Abstract
Background
Acute myeloid leukemia (AML) stem cells (LSCs) are capable of surviving current standard chemotherapy and are the likely source of deadly, relapsed disease. While stem cell transplant serves as proof-of-principle that AML LSCs can be eliminated by the immune system, the translation of existing immunotherapies to AML has been met with limited success. Consequently, understanding and exploiting the unique immune-evasive mechanisms of AML LSCs is critical.
Methods
Analysis of stem cell datasets and primary patient samples revealed CD200 as a putative stem cell–specific immune checkpoint overexpressed in AML LSCs. Isogenic cell line models of CD200 expression were employed to characterize the interaction of CD200+ AML with various immune cell subsets both in vitro and in peripheral blood mononuclear cell (PBMC)–humanized mouse models. CyTOF and RNA-sequencing were performed on humanized mice to identify novel mechanisms of CD200-mediated immunosuppression. To clinically translate these findings, we developed a fully humanized CD200 antibody (IgG1) that removed the immunosuppressive signal by blocking interaction with the CD200 receptor while also inducing a potent Fc-mediated response. Therapeutic efficacy of the CD200 antibody was evaluated using both humanized mice and patient-derived xenograft models.
Results
Our results demonstrate that CD200 is selectively overexpressed in AML LSCs and is broadly immunosuppressive by impairing cytokine secretion in both innate and adaptive immune cell subsets. In a PBMC-humanized mouse model, CD200+ leukemia progressed rapidly, escaping elimination by T cells, compared with CD200− AML. T cells from mice with CD200+ AML were characterized by an abundance of metabolically quiescent CD8+ central and effector memory cells. Mechanistically, CD200 expression on AML cells significantly impaired OXPHOS metabolic activity in T cells from healthy donors. Importantly, CD200 antibody therapy could eliminate disease in the presence of graft-versus-leukemia in immune competent mice and could significantly improve the efficacy of low-intensity azacitidine/venetoclax chemotherapy in immunodeficient hosts.
Conclusions
Overexpression of CD200 is a stem cell–specific marker that contributes to immunosuppression in AML by impairing effector cell metabolism and function. CD200 antibody therapy is capable of simultaneously reducing CD200-mediated suppression while also engaging macrophage activity. This study lays the groundwork for CD200-targeted therapeutic strategies to eliminate LSCs and prevent AML relapse.
Keywords: tumor escape, metabolic networks and pathways, lymphocyte activation, immunomodulation
Background
The primary obstacle to curing acute myeloid leukemia (AML) is the elimination of leukemia stem cells (LSCs), therapy-resistant cells capable of long-term self-renewal and deemed the source of relapse.1 In allogeneic stem cell transplant, immune cells reconstituted from donor bone marrow are capable of destroying residual leukemia cells and curing patients.2 This graft-versus-leukemia (GvL) response is proof-of-principle that LSCs are vulnerable to destruction via immune response. Besides transplant, pharmacologic immunotherapy has not been optimally exploited for AML. Checkpoint inhibitors in patients with AML have had limited success.3 4 Other immunotherapy clinical trials in AML consist of antigen-specific approaches, including drug-conjugated and naked monoclonal antibodies, bispecific engager antibodies, and chimeric antigen receptor (CAR)-T and NK cells.5 However, many of these antigen-specific immunotherapies are insufficient because they are focused on bulk AML cells and are not directed against LSC-specific antigens. Thus, a prominent knowledge gap is identifying LSC-specific immune targets.
Here, we identify CD200 as one such candidate of LSC-specific immunosuppression. It was recently demonstrated that CD200 is expressed on both healthy and leukemic stem cells and can distinguish functional LSCs in xenotransplantation assays.6 In our meta-analysis of AML LSC datasets, we found CD200 is not only a marker of stem cells but is also consistently overexpressed in AML LSCs when compared with healthy hematopoietic stem cells and their corresponding blast cells. This insinuates CD200 may provide a selective advantage for LSCs. While CD200 has no well-established intrinsic function, it has been implicated as a local regulator of immune response through interaction with the CD200 receptor, CD200R1, expressed on subsets of myeloid, T, and NK cells, and may be exploited by LSCs to provide immune privilege.7
CD200 expression on bulk AML disease has been correlated with suppressed NK and memory CD4+ T cell function. Specifically, patients with high bulk CD200 had half the frequency of activated NK cells, and co-culture of these cells with a CD200+ AML cell line could reduce the IFNγ response and impair degranulation.8 Similarly, CD4+ memory T cells from patients with high bulk levels of CD200 leukemia were less active upon ex vivo stimulation and co-culture with a CD200+ AML cell line and could reduce the number of TNFα-producing cells only after CD3/CD28 T cell activation.9 CD200 antibodies treatment was shown to enhance NK cell activity in vitro and marginally improve cytokine-induced killer cell therapy efficacy against CD200+ AML in vivo.10 Clinically, high CD200 expression on bulk AML disease has been associated with a 50% reduction in odds of complete remission and significantly worse overall survival.11–13 Taken together, this suggests that CD200 plays an advantageous role in the regulation of immune cell function in AML; however, the mechanisms of CD200 immunosuppression and CD200 antibody therapy remain unknown, specifically in the context of AML LSCs.
Here, we demonstrate that CD200+ AML could broadly suppress cytokine secretion from T cells and macrophages. In a PBMC-humanized mouse model that faithfully recapitulates GvL, CD200 expression was sufficient to protect AML cells from T cell–mediated elimination. This AML-induced immunosuppression was associated with the expansion of inactive memory CD8 cells and metabolicaly quiescent T cells. Finally, we found that CD200 antibody therapy could eliminate AML in the presence of human immune effector cells and significantly improve the efficacy of low-intensity azacitidine/venetoclax chemotherapy in mice without adaptive immune cells.
Methods
Cell collection and sorting
Primary AML samples (bone marrow or peripheral blood) were collected under MD Anderson IRB protocol PA12-0173. Briefly, whole blood samples were mixed 1:1 with PBS without calcium and magnesium and added to 15 mL Lymphocyte Separation Medium. Cells were centrifuged (1800 rpm for 20 min) and lymphocyte layer extracted. Ammonium chloride solution (5 mL) was used to lyse residual red blood cells for 5 min while shaking. Immune cell subsets were isolated using the selection kits in online supplemental table 1.
jitc-2021-002968supp001.pdf (1.8MB, pdf)
Flow cytometry
Cells (3×105) were then washed twice with staining buffer (PBS+0.5% BSA) and treated with anti-human Fc receptor binding inhibitor for 15 min prior to addition of surface antibodies (online supplemental table 2) for 30 min at room temperature with agitation. DAPI (2 µg/mL) was used for live/dead cell discrimination for collection on a LSRII machine. Cell sorting followed the same staining protocol with larger cell numbers and was run on MoFlo Astrios cell sorter. For phagocytosis, target cells were first labeled with 5 µM CFSE for 5 min at 37°C then quenched with complete media and then co-cultured at a 2:1 ratio with macrophages for 4 hours at 37°C with or without 10 µg/mL anti-CD200 or an isotype matched irrelevant antibody. Cells were detached using StemPro Accutase Cell Dissociation Reagent on ice for 20 min. Percentage of activated macrophages was calculated as the fraction of CFSE+/CD206+ to total CD206+ cells. The isotype control employed in all studies was a fully humanized anti-HER2 IgG1 antibody. All AML cell lines and healthy T cells and macrophages were confirmed to lack HER2 expression.
Mice
NOD-SCID IL2Rgnull (NSG) and CD45.1 C57BL/6 mice were purchased from Jackson Laboratory and bred in house. CD45.2 C57BL/6 mice were purchased from MD Anderson Experimental Radiation Oncology department. All mice were housed in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care and NIH standards. Experiments were conducted according to protocols 00001146-RN02 and 00001446-RN01 and approved by the University of Texas MD Anderson Cancer Center Institutional Animal Care and Use Committee.
