Abstract
The coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) poses innumerous challenges, like understanding what triggered the emergence of this new human virus, how this RNA virus is evolving or how the variability of viral genome may impact the primary structure of proteins that are targets for vaccine. We analyzed 19471 SARS-CoV-2 genomes available at the GISAID database from all over the world and 3335 genomes of other Coronoviridae family members available at GenBank, collecting SARS-CoV-2 high-quality genomes and distinct Coronoviridae family genomes. Additionally, we analyzed 199,984 spike glycoprotein sequences. Here, we identify a SARS-CoV-2 emerging cluster containing 13 closely related genomes isolated from bat and pangolin that showed evidence of recombination, which may have contributed to the emergence of SARS-CoV-2. The analyzed SARS-CoV-2 genomes presented 9632 single nucleotide variants (SNVs) corresponding to a variant density of 0.3 over the genome, and a clear geographic distribution. SNVs are unevenly distributed throughout the genome and hotspots for mutations were found for the spike gene and ORF 1ab. We describe a set of predicted spike protein epitopes whose variability is negligible. Additionally, all predicted epitopes for the structural E, M and N proteins are highly conserved. The amino acid changes present in the spike glycoprotein of variables of concern (VOCs) comprise between 3.4% and 20.7% of the predicted epitopes of this protein. These results favors the continuous efficacy of the available vaccines targeting the spike protein, and other structural proteins. Multiple epitopes vaccines should sustain vaccine efficacy since at least some of the epitopes present in variability regions of VOCs are conserved and thus recognizable by antibodies.
Keywords: COVID-19, SARS-CoV-2 genomics, Spike protein, Epitope prediction, Coronavirus comparative genomics
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) rapidly spread throughout the world after an initial burst first reported in December 2019 at Wuhan, China, presumably after a host jump from animal to human (Lai et al., 2020; Nakagawa and Miyazawa, 2020; Lu et al., 2020).
Coronaviruses are non-segmented positive-sense single-stranded RNA viruses ranging from 26 to 32 Kb in length that belong to the family Coronaviridae, which is sub-divided into four major genera: Alpha, Beta, Gamma and Delta-coronavirus (Wang et al., 2020). Human coronaviruses were initially described in the 1960s associated with the common cold. There are seven coronaviruses that infect humans: two belong to the Alphacoronavirus genus and are responsible for non-severe disease (229E and NL63); the remaining five belong to the Betacoronavirus genus, two of them also causing mild, self-limited respiratory infections (OC43 and HKU1), and three associated with potentially lethal human respiratory infectious (SARS-CoV, MERS-CoV and SARS-CoV-2) (Su et al., 2016). While 229E, OC43, NL63, and HKU1 are well adapted to humans without an animal reservoir, SARS-CoV and MERS-CoV were not well adapted to humans in terms of transmission and have likely jumped from animal (bat, civet and camel) reservoirs (Su et al., 2016). Notably, SARS-CoV-2 has efficiently adapted to humans after a probable recent zoonotic event and is highly transmissible. Close contact with infecting animals provides the opportunity for a host jump, like the two recent epidemics by coronavirus, SARS-CoV (China) and MERS-CoV (Middle East) that had bats as reservoir species and that could be transmitted to humans also from secondary hosts or bridge species like civets and camels, respectively (Su et al., 2016). Indeed, bat SARS-related coronavirus presented sequence similarity and the same cell receptor as SARS-CoV-2, the angiotensin converting enzyme 2 (ACE2) (Fehr and Perlman, 2015). The most probable scenarios for the origin of SARS-CoV-2 are those typical of a zoonosis and include natural selection in an animal reservoir host before zoonotic transfer, or natural selection in humans following zoonotic transfer, during undetected human-to-human transmission (Andersen et al., 2020). The bat and pangolin related coronavirus are the closest relative coronavirus to SARS-CoV-2 (Dos Santos Bezerra et al., 2020; Xiao et al., 2020). Namely, SARS-CoV-2 has high sequence identity with structural proteins of the recent isolated Malayan pangolin coronavirus, which led to the suggestion that pangolins may had been an intermediate host of SARS-CoV-2 (Xiao et al., 2020; Lam et al., 2020).
Importantly, there is a panoply of coronavirus able to infect a large variety of animals, including for instance livestock, exotic and companion animals and wildlife, allowing for the opportunity for genetic recombination resulting in novel viruses (Su et al., 2016). Additionally, the high mutation rate of RNA viruses, yielding offsprings that differ by 1–2 mutations from their parents (Vignuzzi and Andino, 2012), is correlated with enhanced virulence (Duffy, 2018) and favors zoonotic events and epidemic spread, making RNA viruses such as Coronaviruses the most common found in new causes of human disease (Rosenberg, 2015), like COVID-19. Accordingly, closely related coronavirus circulating in the wet animal markets or other places of close contact with humans may allow the cross-species spillover (Fehr and Perlman, 2015). The high mutation rate also provides a means of escaping vaccine-induced immunity and treatment resistance (Duffy, 2018). Despite coronavirus encoding a proofreading exoribonuclease in the NSP14 gene that mediates high-fidelity RNA genome replication (Graepel et al., 2017), the impact of the proof-reading in genome variability is not completely established. Thus, it is important to analyze the level of mutations in a large collection of genomes and evaluate their impact for the development of vaccine or diagnosis methods based on the detection of antibodies. For both, the spike gene is the major target, since the spike glycoprotein (S) is responsible for viral attachment and fusion with the host cell. The S glycoprotein contains a receptor-binding domain (RBD) that specifically binds to ACE2 receptors, starting cell entry. Next, the cleavage of the S glycoprotein by cellular proteases leads to fusion and endocytosis (Pillay, 2020).
To combat the epidemic with a vaccine or with a drug it is vital to understand the genetic variability of SARS-CoV-2. Thus, the aim of the present work is to understand the probable origin of SARS-CoV-2 through sequence comparison with other coronavirus sequences available in public databases; and to contribute to the understanding of the variability of SARS-CoV-2 genomes and its impact on vaccines and diagnostic tests efficacy by analyzing nearly 20,000 SARS-CoV-2 high quality genomes and 200000 spike protein sequences available at GISAID database. The use of a large dataset allowed us to confirm that bats appear to be the main reservoir of diversity of SARS-like coronaviruses, and that SARS-CoV-2 genomes clusters according to geography, presenting hotspots for recombination and mutation accumulation, whose impact in proteins that are used in vaccines is for now negligible.
2. Material and methods
2.1. Coronavirus genome sequences
All high coverage complete sequenced SARS-CoV-2, i.e. genomes sequences with < 1% Ns and < 0.05% unique amino acid mutations, deposited at GISAID were retrieved on May 2020 for analysis comprehending 19471 worldwide genome sequences. The SARS-CoV-2 NC_045512.2 (corresponding to reference EPI_ISL_402125 at GISAID database) was used as reference genome.
Genomes of Coronoviridae family available at NCBI were retrieved, totaling 3335 genomes (collected in June 2020). When available the natural host species was collected using an in-house Python script. Other human coronavirus genomes, including SARS-CoV (58 genomes), MERS-CoV (599 genomes), 229E (43 genomes), NL63 (82 genomes), HKU1 (48 genomes) and OC43 (178 genomes), were retrieved from NCBI, totaling 1013 genome sequences.
