Abstract
Stringent spatiotemporal regulation of the wound healing process involving multiple cell types is associated with epigenetic mechanisms of gene regulation, such as DNA methylation, histone modification and chromatin remodeling, as well as non-coding RNAs. Here we discuss the epigenetic changes that occur during wound healing and the rapidly expanding understanding of how these mechanisms affect healing resolution in both acute and chronic wound milieu. We provide a focused overview of current research into epigenetic regulators that contribute to wound healing by specific cell type. We highlight the role of epigenetic regulators in the molecular pathophysiology of chronic wound conditions. The understanding of how epigenetic regulators can affect cellular functions during normal and impaired wound healing could lead to novel therapeutic approaches, and we outline questions that can provide guidance for future research on epigenetic-based interventions to promote healing. Dissecting the dynamic interplay between cellular subtypes involved in wound healing and epigenetic parameters during barrier repair will deepen our understanding of how to improve healing outcomes in patients affected by chronic non-healing wounds.
Keywords: histone modification, DNA methylation, circRNA, lncRNA, miRNA, skin wound healing
Introduction
Cutaneous wound healing is a complex process involving numerous cell types to accomplish sequential, yet overlapping phases of inflammation, proliferation and tissue remodeling1,2. Immediately after injury, blood components are released into the wound, forming a clot which provides a matrix for the influx of inflammatory cells. The inflammatory phase is characterized by leukocyte migration to the wound. Neutrophils primarily remove bacteria, followed by monocytes which further differentiate into macrophages that exert early pro-inflammatory and late anti-inflammatory functions during the healing process. Deposition of the newly synthesized fibrin matrix and granulation tissue formation follow; these are subsequently replaced by collagen and scar tissue during the final stages of wound healing. The proliferative phase of wound healing is characterized by re-epithelialization, neovascularization, and extracellular matrix deposition1,3.
Historically, exploration of the molecular basis of wound healing has included a primary focus on its spatiotemporal regulation. Given the complexity of the wound healing process and its requirement for stringent regulation, epigenetic regulation including histone modifications and DNA methylation is highly likely to play a role4,5. Indeed, recent discoveries in the field of non-coding RNAs have identified roles for microRNAs (miRs), circular RNAs (circRNA) and long non-coding RNAs (lncRNA) as global gene expression regulators involved in an array of processes important for successful wound healing6–9. While the primary focus of previous reviews has been on the role of epigenetic modifications in acute wound healing4–8, herein we highlight the importance of epigenetic modifiers in the pathophysiology of chronic wound healing disorders, with an emphasis on diabetic foot ulcers (DFU) and venous leg ulcers (VLU). Additionally, the roles of non-coding RNAs, histone modifications and DNA methylation in animal models of delayed healing are reviewed. Future perspectives regarding utilization of epigenetic modifiers as diagnostic and prognostic biomarkers as well as therapeutic targets are also discussed.
1. The role of non-coding RNAs in wound healing pathology
1.1. Long noncoding RNAs in wound healing
While a small portion of the mammalian genome is transcribed into protein coding mRNAs, a majority of the genome is transcribed into lncRNAs10,11 which are classified as RNAs longer than 200 nucleotides not translated into proteins12,13. The human genome contains more than 90,000 lncRNAs14, but only a small subset have a known function. Biogenesis of lncRNAs is similar to that of protein-coding genes, with similar histone modification profiles, splicing signals, and exon/intron lengths10. However, lncRNAs are less abundant and less evolutionarily conserved than mRNAs, are predominantly localized in the chromatin and nucleus, and show cell type-specific expression patterns10,15,16. A growing body of evidence indicates that lncRNAs are regulators of most global cellular processes, including nuclear chromatin organization, mRNA stability, transcription, translation, and cytoplasmic post-translational modifications11,17. As such, lncRNAs have demonstrated roles in human physiology as well as in the pathology of many diseases, including cancer, cutaneous disorders, and diabetes mellitus (DM)12,18–23.
Studies focused on the role of lncRNAs in chronic cutaneous wound healing are still in their early stages. Our group demonstrated that lncRNA GAS5 promoted wound healing by inhibiting c-myc, a biomarker of a hyperproliferative, non-migratory epidermis in chronic wounds24. Ectopic over-expression of lncRNA GAS5 suppressed c-myc even in the presence of dexamethasone, an inducer of c-myc expression, in human keratinocytes. Moreover, topical mevastatin treatment promoted wound healing due to induction of lncRNA GAS5 and the downstream suppression of c-myc24. The evaluation of GAS5 in a diabetic murine wound model revealed a correlation with a proinflammatory M1 macrophage phenotype, indicating distinct roles of this lncRNA in the inflammatory phase of wound healing25. Additionally, lncRNA Lethe deregulation was found involved in the modulation of macrophages under hyperglycemic conditions, resulting in increased reactive oxygen species production via NF-κB signaling26. As such, lncRNAs show cell type-specific functions during wound healing.
LncRNA WAKMAR1 (wound and keratinocyte migration-associated lncRNA 1) was found up-regulated during acute wound healing by TGF-β, while suppressed in DFUs and VLUs27. Knockdown of lncRNA WAKMAR1 decreased cell migration and suppressed re-epithelialization in a human ex vivo skin wound model. LncRNA WAKMAR1 interacted with DNA methyltransferases, resulting in reduced methylation of E2F1 (E2F Transcription Factor 1) promoter and induced expression of the downstream gene network involved in cell migration27. It was also found downregulated in wound edge keratinocytes of VLUs and DFUs when compared to acute wounds28. Silencing of lncRNA WAKMAR2 suppressed the production of inflammatory cytokines, decreased cell migration, and impaired re-epithelialization in human wounds ex vivo28.
LncRNA MALAT1 (Metastasis-associated lung adenocarcinoma transcript 1) was suppressed specifically in infected DFUs, and not regulated in uninfected ulcers29. The expression of lncRNA MALAT1 correlated with downregulation of NRF2 (Nuclear Factor Erythroid 2 Like 2), HIF1 (Hypoxia Inducible Factor 1), and VEGF (Vascular Endothelial Growth Factor) in infected DFUs. Knockdown of lncRNA MALAT1 in endothelial cells reduced expression of pro-angiogenic factors HIF1 and VEGF and pro-inflammatory TNF-α and IL-6, while inducing expression of anti-inflammatory IL-1029, suggesting the potential of lncRNAs for therapeutic intervention in infected ulcers.
Fibroblasts isolated from DFUs expressed low levels of lncRNA H1930. When lncRNA H19 was delivered by exosomes derived from mesenchymal stem cells (MSC), it promoted fibroblasts proliferation and migration, while suppressing apoptosis and inflammation. Mechanistically, lncRNA H19 binds to and suppresses miR-152-3p, which is up-regulated in DFU fibroblasts. This led to an increased level of miR-152-3p target PTEN (Phosphatase and Tensin Homolog), and downstream activation of PI3K/AKT1 signaling. Moreover, ectopic over-expression of lncRNA H19 in DFU fibroblasts reduced miR-29b level, resulting in up-regulation of FBN1 (Fibrillin 1)31. This data supports therapeutic application of MSC-derived exosomes carrying lncRNA H19 for improved diabetic fibroblast function30.
A study by Hu et al.32 showed that lncRNA URIDS (Up-Regulated in Diabetic Skin) is highly expressed in diabetic skin and upregulated in dermal fibroblasts treated with advanced glycation end products (AGE). Silencing of lncRNA URIDS promoted migration of diabetic fibroblasts despite AGE treatment and accelerated wound closure in vivo. The lncRNA URIDS regulates wound healing through interaction with PLOD1 (Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1), which results in decreased PLOD1 protein stability and deregulation of collagen production and ultimately delayed healing32. Another lncRNA TETILA (TET2-interacting long noncoding RNA) was found up-regulated in human diabetic skin33, where it activates transcription of MMP-9 (Matrix Metalloprotease 9), an indicator of poor wound healing in DFUs34,35. Silencing of TETILA increased migration of diabetic keratinocytes even in the presence of AGE. In diabetic skin, lncRNA TETILA recruits TET2 (Ten-eleven translocation 2) and thymine-DNA glycosylase to form a demethylation complex at the MMP-9 promoter, leading to MMP-9 up-regulation and impaired healing33. As DNA demethylation occurs frequently in diabetic tissues and cells36–38, TET2-mediated DNA demethylation may be relevant to global epigenetic changes during diabetic wound healing and will be further discussed in this review.
1.2. The role of microRNAs in wound healing
MicroRNAs (miRs) are short, highly conserved non-coding RNA molecules (~22 bp) that regulate gene expression at a post-transcriptional level39,40. The human genome contains more than 2,000 mature miRs41 which are predicted to target more than 60% of human protein-coding genes42. miRs are transcribed from DNA into primary miRs (pri-miRs) and processed into precursor miRs (pre-miRs) and mature miRs39,40,43. The target recognition domain, named the seed domain, is located at the 5’ end of each miR; miRs that share identical seed sequences are classified into miR families44,45. Most miRs bind to and interact with the 3′ UTR of target mRNAs resulting in target gene silencing43,45, though miRs can stimulate gene expression under certain conditions46. Interaction of miRs with 5′ UTRs, coding sequence regions, and gene promoters is also recognized45. miRs regulate a variety of cellular process and are critical for development, cell differentiation and homeostasis39, while deregulation of miRs is associated with many diseases, including DM and cutaneous disorders47–50. miRs play key roles in all phases of the acute wound healing process, as described in several comprehensive reviews6,7,51–54. Thus, we focus here on the roles of miRNAs in chronic non-healing wounds and animal models of delayed wound healing (Table 1).
