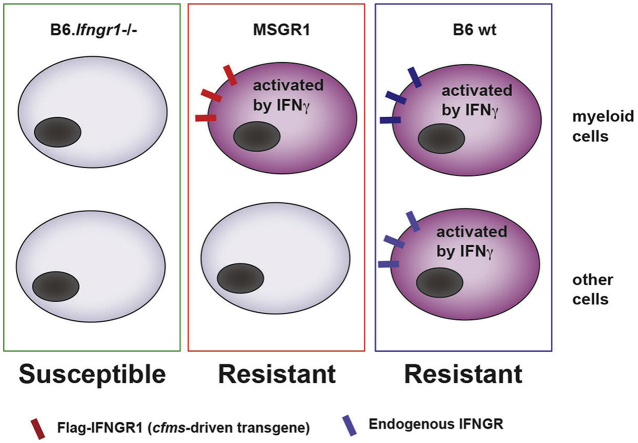
Abstract
The type II interferon (IFNγ) promotes resistance to intracellular pathogens. Most immune and somatic cells also express the IFNγ receptor (IFNGR) and respond to IFNγ. While myeloid cell have been implicated as important targets of IFNγ, it remains unknown if IFNγ signaling to myeloid cell types suffices for resistance to infection. Here, we addressed this question by generating mice in which IFNGR1 is selectively expressed by myeloid cells. These “MSGR1” (myeloid selective IFNGR1) mice express an epitope-tagged Ifngr1 transgene (fGR1) from the myeloid-specific c-fms promoter in a background lacking endogenous Ifngr1. IFNGR staining was selectively observed on myeloid cells in the MSGR1 mice and correlated with responsiveness of these cells to IFNγ. During systemic infection by the bacterium Listeria monocytogenes, activation marker staining was comparable on monocytes from MSGR1 and control B6 mice. Bacterial burdens and survival were also equivalent in MSGR1 and wildtype B6 animals at a timepoint when B6.Ifngr1−/− mice began to succumb. These data confirm that activation of inflammatory monocytes and neutrophils is a key mechanism by which IFNγ promotes innate anti-bacterial immunity and suggest that IFNγ targeting of myeloid cells is largely sufficient to mediate protection against systemic L. monocytogenes.
Keywords: IFNγ, IFNGR, Myeloid cells, Infection, Listeria monocytogenes
Graphical abstract
Highlights
-
•
Expression of IFNGR1 is restricted to monocytes and neutrophils in “MSGR1” (myeloid selective IFNGR1) mice.
-
•
Myeloid cells from MSGR1 mice are responsive to IFNγ and show elevated activation compared to cells from B6.Ifngr1−/− mice.
-
•
MSGR1 myeloid cells respond to Listeria monocytogenes infection and promote early resistance.
-
•
IFNγ stimulation of myeloid cells can thus protect against infection independent of effects on other hematopoietic and non-hematopoietic cell populations.
-
•
Particularly in female mice, IFNγ stimulation of non-myeloid cells may also contribute to improved survival.
Introduction
At steady-state, tissue macrophages, blood monocytes, and granulocytic neutrophils promote tissue homeostasis and surveille for the presence of invasive microbes. The appearance of microbial and inflammatory stimuli induces rapid differentiation of these “resting” myeloid cells towards an anti-microbial and pro-inflammatory state of activation often referred to as M1 “activation” (Mills et al., 2014). The host-derived type II IFN, IFNγ, drives this activation and is required for optimal resistance to a variety of intracellular pathogens (Schroder et al., 2004, Kearney et al., 2013a, Bustamante et al., 2014). IFNγ stimulation induces myeloid cells to upregulate expression of numerous gene products, some of which have been shown to directly interfere with survival or replication of ingested microbes (Schneider et al., 2014, Coers et al., 2018). IFNγ also induces elevated myeloid cell expression of immune-stimulatory gene products such as class II major histocompatibility complex molecules (MHCII), CD54 (ICAM1), and CD64 (Schroder et al., 2004). IFNγ stimulation of non-myeloid cells can impact their differentiation and survival as well as the production of tissue protective factors such as indoleamine 2,3-dioxygenase (IDO) (Desvignes and Ernst, 2009, Lee et al., 2017).
Listeria monocytogenes is an intracellular bacterial pathogen that can replicate within diverse myeloid and non-myeloid cell types to cause severe systemic infections. A systemic model of L. monocytogenes infection has been widely used in immunological studies. In this model, IFNγ plays a critical role in mediating host resistance. Specifically, deficiency in IFNγ or either subunit of the IFNGR significantly increases bacterial burdens and reduces host survival within the first few days of infection (Buchmeier and Schreiber, 1985, Dalton et al., 1993, Huang et al., 1993, Harty and Bevant, 1995, Lu et al., 1998). Further, mice deficient for the IFNGR-stimulated signaling component, STAT1 are unable to clear L. monocytogenes infection (Meraz et al., 1996, Kernbauer et al., 2012). Most immune and non-hematopoietic cell types express IFNGR and can respond to IFNγ (Schroder et al., 2004, Kearney et al., 2013a, Bach et al., 1997). Thus, the protective effects of IFNγ could be due to its activity in some or all of these cells. Studies using mice with transgenic expression of a dominant negative IFNGR1 suggested myeloid cells are an important target of IFNγ during the immune response to L. monocytogenes (Dighe et al., 1995, Lykens et al., 2010). More recent studies using mice with a floxed Ifngr1 or stat1 allele also confirmed that hematopoietic cells are vital targets of protective IFNγ early after infection (Kernbauer et al., 2012, Lee et al., 2013). However, the extent to which IFNγ signaling is protective due to its effects on myeloid cells versus other hematopoietic and non-hematopoietic cells has not been previously determined.
Here, we sought to investigate how myeloid cell-restricted responsiveness to IFNγ impacts host resistance to systemic L. monocytogenes infection. To this end, we generated a novel mouse strain in which only a myeloid cell-restricted flag epitope-tagged IFNGR1 is expressed. These mice utilized a previously reported transgenic strain that expresses flag-IFNGR1 from the myeloid-specific c-fms promoter (Eshleman et al., 2017). To eliminate expression of the endogenous Ifngr1, we crossed the fGR1 transgene to a B6.Ifngr1−/− background. Our studies with these MSGR1 (myeloid selective IFNGR1) mice are reported here. We observed restricted IFNGR1 expression and responsiveness to endogenously produced IFNγ in myeloid cells of infected MSGR1 mice. Further, we determined that the myeloid cell-restricted fGR1 expression in these animals sufficed to mediate host resistance following systemic L. monocytogenes infection. Our results confirm that myeloid cells are a key target of IFNγ and demonstrate for the first time that IFNγ stimulation of myeloid cells is sufficient to promote host resistance to infection in the absence of other IFNγ-responsive cells.
Materials and methods
Mice
Adult male and female mice were used at 8–12 weeks of age. MSGR1 mice were generated by crossing fGR1 transgenic mice (Eshleman et al., 2017) to B6.129S7-Ifngr1tm1Agt/J (B6.Ifngr1−/−) animals (Huang et al., 1993). WT C57BL6/J control mice and B6.Ifngr1−/− mice were purchased from Jackson Laboratories and maintained in our specific pathogen free (SPF) colony at the University of Colorado Medical Campus. All studies were conducted with approval by the Animal Care and Use Committees at the University of Colorado School of Medicine (protocol #00313). Animal studies follow standards enacted by the United States Public Health Service and Department of Agriculture.
