Abstract
Exosomal miRNAs (EmiRs) can be used for prediction of gastric cancer (GC) development. Supposedly, both plasma and urinary microRNAs can also be potential biomarkers for screening, but the diagnostic values of EmiRs in blood and urine are not fully studied. We here collected both types of samples from GC patients and healthy individuals and conducted miRNA sequencing to identify key members of EmiRs in GC. The exosomes samples derived from blood and urine were collected from 3 healthy individuals and 7 GC patients. Differentially expressed miRNAs (DEmiRNAs) were acquired, ontology enrichment analysis and Protein-protein Interaction (PPI) enrichment analysis were performed. There were 8 DEmiRNAs in the serum and 3 DEmiRNAs in the urine. For GC patients, there were three up-regulated DEmiRNAs (hsa-miR-130b-3p, hsa-miR-151a-3p and hsa-miR-15b-3p) in the serum exosomes, and one up-regulated DEmiRNA (hsa-miR-1246) in the urinary exosomes. Using miRNA target prediction databases, we found 418 common targets of hsa-miR-15b-3p, 35 common targets of hsa-miR-151a-3p, 117 common targets of hsa-miR-130b-3p, and 357 common targets of hsa-miR-1246. Some commonly enriched ontology terms were found, including GO BP terms like cell surface receptor signaling pathway involved in cell-cell signaling, positive regulation of catabolic process, morphogenesis of an epithelium, and GO CC terms perinuclear region of cytoplasm. The PPI network show some key nodes, including TAOK1, CMTM6, SCN3A, WASF3, IGF1, CNOT7, GABRG1, PRKD1. Together, this study provided an integrative analysis of expression profile of key circulating exosomal microRNAs. Four key exosomal miRNAs (hsa-miR-130b-3p, hsa-miR-151a-3p and hsa-miR-15b-3p) and the interaction network or enrichments based on their targets (TAOK1, CMTM6, SCN3A, WASF3, IGF1, CNOT7, GABRG1, PRKD1) may provide a reference of the molecular mechanisms in the GC development.
Keywords: gastric cancer, exosome, miRNA, plasma, bioinformatics analysis
Introduction
Gastric cancer (GC) is the fourth most common malignance worldwide (1). An exact detection based on GC biomarkers has a high clinical significance. For GC patients, the exosomal microRNA (miRNA or miR) may have potential clinical application value in diagnosis and status evaluation (2, 3). MicroRNA is a short-chain noncoding RNA molecule (around 22 nucleotides in length). It regulates protein expression of a particular mRNA (by incomplete base pairing), causing inhibition of protein translation of its target genes. Exosomes are a subset of extracellular vesicles containing a variety of bioactive molecules including proteins, lipids, and RNAs. It can play a role in intercellular crosstalk. During tumor development, exosomes can facilitate construction of the specific microenvironment; besides, it strongly affects the occurrence, metastasis, and even drug resistance. Tumor secreted exosomes are sharply different among cancers, and between healthy individuals vs. cancer patients. Theoretically, the contents of exosomes include different tumor-related biomarkers. Commonly, previous studies observed the samples of plasma exosomal miRNAs (EmiRs) for prediction of GC development, progression, and treatment outcomes. Although the development of new biomarkers in blood tests has shown great potential, limited reports have applied multi-biomarkers based on the strategy of exosomal miRNA detection. Supposedly, urinary microRNA can also be a potential biomarker for tumor screening. For example, a lower urine level of miR-30a-5p was found in gastric cancer and colon carcinoma patients when compared to ovarian serous adenocarcinoma, and urinary miR-30a-5p from ovarian cancer patients was notably reduced following the surgical removal of ovarian serous adenocarcinoma (4). However, the diagnostic value of EmiRs in urine for GC is unknown, and that of plasma EmiRs is also not fully studied. We here collected both plasma samples and urine samples from GC patients and healthy control individuals for miRNA sequencing, and some key members were identified.
Methods
Exosomal miRNA Sequencing
The exosomes samples derived from blood and urine were collected from 3 healthy individuals and 7 GC patients (Stage I-II), including 6 blood samples (3 GC vs 3 controls) and 10 urine samples (7 GC vs 3 controls). The median age of the 7 patients was 52 years, and that of the healthy controls was 45 years. All GC patients were confirmed by histopathology. The frozen samples were thawed and centrifuged at 2000 g for 30 min to remove cells/debris. Next, 0.2 volumes of the Total Exosome Isolation reagent (Thermo Fisher, Inc.) were added, the solution was thoroughly mixed and incubated for 30 min at 4°C. Next, the samples were centrifuged at 10000 g for 10 min and the exosome pellet was resuspended in PBS. Next, the exosome-derived RNA was extracted from exosomes using the Total Exosome RNA and Protein Isolation Kit, the total amount of RNA was detected. Subsequently, the Illumina NextSeq 500 SE50 (20M) sequencing was performed. After sequencing, the FastQ-format data were stored and cleaned for further analysis.
Differential miRNAs Analysis
First, miRNA expression was analyzed by miRExpress (http://mirexpress.mbc.nctu.edu.tw), which works based on the miRNA database (http://www.mirbase.org) for quantitative analysis of miRNA. After the second-generation sequencing raw data in FastQ format are imported, the software compares the data with the known miRNA sequence in miRBase and then acquires the read count of the same sequence fragments as Counts, which is used to measure the miRNA levels in a sample. According to the grouping information, the sample expression data was imported into the edgeR R package to compare the expression differences between samples. The Fold Change and p-value were obtained. Then, according to the threshold of p-value<0.05 and Log(Foldchange) >2 or <-2, the DEmiRNAs was acquired. The volcanic plots were produced to present DEmiRNAs using the ggplot2 R package. And a principal component analysis (PCA) was performed using the ggfortify R package based on all detected miRNA levels.
