Abstract
Cryptococcus neoformans is an opportunistic fungal pathogen primarily targeting immunosuppressed populations in both resource-rich and resource-limited nations. Successful treatment is limited to a few antifungals that have become compromised by cryptococcal resistance leading to intensive research for new drug candidates. Two distinguishing hallmarks of this species are the ability to develop a polysaccharide capsule and melanization of the fungal cells. These also act as virulence factors, protecting this pathogen in the host as well as in the environment. Here we describe two classic methods to document capsule and melanin. Although initially described and documented over several decades ago, these methods remain relevant in spite of more sophisticated methodology due in part to their simplicity and cost efficiency.
Keywords: Cryptococcus neoformans, fungal pathogen, melanin, polysaccharide capsule, virulence
INTRODUCTION:
Cryptococcus neoformans is an opportunistic fungal pathogen that manifests primarily as meningoencephalitis in immunosuppressed individuals. In the HIV era, cases of cryptococcosis have become more frequent in this new population with defective CD4-mediated immunity (Rajasingham et al., 2017). In more resource-limited regions of the world, death from HIV-associated cryptococcosis is complicated by delayed diagnosis and limited availability of effective antifungal therapy.
There are several well-characterized virulence factors associated with the pathogenic potential of C. neoformans. Two of these, a polysaccharide capsule and a polyphenol melanin complex, are produced by cryptococcal cells in vivo and can be induced in vitro by specific media and growth conditions (Alspaugh, 2015; Zaragoza, 2019). The complex pathways that control C. neoformans capsule and melanin have been well characterized, and there are dozens of strains and mutants with defective capsule and/or melanin production (Perfect, 2005; Matsumoto et al., 2019). Presented here are simple laboratory assessment protocols to visualize and measure capsule (Basic Protocol 1) and melanin (Basic Protocol 2 and Alternative Protocol 1).
CAUTION:
Cryptococcus neoformans is a Biosafety Level 2 (BSL-2) pathogen. Follow all appropriate guidelines for the use of handling of pathogenic microorganisms.
BASIC PROTOCOL TITLE 1
CAPSULE VISUALIZATION BY INDIA INK COUNTERSTAINING
Introductory paragraph:
C. neoformans cells produce a polysaccharide capsule within the host that is required for its pathogenicity; acapsular strains are rarely recovered from clinical samples, and strains with severe capsule defects are often avirulent in animal models of infection (Perfect, 2005). The capsule protects C. neoformans cells against stress conditions and prevents phagocytosis by immune cells, thus playing a key role in the interaction between cryptococcal cells and the immune system (Campuzano & Wormley, 2018).
In addition to attaching to the cell surface, capsular polysaccharide is also actively secreted from the cryptococcal cell in extracellular vesicles (EVs) (de Oliveira, Castelli, Reis, Rizzo, & Rodrigues, 2020). This exopolysaccharide may serve additional roles in manipulating the external immune environment (Denham et al., 2020). The capsule is primarily composed of glucuronoxylomannan (GXM) and glucuronoxylomannogalactan (GXMGal) with trace amounts of mannoproteins (Zaragoza et al., 2009; O’Meara & Alspaugh, 2012; Z. A. Wang, Li, & Doering, 2018).
C. neoformans cells do not produce detectable surface capsule when grown in rich laboratory medium, but the capsule is induced within the host and by culture in specific nutrient-limiting media. The capsule composition is highly hydrophilic and renders it undetectable under normal light microscopy (Zaragoza et al., 2009). The simplest and most time-efficient method to detect capsule is by India ink counterstaining. The water-laden polysaccharide capsule excludes India ink, creating a discernable transparent halo around the cryptococcal cell that can be visualized by light microscopy. This technique has been used in both research and clinical laboratories.
There are several different types of media that induce the production of capsule including CO2-independent medium, 10% Sabourand-MOPs, DMEM with 5% CO2, and limited iron medium (LIM). In our lab, we primarily use CO2-independent tissue culture medium to mimic host conditions without the necessity of a CO2 incubator. However, this medium may require several days of incubation to achieve growth and capsule induction. Each of these media produce capsule to differing degrees; the key is to include a wild type, capsule-producing strain to access capsule production as a positive control and/or visualize the degree of encapsulation in a non-inducing medium such as YPD (negative control).
Materials:
Cryptococcus neoformans strain(s) (incubated on routine, solid growth medium, such as Sabouraud’s agar or YPD medium. If using potential capsule-defective strains, include a wild-type strain such as H99 (ATCC® 208821™) as a positive control (Perfect, Lang, & Durack, 1980).
YPD liquid medium and/or Sabouraud’s liquid medium
Capsule inducing media (see recipe in Reagents and Solutions)
CO2 independent medium (Gibco 18045088)
Sabouraud-PBS, (see recipe in Reagents and Solutions)
DMEM (Corning 10–013-CM)
Limited Iron Medium (LIM) (see recipe in Reagents and Solutions)
India ink (Remel BactiDrop India Ink, Thermo Scientific R21518)
Test tubes or 250-ml flasks (sterile)
Inoculation sticks (sterile)
Microscope slides and cover slips
Pipette that can measure 2–5 μl
Sterile pipette tips
Shaking incubator set to 30°C or 37°C and 150 rpm
CO2 incubator set to 5% (if using DMEM to induce capsule)
Light microscope equipped with software to capture images. A microscope that includes differential interference contrast (DIC) optics is preferred. DIC enhances the contrast in transparent samples, such as the unstained cryptococcal cell, allowing visualization of internal structures such as nuclei and vacuoles.
Software to measure images; microscope imaging software or a stand-alone software such as Fiji.