Bacterial infections
Male and female sex and aged-matched mice were infected with WT mouse-passaged L. monocytogenes (Lm), strain 10403s. Frozen aliquots of mouse-passaged bacteria were thawed and grown to log phase (Optical Density (OD) of 0.1) in Tryptic Soy Broth (TSB) supplemented with 50 μg/mL of streptomycin at 37 °C with shaking. Lm was diluted in PBS prior to inject via the lateral tail vein (I.V.) at a sublethal dose of 1–1.5 × 104 CFUs/mouse. Livers were harvested into 0.02% NP-40 and homogenized for 1 min with a tissue homogenizer (IKA Work, Inc.). Spleens were processed into single-cell suspensions as previously described (Eshleman et al., 2017) and 2 × 106 splenocytes were lysed in 0.02% NP-40. Serial dilutions were plated on TSB agar plates containing 50 μg/mL of streptomycin. Agar plates were incubated overnight at 37 °C and CFUs were counted. To estimate bacterial burdens in the whole spleen, the number of counted CFUs was multiplied by a ratio of the total spleen cell number divided by the plated cell number (2 × 106).
Cell culture and cytokine stimulations
To culture bone marrow derived macrophages (BMDM), bone marrow was flushed from the femurs and tibias of mice with DM10 media (DMEM (Gibco) supplemented with 10% FBS, 1% sodium pyruvate, 1% l-glutamine, 1% penicillin/streptomycin). The bone marrow cells were cultured for 6 days in BM macrophage media containing 1% P/S (DMEM media supplemented with 10% Bovine Growth Serum, 1% sodium pyruvate, 1% l-glutamine, 50 μM 2-mercaptoethanol, 10% L-cell conditioned media). Cells were provided fresh media on Day 3 and on Day 6 BMDMs were harvested and plated without P/S for experiments to be conducted on Day 7. Peritoneal cells were isolated from naïve mice by injecting 10-mL of ice-cold PBS into the peritoneal cavity and were cultured in DM10 media. For flow cytometric analysis, peritoneal cells were cultured in non-tissue culture treated suspension plates (Gibco) to minimize cell adherence. Spleens were digested and disrupted into single cell suspension as previously described (Eshleman et al., 2017). To determine MHCII expression, cells were stimulated with 100 U/mL murine IFNγ (Life Technology, #PMC-4031) for 18–24 h. All cell cultures were maintained at 37 °C with 7.5% CO2.
Flow cytometry
Murine Fc receptors were blocked before staining using supernatant from hybridoma 2.4G2 (rat anti-CD16/32). Monoclonal antibodies (mAbs) were diluted in FACS Buffer (1% BSA, 0.01% NaN3, PBS) to final concentrations that were empirically determined by pilot staining experiments. The following mAbs clones were used: CD11b (M1/70, eBioscience, 1:400), CD11c (N418, Biolegend, 1:200), Ly6C (Hk1.4, eBioscience, 1:200), Ly6G (1A8, BioLegend, 1:200), CD90.2 (53-2.1, eBioscience, 1:200), IgM (11/41, eBioscience, 1:200), CD64 (X54/5/7.1, BioLegend, 1:200), CD80 (16-10A1, eBioscience, 1:200), MHCII (I-A/I-E) (M5/114.15.2, eBioscience, 1:300), F480 (CL-A3-1, BioRad, 1:200), IFNGR1/CD119 (2E2, BD Bioscience, 1:100), ICAM-1 (CD54) (YN1/1.7.4,eBioscience, 1:100). IFNGR1 (CD119) was stained using a secondary Streptavidin-APC (eBioscience). Cells were analyzed on a BD LSR Fortessa (BD Biosciences) and data were processed with FlowJo software (Treestar).
Statistical analysis
All experiments were repeated at least three times unless otherwise noted. For statistical analysis of flow cytometry data, the geometric mean fluorescent intensity (MFI) was measured for individual samples. For experiments where cultured cells were stimulated in vitro, MFI values were normalized across experiments prior to statistical analysis, as previously described (Kearney et al., 2013b). For studies using cells from infected animals, raw MFIs were pooled from at least 2 experiments using 2–4 mice per group prior to analysis, except where otherwise noted. Error bars represent the mean ± SEM. Statistical calculations were performed using GraphPad Prism software. Significance was determined by one-way ANOVA with Tukey's Multiple Comparisons Test, paired two-tailed t-tests, or log-rank Mantel-Cox test as indicated in figure legends. A p-value of <0.05 was considered significant. Approximate p values are indicated as follows: *p = 0.05-0.01; **p = 0.01-0.005; ***p = 0.005-0.0001, ****p < 0.0001.
Results
IFNGR1 staining is selectively restored on monocytes and macrophages from MSGR1 mice
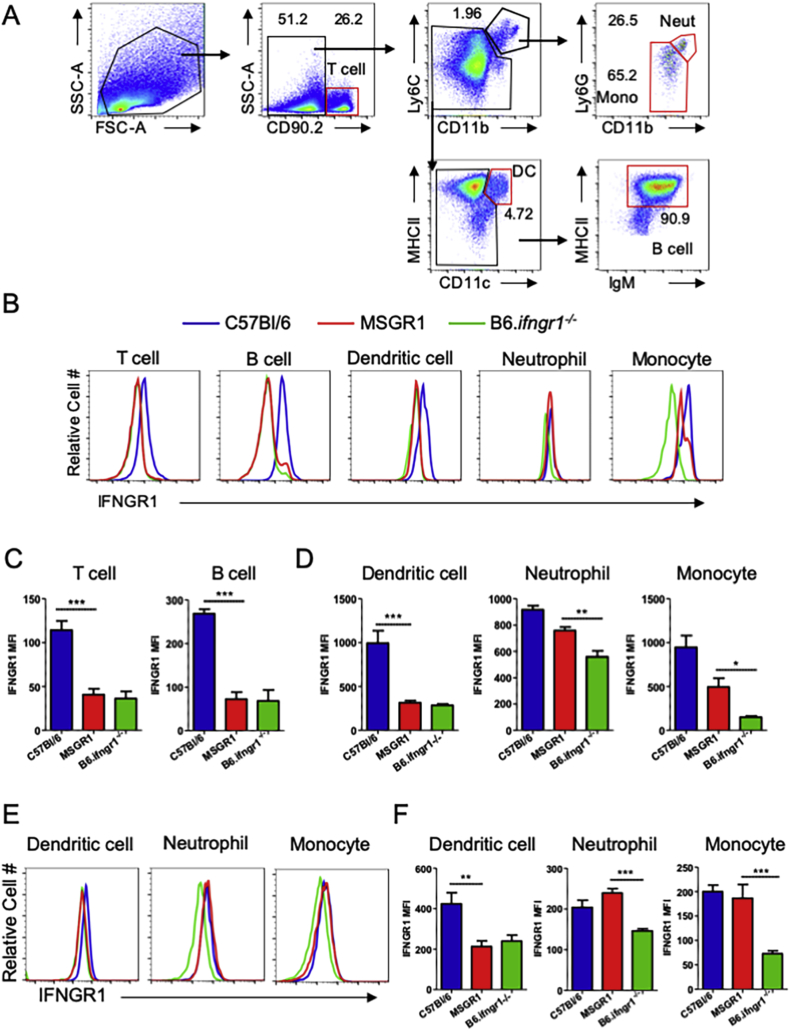
Upon generating fGR1 x B6.ifngr1−/− (MSGR1) mice, we evaluated splenocyte subsets (see Fig. 1A for gating strategy) including inflammatory monocytes (CD11bhi, Ly6Chi, Ly6Glo), neutrophils (CD11bhi, Ly6Chi, Ly6Ghi), dendritic cells (DCs) (CD11chi, MHCIIhi), T cells (Thy1/CD90.2+), and B cells (CD90.2−MHCIIhiIgM+) in naïve mice for IFNGR1 (CD119) expression by flow cytometry. Each of these cell populations stained positive for IFNGR1 in samples from control B6 mice while all cell populations from B6.Ifngr1−/− mice showed negligible IFNGR1 staining (Fig. 1B–D). In the MSGR1 mice, specific IFNGR1 staining was observed on monocytes and neutrophils (Fig. 1B, D), but not on DCs, T cells or B cells (Fig. 1B–D). A comparison of geometric mean fluorescence intensity (MFI) confirmed that the comparable cell surface IFNGR1 staining on monocytes and neutrophils from MSGR1 and B6 mice was significantly higher than that seen on B6.Ifngr1−/− cells (Fig. 1D). Likewise, the lack of staining for IFNGR1 was comparable on DCs, T cells and B cells from MSGR1 and B6.Ifngr1−/− mice and significantly lower than that observed on B6 cells (Fig. 1C and D). After L. monocytogenes infection, splenic neutrophils and inflammatory monocytes, but not DCs, from MSGR1 mice retained IFNGR1 staining that was comparable to cells from infected B6 mice and significantly greater than that observed on B6.Ifngr1−/− cells (Fig. 1E and F).
