Abstract
Communal bacterial processes require intercellular communication mediated by secretion systems to coordinate appropriate molecular responses. Intercellular communication has not been described previously in mycobacteria. Here we show that the ESX secretion-system family member ESX-4 is essential for conjugal recipient activity in Mycobacterium smegmatis. Transcription of esx4 genes in the recipient requires coculture with a donor strain and a functional ESX-1 apparatus in the recipient. Conversely, mutation of the donor ESX-1 apparatus amplifies the esx4 transcriptional response in the recipient. The effect of ESX-1 on esx4 transcription correlates with conjugal DNA transfer efficiencies. Our data show that intercellular communication via ESX-1 controls the expression of its evolutionary progenitor, ESX-4, to promote conjugation between mycobacteria.
Mycobacteria have elaborate cell envelopes and use ESX secretion systems to transport substrates across their diderm cell structure (1). Mycobacteria encode as many as five paralogous esx loci (2). Each esx locus encodes components for its own membrane transporter, secretion substrates, powering adenosine triphosphatase (ATPase), and other proteins that contribute structural or regulatory functions. Although they are homologous, the ESX conserved components (ecc) encoded by each locus are specific to their individual secretory apparatus (3) and are not functionally redundant, as phenotypes arise from mutations in individual paralogs. ESX secretion activity is required for virulence in pathogenic mycobacteria (4–6). However, the full range of functions of the various esx loci remains unknown.
Mycobacterium smegmatis is a fast-growing saprophytic and nonpathogenic species that has been used as a model for slow-growing pathogenic species (7). M. smegmatis has three esx loci that encode the ESX-1, ESX-3, and ESX-4 secretion apparatuses. In M. smegmatis, the ESX-1 apparatus is required for distributive conjugal transfer (DCT), a distinct gene transfer process that occurs between independent and genetically distinct donor and recipient strains and results in progeny with mosaic genomes (fig. S1) (8–11). Mutations that inactivate ESX-1 in either the donor or recipient strains of M. smegmatis alter conjugal DNA transfer efficiencies. A direct model, in which ESX-1 is proposed to serve as the conduit for DNA traversing from the donor strain into the recipient, is ruled out by the finding that esx1 mutations in the donor strain increase DNA transfer efficiencies up to 100-fold (9). Conversely, ESX-1 mutations in the recipient reduce DNA transfer to undetectable levels (8). Recent findings further show that genes determining donor or recipient mating identity in DCT have been mapped to a cluster of six of the 25 esx1 genes (11). The disparate roles for the various ESX secretion apparatuses, in both slow- and fast-growing mycobacteria, indicate that they mediate secretion of substrates that function in diverse pathways.
The esx4 loci appear to encode only the essential core components of a functional ESX apparatus and lack eccA and espG genes that are functionally important for substrate secretion and processing in other ESX systems (12, 13). This observation and the absence of any identified activity for ESX-4 led to the speculation that it is a vestigial locus (5). Loci encoding ESX-4 secretion systems are also found in other Actinobacteria and Firmicutes, which suggests that ESX-4 is the progenitor ESX (2, 3). In spite of its ancestral status, conserved composition, and broad distribution, a functional role for ESX-4 has not been identified.
Here we report mapping of a transposon insertion in the recipient esx4 locus that abolished conjugation (Fig. 1). The transposon inserted into the recipient ortholog of Msmeg_1536, encoding a dedicated Ftsk-SpoIIIE-ATPase, EccC4 (the subscript “4” indicates the associated esx locus), whose paralogs in other esx loci are required for function of their respective ESX apparatus (1, 14). To ensure that the transposon insertion resulted in a null phenotype, we created a precise deletion of eccC4 in the recipient, and it too was defective for DCT (Fig. 1). To formally rule out any possibility of ESX-4 secretion activity, we created a precise deletion of Msmeg_1535, encoding the ESX-4 transporter, EccD4. The recipient EccD4 mutant was also transfer-defective (Fig. 1). Deletion of eccC4 or eccD4 from the donor strain, however, did not abrogate conjugation (Fig. 1). This recipient-specific requirement for ESX-4 is also seen with esx1 mutants in DCT, although the increase in transfer efficiency seen with ESX-1 donor mutants was not evident with the loss of ESX-4 in the donor.