2.2. Phylogenetic analysis and allele diversity of SARS-Cov-2
The 19471 SARS-CoV-2 genomes were aligned with the reference genome using MAFFT version 7 (Katoh and Standley, 2013) default options. Maximum-likelihood phylogenetic trees from alignments of nucleotide were produced using fasttreeMP 2.1.11 (Price et al., 2010). To visualize and annotate produced trees the Interactive Tree Of Life (iTOL) v4 (Letunic and Bork, 2019) was used. For better readability of the phylogenetic tree and to reduce their complexity by eliminating leaves that contribute the least to the tree diversity a smaller dataset with a more even representation of the different phylogenetic groups was obtain after pruning the tree with Treemmer v0.2 (Menardo et al., 2018), using the options -mc 100, to protect from pruning 100 genomes from each continent, keeping 1000 representative leafs. A similar tree pruning with the option -mc 10 to protect 10 genomes from each continent was used to select 100 representative genomes from the large phylogenetic tree. These genomes were used for a comparative genomic analysis with other coronaviruses.
SNVs were extracted from multiple alignments using SNP-sites (Page et al., 2016) producing a vcf file which was processed by the vcftools suite (Danecek et al., 2011) to determine the allele frequency from SARS-CoV-2 genomes. A plot of variant density was produced to show how many SNVs there are and how they are distributed along the genome using a Python script (available at http://alimanfoo.github.io/2016/06/10/scikit-allel-tour.html).
2.3. Comparative genomics and genomic diversity among Coronoviridae and human coronavirus
The 100 SARS-CoV-2 representative genomes and 3335 genomes from Coronoviridae family members were aligned using MAFFT version 7 (Katoh and Standley, 2013). A phylogenetic tree was produced from the nucleotide alignments using fasttreeMP 2.1.11 (Price et al., 2010) and was visualized with iTOL v4 (Letunic and Bork, 2019), as described above. A phylogenetic network was also build using the Neighbor Net algorithm (Bryant and Moulton, 2004) implemented in the software SplitsTree 4.10 (Huson and Bryant, 2006), which is a powerful tool for visualization conflicting and consistent information present in a dataset. The filter taxa option was applied to show only the reference genome of SARS-CoV-2, a genome of each human coronavirus, as well as coronavirus infecting other species clustering with the SARS-CoV-2 reference genome, to evaluate origin and potential relationships between them.
Each group of genomes of human coronavirus retrieved from NCBI (229E, HKU1, MERS-CoV, NL63, OC43, SARS-CoV) was aligned using MAFFT version 7 (Katoh and Standley, 2013), a tree was produced and pruned so that 30 representative genomes of each group could be selected. The 100 representative genomes of SARS-CoV-2 and 30 genomes of each group of human coronavirus were aligned and a network was produce using SplitsTree4 (Huson and Bryant, 2006), since networks may generate more effective presentations of intraspecific evolution. Indeed, a phylogenetic networks allows to observe reticulate events like hybridization, horizontal gene transfer, recombination, or gene duplication and loss (Bryant and Moulton, 2004; Huson and Bryant, 2006).
Additionally, the phylogenetic tree of 100 SARS-Cov-2 plus 3335 genomes from Coronoviridae family allowed to retrieve the group B coronavirus genomes that cluster with SARS-CoV-2. This group of 15 genomes plus the 100 SARS-CoV-2 representative genomes are hereinafter referred to as SARS-Cov-2 emerging cluster. The SARS-CoV-2 emerging cluster was aligned using MAFFT version 7 (Katoh and Standley, 2013), after which SNP-sites (Page et al., 2016) and vcftools suite (Danecek et al., 2011) was used as described above. The SARS-CoV-2 emerging cluster is formed by two subgroups, the SARS-CoV-2 genomes and the Betacoronavirus genus genomes that cluster with SARS-CoV-2. The genetic diversity between each of the groups was done using the PopGenome package (Pfeifer et al., 2014) in R, namely determine FST (fixation index), which tests whether there is genetic structure in the population and quantifies the proportion of genetic variation that lies between subpopulations within the total population; nucleotide diversity to measure the degree of polymorphism in the two groups; and Tajima's D statistics to detect departures from neutrality. Additionally, a principal component analysis (PCA) was done using the R package adegenet (Jombart and Ahmed, 2011). Moreover, a similarity plot and a bootscan plot was build using Simplot v3.5.1 - program (Lole et al., 1999) using a window of 500 nucleotides, which was moved along the SARS-CoV-2 reference genome in steps of 50 nucleotides. This analysis allowed to evaluate possible recombination events in the SARS-CoV-2 emerging cluster and the similarity of non-SARS-CoV-2 genomes to SARS-CoV-2 genome. The sliding window partitions along the alignment of the SARS-CoV-2 emerging cluster method involves the construction of bootstrapped neighbor joining trees. Recombination is detected when a SARS-CoV-2 genome jumps between different clusters in trees constructed from adjacent alignment partitions.
2.4. Tracing epitope conservation of SARS-CoV-2 spike protein (S glycoprotein) and other structural proteins
B-cell epitope prediction of the S glycoprotein (Accession number: YP_009724390.1) was done using BepiPred-2.0 (Jespersen et al., 2017) using default settings. For the 19471 SARS-CoV-2 genome sequences worldwide, the spike gene was extracted and translated using in-house Python scripts. The S glycoprotein sequences were aligned using MAFFT version 7 (Katoh and Standley, 2013) and the positions of the identified epitopes with BepiPred-2.0 (Jespersen et al., 2017) with > 5 amino acid residues in length were extracted with an in-house Python script. Next, a sequence logo graphical representation (Schneider and Stephens, 1990) of the amino acid residues multiple sequence alignment was created with WebLogo 3 (Crooks et al., 2004). A similar analysis was done for the other structural proteins of SARS-CoV-2, i.e., E (envelop protein, accession number: YP_009724392.1), M (membrane glycoprotein, accession number: YP_009724393.1) and N (nucleocapsid phosphoprotein, accession number: YP_009724397.2) proteins. Additionally, all the near 200000 S glycoprotein sequences available at GISAID were collected in November 2020 and the analysis was repeated to check if the conservation of amino acids hold. Thus, for 199984 S glycoprotein sequences (all greater than 1250 amino acids and from SARS-CoV-2 isolates from human hosts) the percent of conservation worldwide and by continent was determined for each of the predicted epitopes. For sequence logo determination a multiple alignment is needed, but performing an alignment of almost 200 thousand sequences can require huge computer power. Thus, we have previously select S glycoprotein unique sequences, than conduct the alignment using MAFFT version 7 (Katoh and Standley, 2013) and then determining the sequence logos with WebLogo 3 (Crooks et al., 2004). Using PyMOL Molecular Graphics System, Version 2.0 (Schrodinger LLC, 2015) it was verified if the predicted epitopes are displayed at the surface of the proteins, whenever the 3D structure of the proteins was available in the protein data bank (PDB). The 3D structures with the accession numbers 6vxx, 5 × 29 and 6VYO for S glycoprotein, E protein and N phosphoprotein were used, respectively.
