell-to-cell signaling.16
Numerous RNA modifications are essential for normal function of the central nervous system (CNS),17−22 but little is known about whether dynamic regulation of these epitranscriptomic marks contributes to its remarkable plasticity. Modification-specific antibodies have revealed region-dependent changes in the levels of N6-methyladenosine (m6A) during oligodendrocyte differentiation,23 acute restraint stress exposure,24 and contextual fear conditioning tasks.25,26 These studies also highlight a broad range of time scales over which post-transcriptional modifications are controlled: on the order of days for cell differentiation, to just minutes for learning-related changes in modified RNAs. Despite a general consensus that such epitranscriptomic regulation may be an understudied component in the molecular underpinnings of learning and memory,27−29 the covariation of multiple RNA modifications during activity-dependent plasticity has not been explored due to the complexity of vertebrate models.
One successful approach to understanding the mechanisms that underlie learning and memory has been to characterize neuronal correlates of simple defensive reflexes in the invertebrate model Aplysia californica (Aplysia).30 Non-associative learning in this animal is governed by a host of biophysical changes initiated by the monoamine neurotransmitter serotonin (5-HT), including post-translational modifications31−33 and pre- and postsynaptic protein synthesis occurring minutes after the application of sensitizing stimuli.34,35Aplysia cytoplasmic polyadenylation element binding protein (ApCPEB) is one of the few known products of 5-HT-induced translation that is required for maintenance of long-term facilitation and possesses an unusual poly-Gln tract at its N-terminus.36 Given that RNA modifications play a newly appreciated role in enhancing translation rates for proteins with poly-Xaa motifs,37−39 it is conceivable that the landscape of post-transcriptional modifications in Aplysia neurons contributes to rapid translation of learning-related mRNAs, resulting in facilitation/behavioral sensitization. However, only a single modified nucleoside has been reported in the Aplysia CNS40 where the abundance, distribution, and learning-related functions of neuronal RNA modifications have yet to be characterized.
Here, we demonstrate that non-associative learning in Aplysia coincides with characteristic time- and region-dependent rearrangements of neuronal RNA modification profiles. Using a liquid chromatography (LC)–tandem mass spectrometry (MS/MS) approach optimized for RNA modifications,41 we identified higher levels of 7-methylguanosine (m7G) in total RNA extracts as well as 1-methyladenosine (m1A) and 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) in tRNA during behavioral sensitization of the tail-elicited siphon withdrawal reflex (TSWR). Treatment of ganglia with mcm5s2U-rich tRNA isolated from sensitized donors increased levels of poly-Q proteins and produced electrophysiological hallmarks of increased excitability in neurons. Together these results provide the first evidence linking tRNA modification profiles to learning and memory and suggest that RNA modification constitutes an additional regulatory layer for protein synthesis that contributes to neuron excitability and ultimately, behavioral changes in Aplysia.
Results
RNA Modifications Heterogeneously Distributed Across the Aplysia CNS
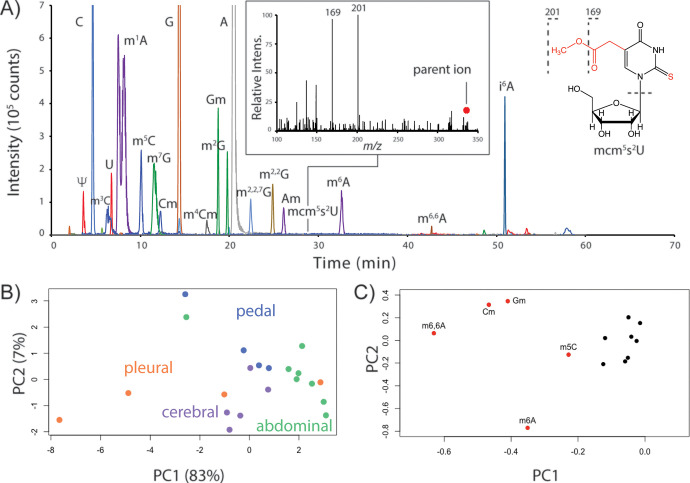
We first established an inventory of RNA modifications in the Aplysia CNS using LC-MS/MS, identifying a total of 26 unique modified nucleosides in the major ganglia (Table S1 of the Supporting Information (SI) and Figure 1A). Technical replicates for RNA digests from the pedal ganglion, which contains neurons involved in locomotion and defensive reflex behaviors, showed that peak areas for modified nucleosides had an average relative standard deviation (RSD) of 7.6 ± 3.6% (n = 3); average RSD values for biological replicates (n = 4) were 21.0 ± 13.0% (Table S2). RNA digests from the abdominal and pleural ganglia showed the highest between-animal variance with average RSD values of 38.8 ± 9.2 and 45.4 ± 17.0%, respectively. To determine whether RNA extraction contributed to variation in RNA modification abundances, we spiked isotopically labeled RNA into ganglia homogenates and performed modification analysis. Stable isotopically labeled canonical nucleosides were detected (Figure S1), but no isotopologues of modified nucleosides were observed. These data show that the variation in RNA modification profiles between animals is ganglion-specific.
Figure 1.
