Abstract
The RNA-dependent RNA polymerase (RdRp) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a promising drug target for coronavirus disease 2019 (COVID-19) because it plays the most important role in the replication of the RNA genome. Nucleotide analogs such as remdesivir and favipiravir are thought to interfere with the RNA replication by RdRp. More specifically, they are expected to compete with nucleoside triphosphates, such as ATP. However, the process in which these drug molecules and nucleoside triphosphates are taken up by RdRp remains unknown. In this study, we performed all-atom molecular dynamics simulations to clarify the recognition mechanism of RdRp for these drug molecules and ATP that were at a distance. The ligand recognition ability of RdRp decreased in the order of remdesivir, favipiravir, and ATP. We also identified six recognition paths. Three of them were commonly found in all ligands, and the remaining three paths were ligand-dependent ones. In the common two paths, it was observed that the multiple lysine residues of RdRp carried the ligands to the binding site like a “bucket brigade.” In the remaining common path, the ligands directly reached the binding site. Our findings contribute to the understanding of the efficient ligand recognition by RdRp at the atomic level.
Significance
Remdesivir and favipiravir are expected to be recognized by the RNA-dependent RNA polymerase (RdRp) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and inhibit the RNA replication. However, the recognition process of these molecules has been unknown. To investigate the recognition process, all-atom molecular dynamics simulations were performed for RdRp with these molecules and ATP located far away. As a result, the behavior of multiple lysine residues of RdRp carrying the ligands to the binding site like a “bucket brigade” was observed. The role of lysine residues arranged in a straight line toward the binding site of RdRp was clarified. We expect our results to contribute to the understanding of the efficient ligand recognition by RdRp.
Introduction
Coronavirus disease 2019 (COVID-19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (1). It causes pathogenic symptoms such as cough, fever, dysgeusia, and olfactory dysfunction (2, 3, 4). Severely ill patients (2) may develop pneumonia and a cytokine storm syndrome (5). In addition, it is human-to-human infectious and has a higher reproduction rate than severe acute respiratory syndrome coronavirus (SARS-CoV), which also increases the risk of the virus spread (6). COVID-19 was first reported in December 2019 in Wuhan, Hubei Province, China, and rapidly spread over the world, causing the declaration of a pandemic by the World Health Organization in March 2020 (7). According to the World Health Organization, as of July 11, 2021, more than 186 million confirmed cases and 4.0 million deaths have been reported worldwide (8). Given the current spread of infection worldwide, drug repositioning of existing therapeutic agents is a promising approach because it could take years to develop new scientifically established therapeutic treatments.
SARS-CoV-2 is classified in the Betacoronavirus belonging to the family Coronaviridae, similar to other highly pathogenic viruses such as SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) (9). It has a large, single-stranded, positive-sense RNA genome composed of ∼30 kilobases, which codes for more than 20 structural and nonstructural proteins (nsps) (10, 11, 12). After the invasion of the virus into the host cell cytoplasm, the RNA genome is released for replication (13, 14, 15). The translation of open reading frames 1a and 1b produces two large viral polyproteins (polyprotein 1a and polyprotein 1ab), which are further cleaved by proteolysis to produce the 16 nsps (nsp1–16) (10,16). Among these nsps, nsp12, known as an RNA-dependent RNA polymerase (RdRp), plays the most important role in the viral replication and transcription processes. This is because it catalyzes the synthesis of the nascent RNA with the help of nsp7 and nsp8, acting as cofactors (Fig. 1) (17, 18, 19, 20, 21, 22, 23). Therefore, RdRp attracts much attention as a primary target for drugs because the replication cycle of the virus can be terminated by inhibiting the function of RdRp (17,24).
Figure 1.
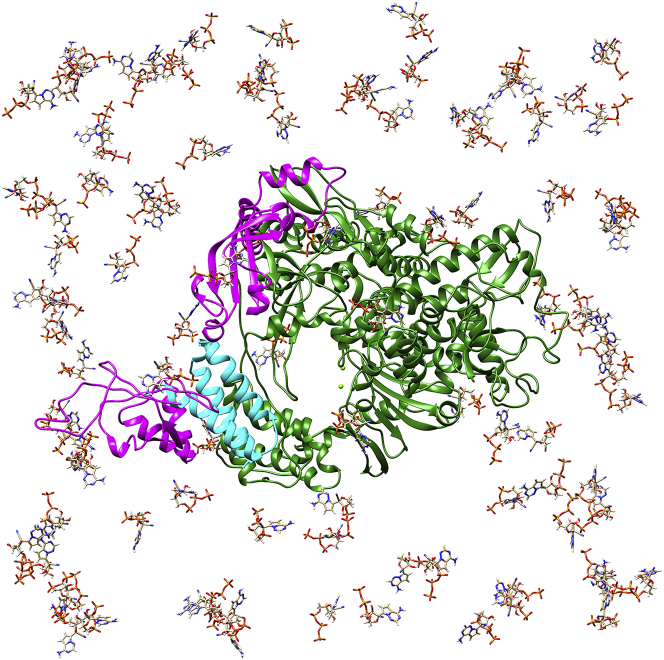
The initial structure of RdRp with 100 RemTPs. Water molecules and counterions are omitted for clarity. Shown are nsp12, two nsp8s, and nsp7 in dark green, magenta, and cyan, respectively. Two small light green spheres indicate Mg2+ ions. To see this figure in color, go online.
RdRp mainly comprises of four parts, an N-terminal β-hairpin (residues D29 to K50), nidovirus RdRp-associated nucleotidyltransferase domain (residues T51 to R249), interface domain (residues A250 to R365), and conserved polymerase domain (residues L366 to Q932) (19). The polymerase domain of RdRp adopts a common architecture among many RNA viruses that resembles a right hand composed of three subdomains (fingers, palm, and thumb). The domain also has seven conserved motifs (A–G) composed of residues that are essentially important for RdRp function at the binding site. The motif C contains the residues forming catalytic active site (residues S759 to D761), which are also conserved in many viral RdRps. The motifs E and F play an important role in primer binding, and motif G is mainly involved in RNA template binding. In addition, RdRp sequence shares a high degree of identity with those of other coronaviruses. In particular, SARS-CoV-2 and SARS-CoV nsp12s show more than 96% sequence identity, and the tertiary structure of SARS-CoV-2 nsp12 is similar to that of SARS-CoV (19,25). These data on conservation and homology of SARS-CoV-2 are useful in applying therapeutic agents tested on other RNA viruses.
Nucleotide analogs such as remdesivir and favipiravir (trade name: Avigan) are promising drug candidates targeting the viral RdRps and have shown potential therapeutic effects. Remdesivir was created by Gilead Sciences (Foster City, CA) as a treatment for Ebola virus disease (26). Favipiravir was created by Toyama Chemical (Tokyo, Japan) as an antiinfluenza virus agent (27). These drugs are expected to inhibit replication of RNA. RdRp usually recognizes nucleoside triphosphates (NTPs) such as ATP to replicate RNA. In cells, remdesivir that is triphosphorylated (RemTP) and favipiravir that is ribosylated and triphosphorylated (FavTP) to become active metabolite form (Fig. S1) are thought to interfere with the RNA replication by RdRp (20,28, 29, 30, 31).
Several experimental and computational studies on the activity of these nucleotide analogs have been reported. In vitro and in vivo experiments showed that remdesivir and favipiravir suppress the replication of SARS-CoV, MERS-CoV, and SARS-CoV-2 with the high potential to inhibit the RdRp function (32, 33, 34, 35, 36, 37, 38). The tertiary structure of an RNA-RdRp complex with remdesivir monophosphate or FavTP was determined by using cryogenic electron microscopy (cryo-EM) (20,28, 29, 30, 31). Several molecular docking and free-energy perturbation studies reported the tight binding of RemTP and FavTP to the binding site of RdRp and the estimated binding poses (24,39, 40, 41). The binding mechanisms of remdesivir were investigated using steered molecular dynamics (MD) and umbrella sampling (42) and MD simulation in implicit solvent (43,44). Quantum mechanics/molecular mechanics (QM/MM) calculations and MD simulations were performed to investigate the inhibition of RdRp activity by RemTP (45,46). We recently compared the dynamic properties of RdRps without ligands in SARS-CoV and SARS-CoV-2 using MD simulations (47).
In this study, we perform MD simulations to clarify how SARS-CoV-2 RdRp takes up and recognizes RemTP, FavTP, and ATP that were at a distance. The MD simulation is a powerful theoretical approach to study the dynamics of drugs (48,49) and disease-related biomolecules, such as proteins (50) and glycans (51). Previous studies on the RdRp systems, including ligands, have focused on the RdRp structure after the drug molecule was recognized (39,46). No studies have been reported for the process in which the drug molecules and NTPs are recognized. This study is the first work to reveal the recognition process of the drug molecules and ATP by RdRp.
