Abstract
Phosphatidic acids (PAs) are glycerophospholipids that regulate key cell signaling pathways governing cell growth and proliferation, including the mTOR and Hippo pathways. Their acyl chains vary in tail length and degree of saturation, leading to marked differences in the signaling functions of different PA species. For example, in mTOR signaling, saturated forms of PA are inhibitory, whereas unsaturated forms are activating. To enable rapid control over PA signaling, we describe here the development of photoswitchable analogues of PA, termed AzoPA and dAzoPA, that contain azobenzene groups in one or both lipid tails, respectively. These photolipids enable optical control of their tail structure and can be reversibly switched between a straight trans form and a relatively bent cis form. We found that cis-dAzoPA selectively activates mTOR signaling, mimicking the bioactivity of unsaturated forms of PA. Further, in the context of Hippo signaling, whose growth-suppressing activity is blocked by PA, we found that the cis forms of both AzoPA and dAzoPA selectively inhibit this pathway. Collectively, these photoswitchable PA analogues enable optical control of mTOR and Hippo signaling, and we envision future applications of these probes to dissect the pleiotropic effects of physiological and pathological PA signaling.
Short abstract
Photoswitchable analogues of the lipid second messenger phosphatidic acid enable control of mitogenic mTOR and Hippo signaling pathways in mammalian cells using light stimulation.
Introduction
Phosphatidic acids (PA) are a class of signaling lipids that mediate diverse and distinct physiological processes in mammalian cells.1−3 Production of PA is elevated in many cancers, likely due to its pleiotropic effects in promoting cell proliferation.4,5 In particular, PA regulates two major pathways that control cell growth and organ size, mammalian target of rapamycin (mTOR) and Hippo signaling.6−8 mTOR signaling is a central pathway that regulates cell growth, and its activation enhances cell proliferation by promoting biosynthesis and suppressing autophagy.9 By contrast, activation of Hippo signaling restrains organ size by inhibiting cell proliferation and stimulating apoptosis.10 Notably, the reported activities of PA include both activation of mTOR and inhibition of Hippo signaling, thus resulting in progrowth effects in both pathways, yet via independent mechanisms.8 However, it is unclear if, in all contexts, these pathways are directly regulated by PA or if they involve indirect mechanisms that are dependent on PA being metabolized to other lipids.11,12
PAs form a central hub in glycerophospholipid metabolism and are potent signaling agents. Their biosynthesis occurs via at least three major routes and on multiple organelle membranes, and they can be rapidly interconverted with other lipids.13 Further, though all PAs contain a signature phosphate monoester headgroup, the acyl tail composition can vary, and not all PA species have equivalent bioactivities. This dynamic metabolic network and structural diversity complicate efforts to understand how individual PA species control specific signaling outcomes.
Frontier approaches that allow for high spatiotemporal and molecular control could therefore facilitate the study of PA physiology. Our laboratory has recently developed an optogenetic approach that features controlled PA synthesis by engineered phospholipase Ds at organelle-level resolution and with the temporal precision of light.14 This approach, which enables the dissection of PA functions at different cellular locations, revealed that PA pools specifically generated at the plasma membrane attenuate Hippo signaling. Yet, optogenetics tools relying on a phospholipase D have limited control over PA acyl tail composition and rely on genetic manipulations, potentially restricting the scope of in vivo applications.
A photopharmacological strategy15−17 to optically control PA signaling could potentially complement this approach and even offer certain advantages. Such tools would retain the exquisite spatiotemporal control afforded by light activation. Additionally, because exogenously added photopharmacological agents would not rely on endogenous metabolism, they would afford complete control over the molecular composition of the target lipid species. Photolipids would also not require genetic perturbations and could thus be used in a broader set of biological systems.
Photocaged lipids have emerged as attractive strategy for the photoactivation of lipids through cleavage of a photosensitive protection group to reveal the native lipid.18 Photocaged versions of phosphatidic acid19,20 have been used to control the activation of matrix metalloproteinase 221 and flagellar excision.22 Chemical modifications of the photosensitive groups also allow for subcellular targeting.23−26
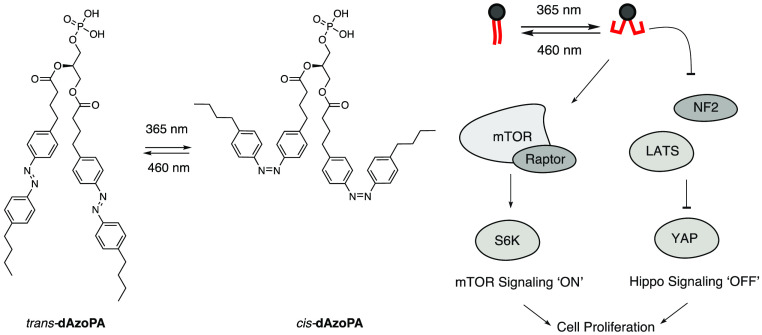
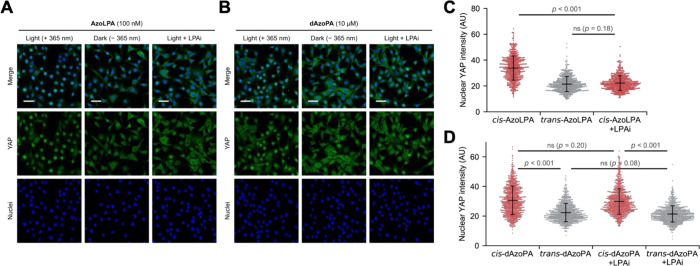
As an alternative strategy to precisely control lipid functions in a cellular context, we have begun to systematically explore photoswitchable lipids. These lipid analogues exhibit a hydrophobic azobenzene photoswitch in their lipid tail, allowing for retention of the headgroup and biological function of lipids while often enabling optical control of that function through differences in the bioactivities of the trans and cis forms.27 Photoswitchable lipids have yielded optical control of several biological targets, including ion channels,28−30 G-protein-coupled receptors (GPCRs),31−33 enzymes,34,35 nuclear hormone receptors,36−38 immune receptors,39,40 as well as membrane properties.41−46 Among the photolipids developed to date several are analogues of lipids implicated in PA metabolism, which include fatty acids (FAAzo4),28 diacylglycerol (PhoDAG and OptoDArG),29,34 lysophosphatidic acid (AzoLPA),33 and phosphatidylcholine (AzoPC)44 (Figure 1A,B).
Figure 1.
Design and synthesis of the photoswitchable PA variants AzoPA and dAzoPA. (A) Chemical structures of PA and previously developed photoswitchable lipids, including PhoDAG, OptoDArG, AzoLPA, AzoPC, FAAzo4, and PhotoS1P. (B) Photoisomerization of an azobenzene-containing photolipid (shown: FAAzo4). (C) Synthesis of AzoPA from PhoDAG-1. (D) Synthesis of dAzoPA from a protected form of AzoLPA.
Yet, a major omission in the growing compendium of photoswitchable lipids are analogues of PA itself, which we reasoned would represent powerful tools to manipulate and understand PA signaling. Herein, we report the development of photoswitchable analogues of PA and demonstrate that they are versatile tools for the optical control of PA-dependent signaling in both the mTOR and the Hippo pathways. Interestingly, though the cis forms of the two photoswitchable PA analogues are more active than their corresponding trans forms, we found differences in abilities of these probes to regulate the mTOR and Hippo pathways. These results provide a roadmap for using these photolipids to control selective outputs of PA signaling, suggesting their use to dissect the pleiotropic effects of this lipid signaling agent.