Optional: Microcentrifuge and 1.5-ml sterile microcentrifuge tubes to concentrate cells.
Protocol steps — Step annotations:
Step 1. Inoculate cryptococcal strain(s).
For each strain to be tested, add 5 to 50-ml capsule-inducing medium to a sterile test tube or sterile flask. As a positive control, include a wild-type strain such as H99.
Optional- As a negative control for each strain to be tested, add 5 to 50-ml of non-capsule inducing liquid medium (YPD or Sabouraud’s) to a sterile test tube or sterile flask.
Use a sterile inoculation stick to transfer a small amount of cells to each test tube/flask. Incubate in a shaking incubator at 37°C overnight or for several days, depending on strain growth rate and medium, until sufficient (visible) growth is achieved. Note that optimal capsule production occurs at 37°C, however 30°C is acceptable if your strains are temperature-sensitive.
Step 2. Prepare sample for visualization.
Pipette 2–5 μl of cell suspension onto a clean, dust-free microscope slide.
Add an equal amount of ink directly onto cells. Using the pipette tip, gently swirl the cells and India ink together.
Place a cover slip over the sample by placing the edge of the cover slip at a right angle to the slide and sample so that some of the liquid draws up onto the cover slip. Then quickly drop cover slip onto slide. This will prevent bubbles and disperse the cells evenly.
Step 3: Microscopic visualization.
Turn on microscope according to brand and model. Place prepared microscope slide on stage. Use a lower power objective (10–20x) to visualize the cells. The cryptococcal cells with surface capsule will stand out against the blackness of the India ink. After locating cells, adjust objective power to best visualize individual cells. Cells without significant surface capsule will be more difficult to distinguish, so it is often helpful to use a capsule-positive strain, such as C. neoformans H99, to ensure adequate capsule induction. If using DIC optics, follow the microscope instruction guide to adjust DIC slider settings to produce the best contrast. Note that the India ink:cell mixture may not be evenly dispersed (i.e., darker in some areas in the sample). This allows you to find the best contrast in any one sample to image. Use the microscope software to acquire and save images. Capture multiple images if the goal is to measure the capsule. Figure 1 illustrates the expected images of a wild-type and a capsule-defective strain grown in CO2-independent medium.
Figure 1. India ink counterstaining of a C. neoformans wild type (H99) and a capsule-defective mutant strain.
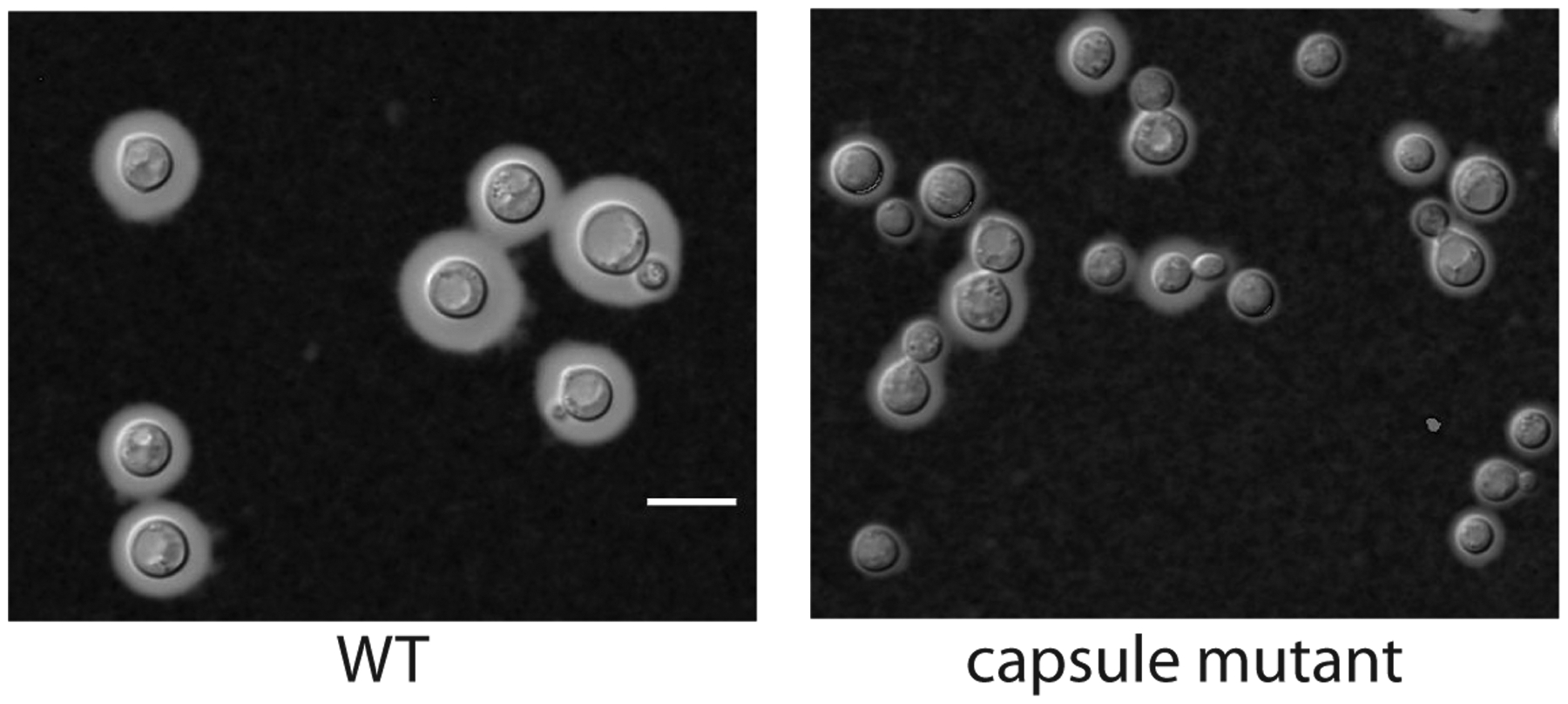
Strains were grown for 48 h in CO2-independent medium then prepared for microscopy by counterstaining with India ink. Capsule is shown as the clear area between the cell body and the surrounding India ink. Note that the cells from the capsule defective strain have much less capsule when compared to the wild-type cells. Also note the heterogeneity of capsule between individual cells from the same strain. The images were captured using a Zeiss Axio Imager A.1 at 1000x magnification using DIC optics and processed with Zeiss Zen software. Scale bar = 10 microns.