3. Results
3.1. SARS-CoV-2 genome variability
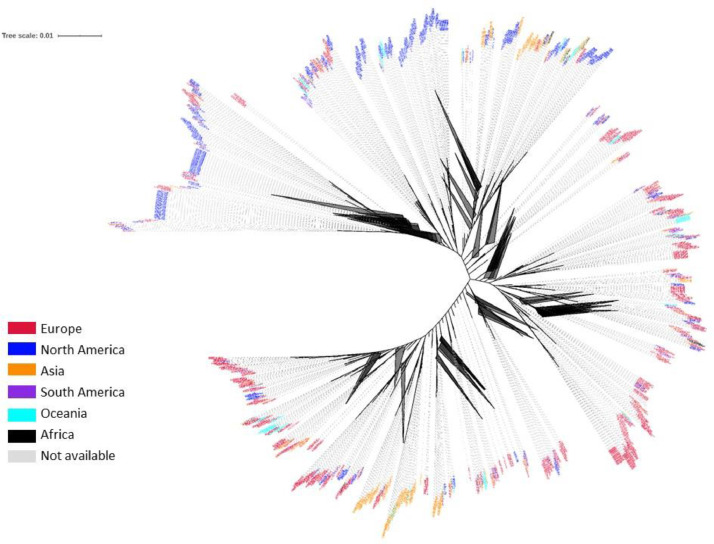
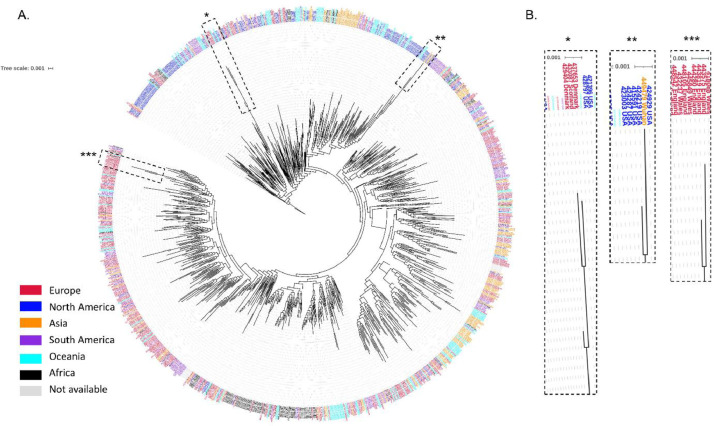
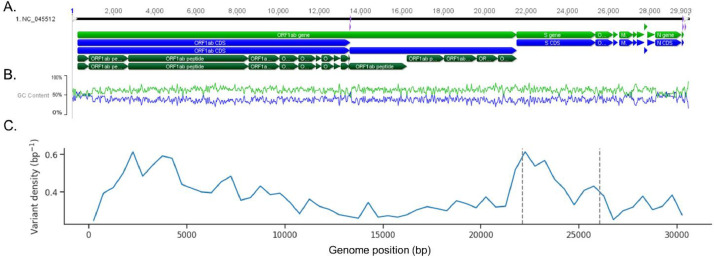
The Maximum-likelihood phylogenetic tree (Fig. 1 ) of 19471 SARS-CoV-2 genomes created with fasttreeMP 2.1.11 (Price et al., 2010), demonstrates the presence of clades associated with the geographic area of isolation. American and European isolates make up the majority of these genomes. Highly similar genomes are shown collapsed in the phylogenetic tree (Fig. 1), most probably representing isolates within the same transmission chain. Tree pruning helps visualization of these clusters after removing similar genomes that correspond to proximal tree nodes. Thus, the phylogenetic tree was pruned using Treemmer v0.2 (Menardo et al., 2018) to increase its readability (Fig. 2 ). Focusing on 1000 genomes in the pruned phylogenetic tree that represent worldwide diversity shows that each region contains multiple clades although for each world region there are dominant spreading clades. The existence of clades reveals a high genome variability, which is typical of RNA viruses, as evidenced by 9632 SNVs among the 19471 SARS-CoV-2 genomes. The variant density is around 0.3 over the genome, that is the raw data contains a SNV approximately every 3 bases of the genome, unevenly distributed along the genome (Fig. 3 and Table S1). Indeed, the variant density is higher for the first two mature peptides of orf1ab/orfa coding for the leader protein and nsp2, and from open reading frame 6 to 10. Importantly, although generally the spike glycoprotein presents a variant density of 0.31, some of their conserved domains present a higher variant density, towards the N-terminal domain of the coronavirus spike glycoprotein that functions as a receptor binding domain (Table S1).
Fig. 1.
Maximum-likelihood phylogenetic tree of 19471 SARS-CoV-2 genomes, created with fasttreeMP 2.1.11 and visualized with iTOL v4. Black triangles represent collapsed nodes of highly similar nodes (genomes). Each genome is colored coded by continent of origin.
Fig. 2.
(A) Trimmed phylogenetic tree of 1000 SARS-CoV-2 genomes representing the worldwide diversity, created with fasttreeMP 2.1.11 and visualized with iTOL v4. Each genome is colored coded by continent of origin. (B) Magnified detail view of topology of the phylogenetic tree evidencing strains in long branches.
Fig. 3.
SNVs positions across SARS-CoV-2 genome. (A) Genome map of SARS-CoV-2. (B). GC content across the SARS-CoV-2 genome. Blue - GC % content; Green – AT % content. (C) Plot exhibiting SNVs distribution along the genome using a window size 500 bp; dashed grey bars indicate S gene position. (Figure caption using Geneious 8).