Results
Design, Synthesis, and Photophysical Characterization of Photoswitchable PA Analogues
The two acyl chains of phosphatidic acid can exist in saturated or unsaturated forms. Notably, the signaling function of PA is strongly altered by lipid saturation. In mTOR signaling, PA with two saturated acyl chains (16:0/16:0) inhibits mTOR signaling,47,48 whereas forms of PA with monounsaturated acyl chains in one or two positions stimulate mTOR signaling. Thus, we envisioned the development of photoswitchable analogues of PA in which the azobenzene N=N double bond is near the middle of the lipid tail, which corresponds to the C=C cis double bond in unsaturated forms of PA. In previous work, we and others have found that cis forms of azobenzene-containing lipids exhibit bioactivities corresponding to mono- and/or polyunsaturated lipids.28,29,33,34
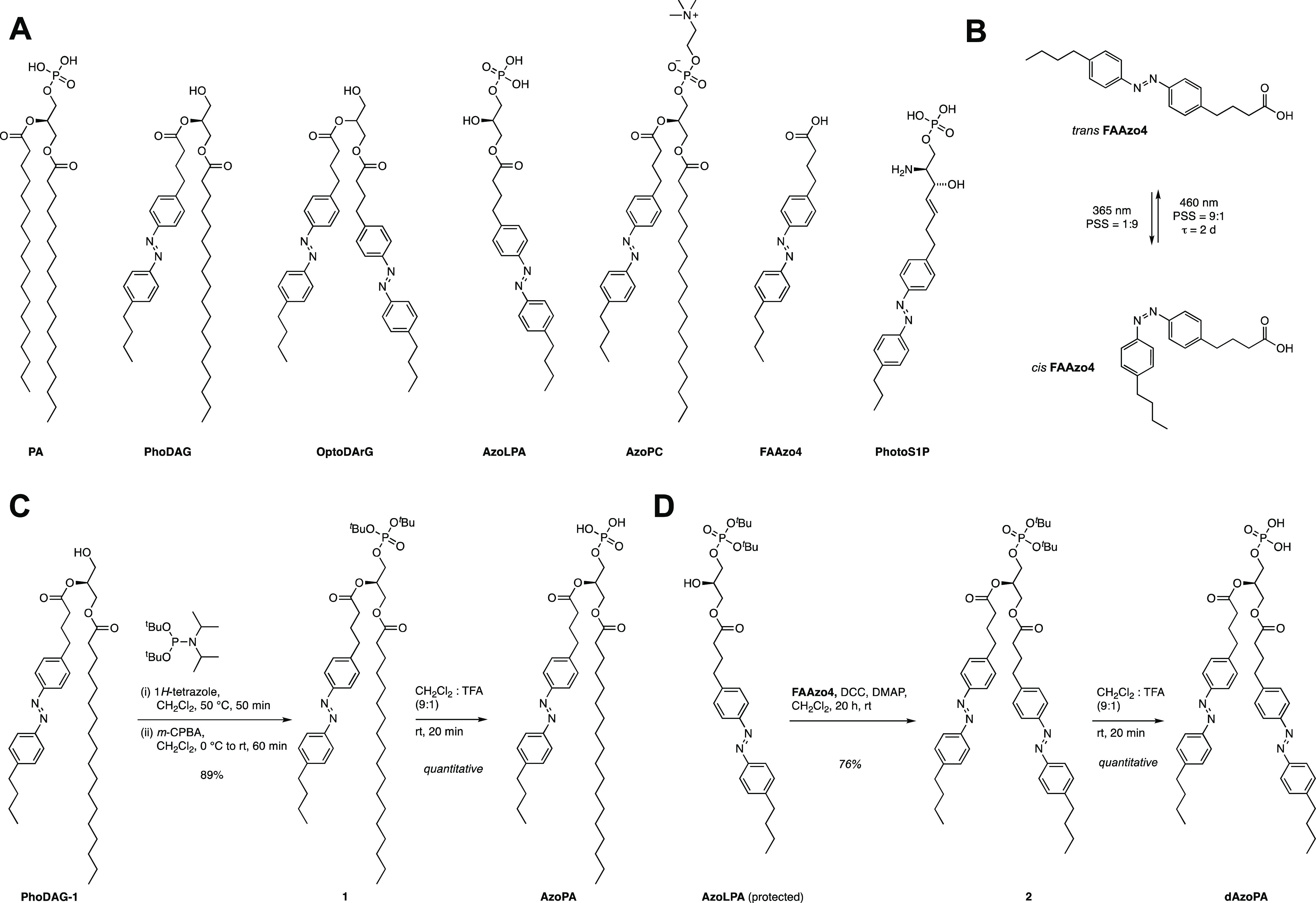
Two photoswitchable analogues of PA were prepared: AzoPA, which bears FAAzo4 at the sn-2 position and stearic acid at the sn-1 position, and dAzoPA, which contains FAAzo4 at both positions. The synthesis of AzoPA began from the previously published photoswitchable analogue of diacylglycerol, termed PhoDAG-1(34) (Figure 1C). Phosphorylation of PhoDAG-1 with di-tert-butyl-N,N-diisopropyl phosphoramidite and subsequent oxidation yielded compound 1. AzoPA was obtained through deprotection of 1 with TFA. The synthesis of dAzoPA was achieved starting from di-tert-butyl-protected AzoLPA(33) through acylation with FAAzo4 and subsequent deprotection with TFA (Figure 1D).
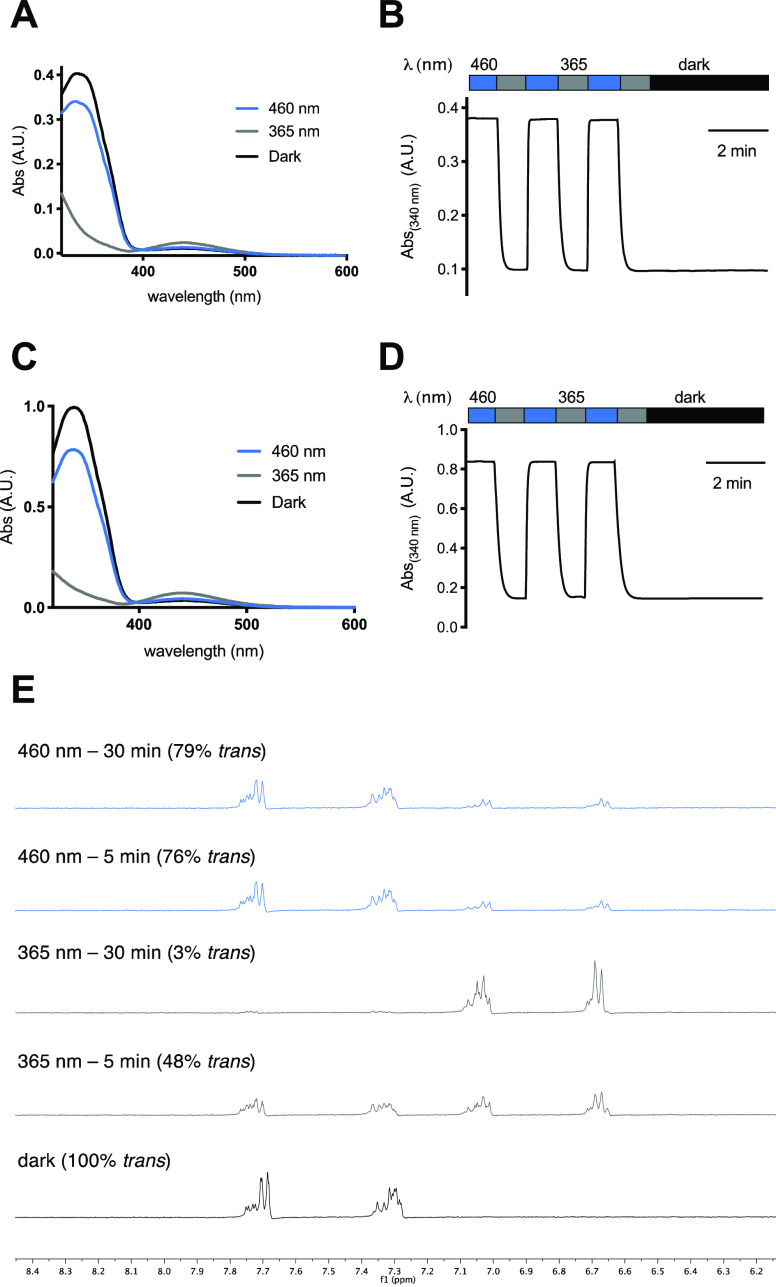
The photophysical characterization of AzoPA and dAzoPA revealed similar properties to classical azobenzenes and other photoswitchable lipids (Figure 2). Both photolipids could be reversibly switched to their respective cis and trans forms with UV-A (365 nm) and blue light (460 nm) and underwent slow thermal relaxation (t1/2 = 678 min in PBS) (Figures 2A–D and S1). Interestingly, the molar absorption of dAzoPA at 340 nm is approximately 2 times higher than that of AzoPA but almost equally high after switching to the cis form, suggesting that both azobenzenes are effectively switched to cis upon irradiation. To quantify the absolute amount of the trans and cis forms of dAzoPA after irradiation, we performed NMR studies (Figure 2E). These studies confirmed that both azobenzenes can be effectively switched from their trans form (100% total trans isomer) to their cis form (97% total cis isomer) upon irradiation with 365 nm light.
Figure 2.
Photophysical characterization of AzoPA (A,B) and dAzoPA (C,D). (A,C) UV–vis spectra of AzoPA (A) and dAzoPA (C) in the dark-adapted (black, trans), 365-nm-adapted (gray, cis), and 460-nm-adapted (blue, trans) photostationary states (50 μM, DMSO). (B,D) Reversible cycling between photoisomers with alternating illumination at 365/460 nm for AzoPA (B) and dAzoPA (D). (E) Nuclear magnetic resonance (NMR) study (performed in DMSO-d6) of the aromatic region of dAzoPA before and after irradiation with 365 or 460 nm and quantification of the trans form relative to the cis form.
Cellular Uptake and Metabolism of AzoPA and dAzoPA
PA has a complex metabolic pathway in mammalian cells (Figure 3A), motivating us to investigate the cellular uptake and metabolism of AzoPA and dAzoPA. We treated NIH 3T3 mouse embryonic fibroblast cells with these lipids in their cis or trans forms, precomplexed to fatty-acid-free bovine serum albumin (BSA), for 1 h and then analyzed cellular lipid extracts by HPLC. Because photoswitchable lipids contain a unique azobenzene moiety with a strong absorption at 350 nm in its trans form, they are easily distinguishable from endogenous lipids via HPLC. This analysis revealed that cis and trans forms of both AzoPA and dAzoPA were taken up to similar extents (Figure 3B,C), corresponding to approximately 1 × 108 molecules/cell when cells were treated with 10 μM photolipid (Figure S2A).
Figure 3.
