Abstract
Background
Stomata are adjustable pores on the surface of plant shoots for efficient gas exchange and water control. The presence of stomata is essential for plant growth and survival, and the evolution of stomata is considered as a key developmental innovation of the land plants, allowing colonization on land from aquatic environments some 450 million years ago. In the past two decades, molecular genetic studies using the model plant Arabidopsis thaliana identified key genes and signalling modules that regulate stomatal development: master regulatory transcription factors that orchestrate cell state transitions and peptide–receptor signal transduction pathways, which, together, enforce proper patterning of stomata within the epidermis. Studies in diverse plant species, ranging from bryophytes to angiosperm grasses, have begun to unravel the conservation and uniqueness of the core modules in stomatal development.
Scope
Here, I review the mechanisms of stomatal development in the context of epidermal tissue patterning. First, I introduce the core regulatory mechanisms of stomatal patterning and differentiation in the model species A. thaliana. Subsequently, experimental evidence is presented supporting the idea that different cell types within the leaf epidermis, namely stomata, hydathodes pores, pavement cells and trichomes, either share developmental origins or mutually influence each other’s gene regulatory circuits during development. Emphasis is placed on extrinsic and intrinsic signals regulating the balance between stomata and pavement cells, specifically by controlling the fate of stomatal-lineage ground cells (SLGCs) to remain within the stomatal cell lineage or differentiate into pavement cells. Finally, I discuss the influence of intertissue layer communication between the epidermis and underlying mesophyll/vascular tissues on stomatal differentiation. Understanding the dynamic behaviours of stomatal precursor cells and their differentiation in the broader context of tissue and organ development may help design plants tailored for optimal growth and productivity in specific agricultural applications and a changing environment.
Keywords: Stomata, peptide hormones, auxin, receptor kinase, signal transduction, bHLH proteins, meristemoid, stomatal-lineage ground cell, guard cell, pavement cell, trichome, hydathode water pore, hypocotyl epidermal cell files, root hair patterning
Introduction
Stomata, turgor-driven cellular valves on the plant aerial epidermis, serve as an interface between a plant and its environment. By dynamically adjusting stomatal pore apertures, stomata facilitate gas exchange for photosynthesis while minimizing water loss via transpiration. The acquisition of both stomata and epidermal pavement cells that protect plants from dry atmospheric conditions are considered key developmental innovations that enabled green plants to conquer the land some 450 million years ago (Peterson et al., 2010; Chater et al., 2017). Stomata exist in land plants including basal land plants, with the exception of liverworts (Chater et al., 2016; Bowman et al., 2017). Increasing evidence suggests that the core genes and signalling mechanisms governing stomatal development, which were discovered in the model angiosperm Arabidopsis thaliana, are conserved across the land plant species, with unique variations signifying diversity in morphology and function of stomatal complexes (Chater et al., 2017; Nunes et al., 2020). A corollary to this hypothesis is a striking finding from the genome of an aquatic grass plant, Eelgrass Zostera marina, which revealed that the loss of genes regulating stomatal development and epicuticular wax synthesis signifies the adaptation of the land-based angiosperm to the sea (Olsen et al., 2016). The evolutionary history of land plants and their adaptation to aquatic environments imply that development of stomata and epidermal pavement cells is coupled to generate functional epidermis. However, studies of stomatal development, including the influence of environmental and hormonal signals, have so far been focused within the context of cell lineages, rather than at a tissue level (Lau and Bergmann, 2012; Qi et al., 2020).
In addition to stomata, the leaf epidermis of arabidopsis differentiates three additional primary cell types, epidermal pavement cells, trichomes and hydathode water pores, each responsible for specific functions (Fig. 1). Interlocking jigsaw puzzle-shaped pavement cells not only provide protection from numerous environmental insults, but also guide plant growth through localized elongation and expansion (Javelle et al., 2011). Trichomes are branched, rigid hair-like structures that provide protection against herbivores (Schellmann and Hulskamp, 2005). Lastly, hydathode water pores located at the leaf margin function as a site of guttation to release water pressure (Cerutti et al., 2019). We are beginning to understand how differentiation programmes of these different cell types are mutually influenced and co-ordinated at the level of gene regulatory circuits and cell–cell signalling. In this review, I first describe the basics of arabidopsis stomatal development, and explain the core genes and regulators of stomatal differentiation and patterning. In addition, some variations on a theme of stomatal development in distant plant species are introduced. Subsequently, I describe the core regulatory mechanisms underpinning the development of hydathode water pores, trichomes and pavement cells, and discuss how the core molecular mechanisms of stomatal development are integrated with other epidermal cell differentiation programmes. Specific emphasis will be given to stomatal-lineage ground cells (SLGCs), bipotent cells that can produce a stoma or pavement cell. Lastly, further roles of intertissue layer communication will be discussed. Through synthesis, I aim to bridge the gap in our knowledge of stomatal development in the broader context of tissues and organs, toward gaining a holistic view of its importance for plant growth and sustenance.
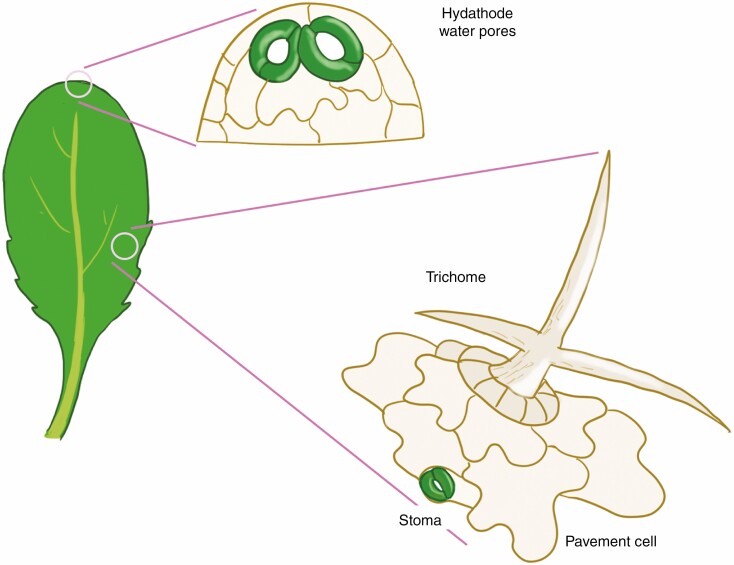
Fig. 1.
Epidermal cell types in arabidopsis rosette leaf. A schematic diagram of an arabidopsis rosette leaf (left). Hydathode water pores are found at/near the tip of the leaf edges and are typically paired large stomata that remain open (top right). Epidermal pavement cells, trichomes and stomata (right) are found on the leaf blade throughout. Trichomes are only found on the adaxial side of the leaf blade, whereas more stomata are located on the abaxial side. Neither stomata nor trichomes differentiate from the epidermis immediately above the midvein.
THE CORE MECHANISMS OF STOMATAL DEVELOPMENT
Stomatal cell lineages
Stomatal development has been extensively studied in arabidopsis, where a series of cell divisions and cell state transitions occur in a stereotypical manner to produce anisocytic stomatal complexes consisting of a two-cell stoma surrounded by up to three non-stomatal epidermal pavement cells (Nadeau and Sack, 2002a) (Fig. 2). Stomatal development initiates within a sub-population of protodermal cells, adopting an identity termed meristemoid mother cells (MMCs), which then initiate asymmetric division (asymmetric ‘entry’ division) that produces two daughter cells with different characteristics: a meristemoid cell and an SLGC (Fig. 2). The stem-like meristemoid reiterates asymmetric divisions (asymmetric ‘amplifying’ divisions), renewing itself while amplifying SLGCs. The SLGC can then re-adopt the MMC identity to give rise to a satellite meristemoid or, alternatively, terminally differentiate into a pavement cell (Fig. 2). The meristemoid eventually differentiates into a round guard mother cell (GMC), which subsequently divides symmetrically and terminally differentiates into a pair of guard cells enclosing a pore (Bergmann and Sack, 2007; Lau and Bergmann, 2012; Pillitteri and Torii, 2012; Herrmann and Torii, 2021) (Fig. 2).