Table 1.
miRs role in chronic wounds and animal models of impaired wound healing.
| microRNA | Effect on wound healing | Dysregulation in chronic wounds | Ref. |
|---|---|---|---|
| Tissue miRs in patients | |||
| miR-15b | Suppresses DNA repair and inflammatory response; impairs angiogenesis | Up-regulated in DFU and diabetic murine wounds | 64,65 |
| miR19a/b and miR-20a | Anti-inflammatory; promote wound healing | Down-regulated in DFU, VLU and pressure ulcers | 59 |
| miR-21 | Suppresses granulation tissue formation; impairs proliferation and migration, and induces senescence | Up-regulated in VLU and fibroblasts isolated from DFU | 55,56 |
| miR-34 family | Pro-inflammatory; impairs proliferation and migration, inhibits healing | Up-regulated in VLUs and fibroblasts isolated from DFU | 56,58 |
| miR-92a | Suppresses cell proliferation and angiogenesis; inhibits wound healing | Up-regulated in VLU and diabetic murine wounds | 60,61 |
| miR-132 | Anti-inflammatory; accelerates wound closure | Down-regulated in DFU and diabetic murine wounds | 63 |
| miR-152-3p | Inhibits proliferation and migration | Up-regulated in DFU fibroblasts | 30 |
| miR-296-5p | Promotes wound healing | Down-regulated in amputated tissue | 217 |
| miRs in diabetic animal models of impaired healing | |||
| miR-25 &miR-29a | Dysregulation of collagen production | Up-regulated in diabetic murine wounds | 218 |
| miR-26a | Impairs angiogenesis, decreases granulation tissue; inhibits healing | Up-regulated in diabetic murine wounds | 219 |
| miR-146a | Anti-inflammatory | Down-regulated in diabetic murine wounds | 220 |
| miR-155 | Pro-inflammatory; impairs wound closure | Up-regulated in diabetic murine wounds | 221 |
| miR-497 | Anti-inflammatory; improves wound healing | Down-regulated in diabetic murine wounds | 222 |
| miR-146a and miR-106 | Compromise tight junction function | Up-regulated in wounds infected with bacterial biofilm | 66 |
| Circulating miRs | |||
| miR-15a-3p | Reduces ROS production; impairs wound closure | Up-regulated in exosomes isolated from blood of DFU patients | 68 |
| miR-24 | Negatively corelated with the amputation rate in DFU | Down-regulated in peripheral plasma from DFU with osteomyelitis | 69 |
| miR-126 | Pro-angiogenic, promotes collagen maturation and healing | Down-regulated in the peripheral blood from the patients with DFU | 223,224 |
| miR-129 and miR-335 | Accelerate wound closure | Down-regulated in serum and tissue from DFU patients | 34 |
| miR-191 and miR-200b | Pro-inflammatory, suppress angiogenesis and migration | Up-regulated in plasma samples from DFU patients | 70 |
| miR-217 | Suppresses angiogenesis, pro-inflammatory; inhibits healing | Up-regulated in serum from DFU patients | 225 |
In early studies, we identified miR-16, -20a, -21, -106, -130a, -203 upregulation in the epidermis of non-healing VLUs55. Overexpression of miR-21 and miR-130a, the most induced miRs, led to suppression of epithelialization in an acute human skin ex vivo wound model. Induction of miR-21 inhibited granulation tissue formation, reduced epithelialization, and prolonged inflammation in an in vivo rat wound model. The mechanism of miR-21 and miR-130 in healing inhibition was mediated by the inhibition of leptin receptor and early growth response factor 355. Moreover, we found miR-21-5p upregulated in fibroblasts isolated from DFUs, contributing to reduced cell proliferation and migration, and induced cell senescence in DFUs56. On the other hand, another member of the miR-21 family, miR-21-3p, promoted fibroblast proliferation and wound closure in a diabetic murine wound model57.
We also identified miR-34a-5p and miR-145-5p upregulation in fibroblasts isolated from DFUs56. Upregulation of miR-34a-5p and miR-145-5p together with suppression of its targets contributes to impaired cell proliferation and migration, as well as induced differentiation and cell senescence in DFU fibroblasts56. In line with this, the miR-34 family was found upregulated in VLUs where it enhanced inflammation and suppressed healing58. The underlying mechanism of miR-34 activity is mediated through LGR4 (leucine rich repeat containing G protein-coupled receptor 4) and silencing of NF-κBsignaling58.
A study by Li et al.59 showed that the miR-17~92 cluster (miR-17, miR-18a, miR-19a, miR-19b, and miR-20a) is upregulated during acute wound healing and downregulated in VLUs, DFUs and pressure ulcers. Knockdown of the miR-17~92 cluster led to impaired wound closure in the murine model under normal and diabetic conditions. Specifically, suppression of miR-19a/b and miR-20a contributed to increased inflammation through toll like receptor-3 (TLR3) mediated NF-κBsignaling59. A member of the same miR-17~92 cluster, miR-92a, has been found to inhibit angiogenesis60,61. A synthetic miR-92a inhibitor, shown to promote angiogenesis in both diabetic and non-diabetic wound models, is currently under clinical investigation as a novel wound healing therapeutic62. miR-132 is found suppressed in DFUs and a diabetic murine model63. Overexpression of miR-132 accelerated re-epithelialization of human ex vivo skin wounds, increased murine wound closure in vivo, and suppressed inflammation in part through inhibition of NF-κBsignaling63.
We have shown that miR-15b-5p is upregulated in DFUs, where it suppresses DNA repair and inflammatory response through downregulation of multiple target genes including IKBKB, WEE1, FGF2, RAD50, MSH2, and KIT64. Moreover, infection of acute human wounds with Staphylococcus aureus, a frequent colonizer of DFUs, triggered miR-15b-5p expression and consequently suppressed target genes involved in inflammation and DNA repair, leading to the accumulation of double strand DNA breaks64. In addition, miR-15b was found upregulated in diabetic murine wounds where it was associated with impaired angiogenic response65. A majority of chronic wounds are colonized with bacteria that form biofilm66,67. Wound biofilm infection led to induction of miR-146a and -106, and downregulation of their targets ZO-1 and ZO-2 (Zonula Occludens Protein-1 and -2) resulting in failed cutaneous barrier repair and increased transepithelial water loss even upon wound closure66.
In addition to tissue miRs, recent research focused on identifying circulating miRs as potential diagnostic and therapeutic targets (Table 1). miR-15a-3p was found upregulated in exosomes isolated from the blood of DFU patients (DFU-exo)68. Treatment of murine wounds with DFU-exo impaired wound closure, while co-treatment with antagomiR-15a-3p accelerated wound closure. The mechanism of miR-15a-3p activity is through suppression of NADPH oxidase 5 and subsequent reduction of reactive oxygen species release68.
miR-24 was found downregulated in the peripheral plasma from patients with DFUs69. This miR was significantly decreased in DFUs that developed osteomyelitis compared to DFUs without osteomyelitis. Further, miR-24 was negatively correlated with amputation rate in DFUs and positively correlated with healing rate, with low miR-24 expression levels serving as an independent risk factor for DFUs69.
A study by Dangwal et al.70 identified miR-191 and miR-200b upregulation in plasma samples from patients with DM and chronic wounds. Increased circulating levels of miR-191 and miR-200b correlated with inflammatory markers and chronic wound size. Overexpression of both miRNAs inhibited tube formation and migration of endothelial cells. miR-191 secreted by vascular endothelial cells was either uptaken by dermal endothelial cells or by dermal fibroblasts, leading to suppression of angiogenesis and fibroblast migration, respectively70. Moreover, miR-200b is upregulated in endothelial tissue elements of the DFU wound edge, whereas its promotor is hypomethylated71. Treatment of diabetic murine wounds with S-adenosyl-L-methionine (SAM) hypermethylated the miR-200b promotor, down-regulated miR-200b expression, and consequently improved vascularization and closure of murine wounds71. A study by Wang et al.34 identified miR-129 and miR-335 downregulation in both serum and tissue samples from patients with DFUs. A direct target of both miRs is Specificity Protein-1 (SP1) gene which was found upregulated in DFUs. SP1 binds directly to the MMP-9 promoter and induces its expression. Ectopic overexpression of miR-129 and miR-335 accelerated wound closure, and downregulated SP1 and MMP-9 expression in a diabetic rat model34.
miRs are not only important post-transcriptional epigenetic regulators of cellular response in acute and chronic wounds, but their expression can also be regulated by epigenetic modifications, including DNA methylation and histone modifications, and vice versa in which miRs can regulate other epigenetic regulators72,73.
1.3. Circular RNAs in wound healing
CircRNAs are a class of non-coding RNA molecules with a covalent closed continuous loop lacking a poly-adenylated tail74,75. Unlike mRNAs that are transcribed in a linear mode, circRNAs are produced by a non-canonical splicing termed “backsplicing” where 3′ and 5′ ends of an RNA molecule are joined together74. Until recently, circular RNA isoforms were thought to be non-functional consequences of rare mistakes in the splicing machinery76. However, circRNAs are present and conserved across most organisms, supporting an evolutionary mechanism for generating circular transcripts77–82.