Optional: Concentrate cells using microcentrifuge and microcentrifuge tubes if the cultures did not grow well or if the cells are too dilute to locate microscopically.
Step 4. Capsule and cell measurement
To determine capsule thickness, the diameter of the capsule is measured along with cell body size. We typically use a Zeiss microscope (Axio Imager A.1) and software (Zen) to capture images but obtain measurements using Fiji, a JAVA-based free software program (Schneider et al., 2012). This program can open up image file formats from different microscope manufacturers (Zeiss, Nikon, Leica, etc.). First, open the image file in Fiji. Before measuring, confirm that the image has retained scale (Image>Properties). The unit of length should be in microns and the pixel width corresponding to the objective used during acquisition (i.e. 63X objective = 0.163). This information should be imported as part of the image file metadata. Next, open the ROI manager (Anazlye>Tools>ROI Manager). To begin measuring, select the line tool from the selection tool bar then draw a straight line through the widest point of the cell, beginning and ending at the outer edge of the capsule. This is the total cell diameter. Press the T key on the keyboard or select “Add (t)” in the ROI manager tool box. This will add the measurement to the ROI Manager. Next, draw a straight line through the same cell, starting and ending with the cell body. Similarity, add this measurement to the ROI Manager. This is the cell body diameter. Continue measuring cells until 50 cells have been measured or all of the cells in the current image have been measured. Multiple images can be opened and cells measured during a single ROI manager session. A minimum of 50 cells should be measured per sample. When done measuring the cells in the image, select “Measure” in the ROI toolbox. This will produce a data chart that includes multiple columns of data; the last being length which represents the values needed to calculate capsule thickness. The results are numbered so that the 1st and 2nd entries will represent the first cell measured (total cell diameter and body diameter) followed by the second cell measured (entries 3 and 4), etc. Copy and paste the data chart into a spreadsheet such as Excel or manually acquire the data. Calculate the thickness of the capsule for each cell by subtracting the cell body diameter from the total cell diameter then divide by 2. This value will represent the thickness of capsule. Finally, average the results.
BASIC PROTOCOL 2 TITLE
ASSESSMENT OF MELANIN PRODUCTION ON SOLID MEDIA
Introductory paragraph:
Similar to capsule production, C. neoformans strains that are unable to produce melanin are correspondingly avirulent in animal models of infection. Melanin is a dark brown to black pigment produced by the oxidation of imported diphenolic compounds such as dopamine. This phenol oxidation reaction is the rate-limiting step of melanin production, mediated by laccase enzymes (Williamson, 1997). Studies suggest that melanin acts as a free radical scavenger to protect C. neoformans cells from reactive oxygen and nitrogen species produced in the environment and in vivo by host immune cells (Y. Wang & Casadevall, 1994). Melanin also acts to prevent cells from phagocytosis in vivo (Y. Wang, Aisen, & Casadevall, 1995).
Investigators have found several different media that reproducibly induce melanin on agar plates and liquid medium. These include media containing Nyjer seed, caffeic acid, L-dopamine, and epinephrine. In our lab, we primarily use Nyjer seed agar to induce and visualize melanin. The key is to include a wild-type, melanin-producing strain as a positive control and/or visualize the degree of melanin produced in a non-inducing medium such as YPD (negative control). Described here is a simple assay to image melanin production in C. neoformans.
Materials:
Cryptococcus neoformans strain(s) (incubated on routine, solid growth medium, such as Sabouraud’s agar or YPD agar. If using potential melanin-defective strains, include a wild-type strain such as H99 (ATCC® 208821™) as a positive control (Perfect, Lang, & Durack, 1980).
Melanin-inducing agar medium (Nyjer seed, caffeic acid, L-dopamine, or epinephrine) (see recipe in Reagents and Solutions)
YPD or Sabouraud’s agar (see recipe in Reagents and Solutions)
YPD liquid medium and/or Sabouraud’s liquid medium (see recipe in Reagents and Solutions)
PBS
Sterile dH2O
Test tubes or 250-ml flasks (sterile)
Inoculation sticks (sterile)
Pipettes to measure 5 μl and 1 ml
Table top centrifuge
Sterile pipette tips
Spectrometer
Spectrometer cuvettes
Shaking incubator set to 30°C and 150 rpm
Protocol steps—Step annotations:
Step 1. Inoculate Cryptococcus strain(s)
Add 5 ml of YPD or Sabouraud’s liquid medium or to a sterile test tube.
Use a sterile inoculation stick to transfer a small amount of cells to the test tube/flask.
Incubate overnight in a shaking incubator set to at 30°C and 150 rpm.
Step 2. Cell Preparation
Pellet cells of each sample by centrifuging each test tube for 5 min at 1900 RCF in a tabletop centrifuge. Discard supernatant and resuspend in 5 ml of sterile PBS or sterile dH2O. Repeat twice.
Measure OD600 of each sample.
Dilute cells to final OD600 0.2–0.3 in sterile PBS or sterile water.
