Abstract
Background and Aims
It is postulated that molecular methods along with mathematical modeling can provide critical inference regarding epidemiological parameters, transmission dynamics, spatiotemporal characteristics, and intervention efficacy. Hence, studying molecular epidemiology of human immunodeficiency virus (HIV)‐1 infection, especially in resource‐limited settings and with a large diaspora of the migrant population such as that of Bangladesh, is of paramount importance. The purpose of this systematic review was to concisely present and discuss the findings from previous studies conducted in Bangladesh regarding HIV‐1 subtype prevalence.
Methods
Articles were retrieved from six publicly available databases regarding HIV‐1 molecular epidemiology using keywords HIV, HIV‐1, subtype(s), Bangladesh, and any combination of aforementioned keywords using Boolean operators. A total of 14 articles were downloaded and screened for suitability. Finally, five studies, containing pooled sequences from 317 individuals, were included in this systematic review.
Results
Results revealed a preponderance of subtype C among HIV‐1 infected population (51.10%), followed by circulating recombinant form (CRF)_07BC (15.46%), CRF_01AE (5.68%), A1 (4.73%), CRF_02AG (3.47%), G (3.15%), CRF_62BC (2.84%), B (2.21%), and other subtypes and recombinant forms in small percentages. Subtype C was largely predominant in intravenous drug users as well as female sex workers, whereas the migrant population exhibited a diverse subtype including rare recombinant forms, largely due to their travel in the Middle East and other South East Asian countries.
Conclusion
With the number of HIV‐1 infections increasing among the general population and a steady increase in the migrant population, molecular epidemiological data are required to curb the progression of the HIV‐1 epidemic in Bangladesh.
Keywords: Bangladesh, HIV‐1, molecular epidemiology, prevalence, subtypes, systematic review
1. INTRODUCTION
Since the first case detection in the early 1980s among a group of homosexual individuals, human immunodeficiency virus (HIV) infection and its subsequent transformation to acquired immune deficiency syndrome (AIDS) has become a major global health concern. 1 Belonging to the Retriviridae family and Lentivirinae genus, the virus is enveloped, spherical in shape, and with a diameter of 120 nm. Like most other retroviruses, HIV is composed of three major structural genes, namely group‐specific antigen (gag), polymerase (pol), and envelope (env) genes. In addition to that several other genes, such as transactivator of viral transcription (tat), regulation of RNA transport (rev), viral protein R (vpr), negative factor (nef), viral infectivity factor (vif), and viral protein U (vpu) make up the viral genome. 2
More than 75.9 million people have been diagnosed with HIV infection since the start of the epidemic, and of them, 32.7 million people have died due to various AIDS‐related complications. 3 In 2019, 1.7 million people worldwide have gotten infected with the virus, marking a 23% reduction in new cases since 2010. 3 Still, the progress achieved during the past decade is way below the expectation set forth by the Joint United Nations Program on HIV/AIDS (UNAIDS), with three times more new infections reported compared to the UNAIDS estimation. Efforts regarding curbing HIV transmission are still lacking, and more so prevalent in countries like Bangladesh—a low middle‐income country with a large diaspora of migrant workers. 4
The first case of HIV in Bangladesh was detected in 1989. 5 It is estimated that more than 14 000 people are currently living with HIV in the country with 1800 newly diagnosed cases in 2018. Unlike most countries, Bangladesh is on an upward trend with the number of newly diagnosed cases each year increased by 600 since 2012. 4 Despite being a low prevalence country and the percentage of HIV infection among key population groups still being <1%, there is evidence of the infection being spread to housewives and pregnant women—a sign of feminization and an early sign of epidemic. 6 While the government has taken numerous initiatives to diagnose and treat HIV cases, people at risk are still reluctant to get tested and treated due to the prevalence of stigma around the infection. As such, more than 70% of people are still beyond the reach of available testing and treatment programs. 6