Fig. 1.
IFNGR1 is selectively expressed on splenic monocytes and neutrophils from MSGR1 mice. A) Gating strategy for different immune cell populations, red gates indicate the final gate for each population. B) Histogram overlays for IFNGR1 expression for each of the cell populations gated in panel A. C) IFNGR1 MFI in splenic lymphocytes and D) indicated myeloid cell populations. Naïve mice were used for staining in A-D. E) Histogram overlays and F) plots of IFNGR1 MFIs for indicated splenic myeloid cell populations at 72 hpi with L. monocytogenes. For all panels, C57Bl/6-blue, MSGR1-red, B6.Ifngr1−/−-green. Geometric MFIs were pooled from at least 2-independent experiments with 3–4 mice per group per experiment. (*two-tailed t-test).
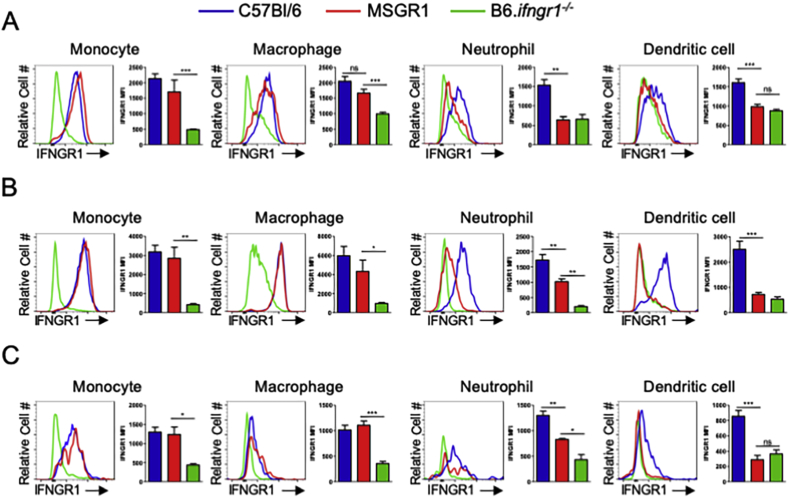
We next determined the expression of IFNGR1 in myeloid cell populations present in other organs including the liver (Fig. 2A), lung (Fig. 2B), and mesenteric lymph nodes (Fig. 2C). Consistent with the spleen, IFNGR1 was only detected in inflammatory monocyte (CD11bhi, Ly6Chi, Ly6Glo) and macrophage (CD11bhi, F480hi) populations isolated from these organs, including alveolar macrophages (gated as CD11c+, F480hi) in the lung (Fig. 2B). DC (CD11chi, MHCIIhi) populations evaluated in these organs also failed the express IFNGR1 in MSGR1 mice. Interestingly, MSGR1 neutrophils (CD11bhi, Ly6Chi, Ly6Ghi) present in the lung (Fig. 2B) and mesenteric lymph nodes (Fig. 2C) have higher IFNGR1 than B6.ifngr1−/− cells, but lower than B6 mice, while MSGR1 neutrophils from the liver are devoid of IFNGR1 (Fig. 2A). Together, these data support the conclusion that fGR1 expression sufficed to selectively restore cell surface IFNGR1 on monocyte/macrophage cell populations in MSGR1 mice.
Fig. 2.
IFNGR1 expression is restricted to macrophage and monocytes. Myeloid cell populations were assessed 72 hpi in the A) liver, B) lung, and C) mesenteric lymph nodes. C57Bl/6-blue, MSGR1-red, B6.Ifngr1−/−-green. Geometric MFIs are from two experiments 3 mice per group per experiment. (*one-way ANOVA).
MSGR1 and WT myeloid cells respond comparably to IFNγ stimulation
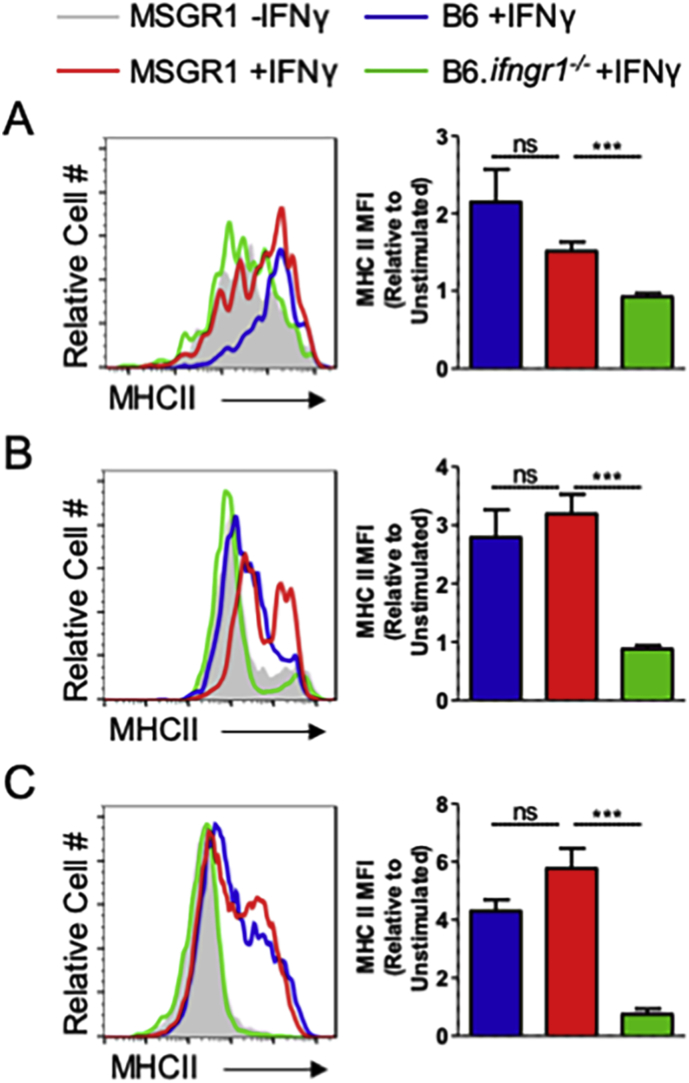
To evaluate the responsiveness of MSGR1 myeloid cells to IFNγ, splenocytes, peritoneal cells, and BMDMs were isolated and cultured with IFNγ (100 U/ml). After 24 h cells were stained and analyzed by flow cytometry. When evaluated for cell surface MHC class II (MHCII) we observed that IFNγ treatment significantly increased staining for MHCII on gated splenic monocytes (Fig. 3A), peritoneal macrophages (Fig. 3B), and BMDMs (Fig. 3C) from both B6 and MSGR1 mice. By comparison and as expected, staining for MHCII was not increased in the IFNγ-treated B6.Ifngr1−/− cells. These results demonstrate that expression of fGR1 in the MSGR1 animals restores responsiveness of macrophages and monocytes to IFNγ. Moreover, there was no significant difference in the responses of myeloid cells from B6 and MSGR1 mice.