Materials and methods
System setup
We employed the cryo-EM structure of SARS-CoV-2 RdRp (Protein Data Bank (PDB) entry: 7bv2) (20) as the initial structure. Because one of the two nsp8s in this cryo-EM structure was missing, the nsp8 corresponding to the chain D in the apo RdRp (PDB entry: 7bv1) (20) was added. The missing nonterminal residues in the cryo-EM structure were complemented using the modeller plugin (52) in UCSF Chimera (53). Hydrogen atoms were added using the LEaP and reduce plugin (54) in AMBER. Proteins were described with Amber ff14SB force field (55). TIP3P parameters (56) were employed for water molecules. The geometries of the ligand molecules (RemTP, FavTP, and ATP) were optimized at Hartree-Fock/6-31+G(d) in vacuo. The restraint electrostatic potential method (57) was used to determine the atomic partial charges of the ligand molecules. The geometrical optimization and restraint electrostatic potential calculations of the ligand molecules were performed using the Gaussian 16 Revision C.01 package (58). The Lennard-Jones (LJ) parameters for the ligand molecules were obtained from the general Amber force field parameter version 2 set (59) assigned by Antechamber (60). The LJ parameters for the divalent and monovalent metal ions were taken from references (61) and (62), respectively. Ligand molecules were distributed using the generalized-ensemble molecular biophysics (GEMB) program, which was developed by one of the authors (H.O.) (63). Initial conformations of several protein systems have been prepared, and their simulations have been performed using this program so far (64, 65, 66). 100 ligand molecules were randomly placed around RdRp at a distance of more than 50 Å from one of the two Mg2+ ions (residue: 1193) at the binding site (Fig. 1). The distance between the centers of mass of any two ligands was set to 17 Å or more. Using different seeds of random numbers for the ligand arrangement, five initial conformations were prepared for each system. 415 Na+ ions were added to neutralize the system, and the total system consisted of ∼400,000 atoms (402,188 atoms for the RemTP system, 402,489 for the FavTP system, and 402,465 for the ATP system). The entire system was immersed in a cubic box with a volume of 1603 Å3. The ligand concentration in each system was set to 40.5 mM to observe the spontaneous ligand recognition by RdRp during the simulation time. Note that this concentration is higher than the physiological concentration of ATP in vivo (∼2–8 mM) (67) and the 50% cytotoxic concentration of remdesivir and favipiravir in vitro (∼100–400 μM) (68).
MD simulations
All MD simulations were performed using the program AMBER18 (69). The system was first minimized with 5000 steps of the steepest descent method and then with 5000 steps of the conjugate gradient method. The system with minimized energy was gradually heated from 0 to 310 K in 310 ps. After the heating phase, short equilibration at 310 K lasted for 690 ps. A production run simulation was then performed for 110 ns. 50 independent MD simulations were conducted for each system with the combinations of the 5 initial conformations and 10 initial velocities. All MD simulations were performed in the isothermal-isochoric (NVT) ensemble at T = 310 K, except for the heating phase. Langevin heat bath with a damping coefficient of 2 ps−1 was used to keep the temperature constant. The LJ potential cutoff was 12 Å. Long-range electrostatic interactions were calculated using the Particle Mesh Ewald method (70) with an Ewald coefficient of 0.305 Å−1, and the grid size was set to 1603. Periodic boundary conditions were used. Bonds involving hydrogen atoms were constrained using the SHAKE method (71). The equations of motion were integrated using the leapfrog algorithm with a time step of 2 fs except for the heating phase, in which a time step of 1 fs was used. The cpptraj program (72) implemented in AMBER was used for trajectory analysis. The trajectories and conformations were visualized by UCSF Chimera (53).
Results and discussion
Ligand recognition probability of RdRp
We performed MD simulations for three systems. Each system consists of RdRp with either RemTPs, FavTPs, or ATPs. We observed that the ligands were recognized by RdRp in all three systems. The number of trajectories, 50, and simulation time, 110 ns, in this study are more than those in similar simulations for other proteins with many ligands (49), and ligand recognition behavior was sufficiently observed within 110 ns. More specifically, the ligands were taken up at the binding site of RdRp during the simulations, as shown in Video S1. To investigate the ligand dependence of the ligand recognition of RdRp, we calculated ligand recognition probabilities. Here, when the closest distance between the Mg2+ ions at the binding site and any atoms of the ligand was less than 3.5 Å, it was considered as a “contact” event. Furthermore, when the contact events were observed in more than 10 snapshots out of the last 1000 snapshots (corresponding to the last 10 ns) in the simulations, it was considered as a “ligand recognition” event. The ligand recognition probability was obtained by dividing the number of the MD simulations with the ligand recognition events by the total number of the MD simulations (50 MD simulations), as listed in Table 1. Although within the statistical errors, RemTP shows the highest probability, and FavTP has the second-highest probability, followed by ATP. These results are qualitatively consistent with previous experimental studies (20,36). In addition, our results agree with the MD simulations using the free-energy perturbation method for the RdRp-RemTP complex, in which it was shown that RemTP binds much stronger to RdRp than ATP (39). We remark that the ligands sometimes aggregated because of a large number of the counterions (415 Na+) in the systems. These aggregates were stabilized by the interaction between the phosphate groups of the ligands and the counterions. To eliminate this effect, even if the ligand aggregates were recognized by RdRp, it was not considered as a ligand recognition event. Here, the number of trajectories with aggregation events were only one for RemTP, three for FavTP, and two for ATP.
Table 1.
The number of MD simulations in which ligand recognition events occurred out of the total 50 MD simulations for each ligand system
| Ligand | Ligand recognition/total | Probability |
|---|---|---|
| RemTP | 12/50 | 0.24 ± 0.07 |
| FavTP | 9/50 | 0.18 ± 0.06 |
| ATP | 7/50 | 0.14 ± 0.06 |
The ligand recognition probabilities by RdRp are also listed. The errors were estimated using the bootstrap method.
Ligand recognition path by RdRp
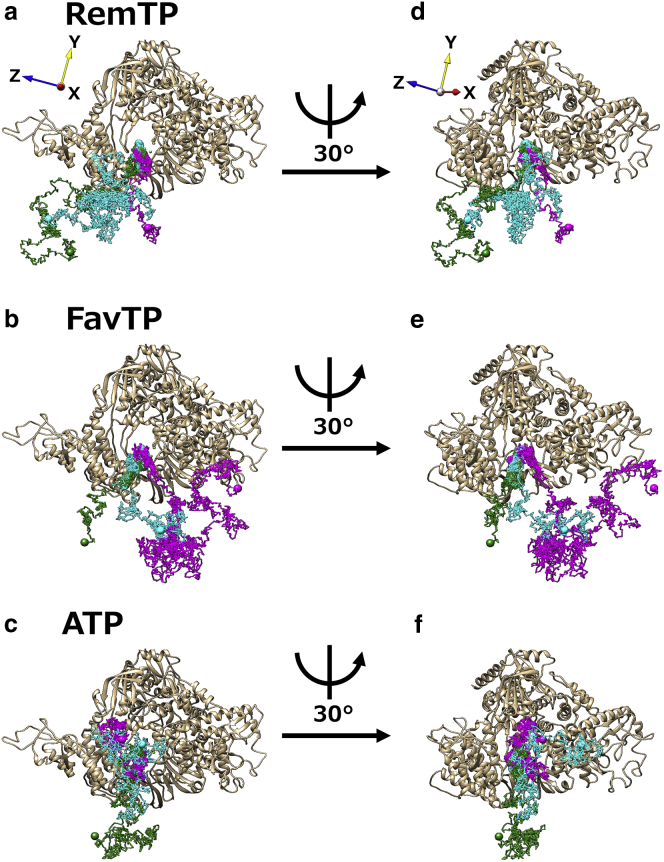
To understand the ligand recognition mechanism by RdRp, trajectories of the recognized ligands were investigated. We found that the ligand recognition paths can be classified into six types, paths 1–6, based on the time series of contact probabilities between the recognized ligand and each residue of RdRp. Paths 1–3 were commonly observed for all ligands. The representative trajectories for paths 1–3 are shown in Fig. 2 (all paths are shown in Fig. S2, a–c). Here, the trajectories of the center of mass of the ligands are represented by lines with different colors. In path 1 (dark green line in Fig. 2), the ligands reach the binding site (Mg2+) by interacting sequentially with multiple lysine residues in nsp7 and nsp12, which is shown on the left side of the binding site in this figure. In path 2 (magenta line in Fig. 2), the ligands are in frequent contact with any residues of D155 to E167 (especially K160) of nsp12, which is shown on the right side of the binding site. In path 3 (cyan line in Fig. 2), the ligands enter the binding site from the front and bind to the Mg2+ ions with almost no interaction with any residues of RdRp. In addition to these common paths, the following three paths (paths 4–6) were also found in one or two of the systems; namely, they are ligand-specific paths. In path 4, which was observed in the RemTP and FavTP systems, the ligands interacted with the histidine residues (H810 and H816 of nsp12) in the vicinity of the binding site. In path 5, which was found only in the FavTP system, FavTP reached the binding site after interacting with Y826 of nsp12. In path 6, which was found only in the ATP system, ATP bound to Mg2+ after ATP was trapped in the gap between nsp8 and nsp7. We individually discuss the details of these paths below.
