Abstract
Microfluidics-enhanced bioprinting holds great promise in the field of biofabrication as it enables the fabrication of complex constructs with high shape fidelity and utilization of a broad range of bioinks with varying viscosities. Microfluidic systems contain channels on the micrometer-scale, causing a change in fluid behaviors, enabling unconventional bioprinting applications such as facilitating the precise spatial positioning and switching between bioinks with higher accuracy compared to traditional approaches. These systems can roughly be divided into three groups: microfluidic chips, co- and triaxial printheads, and printheads combining both. Although several aspects and parameters remain to be improved, this technology is promising as it is a step toward recapitulating the complex native histoarchitecture of human tissues more precisely. In this Perspective, key research on these different systems will be discussed before moving onto the limitations and outlook of microfluidics-enhanced bioprinting as a whole.
I. INTRODUCTION
Three-dimensional (3D) bioprinting is a form of additive manufacturing, defined as the spatial patterning of living cells, biomaterials, or their combinations in a precisely controlled fashion.1 The aim is to engineer constructs that closely mimic the functionality of specific human tissues for fundamental research, drug discovery, and ultimately regenerative purposes.2–4 Recreating human tissues outside of the body, however, is a considerable challenge. To do so, three core elements must be combined in intricate spatial patterns and architectures, namely, cells, extracellular matrix (ECM), and vasculature.5
Current bioprinting techniques are classified as either inkjet bioprinting,6 vat-polymerization bioprinting,7 laser-assisted bioprinting,8 or extrusion-based bioprinting (EBB).4 Due to the relatively low cost, compatibility with different bioinks, and versatility, EBB has been the main focus of bioprinting research over the past decades.2 In the EBB process, a bioink often consists of a cell-laden hydrogel that possesses shear-thinning properties. As the bioink passes through the nozzle, it is subjected to shear forces resulting in a more fluid-like behavior as the material viscosity decreases. A change in viscosity upon applied shear rate is a key characteristic of non-Newtonian fluids.9 The shear stress and low viscosity remain until the extrusion point, where the bioink quickly increases viscosity and rapidly gelates, stabilizing the cylindrical fiber-shape. The printer follows a computer-aided design (CAD) file and manufactures the construct layer-by-layer as the filament is extruded via pneumatic or mechanical force. Although this technique is promising, there are constraints such as the strong dependence of the printing process on bioink rheology, affecting bioink options, shear stress-induced cell death, as well as the resolution of printed features, which is typically limited to approximately 100 μm.4
Technical advances have been made by enhancing the EBB technique with microfluidic systems. Microfluidics is a multidisciplinary field that intersects with engineering, physics, chemistry, nanotechnology, and biotechnology.10 It has been an asset to a multitude of fields, including but not limited to disease diagnostics,11 drug delivery,12 and biosensing.13 In microfluidic systems, fluid passes through channels with diameters in the range from 100 nm to several hundred micrometers. Harnessing the distinct fluid properties that arise on this scale allows for often unexpected applications.10 Microfluidic systems add a layer of control to the bioprinting process, using either a chip with embedded microchannels or concentric nozzles, otherwise known as coaxial dispensing. In this Perspective, we will first discuss how microfluidic chips have been used to enhance bioprinting, highlighting their potentials. We will then focus on coaxial printheads and how they are used to create solid and hollow fibers while shedding light on their applications. Next, we will describe how these two microfluidic approaches have been combined in single printheads before moving onto the limitations, providing insights and a future perspective on microfluidics-enhanced bioprinting.
II. THE MICROFLUIDIC CHIPS
The microfluidic chip consists of a set of microchannels that are patterned in diverse geometries. Due to the easy customization and flexibility of the design, these chips can have many added benefits within biofabrication.14 A fundamental asset of incorporating a microfluidic chip lies in the ease and simplicity of creating zonal heterogeneity within a single construct. As human tissue is rarely made up of a homogenous mixture of cells and ECM, zonal heterogeneity is crucial to accurately recapitulate native tissue complexity and, therefore, functionality.2 Microfluidic chips allow for easy automated and smooth switching between different materials.
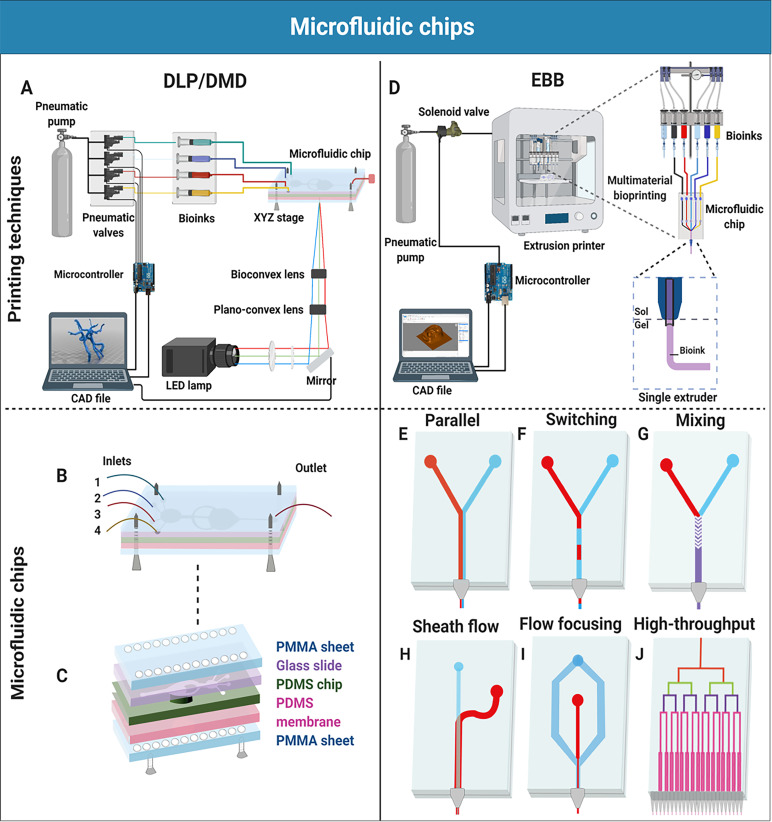
Zonally heterogeneous constructs have been created with different bioprinting techniques. For example, microfluidic chips have been used in digital light processing (DLP) bioprinting [Figs. 1(a)–1(c)], although they are most commonly incorporated into EBB [Figs. 1(d)–1(j)]. DLP bioprinting is a light-based method, where a bath containing a bioink and a photo-excitable cross-linking-initiating agent is illuminated by a light pattern.7 When a threshold level of light intensity is reached, the cross-linking is initiated to cross-link the bioink, trapping the cells inside the mesh. Depending on the design, the stage moves either up or down to cover the previous layer with additional bioink before repeating this process for the subsequent layers. We previously created a DLP bioprinting platform based on a multi-material digital micromirror device (DMD) for rapid bioink switching [Fig. 1(a)].15 Instead of using the conventional open vat, a microfluidic chip was covered by an elastomeric membrane, creating the closed chamber. The membrane was moved by a piston, enabling a layer-by-layer fabrication process. Different layers could be bioprinted with different bioinks with washing steps in between to minimize the cross-bioink contamination. This technology allowed printing materials including poly(ethylene glycol) diacrylate (PEGDA) and gelatin methacryloyl (GelMA) with different viscosities. In the range of Reynolds numbers 10–100, a smooth transition between biomaterials was achieved due to this laminar flow regime.15 The assembly of the microfluidic chip consisted of different layers of the materials polymethylmethacrylate (PMMA), polydimethylsiloxane (PDMS), and glass [Figs. 1(b) and 1(c)]. Although further research is necessary to improve this approach in terms of printing speed and versatility, we were able to successfully generate multi-layered structures and multi-material tissue patterns to model tumor angiogenesis and musculoskeletal systems.15 Considering that the implementation of a microfluidic chip in the biofabrication field is still in its infancy, this study on microfluidics-enhanced DLP demonstrates that the incorporation of microfluidics is not limited to the EBB technology.