3.2. Comparative genomics of SARS-CoV-2 and other Coronaviridae
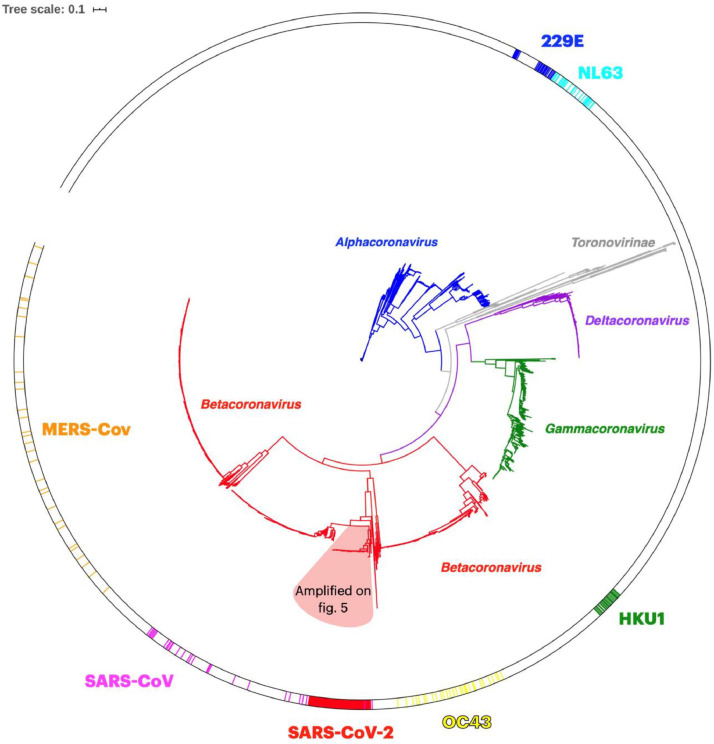
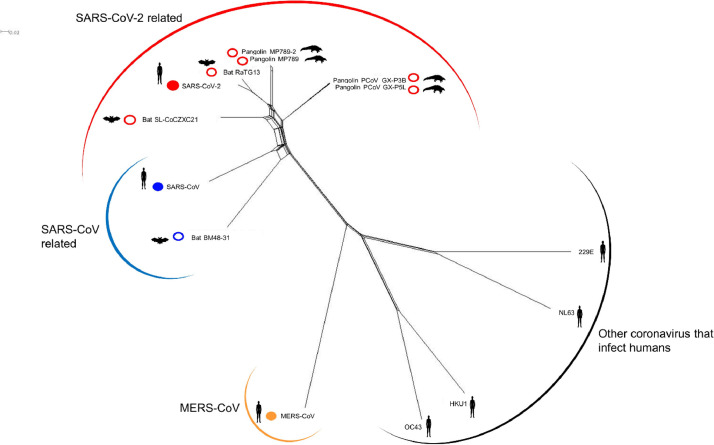
Tree pruning was also used to select 100 worldwide representative genomes from the large phylogenetic tree. Then, a comparison of a 100 representative SARS-CoV-2 genomes with 3335 genomes from other Coronoviridae family members showed, as expected, that the genomes cluster according to the coronavirus genera: Alpha, Beta, Delta and Gammacoronavirus and the more distant Toronovirinae subfamily (Fig. 4 ). Coronavirus capable of infecting humans belong to distinct groups and those associated with milder disease outcomes are in Alphacoronavirus (229E-CoV and NL63-CoV), and Betacoronavirus (HKU1-CoV and OC43-CoV) genera. Betacoronavirus genus harbors all human coronavirus that have been provoking serious epidemic episodes (SARS-CoV, MERS-CoV and SARS-CoV-2) (Fig. 4). A closer inspection of genomes clustering together with 100 representative SARS-CoV-2 genomes reveals 13 betacoronavirus genomes whose host is the bat or the pangolin (the remaining 2 genomes to complete the group of 115 correspond to SARS-CoV-2 genomes retrieved from GenBank) (This detail of Fig. 4 is zoom-in in Fig. 5 ). The phylogenetic network analysis (Fig. 6 ) presented using the filtering option of SplitsTree (Huson and Bryant, 2006) to show only the SARS-CoV-2 reference genome and the 6 genomes of the bat and pangolin coronavirus that clusters together (the remaining 5 of the group of 13 coronavirus (Table 1 ) that cluster with the 100 representative SARS-CoV-2 genomes are similar to the ones showed above and for better readability are not presented) shows short inner branches and long terminal branch lengths leading to the tips, i.e. showing deep divergence between strain lineages. A similar observation occurs for the SARS-CoV genome from the 2003 epidemics and a close related bat genome (the bat coronavirus BM48-31) (Fig. 6). The inner reticulation branching pattern observable is indicative of extensive recombination (Lassalle et al., 2020). However, while distinct human coronavirus are in different clusters, genomes of each group of human coronavirus cluster together (Fig. S1).
Fig. 4.
Maximum-likelihood phylogenetic tree of 3435 genomes of Coronaviridae family, created with fasttreeMP 2.1.11 and visualized with iTOL v4. Blue clade – Alphacoronavirus; Grey clade - Toronovirinae subfamily; Violet clade – Deltacoronavirus; Green clade – Gammacoronavirus; Red clade – Betacoronavirus (includes 100 genomes of SARS-CoV-2 representing worldwide variability). Reddish cone represents the SARS-CoV-2 emerging cluster which is detailed in Fig. 5. The circular strip highlights coronavirus capable of infecting humans, colored clockwise as: Blue – 229E; Cyan – NL63; Green – HKU1; Yellow – OC43; Red – SARS-CoV-2 (detailed in Fig. 5); Magenta – SARS-CoV; Orange – MERS-CoV.
Fig. 5.
Magnified detail view of topology of the phylogenetic tree cluster from which SARS-CoV-2 emerged (corresponding to the reddish cone in Fig. 4), evidencing all non-human SARS-CoV-2 emerging cluster and part of the human-SARS-CoV-2 emerging cluster. For SARS-CoV-2 genomes belonging to the human-SARS-CoV-2 emerging cluster just a few ones are showed to better readability (genome codes from GISAID identify the genomes). For other species the isolate name is presented. The host of the coronavirus is represented by human, bat and pangolin cartoons.
Fig. 6.
Filter of the SplitsTree network of the coronavirus family evidencing the reference SARS-CoV-2 genome (filled red circle) and other coronavirus belonging to the same cluster (unfilled red circle). SARS-CoV genome (filled blue circle) and a closer bat genome (unfilled blue circle) is showed. MERS-CoV genome is showed (filled orange circle) as well as one genome of each coronavirus group that infects humans. The host of the coronavirus is represented by human, bat and pangolin cartoons.
Table 1.
Non-human-SARS-CoV-2 emerging cluster data.
| Strain | Host | Isolation country | Collection date | Publication date | % similarity SARS-CoV-2 | Accession number | Reference |
| RaTG13 | Rhinolophus affinis | China | 2013 | 2020 | 96.114 | MN996532.1 | (Zhou et al., 2020) |
| bat-SL-CoVZXC21 | Rhinolophus sinicus bat | China | 2015 | 2018 | 87.410 | MG772934.1 | (Hu et al., 2018) |
| bat-SL-CoVZC45 | Rhinolophus sinicus bat | China | 2015 | 2018 | 87.640 | MG772933.1 | (Hu et al., 2018) |
| MP789-2 | SARS-CoV pangolin | China | 2019 | 2020 | 89.926 | MT121216.1 | (Liu et al., 2020) |
| MP789 | Manis javanica | China | 2019 | 2020 | 78.523 | MT084071.1 | (Liu et al., 2020) |
| PCoV_GX-P4L | Manis javanica Malayan pangolin | China | 2017 | 2020 | 85.235 | MT040333.1 | (Lam et al., 2020) |
| PCoV_GX-P3B | pangolin | China | 2017 | 2020 | 80.234 | MT072865.1 | (Lam et al., 2020) |
| PCoV_GX-P5L | Manis javanica Malayan pangolin | China | 2017 | 2020 | 85.245 | MT040335.1 | (Lam et al., 2020) |
| PCoV_GX-P1E | Manis javanica Malayan pangolin | China | 2017 | 2020 | 85.211 | MT040334.1 | (Lam et al., 2020) |
| PCoV_GX-P5E | Manis javanica Malayan pangolin | China | 2017 | 2020 | 85.208 | MT040336.1 | (Lam et al., 2020) |
| PCoV_GX-P2V | pangolin | China | 2017 | 2020 | 85.211 | MT072864.1 | (Lam et al., 2020) |
| BtKY72 | Rhinolophus sp. bat | Kenya | 2007 | 2019 | 74.654 | KY352407.1 | (Tao and Tong, 2019) |
| BM48-31/BGR/2008 | Rhinolophus blasii bat | Bulgaria | 2008 | 2010 | 74.638 | NC_014470.1 | (Drexler et al., 2010) |
Note: This table just presents the subgroup of non-human-SARS-CoV-2 emerging cluster. The SARS-CoV-2 emerging cluster is composed of 115 genomes, of which 13 genomes belong to the non-human-SARS-CoV-2 emerging cluster and the remaining to the human-SARS-CoV-2 emerging cluster.