LC-MS/MS characterization of numerous RNA modifications in the Aplysia CNS. (A) Extracted ion chromatograms for modified nucleosides (±0.01 m/z) in the pedal ganglion: m1A, Am, m6A (m/z 282.12); m3C, m5C, Cm (m/z 258.11); m7G, Gm, m2G (m/z 298.12); m4Cm (m/z 272.13); m2,2,7G (m/z 326.15); m2,2G (m/z 312.13), m6,6A (m/z 296.14); mcm5s2U (m/z 333.08); and i6A (m/z 336.17). The inset shows the MS/MS spectrum used to positively identify mcm5s2U. (B) PCA score and (C) loading plots using 13 total RNA modifications in Aplysia ganglia.
We then determined whether RNA modification profiles were spatially distinct across the Aplysia CNS. Unsupervised principal component analysis (PCA) of 13 RNA modifications showed that ganglia generally clustered together (Figure 1B). Pleural ganglia showed the weakest clustering, presumably due to higher animal-to-animal variation (vide supra). Heterogeneity between ganglia was driven primarily by N6,6-dimethyladenosine (m6,6A), m6A, and 5-methylcytidine (m5C), as well as 2′-O-methylation of guanosine (Gm) and cytidine (Cm) (Figure 1C). These results highlight the influence of noncoding RNAs (e.g., m6,6A) in establishing ganglia-specific RNA modification patterns.
Non-associative Learning in Aplysia Accompanied by Global Changes in Total Neuronal RNA Modification Profiles
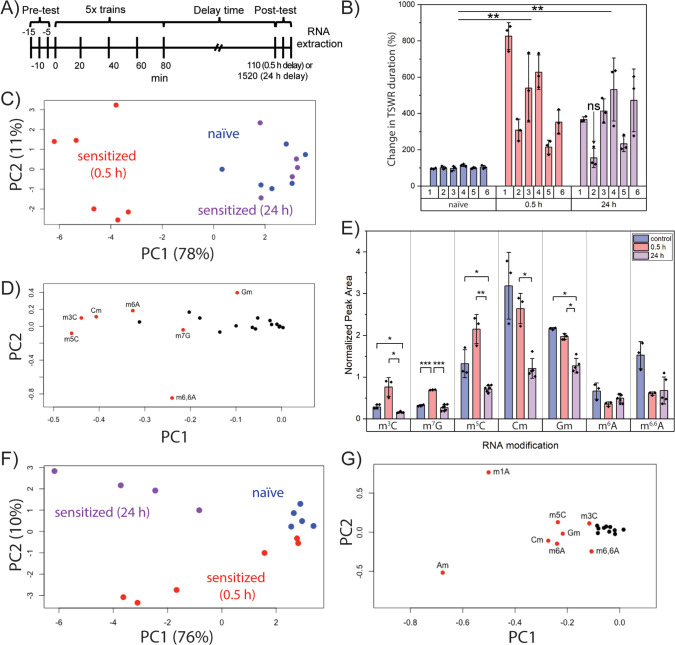
Sensitization of the TSWR in Aplysia involves heightened response to innocuous stimuli as a result of successive applications of a noxious stimulus (e.g., electrical shock). To determine whether neuronal RNA modification profiles were altered at different time points during sensitization training, we subjected animals to behavioral training and either a 0.5 or 24 h delay before the TSWR test (Figure 2A). TSWR times were significantly longer for both the 0.5 and 24 h groups (Figure 2B), indicating sensitization. Animals were excluded if they did not display a significantly longer TSWR time.
Figure 2.
Unique RNA modification profiles in the pedal ganglia for naive and sensitized animals. (A) Behavioral timeline for sensitization of Aplysia tail-elicited siphon withdrawal reflex (TSWR). (B) Comparison of TSWR duration for representative cohorts, each consisting of six animals that were assayed 0.5 or 24 h post-training or were naive to electrical shock. Error bars are ±1 SD, (ns) indicates an animal that did not show a statistically different mean TSWR duration. ** indicates a significantly different (p < 0.005) mean TSWR duration compared to naive Aplysia (unpaired t test). (C) PCA score and (D) loading plots of 20 RNA modifications in the pedal ganglia of naive or sensitized animals (0.5 or 24 h post-training). (E) Quantities of select RNA modifications in the pedal ganglia of sensitized and naive Aplysia. Error bars are ±1 SD. Unpaired t test, Bonferroni correction: * p < 0.05, ** p < 0.005, and *** p < 5 × 10–4. (F) PCA score and (G) loading plots of 18 RNA modifications in the cerebral ganglia of trained and untrained animals.