FIG. 1.
A schematic illustration of microfluidic chips used in biofabrication. (a) A schematic illustration of a DMD-based DLP microfluidic bioprinting platform setup. A CAD file is converted into a set of images, projected by a DMD before passing through the optical lenses/mirror and reaching the reservoir (microfluidic chip), while the pneumatic valves control the dispensation of the bioinks into the microfluidic chip. (b) Microfluidic chip design for the DMD/DLP bioprinting technology with four inlets and one outlet. (c) The assembly of the microfluidic chip consisting of multiple layers [i.e., poly(methyl methacrylate) (PMMA) sheets, glass slide, PDMS chip, PDMS membrane]. (d) A schematic illustration of a microfluidics-enhanced EBB setup. The CAD file is converted into G codes that help the microcontroller control the dispensation of the bioinks using solenoid valves for multi-material bioprinting. The bioinks flow through the microfluidic chip and are dispensed through a single extruder or multiple extruders. (e)–(j) Microfluidic chips with one or more inlets for distinct purposes. (e) Parallel deposition of two bioinks. (f) Switching between two bioinks for an alternating deposition. (g) Mixing of two bioinks with a herringbone micromixer. (h) Sheath flow ensures to keep the cells in the center of the flow for protection. (i) Flow-focusing controls the fiber diameter by alternating the ratio between the core flow (red) and sheath flow (blue). (j) High-throughput bioprinting using multiple outlets to increase scalability.
Coming back to EBB, switching between bioinks is traditionally done by using multiple cartridges in different printheads, making switching a rather slow and tedious process.16 Instead, a Y or T-shaped microfluidic chip can be used to print with different bioinks in either a concurrent [Fig. 1(e)] or alternating [Fig. 1(f)] fashion. Serex and co-workers17 showed that by using a chip with multiple inlets and channels that converged into a single outlet, material-switching could be achieved without slowing the printing down. Each inlet was connected to a bioink reservoir controlled by an actuated syringe pump, allowing material-switching in a mere 500 ms. This approach can be further refined by replacing the actuated syringe pump for actuated virtual valves, enabling faster switching with higher accuracy.17 Lewis and co-workers18 specifically designed a T-shaped chip to accommodate viscoelastic materials and used it to print different PDMS patterns. The PDMS precursor exhibited shear-thinning properties with a viscosity of 91 Pa s. Unfortunately, PDMS does not allow for the presence of cells and is therefore not well-suited for bioprinting. Still, this work indicates that the design of the chip can be adjusted to accommodate a wide range of materials.
A clear and clean transition between the bioinks in multi-material bioprinting is made possible by the dominantly laminar flow and low diffusion rate of fluid flowing through microscale channels, ensuring minimal bioink mixing. However, in some cases, the mixing of materials is desired. Rather than pre-mixing solutions, a mixing unit can be introduced into the printhead itself [Fig. 1(g)].19 In this scenario, the dominantly laminar flow poses a challenge to efficiently mix fluids in a microfluidic chip. Therefore, various methods have been proposed to tackle this issue, which can be divided into two main categories: passive and active mixing.19 Lee and co-workers20 stated that the passive micromixers employ a pressure head for the flow of fluids at a constant rate and are predominantly dependent on chaotic advection effects deployed by geometrical constraints, controlling the flow. Mixing materials with high viscosities using a passive mixer is considered challenging, as these mixers are typically more suitable for low-viscosity materials due to the presence of diffusive species.19 However, Serex and co-workers17 demonstrated the adequate mixing of two materials within several millimeters by adding a passive herringbone micromixer, in which one was the relatively high-viscosity material glycerol with a viscosity of 1.412 Pa s. Active mixers require external energy for either stirring or mixing using a continuous force. They are generally more controllable and efficient than passive mixers, as mixing intensity is decoupled from the flow rate and volume but also significantly more complex with higher working volumes.19,20 Lewis and co-workers19 also incorporated an impeller-based active mixer into the EBB printhead for optimal bioink homogenization of complex materials such as Newtonian fluids. Ratios between fluids could be controlled on-demand, and intricate gradients of materials and mechanical properties were formed, contributing to the constructs' inter-zonal heterogeneity. Micromixers also facilitate last-minute reactions, opening the door to the use of new materials not traditionally used in bioprinting, such as polymer systems initiated by peroxides by inducing a reaction right before printing.17
Shear stress is one of the limiting factors of EBB, as it takes a toll on extruded cells, which can result in severe damage or even cell death. However, cells do vary in robustness and how susceptible they are to external stressors.21 As an example, neuronal cells are notoriously hard to print due to their high sensitivity. Therefore, Willerth and co-workers22 utilized a microfluidic printhead to reduce the shear stress on the cells using a protective sheath layer, resulting in high cell viability in the printed constructs. Shear stress is most abundant around the outer edges of a flow, where friction occurs between the fluid and the nozzle wall.23 A sheath flow was introduced to hydrodynamically focus cells to the center of the flow, resulting in reduced force exertion on the cells, thus less damage and higher viability [Fig. 1(h)]. Bioink flow-focusing is not only useful in avoiding shear stress but as Serex and co-workers17 demonstrated, it could be utilized to decrease the fiber diameter as well, consequently increasing the resolution [Fig. 1(i)]. Fiber diameter decreased from 800 μm to 200 μm while being controlled by fluid pressure, allowing for real-time adjustments.