Fig. 3.
MSGR1 macrophages respond to IFNγ similar to WT cells. Naïve A) splenic monocytes (CD11bhi, Ly6Chi, Ly6Glo), B) peritoneal macrophages (CD11bhi, F480hi), and C) BMDM were stimulated with 100 U/mL of IFNγ for 24 h then analyzed by flow cytometry for MHCII expression. Geometric MFIs were normalized to unstimulated controls and pooled from 2-independent experiments with 1–3 mice per group per experiment. (*two-tailed t-test).
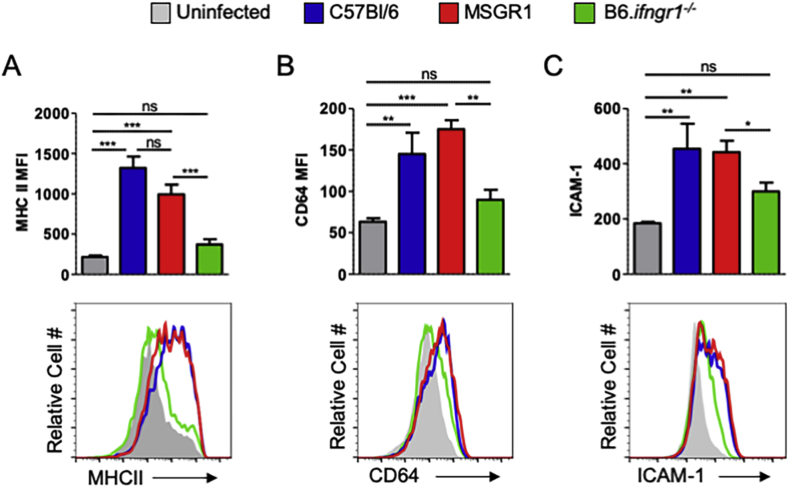
Activation of inflammatory monocytes is comparable following infection of WT B6 and MSGR1 mice
Given evidence that IFNγ responsiveness was restored in cultured myeloid cells from MSGR1 mice, we evaluated upregulation of several cell surface markers indicative of IFNγ responsiveness and “M1” activation on myeloid cells during L. monocytogenes infection. Spleens were harvested from control mice and at 72 hpi, stained, and analyzed by flow cytometry. Gating on inflammatory monocytes, we observed that in B6 and MSGR1 mice the infection significantly increased staining for MHCII (Fig. 4A), CD64 (Fig. 4B) and ICAM-1 (Fig. 4C). In contrast, there was no significant increase in staining for these IFNγ-responsive activation markers on inflammatory monocytes from the infected B6.ifngr1−/− mice. Together with the results from cultured cells in Fig. 3 these findings indicate that myeloid cells from MSGR1 mice are as responsive as B6 cells to both exogenous IFNγ and endogenous IFNγ produced in response to systemic L. monocytogenes infection.
Fig. 4.
L. monocytogenes infection similarly induces IFNγ-dependent activation markers in splenic MSGR1 and WT myeloid cells. Inflammatory monocytes (as gated in Fig. 1A) were analyzed from spleens 72 hpi and assessed for expression of A) MHCII, B) CD64, and C) ICAM-1. Uninfected controls-grey, C57Bl/6-blue, MSGR1-red, B6.Ifngr1−/−-green. Geometric MFI values were pooled from 3-independent experiments with 3–4 mice per group per experiment, (*one-way ANOVA).
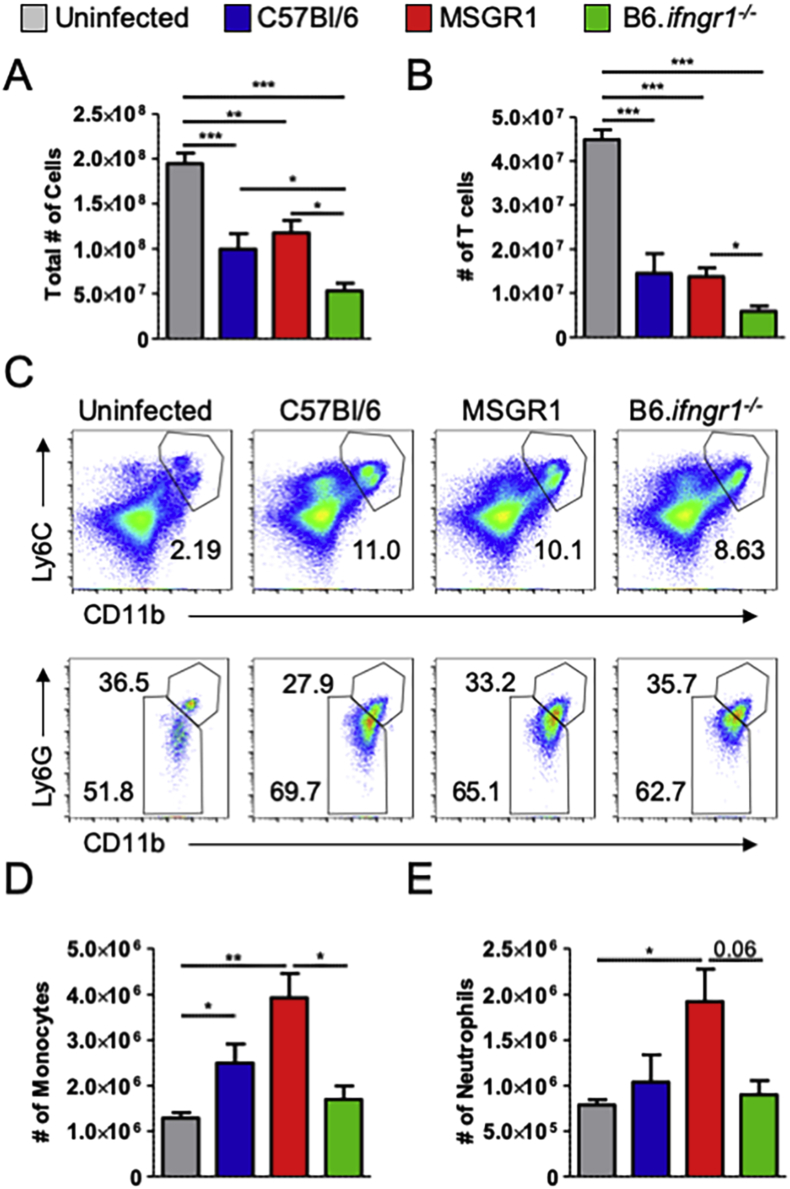
Survival/retention of T cells and inflammatory monocytes is increased in spleens of MSGR1 mice
Adhesion molecules such as ICAM-1 on immune cells interact with LFA-1 on endothelial cells to increase tethering and extravasation of immune cells to sites of infection (Kolaczkowska and Kubes, 2013). Hence we evaluated spleen immune cell numbers in control infected B6, B6.ifngr1−/−, and MSGR1 mice. Systemic L. monocytogenes infection was previously shown to trigger apoptosis of splenic lymphocytes in a manner that is independent of IFNγ (Merrick et al., 1997). Consistent with this the total number of splenocytes was significantly reduced by the infection in all three groups of mice (Fig. 5A). In particular, T cell numbers were reduced by ∼60–75% (Fig. 5B). The reduction in overall splenocyte and T cell numbers were comparable in B6 and MSGR1 mice (Fig. 5A and B). Interestingly, the numbers of total splenocytes and T lymphocytes were reduced further in infected B6.Ifngr1−/− animals. These data suggest myeloid cell responsiveness to IFNGR1 improved the survival of IFNγ-non-responsive T cells in the infected spleens.