AML assays in vivo
NSG or CD45.1 C57BL/6 mice were sublethally irradiated (250 cGy and 450 cGy, respectively) 24 hours before injection. Then 2.5×105 murine AML or 5×104 OCI-AML3 cells/mouse were injected intravenously. Engraftment was tracked weekly by flow cytometry assessment of peripheral blood. Firefly luciferase was used to track engraftment of OCI-AML3 cells. D-Luciferin (30 mg/mL) was injected intraperitoneally 5 min prior to IVIS imaging (IVIS-Xenogen 100 system). For PBMC-humanized mice, 1×107 PBMCs were injected intravenously into unconditioned NSG mice. After 1 week, engraftment was confirmed by measuring the proportion of human CD3+/CD45+ present in peripheral blood by flow cytometry and mice were randomized into treatment groups. NSG and PBMC-humanized mice were treated with 10 mg/kg intraperitoneally 3 times a week for 2 weeks beginning on day 7 after leukemia injection. In PDX mice, when disease in the peripheral blood reached 1%–3%, mice were randomized to receive AZA/VEN (AZA: 2.5 mg/kg/day, intraperitoneally once a day, 7 days; VEN: 50 mg/kg/day, orally once a day, 14 days; beginning day 84), anti-CD200 (10 mg/kg/day, intraperitoneally 3 times a week for 3 weeks; beginning day 84), or the combination.
Cytokine profiling
Effector cells (2×105) were co-cultured at a 1:1 ratio with desired target cells with and without anti-CD200 or isotype matched irrelevant antibodies (10 µg/mL) and incubated for 16–24 hours prior to collecting supernatant. Secreted cytokine abundance was determined by flow cytometry using Legendplex kits (online supplemental table 3) according to manufacturer’s instructions.
Statistics
Unless otherwise specified, data analysis performed using GraphPad Prism V.8. Statistical tests and methods for correcting for multiple testing are reported in-line.
Results
CD200 is overexpressed in AML LSCs in human and murine disease
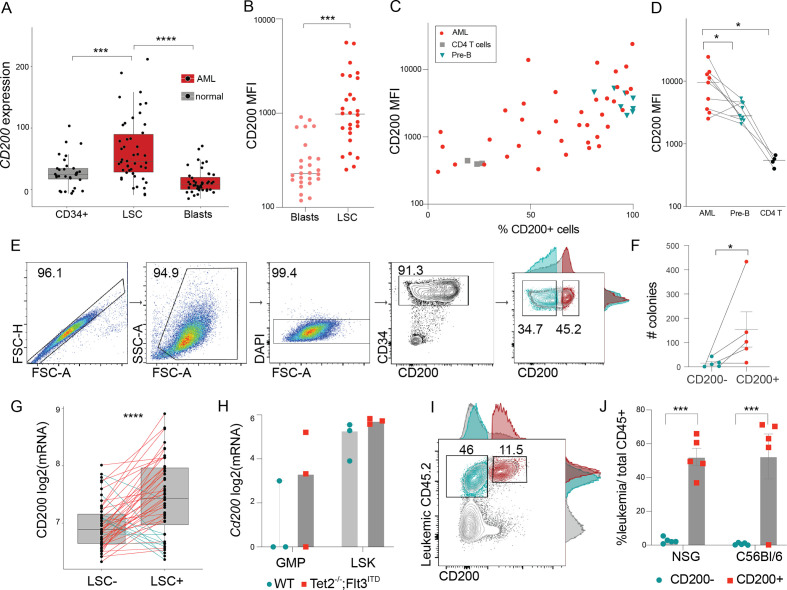
To identify putative LSC markers, we mined three large, publicly available gene expression datasets14–16 of immunophenotypically sorted AML and normal hematopoietic cells and identified CD200 as being significantly overexpressed in LSCs compared with corresponding blast cells or healthy HSCs (figure 1A, S1A–C). We screened primary AML patient samples by flow cytometry and confirmed at the protein level that CD200 was significantly higher in the LSC fraction of disease compared with paired CD34− blast cells (figure 1B). Because healthy HSCs are rare and difficult to distinguish in AML samples, we mined publicly available single-cell CITE-seq of normal bone marrow17 (online supplemental figure 1D) to identify more readily available cell types for comparison. We found that normal HSCs have CD200 protein expression similar to that of CD4 T cells and roughly threefold less than naive B cells (online supplemental figure 1E). In our data, MFI of CD200+ AML was comparable with paired naive B cells (n=8) and significantly greater than paired CD4 T cells (n=4; figure 1C–D). This strongly suggests CD200 protein is also overexpressed in AML LSCs, consistent with our in silico findings. Using the Leucegene18 AML cohort, CD200 expression was also found to be significantly higher in patients with complex karyotype (online supplemental figure 1G) and in relapsed disease (online supplemental figure 1F), both poor prognostic categories with significantly worse survival.19 20
Figure 1.
CD200 is overexpressed in functional leukemia stem cells. (A) CD200 mRNA expression across immunophenotypically sorted healthy (gray) and leukemic (red) cell populations; mined from de Jonge et al15 [GSE74246] (mean±SD; two-sample t-test). (B) CD200 MFI of paired blast (CD34−) and LSCs (CD34+) (n=28; two-sample t-test). (C) Frequency of CD200+ cells against CD200 MFI in 38 primary patients with AML (red), naive B cells (n=8; green), and CD4+ T cells (n=4; gray). (D) CD200 MFI summary for patients with paired naive B and T cell data (Wilcoxon paired test). (E) Representative gating strategy for FACS sorting CD200+ and CD200− cells from CD34+ AML samples. (F) Number of colonies formed in the CD34+CD200 and CD34+CD200+ fractions (mean±SD; Wilcoxon paired test). (G) CD200 mRNA expression in cell fractions that either did (LSC+) or did not (LSC−) engraft in NSG mice from Ng et al21 [GSE76009] (lines connect cells from the same AML patient; paired t-test). (H) RNA-seq gene expression data for sorted progenitor (GMP) and stem (LSK) cells derived from wild type (WT) and the Tet2–/–;Flt3ITD murine leukemia model [GSE57244].25 (I) Representative gating strategy for CD200+ leukemia (CD45.2) stem cells. (J) Combined leukemic burden for NSG and C57Bl/6 mice from 2 experiments (mean±SD; Wilcoxon). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
In most samples, CD200 was co-expressed with CD34, a canonical marker of HSCs/LSCs. To determine whether CD200 further refined the detection of functional LSCs within the CD34+ subset, colony formation potential was compared between paired CD34+CD200+ and CD34+CD200 cells from five primary AML patient samples (figure 1E). Within a given patient sample, CD34+CD200+ cells formed significantly more colonies compared with the CD34+CD200− subset (figure 1F). To further validate CD200 as a functional LSC marker, we examined a functional LSC xenotransplantation dataset (Ng et al21) using our previously described method22 and found that functional LSCs had significantly higher CD200 gene expression within a given patient sample (figure 1G). Further, using the Tet2−/−;Flt3ITD murine AML model, CD200 expression was higher in progenitor and stem cells from AML-primed mice, and only CD200+ leukemia cells were capable of engrafting and repopulating disease in both immunodeficient and immunocompetent mice (figure 1H–J).23–25 Together, these data suggest that CD200 is preferentially overexpressed by LSCs, upregulated on poor-risk AML, and may serve as a clinically important therapeutic target.