Figure 2.
Representative trajectories of the ligands in paths 1–3 for (a and d) RemTP, (b and e) FavTP, and (c and f) ATP. RdRp is displayed as a ribbon model. A trajectory in each path is displayed as a line of a different color. Dark green, magenta, and cyan represent path 1, path 2, and path 3, respectively. The large spheres represent the start and end points of each trajectory. Small light green spheres indicate Mg2+ ions. Coordinate axes are also shown in (a) and (d). To see this figure in color, go online.
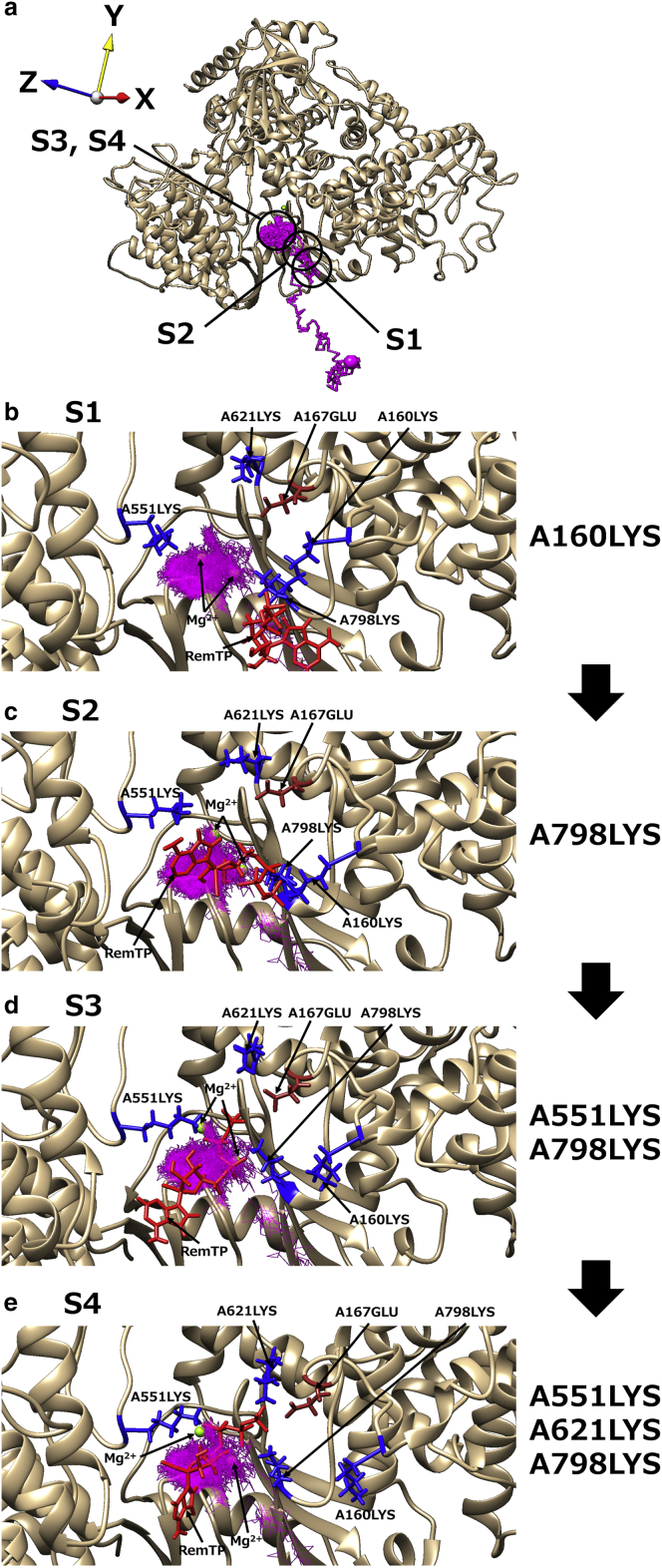
Path 1
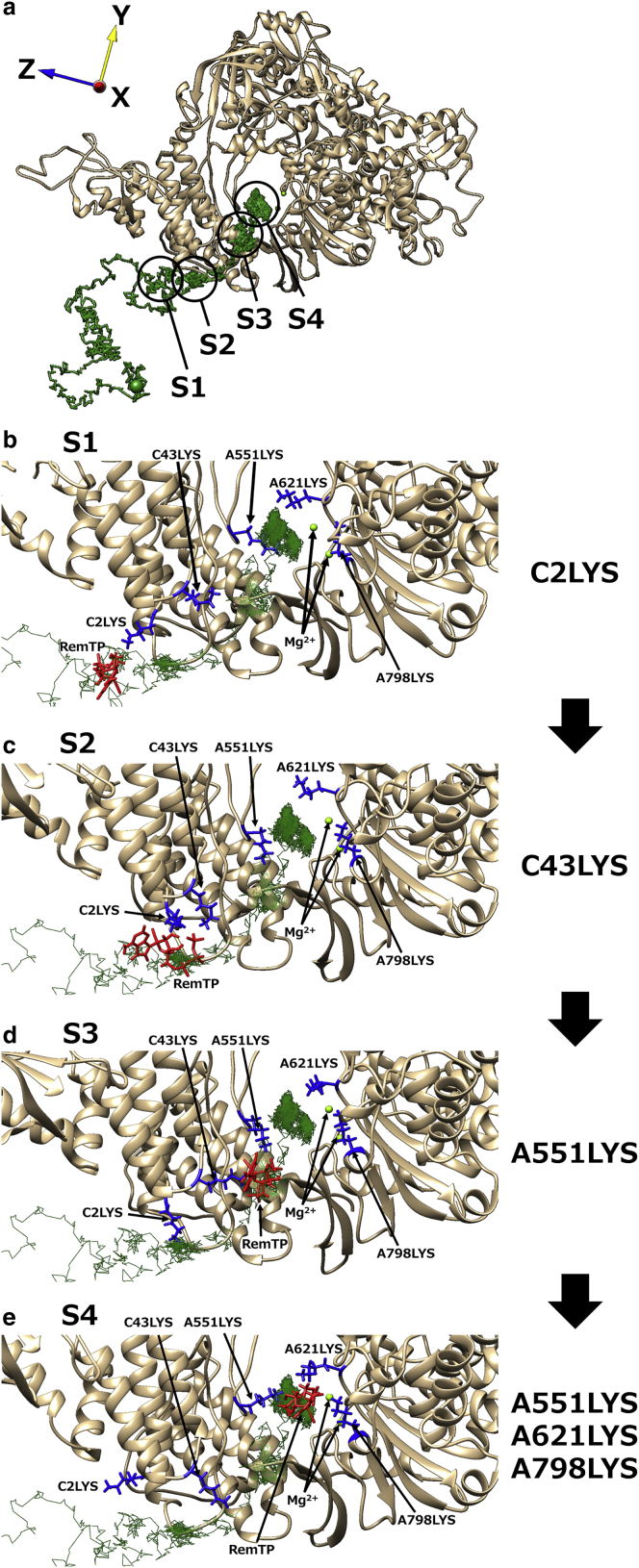
We classified the trajectories in which the phosphate group of the ligand had contact with K43 of nsp7 and K438, K551, and K798 of nsp12 before the ligand recognition as path 1. To understand the ligand recognition process in path 1, we analyzed how the ligands interacted with residues of RdRp. The representative trajectory of RemTP in path 1 is shown in Fig. 3. Snapshots in which RemTP has contact with specific residues are also presented in Fig. 3. Here, when the closest distance between any atoms of the ligand and those of a residue of RdRp was less than 3.5 Å, it was regarded as a “contact” between the ligand and residue. In Fig. 3, b–e, the capital letters at the beginnings of the residues (A or C) correspond to the chain labels in the cryo-EM structure of the original PDB (chains A and C are nsp12 and nsp7, respectively). From now on, the residue of RdRp is described as “chain label + residue number + residue name” for convenience.
Figure 3.
(a) The representative trajectory of RemTP recognized by RdRp in path 1 (the same as Fig. 2a). The black circles show the positions at which RemTP is in stable contact with residues. (b)–(e) Characteristic structure at each state (S1–S4). In (b)–(e), RemTP and the lysine residues involved in the ligand recognition are represented as red and blue stick models, respectively. The first capital letters of residues (A and C) represent the chain labels in the original PDB (residues with “A” and “C” are residues of nsp12 and nsp7, respectively). To see this figure in color, go online.