Step 3. Incubation on melanin-inducing media and imaging
Gently pipette a 5 μl aliquot of each strain being tested onto the melanin-inducing agar medium. Allow the aliquot of cells to dry as a small spot. Remember to include a wild-type strain as well as include non-inducing media (YPD or Sabouraud’s) as positive and negative controls. Incubate at 30°C for several days, monitoring the production of pigment each day until the wild type strain becomes dark brown. Melanin production in vitro is best visualized after incubation at 30°C compared to higher temperatures. Several strains can be included on each agar plate but be sure to space each sample equally since melanin production is influenced by carbohydrate deprivation. The amount of melanin produced by each sample is compared to a positive control (H99) and/or negative controls (strains spotted onto YPD or Sabouraud’s agar plate). Figure 2 illustrates a variety of wild-type and melanin-deficient strains incubated on Nyjer seed agar.
Figure 2. C. neoformans wild-type and mutant strains screened for melanin on Nyjer seed agar.
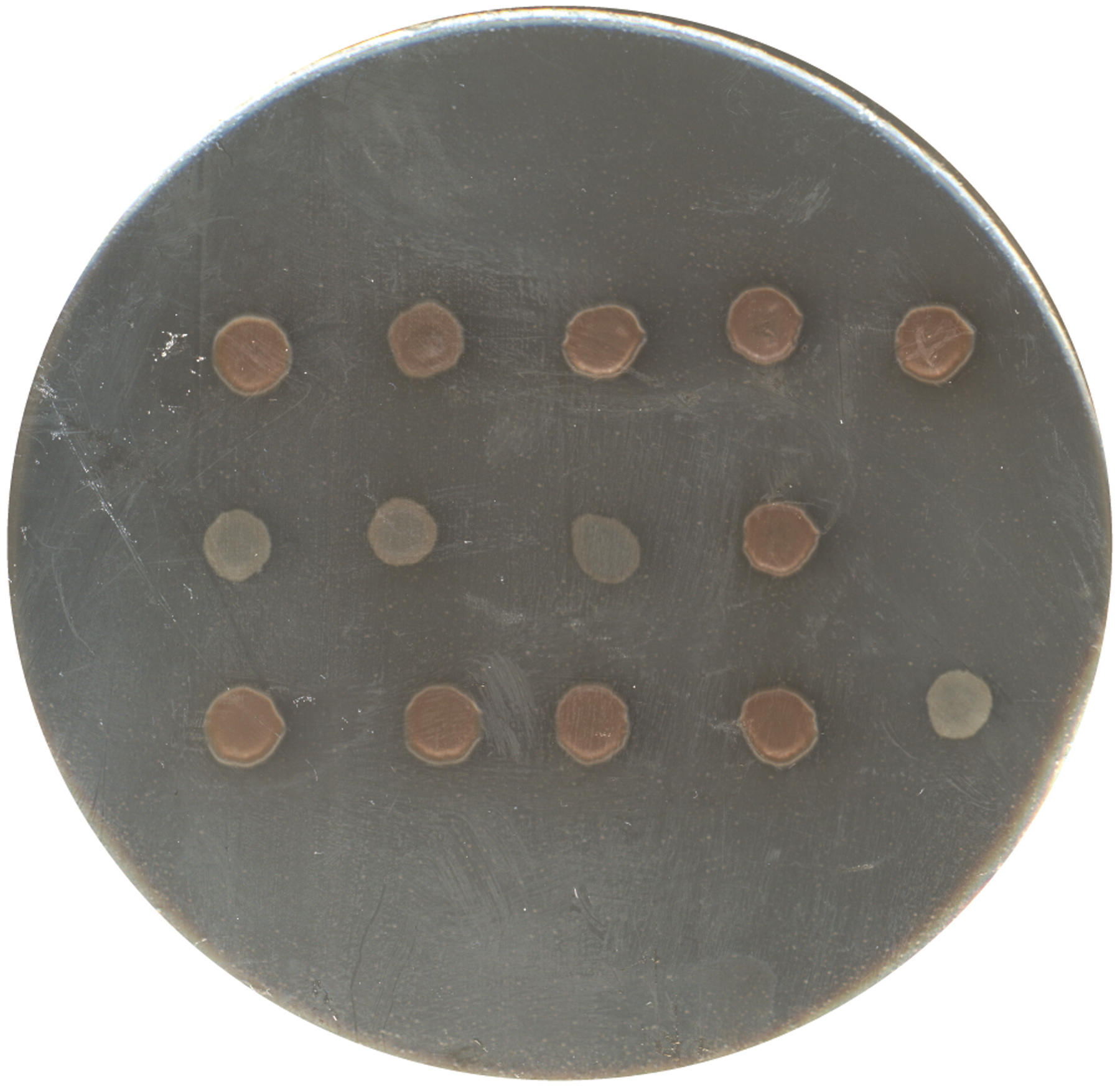
Wild type (H99) and putative melanin-defective strains were grown 24 h in YPD media. Strains were washed with PBS and then spotted (5 μl) onto a Nyjer seed agar plate. The plate was incubated for 3 days at 30°C prior to imaging with camera equipped smart phone. The upper left spot is H99 showing well-developed brown pigmentation (melanin). The remaining spots are various mutant strains ranging from no pigmentation (white) to wild type levels of pigmentation.