Fig. 1. Recipient ESX-4 is required for mycobacterial conjugation.
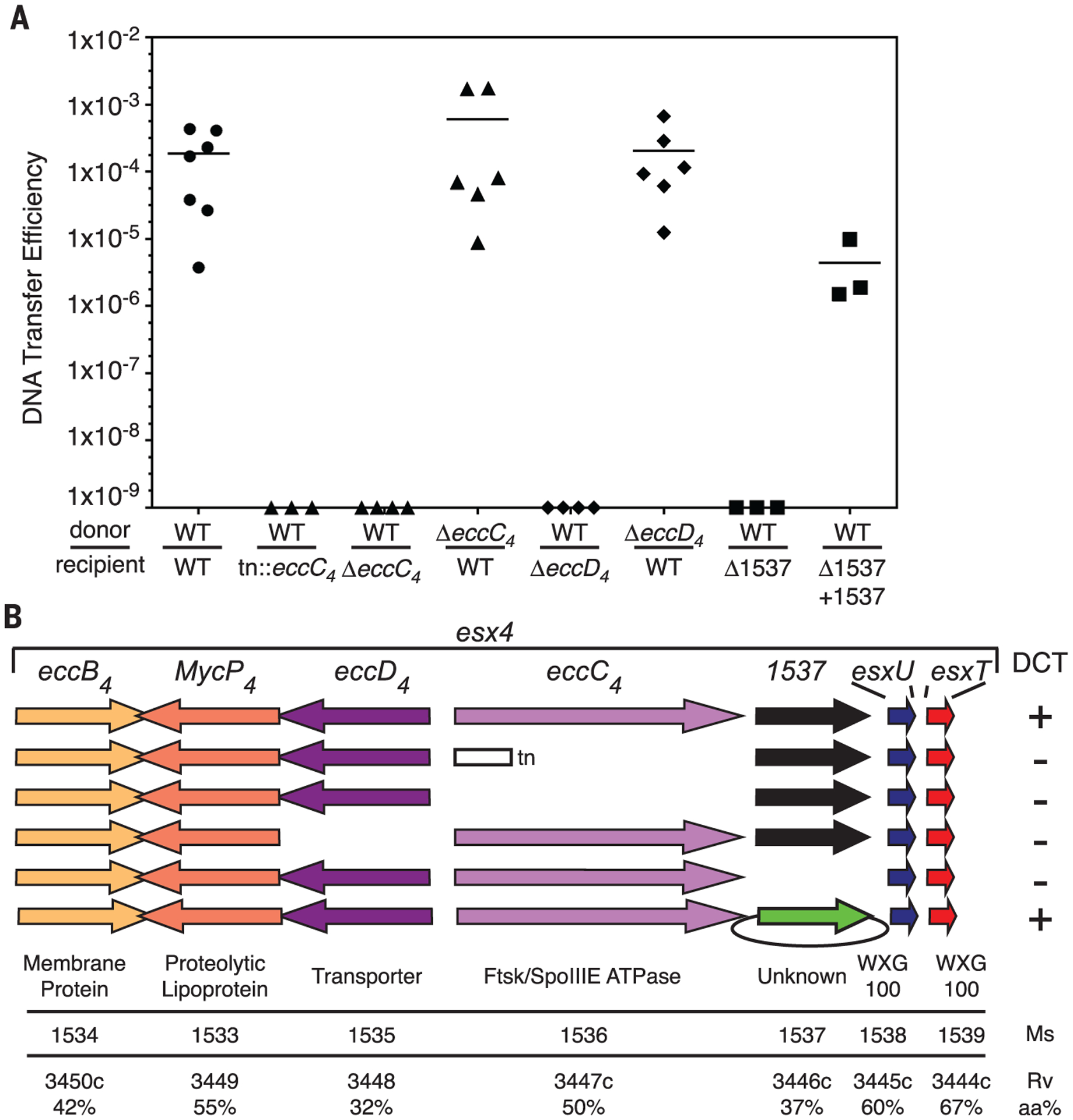
(A) DNA transfer efficiencies (transconjugants per donor cell) show that DCT requires an intact esx4 locus in the recipient strain. Transconjugants were not recovered in matings that lacked a recipient ESX-4 component. Mating-pair genotypes are indicated for donor and recipient, above and below the line, respectively. (B) Schematic showing the conserved gene content and order of esx4 loci. ESX nomenclature of the encoded proteins is shown above the arrows (ecc designates an ESX essential core protein). The gene specific to esx4 is indicated by its M. smegmatis gene name, Msmeg_1537. Recipient locus mutations are summarized by a transposon insertion (tn), a deletion (the absence of an arrow), or a complementing plasmid (green arrow with oval). M. smegmatis (Ms) and M. tuberculosis (Rv) gene numbers and amino acid identities (aa%) and the putative function of the encoded proteins are shown below each gene.
Only one gene is exclusive to esx4 (Fig. 1B). Msmeg_1537 is of unknown function and is conserved in position in all esx4 loci, but homologs are not found in other paralogous esx loci. The conserved presence of this gene within the esx4 locus led us to hypothesize that it is necessary for ESX-4 function. We created a deletion mutant of Msmeg_1537 in the recipient strain and found that this mutant strain was also defective for DCT, producing no transconjugants. Complementation by ectopic expression of Msmeg_1537 restored conjugation (Fig. 1).