In this path, interesting behavior of RdRp was observed in which lysine residues transported the ligand to the binding site as if it were a “bucket brigade” (Video S2). These lysine residues (C2LYS, C43LYS, A438LYS, and A551LYS) have a positive charge and are arranged in a straight line toward the binding site. In this ligand-transport process, the phosphate group of RemTP first interacted with the side chain of C2LYS (state 1 (S1); Fig. 3 b). Subsequently, C2LYS passed it to C43LYS, which was spatially close (state 2 (S2); Fig. 3 c). C43LYS then passed RemTP to A551LYS (state 3 (S3); Fig. 3 d), and finally, the phosphate group of RemTP reached the binding site (state 4 (S4); Fig. 3 e). At the binding site, the ligand electrostatically interacted with A621LYS and A798LYS, which was located in the vicinity of the binding site. The same ligand-transport process was observed in the FavTP and ATP systems, except for minor differences (Videos S3 and S4). In the FavTP and ATP systems, C43LYS instead of C2LYS first contacted with the phosphate group of the ligands. In the FavTP system, A438LYS between C43LYS and A551LYS also participated in the ligand transport.
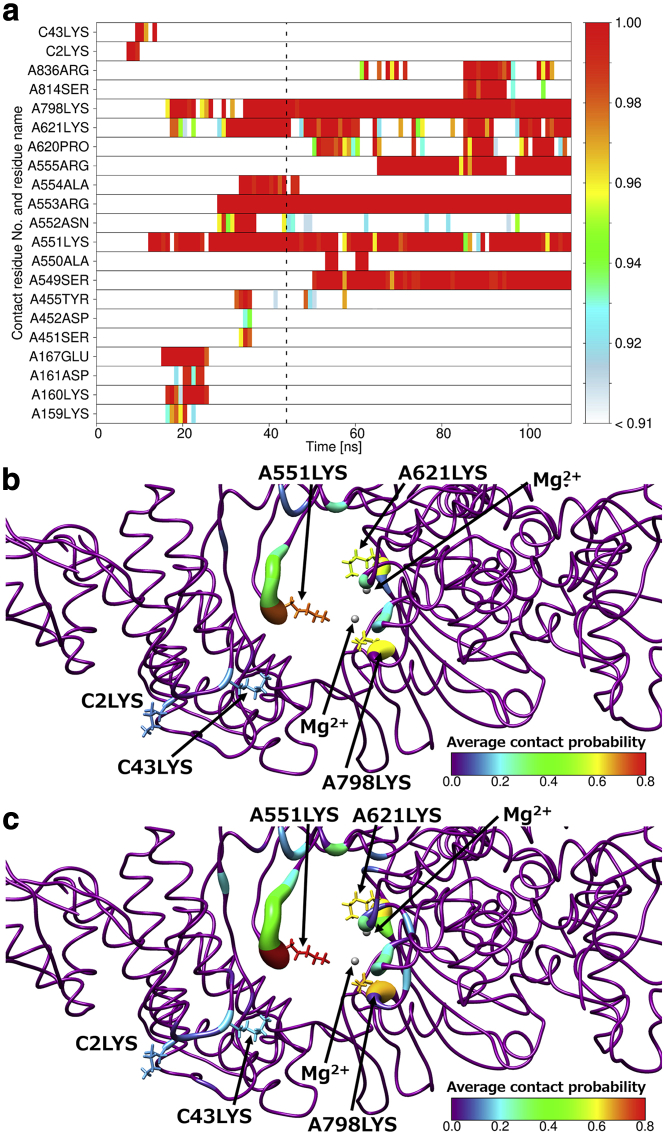
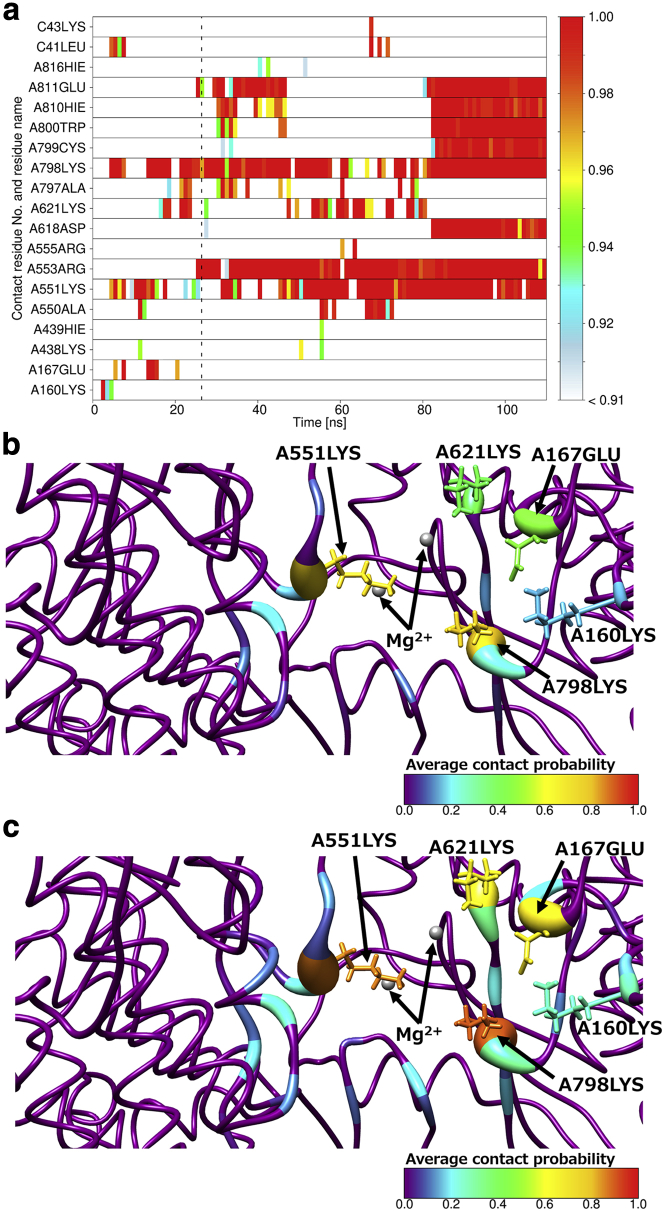
For quantitative analysis, we calculated the time series of contact probabilities between ligands and each residue of RdRp in all MD simulations. The time series that correspond to Fig. 3 are shown in Fig. 4 a. We show here the time series for one typical RemTP trajectory because those obtained in the FavTP and ATP systems are essentially the same. Here, the trajectory for 110 ns was divided into 110 bins, and the contact probability was calculated every 1 ns. In this figure, we show residues with the contact probability of more than 0.9 even once during the simulation time on the vertical axis. The dotted line in the figure shows the time when the first contact event between RemTP and Mg2+ occurred in this trajectory. Fig. 4 a shows that the contact probabilities between RemTP and C2LYS, C43LYS, and A551LYS at the initial stage in path 1 were particularly high. This result also suggests that the electrostatic interaction between the phosphate group of RemTP and these lysine residues plays an important role in the ligand recognition process in path 1. In addition, the probability of contact with C43LYS exceeded 0.9 just before that with C2LYS became less than 0.9. Similarly, the probability of contact with A551LYS increased when that with C43LYS decreased. After the probability of contact with A551LYS exceeded 0.9, that with A621LYS and A798LYS exceeded 0.9. These results also indicate that RemTP reached the binding site through these lysine residues in sequence. Several experimental studies have reported that the NTP entry channel of RdRp has positively charged residues placed on a line (18,20,73). However, no experimental work has been reported that the lysine residues of RdRp transport the ligands to the binding site. This is the first study to show the ligand-transport mechanism of RdRp via multiple lysine residues.
Figure 4.
(a) Representative time series of contact probabilities for each residue with RemTP in path 1. Residues with contact probabilities more than 0.9 are shown. The vertical dashed line represents the simulation time when the first contact event between Mg2+ and RemTP was observed. Shown is a visualizing of the average contact probabilities on the RdRp structure by color map using thresholds of (b) 3.5 and (c) 5.0 Å. Small gray spheres indicate Mg2+ ions. To see this figure in color, go online.
To identify the residues involved in the ligand recognition, we set the threshold of the distance between the ligand and RdRp as 3.5 Å. To examine the threshold dependence, in addition to using this threshold, we calculated the average contact probability between RemTP and each residue before the first contact event between RemTP and Mg2+ using the threshold set to 5.0 Å, as shown in Fig. 4, b and c. We can see that the average contact probabilities of the lysine residues, arranged in a straight line toward the binding site, are higher than those of the surrounding residues regardless of the thresholds. Therefore, it is considered that the threshold dependence is small in identifying the residues involved in the ligand recognition.
It has been reported that these positively charged basic residues, especially those at motif F (residues A544LEU to A555ARG) in nsp12, favor NTP uptake (19,20). Moreover, the above lysine residues (A438LYS, A551LYS, A621LYS, A798LYS, C2LYS, and C43LYS) are highly conserved in RdRp of SARS-CoV (21), as shown in Fig. S3. Therefore, it is assumed that the NTP recognition ability of RdRp is enhanced by carrying NTP to the binding site by these linearly arranged lysine residues for SARS-CoV-2 and SARS-CoV.