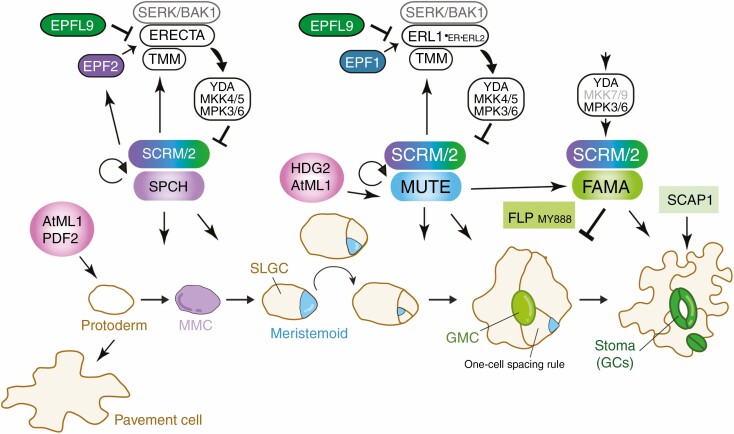
Fig. 2.
Core regulatory mechanism of stomatal development. In the typical dicot plant, arabidopsis, protodermal fate is specified by two HD-ZIP IV transcription factors, AtML1 and PDF2. Stomatal development initiates on a sub-set of protodermal cells termed meristemoid mother cells (MMCs: lilac) with high accumulation of the bHLH heterodimer module of SPCH and SCRMs (SPCH–SCRMs). An MMC undergoes an asymmetric entry division and gives rise to two daughter cells, meristemoid (cyan) and stomatal-lineage ground cell (SLGC: ivory). The meristemoids reiterate asymmetric amplifying division, which is maintained by high SPCH activity. A switch from the proliferation to differentiation state is regulated by the MUTE–SCRM module. MUTE promotes differentiation of the guard mother cell (GMC), and orchestrates the single symmetric cell division. A FAMA–SCRM bHLH module and Myb proteins FLP and MYB88 restrict the single symmetric cell division. FAMA and SCAP1 promote guard cell (GC) morphogenesis. Peptide–receptor kinase signalling pathways mediated by the EPF2–ERECTA–TMM and EPF1–ERL1–TMM ligand–receptor module restricts the entry into stomatal cell lineages and enforces proper asymmetric spacing division, respectively. EPFL9 (Stomagen) antagonizes the signalling by competitively binding to the receptors. The activated receptor associates with SERK/BAK1 co-receptor, and the signal is transduced via an MAPK cascade consisting of YODA–MKK4/5–MPK3/6. MPK3/6 is recruited by SCRM to downregulate SPCH and probably MUTE. The stomatal bHLH proteins directly regulate the cell–cell signalling components, and this negative feedback loop maintains proper cell fate specification.
Transcription factor control of stomatal differentiation
Key molecular–genetic studies over the past two decades have unravelled core regulatory mechanisms of stomatal development (Bergmann and Sack, 2007; Lau and Bergmann, 2012; Pillitteri and Torii, 2012; Herrmann and Torii, 2021). Cell state transitions leading to stomatal differentiation are directed by sequential actions of three basic–helix–loop (bHLH) proteins, SPEECHLESS (SPCH), MUTE and FAMA, as well as their heterodimeric partners, SCREAM (SCRM; also known as ICE1) and SCRM2, here referred to as SCRMs (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007; Kanaoka et al., 2008) (Fig. 2). The activity of SPCH–SCRMs in the nucleus drives the initiation of stomatal cell lineages and promotes asymmetric amplifying divisions of meristemoids and MMCs; spch or scrm scrm2 double mutants confer an epidermis solely composed of (non-stomatal) pavement cells. Next, MUTE–SCRMs orchestrate the cell state switch in meristemoids from proliferation to differentiation into GMCs (Pillitteri et al., 2007; Kanaoka et al., 2008) (Fig. 2). FAMA–SCRMs regulate the symmetric division of GMCs and trigger their terminal differentiation into guard cells. In concert with FAMA, the Myb protein FOUR LIPS (FLP) and its redundant paralogue MYB88 restrict the symmetric division of the GMC (Ohashi-Ito and Bergmann, 2006; Kanaoka et al., 2008) (Fig. 2). Recent experimental evidence and modelling studies have shown that MUTE orchestrates a single symmetric division of GMCs by directly inducing the expression of cell cycle regulators to initiate division, as well as FAMA and FLP, which directly repress the cell cycle regulators to end the division cycle (Lai et al., 2005). Additional transcription factors, including a C2H2 Dof transcription factor STOMATA CARPENTER1 (SCAP1), are required for proper guard cell maturation (Negi et al., 2013) (Fig. 2).
Cell–cell signalling enforcing stomatal patterning
Stomatal mechanics rely on rapid water and ion movement between stomatal guard cells and neighbouring non-stomatal epidermal cells. Stomata are patterned in such a way that two stomata will not form immediately adjacent to each other (Nadeau and Sack, 2002a). This probably reflects the underlying physiology for optimal solute exchange between the stomatal guard cells and neighbouring non-stomatal cells (such as subsidiary cells) for efficient stomatal movement as well as for effective water vapour and carbon dioxide diffusion through stomata (Roth-Nebelsick, 2007). This so-called ‘one-cell spacing rule’ is enforced by local peptide–receptor kinase signalling. In arabidopsis, a family of secreted cysteine-rich peptides EPIDERMAL PATTERNING FACTORS (EPF)/EPF-LIKES (EPFL) act as paracrine signals to enforce proper stomatal patterning (Hara et al., 2007; Hunt and Gray, 2009; Rychel et al., 2010) (Fig. 2). Initial spatial patterning of stomatal progenitor cells is controlled by EPF2, which is secreted from MMCs and inhibits neighbouring cells from adopting stomatal lineage identity. EPF2 peptide is primarily perceived by ERECTA-family receptor kinases and its partner TOO MANY MOUTHS (TMMs) (Nadeau and Sack, 2002b; Shpak et al., 2005; Hara et al., 2009; Hunt and Gray, 2009; Lee et al., 2012, 2015). Upon ligand perception, the ERECTA–TMM receptor complex with co-receptor SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES (SERKs)/BAK1 trigger activation of a mitogen-activated protein kinase (MAPK) signalling cascade composed of YODA MAPKKK, MKK4/5 MAPKKs and MPK3/6 MAPKs (Bergmann et al., 2004; Wang et al., 2007; Bemis et al., 2013; Meng et al., 2015) (Fig. 2). EPF2 is directly induced by SPCH–SCRMs, and in turn EPF2 signalling leads to inhibition of SPCH–SCRMs via MPK3/6-dependent phosphorylation and degradation (Lampard et al., 2008; Lau et al., 2014; Horst et al., 2015; Putarjunan et al., 2019) (Fig. 2). Differentiation of meristemoids as well as the enforcement of the ‘one-cell spacing rule’ are mediated by EPF1, which is primarily perceived by ERECTA-LIKE1 (ERL1), forming a receptor complex with TMM and SERKs/BAK1 (Lee et al., 2012; Meng et al., 2015). EPF1 signalling inhibits MUTE activity, while MUTE–SCRM modules, in turn, directly induce ERL1 and TMM gene expression, thereby forming a negative feedback loop to balance signal strengths (Qi et al., 2017; Han et al., 2018) (Fig. 2).
Conservation of core regulators of stomatal development
Thus far, research in other plant species strongly supports the hypothesis that the core mechanisms of stomatal development are conserved throughout land plants that possess stomata. For example, recent studies showed that in the moss Physcomitrella patens, which generates a single row of stomata on its sporophyte (Peterson et al., 2010), ancient SPCH-MUTE-FAMA (PpSMF) bHLH genes as well as EPF1-TMM (and probably ERECTA) peptide receptor genes (PpEPF1, PpERECTA and PpTMM) regulate stomatal development and patterning (Chater et al., 2016; Caine et al., 2020). Likewise, the differentiation of highly derived and efficient stomatal complexes in monocot cereal species is regulated by the same conserved set of stomatal bHLH proteins and cell–cell signalling components, with some acquiring unique novel functions (Liu et al., 2009; Raissig et al., 2016, 2017). The most striking example is Brachypodium MUTE (BdMUTE), which acquired a novel function to promote subsidiary cell development (Raissig et al., 2017). Unlike BdMUTE, the loss-of-function mutation in MUTE (OsMUTE) confers excessive asymmetric division of a meristemoid and its eventual arrest, a phenotype equivalent to the arabidopsis mute mutant (Wu et al., 2019). Instead of OsMUTE, OsFAMA appears to influence subsidiary cell division (Pillitteri et al., 2007; Wu et al., 2019), emphasizing that the precise functions of stomatal bHLHs may be co-opted differently amongst the grass species. Among the orthologues of cell–cell signalling components, EPFs and YODA MAPKKK exhibit clear functions in grass family stomatal development, including in wheat, barley and Brachypodium (Hughes et al., 2017; Abrash et al., 2018; Caine et al., 2019; Dunn et al., 2019). On the other hand, specific roles for the peptide receptors, including the ERECTA family, TMM and SERKs/BAK1, in stomatal patterning have not been demonstrated in grass species. Excellent review articles on grass stomatal development are available for further reading (Hepworth et al., 2018; McKown and Bergmann, 2020; Nunes et al., 2020).
