Summary
Study of evolution and selection pressure on HIV-1 in fetuses will lead to a better understanding of the role of immune responses in shaping virus evolution and vertical transmission. Detailed genetic analyses of HIV-1 env gene from 12 in utero transmission pairs show that most infections (67%) occur within 2 months of childbirth. In addition, the env sequences from long-term-infected fetuses are highly divergent and form separate phylogenetic lineages from their cognate maternal viruses. Host-selection sites unique to neonate viruses are identified in regions frequently targeted by neutralizing antibodies and T cell immune responses. Identification of unique selection sites in the env gene of fetal viruses indicates that the immune system in fetuses is capable of exerting selection pressure on viral evolution. Studying selection and evolution of HIV-1 or other viruses in fetuses can be an alternative approach to investigate adaptive immunity in fetuses.
Keywords: HIV-1, mother to child transmission, fetus, neonate, in utero transmission, immune selection, signature
Graphical abstract

Highlights
Most in utero HIV-1 transmission occurs within 2 months of childbirth
The HIV-1 env gene sequences from long-term-infected fetuses are highly divergent
Selection sites unique to fetal viruses are found in regions targeted by immunity
Presence of selection sites indicates active adaptive immune responses in fetuses
Marichannegowda et al. find that in utero transmission of HIV-1 usually occurs within 2 months of childbirth. Analysis of the env gene sequences from long-term-infected fetuses identifies host-selection signatures that are unique to viruses in infants, suggesting that the fetal immune system can exert selection pressure on viral evolution.
Introduction
Among 38 million people currently living with HIV/AIDS, 1.8 million are children infected through mother-to-child transmission (MTCT).1 Perinatal HIV-1 infection can be acquired through antepartum (in utero), intrapartum (around delivery), or postpartum (through breast feeding). Prior to the availability of antiretroviral therapy (ART), the rate of HIV-1 MTCT among pregnant women was about 30%–40%,2 and it is estimated that each transmission route accounted for about one-third of the total MTCT transmissions. After the introduction of ART, the MTCT rate was significantly reduced to ∼2%.3,4 However, MTCT still leads to well over 100,000 new infant infections annually. With the “90-90-90” campaign by the United Nations Program on HIV/AIDS (UNAIDS), it was expected that pediatric HIV-1 infections would be reduced to fewer than 20,000 by 2020.5 However, because HIV testing and ART are not widely accessible to all pregnant women in resource-limited countries, this goal will not be achieved.6,7 Further, as ART is often initiated upon diagnosis of HIV-1 during prenatal visits or around the time of delivery and will reduce the number of infection through intrapartum and postpartum, in utero infections are responsible for a higher portion of the ongoing cases of MTCT than would have been the case in the pre-ART era.
During pregnancy, passive transfer of maternal immunoglobulin G (IgG) antibodies (Abs) to fetuses may play a role in blocking MTCT and exert immune selection pressure on viruses in fetuses,8, 9, 10, 11 but the mechanism is still poorly understood.11, 12, 13, 14 The relatively high rate of in utero MTCT suggests that transferred maternal Abs cannot fully prevent HIV-1 infection of fetuses, possibly due to the presence of escape variants resistant to maternal neutralizing antibodies (nAbs). Our previous studies showed that the transmitted viruses in infants are often resistant to neutralization by cognate maternal plasma,8,10,15, 16, 17 yet it remains unclear whether maternal antibodies contribute to blocking placental HIV transmission.
Innate immune responses and antigen-specific T cell responses to pathogens are detectable in fetuses, suggesting exposure to maternal infection in utero may prime the developing immune system.18 In fact, aberrant fetal immune responses to maternal antigens and bacterial, viral, and fungal infections may play an important role in the pathogenesis of preterm labor.19, 20, 21 A recent study demonstrated that fetal antigen-presenting cells (APCs), such as conventional dendritic cells (DCs), can respond to toll-like receptors (TLRs) as early as the second trimester.22 However, a comparison of immune responses between fetuses and infants shows that the immune responses mediated by macrophages are more geared toward immune tolerance in fetuses. Little is known about the function of fetal B lymphocytes in response to infectious agents.23 Fetal exposure to HIV-1 primes the immune system to enhance immune activation and alter T cell homing in fetuses.24 These studies indicate that innate and adaptive immune responses are functioning in the fetus, but none have demonstrated that immune responses can exert selection pressure on viral evolution during in utero infection.
HIV-1 infection of fetuses can occur in pregnant women without ART, but the timing and frequency of infection as well as viral evolution in fetuses remain unknown. Moreover, the role of immune selection on the viruses in fetuses has not yet been established. To address these questions, we have studied a total of 721 HIV-1 env gene sequences from 12 pairs of mothers and neonates who were infected in utero from two cohorts. We found that most in utero infections occur late in pregnancy, and unique strong selection on viral evolution during long-term in utero infection is different between fetuses and their cognate mothers, suggesting the presence of adaptive immune responses against HIV-1 in fetuses.
Results
Most in utero MTCTs occur during the late stage of pregnancy
Twelve in utero mother-neonate transmission pairs were selected from two cohorts (10 from Women and Infant Transmission Study [WITS] and 2 from Center for HIV/AIDS Vaccine Immunology protocol 9 [CHAVI009]). All neonates were infected in utero, as determined by positive PCR with HIV-1 DNA genome extracted from neonate blood samples at birth. All but one mother (0842 m) had plasma samples available at delivery. Neonate plasma samples were available from various time points after delivery (3 at delivery, 7 at ∼1 month after birth, and 2 at ∼2 months after birth; Table 1). A total of 721 HIV-1 env gene sequences (317 and 404 from mothers and neonates, respectively) were obtained by single-genome amplification (SGA). All sequences from each mother-neonate pair were clustered together in the overall phylogenetic tree, and sequences from different mother-neonate pairs were phylogenetically distinctive from each other (Figure S1). Phylogenetic tree and Highlighter plot (https://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html) analyses showed that the env sequences from five neonates (pair 1_0782n, pair 2_1730n, pair 3_0489n, pair 4_1348n, and pair 5_9112n) were highly homogeneous, and a single transmitted/founder (T/F) viral sequence was inferred for each neonate (Figures 1 and S2). The env sequences from three neonates were heterogeneous, consisting of two or three homogeneous sub-populations. Among them, two T/Fs were inferred for two neonates (pair 6_1744n and pair 7_0842n) and three T/Fs for the third neonate (pair 8_3915n). The env sequences from four other neonates (pair 9_1039n, pair 10_2093n, pair 11_1580n, and pair 12_2564n) were highly heterogeneous, and no T/F sequences could be reliably inferred (Figures 1 and S2). Of note, these four neonates were on average sampled at an older age, when the evolving viral populations likely had already replaced the original infecting strains: the mean number of days after birth was 43 days, compared to 17 days for the other 8 neonates.
Table 1.
Demographic and genetic characteristics of mother-neonate transmission pairs
| Transmission pair |
Mother |
Viral load (copies/mL) |
No. of SGAs |
No. of T/Fs | Neonate age (days) | Estimated days of infection (95%, CI) | Transmission days from birth | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | ID | Age (years) | Gestation (weeks) | Mother | Neonate | Mother | Neonate | ||||
| 1 | 0782 | 16 | 34 | 8,224 | 2,716 | 32 | 7 | 1 | 6 | 9 (4, 15) | −3 |
| 2 | 1730 | 19 | 34 | 45,924 | 716,003 | 28 | 29 | 1 | 27 | 44 (33, 54) | −17 |
| 3 | 0489 | 31 | 34 | 30,067 | 779,602 | 35 | 26 | 1 | 20 | 32 (21, 44) | −12 |
| 4 | 1348 | 21 | 34 | 4,458 | 332,727 | 19 | 46 | 1 | 40 | 61 (52, 70) | −21 |
| 5 | 9112 | 25 | 34 | N/A | N/A | 34 | 21 | 1 | 1 | 20 (14, 26) | −19 |
| 6 | 1744 | 27 | 34 | 276,955 | N/A | 21 | 35 | 2 | 21 | 49 (29, 69) | −28 |
| 7 | 0842 | 21 | 34 | 6,057 | 216,000 | N/A | 37 | 2 | 20 | 27 (23, 32) | −7 |
| 8 | 3915 | 28 | 30 | N/A | N/A | 41 | 51 | 3 | 1 | 38 (35, 41) | −37 |
| 9 | 1039 | 24 | 34 | 25,049 | 1,957,418 | 24 | 30 | chronic | 39 | ND | |
| 10 | 2093 | 20 | 34 | 3,101,258 | 1,119,972 | 38 | 45 | chronic | 54 | ND | |
| 11 | 1580 | 30 | 34 | 78,174 | N/A | 32 | 53 | chronic | 51 | ND | |
| 12 | 2564 | 21 | 34 | 26,640 | N/A | 13 | 24 | chronic | 28 | ND | |
See also Figure S3. ND: not determined.
Figure 1.
Phylogenetic trees of the env sequences from each mother-neonate transmission pair
Phylogenetic trees were constructed by the neighbor-joining method with the Kimura 2-parameter model, and their reliability was estimated by 1,000 bootstrap replicates. The nodes that are supported by more than 70% of bootstrap replicates are indicated by asterisks. Sequences from neonates and mothers are indicated in blue and red dots, respectively. See also Figures S1–S3.
Using the Poisson Fitter tool,25 we were able to determine the infection time for the eight neonate samples with well-defined T/F lineages, which ranged from 9 to 61 days, corresponding to 3–37 days before birth (Table 1). For neonate pair 6_1744n, pair 7_0842n, and pair 8_3915n, who were infected with 2 or 3 distinct T/F viruses, the overall time since infection was calculated by taking the harmonic mean of the time estimates from each separate lineage, as previously described.25 Their infection times were 49, 27, and 38 days, respectively, corresponding to 28, 7, and 37 days before birth. The estimated in utero infection times for pair 1_0782n and pair 7_0842n were the shortest, only 3 and 7 days before birth, respectively. Importantly, all estimated times of infection were before their corresponding birth dates, further confirming that all neonates had been infected in utero, within 40 days before birth (Table 1).
The env sequences in four neonates (pair 9_1039n, pair 10_2093n, pair 11_1580n, and pair 12_2564n) were distinctive from all other neonates in that their genetic diversity was far more divergent (Figures 1 and S2), indicating that these infections were too far out to allow the use of the Poisson Fitter tool, which is based on the assumption of random accumulation of mutations prior to the onset of selection. The BEAST (Bayesian evolutionary analysis by sampling trees) software package has been widely used to estimate the time of the most recent common ancestor (MRCA) or infection time.26 However, in this context, BEAST is also limited for two reasons. First, any timing inference on the neonate sequences yields the time since the MRCA, which likely took place in the mother, not the neonate. Indeed, when we ran BEAST on viral sequences from these four neonates, we obtained timing estimates far longer than the 40-week gestational time. Second, further analyses using the recombination detection tool Recombination Analysis PRogram (RAPR)27 reveal extensive recombination in all four neonate samples (Figure S3A). This degree of pervasive recombination can cause branch length artifacts in the phylogenetic reconstructions upon which the BEAST methods are based, potentially confounding the time estimates.28
Indeed, the RAPR analysis revealed that, for two neonates in particular, pair 10_2093n and pair 12_2564n, all but 3 or less sequences had derived from recombination events. In our previous study,27 we showed that, in heterogeneous infections (multiple T/Fs), recombinants rapidly replace the lineages evolving from the initial infecting strains, with a median half-time of 27 days. This result is consistent with the two neonates being infected in utero with multiple T/Fs.
For the other two neonates (pair 9_1039n and pair 11_1580n), after removing all recombinants and their descendants, there were enough sequences left to discern lineages (Figure S3B) and infer infection times using our previous methods. Although we cannot discern with certainty whether these lineages are the original T/Fs or whether, more likely given the level of accumulated diversity in the original sample, they are in fact subclades that originated from subsequent bottlenecks, the timing of these sequences provides useful information on the least number of days since the MRCA. The minimum number of days since infection were 104 (95% confidence interval [CI] 84–123 days) for neonate pair 9_1039n after removing recombinants and selection epitopes and 53 (95% CI 29–76 days) for neonate pair 11_1580n based on the “oldest” lineage (see STAR Methods).
Taken together, these results show that the majority of in utero infections (67%) occur during the last 2 months of the pregnancy in this study.
High genetic diversity of viral populations during long-term in utero infection
To determine the genetic diversity accumulated during in utero infections, we calculated the genetic p distances for the viral population in each neonate and compared the diversity levels of viruses in the neonates to their cognate mothers. The within-lineage viral populations in eight neonates from whom the T/F viral sequences could be inferred were highly homogeneous. As expected, the average diversity of the within-lineage viral populations from each T/F virus was much lower in the neonates than in the mothers (0.12% and 1.56%, respectively; p = 9.8 × 10−4 by paired Wilcoxon test; Figure 2). The p diversity values in the short-term infections were calculated within lineages after removing all detected recombinants. However, the genetic diversity levels were much higher in viral populations of the four neonates from whom the T/F viral sequences could not be inferred, which is also to be expected given the high level of recombination detected in these samples. In these neonates, the viral population average diversity was 0.62% (0.27%–1.13%), compared to the 3-fold higher diversity observed in their cognate maternal viral populations (1.68%; 1.3%–2.39%). Although the diversity levels were similar among the maternal viruses from both groups (1.56% versus 1.68%), the diversity of the viral population in long-term infection neonates was 5 times higher than that in short-term infection neonates (p = 0.006 by 2-sided Wilcoxon test). In pair 12_2564, the average diversity (0.75%) in 2564n was less than 2-fold different of that (1.3%) in 2564m (Figure 2). Lineages in two of the long-term-infected neonates were no longer detectable, but, for the two neonates for which tentative lineages were identified after removing all possible recombinant sequences, the average p diversity was 0.38% and 0.35%, which was a little less than half of diversities when recombinant sequences were included for analysis but still 3-fold higher than the mean diversity in the neonates with clearly defined T/F sequences. This indicates that recombination played an important role in increasing diversity in these long-term-infected neonates.
Figure 2.
Comparison of genetic diversity of the viral populations of mother and neonate from each transmission pair
Genetic diversity was measured by calculating within-lineage pairwise genetic distances (p-distance) of neonate sequences at different time points and maternal sequences. P distances are plotted as black dots. The middle blue line indicates the median, the box shows 25–75 percentiles of the diversity, and whisker shows 10–90 percentiles of the diversity. Neonate versus maternal sequence diversity was statistically significant (p = 9.8 × 10−4; paired Wilcoxon test), and the diversity of sequences from the short-term-infected infants versus the long-term-infected infants are statistically significant (p = 0.006; paired Wilcoxon test). See also Figures S4 and S5.
One caveat with the higher diversity observed in the four long-term-infected neonates is that they were on average sampled at a later time (on average 43 days after birth) than the other eight neonates (on average 17 days after birth). The one exception is neonate pair 4_1348n, sampled at 40 days after birth, whose infection was clearly initiated by a single T/F virus. Although recombination is pervasive in all HIV infections, due to the homogeneity of the sequences, it takes longer for recombinants to be detectable in a single T/F infection.27 Therefore, it is likely that the long in utero infection time allows transmission of multiple T/F viruses to the fetuses and extensive viral recombination to take place, so that, by the time these neonates were sampled, recombinants had completely replaced the infecting viral populations.
Additionally, neonate pair 4_1348n, together with three long-term infected neonates (pair 9_1039n, pair 11_1580n, and pair 12_2564n), were all sampled a second time (Figure S4A), 1–3 months later (35 days later for pair 4_1348n; 26, 51, and 85 days later for pair 9_1039n, pair 11_1580n, and pair 12_2564n, respectively). All second time point sequences showed no significant increase in p diversity from the first time point (p = 0.9 by paired Wilcoxon test; Figure 2) and were phylogenetically indistinguishable from their respective first time point sets (Figure S4B). These results indicate that the viral genetic diversity generally remained similar in neonates during 1–3 months of infection after birth, demonstrating that the mutations accumulated after birth only marginally contributed to the high levels of genetic variation of viruses in those long-term in utero infections. This suggests that these viruses are under host-selection pressure while replicating in utero.
Transmission of multiple T/F viruses in long-term in utero infections
The amino acid sequences in the neonate viral populations present many unique divergent sequence motifs in the env genes. We then sought to investigate whether the sequence motifs observed in the neonates had originated from their cognate mothers or evolved de novo in fetuses. Three unique sequence motifs (A, B, and C) were detected in variable loop 1 (V1), while one major (D) and one minor (E) population, as well as some recombinants, were found in the V3/C4/V4 region in pair 10_2093n (Figure 3). Sequences with identical motif A and nearly identical motif B in the neonate were also found in the cognate maternal viruses (represented by 1 and 3 sequences, respectively). Motif C, represented by a single sequence in pair 10_2093n.5, was found in two of the mother’s viral sequences. In the V3/C3/V4 region, there were two viral populations (D and E) in the neonate (Figure 3). The minority motif E was found in three neonate sequences, but only one similar sequence was found among the maternal sequences (pair 10_2093m.20). Additionally, two more unique motifs were found in the neonate sequences (2093n.18 and 2093n.5a, highlighted in red and gray in Figure 3). Interestingly, a motif identical to the one found in pair 10_2093n.18 (highlighted in red in Figure 3) was also found in one maternal viral sequence (pair 10_2093m.26), and motifs similar to the ones in pair 10_2093n.5a (highlighted in gray) were detected in four maternal viral sequences (pair 10_2093m.5, 2093 m.31, 2093 m.38 s, and 2093 m.42). No sequences in the mother were found to be identical to the predominant D sequence in the neonate, although one maternal viral sequence (pair 10_2093m.3) was similar. Neonate sequences carrying motifs similar to the ones observed in the maternal viral population indicate that these viruses were most likely derived from the viruses transmitted from the cognate, rather than emerging de novo in the neonate.
Figure 3.
Identification of unique sequence motifs in the env gene from the mother-neonate pair 10_2093
The sequences from the neonate and mother transmission pair 10_2093n are compared to the consensus sequence of the viruses in neonate (the top line). The different amino acids are indicated as they are, while identical amino acids are shown as dashes. Distinct motif sequences in the viral population in the neonate (noted as A through E) are indicated by different colors, while the identical or similar motif sequences found in the maternal virus sequences are indicated with the same colors. See also Figure S3.
When the sequence motifs in both V1 and V3C3V4 regions are analyzed together, none of the sequence patterns in the neonate were found in the maternal viral sequences (Figure 3), indicating that they were recombinants as shown in Figure S3A. The recombination was highly significant with the A/D motif recombinants (p = 10 × 10−6), the C/D motif recombinants (p = 6.5 × 10−11), the B/D motif recombinants (p = 0.03), and the A/E motif recombinants (p = 0.007). Because they were not found in the maternal viruses, they were most likely generated in the neonate after transmission. For example, four recombinant sequences (1, 18, 5a, and 2a) all carry motif B in V1 and distinct motifs in the V3/C3/V4 region, and this pairing of the two motifs in the same sequence is not found in any of the maternal sequences (Figure 3). All but two neonate sequences (6 and 14) had pairings of unique motif patterns that were not present in the sampled maternal viral sequences (Figure 3). Because the maternal sample consisted of a total of 38 sequences, we are 90% confident that we sampled all circulating variants that were 10% or more prevalent in the viral population. These results are consistent with our previous findings that the majority of the viruses in the long-term-infected people are recombinants,27 as shown in Figure S3. It is most likely that, after the different viral variants were transmitted from the mother to the fetus, these viruses recombined among themselves and only parts of transmitted maternal viral genomes were detectable later in the neonate virus genomes. Similar results were also observed in pair 11_1580n (Figure S5). Taken together, the analysis of unique motif sequences of the neonate env genes in their cognate maternal viral populations shows that multiple viral variants are often transmitted to the fetuses during pregnancy, and frequent recombination further increases the levels of genetic complexity of the viral population in fetuses.
Host selection on viruses in fetuses
The high levels of genetic diversity in the viral populations in the four long-term-infected neonates (pair 9_1039n, pair 10_2093n, pair 11_1580n, and pair 12_2564n) prompted us to investigate whether specific loci within the env gene were under host selection pressure. We used the Synonymous Non-synonymous Analysis Program (SNAP) tool from the Los Alamos National Laboratory (LANL) database (https://www.hiv.lanl.gov/content/sequence/SNAP/SNAP.html) to determine the average pairwise cumulative codon behavior for synonymous (syn), nonsynonymous (non-syn), and insertions/deletion (indel) mutations across the entire env gene. For all four transmission pairs, non-syn plots were higher than syn ones in both the neonates and their cognate mothers (Figure 4). The average ratio of the rate of non-syn substitutions per non-syn site (dS/dN or ω) in neonate pair 9_1039n and pair 11_1580n were 64% and 50% higher than their cognate mothers, respectively. However, overall, there was no statistical difference in ω values between neonates and mothers in the four pairs (p = 0.38 by paired Wilcoxon test). Similar analyses of viral populations in the other eight short-term infected neonates showed similar accumulation of syn and non-syn mutations across the entire env gene in all but neonate pair 4_1348n, who was the oldest neonate (40 days) and had been infected for the longest time (61 days) among the eight short-term-infected neonates (Figure S6).
Figure 4.
Identification of selection sites by analyzing the accumulation of mutations across the env gene in the neonate viruses
Cumulative plots of each codon average behavior for all sequences pairwise compared for the neonate and maternal viruses for synonymous (blue), non-synonymous (red) mutations, and indels (gray). Sites found under statistically significant diversifying selection by the MEME analysis are marked by arrows, where solid arrows indicate p < 0.01 and dashed arrows p = 0.01. Magenta arrows show sites under diversifying selection in both the mother and the neonate. Values of ω denote average ratios of the rate of non-synonymous substitutions per non-synonymous site (dN/dS) for each sample. See also Figure S6.
In order to identify sites under positive diversifying selection, we analyzed the env sequences from the four long-term-infected mother-neonate pairs using the online Mixed Effects Model of Evolution (MEME) tool from the datamonkey server.29 With a significance threshold of p = 0.01 or lower, MEME identified a total of 23 sites significantly under diversifying selection among the four neonates and 22 among the four mothers, with only two sites overlapping between mother and neonate in two transmission pairs at position 281 (based on the position in HXB2) in pair 2093 and position 304 in pair 11_1580 (Figure 4). Notably, both sites have been found in previous studies to be contact sites for broadly neutralizing antibodies (bnAbs) that target CD4bs30 and V3-glycan, respectively.31,32 Positively selected sites were overwhelmingly concentrated in gp120 (20 out of 23 in the neonates and 17 out of 22 in the mothers) and, in particular, in the V3/C3/V4 region (14 out of 42 unique sites). Site 350 in particular, located in C3, was significant in two mothers (pair 11_2093m and pair 10_1580m; Figure 4).
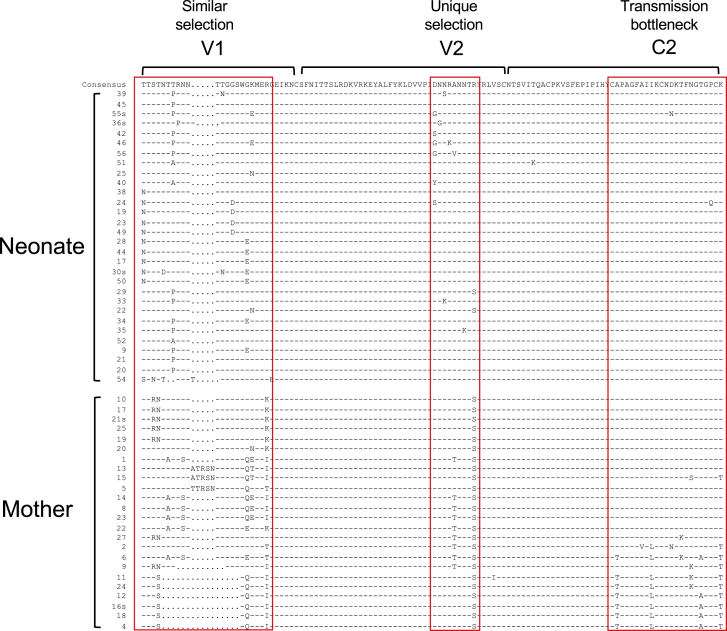
Examination of clustered mutations in the V1V2C2 region in neonate pair 9_1039n (the first consecutive two vertical steep increases of non-syn mutations) showed that different regions were under disparate selection (Figure 5). The C2 sequences were highly homogeneous in pair 9_1039n and similar to half of those sequences in the mother. However, nearly half of the viral population in pair 9_1039m had variable C2 sequences. This suggests that the viral sequences in the region in pair 9_1039n were a result of bottleneck selection of the complex maternal viral population. In contrast, the V2 sequences were very homogeneous in 1039 m, except that one-third of the sequences had one A186T mutation, but about one-third of the sequences had various distinct mutations in the neonate. This suggests that this region was under positive selection in the neonate, but not in the cognate mother. The V1 sequences were highly variable in both neonate and mother. However, the sequences in the neonate were different from those in the mother in this region (Figure 5). Phylogenetic tree analysis confirmed that the neonate and maternal sequences were distinct from each other (Figure S7). Only one maternal virus sequence (1039 m.20) clustered with two neonate sequences (1039n.25 and 1039n.22). They differed from each other by one amino acid. These results indicate that V1 was under selection pressure in both neonate and mother, but the selection was different in the neonate and the mother. As noted before for other maternal samples, one caveat of this analysis is that, with a sample size of 24 maternal sequences, we are 80% confident that we have sampled all variants that are at least 10% prevalent in the viral population and that we may have missed variants closer to the neonate viruses if they happened to be less than 10% prevalent in the maternal circulating viral population.
Figure 5.
Selection signatures in env sequences from mother-neonate pair 9_1039
The V1V2C2 region sequences from the neonate and mother transmission pair 9_1039n are compared to the consensus sequence of the viruses in neonate (the top line). The different amino acids are indicated as they are, while identical amino acids are shown as dashes. Distinct selection signatures at different sites in the neonate and mother viruses are indicated by red boxes; similar selection in V1 for both neonate and mother viruses, unique selection in V2 for the neonate viruses, and transmission bottleneck (or purifying selection) in C2 for neonate viruses are shown. See also Figure S7 and Table S1.
Evidence of similar selection pressure at the same region of the env gene in both neonates and mothers was also found for pair 10_2093 and pair 12_2564. Divergent viral populations in both neonates and mothers were observed in the cytoplasmic domain of gp41 for the pair 10_2093 (Figure S8A) and in V5C5 for pair 12_2564 (Figure S8B). New selection sites in the neonate viral population were also detected in the V3 sequences in pair 12_2564n (Figure S8B) and in the gp41 sequences in pair 11_1580n (Figure S8C). In contrast, the viral populations were highly homogeneous at C3 in pair 12_2564n (Figure S8B) and V1 in pair 11_1580n (Figure S8C), where the viruses were highly heterogeneous in their cognate mothers. Taken together, detection of selection sites unique for the viruses in all four long-term in utero infected neonates indicates that viruses in fetuses are under host-specific selection pressure that is different from those in their cognate mothers.
Nine strong selection regions (Figures 5 and S8) were identified among the four long-term in utero infected cases. Five of them were found in variable regions that are often targeted by neutralizing antibodies,33,34 indicating that selection pressure might be due to neutralizing antibodies (Table S1). The other four sites were found in the more conserved gp41 regions or the signal peptide, where many T cell epitopes have been identified, as reported in the HIV Immunology Database (https://www.hiv.lanl.gov/content/immunology), suggesting that these sites might have been targeted by T cell immune responses in fetuses (Table S1). These mutations were not random but rather concentrated in regions usually targeted by neutralizing antibodies or T cell responses, suggesting that these sites were most likely selected by host immune responses in the fetuses.
Because no viable cells were available from fetuses or neonates from birth, we could not determine whether these sites are targeted by the T cells in the neonates. However, some residual plasma samples were available from both the neonate and the mother form pair 11_1580 for us to determine autologous neutralization activity. We chemically synthesized two env genes that represent two sub-clusters in the neonate viral population and one representative env gene in the maternal viral population and generated Env pseudovirus for each env clone. Both neonate pseudoviruses and the maternal pseudovirus were not or weakly neutralized by the neonate or maternal plasmas (Table S2)3, indicating that the neonate and maternal viruses are resistant to the plasma from the neonate or the mother.
Higher neutralization activity in placenta plasma than systemic blood from the same HIV-1-infected mothers
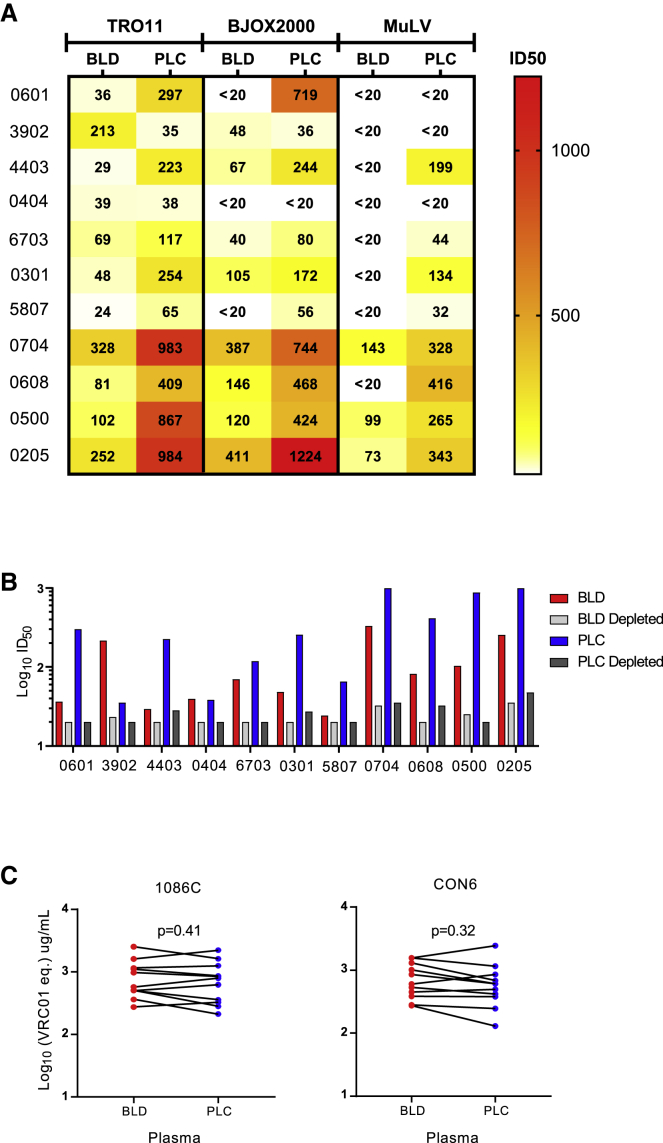
To determine whether viruses in the fetus may be exposed to differential neutralizing antibody pressure than those found in maternal plasma, we next sought to determine whether neutralizing activities were different in plasma samples from systemic blood compared to placental blood from an additional group of chronically infected pregnant women. The plasma samples were collected from the placenta and peripheral blood of HIV-1-infected pregnant women. Interestingly, placental plasma neutralized tier-2 pseudoviruses TRO11 and BJOX2000 at higher titers than paired systemic blood plasma (Figure 6A), although placental plasma also showed higher non-specific antiviral properties against the pseudovirus generated with Env glycoprotein from murine leukemia virus (MuLV) than systemic blood plasma from the same mothers, indicating more non-specific neutralizing activity in placental plasma. However, neutralization titers of placental plasma were 2.97- to 11.98-fold higher than those of the systemic blood plasma from the same mothers. To determine whether the neutralization activity was mediated by IgG, we depleted IgG from placental or systemic blood plasma samples and then determined their neutralization activity against TRO11. After IgG depletion, the neutralization activities were reduced to undetectable levels in over half of systemic and placental plasma samples (54%) or significantly reduced by 8.26- to 28.09-fold for the remaining samples (Figure 6B), suggesting that IgG was responsible for the high levels of neutralization activity in placenta plasma. After IgG depleting, the non-specific neutralization activity against MuLV was also dramatically reduced in both systemic and placental plasma samples (Figure S9). To determine whether the high titers of neutralization activities in placental plasma were due to high levels of HIV-1 Env-specific IgG, we compared binding Ab titers to two different HIV-1 gp120s (1086C and CON6) with paired systemic and placental plasma samples from the same mothers. The IgG titers to both 1086C and CON6 Env were similar between placental and systemic blood plasma from the same mothers (Figure 6C). These results show that the IgG was responsible for the higher HIV-1 neutralization activity in the placental plasma compared to peripheral blood plasma from the mothers, indicating the potential for selective transfer of potently neutralizing IgG antibodies across the placenta,12 and then contributes to selective pressure on fetal HIV-1 variants.
Figure 6.
Higher neutralization activity of placental plasma than blood plasma from the same mothers
(A) Heatmap analysis of neutralizing titers of placental (PLC) plasma than systemic blood (BLD) plasma from the same mothers. The neutralization titers are color coded from dark to light, where the darker color indicates higher neutralization titers.
(B) Loss of neutralization activity after IgG depletion by protein G column in both placental plasma and systemic blood plasma. The dotted line indicates the threshold for detection.
(C) Similar IgG concentrations in placental plasma and blood plasma from the same mothers. HIV-1-specific IgG concentrations were determined by measuring binding of blood and placental plasma to 1086C and CON6 gp120, respectively.
See also Figure S9.
Discussion
The placenta is a physical barrier that protects fetuses from infections by pathogens,23 but HIV-1 still can pass through the placenta and infect fetuses in 5%–10% of untreated HIV-1 pregnant women.2 However, the time during pregnancy at which HIV-1 is more likely to cross the placenta and infect fetuses is not well understood. By analyzing the viral population using the SGA method, we found that T/F viruses could be well defined in eight out of 12 in utero infected neonates. The median of days of infection was 18 (3–37) days before birth. The majority (67%) of in utero infections happened within the last 40 days before delivery during pregnancy in this cohort where the timing of infection could be discerned. Because the sample size in this study is small (12 pairs), additional samples are warranted to further confirm this finding. The placental barrier changes dynamically during pregnancy. In the first trimester, it consists of the syncytiotrophoblast, the cytotrophoblast, the villus mesenchyma, and the fetal capillary walls. During the last trimester, the thickness of the barrier significantly decreases as the cytotrophoblast disappears and fetal vessels get closer to the villus surface. The exchange zones of the barrier consist of only membranes of syncytiotrophoblast and adjacent capillaries walls.35, 36, 37 Conventional T lymphocytes can be detected in fetal spleen and lymph nodes only by 14 weeks of gestation.23 Thus, HIV-1 infection may be less likely to occur in fetuses during early pregnancy, when T cells have not developed yet. Although it is unclear how HIV-1 viruses cross the placenta, the risk of transmission is increased during the third trimester, when the placental barrier becomes thinner. This is consistent with our finding that most of in utero MTCTs occur during the late stage of pregnancy.
The genetic diversity levels of the viral populations in neonates with long-term in utero infection were high, only about 3-fold lower than those in their cognate mothers, while the difference was as high as 13-fold lower when the genetic diversity in the other eight neonates infected only for a short time were compared to their mothers. The diversity of the viruses in neonate pair 12_2564n was only slightly lower than that of the cognate maternal viruses. Importantly, the neonate viruses formed independent lineages that are separate from their cognate maternal viruses, strongly suggesting that the neonate viruses have different evolutionary pathways from the viruses in their cognate mothers during in utero infection. Sequence analysis showed that many env gene regions were highly homogeneous in four long-term in utero infected neonates while the sequences were highly divergent at the same region in their cognate mothers, suggesting a transmission bottleneck. However, neonate viral sequences were divergent at some regions of the env gene where the cognate maternal sequences were homogeneous or similarly divergent but with different sequences. These results indicate that the viruses are under different selection pressures in the fetuses compared to the maternal viruses.
Although fetuses are generally thought to reside in a mostly sterile condition, they can be exposed to a wide range of immuno-stimulatory molecules, such as ingested amniotic-fluid substances,38,39 semi-allogenic antigens from maternal cells,40,41 microbes,42 and food antigens.43 Innate immune responses and antigen-specific T cell responses to pathogens are detectable in fetuses, suggesting that the exposure to maternal infection in utero may prime the developing immune system.18 Aberrant fetal immune responses to maternal antigens are found to play an important role in the pathogenesis of preterm labor.19, 20, 21 A recent study showed that, as early as the second trimester, fetal APC subset, such as conventional DC, can respond to TLRs.22 However, when compared to neonate immune responses, fetus immune responses mediated by macrophages are more geared toward immune tolerance. Precursors of mature B lymphocytes (pre-B cells) are detected in the fetal bone marrow by 13 weeks of gestation,23 although the diversity of the immunoglobulin repertoire is limited during the initial stages of B cell development but appears to be similar to that of adults by the third trimester of gestation.44 However, the role of antibody responses in preventing in utero HIV-1 infections is still unclear.23,45
HIV-1-specific CD8+ T cell activity is detectable from birth in up to 70% of in utero infected neonates,46 suggesting that T cell responses may exert selection pressure on HIV-1. Fetal exposure to HIV-1 primes the immune system to enhanced immune activation and alters T cell homing in fetuses.24 Although these studies suggest immune responses in fetuses, no data show that adaptive immune responses play a role in shaping pathogen evolution. In this study, we now show that more than half of the selected sites in the env gene in fetuses were found in variable regions targeted by neutralizing antibodies,33,34 indicating selection pressure possibly triggered by neutralizing antibodies. The other half of the sites were found in the conserved gp41 regions or in the signal peptide, both of which are frequently targeted by T cell responses (https://www.hiv.lanl.gov/content/immunology). Because there were no cells or sufficient amount of plasma samples available from these studied subjects, we could not determine histocompatibility leukocyte antigen (HLA) alleles in these transmission pairs or whether those sites in the env gene were selected by T cell responses. However, the detection of different amino acids in the neonate viruses at the previously well-defined T cell epitopes compared to the cognate maternal viruses indicates that T cell responses may also play a role in selecting these mutations. Taken together, these data suggest that host selection pressure has played an important role in selecting escape mutations and shaping the viral evolutionary pathway in fetuses.
The immune responses in fetuses are difficult to study, because it is nearly impossible to obtain adequate blood samples from fetuses during pregnancy. In addition, maternal antibodies and half of HLA alleles are transferred from the mother to the fetus, making it a challenge to determine whether the immune responses are from the infected fetuses or their cognate mothers. However, the detection of stronger IgG-mediated neutralization activity from placental plasma compared to the systemic plasma in the same mothers at birth indicates that placental selection of antibodies for transfer to the neonates may play a role in blocking MTCT transmission and driving these mutations in variable loops in the env gene. It is also plausible that paternal HLA alleles may contribute to the selection of mutations at those well-defined T cell epitope sites, which are not likely to be associated with neutralizing antibodies. The presence of distinct mutation clusters in the env gene of neonate viruses compared to the same sites in cognate mother viruses clearly shows that the selection at these sites is due to unique immune response in fetuses.
In summary, we performed detailed genetic analyses of HIV-1 env gene sequences from perinatal transmission pairs and found that most in utero transmissions occur during the last 2 months of pregnancy. Moreover, host selection can be detected during long-term in utero infection of the fetuses. Knowing the timing of in utero infection during pregnancy will be important for the development of more effective approaches to prevent perinatal MTCT. The identification of strong selection sites potentially mediated by neutralizing Abs and T cell immune responses indicates that the immune system in fetuses is capable of exerting selection pressure on viral evolution during in utero infection. A similar approach can also be applied to other viral infections. Studying the selection and evolution of HIV-1 and other viruses in fetuses can be an ideal alternative approach to study the development of adaptive immunity in fetuses.
Limitations of the study
Potential limitations of this study are that we may have missed rare (<10% frequency) variants in the maternal or infant virus populations due to sequence sampling. Additionally, the immune responses in fetuses are challenging to study because of the difficulty to obtain adequate blood samples from fetuses during pregnancy.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Horseradish peroxidase (HRP)-conjugated goat anti-human IgG | Jackson ImmunoResearcsh | Cat#109-035-008; RRID:AB_90720 |
| CH103 | (Liao et al., 2013) | RRID:AB_2491065 |
| VRC01 IgG | NIH-ARP | Cat#12033; RRID:AB_2491019 |
| Virus strains | ||
| TR011 | (Decamp et al., 2014) | N/A |
| SF162 | (Decamp et al., 2014) | N/A |
| MulV | (Decamp et al., 2014) | N/A |
| BJOX2000 | (Decamp et al., 2014) | N/A |
| SVA.MulV | (Decamp et al., 2014) | N/A |
| Biological samples | ||
| Human: Plasma | (Moody et al.47; Permar et al.8) | N/A |
| Human: Peripheral blood mononuclear cells | (Moody et al.47; Permar et al.8) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Platinum Taq DNA polymerase High Fidelity | Invitrogen | Cat#11304102 |
| Tween 20 | Sigma Aldrich | Cat#P2287 |
| Dulbecco’s modified Eagle’s medium (DMEM) | GIBCO | Cat#11995-065 |
| Fetal Bovine Sera (FBS) | Sigma Aldrich | Cat#12106C |
| 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) | Sigma Aldrich | Cat# 83264-100ML-F |
| Gentamycin | Sigma Aldrich | Cat#G1397 |
| DEAE | Sigma Aldrich | Cat# D9885 |
| SureBlue reserve TMB substrate | KPL | Cat#95059-294 |
| FuGene 6 | Promega | Cat#E2691 |
| Bright-Glo | Promega | Cat#E2610 |
| 1086Cgp120 | NIH-ARP | Cat#12582 |
| CON6 gp120 | (Gao et al., 2005) | N/A |
| Critical commercial assays | ||
| PureLink Viral RNA/DNA mini kit | Invitrogen | Cat#12280-050 |
| SuperScript III reverse transcription mix | Invitrogen | Cat#18080085 |
| Deposited data | ||
| HIV-1 sequences | This paper | GenBank: MT853116 to MT853181 and MT861212 -MT861991 |
| Experimental models: cell lines | ||
| Human TZM-bl cells | NIH-ARP | Cat#8129 |
| Human HEK293T cells | ATCC | Cat#CRL-3216 |
| Oligonucleotides | ||
| cDNA antisense primer R3.B3R: ACTACTTGAAGCAC TCAAGGCAAGCTTTATTG |
IDT | N/A |
| PCR primer 07For7: AAATTAYAAAAATTCAAAATTTT CGGGTTTATTACAG |
IDT | N/A |
| PCR primer 2.R3.B6R: TGAAGCACTCAAGGCAAGCT TTATTGAGGC |
IDT | N/A |
| PCR primer VIF1: GGGTTTATTACAGGGACAGCAGAG | IDT | N/A |
| PCR primer Low2c: TGAGGCTTAAGCAGTGGGTTCC | IDT | N/A |
| Recombinant DNA | ||
| pcDNA3.1(+)-1580.env.cons1 | Thermo Fisher/This paper | N/A |
| pcDNA3.1(+)-1580.env.cons2 | Thermo Fisher/This paper | N/A |
| pcDNA3.1(+)-1580.env.M2 | Thermo Fisher/This paper | N/A |
| Software and algorithms | ||
| Sequencher 4.5 | Gene Codes | https://www.genecodes.com/ |
| MEGA7 | MEGA | https://www.megasoftware.net/ |
| Gene Cutter tool | HIV database | https://www.hiv.lanl.gov/content/sequence/GENE_CUTTER/cutter.html |
| Highlighter tool | (Keele et al.48) | https://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html |
| Hypermut tool | (Rose and Korber, 2000) | https://www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html |
| RAPR tool | (Song et al.27) | https://www.hiv.lanl.gov/content/sequence/RAP2017/rap.html |
| Poisson-Fitter tool | (Giorgi et al.25) | http://www.hiv.lanl.gov/content/sequence/POISSONFITTER/pfitter.html |
| SNAP | (Korber, 2000) | https://www.hiv.lanl.gov/content/sequence/SNAP/SNAP.html |
| MEME | (Murrell et al.29) | https://www.datamonkey.org/ |
| R | (R Development Core Team49) | http://www.R-project.org |
| Other | ||
| ABI3730xl genetic analyzer | Applied Biosystems | https://www.thermofisher.com/us/en/home.html |
| Spin-X centrifuge tube filters | Corning | Cat#29442-752 |
| 96-well black solid plate | PerkinElmer | Cat#6002270 |
| Protein G High Performance MultiTrap 96-well plate | GE Healthcare | Cat#28903135 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Feng Gao (fgao@duke.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate new datasets or code.
Experimental model and subject details
Sample collection
Plasma samples were collected from 10 pairs of mothers and neonates from the Women Infant Transmission Study (WITS) cohort8,10 and 2 pairs from the Center for HIV/AIDS Vaccine Immunology protocol (CHAVI009) cohort47. All neonates were diagnosed as in utero infection by positive PCR at birth using genomic DNA extracted from peripheral blood mononuclear cells from the newborn babies. Plasma samples were collected from neonates between delivery and 54 days after birth and from mothers at delivery. Samples from two different time points were collected from four neonates (pair 9_1039n, pair 12_2564n, pair 4_1348n and pair 11_1580n). Two mothers (pair 5_9112m and pair 8_3915m) from the CHAVI009 cohort received 1 dose of nevirapine at delivery. No other individual was on antiretroviral therapy. No neonates had complications at birth and no gender information of the neonates is available.
Placental plasma was obtained as described previously from the CHAVI009 cohort50. Briefly, an incision was made at the cleaned maternal surface of the placenta, and blood from the incision was aspirated into tubes containing EDTA (Ethylenediaminetetraacetic acid), sodium citrate and heparin. The samples were then separated and the plasma was stored at −80°C.
Ethics statement
All samples in the study used were received from the existing cohorts WITS and CHAVI09, which are deidentified and deemed as research not involving human subjects by Duke University Institutional Review Board. The protocol number is Pro00016627.
Cell lines
Human HEK293T and human TZM-bl cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) medium (GIBCO, Thermo Fisher Scientific, MD) supplemented with 10%FBS (Sigma Aldrich, St. Louis, MO) containing HEPES (Sigma Aldrich, St. Louis, MO) and Gentamycin (Sigma Aldrich, St. Louis, MO) at 37°C.
Method details
Analysis of the env gene sequences by single genome amplification
Viral RNA was extracted from plasma samples using PureLink Viral RNA/DNA mini kit (Invitrogen, Carlsbad, CA) and used to generate complementary DNA (cDNA) using SuperScript III reverse transcription mix (Invitrogen) and antisense primer R3.B3R (5′-ACTACTTGAAGCACTCAAGGCAAGCTTTATTG-3′; according to nucleotide [nt] position 9611-9642 in HXB2) for the 3′-half genome. The synthesized cDNA was subjected to single genome amplification (SGA) analysis using Platinum Taq DNA polymerase High Fidelity (Invitrogen)17. The first round PCR was performed using the primers 07For7 (5′-AAATTAYAAAAATTCAAAATTTTCGGGTTTATTACAG-3′; nt 4875-4912) and 2.R3.B6R (5′-TGAAGCACTCAAGGCAAGCTTTATTGAGGC-3′; nt 9636–9607), and second round PCR was carried out using 2 μL of the first round product with primers VIF1 5′-GGGTTTATTACAGGGACAGCAGAG-3′; nt 4900–4923) and Low2c 5′-TGAGGCTTAAGCAGTGGGTTCC-3′; nt 9612-9591). The first round PCR conditions were as follows: one cycle at 94°C for 2 min; 35 cycles of a denaturing step at 94°C for 15 s, an annealing step at 58°C for 30 s, an extension step at 68°C for 4 min; and one cycle of an additional extension at 68°C for 10 min. The second round PCR conditions were the same as for the first round PCR, except that 45 thermocycling cycles were used. The final PCR products were visualized by agarose gel electrophoresis, purified and sequenced for the entire env gene using ABI3730xl genetic analyzer (Applied Biosystems, Foster City, CA).
Sequence analysis
Each SGA amplicon was sequenced using the primer walking method and all sequences were assembled and edited using Sequencher 4.5 (Gene Codes, Ann Arbor MI). The final assembled sequences from each neonate and mother transmission pair were aligned using the Gene Cutter tool (https://www.hiv.lanl.gov/content/sequence/GENE_CUTTER/cutter.html), and the alignments were optimized manually using Seaview51. The Highlighter plots were generated using the Highlighter tool (https://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html). APOBEC-mediated enrichment of G-to-A substitutions was screened using the Hypermut tool (https://www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html) and sequences with significant hypermutation (p < 0.1) were excluded from further analysis. Recombinants were detected using the RAPR tool (https://www.hiv.lanl.gov/content/sequence/RAP2017/rap.html). Recombinants and their putative descendants were excluded from the timing analysis. Neighbor-joining trees were constructed using the Kimura 2-parameter model with 1000 bootstrap replications. Phylogenetic trees and highlighter plots were used to infer the neonate T/F env gene sequences as previously described48. Infection time was estimated using the Poisson Fitter tool (https://www.hiv.lanl.gov/content/sequence/POISSON_FITTER/pfitter.html) after excluding all hypermutated and recombinant sequences25. Time estimates for neonates with multiple infecting strains were obtained on each individual lineage and then averaged using a harmonic mean. For two long-term infected neonates (pair 9_1039n and pair 11_1580n) we obtained a lower boundary on the number of days since the most recent common ancestor (MRCA) by separating into sublineages for the remaining viral population after removing all detected recombinants. In neonate pair 9_1039n, the remaining population formed a single lineage that showed evidence of non-random accumulation of mutations at three sites in particular, 404, 421, and 566. Therefore, in order to infer the time since the selection bottleneck, those sites were masked for the Poisson Fitter analysis. Neonate pair 11_1580n, on the other hand, did not show any evidence of selection sites after removing all recombinants. The remaining viral population could be divided into three sub lineages, of which two had originated through recombination events identified by the RAPR analysis.
Within-lineage genetic pairwise p-distances were calculated using sequence data from each sample, after removing recombinants and hypermutated sequences, and compared between neonate and maternal viral populations. Statistical tests were conducted using the Wilcoxon test function in R. For the paired comparison between maternal p-distances and neonate p-distances, values for neonates with either multiple T/Fs or multiple time points, were first averaged within time point and/or lineage and then averaged overall in order to compare to the maternal mean p-distance. The accumulation of synonymous and non-synonymous mutations across the entire env gene was determined by pairwise comparison of all the env sequences in neonates or mothers according to the method developed by Nei and Gojobori52 using SNAP (https://www.hiv.lanl.gov/content/sequence/SNAP/SNAP.html). The accession numbers for the sequences from this study are GenBank: MT853116 to MT853181 and MT861212 -MT861991.
Generation of Env-pseudoviruses
Two env genes that represent the consensus sequences (neonate 1 and 2) of two sub-clusters in the viral population in neonate pair 11_1580n and one env gene (M.2) that represents one major cluster viruses in mother pair 11_1580m were codon optimized and chemically synthesized (Thermo Fisher, Waltham, MA, USA). The env genes were cloned into pcDNA3.1(+) at the Hind III and EcoR I sites. The selection of MTCT pair 11_1580 is because it is the only pair from which the plasma samples from both the neonate and the mother were available for autologous neutralization analysis. The env gene clones together with the env-deficient HIV-1 plasmid DNA (SG3Δenv) were co-transfected into HEK293T cells (ATCC, Manassas, VA, USA) using the FuGene 6 transfection reagent (Promega, Madison, WI, USA). Two days after transfection, the culture supernatant containing pseudoviruses was harvested, aliquoted, and stored at −80°C.
Neutralization assay
Neutralization activities of the plasma samples were determined by the single-round infection of HIV-1 Env-pseudoviruses in TZM-bl cells as described previously53. Briefly, plasma samples were first heat-inactivated by incubating at 56°C for 60 min. After the 1:3 serial diluted plasma samples were incubated with Env-pseudoviruses at 37°C for 1 hour, the mixtures were used to infect TZM-bl cells. Plates were read after incubating at 37°C for 48 hours. The 50% inhibitory dose (ID50) was defined as plasma reciprocal dilution at which relative luminescence units (RLU) were reduced by 50% compared with RLU in virus control wells after subtraction of background RLU in cell control wells. A response was considered positive if the neutralization titer was higher than 1:30.
Detection of HIV-1 specific antibodies by ELISA
HIV-1 specific binding antibody titers were performed by coating high-binding 384 well plates (Corning, Corning, NY) overnight at 4°C with 1086C and Con6 gp120s. Plates were washed once and blocked for 1 hour at room temperature (RT) with SuperBlock (4% whey protein, 15% goat serum, and 0.5% Tween 20 diluted in 1X phosphate-buffered saline [PBS]). Plates were washed and 10 μl of plasma in 3-fold serial dilutions were added in duplicate and incubated at room temperature for 1 hour. Plates were washed two times and horseradish peroxidase (HRP)-conjugated goat anti-human IgG antibody (Sigma Aldrich, St. Louis, MO) was used at a 1:5,000 dilution and incubated at room temperature for 1 hour. Plates were washed four times and SureBlue reserve TMB substrate (KPL, Gaithersburg, MD) was added. Reactions were stopped by stop solution (KPL, Gaithersburg, MD) and optical densities (OD) were measured at 450nm. Concentrations were calculated using a VRC01 IgG standard curve (at a 2-fold series from 0.0005 to 1 μg/mL).
Depletion of IgG in plasma
IgG depletion was performed by diluting systemic blood and placenta plasma by 2-fold with Tris-buffered saline (TBS) and filtering in Spin-X centrifuge tube filters (Corning). Diluted and filtered samples were then added to a Protein G High Performance MultiTrap 96-well plate (GE Healthcare, Chicago, IL) and shaken at 1,100 RPM for one hour at room temperature. IgG-depleted plasma samples were obtained by centrifugation at 700 g for 3 minutes onto a receiving plate.
Quantification and statistical analysis
Sites under significant diversifying selection pressure were identified using the MEME tool, available on the datamonkey server29. All statistical comparisons of p-distances and the rate of nonsynonymous substitutions per nonsynonymous site (dN/dS) were conducted in R49.
Acknowledgments
We thank Bette Korber for the helpful and insightful discussions. We would like to acknowledge the support of Pediatric HIV/AIDS Cohort Study (PHACS) team for their management of the Women and Infant Transmission study (WITS) cohort samples (U01 HD052102-04 and U01 HD052104-01), the CHAVI009 team, and all study participants. David R. Martinez was supported by an American Society for Microbiology Robert D. Watkins Graduate Research Fellowship, a Burroughs Wellcome Graduate Diversity Fellowship, and an NIH National Institute of Allergy and Infectious Diseases (NIAID) Ruth L. Kirschstein National Research Service Award F31 F31AI127303. This work was supported by a grant (1R01AI22909) from National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID). Elena E. Giorgi was also supported in part by the Laboratory Directed Research and Development program of Los Alamos National Laboratory under project number 20200554ECR.
Author contributions
Conceptualization, F.G. and S.R.P.; data collection, M.H.M., M.M., A.K., J.J.T., D.R.M., and X.L.; data analysis, M.H.M., M.M., A.K., E.E.G., J.J.T., E.O.R.-S., L.F., S.R.P., and F.G.; writing – original draft, M.H.M. and F.G.; writing – review & editing, M.H.M., M.M., A.K., J.J.T., D.R.M., E.E.G., L.F., S.R.P., and F.G.; funding acquisition, S.R.P. and F.G.
Declaration of interests
The authors declare no competing interests.
Published: June 16, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100315.
Supplemental information
References
- 1.UNAIDS Global HIV & AIDS statistics — 2020 fact sheet. https://www.unaids.org/en/resources/fact-sheet.
- 2.Wolinsky S.M., Wike C.M., Korber B.T., Hutto C., Parks W.P., Rosenblum L.L., Kunstman K.J., Furtado M.R., Muñoz J.L. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science. 1992;255:1134–1137. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- 3.Mahy M., Stover J., Kiragu K., Hayashi C., Akwara P., Luo C., Stanecki K., Ekpini R., Shaffer N. What will it take to achieve virtual elimination of mother-to-child transmission of HIV? An assessment of current progress and future needs. Sex. Transm. Infect. 2010;86(Suppl 2):ii48–ii55. doi: 10.1136/sti.2010.045989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorenbaum A., Cunningham C.K., Gelber R.D., Culnane M., Mofenson L., Britto P., Rekacewicz C., Newell M.L., Delfraissy J.F., Cunningham-Schrader B., International PACTG 316 Team Two-dose intrapartum/newborn nevirapine and standard antiretroviral therapy to reduce perinatal HIV transmission: a randomized trial. JAMA. 2002;288:189–198. doi: 10.1001/jama.288.2.189. [DOI] [PubMed] [Google Scholar]
- 5.Bailey H., Zash R., Rasi V., Thorne C. HIV treatment in pregnancy. Lancet HIV. 2018;5:e457–e467. doi: 10.1016/S2352-3018(18)30059-6. [DOI] [PubMed] [Google Scholar]
- 6.Kellerman S., Essajee S. HIV testing for children in resource-limited settings: what are we waiting for? PLoS Med. 2010;7:e1000285. doi: 10.1371/journal.pmed.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang J., Nour N.M. HIV and pregnancy in resource-poor settings. Rev. Obstet. Gynecol. 2010;3:66–71. [PMC free article] [PubMed] [Google Scholar]
- 8.Permar S.R., Fong Y., Vandergrift N., Fouda G.G., Gilbert P., Parks R., Jaeger F.H., Pollara J., Martelli A., Liebl B.E. Maternal HIV-1 envelope-specific antibody responses and reduced risk of perinatal transmission. J. Clin. Invest. 2015;125:2702–2706. doi: 10.1172/JCI81593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fouda G.G., Martinez D.R., Swamy G.K., Permar S.R. The impact of IgG transplacental transfer on early life immunity. Immunohorizons. 2018;2:14–25. doi: 10.4049/immunohorizons.1700057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez D.R., Vandergrift N., Douglas A.O., McGuire E., Bainbridge J., Nicely N.I., Montefiori D.C., Tomaras G.D., Fouda G.G., Permar S.R. Maternal binding and neutralizing IgG responses targeting the C-terminal region of the V3 loop are predictive of reduced peripartum HIV-1 transmission risk. J. Virol. 2017;91:e02422-16. doi: 10.1128/JVI.02422-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albrecht M., Arck P.C. Vertically transferred immunity in neonates: mothers, mechanisms and mediators. Front. Immunol. 2020;11:555. doi: 10.3389/fimmu.2020.00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez D.R., Fong Y., Li S.H., Yang F., Jennewein M.F., Weiner J.A., Harrell E.A., Mangold J.F., Goswami R., Seage G.R., 3rd Fc characteristics mediate selective placental transfer of IgG in HIV-infected women. Cell. 2019;178:190–201.e11. doi: 10.1016/j.cell.2019.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilcox C.R., Holder B., Jones C.E. Factors affecting the FcRn-mediated transplacental transfer of antibodies and implications for vaccination in pregnancy. Front. Immunol. 2017;8:1294. doi: 10.3389/fimmu.2017.01294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmeira P., Quinello C., Silveira-Lessa A.L., Zago C.A., Carneiro-Sampaio M. IgG placental transfer in healthy and pathological pregnancies. Clin. Dev. Immunol. 2012;2012:985646. doi: 10.1155/2012/985646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez D.R., Tu J.J., Kumar A., Mangold J.F., Mangan R.J., Goswami R., Giorgi E.E., Chen J., Mengual M., Douglas A.O. Maternal broadly neutralizing antibodies can select for neutralization-resistant, infant-transmitted/founder HIV variants. MBio. 2020;11:e00176-20. doi: 10.1128/mBio.00176-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moody M.A., Gao F., Gurley T.C., Amos J.D., Kumar A., Hora B., Marshall D.J., Whitesides J.F., Xia S.M., Parks R. Strain-specific V3 and CD4 binding site autologous HIV-1 neutralizing antibodies select neutralization-resistant viruses. Cell Host Microbe. 2015;18:354–362. doi: 10.1016/j.chom.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar A., Smith C.E.P., Giorgi E.E., Eudailey J., Martinez D.R., Yusim K., Douglas A.O., Stamper L., McGuire E., LaBranche C.C. Infant transmitted/founder HIV-1 viruses from peripartum transmission are neutralization resistant to paired maternal plasma. PLoS Pathog. 2018;14:e1006944. doi: 10.1371/journal.ppat.1006944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilcox C.R., Jones C.E. Beyond passive immunity: is there priming of the fetal immune system following vaccination in pregnancy and what are the potential clinical implications? Front. Immunol. 2018;9:1548. doi: 10.3389/fimmu.2018.01548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frascoli M., Coniglio L., Witt R., Jeanty C., Fleck-Derderian S., Myers D.E., Lee T.H., Keating S., Busch M.P., Norris P.J. Alloreactive fetal T cells promote uterine contractility in preterm labor via IFN-γ and TNF-α. Sci. Transl. Med. 2018;10:eaan2263. doi: 10.1126/scitranslmed.aan2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agrawal V., Hirsch E. Intrauterine infection and preterm labor. Semin. Fetal Neonatal Med. 2012;17:12–19. doi: 10.1016/j.siny.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neta G.I., von Ehrenstein O.S., Goldman L.R., Lum K., Sundaram R., Andrews W., Zhang J. Umbilical cord serum cytokine levels and risks of small-for-gestational-age and preterm birth. Am. J. Epidemiol. 2010;171:859–867. doi: 10.1093/aje/kwq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGovern N., Shin A., Low G., Low D., Duan K., Yao L.J., Msallam R., Low I., Shadan N.B., Sumatoh H.R. Human fetal dendritic cells promote prenatal T-cell immune suppression through arginase-2. Nature. 2017;546:662–666. doi: 10.1038/nature22795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dauby N., Marchant A. Fetal infections: immune response to infections during fetal life. In: Johnson A., Oepkes D., Kilby M.D., editors. Fetal Therapy: Scientific Basis and Critical Appraisal of Clinical Benefits. Cambridge University; 2020. pp. 215–223. [Google Scholar]
- 24.Bunders M.J., van Hamme J.L., Jansen M.H., Boer K., Kootstra N.A., Kuijpers T.W. Fetal exposure to HIV-1 alters chemokine receptor expression by CD4+T cells and increases susceptibility to HIV-1. Sci. Rep. 2014;4:6690. doi: 10.1038/srep06690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giorgi E.E., Funkhouser B., Athreya G., Perelson A.S., Korber B.T., Bhattacharya T. Estimating time since infection in early homogeneous HIV-1 samples using a poisson model. BMC Bioinformatics. 2010;11:532. doi: 10.1186/1471-2105-11-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suchard M.A., Lemey P., Baele G., Ayres D.L., Drummond A.J., Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song H., Giorgi E.E., Ganusov V.V., Cai F., Athreya G., Yoon H., Carja O., Hora B., Hraber P., Romero-Severson E. Tracking HIV-1 recombination to resolve its contribution to HIV-1 evolution in natural infection. Nat. Commun. 2018;9:1928. doi: 10.1038/s41467-018-04217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giorgi E.E., Li H., Bhattacharya T., Shaw G.M., Korber B. Estimating the timing of early simian-human immunodeficiency virus infections: a comparison between Poisson Fitter and BEAST. MBio. 2020;11:e00324-20. doi: 10.1128/mBio.00324-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murrell B., Wertheim J.O., Moola S., Weighill T., Scheffler K., Kosakovsky Pond S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou T., Xu L., Dey B., Hessell A.J., Van Ryk D., Xiang S.H., Yang X., Zhang M.Y., Zwick M.B., Arthos J. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore P.L., Gray E.S., Wibmer C.K., Bhiman J.N., Nonyane M., Sheward D.J., Hermanus T., Bajimaya S., Tumba N.L., Abrahams M.R. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat. Med. 2012;18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pejchal R., Walker L.M., Stanfield R.L., Phogat S.K., Koff W.C., Poignard P., Burton D.R., Wilson I.A. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc. Natl. Acad. Sci. USA. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwong P.D., Mascola J.R. HIV-1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity. 2018;48:855–871. doi: 10.1016/j.immuni.2018.04.029. [DOI] [PubMed] [Google Scholar]
- 34.Sok D., Burton D.R. Recent progress in broadly neutralizing antibodies to HIV. Nat. Immunol. 2018;19:1179–1188. doi: 10.1038/s41590-018-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schatten H., Constantinescu G.M. Blackwell; 2007. Comparative Reproductive Biology. [Google Scholar]
- 36.Codaccioni M., Bois F., Brochot C. Placental transfer of xenobiotics in pregnancy physiologically-based pharmacokinetic models: structure and data. Comput. Toxicol. 2019;12:100111. doi: 10.1016/j.taap.2020.115318. [DOI] [PubMed] [Google Scholar]
- 37.Chatuphonprasert W., Jarukamjorn K., Ellinger I. Physiology and pathophysiology of steroid biosynthesis, transport and metabolism in the human placenta. Front. Pharmacol. 2018;9:1027. doi: 10.3389/fphar.2018.01027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Underwood M.A., Gilbert W.M., Sherman M.P. Amniotic fluid: not just fetal urine anymore. J. Perinatol. 2005;25:341–348. doi: 10.1038/sj.jp.7211290. [DOI] [PubMed] [Google Scholar]
- 39.Mold J.E., Michaëlsson J., Burt T.D., Muench M.O., Beckerman K.P., Busch M.P., Lee T.H., Nixon D.F., McCune J.M. Maternal alloantigens promote the development of tolerogenic fetal regulatory T cells in utero. Science. 2008;322:1562–1565. doi: 10.1126/science.1164511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Claas F.H., Gijbels Y., van der Velden-de Munck J., van Rood J.J. Induction of B cell unresponsiveness to noninherited maternal HLA antigens during fetal life. Science. 1988;241:1815–1817. doi: 10.1126/science.3051377. [DOI] [PubMed] [Google Scholar]
- 41.Aagaard K., Ma J., Antony K.M., Ganu R., Petrosino J., Versalovic J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014;6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell D.E., Boyle R.J., Thornton C.A., Prescott S.L. Mechanisms of allergic disease - environmental and genetic determinants for the development of allergy. Clin. Exp. Allergy. 2015;45:844–858. doi: 10.1111/cea.12531. [DOI] [PubMed] [Google Scholar]
- 43.Berry S.M., Fine N., Bichalski J.A., Cotton D.B., Dombrowski M.P., Kaplan J. Circulating lymphocyte subsets in second- and third-trimester fetuses: comparison with newborns and adults. Am. J. Obstet. Gynecol. 1992;167:895–900. doi: 10.1016/s0002-9378(12)80008-1. [DOI] [PubMed] [Google Scholar]
- 44.Rechavi E., Lev A., Lee Y.N., Simon A.J., Yinon Y., Lipitz S., Amariglio N., Weisz B., Notarangelo L.D., Somech R. Timely and spatially regulated maturation of B and T cell repertoire during human fetal development. Sci. Transl. Med. 2015;7:276ra25. doi: 10.1126/scitranslmed.aaa0072. [DOI] [PubMed] [Google Scholar]
- 45.Douglas A.O., Martinez D.R., Permar S.R. The role of maternal HIV envelope-specific antibodies and mother-to-child transmission risk. Front. Immunol. 2017;8:1091. doi: 10.3389/fimmu.2017.01091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thobakgale C.F., Ramduth D., Reddy S., Mkhwanazi N., de Pierres C., Moodley E., Mphatswe W., Blanckenberg N., Cengimbo A., Prendergast A. Human immunodeficiency virus-specific CD8+ T-cell activity is detectable from birth in the majority of in utero-infected infants. J. Virol. 2007;81:12775–12784. doi: 10.1128/JVI.00624-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moody M.A., Pedroza-Pacheco I., Vandergrift N.A., Chui C., Lloyd K.E., Parks R., Soderberg K.A., Ogbe A.T., Cohen M.S., Liao H.X. Immune perturbations in HIV-1-infected individuals who make broadly neutralizing antibodies. Sci. Immunol. 2016;1:aag0851. doi: 10.1126/sciimmunol.aag0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keele B.F., Giorgi E.E., Salazar-Gonzalez J.F., Decker J.M., Pham K.T., Salazar M.G., Sun C., Grayson T., Wang S., Li H. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R Development Core Team . R Foundation for Statistical Computing; 2009. R: A language and environment for statistical computing. [Google Scholar]
- 50.Anderson V.M. The placental barrier to maternal HIV infection. Obstet. Gynecol. Clin. North Am. 1997;24:797–820. doi: 10.1016/s0889-8545(05)70345-4. [DOI] [PubMed] [Google Scholar]
- 51.Gouy M., Guindon S., Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 52.Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 53.Montefiori D.C. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 2009;485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate new datasets or code.