There are multiple ways to take simple images to document melanin production including microscopy at low power (4x objective), imaging systems, document scanners, and smart phones. In our lab we use camera-equipped smart phones to document melanin. If you have access to a gel-doc imaging system with spot densitometry software, it is possible to get a quantitative measurement of melanin production based on the amount of light reflected from each melanin spot (Samarasinghe et al., 2018)
ALTERNATE PROTOCOL 1 TITLE:
QUANTIFICATION OF MELANIN PRODUCTION IN LIQUID MEDIUM
Introductory paragraph:
This protocol uses spectrophotometry to measure the absorbance of melanin secreted by strains incubated in liquid melanin-inducing media and provides both a quantitative and temporal measure of melanin (Pukkila-Worley et al., 2005)
Additional Materials:
Melanin-inducing liquid medium (L-dopamine or epinephrine)
Protocol steps—
Step 1. Inoculate Cryptococcus strain(s).
Add 5 ml of YPD or Sabouraud’s to a sterile test tube.
Use a sterile inoculation stick to transfer a small amount of cells to the test tube/flask.
Incubate overnight in a shaking incubator at 150 rpm at 30°C.
Step 2. Cell Preparation
Pellet cells of each sample by centrifuging each test tube for 5 min at 1900 RCF in a tabletop centrifuge. Discard supernatant and add 5 ml of sterile PBS or sterile dH2O. Resuspend and repeat 2X. Resuspend in 5 ml of sterile PBS or sterile dH2O.
Measure OD600 of each sample.
Resuspend cells to OD600 0.2–0.3 in 20–50 ml in melanin inducing medium and YPD or Sabouraud’s (negative controls).
Step 3. Spectrometric measurement of samples
At regular intervals (24-, 48-, and 72 h post inoculation) remove 2 ml from each sample to a new sterile test tube.
Pellet cells of each sample by centrifuging each test tube for 5 min at 1900 RCF in a table top centrifuge.
Gently pipette 1 ml of the supernatant to a spectrometer cuvette without disturbing the cell pellet.
Measure absorbance at 475 nm in a spectrometer. Blank each sample against the initial sterile growth medium. Include a wild-type strain as a positive control and/or include measurements of samples grown in non-melanin inducing media (YPD or Sabouraud’s).
Create a graph of choice using the absorbance measurements (absorbance vs. time, etc.).
REAGENTS AND SOLUTIONS:
Reagents and Solutions
20% sterile glucose
200 g Glucose (D+ Glucose, CAS 50-99-7, Sigma-Aldrich 8270)
1 L dH2O
Scale
2 L beaker
Magnetic stir heat plate
Magnetic stir bar
1 L graduated cylinder
500 ml bottles, 2
Autoclave
Combine glucose with 600 ml dH2O in a 2 L beaker. Add magnetic stir bar and place on magnetic stir plate. Stir until dissolved with low heat. Bring up to 1 L total volume with dH2O. Split between two 500 ml bottles. Autoclave for 20 min. Store at room temperature.
Yeast peptone dextrose (YPD) liquid medium or agar plates
900 ml dH2O
20 g BD Bacto™ Peptone (Gibco 211677)
10 g BD Bacto™ Yeast Extract (Gibco 212750)
20 g BD Bacto™ granulated agar (for solid agar medium) (Gibco 214050)
100 ml sterile 20% glucose
Scale
1 L graduated cylinder
2 L Erlenmeyer flask
Magnetic stir plate
Magnetic stir bar
500 ml bottles (2 for liquid medium)
Autoclave
Sterile petri dishes (for solid agar medium)
100 ml graduated cylinder, sterile
Combine peptone, yeast extract, and dH2O into 2 L Erlenmeyer flask. Add magnetic stir bar and mix on magnetic stir plate until dissolved.
For liquid medium divide between two 500 ml bottles, autoclave 20 min, then aseptically add 50 ml of 20% sterile glucose to each bottle. Store at room temperature.
For agar add 20 g granulated agar to the mixture and stir until dissolved. Autoclave 20 min and allow to cool to touch (55°C). Aseptically add 100 ml of 20% sterile glucose, gently mix by stirring, then pour 20–25 ml into sterile petri dishes. Allow agar to harden then store inverted at 4°C.
Limited Iron Medium
5 g Glucose (D+ Glucose, CAS 50-99-7, Sigma-Aldrich G8270)
400 mg K2HPO4 (CAS 7758-11-4, Sigma-Aldrich P3786)
5 g L-Asparagine (CAS 70-47-3, Sigma-Aldrich A0884)
250 mg CaCl2-H2O (CAS 10035-04-08, Sigma-Aldrich C7902)
0.4 mg Thiamine hydrochloride (CAS 67-03-08, Sigma-Aldrich T4625)
5 mg CuSO4 (CAS 7758-98-07, Sigma-Aldrich C1297)
2 mg ZnSO4-7 H2O (CAS 7446-20-0, Sigma-Aldrich Z0251)
0.01 mg MnCl2-4 H2O (CAS 13446-34-9, Sigma-Aldrich M3634)
80 mg MgSO4-7 H2O (CAS 10034-99-8, Sigma-Aldrich M1880)
0.46 mg Na2MoO4 (CAS 10102-40-6, Sigma-Aldrich M1003)
0.057 mg Boric acid (CAS 10043-35-3, Sigma-Aldrich B0252)
1 L dH2O
Scale
2 L beaker
1 L graduated cylinder
Magnetic stir plate
Magnetic stir bar
1 L filter sterilizing unit with receptacle
Combine components with 700 ml dH2O in a 2 L beaker. Add a magnetic stir bar and mix on a magnetic stir plate until dissolved. Adjust volume to 1000 ml. Filter sterilize. Store at room temperature.
Sabouraud liquid medium or agar plates
30 g Sabouraud (Difco 238230)
20 g BD Bacto™ granulated agar (for solid agar medium) (Difco 214050)
1 L dH2O
1 L graduated cylinder
Scale
Magnetic stir plate
Magnetic stir bar
500 ml bottles, 2 (for liquid medium)
Autoclave
Sterile petri dishes (for solid agar medium)
Follow manufacturer’s instructions and combine 30 g in 1000 ml dH2O in a 2 L Erlenmeyer flask. Add magnetic stir bar and mix until boiling on magnetic stir plate with heat.