Fig. 5.
Myeloid cell-restricted IFNGR1 expression restores inflammatory cell accumulation to L. monocytogenes-infected spleens. Mice were infected with L. monocytogenes and splenocytes harvested 72 hpi. A) Total number of splenocytes and B) T cells (CD90.2+). C) The frequency of inflammatory monocytes (CD11bhi, Ly6Chi, Ly6Glo) and neutrophils (CD11bhi, Ly6Chi, Ly6Ghi). D) Total numbers of monocytes and E) neutrophils. Data were pooled from 3 independent experiments with 3–4 mice per group per experiment. (*one-way ANOVA).
In contrast to the reductions in T cell numbers, the proportion and numbers of splenic inflammatory monocytes and neutrophils increase 2–4 d after systemic L. monocytogenes infection (Eshleman et al., 2017, Serbina et al., 2003). We thus observed a large population of CD11b+Ly6C+ inflammatory myeloid cells in spleens of the infected mice (Fig. 5C). The increase in inflammatory myeloid cells was more pronounced in the B6 and MSGR1 spleens when compared to spleens of infected B6.ifngr1−/− animals. Similar numbers of Ly6G− inflammatory monocytes and Ly6G+ neutrophils were recovered from spleens of the infected MSGR1 and B6 mice (Fig. 5D and E). Fewer monocytes were recovered from the B6.Ifngr1−/− mice, but there were comparable numbers of neutrophils in the B6 and B6.Ifngr1−/− spleens. These data suggest that expression of IFNGR1 on myeloid cells improves survival and/or accumulation of inflammatory monocytes and T cells in infected tissues.
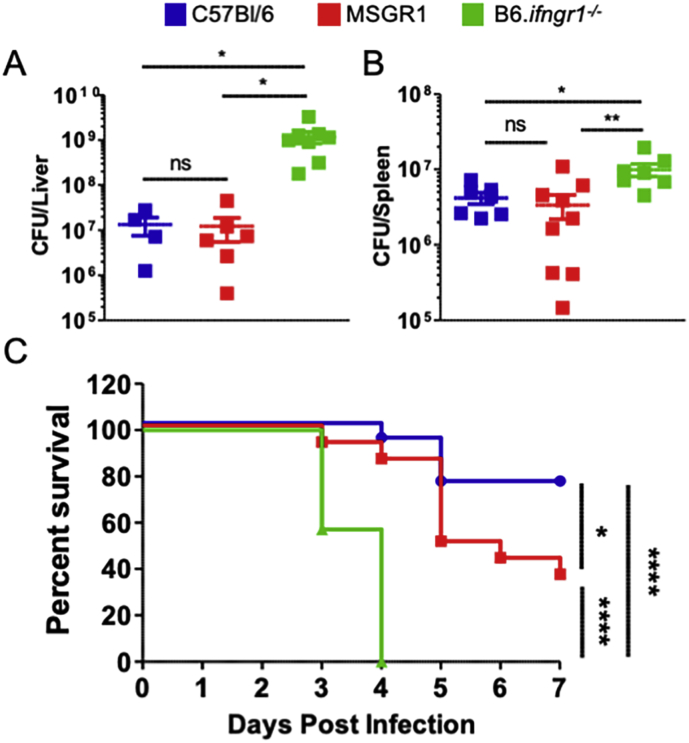
Myeloid IFNGR1 suffices for resistance to systemic L. monocytogenes infection
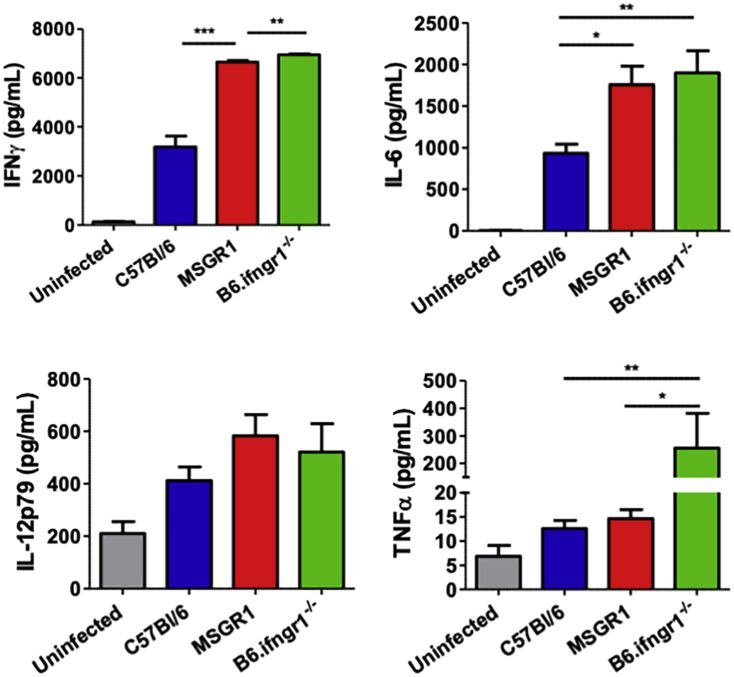
To determine if the restoration of myeloid cell IFNγ responsiveness in MSGR1 mice sufficed to enhance host resistance, we evaluated bacterial burdens from livers and spleens of B6, B6.Ifngr1−/− and MSGR1 mice at 72 h after systemic L. monocytogenes infection. A dose of 104 bacteria (∼1/2 LD50) was used in these experiments. Quantification of bacterial burdens confirmed that universal IFNγ responsiveness of cells significantly reduced bacterial burdens in spleens and livers of B6 mice when compared to B6.Ifngr1−/− mice (Fig. 6A and B). Bacterial burdens in these organs were also lower in tissues from MSGR1 mice versus those from B6.ifngr1−/− mice. Most strikingly, the burdens recovered from both organs were nearly identical in the MSGR1 and WT B6 mice (Fig. 6A and B). These results indicate that at least at this early timepoint after infection the selective restoration of IFNγ responsiveness in myeloid cells enabled mice to limit bacterial replication equivalently as mice in which all cell types respond to IFNγ. Interestingly, serum IFNγ concentrations were similarly elevated at 72 hpi in MSGR1 and B6.Ifngr1−/− mice. This suggests that IFNGR expression by non-myeloid cells (presumably T or NK cells) regulates early serum concentrations of this cytokine (Fig. S1). Data in this figure further show that serum IL-6 was similarly elevated in the infected MSGR1 and B6.Ifngr1−/− mice, whereas IL-12p70 was not affected by IFNGR1 deficiency and TNFα was selectively elevated in the mice that lack IFNGR1 on myeloid cells. These data demonstrate distinct effects of myeloid and non-myeloid cell IFNGR1 on the overall serum cytokine abundance and resistance to the systemic infection.