Path 2
Path 2 includes trajectories in which the phosphate group of the ligand was in frequent contact with any residues between D155 and E167 (especially K160) of nsp12 before the ligand was taken up. The analyzed results of the RemTP trajectory are shown in Figs. 5 and 6. The behavior of the lysine residues that carry RemTP to the binding site like a “bucket brigade” was again observed in path 2 (Video S5). First, the phosphate group of RemTP interacted with A160LYS (S1; Fig. 5 b). The ligand was passed to A798LYS (S2; Fig. 5 c). After that, the phosphate group electrostatically interacted with the side chain of A551LYS (S3; Fig. 5 d), then with that of A621LYS (S4; Fig. 5 e) before reaching the binding site. Such ligand uptake was also observed in the FavTP and ATP systems (Videos S6 and S7).
Figure 5.
(a) The representative trajectory of RemTP recognized by RdRp in path 2 (the same as Fig. 2d). The black circles showed the positions at which RemTP was in stable contact with residues. (b)–(e) Characteristic structure at each state (S1–S4). In (b)–(e), RemTP, the lysine residues involved in the ligand recognition, and A167GLU are shown in red, blue, and brown, respectively. To see this figure in color, go online.
Figure 6.
(a) Representative time series of contact probabilities for each residue with RemTP in path 2. Residues with contact probabilities more than 0.9 are shown. The vertical dashed line represents the simulation time when the first contact event between Mg2+ and RemTP was observed. Shown is a visualizing of the average contact probabilities on the RdRp structure by color map using thresholds of (b) 3.5 and (c) 5.0 Å. Small gray spheres indicate Mg2+ ions. To see this figure in color, go online.
The time series of contact probabilities between the ligand and each residue in path 2 are shown in Fig. 6 a. We again focused on the results of RemTP because similar results were obtained in the FavTP and ATP systems. As can be seen in Fig. 6 a, A160LYS and the ligand were in frequent contact at the beginning of the simulation. It suggests that for this path, the electrostatic interaction between the phosphate group of the ligand and this lysine plays a particularly important role in ligand recognition. Furthermore, as in path 1, when the probability of contact with A160LYS became less than 0.9, those with A551LYS and A798LYS exceeded 0.9. More specifically, RemTP was passed between these lysine residues and approached the binding site. Because A160LYS is highly conserved in RdRp of SARS-CoV, as shown in Fig. S3, it is speculated that this lysine residue also contributes to the NTP recognition of RdRp in SARS-CoV.
We calculated the average contact probability between RemTP and each residue of RdRp before the first contact event between RemTP and Mg2+ again using two thresholds of 3.5 and 5.0 Å, as shown in Fig. 6, b and c. We can see that the average contact probabilities of the lysine residues mainly involved in the ligand recognition in path 2 are higher than those of the surrounding residues regardless of the thresholds. Therefore, it is considered that the threshold dependence is also small in identifying the residues involved in the ligand recognition in path 2.
It is worth mentioning that the effect of counterions was seen in path 2. For example, the phosphate group of RemTP and A167GLU near the binding site interacted with each other via Na+ (Fig. S4). Such a counterion effect was observed because three acidic residues (A161ASP, A164ASP, and A167GLU) are located between A160LYS and the binding site. Na+ binds to these negatively charged acidic residues, neutralizing the electrostatic repulsion between the phosphate groups of the ligands and these residues. Such interaction was also observed in the other ligand systems.
Path 3
This path is the simplest ligand recognition process. The ligands entered the binding site with almost no interaction with any residues of RdRp because of the electrostatic interaction with the two Mg2+ ions at the binding site. As seen in the time series of the contact probabilities between RemTP and each residue in Fig. S5, little contact was seen until the ligand reached the RdRp binding site (Video S8). Because this path was observed in all ligand systems as well as paths 1 and 2, it is also one of the main ligand recognition processes.
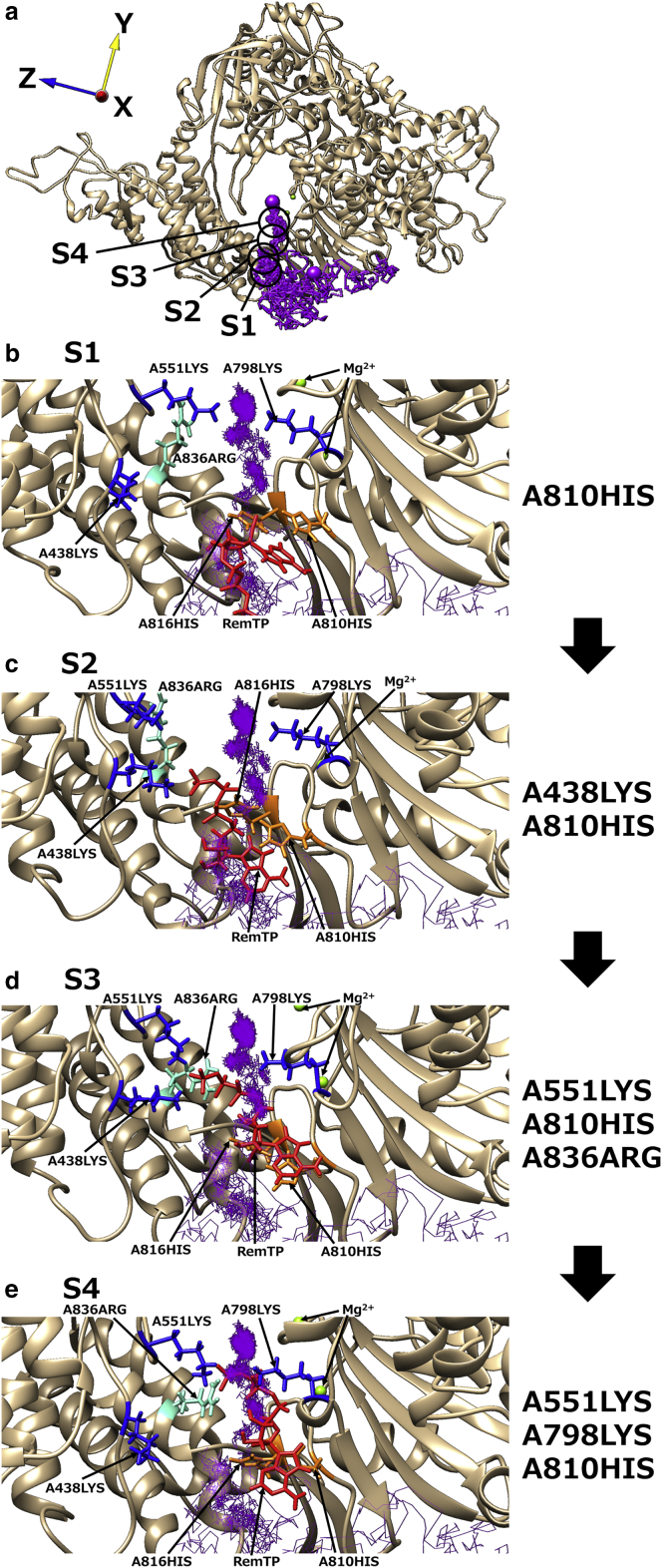
Path 4
Path 4 was found only in the RemTP and FavTP systems. The analysis results of the RemTP trajectory in path 4 are shown in Fig. 7 and Fig. S6. As a feature of this path, histidine residues in the vicinity of the binding site (A810HIS and A816HIS) mainly contributed to the ligand recognition. As shown in Fig. 7, the base moiety of RemTP first formed a π-π stacking with the side chains of A810HIS and A816HIS (S1; Fig. 7 b). Subsequently, the phosphate group of RemTP electrostatically interacted with A438LYS (S2; Fig. 7 c). RemTP was passed from A438LYS to A551LYS and A836ARG (S3; Fig. 7 d) and then interacted with A551LYS and A798LYS (S4; Fig. 7 e). RemTP finally reached the binding site while maintaining interaction with A551LYS. During these processes (S1–S4), the π-π stacking between the base moiety of RemTP and A810HIS was almost maintained, and the π-π stacking was broken when RemTP reached the binding site (Video S9). Fig. S6 also shows that before reaching the binding site, RemTP was in contact with A810HIS and A816HIS for a longer period of time compared with other residues. In addition, similar to paths 1 and 2, the basic residues sequentially carried the phosphate group of RemTP to the binding site. As for FavTP, although sequential delivery of the ligand between these basic residues was not observed, the ligand approached the binding site while maintaining π-π stacking with A810HIS in the same way as RemTP.
Figure 7.
(a) The representative trajectory of RemTP recognized by RdRp in path 4. The black circles showed positions at which RemTP was in stable contact with residues. (b)–(e) Characteristic structure at each state (S1–S4). In (b)–(e), RemTP, the lysine residues involved in the ligand recognition, histidine residues, and A836ARG are shown in red, blue, orange, and light blue, respectively. To see this figure in color, go online.
