Abstract
Background
Our understanding of adaptive immune responses in patients with coronavirus disease 2019 (COVID-19) is rapidly evolving, but information on the innate immune responses by natural killer (NK) cells is still insufficient.
Objective
We aimed to examine the phenotypic and functional status of NK cells and their changes during the course of mild and severe COVID-19.
Methods
We performed RNA sequencing and flow cytometric analysis of NK cells from patients with mild and severe COVID-19 at multiple time points in the course of the disease using cryopreserved PBMCs.
Results
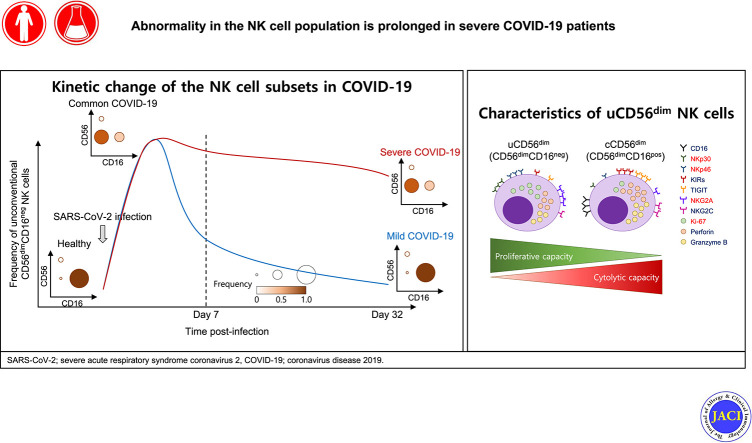
In RNA-sequencing analysis, the NK cells exhibited distinctive features compared with healthy donors, with significant enrichment of proinflammatory cytokine-mediated signaling pathways. Intriguingly, we found that the unconventional CD56dimCD16neg NK-cell population expanded in cryopreserved PBMCs from patients with COVID-19 regardless of disease severity, accompanied by decreased NK-cell cytotoxicity. The NK-cell population was rapidly normalized alongside the disappearance of unconventional CD56dimCD16neg NK cells and the recovery of NK-cell cytotoxicity in patients with mild COVID-19, but this occurred slowly in patients with severe COVID-19.
Conclusions
The current longitudinal study provides a deep understanding of the NK-cell biology in COVID-19.
Key words: SARS-CoV-2, COVID-19, innate immunity, NK cells, cytotoxicity, unconventional CD56dimCD16neg (uCD56dim) NK cell
Abbreviations used: cCD56bright, Conventional CD56brightCD16neg; cCD56dim, Conventional CD56dimCD16pos; COVID-19, Coronavirus disease 2019; HLH, Hemophagocytic lymphohistiocytosis; NK, Natural killer; RNA-seq, RNA sequencing; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; uCD56dim, Unconventional CD56dimCD16neg
Graphical abstract
A worldwide pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has continued unceasingly. Since a cluster of atypical pneumonia cases of unknown origin, now identified as being caused by SARS-CoV-2,1 , 2 were reported in Wuhan city, Hubei province, China, in December 2019, more than 90 million confirmed cases and more than 2 million deaths have been reported globally as of January 19, 2021.3 Most patients with coronavirus disease 2019 (COVID-19), which is caused by SARS-CoV-2 infection, recover spontaneously, complaining of only mild symptoms, but some patients, especially elderly patients, develop severe manifestations, including acute respiratory distress syndrome, multiorgan failure, and death.4 Our understanding of the immune responses against SARS-CoV-2 has deepened significantly,5, 6, 7, 8 but the responses of and changes to the innate lymphocytes are still unclear.
Natural killer (NK) cells are a major subset of innate lymphocytes and play an important role in early protection against viruses and the regulation of the cellular and humoral adaptive immune responses.9, 10, 11 NK cells are conventionally subdivided into 2 subsets on the basis of relative surface expression of CD56 and CD16: the CD56dimCD16pos NK-cell subset, which is predominant in peripheral blood and considered to be the cytolytic NK-cell subset, and the CD56brightCD16neg NK-cell subset, which is predominant in the secondary lymphoid tissues and considered to be the cytokine-producing NK-cell subset.11 , 12 Recently, several studies have reported the presence of the unconventional CD56dimCD16neg (uCD56dim) NK-cell population in the peripheral blood of healthy donors and showed that the population can be expanded in several clinical conditions.13 , 14 However, little is currently known about the role of this uCD56dim NK-cell population in acute viral infection.
In patients with COVID-19, the absolute number of NK cells is reduced in line with general lymphopenia.15 , 16 The Karolinska COVID-19 Study Group reported that NK cells in patients with COVID-19 exhibit activated phenotypes (higher expression of Ki-67, HLA-DR, and CD69 than in healthy controls) and are enriched with “activation/proliferation signatures” and an “effector function gene signature,”17 whereas others have reported that NK cells present exhausted phenotypes (higher expression of T cell immunoglobulin and mucin domain-containing protein 3 (Tim-3) and programmed cell death protein 1 (PD-1) than in healthy donors) and are functionally impaired.18 , 19
In the present study, we examined the phenotypic and functional status of NK cells and their changes during the course of mild and severe COVID-19. We performed flow cytometric analysis and RNA sequencing (RNA-seq) using cryopreserved PBMCs from patients with mild and severe COVID-19 at several serial time points. We found that a unique subset of NK cells, uCD56dim NK cells, is characteristically expanded in patients with COVID-19. In addition, we demonstrate that the cytotoxicity of NK cells is decreased in patients with COVID-19. During the course of COVID-19, uCD56dim NK cells disappear rapidly in mild patients but slowly in severe patients, and the cytotoxicity of NK cells recovers accordingly.
Methods
Study design
We prospectively enrolled patients diagnosed with COVID-19 from Chungnam National University Hospital, Daejeon, Korea. Fresh blood and plasma samples were obtained at several time points during hospitalization. Table E1 in this article’s Online Repository at www.jacionline.org presents the clinical characteristics of the enrolled patients and healthy donors. Table E2 in this article’s Online Repository at www.jacionline.org presents the sampling time points and the assessment assays in which the samples were used. The time was counted from the day of diagnosis (day 1). The patients with COVID-19 were diagnosed an average 9.9 days from symptom onset. Table E3 in this article’s Online Repository at www.jacionline.org presents the initial laboratory test results of the enrolled patients with COVID-19. SARS-CoV-2 RNA was detected in patients’ nasopharyngeal swab and sputum specimens by real-time PCR using the PowerChek 2019-nCoV Real-time PCR Kit (Kogenebiotech, Seoul, Korea). Hospitalized patients diagnosed with acute influenza A virus infection by a rapid antigen test of a nasopharyngeal swab were also enrolled from Chungbuk National University Hospital from December 2015 to April 2016 before the emergence of COVID-19. We collected samples from healthy donors and patients with influenza before the emergence of COVID-19, so we reasonably assumed that they were free from SARS-CoV-2 infection.
All patients provided written informed consent. This study was conducted in accordance with the Declaration of Helsinki (1996) and approved by the institutional review boards of all participating institutions.
All resources including antibodies and agents used in this study are presented in Table E4 in this article’s Online Repository at www.jacionline.org.
Isolation of PBMCs and NK cells
Using whole blood samples obtained from patients with COVID-19, patients with influenza A, and healthy donors, we isolated PBMCs by Ficoll-Paque density gradient centrifugation using lymphocyte separation medium (Corning, New York, NY). PBMCs were cryopreserved until use.
After thawing cryopreserved PBMCs, the cell viability was 86.88% ± 7.35% (healthy donors), 69.21% ± 13.77% (patients with influenza), 79.75% ± 12.11% (patients with mild COVID-19), and 75.22% ± 10.19% (patients with severe COVID-19) (see Table E5 in this article’s Online Repository at www.jacionline.org). Thawed PBMCs were directly analyzed without in vitro incubation.
NK cells were isolated 2 ways. For RNA-seq, we stained PBMCs with antibodies against 7-AAD, CD3, and CD56, and sorted NK cells on the basis of surface expression of CD3 and CD56 (CD3–CD56+) using an ARIA II cell sorter. The gating strategies for NK-cell sorting are presented in Fig E1 in this article’s Online Repository at www.jacionline.org. For the NK-cell cytotoxicity assay, to avoid affecting NK cells as much as possible, we negatively isolated NK cells using the NK-cell isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany). We stained other cells except NK cells in the PBMCs according to the manufacturer’s instructions and obtained NK cells by removing magnetically labeled cells using a magnetic-activated cell sorter (Miltenyi Biotec).
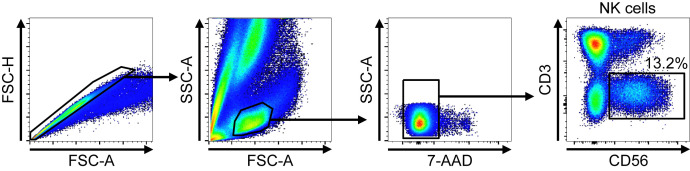
Fig E1.
Gating strategies for NK-cell sorting before RNA-seq. After singlets and lymphocytes were gated by FSC and SSC, live cells were gated by 7-AAD staining. The CD3–CD56+ cells were then gated for NK cells. FSC-A, Forward scatter-area; FSC-H, forward scatter-height; SSC-A, side scatter-area.
RNA-sequencing
RNA-seq was performed using sorted CD3–CD56+ NK cells from cryopreserved/thawed PBMCs obtained from patients with COVID-19 and healthy donors as described above. The sorted CD3–CD56+ NK cells were immediately dissolved in TRIzol LS reagent (Invitrogen, Waltham, Mass), and RNA extraction was performed following the manufacturer’s instructions. RNA quality was assessed by a bioanalyzer system (Agilent 2100, Agilent Scientific Instrument, Santa Clara, Calif), and an RNA library was constructed using the QuantSeq 3′ mRNA-Seq Library Prep Kit (Lexogen, Vienna, Austria). Sequencing was performed using a NextSeq 550; the sequencing reads were aligned to the human reference genome GRCh38 using STAR (v 2.7.2)20 and normalized for effective library size. We analyzed differentially expressed genes using the DESeq2 algorithm21 and defined them on the basis of a P value of less than .05 and log2 fold change of more than 1.
Gene set enrichment analysis was performed using web-based software Enrichr.22 , 23 The access date for the Kyoto Encyclopedia of Genes and Genomes and gene ontology analyses was January 4, 2021.
The RNA sequencing data have been uploaded to the Gene Expression Omnibus database under Gene Expression Omnibus accession number GSE165461.
Flow cytometry and intracellular cytokine staining
For surface protein staining, cryopreserved PBMCs were thawed and stained first using the LIVE/DEAD Fixable Dead Cell Stain Kit (Invitrogen) for 5 minutes at room temperature in the dark. Without washing, the LIVE/DEAD-stained cells were stained with multiple fluorochrome-conjugated antibodies against surface proteins for 20 minutes at 4°C, and then washed with fluorescence-activated cell sorting buffer (PBS + 1% FBS + 0.05% sodium azide). For intracellular cytokine staining, the cells stained with LIVE/DEAD and antibodies against surface proteins were fixed and permeabilized using a Foxp3/Transcription Factor Staining Buffer Set (Invitrogen), and then further stained with antibodies against intracellular proteins.