Throughput and scalability are essential factors in determining the feasibility of widespread acceptance and usage of bioprinted constructs in a medical setting. For research purposes, small constructs will suffice, but for patient-oriented tissue engineering, it will be oftentimes necessary to generate larger constructs at higher quantities in a short amount of time. For this reason, Lewis and co-workers24,25 created microfluidic chip-based printheads with a multitude of outlets for the simultaneous deposition of identical structures featuring multiple materials in close proximity to each other, which could be easily incorporated in EBB technologies for biomedical applications [Fig. 1(j)]. They demonstrated that materials with varying viscosities could be printed simultaneously by reaching uniform ink flow rates despite their distinct flow characteristics. A viscoelastic wax ink and a Newtonian fluid were co-printed in which the latter ink exhibited a viscosity of 0.3 Pa s. Although both studies used several viscoelastic materials with multi-nozzle systems to create complex structures in a fraction of time, the incorporation of cells has not yet been demonstrated. The usage of bioinks will require the precise tuning of ink rheological properties and printing parameters. Even inks with varying viscosities could be used simultaneously, for example, by adapting the lengths of channels delivering the lower-viscosity inks to match the printing speeds.25 As human tissue often consists of complex compositions of different cells and ECM molecules ordered in small repetitive units, these multi-nozzle systems create an immense increase in throughput, making microfluidics-enhanced EBB an attractive technique for the creation of tissue constructs on a large scale at an increased rate.
III. COAXIAL EXTRUSION PRINTHEADS
The fabrication of both solid and hollow fibers can be achieved using another microfluidic system commonly used in EBB, namely, co- and triaxial nozzle systems (Fig. 2).26,27 A coaxial printhead extrudes the bioink and the cross-linking agent from the nozzle simultaneously, directly cross-linking the fibers at the very bottom-tip of the printhead. In coaxial systems, the printhead configuration consists of two needles in a coaxial arrangement allowing for two entirely separated fluid flows before coming into contact with one another at the moment of extrusion. By generating a core flow of bioink and adding a sheath flow of cross-linking agent, the bioink is rapidly crosslinked upon contact from the outside inward to form a solid fiber. With an inverted configuration in the same coaxial dispenser, the shell of the bioink is crosslinked from the inside out, leaving behind a hollow and perfusable structure. The fiber is deposited with spatial precision by moving the printhead in pre-defined directions, sequentially creating all layers of the desired construct [Fig. 2(a)].26 The appeal of this approach partly lies in the decoupling of rheological properties of the bioink from the printing process. In coaxial EBB, cross-linking does not rely on the shear-thinning properties of the bioink, as often as in conventional EBB. Instead, it solely relies on the contact with the cross-linking agent, which allows the use of a larger variety of bioinks, including the ones with very low-viscosity values.4,26
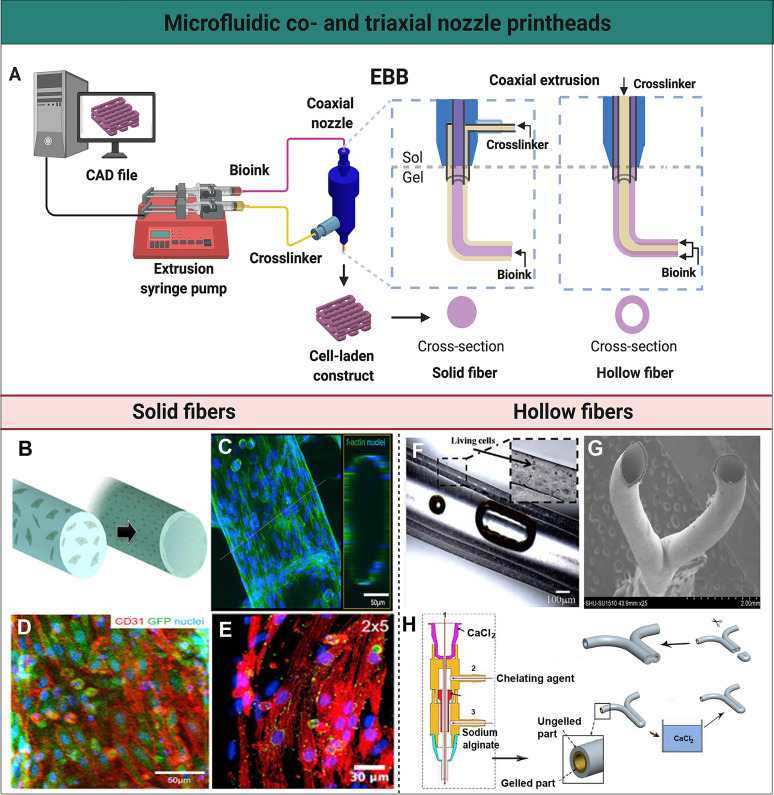
FIG. 2.
Microfluidic co- and triaxial printheads for the fabrication of solid and hollow fibers. (a) A schematic illustration of a microfluidics-enhanced EBB setup with a coaxial extruder. The mechanical-based extrusion syringe pump controls the dispensation of the bioink to create solid or hollow fibers, or their spatial patterns, in a controlled manner based on the CAD design. (b) A schematic representation of solid fibers encapsulating HUVECs that migrate to the periphery to form a lumen-like structure. (c) High-resolution confocal fluorescence micrograph visualizing the distribution of HUVECs on the endothelialized solid fiber. (d) Confocal fluorescence micrograph demonstrating green fluorescent protein (GFP)-HUVECs expressing the surface marker CD31 (red) indicating tight junction-formation. (e) A micrograph showing immunofluorescence staining of the post-seeded directionally aligned cardiomyocytes expressing sarcomeric α-actin (red) and connexin-43 (green). (b)–(e) Reprinted with permission from Zhang et al., Biomaterials 110, 45 (2016). Copyright 2016 Elsevier. (f) Perfusion of the medium through a hollow channel containing living cells. Reprinted with permission from Zhang et al., Nanotechnol. Eng. Med. 4, 0202902 (2013). Copyright 2013 American Society of Mechanical Engineers. (g) Scanning electron microscopy image of a Y-shaped hollow bifurcation. (h) Process of the fabrication of a Y-shaped hollow structure with a triaxial nozzle containing (1) a CaCl2 solution, (2) a chelating agent, and (3) sodium alginate. A Y-shaped hollow fiber was fabricated with this triaxial nozzle, subsequently crosslinked in a CaCl2 bath, and cut to form a branched structure. (g) and (h) Reprinted with permission from Li et al., Biomicrofluidics 10, 64104 (2016). Copyright 2016 IOP Publishing Ltd.