This path suggests that residues other than the positively charged basic ones near the binding site also promote the ligand recognition of RdRp. The location of the aromatic residues such as histidine residues in the vicinity of the binding site would make a beneficial contribution to the transport of the NTP and NTP-like inhibitors to the binding site. Although this recognition process mediated by the histidine residues was not observed for ATP, it is expected that a similar path will be observed in the ATP system by improving the statistics. Furthermore, these histidine residues are also conserved in RdRp of SARS-CoV (Fig. S3) and can also play an important role in the NTP recognition of RdRp in SARS-CoV.
Path 5 and path 6
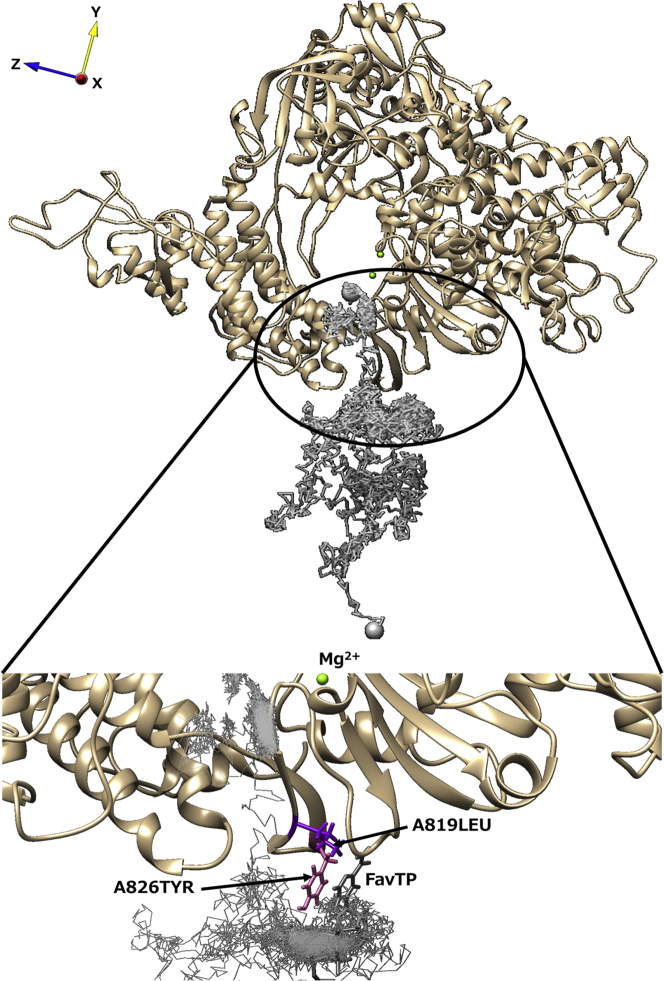
Path 5 and path 6 were observed only in the FavTP system and the ATP system, respectively. Fig. 8 shows the trajectory of FavTP in path 5. The time series of contact probabilities between FavTP and each residue are presented in Fig. S7. As a noticeable difference from the other paths, path 5 is characterized by long-term contact with A819LEU and A826TYR (Fig. S7). Because these residues do not have a net electrostatic charge, it is expected that a moiety of the ligand other than the phosphate group interacts with these residues. The analysis of the trajectory in path 5 revealed that the base moiety of FavTP and the side chain of A826TYR formed π-π stacking (Video S10), resulting in high probabilities of contact with these residues.
Figure 8.
Representative trajectory and the characteristic structure in path 5 observed only for FavTP. The ligand trajectory is displayed as a gray line. The large gray spheres represent the start and end points of this trajectory. The black circle showed a position at which FavTP was in stable contact with some residues. The enlarged view is shown as an inset. In the inset, FavTP, A819LEU, and A826TYR are shown in dark gray, purple, and pink, respectively. To see this figure in color, go online.
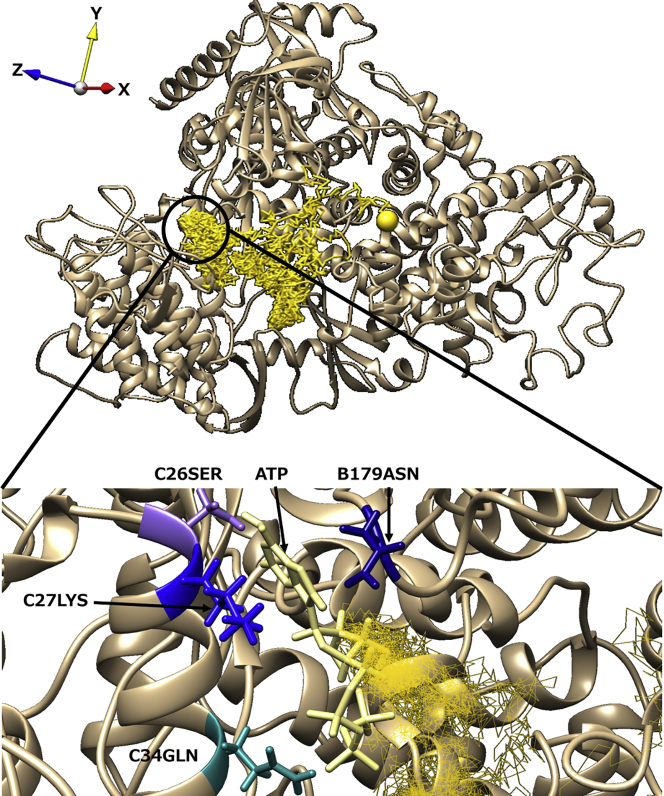
Similarly, we show the trajectory of ATP in path 6 and the time series of probabilities of contact with each residue in Fig. 9 and Fig. S8, respectively. A distinct feature of this path is that the long-term contact of ATP with multiple residues in chains B (nsp8) and C (nsp7) was observed (Fig. S8). It was attributed to the base moiety of ATP being trapped in the gap between chain B and chain C (see Fig. 9; Video S11).
Figure 9.
Representative trajectory and the characteristic structure in path 6 observed only for ATP. The ligand trajectory is displayed as a yellow line. The large yellow spheres represent the start and end points of the trajectory. The black circle showed a position at which ATP was in stable contact with some residues. The enlarged view is shown as an inset. In the inset, ATP, B179ASN, C26SER, C27LYS, and C34GLN are shown in a light yellow, navy blue, light purple, blue, and dark cyan, respectively. To see this figure in color, go online.
Contact probabilities between the ligand and RdRp residues before recognition by RdRp
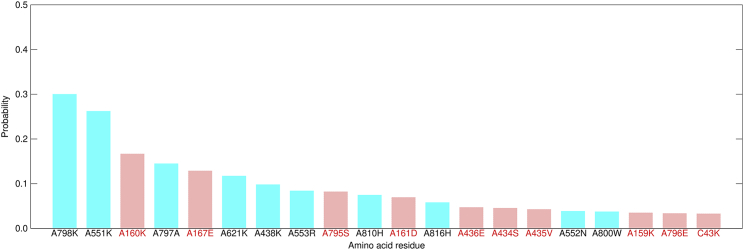
To investigate the key residues of the ligand recognition, we calculated probabilities with which residues were in contact with the ligands before and after the first contact event between the ligands and Mg2+. The contact probabilities in the RemTP system were averaged for all 12 trajectories in which the ligand recognition events were observed. Fig. 10 shows the average contact probabilities of the top 20 residues in contact with RemTP before the first contact event between Mg2+ and RemTP. The average contact probabilities for FavTP and ATP were also calculated in the same way, as shown in Fig. S9. In Fig. 10 and Fig. S9, the red letters and pink vertical bars showed the residues that were not included in the top 20 ones after the first contact between the ligands and Mg2+ (Fig. S10). In other words, the residues shown in red letters indicate that they have been in contact with the ligand only before the ligand recognition.
Figure 10.
The average contact probabilities of the top 20 residues in contact with RemTP before the first contact event between Mg2+ ions and RemTP. Residues that are not included in the top 20 of the average contact probabilities after the first contact event (Document S1. Figs. S1–S13 and Table S1, Video S1. Whole view of the RdRp and RemTP system, Video S2. The RemTP recognition process in path 1, Video S3. The FavTP recognition process in path 1, Video S4. The ATP recognition process in path 1, Video S5. The RemTP recognition process in path 2, Video S6. The FavTP recognition process in path 2, Video S7. The ATP recognition process in path 2, Video S8. The RemTP recognition process in path 3, Video S9. The RemTP recognition process in path 4, Video S10. The base moiety of FavTP and the side chain of A826TYR of RdRp formed π-π stacking in path 5, Video S11. ATP was trapped in the gap between chain B and chain C of RdRp in path 6, Document S2. Article plus supporting materiala) are shown in red letters and pink vertical bars. To see this figure in color, go online.