STOMATAL DEVELOPMENT IN THE CONTEXT OF EPIDERMAL PATTERNING
Role of protoderms in stomatal development
Whereas the core mechanism of stomatal development has been extensively studied, stomatal development in leaves occurs in the context of epidermal tissues and, more broadly, the whole leaf. In addition to stomata, specialized cell types in the leaf epidermis, namely epidermal pavement cells, trichomes and hydathode water pores, all originate from protodermal cells. The protodermal fate is specified by the family of homeodomain-leucine zipper IV (HD-ZIP IV) proteins, AtML1 and its paralogue PDF2 (Lu et al., 1996; Abe et al., 2003) (Fig. 2). These transcription factors are sufficient to direct stomata and trichome differentiation in leaf cells of non-protodermal origin. For instance, ectopic AtML1 overexpression triggers the differentiation of aberrant trichomes and stomata in internal mesophyll tissues (Takada et al., 2013). During stomatal development, the HD-ZIP IV protein HDG2 (and, to a lesser extent, AtML1) is expressed in the meristemoids and promotes stomatal differentiation via upregulating MUTE expression (Fig. 2). Consistently, ectopic overexpression of HDG2 confers ectopic stomata (but not ectopic trichomes) in the internal mesophyll tissues (Peterson et al., 2013). It is still not clear how HD-ZIP IV proteins trigger the initiation of different leaf epidermal cell types or whether the regulatory programme driving the differentiation of a single epidermal cell type interferes (or co-operates) with any other. Recent studies are shedding light on the relationships between the core stomatal development mechanisms and those of other epidermal cell types.
Stomata and hydathode water pores
The hydathode is a special organ on the leaf tips responsible for guttation – releasing water droplets from the vasculature to reduce hydrostatic pressure (Cerutti et al., 2019). It is composed of vasculature, small parenchyma cells called epithum, and water pores from which the water droplets (dew drops) are released. The water pores are considered to be modified stomata: closely resembling stomata but with some notable differences. Unlike regular stomata, which rarely form next to another stoma (due to the ‘one-cell spacing rule), the water pores in arabidopsis are composed of a pair of large stomata in contact, and which remain constitutively open (Pillitteri et al., 2008) (Fig. 2). The water pores in the grass species, such as Brachypodium, rice and maize, are aligned along the leaf tip, tend to remain open and seemingly lack proper subsidiary cells (Jauneau et al., 2020).
Although the specific molecular mechanisms generating hydathode water pores have not yet been explored, at least one of the canonical stomatal transcription factor genes, MUTE, is required for water pore differentiation, since water pores are absent in mute cotyledons and leaves (Pillitteri et al., 2008). It has been proposed that hydathode differentiation occurs at the site of auxin synthesis during leaf morphogenesis (Aloni, 2001). Consistent with this hypothesis, the initial expression of the auxin response reporter DR5::GFP is visible at the leaf tip where initial MUTE expression is also detected. Whether the differentiation of water pores requires high levels of auxin is an interesting question. Whereas mute cotyledons and leaves lack water pores, they still exhibit high activity of the auxin reporter DR5:GFP at the leaf tips. Likewise, treatment with the auxin transport inhibitor N-1-naphthylphthalamic acid (NPA) did not disrupt MUTE promoter activity at the leaf tip (Pillitteri et al., 2008). Thus, the tip-localized auxin may act independently from MUTE or, alternatively, auxin promotes the water pore initiation before MUTE acts (e.g. via SPCH). Very recently, RNA-seq analysis of Arabidopsis hydathode regions was performed in an attempt to identify genes specifically expressed in hydathodes, which include several auxin biosynthesis and auxin-related genes (Yagi et al., 2020). Such an analysis may reveal genes and pathways uniquely associated with water pore differentiation and additional overlapping features with stomata development.
Patterning of stomata and trichomes
Yet another specialized cell type on the leaf epidermis is the trichome, which serves as a mechanical protection against herbivores (Mauricio and Rausher, 1997) among other functions, such as production of essential oils and other metabolic products in glandular trichomes (Glas et al., 2012; Huchelmann et al., 2017). In arabidopsis, a trichome is an extremely enlarged, polarized unicellular non-glandular structure formed on the adaxial side of the leaf (Schellmann and Hulskamp, 2005). Like stomata, leaf trichomes are formed de novo in an evenly distributed manner, and clonal analysis has shown that cell–cell communication also specifies the trichome initials. However, the 2-D spatial patterns of stomata and trichome distribution are governed by distinct molecular and signalling modules. Trichome fate is specified by a transcription factor complex consisting of a MYBR2R3 protein, GLABRA1 (GL1), a bHLH protein, GLABRA3 (GL3), and its paralogues ENHANCER OF GL3 (EGL3) and MYC1, as well a WD40-repeat protein, TRANSPARENT TESTA GL1 (TTG1) (Larkin et al., 1994; Walker et al., 1999; Payne et al., 2000; Schellmann and Hulskamp, 2005; Zhao et al., 2012) (Fig. 3A). The Myb–bHLH–WD40 transcription factor complex induces GL2, an HD-ZIP transcription factor directing trichome differentiation (Rerie et al., 1994). The 2-D spatial patterns of trichome initial cells are maintained by lateral inhibition, which involves a family of short MYBR3 proteins, TRIPTYCON (TRY) CAPRICE (CPC), and redundant paralogues, ENHANCER OF TRY AND CPC2/3 (ETC2/3) (Schellmann et al., 2002) (Fig. 3A). These short MBR3 proteins associate with GL3/EGL3 but fail to bind to the target promoter regions. The GL1–GL3/EGL3–TTG1 complex induces TRY/CPC expression, which is capable of moving to the neighbouring cells via plasmodesmata and inhibiting the activity of GL1–GFL3/EGL3–TTG1 by competitively displacing the transcriptional activator GL1 (Fig. 3A). This switches the Myb–bHLH–WD40 complex from an activator to a repressor of trichome fate (Schellmann and Hulskamp, 2005).