CircRNAs show high stability and specificity74,75. Their stability is attributed to the unique closed loop structure; by evading RNA degradation machineries that recognize linear RNA ends, circRNAs can accumulate inside and outside of the cell80,83–86. The average half-life of circRNAs in the cell is over 48 h, well above that of linear mRNAs (~10 h)84. circRNAs have tissue-specific and developmental stage-specific expression patterns82,87–89. The best-described role of circRNAs is their activity as sponges for miRNAs. By binding and sequestering transcriptionally inhibitory miRs, they “block the blockers,” introducing a new level of gene expression regulation77,83. In contrast to other miR sponges (e.g. competing endogenous RNAs (ceRNAs); pseudogenes90–92), circular RNAs are more abundant and contain more miRNA binding sites, with increased potency. CircRNAs can also act as protein sponges, enhance protein function, mediate protein scaffolding and recruitment, and function as templates for translation74.
circRNAs are involved in development and progression of several human diseases including DM, cancers, neurological disorders, cardiovascular diseases, and chronic inflammatory diseases85,93–96. A growing body of evidence implicates circRNA in the pathogenesis of skin diseases97–103, including impaired wound healing73,97,104–106. Earlier studies modeled circRNA roles in chronic wound healing by employing a bioinformatics-based approach to highlight studies that confirmed the role and function of circRNA in chronic wounds in vivo.
Work by Yang et al.73 laid the groundwork for analysis of circRNAs in wound healing by showing that circ-Amotl1 accelerates healing in a murine wound model. Additionally, overexpression of circ-Amotl1 increased proliferation and migration of murine fibroblasts73. The mechanism of circ-Amotl1 pro-healing activity is mediated by the STAT3-DNMT3-miR-17-5p axis. Upregulation of circ-Amotl1 was accompanied by an increased level of STAT3 (signal transducer and activator of transcription 3) and its nuclear translocation. Once in the nucleus, STAT3 binds to the DNMT3 (DNA methyltransferase 3) promoter and increases DNMT3 expression. DNMT3 methylated the promotor of miR-17-5p, leading to suppression of miR-17-5p expression, subsequent up-regulation of its targets STAT3, DNMT3, and fibronectin, and resulting in promotion of cell proliferation, migration, adhesion, survival and wound closure73. This regulatory loop highlights the interaction of non-coding RNAs and DNA methylation in regulation of the wound healing process.
Specific circRNA were also found elevated with keloid scars107, namely hsa_circ_0057452, hsa_circ_0007482, hsa_circ_0020792, hsa_circ_0057342, and hsa_circ_0043688. circRNA-miRNA interaction networks show that hsa_circRNA_0057342 and hsa_circ_0020792 can sponge miR-29a, -23a-5p and -1976. While it is known that miR-23a and -29a have important functions in keloids, by sponging them, hsa_circ_0020792 and hsa_circRNA_0057342 may participate in keloid development107.
A recent study by Wang et al.104 associated induction of hsa_circ_0084443 with poor wound healing in DFUs. Hsa_circ_0084443 was found downregulated during normal wound healing, in contrast to DFUs. Overexpression of hsa_circ_0084443 increased proliferation and suppressed migration of human keratinocytes, most likely through modulation of PI3K, EFGR, and ERK signaling pathways. Moreover, hsa_circ_0084443 regulated expression of HBEGF (heparin binding EGF like growth factor) and HIF-1α (hypoxia inducible factor 1 subunit alpha)104.
Given its abundance, evolutionary conservation, and miRNA regulatory function, further characterization of circRNAs can expand our understanding of regulation of the wound healing process, as well as the role of circRNA in pathogenesis of wound healing disorders. Future research focusing on identification and functional characterization of circRNA holds a potential for novel “sponge-based” treatments for various cutaneous diseases associated with aberrant miRNA expression. Furthermore, due to their high stability both circRNA and miRs have been considered as biomarkers for various disorders including non-healing ulcers35,108.
2. Histone Modifications and Chromatin Remodeling in Wound Healing
Within the cell, DNA is packaged in a hierarchical structure with the base unit of a double-stranded DNA helix. Approximately 146 base pairs of DNA are wrapped around a hetero-octameric histone (H2A, H2B, H3 and H4) protein complex and together the DNA-histone complexes are termed nucleosomes109. Nucleosomes are interspersed along the DNA with short sequences between them allowing for the interactions between separate nucleosomes and subsequent chromatin fiber formation. Chromatin is differentially packed from compact (heterochromatin) to loose (euchromatin) structures which serves as a form of gene regulation, as loosely packaged DNA is more accessible for protein binding and transcription109. To modify DNA packaging structure, cells utilize post-translational modification of specific histone sites, generally the N-terminal or C-terminal histone tail regions, as well as large ATP-dependent protein complexes to modify nucleosome positioning in order to activate or repress gene expression110,111.
A variety of histone modifiers have been identified, including acetylation, methylation, phosphorylation, and ubiquitination110. Generally, histone methylation and ubiquitination can be associated with gene activation and repression, while histone acetylation is typically only associated with gene activation. In particular, modifications on Lysine (K) such as H3K4me3, H3K36me3, H3K79me3, and H2BK120ub1 are found on activated genes110, while common repression-associated modifications include H3K9me3, H3K27me3, and H2AK119ub1110. In addition, phosphorylation of serine, threonine and/or tyrosine histone residues has important roles in DNA damage repair, mitosis, apoptosis, and transcriptional activation112. Ubiquitylation of H2A and H2B are also associated with DNA damage response and transcriptional repression113. Additionally, distinct families of chromatin remodelers have been identified including SWI/SNF, ISWI, CHD, and INO80, with all families containing DNA-dependent ATPase domains111. Though functional roles for histone modifications have been demonstrated in many pathophysiologic contexts, few studies have examined histone modifications and chromatin remodeling in the skin and their consequences in wound healing5. Here we review current research as demonstrated in different cell lineages within wounded skin, with a focus on immune cells, keratinocytes, and fibroblasts, as well as outline the role of histone modification in angiogenesis (Table 2).
Table 2. The role of histone modifications in wound healing.
HDAC= Histone Deacetylase; HAC = Histone Acetyltransferase; HMT=Histone methyltransferase; HDM = Histone demethylase N/A= function or target not available.
| Enzyme | Target modification | Effect on wound healing | Dysregulation in pathologic conditions | Ref. |
|---|---|---|---|---|
| Macrophages | ||||
| Class I HDACs (HDAC1, 2, 3, 8) | H4K16 | M1 macrophage polarization; inhibition improves healing | Induced HDAC3 in DM and HDAC1 in DFU; suppression of HDAC2/ HDAC8 in DFU | 127–130 |
| HDAC6 | cytoplasmic proteins | Pro-inflammatory | Elevated in diabetic mice; inhibition improves healing | 133,134 |
| MOF (HAC) | H4K16ac | Pro-inflammatory | Elevated in diabetic mouse models; inhibition improves healing | 135 |
| Setdb2 (HMT) | H3K9me3 | Anti-inflammatory | Decreased in patients with type 2 DM | 136 |
| MLL1 (HMT) | H3K4me3 | Pro-inflammatory; inhibits healing | Elevated in monocytes of patients with type 2 DM | 137–139 |
| Ash1l (HMT) | H3K4me3, H3K36me3 | Anti-inflammatory | Human genetic polymorphisms associated with SLE/RA and RA | 140,141,226,227 |
| Jmjd3 (HDM) | H3K27, H3K4 | Pro-inflammatory, promotes M2 polarization | Increased in DIO mice and type 2 DM; inhibition improves healing | 142–145 |
| Neutrophils | ||||
| Peptidylarginine deiminase 4 (PAD-4) | H3 citrullination | NET formation; inhibition improves healing | Elevated PAD4 in patients with DM increased NETosis in DFU | 114,115,117 |
| unknown | H4 acetylation | NET formation (low levels), and neutrophil apoptosis (high levels) | 119–122 | |
| Keratinocytes | ||||
| HDAC2 | H4K16 | Inhibits wound healing | Downregulated in DFU | 128,130,168,228 |
| Sirtuins aka Class III HDACs | H3K9 | Improves proliferation&healing | N/A | 168 |
| PCAF (HAC) | Rb-acH3K14ac/K9ac/K8ac | Promotes keratinocyte differentiation & improves healing | Increased in patients with Type 1 or 2 DM | 163–166 |
| Ash1l (HMT) | H3K4me3, H3K36me3 | Induces keratinocyte terminal differentiation | Aged Ash1l knockout mice show epidermal hyperplasia; deficiency impairs wound healing | 172 |
| Jmjd3 (HDM) | H3K27, H3K4 | Promotes keratinocyte migration and differentiation | Increased in DIO mice and patients with type 2 DM | 142,143,169–171 |
| Brg1 | SWI2/SNF2 remodeling complex | Induces keratinocyte terminal differentiation & improves healing | N/A | 148,175 |
| Mi-2β or Chd4 | nucleosome remodeling complex | Inhibits keratinocyte differentiation and proliferation | Deficiency impairs barrier repair | 174 |
| Fibroblasts | ||||
| HDAC1 | Fli1 & COL1A2 | Regulation of collagen production | Upregulated in DFU | 130,160 |
| HDAC 4, 6, and 8 | N/A | Promote myofibroblast differentiation; HDAC6 delays diabetic murine wound healing | HDAC4 - upregulated in DFU HDAC8 - downregulated in DFU HDAC6 - upregulated in diabetic mouse |
130,134,158 |
2.1. Histone modification in neutrophils
Neutrophils are recruited to the wound site during the early inflammatory phase and play an important role in first-line defense of clearing foreign debris and bacteria. One mechanism of extracellular bacterial killing is producing neutrophil extracellular traps (NETs), an extracellular matrix involving decondensed chromatin lined with cytotoxic proteins114. During NET formation and release, known as NETosis, activated neutrophils undergo histone citrullination by peptidylarginine deiminase 4 (PAD4) to induce chromatin decondensation114,115. Therefore, commonly used markers for NETosis are citrullinated H3 and PAD4. NETs, however, can damage the surrounding host tissue which may impair wound healing and contribute to the progression of many inflammatory diseases116. DM primes neutrophils to undergo NETosis114, with elevated PAD4 observed in neutrophils of diabetic patients114 and NETs overproduction in human DFU115,117. Interestingly, increased level of NET components in the wound corresponds with infection and worsened state of the DFU115, while healing improved after inhibition of NETs in diabetic murine models114,117,118. PAD4 deficiency also improved wound repair in normoglycemic mice114, suggesting a role of NETosis in delaying both diabetic and normal wound healing processes (Table 2).