Fig. 10 shows that A160LYS and C43LYS, which mainly contribute to the uptake of RemTP in paths 1 and 2, appear only before the ligand recognition. These two residues also appear in the FavTP and ATP systems only before the recognition (Fig. S9). This suggests that these residues are essential in the NTP recognition by RdRp, as discussed in Path 1 and Path 2. Other lysine residues (A438LYS, A551LYS, A621LYS, and A798LYS) identified in paths 1 and 2 also appear in all ligand systems. Furthermore, in other paths (except for path 3), there are also high contact probabilities between the ligand and these lysine residues before the first contact between the ligand and Mg2+ (Figs. S6–S8). Therefore, the sequential electrostatic interaction between the phosphate group of the ligand and these lysine residues is the main driving force for the ligand recognition in the paths other than path 3.
Because these lysine residues (A160LYS, A438LYS, A551LYS, A621LYS, A798LYS, and C43LYS) have higher contact probabilities for all ligands, paths 1 and 2 are considered as the main common paths for the NTP recognition by RdRp. Conversely, the residues identified in path 4 (A810HIS and A816HIS) appear only for RemTP and FavTP (Fig. 10; Fig. S9 a). In addition, the residues identified in path 5 (A819LEU and A826TYR) and those in path 6 (C27LYS and C34GLN) appear only for FavTP and ATP, respectively. Therefore, these results show that paths 4–6 are relatively minor paths.
According to the literature (74), even divalent ions such as Mg2+ have an interaction range of only ∼10 Å in water at most. Therefore, it is considered that when the ligand happens to approach the binding site while floating in solution, it is attracted to the Mg2+ ions, rather than a distant ligand is attracted directly to the Mg2+ ions. In fact, the ligand was suddenly attracted to the Mg2+ ions when it came near the binding site in path 3 (Video S8). In addition to the direct interaction with the Mg2+ ions, the presence of the lysine residues, such as C2LYS (34–36 Å away from the Mg2+ ions) and A160LYS (21–25 Å away from the Mg2+ ions), makes it possible to capture ligands that are further distant. We consider that transporting the ligands to the binding site like a “bucket brigade” by these lysine residues also contributes to the recognition of distant ligands by RdRp.
The MD simulations in this study were performed without RNA strands. To date, some structures of the RdRp bound to an RNA duplex were experimentally solved (20,28, 29, 30,75, 76, 77, 78). From these structures, it is clarified that the nascent RNA extends on the opposite side of the ligand entry side. It is also shown that the RNA template strand enters the binding site from the opposite side of the ligand entry side and translates in the same direction as the nascent RNA elongation. Therefore, we consider that the effect of the presence of these RNA strands on the ligand recognition mechanism of RdRp revealed in this study is small.
In this study, we employed a simple model for the divalent metal ions, which is represented only by the Coulombic and LJ interactions. Other models such as cationic dummy atom models (79) and polarizable models (80) are useful for considering charge transfer, polarization, and covalent interactions. However, our study focuses on the ligand recognition paths before the ligand binding with the metal ions. Therefore, the force field employed in this study is sufficient for our discussion on the ligand recognition of RdRp.
Directivity in the ligand uptake of RdRp
The structure of RdRp was used without RNA as the initial structure in our MD simulations. The binding site is a cavity containing only two catalytic Mg2+ ions (Fig. S11). Therefore, the ligands can enter the binding site from either front (“Front” in Fig. S11) or back (“Back” in Fig. S11) in these MD simulations, although the back side is inherently blocked by the extended RNA. One might expect that the frequency of the ligand entry from the front is the same as that from the back. However, as shown in Table S1, the number of trajectories approaching from the correct direction (front entry) was more than that from the opposite direction (back entry). Note that the paths we discussed in the previous subsections are only those from the front.
To understand the difference in the spatial accessibility to the binding site, we calculated the solid angle with which outside is seen from the two Mg2+ ions without being disturbed by any residues of RdRp. Figs. S12 and S13 show the parts of the spherical surface created by these solid angles whose vertices are the two Mg2+ ions. Here, the initial structure of RdRp was used. The solid angle at the front side is larger than that at the back side for both Mg2+ ions (Table 2). For Mg2+(1) in Fig. S11, particularly, the solid angle at the front side is approximately twice as large as that at the back side. This indicates that the ligand is more accessible from the correct direction when it reaches the binding site with little contact with the residues as in path 3. In addition, it is considered that, as seen in other paths, the intermolecular interaction between the ligand and residues, especially the positively charged basic residues, contributes to the ligand recognition of RdRp from the correct direction.
Table 2.
Solid angles with which the outside is seen from the two Mg2+ ions without being disturbed by any residues of RdRp
| Front | Back | |
|---|---|---|
| Mg2+(1) | 0.96 | 0.54 |
| Mg2+(2) | 0.94 | 0.82 |
They were calculated for both front and back sides and from both two Mg2+ ions. See Fig. S11 for the definitions of the front and back sides and the numbers in parentheses for the Mg2+ ions. The solid angles were measured in steradians
Conclusions
In this study, we performed the MD simulations of SARS-CoV-2 RdRp with the three kinds of ligands, RemTP, FavTP, and ATP, to clarify the recognition mechanism for the drug molecules. It was found that the recognition probability for RemTP was the highest, that for FavTP was intermediate, and that for ATP was the lowest.
We identified six recognition paths (paths 1–6) in this study. Paths 1–3 were commonly found in all ligands, and paths 4–6 were ligand-dependent paths. In path 1, “bucket brigade” behavior was observed, in which the ligands were carried to the binding site by multiple lysine residues. The lysine residues interacting with the phosphate group of the ligand in this path were K2 and K43 of nsp7 and K438, K551, K621, and K798 of nsp12. In path 2, the transport of the ligands from K160 of nsp12 to the lysine residues around the binding site was also observed. These paths are considered as the main ones in the NTP recognition by RdRp together with path 3, in which the ligands directly reach the binding site. Conversely, in paths 4–6, the residues other than the positively charged basic ones near the binding site were mainly involved in the ligand recognition of RdRp. In our simulations, these paths were not common to all ligands and thus may be relatively minor paths.
We have clarified the NTP recognition processes in SARS-CoV-2 RdRp at the atomic level for the first time. The results obtained in this study fill the gap between the apo form of RdRp and the structure of the RdRp-NTP complex. We expect our results to contribute to the understanding of the efficient NTP uptake by RdRp and the development of drug molecules for suppressing the RdRp function. In addition, the residues identified in simulation as promoting the NTP recognition are highly conserved in RdRp of SARS-CoV. Thus, our findings can be extended to the NTP recognition mechanism of other RNA viruses that have RdRp similar to SARS-CoV-2 RdRp.
We performed the MD simulations for the ligand recognition by RdRp before the RNA replication. Future studies should explore the NTP recognition mechanism in the complex of RdRp containing the template and nascent RNA strands to develop more effective therapeutic drugs.
Author contributions
S.G.I. and H.O. designed the research. S.T. and H.O. modeled the initial conformations. S.T. performed simulations, analyzed the results, and wrote the article. All authors discussed the results and revised the article.
Acknowledgments
This work used supercomputers at the Research Center for Computational Science, Okazaki Research Facilities, National Institutes of Natural Sciences, Japan, and computational resources of the TSUBAME3.0 provided by Tokyo Institute of Technology through the HPCI System Research Project (Project ID: hp200142). Molecular graphics were depicted with the UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (UCSF) (53).
Editor: Monika Fuxreiter.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.07.026.