RNA modification profiles were then established for the behavioral groups. PCA showed no obvious differences between groups for the abdominal or pleural ganglia (Figure S2). However, RNA modification profiles in the pedal ganglia of sensitized animals (0.5 h delay) were readily distinguished from naive and sensitized animals (24 h delay) (Figure 2C). The naive animals and 24 h postsensitization group coclustered, indicating similar neuronal RNA modification profiles. Figure 2D highlights seven RNA modifications that contributed to the separation of sensitized animals (0.5 h) from other animals of the cohort: methylcytidine positional isomers (m3C, m5C, Cm), m6A, m6,6A, m7G, and Gm. We then subjected a new cohort to behavioral training and performed targeted analysis of these seven modifications. Levels of m3C and m5C trended higher for the 0.5 h group compared to naive animals and were significantly higher compared to the 24 h group (Figure 2E). Significantly higher amounts of m7G (p < 5 × 10–4) were detected in the 0.5 h group compared to both naive and 24 h postsensitization groups. Cerebral ganglia were also distinguishable on the basis of behavioral status (Figure 2F), where m1A and 2′-O-methyladenosine (Am) contributed to group separation (Figure 2G). These results reveal time- and ganglia-specific changes in RNA modification patterns during behavioral change.
mcm5s2U and m1A Modifications in Neuronal tRNAs Correlation with Sensitization
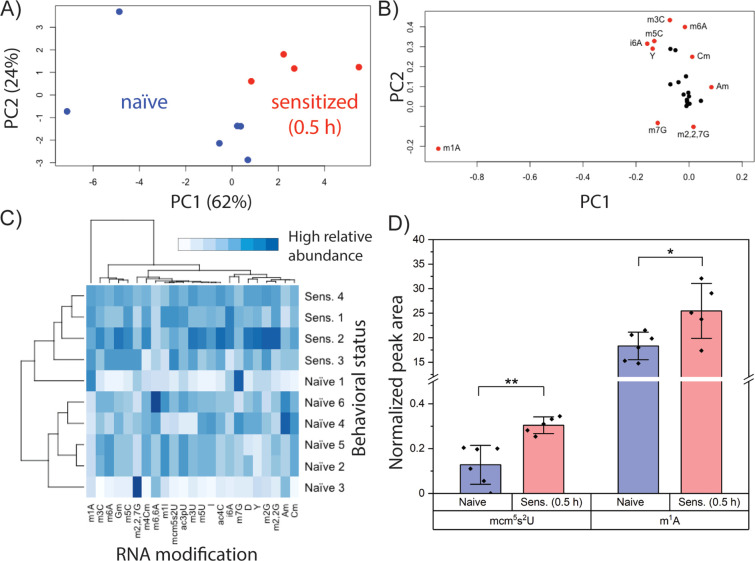
Given the heterogeneity of noncoding RNA modification distribution in the Aplysia CNS, we then asked whether tRNA modification profiles responded to behavioral training. Subtle clustering of behavioral status was observed for tRNA modifications in the cerebral ganglia (Figure S3), but an obvious distinction between naive and sensitized animals was observed in the pedal ganglia (Figure 3A). Clustering was driven by m1A and, to a lesser extent, m7G, m3C, and m6A (Figure 3B), which were also important in total RNA fractions for distinguishing naive and sensitized animals (Figure 2D,G). Hierarchical clustering analysis recapitulated the results of the behavioral training, providing a strong link between pedal tRNA modifications and non-associative learning. Inspection of the heat map (Figure 3C) shows m1A, m5C, mcm5s2U, N4-acetylcytidine (ac4C), and N2-methylguanosine (m2G) at higher levels in nearly all sensitized animals relative to their naive counterparts. A trend toward lower Cm amounts in sensitized animals was also observed. While these modifications exist in multiple tRNAs, mcm5s2U was the only modification unambiguously involved in translation since it is positioned solely in the anticodon of the three eukaryotic tRNAs in which it is present: tRNALysUUU (tKUUU), tRNAGluUUC (tEUUC), and tRNAGlnUUG (tQUUG).37 We therefore examined mcm5s2U levels in a new cohort of animals, as well as m1A levels given their strong influence on clustering of naive and sensitized groups. Both mcm5s2U and m1A were again observed at higher abundance (p < 0.005 and p < 0.05, respectively) in sensitized compared to naive animals (Figures 3D and S4). These results highlight differential tRNA modification in naive and sensitized animals and implicate an anticodon modification (mcm5s2U) in the early stages of non-associative learning, raising the question of whether select tRNA modifications promote the synthesis of learning-related proteins.
Figure 3.
tRNA modification profiles in the pedal ganglion distinguish naive and sensitized animals. (A) PCA of 25 neuronal tRNA modifications for naive and sensitized Aplysia (pedal ganglia pooled from four animals). (B) Loading plot for panel A. (C) Heat map diagram showing hierarchical clustering analysis of tRNA modifications in pedal ganglia of sensitized and naive animals. (D) Comparison of mcm5s2U and m1A levels in the tRNA fraction of pedal ganglia from naive and sensitized animals. Error bars are ±1 SD. Unpaired t test, Bonferroni correction: * p < 0.05 and ** p < 0.005.