Together, these data show that ESX-4 is essential for DCT in the recipient but not the donor and that DCT is a sensitive and reliable assay for ESX-4 function. Thus, ESX-1 and ESX-4 have non-redundant roles in the same biological pathway. ESX-4 function cannot compensate for ESX-1 mutations and vice versa. Although the traditional oriT-based conjugation systems have evolved as a donor function encoded by a specific mobile element for self-propagation, all of the genes that have thus far been identified as necessary for mycobacterial DCT are recipient-specific.
Conjugation is a tightly regulated biological process that requires coordinated gene expression (15–17). We hypothesized that a subset of genes involved in DCT would respond to the presence of the opposite mating type. We used RNA profiling to detect key transcriptional programs that were activated or silenced upon coculture of donor and recipient strains. The many single-nucleotide polymorphisms (SNPs) between donor and recipient genomes act as strain-specific identifiers for the mRNAs and allowed us to perform strain-specific expression profiling (Fig. 2A). cDNA libraries were prepared from mRNA isolated from donor and recipient cells grown under mating conditions in either monoculture or coculture (fig. S2A). After library sequencing, reads were mapped back to each reference genome, and the embedded SNPs were used to identify which strain produced the mapped read. All genes were evaluated for their transcript levels under mating conditions relative to their levels from monoculture to identify the transcripts that responded to the presence of the opposite mating type (fig. S3).
Fig. 2. Coculture with donor induces recipient esx4 transcript levels and requires recipient ESX-1 activity.