Due to great mechanical properties, biocompatibility, and ease of gelation, the most commonly used bioinks are alginate-based and are physically crosslinked by a cytocompatible calcium chloride (CaCl2) solution. The natural polysaccharide alginate lacks inherent bioactivity, and therefore, functionalization with peptide ligands such as the arginine-glycine-aspartic acid (RGD)-motif is crucial to enhance cell adhesion, migration, and ultimately functionality of the bioprinted constructs.28 Biomaterials that do possess these intrinsic bioactive qualities, such as ECM-like gelatin and collagen, exhibit poor printability by themselves due to their inherent low mechanical properties at lower concentrations causing mechanical instability in constructs. In an attempt to mitigate this limitation, alginate has been blended with ECM-like hydrogels to produce the bioinks.29–31
Our group blended alginate and GelMA, in which human umbilical vein endothelial cells (HUVECs) were encapsulated to generate endothelialized networks [Figs. 2(b)–2(e)].30,32 We extruded the bioink through a coaxial nozzle, ionically cross-linking alginate, which was critical to the scaffold's structural integrity, before chemical photo-cross-linking of the GelMA component post-printing. Also, the continuous sheath flow of CaCl2 resulted in the scaffold's continuous wetting, avoiding dehydration while ensuring cross-linking of the microfibers in adjacent layers. The construct consisted of solid fibers with encapsulated HUVECs that were evenly distributed [Fig. 2(c)]. Cell migration toward the peripheries and complete endothelialization were generally seen after 15 days in culture, and maturation was confirmed by the presence of tight junctions between adjacent cells [Fig. 2(d)]. After the formation of a confluent endothelial layer, cardiomyocytes were seeded onto the construct. Macroscale anisotropic structures were created by varying the distance between fibers deposited on the y axis while maintaining the same distance on the x axis. The anisotropic solid fibers ensured that the seeded cardiomyocytes were locally aligned and expressed sarcomeric α-actin and connexin-43, which are important for the contractile function, resembling the native myocardium [Fig. 2(e)]. Functionality was demonstrated by increased cellular alignment and subsequent beating of the construct for up to 28 days in culture.
In another relevant work, we used a bioink blend of alginate (2% w/v) with not only GelMA but also gelatin or type I collagen.29 GelMA (7% w/v)-alginate, gelatin (10% w/v)-alginate, and collagen (1 mg ml−1)-alginate blend bionks all exhibited non-Newtonian behavior and shear-thinning properties. Alginate merely served as a template, providing temporary structural support during and directly after printing. First, the bioink was extruded through a coaxial printhead, physically cross-linking alginate with a sheath flow of CaCl2. After printing, the second round of cross-linking was induced to either photochemically, chemically, or physically cross-link the GelMA, gelatin, or type I collagen component, respectively. The construct was then moved into a bath containing a Ca2+-chelator, selectively removing the alginate, leaving only the desired biomacromolecules supporting cell adherence, migration, and proliferation. Using this templating method, it becomes possible to create constructs of polymers with bioactive properties with coaxial bioprinting without being hampered by their low printability. This study also highlights the potential of extrusion bioprinting using bioinks with a broad range of viscosities.
Coaxially printed hollow fibers can be used for emulating many tissues with microtubular structures but are particularly promising in the vascularization of larger scaffolds.2 This is a crucial aspect in tissue engineering since the vascular network supplies all cells with necessary oxygen and nutrients and disposes of waste products, and only gains importance as the size of the construct increases, as oxygen and nutrients have a typical diffusion limit of 100–200 μm.33 There is a tremendous need for prevascularized tissue-engineered constructs, as the incorporation of functional blood vessel networks in organ constructs remains one of the most profound challenges researchers are currently facing.
Ozbolat and co-workers34 were one of the first to utilize a coaxial printhead for direct hollow fiber fabrication, eliminating any processing steps crucial in other methods.34–36 Hollow fibers consisting of either cell-laden chitosan or alginate hydrogels were bioprinted with a lumen diameter of fewer than 200 μm. Alginate was superior to chitosan in terms of structural and mechanical integrity. Alginate hydrogel also demonstrated good perfusability as there was no breaking or leaking observed upon oxygenized medium perfusion [Fig. 2(f)]. However, human vasculature does not run in a straight line and contains countless bifurcations. Hu and co-workers37 made the first attempt at fabricating such a bifurcated structure using a triaxial nozzle system to generate a branched hollow fiber [Fig. 2(g)]. The core consisted of a CaCl2 flow, surrounded by an alginate shell. This system was used to print the Y-shaped hollow fiber and was moved into a CaCl2 bath to fully cross-link the outer layer, allowing the fiber to fuse with itself in the lower part of the Y-shape. The barrier wall was removed, unifying the two fused fibers and subsequently creating a bifurcated structure [Fig. 2(h)]. In this particular approach, the ungelled outer layer of the hollow fiber was important for inter-layer attachment. After the printing is completed, the extruder is more prone to clogging. The third nozzle was, therefore, used to swiftly clear the nozzle upon clogging by extruding a chelating solution. Further research is required to optimize this technique to generate multiple bifurcations as the need for a single-step process to fabricate intricate vascular networks containing bifurcations remains.
IV. COMBINING MICROFLUIDIC CHIPS WITH COAXIAL EXTRUSION
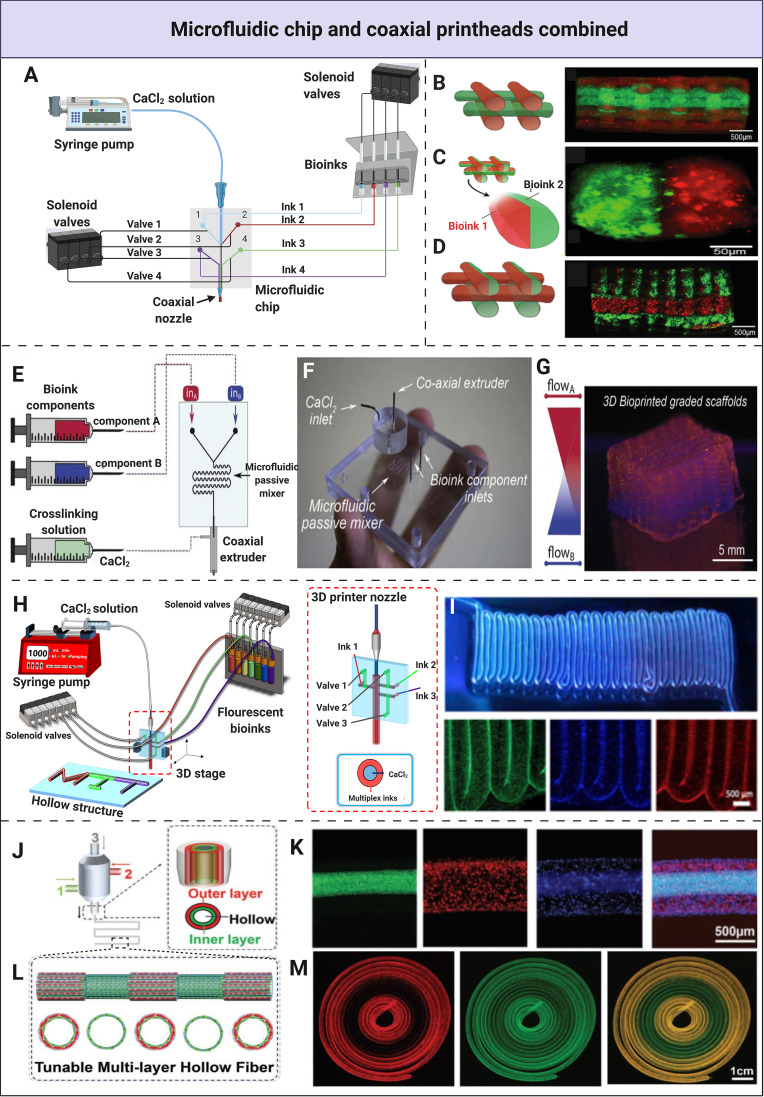
By combining the two microfluidic approaches, the advantages of both microfluidic chips and coaxial printheads can be leveraged. The chip can be mounted upstream of the coaxial dispenser, making it possible to print with low-viscosity bioinks while achieving a higher level of histoarchitectural complexity [Fig. 3(a)]. Depending on the desired construct and its final purpose, a microfluidic chip with a specific function can be designed and incorporated in a printhead with a co-, tri-, or multiaxial extruder. Khademhosseini and co-workers38 used a simple Y-shaped microfluidic chip and a coaxial mechanism to create a heterogeneous cell-laden construct made of solid fibers, using bioink blends of alginate (4%) and GelMA (4.5%) containing either red or green fluorescent beads. After extrusion and initial cross-linking with CaCl2, GelMA was photocrosslinked by UV light, ensuring inter-layer adherence, as well as shape fidelity upon removal of alginate. The relatively low viscosity of alginate/GelMA mixture allowed for the use of a thinner nozzle resulting in a higher resolution and high dispensing speed. This alginate/GelMA blend exhibited Newtonian behavior in the studied range, allowing for the adjustment of dispensing speed without compromising resolution. The system enabled rapid switching between two bioinks and demonstrated versatile deposition patterns [Figs. 3(b)–3(d)]. By alternating between the bioinks, longitudinally diverse fibers were created [Fig. 3(b)], while by simultaneous deposition, miscellaneous fibers were created in predetermined parallel patterns creating transversal heterogeneity [Fig. 3(c)]. Both methods were combined within a single construct, alternating between longitudinal and transversal diversity on-demand [Fig. 3(d)].