CD200+ AML broadly suppresses T cell cytokine production
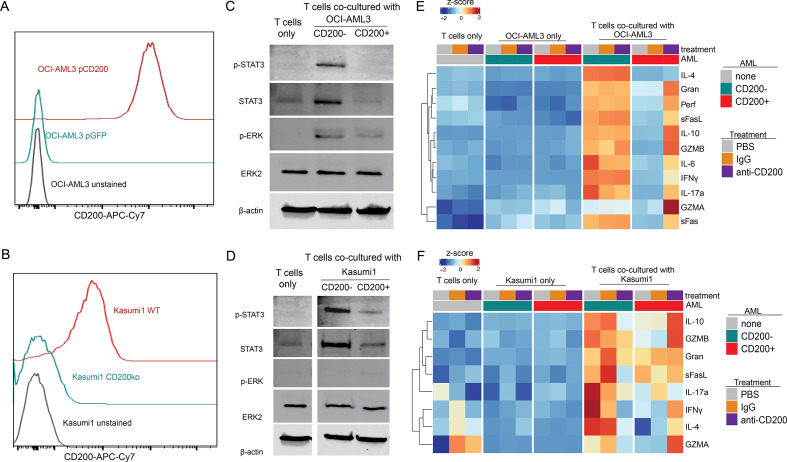
To study the interaction of CD200+ AML with immune cells, we established and characterized two isogenic cell line model systems: CD200 knockout in the Kasumi1 cell line (low basal CD200 expression) and CD200 overexpression in OCI-AML3 cells (no baseline CD200 expression; figure 2A–B). Manipulation of CD200 expression in these cells had no impact on AML cell metabolism, proliferation, or survival (online supplemental figure 2). While it was previously shown that CD200-expressing AML could reduce the frequency of activated CD4 T cells capable of producing TNFα, the mechanism and extent of suppression remains unclear.9 So, we co-cultured our isogenic cell lines with healthy, sorted CD3 T cells and assessed signaling pathways by immunoblotting (figure 2C–D). CD200R engagement results in recruitment of RasGAP and subsequent inhibition of the Ras/MAPK pathway,26 a pattern of inhibition corroborated in OCI-AML3 co-culture, where T cell p-ERK was activated in response to AML, but significantly attenuated in the presence of CD200 (figure 2C). We further discovered a CD200-dependent suppression of STAT3 signaling in T cells invoked by both OCI-AML3 and Kasumi1 models (figure 2C–D). Because STAT3 is a regulator of cytokine response, we evaluated T cell cytokine production in vitro. In the presence of CD200+ OCI-AML3 or Kasumi1 cells, T cells had significantly impaired cytokine secretion (figure 2E–F), consistent with the decreased pSTAT3 seen by western blot. Importantly, this inflammatory cytokine response could be partially restored by blocking CD200–CD200R interaction using anti-CD200 antibody. The alterations in T-cell function did not correlate with significant differences in either peptide-specific or allogeneic T cell–mediated AML cell killing (online supplemental figure 3). These data indicate that CD200+ AML cells directly suppress effector T cell cytokine production through dampening the MAPK and STAT3 signaling pathways, thereby nominating CD200 as an immunosuppressive molecule.
Figure 2.
CD200+ AML cells suppresses T cell cytokine production in vitro. (A–B) Cell surface flow cytometry of CD200 in the OCI-AML3 (A) and Kasumi1 (B) engineered cell line models. (C–D) Representative western blot analysis of healthy T cells purified after 24-hour co-culture at a 1:1 ratio with OCI-AML3 (C) or Kasumi1 (D) cells. (E–F) Relative abundance of cytokines secreted from T cells alone, AML cells alone, and T cells in response to co-culture with CD200− or CD200+ OCI-AML3 (E) or Kasumi1 (F) cells. All conditions were assessed after treatment with a blocking CD200 antibody (anti-CD200), IgG1 isotype matched control (IgG), or PBS alone (untreated). Heatmap represents the average abundance of 3 technical replicates.
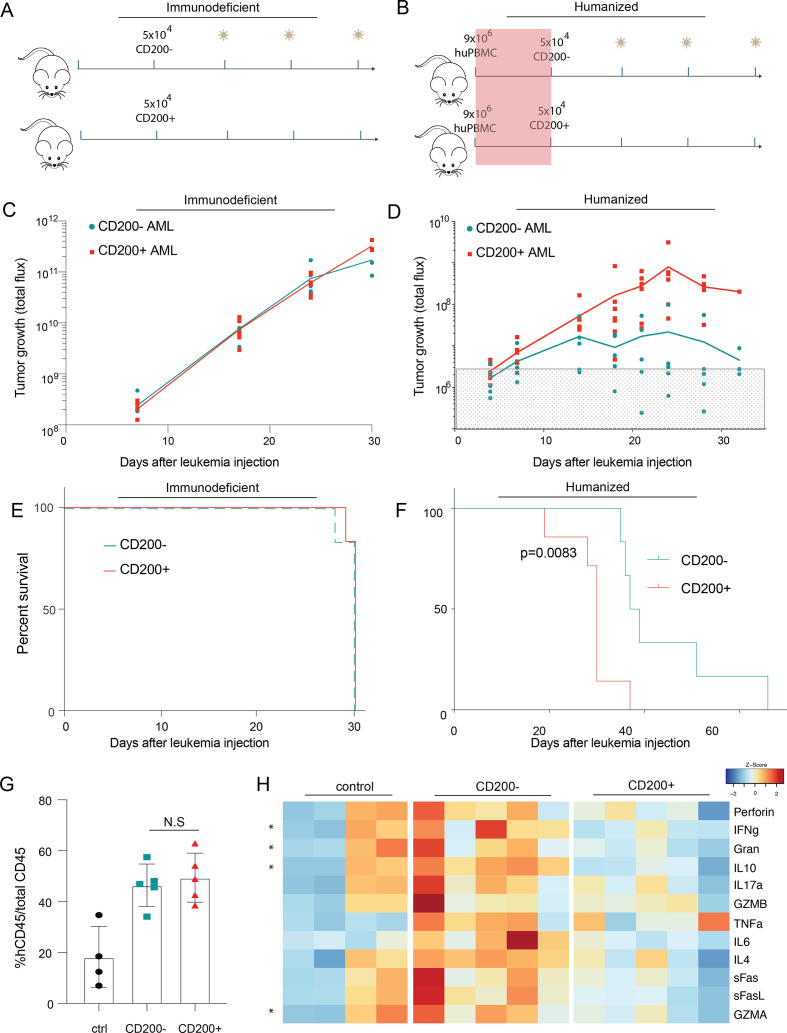
CD200 prevents T cell–mediated leukemia rejection in vivo
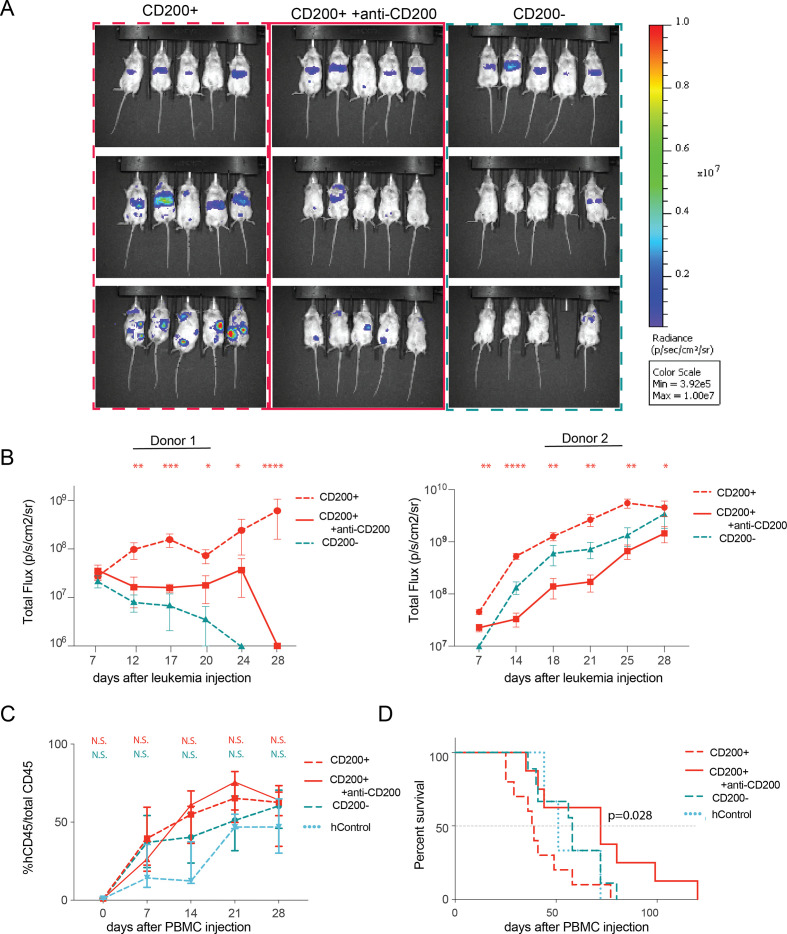
To explore this possibility, we next asked whether CD200 could suppress T cells and protect AML cells in vivo. In immunodeficient NSG mice with AML, CD200 expression did not impact disease progression (figure 3C) or overall survival (figure 3E). However, when mice were humanized with healthy human PBMCs, prior to leukemia introduction (figure 3B), we observed significant and expeditious progression of CD200+ leukemia (figure 3D). While both lines were capable of engraftment, only the CD200+ persisted, while CD200− cells were eliminated by T cells. Indeed, all mice injected with CD200− leukemia died without detectable disease, translating to significantly prolonged survival (figure 3F); however, all mice (regardless of CD200 leukemia status) eventually succumbed to histologically confirmed graft-versus-host disease (GVHD; online supplemental figure 4). There was no significant difference in the fraction of human non-leukemic CD45+ (>90% CD3+ human T cells) cells between the two groups (online supplemental figure 5).