With an epidemic looming large and the unavailability of widespread testing and treatment facilities, HIV prevention, for the time being, is the best bet for Bangladesh. Observing trends and tracking individuals through molecular epidemiology could be an effective strategy for health authorities, especially because of the goal of eliminating the disease by 2030. 7 Besides, viral subtypes could be potentially determining factors regarding epidemiological and therapeutic outcomes in infected individuals, as previous studies suggest differences between clades can explain several determining factors of infection and disease progression, including transmission rates, CD4 cells decline rate, natural history, response to antiretroviral therapy (ART) drugs, and immune evasion by the virus. 8 For example, people infected with subtype C, the prevalent HIV‐1 subtype in India and Bangladesh, were found to be susceptible to earlier disease progression than people infected with subtype‐B. 9 In addition, CRF_AE, a subtype found among Thai HIV‐infected patients, was found to be responsible for higher plasma viral load and higher transmission rates. 10 Now, to put that into Bangladeshi perspective, genome sequencing prior to starting ART drugs is not performed. Patients have been prescribed a standard three‐drug regimen consisting of two nucleoside reverse transcriptase inhibitor (NRTI) and one nonnucleoside reverse transcriptase inhibitor (non‐NRTI), irrespective of subtype and mutation status. 11 Besides, epidemiological tracking is limited to serological testing at few centers in big cities, with most people around the country still devoid of HIV testing services. 5 , 11
Several studies that tried to fill the knowledge gap regarding HIV‐1 subtypes circulating in Bangladesh have been published with the oldest being published in 2002 and the latest one in 2020. But the time difference and the discrepancies in subtyping results between two studies make it harder for researchers to draw any valid inference by looking into a single study. A concerted effort is required to compile and analyze existing molecular epidemiological studies to construct the prevailing scenario of HIV‐1 transmission patterns in Bangladesh, as well as formulating effective treatment strategies. Hence, this study aimed to perform a systematic review of studies concerning HIV‐1 molecular epidemiology in Bangladesh.
2. METHODS
2.1. Data source
For this systematic review, articles regarding molecular epidemiology of HIV‐1 in Bangladesh were searched in different databases following the guidelines of Preferred Reporting Items for Systematic Reviews and Meta‐Analyses. 12 However, we did not register the systematic review as this is a systematic review of molecular epidemiology.
Articles were retrieved from six publicly available databases on March 5, 2021. The databases are Medline through PubMed, Cochrane database, Web of Science, CINAHL, EMBASE, and Google Scholar. Several primary keywords and their combinations using Boolean operators were used to search articles in databases (Table 1). Besides, we also searched bibliographies of primarily selected articles for suitable articles on our topic.
TABLE 1.
Search strategy for different databases
| Database | Search strategy |
|---|---|
| PubMed (Medline) | HIV AND HIV‐1 AND Subtypes AND Bangladesh |
| Web of Science | HIV AND HIV‐1 AND Subtypes AND Bangladesh |
| Cochrane database | HIV AND HIV‐1 AND Subtypes AND Bangladesh |
2.2. Inclusion and exclusion criteria
Regarding inclusion criteria, articles in English, from the inception of record‐keeping to March 2021, that discussed primary data regarding molecular epidemiology and subtyping of HIV‐1‐infected individuals in Bangladesh were initially included within the review process. Review articles or case series with less than five participants were excluded. Articles discussing genotyping any region of the HIV‐1 genome (env, gag, pol) through sequencing followed by subtyping using an established online tool or manual phylogeny were considered for the systematic review. In the case of multiple articles discussing findings from the same accession numbers of sequences, the oldest article with original data was kept, and the rest were discarded. Additionally, in the case of articles with multiple regions of sequencing (env, gag, pol), for initial reporting of HIV‐1 prevalence in Bangladesh, subtyping results from the region with the highest number of sequenced samples within the article was reported. Grey literature such as conference papers, theses, and dissertations was excluded from the final list as the authors could not validate the authenticity of the reported data. Besides, the authors did not have any reservations regarding the age of included participants, sex, their geographical locations, migration history, length of genome sequenced, and the method of sequencing.
2.3. Screening process
After removing duplicates, articles were screened by titles and abstracts for suitability by two independent authors. If deemed suitable, full‐text articles were downloaded and further explored for suitability. Articles were excluded based on the above‐mentioned inclusion and exclusion criteria. The full‐text screening was also conducted by two independent researchers. In case of disagreement over whether to include the full‐text article for review, an opinion was sought from a third author.