The SARS-CoV-2 emerging cluster is composed by 115 genomes that were separated in two subgroups, the non-SARS-CoV-2 genomes (13 genomes) and SARS-CoV-2 (102 genomes). These two subgroups are hereinafter referred to as non-human-SARS-CoV-2 emerging cluster and human-SARS-CoV-2 emerging cluster, respectively. The R PopGenome package (Pfeifer et al., 2014) allowed to determine several statistics from multiple sequence alignments and single-nucleotide variant (SNV) data of the SARS-CoV-2 emerging cluster. The Tajima-D statistics (a measure of the mutation frequency spectrum) in the sample for these subgroups was -2.894 and 0.070, respectively. The negative value of this statistics reflects recent population expansion after a recent bottleneck, which is in agreement with the recent emerging of SARS-CoV-2 and rapid pandemic expansion. On the other hand, the roughly zero value points to a population with no evidence of selection. The nucleotide diversity was 3.810 and 1201.538, respectively, revealing that the degree of polymorphism in the non-human-SARS-CoV-2 emerging cluster subgroup was > 300 superior than in the human-SARS-CoV-2 emerging cluster subgroup. FST values may vary from 0 (not different) to 1 (completely different with every SNV fixed in each population). Thus, higher FST values are consistent with a considerable degree of differentiation among populations. The observed FST value of 0.475 points to a differentiation among the two subgroups.
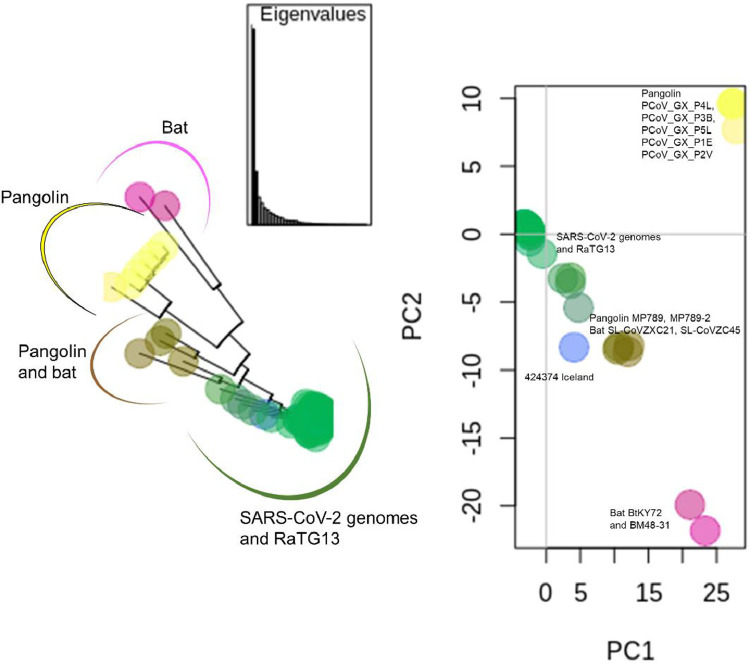
A principal component analysis (PCA), a technique for reducing the dimensionality of large datasets, was carry out in this group of 115 genomes done using the R package adegenet (Jombart and Ahmed, 2011). The first two principal components (Fig. 7 ) explained 47.15% and 12.60% of the total variance in the dataset. The PCA analysis was able to divide the 115 genomes of the SARS-CoV-2 emerging cluster into three groups, namely SARS-CoV-2, a group of bat coronavirus genomes and a mixed group of bat and pangolin coronavirus genomes (Fig. 7). In addition, the PCA confirms that the closest related genome to the SARS-CoV-2 genomes corresponds to the bat coronavirus RaTG13 (Table 1). This distribution is also observable in the phylogenetic tree (Figs. 5 and 7). The similarity plot performed with SimPlot (Lole et al., 1999) along the reference genome of SARS-CoV-2 shows how the genomes from the non-human-SARS-CoV-2 emerging cluster are related with the reference SARS-CoV-2 genome (Figure S2.A). The recombination analysis, also performed with SimPlot (Lole et al., 1999), detected evidence of possible recombination (Figure S2.B), mainly around positions ∼2350 to ∼2400 (region of orf 1ab) and ∼25400 to ∼25500 (region of spike gene) between the bat coronavirus RaTG13 and the group bat-SL-CoVZXC21-bat-SL-CoVZC45 and between the bat coronavirus RaTG13 and the pangolin coronavirus MP789, respectably (consult Table 1 for details).
Fig. 7.
Phylogenetic tree and principal component analysis (PCA) of 115 genomes of the SARS-CoV-2 emerging cluster, done with R package adegenet. Phylogenetic tree exhibiting the same colors as in the scatter plot of the first two principal components. PC1 explains 47.15% of the variance and PC2 12.60% % of the variance. Green dots – SARS-CoV-2 and the bat coronavirus RaTG13; Blue dot – SARS-CoV-2 genome (424374 Iceland); pink dots – bats Rhinolophus sp. (BtKY72) and Rhinolophus blasii (BM48-31); yellow dots – several pangolin Manis javanica; brown dots – pangolin (MP789-2 and MP789) and bat (Rhinolophus sinicus bat-SL-CoVZC45and bat-SL-CoVZXC21) genomes .