Expression Analysis of DEmiRNAs in TCGA and EVmiRNA
Exosomal miRNAs that are up-regulated in the urine or blood exosomes of tumor patients have potential clinical applications, so the differential miRNAs obtained from screening were subjected to expression analysis. From the Cancer Genome Atlas (https://portal.gdc.cancer.gov/) and EVmiRNA (http://bioinfo.life.hust.edu.cn/EVmiRNA#!/) databases, the raw count of tumor and exosomal miRNA expression data were obtained. And box plots were drawn by ggplot2 R software.
Prognostic Analysis of DEmiRNAs in TCGA
Corresponding clinical information were obtained from The TCGA dataset. High and low expression of miRNAs was defined with the median. For the Kaplan-Meier curve, the p value and the hazard ratio (HR) with 95% confidence interval (CI) are obtained by logrank test and univariate Cox proportional hazard regression. p<0.05 was considered statistically significant. And p < 0.05 was considered as statistically significant.
Target Prediction of the DEmiRNAs
First, we focused on the up-regulated DEmiRNAs which may be encapsulated in circulating exosomes. The mRNAs potentially targeted by these miRNAs were predicted using both miRDB and Targetscan databases. When the intersection of two databases regarding the targets of each up-regulated DE-miR was acquired, the overlap set of mRNAs were further used for enrichment analysis.
Predicted Target Gene Enrichment Analysis
The Metascape gene list analysis online tool was used for enrichment analysis of the predicted target genes. All targets were imported into the tool, gene annotation was performed, and pathway and process enrichment analysis were carried out with the following ontology sources: KEGG Pathway, GO Biological Process, GO Molecular Function and GO Cell Component. The Homo Sapiens background was used for enrichment analysis of all predicted target genes. Terms with a p-value < 0.01, a minimum count of 3, and an enrichment factor > 1.5 (the ratio between the observed counts and the counts expected by chance) were collected and grouped into clusters based on their according DEmiRNAs. The statistically enriched terms were presented by accumulative hypergeometric p-values and enrichment factors, and then hierarchically clustered into a tree based on Kappa-statistical similarities.
Protein-Protein Interaction Analysis
Protein-protein interaction analysis was carried out with STRING and BioGrid. And the Molecular Complex Detection (MCODE) was applied to identify densely connected network components. The MCODE networks identified for individual gene lists were gathered and drawn by the Metascape (https://metascape.org/).
Network Hub Gene Expression Analysis
For the analysis of hub genes in the resulting MCODE subnetwork, GEPIA (http://gepia.cancer-pku.cn/) tools were used to analyze their expression profiles in the STAD from TCGA. And p < 0.05 was considered as statistically significant.
Results
Identification of GC Associated DEmiRNAs
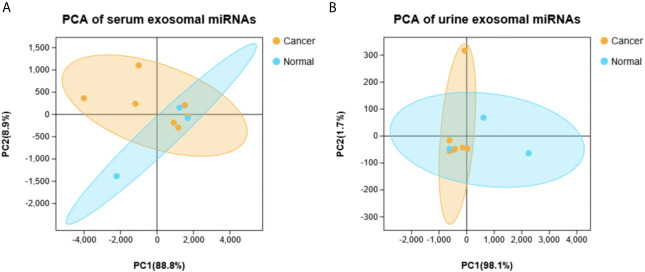
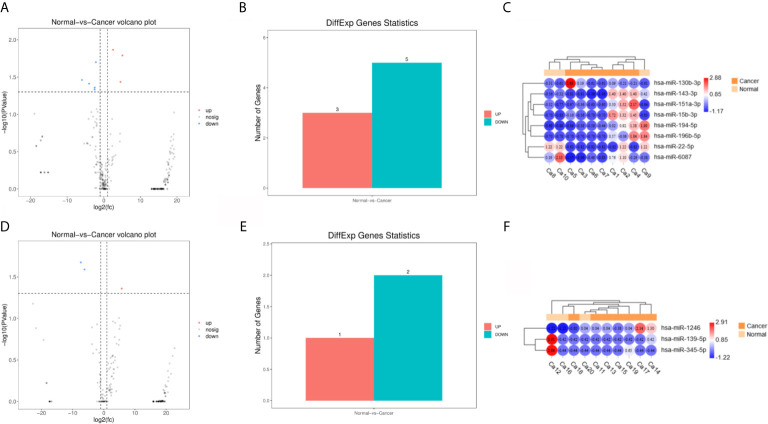
Principal component analysis were presented in Figure 1 , which suggested that there is some heterogeneity in the expression profiles of blood and urine exosomal miRNAs in patients. As Table 1 and Figure 2 shown, there were 8 DEmiRNAs in the serum and 3 DEmiRNAs in the urine. The expression profiles of all differentially expressed miRNAs are shown in the Heatmap. For GC patients, there were 3 up-regulated DEmiRNAs (hsa-miR-130b-3p, hsa-miR-151a-3p and hsa-miR-15b-3p) in the serum exosomes, and one DEmiRNA (hsa-miR-1246) enriched in the urinary exosomes. Theoretically, these up-regulated miRNAs may be transferred to tissues or cells and target specific RNAs, which finally promotes the GC development.
Figure 1.
The principal component analysis (PCA) based on all detected miRNA levels. (A) Serum. (B) Urine.
Table 1.