Another form of histone modification in NETosis besides citrullination is acetylation, notably of H4. Histone acetylation also facilitates chromatin decondensation, promoting baseline NETosis in human neutrophils in addition to both NADPH oxidase (NOX)-dependent and independent NETosis pathways119. While low concentrations of histone deacetylase (HDAC) inhibitors promote NETosis, interestingly higher concentrations promote neutrophil apoptosis120,121. Enriched regions of H4K16 acetylation are associated with DNA damage during neutrophil apoptosis122. The shifts between NETosis and apoptosis suggest that H4 acetylation plays a role in regulating balance between the two processes. Aside from NETosis, acetylation can also modulate other inflammatory functions as HDAC11 represses expression of pro-inflammatory TNFα and IL-6123.
2.2. Histone modification in macrophages
Macrophages are essential to the initiation and resolution of inflammatory response during wound healing1. The transition from inflammation to proliferation in normal acute wound healing corresponds with a shift in macrophage polarization from the classically activated M1 phenotype, characterized by pro-inflammatory cytokines and nitric oxide production, to alternatively activated M2 phenotype, characterized by markers of inflammatory resolution124–126. Deregulated activation and polarization of wound macrophages with predominance of the M1 phenotype can result in persistent inflammation and impaired host defense, observed in the type 2 DM patient population commonly affected with chronic ulcers124. Advances in epigenetics have revealed multiple chromatin remodeling mechanisms involved in regulating transcription of inflammatory genes affecting macrophage polarization and antimicrobial properties important for successful wound healing (Table 2).
In general, histone acetyltransferase (HAT) and deacetylases (HDAC) promote pro-inflammatory monocyte differentiation and macrophage phenotypes. During development of myeloid progenitors, HDACs are required for differentiation into pro-inflammatory monocytes characterized by high Ly6C markers. Upon differentiation, HDACs further promote the M1 phenotype, while class I-IIa HDAC inhibitor trichostatin A promotes elongated shape and potential to transition into a M2 phenotype through chromatin changes specifically involving H4K16127,128. Trichostatin A topically added to murine wounds mirrored the behavior observed in vitro resulting in enhanced wound closure128. Class I HDACs downregulate myeloid plasticity and favor differentiation into “pro-inflammatory” M1 macrophages, while inhibition renews macrophage plasticity128. But HDACs do not all have the same function, even within the same class, as gene profiling of peripheral blood monocytes (PBMCs) in patients with type 2 DM and DFUs demonstrated differential regulation of HDACs in the context of increased pro-inflammatory mediators129,130. Among DFU patients, significant upregulation of class I HDACs 1, 3 and downregulation of class I HDACs 2 and 8 were observed130 while genetic variations of HDAC3 were associated with an increased prevalence of type 2 DM in certain populations131. Unlike other HDACs that are localized in the nucleus, Class IIb HDAC6 acts on cytoplasmic proteins such as microtubules to regulate cytoskeletal dynamics. HDAC6 facilitates lipopolysaccharide (LPS)-induced macrophage activation132, enhances monocyte/macrophage infiltration into tissue, and promotes phagocytic capacity of peritoneal macrophages133. However, HDAC6 exhibited sustained overexpression in the wounds of diabetic mice134. Selective inhibition reduced pro-inflammatory IL-1β and increased anti-inflammatory IL-10 expression in LPS or high glucose stimulated macrophages and accelerated wound healing in diabetic mice134. The histone acetyltransferase “Males absent on the first” (MOF) also targets H4K16 to promote an inflammatory profile in wound macrophages135. TNFα-mediated MOF activity increases NF-κB–mediated transcription of inflammatory genes, peaking in the normal wound healing process, and is comparatively elevated in wound macrophages of diet-induced obese mice and db/db mice135. TNF-α inhibition via etanercept (an FDA-approved inhibitor) reduced MOF levels and improved outcomes of diabetic wound healing135. The overlapping pro-inflammatory results between histone acetyltransferase and deacetylases could be due to action on different genes promoters involved with the H4K16 site: HAT to promote pro-inflammatory genes and HDAC to repress anti-inflammatory genes.
Histone methyltransferases also play an important role in regulating the transition of macrophages from an inflammatory to an anti-inflammatory phenotype. Deletion of histone methyltransferase Setdb2 impairs the transition of macrophages from an inflammatory M1 to anti-inflammatory M2 phenotype, leading to increased NF-κB-mediated transcription of inflammatory cytokines IL-1β, TNF-α, and antimicrobial nitric oxide synthase (NOS2)136. Another histone methyltransferase, mixed-lineage leukemia-1 (MLL1), also promotes a pro-inflammatory phenotype137 and increases NF-κB-mediated transcription of inflammatory cytokines via trimethylation of H3K4, correlating with impaired wound healing in mice138. Peripheral blood monocytes of patients with type 2 DM had elevated levels of MLL-1 compared to control subjects, suggesting a predisposed hyperinflammatory systemic state even before the occurrence of a wound138. MLL1 additionally decreases antimicrobial activity of macrophages. MLL1-deficient macrophages in vitro exhibit increased phagocytosis and bacterial killing activity of Group A Streptococcus, and upregulation of anti-viral related genes such as interferon-induced GTP-binding proteins139. Alternatively, Ash1l histone methyltransferase activity on H3K4 at the TNF-α inducible protein 3 (Tnfaip3) promoter induces an anti-inflammatory phenotype by suppressing NF-κB-mediated IL-6 and TNF-α production in TLR-triggered macrophages140,141. Histone demethylase Jmjd3 removes repressive methylation of H3K27, which promotes NF-κB -mediated transcription of inflammatory genes lL-1β and IL-12142,143. JMJD3 is also important for activation of LPS-induced inflammatory genes, independent of H3K27me3, by targeting H3K4me3 instead. However, JMJD3 is also involved in IL-4 associated activation of M2 polarization in bone marrow–derived macrophages144,145, suggesting the demethylase may differentially regulate macrophage phenotype based on environmental context. Pre-treatment of Jmjd3 specific inhibitor (GSK□J4) reduced inflammatory cytokine expression in circulating monocytes and wound macrophages and improved wound healing in diabetic mice143.
The ATP-dependent chromatin remodeling SWI/SNF complex (also termed BAF) also participates in macrophage development. SWI/SNF interacts with histone acetyltransferase p300 to regulate H3K27ac and control genes important to cell development and differentiation146. The complex uses two alternative ATP-dependent enzymes, brahma-related gene 1 (BRG1) and brahma (BRM), that recognize and replace nucleosomes marked by the coordinated activity between P300 and HDAC1147. Of the two enzymes, chromatin remodeler Brg1 has also been shown to promote arginase-1 gene (Arg1) transcription in M2 polarized macrophages and improve wound healing during treatment with topical all-trans retinoic acid and IL-4148,149. In summary, chromatin dynamics governed by histone modifying enzymes has been extensively studied in macrophages and regulate multiple aspects of macrophage development and effector functions during wound healing.