FIG. 3.
Integration of microfluidic chips and coaxial nozzles into single printheads. (a) A schematic representation of the multi-material fabrication process and the bioprinting setup with the use of a microfluidic chip incorporated in a coaxial printhead. Solenoid valves have been utilized for the dispensation, mixing the bioinks, or both, and the CaCl2 solution. (b) Extrusion of two bioinks with an alternating position. (c) The combination of alternating and parallel deposition. (d) The deposition of homogenous fibers with parallel patterns. (b)–(d) Reprinted with permission from Colosi et al., Adv. Mater. 28, 677 (2016). Copyright 2015 Wiley-VCH Verlag GmbH & Co. KGaA. (e) Schematics of a system to produce the bioprinted graded constructs. (f) Microfluidic extrusion system consisting of the microfluidic printing head, the coaxial adapter, and the mixing index heatmap. (g) A scaffold featuring a continuous gradient consisting of two bioinks was created with the use of a microfluidic chip with a serpentine mixer and a coaxial extruder. (e)–(g) Reprinted with permission Idaszek et al., Biofabrication 11, 44101 (2019). Copyright 2019 IOP Publishing Ltd. (h) Schematics of the microfluidic system used to print multi-material hollow fibers longitudinally. (i) Longitudinal bioprinting of hollow fibers with a gradual transition between distinct bioinks. (h) and (i) Unpublished data by the authors. (j) The fabrication of multi-layered circumferential fibers with a triaxial nozzle system. (k) Fluorescence microscopy images demonstrating a three-layered fiber consisting of green, red, and blue fluorescent beads representing the inner, middle, and outer layers, respectively. (l) The tunable fabrication of multi-layered fibers with regular intervals of single- and double layers. (m) Confocal fluorescence microscopy images demonstrating the dynamical conversion of single- and double-layer circumferential fibers. (j)–(m) Reprinted with permission from Pi et al., Adv. Mater. 30, 1706913 (2018). Copyright 2018 Wiley-VCH Verlag GmbH & Co. KGaA.
As previously mentioned, the ability to create a smooth transient gradient of distinct bioinks has been a challenge with conventional bioprinting approaches. To overcome this, Święszkowski and co-workers39 were able to create an osteochondral analog by mimicking the zonal cartilage architecture, consisting of hyaline articular cartilage and calcified cartilage layers with an intermediate zone. In order to get a transient construct, the mixing efficiencies of several materials with viscosities ranging from 1 × 10−3 Pa s to 1.1 Pa s were assessed with the use of a passive serpentine mixer. They were still able to mix two inks to a certain degree by altering the volumetric ratios, although there was no perfect mixing. For the osteochondral analog, a continuous transition between bioinks was achieved with the passive mixer, which was incorporated into the microfluidic chip and extruded through a coaxial nozzle [Fig. 3(e)]. A continuous gradient within the construct with high shape fidelity was generated that closely recapitulated the intermediate zone and, thus, the in vivo environment. This system will be particularly valuable in fabricating tissue-engineered analogs exhibiting transient characteristics.
For the generation of tissue-engineered human tubular structures recapitulating their native counterparts, the combination of coaxial dispensers and microfluidic chips can be employed in the creation of hollow fibers. Most human tubular structures are composed of multiple layers with distinct biochemical and mechanical properties due to cellular heterogeneity and organized hierarchical architecture. For instance, in our group's unpublished work, we were the first to generate a multi-material hollow fiber by performing longitudinal coding [Figs. 3(h) and 3(i)]. Here, the switching between bioinks was controlled by the on-chip pneumatic valves to achieve highly accurate transitions of distinct materials that were subsequently extruded through a coaxial nozzle to obtain a hollow structure [Fig. 3(h)].
Continuous bioprinting of hollow fibers exhibiting heterogeneity in the transversal plane has also been achieved with accurate transitions between materials. We generated tubular structures such as blood vessels and urethral tissues using a triaxial dispenser, creating double- and triple-layered hollow fibers, marching a step closer to mimicking the in vivo environment [Figs. 3(j)–3(m)].23 This system allowed deposition ranging between one and three layers to create multi-layered hollow fibers by fluidic manipulation upon extrusion in a single step [Figs. 3(j) and 3(k)]. The versatility of this microfluidic extrusion system was shown by tuning single- and double-layered hollow fibers at regular intervals during bioprinting [Fig. 3(l)] with clear margins between the dynamic conversions [Fig. 3(m)]. The bioink was composed of GelMA, alginate, and 8-arm polyethylene glycol-acrylate with a tripentaerythritol core (PEGOA), respectively, ensuring optimized viscosity during printing, satisfactory cell attachment and proliferation, and sustained mechanical stability. Proof-of-concept of recapitulating native blood vessels containing HUVECs and human vascular smooth muscle cells as well as urethral tissue containing both human urothelial cells and bladder smooth muscle cells was demonstrated. The constructs showed high cell viability and decent functionality for up to 2 weeks. Combining microfluidic chips with coaxial nozzles is not only a significant step in recapitulating vasculature more accurately but it could also be a potential approach for the fabrication of other human tissues that present microtubular structures such as the bile duct and the milk duct.