Differentially expressed exosomal microRNAs identified by next generation sequencing.
| miRNA Information | Statistics & Regulation | ||||
|---|---|---|---|---|---|
| Source | Mature-miRNA | sequence | Fold Change | P-value | Regulation |
| Serum | hsa-miR-130b-3p | CAGUGCAAUGAUGAAAGGGCAU | 4.555389982 | 0.036615174 | Up |
| Serum | hsa-miR-143-3p | UGAGAUGAAGCACUGUAGCUC | -2.481915254 | 0.043638531 | Down |
| Serum | hsa-miR-151a-3p | CUAGACUGAAGCUCCUUGAGG | 2.545806244 | 0.013604136 | Up |
| Serum | hsa-miR-15b-3p | CGAAUCAUUAUUUGCUGCUCUA | 5.095125215 | 0.016185555 | Up |
| Serum | hsa-miR-194-5p | UGUAACAGCAACUCCAUGUGGA | -2.508236592 | 0.046425407 | Down |
| Serum | hsa-miR-196b-5p | UAGGUAGUUUCCUGUUGUUGGG | -4.002942023 | 0.038692865 | Down |
| Serum | hsa-miR-22-5p | AGUUCUUCAGUGGCAAGCUUUA | -5.970168823 | 0.034517425 | Down |
| Serum | hsa-miR-6087 | UGAGGCGGGGGGGCGAGC | -2.154113907 | 0.019948851 | Down |
| Urine | hsa-miR-1246 | AAUGGAUUUUUGGAGCAGG | 5.747077844 | 0.043731253 | Up |
| Urine | hsa-miR-139-5p | UCUACAGUGCACGUGUCUCCAGU | -6.184723327 | 0.025724708 | Down |
| Urine | hsa-miR-345-5p | GCUGACUCCUAGUCCAGGGCUC | -7.376130587 | 0.021140159 | Down |
Fold change, Fold change between two groups; P-value, P-value is calculated by paired t-test; Regulation, ‘up’ indicates up-regulation, and ‘down’ indicates down-regulation.
Figure 2.
Differentially expressed miRNA (DEmiRNAs) in plasma and urine samples of the gastric cancer (GC) patients. (A) The volcano plots of DEmiRNAs in serum. (B) Significant ups and downs in serum. (C) Expression heatmap of significantly expressed miRNAs in serum. (D–F) Urine.
Expression of DEmiRNAs in TCGA and EVmiRNA
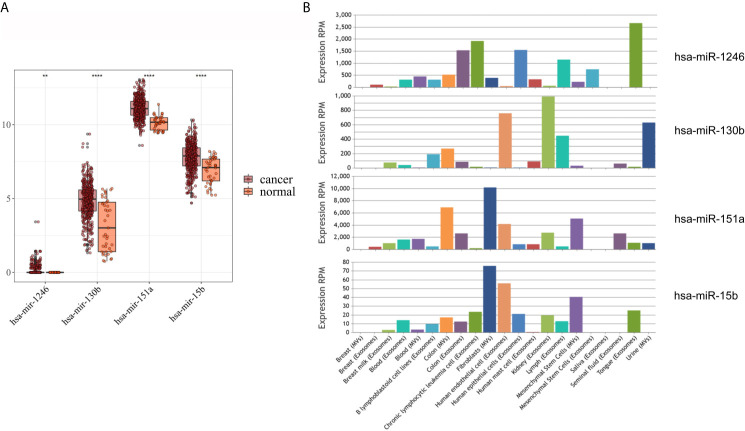
As shown in Figure 3A , hsa-mir-130b-3p, hsa-mir-151a-3p and hsa-mir-15b-3p, which were significantly upregulated in serum exosomes from GC patients, were expressed at lower levels in gastric cancer tissues compared with adjacent noncancerous tissues, and hsa-mir-1246, which was significantly upregulated in urine exosomes, was barely expressed in gastric cancer tissues or adjacent noncancerous tissues. While in the EVmiRNA database, which exclusively exhibits exosomal or microvesicular miRNA expression data, the expression profiles of these four miRNAs in different tissues of origin are shown in Figure 3B , it is worth noting that hsa-mir-1246 has never been previously reported in urine exosomes.
Figure 3.
Expression analysis of DEmiRNAs in TCGA and EVmiRNA. (A) The expression level of DEmiRNAs in TCGA dataset. (B) EVmiRNA dataset.
Prognostic of DEmiRNAs in TCGA
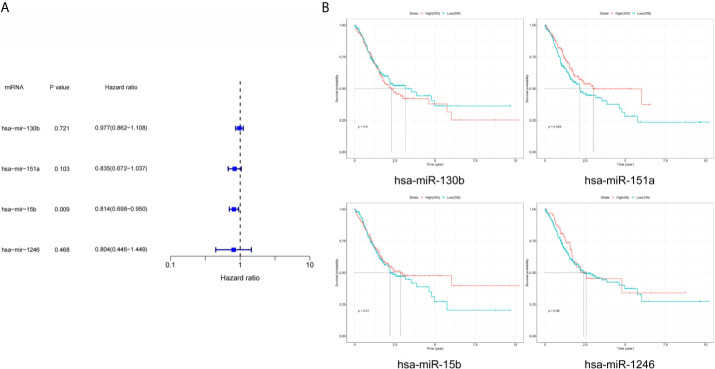
As shown in Figure 4A , the results of univariate Cox analysis regarding these four miRNAs revealed that only one serum exosomal hsa-mir-15b-3p was identified to be associated with GC patient prognosis (HR = 0.814, P < 0.05). None of the other three were related, hsa-mir-130b-3p (HR = 0.977, P > 0.05), hsa-mir-151a-3p (HR = 0.835, P > 0.05), hsa-mir-1246 (HR = 0.804, P > 0.05). While the results of prognostic analysis in Figure 4B were different, high hsa-mir-151a-3p expression was considered to be associated with poor survival outcomes in GC patients (P < 0.05), while the other three miRNAs were not.
Figure 4.
Prognostic analysis of DEmiRNAs in TCGA. (A) The p value, risk coefficient HR and confidence interval of the DEmiRNAs. (B) Kaplan-Meier survival analysis of the DEmiRNAs signature.