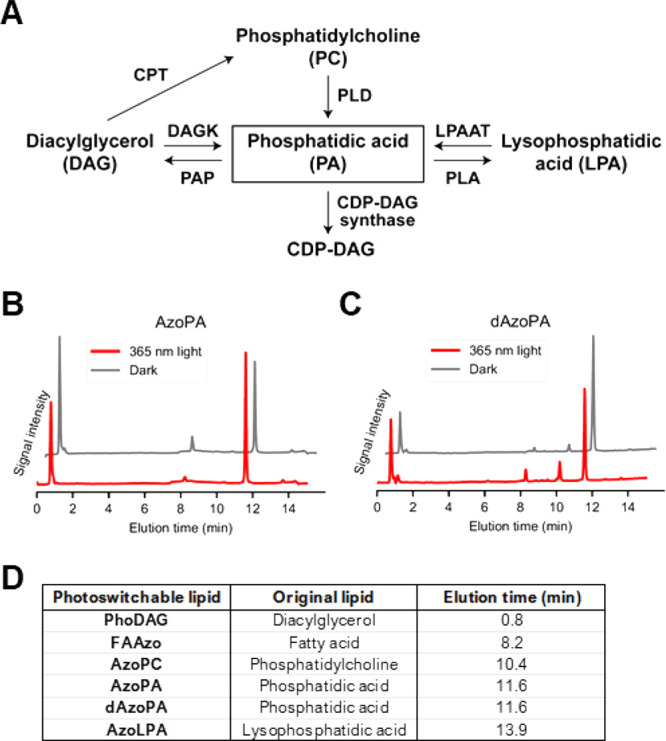
Characterization of cellular uptake and metabolism of AzoPA and dAzoPA. (A) Schematic diagram of mammalian PA metabolism. CDP: Cytidine diphosphate, CPT: cholinephosphotransferase, DAGK: diacylglycerol kinase, LPAAT: lysophosphatidic acid acyl transferase, PAP: phosphatidic acid phosphatase, PLA: phospholipase A, PLD: phospholipase D. (B,C) HPLC trace of lipid extracts from NIH 3T3 cells treated with 20 μM AzoPA (B) or dAzoPA (C) in the light-induced cis form (red) or dark-adapted trans form (gray) for 1 h. Just prior to the analysis, all samples were irradiated with 460 nm light to switch azobenzene moieties into the trans forms and normalize the absorption. (D) Observed elution times of standard photolipid molecules on HPLC.
We observed several other azobenzene-containing species that corresponded to photolipid analogues of diacylglycerol (PhoDAG), lysophosphatidic acid (AzoLPA), fatty acid (FAAzo4), and phosphatidylcholine (AzoPC), suggesting that AzoPA and dAzoPA are metabolized similarly to endogenous PA12 (Figure 3D). The non-PA species were not present in the cell culture media (Figure S2B), suggesting that the conversion occurs inside cells and not in an extracellular environment. Interestingly, these photolipid species, including AzoPA, were also observed in cells treated with AzoLPA and PhoDAG (Figure S2C–E), revealing the interconnectivity of photolipid metabolism. We used deuterium-labeled 16:0(d31)-18:1 PA (d31-POPA) in a pulse-chase experiment to study the rate of turnover in comparison to dAzoPA and found that d31-POPA turnover rates in NIH 3T3 cells were similar to those of the synthetic analogue dAzoPA (Figure S2F). Together, our studies validate that both AzoPA and dAzoPA are effectively incorporated into cells and recognized by endogenous lipid-modifying enzymes.
Our HPLC results show that our PA photolipids can be converted into the corresponding DAG analogues in cells (Figure 2B,C). To evaluate if AzoPA/dAzoPA metabolism leads to formation of sufficiently large amounts of azobenzene-containing DAG analogues to cause activation of PKC signaling,29,34 we performed imaging assays using C1-EGFP, a DAG-binding fluorescent probe. Whereas the exogenous addition of a phorbol ester or PhoDAG itself led to pronounced translocation of C1-EGFP from the cytosol to the plasma membrane, such translocation was not observed upon addition of AzoPA or dAzoPA (Figure S2G). These data suggest that the small amounts of PhoDAG formed by metabolic conversion of AzoPA or dAzoPA have negligible effects on PKC signaling.
Optical Control of mTOR Signaling
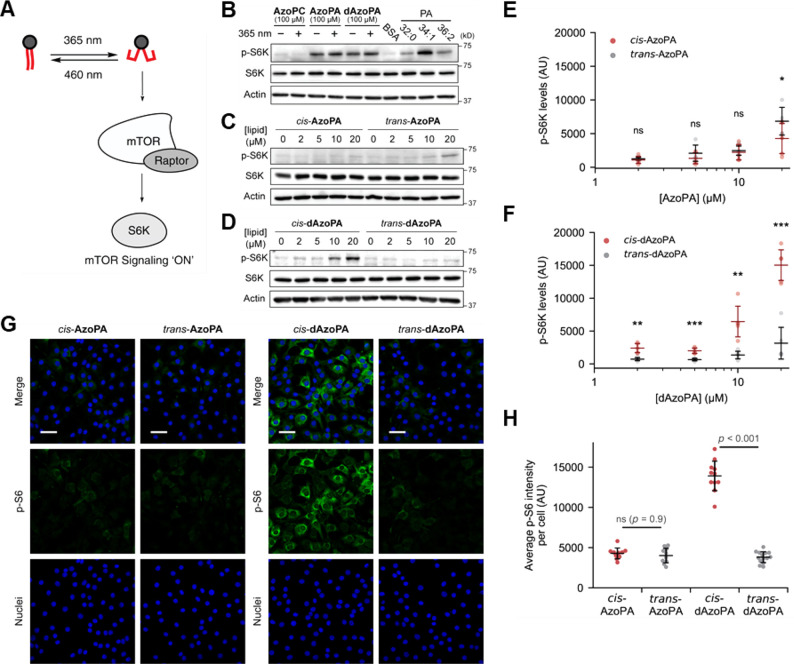
We then studied the bioactivity of AzoPA and dAzoPA and their abilities to enable control of cellular signaling pathways with light. PA species with unsaturated acyl chains activate the mTOR Complex 1 (mTORC1) through binding to the FKBP12 rapamycin binding (FRB) domain and displacing its inhibitor, DEPTOR.7 We first tested if the photoswitchable analogues of PA could also affect mTOR signaling and thus manipulate the activity of a key downstream target of mTORC1 signaling, p70-S6 kinase (S6K) (Figure 4A). To examine mTORC1 activation, we measured the levels of S6K phosphorylated at Thr389 (p-S6K) by Western blot analysis. Unlike most other signaling pathways, mTOR signaling has been previously reported to directly respond to 365 nm light.49 Indeed, we found that intermittent 365 nm light irradiation to cells (5 ms every 15 s) suppressed PA-induced activation of p-S6K (Figure S3A). Thus, to exclude direct effects of light on mTOR signaling, we applied 365 nm light only prior to and not during the incubation with cells to obtain samples with the cis-isomer. Serum-starved NIH 3T3 cells were treated with photolipids (AzoPC, AzoPA, or dAzoPA; at 100 μM, with or without 365 nm light) or natural PA species with different acyl tail compositions of varying degrees of unsaturation (32:0, 34:1, or 36:2) for 1 h. Whereas all the PA species increased p-S6K levels, the unsaturated ones (34:1, 36:2) displayed significantly more activity, in accordance with previous reports (Figure 4B).7 Both AzoPA and dAzoPA, but not AzoPC, were effective in increasing p-S6K levels, suggesting that the photoswitchable analogues of PA can mimic its bioactivity (Figure 4B).
Figure 4.
Optical control of mTOR signaling in NIH 3T3 cells. (A) Schematic depiction of the optical control of mTOR activation induced by PA. (B) Western blot analysis of NIH 3T3 cells treated with BSA (4 mg/mL) or indicated lipids (100 μM) for 1 h, probing for p-S6K, S6K, or actin as a loading control. (C,D) Western blot analysis of cells treated with AzoPA (C) or dAzoPA (D) in their light-induced cis form or dark-adapted trans form at different concentrations (0–20 μM) for 30 min. (E,F) Quantification of p-S6K levels in (C) and (D). Horizontal bars indicate mean (n = 6), and vertical error bars indicate standard deviation. Statistical significance was calculated using an unpaired two-tailed Student’s t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001. (G) Confocal microscopy images of NIH 3T3 cells treated with the indicated lipids (5 μM) for 30 min and immunostained for p-S6. Green, p-S6; blue, DAPI (nuclei). Scale bars: 50 μm. (H) Quantification of immunofluorescence analysis. The plots show average p-S6 intensity per cell in each image. Black horizontal bars indicate mean (n = 10–14), and vertical error bars indicate standard deviation. Statistical significance was calculated using one-way ANOVA, followed by Tukey’s HSD test.