2.4. Data extraction
The following information was extracted from all included articles—the name of the first author, date of article publishing, year of the study conducted, participant demographics such as age, gender, and geographical location, their exposure, and migration history if available, sampling procedure, sample collection method, regions of genome analyzed, sequencing, and subtyping method. Retrieved data were anonymized, and all studies reported approval from the local ethical committee and/or informed written consent from study participants. Any discrepancy in data findings was resolved by discussions among authors. For this systematic review, the risks of bias and quality appraisal were not evaluated due to the absence of a specific tool describing criteria for the evaluation of articles regarding molecular epidemiology.
2.5. Data analysis
Data were extracted in Microsoft Excel and analyzed using native functions of the software. Since there was heterogeneity in the outcome of the included articles, a meta‐analysis could not be performed.
2.6. Ethical consideration
This systematic review of HIV‐1 subtype prevalence in Bangladesh was conducted using publicly available secondary data. No human being was interviewed as part of the process. Hence, ethical permission from local/regional authorities was waived.
3. RESULTS
After searching different databases, a total of 14 articles were included for initial review. After removing five duplicates, nine articles were considered for the title and abstract screening. A total of two articles were deleted (one review article and another article not related to Bangladesh), with seven articles downloaded and further screened for suitability. Two of those articles were removed as those did not meet the inclusion criteria set forth, and finally five articles were included in the systematic review. A detailed explanation for each step is illustrated in Figure 1.
FIGURE 1.
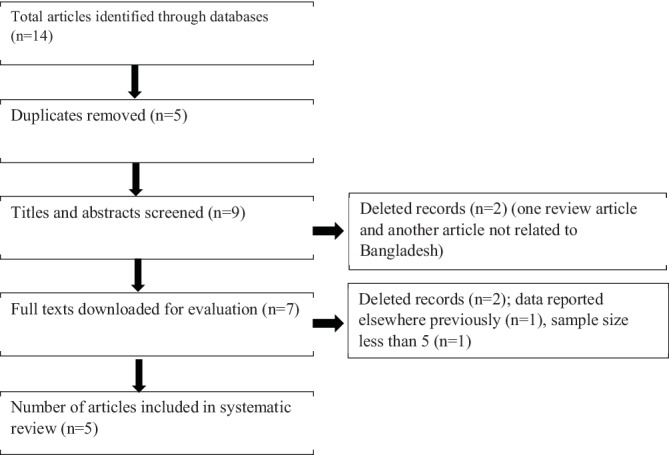
Flow diagram of article screening procedure regarding human immunodeficiency virus (HIV)‐1 subtyping in Bangladesh
A brief description of each study included in this systematic review is provided in Table 2.
TABLE 2.
Characteristics of the included studies
| Author | Published year | Sampling year | Sample size | Gene sequenced | Prevalent subtype |
|---|---|---|---|---|---|
| Ljungberg et al 13 | 2002 | 2001 | 9 | gag/env | Subtype C |
| Azim et al 14 | 2002 | 1999‐2000 | 6 | gag/env | Subtype C |
| Sarker et al 15 | 2008 | 1999‐2005 | 198 | gag | Subtype C |
| Sarker et al 16 | 2020 | 2005–2007 | 40 | gag/pol/env | Subtype C |
| Rahman et al 17 | 2020 | 2016 | 64 | gag/pol/env | Subtype C |
3.1. Demographic characteristics of key populations
A total of five studies conducted between 1999 and 2016 observed that the largest numbers of infections were attributed to injecting drug users (IDUs) and a total of three studies reported HIV in this key population of Bangladesh, which analyzed 129 (40.69%, 129/317) sequences of IDU. Mostly, they were from the southern part of the Dhaka district and a few of them were from different parts of the country. Most of them were male. A total of two studies conducted between 2005‐2007 and 2016 sequenced 43 samples from female sex workers (FSW), transgender, men who have sex with men (MSM), and patients with sexually transmitted infection (STI). These groups attributed to 13.56% (43/317) of total sequencing data. Of these, 28 (65.11%) were female sex workers, 5 (11.63%) were transgender, 5 (11.63%) were MSM, and 5 (11.63%) were patients with STI. In between 2001 and 2007, three studies were conducted, which analyzed sequences from 77 (24.29%, 77/317) migrant workers and their contacts. Among them, 48 (62.33%) were migrant workers, 22 (28.57%) were spouses or sex partners, and 7 (9.09%) were children of HIV‐positive migrants. Of these 48 migrant workers, 44 (91.67%) were male and 4 (8.33%) were female. Out of 77 migrant workers, the residence history of 47 patients was mentioned in previous studies. It was reported that 59.57% (28/47) of this group were from Dhaka, followed by Sylhet (29.78%, 14/47), Chittagong (4.25%, 2/47), Gazipur (4.25%, 2/47), and Faridpur district (2.12%, 1/47). All migrant workers had a history of migration abroad. The majority of the migrants (72.91%, 35/48) had traveled to countries in the Middle East (UAE, Saudi Arabia, Jordan, Bahrain, Kuwait, etc) while 20.83% (10/48) to India and 2.08% (1/48) to Nepal, Malaysia, and Singapore.