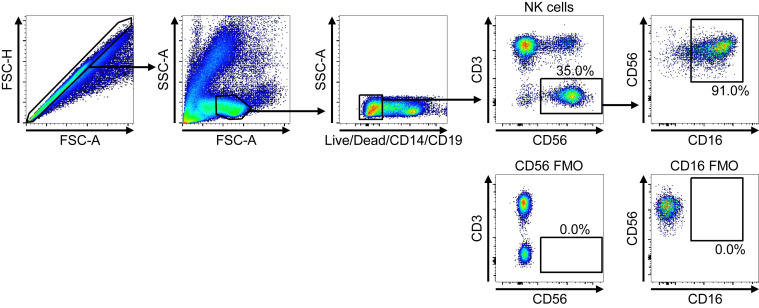
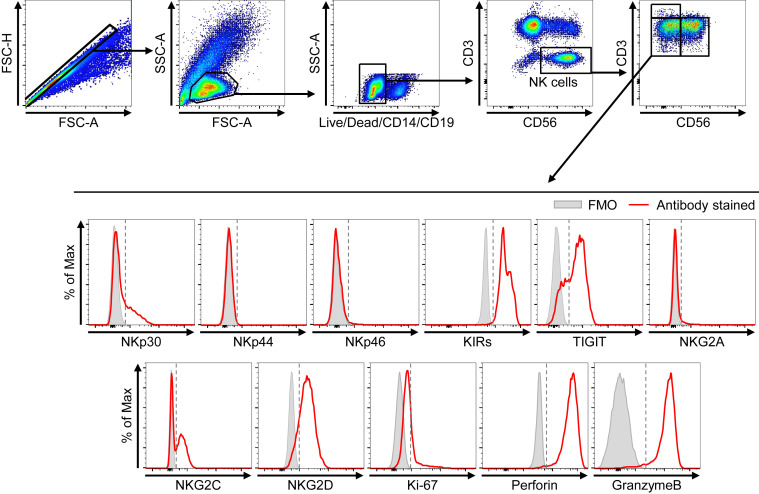
For the analysis of NK-cell subpopulations from PBMCs, singlet cells were gated first, followed by lymphogating. Positive cells for dump colors (Live/Dead, anti-CD14, anti-CD19, PE-Texas Red) were excluded. CD3–CD56+ cells were gated as NK cells and analyzed for the expression of CD16 and CD56. Consequently, the frequencies of conventional CD56brightCD16neg (cCD56bright), conventional CD56dimCD16pos (cCD56dim), and uCD56dim NK cells were determined among CD3–CD56+ NK cells. The expression of NK receptors, proliferation marker, and cytotoxic molecules within each NK subpopulation was also analyzed. Fluorescence minus one samples were used as negative controls. The gating strategies are presented in Fig E3 (NK-cell gating) and Fig E9 (surface markers and intracellular cytokine staining gating) in this article’s Online Repository at www.jacionline.org.
Fig E3.
Gating strategies for flow cytometric analysis of CD56 and CD16. FMO, Fluorescence minus one; FSC-A, forward scatter-area; FSC-H, forward scatter-height; SSC-A, side scatter-area.
Fig E9.
Gating strategies for flow cytometric analysis of surface markers and intracellular cytokine staining. Flow cytometry plots for the staining of each marker (red line) in uCD56dim NK cells. Fluorescence minus one (FMO; gray bar) was used as a negative control. Gray dashed lines are cutoff points determined by FMO staining. FSC-A, forward scatter-area; FSC-H, forward scatter-height; SSC-A, side scatter-area
Flow cytometry was performed using an LSR II instrument and FACSDiva software (BD, Franklin Lakes, NJ), and the data were analyzed using FlowJo software (BD).
NK-cell cytotoxicity assay
NK cells isolated from cryopreserved/thawed PBMCs using the NK-cell isolation kit (Miltenyi Biotec) were used as effector cells. K562 cells were used as target cells; they were labeled with PKH26 (Sigma-Aldrich) and cocultured with isolated NK cells at various effector to target ratios in 96-well U-bottom plates. After 6 hours, cocultured cells were gathered, and TO-PRO-3-iodide (Invitrogen) was added at a final concentration of 0.5 μM. The cells were immediately analyzed by flow cytometry. The background TO-PRO-3-iodide staining was evaluated by staining PKH26-labeled K562 cells without coculture with NK cells after 6-hour incubation in medium. The percent-specific lysis for each coculture was calculated as the % TO-PRO-3-iodide+ cells in background subtracted from the % TO-PRO-3-iodide+ cells in PKH26+ cells in each coculture.
Statistical analysis
Statistical analyses were performed using Prism software version 8.4.3 (GraphPad, GraphPad Software, San Diego, Calif). The Mann-Whitney U test was used in unpaired nonparametric comparisons, and the Wilcoxon matched-pairs signed rank test was used in paired nonparametric comparisons. The nonparametric Spearman rank correlation test was used to evaluate correlations between 2 parameters. The 2-way ANOVA test was used in group comparisons. P less than .05 was considered significant. In the analysis of next-generation sequencing (NGS) data, we used R program to implement a Walt test for significance testing (ie, the shrunken estimate of the logarithmic fold change divided by its SE). The Walt test P values were adjusted for multiple testing using the procedure of Benjamini and Hochberg.24
Results
Transcriptional profiles of NK cells from patients with COVID-19
We recruited 13 healthy donors, 7 patients with influenza, and 30 patients with COVID-19 (mild, n = 22; severe, n = 8) for this study. We used cryopreserved PBMCs from the patients with COVID-19 at several different time points. The clinical characteristics of the enrolled patients with COVID-19 are presented in Table E1, and the time points when samples were obtained are presented in Table E2.
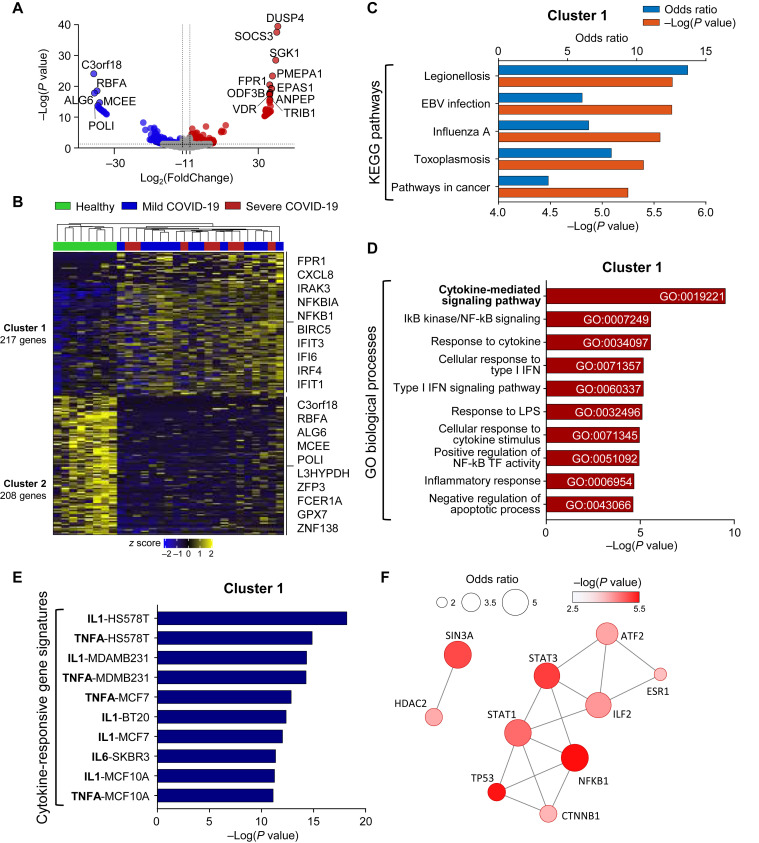
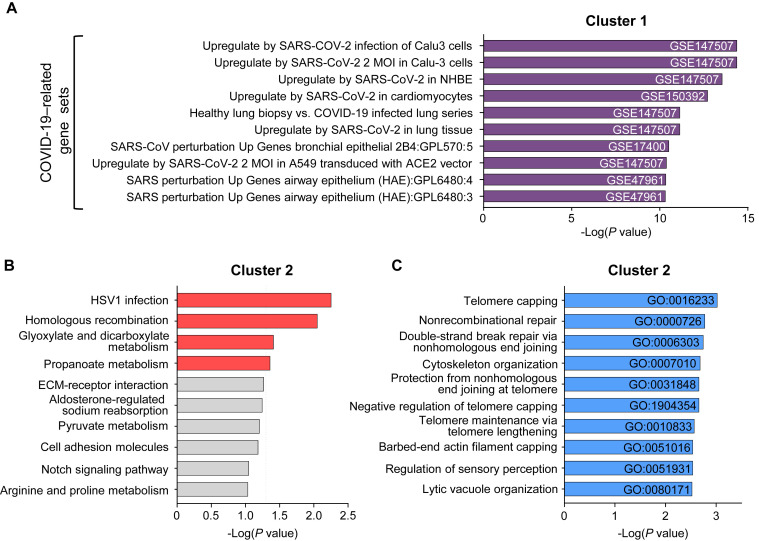
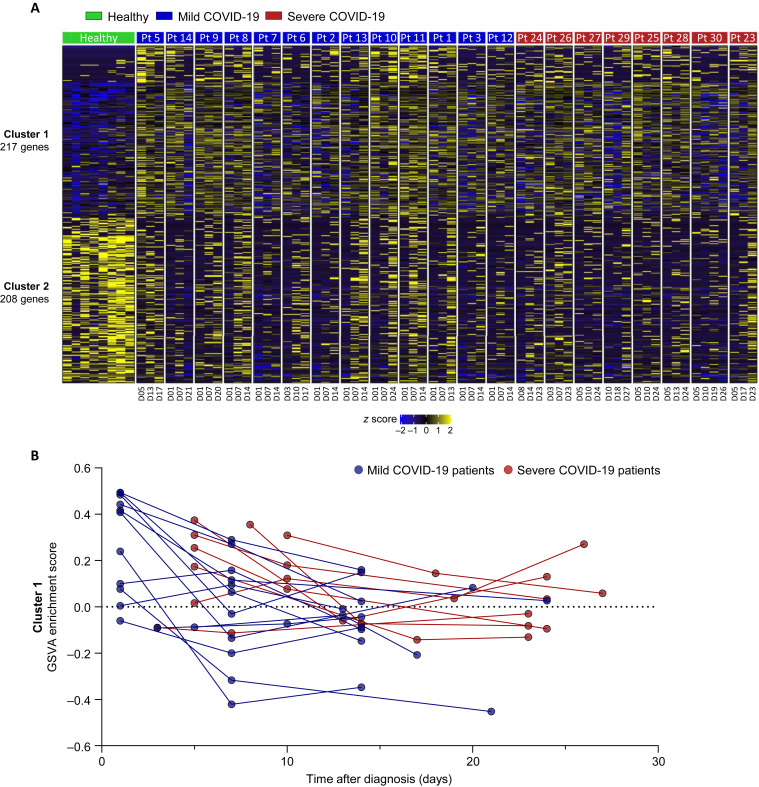
First, we sorted CD3–CD56+ NK cells from cryopreserved PBMCs of healthy donors (n = 8) and patients with COVID-19 (mild, n = 13; severe, n = 8) and performed RNA-seq. When we analyzed the data obtained at the earliest time point for each patient with COVID-19, 217 genes (cluster 1) were upregulated and 208 genes (cluster 2) were downregulated in NK cells compared with the healthy donors (Fig 1 , A and B). Cluster 1 was commonly enriched in previously reported gene sets related to SARS-CoV-2 infection (see Fig E2, A, in this article’s Online Repository at www.jacionline.org). The Kyoto Encyclopedia of Genes and Genomes pathway analysis revealed that cluster 1 was associated with pathways of several other infections, including legionellosis, EBV infection, influenza A, and toxoplasmosis, and pathways in cancer (Fig 1, C). These genes were most significantly associated with “cytokine-mediated signaling pathway,” and also with “IκB kinase/NF-κB signaling,” “response to cytokine,” “cellular response to type I interferon,” and “type I interferon signaling pathway” in the gene ontology analysis of biological processes (Fig 1, D). However, Kyoto Encyclopedia of Genes and Genomes pathway and gene ontology analysis showed that cluster 2 was associated with several gene sets, including “HSV 1 infection,” “homologous recombination,” “telomere capping,” and “nonrecombinant repair” with relatively high P values (Fig E2, B and C).
Fig 1.