No differences between the cis and trans forms were observable at this relatively high concentration (100 μM); thus, we performed dose–response studies with each isomer to quantify their activities. Excitingly, we found that, of the two pairs of isomeric PA analogues, only a single isomeric compound, cis-dAzoPA, induced phosphorylation of S6K at lower concentrations (10–20 μM) (Figure 4C–F). To confirm that this effect was not due to cellular metabolism of AzoPA and dAzoPA, we analyzed p-S6K level in cells treated with AzoLPA or PhoDAG, the major metabolites of AzoPA and dAzoPA. Gratifyingly, we observed no detectable increase in p-S6K levels in these cells, suggesting that AzoLPA and PhoDAG are not involved in this effect (Figure S3B,C).
To further ascertain the activation of p-S6K, we quantified the amount of phosphorylated ribosomal protein S6 (p-S6), which is a downstream target of p-S6K, by immunofluorescence. Indeed, cells treated with cis-dAzoPA exhibited a substantial increase in levels of p-S6 (Figure 4G,H), consistent with the previous Western blot analysis of p-S6K levels. Notably, these effects were selective to dAzoPA and not AzoPA, suggesting a high degree of acyl chain selectivity in PA-induced activation of mTOR signaling. Overall, these studies validate dAzoPA as an effective tool for optical control of mTOR signaling via mimicry of PA species with unsaturated acyl tails and underscore the potency of unsaturated forms of PA as effective activators of mTORC1 signaling.
Optical Control of Hippo Signaling
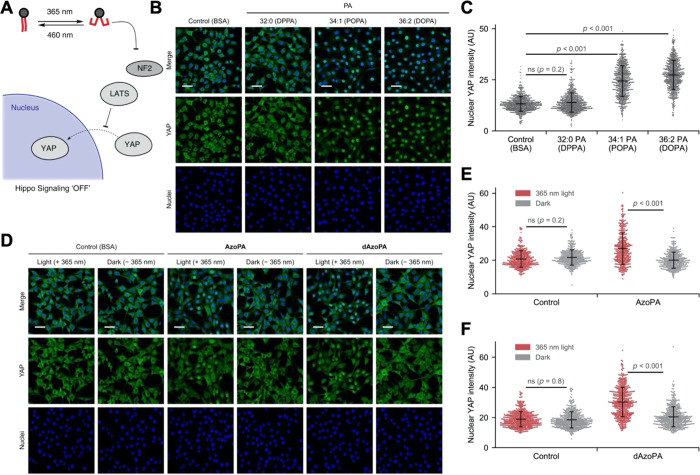
Having demonstrated that the photoswitchable PA analogues could activate a well-established PA-dependent pathway, mTOR signaling, with optical control, we next sought to assess the ability of AzoPA and dAzoPA to modulate Hippo signaling, whose connection to PA signaling was only recently described.50 The Hippo pathway is an important attenuator of cell growth and proliferation.10 When Hippo signaling is on, large tumor suppressor 1/2 (LATS1/2) kinases are active and phosphorylate the transcription factor Yes-associated protein (YAP) to prevent its translocation to the nucleus. PA has recently been discovered to modulate Hippo signaling.8 PA can bind to both LATS1/2 and an upstream factor, NF2, thereby inhibiting Hippo signaling. Using an optogenetic phospholipase D, we have previously demonstrated that the pool of PA that exerts these effects resides at the plasma membrane.14 In different contexts, PA has been proposed to bind to the Rab-like GTPase RAP2 and thereby modulate Hippo signaling.51 The full spectrum of PA’s roles in affecting Hippo signaling remains to be determined, including the relative importance of different PA species in such regulation. These mechanisms are of vital importance to understand, given the established connection of both PA and the Hippo pathway to the control of cell growth and proliferation in normal physiology and in cancer.4,5,52
As a first step toward these goals, we investigated the effects of the photoswitchable PA analogues AzoPA and dAzoPA on the regulation of Hippo signaling, which can be quantified by measuring nuclear translocation of YAP (Figure 5A). For Hippo signaling, the potency of different PA species has not been reported. We treated serum-starved NIH 3T3 cells with 50 μM PA with different acyl tail compositions (32:0, 34:1, or 36:2), complexed to bovine serum albumin (BSA), for 1 h and visualized the subcellular location of YAP by immunofluorescence. Interestingly, we found that PA species with unsaturated acyl chains (34:1, 36:2) triggered YAP translocation into nuclei most strongly (Figure 5B,C). However, the fully saturated DPPA was not incorporated as efficiently into lipid–BSA complexes (Figure S4A), which could contribute to the decreased bioactivity observed compared to unsaturated forms of PA. It is worth noting that photoswitchable analogues of PA have similar cellular uptake in their cis and trans forms (Figure 3B,C), and their ability to be photoswitched in situ enables the study of bioactivity of forms with different acyl tail shapes independent from effects on cellular uptake. We thus performed the same experiment using 10 μM AzoPA or dAzoPA, with or without preirradiation with 365 nm light to obtain their cis or trans forms. Excitingly, both cis-AzoPA and cis-dAzoPA, and not the trans isomers, induced nuclear translocation of YAP (Figure 5D–F). Similar results were obtained when 365 nm light was applied after addition of the photolipids to cells in their inactive form to temporally control the photoconversion in cell culture (Figure S4B,C). We also tested two related photoswitchable lipids with different headgroup structures, AzoPC and PhoDAG. These molecules, in either the cis or trans forms, did not induce any observable increase in YAP localization (Figure S4D,E), confirming the selectivity of this effect. Collectively, these results suggest that both AzoPA and dAzoPA can regulate Hippo signaling in a light-dependent manner.
Figure 5.
Optical control of Hippo signaling in NIH 3T3 cells. (A) Schematic depiction of the optical control of Hippo deactivation induced by PA. (B) Confocal microscopy images of NIH 3T3 cells treated with BSA (4 mg/mL) or the indicated lipids (50 μM) for 1 h and immunostained for YAP. (C) Quantification of nuclear YAP levels in (B). The plots show mean nuclear YAP intensity in each cell. Black horizontal bars indicate mean (n = 645), and vertical error bars indicate standard deviation. (D) Confocal microscopy images of NIH 3T3 cells treated with BSA (4 mg/mL) or indicated lipids (10 μM) for 1 h with or without 365 nm light (5 ms every 15 s) and immunostained for YAP. (E,F) Quantification of nuclear YAP levels in (C) and (D). The plots show mean nuclear YAP intensity in each cell. Black horizontal bars indicate mean (n = 342 (C) and 465 (D)), and vertical error bars indicate standard deviation. Statistical significance was calculated using one-way ANOVA, followed by Tukey’s HSD test. Green, YAP; blue, DAPI (nuclei). Scale bars: 50 μm.
Lysophosphatidic acid (LPA), which is formed from PA by phospholipase A enzymes, can also regulate Hippo signaling via activation of GPCRs from the LPA receptor family.53 Previous work has established that the effects of PA on Hippo signaling are independent of LPA signaling.8 Yet, because the photoswitchable PA analogues are distinct molecular species from natural PA, we sought to determine if conversion of AzoPA to AzoLPA, and subsequent signaling via LPA receptors, was involved in the AzoPA-mediated Hippo regulation. For these studies, we used an LPA receptor inhibitor, Ki16425 (LPAi), which blocks the binding of LPA to two major LPA receptors, LPA1 and LPA3, and thus prevents LPA-induced YAP dephosphorylation.53,54
We visualized the localization of YAP in cells treated with 10 μM LPAi for 1 h, followed by treatment with 100 nM AzoLPA or 10 μM dAzoPA for 1 h, with or without 365 nm light. Interestingly, cis-AzoLPA, but not the trans form, induced the nuclear translocation of YAP, and this effect was completely blocked by treatment with LPAi (Figure 6A,C). This observation is consistent with the ability of AzoLPA to optically control LPA signaling via LPA receptors, as previously assessed by different readouts of LPA receptor activation.33 By contrast, LPAi had a negligible effect on YAP translocation induced by cis-dAzoPA (Figure 6B,D). Taken together, these results demonstrate that, although AzoLPA can indeed affect Hippo signaling through its binding to the cell surface receptors, it is not involved in the process of Hippo regulation that is mediated by photoswitchable PA analogues.
Figure 6.
Optical control of Hippo signaling in NIH 3T3 cells with the inhibition of LPA receptor signaling. (A,B) Confocal microscopy images of NIH 3T3 cells treated with AzoLPA (100 nM) or dAzoPA (10 μM) for 1 h with or without 365 nm light and with or without LPA receptor inhibitor (LPAi: Ki16425, 10 μM) and immunostained for YAP. Green, YAP; blue, DAPI (nuclei). Scale bars: 50 μm. (C,D) Quantification of nuclear YAP levels in (A). The plots show mean nuclear YAP intensity in each cell. Black horizontal bars indicate mean (n = 540 (C) and 811 (D)), and vertical error bars indicate standard deviation. Statistical significance was calculated using one-way ANOVA, followed by Tukey’s HSD test.
Discussion
In this study, we developed photoswitchable analogues of PA and demonstrated that they can enable the optical control of two major PA-dependent pathways, mTOR and Hippo signaling. Our photoswitchable analogues, AzoPA and dAzoPA, were designed to incorporate azobenzene units in their lipid tails, which preserves the integrity of the headgroup and allows these photolipids to effectively mimic the endogenous lipid. Unlike natural PA, however, the photoswitchable PAs allow for optical control of the shape of the acyl tails. We found that the exclusively doubly cis form of dAzoPA mimics the potent bioactivity of unsaturated species of PA in regulation of mTOR signaling, whereas the trans form, and even the cis form of AzoPA, which still contains a saturated acyl tail at the sn-1 position, have much lower activities, mimicking the relatively inactive, saturated PA species. This finding highlights the remarkable selectivity afforded by dAzoPA, which, by virtue of containing two azobenzene photoswitches, undergoes a major light-induced structural rearrangement.