2.3. Histone modification in fibroblasts
During the remodeling phase of wound healing, granulation tissue with a high ratio of collagen III is replaced by mature extracellular matrix (ECM) enriched with collagen I150. Other ECM components include fibronectin, elastin, laminins, proteoglycans, hyaluronic acid, glycoproteins, and matricellular proteins. These ECM components are primarily synthesized by fibroblasts, while epithelial, endothelial, and immune cells contribute as well. As the tensile strength and diameter of collagen fibers increase, fibroblasts are induced to form myofibroblasts151,152. Alpha-smooth muscle actin (αSMA) is produced by myofibroblasts, providing contractility that aids in wound healing153,154. While stimulating fibroblasts may be of interest in wound healing, care must be taken to prevent fibroblast over-activation and fibrotic response associated with impaired wound healing155–157. A critical step for myofibroblast differentiation is regulation by TGF-β1 signaling. Silencing of HDAC 4, 6, and 8 impaired TGF-β1 induction of α-SMA and thus fibroblast differentiation158,159, while suppression of HDAC1 increased collagen production160 (Table 2). Increasing HDAC1 results in the deacetylation of Fli1 (friend leukemia integration 1 transcription factor) and a subsequent rise in Fli1 DNA binding and inhibition of Collagen Type I Alpha 2 (COL1A2). The Fli1-HDAC1 complex release from the COL1A2 promoter is also mediated by TGF-β160. While HDAC inhibitors cause an accumulation of acetylated histones and have been shown to inhibit the growth of keratinocytes, fibroblasts were not growth inhibited161. In a streptozotocin induced diabetic murine model, Guo et. al. found that lncRNA H19 promotes H3K4me3 methylation, resulting in activation of the HIF-1α signaling pathway by recruiting EZH2162. By modulation of histone methylation, lncRNA enhanced wound healing in diabetic mice162. Using primary fibroblasts from DFUs, Park et. al. analyzed DNA methylation patterns compared to site and age-matched normal foot fibroblasts37. Overall, they found that DFU derived fibroblasts had lower global DNA methylation on genes associated with wound healing37. Further studies are needed to decipher the role of epigenetic modifications in fibroblasts from other types of chronic wounds.
2.4. Histone modifications in keratinocytes
The later phases of wound healing involve keratinocytes as key players of successful wound closure1,2. Regulation of keratinocytes proliferation, migration, and differentiation during wound healing is modulated by multiple growth factors and cytokines, non-coding RNAs as well as histone and chromatin modifications5. Histone acetylation of different target sites promote keratinocyte migration and terminal differentiation for overall accelerated wound repair. Activity of acetyltransferase P300/CBP-associated factor (PCAF) on retinoblastoma protein (Rb), an important regulator for cell differentiation, is required for normal keratinocyte differentiation163. Additional acetylation targets for PCAF are H3 and H4164. Topical treatment of a PCAF activator accelerated wound closure in mice, in association with increased H4 acetylation165. High glucose conditions can also increase PCAF action on inflammatory genes, with increased H3K9 acetylation at TNF-α promoters of human blood monocytes in type 1 and 2 DM166. Meanwhile, photobiomodulation therapy-induced histone 3 (H3) acetylation and corresponding NF-κB expression stimulate keratinocyte migration in the early stages of healing while decreased H3 acetylation and NF-κB expression accelerate keratinocyte terminal differentiation in the later stages of healing167. Histone deacetylation can impact keratinocyte activity differently depending on the type of HDAC activated. Class I HDAC2 inhibits expression of growth factors important to keratinocyte proliferation and wound healing, such as IGF-I (insulin growth factor I, FGF-10 (fibroblast growth factor) 10, and EGF (epidermal growth factor)168. Class III HDACs 1-3 (Sirtuins), on the other hand, negatively regulate class I HDACs and hence promote keratinocyte proliferation through increased endothelial cell NO synthesis. When treated with class I HDAC inhibitors or sirtuin activators, mice exhibited increased keratinocyte proliferation and improved wound healing168 (Table 2).
In addition to activity for macrophages, H3K27 demethylase JMJD3 interacts with NF-κB at the wound edge to increase expression of genes encoding inflammatory cytokines and growth factors important to keratinocyte function and wound closure169. JMJD3 activity promotes keratinocyte migration through Notch1 activation169,170 and is also important for keratinocyte differentiation171. Histone methylase Ash1l also promotes wound healing through regulation of keratinocyte proliferation and induced terminal differentiation172. Ash1l trimethylation of H3K36 antagonizes Polycomb group-mediated H3K27 trimethylation, which is involved in epidermal stem cell proliferation and c-myc activation in tumor cells. Ash1l knockdown caused delayed re-epithelialization upon wounding despite hyperproliferation of keratinocytes while leading to epidermal hyperplasia in aged mice, highlighting its role in maintaining epidermal homeostasis172. Using a murine wound model, Shaw and Martin showed that Polycomb group proteins Eed, Ezh2 and Suz12 are down regulated, while finding demethylases Jmjd3 and Utx upregulated during wound healing and an overall reduction of H3K27me3 in the epidermis of healing wounds173.
Aside from histone modification, chromatin remodelers also play a role in regulating keratinocyte functions important in wound healing. Nucleosome remodeler Mi-2β (also known as Chd4) works with HDAC1 and HDAC2 as part of nucleosome remodeling and deacetylase (NuRD) complex and helps to inhibit keratinocyte differentiation and proliferation174. Through H3K27Ac modulation, the complex represses Activator protein 1 (AP1)-dependent transcription of stress response genes in keratinocytes, including gene clusters involved with keratinocyte terminal differentiation, cell proliferation and inflammatory cytokines. Disruption of the skin barrier causes a transient reduction of Mi-2β expression, which allows activation of key stress signaling and keratinocyte proliferation/differentiation during barrier repair174. Brahma-related gene 1 (Brg1) in the SWI2/SNF2 nucleosome remodeling complex is required for keratinocyte terminal differentiation175. Brg1 is also involved in control of hair follicle stem cells (SC) during tissue regeneration176. Activation of Brg1 activates Sonic Hedgehog through NF-κB to keep the hair follicle SC reservoir at full capacity176. In contrast, there is an increase in hair follicle SC apoptosis and an inhibition of SC proliferation and re-epithelialization without Polycomb proteins Ezh1 and Ezh2177. Furthermore, blocking hypomethylation at histone H3 K4me3, K9me3, and K27me3 in hair follicle SCs leads to impaired wound healing through upregulated BMP4 expression178. While we recognize that SC are mobilized to switch fate and contribute to barrier repair in response to wounding178–180 when the epidermal SC niche is depleted in chronic VLUs181, the role of epigenetic modification in regulation of the epidermal SC niche has been extensively reviewed5 and is out of the scope of this manuscript. Despite multiple studies focusing on the epigenetic modifications in keratinocytes during acute wound healing, not many studies have evaluated the effects of the chromatin modifications in the pathology of the chronic wound epidermis. We have recently found accumulation of phosphorylated H2AX in the epidermis of DFUs as a result of inhibition of DNA repair genes and accumulation of DNA breaks64. Further studies are needed to determine the cell-specific role of histone modifications associated with wound healing disorders.
3. The role of DNA methylation in wound healing
The role of DNA methylation in the regulation of the normal wound healing response is in early stages of investigation5. It is well established that DNA methylation is associated with silencing of gene expression and occurs via the action of DNA methyltransferases (DNMTs). Typically, the cytosine 5’ position of CpG dinucleotides is the target, leading to the formation of 5-methylcytosine (5mC)182. DNMT1, DNMT3A and DNMT3B are the primary methyltransferase enzymes that maintain patterned methylation in human cells183; DNMT3A and DNMT3B can also participate in de novo methylation, whereby they target previously unmethylated CpG dinucleotides184. The TET dioxygenase family is also responsible for DNA demethylation, oxidizing 5mC to 5-hydroxymethylcytosine (5hmC), which can then be further processed to unmodified cytosine185. The complex process of gene expression depends on both the density of the methylated sites as well as their relative proximity to the promoter region186. Here we review the role of DNA methylation in cellular subtypes involved in wound healing, with particular reference to immune cells, keratinocytes, and fibroblasts, and discuss potential contributions of DNA methylation to cellular malfunctioning in wound healing disorders.
3.1. DNA methylation in inflammatory cells
The specific effects of DNA methylation during the inflammatory phase of wound healing have not yet been fully elucidated, though it is likely to be an important factor in the healing response. For example, it has been shown that macrophage secretion of certain proinflammatory cytokines is mediated by DNA methylation187. Likewise, patterns of glucocorticoid receptor promoter methylation have been found to vary between individuals, which is potentially significant given that glucocorticoids are synthetized in skin and act as moderators of inflammation during cutaneous wound healing188–190. The Peroxisome Proliferator Activated Receptor (PPAR) family of transcription factors also plays a role in the inflammatory phase of wound healing and differentially methylated sites within the PPAR-α promoter in macrophages have been associated with altered inflammatory cytokine expression, though the significance for the wound healing is yet unclear191.
In murine models of diabetes, bone marrow derived stem cells have shown increased levels of DNMT1 and a pro-inflammatory macrophage phenotype. Subsequent DNMT1 reduction led to increased wound healing192,193. Davis et al.194 have recently shown decreased expression of DNMTs 3a and 3b in diet-induced obese and db/db−/− murine wound monocytes/macrophages. TGF-β-regulated miR-29b inhibition of DNMT3 led to hypomethylation and overexpression of Cox-2, which promoted increased Prostaglandin E2 (PGE2) synthesis. Elevation of this Cox-2/PGE2 pathway caused impaired macrophage function, with increased inflammatory cytokine expression and impaired phagocytosis of bacteria. Of note, inhibition of this pathway resulted in decreased inflammatory cytokine production and improved wound healing194.