Predicted Target Gene of the DEmiRNAs
Next, the targets of above three miRNAs were acquired. In the miRDB database, there were 470 targets of hsa-miR-15b-3p, 220 targets of hsa-miR-151a-3p, 917 targets of hsa-miR-130b-3p, and 407 targets of hsa-miR-1246; in the Targetscan database, there were 3573 targets of hsa-miR-15b-3p, 112 targets of hsa-miR-151a-3p, 183 targets of hsa-miR-130b-3p, and 3031 targets of hsa-miR-1246. The intersect of two databases were calculated and presented in Figure 5 . Between two databases, there were 418 common targets of hsa-miR-15b-3p, 35 common targets of hsa-miR-151a-3p, 117 common targets of hsa-miR-130b-3p, and 357 common targets of hsa-miR-1246.
Figure 5.
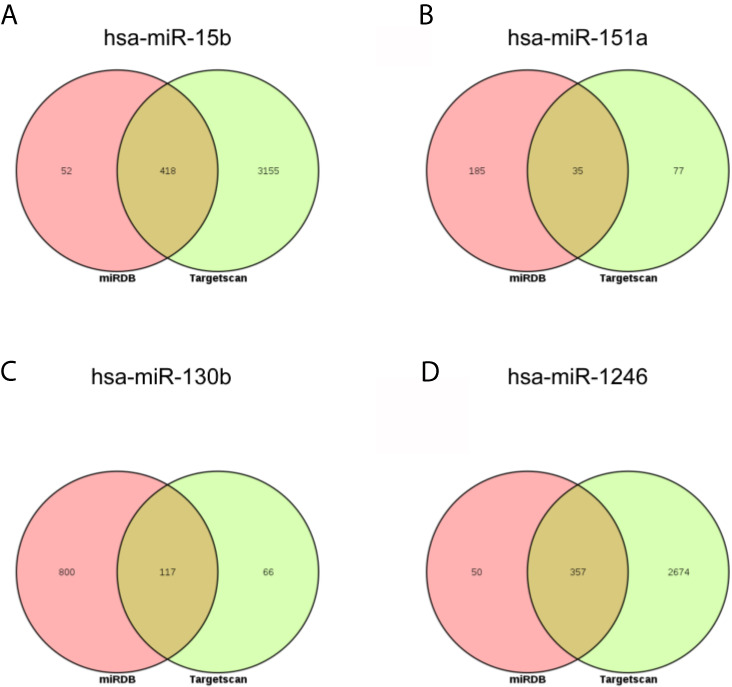
The intersection analysis of predicted target genes of DEmiRNAs. (A) hsa-miR-15b-3p. (B) hsa-miR-151a-3p. (C) hsa-miR-130b-3p. (D) hsa-miR-1246.
Predicted Target Gene Enrichment Analysis
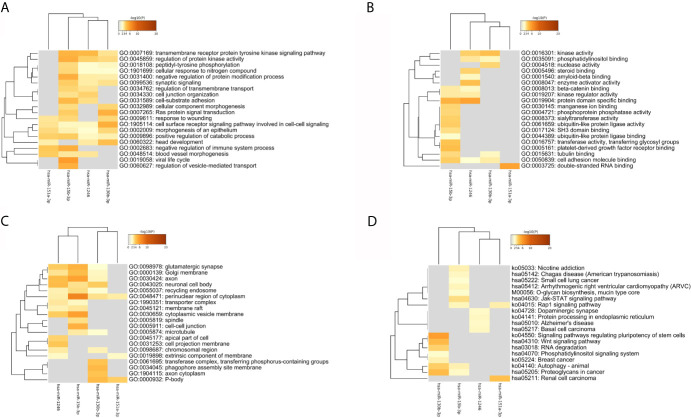
As Figure 6 and Table 2 shown, the enrichment analysis was conducted based on above targets. Some commonly enriched ontology terms were found, including GO BP terms like cell surface receptor signaling pathway involved in cell-cell signaling, positive regulation of catabolic process, morphogenesis of an epithelium, and GO CC terms perinuclear region of cytoplasm.
Figure 6.
The heatmap of the enriched GO terms and KEGG pathways based all the common targets of the four key DEmiRNAs. (A) Biology Process. (B) Molecule Function. (C) Cellular Components. (D) KEGG pathway.
Table 2.
List of enriched terms.
| GO | Category | Description | Count | % | Log10(P) | Log10(q) |
|---|---|---|---|---|---|---|
| GO:0045859 | GO Biological Processes | regulation of protein kinase activity | 63 | 7.13 | -10.64 | -6.45 |
| GO:0007169 | GO Biological Processes | transmembrane receptor protein tyrosine kinase signaling pathway | 59 | 6.68 | -10.17 | -6.45 |
| GO:0031400 | GO Biological Processes | negative regulation of protein modification process | 48 | 5.44 | -9.51 | -5.92 |
| GO:1905114 | GO Biological Processes | cell surface receptor signaling pathway involved in cell-cell signaling | 50 | 5.66 | -9.01 | -5.59 |
| GO:0009896 | GO Biological Processes | positive regulation of catabolic process | 41 | 4.64 | -8.86 | -5.56 |
| GO:0099536 | GO Biological Processes | synaptic signaling | 53 | 6 | -8.09 | -5.04 |
| GO:0007265 | GO Biological Processes | Ras protein signal transduction | 33 | 3.74 | -7.98 | -4.96 |
| GO:0002009 | GO Biological Processes | morphogenesis of an epithelium | 42 | 4.76 | -7.19 | -4.32 |
| GO:0019058 | GO Biological Processes | viral life cycle | 32 | 3.62 | -7.14 | -4.29 |
| GO:0032989 | GO Biological Processes | cellular component morphogenesis | 53 | 6 | -7.11 | -4.28 |
| GO:0031589 | GO Biological Processes | cell-substrate adhesion | 32 | 3.62 | -6.83 | -4.04 |
| GO:0048514 | GO Biological Processes | blood vessel morphogenesis | 48 | 5.44 | -6.79 | -4.02 |
| GO:0009611 | GO Biological Processes | response to wounding | 47 | 5.32 | -6.77 | -4.02 |
| GO:0034330 | GO Biological Processes | cell junction organization | 49 | 5.55 | -6.75 | -4.02 |
| GO:0060627 | GO Biological Processes | regulation of vesicle-mediated transport | 40 | 4.53 | -6.71 | -4.02 |
| GO:1901699 | GO Biological Processes | cellular response to nitrogen compound | 49 | 5.55 | -6.71 | -4.02 |
| GO:0002683 | GO Biological Processes | negative regulation of immune system process | 34 | 3.85 | -6.68 | -4 |