Treatment of Neurons with tRNA from Sensitized Animals Increased Synthesis of Poly-Q Proteins
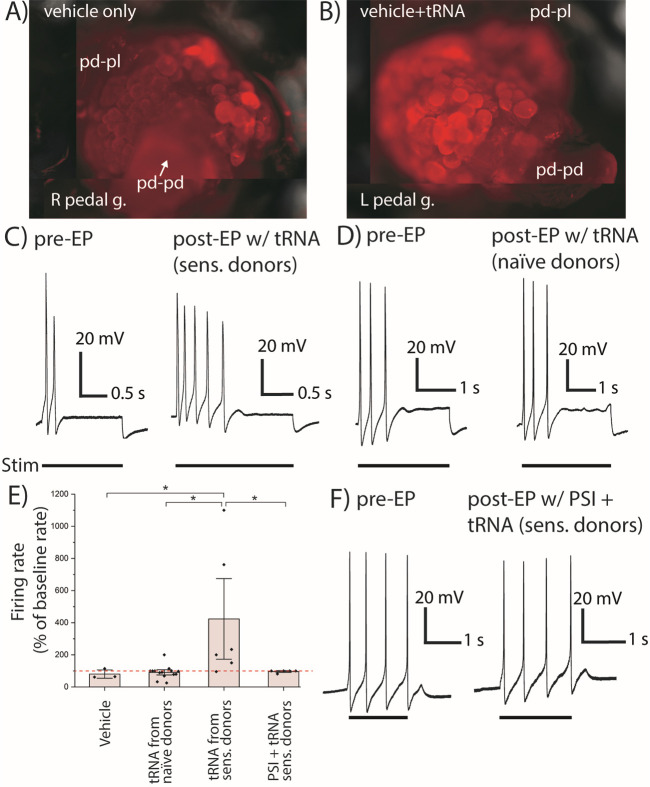
We then considered whether the mcm5s2U modification contributes to synthesis of a particular class of proteins in sensitized animals. The hypermodified uridine at position 34 in the anticodon of tKUUU, tEUUC, and tQUUG is known to modulate translation rates for poly-K, -E, and -Q,37,39 so we asked whether any proteins containing these motifs are involved in sensitization in Aplysia. One such protein, ApCPEB, contains 72 Q residues out of the N-terminal 160 amino acids and is rapidly synthesized following 5-HT stimulation.36,42 To confirm 5-HT induction of ApCPEB, we subjected pedal-pleural ganglia to 5 × 5 min bath applications of 5-HT (15 min wash after each), and immunostained with poly-Q-specific primary antibody MW1.43 Higher fluorescence intensities (119 ± 5% of control, n = 2) were observed for pedal serotonergic (PS) cluster neurons in ganglia treated with 5-HT compared to untreated pedal ganglia (Figure S5A–D), which aligns with previous findings that showed 5-HT increased ApCPEB levels in pedal-pleural homogenates.42 We then asked whether treatment of pedal ganglia with tRNA from sensitized animals would similarly induce translation of poly-Q. For these experiments, we transfected pedal-pleural ganglia with pedal tRNA from sensitized animals, incubated for 30 min, and immunostained for poly-Q. Higher fluorescence intensities (128 ± 9%, p = 0.035, n = 3) were observed in PS neurons of tRNA-treated ganglia relative to controls (Figures 4A,B, and S5E–J). Taken together with our mass spectrometry (MS) data, these findings implicate tRNAs and their modifications in the rapid translation of poly-Q proteins.
Figure 4.
Treatment of pedal ganglia with tRNA from sensitized donors, increasing abundance of poly-Q proteins and spike frequency. Immunostaining of poly-Q in pedal hemiganglia treated with (A) vehicle only or (B) vehicle and pedal tRNA from sensitized donors after 30 min incubation. Representative intracellular recordings from excitability tests for neurons in the pedal serotonergic (PS) cluster (C) before and after electroporation (EP) with pedal tRNA from sensitized donors (n = 6) or (D) naive donors (n = 14). Bar below recording indicates period of electrical stimulation. Cells were stimulated with 0.5–2.5 nA depending on the amount of current needed to elicit spikes. The same amount of depolarizing current was injected for pre- and post-EP tests. Resting potentials ranged from −27 to −55 mV for the studied cells and did not change after EP (paired t test). (E) Baseline normalized stimulation-induced firing rates for neurons post-EP. The red dashed line shows 100% (no change). Error bars are ±1.5 standard error. Mann–Whitney U test, * p < 0.05. (F) Sample electrophysiological recording from excitability tests of neurons in the PS cluster before and after EP of pedal tRNA from sensitized donors and anisomycin treatment (n = 5).
Electroporation with tRNA from Sensitized Animals Increased Neuron Excitability
We then asked whether altering the tRNA content of individual neurons would result in the electrophysiological hallmarks of sensitization. We electroporated ∼10 pg of pedal tRNA from sensitized (0.5 h) or naive animals into PS cluster neurons,44 which project to the tail nerve and are activated by sensitizing stimuli.45,46 After 30 min, neurons that were electroporated with tRNA from sensitized donors showed significantly increased responses to depolarizing current (n = 6, 423 ± 183% of baseline firing rate) as compared to neurons electroporated with either tRNA from naive animals (n = 14, 91 ± 11% of baseline; U = 8.5, p = 0.0204) or vehicle solution (n = 3, 80 ± 21% of baseline; U = 1, p = 0.0476) (Figures 4C–E and S5K). These findings were similar to increased excitability induced by 5-HT (Figure S5L) and previous reports of increased firing rates in PS cluster neurons.46 Neurons electroporated with tRNA from naive animals showed no significant difference from neurons treated with vehicle alone (p = 0.4857) (Figure 4E).
To determine if the tRNA-induced excitability was protein synthesis-dependent, we electroporated neurons with pedal tRNA from sensitized animals while applying a bath of the protein synthesis inhibitor anisomycin. Postelectroporation firing rates (n = 5, 96 ± 4% of baseline) were significantly reduced compared to neurons treated with tRNA from sensitized animals (U = 4; p = 0.0498) and were not significantly different from either tRNA from naive donors (p = 0.5968) or vehicle-treated groups (p = 0.5253) (Figure 4E,F). Baseline firing rates were not significantly different between groups (p = 0.1063, one-way ANOVA). These results indicate that hyperexcitability induced by electroporation with tRNA was protein synthesis-dependent and support a contributing role for neuronal tRNAs and their modifications in neuronal excitability.
Discussion
Rapid translation of select mRNAs is a well-established phenomenon in learning and memory paradigms that involves both pre- and postsynaptic mechanisms to tightly regulate spatiotemporal protein profiles in neurons. Here, we report evidence that neuronal RNA modifications constitute an additional layer of regulation during non-associative learning in Aplysia. Using state-of-the-art MS, we found that RNA modifications not only exhibited spatial heterogeneity (Figure 1) but also temporal dependence in the pedal and cerebral ganglia following behavioral sensitization (Figure 2C,F). Notably, striking differences in m1A and mcm5s2U abundances were observed between naive and sensitized animals (Figure 3). These findings support the existence of heterogeneous pools of modified tRNAs in neurons that, upon activation by sensitizing stimuli, may (i) favor the transcription/stabilization of select tRNA isoacceptors, (ii) reprogram modification statuses of tRNAs, or (iii) a combination of both mechanisms. The results also raise the intriguing possibility that tRNAs are differentially modified to fine-tune translation during sensitization. Indeed, stress-induced changes in tRNA modification profiles have been linked to the translation of codon-biased mRNAs in yeast as well as mycobacteria.11,12,47
In Aplysia, our data implicate changes in tRNA modifications during non-associative learning that facilitate the translation of Q-rich proteins, such as the learning-related protein ApCPEB. Since the absence of mcm5s2U in tQUUG results in ribosome pausing at cognate codons,39 the behavioral training-induced increases in mcm5s2U levels we observed may gate poly-Q translation. This hypothesis is supported by our finding that the treatment of pedal ganglia with tRNA from sensitized donors increases poly-Q abundance (Figure 4A,B). It is possible that introducing excess tRNA to Aplysia neurons may result in the translation of poly-Q proteins not ordinarily synthesized during sensitization. Our tRNA electroporation experiments address this question in part by showing increased PS neuron excitability mediated by protein synthesis (Figure 4C–F). Moreover, electroporation with tRNA from naive donors had no effect on PS neuron excitability, highlighting the importance of training-induced tRNA modification profiles.
Although questions remain about the specific RNA sequences that are responsible for the changes observed in neuron activity, hallmarks of sensitization have also been reported in Aplysia sensory neurons upon administration of RNA. Hyperexcitability of sensory neurons was previously induced by noncoding RNA from the pedal-pleural ganglia of sensitized donors,48 which is consistent with our findings linking electrophysiological changes to tRNA. However, this does not rule out the possibility that other noncoding RNAs are involved since tRNA fractions may contain other RNAs of similar size.
Despite their involvement in the TSWR, we did not detect sensitization-induced changes to RNA modifications in the abdominal or pleural ganglia. This may be due to larger between-animal variation in RNA modifications (Table S2), perhaps stemming from the giant neurons in these ganglia (∼0.5–1 mm diameter) that contain up to 2 μg of RNA.49 This represents ∼15% of the total RNA in a typical ganglionic extract, rendering our whole-ganglia sampling approach less likely to detect subtle changes in these ganglia. With the development of increasingly sensitive analytical methods that simultaneously detect multiple RNA modifications, we anticipate that the characterization of modification profiles of individual neurons within behavioral circuits will further unravel the molecular underpinnings of non-associative learning and memory formation.
Methods
Detailed methods are described in the Supporting Information.
Acknowledgments
This work was funded by the National Institute on Drug Abuse under Award No. P30 DA018310, the National Human Genome Research Institute under Award No. RM1HG010023, and the U.S. Office of Naval Research Award No. N00014-19-1-2373. The National Resource for Aplysia is funded by the PHS Grant P40OD010952. K.D.C. is supported by a Beckman Institute Postdoctoral Fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.1c00351.