For liquid medium divide between two 500 ml bottles and autoclave 20 min. Store at room temperature.
For agar add 20 g granulated agar to the mixture and stir until dissolved. Autoclave 20 min and allow to cool to touch (55°C). Pour 20–25 ml into sterile petri dishes. Allow agar to harden then store inverted at 4°C.
Sabouraud-PBS
100 ml Sabouraud liquid medium
900 ml PBS
1 L graduated cylinder
1 L filter sterilizing unit with receptacle
Dilute Sabourand liquid medium in PBS and filter sterilize.
Store at room temperature.
Caffeic acid agar
334 g Caffeic acid agar (CRITERION C7781, 500 g)
Scale
1 L dH2O
1 L graduated cylinder
2 L Erlenmeyer
Magnetic stir plate with heat
Magnetic stir bar
Autoclave
Sterile petri dishes
Follow manufacturer’s instructions and combine 334 g in 1 L dH2O in a 2 L Erlenmeyer flask. Add magnetic stir bar and mix until boiling on magnetic stir plate with heat. Autoclave 20 min and allow to cool to touch (55°C). Pour 20–25 ml into sterile petri dishes. Allow agar to harden then store inverted at 4°C.
L-dopamine or epinephrine liquid medium or solid agar plates
1 g L-Asparagine (CAS 70-47-3, Sigma-Aldrich A0884)
1 g Glucose (D+ Glucose, CAS 50-99-7, Sigma-Aldrich 8270)
3 g K2HPO4 (CAS 7758-11-4, Sigma-Aldrich 3786) 0.25 g MgSO4-7H2O
100 mg L-DOPA (CAS 59-92-7, Sigma-Aldrich D9628) or 100 mg (−)-Epinephrine (CAS 51-43-4, Sigma-Aldrich E4250)
1 mg Thiamine hydrochloride (CAS 67-03-08, Sigma-Aldrich T4625)
5 μg Biotin (CAS 58-85-5, Sigma-Aldrich B4639)
1 L dH2O
20 g BD Bacto™ granulated agar (for solid agar medium) (Difco 214050)
Scale
1 L graduated cylinder
2 L Erlenmeyer flask
Magnetic stir plate
Magnetic stir bar
Autoclave
500 ml beaker
Filter sterilizing unit with receptacle, 250 ml
pH meter
Sterile petri dishes
For agar plates, mix 20 g Bacto agar with 900 ml dH2O and in a 2 L Erlenmeyer flask. Add magnetic stir bar and mix on magnetic stir plate until dissolved.
Autoclave 40 min then place flask on magnetic stir plate and mix gently until able to touch the side of the flask (50°C).
While agar is cooling, mix together in a beaker: 1 g L-asparagine, 1 g glucose, 3 g KH2PO4, 0.25 g MgSO4-7H2O, 100 mg L-DOPA (or 100 mg epinephrine) with 100 ml distilled water.
Adjust pH to 5.6.
Add 1 mg thiamine-HCl and 5 μg biotin.
Filter sterilize.
Aseptically add mixture to agar and mix gently on magnetic stir plate.
Pour 20–25 ml into sterile petri dishes.
Let agar harden then invert plates for storage at 4°C.
For liquid medium, mix together in a beaker: 1 g L-asparagine, 1 g glucose, 3 g KH2PO4, 0.25 g MgSO4-7H2O, 100 mg L-DOPA (or 100 mg epinephrine) with 600 ml distilled H2O.
Adjust pH to 5.6.
Adjust volume to 1 L.
Add 1 mg thiamine-HCl and 5 μg biotin.
Filter sterilize and store at room temperature.
Nyjer seed agar
70g Nyjer seed (Guizotia abyssinica; seed available from bird or pet stores, also known as thistle seed)
1 L dH2O
1 g Glucose (D+ Glucose, CAS 50-99-7, Sigma-Aldrich 8270)
20 g BD Bacto™ granulated agar (for solid agar medium) (Difco 214050)
Scale
Coffee grinder
Cheesecloth
1 L graduated cylinder
1 L Erlenmeyer flask
2 L Erlenmeyer flask
Magnetic stir plate
Magnetic stir bar
Autoclave
Sterile petri dishes
Grind Nyjer seed into a fine powder using coffee grinder or mortar/pestle.
Combine with 350 ml dH2O into a 1 L Erlenmeyer flask.
Add magnetic stir bar and mix on a magnetic stir plate until all components are either dissolved or well suspended.
Autoclave 15 min.
Remove particulate matter by filtering through cheesecloth into a 2 L Erlenmeyer flask.
Adjust volume to 1000 ml.
Add 1 g glucose.
Add 20 g granulated agar.
Add a magnetic stir bar and mix on a magnetic plate until dissolved.
Autoclave 40 min.
Pour 20–25 ml into sterile petri dishes.
Allow agar to solidify and then invert plates for storage at 4°C.
COMMENTARY
BACKGROUND INFORMATION:
Cryptococcus neoformans is a fungal opportunistic pathogen that can infect a wide variety of hosts, including humans. The resulting disease, cryptococcosis, can manifest from minor skin lesions to meningoencephalitis, which can be fatal. Entry into the body is via inhalation of yeast cells or spores that can either remain dormant or multiply to cause pneumonia in the lungs and disseminate throughout the body with a preference for the central nervous system and brain.