| GO:0060322 | GO Biological Processes | head development | 51 | 5.78 | -6.61 | -3.96 |
| GO:0034762 | GO Biological Processes | regulation of transmembrane transport | 42 | 4.76 | -6.57 | -3.95 |
| GO:0018108 | GO Biological Processes | peptidyl-tyrosine phosphorylation | 32 | 3.62 | -6.57 | -3.95 |
| GO:0019904 | GO Molecular Functions | protein domain specific binding | 58 | 6.57 | -10.91 | -7.24 |
| GO:0016301 | GO Molecular Functions | kinase activity | 50 | 5.66 | -6.36 | -3.17 |
| GO:0050839 | GO Molecular Functions | cell adhesion molecule binding | 39 | 4.42 | -5.82 | -2.75 |
| GO:0019207 | GO Molecular Functions | kinase regulator activity | 22 | 2.49 | -5.73 | -2.75 |
| GO:0003725 | GO Molecular Functions | double-stranded RNA binding | 4 | 11.43 | -5.6 | -2.16 |
| GO:0015631 | GO Molecular Functions | tubulin binding | 29 | 3.28 | -5.16 | -2.34 |
| GO:0017124 | GO Molecular Functions | SH3 domain binding | 14 | 1.59 | -4.29 | -1.56 |
| GO:0008047 | GO Molecular Functions | enzyme activator activity | 35 | 3.96 | -4.28 | -1.56 |
| GO:0005161 | GO Molecular Functions | platelet-derived growth factor receptor binding | 4 | 0.96 | -4.24 | -1.43 |
| GO:0016757 | GO Molecular Functions | transferase activity, transferring glycosyl groups | 22 | 2.49 | -4.04 | -1.41 |
| GO:0035091 | GO Molecular Functions | phosphatidylinositol binding | 20 | 2.27 | -3.8 | -1.36 |
| GO:0061659 | GO Molecular Functions | ubiquitin-like protein ligase activity | 14 | 3.35 | -3.68 | -1.19 |
| GO:0008373 | GO Molecular Functions | sialyltransferase activity | 4 | 0.96 | -3.63 | -1.19 |
| GO:0005496 | GO Molecular Functions | steroid binding | 7 | 1.97 | -3.55 | -1.15 |
| GO:0030145 | GO Molecular Functions | manganese ion binding | 6 | 1.44 | -3.5 | -1.14 |
| GO:0004518 | GO Molecular Functions | nuclease activity | 6 | 5.13 | -3.49 | -1.14 |
| GO:0008013 | GO Molecular Functions | beta-catenin binding | 10 | 1.13 | -3.44 | -1.05 |
| GO:0044389 | GO Molecular Functions | ubiquitin-like protein ligase binding | 22 | 2.49 | -3.42 | -1.05 |
| GO:0004721 | GO Molecular Functions | phosphoprotein phosphatase activity | 10 | 2.39 | -3.38 | -1.09 |
| GO:0001540 | GO Molecular Functions | amyloid-beta binding | 10 | 1.13 | -3.36 | -1.01 |
| GO:0048471 | GO Cellular Components | perinuclear region of cytoplasm | 62 | 7.02 | -12.08 | -8.8 |
| GO:0098978 | GO Cellular Components | glutamatergic synapse | 36 | 4.08 | -10.86 | -8.09 |
| GO:0043025 | GO Cellular Components | neuronal cell body | 46 | 5.21 | -10.83 | -8.09 |
| GO:0030424 | GO Cellular Components | axon | 54 | 6.12 | -10.77 | -8.09 |
| GO:0000139 | GO Cellular Components | Golgi membrane | 56 | 6.34 | -8.15 | -5.73 |
| GO:0030659 | GO Cellular Components | cytoplasmic vesicle membrane | 55 | 6.23 | -7.67 | -5.39 |
| GO:0055037 | GO Cellular Components | recycling endosome | 20 | 2.27 | -5.57 | -3.61 |
| GO:1990351 | GO Cellular Components | transporter complex | 28 | 3.17 | -5.46 | -3.53 |
| GO:0045177 | GO Cellular Components | apical part of cell | 31 | 3.51 | -5.03 | -3.15 |
| GO:0005911 | GO Cellular Components | cell-cell junction | 34 | 3.85 | -4.9 | -3.05 |
| GO:0034045 | GO Cellular Components | phagophore assembly site membrane | 3 | 2.56 | -4.51 | -2.24 |
| GO:0005819 | GO Cellular Components | spindle | 28 | 3.17 | -4.5 | -2.68 |
| GO:0005874 | GO Cellular Components | microtubule | 30 | 3.4 | -4.49 | -2.68 |