Having validated that photoswitchable PA analogues can effectively modulate an established PA-dependent, oncogenic pathway (mTORC1 signaling), we next investigated the effects of AzoPA and dAzoPA on Hippo signaling, a recently discovered target of PA. We found that unsaturated forms of PA may be more potent agents at suppressing the Hippo pathway than the saturated forms. Although we cannot decouple the contribution of reduced efficiency of saturated PA (DPPA) in the formation of lipid–BSA complexes, compared to DOPA and POPA, from this effect, both AzoPA and dAzoPA enabled optical control of this signaling pathway and were more potent in their light-induced cis forms. Notably, they can be delivered to cells in one form and subsequently switched, which could potentially allow the study of bioactivity independent of cellular uptake. These results stand in contrast to effects of our photoswitchable PA analogues on mTOR signaling, which is exclusively activated by cis-dAzoPA. This divergence in activities likely reflects differential lipid-binding requirements of the relevant PA effectors implicated in these two distinct pathways. Nevertheless, our findings highlight an added measure of selectivity afforded by AzoPA, in that its photoisomerization can favor activation of one PA-dependent outcome over another. Such unprecedented selectivity suggests the use of the photolipids reported here to decouple the pleiotropy of PA signaling in additional contexts.
Finally, beyond an ability to control PA-dependent signaling pathways, AzoPA and dAzoPA undergo dynamic metabolism, similar to endogenous PA, as confirmed by HPLC analysis with authentic photolipid standards to detect and assign AzoPA- and dAzoPA-derived photolipid metabolites in cells. Importantly, using a panel of photolipids and pharmacological manipulations, we ruled out potential effects of the observed photolipid metabolites on mTOR and Hippo signaling in this particular context and ascribe the observed bioactivities to our photoswitchable analogues of PA. Yet, the appearance of azobenzene-derived metabolites of AzoPA and dAzoPA indicate additional uses of these photolipids as probes of dynamic PA metabolism. Overall, the photolipid analogues of PA presented here comprise a set of tools to control PA signaling and also aid in determining the multiple effects of PA in the context of complex and dynamic lipid metabolic pathways.
Experimental Section
General Methods
All reagents and solvents were purchased from commercial sources (Sigma-Aldrich, TCI Europe N.V., Strem Chemicals, etc.) and were used without further purification. Solvents were obtained from Fisher Scientific. Reactions were monitored by thin layer chromatography (TLC) on precoated, Merck Silica gel 60 F254 aluminum-backed plates, and the chromatograms were first visualized by UV irradiation at λ = 254 nm. Flash silica gel chromatography was performed using silica gel (SiO2, particle size 40–63 μm) purchased from SiliCycle. NMR spectra were measured on a BRUKER Avance III HD 400 (equipped with a CryoProbe). Multiplicities in the following experimental procedures are abbreviated as follows: s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet. 1H chemical shifts are expressed in parts per million (ppm, δ scale) and are referenced to the residual protium in the NMR solvent (CDCl3: δ = 7.26; THF-d8: δ = 1.72). 13C chemical shifts are expressed in ppm (δ scale) and are referenced to the carbon resonance of the NMR solvent (CDCl3: δ = 77.16, THF-d8: δ = 25.31). NOTE: Due to the trans/cis isomerization of some compounds containing an azobenzene functionality, more signals were observed in the 1H and 13C spectra than would be expected for the pure trans-isomer. Only signals for the major trans-isomer are reported.
Synthesis of 1
PhoDAG-1(34) (50.0 mg, 75.2 mmol, 1.0 equiv) was dissolved in dry CH2Cl2 (2.0 mL) under a nitrogen atmosphere. Di-tert-butyl diisopropylphosphoramidite (37.1 mg, 134 mmol, 1.8 equiv) and 1H-tetrazole (0.45 M solution in MeCN, 500 mL) were added, and the solution was stirred for 50 min at room temperature. The mixture was cooled to 0 °C, and mCPBA (1.20 g, 7.20 mmol, 3.0 equiv) was added. The mixture was stirred for 30 min at 0 °C and 30 min at room temperature, washed with aqueous Na2S2O3 solution (10%) and NaHCO3 solution (saturated), dried, and filtered, and the solvent was evaporated under reduced pressure. The crude mixture was purified by flash column chromatography (4:1 hexanes/ethyl acetate) to yield di-tert-butyl AzoPA (1) as orange oil (57.2 mg, 66.7 mmol, 89%). 1H NMR (400 MHz, CDCl3) δ 7.82 (dd, J = 8.3, 2.9 Hz, 4H), 7.33–7.29 (m, 4H), 5.27–5.21 (m, 1H), 4.38 (dd, J = 12.0, 4.0 Hz, 1H), 4.19–4.05 (m, 3H), 2.71 (dt, J = 18.3, 7.6 Hz, 4H), 2.38 (t, J = 7.4 Hz, 2H), 2.30 (t, J = 7.6 Hz, 2H), 2.04–1.95 (m, 2H), 1.70–1.62 (m, 2H), 1.48–1.46 (m, 18H), 1.42–1.35 (m, 2H), 1.24 (d, J = 4.4 Hz, 30H), 0.94 (t, J = 7.3 Hz, 3H), 0.87 (d, J = 7.0 Hz, 3H). 13C NMR (100 MHz, CDCl3) δ 173.5, 172.5, 151.4, 151.1, 146.5, 144.6, 129.3, 129.2, 123.0, 122.9, 82.9, 70.0, 64.5, 62.1, 53.6, 35.7, 35.0, 34.2, 33.6, 33.6, 32.1, 30.0, 30.0, 29.9, 29.8, 29.8, 29.6, 29.5, 29.4, 29.3, 26.4, 25.0, 22.9, 22.5, 14.3, 14.1. 31P NMR (162 MHz, CDCl3) δ −9.89. HRMS: m/z calcd. for C49H82N2O8P+ ([M + H]+): 857.5803, found: 857.5794.
Synthesis of AzoPA
Di-tert-butyl AzoPA (1) (37.2 mg, 43.4 μmol, 1.0 equiv) was dissolved in CH2Cl2 (10 mL). TFA (1.1 mL) was added, and the solution was stirred for 10 min at room temperature. MeOH (128 μL) was added, and the mixture was stirred for 10 min at room temperature. Toluene (5 mL) was added, and the solvents were removed under reduced pressure. AzoPA was obtained as an orange solid (32.2 mg, 43.2 μmol, quantitative). 1H NMR (400 MHz, CDCl3) δ 7.81 (d, J = 8.0 Hz, 4H), 7.29 (d, J = 8.1 Hz, 4H), 5.25 (s, 1H), 4.34 (d, J = 10.2 Hz, 1H), 4.16 (s, 3H), 2.76–2.63 (m, 4H), 2.38 (t, J = 6.9 Hz, 2H), 2.28 (t, J = 7.5 Hz, 2H), 2.03–1.92 (m, 2H), 1.63 (p, J = 7.6 Hz, 2H), 1.55 (s, 2H), 1.37 (q, J = 7.4 Hz, 2H), 1.23 (d, J = 10.1 Hz, 30H), 0.94 (t, J = 7.3 Hz, 3H), 0.87 (t, J = 6.8 Hz, 4H). 13C NMR (100 MHz, CDCl3) δ 174.1, 173.2, 151.1, 150.8, 146.7, 144.8, 129.4, 129.2, 123.1, 123.0, 70.0, 65.2, 62.2, 35.7, 34.9, 34.2, 33.6, 33.5, 32.1, 29.9, 29.8, 29.8, 29.7, 29.5, 29.4, 29.3, 26.3, 24.9, 22.8, 22.5, 14.3, 14.1. 31P NMR (162 MHz, CDCl3) δ 0.82. HRMS: m/z calcd. for C41H65N2O5+ ([M–PO3]+): 665.4888, found: 665.4897.