Transcriptome analysis of NK cells from healthy donors and patients with COVID-19. RNA-seq of sorted CD3–CD56+ NK cells from cryopreserved/thawed PBMCs obtained from 8 healthy donors, 13 patients with mild COVID-19, and 8 patients with severe COVID-19 at the earliest time point for each. A and B, Volcano plot (Fig 1, A) and heatmap (Fig 1, B) showing upregulated or downregulated genes between healthy donors and patients with COVID-19: 217 genes were upregulated (cluster 1) and 208 genes were downregulated (cluster 2) in patients with COVID-19. Representative genes are annotated. C, KEGG pathway analysis of cluster 1. The top 5 pathways are presented with the odds ratio (blue bar) and −log(P value) (orange bar). D, Bar graphs showing the enrichment P values of the top 10 GO biological processes for cluster 1. GO IDs are annotated. E, Bar graphs showing the enrichment P values of the top 10 cytokine-responsive gene signatures from the L1000 LINCS database for cluster 1. Gene signatures are presented in the order of the cytokines (bold) and the name of the cytokine-treated cell lines. F, Protein-protein interaction network analysis of the top 10 cluster 1–related transcription factors. The size of the circle represents the odds ratio, and the fill color represents the value of −log(P value). GO, Gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; TF, transcription factor.
Fig E2.
Gene set enrichment analysis of clusters 1 and 2. A, Bar graphs showing the top 10 enriched gene sets for cluster 1 from published COVID-19–related gene sets. GSE numbers are indicated. B and C, KEGG pathway analysis (Fig E2, B) and GO biological processes analysis with GO IDs (Fig E2, C) for cluster 2. The dashed line in Fig E2, B, indicates a P value of .05. The enriched pathways with P less than .05 are presented as red bars, and others are presented as gray bars. GO, Gene ontology; GSE, gene set enrichment; KEGG, Kyoto Encyclopedia of Genes and Genomes; MOI, multiplicity of infection; HSV1, herpes simplex virus type 1; ECM, exctracellular matrix.
To determine which cytokine contributed to these changes in the NK cells, we performed gene set enrichment analysis using cytokine-responsive gene signatures from the LINCS L1000 data set. Cluster 1 was highly enriched in proinflammatory cytokine-responsive gene signatures, including IL-1, TNF-α, and IL-6 (Fig 1, E). The protein-protein interaction network analysis revealed that key transcription factors associated with NK cells in COVID-19 are signal transducer and activator of transcription 3, signal transducer and activator of transcription 1, nuclear factor kappa B subunit 1, and tumor protein p53 (Fig 1, F).
Expansion of the uCD56dim NK-cell population in patients with COVID-19
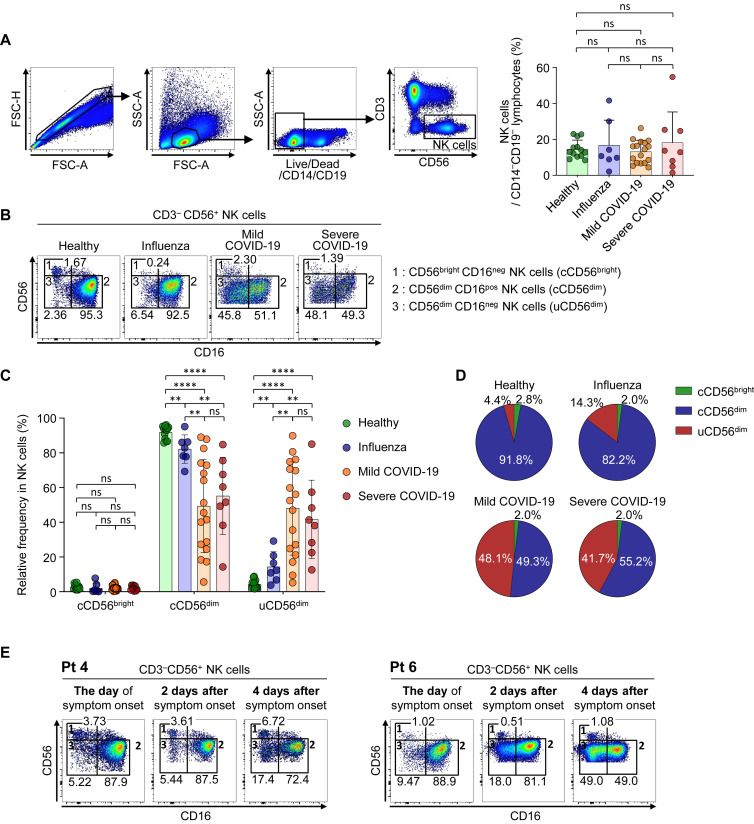
To examine the phenotype of NK cells in patients with COVID-19, we performed flow cytometric analysis using cryopreserved PBMCs from healthy donors (n = 13), patients with severe influenza (n = 7), patients with mild COVID-19 (n = 17), and patients with severe COVID-19 (n = 8). Samples obtained on the day of diagnosis (day 1) were used for patients with influenza and patients with COVID-19. First, we analyzed the frequency of NK cells among CD14–CD19– lymphocytes (Fig 2 , A) and absolute number (see Fig E4, A, in this article’s Online Repository at www.jacionline.org) and found no significant differences between the groups. We also found no significant difference in the frequency of NKG2C+CD57+ adaptive NK cells and CD3+CD56+ T cells between each group (see Figs E5 and E6 in this article’s Online Repository at www.jacionline.org). However, we found that, within the NK-cell population, the uCD56dim NK-cell population was significantly expanded alongside the shrinkage of cCD56dim NK cells in patients with COVID-19 regardless of disease severity, whereas the frequency of cCD56bright NK cells did not change (Fig 2, B-D, and Fig E4, B).
Fig 2.
Identification of NK-cell subsets and phenotypes of each subset using flow cytometry. Data from samples obtained on the day of diagnosis (day 1) are presented for patients with COVID-19 (A-D). A, Representative flow cytometry plots showing the gating strategy for CD3–CD56+ NK cells on the left. Right bar plots show cumulative data regarding the frequency of NK cells among CD14–CD19– lymphocytes in healthy donors (green bar, n = 13), patients with influenza (blue bar, n = 7), patients with mild COVID-19 (orange bar, n = 17), and patients with severe COVID-19 (red bar, n = 8). B, Flow cytometry plots of NK cells from a representative healthy donor, patients with influenza, patients with mild COVID-19, and patients with severe COVID-19 with identification of each NK-cell subset: subset 1 represents cCD56bright NK cells, subset 2 cCD56dim NK cells, and subset 3 uCD56dim NK cells. The frequencies of each subset among NK cells are presented. C, Bar plots showing cumulative data regarding the relative frequencies of each subset among NK cells in healthy donors (green bar), patients with influenza (blue bar), patients with mild COVID-19 (orange bar), and patients with severe COVID-19 (red bar). D, Pie charts showing the average frequency of each NK-cell subset by disease group: green slice for cCD56bright, blue slice for cCD56dim, and red slice for uCD56dim NK cells. E, Expansion of the uCD56dim NK-cell population during the early acute phase of COVID-19. Flow cytometry plots for 2 patients (Pt 4 and Pt 6) on the day of symptom onset, 2 days after symptom onset, and 4 days after symptom onset are presented. FSC-A, Forward scatter-area; FSC-H, forward scatter-height; ns, not significant; Pt, patient; SSC-A, side scatter-area. ∗P < .05. ∗∗P < .01. ∗∗∗P < .001. ∗∗∗∗P < .0001.
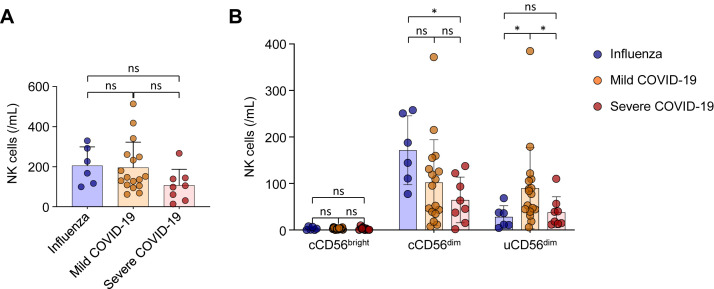
Fig E4.
Comparison of the absolute number of NK cells among patients with influenza and patients with mild and severe COVID-19. All data are presented as absolute counts of NK cells in 1 mL of blood. A, Comparison of the absolute number of NK cells among disease groups. B, Comparison of the absolute number of NK cells in each subset among disease groups. Blue dots represent patients with influenza, orange dots represent patients with mild COVID-19, and red dots represent patients with severe COVID-19. ns, Not significant. ∗P < .05.
Fig E5.
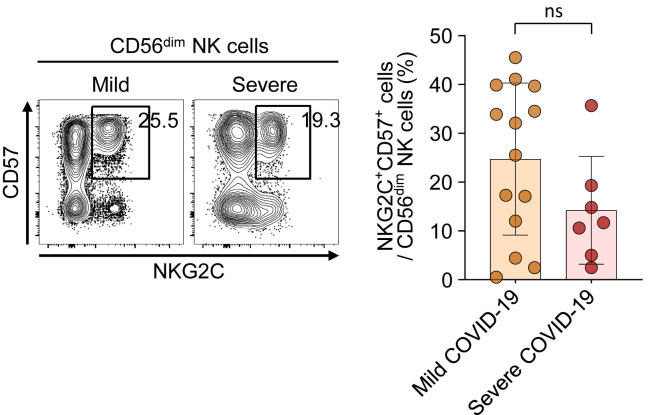
The frequency of adaptive NK cells in patients with mild and severe COVID-19. Left, Representative flow cytometry plots showing the gating strategy for adaptive NK cells (CD57+NKG2C+CD56dim NK cells) in PBMCs. Right, Bar plots showing cumulative data regarding the frequency of adaptive NK cells in patients with mild COVID-19 (orange bar, n = 14) and patients with severe COVID-19 (red bar, n = 7). ns, Not significant.
Fig E6.
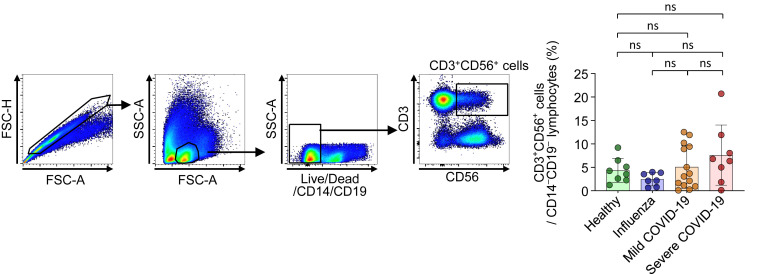
The frequency of CD3+CD56+ cells in each disease group. Left, Representative flow cytometry plots showing the gating strategy for CD3+CD56+ cells in PBMCs. Right, Bar plots showing cumulative data regarding the frequency of CD3+CD56+ cells among CD14–CD19– lymphocytes in healthy donors (green bar, n = 8), patients with influenza (blue bar, n = 7), patients with mild COVID-19 (orange bar, n = 14), and patients with severe COVID-19 (red bar, n = 8). FSC-A, forward scatter-area; FSC-H, forward scatter-height; ns, not significant; SSC-A, side scatter-area.
To demonstrate the expansion of the uCD56dim NK-cell population during the early phase of COVID-19, we analyzed longitudinal PBMC samples obtained from 2 patients during the early acute phase with short intervals from the day of symptom onset. We found that the frequency of uCD56dim NK cells was low on the day of symptom onset and gradually increased during the first 4 days after symptom onset (Fig 2, E), indicating that the uCD56dim NK-cell population expands during the early phase of COVID-19.