3.2. HIV‐1 subtype prevalence
Sequences were retrieved from a total of 317 individuals from all five included studies spanning a duration of 18 years with the earliest study being conducted in 2002 and the latest in 2020. All three major genes of HIV‐1—gag, pol, and env were sequenced, with gag being the most sequenced region (n = 317), followed by env (n = 111) and pol (n = 50). The highest number of HIV‐1 sequences (n = 198) were reported by Sarker et al (2008) and lowest (n = 7) by Azim et al (2002). Among those studied sequences, subtype C was found to be most prevalent (51.10%), followed by CRF_07BC (15.46%), CRF_01AE (5.68%), A1 (4.73%), CRF_02AG (3.47%), G (3.15%), CRF_62BC (2.84%), B (2.21%), and other subtypes in small percentages Table 3.
TABLE 3.
Human immunodeficiency virus (HIV)‐1 subtype distribution in Bangladesh
| Sample Size (n) | A1 | A | B | C | D | G | CRF_01AE | CRF_02AG | CRF_06AGJK | CRF_07BC | CRF_08BC | CRF_09‐02AG,AU | CRF_10CD | CRF_13A,AE | CRF_14BG | CRF_15 A,E,B | U | CRF_09AGU | CRF13cpx_A_AE_G_J_U | CRF_16 A2D | CRF_25 AGU | CRF_43AG | CRF_62BC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 198 | 13 | 0 | 5 | 82 | 4 | 9 | 18 | 5 | 1 | 48 | 5 | 2 | 2 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 2 | 0 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 1 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 40 | 2 | 0 | 1 | 18 | 0 | 0 | 0 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 1 | 3 | 3 | 1 | 0 |
| 64 | 0 | 0 | 0 | 52 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 |
| 317 | 15 | 2 | 7 | 162 | 4 | 10 | 18 | 11 | 2 | 49 | 6 | 2 | 2 | 1 | 1 | 2 | 3 | 3 | 1 | 3 | 3 | 1 | 9 |
3.3. Pattern of subtype distribution among key population groups
Considering partial sequencing of gag or env or both genes and neighbor‐joining trees of the HIV sequences of the 129 available samples from IDU (intravenous drug users) risk group and reference sequences of HIV‐1 group M revealed multiple distinct subtypes and CRFs. Based on the gag gene, 128 samples were successfully amplified, and the predominant subtype was identified as HIV‐1 subtype C, which accounted for 82.03% (105/128) of total infections, followed by CRF07_BC (8.59%, 11/129), CRF62_BC (6.25%, 8/128), subtype B (1.56%, 2/128), CRF08_BC (0.78%, 1/128), and CRF01_AE (0.78%, 1/128), respectively. While considering env gene, only 57 samples were sequenced (one study out of three did not sequence env gene), and CRF08_BC was the most frequent subtype reported, responsible for 59.64% (34/57), followed by subtype C (15.78%, 9/57), CRF64_BC (10.52%, 6/57), CRF07_BC (8.77%, 5/57), subtype E (1.75%, 1/57), CRF61_BC (1.75%, 1/57), and CRF57_BC (1.75%, 1/57).
Data from two studies revealed 43 genome sequences from the sex workers group. The most prevalent subtype was C‐related recombinant forms. Frequency of CRF07_BC was responsible for 44.18% (19/43), followed by subtype C (27.90%, 12/43), subtype‐A‐related recombinant forms (16.27%, 7/43), subtype A1 (6.97%, 3/43), and subtype‐C‐related other recombinant forms (4.65%, 2/43) based on partial sequence of gag gene. Whereas subtype‐C‐related recombinant forms were mostly observed while considering env gene sequence among the five studied samples from this group.