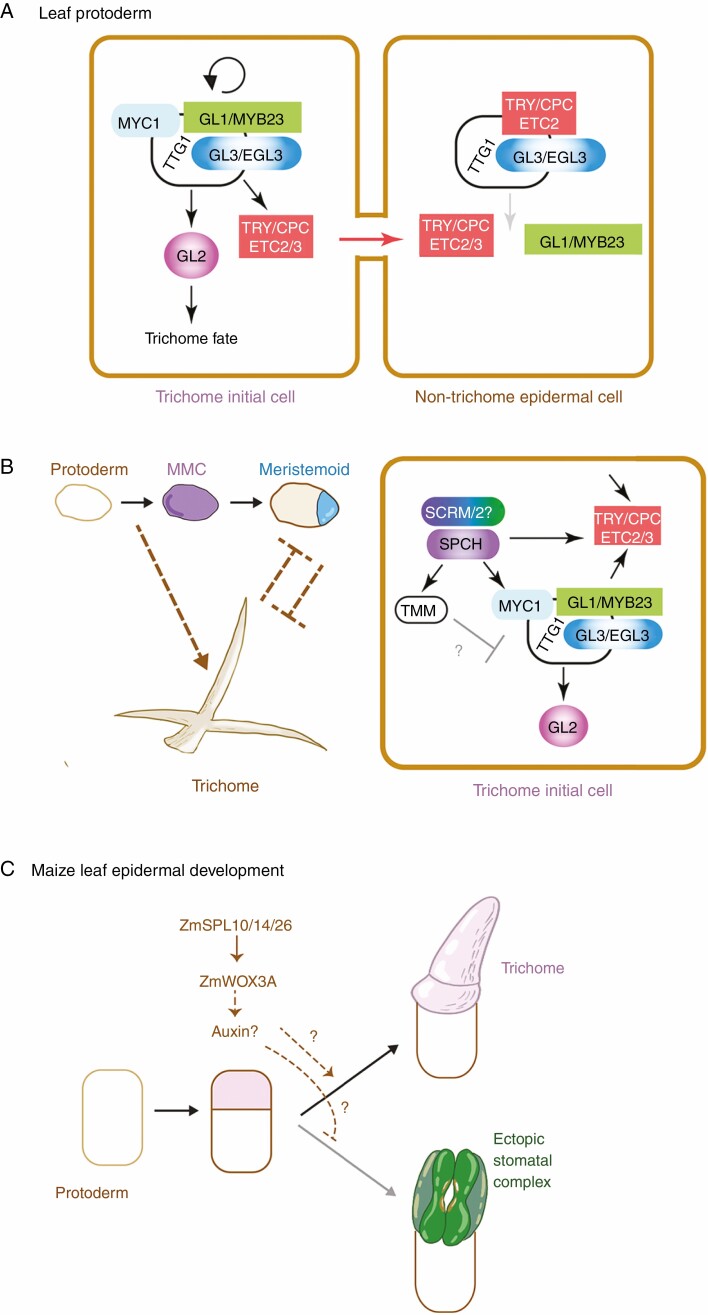
Fig. 3.
Trichome initial patterning and its intersection with the core stomatal developmental programme. (A) Trichome initial fate specification. The Myb–bHLH–WD40 complex containing GL1/MYB23, GL3/EGL3 (and MYC2) and TTG autoactivates itself and induces the expression of the HD-ZIP IV protein GL2, which specifies the trichome fate. The Myb–bHLH–WD40 complex in addition induces the expression of small Myb proteins, TRY/CPC/ETC2/ETC3, which can move from cell to cell via plasmodesmata and repress the activity of the Myb–bHLH–WD40 in the adjacent cell through competitive inhibition. (B) Role of the core stomatal development programme in trichome fate specification. Left: protoderm and MMCs may induce the trichome initiation programme, whereas later in the developmental progression both stomata and trichome precursors may mutually exclude each other’s fate. Right: in the protoderm/MMC, SPCH directly upregulates MYC1. In addition, SPCH directly upregulates TMM, whose overexpression reduces trichome numbers via an unknown mechanism. (C) Regulation of trichome (hair) development in maize. A trichome precursor cell (pink) is generated via an asymmetric cell division of a protodermal cell (white). ZmSPL10/14/26 upregulate the expression of ZmWOX3A, and promote trichome fate of the precursor, probably through auxin. In the absence of these ZmSPL genes, a stomatal complex transdifferentiates from the precursor instead of the trichome. The figure is modified and re-drawn from Kong et al. (2021).
Phenotypic analyses of stomatal mutant and transgenic plants suggest complex regulatory relationships between stomatal development and trichome patterning. For example, the scrm-D mutant, in which SCRM is constitutively active, overproduces stomata seemingly at the expense of trichomes (Kanaoka et al., 2008). Likewise, overexpression of TMM reduces trichome numbers and branching (Yan et al., 2014), suggesting that stomatal development is antagonistic to trichome formation (Fig. 3B). Interestingly, however, a study of direct SPCH targets hints that the story is not that simple (Adrian et al., 2015). The SPCH protein binds to the promoter region of MYC1 and ETC3, and upregulates their transcripts, indicating that SPCH directly activates the genes promoting trichome differentiation (Fig. 3B). In addition, ETC2 is strongly expressed in stomata and stomatal lineages, although loss-of-function etc2 mutant as well as try cpc etc2 triple mutant cotyledons exhibit no defects in stomatal density and patterning (Kirik et al., 2004). These seemingly counterintuitive findings imply that the molecular mechanisms underlying the initiation of stomatal cell lineages also contribute to the initiation of trichomes. However, a rosette leaf of the spch null mutant produces trichomes in apparent proper density (Hara et al., 2009), indicating that SPCH is not required for the initiation or spacing of trichomes. Thus, the biological significance of SPCH activation of trichome genes, such as MYC1 and ETC3, remains unclear.
A very recent study of leaf epidermal development in maize revealed an antagonistic regulatory mechanism between trichome and stomatal development. Maize triple loss-of-function mutants lacking three SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) genes ZmSPL10/14/26 develop glabrous leaf epidermis devoid of trichomes (Kong et al., 2021). In maize, three different types of trichomes (known as macrohairs, prickle hairs and bicellular hairs) exist, but Zmspl10/14/16 triple mutants lack all types of trichomes, but instead transdifferentiate ectopic stomatal complexes. This striking observation suggest that both maize trichomes and stomatal complexes originate from the same precursor cell (generated after the asymmetric division of a protodermal cell), and ZmSPL10/14/26 are required to suppress stomatal fate (Kong et al., 2021) (Fig. 3C). Through a molecular analysis, Kong et al. (2021) propose that ZmSPL10/14/26 activates the expression of ZmWOX3A, which then activates an auxin biosynthesis gene. Perhaps, through the action of auxin, the stomatal precursor fate is suppressed to ensure proper trichome differentiation (Fig. 3C). Whether this pathway is maize specific or more broadly applicable to other species remains to be seen. In any event, the studies in arabidopsis and maize highlight the complex regulatory relationships between the development of stomata and trichomes.
Stomatal patterning in hypocotyls: co-option of root hair patterning programmes
Unlike in cotyledons and leaves, the hypocotyl epidermis of arabidopsis shares characteristics of the root epidermis, with a common set of regulatory modules used in trichome development and root epidermal patterning (Schiefelbein, 2003; Grebe, 2012). During the development of the root epidermis, the Myb–bHLH–WD40 complex composed of WEREWOLF (WER)–GL3/EGL3–TTG1 induces GL2 expression, which subsequently suppresses expression of a series of root hair initiation and elongation genes, including RHD6, to specify non-hair cell fate. CPC/TRY induced by the WER–GL3/EGL3–TTG1 complex moves to a neighbouring cell and outcompetes WER for binding with GL3/EGL3 to inhibit GL2 induction, thereby promoting root hair cell fate (Wada et al., 1997; Lee and Schiefelbein, 1999; Schiefelbein et al., 2009) (Fig. 4A). The hypocotyl epidermis produces elongated, non-stomatal cell files and sunken stomata-producing files with cell division capacity. These cells exhibit no GL2 expression. However, not all cells in stomata-producing cell files in the hypocotyl epidermis develop into stomata, indicating that the stomatal cell fate determination requires additional signals. The elongated non-stomata-producing cell files exhibit strong GL2 expression, therefore sharing molecular characteristics of non-hair-producing root epidermal cell files (Berger et al., 1998; Hung et al., 1998) (Fig. 4A). Further mutant analyses showed that loss-of-function gl2 and ttg mutants ectopically expand stomata-producing cell files in hypocotyls, resulting in ectopic stomata (Fig. 4B). Unlike GL2 and TTG, GL1 and TRY play a role only in the trichome patterning. Loss-of-function gl1 and try mutants did not confer any effects on stomatal patterning in hypocotyls (Berger et al., 1998; Hung et al., 1998). Taken together, the hypocotyl stomatal patterning follows the root hair patterning rule.