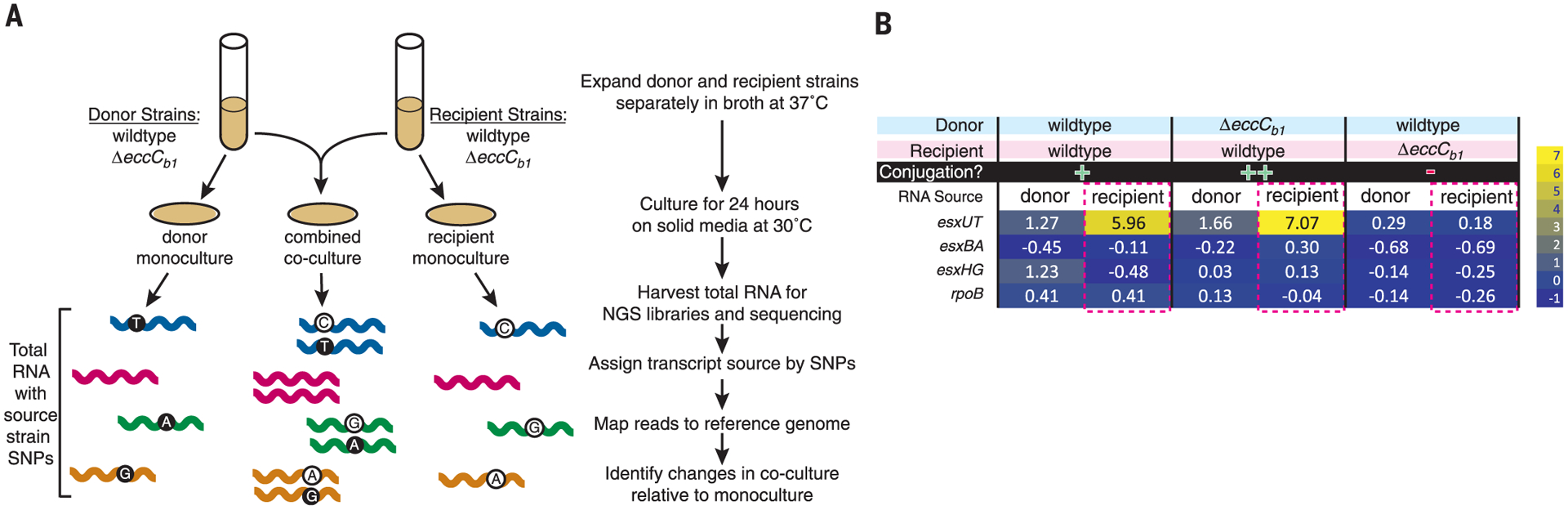
(A) Experimental design for SNP-guided RNA-seq of DCT. The ESX-1mut strains have targeted deletions of eccCb1, encoding the ATPase required for ESX-1 secretion activity and required in the recipient for conjugation. The four strains were grown individually and in mixed cultures of WT × WT, WT × ESX-1mut, or ESX-1mut × WT. Conditions, processing, and analysis are as indicated at right. (B) Heat-map cells of WXG100 genes from esx4 (esxUT), esx1 (esxBA), and esx3 (esxHG), with changes upon coculture shown as log2 insets. For each mating, the donor and recipient genotypes are shown, and the conjugal mating proficiencies are indicated. A housekeeping gene (rpoB) is included as an independent expression control.
During coculture of wild-type (WT) M. smegmatis strains, one of the most highly induced transcripts was from the esx4 locus (Fig. 2B, fig. S2B, and table S1). esx4 transcripts were elevated only in the recipient strain, as can be observed in the heat map of esxUT (Fig. 2B), the tandem gene pair encoding the primary WXG100 secretion substrates of ESX-4. Notably, the paralogous esx1 and esx3 WXG100 genes—esxBA and esxGH, respectively—did not transcriptionally respond in either strain to coculture with their mating partner (Fig. 2B). Therefore, coculture of donor and recipient strains specifically increased transcript levels of a recipient locus required for conjugation. The specific conditions required for the expression of esx4 locus genes (in this case, coculture of a mating pair) may explain why identification of ESX-4 expression and function has been elusive. Overexpression of the sigma factor, SigM, has been associated with increased transcription of esxUT in M. tuberculosis (18, 19), but our M. smegmatis DCT RNA sequencing (RNA-seq) analyses detected no transcriptional change in sigM or conserved genes of its regulon.