V. SUMMARY AND OUTLOOK
Microfluidic systems are a simple, low cost, and versatile addition to existing bioprinting techniques and enable the fabrication of sophisticated and compositionally complex 3D tissues, combining cells, ECM materials, and vasculature. Regardless of the relative simplicity of microfluidic devices, adding a technical feature will inevitably add a layer of complexity to a bioprinting process, increasing the chance of problems arising. Microfluidics-enhanced printheads can be limited by increased shear-stresses,40 flow instabilities,17 and technical issues such as the clogging of channels and leaking of fluids.37 DLP bioprinting has progressed from moving between open resin baths upon bioink switching to a closed-circuit system on a microfluidic chip enabling rapid switching between bioinks and washing, referred to as microfluidics-enhanced DLP bioprinting.15 Although this sounds appealing, it still leaves a need for intra-layer diversity. EBB microfluidic printheads do enable intra-layer diversity, answering the need for continuous single-printhead, multi-material printing. Especially in combination with a mixing unit, it becomes possible to create continuous gradients and also ensure bioink homogeneity, as well as combining patterns of different cells, ECM molecules, bioactive agents, and mechanical properties.
Bioinks and their rheological properties play a pivotal role in the entire bioprinting process. In microfluidics-enhanced EBB, rapid gelation and viscosity of bioinks become even more important due to increasing complexity and the need for accuracy, thus making the selection of proper bioinks crucial. Coaxial dispensing systems allow for printing low-viscosity bioinks with viscosities down to 0.08 Pa s and similar values38 due to rapid gelation at extrusion upon contact with a cross-linking agent, expanding the range of bioprintable materials. Although the use of natural bioinks, such as collagen and fibrin, is preferable in terms of cell-friendliness, they typically require more time to gelate due to their inherent biochemical properties compromising mechanical stability directly post-printing.27 For instance, we successfully bioprinted type I collagen but it required 30 min to be cross-linked.29 Factors such as rheological properties, bioink viscosity, gelation time, bioactivity, and mechanical properties are of utmost importance to the entire printing process, and many currently available bioinks do not satisfy all of these needs. Therefore, the development of novel bioink formulations will be necessary to move the field forward. Researchers have found solutions to increase mechanical stability by blending natural biomaterials with synthetic ones, referred to as blend, hybrid, or composite bioinks, where desirable properties of both worlds have been combined.41,42 Looking one step further, the development of new bioink formulations could lead to more advanced technologies. For instance, the use of smart materials, which are materials that respond to external stimuli and subsequently change in shape and morphology after fabrication, could be used in four-dimensional (4D) bioprinting technologies. The potential of 4D technologies combined with microfluidics can lead to the fabrication of biomimetic tissues with more control during and after printing because of utilizing a new dimension, namely, time. The fourth dimension creates a dynamically changing environment to which cells can adapt.43 On top of that, it will also facilitate more control over shape, size, structure, and transformability.10
Even after scientific efforts that might lead to the development of more sophisticated bioinks, other factors such as shear stress may negatively impact the construct's quality and functionality and should, therefore, be taken into consideration. Thus, to protect the cells, researchers can use a sheath flow to add a protective layer, minimizing the stress exerted on the cells while allowing for on-demand tuning of the fiber diameter by adjusting the flow rates.22 Although a reduction of fiber diameter and, thus, an increase in resolution sounds promising, the difference between flow rates can become too large, creating flow instabilities. These instabilities result in a loss of precision and consistency, limiting the degree to which resolution can be enhanced.17 Also, other printing parameters such as dispensing pressure, ink viscosity, and nozzle tip diameter will affect cell viability indirectly.40
Even though considerable progress has been made over the years, reaching the full complexity of a vascular network is currently out of reach. Current hollow fibers created with a coaxial printhead have a diameter down to a couple of hundred μm.36 The diameter of vasculature in the body ranges from several micrometers in the capillaries to >1 cm in the larger veins and arteries.44 A higher resolution is, therefore, needed to closely mimic the function of small-diameter vessels. Besides diameter, it must be noted that the construct does not achieve the complexity of the native vascular bed, as it is a single straight tube. The lack of a perfusable vascular network in larger tissue-engineered constructs, where diffusion is limited, is problematic and will result in a necrotic center due to the lack of oxygen and nutrients that reach the cells.45
Although Hu and co-workers37 made a step toward a branched vascular system by fusing and cutting fibers to create a single bifurcation, this approach lacked simplicity and flexibility. There is still a need to create multiple bifurcations without any additional processing steps. However, when these challenges are overcome, vessels with digitally tunable layers, cell types, and diameters can be printed, making it possible to combine cells, ECM components, and vasculature in a single construct, bringing the entire biofabrication field forward.
For biofabrication to be relevant in regenerative medicine, speed and throughput are critical. Higher throughput means the technique will be easier to adopt in modern medicine practices and allows quick adjustments when aberrations occur. A multi-nozzle chip allows either the simultaneous creation of multiple different and identical constructs or the simultaneous creation of repetitive structures within a single construct, significantly speeding up the printing process. By adopting a modular approach in designing a microfluidic system and combining different blocks or modules, one can integrate multiple features into a single intricate system allowing for easy reconfiguration, even post-fabrication of the system itself.46 To our knowledge, this concept has not yet been applied to bioprinting, but drawing from the abundance of research into microfluidic systems in general as well as multi-nozzle systems, it can contribute to transforming bioprinting into a more versatile and high(er)-throughput technique.
We anticipate that advances in microfluidics can lead to the optimization of handheld extrusion bioprinters in combination with robotic arms and artificial intelligence, allowing for in situ bioprinting.47,48 Up until now, tissues such as skin48 and cartilage49 have been fabricated using an in situ approach. Most of the in situ approaches used coaxial nozzle systems for multi-material printing since these tissues are complex and heterogeneous. The desirable features of microfluidic chips and multiaxial printheads will aid in the progression of biofabrication as a whole and ultimately to the development of more advanced treatments and innovative technologies. For example, the fabrication of in situ-bioprinted autologous skin substitutes that can mimic the complex skin architecture to a greater extent by printing multiple layers consisting of multiple cell types and materials in a rapid and highly organized manner for greater in vivo functionality. Tissue regeneration can then be turned into a single-step personalized procedure, directly printing the desired substitute with autologous cells at the target site. Advancements in microfluidics may also lead to the printing of other complex multi-material constructs and hollow in situ vessels. Despite these advances, connecting the vessels to host vasculature and long-term graft survival remain obstacles.