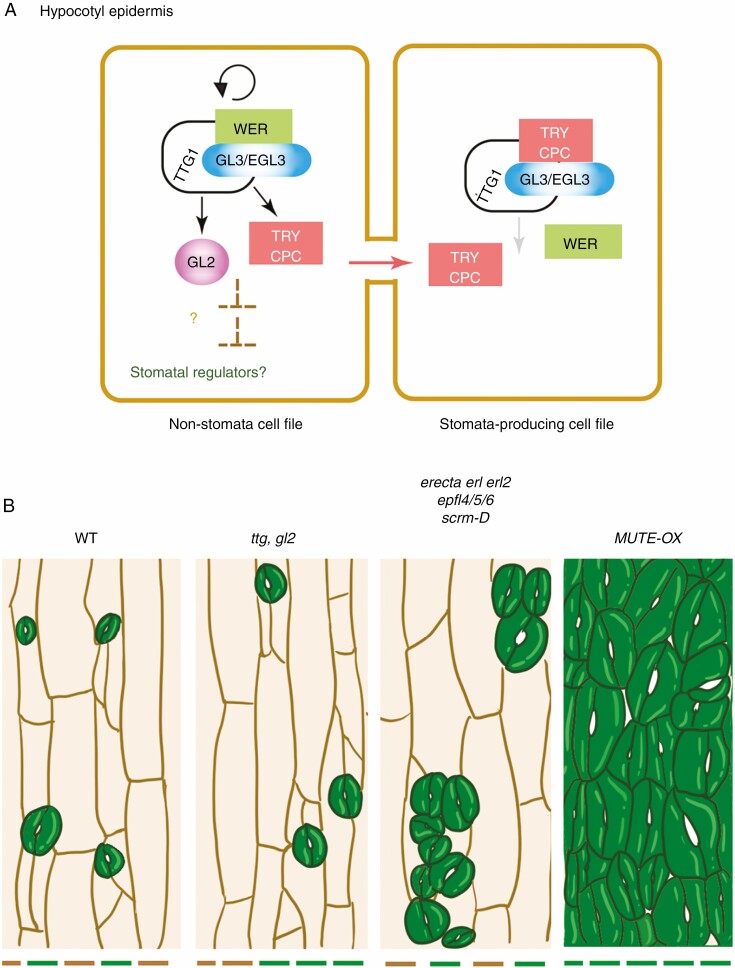
Fig. 4.
Hypocotyl epidermal patterning and its role in stomatal development. (A) The hypocotyl cell file follows the root hair/non-root hair epidermal patterning programme. Similar to the trichome fate specification programme, the Myb–bHLH–WD40 transcriptional complex consisting of WER–GL3/EGL3–TTG1 directly upregulates GL2 expression, which specifies a non-root hair cell file and non-stomatal cell file in the root and hypocotyl epidermis, respectively. The Myb–bHLH–WD40 complex in addition induces inhibitory small Myb proteins, TRY/CPC. TRY/CPC move from cell to cell and repress the Myb–bHLH–WD40 complex by competitively displacing WER, and this leads to eventual differentiation to a root hair cell file and a stomata-producing cell file in root and hypocotyl epidermis, respectively. (B) Hypocotyl epidermis phenotypes. Wild-type (WT) hypocotyl produces ordered columns of stomata-producing (green bars) and non-stomata-producing (brown bars) cell files. The loss-of-function mutants in Myb–bHLH–WD40, such as ttg and gl2 mutants, disrupt cell files. Core stomatal development mutants, such as erecta erl1 erl2, epfl4/5/6 and scrm-D, produce stomatal clusters only within the stomata-producing cell files. In contrast, the MUTE overexpressor produces hypocotyls solely composed of stomata. These ectopic stomata are very large and align in files, suggestive of direct reprogramming of protodermal cells into stomata.
Now that the underlying molecular players that potentiate the hypocotyl epidermis to make stomata have been elucidated, the important next question is to address their regulatory relationships with the core stomatal transcription factors (e.g. SPCH, MUTE, FAMA and SCRMs) and cell–cell signalling modules (e.g. EPF–ERECTA/TMM–MAPK). Indeed, phenotypic analysis demonstrated that the core regulators of stomatal development operate only in the ‘pre-patterned’ stomata-producing cell files: erecta erl1 erl2, epfl4/epfl5/epfl6 (also known as challah/chll1/chll2) and scrm-D mutants all produce dense stomatal clusters in hypocotyls, but only within the stomata-producing cell files (Kanaoka et al., 2008; Pillitteri et al., 2008; Abrash et al., 2011) (Fig. 4B). Likewise, mute mutant hypocotyls produce arrested meristemoids only in the stomata-producing cell files, but additional gl2 or ttg mutations confer production of ectopic cell files with meristemoids (Pillitteri et al., 2008).
Can the ectopic activation of stomatal development over-ride the hypocotyl cell file positional information? Indeed, ectopic overexpression of MUTE converted all hypocotyl epidermal cells into stomata, regardless of the original cell files (Pillitteri et al., 2008). The large size and block wall-like arrangements of these ectopic stomata in MUTE overexpressors suggest that all hypocotyl epidermal cells underwent direct reprogramming to differentiate stomata (Pillitteri et al., 2008) (Fig. 4B). Despite this observation, the molecular connection of the Myb–bHLH–WD40 (WER–GL3/EGL3–TTG1) transcription factor complex to the core stomatal development programme is not known. One possibility is that the transcription factor complex or its downstream effector, GL2, represses SPCH expression. Thus far, however, ChIP on chip experiments have not identified SPCH as a direct target of GL1 or GL3 (Morohashi and Grotewold, 2009). Very recently, the C2H2-Zinc Finger protein ZP1 was identified as a negative regulator of root hair development (Han et al., 2020). ZP1 acts downstream of GL2 and directly represses the expression of a set of ‘root hair’ bHLH genes: ROOT HAIR DEFECTIVE 6 (RHD6), which initiates root hair formation, and subsequently RHD6-LIKE2 (RSL2) and RSL4, which promote root hair elongation (Han et al., 2020). By analogy, it is tempting to speculate that a similar gene regulatory network may specify non-stomatal cell files in hypocotyls. Alternatively, the elongated and protruded non-stomatal-producing (and GL2-positive) cell files may harbour a specific epigenetic state that is incompatible with stomatal differentiation programmes. In this regard, it is worth mentioning that certain regulators of histone modifications, specifically HISTONE DEACETYLASE18 (HDA18), a negative regulator of histone acetylation, and GEM, a negative regulator of histone H3 K9 trimethylation (H3K9me3), are required for proper GL2 expression and proper patterning of root hair cell files (Xu et al., 2005; Caro et al., 2007). Future studies may reveal the exact molecular connection of gene regulatory networks specifying hypocotyl epidermal patterning and stomatal development.
Relationships between epidermal pavement cells and stomatal cell lineages
In addition to protecting plants from numerous environmental stresses, epidermal pavement cells provide a mechanical strength for plant growth and help guide organ morphogenesis (Bemis and Torii, 2007; Savaldi-Goldstein et al., 2007). The arabidopsis epidermal pavement cells have been characterized extensively for their unique jigsaw-puzzle shapes with characteristic lobes and sinuses. Numerous empirical and mathematical modelling studies have been performed to address how pavement cell shapes emerge and how mechanical stress influences the pavement cell geometry through dynamic behaviours of microtubules, cytoskeletons and cell walls (Sampathkumar et al., 2014; Armour et al., 2015; Elsner et al., 2018; Altartouri et al., 2019; Haas et al., 2020). Moreover, the plant hormone auxin co-ordinates pavement cell interdigitation via specific activation of two antagonistic small GTPase proteins: ROP2/4 determines the site of lobe expansion via local F-actin accumulation and, conversely, ROP6 specifies the site of the sinus via cortical microtubule organization through localized bundling (Fu et al., 2002, 2009; Xu et al., 2010). The auxin efflux carrier PIN1 is implicated in orchestrating the process by supplying auxin flow (Xu et al., 2010), although there is a contradictory observation refuting the role of PIN proteins in pavement cell morphogenesis (Belteton et al., 2018).
The leaf epidermal pavement cell differentiation does not require other epidermal cell types, including stomatal cell lineages. The most extreme example is the spch mutant, which produces an epidermis composed solely of perfectly interdigitated pavement cells (MacAlister et al., 2007; Pillitteri et al., 2007). In fact, researchers took advantage of the spch mutant to describe and model the pavement cell morphogenesis without the confounding effects of ‘irregularly shaped’ stomatal lineage cells (Carter et al., 2017). Although pavement cells can differentiate directly from protodermal cells, it is estimated that the vast majority of pavement cells in wild-type epidermis are derived from the stomatal lineage (approx. 67 % of all pavement cells in cotyledons and approx. 48 % in leaves) through the proliferative activities of MMCs and meristemoids (Geisler et al., 2000). Thus, stomatal lineage cells not only produce stomata, but are also a major source for pavement cells.
The key cell type supplying new pavement cells is the SLGC, which is defined as the larger daughter cell generated by the asymmetric division of an MMC or meristemoid (Shpak et al., 2005). The SLGC has interesting bi-potency: it could re-enter the stomatal initial state (MMC state) and undergo secondary asymmetric cell division to generate a satellite stoma, or it could lose the asymmetric cell division potential and differentiate into a pavement cell. Maintenance of the MMC state requires high amounts of SPCH–SCRMs (Horst et al., 2015) and, as such, the mechanisms leading to the loss of expression of SPCH–SCRMs drive the eventual differentiation of SLGCs into pavement cells. In arabidopsis, both extrinsic EPF peptide signalling and intrinsic polarity pathways downregulate SPCH–SCRMs via the activation of MAPK cascades (Herrmann and Torii, 2021). Indeed, a tissue mosaic study using Cre–Lox recombination-dependent EPF1 overexpression sectors showed that secreted extrinsic signals influence adjacent cells as well as those within a range of a few cells (Zeng et al., 2020). The intrinsic polarity component, BASL (BREAKING OF ASSYMMETRY IN THE STOMATAL LINEAGE), specifies the SLGC fate to one of the two daughter cells generated by the asymmetric cell division of a meristemoid (Dong et al., 2009). In newly formed SLGCs, BASL localizes at the cellular cortex and recruits YODA, and in return MPK3/6 phosphorylates BASL in the nucleus (Zhang et al., 2015). This BASL–MAPK positive feedback loop ensures the inhibition of SPCH accumulation, thereby preventing the SLGC from becoming a stomatal precursor cell (Zhang et al., 2016).
SLGCs that are transitioning into pavement cells begin to exhibit the characteristic jigsaw-puzzle shape with lobes and sinuses. The timing of this transition appears to coincide with the differentiation of its sister cell (now a GMC) into guard cells. Is the stomatal differentiation somehow coupled with the pavement cell differentiation? If so, what is the underlying mechanism? Very recently, this question was addressed by Grones et al. (2020), who observed that the position of first lobe outgrowth in SLGCs is distal to the neighbouring stoma, implying the presence of signals emanating from the differentiating stoma. Indeed, observation using the auxin response reporter DR5::NLS-Venus revealed a gradient of auxin response in a spiral manner within each stomatal complex, with the GMC exhibiting the lowest auxin response, followed by the youngest SLGC, and the highest response in the oldest SLGC (Grones et al., 2020). A previous study showed that the auxin efflux carrier PIN3 accumulates in late meristemoids transitioning to GMCs (Le et al., 2014). Characterization of PIN3 localization patterns and mutant phenotypes of higher order pin3/4/7 triple mutants led to the conclusion that timely auxin efflux from GMCs is critical for stomatal differentiation (Le et al., 2014). Further analysis on the spatiotemporal dynamics of auxin efflux carriers, influx carriers and transporters (e.g. PIN3, PIN7, AUX and ABCB1) supported the hypothesis that the directed auxin flow from GMC to SLGC promotes lobe formation, driving pavement cell morphogenesis (Grones et al., 2020). Thus, dynamic cell–cell auxin flow co-ordinates the differentiation of both stomata and pavement cells, and thus a deeper understanding of epidermal patterning is achieved only through characterizing the stomatal complex (stomatal precursors and SLGCs) as a unit. How the core regulators of stomatal development shape the dynamic auxin flow remains an open question.
Stomatal development in the context of intertissue communication
While this review focused on how developing stomata and other epidermal cell types influence each other, co-ordination between stomata and underlying mesophyll and vascular tissues must also be considered. For example, EPFL9 peptide, also known as Stomagen, is secreted from internal immature mesophyll cells and promotes stomatal development via antagonizing the EPF–ERECTA/TMM receptor module in the protoderm (Kondo et al., 2010; Sugano et al., 2010; Shimada et al., 2011; Lee et al., 2015). The intertissue layer communication is not unidirectional; developing epidermis also influences mesophyll cell organization, whereby each stoma is subtended by a mesophyll air space immediately underneath. In a wheat EPF1 (TaEPF1) overexpression line, the mesophyll air space was not produced underneath arrested meristemoids (Lundgren et al., 2019), suggesting that mature stomata are required in the epidermal layer for mesophyll air cavity formation. Therefore, physical (e.g. air flow and the humidity gradient from open stomata) and biochemical (e.g. peptides and hormones from developing guard cells) may actively influence mesophyll development. The presence of the air cavity below stomata is not unique to arabidopsis or wheat, and the connection between mature stomata and the air cavity underneath may be universal in land plants with stomata, including the moss Physcomitrella (Caine et al., 2020). Moreover, astomatous land plants, such as liverwort (Marcanthia polymorpha), develop air cavities underneath their multicellular air pores (Kohchi et al., 2021). While it is not known whether the air cavity development in both stomatous and astomatous land plants involves similar physiological, biochemical and/or physical mechanisms, understanding their mechanistic bases is certainly an important future direction.
Similar intertissue layer communication probably exists between vasculature and epidermal stomata development. It is known that the epidermis immediately above major veins does not develop stomata (McKown and Bergmann, 2020; Nunes et al., 2020). This architecture of excluding stomata immediately above the vasculature is likely to maximize efficient transport of photosynthates (sugar) from mesophyll cells. As one might expect, SPCH promoter activity is absent in the epidermis immediately above the midvein, suggesting that, like non-stomatal cell files in the hypocotyls, the mechanism to initiate SPCH expression is absent for this specialized epidermal region. The underlying cell–cell interactions and molecular mechanisms creating this ‘stomata-free’ zone remain unexplored, and are certainly an exciting future direction for investigation.
CONCLUSION AND PERSPECTIVES
Historically, studies of cell type differentiation in the leaf epidermis have focused on specific cell types of interest, whether stomata, trichomes or pavement cells. The identification of key genes, signalling components and regulatory mechanisms underlying the development of stomata and other leaf cell types now enables us to explore how stomatal development occurs in the context of epidermal tissues and integrates with the development of internal mesophyll and vascular tissues. Because stomata and other tissue/cell types are interconnected for generating a functional leaf, understanding how cell–cell interaction and cellular differentiation programmes are elaborated into supra-cellular tissue and organ patterning is necessary to gain a holistic view of plant development. Recent technological advancements now enable the profiling of cellular gene expression characteristics at a single-cell resolution (e.g. scRNA-seq, scATAC-seq) (Liu et al., 2020) and visualization of cell–cell interactions at high spatiotemporal resolution (Seerangan et al., 2020). These integrated approaches can be harnessed to design and adjust the stomatal patterns in a holistic way to maximize plant productivity and sustenance for specific agricultural conditions in a changing climate.
ACKNOWLEDGEMENTS
I thank Drs Daniel Zuch, Arvid Herrmann and Jiyuan Yang for commenting on this review article. I am truly grateful to the Australian Society for Plant Scientists for the honour of being selected as the 2018 Annals of Botany Lecturer. The author declares no conflict of interest.
Note Added in Proof: Very recently, subtractive RNA-sequencing was performed by Ho et al. to profile SLGC-enriched gene expression profiles. The results confirm the bipotent nature of SLGC to stay mitotic or undergo endoreduplication to become a pavement cell.
Ho CMK, Bringmann M, Oshima Y, Mitsuda N, Bergmann DC. 2021. Transcriptional profiling reveals signatures of latent developmental potential in Arabidopsis stomatal lineage ground cells. Proc Natl Acad Sci USA 118: e2021682118
FUNDING
Research in my laboratory is supported by the Howard Hughes Medical Institute, Johnson & Johnson Centennial Chair in Plant Cell Biology, The University of Texas at Austin, Molecular Biosciences, MEXT/JSPS KAKENHI JP26113507, JP16H01237, JP17H06476 and JP19H00990 and Ministry of Education, Culture, Sports, Science and Technology, Japan Society for the Promotion of Sciences.
LITERATURE CITED
- Abe M, Katsumata H, Komeda Y, Takahashi T. 2003. Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130: 635–643. [DOI] [PubMed] [Google Scholar]
- Abrash E, Anleu Gil MX, Matos JL, Bergmann DC. 2018. Conservation and divergence of YODA MAPKKK function in regulation of grass epidermal patterning. Development 145: dev165860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrash EB, Davies KA, Bergmann DC. 2011. Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand–receptor interactions. The Plant Cell 23: 2864–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian J, Chang J, Ballenger CE, et al. 2015. Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self-renewing population. Developmental Cell 33: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloni R. 2001. Foliar and axial aspects of vascular differentiation: hypotheses and evidence. Journal of Plant Growth Regulation 20: 22–34. [Google Scholar]
- Altartouri B, Bidhendi AJ, Tani T, et al. 2019. Pectin chemistry and cellulose crystallinity govern pavement cell morphogenesis in a multi-step mechanism. Plant Physiology 181: 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armour WJ, Barton DA, Law AM, Overall RL. 2015. Differential growth in periclinal and anticlinal walls during lobe formation in arabidopsis cotyledon pavement cells. The Plant Cell 27: 2484–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belteton SA, Sawchuk MG, Donohoe BS, Scarpella E, Szymanski DB. 2018. Reassessing the roles of PIN proteins and anticlinal microtubules during pavement cell morphogenesis. Plant Physiology 176: 432–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis SM, Torii KU. 2007. Autonomy of cell proliferation and developmental programs during Arabidopsis aboveground organ morphogenesis. Developmental Biology 304: 367–381. [DOI] [PubMed] [Google Scholar]
- Bemis SM, Lee JS, Shpak ED, Torii KU. 2013. Regulation of floral patterning and organ identity by Arabidopsis ERECTA-family receptor kinase genes. Journal of Experimental Botany 64: 5323–5333. [DOI] [PubMed] [Google Scholar]
- Berger F, Linstead P, Dolan L, Haseloff J. 1998. Stomata patterning on the hypocotyl of Arabidopsis thaliana is controlled by genes involved in the control of root epidermis patterning. Developmental Biology 194: 226–234. [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Sack FD. 2007. Stomatal development. Annual Review of Plant Biology 58: 163–181. [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Lukowitz W, Somerville CR. 2004. Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Kohchi T, Yamato KT, et al. 2017. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171: 287–304 e15. [DOI] [PubMed] [Google Scholar]
- Caine RS, Yin X, Sloan J, et al. 2019. Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytologist 221: 371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine RS, Chater CCC, Fleming AJ, Gray JE. 2020. Stomata and sporophytes of the model moss Physcomitrium patens. Frontiers in Plant Science 11: 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro E, Castellano MM, Gutierrez C. 2007. A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature 447: 213–217. [DOI] [PubMed] [Google Scholar]
- Carter R, Sánchez-Corrales YE, Hartley M, Grieneisen VA, Marée AFM. 2017. Pavement cells and the topology puzzle. Development 144: 4386–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti A, Jauneau A, Laufs P, et al. 2019. Mangroves in the leaves: anatomy, physiology, and immunity of epithemal hydathodes. Annual Review of Phytopathology 57: 91–116. [DOI] [PubMed] [Google Scholar]
- Chater CC, Caine RS, Tomek M, et al. 2016. Origin and function of stomata in the moss Physcomitrella patens. Nature Plants 2: 16179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater CCC, Caine RS, Fleming AJ, Gray JE. 2017. Origins and evolution of stomatal development. Plant Physiology 174: 624–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, MacAlister CA, Bergmann DC. 2009. BASL controls asymmetric cell division in Arabidopsis. Cell 137: 1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J, Hunt L, Afsharinafar M, et al. 2019. Reduced stomatal density in bread wheat leads to increased water-use efficiency. Journal of Experimental Botany 70: 4737–4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsner J, Lipowczan M, Kwiatkowska D. 2018. Differential growth of pavement cells of Arabidopsis thaliana leaf epidermis as revealed by microbead labeling. American Journal of Botany 105: 257–265. [DOI] [PubMed] [Google Scholar]
- Fu Y, Li H, Yang Z. 2002. The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. The Plant Cell 14: 777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Xu T, Zhu L, Wen M, Yang Z. 2009. A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Current Biology: CB 19: 1827–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Nadeau J, Sack FD. 2000. Oriented asymmetric divisions that generate the stomatal spacing pattern in arabidopsis are disrupted by the too many mouths mutation. The Plant Cell 12: 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glas JJ, Schimmel BC, Alba JM, Escobar-Bravo R, Schuurink RC, Kant MR. 2012. Plant glandular trichomes as targets for breeding or engineering of resistance to herbivores. International Journal of Molecular Sciences 13: 17077–17103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe M. 2012. The patterning of epidermal hairs in Arabidopsis – updated. Current Opinion in Plant Biology 15: 31–37. [DOI] [PubMed] [Google Scholar]
- Grones P, Majda M, Doyle SM, Van Damme D, Robert S. 2020. Fluctuating auxin response gradients determine pavement cell-shape acquisition. Proceedings of the National Academy of Sciences, USA 117: 16027–16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas KT, Wightman R, Meyerowitz EM, Peaucelle A. 2020. Pectin homogalacturonan nanofilament expansion drives morphogenesis in plant epidermal cells. Science 367: 1003–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G, Wei X, Dong X, et al. 2020. Arabidopsis ZINC FINGER PROTEIN1 acts downstream of GL2 to repress root hair initiation and elongation by directly suppressing bHLH genes. The Plant Cell 32: 206–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SK, Qi X, Sugihara K, et al. 2018. MUTE directly orchestrates cell-state switch and the single symmetric division to create stomata. Developmental Cell 45: 303–315 e5. [DOI] [PubMed] [Google Scholar]
- Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. 2007. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes & Development 21: 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yokoo T, Kajita R, et al. 2009. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant & Cell Physiology 50: 1019–1031. [DOI] [PubMed] [Google Scholar]
- Hepworth C, Caine RS, Harrison EL, Sloan J, Gray JE. 2018. Stomatal development: focusing on the grasses. Current Opinion in Plant Biology 41: 1–7. [DOI] [PubMed] [Google Scholar]
- Herrmann A, Torii KU. 2021. Shouting out loud: signaling modules in the regulation of stomatal development. Plant Physiology 185: 765–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst RJ, Fujita H, Lee JS, et al. 2015. Molecular framework of a regulatory circuit initiating two-dimensional spatial patterning of stomatal lineage. PLoS Genetics 11: e1005374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huchelmann A, Boutry M, Hachez C. 2017. Plant glandular trichomes: natural cell factories of high biotechnological interest. Plant Physiology 175: 6–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J, Hepworth C, Dutton C, et al. 2017. Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiology 174: 776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CY, Lin Y, Zhang M, Pollock S, Marks MD, Schiefelbein J. 1998. A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiology 117: 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L, Gray JE. 2009. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Current Biology 19: 864–869. [DOI] [PubMed] [Google Scholar]
- Jauneau A, Cerutti A, Auriac MC, Noël LD. 2020. Anatomy of leaf apical hydathodes in four monocotyledon plants of economic and academic relevance. PLoS One 15: e0232566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelle M, Vernoud V, Rogowsky PM, Ingram GC. 2011. Epidermis: the formation and functions of a fundamental plant tissue. New Phytologist 189: 17–39. [DOI] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, et al. 2008. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. The Plant Cell 20: 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Simon M, Wester K, Schiefelbein J, Hulskamp M. 2004. ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Molecular Biology 55: 389–398. [DOI] [PubMed] [Google Scholar]
- Kohchi T, Yamato KT, Ishizaki K, Yamaoka S, Nishihama R. 2021. Development and molecular genetics of Marchantia polymorpha. Annual Review of Plant Biology 10.1146/annurev-arplant-082520-094256. [DOI] [PubMed] [Google Scholar]
- Kondo T, Kajita R, Miyazaki A, et al. 2010. Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant & Cell Physiology 51: 1–8. [DOI] [PubMed] [Google Scholar]
- Kong D, Pan X, Jing Y, et al. 2021. ZmSPL10/14/26 are required for epidermal hair cell fate specification in maize leaf. New Phytologist 24: 17293. [DOI] [PubMed] [Google Scholar]
- Lai LB, Nadeau JA, Lucas J, et al. 2005. The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. The Plant Cell 17: 2754–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard GR, Macalister CA, Bergmann DC. 2008. Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Larkin JC, Oppenheimer DG, Lloyd AM, Paparozzi ET, Marks MD. 1994. Roles of the GLABROUS1 and TRANSPARENT TESTA GLABRA genes in Arabidopsis trichome development. The Plant Cell 6: 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Bergmann DC. 2012. Stomatal development: a plant’s perspective on cell polarity, cell fate transitions and intercellular communication. Development 139: 3683–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Davies KA, Chang J, et al. 2014. Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 345: 1605–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J, Liu XG, Yang KZ, et al. 2014. Auxin transport and activity regulate stomatal patterning and development. Nature Communications 5: 3090. [DOI] [PubMed] [Google Scholar]