3.3. Glycoprotein S–glycoprotein epitope conservation
This study focused on predicted antibody-epitope interactions of the spike glycoprotein. B-cells play an important role in the adaptive immune system due to the production of antibodies that recognize target antigens by binding to a specific epitope in the antigen. Vaccines rely on the humoral immune response and attenuated or subunit vaccines that mimic the presentation of antigens to stimulate antibody production (Jespersen et al., 2017). The SARS-CoV-2 S glycoprotein (a spike protein) is a good target for vaccine development: first, because of the role of this structural protein in viral attachment, fusion, and entry into the host cell (Samrat et al., 2020); secondly, because the generation of neutralizing antibodies to the spike protein should certainly block virus entry. Using the S glycoprotein sequence of the SARS-CoV-2 genome, 29 epitopes with more than 5 amino acids were predicted, ranging from 6 to 26 amino acid residues (Figure S3, Table S2 Movies S1 and S2) using BepiPred-2.0, a sequence-based epitope prediction tool based on based on a random forest algorithm trained on epitope data from crystal structures, improving the algorithm predictive power (Jespersen et al., 2017). Conserved epitopes are likely to provide broader protection across multiple virus strains, than those derived from highly variable genome regions. The degree of the predicted epitopes conservation was evaluated. The impact in epitope sequence conservation was residual. In fact, the 29 predicted epitopes appear to be conserved across the 19471 SARS-CoV-2 genomes, as observed by the epitopes’ sequence logos (Table S2). In a logo the height of the stack represents the sequence conservation at each position and the height of the amino acid symbol within the stack represents its relative frequency at that position (Crooks et al., 2004). The conservation of S glycoprotein predicted epitopes is high considering that the height of the stack is close to maximal for the majority of the residues and each stack has a clear predominant amino acid residue at every position in the epitope. Since the pdb 3D structure of the S glycoprotein is available (pdb: 6vxx), we have checked which of the predicted epitopes have their amino acids exposed in the 3D structure, verifying that about half are presumably entirely exposed (Table S2), favoring their application for vaccine development. Additionally, we have determined the percentage of conservation of the predicted epitopes among the 19471 SARS-CoV-2 S glycoprotein sequences. In agreement with the sequence logos, the predicted epitopes sequences were conserved between 92.6% and 95.6% of the S glycoprotein sequences (Table S2). In an unprecedented sequencing effort, new SARS-CoV-2 genomes are deposit at a daily basis at GISAID database. Thus, to verify the conservation of the S glycoprotein predicted epitopes, we have repeated the analysis for 199984 spike glycoprotein sequences (Table S3). We found that the predicted epitopes maintained their high percentage of conservation worldwide (the predicted epitope sequence is found with 100% identity in a high percentage of analyzed sequences), varying from 85.7% to 99.8%. The percentage of conservation determined by continent showed that the predicted epitopes are conserved across continents, with a punctual exception for one of the predicted epitopes in Oceania (Table S3). Of the 199984 S glycoprotein sequences, 31323 are unique sequences. This conservation is also observable in the sequence logos obtain from the multiple alignment of the 31323 S glycoprotein unique sequences, where the sequence logo is almost always represented by a single amino acid (Table S2). Besides well conserved as observed by the sequences logos, in the 31323 S glycoprotein sequences the proportion of each unique sequence is not identical for all sequences. The most prevalent S glycoprotein sequence is present in 37% (73997/199984) of the total S glycoprotein sequences, and the second and third most frequent sequences in 6.2% and 4.1%, respectively.
The current variants of concern (VOCs) established by WHO and/or CDC named alpha, beta, gamma, delta and epsilon, which may be associated with increased transmissibility or virulence, and decreased effectiveness of public health measures, present a defined set of amino acid changes in the S glycoprotein, mainly amino acid substitutions, but also few deletions. Whenever these amino acids that are changed or deleted in each VOC are present in the predicted epitopes they were highlighted (Table S2). The majority of the predicted epitopes (56.6%, 17/29) is not affected in any amino acid that is altered in VOCs. Additionally, for each VOC the amino acid substitutions or deletions are very disperse along the S glycoprotein, affecting few of the predicted epitopes. For VOC alpha, 20.7% (6/29) of the predicted epitopes have at least one amino acid change; VOC beta has 20.7% (6/29); VOC gamma has 17.2% (5/29); VOC delta has 13.8% (4/29) and VOC epsilon has 3.4% (1/29).
3.4. Other structural proteins (E, M and N) epitope conservation
For the other three structural proteins of SARS-CoV-2, the E, M and N proteins the number of predicted epitopes using BepiPred-2.0 (Jespersen et al., 2017) with more than 5 amino acids was 1 (length of 15 amino acid residues, not predicted to be entirely exposed on the 3D structure of E protein), 4 (ranging from 6 to 21 amino acid residues) and 9 (ranging from 6 to 59 amino acid residues, half of each exposed at the 3D M protein surface), respectively (Tables S4, S5 and S6). In general, the predicted epitopes were found to have their sequences conserved in about 99% of the sequence for E, M and N proteins (Tables S4, S5 and S6). One of the predicted epitopes from the N phosphoprotein was found to be conserved only in about 70% of the sequences (Table S6).
4. Discussion
We applied phylogenetic and sequence analyses to address these pressing issues. The phylogenetic clustering is a powerful technique to understand how SARS-CoV-2 genomes are related to each other and to other coronavirus that infect humans or animals, while sequence comparisons can identify which epitopes are stable versus those that are hotspots for mutation and are thus unsuitable as vaccine or diagnostic targets.
Applying phylogenetic analysis to 100 representative SARS-CoV-2 genomes plus 3335 genomes of other members of the Coronoviridae family demonstrated that they clustered into the 4 known Coronoviridae genera and the more distal Torovirinae genus. Focusing on the SARS-CoV-2 genomes confirmed their presence in the Betacoronavirus genus (Fig. 4). Moreover, it identified a SARS-CoV-2 emerging cluster containing the 100 representative SARS-CoV-2 genomes and 13 genomes from bat and pangolin hosts (Figs. 5 and 6). These findings suggest a likely link to viruses infecting these animal hosts.
The current knowledge on viral biodiversity is biased due to the limited number of closely related genomes available in public databases. The true betacoronavirus diversity is certainly far from being completely described, as databases represent mainly samples from human virus outbreaks (Kitchen et al., 2011), rather than non-human sources. This imposes a huge constraint and limitation in deciphering the origin of SARS-CoV-2. Continued sampling in areas where humans are in close contact with bats and pangolins may lead to the identification of closer relatives of SARS-CoV-2 (Zhang and Holmes, 2020). Nonetheless, SARS-CoV-2 presents an average whole genome similarity of 96.1% with the bat virus RaTG13 strain isolated in China from Rhinolophus affinis (Table 1), making this genome the closest relative to SARS-CoV-2 so far (Zhou et al., 2020). This observation is in agreement with bats being a significant reservoir for coronavirus from which spillovers infecting other species appear to routinely emerge (Fehr and Perlman, 2015). In general, the non-human-SARS-CoV-2 emerging cluster presents less similarity to the reference SARS-CoV-2 genome in the regions coding for the ORF1ab polyprotein and the spike glycoprotein (Figure S2), which are precisely the regions of greatest variability among the SARS-CoV-2 genomes (Fig. 3), as discussed below. Genetic recombination within positive-strand RNA viruses is an important evolutionary mechanism increasing viral diversity through the formation of novel chimeric genomes (Bentley and Evans, 2018). The present work showed evidence of recombination among the SARS-CoV-2 emerging cluster, which may have contributed to more efficient transmission and wider host range (Figs. 2 and S2). Importantly, one of the regions where recombination was detected is precisely the spike gene, coding the S glycoprotein responsible for initial attachment of the virus to the host cell (Zhang and Holmes, 2020). The existence of an intermediate host, namely the pangolin, has been suggested (Dos Santos Bezerra et al., 2020; Lam et al., 2020; Liu et al., 2020). This theory is supported by the observation that the E, M, N and S proteins of coronavirus isolated from pangolins showed > 90% amino acid identity and infected pangolins presented antibodies that reacted with the spike glycoprotein of SARS-CoV-2 (Xiao et al., 2020). The possible recombination detected between bat and pangolin coronavirus in the region of the spike glycoprotein, more specifically between bat genome RaTG13 isolated from Rhinolophus affinis and the pangolin genome MP789 isolated from Manis javanica, contribute to the theory that the pangolin was an intermediate host (Table 1). The PCA analysis confirmed the phylogenetic analysis of the SARS-CoV-2 emerging cluster, pointing to a genomic divergence from other betacoronaviruses. The related bat genome RaTG13 isolate in China from Rhinolophus affinis (Table 1) cluster together with SARS-CoV-2 genomes constituting a tight cluster, except for only one genome (from an Iceland SARS-CoV-2 isolate – GISAID EPI_ISL_424374). The Tajima D statistics may be computed either from within-species or among-species polymorphisms to test for neutrality (Bhatt et al., 2010). The observed Tajima's D values < 0 for SARS-CoV-2 is consistent with population expansion after a bottleneck, which is in agreement with others (Fang et al., 2020; Laskar and Ali, 2020). In opposition, the inter-species Tajima D near zero is compatible with absence of selection and neutral evolution. However, the nucleotide diversity among-species of the SARS-CoV-2 emerging cluster is an order of magnitude higher (>300) than that observed within the species, which in combination with the recombination potential makes the SARS-CoV-2 emerging cluster a pool for potential emergence of novel coronavirus strains capable of infecting new hosts, like the SARS-CoV-2.