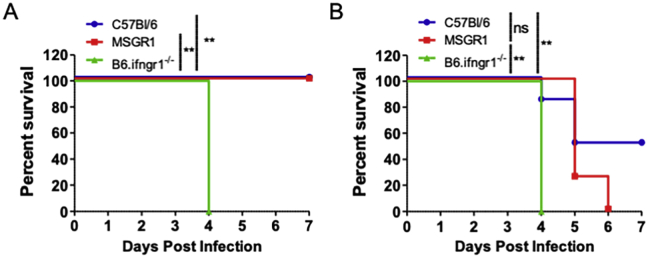
Fig. 6.
Myeloid IFNGR1 suffices for early resistance to L. monocytogenes. Mice were infected with L. monocytogenes and bacterial burdens were determined in the A) liver and B) spleen 72 hpi. CFUs were pooled from 3-independent experiments with 3–4 mice per group per experiment. (*one-way ANOVA). C) Survival of mice infected with L. monocytogenes. Data pooled from 2-independent experiments with 8–9 mice per group per experiment. (*log-rank Mantel-Cox test).
To further explore the longer-term consequences of myeloid cell-selective responsiveness to IFNγ, we evaluated survival of the B6, B6.Ifngr1−/−, and MSGR1 over the course of one week after initiating systemic L. monocytogenes infection. Consistent with the known importance of IFNγ, the infection dose used (1.5 × 104 CFU) caused severe moribundity in all B6.Ifngr1−/− mice by 4 dpi (Fig. 6C). However, most MSGR1 mice remained healthy at 4–5 dpi and over 35% survived until termination of the experiment on day 7. There was nonetheless a difference in survival of the B6 and MSGR1 strains, suggesting an important protective role for IFNγ signaling in one or more cell type that does not express fGR1. Data from animals of both sexes are included in Fig. 6C. When broken down by sex, we found that male MSGR1 mice were much more resistant than females and in fact showed equivalent survival as B6 males (Fig. S2A). By contrast, female B6 and MSGR1 mice showed increased morbidity and reduced survival (Fig. S2B). The difference in survival of B6 and MSGR1 mice was not significant by the test used (p∼0.052) but may suggest non-myeloid cell IFNγ signaling has increased importance in female mice. Nevertheless, both sexes of MSGR1 mice showed significantly improved survival when compared to sex-matched B6.Ifngr1−/− animals completely lacking IFNGR1. Hence, myeloid cell activation by IFNγ is largely responsible for the protective effects of this cytokine at early stages of the bacterial infection.
Discussion
The goal of these studies was to improve understanding of the respective roles for IFNγ stimulation in myeloid versus non-myeloid cells during innate resistance to microbial infection. We employed a well-established systemic infection model with the intracellular bacterial pathogen Listeria monocytogenes to build on results from several prior studies that used this model to demonstrate IFNγ -dependent control of infection (Buchmeier and Schreiber, 1985, Kernbauer et al., 2012, Dighe et al., 1995, Lykens et al., 2010, Eshleman et al., 2017, Portnoy et al., 1989). By developing and utilizing a murine model (MSGR1) in which only myeloid cells could respond to endogenous IFNγ, we were for the first time able to directly test if such activation suffices to limit bacterial replication and increase host survival in the context of a systemic L. monocytogenes infection. Our results confirmed that bacterial burdens were significantly lower in MSGR1 mice than in B6.Ifngr1−/− animals. Further, the MSGR1 mice showed improved survival following a high-dose challenge infection. The finding that bacterial burdens in MSGR1 mice were similar to those in infected wildtype B6 mice is suggestive that IFNγ activation of myeloid cells is the major mechanism by which IFNγ promotes early bacterial containment in wildtype animals. Thus, our findings confirm that myeloid cell activation by IFNγ is important for resistance to infection and further argue that the targeting of these cells is the key mechanism by which IFNγ promotes innate resistance.
Despite the evidence here indicating myeloid cells are a vital and sufficient target for IFNγ-driven protective immunity, the receptor for this cytokine (IFNGR) is expressed ubiquitously and IFNγ-stimulation induces diverse gene expression changes in non-myeloid cell types. What are the benefits of targeting non-myeloid cells? Certainly IFNγ triggers antiviral gene expression in non-myeloid populations and can also mediate direct anti-tumor effects through the induction of apoptosis. In T cells, IFNγ-stimulation is also instrumental for enforcing differentiation of type 1 responses. Our focus here on early events after an acute infection would not be expected to reveal evidence for these non-myeloid cell effects of IFNγ. However, our data here do lend support for another beneficial role associated with IFNγ targeting of non-myeloid cells. Namely, we observed that despite harboring bacterial burdens equivalent to those seen in B6 mice, the overall survival of MSGR1 mice was significantly lower than that seen in B6 animals. Thus, IFNγ targeting of myeloid cells permits killing of bacteria but overall host survival appears to require targeting of a non-myeloid cell type(s) – particularly in female mice. While our experiments did not specifically discern whether IFNγ−responsiveness alters cell death or accumulation, the differences in survival of B6 and MSGR1 mice correlated with a (non-significant) trend towards higher numbers of neutrophils and monocytes in spleens of the infected MSGR1 mice. Hence, IFNγ effects on non-myeloid cells may improve survival by reducing inflammatory cell infiltration that damages host tissues. These data are consistent with previous work showing that IFNGR1 expression by stromal cells is required for production of indoleamine-2,3-dioxygenase (IDO) that reduces neutrophil influx and lung damage during Mycobacterium tuberculosis infection (Desvignes and Ernst, 2009). We did not observe any significant difference in overall T cell numbers between infected MSGR1 and wildtype animals, but another model worth investigating further in future studies is the possibility that the lack of IFNγ signaling to T cells favors over-active T cell responses in the MSGR1 mice. IFNγ signaling to T cells has been shown to promote death of these cells in chronic disease settings (Dalton et al., 2000, Berner et al., 2007). CD4+ T cells thus down regulate IFNGR2 to avoid this IFNγ-driven cell death (Bach et al., 1995, Pernis et al., 1995).