Apart from this, we also anticipate that future advancements in microfluidics-enhanced bioprinting will contribute to further advancing current technologies where microfluidic bioprinting and other strategies such as organ-on-a-chip modeling and the fabrication of micro-tissues, cell-laden microspheres, and organoids are synergistically combined.50–52 Future applications are diverse and range from facilitating the development of novel delivery mechanisms as well as novel drug discovery to disrupting the status quo of in vitro drug testing by aiding the process of clinical trials.53,54
There is a need for smarter dispensing tools as bioprinting aims to contribute to regenerative medicine. This novel bioprinting paradigm has the potential to make this a reality. The combinations of microfluidic chips with coaxial nozzles offer many functions that may vary from the incorporation of micromixers to homogenize the bioink and form gradients in hollow fibers to printing with multiple bioinks with diverse viscosities and accurately switching between them by using automated valves. We believe that these smarter dispensing tools can enable researchers to create complex constructs using multiple materials with distinct rheological properties, cell types, cell densities, and biological stimulants, thereby generating distinct structural and mechanical integrities resembling the target tissue.17 Microfluidic printheads are a way of automating and simplifying the creation of complex constructs, increasing accuracy, reproducibility, and ease of use. Even though more extensive research into the development of new bioink formulations and optimization of printheads are required, we believe that microfluidic technology has the potential to disrupt the norm in bioprinting in a multitude of ways, taking bioprinting to the next level in ways that exceed our current imagination, potentially changing the world of medicine.
AUTHORS’ CONTRIBUTIONS
D.N.d.C., K.P.F., and P.A. contributed equally to this work.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (Nos. R21EB025270, R21EB026175, R00CA201603, R01EB028143, R01HL153857, and R21EB030257), the National Science Foundation (No. 1936105), and Brigham Research Institute.
Y.S.Z. sits on the Scientific Advisory Board of Allevi, Inc., which however, did not participate or sponsor this work.
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1.Heinrich M. A., Liu W., Jimenez A., Yang J., Akpek A., Liu X., Pi Q., Mu X., Hu N., Schiffelers R. M., Prakash J., Xie J., and Zhang Y. S., Small 15, 1805510 (2019). 10.1002/smll.201805510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levato R., Jungst T., Scheuring R. G., Blunk T., Groll J., and Malda J., Adv. Mater. 32, 1906423 (2020). 10.1002/adma.201906423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy S. V. and Atala A., Nat. Biotechnol. 32, 773 (2014). 10.1038/nbt.2958 [DOI] [PubMed] [Google Scholar]
- 4.Ozbolat I. T. and Hospodiuk M., Biomaterials 76, 321 (2016). 10.1016/j.biomaterials.2015.10.076 [DOI] [PubMed] [Google Scholar]
- 5.Kolesky D. B., Homan K. A., Skylar-Scott M., and Lewis J. A., Altern. Lab. Anim. 46, 209 (2018). 10.1177/026119291804600404 [DOI] [PubMed] [Google Scholar]
- 6.Compaan A. M., Christensen K., and Huang Y., ACS Biomater. Sci. Eng. 3, 1519 (2017). 10.1021/acsbiomaterials.6b00432 [DOI] [PubMed] [Google Scholar]
- 7.Li W., Mille L. S., Robledo J. A., Uribe T., Huerta V., and Zhang Y. S., Adv. Healthcare Mater. 9, 2000156 (2020). 10.1002/adhm.202000156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vinson B. T., Sklare S. C., and Chrisey D. B., Curr. Opin. Biomed. Eng. 2, 14 (2017). 10.1016/j.cobme.2017.05.005 [DOI] [Google Scholar]
- 9.Guvendiren M., Lu H. D., and Burdick J. A., Soft Matter 8, 260 (2012). 10.1039/C1SM06513K [DOI] [Google Scholar]
- 10.Hou X., Zhang Y. S., Trujillo-de Santiago G., Alvarez M. M., Ribas J., Jonas S. J., Weiss P. S., Andrews A. M., Aizenberg J., and Khademhosseini A., Nat. Rev. Mater. 2, 17016 (2017). 10.1038/natrevmats.2017.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park S., Zhang Y., Lin S., Wang T.-H., and Yang S., Biotechnol. Adv. 29, 830 (2011). 10.1016/j.biotechadv.2011.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Q., Hashimoto M., Dang T. T., Hoare T., Kohane D. S., Whitesides G. M., Langer R., and Anderson D. G., Small 5, 1575 (2009). 10.1002/smll.200801855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giouroudi I. and Keplinger F., Int. J. Mol. Sci. 14, 18535 (2013). 10.3390/ijms140918535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nie M. and Takeuchi S., Biofabrication 10, 44103 (2018). 10.1088/1758-5090/aadef9 [DOI] [PubMed] [Google Scholar]
- 15.Miri A. K., Nieto D., Iglesias L., Goodarzi Hosseinabadi H., Maharjan S., Ruiz-Esparza G. U., Khoshakhlagh P., Manbachi A., Dokmeci M. R., Chen S., Shin S. R., Zhang Y. S., and Khademhosseini A., Adv. Mater. 30, 1800242 (2018). 10.1002/adma.201800242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richard C., Richard C., Neild A., Cadarso V. J., and Cadarso V. J., Lab Chip 20, 2044 (2020). 10.1039/C9LC01184F [DOI] [PubMed] [Google Scholar]
- 17.Serex L., Bertsch A., and Renaud P., Micromachines 9, 86 (2018). 10.3390/mi9020086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardin J. O., Ober T. J., Valentine A. D., and Lewis J. A., Adv. Mater. 27, 3279 (2015). 10.1002/adma.201500222 [DOI] [PubMed] [Google Scholar]
- 19.Ober T. J., Foresti D., and Lewis J. A., Proc. Natl. Acad. Sci. U.S.A. 112, 12293 (2015). 10.1073/pnas.1509224112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee C.-Y., Chang C.-L., Wang Y.-N., and Fu L.-M., Int. J. Mol. Sci. 12, 3263 (2011). 10.3390/ijms12053263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pati F., Jang J., Lee J.-W., and Cho D.-W., in Essentials of 3D Biofabrication and Translation (Academic Press, 2015), pp. 123–152. 10.1016/B978-0-12-800972-7.00007-4 [DOI] [Google Scholar]