When compared with other coronaviruses, SARS-CoV-2 forms a tight cluster. However, this does not mean that SARS-CoV-2 genomes are free of variation. On the contrary, when analyzing the world variability of nearly 20000 genomes a geographic distribution is clear, pointing to the ways of spread of the pandemic virus in each country and aggregating countries by continent. Interestingly, there are dominant virus spreading in each region (collapsed nodes of highly similar genomes, Fig. 1). The current analysis is in agreement with others showing that the virus is evolving and that strains from different continents exhibit different mutation patterns (Pachetti et al., 2020; Forster et al., 2020). The genomes collected from the GISAID database included isolates from the mint and tiger non-human hosts, and they cluster together with SARS-CoV-2, which points to a transmission from human to animal, demonstrating that SARS-CoV-2 has a host range larger than humans.
In the analyzed SARS-CoV-2 genomes one third of the genome has mutated in at least one of the analyzed genomes, totaling 9632 SNVs. However, these SNVs are not equally distributed along the SARS-CoV-2 genome and accumulate in hot spots for mutations, i.e., accumulating in the spike gene and ORF 1ab (Fig. 3). These regions are precisely the ones showing detectable recombination (Figure S2) and where SARS-CoV-2 exhibits in general less similarity with the non-human-SARS-CoV-2 emerging cluster, suggesting that these genome regions are hypervariable. Most vaccines target the spike glycoprotein (Funk et al., 2020), because of the essential role of the S protein in virus binding and uptake into the host cell allowing the replicative infection cycle to start. Certainly, the role of the spike protein in binding with host receptors makes it a perfect target for vaccine and antiviral therapeutic development (Samrat et al., 2020). The finding that the spike gene is a hotspot of variability in the SARS-CoV-2 genome might pose a problem for vaccine effectiveness as well as diagnostics and therapeutic targeting. However, a careful analysis of the impact of this variability in a set of predicted spike glycoprotein epitopes showed that presently this variability is negligible, which is a good predictor for the continuous success of a vaccine targeting the spike glycoprotein. Therefore, although the increased variability found for the spike gene (Fig. 3C), this is not reflected in the amino-acids residues of the epitopes found (Table S2), which is in agreement with others (Dearlove et al., 2020). Additionally, the amino acids alterations of VOCs over the predicted epitopes are limited, reinforcing the effectiveness of vaccines targeting the S glycoprotein against current VOCs. The high degree of epitope conservation found in a large group of SARS-CoV-2 genomes confirms that this glycoprotein is a good target for vaccine development, especially if they rely on multiple epitope presentation. The conservation found for the epitopes may be related to the fact that most of the variants of the ACE2 human receptor are rare (Fujikura and Uesaka, 2021). Even that a certain S protein presents for some epitope a sequence that differs from the epitope consensus sequence (Table S2), multiple epitope vaccines continue to stimulate the production of antibodies that still are capable of recognizing if not all at least some of the epitopes. Nonetheless, due to the fact that this is a hypervariable region a constant monitoring of the evolution of the sequence and its impact on epitope stability is mandatory. Accordingly, the predicted epitopes conservation analysis of the S glycoprotein for the 199984 sequences showed that they are worldwide conserved, keeping this conservation across each continent (Table S3) and over a time interval (Table S2). Even though most vaccines target the S protein, other structural proteins have been proposed as vaccine targets, for being associated with viral envelope: M and E; or for being highly immunogenic and abundantly expressed during infection by coronaviruses: N protein (Funk et al., 2020; Dutta et al., 2020; Florindo et al., 2020). The S glycoprotein plays a crucial role in both viral replication and neutralization potential. The E protein has been associated with the pathogenesis of the cytokine storm observed in some patients with severe COVID-19 (Schoeman and Fielding, 2020) and M protein has a major role in virion self-assembly. Furthermore, if these proteins are immunogenic and target for host antibodies, binding of these antibodies could block virus-cell interaction, precluding binding and/or fusion events through a mechanism of steric hindrance. We have thus succeed to predict epitopes for E, M and N proteins, which are less abundant than in the S glycoprotein (due to the smaller size of these proteins), but well conserved in nearly 20000 SARS-CoV-2 genomes (Tables S4, S5 and S6), supporting their application as vaccine targets.
5. Conclusion
In the current century this is the third emergency caused by coronavirus, and it is highly probable that new viruses will continue to emerge causing outbreaks due to their ability to mutate, recombine, and infect multiple species. The current study points to bats as the main reservoir of diversity of SARS-like coronaviruses, evidencing their ability to change their genomes which may in turn trigger the capacity of emerge in novel hosts and escape vaccine. Indeed, the present analysis evidenced the existence of all these properties typical of RNA virus, namely existence of recombination events and high mutational rate in SARS-CoV-2, that accumulate in genome hotspots, for the time being without an impact in the conservation of epitope sequences.
Data availability
The data underlying this article are available in the article and in its online supplementary material files named DataInfo.pdf. The in-house python scripts are available at https://github.com/v888888/programing/releases/tag/scriptsGenomics
CRediT authorship contribution statement
Filipa F. Vale: Conceptualization, Formal analysis, Methodology, Writing – original draft. Jorge M.B. Vítor: Conceptualization, Validation, Writing – review & editing. Andreia T. Marques: Methodology, Validation, Writing – review & editing. José Miguel Azevedo-Pereira: Conceptualization, Validation, Writing – review & editing. Elsa Anes: Conceptualization, Validation, Writing – review & editing. Joao Goncalves: Conceptualization, Funding acquisition, Validation, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work is based on SARS-CoV-2 genomes available in the GISAID database, so we would like to express our appreciation to all the researchers who contributed to the effort of sequencing and make genomes available. The authors would like to thank Francine Perler for valuable comments and suggestions. This work was funded by national funds from FCT – Fundação para a Ciência e a Tecnologia, I.P., projects UIDB/04138/2020 and UIDP/04138/2020. Filipa F. Vale is financed by FCT through Assistant Researcher grant CEECIND/03023/2017.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2021.198526.
