Abstract
Objective
The United States is experiencing an opioid epidemic. In recent years, there were more than 10 million opioid misusers aged 12 years or older annually. Identifying patients at high risk of opioid use disorder (OUD) can help to make early clinical interventions to reduce the risk of OUD. Our goal is to develop and evaluate models to predict OUD for patients on opioid medications using electronic health records and deep learning methods. The resulting models help us to better understand OUD, providing new insights on the opioid epidemic. Further, these models provide a foundation for clinical tools to predict OUD before it occurs, permitting early interventions.
Methods
Electronic health records of patients who have been prescribed with medications containing active opioid ingredients were extracted from Cerner’s Health Facts database for encounters between January 1, 2008, and December 31, 2017. Long short-term memory models were applied to predict OUD risk based on five recent prior encounters before the target encounter and compared with logistic regression, random forest, decision tree, and dense neural network. Prediction performance was assessed using F1 score, precision, recall, and area under the receiver-operating characteristic curve.
Results
The long short-term memory (LSTM) model provided promising prediction results which outperformed other methods, with an F1 score of 0.8023 (about 0.016 higher than dense neural network (DNN)) and an area under the receiver-operating characteristic curve (AUROC) of 0.9369 (about 0.145 higher than DNN).
Conclusions
LSTM–based sequential deep learning models can accurately predict OUD using a patient’s history of electronic health records, with minimal prior domain knowledge. This tool has the potential to improve clinical decision support for early intervention and prevention to combat the opioid epidemic.
Keywords: opioid use disorder, machine learning, deep learning, electronic health records
INTRODUCTION
Opioid use disorder (OUD), including opioid dependence and opioid addiction, is a physical or psychological reliance on opioids, which are a class of substances found in certain prescription pain medications and illegal drugs such as heroin.1 Misuse and abuse of opioids are attributable to the deaths of more than 130 Americans daily,2 making them a leading cause of accidental death in the United States.3 Opioid prescribing has increased due to its effectiveness in treating acute pain.4 According to Han et al,5 91.8 million (37.8%) civilian noninstitutionalized adults in the United States consumed prescription opioids in 2015. Among them, 11.5 million (4.7%) misused them and 1.9 million (0.8%) had a use disorder. Overdose from prescription opioids has risen from over 4000 to over 16 000 in 2010, making it the fastest-growing cause of overdose deaths.6 OUD among individuals on prescription opioids has become a significant public health concern.
Early intervention in the developmental trajectory of OUD has the potential to reduce morbidity and mortality. Interventions, such as reducing opioid dosage or suggesting alternative options for chronic pain management, can potentially reduce the risk of OUD. The Centers for Disease Control and Prevention has also provided recommendations for safer use of opioid prescriptions in chronic pain care.7 Predicting OUD risk for specific patients or patient groups can help target these interventions.
Electronic health records (EHRs) have been widely adopted with the introduction of the Health Information Technology for Economic and Clinical Health Act of 2009.8 Besides EHR data managed by healthcare providers, large-scale EHR data are also made available through commercial EHR vendors for research purpose. For example, Cerner’s Health Facts9 is a large multi-institutional de-identified database derived from EHRs and administrative systems. Given the availability of vast EHR data, we can build predictive models, which assess the risk of OUD and provide top risk factors as explanations. Therefore, meaningful clinical decision support tools are possible.10
Traditional statistical and machine learning based models for OUD prediction have been proposed in previous works.11–14 For example, the Cox regression method was applied to extract the most relevant features and then to build a multivariate regression model to fit those features to predict 2-year risk of opioid overdose.15 Ellis et al16 studied Gini importance, effect size, and the Wilcoxon rank sum test to measure the importance of different features and applied a random forest classifier to predict opioid dependence. In addition to random forests, decision trees and logistic regression were also proposed by Wadekar et al17 for OUD prediction using demographic, socioeconomic, physical, and psychological features, which revealed first use of marijuana before 18 years of age as the greatest risk factor. Lo-Ciganic et al18,19 applied gradient boosting machine (GBM) and dense neural network models for opioid overdose prediction using a set of handcrafted features, including demographics, medical codes, and other aggregated features such as daily morphine milligram equivalents (MME). Calcaterra et al20 applied logistic regression on laboratory tests, demographics, and diagnosis codes to predict future chronic opioid use in 30 days.
Recently, deep learning methods have gained popularity in EHR based predictive modeling. For instance, Rajkomar et al21 performed a large-scale study on multiple medical event prediction based on EHR data using deep learning, which achieved high prediction accuracy. Another study employed a fully connected deep neural network to suggest candidates for palliative care.22 Sequential deep learning models were also applied to tackle problems relevant to opioids. For instance, recurrent neural networks (RNNs) were proposed by Che et al23 to classify opioid users into long-term users, short-term users, and opioid-dependent patients, with diagnoses, procedures and medications as features. Another study explored the application of RNNs for chronic disease prediction using medical notes.24 Our recent work also applied fully connected networks for predicting diseases and improving coding25,26 and opioid overdose prediction.27
In this article, we propose a sequential deep learning model built on long short-term memory (LSTM) to predict OUD among patients prescribed with opioid medications in their past health records. This sequential model can better represent the progression of diseases and identify the most important features as potential risk factors for the diseases. We used patients’ past medical history including diagnosis codes, procedure codes, laboratory test results, medications, clinical events, and demographics to train the model. We also compared our method with traditional machine learning algorithms and dense neural networks. Our results demonstrate that with comprehensive EHR data, our sequential deep learning model can provide highly accurate predictions, with a higher F1 score than the other methods. We also identified OUD-related diagnoses and medications as important features for prediction.
MATERIALS AND METHODS
Data source
Cerner’s Health Facts database includes de-identified EHR data from over 600 participating Cerner client hospitals and clinics in the United States. In addition to encounters, diagnoses, procedures, and patients’ demographics that are typically available in claims data, Health Facts also includes medication dosage and administration information, vital signs, laboratory test results, surgical case information, other clinical observations, and health systems attributes.28
Data selection
As patients with opioid prescriptions were the target cohort of this study, we extracted all the patients who had been prescribed with medications containing opioid related ingredients according to their medical records. For retrieval of those ingredients, we used the Anatomical Therapeutic Chemical (ATC) level 3 code “N02A” and categories description ‘opioid’ to retrieve all relevant active ingredients from DrugBank 5.1.4.29 Selected opioid related ingredients include butorphanol, diamorphine, eluxadoline, oxycodone, oxymorphone, naloxone, tramadol, levacetylmethadol, pentazocine, hydromorphone, levorphanol, remifentanil, normethadone, opium, sufentanil, piritramide, tapentadol, morphine, codeine, dezocine, fentanyl, nalbuphine, meperidine, naltrexone, buprenorphine, methadone, hydrocodone, alfentanil, dihydrocodeine, and diphenoxylate.