A total of 77 sequences from migrant workers and their contacts were reported in three previous studies. Based on two studies that reported 47 sequences out of 77, the majority of migrant workers and their contacts were infected with subtype‐C while using a partial sequence of gag, env, and/or pol gene. Subtype‐C was identified in 22 individuals out of 47 study populations (46.80%). The next predominant subtype was G‐related recombinant forms, accounting for 29.78% (14/47), followed by CFR16_A2D (6.38%, 3/47), and subtype A, B, G, and unique recombinant form together responsible for 17.02% (8/47) of total infections in this group. Another study described the sequences of 96 patients from the voluntary counseling and testing unit (VCT), and among them, 30 had a history of migration or relation with a migrant worker. As this study did not delineate the individual sequence data of these 96 VCT patients, separately, we could not identify the exact frequency of different subtypes among 30 migrant workers. It can only be stated that subtype C was responsible for 21.87% (21/96), whereas diverse recombinant forms together attributed to 63.54% (61/96) of total VCT strains. CRF07_BC was the prevailing subtype among recombinant forms (19.79%, 19/96) and next predominant subtype was CRF01_AE (14.58%, 14/96), subtype A1 (10.41%, 10/96), subtype G (9.37%, 9/96), subtype B (4.16%, 4/96), subtype D (4.16%, 4/96), and other recombinant forms (CRF02_AG; CRF08_BC; CRF09‐02_AG, AU; CRF10_CD; CRF13_A, AE; CRF14_BG; and CRF15_ A, E, B) together accounted for 14.58% (14/96) of VCT clients.
4. DISCUSSION
To our knowledge, this is the first systematic review discussing the prevalent HIV‐1 subtypes in Bangladesh. This was necessitated due to the long interval between each published study, a large number of undetected cases, a large diaspora of migrants, and the recent surge in HIV‐1 infection in Bangladesh. But, concluding this systematic review was not straightforward for us. The low number of studies available along with the long interval between studies and small sample size in two of those included studies might not reflect the actual scenario of HIV‐1 molecular epidemiology in Bangladesh. Despite such challenges, we tried to construct the pattern of distributions of HIV‐1 subtypes in Bangladesh. In the following sections, we will be providing an account of HIV‐1 transmission patterns among key population groups based on phylogenetic analysis along with our recommendations for future research.
4.1. Transmission links by phylogenetic analysis
Injecting drug use represents the main mode of HIV transmission in Bangladesh as indicated by previous studies. Subtype C is the predominant clade driving the epidemic among IDU. Phylogenetic analysis which includes gag sequences from HIV strains of IDU samples showed clustering of IDU strains and separation from other risk group populations. Initial studies were conducted from 1999 to 2005, and 36 samples were used to draw dendrogram using only predominant strain subtype C and CRF07_BC. Among them, 31 Bangladeshi IDU samples were clustered with Indian strains, and five of them were similar to Ethiopian and South African strains, whereas a study conducted in 2016 exhibited that IDU strains clustered in a monophyletic branch, and most of them were clustered with previous Bangladeshi IDU strains. These data suggest that the initial introduction of these strains into the IDU may have been from India and other foreign countries, but the present epidemic has been propagating locally within the Bangladeshi strain for at least several years, and there is a little epidemiologic linkage between IDU and FSW—two of the most important risk groups in Bangladesh. Strains isolated from commercial sex workers and transgender were closely related to each other. The isolates from the sex workers were placed in different branches with Indian, Chinese, Zimbabwean, and South African isolates. Transgender isolates clustered with either Chinese or Indian strains.
Sequence analysis of migrant workers indicated multiple subtypes, suggesting multiple travels related to the exogenous entry of genetically diverse HIV‐1 subtypes rather than endemic transmission. Subtype C isolated from migrants mostly clustered with Indian strains and a few of them with strains from Nepal, South Africa, and the Middle East. The majority of subtype A1‐, G‐, and G‐related recombinant forms were clustered with strains from South Africa whereas predominant VCT (subtype C and CRF07_BC) strains, which included migrant workers were clustered with Indian, Myanmar, Zimbabwean, and Ethiopian strains. HIV strains of these migrant populations dispersed all through the tree and shared multiple branches with different global strains indicating extreme polyphyletic origin. Bangladeshi migrants usually travel to Middle East, Europe, and other South East Asian countries, and previous reports suggest many of them purchase sex from commercial sex workers, with some of them admitting sexual interaction with male partners. Subtype C is predominant in India, subtype B, and CRF_01AE are prevalent in Thailand, and other subtypes and recombinant forms in countries such as Malaysia, Saudi Arabia, United Arab Emirates, and Qatar—where Bangladeshis mostly travel for work. 18 This may justify the diverse subtypes of HIV‐1 found in migrants. Besides, this may be the reason why the strains isolated from migrants were not clustered with IDU and sexual workers' strain, since the epidemic is not concentrated within any specific population and spread is usually limited to immediate sexual partners and their children.