The regulation of cellular IFNγ responsiveness impacts host resistance to a number of infectious agents (Crisler and Lenz, 2018). Many questions remain unanswered regarding the regulation of myeloid cell IFNGR expression and activity. Our lab and others previously demonstrated that responsiveness of myeloid cells to IFNγ can also be attenuated in response to type I IFNs and infections that elicit production of these cytokines (Rayamajhi et al., 2010, Yoshida et al., 1988, Ling et al., 1985). These effects are associated with transcriptional silencing of Ifngr1 (Kearney et al., 2013b). This downregulation of myeloid cell IFNGR1 is conserved in mice and humans, despite evolutionary divergence of the IFN receptors and ligands, and thus likely plays an important role in controlling myeloid cell activity. The fGR1 transgenic mice were developed to overcome the effects of Ifngr1 silencing (Eshleman et al., 2017). Hence, fGR1 expression in myeloid cells from MSGR1 mice might be expected to result in elevated cell surface IFNGR1 (vs. that in B6 cells) in the presence of type I IFNs or other stimuli that normally drive Ifngr1 silencing. However, we failed to observe obvious differences in the intensity of myeloid cell surface IFNGR1 staining in L. monocytogenes infected B6 and MSGR1 mice. Moreover, IFNγ stimulation induced similar expression levels of MHCII on monocytes and macrophages from naïve B6 and MSGR1 mice, and activation marker expression was comparable on monocytes from these mice after L. monocytogenes infection. The similar IFNGR1 staining and IFNγ responsiveness in myeloid cells from infected B6 and MSGR1 mice may be a fortuitous consequence of the low surface expression of the transgenic FLAG-tagged IFNGR1. Regardless, the available data argue that the amounts of IFNGR1 available at the myeloid cell surface and the responsiveness of myeloid cells to IFNγ is comparable in the infected B6 and MSGR1 myeloid cells.
Which myeloid cell populations must respond to IFNγ to support resistance? Results of prior studies using Lyz2 or Cd68 promoters to drive expression of dominant negative IFNGR1 support the conclusion that macrophages, monocytes, and/or neutrophils are key targets of IFNγ during L. monocytogenes and Toxoplasma gondii infection (Dighe et al., 1995, Lykens et al., 2010). A complementary study using Vav-cre or Itgax-cre to delete a floxed Ifngr1 allele implicated hematopoietic cells and suggested an important role for IFNγ signaling to CD8α+ DC (Lee et al., 2013). In our studies, we failed to observe a significant increase in IFNGR1 staining on DCs (Fig. 1, Fig. 2). However, IFNGR1 staining and IFNγ responsiveness was observed in macrophages, neutrophils, and inflammatory monocytes from infected MSGR1 animals. Inflammatory monocytes and neutrophils are recruited rapidly to sites of microbial infection and within 2–3 days are the most abundant splenic myeloid cell populations (V Serbina et al., 2003, Clark et al., 2016, Clark et al., 2018). Further, we recently showed that these cells are heavily associated with L. monocytogenes and that IFNγ enhances their ability to inactivate or resist productive infection by this bacterium (Eshleman et al., 2017). Together with the findings from our studies here, these data suggest innate IFNγ-dependent resistance to systemic L. monocytogenes largely results from the activation of these accumulating inflammatory monocytes and neutrophils.
Over 35 years have passed since IFNγ was identified as a factor that stimulates M1-type antimicrobial macrophage activation (Nathan et al., 1983). In the interim, this cytokine has been firmly-established as a vital mediator of host resistance to pathogenic intracellular microbes. The efforts here establish for the first time that IFNγ responsiveness in myeloid cells suffices to mediate early host protection in the absence of responsiveness in other hematopoietic and non-hematopoietic cell populations. This information furthers our understanding of how IFNγ mediates protection and underscores the importance of further defining how IFNγ responsiveness is regulated in myeloid cells. Future efforts to further dissect the mechanisms by which IFNγ promotes resistance of MSGR1 inflammatory monocytes and neutrophils to L. monocytogenes and other pathogens could reveal new approaches to exploit or mimic the protective effects of IFNγ targeting in these myeloid cell populations for host-directed antimicrobial therapies.
Author contributions
EME and LLL conceived, wrote, and edited the manuscript. EME analyzed the data and prepared figures. NB, DSM, and WJC performed specific experiments and contributed to the curation of data. LLL was responsible for funding acquisition and project administration.
Conflict of interest
The authors declare that there are no conflict of interest.
Acknowledgements
We thank current and past members of the Lenz laboratory for helpful discussion and input into these studies. This work was supported by National Institute of Allergy and Infectious Diseases research grants to LLL: R33AI102264, R21AI103782, R01AI65638, and R21AI140499. EME and WJC received support from NIH training grant T32AI052066. DSM received support from T32AI007405. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crimmu.2020.01.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1.
Fig. S2.
References
- Bach E.A., Szabo S.J., Dighe A.S., Ashkenazi A., Aguet M., Murphy K.M., Schreiber R.D. Ligand-induced autoregulation of IFN-γ receptor β chain expression in T helper cell subsets. Science. 1995;270:1215–1218. doi: 10.1126/science.270.5239.1215. [DOI] [PubMed] [Google Scholar]
- Bach E.A., Aguet M., Schreiber R.D. The IFNγ receptor: a paradigm for cytokine receptor signaling. Annu. Rev. Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- Berner V., Liu H., Zhou Q., Alderson K.L., Sun K., Weiss J.M., Back T.C., Longo D.L., Blazar B.R., Wiltrout R.H., Welniak L.A., Redelman D., Murphy W.J. IFN-γ mediates CD4+ T-cell loss and impairs secondary antitumor responses after successful initial immunotherapy. Nat. Med. 2007;13:354–360. doi: 10.1038/nm1554. [DOI] [PubMed] [Google Scholar]
- Buchmeier N.A., Schreiber R.D. Requirement of endogenous interferon-gamma production for resolution of Listeria monocytogenes infection. Proc. Natl. Acad. Sci. U. S. A. 1985;82:7404–7408. doi: 10.1073/pnas.82.21.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante J., Boisson-Dupuis S., Abel L., Casanova J.L. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-γ immunity. Semin. Immunol. 2014;26:454–470. doi: 10.1016/j.smim.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Filak H.C., Guthrie B.S., Schmidt R.L., Jamieson A., Merkel P., Knight V., Cole C.M., Raulet D.H., Lenz L.L. Bacterial manipulation of NK cell regulatory activity increases susceptibility to Listeria monocytogenes infection. PLoS Pathog. 2016;12:1–21. doi: 10.1371/journal.ppat.1005708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Schmidt R.L., McDermott D.S., Lenz L.L. A Batf3/Nlrp3/IL-18 Axis promotes Natural killer cell IL-10 production during Listeria monocytogenes infection. Cell Rep. 2018;23:2582–2594. doi: 10.1016/j.celrep.2018.04.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers J., Brown H.M., Hwang S., Taylor G.A. Partners in anti-crime: how interferon-inducible GTPases and autophagy proteins team up in cell-intrinsic host defense. Curr. Opin. Immunol. 