Figure 3.
CD200 protects AML cells from T cell–mediated clearance in vivo. (A, B) Schematic for immunodeficient (A) and PBMC-humanized (B) mouse model systems. (C, D) Tumor growth measured by luciferase imaging of CD200± OCI-AML3 cells in immunodeficient (C) and PBMC-humanized (D) mice. Gray box indicates background flux levels. (E, F) Overall survival of mice with CD200± leukemia measured from the day of leukemia injection in immunodeficient (E) and PBMC-humanized (F) models. (G) Fraction of circulating human PBMCs in humanized mice with either CD200± leukemia or humanized-only controls at day 7 after OCI-AML3 injection. (H) Normalized serum cytokine abundance for each mouse in (G). Multiple t-tests compared cytokine production in CD200− vs CD200+ mice. *q<0.05.
When mice from both the CD200− and CD200+ models were confirmed to have equal engraftment and comparable human CD45+ abundance (figure 3G), serum was collected and profiled for human inflammatory cytokines traditionally secreted by effector T cells (figure 3H). Mice with CD200+ leukemia had significantly reduced IFNγ, granzyme A (GZMA), granulysin (Gran), and IL-10 abundance compared with mice with CD200− leukemia. These data corroborate that AML-expressing CD200 provides a survival advantage in vivo exclusively in the presence of activated human immune cells by impairing cytokine secretion from T cells.
CD200+ AML alters T cell composition and cell cycle
Patients with high levels of bulk AML CD200 expression have been reported to have altered T cell composition.27 To further characterize this descriptive observation, and determine whether CD200+ AML significantly altered the composition or fitness of the immune microenvironment in vivo, we performed CyTOF on the humanized immune system of mice injected with CD200+ or CD200− leukemia (figure 4A). At this timepoint, mice from both leukemia cohorts had comparable disease burden, white blood cell counts (WBC), and healthy human CD45+ abundance (online supplemental figure 5). On average, >87% of human CD45 immune cells were CD3+ T cells. Mice injected with CD200− leukemia had significantly higher fractions of central memory T cells (CD4+CD45RA−CCR7+). Interestingly, mice with CD200+ AML had significant enrichments of the CD8 effector memory cluster 1 (CD8 EM 1; CD8+CD45RA−CCR7−) and CD8 central memory cluster 2 (CD8 CM 2; CD8+CD45RA−CCR7+) (figure 4B–C). These specific clusters of CD8 memory T cells appeared to be immunophenotypically inactive, distinguished by the absence of markers indicative of T-cell activation, including CD69 and 4-1BB (CD127) (figure 4D). Furthermore, these CD8 clusters contained significantly fewer actively cycling cells (Ki67+) in mice with CD200+ disease (figure 4E). This suggests that the presence of CD200 on leukemia cells is sufficient to produce a more immunosuppressed microenvironment characterized by functionally impaired CD8 T cells and reduced central memory CD4 cells. These CD200-mediated differences in T cell composition may partly explain the expedited leukemia progression in humanized mice, further implicating CD200 as a critical immunosuppressive molecule in AML.
Figure 4.
CD200+ alters T cell composition in vivo. (A) CyTOF summary UMAP of healthy human immune cells derived from humanized mice with cluster definitions. (B) Cluster proportions for each mouse. (C) Clusters with significantly different frequency between mice with CD200+ and CD200− disease (Wilcoxon test). (D) Heatmap of average protein intensity for all markers by cluster and disease status. (E) Density plots of Ki67 expression in cells from CD8 CM 2 and EM 1 clusters for each mouse evaluated (left) and summarized for all cells from a specific disease group (right; Wilcoxon test). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
CD200+ AML impairs T cell metabolism
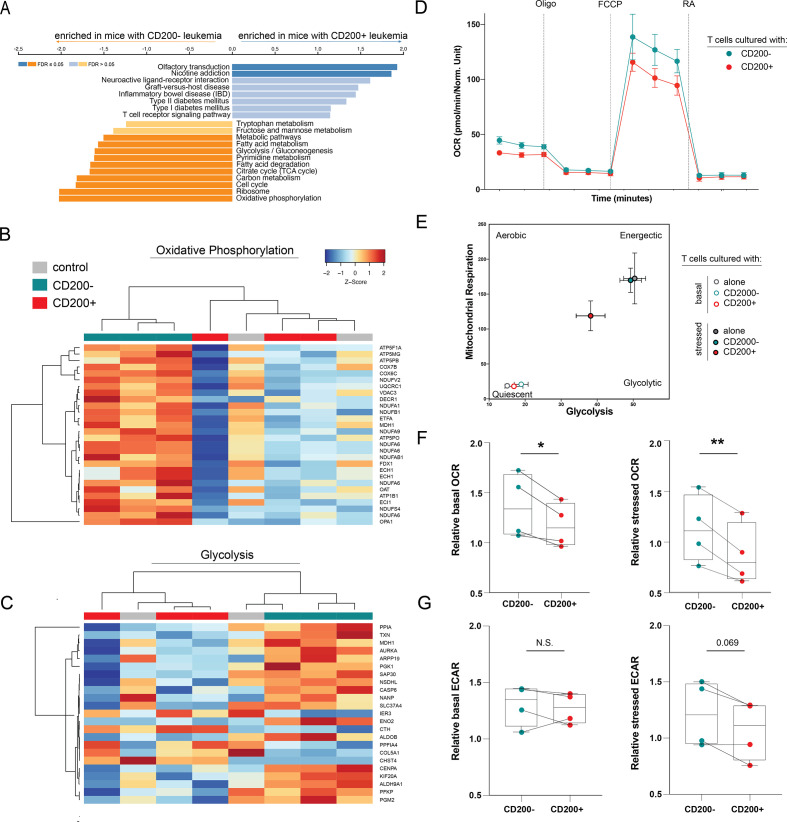
The differences in cytokine expression and T cell phenotypes inspired us to perform RNA-sequencing to evaluate specific transcriptional pathways altered by CD200± leukemia. Purified CD3+ T cells from mice with CD200+ leukemia were enriched in genes involved in T cell–mediated inflammation (figure 5A). Surprisingly, these T cells had significantly downregulated genes across multiple critical metabolic pathways, providing a possible explanation for the fewer observed actively replicating T cells in CyTOF and reduced serum cytokine abundance. Based on gene set enrichment analysis, these cells were significantly devoid of active metabolic signaling through glycolysis and oxidative phosphorylation (OXPHOS) when compared with T cells exposed to CD200− AML (figure 5B–C).
Figure 5.