Historically, disease resulting from C. neoformans was first described in the late 1800’s, but there was a dramatic increase in cryptococcosis during the AIDS epidemic in the 1980’s (Srikanta, Santiago-Tirado, & Doering, 2014). It was during this time that an association between C. neoformans and CD4-mediated immunity was realized. Globally it remains one of the leading fungal pathogens with an estimated 220,000 cases per year, targeting HIV-associated populations in resource-limited countries and other immunosuppressed populations in developed countries (Rajasingham et al., 2017). It is one of the top three fungal pathogens that infects solid organ transplant patients, ranging from months to years to manifest post-transplantation (Baddley & Forrest, 2019).
In more economically developed countries, optimal treatment of cryptococcosis with antifungals includes agents such as fluconazole and amphotericin B in combination with flucytosine. In more resource-limited countries, treatment options are suboptimal due to the limited availability of amphotericin B and flucytosine. Limited antifungal choice and emerging resistance have created a need for new anti-cryptococcal drugs (Bermas & Geddes-McAlister, 2020) (Mourad & Perfect, 2018). Analysis of patient and environmental isolates as well as analysis of lab-generated mutant strains for avirulent phenotypes is one method of exploring new pathways that may lead to new drug discovery.
The ability to produce capsule and melanin as well as growth at 37°C comprise the three major virulence factors of C. neoformans. Understanding the pathways that control the production of capsule and melanin are important for further drug development making simple assays described here a critical component of our arsenal of techniques characterizing C. neoformans.
The techniques described here are the classic techniques to visualize capsule and melanin but they are certainly not the only protocols available. In addition to India ink counterstaining, there are fluorescent labelling techniques to visualize capsule using antibodies directed against capsule constituents. Automated imaging software is now available to more accurately measure the capsule revealed by fluorescent labeling and by India ink counterstaining (Casadevall et al., 1998; Dragotakes & Casadevall; 2018; Guess et al., 2018). Recently a technique using Percoll and refractive index has been described to visualize the capsule in real time (Paes et al., 2018; Paes et al., 2019), Another powerful visualization technique is scanning electron microscopy (SEM) (Cleare & Casadevall, 1999). However, all of these techniques require specialized equipment, special fixatives in the case of SEM, time, and/or expensive reagents while capsule visualization by India ink only requires a light microscope.
In contrast to capsule, alternative visualization protocols for melanin activity are more limited. In depth visual analysis can be determined by transmission electron microscopy (TEM) but this technique requires the isolation of melanin from cryptococcal cells (melanin ghosts) as well as specialized equipment (Eisenman et al., 2005). However, there are biochemical assays to analyze melanin activity and components including a quantitative laccase activity assay that was first described by Rhodes et al. in 1982 but is still utilized with modifications (Rhodes, Polacheck, & Kwon-Chung, 1982; Ikeda et al., 2002; Eisenman et al., 2007).
CRITICAL PARAMETERS & TROUBLESHOOTING:
The most relevant critical parameter and troubleshooting is to explore the different media listed in this protocol and to consistently compare the phenotype of your strain of interest to strains known to produce abundant capsule and melanin. A well characterized wild type strain that produces abundant capsule and melanin is C. neoformans strain H99. In the event that your wild type strain is not producing capsule and melanin in the different inducing media it is possible that the strain has mutated or been contaminated by another strain or fungi (budding yeasts Saccharomyces cerevisiae and Candida species have cell morphologies similar to C. neoformans). Fungi contamination is less common than bacteria contamination. Both can be prevented by maintaining sterile technique. There have been instances of mutation in C. neoformans strain H99 during laboratory passage and storage. If this is suspected, H99 can be purchased from the ATCC or acquired from another laboratory.
UNDERSTANDING RESULTS:
There is quite a bit of range in C. neoformans cell size and capsule. When cultured in capsule-inducing conditions, we typically observe 3.5–8 μm body sizes and 1–5 μm capsule thickness using the technique described here. Although some capsule mutant strains produce no capsule, other capsule mutant strains will produce reduced capsule as compared to wild type. For example, the rim101 mutant strain has a capsule attachment defect and the capsule that remains attached is reduced compared to wild type, ranging in thickness from 0.45 to 2 μm (personal data; O’Meara and Alspaugh, 2012). To avoid artifacts from imaging and analysis, it is important to have some type of positive control for capsule such as a wild type strain and also to acquire the images (control and experimental sample) during the same imaging session.
Similarly, there is also a range of C. neoformans melanization. This is easily discerned when viewing melanin produced from cells cultured on melanin inducing agar over several days. While wild type strains such as H99 will accumulate melanin and become dark brown in color, mutant strains deficient in melanin production will remain white in color or become lighter shades of brown compared to wild type. When quantifying melanin accumulation in a culture supernatant, a typical wild type measurement will be around OD475 0.5 at 48 h and increasing to 3 by 72 h. In comparison, the lac2 mutant strain has an initial delay in melanin accumulation and starts low (i.e. OD475 0.2 at 48 h) but increases to wild type levels by 72 h. In contrast, the lac1 mutant strain is unable to accumulate high amounts of melanin (i.e. OD475 less than 0.1 at 48 h and 1.0 at 72 h) (Pukkila-Worley et al., 2005).
TIME CONSIDERATIONS:
Using these two simple assays the investigator can quickly assess two of the three major virulence factors associated with C. neoformans in strains isolated from the environment, clinical samples, or laboratory-generated mutant strains in a short period of time. Starting with a cultured strain or isolate, results can be obtained from as early as overnight (capsule staining) to a few days.
ACKNOWLEDGEMENTS:
This work was supported by NIH R01 grant AI074677.