Following procedures from Moore et al,30 we selected a group of International Classification of Diseases–Ninth Revision (ICD-9) and ICD–Tenth Revision (ICD-10) codes to define OUD diagnosis. We excluded opioid poisoning ICD codes for OUD patient identification, as opioid poisoning can be fatal and prompt medication interventions are needed. Our work targets OUD patients for potential early intervention at the trajectory from OUD to overdose, with a similar idea as in Lo-Ciganic et al.18 The summary of the codes can be found in Supplementary Appendix S1. Patients with one or more of these codes were considered OUD patients.
Because opioid medications have proven successful in treatment of cancer pain,31 cancer patients may receive many more opioid prescriptions than other patients. This fact may lead the model to misclassify these patients as having OUD, so we removed all patients with cancer diagnosis. To identify patients with cancer, we used the ICD-932 and ICD-10 codes.33 The summary of these ICD codes can be found in Supplementary Appendix S2.
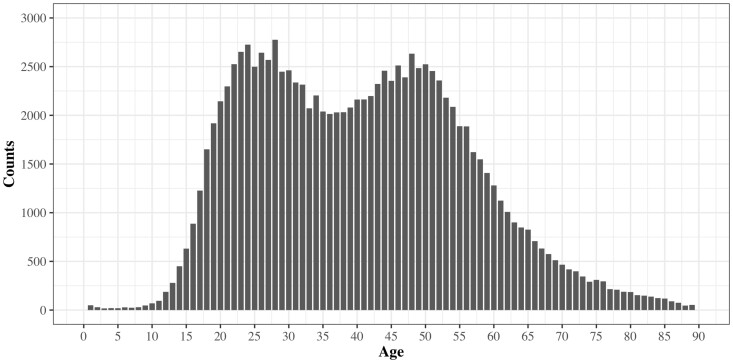
The majority of OUD patients (91.08%) were between 18 and 66 years of age. The portion of patients older than 66 years of age and younger than 18 years of age between OUD and non-OUD patients differed significantly. To make positive and negative cases consistent and prevent potential bias on age, we extracted both OUD and non-OUD patients based on their age of first exposure to opioid medications between 18 and 66 years of age. The age distribution of first opioid medication exposure for OUD patients is shown in Figure 1.
Figure 1.
Age distribution of first opioid medication exposure for patients with opioid use disorder.
The patient selection process is illustrated in Supplementary Figure 1S. After age filtering, there were 111 456 positive (OUD) patients and 5 072 110 negative patients. For positive patients, we identified the target encounter as the encounter with the earliest diagnosis of OUD (label “1”). We then selected the five encounters prior to the target encounter to use as the input features (Supplementary Figure S2a). For negative patients, we selected the last available encounter as the target encounter (label “0”) and then selected the 5 prior encounters as the input data (Supplementary Figure S2b).
Encounter selection
We used the 5 prior encounters before the target encounter to build the feature matrix. The choice of 5 encounters was based on two considerations. First, the median number of encounters for all patients was close to 5. Second, we tested the model with 5 to 10 encounters and the result showed that the prediction with 5 prior encounters had optimal performance. Note that some patients had fewer than 5 prior encounters. If a patient did not have 5 prior encounters, we replicated the last encounter before the target encounter to fill the gap. For example, if a patient had 7 prior encounters (1-7), encounters 3 to 7 would be used; if a patient had 3 prior encounters (1-3), encounter 3 would be replicated to generate a sequence of 5 encounters (1,2,3,3,3). The selection of encounters is illustrated in (Supplementary Figure S2a).
Feature selection
Information used for the feature matrix included diagnosis codes, procedure codes, laboratory tests, medications, clinical events, and demographic information.
Diagnosis codes specify patients’ diseases and symptoms. This history of diseases is critical information for predicting the future. In Health Facts, both ICD-9 codes (before October 1, 2015) and ICD-10 codes (after October 1, 2015) exist. We converted all ICD-9 codes to ICD-10 codes to avoid dispersion of predictability for each diagnosis feature34 and used the first 3 digits of the ICD-10 codes to reduce granularity and accelerate the training process.
Medications are recorded by National Drug Code (NDC) codes in Health Facts, which give labeler, product, and package information. For a more clinically meaningful representation, we converted all NDC codes to ATC codes. ATC codes indicate active ingredients and are organized hierarchically by the system/organ they act on and their therapeutic or pharmacological class. Moreover, by using ATC codes, we reduced the number of medication features while retaining meaningful information about the active ingredients. ATC level 3 codes were chosen35 to represent all medications. For each medication, the total medication quantity prescribed to each patient was taken as a feature for the medication. In addition to the total medication quantity, we also calculated the amount of opioid ingredients contained in each medication and converted it to MME as an aggregate feature, which represents the dosing strengths of prescription opioids based on their relative potencies compared with morphine.36,37 MME was then used to determine a patient’s cumulative intake of opioids.
The numeric value for each laboratory test is recorded in Health Facts as well as the standardized interpretation of the value (i.e., indicating whether it is high, low, or normal). We calculated the number of high, low, and normal values that a patient received for each test, as well as the total number of laboratory tests that the patient received.
Clinical events are related symptoms, procedures, and personal situations that are not formally classified into any of the previous codes, for instance, the pain level of patients, smoking history, height, weight, and travel information. Since 79.21% of hospitals in Health Facts report clinical events, we included clinical events in the feature space.
Demographic information includes age, gender, and race or ethnicity. These variables were also included in the feature space.
We extracted 1468 features to predict future OUD based on a patient’s EHR history, including 457 diagnosis features, 530 laboratory test features, 3 demographic features, 251 clinical event features, and 227 medication features, as summarized in Table 1. Features of low co-occurrence with OUD were removed to prevent overfitting and sparsity problems; if a feature was present in less than 1% of all OUD patients, that feature was not included in the feature space.
Table 1.