| GO:0045121 | GO Cellular Components | membrane raft | 25 | 2.83 | -4.41 | -2.64 |
| GO:0000932 | GO Cellular Components | P-body | 5 | 4.27 | -4.4 | -2.24 |
| GO:0061695 | GO Cellular Components | transferase complex, transferring phosphorus-containing groups | 21 | 2.38 | -4.27 | -2.53 |
| GO:0031253 | GO Cellular Components | cell projection membrane | 25 | 2.83 | -4.13 | -2.42 |
| GO:1904115 | GO Cellular Components | axon cytoplasm | 4 | 3.42 | -4 | -2.02 |
| GO:0019898 | GO Cellular Components | extrinsic component of membrane | 23 | 2.6 | -3.99 | -2.34 |
| GO:0098687 | GO Cellular Components | chromosomal region | 25 | 2.83 | -3.99 | -2.34 |
| ko04550 | KEGG Pathway | Signaling pathways regulating pluripotency of stem cells | 7 | 5.98 | -5.71 | -2.84 |
| ko04140 | KEGG Pathway | Autophagy - animal | 15 | 1.7 | -4.92 | -2.72 |
| ko05224 | KEGG Pathway | Breast cancer | 16 | 1.81 | -4.91 | -2.72 |
| hsa04310 | KEGG Pathway | Wnt signaling pathway | 6 | 5.13 | -4.52 | -2.39 |
| hsa05211 | KEGG Pathway | Renal cell carcinoma | 3 | 8.57 | -4.14 | -2.34 |
| hsa05217 | KEGG Pathway | Basal cell carcinoma | 8 | 0.91 | -3.52 | -1.76 |
| hsa03018 | KEGG Pathway | RNA degradation | 4 | 3.42 | -3.52 | -1.91 |
| ko04015 | KEGG Pathway | Rap1 signaling pathway | 17 | 1.93 | -3.44 | -1.75 |
| hsa04630 | KEGG Pathway | Jak-STAT signaling pathway | 14 | 1.59 | -3.38 | -1.74 |
| hsa05142 | KEGG Pathway | Chagas disease (American trypanosomiasis) | 11 | 1.25 | -3.27 | -1.67 |
| ko04728 | KEGG Pathway | Dopaminergic synapse | 12 | 1.36 | -3.09 | -1.53 |
| hsa04070 | KEGG Pathway | Phosphatidylinositol signaling system | 10 | 1.13 | -3.03 | -1.53 |
| hsa05412 | KEGG Pathway | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 8 | 0.91 | -2.73 | -1.35 |
| ko05033 | KEGG Pathway | Nicotine addiction | 4 | 0.96 | -2.54 | -1.23 |
| ko04141 | KEGG Pathway | Protein processing in endoplasmic reticulum | 7 | 1.97 | -2.27 | -1.15 |
| hsa05010 | KEGG Pathway | Alzheimer’s disease | 7 | 1.97 | -2.2 | -1.13 |
| M00056 | KEGG Pathway | O-glycan biosynthesis, mucin type core | 3 | 0.72 | -2.14 | -1.1 |
| hsa05222 | KEGG Pathway | Small cell lung cancer | 5 | 1.2 | -2.08 | -1.1 |
PPI Network
Applying the STRING and BioGrid data, the PPI network was generated. Meanwhile, each MCODE network was drawn assigned by a unique color ( Figures 7B, D ) or by the matched DEmiRNAs ( Figures 7A, C ). Some genes were located at key nodes, such as TAOK1, CMTM6, SCN3A, WASF3, IGF1, CNOT7, GABRG1 and PRKD1. And the information of these key genes from mcode screening was exhibited in Table S1 .
Figure 7.
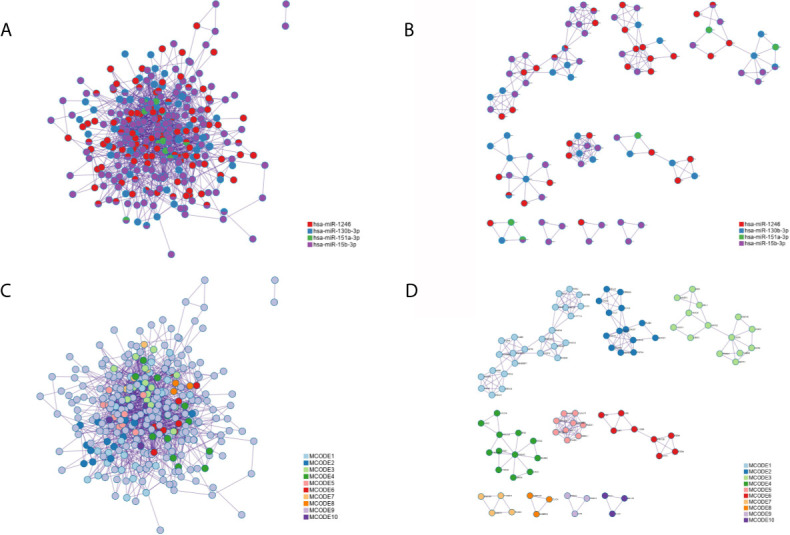
The protein-protein interaction (PPI) network of all common targets. (A) Network colored by DEmiRNAs group. (B) MCODE subnetworks colored by DEmiRNAs group. (C) Network colored by MCODE clusters. (D) MCODE subnetworks colored by MCODE clusters.
Network Hub Gene Expression Analysis
As shown in Figure 8 , the expression levels of eight key genes screened by MCODE in STAD tissues of TCGA were analyzed. The results showed that the expression of TAOK1, CMTM6 and CNOT7 were significantly lower in the adjacent cancers, while the expression of WASF3 was significantly higher in the adjacent cancers. SCN3A, IGF1, GABRG1 and PRKD1 were not significantly expressed.
Figure 8.
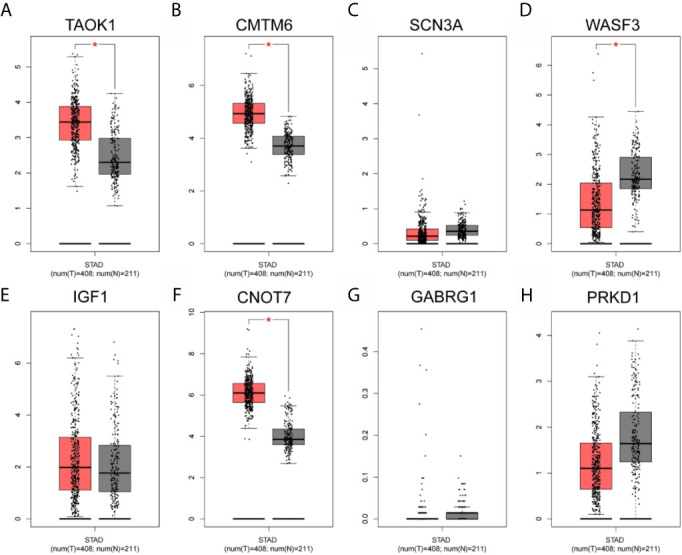
Network hub gene expression analysis. (A) TAOK1. (B) CMTM6. (C) SCN3A. (D) WASF3. (E) IGF1. (F) CNOT7. (G) GABRG1. (H) PRKD1.