Materials, methods, and complete experimental details; nucleoside isotopologues and extracted ion chromatograms (Figure S1), total RNA modification profiles comparison (Figure S2), tRNA purity assessment and hierarchical clustering (Figure S3), EICs (Figure S4), immunostaining images and intracellular recordings (Figure S5); all modified nucleosides identified (Table S1) and normalized peak areas for neuronal RNA modifications (Table S2) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Cantara W. A.; Crain P. F.; Rozenski J.; McCloskey J. A.; Harris K. A.; Zhang X.; Vendeix F. A. P.; Fabris D.; Agris P. F. The RNA Modification Database, RNAMDB: 2011 Update. Nucleic Acids Res. 2011, 39 (Suppl_1), D195–D201. 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P.; Machnicka M. A.; Purta E.; Piątkowski P.; Bagiński B.; Wirecki T. K.; de Crécy-Lagard V.; Ross R.; Limbach P. A.; Kotter A.; Helm M.; Bujnicki J. M. MODOMICS: A Database of RNA Modification Pathways. 2017 Update. Nucleic Acids Res. 2018, 46 (D1), D303–D307. 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Magro C.; Keller P.; Kotter A.; Werner S.; Duarte V.; Marchand V.; Ignarski M.; Freiwald A.; Müller R.-U.; Dieterich C.; Motorin Y.; Butter F.; Atta M.; Helm M. A Vastly Increased Chemical Variety of RNA Modifications Containing a Thioacetal Structure. Angew. Chem., Int. Ed. 2018, 57 (26), 7893–7897. 10.1002/anie.201713188. [DOI] [PubMed] [Google Scholar]
- Helm M. Post-Transcriptional Nucleotide Modification and Alternative Folding of RNA. Nucleic Acids Res. 2006, 34 (2), 721–733. 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S.; Waldor M. K. The RNA Degradosome Promotes tRNA Quality Control through Clearance of Hypomodified tRNA. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (4), 1394–1403. 10.1073/pnas.1814130116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Miao W.; Williams P.; Guo C.; Wang Y. SLIRP Interacts with Helicases to Facilitate 2′-O-Methylation of rRNA and to Promote Translation. J. Am. Chem. Soc. 2019, 141 (28), 10958–10961. 10.1021/jacs.9b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G.; Fu Y.; Zhao X.; Dai Q.; Zheng G.; Yang Y.; Yi C.; Lindahl T.; Pan T.; Yang Y.-G.; He C. N6-methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 2011, 7 (12), 885–887. 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Xiong X.; Wang K.; Wang L.; Shu X.; Ma S.; Yi C. Transcriptome-Wide Mapping Reveals Reversible and Dynamic N1-methyladenosine Methylome. Nat. Chem. Biol. 2016, 12 (5), 311–316. 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- Roundtree I. A.; Evans M. E.; Pan T.; He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169 (7), 1187–1200. 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaian A.; Rothe K.; Girodat D.; Minia I.; Djondovic S.; Milek M.; Spencer Miko S. E.; Wieden H.-J.; Landthaler M.; Morin G. B.; Mager D. L. Loss of m1acp3Ψ Ribosomal RNA Modification Is a Major Feature of Cancer. Cell Rep. 2020, 31 (5), 107611. 10.1016/j.celrep.2020.107611. [DOI] [PubMed] [Google Scholar]
- Chan C. T. Y.; Dyavaiah M.; DeMott M. S.; Taghizadeh K.; Dedon P. C.; Begley T. J. A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular Stress. PLoS Genet. 2010, 6 (12), e1001247. 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. T. Y.; Pang Y. L. J.; Deng W.; Babu I. R.; Dyavaiah M.; Begley T. J.; Dedon P. C. Reprogramming of tRNA Modifications Controls the Oxidative Stress Response by Codon-Biased Translation of Proteins. Nat. Commun. 2012, 3 (1), 1–9. 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkatza N. A.; Castro C.; Harvey R. F.; Heiß M.; Popis M. C.; Blanco S.; Bornelöv S.; Sajini A. A.; Gleeson J. G.; Griffin J. L.; West J. A.; Kellner S.; Willis A. E.; Dietmann S.; Frye M. Cytosine-5 RNA Methylation Links Protein Synthesis to Cell Metabolism. PLoS Biol. 2019, 17 (6), e3000297. 10.1371/journal.pbio.3000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng C. S.; Sinha A.; Aniweh Y.; Nah Q.; Babu I. R.; Gu C.; Chionh Y. H.; Dedon P. C.; Preiser P. R. tRNA Epitranscriptomics and Biased Codon Are Linked to Proteome Expression in Plasmodium Falciparum. Mol. Syst. Biol. 2018, 14 (10), e8009. 10.15252/msb.20178009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basanta-Sanchez M.; Temple S.; Ansari S. A.; D’Amico A.; Agris P. F. Attomole Quantification and Global Profile of RNA Modifications: Epitranscriptome of Human Neural Stem Cells. Nucleic Acids Res. 2016, 44 (3), e26–e26. 10.1093/nar/gkv971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gámbaro F.; Li Calzi M.; Fagúndez P.; Costa B.; Greif G.; Mallick E.; Lyons S.; Ivanov P.; Witwer K.; Cayota A.; Tosar J. P. Stable tRNA Halves Can Be Sorted into Extracellular Vesicles and Delivered to Recipient Cells in a Concentration-Dependent Manner. RNA Biol. 2020, 17 (8), 1168–1182. 