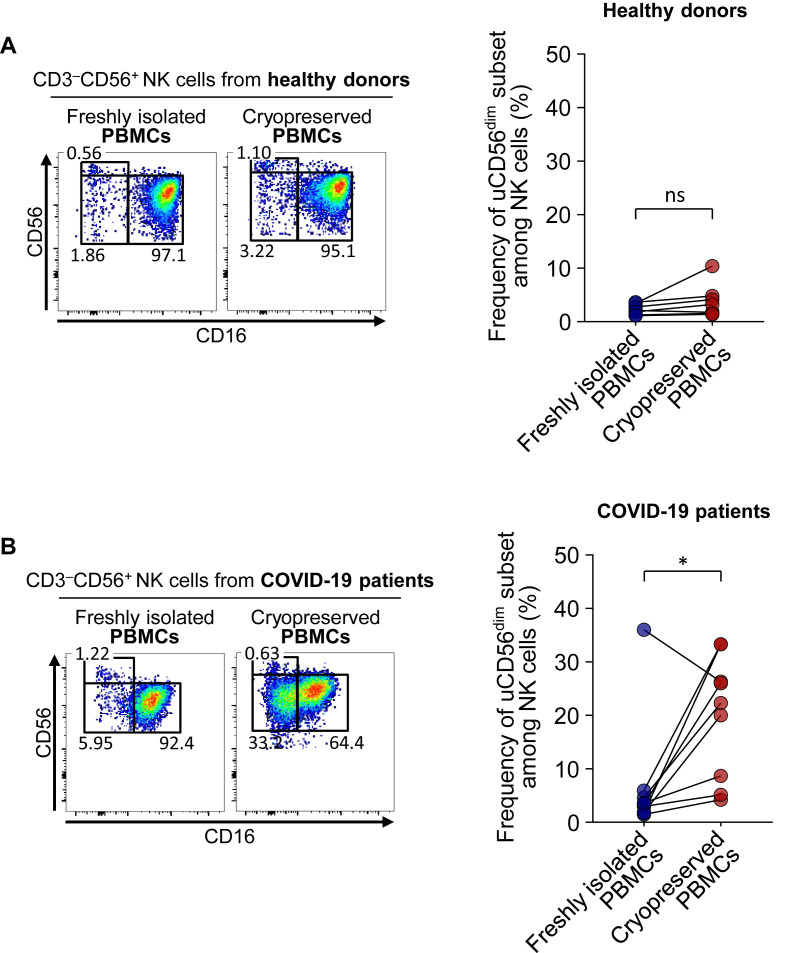
We analyzed the NK-cell subpopulations using freshly isolated PBMC samples and matched cryopreserved PBMC samples and found that the uCD56dim NK-cell population was a minor population in both fresh and cryopreserved PBMC samples from healthy donors (see Fig E7, A, in this article’s Online Repository at www.jacionline.org). In patients with COVID-19, the uCD56dim NK-cell population was prominently observed in cryopreserved samples, but not in freshly isolated samples (Fig E7, B), indicating that the frequency of the uCD56dim NK-cell population was significantly increased by cryopreservation and thawing only in patients with COVID-19. These data show that NK cells from patients with COVID-19 easily lose their CD16 expression via cellular stress.
Fig E7.
Comparison of NK-cell subsets between freshly isolated PBMCs and cryopreserved PBMCs from healthy donors (A) and patients with COVID-19 (B). Left, Representative flow cytometry plots. Right, Bar graph showing cumulative data regarding the paired comparison of the frequency of the uCD56dim subset among NK cells in healthy donors (Fig E7, A) and patients with COVID-19 (Fig E7, B). ns, Not significant. ∗P < .05.
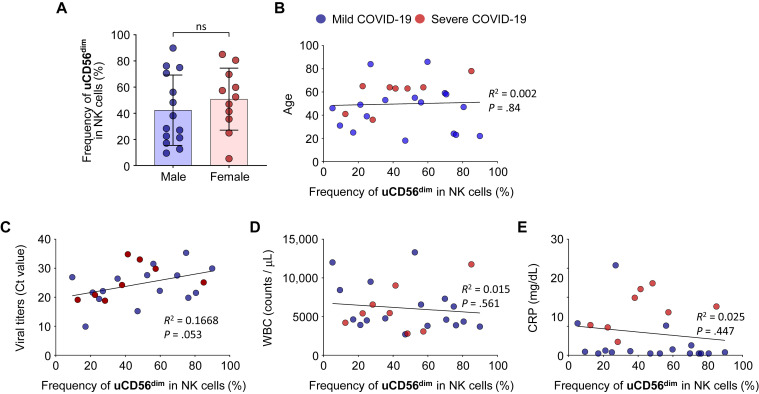
The expansion of uCD56dim NK-cell population was common in patients with COVID-19 regardless of clinical parameters, including sex, age, viral titers, white blood cell count, and C-reactive protein levels (see Fig E8 in this article’s Online Repository at www.jacionline.org).
Fig E8.
Correlation between the expansion of uCD56dim NK-cell population and clinical parameters. Each bar plot and dot plot represents the correlation between the frequency of uCD56dim NK cells in patients with COVID-19 and clinical parameters, including sex (A), age (B), viral titers (C), WBC counts (D), and CRP (E). N = 25 (17 patients with mild COVID-19 and 8 patients with severe COVID-19). CRP, C-reactive protein; WBC, white blood cell.
uCD56dim NK cells as a phenotypically and functionally distinct NK-cell subset
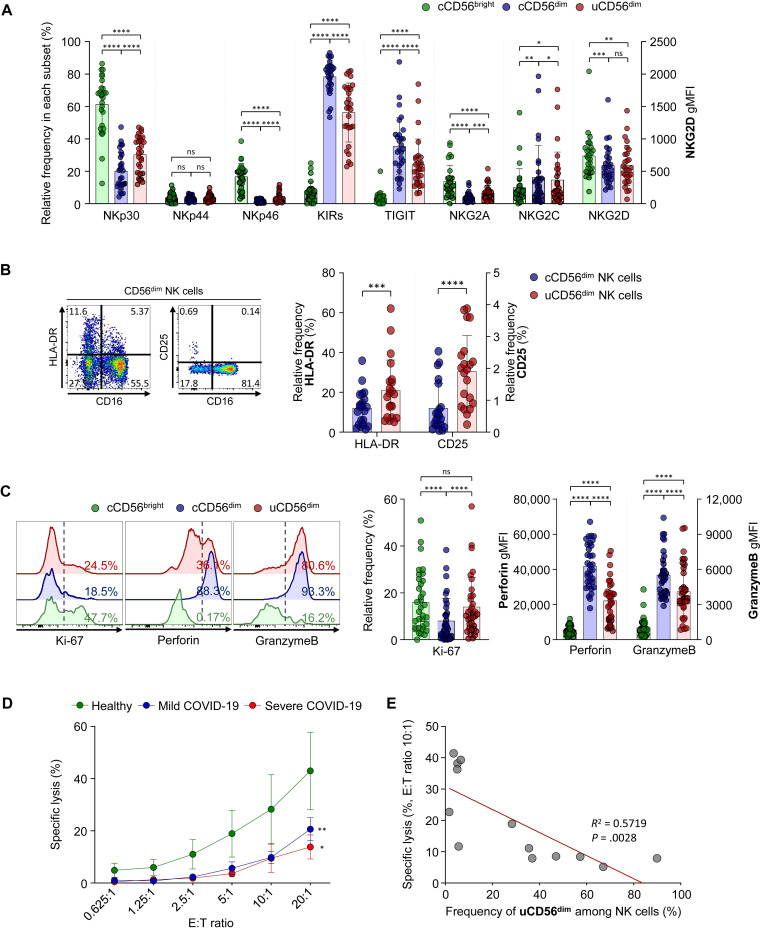
To identify the characteristics of the uCD56dim NK-cell population, we compared the expression of activating receptors, including NKp30, NKp44, NKp46, NKG2C, and NKG2D, and inhibitory receptors, including inhibitory killer-cell immunoglobulin-like receptors, T-cell immunoglobulin and ITIM domain, and NKG2A, using flow cytometry. The uCD56dim NK cells resembled cCD56dim more than cCD56bright with low expression of NKp30, NKp46, NKG2A, and NKG2D and high expression of inhibitory killer-cell immunoglobulin-like receptors, T-cell immunoglobulin and ITIM domain, and NKG2C (Fig 3 , A). However, uCD56dim NK cells were distinguished from cCD56dim NK cells by higher expression of NKp30, NKp46, and NKG2A and lower expression of inhibitory killer-cell immunoglobulin-like receptors, T-cell immunoglobulin and ITIM domain, and NKG2C (Fig 3, A).
Fig 3.
Intracellular cytokine staining to evaluate the proliferating and cytolytic capacities of each NK-cell subset and assessment of NK-cell cytotoxicity the first week after diagnosis. A, Bar plots showing the cumulative data regarding the expression of NK-cell receptors in each NK-cell subset: cCD56bright (green bar), cCD56dim (blue bar), and uCD56dim NK cells (red bar). Data from 9 healthy donors, 7 patients with influenza, 6 patients with mild COVID-19, and 7 patients with severe COVID-19 are presented. The expression of NKp30, NKp44, NKp46, KIRs (KIR2D and KIR3DL1/2), TIGIT, NKG2A, and NKG2C is presented as the frequency among each NK-cell subset, and the expression of NKG2D is presented as the geometric MFI (gMFI). B, The expression of activation markers, including HLA-DR and CD25, was compared between cCD56dim (blue bar) and uCD56dim (red bar) NK cells from 21 patients with COVID-19. Left, Representative flow cytometry plots. Right, Bar plots showing cumulative data regarding the relative frequencies of HLA-DR and CD25 in each NK-cell subset. C, Representative half-offset flow cytometry histograms (left) showing Ki-67, perforin, and granzyme B expression in each NK-cell subset. Bar plots (right) are cumulative data regarding Ki-67 expression (%), perforin, and granzyme B (gMFI) in each NK-cell subset (green for cCD56bright, blue for cCD56dim, and red for uCD56dim NK cells). Data from 8 healthy donors, 7 patients with influenza, 12 patients with mild COVID-19, and 7 patients with severe COVID-19 are presented. D, Assessment of NK-cell cytotoxicity the first week after diagnosis (9 healthy donors, 7 patients with mild COVID-19, and 5 patients with severe COVID-19). E, Correlation of the percentage of specific lysis at an E:T ratio of 10:1 and the frequency of uCD56dim NK cells. Data from 6 healthy donors, 3 patients with mild COVID-19, and 4 patients with severe COVID-19 are presented. E:T, Effector to target; KIR, inhibitory killer-cell immunoglobulin-like receptor; ns, not significant; TIGIT, T-cell immunoglobulin and ITIM domain. Error bars indicate SD. ∗P < .05. ∗∗P < .01. ∗∗∗∗P < .0001.
We also examined the expression of activation markers and found that HLA-DR and CD25 are upregulated in uCD56dim NK cells compared with cCD56dim NK cells, indicating that uCD56dim NK cells were recently activated (Fig 3, B).
The cCD56dim NK cells are cytolytic NK cells, and cCD56bright NK cells are highly proliferating and cytokine-producing NK cells.11 , 25 Consistent with previous studies, cCD56dim NK cells expressed a lower level of Ki-67 and higher levels of perforin and granzyme B compared with the cCD56bright NK cells (Fig 3, C). However, uCD56dim NK cells exhibited a distinctive functional phenotype from both cCD56dim and cCD56bright NK cells, with similar expression of Ki-67 but higher expression of perforin and granzyme B than in cCD56bright NK cells, and higher expression of Ki-67 but lower expression of perforin and granzyme B than in cCD56dim NK cells (Fig 3, C), indicating that uCD56dim NK cells exhibit a higher proliferating capacity but lower cytotoxic capacity compared with cCD56dim NK cells.
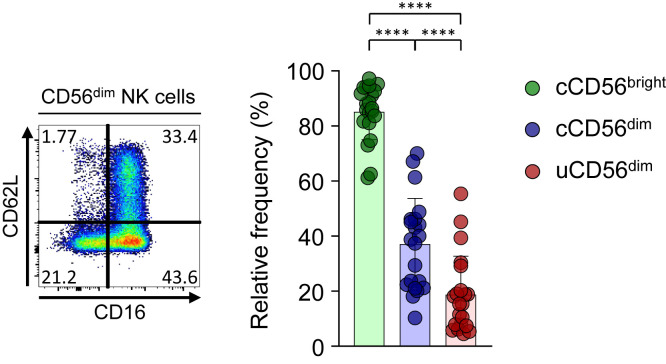
In addition, we examined the expression of CD62L, which is known to be cleaved by ADAM17 in activated NK cells,26 and found that the frequency of CD62L+ cells was significantly lower among uCD56dim NK cells than among cCD56dim NK cells (see Fig E10 in this article’s Online Repository at www.jacionline.org), indicating that ADAM17 activity is increased in uCD56dim NK cells. Given that CD16 can also be cleaved by ADAM17,26 , 27 these results suggest that NK cells from patients with COVID-19 easily lose CD16 expression via increased ADAM17 activity.
Fig E10.