While neutrophils play a critical role in the early wound healing response, little is specifically known about the role of DNA methylation in neutrophil function. We have recently shown that suppression of FOXM1 in DFU tissue and a diabetic mouse model caused decreased neutrophil and macrophage recruitment to the wound as well as delayed wound healing195. Furthermore, release of neutrophil extracellular traps (NETs) and death by NETosis is found dysregulated in diabetic wound healing, contributing to delayed wound healing in db/db−/− mice115. A recent in vitro study established a relationship between NETosis and DNA methylation, demonstrating increased NETosis via DNMT inhibition196 and warranting further in vivo studies. Taken together, these findings suggest that DNA methylation may be an important epigenetic regulator in the inflammatory phase of wound healing acting through modulation of neutrophils and macrophages.
3.2. DNA methylation in keratinocytes
DNA methylation is also an essential regulator of epidermal homeostasis197. As the skin ages, in concert with accumulated UV radiation exposure, epidermal DNA methylation patterns among individuals change and become more heterogeneous, a process referred to as epigenetic drift. In general, this phenomenon leads to promoter hypermethylation and lamina-associated domain hypomethylation197. As aging has long been associated with impaired wound healing198,199 the role of DNA methylation in the pathogenesis of wound healing disorders can’t be overlooked.
It has been shown that DNMT1 expression plays a critical role in maintaining the undifferentiated state and capacity for self-renewal in epidermal progenitor cells. Relative hypomethylation occurs during the transition to differentiation200. DNMT1 is expressed in the basal epidermis and outer root sheath of hair follicle stem cells. Loss of DNMT1 leads to defects in stem cell proliferation and maintenance200,201. DNA hypermethylation and increased DNMT expression have been associated with wound re-epithelialization and regeneration in proliferating tissue5,202. Luo et al.203 found that corneal keratinocytes in mice show global DNA hypermethylation during corneal epithelial wound healing, which was also associated with increased DNMT1 and DNMT3B expression. Moreover, selective silencing of DNMT1, but not DNMT3B, inhibited corneal wound healing. This effect was mediated by hypermethylation of the promoters for miR-200a and Cdkn2b, with subsequent decreased expression of these genes203. Similar mechanisms may be involved in cutaneous wound healing.
DNA demethylation, a relatively newly recognized phenomenon, is also increasingly recognized as a fundamental process regulating wound healing. Zhang et al.204 found that the demethylating enzyme TET2 mediated increased expression of MMP-9 in human keratinocytes cultured in the presence of AGEs. AGEs induced keratinocyte expression of TET2 in vitro, leading to hypomethylation of the MMP-9 promoter204. Furthermore, Ling et al.205 demonstrated that TNFα-treated keratinocytes showed site-specific DNA demethylation in the MMP-9 promoter, which was correlated with increased MMP-9 expression. Such results may guide future novel therapeutic strategies targeting DNA demethylation in chronic wounds associated with increased levels of MMP-935,108.
3.3. DNA methylation in fibroblasts
The study of fibrotic disorders has also shed some light on the importance of DNA methylation in cutaneous wound healing. In systemic sclerosis (SSc), myofibroblasts show promoter hypermethylation at DKK1 and SFRP1, both Wnt antagonists206. Treatment with 5-aza-2′-deoxycytidine (decitabine), a DNMT inhibitor, reversed this hypermethylation and lessened fibrosis in a mouse model207. RUNX1 and RUNX2, regulators of tissue inhibitor of metalloproteinases 1 (TIMP-1), are also hypomethylated in dermal fibroblasts from patients with SSc208. FLI1, a negative regulator of collagen production and fibrosis, is hypermethylated and suppressed in SSc myofibroblasts209, while inhibition of FLI1 led to excessive production of ECM in normal cells210. Additionally, global methylation patterns of dermal SSc fibroblasts as well as keloid fibroblasts were altered, with a predominance of hypomethylated sites208,211. Expression of αSMA has been shown to be regulated in part via DNA methylation in lung and hepatic tissue212,213. The αSMA gene locus is hypomethylated in a mouse model of early stage liver fibrosis214.
While current efforts were focused on the role of DNA methylation in fibrotic disorders, we also recognize the importance of fibrosis in the pathogenesis of non-healing VLUs157, warranting further investigation on the role of this epigenetic modification associated with the aberrant upregulation of fibrogenic pathways in chronic wounds155,157.
Conclusions and future perspective
The epigenetic landscape emerges as a fine-tuned regulator of cellular responses during wound healing. Given the abundance, stability, evolutionary conservation and regulatory functions of non-coding RNAs, further characterization of circRNAs, miRs, and lncRNAs and their interaction with histone modifications and DNA methylation in wounded skin over time provides new opportunities to predict and ultimately halt the transition of acute wounds into their devastating pathologic counterpart, chronic non-healing wounds (Figure 1). While the field has advanced in identifying epigenetic modifiers as potential therapeutic targets in animal models of delayed wound healing4,5, future efforts focusing on aberrant epigenetic modification in patients affected with chronic wounds will provide directions for identification of novel therapeutic targets. Patients suffering from chronic DFU and VLU are mostly elderly and burdened by comorbidities, including metabolic disorders, diabetes and cardiovascular disease1,108 while it is established that metabolism and epigenetics are intricately linked and work together to influence the ability to respond to aging and injury215,216 . It is also well known that hyperglycemic conditions cause an adverse effect on the DNA 5-hydroxymethylome, while metformin can prevent global changes in 5-hydroxymethylcytosine levels216, potentially offering additional beneficial effects in the diabetic population at risk of DFU. Although various anti-aging compounds including metformin, quercetin, and even aspirin215 have been linked to an improved ‘younger’ chromatin architecture, further studies are needed to elucidate how these drugs target epigenetic networks to affect the cutaneous wound healing process and potentially prevent development of chronic wounds. The utilization of non-coding RNAs as therapeutic targets for wound healing disorders is also rapidly developing60,72, however caution should be taken as the pathology of impaired wound healing results from a complex etiology and malfunction of multiple cell types, often involving more than a single non-coding RNA.
Figure 1.
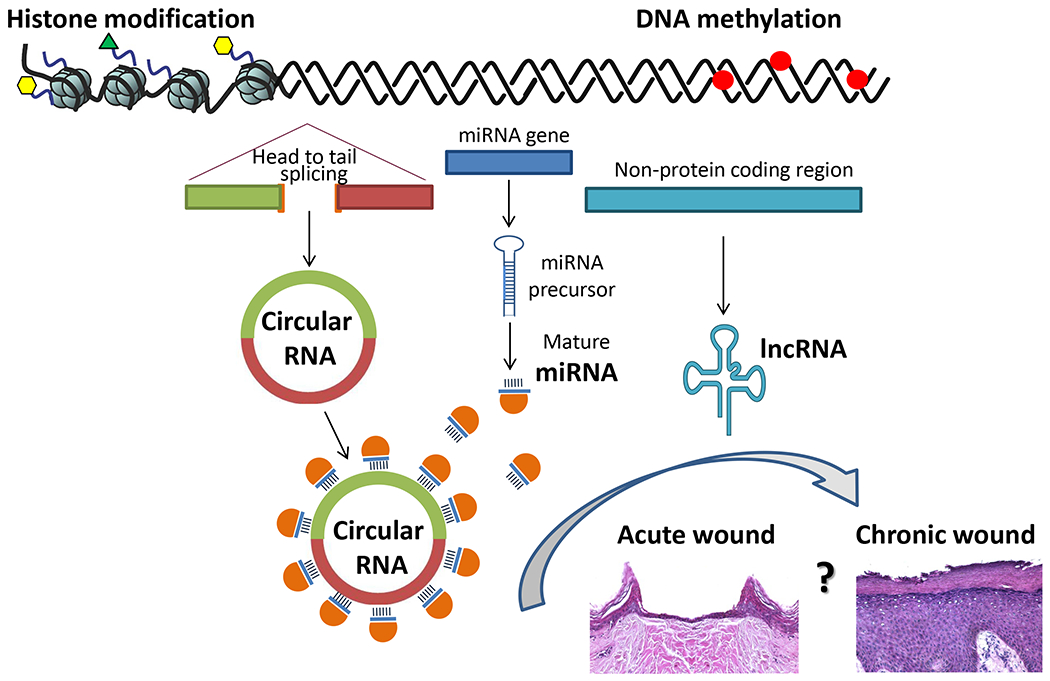
Histone modifications, DNA methylation and non-coding RNAs (circRNAs, miRs and lncRNAs) play a role as epigenetic regulators of wound healing. Their role is balanced in acute wound healing, while deregulation of the epigenetic regulators contributes to molecular pathology of chronic wounds. However, how changes in epigenetic makeup contribute to transition of an acute wound to a chronic wound and vice versa remains to be elucidated.
The molecular features of non-coding RNAs reviewed here support their potential application as novel diagnostic and prognostic markers for wound healing disorders. In particular circRNA and miRs have been considered biomarkers due to their high stability and abundance. Utilization of circRNA/miRs as predictive and diagnostic biomarkers has a potential to improve outcomes, maximize treatment outcomes, and reduce risks associated with chronic ulcers.
Better understanding of the underlying causes of cellular malfunction in chronic wounds, as well as the epigenetic targets of healing interventions, is essential for developing more effective strategies to ameliorate chronic wound disorders and to identify novel therapeutic and diagnostic targets.
ACKNOWLEDGEMENTS
We are grateful to current and past members of our collaborative clinical and research teams as well as our colleagues in the field for their continuous inspiration.
Funding information
This work was supported by NIH R01NR015649, U01DK119085, R01NR01388, R01AR073614, NIH Bench-to-Bedside award made possible by the NIH Office of Clinical Research (all to MTC); U01DK119085-02S1 (to MTC and RCS); and University of Miami SAC-2016-9R1 award (to RCS.)