10.1080/15476286.2019.1708548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte B.; Wang F.; Ngoc L. V.; Collignon E.; Bonvin E.; Deplus R.; Calonne E.; Hassabi B.; Putmans P.; Awe S.; Wetzel C.; Kreher J.; Soin R.; Creppe C.; Limbach P. A.; Gueydan C.; Kruys V.; Brehm A.; Minakhina S.; Defrance M.; Steward R.; Fuks F. Transcriptome-Wide Distribution and Function of RNA Hydroxymethylcytosine. Science 2016, 351 (6270), 282–285. 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- Satterlee J. S.; Basanta-Sanchez M.; Blanco S.; Li J. B.; Meyer K.; Pollock J.; Sadri-Vakili G.; Rybak-Wolf A. Novel RNA Modifications in the Nervous System: Form and Function. J. Neurosci. 2014, 34 (46), 15170–15177. 10.1523/JNEUROSCI.3236-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.; Tuck S.; Byström A. S. Defects in tRNA Modification Associated with Neurological and Developmental Dysfunctions in Caenorhabditis elegans Elongator Mutants. PLoS Genet. 2009, 5 (7), e1000561. 10.1371/journal.pgen.1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M.; Kataoka N.; Miyauchi K.; Ohe K.; Iida K.; Yoshida S.; Nojima T.; Okuno Y.; Onogi H.; Usui T.; Takeuchi A.; Hosoya T.; Suzuki T.; Hagiwara M. Rectifier of Aberrant mRNA Splicing Recovers tRNA Modification in Familial Dysautonomia. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (9), 2764–2769. 10.1073/pnas.1415525112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal J. J. C.; Seeburg P. H. A-to-I RNA Editing: Effects on Proteins Key to Neural Excitability. Neuron 2012, 74 (3), 432–439. 10.1016/j.neuron.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S.; Dietmann S.; Flores J. V.; Hussain S.; Kutter C.; Humphreys P.; Lukk M.; Lombard P.; Treps L.; Popis M.; Kellner S.; Hölter S. M.; Garrett L.; Wurst W.; Becker L.; Klopstock T.; Fuchs H.; Gailus-Durner V.; Hrabě de Angelis M.; Káradóttir R. T.; Helm M.; Ule J.; Gleeson J. G.; Odom D. T.; Frye M. Aberrant Methylation of tRNAs Links Cellular Stress to Neuro-Developmental Disorders. EMBO J. 2014, 33 (18), 2020–2039. 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.; Dzhashiashvili Y.; Shah A.; Kunjamma R. B.; Weng Y.; Elbaz B.; Fei Q.; Jones J. S.; Li Y. I.; Zhuang X.; Ming G.; He C.; Popko B. m6A mRNA Methylation Is Essential for Oligodendrocyte Maturation and CNS Myelination. Neuron 2020, 105 (2), 293–309.e5. 10.1016/j.neuron.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel M.; Eggert C.; Kaplick P. M.; Eder M.; Röh S.; Tietze L.; Namendorf C.; Arloth J.; Weber P.; Rex-Haffner M.; Geula S.; Jakovcevski M.; Hanna J. H.; Leshkowitz D.; Uhr M.; Wotjak C. T.; Schmidt M. V.; Deussing J. M.; Binder E. B.; Chen A. The Role of m6A/m-RNA Methylation in Stress Response Regulation. Neuron 2018, 99 (2), 389–403.e9. 10.1016/j.neuron.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Wang M.; Xie D.; Huang Z.; Zhang L.; Yang Y.; Ma D.; Li W.; Zhou Q.; Yang Y.-G.; Wang X.-J. METTL3-Mediated N6-methyladenosine mRNA Modification Enhances Long-Term Memory Consolidation. Cell Res. 2018, 28 (11), 1050–1061. 10.1038/s41422-018-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H.; Zhang X.; Weng Y.-L.; Lu Z.; Liu Y.; Lu Z.; Li J.; Hao P.; Zhang Y.; Zhang F.; Wu Y.; Delgado J. Y.; Su Y.; Patel M. J.; Cao X.; Shen B.; Huang X.; Ming G.; Zhuang X.; Song H.; He C.; Zhou T. m6A Facilitates Hippocampus-Dependent Learning and Memory through YTHDF1. Nature 2018, 563 (7730), 249–253. 10.1038/s41586-018-0666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S.; Mehler M. F. RNA Editing, DNA Recoding and the Evolution of Human Cognition. Trends Neurosci. 2008, 31 (5), 227–233. 10.1016/j.tins.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Marshall P.; Bredy T. W. Cognitive Neuroepigenetics: The next Evolution in Our Understanding of the Molecular Mechanisms Underlying Learning and Memory?. Npj Sci. Learn. 2016, 1 (1), 1–8. 10.1038/npjscilearn.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nainar S.; Marshall P. R.; Tyler C. R.; Spitale R. C.; Bredy T. W. Evolving Insights into RNA Modifications and Their Functional Diversity in the Brain. Nat. Neurosci. 2016, 19 (10), 1292–1298. 10.1038/nn.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel E. R. The Molecular Biology of Memory Storage: A Dialogue Between Genes and Synapses. Science 2001, 294 (5544), 1030–1038. 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Sweatt J. D.; Kandel E. R. Persistent and Transcriptionally-Dependent Increase in Protein Phosphorylation in Long-Term Facilitation of Aplysia Sensory Neurons. Nature 1989, 339 (6219), 51–54. 10.1038/339051a0. [DOI] [PubMed] [Google Scholar]