Summary of features
| Category | Number of features | Description |
|---|---|---|
| Diagnoses | 457 | First three digits of ICD-10 codes (ICD-19 codes were first converted to ICD-10 codes) |
| Medications | 227 | The total quantity of a medication a patient received |
| Clinical events | 251 | The highest, lowest and median values for each event if there are multiple values in one encounter |
| Laboratory tests | 530 | The numbers of high, low and normal values and the total number for each test |
| Demographics | 3 | Gender, age, race/ethnicity |
ICD-9: International Classification of Diseases–Ninth Revision; ICD-10: International Classification of Diseases–Tenth Revision.
Feature matrix construction
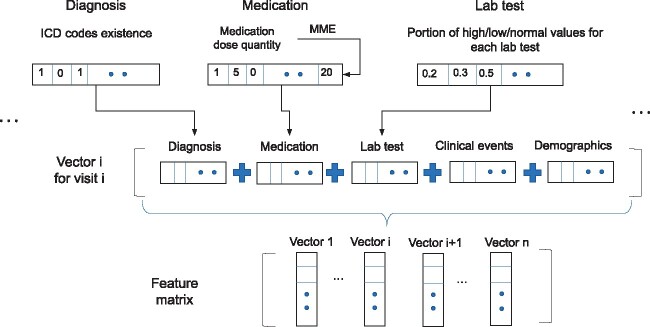
We used a binary representation to denote the presence or absence of a diagnosis code in a patient's EHR history. Ages were segmented into groups: the first age group was 18 to 27 years of age, followed by groups spanning 10 years each (28-37 years of age and so on). Race or ethnicity was encoded using one-hot encoding, the most common coding scheme for categorical variables. One-hot encoding transforms a single variable with n distinct possible values into n binary variables indicating presence (1) or absence (0). Numeric features, such as medication dosage, blood pressure, height, and pain score, were assigned with numeric values in corresponding units in the database, such as height measured in cm, and blood pressure measured in mm Hg. For patients without any value for a given laboratory test, we imputed a value. In addition, for some clinical events, there were multiple values in one encounter, such as body temperature taken multiple times in the encounter. We therefore recorded the highest, lowest and median values of the feature in that encounter. We replaced missing values with median values for numeric features, or majority values for binary features. Figure 2 shows feature matrix construction from feature vectors.
Figure 2.
Construction of the feature matrix. ICD: International Classification of Diseases; MME: morphine milligram equivalents.
Missing values
Missing values may impact prediction tasks. In our work, missing values were more challenging to address in the numeric data types in the clinical events and laboratory tests. Table 2 shows the basic statistics about the missing value proportion in laboratory tests and clinical events. The proportion of missing values was higher than 10% in both cases, so the issue is considerable. In our model, we used median imputation methods to handle missing values. We also compared median imputation with other common imputation methods including mean imputation, KNN38 and MICE.39 For patients with a missing value, KNN imputation finds the k patients with closest values to that patient based on known values, and then imputes the missing value with the weighted average value of that feature for all the k patients. We applied KNN imputation with a k value of 100. MICE imputation models each feature as a function of other features, trains a regressor based on known values, and then imputes missing values with the trained regressor. We used scikit-learn to implement these methods.
Table 2.
Statistics on missing values in the datasets
| Non-OUD patients | OUD patients | P value | |
|---|---|---|---|
| Total number of patients | 5 072 110 | 111 456 | |
| Clinical events | |||
| Average number of clinical events per patient | 335.46 | 539.28 | .0046 |
| Portion of clinical events with missing values | 37.89 (11.30) | 71.27 (13.21) | |
| Laboratory tests | |||
| Average number of laboratory tests per patient | 184.02 | 356.65 | .0696 |
| Portion of laboratory tests with missing values | 40.57 (22.05) | 85.37 (23.94) | |
Values are n (%), unless otherwise indicated.
OUD: opioid use disorder.
Dense neural networks baseline
Deep neural networks (DNNs) have been proven effective in many healthcare prediction applications21–24 due to their ability to handle large numbers of features, making them well suited for our problem. We took the DNN as a baseline for comparison. Since the performance of the DNN may vary due to its flexible structure, for a fair comparison, we tuned the DNN for optimal performance. We tested different structures of DNNs, with different numbers of layers (2, 3, 4, 5, 6), different dimensions (64, 128, 256, 512) and different dropout rates (0.2, 0.25, 0.3, 0.35, 0.4, 0.45, 0.5). We also took complexity into consideration. The optimized DNN model for comparison was composed of 6 fully connected layers, with each of the first 5 layers having a dimension of 512, with ReLU as the activation function, and with a dropout of 0.3. The last layer had a dimension of 8 and was connected to the output layer with binary cross-entropy loss function and Adam optimizer, whose learning rate was 0.01. The dropout layer randomly drops out a portion of outputs from the previous fully connected layer at each training epoch to prevent overfitting. The framework of our network is illustrated in Figure 3.
Figure 3.
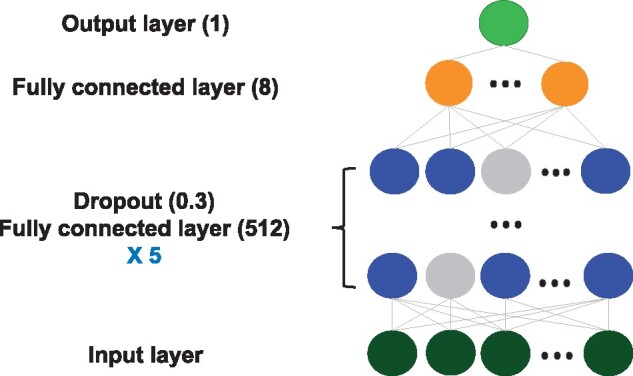
Structure of the dense neural network model.