Footnotes
CONFLICT OF INTEREST
All authors have declared no conflicting interests.
References
- 1.Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: mechanisms, signaling, and translation. Sci Transl Med 2014;6:265sr6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pastar I, Stojadinovic O, Yin NC, Ramirez H, Nusbaum AG, Sawaya A, Patel SB, Khalid L, Isseroff RR, Tomic-Canic M. Epithelialization in Wound Healing: A Comprehensive Review. Adv Wound Care (New Rochelle) 2014;3:445–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brem H, Tomic-Canic M. Cellular and molecular basis of wound healing in diabetes. J Clin Invest 2007;117:1219–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis CJ, Mardaryev AN, Sharov AA, Fessing MY, Botchkarev VA. The Epigenetic Regulation of Wound Healing. Adv Wound Care (New Rochelle) 2014;3:468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis CJ, Stevenson A, Fear MW, Wood FM. A review of epigenetic regulation in wound healing: Implications for the future of wound care. Wound Repair Regen 2020;28:710–718. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee J, Sen CK. MicroRNAs in skin and wound healing. Methods Mol Biol 2013;936:343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li D, Landen NX. MicroRNAs in skin wound healing. Eur J Dermatol 2017;27:12–14. [DOI] [PubMed] [Google Scholar]
- 8.Pastar I, Ramirez H, Stojadinovic O, Brem H, Kirsner RS, Tomic-Canic M. Micro-RNAs: New Regulators of Wound Healing. Surg Technol Int 2011;21:51–60. [PubMed] [Google Scholar]
- 9.Herter EK, Xu Landen N. Non-Coding RNAs: New Players in Skin Wound Healing. Adv Wound Care (New Rochelle) 2017;6:93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol 2019;21:542–551. [DOI] [PubMed] [Google Scholar]
- 12.Fang Y, Fullwood MJ. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics Proteomics Bioinformatics 2016;14:42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013;193:651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J, Mestdagh P. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res 2015;43:D174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature 2012;489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011;25:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 2009;23:1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev 2016;96:1297–1325. [DOI] [PubMed] [Google Scholar]
- 19.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci 2016;73:2491–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152:1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan DC, Wang KC. Long noncoding RNA: significance and potential in skin biology. Cold Spring Harb Perspect Med 2014;4:a015404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hulstaert E, Brochez L, Volders PJ, Vandesompele J, Mestdagh P. Long non-coding RNAs in cutaneous melanoma: clinical perspectives. Oncotarget 2017;8:43470–43480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He X, Ou C, Xiao Y, Han Q, Li H, Zhou S. LncRNAs: key players and novel insights into diabetes mellitus. Oncotarget 2017;8:71325–71341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawaya AP, Pastar I, Stojadinovic O, Lazovic S, Davis SC, Gil J, Kirsner RS, Tomic-Canic M. Topical mevastatin promotes wound healing by inhibiting the transcription factor c-Myc via the glucocorticoid receptor and the long non-coding RNA Gas5. J Biol Chem 2018;293:1439–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J, Zhang L, Liechty C, Zgheib C, Hodges MM, Liechty KW, Xu J. Long Noncoding RNA GAS5 Regulates Macrophage Polarization and Diabetic Wound Healing. J Invest Dermatol 2020;140:1629–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zgheib C, Hodges MM, Hu J, Liechty KW, Xu J. Long non-coding RNA Lethe regulates hyperglycemia-induced reactive oxygen species production in macrophages. PLoS One 2017;12:e0177453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li D, Kular L, Vij M, Herter EK, Li X, Wang A, Chu T, Toma MA, Zhang L, Liapi E, Mota A, Blomqvist L, Gallais Serezal I, Rollman O, Wikstrom JD, Bienko M, Berglund D, Stahle M, Sommar P, Jagodic M, Landen NX. Human skin long noncoding RNA WAKMAR1 regulates wound healing by enhancing keratinocyte migration. Proc Natl Acad Sci U S A 2019;116:9443–9452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herter EK, Li D, Toma MA, Vij M, Li X, Visscher D, Wang A, Chu T, Sommar P, Blomqvist L, Berglund D, Stahle M, Wikstrom JD, Xu Landen N. WAKMAR2, a Long Noncoding RNA Downregulated in Human Chronic Wounds, Modulates Keratinocyte Motility and Production of Inflammatory Chemokines. J Invest Dermatol 2019;139:1373–1384. [DOI] [PubMed] [Google Scholar]
- 29.Jayasuriya R, Dhamodharan U, Karan AN, Anandharaj A, Rajesh K, Ramkumar KM. Role of Nrf2 in MALAT1/ HIF-1alpha loop on the regulation of angiogenesis in diabetic foot ulcer. Free Radic Biol Med 2020;156:168–175. [DOI] [PubMed] [Google Scholar]
- 30.Li B, Luan S, Chen J, Zhou Y, Wang T, Li Z, Fu Y, Zhai A, Bi C. The MSC-Derived Exosomal lncRNA H19 Promotes Wound Healing in Diabetic Foot Ulcers by Upregulating PTEN via MicroRNA-152-3p. Mol Ther Nucleic Acids 2020;19:814–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B, Zhou Y, Chen J, Wang T, Li Z, Fu Y, Zhai A, Bi C. Long noncoding RNA H19 acts as a miR-29b sponge to promote wound healing in diabetic foot ulcer. FASEB J 2021;35:e20526. [DOI] [PubMed] [Google Scholar]
- 32.Hu M, Wu Y, Yang C, Wang X, Wang W, Zhou L, Zeng T, Zhou J, Wang C, Lao G, Yan L, Ren M. Novel Long Noncoding RNA lnc-URIDS Delays Diabetic Wound Healing by Targeting Plod1. Diabetes 2020;69:2144–2156. [DOI] [PubMed] [Google Scholar]
- 33.Zhou L, Ren M, Zeng T, Wang W, Wang X, Hu M, Su S, Sun K, Wang C, Liu J, Yang C, Yan L. TET2-interacting long noncoding RNA promotes active DNA demethylation of the MMP-9 promoter in diabetic wound healing. Cell Death Dis 2019;10:813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W, Yang C, Wang XY, Zhou LY, Lao GJ, Liu D, Wang C, Hu MD, Zeng TT, Yan L, Ren M. MicroRNA-129 and -335 Promote Diabetic Wound Healing by Inhibiting Sp1-Mediated MMP-9 Expression. Diabetes 2018;67:1627–1638. [DOI] [PubMed] [Google Scholar]
- 35.Lindley LE, Stojadinovic O, Pastar I, Tomic-Canic M. Biology and Biomarkers for Wound Healing. Plast Reconstr Surg 2016;138:18S–28S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volkmar M, Dedeurwaerder S, Cunha DA, Ndlovu MN, Defrance M, Deplus R, Calonne E, Volkmar U, Igoillo-Esteve M, Naamane N, Del Guerra S, Masini M, Bugliani M, Marchetti P, Cnop M, Eizirik DL, Fuks F. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J 2012;31:1405–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park LK, Maione AG, Smith A, Gerami-Naini B, Iyer LK, Mooney DJ, Veves A, Garlick JA. Genome-wide DNA methylation analysis identifies a metabolic memory profile in patient-derived diabetic foot ulcer fibroblasts. Epigenetics 2014;9:1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams KT, Garrow TA, Schalinske KL. Type I diabetes leads to tissue-specific DNA hypomethylation in male rats. J Nutr 2008;138:2064–2069. [DOI] [PubMed] [Google Scholar]
- 39.Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol 2019;20:21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics 2010;11:537–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014;42:D68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014;15:509–524. [DOI] [PubMed] [Google Scholar]
- 44.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell 2003;115:787–798. [DOI] [PubMed] [Google Scholar]
- 45.Broughton JP, Lovci MT, Huang JL, Yeo GW, Pasquinelli AE. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol Cell 2016;64:320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vasudevan S Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA 2012;3:311–330. [DOI] [PubMed] [Google Scholar]
- 47.Esteller M Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861–874. [DOI] [PubMed] [Google Scholar]
- 48.Feng J, Xing W, Xie L. Regulatory Roles of MicroRNAs in Diabetes. Int J Mol Sci 2016;17:1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Q, Wu DH, Han L, Deng JW, Zhou L, He R, Lu CJ, Mi QS. Roles of microRNAs in psoriasis: Immunological functions and potential biomarkers. Exp Dermatol 2017;26:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jinnin M Various applications of microRNAs in skin diseases. J Dermatol Sci 2014;74:3–8. [DOI] [PubMed] [Google Scholar]