4.2. Future recommendations
The prevalence of HIV‐1 in Bangladesh among the general population is still below <0.1% and reaching 1% in high‐risk groups. But considering recent trends and the introduction of diverse subtypes, especially through migrant populations, Bangladesh should strengthen their overall surveillance system and molecular epidemiology, despite being cost‐prohibitive and currently unavailable barring few centers in Dhaka, could be an important tool for future epidemic management in Bangladesh. Our research illustrated the concentrated prevalence of HIV‐1, especially subtype C in high‐risk groups such as intravenous drug users and female sex workers. We would suggest the authorities including the migrant population as high‐risk groups and introduce a regular screening and, if found positive, carry out molecular epidemiological tracking in order to contain the spread.
To our knowledge, very few laboratories in Bangladesh currently provide facilities for HIV‐1 genome sequencing. Moreover, all the studies included in our systematic review were conducted by a single group of researchers. We would advise the health authorities to consider opening laboratories in each divisional cities, especially border city, with a high percentage of risk group populations. Otherwise, the goal of eliminating the virus by 2030 would be hard to achieve for Bangladesh.
There are several limitations to our study. First of all, the number of eligible studies was highly limited. Moreover, two of those included studies had a low sample size (<10). We could not perform a meta‐analysis due to the heterogeneity of the included articles. Besides, samples were stored in the freezer and analyzed decades later in one of the included studies, further undermining the credibility of published results. Besides, analyzing stored samples from the past might not reflect the true transmission pattern of the infection and could simply refer to an epidemic occurring in the past. Hence, we would advise future research to be conducted with samples from newly diagnosed cases in order to avoid uncertainty regarding infection patterns and to portray the true dynamics of HIV‐1 subtypes circulating in Bangladesh.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
Conceptualization: Maruf Ahmed Molla, K.M. Saif‐Ur‐Rahman.
Data Curation: Asish Kumar Ghosh, Tasnim Nafisa.
First Draft Preparation: Maruf Ahmed Molla, Mahmuda Yeasmin.
Methodology: Maruf Ahmed Molla, K.M. Saif‐Ur‐Rahman, Mahmuda Yeasmin.
Review and Editing: K.M. Saif‐Ur‐Rahman, Khairul Islam.
Supervision: K.M. Saif‐Ur‐Rahman.
All authors have read and approved the final version of the manuscript.
K.M. Saif‐Ur‐Rahman had full access to all of the data in this study and takes complete responsibility for the integrity of the data and the accuracy of the data analysis.
TRANSPARENCY STATEMENT
K.M. Saif‐Ur‐Rahman affirms that this manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained.
ACKNOWLEDGEMENTS
We would like to thank all the healthcare workers in Bangladesh tasked with testing, treating, and caring for HIV‐infected individuals. icddr,b is also grateful to the Governments of Bangladesh, Canada, Sweden, and the United Kingdom for providing core/unrestricted support.
Molla MMA, Yeasmin M, Ghosh AK, Nafisa T, Islam MK, Saif‐Ur‐Rahman KM. HIV‐1 molecular epidemiology in Bangladesh: A systematic review. Health Sci Rep. 2021;4:e344. 10.1002/hsr2.344
Maruf Ahmed Molla and Mahmuda Yeasmin contributed equally to this study.
Funding information The authors did not receive any funding for this study
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Sharp PM, Beatrice HH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1(1):a006841. 10.1101/cshperspect.a006841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. German Advisory Committee Blood (Arbeitskreis Blut), Subgroup ‘Assessment of Pathogens Transmissible by Blood’ . Human immunodeficiency virus (HIV). Transfus Med Hemother. 2016;43(3):203‐222. 10.1159/000445852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. WHO . HIV data and statistics. 2020. https://www.who.int/teams/global‐hiv‐hepatitis‐and‐stis‐programmes/hiv‐programme/strategic‐information/hiv‐data‐and‐statistics. Accessed March 25, 2021.