2018;54:93–101. doi: 10.1016/j.coi.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisler W.J., Lenz L.L. Crosstalk between type I and II interferons in regulation of myeloid cell responses during bacterial infection. Curr. Opin. Immunol. 2018;54:35–41. doi: 10.1016/j.coi.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton D.K., Pitts-Meek S., Keshav S., Figari I.S., Bradley A., Stewart T.A. Multiple defects of immune cell function in mice with disrupted interferon-γ genes. Science. 1993;259:1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- Dalton D.K., Haynes L., Chu C.Q., Swain S.L., Wittmer S. Interferon γ eliminates responding CD4 T cells during mycobacterial infection by inducing apoptosis of activated CD4 T cells. J. Exp. Med. 2000;192:117–122. doi: 10.1084/jem.192.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvignes L., Ernst J.D. Interferon-γ-Responsive Nonhematopoietic cells regulate the immune response to Mycobacterium tuberculosis. Immunity. 2009;31:974–985. doi: 10.1016/j.immuni.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dighe A.S., Campbell D., Hsieh C.S., Clarke S., Greaves D.R., Gordon S., Murphy K.M., Schreiber R.D. Tissue-specific targeting of gytokine unresponsiveness in transgenic mice. Immunity. 1995;3:657–666. doi: 10.1016/1074-7613(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Eshleman E.M., Delgado C., Kearney S.J., Friedman R.S., Lenz L.L. Down regulation of macrophage IFNGR1 exacerbates systemic L. monocytogenes infection. PLoS Pathog. 2017;13:1–22. doi: 10.1371/journal.ppat.1006388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty J.T., Bevant M.J. Specific immunity to listeria monocytogenes in the absence of IFNγ. Immunity. 1995;3:109–117. doi: 10.1016/1074-7613(95)90163-9. [DOI] [PubMed] [Google Scholar]
- Huang S., Hendriks W., Althage A., Hemmi S., Bluethmann H., Kamijo R., Vilček J., Zinkernagel R.M., Aguet M. Immune response in mice that lack the interferon-γ receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- Kearney S., Delgado C., Lenz L.L. Differential effects of type i and II interferons on myeloid cells and resistance to intracellular bacterial infections. Immunol. Res. 2013;55:187–200. doi: 10.1007/s12026-012-8362-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney S.J., Delgado C., Eshleman E.M., Hill K.K., O'Connor B.P., Lenz L.L. Type I IFNs downregulate myeloid cell IFN-γ receptor by inducing recruitment of an early Growth response 3/NGFI-A Binding protein 1 complex that silences ifngr1 transcription. J. Immunol. 2013;191:3384–3392. doi: 10.4049/jimmunol.1203510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernbauer E., Maier V., Stoiber D., Strobl B., Schneckenleithner C., Sexl V., Reichart U., Reizis B., Kalinke U., Jamieson A., Müller M., Decker T. Conditional stat1 ablation reveals the importance of interferon signaling for immunity to Listeria monocytogenes infection. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowska E., Kubes P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 2013;13:159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- Lee S.H., Carrero J.A., Uppaluri R., White J.M., Archambault J.M., Lai K.S., Chan S.R., Sheehan K.C.F., Unanue E.R., Schreiber R.D. Identifying the initiating events of anti- Listeria responses using mice with conditional loss of IFN-γ receptor subunit 1 (IFNGR1) J. Immunol. 2013;191:4223–4234. doi: 10.4049/jimmunol.1300910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.M., Park H.Y., Suh Y.S., Yoon E.H., Kim J., Jang W.H., Lee W.S., Park S.G., Choi I.W., Choi I., Kang S.W., Yun H., Teshima T., Kwon B., Seo S.K. Inhibition of acute lethal pulmonary inflammation by the Ido–AhR pathway. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E5881–E5890. doi: 10.1073/pnas.1615280114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling P.D., Warren M.K., Vogel S.N. Antagonistic effect on interferon-β on the interferon-γ-induced expression of Ia antigen in murine macrophages. J. Immunol. 1985;135:1857–1863. [PubMed] [Google Scholar]
- Lu B., Ebensperger C., Dembic Z., Wang Y., Kvatyuk M., Lu T., Coffman R.L., Pestka S., Rothman P.B. Targeted disruption of the interferon-γ receptor 2 gene results in severe immune defects in mice. Proc. Natl. Acad. Sci. U. S. A. 1998;95:8233–8238. doi: 10.1073/pnas.95.14.8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykens J.E., Terrell C.E., Zoller E.E., Divanovic S., Trompette A., Karp C.L., Aliberti J., Flick M.J., Jordan M.B. Mice with a selective impairment of IFN-γ signaling in macrophage lineage cells demonstrate the critical role of IFN-γ–Activated macrophages for the control of Protozoan parasitic infections in vivo. J. Immunol. 2010;184:877–885. doi: 10.4049/jimmunol.0902346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraz M.A., White J.M., Sheehan K.C.F., Bach E.A., Rodig S.J., Dighe A.S., Kaplan D.H., Riley J.K., Greenlund A.C., Campbell D., Carver-Moore K., DuBois R.N., Clark R., Aguet M., Schreiber R.D. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/S0092-8674(00)81288-X. [DOI] [PubMed] [Google Scholar]
- Merrick J.C., Edelson B.T., Bhardwaj V., Swanson P.E., Unanue E.R. Lymphocyte apoptosis during early phase of Listeria infection in mice. Am. J. Pathol. 1997;151:785–792. [PMC free article] [PubMed] [Google Scholar]
- Mills C.D., Thomas A.C., Lenz L.L., Munder M. Macrophage: SHIP of immunity. Front. Immunol. 2014;5:1–5. doi: 10.3389/fimmu.2014.00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C.F., Murray H.W., Wlebe I.E., Rubin B.Y. Identification of interferon-γ, as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J. Exp. Med. 1983;158:670–689. doi: 10.1084/jem.158.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernis A., Gupta S., Gollob K.J., Garfein E., Coffman R.L., Schindler C., Rothman P. Lack of interferon γ receptor β chain and the prevention of interferon γ signaling in TH1 Cells. Science. 1995;269:245–247. doi: 10.1126/science.7618088. [DOI] [PubMed] [Google Scholar]
- Portnoy D.A., Schreiber R.D., Connelly P., Tilney L.G. Gamma-Interferon limits access of Listeria monocytogenes to the macrophage cytoplasm. J. Exp. Med. 1989;170:2141–2146. doi: 10.1084/jem.170.6.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi M., Humann J., Penheiter K., Andreasen K., Lenz L.L. Induction of IFN-αβ enables Listeria monocytogenes to suppress macrophage activation by IFN-γ. J. Exp. Med. 2010;207:327–337. doi: 10.1084/jem.20091746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K., Hertzog P.J., Ravasi T., Hume D.A. IFN Gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252.Journal. [DOI] [PubMed] [Google Scholar]
- Serbina N.V., Salazar-Mather T.P., Biron C.A., Kuziel W.A., Pamer E.G. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003;19:59–70. doi: 10.1016/S1074-7613(03)00171-7. [DOI] [PubMed] [Google Scholar]
- Serbina V.N., Kuziel W., Flavell R., Akira S., Rollins B., Pamer E.G. Sequential MyD88-independent and -dependent activation of innate immune responses to intracellular bacterial infection. Immunity. 2003;19:891–901. doi: 10.1016/S1074-7613(03)00330-3. [DOI] [PubMed] [Google Scholar]
- Yoshida R., Murray H.W., Nathan C.F. Agonist and antagonist effects of gabaa modulators. J. Exp. Med. 1988;167:1171–1185. doi: 10.1097/00008877-199812001-00159. [DOI] [PMC free article] [PubMed] [Google Scholar]