The expression of CD62L in CD56dim NK-cell subpopulations. Left, Representative flow cytometry plot from 1 patient. Right, Bar plots showing cumulative data regarding the relative frequency of CD62L in each NK-cell subset (green bar for cCD56bright, blue bar for cCD56dim, and red bar for uCD56dim; n = 21). Error bars indicate SD. ∗∗∗∗P < .0001.
Impaired NK-cell cytotoxicity in patients with COVID-19
Next, we evaluated the cytotoxic function of NK cells. We isolated NK cells from the PBMCs of healthy donors and patients with COVID-19 the first week after diagnosis and assessed the NK-cell cytotoxicity against K562 cells. We found that NK-cell cytotoxicity was significantly impaired in patients with COVID-19 compared with healthy donors regardless of disease severity (Fig 3, D). Furthermore, the NK-cell cytotoxicity negatively correlated with the relative frequency of uCD56dim NK cells in the NK-cell population (Fig 3, E). Taken together, these results indicate that the expansion of the uCD56dim subset, which had decreased expression of perforin and granzyme B compared with cCD56dim NK cells, leads to impaired cytotoxicity of the NK-cell population in patients with COVID-19.
Recovery of impaired NK-cell cytotoxicity and the cCD56dim NK-cell population depending on disease severity
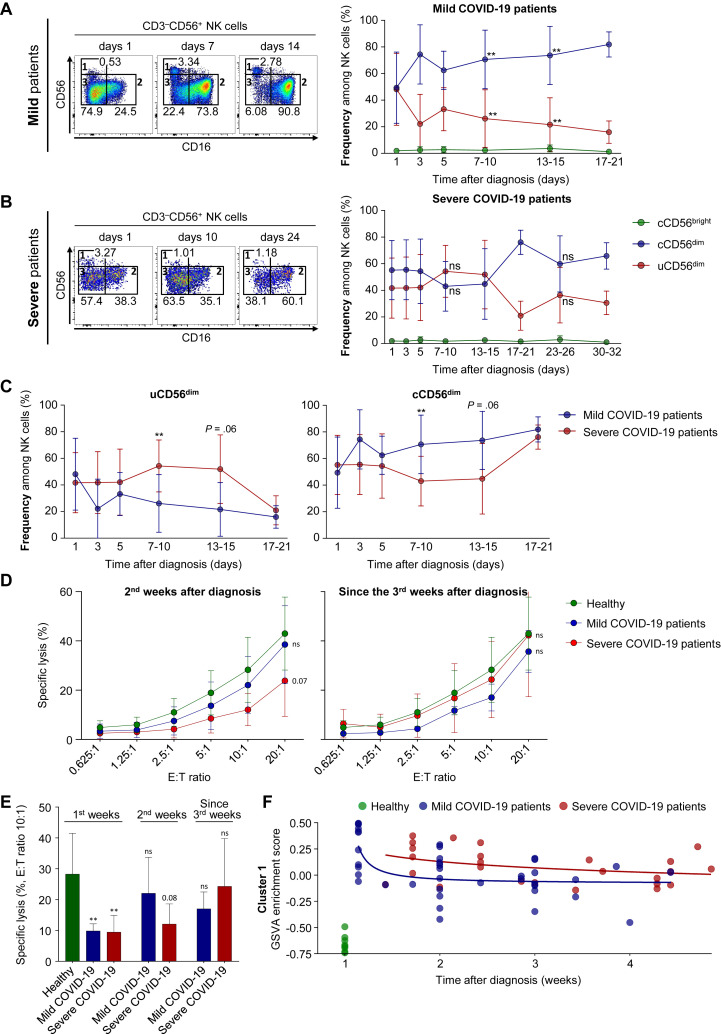
Finally, we investigated how the expansion of uCD56dim NK cells and impaired NK-cell cytotoxicity in patients with COVID-19 changes during the course of disease. We examined PBMCs collected from patients with COVID-19 at serial time points (indicated in Table E2) by flow cytometry and found that the frequency of uCD56dim NK cells decreased more rapidly in mild patients than in severe patients (Fig 4 , A and B). Until 7 days after diagnosis, we found no differences in the frequency of uCD56dim and cCD56dim NK cells between the 2 groups (Fig 4, C), but from 7 days after diagnosis, the frequency of uCD56dim NK cells was significantly higher and the frequency of cCD56dim NK cells was significantly lower in severe patients compared with mild patients (Fig 4, C). After 15 days from diagnosis, the differences disappeared, as the frequency of uCD56dim NK cells decreased and the frequency of cCD56dim NK cells increased in patients with severe COVID-19 (Fig 4, C).
Fig 4.
The longitudinal change in NK cells. A and B, The longitudinal change in the frequencies of NK-cell subsets in 17 mild (Fig 4, A) and 8 severe (Fig 4, B) patients. Each representative flow cytometry plot of NK cells on the left is from 1 patient at 3 different time points after diagnosis. Right graphs represent cumulative data regarding the frequencies of each NK-cell subset (green for cCD56bright, blue for cCD56dim, and red for uCD56dim NK cells). Statistical analysis at each time point was a comparison to day-1 samples. C, Frequency of uCD56dim (left) and cCD56dim (right) at each time point between mild patients (blue line) and severe patients (red line). The statistical analysis at each time point was a comparison of mild patients with severe patients. D, The NK-cell cytotoxicity against K562 cells was assessed the second week after diagnosis (left, 8 mild patients and 5 severe patients) and since the third week after diagnosis (right, 4 mild patients and 5 severe patients). The green line represents the NK-cell cytotoxicity of healthy donors, the blue line represents mild patients, and the red line represents severe patients. E, Bar graph showing the specific lysis of K562 cells by NK cells at an E:T ratio of 10:1 at different time points. The statistical analysis was a comparison to healthy donors. F, The longitudinal GSVA enrichment score for cluster 1. E:T, Effector to target; GSVA, gene set variation analysis; ns, not significant. Error bar indicates SD. ∗∗P < .01. ∗∗∗P < .001. ∗∗∗∗P < .0001.
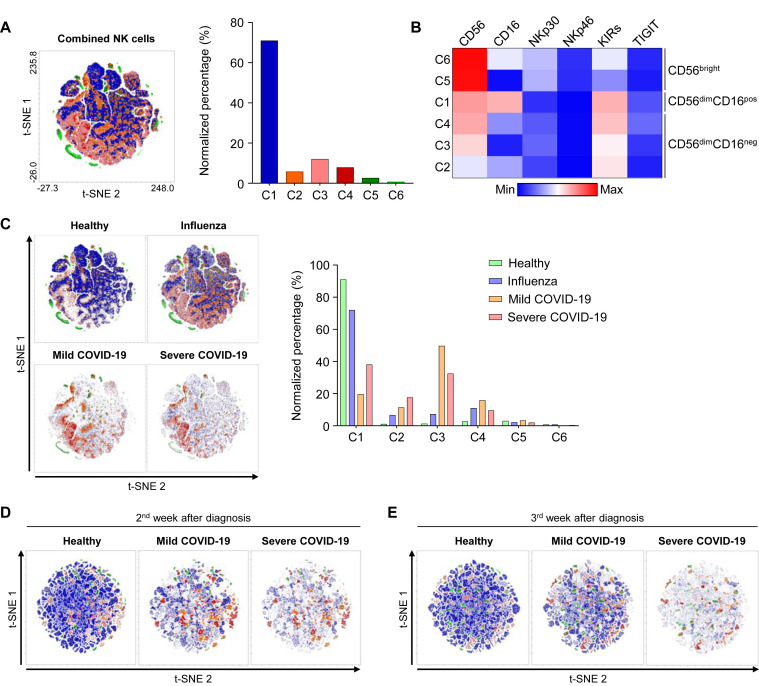
We also performed combined t-distributed stochastic neighbor embedding analysis of NK cells from healthy donors, patients with influenza, and patients with mild and severe COVID-19 using flow cytometry data to examine the NK-cell population clusters and changes during the disease course (see Fig E11 in this article’s Online Repository at www.jacionline.org). The t-distributed stochastic neighbor embedding analysis revealed 6 different clusters of NK cells, and they could be grouped into 3 major populations: CD56bright NK cells, CD56dimCD16pos NK cells, and CD56dimCD16neg NK cells (Fig E11, A and B). Further analysis demonstrated that the CD56dimCD16neg NK-cell population was exclusively expanded in patients with COVID-19, and it normalized rapidly in patients with mild COVID-19 (Fig E11, C-E).
Fig E11.
t-SNE analysis of NK cells. A-C, The t-SNE analysis of NK cells from 8 healthy donors, 7 patients with influenza, 8 patients with mild COVID-19, and 7 patients with severe COVID-19 on the day of diagnosis. A, The left t-SNE plot shows 6 different clusters, and the bar graph shows the normalized percentage of each cluster. B, Heatmap showing the differential expression of CD56, CD16, NKp30, NKp46, KIRs, and TIGIT in each group. C, The t-SNE analysis performed separately by disease group. The bar graph shows the normalized percentage of each cluster by group (green box for healthy donors, blue box for patients with influenza, orange box for patients with mild COVID-19, and red box for patients with severe COVID-19). D and E, The t-SNE analyses were performed separately by group (6 healthy donors, 5 patients with mild COVID-19, and 6 patients with severe COVID-19) 1 week after diagnosis (Fig E11, D) and 2 weeks after diagnosis (Fig E11, E). KIR, Inhibitory killer-cell immunoglobulin-like receptor; TIGIT, T-cell immunoglobulin and ITIM domain; t-SNE, t-distributed stochastic neighbor embedding.
In addition, we evaluated the NK-cell cytotoxicity during the course of disease. The second week after diagnosis, the impaired NK-cell cytotoxicity recovered in mild patients but not in severe patients (Fig 4, D and E). Since the third week after diagnosis, the impaired NK-cell cytotoxicity also recovered in severe patients, and the differences among disease groups disappeared (Fig 4, D and E). The results from the NK-cell cytotoxicity assays are compatible with the faster disappearance of uCD56dim NK cells and faster reemergence of cCD56dim NK cells in mild patients than in severe patients. We also analyzed RNA-seq data obtained from patients with COVID-19 (mild, n = 13; severe, n = 8) at serial time points. The gene set variation analysis enrichment score for cluster 1, upregulated genes in patients with COVID-19, decreased earlier in mild patients than in severe patients (Fig 4, F; see Fig E12 in this article’s Online Repository at www.jacionline.org), confirming the rapid and slow recovery of the NK-cell population in patients with mild and severe COVID-19, respectively.
Fig E12.
Transcriptomic profiles of NK cells during the disease course by patient. A, Heatmap showing the dynamic changes in the expression of cluster 1 and cluster 2 genes during the course of the disease by patient. Patient ID is annotated in green, blue, and red boxes, and the time points are annotated below. B, The longitudinal GSVA enrichment score for cluster 1. The same patients are connected by a blue (patients with mild COVID-19) or red line (patients with severe COVID-19). GSVA, Gene set variation analysis.
Discussion
Previous studies have reported that the absolute number of NK cells in patients with COVID-19 is reduced alongside lymphopenia, but the frequency of NK cells in lymphocytes is similar to that of healthy donors.17, 18, 19 Among NK cells, the frequency of CD56brightCD16neg NK cells has been reported to decrease in patients with COVID-19, especially those with severe disease, and the frequency of CD56dimCD16pos NK cells has been reported to slightly increase in patients with COVID-19. In the current study using cryopreserved PBMC samples, we found no difference in the frequency of NK cells among lymphocytes between healthy donors, patients with influenza, and patients with mild and severe COVID-19, and there was no difference in the frequency of cCD56bright NK cells. Interestingly, the cCD56dim NK-cell population was significantly reduced in patients with COVID-19 and the uCD56dim NK-cell population was significantly expanded in patients with COVID-19 on the day of diagnosis regardless of severity, which was scarcely found in healthy donors and patients with influenza. The uCD56dim NK cells had a higher proliferating capacity but lower cytolytic capacities than cCD56dim NK cells, which accounted for most of the NK cells in healthy donors and patients with influenza. The expansion of uCD56dim NK cells was associated with impaired NK-cell cytotoxicity in patients with COVID-19.