CD200+ leukemia directly suppresses T cell metabolism. (A) GSEA analysis of genes enriched in T cells from mice CD200+ leukemia compared with T cells from mice with CD200− leukemia. (B–C) Heatmap of differentially expressed genes in the oxidative phosphorylation (B) and glycolysis (C) pathways between human T cells from mice with CD200+ or CD200− OCI-AML3 leukemia or humanized-only controls. Only regulated genes with p<0.05 are displayed. The color scale corresponds to the row-wise Z score of gene expression. (D) Representative OCR measured at baseline and in response to oligomycin (Oligo), carbonyl cyanide p-trifluoromethoxy-phenylhydrazone (FCCP), and rotenone+antimycin A (RA) in human CD3+ T cells after co-culture with CD200+ or CD200− OCI-AML3 leukemia. (E) Representative XF cell energy phenotype of human CD3+ T cells alone or after co-culture with CD200+ or CD200− OCI-AML3 leukemia. (F–G) Summary basal and stressed OCR (F) and ECAR (G) for 4 healthy T cell donors. To account for donor variability, OCR and ECAR are reported relative to donor T cells without co-culture. Paired two-sample t-tests were used. *p<0.05, **p<0.01.
To functionally validate the effect of CD200+ leukemia on the metabolic activity of the immune microenvironment, we performed mitochondrial stress tests on T cells isolated from four healthy human donors after exposure to either CD200+ or CD200− OCI-AML3 cells. T cells co-cultured with CD200+ AML had significantly impaired oxygen consumption rates (OCR), a surrogate marker of OXPHOS, when compared with T cells from the same donor cultured with CD200− AML (figure 5D–F). We also observed that the extracellular acidification rate (ECAR), an indicator of glycolytic activity, under stressed conditions tended to be reduced in the presence of CD200+ leukemia (figure 5G). These data indicate that CD200+ AML exerts an immunosuppressive phenotype by metabolically hampering neighboring T cells.
CD200 antibody monotherapy can eliminate AML in the presence of immune effectors
Our data indicate that CD200 suppresses T cell–mediated response to AML in a mouse model. To exploit these findings therapeutically, a fully humanized IgG1 antibody against CD200 (anti-CD200) was developed to specifically reverse the immunosuppressive effects of CD200-expressing AML by blocking interaction with CD200R (online supplemental figure 6). We assessed the efficacy of single-agent antibody therapy in 2 PBMC-humanized NSG mouse models with varying degrees of GvHD. In mice from the first donor, which had substantial GVHD and therefore potent GvL potential, anti-CD200 therapy resulted in significant disease burden reduction (figure 6A–B) and ultimate clearance of leukemia. While strong GvL/GvHD was beneficial in anti-leukemia activity in this model, it made overall survival uninterpretable, as mice died from lethal GvHD almost immediately on disease elimination. In a second humanized model with significantly less GvHD, we found anti-CD200 single-agent therapy similarly reduced disease burden (figure 6B). This translated to a significant improvement in overall survival (p=0.028) with an increase in median survival of 33.5 days (+117.5%) (figure 6D). Interestingly, the leukemia-treated mice also survived significantly longer than the humanized-only controls, possibly indicating that CD200-expressing leukemia could also mitigate the lethal GvHD component of T cell response. Using this model, it was also confirmed that the anti-CD200 treatment had no significant effect on the expansion of human immune cells in these mice (figure 6C). These results suggest that anti-CD200 therapy is effective in the presence of activated T cells.
Figure 6.
Anti-CD200 monotherapy significantly reduces leukemia in humanized mice. (A) Bioluminescent images of humanized mice (donor 1) over the course of anti-CD200 treatment. (B) Total leukemia burden in 2 humanized models (ANOVA with Dunnett’s test for multiple comparisons). (C) Flow cytometry of human PBMC expansion in humanized mice (ANOVA with Dunnett’s). (D) Overall survival for mice humanized with PBMCs from donor 2 (Mantel-Cox log-rank test). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
CD200 attenuates the macrophage response to AML
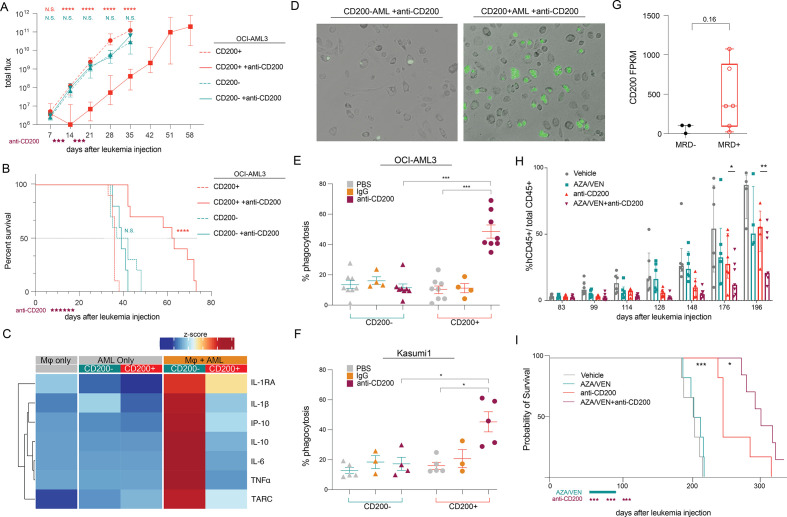
We also found a transient effect of CD200 antibody monotherapy in unmanipulated NSG mice—which are reported to have functional phagocytes.28 After three doses, there was a significant decrease in the disease burden of CD200+ anti-CD200-treated mice. However, the remaining three doses appeared largely ineffective, as the disease burden began to increase at the same rate as the control-treated mice (figure 7A). This transient leukemia suppression translated to significantly prolonged survival (p<0.0001), with a 74% increase in median survival, but did not cure any mice of disease (figure 7B). Quite interestingly, CD200 was originally implicated as a regulator of macrophage function; however, the relationship between this innate immune subset and AML through the CD200/CD200R signaling axis has not been reported.7 29 Similar to our T cell findings, healthy macrophages secreted important inflammatory cytokines when co-cultured with CD200− OCI-AML3 cells in vitro—a phenomenon markedly abrogated by ectopic CD200 expression (figure 7C). This suggests that CD200+ AML can also exert an immunosuppressive effect on the innate immune response.
Figure 7.
CD200+ AML regulates macrophage function. (A) NSG mice with CD200+ or CD200− OCI-AML3 disease were treated with 3 times a week for 2 weeks (6 total doses) with 10 mg/kg anti-CD200 or PBS. Total leukemia burden was tracked by bioluminescence imaging (ANOVA with Dunnett’s test for multiple comparisons). (B) Overall survival of mice with CD200+/CD200− AML after anti-CD200 therapy (Mantel-Cox log-rank test). (C) Relative abundance of cytokines secreted from human macrophages or CD200−/CD200+ OCI-AML3 cells and after co-culture. Heatmap represents the average abundance of 3 technical replicates. (D) Representative images of phagocytosed CFSE-labeled CD200− (left) and CD200+ (right) OCI-AML3 treated with anti-CD200 after 4-hour co-incubation with healthy macrophages. (E–F) Flow cytometry analysis of phagocytosis of CD200−/CD200+ OCI-AML3 (E) or Kasumi1 (F) cells treated with anti-CD200, IgG-control, or PBS alone (Kruskal-Wallis with Dunn’s multiple comparison test). (G) CD200 RNA expression in LSCs from patients with (n=6) and without (n=3) minimal residual disease (MRD) after treatment with AZA/VEN [GSE155431].30 (H–I) Leukemic burden as measured by the fraction of human CD45+ leukemic cells (F) and overall survival (G) in mice harboring a CD200+ AML PDX (46, XY) treated with either vehicle, AZA/VEN (AZA: 2.5 mg/kg/day, intraperitoneally once a day, 7 days; VEN: 50 mg/kg/day, orally once a day, 14 days; beginning day 84), anti-CD200 (10 mg/kg/day, intraperitoneally 3× week for 3 weeks; beginning day 84), or the combination. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
We next examined the functional response of macrophages in the presence of AML cells treated with anti-CD200. A primary mechanism of action of monoclonal antibodies is to engage macrophages and induce antibody-dependent cell phagocytosis (ADCP). To test whether the anti-CD200 antibodies could induce specific ADCP, CFSE-labeled CD200+ and CD200− leukemia cells were co-cultured with mature macrophages and treated with either anti-CD200, an isotype control, or PBS alone. Phagocytosis was visualized using fluorescent microscopy to manually inspect evidence of cell engulfment after co-incubation. In the presence of CD200 antibody, only CD200+ leukemia cells were observed within the perimeter of a given macrophage, suggesting macrophages were capable of engulfing AML cells (figure 7D). High-throughput flow cytometry was subsequently employed to assess phagocytosis under various conditions. Anti-CD200 treatment significantly increased the fraction of phagocytosing macrophages exclusively with CD200+ OCI-AML3 (figure 7E) and CD200+ Kasum-1 (figure 7F) cells. Together, these data demonstrate that CD200 antibody treatment potently and specifically facilitates macrophage-mediated ADCP of CD200+ AML cells.