Synthesis of 2
To a solution of di-tert-butyl AzoLPA33 (20.8 mg, 35.2 μmol, 1.0 equiv) in dry CH2Cl2 under nitrogen was added FAAzo-4(28) (30.3 mg, 93.3 μmol, 2.7 equiv), followed by DCC (19.4 mg, 94.0 μmol, 2.7 equiv) and DMAP (70.0 μg, 5.60 μmol, 0.2 equiv). The resulting mixture was stirred for 20 h at room temperature. After this time, the reaction mixture was filtered through a pad of Celite, and the filtrate was concentrated under reduced pressure. The crude product was purified by flash column chromatography (hexane/EtOAc/Et3N, 80:20:0.1 to 70:30:0.1) to yield di-tert-butyl dAzoPA (2) as an orange oil (24.0 mg, 26.8 μmol, 76%). 1H NMR (400 MHz, CDCl3) δ 7.82 (d, J = 8.0 Hz, 8H), 7.30 (dd, J = 8.1, 4.2 Hz, 8H), 5.28–5.23 (m, 1H), 4.40 (dd, J = 12.0, 4.1 Hz, 1H), 4.19 (dd, J = 12.1, 6.1 Hz,1H), 4.13–4.03 (m, 2H), 2.74–2.63 (m, 7H), 2.40–2.34 (m, 3H), 2.11–1.93 (m, 5H), 1.76–1.62 (m, 6H), 1.46 (s, 18H), 1.42–1.34 (m, 4H), 0.94 (t, J = 7.6 Hz, 6H). 13C NMR (100 MHz, CDCl3) δ 172.9, 172.5, 151.4, 151.1, 146.4, 144.5, 144.4, 129.3, 129.2, 129.2, 129.2, 123.0, 122.89, 83.0, 82.9, 82.9, 70.0, 69.9, 64.5, 64.4, 62.3, 49.3, 36.8, 35.7, 35.0, 35.0, 34.1, 33.6, 33.5, 33.4, 33.4, 30.0, 29.9, 29.8, 26.4, 26.3, 25.7, 25.1, 24.8, 22.5, 22.4, 14.1, 14.0. 31P NMR (162 MHz, CDCl3) δ −9.92.
Synthesis of dAzoPA
Di-tert-butyl dAzoPA (2) (24.0 mg, 26.8 μmol, 1.0 equiv) was dissolved in CH2Cl2 (5.2 mL). TFA (1.3 mL) was added, and the solution was stirred for 10 min at room temperature. MeOH (75 μL) was added, and the mixture was stirred for 10 min at room temperature. Toluene (15 mL) was added, and the solvents were removed under reduced pressure. dAzoPA was obtained as an orange solid (20.8 mg, 26.5 μmol, quantitative). 1H NMR (400 MHz, THF-d8) δ 7.80 (d, J = 6.2 Hz, 8H), 7.31 (d, J = 7.9 Hz, 8H), 5.26 (br. s, 1H), 4.40 (dd, J = 11.8, 3.6 Hz, 1H), 4.20 (dd, J = 12.4, 6.3 Hz,1H), 4.16–4.08 (m, 2H), 2.75–2.65 (m, 7H), 2.41–2.31 (m, 3H), 2.01–1.92 (m, 3H), 1.69–1.61 (m, 5H), 1.44–1.34 (m, 4H), 0.94 (t, J = 7.4 Hz, 6H). 13C NMR (100 MHz, THF-d8) δ 173.2, 173.0, 152.3, 152.1, 147.2, 146.3, 146.2, 130.2, 130.1, 130.1, 130.0, 123.8, 123.7, 123.7, 63.1, 55.1, 49.3, 36.4, 35.8, 35.8, 34.9, 34.6, 33.9, 27.5, 27.4, 26.0, 23.4, 14.4. 31P NMR (162 MHz, THF-d8) δ 1.07. HRMS: m/z calcd. for C43H53N4O5 ([M–PO3]+): 705.4010, found: 705.3994.
Photophysical Characterization
UV–vis spectra were recorded using a Varian Cary 50 Bio UV–visible Spectrophotometer. Photoswitching was achieved using 365 or 460 nm LED light sources. The LEDs were pointed directly onto the top of the sample cuvette with AzoPA (50 μM in DMSO). An initial spectrum was recorded (dark-adapted state) and then again following illumination at 365 nm for 30 s (cis-adapted state). A third spectrum was recorded after irradiation at 460 nm for 30 s (trans-adapted state). Absorption at 340 nm was recorded over several switching cycles while alternating illumination at 365 and 460 nm with. The light source was directly pointed onto the top of the sample cuvette.
Mammalian Cell Culture
NIH 3T3 mouse fibroblast cells were grown in DMEM (Corning) supplemented with 10% bovine calf serum (Corning) and 1% penicillin/streptomycin (Corning) at 37 °C in a 5% CO2 atmosphere. For serum starvation, NIH 3T3 cells were cultured in DMEM supplemented with 1% penicillin/streptomycin (serum-free medium). For experiments, cells were seeded in 12-well plates for Western blot analysis, 35 mm glass-bottom imaging dishes (MatTek Corporation) for immunofluorescence, and 35 mm dishes for HPLC and LC–MS analysis. For C1-EGFP translocation assays, cells seeded on 35 mm imaging dishes were transfected using 1 μg of C1-EGFP plasmid (a gift from the De Camilli laboratory, Yale University, New Haven, CT) and 3 μL of Lipofectamine 2000 (Invitrogen) per dish, 24 h prior to the experiment.
Preparation of Lipid–BSA Complexes
Photoswitchable lipid analogues were synthesized in the lab, and PA was purchased (DPPA, Cayman, 15082; POPA, Avanti Polar Lipids, 840857; DOPA, Avanti Polar Lipids, 840875). The lipid was dissolved in chloroform and dried under a stream of N2 gas to form a lipid film. The lipid film was resuspended, with sonication, in 4 mg/mL of fatty-acid-free BSA (Sigma-Aldrich, A8806) in DPBS (Corning) to a final concentration of 1 mM lipid to generate a stock solution. For PhoDAG, because of its low solubility in aqueous solution,30 the lipid film was resuspended in DMSO to a final concentration of 10 mM. For isomerization to the cis form, the lipid solution was treated with 365 nm light for 10 min using a UV lamp (Thermo Fisher, # UVP95004207) prior to addition to cells.
HPLC and LC–MS Analysis of Lipid Extracts
NIH 3T3 cells were starved in serum-free medium for 20 h, and cells were treated with the indicated lipids (5–20 μM lipid in the form of lipid–BSA complexes, with or without pretreatment of 5 min illumination with 365 nm light) for 30 min or 1 h. After the incubation, cells were rinsed with PBS for three times, and lipid extracts were obtained using a modified Bligh–Dyer extraction as reported previously.14 Briefly, cells were scraped in 250 μL of methanol, 125 μL of acetic acid (20 mM in water), and 100 μL of PBS using cell scrapers (Corning), and the suspension was transferred into 1.5 mL microcentrifuge tubes. After the addition of 500 μL of chloroform, the mixture was vortexed vigorously for 5 min and centrifuged at 17 000g for 1 min. The organic layer (bottom) was collected in a new tube and dried under a stream of N2 gas to obtain a lipid film, which was then resuspended in chloroform. For HPLC analysis, the lipid solution was irradiated with blue light (488 nm laser) for 10 s and quickly injected into a Shimadzu Prominence HPLC equipped with an in-line fluorescence detector. Separation was achieved using a Luna 3 μm Silica LC Column (Phenomenex; 150 × 4.6 mm) with a binary gradient elution system where solvent A was chloroform/methanol/ammonium hydroxide (95:7:0.5), and solvent B was chloroform/methanol/water/ammonium hydroxide (60:34:5:0.5). For LC–MS analysis, the lipid solution was analyzed on an Agilent 6230 electrospray ionization–time-of-flight MS coupled to an Agilent 1260 HPLC equipped with a Luna 3 μm Silica LC Column (Phenomenex; 50 × 2 mm) using a binary gradient elution system where solvent A was chloroform/methanol/ammonium hydroxide (85:15:0.5), and solvent B was chloroform/methanol/water/ammonium hydroxide (60:34:5:0.5).
Quantification of Lipids in Lipid–BSA Complexes
Lipid–BSA complexes were separated from free lipids by size-exclusion chromatography using an ÄKTA pure system equipped with a Superdex 200 Increase 10/300 GL column in PBS. The fractions containing BSA were subjected to Bligh–Dyer lipid extraction as described above, and the obtained lipid extracts were analyzed by LC–MS as described above.
Evaluation of mTORC1 Activity
NIH 3T3 cells were starved in serum-free medium for 20 h, and cells were treated with the indicated lipids (10 μM lipid in the form of lipid–BSA complexes with or without pretreatment of 5 min illumination with 365 nm light) for 30 min or 1 h. For Western blot analysis, cells were lysed in RIPA lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton-X, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 1× cOmplete Protease Inhibitor, 1 mM sodium orthovanadate). The membrane was blotted with antibodies for phospho-p70 S6 kinase (Thr389) (Cell Signaling Technology, #9205), p70 S6 kinase (Santa Cruz Biotechnology, sc-8418), or actin (MP Biomedicals, 08691001), with detection by chemiluminescence using the Clarity Western ECL Substrate (Bio-Rad) and acquisition on a Bio-Rad ChemiDoc MP System. For immunofluorescence, sample preparation and confocal microscopy were performed as reported previously.14 Briefly, cells were fixed with 4% formaldehyde for 10 min, permeabilized with 0.5% Triton X-100 in PBS for 5 min, and blocked with 1% BSA and 0.1% Tween-20 in PBS (blocking buffer) for 30 min. Primary antibody incubation was performed with a 1:250 dilution of a phospho-S6 (Ser240/244) antibody (Cell Signaling Technology, #5364), and secondary antibody incubation was performed with a 1:500 dilution of antimouse–Alexa Fluor 488 (Invitrogen, A-21202). Image acquisition by laser-scanning confocal microscopy was performed as described before by using solid-state lasers (405, 488 nm) to excite DAPI and Alexa Fluor 488, respectively.14 For calculation of the average p-S6 intensity per cell, the total integrated density of p-S6 signal in the frame was measured and divided by the number of cells in the frame using FIJI and a Python script: “Intensity_per_cell.py”.