NK cells are a major subtype of type 1 innate lymphoid cells and play important roles against viral infections.9, 10, 11 Therefore, the study of NK-cell biology in viral diseases is important for understanding the natural disease course. However, because of the urgent need for the development of vaccines and treatments for COVID-19, more research and attention is focused on the adaptive immune responses, and understanding of the NK-cell biology in COVID-19 is lacking. In the present study, we aimed to identify the phenotypic and functional changes in NK cells during COVID-19 and how NK cells change during the course of the disease. We found that NK cells in COVID-19, regardless of disease severity, are characterized by the expansion of uCD56dim NK cells, which exhibit lower cytolytic capacities than do cCD56dim NK cells. This characteristic change in NK cells during COVID-19 was accompanied by impaired NK-cell cytotoxicity. Both the expansion of uCD56dim NK cells and impaired NK-cell cytotoxicity recovered earlier in mild patients compared with severe patients. A few studies have reported that the natural cytotoxicity of PBMCs from patients with COVID-19 is impaired,18 , 19 but to our knowledge, our study is the first to show the impaired function of NK cells directly using sorted CD3–CD56+ NK cells and how their function recovered during the course of the disease.
The expansion of the uCD56dim NK-cell population has been reported in a few clinical conditions, including malignancies, infectious mononucleosis, haploidentical hematopoietic stem cell transplantation, and hemophagocytic lymphohistiocytosis (HLH).14 , 28, 29, 30, 31, 32, 33 However, the mechanism underlying the expansion of the uCD56dim NK-cell population has not been clarified. Previously, uCD56dim NK cells were considered to be the precursors of cCD56dim NK cells. However, several studies have shown that uCD56dim NK cells are bona fide differentiated NK cells.32 , 34 , 35 Interestingly, the loss of CD16 expression on NK cells by cryopreservation/thawing of human PBMCs has been reported in previous studies.14 , 34 , 36 In the current study, the uCD56dim NK-cell population was prominently observed in cryopreserved PBMCs from patients with COVID-19, but not healthy donors, indicating that NK cells become abnormally labile to ex vivo cellular stress, such as cryopreservation/thawing, during COVID-19. In the present study, we also showed that the frequency of CD62L+ cells was significantly lower among uCD56dim NK cells than among cCD56dim NK cells (Fig E10), indicating that the ADAM17 activity is increased in uCD56dim NK cells. Given that CD16 can also be cleaved by ADAM17,26 , 27 these results suggest that NK cells from patients with COVID-19 easily lose CD16 expression via increased ADAM17 activity. In this regard, we hypothesize that ADAM17 activity is involved in the cleavage and loss of CD16 on NK cells in patients with COVID-19.
In addition, considering that COVID-19–specific upregulated genes were significantly enriched in gene sets regulated by proinflammatory cytokines, such as IL-1, TNF-α, and IL-6, we hypothesize that high levels of proinflammatory cytokines can be one of the mechanisms inducing the expansion of uCD56dim NK-cell population. The clinical conditions previously reported for the expansion of uCD56dim NK cells are commonly characterized by overwhelming inflammation and hypersecretion of various cytokines.30, 31, 32, 33 , 37 However, the frequency of uCD56dim NK cells did not correlate with C-reactive protein levels in the current study. Further studies are required to reveal the origin and fate of uCD56dim NK cells.
HLH is a fatal systemic inflammatory syndrome and can be caused by several medical conditions, inducing strong activation of the immune system, such as viral infection or cancer (secondary HLH).38, 39, 40 HLH is typically accompanied by impaired NK-cell function.41, 42, 43 Recently, several studies have reported that patients with severe COVID-19 undergo clinical conditions reminiscent of secondary HLH.37 , 44, 45, 46 In the current study, the expansion of uCD56dim NK cells lasted for a longer period in patients with severe COVID-19 than in patients with mild COVID-19. Particularly, in a mortality case, the frequency of uCD56dim NK cells remained elevated (>30%) during the course of the disease. Secondary HLH needs to be suspected in patients with COVID-19, particularly when the high frequency of uCD56dim NK cells is sustained.
Conclusions
We found that a unique NK-cell subset, uCD56dim NK cells, is expanded in COVID-19. We demonstrated that the NK-cell cytotoxicity was impaired in COVID-19 alongside the expansion of uCD56dim NK cells. Furthermore, we longitudinally showed that the rapid disappearance of uCD56dim NK cells and reemergence of cCD56dim NK cells in mild patients was accompanied by rapid recovery of NK-cell cytotoxicity. However, the recovery of the NK-cell population occurred in patients with severe COVID-19 in a delayed manner. To our knowledge, this was the first longitudinal study assessing NK cells from patients with COVID-19 in regard to phenotypic and functional changes. Further studies are needed on how these characteristic uCD56dim NK cells play a role in the protection or pathology of patients with COVID-19.
Key messages.
-
•
The uCD56dim NK-cell population expands in cryopreserved PBMCs from patients with COVID-19 regardless of disease severity.
-
•
The expansion of uCD56dim NK cells is accompanied by decreased NK-cell cytotoxicity.
-
•
The expansion of uCD56dim NK cells lasts longer in severe COVID-19 than in mild COVID-19.
Footnotes
This research was supported by the 2020 Joint Research Project of Institutes of Science and Technology.
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Data and materials availability: RNA sequencing data have been uploaded in the Gene Expression Omnibus (GEO) database under GEO accession number GSE165461. All other data that support the findings of this study and materials are available on request.
Appendix
Table E1.
Clinical characteristics of enrolled healthy donors and patients
| ID | Age (y) | Sex | Disease severity∗ | Mortality† | Underlying diseases | Oxygen demand (L/min or oxygen % for MV) | Systoloic BP | Respiratory rate (/min) | Mental status‡ | ICU admission |
|---|---|---|---|---|---|---|---|---|---|---|
| Patients with mild COVID-19§ | ||||||||||
| 1 | 46 | F | 3 | 0 | DM, SCZ | 0 | 96 | 22 | A | No |
| 2 | 39 | F | 3 | 0 | None | 0 | 111 | 20 | A | No |
| 3 | 31 | M | 2 | 0 | None | 0 | 151 | 18 | A | No |
| 4 | 47 | F | 2 | 0 | None | 0 | 111 | 20 | A | No |
| 5 | 53 | F | 3 | 0 | None | 2 | 143 | 26 | A | No |
| 6 | 49 | M | 3 | 0 | None | 0 | 146 | 20 | A | No |
| 7 | 55 | F | 2 | 0 | HTN, DM | 0 | 158 | 20 | A | No |
| 8 | 86 | F | 1 | 0 | HTN, A-fib, dementia | 0 | 107 | 20 | A | No |
| 9 | 84 | M | 3 | 0 | HTN, DM, dementia | 4 | 129 | 20 | A | No |
| 10 | 58 | M | 3 | 0 | None | 0 | 123 | 20 | A | No |
| 11 | 59 | F | 3 | 0 | None | 0 | 121 | 20 | A | No |
| 12 | 23 | M | 2 | 0 | None | 0 | 108 | 20 | A | No |
| 13 | 51 | M | 3 | 0 | None | 0 | 114 | 18 | A | No |
| 14 | 25 | M | 2 | 0 | None | 0 | 118 | 20 | A | No |
| 15 | 49 | F | 3 | 0 | HTN | 0 | 111 | 20 | A | No |
| 16 | 22 | M | 1 | 0 | None | 0 | 115 | 20 | A | No |
| 17 | 18 | F | 1 | 0 | None | 0 | 96 | 18 | A | No |
| 18 | 24 | M | 2 | 0 | None | 0 | 101 | 18 | A | No |
| 19 | 24 | F | 1 | 0 | None | 0 | 116 | 18 | A | No |
| 20 | 61 | M | 2 | 0 | None | 0 | 118 | 20 | A | No |
| 21 | 49 | M | 1 | 0 | None | 0 | 100 | 20 | A | No |
| 22 | 29 | M | 1 | 0 | None | 0 | 112 | 20 | A | No |
| Patients with severe COVID-19§ | ||||||||||
| 23 | 65 | M | 5 | 0 | HTN, DM | MV 70% | 127 | 20 | V | Yes |
| 24 | 63 | M | 5 | 0 | SCZ | MV 40% | 106 | 19 | P | Yes |
| 25 | 64 | M | 5 | 0 | HTN, SCZ | MV 50% | 127 | 20 | P | Yes |
| 26 | 36 | M | 5 | 0 | None | MV 40% | 143 | 20 | A | Yes |
| 27 | 64 | F | 5 | 0 | None | MV 50% | 102 | 26 | U | Yes |
| 28 | 63 | F | 5 | 0 | None | MV 60% | 96 | 28 | A | Yes |
| 29 | 78 | F | 5 | 1 | HTN | MV 100% | 89 | 30 | P | Yes |
| 30 | 41 | M | 5 | 0 | None | MV 60% | 128 | 22 | U | Yes |
| Healthy donors | ||||||||||
| H1 | 31 | M | — | — | None | — | — | — | — | — |
| H2 | 31 | M | — | — | None | — | — | — | — | — |
| H3 | 38 | M | — | — | None | — | — | — | — | — |
| H4 | 38 | M | — | — | None | — | — | — | — | — |
| H5 | 30 | M | — | — | None | — | — | — | — | — |
| H6 | 27 | M | — | — | None | — | — | — | — | — |
| H7 | 28 | F | — | — | None | — | — | — | — | — |
| H8 | 36 | M | — | — | None | — | — | — | — | — |
| H9 | 35 | M | — | — | None | — | — | — | — | — |
| H10 | 45 | M | — | — | None | — | — | — | — | — |
| H11 | 55 | M | — | — | None | — | — | — | — | — |
| H12 | 27 | M | — | — | None | — | — | — | — | — |
| H13 | 36 | M | — | — | None | — | — | — | — | — |
| Patients with influenza | ||||||||||
| I1 | 89 | M | 4 | 1 | Prostate cancer | 2 | 98 | 18 | A | Yes |
| I2 | 66 | M | 5 | 0 | MM, HTN | 10 | 103 | 24 | V | Yes |
| I3 | 75 | F | 2 | 0 | HTN, DM | 1 | 130 | 24 | A | No |
| I4 | 70 | M | 3 | 1 | HTN, DM | 0 | 120 | 16 | A | Yes |
| I5 | 33 | M | 2 | 0 | PCOS | 0 | 134 | 20 | A | No |
| I6 | 59 | M | 3 | 0 | ESRD | 0 | 144 | 18 | A | No |
| I7 | 59 | F | 3 | 0 | BLL | 0 | 118 | 18 | A | No |
A-fib, Atrial fibrillation; BLL, B-cell lymphocytic leukemia; BP, blood pressure; RR, respiratory rate; DM, diabetes mellitus; ESRD, end-stage renal disease; F, female; HTN, hypertension; ICU, intensive care unit; M, male; MM, multiple myeloma; MV, mechanical ventilation; NIH, National Institutes of Health; PCOS, polycystic ovary syndrome; SCZ, schizophrenia.
Disease severity was calculated on the basis of NIH severity score (1 = asymptomatic, 2 = mild, 3 = moderate [pneumonia saturation ≥ 94%], 4 = severe [pneumonia with RR > 30, saturation < 94%, PaO2/FiO2 < 300, lung infiltration > 50%], 5 = critical [respiratory failure, shock, multiple organ dysfunction]).
Mortality is presented as 0 = alive and 1 = death.
Mental status is presented as A = alert, V = voice, P = pain, and U = unresponsive.
Mild and severe patients were classified by the NIH severity score (1-3 = mild, 4-5 = severe).
Table E2.