Supporting material
References
- 1.Gorbalenya A.E., Baker S.C., Ziebuhr J., Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai C.-C., Shih T.-P., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehta P., McAuley D.F., Manson J.J., HLH Across Speciality Collaboration, UK COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacLaren G., Fisher D., Brodie D. Preparing for the most critically ill patients with COVID-19: the potential role of extracorporeal membrane oxygenation. JAMA. 2020;323:1245–1246. doi: 10.1001/jama.2020.2342. [DOI] [PubMed] [Google Scholar]
- 5.Cook D.J., Marshall J.C., Fowler R.A. Critical illness in patients with COVID-19: mounting an effective clinical and research response. JAMA. 2020;323:1559–1560. doi: 10.1001/jama.2020.5775. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y., Gayle A.A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020;27:1–4. doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P., Yang X.-L., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization (WHO) 2021. Weekly operational update on COVID-19–12 July 2021. https://www.who.int/publications/m/item/weekly-operational-update-on-covid-19—12-july-2021. [Google Scholar]
- 9.Zhu N., Zhang D., Tan W., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim D., Lee J.-Y., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu F., Zhao S., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu R., Zhao X., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders J.M., Monogue M.L., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;323:1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 14.Wu C., Liu Y., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon D.E., Jang G.M., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alanagreh L., Alzoughool F., Atoum M. The human coronavirus disease COVID-19: its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens. 2020;9:331. doi: 10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., Anirudhan V., Rong L. RNA-dependent RNA polymerase of SARS-CoV-2 as a therapeutic target. J. Med. Virol. 2021;93:300–310. doi: 10.1002/jmv.26264. [DOI] [PubMed] [Google Scholar]
- 18.Venkataraman S., Prasad B.V.L.S., Selvarajan R. RNA dependent RNA polymerases: insights from structure, function and evolution. Viruses. 2018;10:76. doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Y., Yan L., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin W., Mao C., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subissi L., Posthuma C.C., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romano M., Ruggiero A., Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elfiky A.A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silico perspective. J. Biomol. Struct. Dyn. 2021;39:3204–3212. doi: 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W.-F., Stephen P., Lin S.-X. Novel coronavirus polymerase and nucleotidyl-transferase structures: potential to target new outbreaks. J. Phys. Chem. Lett. 2020;11:4430–4435. doi: 10.1021/acs.jpclett.0c00571. [DOI] [PubMed] [Google Scholar]
- 26.Warren T.K., Jordan R., Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furuta Y., Takahashi K., Shiraki K. Mechanism of action of T-705 against influenza virus. Antimicrob. Agents Chemother. 2005;49:981–986. doi: 10.1128/AAC.49.3.981-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q., Wu J., Rao Z. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428.e13. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kokic G., Hillen H.S., Cramer P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021;12:279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naydenova K., Muir K.W., Russo C.J. Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2021946118. e2021946118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng Q., Peng R., Shi Y. Structural basis of SARS-CoV-2 polymerase inhibition by Favipiravir. Innovation. 2021;2:100080. doi: 10.1016/j.xinn.2021.100080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agostini M.L., Andres E.L., Denison M.R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 doi: 10.1128/mBio.00221-18. e00221–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheahan T.P., Sims A.C., Baric R.S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheahan T.P., Sims A.C., Baric R.S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon C.J., Tchesnokov E.P., Götte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon C.J., Tchesnokov E.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shannon A., Selisko B., Canard B. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat. Commun. 2020;11:4682. doi: 10.1038/s41467-020-18463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaptein S.J.F., Jacobs S., Delang L. Favipiravir at high doses has potent antiviral activity in SARS-CoV-2-infected hamsters, whereas hydroxychloroquine lacks activity. Proc. Natl. Acad. Sci. USA. 2020;117:26955–26965. doi: 10.1073/pnas.2014441117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L., Zhou R. Structural basis of the potential binding mechanism of remdesivir to SARS-CoV-2 RNA-dependent RNA polymerase. J. Phys. Chem. B. 2020;124:6955–6962. doi: 10.1021/acs.jpcb.0c04198. [DOI] [PubMed] [Google Scholar]
- 40.Elfiky A.A. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen H.L., Thai N.Q., Li M.S. Remdesivir strongly binds to both RNA-dependent RNA polymerase and main protease of SARS-CoV-2: evidence from molecular simulations. J. Phys. Chem. B. 2020;124:11337–11348. doi: 10.1021/acs.jpcb.0c07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koulgi S., Jani V., Joshi R. Remdesivir-bound and ligand-free simulations reveal the probable mechanism of inhibiting the RNA dependent RNA polymerase of severe acute respiratory syndrome coronavirus 2. RSC Adv. 2020;10:26792–26803. doi: 10.1039/d0ra04743k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wakchaure P.D., Ghosh S., Ganguly B. Revealing the inhibition mechanism of RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2 by remdesivir and nucleotide analogues: a molecular dynamics simulation study. J. Phys. Chem. B. 2020;124:10641–10652. doi: 10.1021/acs.jpcb.0c06747. [DOI] [PubMed] [Google Scholar]
- 45.Aranda J., Orozco M. RNA-dependent RNA polymerase from SARS-COV-2. Mechanism of reaction and inhibition by remdesivir. bioRxiv. 2020 doi: 10.1101/2020.06.21.163592. [DOI] [Google Scholar]
- 46.Zhang L., Zhang D., Huang X. 1′-ribose cyano substitution allows remdesivir to effectively inhibit nucleotide addition and proofreading during SARS-CoV-2 viral RNA replication. Phys. Chem. Chem. Phys. 2021;23:5852–5863. doi: 10.1039/d0cp05948j. [DOI] [PubMed] [Google Scholar]
- 47.Itoh S.G., Tanimoto S., Okumura H. Dynamic properties of SARS-CoV and SARS-CoV-2 RNA-dependent RNA polymerases studied by molecular dynamics simulations. Chem. Phys. Lett. 2021;778:138819. doi: 10.1016/j.cplett.2021.138819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ngoc L.L.N., Itoh S.G., Okumura H. Replica-permutation molecular dynamics simulations of an amyloid-β(16–22) peptide and polyphenols. Chem. Phys. Lett. 2020;758:137913. [Google Scholar]
- 49.Takemura K., Sato C., Kitao A. ColDock: concentrated ligand docking with all-atom molecular dynamics simulation. J. Phys. Chem. B. 2018;122:7191–7200. doi: 10.1021/acs.jpcb.8b02756. [DOI] [PubMed] [Google Scholar]
- 50.Okumura H., Itoh S.G. Amyloid fibril disruption by ultrasonic cavitation: nonequilibrium molecular dynamics simulations. J. Am. Chem. Soc. 2014;136:10549–10552. doi: 10.1021/ja502749f. [DOI] [PubMed] [Google Scholar]
- 51.Tachi Y., Okamoto Y., Okumura H. Conformational change of amyloid-β 40 in association with binding to GM1-glycan cluster. Sci. Rep. 2019;9:6853. doi: 10.1038/s41598-019-43117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Šali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 53.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 54.Word J.M., Lovell S.C., Richardson D.C. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 55.Maier J.A., Martinez C., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 57.Bayly C.I., Cieplak P., Kollman P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 58.Frisch M.J., Trucks G.W., Fox D.J. Gaussian, Inc.; Wallingford, CT: 2016. Gaussian 16, revision C.01. [Google Scholar]
- 59.Case D.A., Betz R.M., Kollman P.A. University of California; San Francisco, CA: 2016. AMBER 2016. [Google Scholar]
- 60.Wang J., Wang W., Case D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 61.Li P., Roberts B.P., Merz K.M., Jr. Rational design of particle mesh ewald compatible Lennard-Jones parameters for +2 metal cations in explicit solvent. J. Chem. Theory Comput. 2013;9:2733–2748. doi: 10.1021/ct400146w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joung I.S., Cheatham T.E., III Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okumura H. Temperature and pressure denaturation of chignolin: folding and unfolding simulation by multibaric-multithermal molecular dynamics method. Proteins. 2012;80:2397–2416. doi: 10.1002/prot.24125. [DOI] [PubMed] [Google Scholar]
- 64.Chiang H.-L., Chen C.-J., Hu C.-K. Transformation between α-helix and β-sheet structures of one and two polyglutamine peptides in explicit water molecules by replica-exchange molecular dynamics simulations. J. Comput. Chem. 2014;35:1430–1437. doi: 10.1002/jcc.23633. [DOI] [PubMed] [Google Scholar]
- 65.Okumura H., Itoh S.G. Molecular dynamics simulations of amyloid-β(16-22) peptide aggregation at air-water interfaces. J. Chem. Phys. 2020;152:095101. doi: 10.1063/1.5131848. [DOI] [PubMed] [Google Scholar]
- 66.Okumura H., Itoh S.G. Structural and fluctuational difference between two ends of Aβ amyloid fibril: MD simulations predict only one end has open conformations. Sci. Rep. 2016;6:38422. doi: 10.1038/srep38422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 68.Wang M., Cao R., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Case D.A., Ben-Shalom I.Y., Kollman P.A. University of California; San Francisco, CA: 2019. AMBER 2019. [Google Scholar]
- 70.Darden T., York D., Pedersen L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 71.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 72.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 73.Jiang Y., Yin W., Xu H.E. RNA-dependent RNA polymerase: structure, mechanism, and drug discovery for COVID-19. Biochem. Biophys. Res. Commun. 2021;538:47–53. doi: 10.1016/j.bbrc.2020.08.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bruni F., Imberti S., Ricci M.A. Aqueous solutions of divalent chlorides: ions hydration shell and water structure. J. Chem. Phys. 2012;136:064520. doi: 10.1063/1.3684633. [DOI] [PubMed] [Google Scholar]
- 75.Chen J., Malone B., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573.e13. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hillen H.S., Kokic G., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 77.Yan L., Zhang Y., Lou Z. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat. Commun. 2020;11:5874. doi: 10.1038/s41467-020-19770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bravo J.P.K., Dangerfield T.L., Johnson K.A. Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol. Cell. 2021;81:1548–1552.e4. doi: 10.1016/j.molcel.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oelschlaeger P., Klahn M., Warshel A. Magnesium-cationic dummy atom molecules enhance representation of DNA polymerase β in molecular dynamics simulations: improved accuracy in studies of structural features and mutational effects. J. Mol. Biol. 2007;366:687–701. doi: 10.1016/j.jmb.2006.10.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jing Z., Liu C., Ren P. Many-body effect determines the selectivity for Ca2+ and Mg2+ in proteins. Proc. Natl. Acad. Sci. USA. 2018;115:E7495–E7501. doi: 10.1073/pnas.1805049115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.