Evaluation of Nuclear YAP Localization
NIH 3T3 cells were starved in serum-free medium for 6 h, and cells were treated with the indicated lipids (10 μM lipid in the form of a lipid–BSA complex, with or without pretreatment of 5 min illumination with 365 nm light) for 1 h with or without 365 nm light (5 ms of illumination at 15 s intervals). For experiments with LPA receptor inhibitor, cells were pretreated with 10 μM Ki16425 (Cayman Chemical Company, 10012659) for 1 h. Immunofluorescence analysis was performed as described above, and primary antibody incubation was done with 1:200 dilution of YAP antibody (Santa Cruz Biotechnology, sc-101199). For statistical analysis of YAP nuclear intensity, the mean of YAP signal in each nucleus was collected using FIJI and a Python script: “Nuclear_intensity.py”.
Safety Statement
No unexpected or unusually high safety hazards were encountered.
Acknowledgments
J.M.B. acknowledges support from a Beckman Young Investigator award, a Sloan Research Fellowship, and the NSF (CAREER CHE-1749919). D.T. acknowledges support from NYU. R.T. was supported by Honjo International, Funai Overseas, and Cornell Fellowships. J.M. thanks the German Academic Scholarship Foundation for a fellowship, the New York University for a MacCracken fellowship and a Margaret and Herman Sokol fellowship, and the NCI for an F99/K00 award (1F99CA253758-01). We thank the Fromme lab for use of equipment.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.1c00444.
1H and 13C NMR spectra, Python scripts, and Supplementary Figures (PDF)
Author Contributions
# R.T. and J.M. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Liu Y.; Su Y.; Wang X. Phosphatidic Acid-Mediated Signaling. Lipid-mediated Protein Signaling 2013, 991, 159–176. 10.1007/978-94-007-6331-9_9. [DOI] [PubMed] [Google Scholar]
- Kooijman E. E.; Burger K. N. J. Biophysics and Function of Phosphatidic Acid: A Molecular Perspective. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2009, 1791 (9), 881–888. 10.1016/j.bbalip.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Tanguy E.; Wang Q.; Moine H.; Vitale N. Phosphatidic Acid: From Pleiotropic Functions to Neuronal Pathology. Front. Cell. Neurosci. 2019, 13, 2. 10.3389/fncel.2019.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cambronero J. Phosphatidic Acid, Phospholipase D and Tumorigenesis. Adv. Biol. Regul. 2014, 54, 197–206. 10.1016/j.jbior.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruntz R. C.; Lindsley C. W.; Brown H. A. Phospholipase D Signaling Pathways and Phosphatidic Acid as Therapeutic Targets in Cancer. Pharmacol. Rev. 2014, 66 (4), 1033–1079. 10.1124/pr.114.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y.; Vilella-Bach M.; Bachmann R.; Flanigan A.; Chen J. Phosphatidic Acid-Mediated Mitogenic Activation of MTOR Signaling. Science 2001, 294 (5548), 1942–1945. 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- Yoon M.-S.; Rosenberger C. L.; Wu C.; Truong N.; Sweedler J. V.; Chen J. Rapid Mitogenic Regulation of the MTORC1 Inhibitor, DEPTOR, by Phosphatidic Acid. Mol. Cell 2015, 58 (3), 549–556. 10.1016/j.molcel.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H.; Qi R.; Zhou J. J.; Ta A. P.; Yang B.; Nakaoka H. J.; Seo G.; Guan K.-L.; Luo R.; Wang W. Regulation of the Hippo Pathway by Phosphatidic Acid-Mediated Lipid-Protein Interaction. Mol. Cell 2018, 72 (2), 328–340.e8. 10.1016/j.molcel.2018.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R. A.; Sabatini D. M. MTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168 (6), 960–976. 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.-X.; Zhao B.; Guan K.-L. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015, 163 (4), 811–828. 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J. N.; Fox T. E.; Kester M.; Jefferson L. S.; Kimball S. R. Phosphatidic Acid Mediates Activation of MTORC1 through the ERK Signaling Pathway. Am. J. Physiol.-Cell Physiol. 2010, 299 (2), C335–C344. 10.1152/ajpcell.00039.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond P. Phosphatidic Acid: Biosynthesis, Pharmacokinetics, Mechanisms of Action and Effect on Strength and Body Composition in Resistance-Trained Individuals. Nutr. Metab. 2017, 14 (1), 12. 10.1186/s12986-017-0166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athenstaedt K.; Daum G. Phosphatidic Acid, a Key Intermediate in Lipid Metabolism. Eur. J. Biochem. 1999, 266 (1), 1–16. 10.1046/j.1432-1327.1999.00822.x. [DOI] [PubMed] [Google Scholar]
- Tei R.; Baskin J. M. Spatiotemporal Control of Phosphatidic Acid Signaling with Optogenetic, Engineered Phospholipase Ds. J. Cell Biol. 2020, 219, e201907013 10.1083/jcb.201907013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beharry A. A.; Woolley G. A. Azobenzene Photoswitches for Biomolecules. Chem. Soc. Rev. 2011, 40 (8), 4422–4437. 10.1039/c1cs15023e. [DOI] [PubMed] [Google Scholar]
- Lerch M. M.; Hansen M. J.; van Dam G. M.; Szymanski W.; Feringa B. L. Emerging Targets in Photopharmacology. Angew. Chem., Int. Ed. 2016, 55 (37), 10978–10999. 10.1002/anie.201601931. [DOI] [PubMed] [Google Scholar]
- Hüll K.; Morstein J.; Trauner D. In Vivo Photopharmacology. Chem. Rev. 2018, 118 (21), 10710–10747. 10.1021/acs.chemrev.8b00037. [DOI] [PubMed] [Google Scholar]
- Höglinger D.; Nadler A.; Schultz C. Caged Lipids as Tools for Investigating Cellular Signaling. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2014, 1841 (8), 1085–1096. 10.1016/j.bbalip.2014.03.012. [DOI] [PubMed] [Google Scholar]
- Dinkel C.; Wichmann O.; Schultz C. Versatile Reagents to Introduce Caged Phosphates. Tetrahedron Lett. 2003, 44 (6), 1153–1155. 10.1016/S0040-4039(02)02832-0. [DOI] [Google Scholar]
- Wagner N.; Schuhmacher M.; Lohmann A.; Nadler A. A Coumarin Triflate Reagent Enables One-Step Synthesis of Photo-Caged Lipid Metabolites for Studying Cell Signaling. Chem. - Eur. J. 2019, 25 (68), 15483–15487. 10.1002/chem.201903909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williger B.-T.; Reich R.; Neeman M.; Bercovici T.; Liscovitch M. Release of Gelatinase A (Matrix Metalloproteinase 2) Induced by Photolysis of Caged Phosphatidic Acid in HT 1080 Metastatic Fibrosarcoma Cells. J. Biol. Chem. 1995, 270 (50), 29656–29659. 10.1074/jbc.270.50.29656. [DOI] [PubMed] [Google Scholar]
- Goedhart J.; Gadella T. W. J. Photolysis of Caged Phosphatidic Acid Induces Flagellar Excision in Chlamydomonas. Biochemistry 2004, 43 (14), 4263–4271. 10.1021/bi0351460. [DOI] [PubMed] [Google Scholar]
- Nadler A.; Yushchenko D. A.; Müller R.; Stein F.; Feng S.; Mulle C.; Carta M.; Schultz C. Exclusive Photorelease of Signalling Lipids at the Plasma Membrane. Nat. Commun. 2015, 6, 10056. 10.1038/ncomms10056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner N.; Stephan M.; Höglinger D.; Nadler A. A Click Cage: Organelle-Specific Uncaging of Lipid Messengers. Angew. Chem., Int. Ed. 2018, 57 (40), 13339–13343. 10.1002/anie.201807497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S.; Harayama T.; Montessuit S.; David F. P.; Winssinger N.; Martinou J.-C.; Riezman H. Mitochondria-Specific Photoactivation to Monitor Local Sphingosine Metabolism and Function. eLife 2018, 7, e34555 10.7554/eLife.34555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S.; Harayama T.; Chang D.; Hannich J. T.; Winssinger N.; Riezman H. Lysosome Targeted Photoactivation Reveals Local Sphingosine Metabolism Signature. Chem. Sci. 2019, 10, 2253. 10.1039/C8SC03614D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morstein J.; Impastato A. C.; Trauner D. Photoswitchable Lipids. ChemBioChem 2021, 22, 73. 10.1002/cbic.202000449. [DOI] [PubMed] [Google Scholar]