Proposed LSTM based model
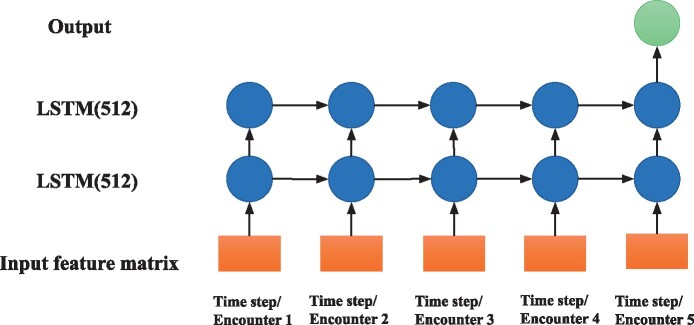
Our proposed model is based on the LSTM network, a class of RNN architecture in which connections between nodes form a directed graph along a temporal sequence. This structure makes the RNN model well suited to make predictions from time series data. LSTM networks are a version of RNNs, modified to better store past data in memory. Additionally, LSTMs can solve the vanishing gradient problem common in RNN models.40 We implemented an LSTM based network composed of 2 LSTM layers of 512 units. The framework of our LSTM based neural network is illustrated in Figure 4. The feature vector for each encounter was treated as a time step for LSTM, and the number of time steps equaled the number of encounters (5). Our LSTM was trained with a binary cross-entropy loss function and Adam optimizer whose learning rate was 0.01.
Figure 4.
Structure of the LSTM model.
Variants of the LSTM model
There are various techniques that can be employed by an LSTM model for potentially enhancing the performance. For comparison, we implemented widely used techniques including attention mechanism41 and bidirectional structure42 and compared their performance experimentally. The attention mechanism is an input processing technique that allows a model to focus on a specific aspect of a complex input, mimicking how humans solve problems. Bidirectional structure is commonly applied together with RNNs. The principle is to train a regular RNN model in 2 directions, so that the output layer can gain information from backward and forward states simultaneously. It has the advantage of capturing long-term dependencies. We compared performance across the LSTM model, the LSTM with the attention model (LSTM+Attention), the LSTM with bidirectional structure (Bi-LSTM), and the LSTM with both mechanisms (Bi-LSTM+Attention).
Further, we also considered the BERT model43 for our problem. BERT is a transformer-based44 machine learning technique for natural language processing application, which gained popularity recently in sequential model applications. To adapt the BERT model to our problem, we constructed the number of encounters, feature name, and value together as a token for BERT. Our model was implemented with keras-bert45 in Python (Python Software Foundation, Wilmington, DE). Because there was no pretrained model for similar problems, we trained the model from scratch.
Other baseline methods
In addition to DNN and LSTM models, we also applied traditional machine learning methods including decision tree, random forest, and logistic regression. Another popular model for consideration is the convolutional neural network (CNN). However, for CNN models to have an advantage, the elements in the input matrix should be related to their surrounding elements. In our problem, the order of our features is interconvertible, therefore CNNs may not have an advantage over other models. Nevertheless, we tested CNNs with different settings, including different numbers of layers (2, 3, 4), sizes of kernel (3 × 3, 2 × 2) and pooling (max, min, average). Even with the best settings, the prediction performance was poor, with an F1 score around 0.4. Thus, CNN was not included for further analysis in the following experiments. Detailed description of those methods can be found in Supplementary Table S6.
We implemented our models and performed experiments using the programming language Python (version 2.7). Traditional machine learning methods were implemented with the Python Scikit-Learn package.46 Deep learning was implemented with Python TensorFlow47 and Python Keras.48 Other libraries used include Python NumPy49 and Python Pandas.50 The training was performed on one NVIDIA Tesla V100 GPU (16GB RAM; NVIDIA, Santa Clara, CA).
Data availability statement
The original data underlying this article were provided by Cerner (https://www.cerner.com/) under an institutional agreement. Data sharing of the original data is prohibited. However, all result data, models, and codes are publicly available (https://github.com/StonyBrookDB/oudprediction). The Stony Brook University Institutional Review Board determined that the de-identified Cerner Health Facts database is not human subjects data (#170753_MODCR001).
RESULTS
In our experiments, we randomly assigned 80% of patients to the training set and the rest to the test set. For each method, we repeated the training process 10 times to calculate an average value for each performance metric. In our experiments, we portioned the total cohort of negative patients into 10 parts of 507 211 negative patients each. We trained each part with positive patients separately and randomly selected 80% as a training set and the remaining 20% as the test set. For each metric to evaluate a model’s performance, we calculated the average for all 10 parts.
To comprehensively evaluate the models, we computed all common metrics including precision, recall, F1 score, and area under the receiver-operating characteristic curve (AUROC). We note that for imbalanced datasets, AUROC can be misleading.51 Recall is a critical factor, as it indicates the fraction of future OUD patients we can identify, but high recall is not useful without high precision (for instance, if all patients were marked positive, recall would be perfect, but the model would not be informative). Because the F1 score considers both precision and recall, we regard it as the best aggregated assessment of the overall prediction performance. The average and standard deviation of each metric for each method are shown in Table 3. The best results for each metric are highlighted.
Table 3.
Summary of prediction performance of different models
| Model | Precision | Recall | F1 score | AUROC |
|---|---|---|---|---|
| Random forest | 0.8565 ± 0.0014a | 0.6871 ± 0.0027 | 0.7545 ± 0.0022 | 0.9112 ± 0.0014 |
| Decision tree | 0.7592 ± 0.0084 | 0.7281 ± 0.0059 | 0.7453 ± 0.0030 | 0.8823 ± 0.0019 |
| Logistic regression | 0.7507 ± 0.0095 | 0.6020 ± 0.0089 | 0.6722 ± 0.0035 | 0.7933 ± 0.0036 |
| Dense neural network | 0.8019 ± 0.0108 | 0.7694 ± 0.0027 | 0.7855 ± 0.0049 | 0.9224 ± 0.012 |
| LSTM | 0.8184 ± 0.0085 | 0.7865 ± 0.0058a | 0.8023 ± 0.0020a | 0.9369 ± 0.0038 |
| Bi-LSTM | 0.7779 ± 0.0013 | 0.7615 ± 0.0012 | 0.7696 ± 0.0012 | 0.9377 ± 0.0065 |
| LSTM+Attention | 0.8131 ± 0.0081 | 0.7814 ± 0.0071 | 0.7969 ± 0.0035 | 0.9491 ± 0.0023a |
| Bi-LSTM+Attention | 0.7710 ± 0.0086 | 0.7804 ± 0.0109 | 0.7759 ± 0.0019 | 0.9463 ± 0.0006 |
| BERT | 0.7709 ± 0.0123 | 0.6709 ± 0.0056 | 0.7174 ± 0.0079 | 0.8687 ± 0.0060 |
AUROC: area under the receiver-operating characteristic curve; Bi-LSTM: bidirectional long short-term memory; LSTM: long short-term memory.
Best result for the metric.
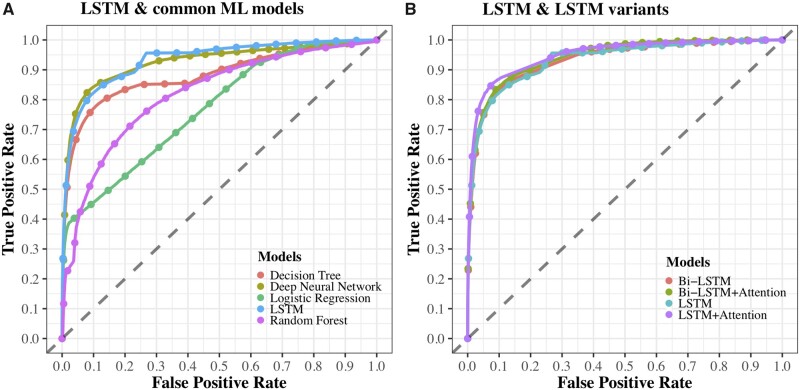
Across the models, the LSTM model achieved the highest F1 score and the highest recall. The random forest achieved the highest precision. The LSTM with attention mechanism achieved the highest AUROC, with a score of 0.9369, which indicates a good performance for clinical psychology applications, for which AUROC scores are recommended to exceed 0.747.52Figure 5 shows the ROC curves for all 8 methods. Curves of random forest, logistic regression, decision tree, DNN, and LSTM are shown in Figure 5A, and curves of the LSTM model and the 3 LSTM variants are shown in Figure 5B. We used a t test to compare the performance of the DNN and the LSTM and found that the LSTM significantly outperformed the DNN in each metric (2-tailed P < .0001). Details are shown in Supplementary Table S3.
Figure 5.
Receiver-operating characteristic (ROC) curves for each method. (A) ROC curves for LSTM and common machine learning models; (B) ROC curves for LSTM and LSTM variants. Bi-LSTM: bidirectional long short-term memory; LSTM: long short-term memory; ML: machine learning.
Compared with the LSTM models, the BERT model performed only modestly. One explanation may be that the size of our dataset was not big enough for BERT’s complex architecture. The two original versions of BERT were trained with 800 million and 2500 million words accordingly and had more than 100 million parameters. Without a pretrained model applicable to our work, BERT’s performance may have been suboptimal, facing issues such as overfitting. While LSTM variants outperformed BERT, they also suffered from the overfitting problem.
In order to evaluate various imputation methods, we compared performance of the best-performing model, the LSTM model, with different imputation methods. The results are shown in Table 4.
Table 4.
Summary of LSTM prediction performance on different imputation methods
| Precision | Recall | F1 score | AUROC | |
|---|---|---|---|---|
| Median | 0.8184 ± 0.0085a | 0.7865 ± 0.0058 | 0.8023 ± 0.0020 | 0.9369 ± 0.0038a |
| Mean | 0.8183 ± 0.0073 | 0.7851 ± 0.0036 | 0.8014 ± 0.0031 | 0.8977 ± 0.0048 |
| MICE | 0.8128 ± 0.0082 | 0.8017 ± 0.0070 | 0.8072 ± 0.0037a | 0.9002 ± 0.0042 |
| KNN | 0.7984 ± 0.0094 | 0.8034 ± 0.0073a | 0.8009 ± 0.0039 | 0.8854 ± 0.0032 |
AUROC: area under the receiver-operating characteristic curve.
Best result for the metric.
The results showed that the imputation methods did not make a tangible difference on the performance. This is in concert with similar studies, which found no significant differences in results when comparing different imputation methods in noisy and large-scale EHR data.25,26
Ablation study
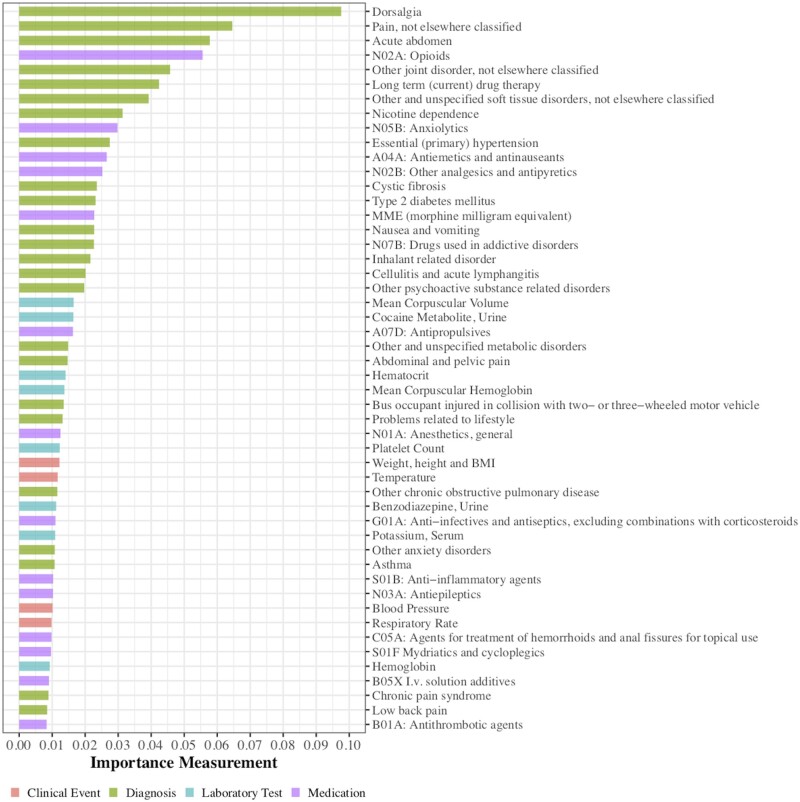
To support researchers or clinicians to exploit potential causes or trajectories of diseases, it is necessary to understand the importance of different features for prediction. Deep learning models are especially difficult to interpret because of their complex structures and millions of parameters. To evaluate the importance of features in our deep learning models, we employed the permutation importance method. It measures the importance of a feature as the size of the decrease in performance after blinding the model to that feature.53 We used the AUROC as the performance metric. This method can be applied to a wide variety of models, including both traditional machine learning models and deep learning methods. We used the Python package eli5 to perform the permutations.54 We report the 50 most important features for our best-performing method, the LSTM model, in Figure 6 and Supplementary Table S4. We also report the top 20 features for four different models in Supplementary Table S5.
Figure 6.
Top 50 important predictors for opioid use disorder selected by the long short-term memory model.
We found some patterns among the top 50 features. Opioid related medications had high rankings, including dose quantity of opioid medications (ATC Level 3 code N02A*) and MME. Other pain treatment related medications (N02B: Other analgesics and antipyretics; N01A: Anesthetics, general; N01B: Anesthetics, local) were also among the top features across different methods. Several highly ranked diagnosis features were also related to pain, such as dorsalgia (ICD-10 codes M54.*), which includes chronic back pain, as well as pain not elsewhere classified (ICD-10 codes G89.*), acute abdominal and pelvic pain (ICD-10 codes R10.*), and joint or tissue disorder (ICD-10 codes M25.* and M79.*). Pain disorders could represent the cause of opioid use initiation. Other substances also appeared as highly ranked features, including tobacco use and alcohol use (recorded as clinical events), and administration of anxiolytics (ATC code N05B). One explanation for the relevance of anxiolytics is that anxiety is common in patients with OUD, and anxiety sensitivity is a significant predictor for addiction severity.55 In summary, most of the top 50 top features seemed conceptually related to OUD, which indicates that our LSTM based model captured meaningful relationships between the features and OUD.
DISCUSSION
Data-driven studies hold high potential for studying the opioid epidemic in the United States. With the wide availability of EHR, predictive modeling provides a powerful approach to automatically predict the risks of OUD for patients who used prescription opioids.
The LSTM model achieved a promising result with the best F1 score. Top important features generated from our model by permutation importance methods also revealed interesting relationships. Chronic pain management and treatment of acute pain are among the top factors leading to OUD.4 While we focused on a specific disease case (OUD), the methodology and pipeline are general and can be applied to other chronic diseases for early detection through a sequence based predictive model. The pipeline can easily plug into other variants of sequential based models, as demonstrated in our experiments.
Comparison with previous work
In Wadekar’s work17 for OUD prediction, they applied random forest and other traditional methods on hand-crafted features. Our AUROC scores represent a significant improvement from their work, which achieved an AUROC score of 0.8938. Lo-Ciganic et al18,19 applied GBMs and dense neural network models to predict opioid overdose using a set of 268 handcrafted features, achieving an AUROC score around 0.90, which is also lower than our best result. These methods used a limited set of features and lacked the modeling of temporal progression with state-of-the-art methods. Our approach can take advantage of as much information as possible to discover hidden relationships.
There are also sequential deep learning models designed for disease prediction like Dipole56 and BHERT.57 Dipole is an attention based bidirectional recurrent neural network that aims to overcome the problem of performance loss when the length of sequences is large. The Dipole model employs a bidirectional RNN with attention mechanism, which is similar to our proposed Bi-LSTM+Attention model. The main difference is that we applied a more advanced LSTM instead of a traditional RNN cell. As for input, the Dipole work only included medical code features that can be encoded as binary values (presence or absence), while our model also processes numeric value features like laboratory tests and clinical events. BHERT57 is a sequence transduction model for EHR data with multitask prediction and disease trajectory mapping. BHERT has an architecture built with a transformer like BERT. It is powerful in multiple disease prediction, but performance varies on different diseases, with the average precision score ranging from 0.1 to 0.7. Thus, for the prediction of a single disease like OUD, we consider it preferable to use designed features processing.
Benefits of the model
Compared with previous works, our study has several advantages. First, our study employed more comprehensive information, in which previous studies only included a limited set of features—diagnoses, medications, or demographics, all of which are included in our model. Second, many previous works required clinical knowledge to make hand-crafted features. Our models require minimal domain knowledge, making them more generalizable. Third, while a disease is often a progressive process, traditional methods do not model time series data. Our LSTM-based model can use temporal relationships to capture meaningful patterns in trajectory. Fourth, the LSTM model has an advantage over basic RNN models, which can face vanishing gradient problems and insensitivity to gap length.40
Clinical significance
Understanding what is involved in the development of OUD is critical for understanding how to construct a prevention response that may curtail the progression from incidental nonmedical use of prescription opioids to habitual or compulsive use in OUD. In many studies, pain is reported as the most common motivation for opioid use in adults who developed OUD. Such pain treatment may involve prescribed or nonprescribed opioids. Nonprescribed opioid use history is hard to track, but related symptoms and risk factors may still appear in the EHR data. Our model can identify such factors not only to help find patients at risk of OUD, but also to assist the development of knowledge and understanding of such relationships.
Limitations
The population of the study is derived from patients based on structured EHR records, and it does not capture nonprescribed opioids or unrecorded use disorders. The Health Facts database does not differentiate between primary and secondary diagnoses, and the implication to the results from the choice of primary diagnosis versus all diagnoses cannot be evaluated. Interpretation of deep learning models is a challenging task. While our work on feature ranking provides important knowledge for clinical decision support, further research on understanding LSTM models is still needed.
Future work
One future work is to include clinical notes as additional knowledge to improve the model, as notes may provide additional information that may not be captured in structured EHR records. LSTM models have potential limitations such as use of extensive resources and overfitting. More advanced deep learning techniques will be explored to work with sequential models to overcome these limitations. Deep learning visualization tools will be explored to help improve interpretability of results for clinical decision support.
CONCLUSION
The opioid epidemic has become a national emergency for public health in the United States. Predicting risk of OUD for patients taking prescription opioids can provide targeted, focused early interventions for smarter and safer clinical decision support. Our LSTM-based deep learning predictive model of OUD using the history of EHR data demonstrates promising results. The sequential deep learning model is capable of identifying patients who will develop OUD in the future and can provide critical insight on risk factors. Our approach can potentially reduce OUD through earlier intervention in the developmental trajectory.
FUNDING
This work was supported by a Stony Brook University OVPR Seed Grant.
AUTHOR CONTRIBUTIONS
FW, RNR, MS, and JS conceived the study. FW directed the project. XD designed and implemented the study. WH performed statistical design. JD and SR contributed to data extraction and normalization. XD wrote the article. All authors reviewed the manuscript and contributed to revisions.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Janos Hajagos for providing the dataset.
DATA AVAILABILITY STATEMENT
The original data underlying this article were provided by Cerner (https://www.cerner.com/) under institutional agreement. Data sharing of the original data is prohibited. However, all result data, models and codes will be made publicly available at: https://github.com/StonyBrookDB/odprediction. Stony Brook University Institutional Review Board determined that the de-identified Cerner Health Facts database is not human subjects data (#170753_MODCR001).
CONFLICT OF INTEREST STATEMENT
The authors have no competing interests to declare.
REFERENCES
- 1.Centers for Disease Control and Prevention. Assessing and Addressing Opioid Use Disorder (OUD). 2020. https://www.cdc.gov/drugoverdose/training/oud/index.html. Accessed March 28, 2021.
- 2.Centers for Disease Control and Prevention/National Center for Health Statistics. National Vital Statistics System, Mortality. 2018. https://wonder.cdc.gov. Accessed March 28, 2021.
- 3. Schiller EY, Goyal A, Mechanic OJ.. Opioid overdose. In: StatPearls. Treasure Island, FL: StatPearls Publishing; 2020. PMID: 29262202. [PubMed] [Google Scholar]
- 4. Gilson AM, Ryan KM, Joranson DE, et al. A reassessment of trends in the medical use and abuse of opioid analgesics and implications for diversion control: 1997–2002. J Pain Symptom Manage 2004; 28 (2): 176–88. [DOI] [PubMed] [Google Scholar]
- 5. Han B, Compton WM, Blanco C, et al. Prescription opioid use, misuse, and use disorders in U.S. adults: 2015 National Survey on Drug Use and Health. Ann Intern Med 2017; 167 (5): 293–301. [DOI] [PubMed] [Google Scholar]
- 6. Compton WM, Volkow NDJD, Dependence A.. Major increases in opioid analgesic abuse in the United States: concerns and strategies. Drug Alcohol Depend 2006; 81 (2): 103–7. [DOI] [PubMed] [Google Scholar]
- 7. Dowell D, Haegerich TM, Chou RJJ.. CDC guideline for prescribing opioids for chronic pain—United States, 2016. JAMA 2016; 315 (15): 1624–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henry J, Pylypchuk Y, Searcy T, Patel V.. Adoption of Electronic Health Record Systems among US Non-Federal Acute Care Hospitals: 2008-2015. ONC Data Brief 35. 2016. https://dashboard.healthit.gov/evaluations/data-briefs/non-federal-acute-care-hospital-ehr-adoption-2008-2015.php. Accessed March 28, 2021. [Google Scholar]
- 9. DeShazo JP, Hoffman MA.. A comparison of a multistate inpatient EHR database to the HCUP Nationwide Inpatient Sample. BMC Health Serv Res 2015; 15 (1): 384. Jun [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bruneau J, Ahamad K, Goyer MÈ, et al. ; CIHR Canadian Research Initiative in Substance Misuse. Management of opioid use disorders: a national clinical practice guideline. CMAJ 2018; 190 (9): E247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng Y, Wang F, Zhang P, Hu J. Risk prediction with electronic health records: A deep learning approach. In: Proceedings of the 2016 SIAM International Conference on Data Mining; 2016: 432–40.
- 12. Miotto R, Wang F, Wang S, Jiang X, Dudley JT.. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform 2018; 19 (6): 1236–46. Nov [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shickel B, Tighe PJ, Bhorac A, Rashidi P.. Deep EHR: a survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. IEEE J Biomed Health Inform 2017; 22 (5): 1589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang F, Casalino LP, Khullar D.. Deep learning in medicine—promise, progress, and challenges. JAMA Intern Med 2018; 179 (3): 293–4. [DOI] [PubMed] [Google Scholar]
- 15. Glanz JM, Narwaney KJ, Mueller SR, et al. Prediction model for two-year risk of opioid overdose among patients prescribed chronic opioid therapy. J Gen Intern Med 2018; 33 (10): 1646–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ellis RJ, Wang Z, Genes N, et al. Predicting opioid dependence from electronic health records with machine learning. BioData Min 2019; 12 (1): 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wadekar AS. Understanding opioid use disorder (OUD) using tree-based classifiers. Drug Alcohol Depend 2020; 208: 107839. [DOI] [PubMed] [Google Scholar]
- 18. Lo-Ciganic WH, Huang JL, Zhang HH, Weiss JC, et al. Using machine learning to predict risk of incident opioid use disorder among fee-for-service Medicare beneficiaries: A prognostic study. PLoS One 2020; 15 (7): e0235981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lo-Ciganic W-H, Huang JL, Zhang HH, et al. Evaluation of machine-learning algorithms for predicting opioid overdose risk among medicare beneficiaries with opioid prescriptions. JAMA Netw Open 2019; 2 (3): e190968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Calcaterra SL, Scarbro S, Hull ML, Forber AD, Binswanger IA, Colborn KL.. Prediction of future chronic opioid use among hospitalized patients. J Gen Intern Med 2018; 33 (6): 898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med 2018; 1 (1): 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Avati A, Jung K, Harman S, Downing L, Ng A, Shah NH.. Improving palliative care with deep learning. BMC Med Inform Decis Mak 2018; 18 (S4): 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Che Z, Sauver JS, Liu H, Liu Y.. Deep learning solutions for classifying patients on opioid use. AMIA Annu Symp Proc 2017; 2017: 525–34. [PMC free article] [PubMed] [Google Scholar]
- 24. Liu J, Zhang Z, Razavian N.. Deep EHR: Chronic disease prediction using medical notes. Proc Mach Learn Res 2018; 85: 440–64. [Google Scholar]
- 25. Rashidian S, Hajagos J, Moffitt R, et al. Disease phenotyping using deep learning: A diabetes case study. arXiv, doi: https://arxiv.org/abs/1811.11818, 28 Nov 2018, preprint: not peer reviewed
- 26. Rashidian S, Hajagos J, Moffitt RA, et al. Deep learning on electronic health records to improve disease coding accuracy. AMIA Summits Transl Sci Proc 2019; 2019: 620–9. [PMC free article] [PubMed] [Google Scholar]
- 27. Dong X, Rashidian S, Wang Y, et al. Machine learning based opioid overdose prediction using electronic health records. AMIA Annu Symp Proc 2019; 2019: 389–98. [PMC free article] [PubMed] [Google Scholar]
- 28. Hughes A, Williams MR, Lipari RN, Bose J, Copello EA, Kroutil LA.. Prescription drug use and misuse in the United States: Results from the 2015 National Survey on Drug Use and Health. NSDUH Data Review 2016; A1–24. https://www.samhsa.gov/data/sites/default/files/NSDUH-FFR2-2015/NSDUH-FFR2-2015.htm. Accessed March 28, 2021. [Google Scholar]
- 29. Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018; 46 (D1): D1074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moore B. M. Barrett Case Study: Exploring How Opioid-Related Diagnosis Codes Translate From ICD-9-CM to ICD-10-CM. 2017. https://www.hcup-us.ahrq.gov/datainnovations/ICD-10CaseStudyonOpioid-RelatedIPStays042417.pdf. Accessed March 28, 2021.
- 31. Portenoy RK, Foley KM.. Chronic use of opioid analgesics in non-malignant pain: report of 38 cases. Pain 1986; 25 (2): 171–86. [DOI] [PubMed] [Google Scholar]
- 32.Centers for Disease Control and Prevention. SCREENING LIST OF ICD-9-CM CODES FOR CASEFINDING. https://www.cdc.gov/cancer/apps/ccr/icd9cm_codes.pdf. Accessed September 22, 2020.
- 33.Centers for Disease Control and Prevention. ICD-10-CM Table of NEOPLASMS. https://ftp.cdc.gov/pub/Health_Statistics/NCHS/Publications/ICD10CM/2019/icd10cm_neoplasm_2019.pdf. Accessed September 22, 2020.
- 34.General Equivalence Mappings ICD-9-CM to and from ICD-10-CM and ICD-10-PCS. https://www.cms.gov/Medicare/Coding/ICD10/downloads/ICD-10_GEM_fact_sheet.pdf. Accessed March 28, 2021.
- 35. Miller GC, Britt H.. A new drug classification for computer systems: the ATC extension code. Int J Biomed Comput 1995; 40 (2): 121–4. [DOI] [PubMed] [Google Scholar]
- 36.Centers for Disease Control and Prevention. Calculating total daily dose of opioids for safer dosage. https://www.cdc.gov/drugoverdose/pdf/calculating_total_daily_dose-a.pdf. Accessed January 17, 2018.
- 37.Centers for Medicare and Medicaid Services. Opioid oral morphine milligram equivalent (MME) conversion factors. https://www.cms.gov/Medicare/Prescription-Drug-Coverage/PrescriptionDrugCovContra/Downloads/Oral-MME-CFs-vFeb-2018.pdf. Accessed September 22, 2020.
- 38. Raghunathan TE, Lepkowski JM, Van Hoewyk J, Solenberger P.. A multivariate technique for multiply imputing missing values using a sequence of regression models. Surv Methodol 2001; 27 (1): 85–96. [Google Scholar]
- 39. Buuren SV, Groothuis-Oudshoorn K.. mice: Multivariate imputation by chained equations in R. J Stat Soft 2011; 45 (3): 1–68. [Google Scholar]
- 40. Hochreiter S, Schmidhuber J.. Long short-term memory. J Neural Comput 1997; 9 (8): 1735–80. [DOI] [PubMed] [Google Scholar]
- 41. Bahdanau D, Cho K, Bengio Y. Neural machine translation by jointly learning to align and translate. arXiv, doi: https://arxiv.org/abs/1409.0473, 1 Sep 2014, preprint: not peer reviewed.
- 42. Schuster M, Paliwal KK.. Bidirectional recurrent neural networks. IEEE Trans Signal Process 1997. Nov; 45 (11): 2673–81. [Google Scholar]
- 43. Devlin J, Chang MW, Lee K, Bert TK. Pre-training of deep bidirectional transformers for language understanding. arXiv, doi: https://arxiv.org/abs/1810.04805, 11 Oct 2018, preprint: not peer reviewed.
- 44. Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. Adv Neural Inf Process Syst 2017; 30: 5998–6008. [Google Scholar]
- 45.CyberZHG. Keras-Bert. https://github.com/CyberZHG/keras-bert. Accessed March 28, 2021.
- 46. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res 2011; 12: 2825–30. [Google Scholar]
- 47. Abadi M, Barham P, Chen J, et al. Tensorflow: A system for large-scale machine learning. In: 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI 16); 2016: 265–83.
- 48. Gulli A, Pal S.. Deep Learning with Keras. Birmingham, United Kingdom: Packt; 2017. [Google Scholar]
- 49. Bressert E. SciPy and NumPy: An Overview for Developers. Sebastopol, CA: O'Reilly Media; 2012. [Google Scholar]
- 50. McKinney W. pandas: a foundational Python library for data analysis and statistics. In: Python for High Performance and Scientific Computing; 2011; 14 (9): 1–9. [Google Scholar]
- 51. Saito T, Rehmsmeier M.. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One 2015; 10 (3): e0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rice ME, Harris GT.. Comparing effect sizes in follow-up studies: ROC Area, Cohen's d, and r. Law Hum Behav 2005; 29 (5): 615–20. [DOI] [PubMed] [Google Scholar]
- 53. Breiman L. Random forests. Mach Learn 2001; 45 (1): 5–32. [Google Scholar]
- 54. Fan A, Jernite Y, Perez E, Grangier D, Weston J, Eli5 AM. Long form question answering. arXiv, doi: https://arxiv.org/abs/1907.09190, 22 Jul 2019, preprint: not peer reviewed.
- 55. Stathopoulou G, Gold AK, Hoyt DL, Milligan M, Hearon BA, Otto MW.. Does anxiety sensitivity predict addiction severity in opioid use disorder? Addict Behav 2020; 112: 106644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ma F, Chitta R, Zhou J, You Q, Sun T, Gao J. Dipole: diagnosis prediction in healthcare via attention-based bidirectional recurrent neural networks. In: Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2017: 1903–11.
- 57. Li Y, Rao S, Solares JR, et al. BeHRt: transformer for electronic health records. Sci Rep 2020; 10 (1): 7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original data underlying this article were provided by Cerner (https://www.cerner.com/) under an institutional agreement. Data sharing of the original data is prohibited. However, all result data, models, and codes are publicly available (https://github.com/StonyBrookDB/oudprediction). The Stony Brook University Institutional Review Board determined that the de-identified Cerner Health Facts database is not human subjects data (#170753_MODCR001).
The original data underlying this article were provided by Cerner (https://www.cerner.com/) under institutional agreement. Data sharing of the original data is prohibited. However, all result data, models and codes will be made publicly available at: https://github.com/StonyBrookDB/odprediction. Stony Brook University Institutional Review Board determined that the de-identified Cerner Health Facts database is not human subjects data (#170753_MODCR001).