Sample time points and assessment assays
| Patient ID | Sampling time points∗ | Assessment assays† |
||
|---|---|---|---|---|
| RNA-seq | Immunophenotyping/ICS | Cytotoxicity assay | ||
| Mild patients‡ | ||||
| 1 | 1, 3, 5, 7, 13 | 1, 7, 13 | 1, 3, 5, 7, 13 | |
| 2 | 1, 3, 5, 7, 14 | 1, 7, 14 | 1, 3, 5, 7, 14 | |
| 3 | 1, 3, 5, 7, 15 | 1, 7, 15 | 1, 3, 5, 7, 15 | |
| 4 | 1, 3, 5, 7, 15 | 1, 3, 5, 7, 15 | ||
| 5 | 1, 3, 5, 13, 17 | 5, 13, 17 | 1, 3, 13, 17 | 1 |
| 6 | 1, 3, 10, 17 | 3, 10, 17 | 1, 3, 10, 17 | 17 |
| 7 | 1, 3, 7, 14, 21 | 1, 7, 14 | 1, 3, 7, 14, 21 | |
| 8 | 1, 3, 7, 14 | 1, 7, 14 | 1, 3, 7, 14 | |
| 9 | 1, 5, 7, 20 | 1, 7, 20 | 1, 5, 7, 20 | |
| 10 | 1, 3, 7 | 1, 7, 24 | 1, 3, 7 | |
| 11 | 1, 3, 7, 14 | 1, 7, 14 | 1, 3, 7, 14 | |
| 12 | 1, 3, 7, 14 | 1, 7, 14 | 1, 3, 7, 14 | |
| 13 | 1, 7, 14 | 1, 7, 14 | 1, 7, 14 | |
| 14 | 1, 7, 21 | 1, 7, 21 | 1, 7, 21 | |
| 15 | 7, 21 | 7, 21 | ||
| 16 | 1, 7, 14 | 1, 7, 14 | 1, 7 | |
| 17 | 1, 7, 14 | 1, 7, 14 | 1, 7 | |
| 18 | 1, 7, 14 | 1, 7, 14 | 1, 7 | |
| 19 | 1, 7 | 1, 7 | ||
| 20 | 1, 7, 21 | 1, 7, 21 | ||
| 21 | 1, 7, 21 | 1, 7, 21 | ||
| 22 | 1, 7 | 1, 7 | ||
| Severe patients‡ | ||||
| 23 | 1, 3, 5, 14, 17, 23 | 5, 17, 23 | 1, 5, 17, 23 | 3, 14, 23 |
| 24 | 1, 5, 8, 14, 23, 26 | 8, 14, 23 | 1, 5, 8, 14, 23, 26 | 5 |
| 25 | 1, 5, 7, 10, 24, 31 | 5, 10, 24 | 1, 5, 10, 24 | 7, 31 |
| 26 | 1, 3, 7, 14, 23 | 3, 7, 23 | 1, 3, 7, 14, 23 | 3, 7, 23 |
| 27 | 1, 3, 5, 10, 20, 24 | 5, 10, 24 | 1, 5, 10, 20, 24 | 3, 24 |
| 28 | 1, 3, 5, 7, 13, 17, 24 | 5, 13, 24 | 1, 3, 5, 7, 13, 17, 24 | 3 |
| 29 | 1, 7, 10, 14, 18, 27, 32 | 10, 18, 27 | 1, 7, 14, 32 | 7 |
| 30 | 1, 3, 5, 7, 10, 19, 26, 30 | 5, 10, 19, 26 | 1, 3, 5, 7, 10, 19, 26, 30 | 7, 26 |
ICS, Intracellular cytokine staining; NIH, National Institutes of Health.
Sampling time points are presented as days after hospitalization.
Assessment assays are presented as number of sampling time points.
Mild and severe patients are classified by NIH severity score (score 1-3: mild, 4-5: severe).
Table E3.
Initial laboratory results for patients with COVID-19 and patients with influenza
| Patient’s ID | Initial laboratory results |
|||||
|---|---|---|---|---|---|---|
| Viral titer (Ct value) | WBC count (/μL) | Neutrophil (%) | Lymphocyte (%) | CRP (mg/dL) | Chest X-ray abnormalities∗ | |
| Mild patients† | ||||||
| 1 | Negative | 12,000 | 83.4 | 10.5 | 5.5 | 1 |
| 2 | 19.4 | 4,500 | 82.7 | 13.2 | 0.5 | 0 |
| 3 | 26.9 | 8,430 | 59.8 | 26.5 | 0.6 | 0 |
| 4 | 21.5 | 4,360 | 72.2 | 19.3 | 0.3 | 0 |
| 5 | 26.4 | 4,770 | 65.5 | 24.7 | 0.7 | 0 |
| 6 | 21.6 | 3,920 | 63.2 | 23.5 | 0.8 | 1 |
| 7 | 27.6 | 13,300 | 67.2 | 24.9 | 0.3 | 0 |
| 8 | 22.2 | 3,800 | 51.4 | 36.7 | 1 | 0 |
| 9 | 22.1 | 9,500 | 76.2 | 10.8 | 15.5 | 1 |
| 10 | Negative | 4,600 | 77.3 | 15.4 | 1.7 | 1 |
| 11 | 27.5 | 7,300 | 82.9 | 14.3 | 0.3 | 0 |
| 12 | 19.8 | 3,880 | 69 | 23.5 | 0.3 | 0 |
| 13 | 31.5 | 6,560 | 57.4 | 32.8 | 5.1 | 1 |
| 14 | 9.87 | 4,630 | 48.6 | 35 | 0.3 | 0 |
| 15 | 32.2 | 5,540 | 70.3 | 23.6 | 0.3 | 0 |
| 16 | 29.9 | 3,710 | 51.8 | 39.1 | 0.5 | 0 |
| 17 | 15.2 | 2,700 | 39.3 | 52.6 | 0.3 | 0 |
| 18 | 35.3 | 6,310 | 46.4 | 39.9 | 0.3 | 0 |
| 19 | 32.3 | 5,450 | 66.9 | 26.5 | 0.5 | 0 |
| 20 | 28.7 | 8,400 | 79.2 | 12.4 | 2.1 | 0 |
| 21 | Negative | 5,600 | 64.1 | 28.7 | 0.6 | 0 |
| 22 | 35.3 | 6,230 | 51.9 | 37.9 | 0.8 | 0 |
| Severe patients† | ||||||
| 23 | 20.8 | 5,420 | 62.9 | 25.3 | 4.8 | 1 |
| 24 | 33.0 | 2,800 | 74.9 | 20.6 | 12.4 | 1 |
| 25 | 24.2 | 5,460 | 77.2 | 12.5 | 9.9 | 1 |
| 26 | 18.8 | 6,560 | 67.2 | 16.9 | 2.3 | 1 |
| 27 | 29.8 | 3,080 | 68.9 | 24 | 7.4 | 1 |
| 28 | 34.8 | 9,020 | 71.4 | 21.4 | 11.4 | 1 |
| 29 | 25.1 | 11,760 | 88.7 | 8.1 | 8.4 | 1 |
| 30 | 19.1 | 4,200 | 64.5 | 29.3 | 5.2 | 1 |
| Patients with influenza | ||||||
| I1 | NA | 11,400 | 75.6 | 10.5 | 9.6 | 1 |
| I2 | NA | 2,700 | 80 | 8.9 | 1.1 | 1 |
| I3 | NA | 9,000 | 69.4 | 25.8 | 0.1 | 0 |
| I4 | NA | 15,400 | 83.2 | 4.9 | 6.7 | 1 |
| I5 | NA | 2,300 | 55.8 | 30.5 | 3.0 | 0 |
| I6 | NA | 5,500 | 74.4 | 16.9 | 0.2 | 1 |
| I7 | NA | 3,700 | 35.3 | 51.1 | 1.2 | 1 |
CRP, C-reactive protein; NA, not available; NIH, National Institutes of Health; WBC, white blood cell.
Chest abnormalities are presented as 0 = normal and 1 = abnormal.
Mild and severe patients are classified by NIH severity score (1-3 = mild, 4-5 = severe).
Table E4.
Key resources
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| BV421 anti-NKG2D (Clone: 1D11) | Biolegend | Cat# 320822 |
| BV510 anti-CD3 (Clone: HIT3a) | BD Biosciences | Cat# 564713 |
| BV605 anti-NKp44 (Clone: p44-8) | BD Biosciences | Cat# 744301 |
| BV605 anti-Ki-67 (Clone: Ki-67) | Biolegend | Cat# 350522 |
| BV650 anti-NKG2A (Clone: 131411) | BD Biosciences | Cat# 747920 |
| BV711 anti-NKG2C (Clone: 134591) | BD Biosciences | Cat# 748164 |
| FITC anti-KIR2D (Clone: REA1042) | Miltenyi Biotec | Cat# 130-117-477 |
| FITC anti-KIR3DL1/DL2 (Clone: REA970) | Miltenyi Biotec | Cat# 130-116-177 |
| FITC anti-Perforin (Clone: dG9) | Biolegend | Cat# 308104 |
| FITC anti-CD3 (Clone: HIT3a) | BD Biosciences | Cat# 555339 |
| PerCP-cy5.5 anti–Granzyme B (Clone: QA16A02) | Biolegend | Cat# 372212 |
| PerCP-cy5.5 7-AAD (Clone: 8-AAD) | BD Biosciences | Cat# 51-68981E |
| PE anti-CD56 (Clone: HCD56) | Biolegend | Cat# 318306 |
| PE-TR anti-CD14 (Clone: MOP9) | BD Biosciences | Cat# 562335 |
| PE-TR anti-CD19 (Clone: HIB19) | BD Biosciences | Cat# 562294 |
| PE-Cy7 anti-TIGIT (Clone: MBSA43) | eBioscience | Cat# 25-9500-42 |
| AF647 anti-NKp30 (Clone: p30-15) | BD Biosciences | Cat# 558408 |
| AF700 anti-NKp46 (Clone: 900) | Biolegend | Cat# 331932 |
| APC-H7 anti-CD16 (Clone: 3G8) | BD Biosciences | Cat# 560195 |
| Critical commercial assays | ||
| LIVE/DEAD Fixable Red Dead Cell Stain Kit | Invitrogen | Cat# L34972 |
| Foxp3/Transcription Factor Staining Buffer Set | Invitrogen | Cat# 00-5523-00 |
| PKH26 Red Fluorescent Cell Linker Kit | Sigma-Aldrich | Cat# PKH26GL |
| TO-PRO-3 Iodide | Invitrogen | Cat# T3605 |
| TRIzol LS reagent | Invitrogen | Cat# 10296028 |
| Lymphocyte Separation Medium | Corning | Cat# 25-072-CV |
| NK Cell Isolation Kit, human | Miltenyi Biotec | Cat# 130-092-657 |
| Deposited data | ||
| Raw and analyzed data | This article | GEO accession number |
| Human reference genome GRCh38 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Experimental models: Cell lines | ||
| K562 | American Type Culture Collection (ATCC) | ATCC CCL243 |
| Software and algorithms | ||
| STAR | Dobin et al,E1 2013 | http://doi:10.1093/bioinformatics/bts635 |
| DESeq2 | Love et al,E2 2014 | http://doi:10.1186/s13059-014-0550-8 |
| Prism software version 8.4.3 | GraphPad | NA |
| FlowJo software version 10.7.1 | Treestar | https://www.flowjo.com/solutions/flowjo/downloads |
| FACSDiva software | BD Biosciences | NA |
FITC, Fluorescein isothiocyanate; GEO, Gene Expression Omnibus; KIR, inhibitory killer-cell immunoglobulin-like receptor; NA, not available; TIGIT, T-cell immunoglobulin and ITIM domain.
Table E5.
Viability information for all samples after thawing
| Healthy∗ | Patients with influenza† | Patients with mild COVID-19‡ | Patients with severe COVID-19§ | |
|---|---|---|---|---|
| Viability, mean ± SD | 86.88 ± 7.35 | 69.21 ± 13.77 | 79.75 ± 12.11 | 75.22 ± 10.19 |
Separate information about the viability of samples is presented below (presented as Pt ID, time point, viability [%]).
Pt, Patient.
Healthy: H1, 1, 85.1; H2, 1, 77.0; H3, 1, 84.6; H4, 1, 77.6; H5, 1, 85.3; H6, 1, 83.0; H7, 1, 86.7; H8, 1, 83.0; H9, 1, 99.4; H10, 1, 99.2; H11, 1, 99.1; H12, 1, 82.5; H13, 1, 86.9.
Patients with influenza: I1, 1, 92.2; I2, 1, 72.9; I3, 1, 61.0; I4, 1, 79.5; I5, 1, 72.3; I6, 1, 60.2; I7, 1, 46.4.