We then tested whether ESX-1 function affected transcriptional profiles in DCT mating conditions. The esx1-encoded Ftsk-SpoIIIE-ATPase ortholog of eccC4 (1), eccCb1, was deleted in the donor and recipient strains for use as mating partners with WT strains. Coculture of the ΔeccCb1 donor with a WT recipient induced transcription of the recipient WT esx4 locus (Fig. 2B), as expected for mutated ESX-1 donors, which are known to perform DCT (9). In contrast, the pairing of the WT donor with the ΔeccCb1 recipient failed to induce esx4 transcription in this recipient strain (Fig. 2B). ESX-1 function in the recipient strain is required for DCT (8). Therefore, the recipient esx4 locus transcriptionally responds only in conjugation-proficient mating pairs. Our RNA-seq analyses show that recipient esx4 gene transcripts are induced only upon coculture with the donor strain and that the induction requires a functional recipient ESX-1 secretion system.
We used the highly responsive bicistronic esxUT transcript in quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays to independently validate and quantify our global SNP-specific RNA-seq profiles (fig. S2B). DCT mating assays were repeated, with duplicate samples being processed for conjugal DNA transfer efficiency (Fig. 3A) and qRT-PCR (Fig. 3B). We used a polymorphic BstUI cleavage site to show that the induced esxUT transcript is exclusively from the recipient strain (Fig. 3C). The WT mating pair showed a 30-fold increase of the esxUT transcript relative to parental monocultures (Fig. 3B). The ESX-1 dependence in the recipient was corroborated, as esxUT was increased less than sixfold in the ΔeccCb1 mutant. This level was slightly elevated relative to our RNA-seq data (Fig. 2B), although