- Lee JS, Kuroha T, Hnilova M, et al. 2012. Direct interaction of ligand–receptor pairs specifying stomatal patterning. Genes & Development 26: 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Hnilova M, Maes M, et al. 2015. Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522: 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. 1999. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483. [DOI] [PubMed] [Google Scholar]
- Liu T, Ohashi-Ito K, Bergmann DC. 2009. Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136: 2265–2276. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhou Y, Guo J, et al. 2020. Global dynamic molecular profiling of stomatal lineage cell development by single-cell RNA sequencing. Molecular Plant 13: 1178–1193. [DOI] [PubMed] [Google Scholar]
- Lu P, Porat R, Nadeau JA, O’Neill SD. 1996. Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. The Plant Cell 8: 2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren MR, Mathers A, Baillie AL, et al. 2019. Mesophyll porosity is modulated by the presence of functional stomata. Nature Communications 10: 2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister CA, Ohashi-Ito K, Bergmann DC. 2007. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445: 537–540. [DOI] [PubMed] [Google Scholar]
- Mauricio R, Rausher MD. 1997. Experimental manipulation of putative selective agents provides evidence for the role of natural enemies in the evolution of plant defense. Evolution 51: 1435–1444. [DOI] [PubMed] [Google Scholar]
- McKown KH, Bergmann DC. 2020. Stomatal development in the grasses: lessons from models and crops (and crop models). New Phytologist 227: 1636–1648. [DOI] [PubMed] [Google Scholar]
- Meng X, Chen X, Mang H, et al. 2015. Differential fuction of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Current Biology 25: 2361–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K, Grotewold E. 2009. A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genetics 5: e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD. 2002a. Stomatal development in Arabidopsis. Rockville, MD: American Society of Plant Biologists. [Google Scholar]
- Nadeau JA, Sack FD. 2002b. Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700. [DOI] [PubMed] [Google Scholar]
- Negi J, Moriwaki K, Konishi M, et al. 2013. A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Current Biology 23: 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes TDG, Zhang D, Raissig MT. 2020. Form, development and function of grass stomata. The Plant Journal 101: 780–799. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC. 2006. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. The Plant Cell 18: 2493–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JL, Rouzé P, Verhelst B, et al. 2016. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530: 331–335. [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. 2000. GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KM, Rychel AL, Torii KU. 2010. Out of the mouths of plants: the molecular basis of the evolution and diversity of stomatal development. The Plant Cell 22: 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KM, Shyu C, Burr CA, et al. 2013. Arabidopsis homeodomain-leucine zipper IV proteins promote stomatal development and ectopically induce stomata beyond the epidermis. Development 140: 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri LJ, Torii KU. 2012. Mechanisms of stomatal development. Annual Review of Plant Biology 63: 591–614. [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. 2007. Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505. [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Bogenschutz NL, Torii KU. 2008. The bHLH protein, MUTE, controls differentiation of stomata and the hydathode pore in Arabidopsis. Plant & Cell Physiology 49: 934–943. [DOI] [PubMed] [Google Scholar]
- Putarjunan A, Ruble J, Srivastava A, et al. 2019. Bipartite anchoring of SCREAM enforces stomatal initiation by coupling MAP kinases to SPEECHLESS. Nature Plants 5: 742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Han SK, Dang JH, et al. 2017. Autocrine regulation of stomatal differentiation potential by EPF1 and ERECTA-LIKE1 ligand–receptor signaling. eLife 6: e24102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Yoshinari A, Bai P, Maes M, Zeng SM, Torii KU. 2020. The manifold actions of signaling peptides on subcellular dynamics of a receptor specify stomatal cell fate. eLife 9: e58097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig MT, Abrash E, Bettadapur A, Vogel JP, Bergmann DC. 2016. Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proceedings of the National Academy of Sciences, USA 113: 8326–8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig MT, Matos JL, Anleu Gil MX, et al. 2017. Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science 355: 1215–1218. [DOI] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD. 1994. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes & Development 8: 1388–1399. [DOI] [PubMed] [Google Scholar]
- Roth-Nebelsick A. 2007. Computer-based studies of diffusion through stomata of different architecture. Annals of Botany 100: 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychel AL, Peterson KM, Torii KU. 2010. Plant twitter: ligands under 140 amino acids enforcing stomatal patterning. Journal of Plant Research 123: 275–280. [DOI] [PubMed] [Google Scholar]
- Sampathkumar A, Krupinski P, Wightman R, et al. 2014. Subcellular and supracellular mechanical stress prescribes cytoskeleton behavior in Arabidopsis cotyledon pavement cells. eLife 3: e01967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Peto C, Chory J. 2007. The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202. [DOI] [PubMed] [Google Scholar]
- Schellmann S, Hülskamp M. 2005. Epidermal differentiation: trichomes in Arabidopsis as a model system. International Journal of Developmental Biology 49: 579–584. [DOI] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, et al. 2002. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. The EMBO Journal 21: 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J. 2003. Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot. Current Opinion in Plant Biology 6: 74–78. [DOI] [PubMed] [Google Scholar]
- Schiefelbein J, Kwak SH, Wieckowski Y, Barron C, Bruex A. 2009. The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis. Journal of Experimental Botany 60: 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seerangan K, van Spoordonk R, Sampathkumar A, Eng RC. 2020. Long-term live-cell imaging techniques for visualizing pavement cell morphogenesis. Methods in Cell Biology 160: 365–380. [DOI] [PubMed] [Google Scholar]
- Shimada T, Sugano SS, Hara-Nishimura I. 2011. Positive and negative peptide signals control stomatal density. Cellular and Molecular Life Sciences 68: 2081–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU. 2005. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293. [DOI] [PubMed] [Google Scholar]
- Sugano SS, Shimada T, Imai Y, et al. 2010. Stomagen positively regulates stomatal density in Arabidopsis. Nature 463: 241–244. [DOI] [PubMed] [Google Scholar]
- Takada S, Takada N, Yoshida A. 2013. ATML1 promotes epidermal cell differentiation in Arabidopsis shoots. Development 140: 1919–1923. [DOI] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. 1997. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, et al. 1999. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. The Plant Cell 11: 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. 2007. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. The Plant Cell 19: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Chen L, Yu Q, et al. 2019. Multiple transcriptional factors control stomata development in rice. New Phytologist 223: 220–232. [DOI] [PubMed] [Google Scholar]
- Xu CR, Liu C, Wang YL, et al. 2005. Histone acetylation affects expression of cellular patterning genes in the Arabidopsis root epidermis. Proceedings of the National Academy of Sciences, USA 102: 14469–14474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, et al. 2010. Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi H, Nagano AJ, Kim J, et al. 2020. Fluorescent protein-based imaging and tissue-specific RNA-seq analysis of Arabidopsis hydathodes. Journal of Experimental Botany 72: 1260–1270. [DOI] [PubMed] [Google Scholar]
- Yan L, Cheng X, Jia R, et al. 2014. New phenotypic characteristics of three tmm alleles in Arabidopsis thaliana. Plant Cell Reports 33: 719–731. [DOI] [PubMed] [Google Scholar]
- Zeng SM, Lo EKW, Hazelton BJ, Morales MF, Torii KU. 2020. Effective range of non-cell autonomous activator and inhibitor peptides specifying plant stomatal patterning. Development 147: dev192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang P, Shao W, Zhu JK, Dong J. 2015. The BASL polarity protein controls a MAPK signaling feedback loop in asymmetric cell division. Developmental Cell 33: 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Guo X, Dong J. 2016. Phosphorylation of the polarity protein BASL differentiates asymmetric cell fate through MAPKs and SPCH. Current Biology 26: 2957–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Wang X, Zhu D, et al. 2012. A single amino acid substitution in IIIf subfamily of basic helix–loop–helix transcription factor AtMYC1 leads to trichome and root hair patterning defects by abolishing its interaction with partner proteins in Arabidopsis. Journal of Biological Chemistry 287: 14109–14121. [DOI] [PMC free article] [PubMed] [Google Scholar]