Patients with mild COVID-19: 1, 1, 78.7; 1, 3, 81.7; 1, 5, 79.1; 1, 7, 78.9; 1, 13, 76.6; 2, 1, 84.9; 2, 3, 84.9; 2, 5, 78.5; 2, 7, 98.9; 2, 14, 98.9; 3, 1, 83.6; 3, 3, 67.7; 3, 5, 74.0; 3, 7, 83.7; 3, 15, 79.9; 4, 1, 95.0; 4, 3, 67.7; 4, 5, 74.0; 4, 7, 98.6; 4, 15, 97.9; 5, 1, 67.5; 5, 3, 81.1; 5, 5, 97.5; 5, 13, 98.9; 5, 17, 95.5; 6, 1, 98.9; 6, 3, 97.5; 6, 10, 92.6; 6, 17, 99.7; 7, 1, 56.4; 7, 3, 88.1; 7, 7, 71.0; 7, 14, 63.7; 7, 21, 73.6; 8, 1, 72.6; 8, 3, 82.4; 8, 7, 88.1; 8, 14, 82.9; 9, 1, 83.5; 9, 5, 63.6; 9, 7, 93.5; 9, 20, 91.1; 10, 1, 82.1; 10, 3, 89.2; 10, 7, 95.1; 10, 24, 93.9; 11, 1, 64.6; 11, 3, 90.8; 11, 7, 72.5; 11, 14, 51.1; 12, 1, 49.7; 12, 3, 95.1; 12, 7, 71.4; 12, 14, 80.3; 13, 1, 66.8; 13, 7, 81.4; 13, 14, 81.5; 14, 1, 56.0; 14, 7, 91.5; 14, 21, 85.7; 15, 7, 73.8; 15, 21, 79.5; 16, 1, 75.2; 16, 7, 75.2; 16, 14, 77.2; 17, 1, 73.2; 17, 7, 84.1; 17, 14, 81.3; 18, 1, 64.5; 18, 7, 79.4; 19, 1, 61.1; 19, 7, 77.2; 19, 18, 66.4; 20, 1, 57.4; 20, 3, 84.2; 20, 5, 81.1; 20, 7, 53.8; 20, 21, 71.2; 21, 3, 85.5; 22, 1, 83.4; 22, 7, 86.0.
Patients with severe COVID-19: 23, 1, 87.4; 23, 3, 78.8; 23, 5, 56.7; 23, 14, 81.9; 23, 17, 85.7; 23, 23, 63.7; 24, 1, 72.1; 24, 5, 64.2; 24, 8, 83.4; 24, 14, 86.5; 24, 23, 87.2; 24, 26, 57.0; 25, 1, 68.8; 25, 5, 74.9; 25, 7, 77.0; 25, 10, 72.3; 25, 24, 74.5; 25, 31, 63.8; 26, 1, 83.1; 26, 3, 78.3; 26, 7, 80.3; 26, 14, 74.7; 26, 23, 86.0; 27, 1, 59.6; 27, 3, 62.8; 27, 5, 61.8; 27, 10, 69.2; 27, 20, 64.3; 27, 24, 75.2; 28, 1, 85.4; 28, 3, 71.2; 28, 5, 73.3; 28, 7, 71.6; 28, 13, 84.1; 28, 17, 80.1; 28, 24, 79.6; 29, 1, 67.5; 29, 7, 45.4; 29, 14, 84.1; 29, 32, 83.5; 30, 1, 84.5; 30, 3, 91.2; 30, 5, 82.2; 30, 7, 67.7; 30, 10, 86.9; 30, 19, 92.9; 30, 26, 79.3; 30, 30, 81.0.
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization Weekly epidemiological update—19 January 2021. https://www.who.int/publications/m/item/weekly-epidemiological-update---19-january-2021 Available at:
- 4.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferretti A.P., Kula T., Wang Y., Nguyen D.M.V., Weinheimer A., Dunlap G.S., et al. Unbiased screens show CD8(+) T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity. 2020;53:1095–1107.e3. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 9.Abel A.M., Yang C., Thakar M.S., Malarkannan S. Natural killer cells: development, maturation, and clinical utilization. Front Immunol. 2018;9:1869. doi: 10.3389/fimmu.2018.01869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andoniou C.E., Andrews D.M., Degli-Esposti M.A. Natural killer cells in viral infection: more than just killers. Immunol Rev. 2006;214:239–250. doi: 10.1111/j.1600-065X.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 11.Vivier E., Tomasello E., Baratin M., Walzer T., Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 12.Alessandro M., Cristina B., Massimo V., Daniela P., Claudia C., Maria Cristina M., et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 13.Fan Y.Y., Yang B.Y., Wu C.Y. Phenotypically and functionally distinct subsets of natural killer cells in human PBMCs. Cell Biol Int. 2008;32:188–197. doi: 10.1016/j.cellbi.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 14.Amand M., Iserentant G., Poli A., Sleiman M., Fievez V., Sanchez I.P., et al. Human CD56(dim)CD16(dim) cells as an individualized natural killer cell subset. Front Immunol. 2017;8:699. doi: 10.3389/fimmu.2017.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B., Han J., Cheng X., Yu L., Zhang L., Wang W., et al. Reduced numbers of T cells and B cells correlates with persistent SARS-CoV-2 presence in non-severe COVID-19 patients. Sci Rep. 2020;10:17718. doi: 10.1038/s41598-020-73955-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazzoni A., Salvati L., Maggi L., Capone M., Vanni A., Spinicci M., et al. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J Clin Invest. 2020;130:4694–4703. doi: 10.1172/JCI138554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maucourant C., Filipovic I., Ponzetta A., Aleman S., Cornillet M., Hertwig L., et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osman M., Faridi R.M., Sligl W., Shabani-Rad M.T., Dharmani-Khan P., Parker A., et al. Impaired natural killer cell counts and cytolytic activity in patients with severe COVID-19. Blood Adv. 2020;4:5035–5039. doi: 10.1182/bloodadvances.2020002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varchetta S., Mele D., Oliviero B., Mantovani S., Ludovisi S., Cerino A., et al. Unique immunological profile in patients with COVID-19. Cell Mol Immunol. 2021;18:604–612. doi: 10.1038/s41423-020-00557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Vaz Meirelles G., et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 25.Freud Ag, Caligiuri M.A. Human natural killer cell development. Immunol Rev. 2006;214:56–72. doi: 10.1111/j.1600-065X.2006.00451.x. [DOI] [PubMed] [Google Scholar]
- 26.Romee R., Foley B., Lenvik T., Wang Y., Zhang B., Ankarlo D., et al. NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease-17 (ADAM17) Blood. 2013;121:3599–3608. doi: 10.1182/blood-2012-04-425397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J., Mishra H.K., Walcheck B. Role of ADAM17 as a regulatory checkpoint of CD16A in NK cells and as a potential target for cancer immunotherapy. J Leukoc Biol. 2019;105:1297–1303. doi: 10.1002/JLB.2MR1218-501R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moeller M.J., Kammerer R., von Kleist S. A distinct distribution of natural killer cell subgroups in human tissues and blood. Int J Cancer. 1998;78:533–538. doi: 10.1002/(sici)1097-0215(19981123)78:5<533::aid-ijc1>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 29.Penack O., Gentilini C., Fischer L., Asemissen A.M., Scheibenbogen C., Thiel E., et al. CD56dimCD16neg cells are responsible for natural cytotoxicity against tumor targets. Leukemia. 2005;19:835–840. doi: 10.1038/sj.leu.2403704. [DOI] [PubMed] [Google Scholar]
- 30.Azzi T., Lunemann A., Murer A., Ueda S., Beziat V., Malmberg K.J., et al. Role for early-differentiated natural killer cells in infectious mononucleosis. Blood. 2014;124:2533–2543. doi: 10.1182/blood-2014-01-553024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hust M.A., Blechacz B.R.A., Bonilla D.L., Daver N., Rojas-Hernandez C.M. Adult cancer-related hemophagocytic lymphohistiocytosis—a challenging diagnosis: a case report. J Med Case Rep. 2017;11:172. doi: 10.1186/s13256-017-1344-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberto A., Di Vito C., Zaghi E., Mazza E.M.C., Capucetti A., Calvi M., et al. The early expansion of anergic NKG2A(pos)/CD56(dim)/CD16(neg) natural killer represents a therapeutic target in haploidentical hematopoietic stem cell transplantation. Haematologica. 2018;103:1390–1402. doi: 10.3324/haematol.2017.186619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vujanovic L., Chuckran C., Lin Y., Ding F., Sander C.A., Santos P.M., et al. CD56(dim) CD16(−) natural killer cell profiling in melanoma patients receiving a cancer vaccine and interferon-alpha. Front Immunol. 2019;10:14. doi: 10.3389/fimmu.2019.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi E., Kuranaga N., Satoh K., Habu Y., Shinomiya N., Asano T., et al. Induction of CD16+ CD56bright NK cells with antitumour cytotoxicity not only from CD16− CD56bright NK cells but also from CD16− CD56dim NK cells. Scand J Immunol. 2007;65:126–138. doi: 10.1111/j.1365-3083.2006.01883.x. [DOI] [PubMed] [Google Scholar]
- 35.Stabile H., Nisti P., Morrone S., Pagliara D., Bertaina A., Locatelli F., et al. Multifunctional human CD56 low CD16 low natural killer cells are the prominent subset in bone marrow of both healthy pediatric donors and leukemic patients. Haematologica. 2015;100:489–498. doi: 10.3324/haematol.2014.116053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mark C., Czerwinski T., Roessner S., Mainka A., Horsch F., Heublein L., et al. Cryopreservation impairs 3-D migration and cytotoxicity of natural killer cells. Nat Commun. 2020;11:5224. doi: 10.1038/s41467-020-19094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tholin B., Hauge M.T., Aukrust P., Fehrle L., Tvedt T.H. Hemophagocytic lymphohistiocytosis in a patient with COVID-19 treated with tocilizumab: a case report. J Med Case Rep. 2020;14:187. doi: 10.1186/s13256-020-02503-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herman T.E., Siegel M.J. Familial hemophagocytic lymphohistiocytosis. J Perinatol. 2010;30:363–365. doi: 10.1038/jp.2010.25. [DOI] [PubMed] [Google Scholar]
- 39.Ramos-Casals M., Brito-Zerón P., López-Guillermo A., Khamashta M.A., Bosch X. Adult haemophagocytic syndrome. Lancet. 2014;383:1503–1516. doi: 10.1016/S0140-6736(13)61048-X. [DOI] [PubMed] [Google Scholar]
- 40.Hayden A., Park S., Giustini D., Lee A.Y., Chen L.Y. Hemophagocytic syndromes (HPSs) including hemophagocytic lymphohistiocytosis (HLH) in adults: a systematic scoping review. Blood Rev. 2016;30:411–420. doi: 10.1016/j.blre.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Perez N., Virelizier J.L., Arenzana-Seisdedos F., Fischer A., Griscelli C. Impaired natural killer activity in lymphohistiocytosis syndrome. J Pediatr. 1984;104:569. doi: 10.1016/s0022-3476(84)80549-1. [DOI] [PubMed] [Google Scholar]
- 42.Stark B., Cohen I.J., Pecht M., Umiel T., Apte R.N., Friedman E., et al. Immunologic dysregulation in a patient with familial hemophagocytic lymphohistiocytosis. Cancer. 1987;60:2629–2636. doi: 10.1002/1097-0142(19871201)60:11<2629::aid-cncr2820601110>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 43.Grom A.A. Natural killer cell dysfunction: a common pathway in systemic-onset juvenile rheumatoid arthritis, macrophage activation syndrome, and hemophagocytic lymphohistiocytosis? Arthritis Rheum. 2004;50:689–698. doi: 10.1002/art.20198. [DOI] [PubMed] [Google Scholar]
- 44.Clark KEN, Nevin WD, Mahungu T, Lachmann H, Singh A. Assessment of the haemophagocytic lymphohistiocytosis HScore in patients with COVID-19 [published online ahead of print September 28, 2020]. Clin Infect Dis. https://doi.org/10.1093/cid/ciaa1463. [DOI] [PMC free article] [PubMed]
- 45.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White K., Benninger L., Manjarres D.G. Covid-19 and a hemophagocytic lymphohistiocytosis-like syndrome in the absence of ARDS. Chest. 2020;158:A1007. [Google Scholar]
References
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]