it is unclear whether this is normal variation in the assay, the rpoB qRT-PCR internal control, or an unknown factor. The accentuated recipient esxUT transcriptional response to the ESX-1 mutant donor strain indicated by RNA-seq data (Fig. 2B) was corroborated by qRT-PCR, which showed a 221-fold induction (Fig. 3B). Thus, coculture induction of ESX-4 corresponds to the reported effects of ESX-1 on conjugation: Recipient ESX-1 mutants do not induce recipient ESX-4 and do not produce transconjugants (8), whereas donor ESX-1 mutants hyperinduced recipient ESX-4 and are hyperconjugative (9). These results indicate that ESX-1 acts upstream of ESX-4 in DCT, directing the induction of esx4 gene expression that results from mating-pair interactions.
Fig. 3. ESX-1 and contact dependence of conjugal communication.
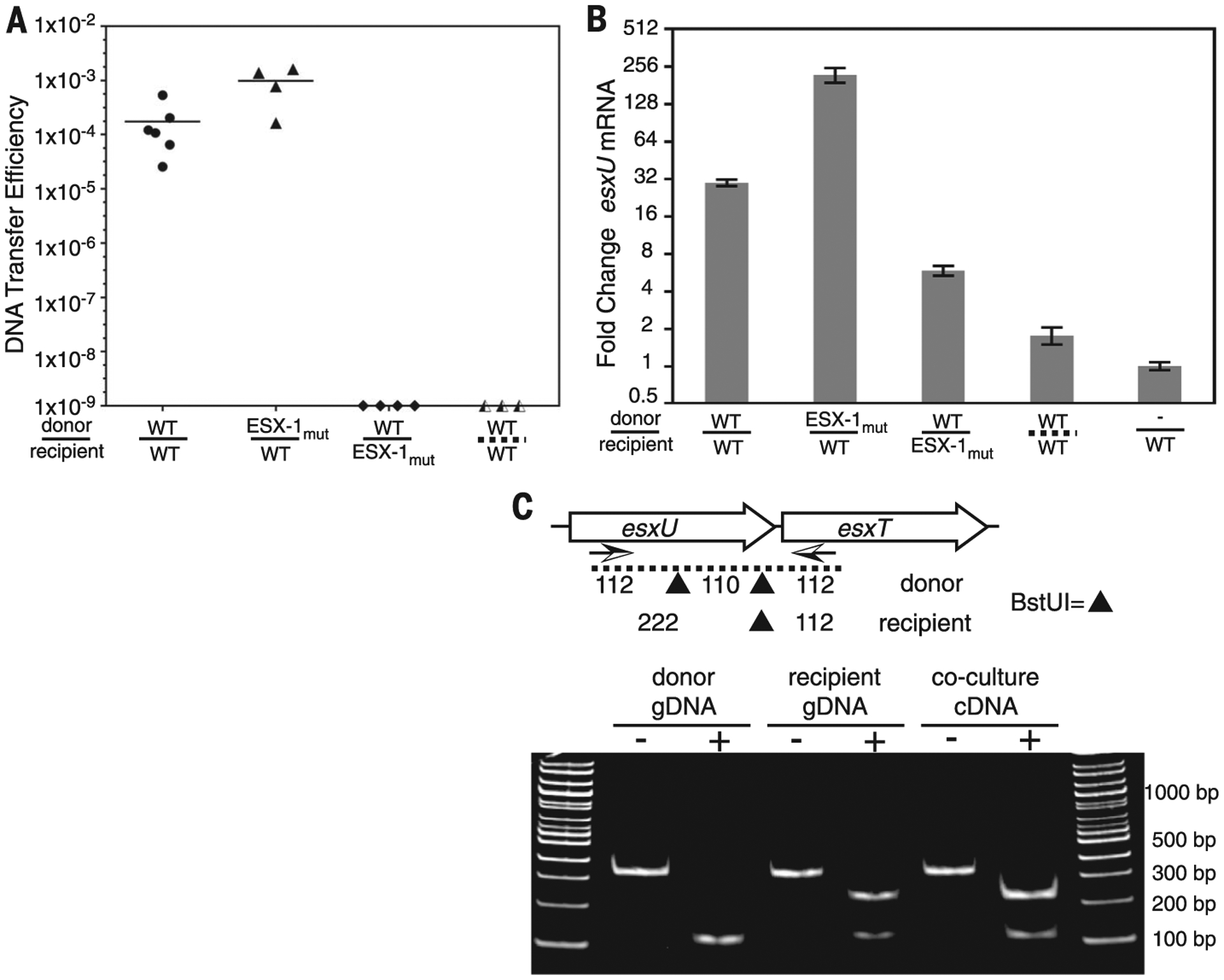
(A) DCT mating efficiencies for conjugal pairs used for RT-PCR. Strains are identified as WT or ESX-1mut (ΔeccCb1) donors (above the line) or recipients (below). The dashed line at bottom right indicates separation of conjugal strains by a porous membrane. (B) qRT-PCR analysis for esxU from cocultures of ESX-1 mutants or physically separated mycobacteria. esxU signals were normalized to rpoB expression. Error bars indicate SD (n = 3). (C) RT-PCR and restriction fragment length polymorphism analysis of BstUI fragments identifies recipient origin of the elevated esxUT transcripts. Arrows below the genetic map indicate the primers used for amplification, and the sites of BstUI cleavage are indicated by triangles. Monoculture genomic DNA (gDNA) controls show the expected parental patterns upon digestion (+) with BstUI. Digestion of coculture-derived cDNA shows a recipient pattern. bp, base pairs.