Venetoclax with azacitidine (AZA/VEN) is a highly effective treatment regimen in AML that is hypothesized to specifically eradicate the AML LSCs by inhibiting amino acid metabolism.30 Analyzing the RNA-seq data generated by Stevens et al,30 we found that LSCs collected at baseline from patients who had minimal residual disease (MRD) after AZA/VEN therapy tended to have higher levels of CD200 than those from patients with no detectable MRD (figure 7G). This suggested CD200 expression on LSCs may contribute to or identify AZA/VEN resistant cells. To investigate the utility of adding anti-CD200 to AZA/VEN therapy in vivo, we employed an established CD200+ patient-derived xenograft model of AML and treated with anti-CD200 monotherapy, (AZA/VEN), or AZA/VEN/anti-CD200. In this AML PDX, known to be refractory to AZA/VEN, anti-CD200 monotherapy impeded disease progression; however, this effect was significantly enhanced in the combination therapy arm (figure 7H). Reduced leukemic burden translated to significantly prolonged survival in mice treated with combination therapy (figure 7I). These data suggest that CD200-antibody therapy is efficacious against primary AML in the presence of functional macrophages in vivo and that anti-CD200 may partially restore sensitivity to lower intensity frontline induction approaches such as AZA/VEN.
Discussion
Most patients with AML achieve remission after induction chemotherapy, but relapse and become unresponsive to traditional treatment. Leukemia and the subsequent relapsed disease are hypothesized to arise from LSCs, implicating LSCs as the critical target for curing AML.1 Stem cell transplantation provides proof-of-principle that AML LSCs can be eliminated by a functional immune system.31 However, the mechanisms by which AML LSCs escape immune surveillance and destruction remain largely unknown. Our work narrows this knowledge gap by identifying and characterizing CD200 as a novel immune target, selectively overexpressed in AML LSCs, capable of negatively regulating both the innate and adaptive immune systems and demonstrate that targeting CD200 is a promising therapeutic strategy for AML.
CD200 has been previously implicated as a marker of certain normal and cancer stem cells. In the healthy setting, CD200 was reported to identify follicular bulge stem cells,32 visceral-fat adipose-derived stem cells,33 and mesenchymal stem cells.34 CD200 was also shown to be co-expressed with CD44 and CD133, markers of solid tumor stem cells.35 Specific to AML, CD200 was found to be a marker of functional leukemic and healthy hematopoietic stem cells.6 Our discoveries build on these observations and further demonstrate that CD200 is a marker of LSCs and that it is significantly overexpressed, both at the transcript and protein level, when compared with healthy hematopoietic stem and progenitor cells as well as blast cells. This suggests that CD200 may provide a selective advantage for these LSCs. Future single-cell sequencing experiments on primary AML bone marrow could provide the granularity to allow for the comparison of CD200 expression in normal and leukemic stem cells within the same patient.
The established mechanism of CD200 immunosuppression via the CD200 receptor involves the downstream to tyrosine kinase (Dok2) adapter protein recruitment to the distal phosphorylation residue where it further employs the SH2 domain containing protein, RAS p21 protein activator (RasGAP). This Dok2/RasGAP complex is sufficient to inhibit Ras activation, which leads to the subsequent inhibition of the MAPK signaling cascade and impaired cytokine response in myeloid cells.36 In the instance of the OCI-AML3 CD200 model system, our data supported this existing signaling paradigm, evident by reduced p-ERK activity in T cells exposed to CD200+ AML and significantly diminished cytokine production. However, we were surprised to see the same pattern of cytokine suppression in the Kasumi1 model where there was no evidence of MAPK signaling in T cells co-cultured with either CD200+ or CD200− AML. In fact, this is the first evidence to suggest that CD200R signaling converges on inhibition of the STAT3 signaling pathway irrespective of MAPK activity. Our data suggest that there may exist either (1) other downstream pathways that are regulated through CD200R1 receptor ligation or (2) the potential of CD200 interaction with another receptor on T cells.
CD200 has been implicated in immunosuppression, in part, through regulation of inflammatory cytokine production. In AML, memory T cells derived from patients with high bulk CD200 expression had impaired IL-2 and IFNγ production on ex vivo stimulation when compared with those from CD200-low patients.9 These observations led to the hypothesis that CD200 could shift the cytokine milieu from Th1 to Th2, in an effort to protect leukemia cells from an inflammatory microenvironment. In alignment with these studies, our data indicated that CD200+ AML could robustly inhibit secretion of critical inflammatory cytokines, including TNFα and IFNγ, from T cells. We also showed, for the first time, that CD200+ AML could also exert the same cytokine suppression on healthy macrophages. In opposition to the existing hypothesis, the immunosuppression seen here was not limited to pro-inflammatory cytokines but also included downregulation of anti-inflammatory Th2 cytokines including IL-4, IL-6, IL-10, and IL-13.37 The broad pattern of cytokine suppression was consistent in our humanized mouse model. This suggests that CD200 signaling to immune cells does not selectively regulate pro-inflammatory cytokines but, rather, may act as a master regulator of all cytokine production in CD200R1+ immune cells.
The role of CD200 in allogeneic immune response has been studied in the transplantation setting where it was originally found that increased expression of CD200 on dendritic cells was functionally important in murine renal allograft survival.38 Similarly, mice with high levels of soluble CD200 in the serum had enhanced graft survival and administration of supplemental CD200-Fc chimeric protein was sufficient to promote survival of allografts and xenografts in mice.39 40 Our PBMC-humanized mouse offers the alternative model where the immune cells are transplanted, instead of grafted tissue. Still, our data are consistent with the role of CD200 in allogeneic immunity in that CD200 expression alone on AML cells was sufficient for protection from allo-induced T cell inflammatory response. It will be important in the future to also elucidate the role of CD200 immunoprotection in a model system without GvL, such as HLA-matched AML in CD34-humanized mice.
Another potential mechanism of CD200-mediated immunosuppression is through the recruitment or expansion of FoxP3+ regulatory T cells (Tregs).40 This is in line with a later clinical observation that high CD200 expression on AML blasts correlated with a higher proportion of FoxP3+ Tregs.27 However, in our humanized mouse models, we saw no significant differences in the frequency of Tregs. In fact, the Tregs present in mice with CD200+ disease appeared to have a less active phenotype as characterized by decreased expression of TIGIT, Lag-3, and Ki67. This discrepancy with the clinical observation may be explained, in part, by potential confounding of various AML subtypes. What we did observe was a significantly reduced proportion of CD4 central memory cells in mice with CD200+ leukemia. This could be explained, in part, by the reliance of CD4 T cell memory cells on effective STAT3 signaling.41 While frequencies of CD8 cell subsets did not substantially vary between groups, we did find significantly more phenotypically inactive CD8 effector and central memory cells in the mice with CD200+ disease which may contribute to the immune-escape mechanism.
Lymphocyte signaling, differentiation, and function are all processes intimately tied to metabolic reprogramming. Adequate T cell activation requires both an increase in mitochondrial OXPHOS production and induction of aerobic glycolysis.42 While T cells from CD200+ leukemia mice appeared to upregulate genes involved in inflammatory response at the transcript level, these cells were surprisingly more metabolically quiescent compared with T cells obtained from mice exposed to CD200− leukemia. Such metabolic quiescence could be the result of T cell anergy; however, our ex vivo metabolic assays demonstrated, for the first time, that CD200+ leukemia could directly suppress OXPHOS in healthy human T cells after only 4 hours of exposure. It is known that tumor cells can also alter T cell function by manipulating pH, nutrient availability, and oxygen abundance in the tumor immune microenvironment.43 Further studies are necessary to understand whether this regulation of T cell metabolism is directly downstream of the CD200 receptor or due to secondary mechanisms.