- Shobe J. L.; Zhao Y.; Stough S.; Ye X.; Hsuan V.; Martin K. C.; Carew T. J. Temporal Phases of Activity-Dependent Plasticity and Memory Are Mediated by Compartmentalized Routing of MAPK Signaling in Aplysia Sensory Neurons. Neuron 2009, 61 (1), 113–125. 10.1016/j.neuron.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougie J. K.; Cai D.; Hastings M.; Farah C. A.; Chen S.; Fan X.; McCamphill P. K.; Glanzman D. L.; Sossin W. S. Serotonin-Induced Cleavage of the Atypical Protein Kinase C Apl III in Aplysia. J. Neurosci. 2012, 32 (42), 14630–14640. 10.1523/JNEUROSCI.3026-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilai A.; Kennedy T. E.; Sweatt J. D.; Kandel E. R. 5-HT Modulates Protein Synthesis and the Expression of Specific Proteins during Long-Term Facilitation in Aplysia Sensory Neurons. Neuron 1989, 2 (6), 1577–1586. 10.1016/0896-6273(89)90046-9. [DOI] [PubMed] [Google Scholar]
- Villareal G.; Li Q.; Cai D.; Glanzman D. L. The Role of Rapid, Local, Postsynaptic Protein Synthesis in Learning-Related Synaptic Facilitation in Aplysia. Curr. Biol. 2007, 17 (23), 2073–2080. 10.1016/j.cub.2007.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K.; Giustetto M.; Etkin A.; Hsu R.; Janisiewicz A. M.; Miniaci M. C.; Kim J.-H.; Zhu H.; Kandel E. R. A Neuronal Isoform of CPEB Regulates Local Protein Synthesis and Stabilizes Synapse-Specific Long-Term Facilitation in Aplysia. Cell 2003, 115 (7), 893–904. 10.1016/S0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- Rezgui V. A. N.; Tyagi K.; Ranjan N.; Konevega A. L.; Mittelstaet J.; Rodnina M. V.; Peter M.; Pedrioli P. G. A. tRNA tKUUU, tQUUG, and tEUUC Wobble Position Modifications Fine-Tune Protein Translation by Promoting Ribosome A-Site Binding. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (30), 12289–12294. 10.1073/pnas.1300781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam R.; Fierer J.; Kaiser S.; Helm M.; Jurkowski T. P.; Jeltsch A. Cytosine Methylation of tRNA-Asp by DNMT2 Has a Role in Translation of Proteins Containing Poly-Asp Sequences. Cell Discovery 2015, 1 (1), 1–10. 10.1038/celldisc.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova D. D.; Leidel S. A. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 2015, 161 (7), 1606–1618. 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.; Antonov I.; Castillejos D.; Nagaraj A.; Bostwick C.; Kohn A.; Moroz L. L.; Hawkins R. D. Intermediate-Term Memory in Aplysia Involves Neurotrophin Signaling, Transcription, and DNA Methylation. Learn. Mem. 2018, 25 (12), 620–628. 10.1101/lm.047977.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. D.; Philip M. C.; Tan Y.; Sweedler J. V. Biphasic Liquid Microjunction Extraction for Profiling Neuronal RNA Modifications by Liquid Chromatography–Tandem Mass Spectrometry. Anal. Chem. 2020, 92 (18), 12647–12655. 10.1021/acs.analchem.0c02830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K.; Choi Y.-B.; White-Grindley E.; Majumdar A.; Kandel E. R. Aplysia CPEB Can Form Prion-like Multimers in Sensory Neurons That Contribute to Long-Term Facilitation. Cell 2010, 140 (3), 421–435. 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Owens G. E.; New D. M.; West A. P.; Bjorkman P. J. Anti-PolyQ Antibodies Recognize a Short PolyQ Stretch in Both Normal and Mutant Huntingtin Exon 1. J. Mol. Biol. 2015, 427 (15), 2507–2519. 10.1016/j.jmb.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas K.; Sin W.-C.; Javaherian A.; Li Z.; Cline H. T. Single-Cell Electroporationfor Gene Transfer In Vivo. Neuron 2001, 29 (3), 583–591. 10.1016/S0896-6273(01)00235-5. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Wainwright M.; Byrne J. H.; Cleary L. J. Quantitation of Contacts Among Sensory, Motor, and Serotonergic Neurons in the Pedal Ganglion of Aplysia. Learn. Mem. 2003, 10 (5), 387–393. 10.1101/lm.63903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinesco S.; Kolkman K. E.; Carew T. J. Serotonergic Modulation in Aplysia. I. Distributed Serotonergic Network Persistently Activated by Sensitizing Stimuli. J. Neurophysiol. 2004, 92 (4), 2468–2486. 10.1152/jn.00209.2004. [DOI] [PubMed] [Google Scholar]
- Chionh Y. H.; McBee M.; Babu I. R.; Hia F.; Lin W.; Zhao W.; Cao J.; Dziergowska A.; Malkiewicz A.; Begley T. J.; Alonso S.; Dedon P. C. tRNA-Mediated Codon-Biased Translation in Mycobacterial Hypoxic Persistence. Nat. Commun. 2016, 7 (1), 13302. 10.1038/ncomms13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bédécarrats A.; Chen S.; Pearce K.; Cai D.; Glanzman D. L. RNA from Trained Aplysia Can Induce an Epigenetic Engram for Long-Term Sensitization in Untrained Aplysia. eNeuro 2018, 5 (3), ENEURO.0038-18-2018. 10.1523/ENEURO.0038-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz L. L.; Kohn A. B. Do Different Neurons Age Differently? Direct Genome-Wide Analysis of Aging in Single Identified Cholinergic Neurons. Front. Aging Neurosci. 2010, 2, 6. 10.3389/neuro.24.006.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.