- 22.Lee C., Abelseth E., De La Vega L., and Willerth S. M., Mater. Today Chem. 12, 78 (2019). 10.1016/j.mtchem.2018.12.005 [DOI] [Google Scholar]
- 23.Pi Q., Maharjan S., Yan X., Liu X., Singh B., van Genderen A. M., Robledo-Padilla F., Parra-Saldivar R., Hu N., and Jia W., Adv. Mater. 30, 1706913 (2018). 10.1002/adma.201706913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen C. J., Saksena R., Kolesky D. B., Vericella J. J., Kranz S. J., Muldowney G. P., Christensen K. T., and Lewis J. A., Adv. Mater. 25, 96 (2013). 10.1002/adma.201203321 [DOI] [PubMed] [Google Scholar]
- 25.Skylar-Scott M. A., Mueller J., Visser C. W., and Lewis J. A., Nature 575, 330 (2019). 10.1038/s41586-019-1736-8 [DOI] [PubMed] [Google Scholar]
- 26.Costantini M., Colosi C., Święszkowski W., and Barbetta A., Biofabrication 11, 12001 (2018). 10.1088/1758-5090/aae605 [DOI] [PubMed] [Google Scholar]
- 27.Onoe H., Okitsu T., Itou A., Kato-Negishi M., Gojo R., Kiriya D., Sato K., Miura S., Iwanaga S., and Kuribayashi-Shigetomi K., Nat. Mater. 12, 584 (2013). 10.1038/nmat3606 [DOI] [PubMed] [Google Scholar]
- 28.Lee K. Y. and Mooney D. J., Prog. Polym. Sci. 37, 106 (2012). 10.1016/j.progpolymsci.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu K., Chen N., Liu X., Mu X., Zhang W., Wang C., and Zhang Y. S., Macromol. Biosci. 18, e1800127 (2018). 10.1002/mabi.201800127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y. S., Arneri A., Bersini S., Shin S.-R., Zhu K., Goli-Malekabadi Z., Aleman J., Colosi C., Busignani F., and Dell’Erba V., Biomaterials 110, 45 (2016). 10.1016/j.biomaterials.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao G., Lee J. H., Jang J., Lee D. H., Kong J.-S., Kim B. S., Choi Y.-J., Jang W. B., Hong Y. J., Kwon S.-M., and Cho D.-W., Adv. Funct. Mater. 27, 1700798 (2017). 10.1002/adfm.201700798 [DOI] [Google Scholar]
- 32.Zhang Y. S., Pi Q., and van Genderen A. M., JoVE 126, e55957 (2017). 10.3791/55957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carmeliet P. and Jain R. K., Nature 407, 249 (2000). 10.1038/35025220 [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y., Yu Y., and Ozbolat I. T., J. Nanotechnol. Eng. Med. 4, 020902 (2013). 10.1115/1.4024398 [DOI] [Google Scholar]
- 35.Liu T., Liu Q., Anaya I., Huang D., Kong W., Mille L. S., and Zhang Y. S., “Investigating lymphangiogenesis in a sacrificially bioprinted volumetric model of breast tumor tissue,” Methods (published online, 2020). 10.1016/j.ymeth.2020.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong J., Schuurmans C. C. L., van Genderen A. M., Cao X., Li W., Cheng F., He J. J., López A., Huerta V., and Manríquez J., Nat. Commun. 11, 1267 (2020). 10.1038/s41467-020-14997-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S., Liu Y., Li Y., Liu C., Sun Y., and Hu Q., Biomicrofluidics 10, 64104 (2016). 10.1063/1.4967456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colosi C., Shin S. R., Manoharan V., Massa S., Costantini M., Barbetta A., Dokmeci M. R., Dentini M., and Khademhosseini A., Adv. Mater. 28, 677 (2016). 10.1002/adma.201503310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Idaszek J., Costantini M., Karlsen T. A., Jaroszewicz J., Colosi C., Testa S., Fornetti E., Bernardini S., Seta M., Kasarełło K., Wrzesień R., Cannata S., Barbetta A., Gargioli C., Brinchman J. E., and Święszkowski W., Biofabrication 11, 44101 (2019). 10.1088/1758-5090/ab2622 [DOI] [PubMed] [Google Scholar]
- 40.Chang R., Nam J., and Sun W., Tissue Eng. A 14, 41 (2008). 10.1089/ten.a.2007.0004 [DOI] [PubMed] [Google Scholar]
- 41.Cui X., Li J., Hartanto Y., Durham M., Tang J., Zhang H., Hooper G., Lim K., and Woodfield T., Adv. Healthcare Mater. 9, 1901648 (2020). 10.1002/adhm.201901648 [DOI] [PubMed] [Google Scholar]
- 42.Gungor-Ozkerim P. S., Inci I., Zhang Y. S., Khademhosseini A., and Dokmeci M. R., Biomater. Sci. 6, 915 (2018). 10.1039/C7BM00765E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y.-C., Zhang Y. S., Akpek A., Shin S. R., and Khademhosseini A., Biofabrication 9, 12001 (2016). 10.1088/1758-5090/9/1/012001 [DOI] [PubMed] [Google Scholar]
- 44.Gao Y., Biology of Vascular Smooth Muscle: Vasoconstriction Dilatation (Springer, Singapore, 2017), pp. 3–12. 10.1007/978-981-10-4810-4_1 [DOI] [Google Scholar]
- 45.Radisic M., Yang L., Boublik J., Cohen R. J., Langer R., Freed L. E., and Vunjak-Novakovic G., Am. J. Physiol. Heart Circ. Physiol. 286, 507 (2004). 10.1152/ajpheart.00171.2003 [DOI] [PubMed] [Google Scholar]
- 46.Lai X., Shi Z., Pu Z., Zhang P., Zhang X., Yu H., and Li D., Microsyst. Nanoeng. 6, 27 (2020). 10.1038/s41378-020-0136-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ozbolat I. T., Trends Biotechnol. 33, 395 (2015). 10.1016/j.tibtech.2015.04.005 [DOI] [PubMed] [Google Scholar]
- 48.Ying G., Manríquez J., Wu D., Zhang J., Jiang N., Maharjan S., Medina D. H. H., and Zhang Y. S., Mater. Today Bio 8, 100074 (2020). 10.1016/j.mtbio.2020.100074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Bella C., Duchi S., O’Connell C. D., Blanchard R., Augustine C., Yue Z., Thompson F., Richards C., Beirne S., and Onofrillo C., J. Tissue Eng. Regener. Med. 12, 611 (2018). 10.1002/term.2476 [DOI] [PubMed] [Google Scholar]
- 50.Yamada M., Utoh R., Ohashi K., Tatsumi K., Yamato M., Okano T., and Seki M., Biomaterials 33, 8304 (2012). 10.1016/j.biomaterials.2012.07.068 [DOI] [PubMed] [Google Scholar]
- 51.Zhao H., Chen Y., Shao L., Xie M., Nie J., Qiu J., Zhao P., Ramezani H., Fu J., and Ouyang H., Small 14, 1802630 (2018). 10.1002/smll.201802630 [DOI] [PubMed] [Google Scholar]
- 52.Sharma R., Smits I. P. M., De La Vega L., Lee C., and Willerth S. M., Front. Bioeng. Biotechnol. 8, 1 (2020). 10.3389/fbioe.2020.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Low L. A., Mummery C., Berridge B. R., Austin C. P., and Tagle D. A., Nat. Rev. Drug Discovery 20, 00793 (2020). 10.1038/s41573-020-0079-3 [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y. S. and Khademhosseini A., Bio-Des. Manuf. 3, 155 (2020). 10.1007/s42242-020-00080-w [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.