- 4. WHO . Bangladesh: HIV Country Profile 2019. 2020. https://cfs.hivci.org/country-factsheet.html. Accessed March 25, 2021.
- 5. Azim T, Khan SI, Haseen F, et al. HIV and AIDS in Bangladesh. J Health Popul Nutr. 2008;26(3):311‐324. 10.3329/jhpn.v26i3.1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Unicef . (2020). Towards ending AIDS. https://www.unicef.org/bangladesh/en/towards‐ending‐aids#:~:text=In%20Bangladesh%2C%20there%20is%20a,cent%20of%20the%20general%20population.&text=The%20pattern%20of%20transmission%20of,workers%20and%20between%20married%20couples. Accessed March 25, 2021.
- 7. Assefa Y, Gliks CF. Ending the epidemic of HIV/AIDS by 2030: will there be an endgame to HIV, or an endemic HIV requiring an integrated health systems response in many countries? Int J Infect Dis. 2020;100:273‐277. 10.1016/j.ijid.2020.09.011 [DOI] [PubMed] [Google Scholar]
- 8. Gatell JM. Antiretroviral therapy for HIV: do subtypes matter? Clin Infect Dis. 2011;53(11):1153‐1155. 10.1093/cid/cir686 [DOI] [PubMed] [Google Scholar]
- 9. Wainberg MA, Brenner BG. Role of HIV subtype diversity in the development of resistance to antiviral drugs. Viruses. 2010;2(11):2493‐2508. 10.3390/v2112493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tovanabutra S, Robison V, Wongtrakul J, et al. Male viral load and heterosexual transmission of HIV‐1 subtype E in northern Thailand. J Acquir Immune Defic Syndr. 2002;29(3):275‐283. 10.1097/00126334-200203010-00008 [DOI] [PubMed] [Google Scholar]
- 11. NASP . (2020). National Anti‐Retroviral Therapy (NRT) Guidelines, Bangladesh. Retrieved from http://asp.portal.gov.bd/sites/default/files/files/asp.portal.gov.bd/page/2d04f70c_c5e5_4d3d_9a13_e8cc7e5af9c9/2020‐10‐13‐14‐47‐6f171c6b8baeb78d8dc2c9e073f894b0.pdf. Accessed July 5, 2021.
- 12. Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372. n–71. 10.1136/bmj.n71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ljungberg K, Hassan MS, Islam MN, et al. Subtypes a, C, G, and recombinant HIV type 1 are circulating in Bangladesh. AIDS Res Hum Retrovir. 2002;18(9):667‐670. 10.1089/088922202760019374 [DOI] [PubMed] [Google Scholar]
- 14. Azim T, Bogaerts J, Yirrell DL, et al. Injecting drug users in Bangladesh: prevalence of syphilis, hepatitis, HIV and HIV subtypes. AIDS. 2002;16(1):121‐123. 10.1097/00002030-200201040-00015 [DOI] [PubMed] [Google Scholar]
- 15. Sarker MS, Rahman M, Yirrell D, et al. Molecular evidence for polyphyletic origin of human immunodeficiency virus type 1 subtype C in Bangladesh. Virus Res. 2008;135(1):89‐94. 10.1016/j.virusres.2008.02.010 [DOI] [PubMed] [Google Scholar]
- 16. Sarker MS, Azim T, Islam LN, Rahman M. Migrant workers play a key role in HIV‐1 strain diversity in Bangladesh. HIV AIDS Rev. 2020;19(4):277‐283. 10.5114/hivar.2020.101794 [DOI] [Google Scholar]
- 17. Rahman M, Rahman S, Reza MM, Khan SI, Sarker MS. HIV‐1 drug resistance and genotypes circulating among HIV‐positive key populations in Bangladesh: 2016 update. Int J Infect Dis. 2020;104:150‐158. 10.1016/j.ijid.2020.12.037 [DOI] [PubMed] [Google Scholar]
- 18. Bbosa N, Kaleebu P, Ssemwanga D. HIV subtype diversity worldwide. Curr Opin HIV AIDS. 2019;14(3):153‐160. 10.1097/COH.0000000000000534 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