LITERATURE CITED:
- Alspaugh JA (2015). Virulence mechanisms and Cryptococcus neoformans pathogenesis. Fungal Genet Biol, 78, 55–58. doi: 10.1016/j.fgb.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddley JW, & Forrest GN (2019). Cryptococcosis in solid organ transplantation-Guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transplant, 33(9), e13543. doi: 10.1111/ctr.13543 [DOI] [PubMed] [Google Scholar]
- Bermas A, & Geddes-McAlister J (2020). Combatting the evolution of antifungal resistance in Cryptococcus neoformans. Mol Microbiol. doi: 10.1111/mmi.14565 [DOI] [PubMed] [Google Scholar]
- Campuzano A, & Wormley FL (2018). Innate Immunity against Cryptococcus, from Recognition to Elimination. J Fungi (Basel), 4(1). doi: 10.3390/jof4010033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Cleare W, Feldmesser M, Glatman-Freedman A, Goldman DL, Kozel TR, … Zhong Z. (1998). Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob Agents Chemother, 42(6), 1437–1446. doi: 10.1128/aac.42.6.1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleare W, & Casadevall A (1999). Scanning electron microscopy of encapsulated and non-encapsulated Cryptococcus neoformans and the effect of glucose on capsular polysaccharide release. Med Mycol, 37(4), 235–243. [PubMed] [Google Scholar]
- Dragotakes Q, & Casadevall A (2018). Automated Measurement of Cryptococcal Species Polysaccaride Capsule and Cell Body. J Vis Exp(131). doi: 10.3791/56957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira HC, Castelli RF, Reis FCG, Rizzo J, & Rodrigues ML (2020). Pathogenic Delivery: The Biological Roles of Cryptococcal Extracellular Vesicles. Pathogens, 9(9). doi: 10.3390/pathogens9090754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman HC, Nosanchuk JD, Webber JB, Emerson RJ, Camesano TA, & Casadevall A (2005). Microstructure of cell wall-associated melanin in the human pathogenic fungus Cryptococcus neoformans. Biochemistry, 44(10), 3683–3693. doi: 10.1021/bi047731m [DOI] [PubMed] [Google Scholar]
- Guess T, Lai H, Smith SE, Sircy L, Cunningham K, Nelson DE, & McClelland EE (2018). Size Matters: Measurement of Capsule Diameter in Cryptococcus neoformans. J Vis Exp(132). doi: 10.3791/57171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Azami S, Shiga H, Nagamachi T, Moriyama H, Yamashita Y, … Sugita T (2019). Induction of signal transduction pathways related to the pathogenicity of Cryptococcus neoformans in the host environment. Drug Discov Ther, 13(4), 177–182. doi: 10.5582/ddt.2019.01047 [DOI] [PubMed] [Google Scholar]
- Mourad A, & Perfect JR (2018). Present and Future Therapy of Cryptococcus Infections. J Fungi (Basel), 4(3). doi: 10.3390/jof4030079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, & Alspaugh JA (2012). The Cryptococcus neoformans capsule: a sword and a shield. Clin Microbiol Rev, 25(3), 387–408. doi: 10.1128/cmr.00001-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect JR (2005). Cryptococcus neoformans: a sugar-coated killer with designer genes. FEMS Immunol Med Microbiol, 45(3), 395–404. doi: 10.1016/j.femsim.2005.06.005 [DOI] [PubMed] [Google Scholar]
- Perfect JR, Lang SD, & Durack DT (1980). Chronic cryptococcal meningitis: a new experimental model in rabbits. Am J Pathol, 101(1), 177–194. Retrieved from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=7004196 [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R, Gerrald QD, Kraus PR, Boily MJ, Davis MJ, Giles SS, … Alspaugh JA (2005). Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot Cell, 4(1), 190–201. doi: 10.1128/ec.4.1.190-201.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, … Boulware DR (2017). Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis, 17(8), 873–881. doi: 10.1016/s1473-3099(17)30243-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes JC, Polacheck I, & Kwon-Chung KJ (1982). Phenoloxidase activity and virulence in isogenic strains of Cryptococcus neoformans. Infect Immun, 36(3), 1175–1184. doi: 10.1128/iai.36.3.1175-1184.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Daynig V, Longair M, Pietzsch T, …Cardona A (2012). “Fiji: an open-source platform for biological-image analysis”. Nature methods, 9(7), 676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanta D, Santiago-Tirado FH, & Doering TL (2014). Cryptococcus neoformans: historical curiosity to modern pathogen. Yeast, 31(2), 47–60. doi: 10.1002/yea.2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Aisen P, & Casadevall A (1995). Cryptococcus neoformans melanin and virulence: mechanism of action. Infect Immun, 63(8), 3131–3136. doi: 10.1128/iai.63.8.3131-3136.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, & Casadevall A (1994). Decreased susceptibility of melanized Cryptococcus neoformans to UV light. Appl Environ Microbiol, 60(10), 3864–3866. doi: 10.1128/aem.60.10.3864-3866.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZA, Li LX, & Doering TL (2018). Unraveling synthesis of the cryptococcal cell wall and capsule. Glycobiology, 28(10), 719–730. doi: 10.1093/glycob/cwy030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson PR (1997). Laccase and melanin in the pathogenesis of Cryptococcus neoformans. Front Biosci, 2, e99–107. doi: 10.2741/a231 [DOI] [PubMed] [Google Scholar]
- Zaragoza O (2019). Basic principles of the virulence of Cryptococcus. Virulence, 10(1), 490–501. doi: 10.1080/21505594.2019.1614383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, & Casadevall A (2009). The capsule of the fungal pathogen Cryptococcus neoformans. Adv Appl Microbiol, 68, 133–216. doi: 10.1016/s0065-2164(09)01204-0 [DOI] [PMC free article] [PubMed] [Google Scholar]