Appendix. Supplementary materials
References
- Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Miyazawa T. Genome evolution of SARS-CoV-2 and its virological characteristics. Inflamm. Regen. 2020;40:17. doi: 10.1186/s41232-020-00126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Li X., Li T., Zhang S., Wang L., Wu X., Liu J. The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2020;39:1629–1635. doi: 10.1007/s10096-020-03899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos Bezerra R., Valença I.N., de Cassia Ruy P., Ximenez J.P.B., da Silva W.A., Jr., Covas D.T., Kashima S., Slavov S.N. The novel coronavirus SARS-CoV-2: from a zoonotic infection to coronavirus disease 2019. J. Med. Virol. 2020 doi: 10.1002/jmv.26072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.-J., Li N., Guo Y., Li X., Shen X., et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- Lam T.T.-Y, Jia N., Zhang Y.-W., Shum M.H.-H., Jiang J.-F., Zhu H.-C., Tong Y.-G., Shi Y.-X., Ni X.-B., Liao Y.-S., et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Vignuzzi M., Andino R. Closing the gap: the challenges in converging theoretical, computational, experimental and real-life studies in virus evolution. Curr. Opin. Virol. 2012;2:515–518. doi: 10.1016/j.coviro.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg R. Detecting the emergence of novel, zoonotic viruses pathogenic to humans. Cell. Mol. Life Sci. 2015;72:1115–1125. doi: 10.1007/s00018-014-1785-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graepel K.W., Lu X., Case J.B., Sexton N.R., Smith E.C., Denison M.R. Proofreading-deficient coronaviruses adapt for increased fitness over long-term passage without reversion of exoribonuclease-inactivating mutations. MBio. 2017;8 doi: 10.1128/mBio.01503-17. e01503-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay T.S. Gene of the month: the 2019-nCoV/SARS-CoV-2 novel coronavirus spike protein. J. Clin. Pathol. 2020;73:366–369. doi: 10.1136/jclinpath-2020-206658. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menardo F., Loiseau C., Brites D., Coscolla M., Gygli S.M., Rutaihwa L.K., Trauner A., Beisel C., Borrell S., Gagneux S. Treemmer: a tool to reduce large phylogenetic datasets with minimal loss of diversity. BMC Bioinform. 2018;19:164. doi: 10.1186/s12859-018-2164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A.J., Taylor B., Delaney A.J., Soares J., Seemann T., Keane J.A., Harris S.R. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genomics. 2016;2 doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D., Moulton V. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Pfeifer B., Wittelsbürger U., Ramos-Onsins S.E., Lercher M.J. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 2014;31:1929–1936. doi: 10.1093/molbev/msu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T., Ahmed I. Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27:3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/JVI.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger LLC . 2015. The PyMOL Molecular Graphics System, Version 1.8. [Google Scholar]
- Lassalle F., Beale M.A., Bharucha T., Williams C.A., Williams R.J., Cudini J., Goldstein R., Haque T., Depledge D.P., Breuer J. Whole genome sequencing of Herpes Simplex Virus 1 directly from human cerebrospinal fluid reveals selective constraints in neurotropic viruses. Virus Evol. 2020;6:veaa012. doi: 10.1093/ve/veaa012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samrat S.K., Tharappel A.M., Li Z., Li H. Prospect of SARS-CoV-2 spike protein: potential role in vaccine and therapeutic development. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen A., Shackelton L.A., Holmes E.C. Family level phylogenies reveal modes of macroevolution in RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 2011;108:238–243. doi: 10.1073/pnas.1011090108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley K., Evans D.J. Mechanisms and consequences of positive-strand RNA virus recombination. J. Gen. Virol. 2018;99:1345–1356. doi: 10.1099/jgv.0.001142. [DOI] [PubMed] [Google Scholar]
- Liu P., Jiang J.Z., Wan X.F., Hua Y., Li L., Zhou J., Wang X., Hou F., Chen J., Zou J., Chen J. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S., Katzourakis A., Pybus O.G. Detecting natural selection in RNA virus populations using sequence summary statistics. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2010;10:421–430. doi: 10.1016/j.meegid.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Fang B., Liu L., Yu X., Li X., Ye G., Xu J., Zhang L., Zhan F., Liu G., Pan T., et al. Genome-wide data inferring the evolution and population demography of the novel pneumonia coronavirus (SARS-CoV-2) bioRxiv. 2020 doi: 10.1101/2020.03.04.976662. [DOI] [Google Scholar]
- Laskar R., Ali S. Phylo-geo-network and haplogroup analysis of 611 novel Coronavirus (nCov-2019) genomes from India. bioRxiv. 2020 doi: 10.1101/2020.09.03.281774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura K., Uesaka K. Genetic variations in the human severe acute respiratory syndrome coronavirus receptor ACE2 and serine protease TMPRSS2. J. Clin. Pathol. 2021;74:307–313. doi: 10.1136/jclinpath-2020-206867. jclinpath- [DOI] [PubMed] [Google Scholar]
- Funk C.D., Laferrière C., Ardakani A. A snapshot of the global race for vaccines targeting SARS-CoV-2 and the COVID-19 pandemic. Front. Pharmacol. 2020;11:937. doi: 10.3389/fphar.2020.00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearlove B., Lewitus E., Bai H., Li Y., Reeves D.B., Joyce M.G., Scott P.T., Amare M.F., Vasan S., Michael N.L., et al. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc. Natl. Acad. Sci. 2020;117 doi: 10.1073/pnas.2008281117. 23652 LP –23662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta N.K., Mazumdar K., Gordy J.T. The nucleocapsid protein of SARS-CoV-2: a target for vaccine development. J. Virol. 2020;94 doi: 10.1128/JVI.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florindo H.F., Kleiner R., Vaskovich-Koubi D., Acúrcio R.C., Carreira B., Yeini E., Tiram G., Liubomirski Y., Satchi-Fainaro R. Immune-mediated approaches against COVID-19. Nat. Nanotechnol. 2020;15:630–645. doi: 10.1038/s41565-020-0732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Is There a link between the pathogenic human coronavirus envelope protein and immunopathology? A review of the literature. Front. Microbiol. 2020;11:2086. doi: 10.3389/fmicb.2020.02086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D., Zhu C., Ai L., He T., Wang Y., Ye F., Yang L., Ding C., Zhu X., Lv R., et al. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg. Microbes Infect. 2018;7:154. doi: 10.1038/s41426-018-0155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Tong S. Complete genome sequence of a severe acute respiratory syndrome-related coronavirus from Kenyan bats. Microbiol. Resour. Announc. 2019;8 doi: 10.1128/MRA.00548-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Gloza-Rausch F., Glende J., Corman V.M., Muth D., Goettsche M., Seebens A., Niedrig M., Pfefferle S., Yordanov S., et al. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J. Virol. 2010;84:11336–11349. doi: 10.1128/JVI.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material files named DataInfo.pdf. The in-house python scripts are available at https://github.com/v888888/programing/releases/tag/scriptsGenomics