Samalizumab is a clinically available, anti-CD200 humanized monoclonal antibody engineered with a cross-subclass IgG2/G4 backbone to minimize Fc-mediated effector functions. In the phase I trial, samalizumab was well tolerated and had a good safety profile.44 TTI‐CD200, a humanized IgG4 CD200 antibody, was recently shown to improve CIK cellular therapy in vivo.10 We hypothesized we could enhance therapeutic efficacy of CD200 antibody by replacing the chimeric Fc region with a potent IgG1 subclass. This would allow specific AML cell targeting by (1) blocking immunosuppressive activity of CD200 while simultaneously (2) engaging Fc-mediated effector cells for AML destruction. In vitro, we confirmed that our novel antibody could largely restore cytokine secretion of effector cells in the presence of CD200+ AML. We also demonstrated that the anti-CD200 antibody could induce potent, specific macrophage-mediated ADCP. While anti-CD200 monotherapy resulted in initial regression of CD200+ disease in NSG mice, all eventually progressed, suggesting that phagocytosis alone was transiently leukemia-suppressive, although insufficient to eliminate AML.28 45 However, in the PBMC-humanized model, which mimics the scenario seen in AML patient samples after allogeneic stem cell transplant, CD200 antibody treatment was capable of eradicating CD200+ disease. This phenomenon appeared to be dependent on the strength of the T cell–mediated response. Together, our findings suggest that clearance of CD200+ AML by targeted antibody therapy requires components of both innate and adaptive immune systems and can be further facilitated by existing AML therapies.
LSCs are widely considered to be resistant to standard AML therapies and have been hypothesized to contribute to MRD and relapse.46 As such, we anticipate a role of CD200 in the identification of, or functional contribution to, MRD. Recently, high CD200 protein expression was recently reported to be associated with MRD in pediatric AML.47 Using the RNA-seq data generated by Stevens et al,30 we were excited to see the highest levels of CD200 expression on LSCs from patients who would go on to have detectable MRD after treatment with AZA/VEN, a regimen designed to specifically eliminate the LSCs. The rationale for combining anti-CD200 with AZA/VEN was to eliminate the CD200+ LSCs that may be potentially resistant to AZA/VEN alone. Indeed, our data from an AZA/VEN-resistant, CD200+ patient-derived xenograft model of AML demonstrated that this combination could profoundly improve efficacy when compared with anti-CD200 therapy or AZA/VEN alone. Further studies are warranted to investigate the role of CD200 in MRD after other frontline therapies.
In summary, we have identified and characterized an AML stem cell immunomodulatory protein, CD200, that acts as a suppressor of metabolic activity, STAT3 and MAPK signaling, and cytokine secretion in innate and adaptive immune cells. In the context of immune competent microenvironment, CD200 expression on AML cells is sufficient for protection against immune-mediated rejection. Lastly, a novel anti-CD200 antibody, capable of restoring cytokine production and inducing a robust Fc-mediated innate immune response, can eliminate CD200+ AML in the presence of immune effectors or low-intensity chemotherapy. Collectively, we offer a new paradigm of LSC-specific immunosuppression via overexpression of CD200 that will facilitate further investigations into exploiting stem cell–specific immunology to eliminate LSCs and cure poor-risk AML subsets by immunotherapy.
Acknowledgments
We kindly thank Dr. Ross Levine for providing the murine AML model. Cell sorting, analytical flow cytometry, and CyTOF were all performed in collaboration with the MD Anderson Flow Cytometry and Cellular Imaging Core Facility.
Footnotes
Twitter: @sherbrich, @maitkencancerhx, @Daver_Leukemia
Correction notice: This article has been corrected since it was first published. Ondrej Havranek has now been added as an author.
Contributors: SH performed the bulk of experiments, analyses, and wrote the manuscript. NB performed and analyzed Seahorse experiments. TC aided in cytokine experiments. CW assisted in the animal studies. MJLA and SMP aided in experimental design and critically revised the manuscript. GA-A helped in cytotoxicity studies. JH, OH and RED designed and assisted with isogenic cell line creation. CS and DZ produced and characterized the anti-CD200 antibody. ND provided clinical immunotherapy insight. GR-C and GS provided and analyzed the AML patient cohort. KB consulted on bioinformatic analyses. MK oversaw this project and edited the manuscript.
Funding: This study was funded by Institutional Research Grant (IRG, MDACC); Moonshot, MDACC.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. Data are available on reasonable request. Raw and processed RNA sequencing data are available through the NCBI GEO database under accession number GSE173194. All other raw data may be requested from SH (SMHerbrich@mdanderson.org).
Ethics statements
Patient consent for publication
Not required.
References
- 1.Thomas D, Majeti R. Biology and relevance of human acute myeloid leukemia stem cells. Blood 2017;129:1577–85. 10.1182/blood-2016-10-696054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powles R. Immunotherapy for acute myelogenous leukaemia. Br J Cancer Suppl 1973;1:365–76. 10.1038/bjc.1973.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albring JC, Inselmann S, Sauer T, et al. Pd-1 checkpoint blockade in patients with relapsed AML after allogeneic stem cell transplantation. Bone Marrow Transplant 2017;52:317–20. 10.1038/bmt.2016.274 [DOI] [PubMed] [Google Scholar]
- 4.Daver N, Garcia-Manero G, Basu S, et al. Efficacy, safety, and biomarkers of response to azacitidine and nivolumab in relapsed/refractory acute myeloid leukemia: a nonrandomized, open-label, phase II study. Cancer Discov 2019;9:370–83. 10.1158/2159-8290.CD-18-0774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masarova L, Kantarjian H, Garcia-Mannero G, et al. Harnessing the immune system against leukemia: monoclonal antibodies and checkpoint strategies for AML. Adv Exp Med Biol 2017;995:73–95. 10.1007/978-3-319-53156-4_4 [DOI] [PubMed] [Google Scholar]
- 6.Ho JM, Dobson SM, Voisin V, et al. Cd200 expression marks leukemia stem cells in human AML. Blood Adv 2020;4:5402–13. 10.1182/bloodadvances.2020001802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoek RM, Ruuls SR, Murphy CA, et al. Down-regulation of the macrophage lineage through interaction with OX2 (CD200). Science 2000;290:1768–71. 10.1126/science.290.5497.1768 [DOI] [PubMed] [Google Scholar]
- 8.Coles SJ, Wang ECY, Man S, et al. Cd200 expression suppresses natural killer cell function and directly inhibits patient anti-tumor response in acute myeloid leukemia. Leukemia 2011;25:792–9. 10.1038/leu.2011.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coles SJ, Hills RK, Wang ECY, et al. Expression of CD200 on AML blasts directly suppresses memory T-cell function. Leukemia 2012;26:2148–51. 10.1038/leu.2012.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rastogi N, Baker S, Man S, et al. Use of an anti-CD200-blocking antibody improves immune responses to AML in vitro and in vivo. Br J Haematol 2021;193:155–9. 10.1111/bjh.17125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiribelli M, Raspadori D, Geromin A, et al. High CD200 expression is associated with poor prognosis in cytogenetically normal acute myeloid leukemia, even in FlT3-ITD-/NPM1+ patients. Leuk Res 2017;58:31–8. 10.1016/j.leukres.2017.04.001 [DOI] [PubMed] [Google Scholar]
- 12.Damiani D, Tiribelli M, Raspadori D, et al. Clinical impact of CD200 expression in patients with acute myeloid leukemia and correlation with other molecular prognostic factors. Oncotarget 2015;6:30212–21. 10.18632/oncotarget.4901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tonks A, Hills R, White P, et al. Cd200 as a prognostic factor in acute myeloid leukaemia. Leukemia 2007;21:566–8. 10.1038/sj.leu.2404559 [DOI] [PubMed] [Google Scholar]