The transfer of DNA between participating cells during DCT clearly requires physical contact, yet the communication might occur by diffusible signals. We performed coculture under typical DCT mating conditions. However, we separated the donor and recipient strains with a 0.45-μm filter membrane intended to allow transit of soluble signaling molecules but prevent cell-cell contact. qRT-PCR analysis revealed that recipient cells cultured with this porous separation did not show an esxUT transcriptional response to the underlying donor strain (Fig. 3B). These data indicated that direct cell-cell contact is needed to initiate the esx4 transcriptional response. We speculate that ESX-1 secretes cell-surface mating identifiers, receptor proteins, and/or tethering scaffolds. One possibility consistent with our data is that ESX-1 secretes a cell surface receptor in both strains: In the recipient, a yet unknown ligand binding to this receptor initiates a signal cascade that induces ESX-4 necessary for DCT (fig. S4). Thus, disabling ESX-1 function in the donor strain would prevent its receptor secretion from competing for ambient ligand, resulting in hyperactivation of the recipient esx4 locus. Candidates for potential involvement are encoded by the subset of esx1 genes that constitute the mating identity locus (mid) (11).
Strain-specific RNA-seq during coculture also identified expression changes in genes that are not involved in DCT. Thus, coculture responses between these mycobacterial strains may not be limited to conjugation. Some of the largest transcriptional changes were dependent on ESX-1. The concept that ESX systems function in intercellular communication among mycobacteria has implications for other mycobacterial species and for intercellular communication in infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Wade for valuable discussions and technical assistance and the Wadsworth Center Core Facilities for Media and Bioinformatics. Primary data can be found in the supplementary materials, and RNA-seq data sets have been deposited at the National Center for Biotechnology Information with accession number SRP080861. This work was supported by National Institute of Allergy and Infectious Diseases grants R01AI097191 and R21AI07258 (to K.M.D. and T.A.G.). R.R.C. generated strains, performed conjugation, and carried out RT-PCR experiments; N.B. identified the initial transposon mutant, constructed mutants, and performed conjugation assays; C.S. prepared RNA-seq libraries; P.L. analyzed RNA-seq data; K.M.D. contributed study design and data analyses; T.A.G. contributed study design, targeted mutations, and data analyses; and K.M.D. and T.A.G. wrote the manuscript.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/354/6310/347/suppl/DC1
Material and Methods
Figs. S1 to S4
Tables S1 and S2
References (20–22)
REFERENCES AND NOTES
- 1.Abdallah AM et al. , Nat. Rev. Microbiol 5, 883–891 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Gey van Pittius NC et al. , Genome Biol 2, research0044.1 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bitter W et al. , PLOS Pathog 5, e1000507 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simeone R, Bottai D, Brosch R, Curr. Opin. Microbiol 12, 4–10 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Stoop EJ, Bitter W, van der Sar AM, Trends Microbiol 20, 477–484 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Tan T, Lee WL, Alexander DC, Grinstein S, Liu J, Cell. Microbiol 8, 1417–1429 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Shiloh MU, DiGiuseppe Champion PA, Curr. Opin. Microbiol 13, 86–92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coros A, Callahan B, Battaglioli E, Derbyshire KM, Mol. Microbiol 69, 794–808 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flint JL, Kowalski JC, Karnati PK, Derbyshire KM, Proc. Natl. Acad. Sci. U.S.A 101, 12598–12603(2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsons LM, Jankowski CS, Derbyshire KM, Mol. Microbiol 28, 571–582 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Gray TA, Krywy JA, Harold J, Palumbo MJ, Derbyshire KM, PLOS Biol 11, e1001602 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiGiuseppe Champion PA, Champion MM, Manzanillo P, Cox JS, Mol. Microbiol 73, 950–962 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houben EN, Korotkov KV, Bitter W, Biochim. Biophys. Acta 1843, 1707–1716 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Converse SE, Cox JS, J. Bacteriol 187, 1238–1245 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koraimann G, Wagner MA, Front. Cell. Infect. Microbiol 4, 54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh PK, Meijer WJ, FEMS Microbiol. Lett 358, 119–128 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Dunny GM, Berntsson RP, J. Bacteriol 198, 1556–1562 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agarwal N, Woolwine SC, Tyagi S, Bishai WR, Infect. Immun 75, 452–461 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raman S et al. , J. Bacteriol 188, 8460–8468 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


