In this issue of Blood, Green et al identify a specific subset of skeletal cells with a unique role in supporting B lymphopoiesis.1
Hematopoiesis is regulated by cell-intrinsic signals and local cell-extrinsic cues from nonhematopoietic cells, which are collectively known as the bone marrow microenvironment, or niche.2 In earlier studies, niche cells have been defined at the “bulk” level as, for example, osteoprogenitors, endothelial, or leptin receptor–positive cells. However, more recent evidence indicates that niche cells are highly heterogeneous3,4 and that a specific cell subset within a “bulk” niche cell population may serve as a predominant source of hematopoiesis-supporting niche factors.5,6 Most of these studies, however, based their conclusions on transcriptional or proteomic profiling of the nonhematopoietic bone marrow compartment. Extending this knowledge toward a translational application relies on the ability to prospectively isolate these small, superspecialized stromal subsets and validate their role in functional assays.
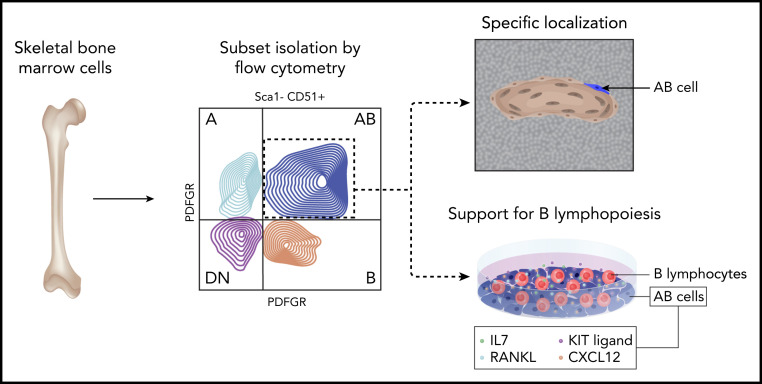
Green et al have undertaken an impressive effort to address this challenge within B lymphopoiesis. Although the role of “bulk” osteolineage cells in B-cell support has been established,7 the molecular identity and localization of the specific skeletal cell subset responsible for this function were not known. In their studies, the authors first used a combination of several skeletal fluorescent reporter lines (Nes:EYFP, Dmp1:EYFP, Osx1:EYFP, Col2a1:EYFP, Prrx1:EYFP) with flow cytometry to dissect the heterogeneity of skeletal cells within mouse bone marrow (see figure). They showed that Sca-1−CD51+ skeletal cells (commonly termed osteoblasts) can be divided into 4 distinct subsets (termed A, B, AB, and double-negative [DN]) based on expression of platelet-derived growth factor receptors α and β (PDFGRα and PDGFRβ). The authors established that although all 4 subsets were found predominantly in the trabecular bone and lining the endosteal surface, they displayed distinct transcriptional signatures and highly diverse behaviors with respect to their ability to form colonies, mineralize, and differentiate into adipocytes in vitro. This suggested that the authors’ prospective isolation strategy revealed a previously unrecognized functional heterogeneity within murine skeletal cells.
Experimental flow for prospective isolation and functional validation of skeletal AB cells in B-lymphopoiesis. Professional illustration by Somersault18:24.
With respect to hematopoiesis-supporting function, the AB subset is of particular interest. Using multiplexed immunofluorescence and spatial analysis, the authors showed that AB cells had a very specific localization to trabecular bone regions, as compared with the A, B, and DN cells and random dots. Notably, the AB subset expressed the highest levels of Flt3l, Cxcl12, kit-ligand, IL7, and tumor necrosis family superfamily member 11 (Tnsf11, encoding RANKL), which have a previously established role in regulating B lymphopoiesis. To functionally test whether this distinct transcriptional profile translates into a superior ability of AB cells to maintain B lymphopoiesis in vitro, the authors cocultured different B-lymphocyte progenitors with A, B, AB, and DN cells, followed by flow cytometry analysis and, for nonadherent pro-B lymphocytes, additional coculture on the B lymphopoiesis–supportive OP9 stromal layer. Importantly, they found that the numbers of pre-B and immunoglobulin M–positive cells were significantly higher following coculture with AB cells, thus functionally validating the unique role of this skeletal cell subset in supporting B lymphopoiesis.
Unexpectedly, the authors found that a higher abundance of AB cells in calvarial bones correlated with a higher number of B-lymphocyte progenitors. This observation raised a possibility that in each individual bone, maintenance of a specific cellular “repertoire” in the bone marrow requires a distinct set of stromal “partners.” To further investigate this intriguing notion, the authors systematically quantified primitive and mature hematopoietic cells across different bones. Surprisingly, they discovered a bone-specific marrow composition that was highly reproducible between the animals: for example, calvarial bones contained more hematopoietic stem and progenitor cells whereas long bones were particularly good at housing erythroid progenitors. Thus, the bone marrow is a highly collaborative “consortium” in which individual “bone factories” uniquely contribute to cumulative blood cell output.
The study by Green et al is another convincing demonstration of functional and transcriptional heterogeneity within the nonhematopoietic bone marrow compartment.8 The niche complexity mirrors that of the hematopoietic system, in which both primitive and mature cell populations have been dissected into more specialized, molecularly distinct fractions.9 We can anticipate that, as these 2 bodies of knowledge expand and merge, a highly accurate “niche cell–hematopoietic cell” pairing in both normal and malignant hematopoiesis will become feasible. This will create new opportunities for selectively manipulating specific hematopoietic subsets and enable design of precision therapies for hematopoietic regeneration and blood cancers.
Supplementary Material
Footnotes
Conflict-of-interest disclosure: The author declares no competing financial interests.
REFERENCES
- 1.Green AC, Tjin G, Lee SC, et al. The characterization of distinct populations of murine skeletal cells that have different roles in B lymphopoiesis. Blood. 2021;138(4):304-317. [DOI] [PubMed] [Google Scholar]
- 2.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505(7483):327-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Severe N, Karabacak NM, Gustafsson K, et al. Stress-induced changes in bone marrow stromal cell populations revealed through single-cell protein expression mapping. Cell Stem Cell. 2019;25(4):570-583.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baryawno N, Przybylski D, Kowalczyk MS, et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell. 2019;177(7):1915-1932.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silberstein L, Goncalves KA, Kharchenko PV, et al. Proximity-based differential single-cell analysis of the niche to identify stem/progenitor cell regulators. Cell Stem Cell. 2016;19(4):530-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crosse EI, Gordon-Keylock S, Rybtsov S, et al. Multi-layered spatial transcriptomics identify secretory factors promoting human hematopoietic stem cell development. Cell Stem Cell. 2020;27(5):822-839.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu JY, Purton LE, Rodda SJ, et al. Osteoblastic regulation of B lymphopoiesis is mediated by Gsalpha-dependent signaling pathways. Proc Natl Acad Sci USA. 2008;105(44):16976-16981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Wu Q, Johnson CB, et al. In situ mapping identifies distinct vascular niches for myelopoiesis. Nature. 2021;590(7846):457-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haas S, Trumpp A, Milsom MD. Causes and consequences of hematopoietic stem cell heterogeneity. Cell Stem Cell. 2018;22(5):627-638. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.