- 14.Corces MR, Buenrostro JD, Wu B, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 2016;48:1193–203. 10.1038/ng.3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Jonge HJM, Woolthuis CM, Vos AZ, et al. Gene expression profiling in the leukemic stem cell-enriched CD34+ fraction identifies target genes that predict prognosis in normal karyotype AML. Leukemia 2011;25:1825–33. 10.1038/leu.2011.172 [DOI] [PubMed] [Google Scholar]
- 16.Jung N, Dai B, Gentles AJ, et al. An Lsc epigenetic signature is largely mutation independent and implicates the HOXA cluster in AML pathogenesis. Nat Commun 2015;6:8489. 10.1038/ncomms9489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemieux S, Sargeant T, Laperrière D, et al. MiSTIC, an integrated platform for the analysis of heterogeneity in large tumour transcriptome datasets. Nucleic Acids Res 2017;45:e122. 10.1093/nar/gkx338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duval M, Klein JP, He W, et al. Hematopoietic stem-cell transplantation for acute leukemia in relapse or primary induction failure. J Clin Oncol 2010;28:3730–8. 10.1200/JCO.2010.28.8852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang M, Storer B, Estey E, et al. Outcome of patients with acute myeloid leukemia with monosomal karyotype who undergo hematopoietic cell transplantation. Blood 2011;118:1490–4. 10.1182/blood-2011-02-339721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng SWK, Mitchell A, Kennedy JA, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016;540:433–7. 10.1038/nature20598 [DOI] [PubMed] [Google Scholar]
- 22.Herbrich SM, Baggerly KA, Konopleva M. Refining statistics clarifies leukaemic stem cell genomics. Br J Haematol 2019;185:1005–7. 10.1111/bjh.15697 [DOI] [PubMed] [Google Scholar]
- 23.Hatherley D, Lea SM, Johnson S, et al. Structures of CD200/CD200 receptor family and implications for topology, regulation, and evolution. Structure 2013;21:820–32. 10.1016/j.str.2013.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright GJ, Cherwinski H, Foster-Cuevas M, et al. Characterization of the CD200 receptor family in mice and humans and their interactions with CD200. J Immunol 2003;171:3034–46. 10.4049/jimmunol.171.6.3034 [DOI] [PubMed] [Google Scholar]
- 25.Shih AH, Jiang Y, Meydan C, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell 2015;27:502–15. 10.1016/j.ccell.2015.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S, Cherwinski H, Sedgwick JD, et al. Molecular mechanisms of CD200 inhibition of mast cell activation. J Immunol 2004;173:6786–93. 10.4049/jimmunol.173.11.6786 [DOI] [PubMed] [Google Scholar]
- 27.Coles SJ, Hills RK, Wang ECY, et al. Increased CD200 expression in acute myeloid leukemia is linked with an increased frequency of Foxp3+ regulatory T cells. Leukemia 2012;26:2146–8. 10.1038/leu.2012.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chao MP, Alizadeh AA, Tang C, et al. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell 2010;142:699–713. 10.1016/j.cell.2010.07.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barclay AN, Wright GJ, Brooke G, et al. Cd200 and membrane protein interactions in the control of myeloid cells. Trends Immunol 2002;23:285–90. 10.1016/S1471-4906(02)02223-8 [DOI] [PubMed] [Google Scholar]
- 30.Stevens BM, Jones CL, Pollyea DA, et al. Fatty acid metabolism underlies venetoclax resistance in acute myeloid leukemia stem cells. Nat Cancer 2020;1:1176–87. 10.1038/s43018-020-00126-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stelljes M, Krug U, Beelen DW, et al. Allogeneic transplantation versus chemotherapy as postremission therapy for acute myeloid leukemia: a prospective matched pairs analysis. J Clin Oncol 2014;32:288–96. 10.1200/JCO.2013.50.5768 [DOI] [PubMed] [Google Scholar]
- 32.Ohyama M, Terunuma A, Tock CL, et al. Characterization and isolation of stem cell-enriched human hair follicle bulge cells. J Clin Invest 2006;116:249–60. 10.1172/JCI26043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ong WK, Tan CS, Chan KL, et al. Identification of specific cell-surface markers of adipose-derived stem cells from subcutaneous and visceral fat depots. Stem Cell Reports 2014;2:171–9. 10.1016/j.stemcr.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delorme B, Ringe J, Gallay N, et al. Specific plasma membrane protein phenotype of culture-amplified and native human bone marrow mesenchymal stem cells. Blood 2008;111:2631–5. 10.1182/blood-2007-07-099622 [DOI] [PubMed] [Google Scholar]
- 35.Kawasaki BT, Mistree T, Hurt EM, et al. Co-Expression of the toleragenic glycoprotein, CD200, with markers for cancer stem cells. Biochem Biophys Res Commun 2007;364:778–82. 10.1016/j.bbrc.2007.10.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mihrshahi R, Barclay AN, Brown MH. Essential roles for Dok2 and RasGAP in CD200 receptor-mediated regulation of human myeloid cells. J Immunol 2009;183:4879–86. 10.4049/jimmunol.0901531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Opal SM, DePalo VA. Anti-Inflammatory cytokines. Chest 2000;117:1162–72. 10.1378/chest.117.4.1162 [DOI] [PubMed] [Google Scholar]
- 38.Gorczynski RM, Chen Z, Fu XM, et al. Increased expression of the novel molecule OX-2 is involved in prolongation of murine renal allograft survival. Transplantation 1998;65:1106–14. 10.1097/00007890-199804270-00016 [DOI] [PubMed] [Google Scholar]
- 39.Gorczynski RM, Cattral MS, Chen Z, et al. An immunoadhesin incorporating the molecule OX-2 is a potent immunosuppressant that prolongs allo- and xenograft survival. J Immunol 1999;163:1654–60. [PubMed] [Google Scholar]
- 40.Gorczynski R, Chen Z, Khatri I, et al. sCD200 present in mice receiving cardiac and skin allografts causes immunosuppression in vitro and induces Tregs. Transplantation 2013;95:442–7. 10.1097/TP.0b013e3182754c30 [DOI] [PubMed] [Google Scholar]
- 41.Siegel AM, Heimall J, Freeman AF, et al. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity 2011;35:806–18. 10.1016/j.immuni.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buck MD, O'Sullivan D, Pearce EL. T cell metabolism drives immunity. J Exp Med 2015;212:1345–60. 10.1084/jem.20151159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin Z, Bai L, Li W, et al. Targeting T cell metabolism in the tumor microenvironment: an anti-cancer therapeutic strategy. J Exp Clin Cancer Res 2019;38:403. 10.1186/s13046-019-1409-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahadevan D, Lanasa MC, Farber C, et al. Phase I study of samalizumab in chronic lymphocytic leukemia and multiple myeloma: blockade of the immune checkpoint CD200. J Immunother Cancer 2019;7:227. 10.1186/s40425-019-0710-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng M, Chen JY, Weissman-Tsukamoto R, et al. Macrophages eat cancer cells using their own calreticulin as a guide: roles of TLR and Btk. Proc Natl Acad Sci U S A 2015;112:2145–50. 10.1073/pnas.1424907112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Felipe Rico J, Hassane DC, Guzman ML. Acute myelogenous leukemia stem cells: from bench to bedside. Cancer Lett 2013;338:4–9. 10.1016/j.canlet.2012.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kandeel EZ, Madney Y, Eldin DN, et al. Overexpression of CD200 and CD123 is a major influential factor in the clinical course of pediatric acute myeloid leukemia. Exp Mol Pathol 2021;118:104597. 10.1016/j.yexmp.2020.104597 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-002968supp001.pdf (1.8MB, pdf)
Data Availability Statement
Data are available in a public, open access repository. Data are available on reasonable request. Raw and processed RNA sequencing data are available through the NCBI GEO database under accession number GSE173194. All other raw data may be requested from SH (SMHerbrich@mdanderson.org).