- Frank J. A.; Moroni M.; Moshourab R.; Sumser M.; Lewin G. R.; Trauner D. Photoswitchable Fatty Acids Enable Optical Control of TRPV1. Nat. Commun. 2015, 6, 7118. 10.1038/ncomms8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenegger M.; Tiapko O.; Svobodova B.; Stockner T.; Glasnov T. N.; Schreibmayer W.; Platzer D.; de la Cruz G. G.; Krenn S.; Schober R.; Shrestha N.; Schindl R.; Romanin C.; Groschner K. An Optically Controlled Probe Identifies Lipid-Gating Fenestrations within the TRPC3 Channel. Nat. Chem. Biol. 2018, 14 (4), 396–404. 10.1038/s41589-018-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinders-Zufall T.; Storch U.; Bleymehl K.; Mederos y Schnitzler M.; Frank J. A.; Konrad D. B.; Trauner D.; Gudermann T.; Zufall F. PhoDAGs Enable Optical Control of Diacylglycerol-Sensitive Transient Receptor Potential Channels. Cell Chem. Biol. 2018, 25 (2), 215–223.e3. 10.1016/j.chembiol.2017.11.008. [DOI] [PubMed] [Google Scholar]
- Frank J. A.; Yushchenko D.; Fine N. H. F.; Duca M.; Citir M.; Broichhagen J.; Hodson D. J.; Schultz C.; Trauner D. Optical Control of GPR40 Signalling in Pancreatic β-Cells. Chem. Sci. 2017, 8, 7604. 10.1039/C7SC01475A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morstein J.; Hill R. Z.; Novak A. J. E.; Feng S.; Norman D. D.; Donthamsetti P. C.; Frank J. A.; Harayama T.; Williams B. M.; Parrill A. L.; Tigyi G. J.; Riezman H.; Isacoff E. Y.; Bautista D. M.; Trauner D. Optical Control of Sphingosine-1-Phosphate Formation and Function. Nat. Chem. Biol. 2019, 15 (6), 623. 10.1038/s41589-019-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morstein J.; Dacheux M. A.; Norman D. D.; Shemet A.; Donthamsetti P. C.; Citir M.; Frank J. A.; Schultz C.; Isacoff E. Y.; Parrill A. L.; Tigyi G. J.; Trauner D. Optical Control of Lysophosphatidic Acid Signaling. J. Am. Chem. Soc. 2020, 142, 10612. 10.1021/jacs.0c02154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. A.; Yushchenko D. A.; Hodson D. J.; Lipstein N.; Nagpal J.; Rutter G. A.; Rhee J.-S.; Gottschalk A.; Brose N.; Schultz C.; Trauner D. Photoswitchable Diacylglycerols Enable Optical Control of Protein Kinase C. Nat. Chem. Biol. 2016, 12 (9), 755–762. 10.1038/nchembio.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kol M.; Williams B.; Toombs-Ruane H.; Franquelim H. G.; Korneev S.; Schroeer C.; Schwille P.; Trauner D.; Holthuis J. C.; Frank J. A. Optical Manipulation of Sphingolipid Biosynthesis Using Photoswitchable Ceramides. eLife 2019, 8, e43230 10.7554/eLife.43230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morstein J.; Awale M.; Reymond J.-L.; Trauner D. Mapping the Azolog Space Enables the Optical Control of New Biological Targets. ACS Cent. Sci. 2019, 5 (4), 607–618. 10.1021/acscentsci.8b00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morstein J.; Trads J. B.; Hinnah K.; Willems S.; Barber D. M.; Trauner M.; Merk D.; Trauner D. Optical Control of the Nuclear Bile Acid Receptor FXR with a Photohormone. Chem. Sci. 2020, 11 (2), 429–434. 10.1039/C9SC02911G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnah K.; Willems S.; Morstein J.; Heering J.; Hartrampf F. W. W.; Broichhagen J.; Leippe P.; Merk D.; Trauner D. Photohormones Enable Optical Control of the Peroxisome Proliferator-Activated Receptor γ (PPARγ). J. Med. Chem. 2020, 63 (19), 10908–10920. 10.1021/acs.jmedchem.0c00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartrampf N.; Seki T.; Baumann A.; Watson P.; Vepřek N. A.; Hetzler B. E.; Hoffmann-Röder A.; Tsuji M.; Trauner D. Optical Control of Cytokine Production Using Photoswitchable Galactosylceramides. Chem. - Eur. J. 2020, 26 (20), 4476–4479. 10.1002/chem.201905279. [DOI] [PubMed] [Google Scholar]
- Hu H.-G.; Chen P.-G.; Wang G.; Wu J.-J.; Zhang B.-D.; Li W.-H.; Davis R. L.; Li Y.-M. Regulation of Immune Activation by Optical Control of TLR1/2 Heterodimerization. ChemBioChem 2020, 21 (8), 1150. 10.1002/cbic.201900591. [DOI] [PubMed] [Google Scholar]
- Anzai J.-I.; Osa T. Photosensitive Artificial Membranes Based on Azobenzene and Spirobenzopyran Derivatives. Tetrahedron 1994, 50 (14), 4039–4070. 10.1016/S0040-4020(01)86704-1. [DOI] [Google Scholar]
- Kinoshita T. Photoresponsive Membrane Systems. J. Photochem. Photobiol., B 1998, 42 (1), 12–19. 10.1016/S1011-1344(97)00099-7. [DOI] [Google Scholar]
- Frank J. A.; Franquelim H. G.; Schwille P.; Trauner D. Optical Control of Lipid Rafts with Photoswitchable Ceramides. J. Am. Chem. Soc. 2016, 138 (39), 12981–12986. 10.1021/jacs.6b07278. [DOI] [PubMed] [Google Scholar]
- Pernpeintner C.; Frank J. A.; Urban P.; Roeske C. R.; Pritzl S. D.; Trauner D.; Lohmüller T. Light-Controlled Membrane Mechanics and Shape Transitions of Photoswitchable Lipid Vesicles. Langmuir 2017, 33 (16), 4083–4089. 10.1021/acs.langmuir.7b01020. [DOI] [PubMed] [Google Scholar]
- Urban P.; Pritzl S. D.; Konrad D. B.; Frank J. A.; Pernpeintner C.; Roeske C. R.; Trauner D.; Lohmüller T. Light-Controlled Lipid Interaction and Membrane Organization in Photolipid Bilayer Vesicles. Langmuir 2018, 34 (44), 13368–13374. 10.1021/acs.langmuir.8b03241. [DOI] [PubMed] [Google Scholar]
- Urban P.; Pritzl S. D.; Ober M. F.; Dirscherl C. F.; Pernpeintner C.; Konrad D. B.; Frank J. A.; Trauner D.; Nickel B.; Lohmueller T. A Lipid Photoswitch Controls Fluidity in Supported Bilayer Membranes. Langmuir 2020, 36 (10), 2629–2634. 10.1021/acs.langmuir.9b02942. [DOI] [PubMed] [Google Scholar]
- Foster D. A. Phosphatidic Acid and Lipid-Sensing by MTOR. Trends Endocrinol. Metab. 2013, 24 (6), 272–278. 10.1016/j.tem.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.; Wendel A. A.; Keogh M. R.; Harris T. E.; Chen J.; Coleman R. A. Glycerolipid Signals Alter MTOR Complex 2 (MTORC2) to Diminish Insulin Signaling. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (5), 1667–1672. 10.1073/pnas.1110730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strozyk E.; Kulms D. The Role of AKT/MTOR Pathway in Stress Response to UV-Irradiation: Implication in Skin Carcinogenesis by Regulation of Apoptosis, Autophagy and Senescence. Int. J. Mol. Sci. 2013, 14 (8), 15260–15285. 10.3390/ijms140815260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totaro A.; Piccolo S. Phosphatidic Acid Enters into the YAP/TAZ Arena. Trends Mol. Med. 2019, 25 (1), 5–7. 10.1016/j.molmed.2018.11.002. [DOI] [PubMed] [Google Scholar]
- Meng Z.; Qiu Y.; Lin K. C.; Kumar A.; Placone J. K.; Fang C.; Wang K.-C.; Lu S.; Pan M.; Hong A. W.; Moroishi T.; Luo M.; Plouffe S. W.; Diao Y.; Ye Z.; Park H. W.; Wang X.; Yu F.-X.; Chien S.; Wang C.-Y.; Ren B.; Engler A. J.; Guan K.-L. RAP2Mediates Mechanoresponses of the Hippo Pathway. Nature 2018, 560 (7720), 655–660. 10.1038/s41586-018-0444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K. C.; Park H. W.; Guan K.-L. Deregulation and Therapeutic Potential of the Hippo Pathway in Cancer. Annu. Rev. Cancer Biol. 2018, 2 (1), 59–79. 10.1146/annurev-cancerbio-030617-050202. [DOI] [Google Scholar]
- Yu F.-X.; Zhao B.; Panupinthu N.; Jewell J. L.; Lian I.; Wang L. H.; Zhao J.; Yuan H.; Tumaneng K.; Li H.; Fu X.-D.; Mills G. B.; Guan K.-L. Regulation of the Hippo-YAP Pathway by G-Protein-Coupled Receptor Signaling. Cell 2012, 150 (4), 780–791. 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H.; Sato K.; Murata N.; Damirin A.; Malchinkhuu E.; Kon J.; Kimura T.; Tobo M.; Yamazaki Y.; Watanabe T.; Yagi M.; Sato M.; Suzuki R.; Murooka H.; Sakai T.; Nishitoba T.; Im D.-S.; Nochi H.; Tamoto K.; Tomura H.; Okajima F. Ki16425, a Subtype-Selective Antagonist for EDG-Family Lysophosphatidic Acid Receptors. Mol. Pharmacol. 2003, 64 (4), 994–1005. 10.1124/mol.64.4.994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.