Discussion
Previous bioinformatic studies have identified amounts of GC related miRNAs (3, 5–8). Some circulating serum exosomal miRNAs were proposed as novel biomarkers for GC diagnosis, such as miR-92a-3p, miR-500a-3p, miR-1246, and miR-423-5p (3, 5, 9–12). Especially, some exosomal miRNA can be even used for GC prognosis, e.g., miR-590-5p and miR-196a-1 (3, 13). In this study, we used collected blood and urine samples to analyze the EmiRs associated with GC development. The sequencing results showed four essential circulating exosomal microRNAs: hsa-miR-130b-3p, hsa-miR-151a-3p, hsa-miR-15b-3p and hsa-miR-1246. The targets of these EmiRs and enriched ontology terms should be paid more attention in GC research.
There is a study has reported that miR-130b-3p was upregulated in GC tissues, and miR-130b-3p promoted survival, metastasis and angiogenesis of GC cells as well as enhanced tumor formation and angiogenesis in GC in vivo (14). Another one has identified exo-miR-15b-3p/DYNLT1/Caspase-3/Caspase-9 axis do promotes GC development and malignant transformation. It means that serum exo-miR-15b-3p may be a potential GC diagnosis and prognosis biomarker, which can be used in precise targeted GC therapy. Studies in vitro revealed that elevated serum miR-1246 was tumor-derived by being packaged into exosomes with the help of ELAVL1 (15). And in a other area, exosomal miR-1246 expressions in serum could differentiate GC patients with TNM stage I from healthy controls and patients with benign diseases (11). Furthermore, there are no correlation between hsa-miR-130b-3p, hsa-miR-151a-3p, hsa-miR-15b-3p and hsa-miR-1246 expression and phenotypic features in the TCGA Stomach database. More clinical studies are to be performed for clarifying the definite roles of above three DEmiRNAs in GC.
A literature search of important hub genes revealed that CKLF like Marvel transmembrane domain 6 (CMTM6) was involved in gene epigenetic regulation and tumorigenesis, and the combined cmtm6 and PD-L1 testing could be used as an indicator to determine the prognosis of patients with gastric cancer (16, 17). Whereas Wiskott Aldrich syndrome protein family member 3 (WASF3) is required for tumor invasion and metastasis. Reports have indicated that WASF3 expression is associated with poor prognosis and is a potential prognostic factor for gastric cancer patients, therefore, targeting WASF3 is a novel potential therapeutic strategy for gastric cancer (18, 19). In addition to this, it has also been documented that mir-218 can inhibit the proliferation, migration and EMT of gastric cancer cells SGC7901 by targeting WASF3 (20). Protein kinase D3 (PRKD3) promotes cancer cell proliferation, growth, migration, and invasion in various tumor types. A growing body of data supports that PRKD3 is a promising therapeutic target for the treatment of cancer (21).
In the PPI network, most of the key nodes were promising candidates that have been fully surveyed, such as TAOK1, CMTM6, SCN3A, WASF3, IGF1, CNOT7, GABRG1 and PRKD1.
Still, this work has some limitations. First, the published studies about hsa-miR-130b-3p, hsa-miR-151a-3p, hsa-miR-15b-3p and hsa-miR-1246 are very few, and known conclusions implied that they are tumor suppressors in all possibility. Again, many targets of our up-regulated DEmiRNAs have been reported to be oncogenic factors or increased in GC (as mentioned above). Their conclusions sharply contradict ours, and the deep reasons need to be further explored. Second, we failed to acquire any common DE-miR in blood and urine, which may be due to that only 3 or 7 samples were included in each group. Theoretically, there may be some enriched circulating exosomal miRNAs in both blood and urine, and more samples should be accumulated in future.
Conclusions
In conclusion, this study provides an integrative analysis of expression profile of key circulating exosomal microRNAs. Four key miRNAs were found: hsa-miR-130b-3p, hsa-miR-151a-3p, hsa-miR-15b-3p and hsa-miR-1246. These risk exosomal microRNAs, as well as the corresponding interaction network or enrichments based on their targets (such as TAOK1, CMTM6, SCN3A, WASF3, IGF1, CNOT7, GABRG1, PRKD1) may provide a better understanding of the molecular mechanisms in the GC development.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .
Ethics Statement
The studies involving human participants were reviewed and approved by The Ethics Committee of Renji Hospital (Shanghai, China). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Conception and design: XQ. Administrative support: DC. Provision of study materials or patients: HW. Collection and assembly of data: FX. Data analysis and interpretation: XQ. Manuscript writing: All authors. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Key Basic Research Program of China (No.2017YFA0205304), Nature Scientific Foundation of China (No.820201080, 81602184, SKLPBS1827), and Medical Engineering Cross Project of Shanghai Jiao Tong University (YG2016ZD10, ZH2018QNA51, ZH2018QNA28). This work was also supported by “the Belt and Road” young scientist exchange program of the Science and Technology Commission of Shanghai (Grant No. 18410741600).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.693360/full#supplementary-material
References
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global Cancer Statistics, 2012. CA Cancer J Clin (2015) 65(2):87–108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2. Zhao G, Zhou A, Li X, Zhu S, Wang Y, Zhang S, et al. The Significance of Exosomal RNAs in the Development, Diagnosis, and Treatment of Gastric Cancer. Genes (Basel) (2021) 12(1):73. 10.3390/genes12010073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zheng GD, Xu ZY, Hu C, Lv H, Xie HX, Huang T, et al. Exosomal miR-590-5p in Serum as a Biomarker for the Diagnosis and Prognosis of Gastric Cancer. Front Mol Biosci (2021) 8:636566. 10.3389/fmolb.2021.636566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou J, Gong G, Tan H, Dai F, Zhu X, Chen Y, et al. Urinary microRNA-30a-5p Is a Potential Biomarker for Ovarian Serous Adenocarcinoma. Oncol Rep (2015) 33(6):2915–23. 10.3892/or.2015.3937 [DOI] [PubMed] [Google Scholar]
- 5. Abe S, Matsuzaki J, Sudo K, Oda I, Katai H, Kato K, et al. A Novel Combination of Serum microRNAs for the Detection of Early Gastric Cancer. Gastric Cancer (2021) 24(4):835–43. 10.1007/s10120-021-01161-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jiao Y, Zhang L, Li J, He Y, Zhang X, Li J. Exosomal miR-122-5p Inhibits Tumorigenicity of Gastric Cancer by Downregulating GIT1. Int J Biol Markers (2021) 36(1):36–46. 10.1177/1724600821990677 [DOI] [PubMed] [Google Scholar]
- 7. Yang R, Liu G, Han L, Qiu Y, Wang L, Wang M. MiR-365a-3p-Mediated Regulation of HELLS/GLUT1 Axis Suppresses Aerobic Glycolysis and Gastric Cancer Growth. Front Oncol (2021) 11:616390. 10.3389/fonc.2021.616390 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8. Zhao Y, Zhang H, Ju Q, Li X, Zheng Y. Comprehensive Analysis of Survival-Related lncRNAs, miRNAs, and mRNAs Forming a Competing Endogenous RNA Network in Gastric Cancer. Front Genet (2021) 12:610501. 10.3389/fgene.2021.610501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu X, Lu J, Wang S, Zhang Y, Ding Y, Shen X, et al. Circulating Serum Exosomal miR-92a-3p as a Novel Biomarker for Early Diagnosis of Gastric Cancer. Future Oncol (2021) 17(8):907–19. 10.2217/fon-2020-0792 [DOI] [PubMed] [Google Scholar]
- 10. Lin H, Zhang L, Zhang C, Liu P. Exosomal MiR-500a-3p Promotes Cisplatin Resistance and Stemness via Negatively Regulating FBXW7 in Gastric Cancer. J Cell Mol Med (2020) 24(16):8930–41. 10.1111/jcmm.15524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shi Y, Wang Z, Zhu X, Chen L, Ma Y, Wang J, et al. Exosomal miR-1246 in Serum as a Potential Biomarker for Early Diagnosis of Gastric Cancer. Int J Clin Oncol (2020) 25(1):89–99. 10.1007/s10147-019-01532-9 [DOI] [PubMed] [Google Scholar]
- 12. Yang H, Fu H, Wang B, Zhang X, Mao J, Li X, et al. Exosomal miR-423-5p Targets SUFU to Promote Cancer Growth and Metastasis and Serves as a Novel Marker for Gastric Cancer. Mol Carcinog (2018) 57(9):1223–36. 10.1002/mc.22838 [DOI] [PubMed] [Google Scholar]
- 13. Feng C, She J, Chen X, Zhang Q, Zhang X, Wang Y, et al. Exosomal miR-196a-1 Promotes Gastric Cancer Cell Invasion and Metastasis by Targeting SFRP1. Nanomed (Lond) (2019) 14(19):2579–93. 10.2217/nnm-2019-0053 [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y, Meng W, Yue P, Li X. M2 Macrophage-Derived Extracellular Vesicles Promote Gastric Cancer Progression via a microRNA-130b-3p/MLL3/GRHL2 Signaling Cascade. J Exp Clin Cancer Res (2020) 39(1):134. 10.1186/s13046-020-01626-7 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15. Wei S, Peng L, Yang J, Sang H, Jin D, Li X, et al. Exosomal Transfer of miR-15b-3p Enhances Tumorigenesis and Malignant Transformation Through the DYNLT1/Caspase-3/Caspase-9 Signaling Pathway in Gastric Cancer. J Exp Clin Cancer Res (2020) 39(1):32. 10.1186/s13046-019-1511-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang C, Zhao S, Wang X. Co-Expression of CMTM6 and PD-L1: A Novel Prognostic Indicator of Gastric Cancer. Cancer Cell Int (2021) 21(1):78. 10.1186/s12935-020-01734-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li X, Chen L, Gu C, Sun Q, Li J. CMTM6 Significantly Relates to PD-L1 and Predicts the Prognosis of Gastric Cancer Patients. PeerJ (2020) 8:e9536. 10.7717/peerj.9536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nie Y, Hu S, Liu S, Fang N, Guo F, Yang L, et al. WASF3 Expression Correlates With Poor Prognosis in Gastric Cancer Patients. Future Oncol (2019) 15(14):1605–15. 10.2217/fon-2018-0516 [DOI] [PubMed] [Google Scholar]
- 19. Nie Y, Liang X, Liu S, Guo F, Fang N, Zhou F. WASF3 Knockdown Sensitizes Gastric Cancer Cells to Oxaliplatin by Inhibiting ATG12-Mediated Autophagy. Am J Med Sci (2020) 359(5):287–95. 10.1016/j.amjms.2020.02.007 [DOI] [PubMed] [Google Scholar]
- 20. Wang G, Fu Y, Liu G, Ye Y, Zhang X. miR-218 Inhibits Proliferation, Migration, and EMT of Gastric Cancer Cells by Targeting Wasf3. Oncol Res (2017) 25(3):355–64. 10.3727/096504016X14738114257367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Y, Song H, Zhou Y, Ma X, Xu J, Yu Z, et al. The Oncogenic Role of Protein Kinase D3 in Cancer. J Cancer (2021) 12(3):735–9. 10.7150/jca.50899 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .