Summary
Epstein-Barr virus (EBV) and related lymphocryptoviruses (LCVs) from nonhuman primates are transmitted through oral secretions, penetrate the mucosal epithelium, and establish persistent infection in B cells. To determine whether neutralizing antibodies against epithelial or B cell infection could block oral transmission and persistent LCV infection, we use rhesus macaques, the most accurate animal model for EBV infection by faithfully reproducing acute and persistent infection in humans. Naive animals are infused with monoclonal antibodies neutralizing epithelial cell infection or B cell infection and then challenged orally with recombinant rhesus LCV. Our data show that high-titer B cell-neutralizing antibodies alone, but not epithelial cell-neutralizing antibodies, can provide complete protection of rhesus macaques from oral LCV challenge, but not in all hosts. Thus, neutralizing antibodies against B cell infection are important targets for EBV vaccine development, but they may not be sufficient.
Keywords: Epstein-Barr virus, lymphocryptovirus, 72A1, E1D1, EBV vaccine
Graphical abstract
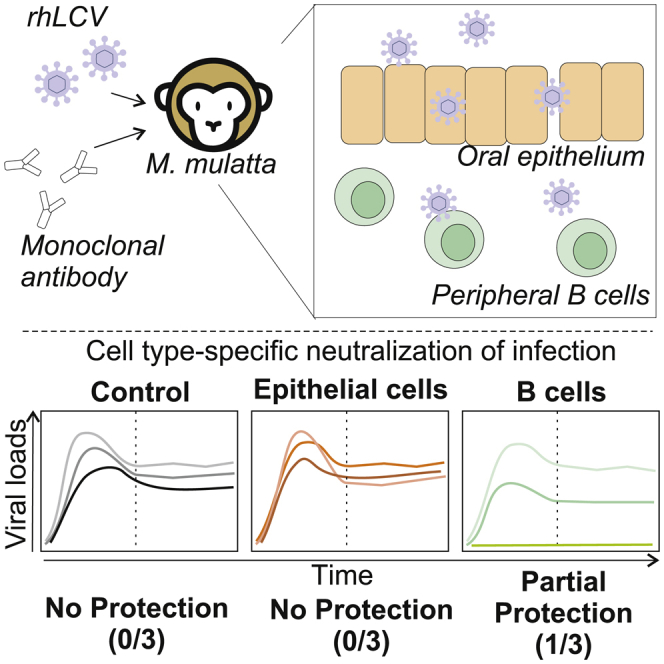
Highlights
mAb infusion leads to high neutralizing titers in nonhuman primates
Protection of epithelial cells does not protect from lymphocryptovirus challenge
Neutralization of B cell infection alone provides partial protection in macaques
Epstein-Barr virus is associated with a variety of diseases and causes a burden for healthcare and the economy. Thus, a vaccine is urgently needed. Mühe et al. provide insights into how protection from natural infection could be achieved and which viral antigens are pivotal for successful vaccine development.
Introduction
Primary Epstein-Barr virus (EBV) infection is the most common cause of infectious mononucleosis and typically occurs by transmission of oral secretions to an EBV-naive host, e.g., during kissing.1 During acute infection, EBV-infected B cells are readily detected in submucosal secondary lymphoid tissues, such as the tonsils, and the virus persists in the host in a small number of latently infected peripheral blood B cells for life.2 Nearly all humans are infected by adulthood.3 Although the vast majority of persistent EBV infections are asymptomatic, they can lead to development of B cell and epithelial cell malignancies.4 There is no vaccine to prevent EBV-associated diseases, such as infectious mononucleosis, post-transplant lymphoproliferative disorder, or cancer.5
Viral targets required for an effective EBV vaccine are not obvious because the early events in acute EBV infection and how EBV penetrates the oral mucosal epithelium remain speculative, and many different viral proteins are involved.6 The classic paradigm postulates that EBV initially infects epithelial cells, where the virus is amplified by lytic replication, and release of the amplified virus goes on to establish latent infection in peripheral blood B cells. Alternatively, EBV may bypass the oral mucosal epithelium (e.g., via microfissures) and infect peripheral blood B cells directly, where it would be amplified by lytic replication and infection of more B cells or through proliferation of latently infected B cells. Blocking the earliest stages of primary EBV infection before viral amplification would likely be the most effective strategy, but that could be EBV infection of epithelial cells, B cells, or both.
B cell-neutralizing antibodies (BnAbs) are specific for the EBV glycoprotein gp350.7,8 BnAbs can block B cell infection but are incapable of blocking EBV infection of epithelial cells.9 The best-characterized BnAb is 72A1, which binds close to the receptor-binding domain on EBV gp350, preventing B cell attachment and infection.8 Epithelial cell-neutralizing antibodies (EnAbs), on the other hand, bind the EBV gH/gL glycoprotein complex and can block epithelial cell infection.9 E1D1 is the prototypic EnAb, and it detects the gH protein when in a complex with gL.10, 11, 12 We tested the role of BnAbs versus EnAbs in primary infection using the rhesus macaque animal model for EBV infection because it is the most authentic animal model for EBV infection in humans.
Rhesus macaques and all other nonhuman primates (NHPs) are naturally infected with gammaherpesviruses, or lymphocryptoviruses (LCVs), closely related to EBV.13 The genomes, proteomes, and biological properties are well conserved among those viruses.14 Notably, LCVs are transmitted orally between NHPs, acute primary infection can result in symptoms similar to infectious mononucleosis, essentially all adult animals harbor life-long persistent LCV infection in their B cells, and LCV infection is associated with development of virus-associated malignancies, e.g., lymphoma.15,16
Experimental infection of rhesus macaques requires use of their endogenous rhesus LCV (rhLCV) because a species-specific mechanism during latent infection prevents EBV from immortalizing rhesus macaque B cells.17,18 The gH/gL amino acid sequence is highly conserved between rhLCV and EBV, and the EnAb E1D1 cross-reacts with rhLCV gH/gL.19 The gp350 sequence in rhLCV and EBV is less conserved, and the BnAb 72A1 does not cross-react with rhLCV gp350.20 We recently demonstrated that a recombinant rhLCV carrying the EBV gp350 in place of rhLCV gp350 could be neutralized with the BnAb 72A1 and could successfully establish acute and persistent infection after experimental oral inoculation of LCV-naive rhesus macaques.20 Therefore, we used this recombinant rhLCV to test whether the epithelial cell or B cell pathway for EBV infection could be inhibited by passive transfer of EnAbs or BnAbs prior to oral viral challenge in naive rhesus macaques.
Results
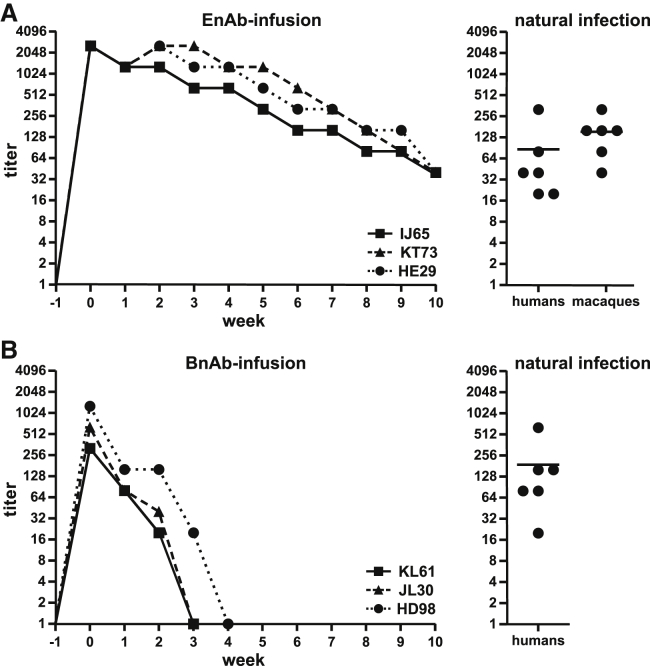
Recombinant EnAbs and BnAbs for use in rhesus macaques were constructed from the immunoglobulin variable regions of E1D1 and 72A1 cloned onto the constant regions of the rhesus IgG1 heavy and kappa light chains, respectively, to reduce potential immunogenicity. No further steps were deemed necessary to reduce the potential immunogenicity because the in vivo effect of these neutralizing antibodies is likely to occur within the first few days after oral inoculation, making prolonged administration unnecessary. Three groups of rhLCV-naive rhesus macaques were infused intravenously with 10 mg/kg of EnAbs (n = 3), BnAbs (n = 3), or control antibodies (n = 4) 1 day before oral virus challenge and boosted with 5 mg/kg intravenously 1 week after virus challenge. The monoclonal antibody (mAb) infusions were well tolerated in all animals. Serum titers of EnAbs and BnAbs were measured by endpoint dilution and flow cytometry on a weekly basis (Figures 1A and 1B, left panels). EnAb infusions achieved slightly higher peak serum titers (1:2,560) than BnAb infusions (1:320–1:1,280). These peak serum titers of mAbs were higher than or comparable to the polyclonal serum titers of gH/gL and gp350 reactivity in naturally infected humans and rhesus macaques (Figure 1, right panels). Only a fraction of the polyclonal serum gH/gL or gp350 reactivity in naturally infected hosts is neutralizing, so experimental infusion of neutralizing mAbs provided much higher serum neutralizing activity in the study animals compared with natural infection. We also observed that EnAb serum titers were more stable than BnAb serum titers and persisted for at least 10 weeks after experimental infusion. Because epithelial or B cell targets are likely to be infected within the first few days after virus inoculation, even 2–3 weeks of high neutralizing titers observed after BnAb infusion should be more than sufficient for relevant biologic activity.
Figure 1.
Serum antibody levels in rhLCV-naive rhesus macaques after infusion of anti-gH/gL mAbs (EnAbs) and anti-gp350 mAbs (BnAbs)
(A and B) Serum endpoint dilution titers for reactivity against recombinant gH/gL or gp350 expressed on EBV-negative B lymphoma cells for three individual animals infused with EnAb (A) or BnAbs (B) (left panels). Serum from three animals infused with a control mAb were non-reactive against gH/gL and gp350 (data not shown). Serum titers for gH/gL and gp350 reactivity in healthy humans and macaques with natural LCV infection using the same assay are shown for comparison (right panels, line indicates mean). Each data point represents at least two technical replicates.
All 10 animals were challenged orally with 106 transforming units of a recombinant rhLCV carrying EBV gp350 in place of rhLCV gp350, i.e., rhLCV-hugp350.20 This rhLCV dose has provided reliable infection of all experimentally inoculated animals to date and is likely to be orders of magnitude higher than what is typically encountered in natural LCV infection of humans and rhesus macaques.
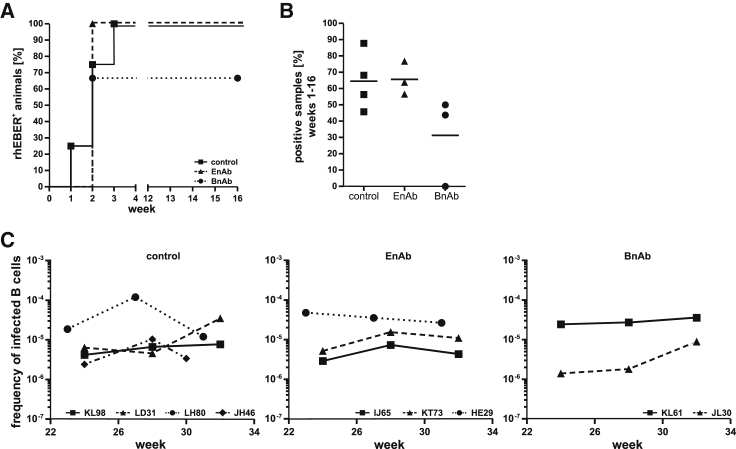
Animals were monitored for acute infection by weekly blood sampling for the first 16 weeks. Multiple aliquots of 5 × 106 peripheral blood mononuclear cells (PBMCs) were prepared from each time point and analyzed by RT-PCR for the presence of the highly abundant small RNAs, rhEBERs, the most sensitive method of detecting LCV infection of the peripheral blood. RhEBERs were detected in PBMCs from a control animal as early as 1 week after challenge and in all 4 control animals by week 3 after oral inoculation (Figure 2A). All EnAb-infused animals were positive for rhEBERs by week 2 as well as 2 of 3 animals infused with BnAbs, indicating that penetration of the oral epithelium by the virus and infection of peripheral B lymphocytes was not delayed by EnAbs or BnAbs in these animals. Similarly, when the results from all PBMC aliquots collected over the first 16 weeks were compared, there was no significant difference in the percentage of aliquots positive for rhEBERs (Figure 2B; 44%–88%), suggesting that there were no obvious differences in viral load during acute infection in these 9 animals. However, in one animal infused with BnAbs, rhEBERs were undetected in any PBMC aliquot during the acute infection period, suggesting that infection was blocked or reduced to undetectable levels during the first 16 weeks in this animal (HD98).
Figure 2.
Evaluation of acute and persistent infection after oral LCV challenge of animals infused with EnAbs, BnAbs, or a control mAb
(A and B) The percentage of animals in each group with at least one PBMC aliquot positive for rhLCV EBERs during the first 16 weeks after oral LCV challenge (A) and the overall percentage of PBMC aliquots positive for rhEBERs during the first 16 weeks (B); line indicates mean. The number of aliquots analyzed during the acute phase of infection (weeks 1–16) was 22–48 for all animals; see STAR Methods for details.
(C) The viral setpoint for individual animals as determined by limiting dilution analysis at three different time points during persistent infection (>16 weeks after viral challenge). Results are shown as the inverse number of peripheral blood B cells containing one LCV-infected B cell.
Persistent infection develops as viral loads during acute infection are controlled over time until a low and stable frequency of virus-infected B cells, or viral setpoint, is established.21 To determine whether neutralizing mAbs might affect establishment of the viral setpoint in persistent infection, larger amounts of peripheral blood were collected at three time points more than 20 weeks after oral challenge, and the viral setpoints were determined by limiting dilution analysis. As shown in Figure 2C, viral setpoints between approximately 1 in 106 to 1 in 104 B cells were detected in 9 of the 10 infected animals. The viral setpoints were relatively stable during the time period measured and persistent infection in 5 of 6 animals infused with EnAbs or BnAbs was established at levels comparable with the 4 animals infused with control mAbs. The virus remained undetectable at all time points beyond 20 weeks in the BnAb-infused animal, HD98, with undetectable viral loads during acute infection.
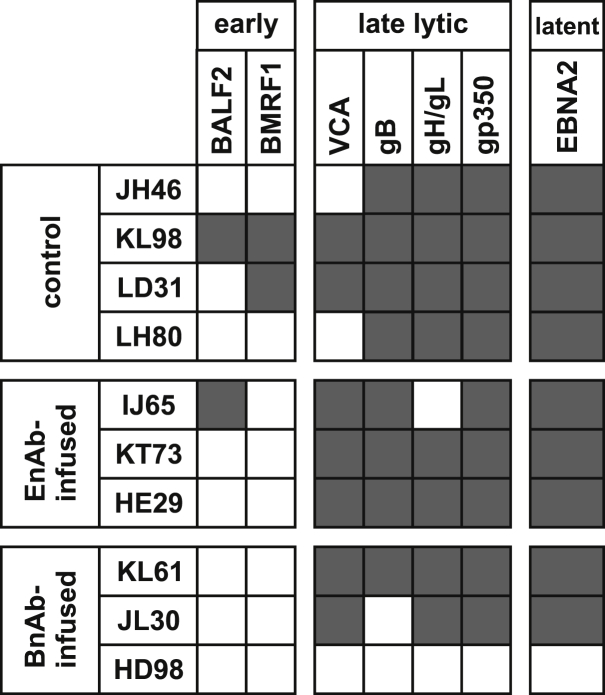
To differentiate complete protection from infection at undetectable levels in HD98, serologic responses to various LCV lytic and latent infection antigens were measured (Figure 3). Serum antibodies against a latent infection protein (EBNA2) and at least 3 of 4 late lytic infection proteins (small viral capsid antigen [sVCA], gB, gH/gL, or gp350) were detected in all animals except HD98. Serum antibodies against early lytic infection proteins (BALF2 and BMRF1) were more variable. Thus, the oral LCV challenge of HD98 resulted in no detectable serologic responses, consistent with sterilizing immunity provided by BnAb infusion, whereas robust responses were readily detected in all other animals.
Figure 3.
Serologic responses to lytic and latent infection LCV antigens after oral rhLCV challenge of macaques infused with control mAbs, EnAbs, or BnAbs
Detection of serum antibodies against early (BALF2 and BMRF1) or late (sVCA, gB, gH/gL, and gp350) lytic infection antigens and a latent infection antigen (EBNA2) during persistent infection are indicated by black boxes. White boxes represent no detection of the antigen-specific antibody response at any of the tested time points. Each time point was tested in duplicate.
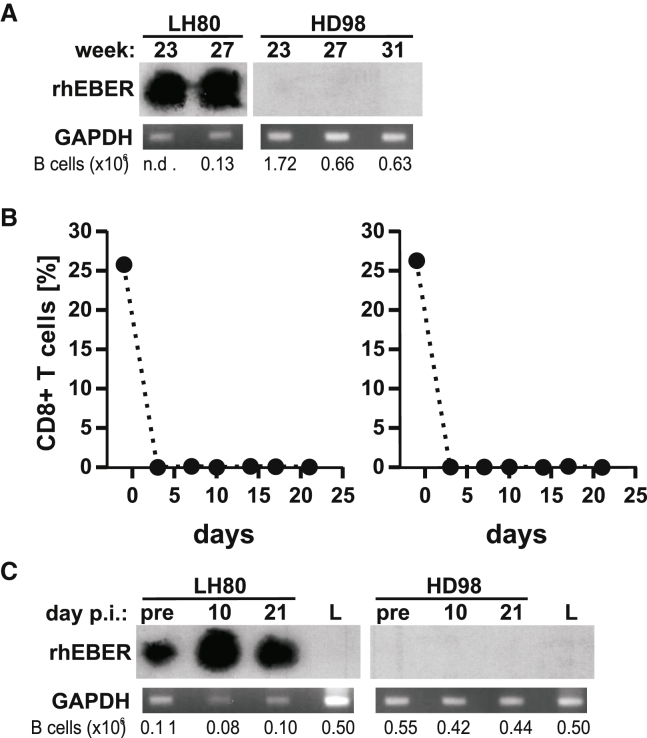
To test whether LCV challenge in the presence of BnAbs resulted in very low-level infection of B cells in HD98, two additional experiments were performed. First, peripheral blood B cells were affinity purified from the peripheral blood at time points more than 20 weeks after viral challenge of HD98 and an animal infused with control mAb, LH80 (Figure 4A). The enriched B cell population contained higher numbers of B cells than in peripheral blood aliquots, increasing the sensitivity of rhEBERs RT-PCR detection by eliminating large amounts of RNA from irrelevant cell types. RhEBERs were still undetectable in purified B cells from three different time points in HD98, whereas they were readily detectable in LH80, where smaller B cell numbers were tested. In a second experiment, HD98 and LH80 were infused with an anti-CD8α mAb to deplete these immune T cells in vivo. CD8+ T cells are important for control of persistent EBV infection in vivo, and we hypothesized that, if persistently infected B cells were present at a low frequency in HD98, they would amplify and become potentially detectable in the absence of CD8+ T cells. As shown in Figure 4B, CD8+ T cells in the peripheral blood were depleted rapidly and markedly in both animals after anti-CD8 mAb infusion. However, rhEBERs remained undetectable in peripheral B lymphocytes from HD98 for 21 days after CD8+ T cell depletion, whereas a modest increase in rhEBERs was detected after 10 days of CD8+ T cell depletion in the LH80 control. In addition, there was no evidence showing that HD98 was resistant to rhLCV infection because peripheral blood B cells from HD98 were readily immortalized in tissue culture with the rhLCV-hugp350 virus. These experiments support the conclusion that administration of BnAbs prevented acute and persistent infection of B cells after oral LCV challenge in a rhesus macaque, HD98.
Figure 4.
Absence of detectable rhLCV infection in enriched B cell populations from HD98 before and after CD8+ T cell depletion
(A–C) Beginning at 23 weeks after oral LCV challenge, B cells were affinity purified from PBMCs at multiple time points from a control animal (LH80) and the BnAb-infused HD98. RNA was isolated and subjected to RT-PCR, with detection of rhEBERs by southern blot hybridization (top panel) or GAPDH by ethidium bromide fluorescence. The total number of purified B cells in each aliquot is listed at the bottom in (A). HD98 and the control animal, LH80, were subsequently infused with an anti-CD8 mAb to deplete CD8+ T cells, as confirmed by flow cytometry analysis of PBMCs (B). RT-PCR detection of rhEBER and GAPDH after CD8+ T cell depletion is shown in (C). RNA from EBV-negative Louckes cells (L) was used as a negative control for the rhEBERs assay.
Discussion
Our results show that high serum titers of BnAbs are sufficient to provide complete protection against oral LCV challenge. Sashihara et al.22 reported that two of four rhesus macaques could be protected from challenge by vaccination with soluble gp350, but the immune mechanism responsible for protection was not defined in that study. Because the total gp350 antibody responses induced by the soluble gp350 vaccine were only one log higher than those in naturally infected animals, one possible hypothesis was that a vaccine capable of inducing more B cell-neutralizing activity might protect all animals. Singh et al.23 showed that infusion of the AMMO1 mAb protected three of four rhesus macaques from oral rhLCV challenge. However, because the AMMO1 mAb can block epithelial and B cell infection, it was not certain which target(s) were important for protection. Our study proves that neutralizing activity against EBV infection of B cells is a key immune mechanism that can be sufficient for immunity against oral transmission. However, even though we achieved extremely high levels of gp350 neutralizing activity by administering mAbs, not all animals could be protected from oral challenge. This suggests that developing EBV vaccines capable of inducing higher B cell-neutralizing activity may not necessarily result in protection from EBV infection in all hosts.
The amount of challenge virus relative to antibody is an important consideration that may have affected the efficacy of the BnAbs. Our protocol is unique in that it uses a very high titer of inoculating virus, 106 transforming units, to ensure reliable infection of all animals in a small study. This titer of virus is likely to be multiple orders of magnitude higher than that typically exchanged naturally among humans through kissing. Our exploratory experiments appropriately used a limited number of animals to screen for complete protection because there were no prior data available to reliably predict a likely outcome. Thus, it is possible that BnAbs might also have blunted the acute viral load or persistent viral setpoint in cases where BnAbs did not provide complete protection. In fact, a second BnAb animal in our study might have had slightly lower acute viral loads and a lower viral setpoint, but our studies were not designed to detect these more subtle quantitative differences. Altering the ratio of challenge virus/mAb to more closely mimic clinical situations may reveal complete protection in a higher percentage of hosts and reveal whether BnAbs can have significant effects on acute viral loads and persistent viral setpoints in the absence of complete protection. However, even with a virus challenge dose of only 50 transforming units, not all rhesus macaques were protected by infusion with 20 mg/kg of mAb AMMO1.23
It is intriguing that in three separate NHP studies, different therapeutic interventions targeting gp350 provided complete protection against oral transmission in some but not all animals. The small numbers of animals used in each study prevent any statistically significant interpretation of whether one treatment was better than another. However, the common finding that not all animals were protected suggests that EBV entry and pathogenesis may be more complicated and may vary between individual hosts. In some individuals, neutralizing B cell infection may be sufficient to provide complete protection, whereas in other individuals, additional pathways may allow successful viral entry and infection. One obvious hypothesis is that infection of epithelial cells with subsequent cell-to-cell transfer to susceptible B cells provides a pathway for EBV infection in some individuals that is resistant to B cell-neutralizing antibodies. However, in our study, administering gH/gL-specific neutralizing antibody E1D1 alone had no effect on oral challenge of rhesus macaques. Despite the higher EnAb titers and longer half-life, no animal was completely protected, and we could not detect any obvious effect on acute or persistent rhLCV infection. Similarly, using AMMO1, which blocks infection of B and epithelial cells, did not result in universal protection either.23,24 Our negative results with EnAbs do not rule out epithelial cell infection as an important potential target for therapeutic intervention. It is possible that, in our study, there were insufficient EnAb levels at the oral mucosal surface to block initial infection of epithelial cells, although the EnAb serum titers we achieved were extremely high (comparable with 100 μg/mL), and serum antibodies can be transduced to the mucosa. Adding gH/gL as a vaccine target to induce EnAbs could be beneficial, but these experiments suggest that vaccine designs that only induce high serum titers of gH/gL-specific antibodies or even high titers of E1D1-like neutralizing activity in the serum are unlikely to be effective against mucosal EBV challenge. We cannot exclude a potential benefit from non-E1D1 EnAbs blocking other functions of gH/gL, high levels of mucosal EnAbs (e.g., immunoglobulin A [IgA] antibodies), or addition of other glycoprotein targets to an EBV vaccine (e.g., the fusion protein gB).
It is possible that an EBV vaccine may be effective at reducing the risk for EBV-induced infectious mononucleosis without providing complete protection against EBV infection because it is unknown why primary EBV infection causes infectious mononucleosis in most individuals but not others. One could speculate that reducing the magnitude of the acute viral infection may prevent induction of infectious mononucleosis without completely preventing EBV infection. Our study proves that EBV neutralizing activity is a key immune parameter for vaccine development, and the rhesus macaque animal model provides a valuable and stringent platform for future investigations to definitively resolve whether it is sufficient to reliably reduce acute viral load in cases without complete protection.
These types of studies will require larger numbers of rhLCV-naive animals than have been used in the past. We used animals from self-sustaining, extended specific pathogen-free breeding colonies because the alternative, obtaining rhLCV-naive animals by separating neonates from their mothers, is labor intensive; costly in terms of resources, space, and time; and somewhat unreliable because of the extremely facile transmission of rhLCV through oral secretions in domestic colonies. Thus, we strongly argue for more ongoing support to expand this powerful research resource because the unique insights into EBV pathogenesis and potential treatments in our and other NHP studies could not be obtained in any other animal model.
Limitations of the study
Rhesus macaques are the most accurate animal model to study natural EBV infection, but they have to be reared free of LCVs in extended specific pathogen-free colonies. Thus, the number of animals is limited, and our study was designed as a proof of concept. Under these circumstances, we could show that passive transfer of BnAbs alone can prevent infection after oral challenge in 1/3 animals, whereas no protection was evident from EnAbs. Why BnAb could not protect all animals will need further investigation of potential biological and technical issues, as discussed above. Similarly, negative results with EnAbs do not rule out a potential role of epithelial cells during oral EBV infection or for vaccine development but suggest that high titers of EnAbs alone will not be sufficient for protection. Additionally, the study design did not allow investigation of whether BnAbs or EnAbs might reduce acute viral loads or persistent viral setpoints because of the limited amount of blood samples available from each animal.
The results presented here provide justification for future experiments with larger numbers of animals that will allow comprehensive statistical analyses, detection of smaller changes, and dissection of the mechanisms important for protection from oral EBV transmission.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BnAb, recombinant rhesus anti-EBV gp350 antibody (chimeric 72A1) | This study and Herrman et al.20 | N/A |
| EnAb, recombinant rhesus anti-EBV gHgL antibody (chimeric E1D1) | This study | N/A |
| rhesus anti-desmipramine IgG1 control antibody | NIH Nonhuman Primate Reagent Resource; MassBiologics | N/A |
| anti-CD8α antibody (MT807R1) | NIH Nonhuman Primate Reagent Resource | RRID: AB_2716320 |
| APC-conjugated anti-CD20 antibody | BD Biosciences | Cat# 559776 |
| goat-anti-Human FITC F (ab’)2 | Jackson ImmunoResearch | Cat# 109-096-008; RRID: AB_2337663 |
| HRP-conjugated anti-human IgG | Jackson ImmunoResearch | Cat# 109-035-003; RRID: AB_2337577 |
| anti-flag antibody | Sigma | Cat# F1804 |
| HRP-conjugated anti-dig-antibody | Jackson ImmunoResearch | Cat# 200-032-156; RRID: AB_2339011 |
| Bacterial and virus strains | ||
| RhLCV-hugp350 | Herrman et al.20 | N/A |
| Vaccinia virus encoding flag-tagged rhLCV BALF2 | Orlova et al.25 | N/A |
| Vaccinia virus encoding flag-tagged rhLCV BMRF1 | Orlova et al.25 | N/A |
| Vaccinia virus encoding flag-tagged rhLCV gB | Orlova et al.25 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RhBFRF3 peptide (AVDTGSGGGAQPQDTSTRGARKKQ) | Peptide/Protein Core Laboratory at Massachusetts General Hospital, Boston | #1971 |
| 3xflag-peptide | Sigma | Cat# F4799 |
| Count Bright counting beads | Life Technologies | Cat# C36950 |
| DYNAL Dynabeads anti-mouse IgG | Thermo Fisher | Cat# 11033 |
| RNA-Bee | Tel-Test, Inc. | CS-501B |
| Experimental models: Cell lines | ||
| hugp350-rhLCV LCL, lymphoblastoid cell line generated from rhesus macaque PBMC using hugp350-rhLCV | Herrman et al.20 | N/A |
| E1D1 hybridoma cells | Lindsey Hutt-Fletcher | N/A |
| BJAB cells expressing EBV gp350 or gH/gL | This manuscript | N/A |
| flag-rhEBNA2-rhLCV LCL | This manuscript | N/A |
| Experimental models: Organisms/strains | ||
| Macaca mulatta | Tulane National Primate Research Center | N/A |
| Oligonucleotides | ||
| rhEBER (173R: AAAACAGGCGGACCACCAG) | Ohashi et al.21 | N/A |
| rhEBER (32F: GGAGGAGATGAGTGTGACTTAAATCA) | Ohashi et al.21 | N/A |
| rhEBER (148R: TGAACCGAAGAGAGCAGAAACC) | Ohashi et al.21 | N/A |
| rhGAPDH (for RT: GTTCACACCCATGACGAACATGG) | Ohashi et al.21 | N/A |
| GAPDH (TF: GCGAGATCCCTCCAAAATCA) | Ohashi et al.21 | N/A |
| GAPDH (TR: CCAGTGGACTCCACGACGTA) | Ohashi et al.21 | N/A |
| Recombinant DNA | ||
| 72A1 heavy chain, CDR grafted (TGTCAGGTGCAG CTGGTGCAGTCTGGGGCCGAAGTGAAGAAGCC AGGCGCCAGCGTGAAGGTGTCTTGCAAGGCTTC CGGCTACACCTTCACAGACTATACAATGAACTG GATGAGACAGGCCCCCGGCCAGAATCTGGAGT GGATCGGCCTGATCAACCCTTACAATGGCGGC ACCAGGTATAACCAGAAGTTTAAGGGCAAGGCC ACCATGACACGGGACACCTCTACATCCACCGCT TACATGGAGCTGTCCAGCCTGCGCAGCGAGGAT ACAGCCGTGTACTATTGTGCTGGCGGCCTGCGT CGCGTGAATTGGTTCGCATATTGGGGGCAGGG GACCCTGGTGACCGTCTCA) |
This study | N/A |
Resource availability
Lead contact
Further communication and requests for resources and reagents should be directed to the lead contact, Fred Wang (fwang@research.bwh.harvard.edu).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
The sequences for recombinant BnAb and EnAb antibodies supporting the current study are available in GenBank: KT211018 (72A1 light chain), GenBank: KX755644 (E1D1 heavy chain), and GenBank: KX755645 (E1D1 light chain).
Experimental model and subject details
Ethics statement
All animal experiments were approved by the Tulane National Primate Research Center Institutional Animal Care and Use Committee (IACUC) and performed in accordance with guidelines from the National Institutes of Health, US Department of Agriculture, and Tulane University.
Animals
RhLCV-naive rhesus macaques from the extended-specific pathogen free colony at the Tulane National Primate Research Center were assigned randomly to the study groups and confirmed to be rhLCV-naive shortly before infection.
The sex and age in years old on day 0 for each animal were as follows: JH46 (M, 6.0); KL98 (M, 5.0); LD31 (M, 4.1); LH80 (M,3.2); IJ65 (M,9.0); KT73 (M, 4.9); HE29 (F, 11.2); KL61 (M, 5.1); JL30 (M 7.1); HD98 (F, 11.2).
Cell lines
RhLCV-hugp350 lymphoblastoid cell line was generated by immortalization of rhesus macaque PBMC with rhLCV-hugp350 virus (as previously described20). E1D1 hybridoma cells were a kind gift from Lindsey Hutt-Fletcher. BJAB cell lines expressing gp350 or gH/gL or were generated by stable transfection of gp350 expression plasmid, or stepwise tranfection of gH- and gL-plasmids. Flag-rhEBNA2-rhLCV LCL is a lymphoblastoid cell line generated from rhesus macaque PBMC using recombinant rhLCV encoding a N-terminally flag-tagged version of EBNA2. Louckes is an EBV-negative human B lymphoma cell line. All B cell lines were cultured in RPMI with 10% FCS, 100 units/ml Penicillin, 100 μg/ml Streptomycin.
Method details
Recombinant antiviral antibodies
To determine E1D1 heavy and light chain sequences, RNA was isolated from E1D1 hybridoma cells (a kind gift from Lindsey Hutt-Fletcher) and used for Rapid Amplification of cDNA Ends (RACE) as previously described for 72A1.20 For production of recombinant antibodies, natural signal peptides were replaced for improved expression as follows: MELGLSWIFLLAILKGVQC for 72A1 heavy chain, MKYLLPTAAAGLLLLAAQPAMA for 72A1 light chain, and osteonectin signal peptide (MRAWIFFLLCLAGRALA) for E1D1 chains.26 The variable regions of the 72A1 heavy chain was CDR grafted with human antibody sequences to improve stability.
Variable regions were fused to rhesus IgG1 heavy chain and kappa light chain and full coding sequences were codon optimized using Gibson assembly with SGI-DNA BioXpTM 3200 System. Optimized ORFs were subcloned into expression vectors. Plasmids encoding corresponding heavy and light chains were subsequently joined to form a single vector containing tandem CMV promoters, i.e., Hu72A1 IgG1 Kappa pYW6 or E1D1 IgG1 Kappa pYW6. Recombinant antibodies were produced in CHO cells, purified from cell supernatants using Protein A column (GE Akta Pure FPLC, ProSep-vA Ultra Chromatography Resin), concentrated, and dialyzed against 20 mM Sodium Citrate, 150nM NaCl, 5% Maltose, pH5 using Pellicon XL Ultrafiltration Module Biomax 30 kDa 0.005 m2 (Millipore). Production of recombinant E1D1 was approximately 10-fold higher than recombinant 72A1 suggesting higher stability. After endotoxin removal final product was also polished by size exclusion chromatography (Superdex 200 Increase 10/300 GL) to confirm the purity of the antibody. Recombinant antibodies were confirmed to have comparable binding affinities as the original hybridoma antibodies by flow cytometry against B lymphoma cells expressing gp350 or gH/gL.
Antibody infusion
For passive immunization, rhesus macaques were infused intravenously with 10 mg/kg of recombinant 72A1, E1D1 or rhesus recombinant IgG1 control antibody (DSPR1 (anti-desmipramine); NIH Nonhuman Primate Reagent Resource) the day before viral challenge and with another i.v. dose of 5 mg/kg one week later.
In week 37 post challenge, two animals were infused subcutaneously with 10 mg/kg anti-CD8α antibody (MT807R1, NIH Nonhuman Primate Reagent Resource) to deplete cytotoxic T cells and NK cells. Additional infusions of 5 mg/kg i.v. followed every three days for 17 days. Blood was sampled every three days to confirm CD8+ T cell depletion by immune cell staining and every 10 days for detection of rhLCV-infected PBMC.
Virus production and cell culture
RhLCV-hugp350 was produced from a lymphoblastoid cell line as previously described.20 Viral replication of large numbers (> 1x109 cells) of rhLCV-hugp350 LCLs was induced by culturing cells with 20 ng/ml phorbol 12-myristate 13-acetate (TPA) and 3 mM butyrate. The following day, cells were resuspended in fresh media at a concentration of 3x106 cells/ml. Cell supernatants were collected 7 days after induction, filtered through a 0.45 μm filter, and concentrated by centrifugation at 13,000 g for 2 hours at 4°C. Virus was resuspended in RPMI (1/300-1/550 of the starting culture volume). Virus stocks were titrated for transforming units by making serial 10-fold dilutions of virus and plating multiple replicates of each virus dilution into microtiter wells with 200,000 rhesus PBMC/well. Wells were scored positive at 8 weeks post infection by visual confirmation of cell outgrowth. Transforming units were calculated as the dilution necessary for outgrowth in 50% of the wells based on the Reed-Muench equation.27
PBMC were isolated from heparinized whole blood by density centrifugation over a Ficoll gradient and washed 3 times with PBS. B cell numbers were determined by staining with APC-conjugated anti-CD20 antibodies (BD Biosciences) while PBMC were counted using Count Bright counting beads (Life Technologies) and analyzed by flow cytometry using a FACScalibur cytometer (BD Biosciences). PBMC used for titration were cultured in RPMI supplemented with 10% FCS, 100 U/ml penicillin, 100 μg/ml streptomycin, 20 mM HEPES, 4 mM Glutamine, 1 mM Oxaloacetic acid, 1.4 μM insulin, 500 μM pyruvic acid, and Cyclosporin A (0.5 μg/ml). PBMC used for detection of rhEBER expression were immediately resuspended in RNA-Bee (Tel-Test, Inc.) and stored at −80°C.
B cells were purified from PBMCs using anti-CD20 antibody and DYNAL Dynabeads anti-mouse IgG (Thermo Fisher) according to manufacturer’s instructions. Purity and quantity of B cells was confirmed by flow cytometry.
Virus challenge
Animals were inoculated with 106 transforming units of virus applied non-traumatically throughout the oral cavity and followed for approximately 40 weeks. Blood samples were taken weekly for the first 16 weeks and biweekly thereafter.
Detection of gp350- and gH/gL-specific antibodies by immune fluorescence
To determine endpoint titers of rhesus plasma, samples were diluted two-fold in FACS buffer (PBS, 1% FCS, 0.1% NaN3) with a starting dilution of 1:10. BJAB-EBVgp350 or BJAB-gH/gL cells were washed and resuspended in plasma dilutions at 1x106/100 μl. Cells were incubated on ice for 45 min, washed and incubated with goat-anti-Human FITC F (ab’)2 (Jackson ImmunoResearch) for 45 min. Stained cells were analyzed by flow cytometry. Three rhLCV-negative plasma samples were run in parallel to determine the threshold, which was defined as the average MFI + 20%. Endpoint titers of test plasma were established as the last plasma dilution generating an MFI above the threshold.
VCA peptide ELISA
RhBFRF3 peptide (AVDTGSGGGAQPQDTSTRGARKKQ, obtained from the Peptide/Protein Core Laboratory at Massachusetts General Hospital) was used to coat ELISA plates at a final concentration of 0.5 μg/well in Bicarbonate buffer (15 mM Na2CO3, 35 mM NaHCO3, 3 mM NaN3, pH 9.6). Plates were coated over night at 4°C and blocked with blocking buffer (PBS, 0.1% Tween-20, 3% BSA). Plasma samples, including known positive and negative samples, were diluted 1:100 in blocking buffer and tested in duplicate. Wells were incubated with 200 μL plasma dilutions for 1h at room temperature, followed by incubation with HRP-conjugated anti-human IgG (Jackson Laboratories). Samples were developed using OPD substrate (Sigma). The threshold was determined as 3x the OD of a known rhLCV-negative serum.
Detection of serum antibodies against BALF2, BMRF1, and gB
Flag-tagged versions of rhLCV BALF2, BMRF1, and gB were produced by Vaccinia virus infection of BHK cells as previously described.25 In brief, BHK cells were infected with VV-rhBALF2/rhBMRF1/rhgB-flag at a multiplicity of infection of ∼1 and incubated for 12-16 h. The next day, cells were washed with PBS, lysed in flag-buffer (50 mM Tris/Hcl, pH7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton-X, Complete protease inhibitor [Sigma]) + 1% NP-40 followed by sonication. Lysates were cleared by centrifugation and incubated with anti-flag antibody (Sigma) over night. RhLCV protein-flag/antibody-complexes were precipitated with protein-G beads, washed and eluted with 3xflag-peptide solution. Protein purification was confirmed by western blot. Purified protein was diluted 1:100 in Bicarbonate buffer and used to coat ELISA plates as described, using PBS, 0.1% Tween-20, and 0.3% I-Block (Applied Biosystems) as blocking buffer.
Detection of EBNA2 antibodies by immunoprecipitation
Cell lysates were prepared from a lymphoblastoid B cell line transformed with rhLCV expressing flag-tagged EBNA2. Cells were lysed in IP buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% sodium deoxycholate, 1% NP-40) and equal volumes of lysate were incubated with 3-10 μL plasma over night at 4°C. Protein G Sepharose beads were preincubated with IP buffer + 1% BSA and then 20 μL of a 50% suspension were used to precipitate immunocomplexes. Beads were incubated with lysates for 30 min at 4°C, washed 3 times with PBS and boiled in protein sample buffer. Samples were separated on a 7.5% SDS-polyacrylamid gel and transferred to nitrocellulose membrane. Immunoprecipitated flag-EBNA2 was detected using HRP-conjugated mouse-anti-flag antibody (Sigma).
Reverse transcriptase mediated PCR detection of rhEBER in PBMC
For rhEBERs detection, total RNA was isolated from aliquots of 5x106 PBMC by phenol:chloroform extraction using RNA-Bee (Tel-Test, Inc.) and converted to cDNA using Mu-MLV reverse transcriptase (NEB) and primers for rhEBER (173R) and rhGAPDH (R). Ninety percent of the cDNA was tested for rhEBER and 10% was tested for GAPDH expression by PCR amplification as previously described.21 PCR products were separated by agarose gel electrophoresis, transferred to a nitrocellulose membrane by Southern blot, hybridized with a digoxigenin-labeled rhEBERs probe, and detected using an HRP-conjugated anti-dig-antibody (Jackson ImmunoResearch).
The number of aliquots analyzed during acute phase of infection (week 1 – 16) was 22-48 for animals in the control group, 22-40 for animals infused with BnAb, and 23-44 for those infused with EnAb. To increase the number of aliquots tested despite low blood availability, up to 11 additional PBMC aliquots with cell numbers between 1 and 4x106 cells were analyzed for each animal.
Frequency analysis for rhLCV infected cells by limiting dilution
To determine the frequency of rhLCV-infected cells in the peripheral blood, serial two-fold dilutions of freshly isolated PBMC were prepared, starting at 1x106 cells, and aliquoted into multiple replicates for each dilution. RNA was then isolated from each replicate and used for the detection of rhEBERs as described above. The frequency of infected cells was calculated by the number of positive and negative replicates in the dilutions using Extreme Limiting Dilution Analysis.28
Quantification and statistical analysis
The frequency of infected cells determined by limiting dilution was calculated by the number of positive and negative replicates in the dilutions using Extreme Limiting Dilution Analysis.28
Acknowledgments
We would like to thank Lindsey Hutt-Fletcher for providing the E1D1 hybridoma cell line. This research was funded by a grant (AI114557 to F.W.) from the National Institute for Allergy and Infectious Diseases, NIH. Core animal services were supported by Tulane National Primate Research Center base grant P51OD011104 and SPF grants U42OD010568 and U42OD024282. Production of recombinant antibodies was supported by NIH R24 OD010976 and U24 AI126683 to MassBiologics, University of Massachusetts Medical School.
Author contributions
This study was conceived by F.W. and designed by J.M., P.P.A., K.E., K.A.R., and F.W. J.M., P.P.A., C.Q., and J.Y.E. collected data. Interpretation of data was performed by J.M., J.Y.E., K.E., K.A.R., and F.W. The figures and first draft were generated by J.M. and F.W. All authors reviewed and approved the final manuscript.
Declaration of interests
F.W. is an inventor on patent WO 2013130565A1 related to the work described in this manuscript.
Published: July 20, 2021
References
- 1.Hoagland R.J. The transmission of infectious mononucleosis. Am. J. Med. Sci. 1955;229:262–272. doi: 10.1097/00000441-195503000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Miyashita E.M., Yang B., Babcock G.J., Thorley-Lawson D.A. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J. Virol. 1997;71:4882–4891. doi: 10.1128/jvi.71.7.4882-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de-Thé G., Day N.E., Geser A., Lavoué M.F., Ho J.H., Simons M.J., Sohier R., Tukei P., Vonka V., Zavadova H. Sero-epidemiology of the Epstein-Barr virus: preliminary analysis of an international study - a review. IARC Sci. Publ. 1975:3–16. [PubMed] [Google Scholar]
- 4.Kutok J.L., Wang F. Spectrum of Epstein-Barr virus-associated diseases. Annu. Rev. Pathol. 2006;1:375–404. doi: 10.1146/annurev.pathol.1.110304.100209. [DOI] [PubMed] [Google Scholar]
- 5.Cohen J.I. Vaccine Development for Epstein-Barr Virus. Adv. Exp. Med. Biol. 2018;1045:477–493. doi: 10.1007/978-981-10-7230-7_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chesnokova L.S., Jiang R., Hutt-Fletcher L.M. Viral Entry. Curr. Top. Microbiol. Immunol. 2015;391:221–235. doi: 10.1007/978-3-319-22834-1_7. [DOI] [PubMed] [Google Scholar]
- 7.Nemerow G.R., Mold C., Schwend V.K., Tollefson V., Cooper N.R. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J. Virol. 1987;61:1416–1420. doi: 10.1128/jvi.61.5.1416-1420.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffman G.J., Lazarowitz S.G., Hayward S.D. Monoclonal antibody against a 250,000-dalton glycoprotein of Epstein-Barr virus identifies a membrane antigen and a neutralizing antigen. Proc. Natl. Acad. Sci. USA. 1980;77:2979–2983. doi: 10.1073/pnas.77.5.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molesworth S.J., Lake C.M., Borza C.M., Turk S.M., Hutt-Fletcher L.M. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 2000;74:6324–6332. doi: 10.1128/jvi.74.14.6324-6332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oba D.E., Hutt-Fletcher L.M. Induction of antibodies to the Epstein-Barr virus glycoprotein gp85 with a synthetic peptide corresponding to a sequence in the BXLF2 open reading frame. J. Virol. 1988;62:1108–1114. doi: 10.1128/jvi.62.4.1108-1114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q., Turk S.M., Hutt-Fletcher L.M. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J. Virol. 1995;69:3987–3994. doi: 10.1128/jvi.69.7.3987-3994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Omerović J., Lev L., Longnecker R. The amino terminus of Epstein-Barr virus glycoprotein gH is important for fusion with epithelial and B cells. J. Virol. 2005;79:12408–12415. doi: 10.1128/JVI.79.19.12408-12415.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mühe J., Wang F. Non-human Primate Lymphocryptoviruses: Past, Present, and Future. Curr. Top. Microbiol. Immunol. 2015;391:385–405. doi: 10.1007/978-3-319-22834-1_13. [DOI] [PubMed] [Google Scholar]
- 14.Rivailler P., Jiang H., Cho Y.G., Quink C., Wang F. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J. Virol. 2002;76:421–426. doi: 10.1128/JVI.76.1.421-426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moghaddam A., Rosenzweig M., Lee-Parritz D., Annis B., Johnson R.P., Wang F. An animal model for acute and persistent Epstein-Barr virus infection. Science. 1997;276:2030–2033. doi: 10.1126/science.276.5321.2030. [DOI] [PubMed] [Google Scholar]
- 16.Rivailler P., Carville A., Kaur A., Rao P., Quink C., Kutok J.L., Westmoreland S., Klumpp S., Simon M., Aster J.C., Wang F. Experimental rhesus lymphocryptovirus infection in immunosuppressed macaques: an animal model for Epstein-Barr virus pathogenesis in the immunosuppressed host. Blood. 2004;104:1482–1489. doi: 10.1182/blood-2004-01-0342. [DOI] [PubMed] [Google Scholar]
- 17.Moghaddam A., Koch J., Annis B., Wang F. Infection of human B lymphocytes with lymphocryptoviruses related to Epstein-Barr virus. J. Virol. 1998;72:3205–3212. doi: 10.1128/jvi.72.4.3205-3212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mühe J., Wang F. Species-specific functions of Epstein-Barr virus nuclear antigen 2 (EBNA2) reveal dual roles for initiation and maintenance of B cell immortalization. PLoS Pathog. 2017;13:e1006772. doi: 10.1371/journal.ppat.1006772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu L., Hutt-Fletcher L.M. Compatibility of the gH homologues of Epstein-Barr virus and related lymphocryptoviruses. J. Gen. Virol. 2007;88:2129–2136. doi: 10.1099/vir.0.82949-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrman M., Mühe J., Quink C., Wang F. Epstein-Barr Virus gp350 Can Functionally Replace the Rhesus Lymphocryptovirus Major Membrane Glycoprotein and Does Not Restrict Infection of Rhesus Macaques. J. Virol. 2015;90:1222–1230. doi: 10.1128/JVI.02531-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohashi M., Fogg M.H., Orlova N., Quink C., Wang F. An Epstein-Barr virus encoded inhibitor of Colony Stimulating Factor-1 signaling is an important determinant for acute and persistent EBV infection. PLoS Pathog. 2012;8:e1003095. doi: 10.1371/journal.ppat.1003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sashihara J., Hoshino Y., Bowman J.J., Krogmann T., Burbelo P.D., Coffield V.M., Kamrud K., Cohen J.I. Soluble rhesus lymphocryptovirus gp350 protects against infection and reduces viral loads in animals that become infected with virus after challenge. PLoS Pathog. 2011;7:e1002308. doi: 10.1371/journal.ppat.1002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh S., Homad L.J., Akins N.R., Stoffers C.M., Lackhar S., Malhi H., Wan Y.H., Rawlings D.J., McGuire A.T. Neutralizing Antibodies Protect against Oral Transmission of Lymphocryptovirus. Cell Rep. Med. 2020;1:100033. doi: 10.1016/j.xcrm.2020.100033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snijder J., Ortego M.S., Weidle C., Stuart A.B., Gray M.D., McElrath M.J., Pancera M., Veesler D., McGuire A.T. An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus. Immunity. 2018;48:799–811.e9. doi: 10.1016/j.immuni.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orlova N., Fogg M.H., Carville A., Wang F. Antibodies to lytic infection proteins in lymphocryptovirus-infected rhesus macaques: a model for humoral immune responses to epstein-barr virus infection. Clin. Vaccine Immunol. 2011;18:1427–1434. doi: 10.1128/CVI.05126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haryadi R., Ho S., Kok Y.J., Pu H.X., Zheng L., Pereira N.A., Li B., Bi X., Goh L.T., Yang Y., Song Z. Optimization of heavy chain and light chain signal peptides for high level expression of therapeutic antibodies in CHO cells. PLoS ONE. 2015;10:e0116878. doi: 10.1371/journal.pone.0116878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- 28.Hu Y., Smyth G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences for recombinant BnAb and EnAb antibodies supporting the current study are available in GenBank: KT211018 (72A1 light chain), GenBank: KX755644 (E1D1 heavy chain), and GenBank: KX755645 (E1D1 light chain).