- 51.Banerjee J, Sen CK. microRNA and Wound Healing. Adv Exp Med Biol 2015;888:291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mulholland EJ, Dunne N, McCarthy HO. MicroRNA as Therapeutic Targets for Chronic Wound Healing. Mol Ther Nucleic Acids 2017;8:46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soliman AM, Das S, Abd Ghafar N, Teoh SL. Role of MicroRNA in Proliferation Phase of Wound Healing. Front Genet 2018;9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fahs F, Bi X, Yu FS, Zhou L, Mi QS. New insights into microRNAs in skin wound healing. IUBMB Life 2015;67:889–896. [DOI] [PubMed] [Google Scholar]
- 55.Pastar I, Khan AA, Stojadinovic O, Lebrun EA, Medina MC, Brem H, Kirsner RS, Jimenez JJ, Leslie C, Tomic-Canic M. Induction of specific microRNAs inhibits cutaneous wound healing. J Biol Chem 2012;287:29324–29335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liang L, Stone RC, Stojadinovic O, Ramirez H, Pastar I, Maione AG, Smith A, Yanez V, Veves A, Kirsner RS, Garlick JA, Tomic-Canic M. Integrative analysis of miRNA and mRNA paired expression profiling of primary fibroblast derived from diabetic foot ulcers reveals multiple impaired cellular functions. Wound Repair Regen 2016;24:943–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu Y, Zhang K, Liu R, Zhang H, Chen D, Yu S, Chen W, Wan S, Zhang Y, Jia Z, Chen R, Ding F. MicroRNA-21-3p accelerates diabetic wound healing in mice by downregulating SPRY1. Aging (Albany NY) 2020;12:15436–15445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu J, Li X, Li D, Ren X, Li Y, Herter EK, Qian M, Toma MA, Wintler AM, Serezal IG, Rollman O, Stahle M, Wikstrom JD, Ye X, Landen NX. MicroRNA-34 Family Enhances Wound Inflammation by Targeting LGR4. J Invest Dermatol 2020;140:465–476 e411. [DOI] [PubMed] [Google Scholar]
- 59.Li D, Peng H, Qu L, Sommar P, Wang A, Chu T, Li X, Bi X, Liu Q, Serezal IG, Rollman O, Lohcharoenkal W, Zheng X, Angelstig SE, Grunler J, Pivarcsi A, Sonkoly E, Catrina SB, Xiao C, Stahle M, Mi QS, Zhou L, Landen NX. miR-19a/b and miR-20a promote wound healing by regulating the inflammatory response of keratinocytes. J Invest Dermatol 2020;141:659–671. [DOI] [PubMed] [Google Scholar]
- 60.Gallant-Behm CL, Piper J, Dickinson BA, Dalby CM, Pestano LA, Jackson AL. A synthetic microRNA-92a inhibitor (MRG-110) accelerates angiogenesis and wound healing in diabetic and nondiabetic wounds. Wound Repair Regen 2018;26:311–323. [DOI] [PubMed] [Google Scholar]
- 61.Lucas T, Schafer F, Muller P, Eming SA, Heckel A, Dimmeler S. Light-inducible antimiR-92a as a therapeutic strategy to promote skin repair in healing-impaired diabetic mice. Nat Commun 2017;8:15162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.U.S. National Library of Medicine ClinicalTrials.gov. Safety, Tolerability, Pharmacokinetics, and Pharmacodynamics of MRG-110 Following Intradermal Injection in Healthy Volunteers. https://ClinicalTrials.gov/show/NCT03603431 Accessed December 15, 2020
- 63.Li X, Li D, Wang A, Chu T, Lohcharoenkal W, Zheng X, Grunler J, Narayanan S, Eliasson S, Herter EK, Wang Y, Ma Y, Ehrstrom M, Eidsmo L, Kasper M, Pivarcsi A, Sonkoly E, Catrina SB, Stahle M, Xu Landen N. MicroRNA-132 with Therapeutic Potential in Chronic Wounds. J Invest Dermatol 2017;137:2630–2638. [DOI] [PubMed] [Google Scholar]
- 64.Ramirez HA, Pastar I, Jozic I, Stojadinovic O, Stone RC, Ojeh N, Gil J, Davis SC, Kirsner RS, Tomic-Canic M. Staphylococcus aureus Triggers Induction of miR-15B-5P to Diminish DNA Repair and Deregulate Inflammatory Response in Diabetic Foot Ulcers. J Invest Dermatol 2018;138:1187–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu J, Zgheib C, Hu J, Wu W, Zhang L, Liechty KW. The role of microRNA-15b in the impaired angiogenesis in diabetic wounds. Wound Repair Regen 2014;22:671–677. [DOI] [PubMed] [Google Scholar]
- 66.Roy S, Elgharably H, Sinha M, Ganesh K, Chaney S, Mann E, Miller C, Khanna S, Bergdall VK, Powell HM, Cook CH, Gordillo GM, Wozniak DJ, Sen CK. Mixed-species biofilm compromises wound healing by disrupting epidermal barrier function. J Pathol 2014;233:331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tomic-Canic M, Burgess JL, O’Neill KE, Strbo N, Pastar I. Skin Microbiota and its Interplay with Wound Healing. Am J Clin Dermatol 2020;21:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiong Y, Chen L, Yu T, Yan C, Zhou W, Cao F, You X, Zhang Y, Sun Y, Liu J, Xue H, Hu Y, Chen D, Mi B, Liu G. Inhibition of circulating exosomal microRNA-15a-3p accelerates diabetic wound repair. Aging (Albany NY) 2020;12:8968–8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X, Tang Y, Jia Z, Zhao X, Chen M. Decreased expression of miR-24 in peripheral plasma of type 2 diabetes mellitus patients associated with diabetic foot ulcer. Wound Repair Regen 2020;28:728–738. [DOI] [PubMed] [Google Scholar]
- 70.Dangwal S, Stratmann B, Bang C, Lorenzen JM, Kumarswamy R, Fiedler J, Falk CS, Scholz CJ, Thum T, Tschoepe D. Impairment of Wound Healing in Patients With Type 2 Diabetes Mellitus Influences Circulating MicroRNA Patterns via Inflammatory Cytokines. Arterioscler Thromb Vasc Biol 2015;35:1480–1488. [DOI] [PubMed] [Google Scholar]
- 71.Singh K, Pal D, Sinha M, Ghatak S, Gnyawali SC, Khanna S, Roy S, Sen CK. Epigenetic Modification of MicroRNA-200b Contributes to Diabetic Vasculopathy. Mol Ther 2017;25:2689–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petkovic M, Sorensen AE, Leal EC, Carvalho E, Dalgaard LT. Mechanistic Actions of microRNAs in Diabetic Wound Healing. Cells 2020;9:2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J, Wu, Gupta S, Yang W, Yang BB. The Circular RNA Interacts with STAT3, Increasing Its Nuclear Translocation and Wound Repair by Modulating Dnmt3a and miR-17 Function. Mol Ther 2017;25:2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675–691. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Z, Yang T, Xiao J. Circular RNAs: Promising Biomarkers for Human Diseases. EBioMedicine 2018;34:267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J 1993;7:155–160. [DOI] [PubMed] [Google Scholar]
- 77.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384–388. [DOI] [PubMed] [Google Scholar]
- 78.Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One 2014;9:e90859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015;10:170–177. [DOI] [PubMed] [Google Scholar]
- 80.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 2014;9:1966–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333–338. [DOI] [PubMed] [Google Scholar]
- 84.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H, Li K, Lai W, Li X, Wang H, Yang J, Chu S, Wang H, Kang C, Qiu Y. Comprehensive circular RNA profiles in plasma reveals that circular RNAs can be used as novel biomarkers for systemic lupus erythematosus. Clin Chim Acta 2018;480:17–25. [DOI] [PubMed] [Google Scholar]
- 86.Hang D, Zhou J, Qin N, Zhou W, Ma H, Jin G, Hu Z, Dai J, Shen H. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med 2018;7:2783–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maass PG, Glazar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, Sauer AV, Toka O, Aiuti A, Luft FC, Rajewsky N. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl) 2017;95:1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Floris G, Zhang L, Follesa P, Sun T. Regulatory Role of Circular RNAs and Neurological Disorders. Mol Neurobiol 2017;54:5156–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014;20:1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011;147:358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Noorbakhsh J, Lang AH, Mehta P. Intrinsic noise of microRNA-regulated genes and the ceRNA hypothesis. PLoS One 2013;8:e72676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fang Y, Wang X, Li W, Han J, Jin J, Su F, Zhang J, Huang W, Xiao F, Pan Q, Zou L. Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus. Int J Mol Med 2018;42:1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hanan M, Soreq H, Kadener S. CircRNAs in the brain. RNA Biol 2017;14:1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 2018;37:555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol 2019;16:503–514. [DOI] [PubMed] [Google Scholar]
- 97.Wu X, Xiao Y, Ma J, Wang A. Circular RNA: A novel potential biomarker for skin diseases. Pharmacol Res 2020;158:104841. [DOI] [PubMed] [Google Scholar]
- 98.Liang J, Wu X, Sun S, Chen P, Liang X, Wang J, Ruan J, Zhang S, Zhang X. Circular RNA expression profile analysis of severe acne by RNA-Seq and bioinformatics. J Eur Acad Dermatol Venereol 2018;32:1986–1992. [DOI] [PubMed] [Google Scholar]
- 99.Moldovan LI, Tsoi LC, Ranjitha U, Hager H, Weidinger S, Gudjonsson JE, Kjems J, Kristensen LS. Characterization of circular RNA transcriptomes in psoriasis and atopic dermatitis reveals disease-specific expression profiles. Exp Dermatol 2020. [DOI] [PubMed] [Google Scholar]
- 100.Moldovan LI, Hansen TB, Veno MT, Okholm TLH, Andersen TL, Hager H, Iversen L, Kjems J, Johansen C, Kristensen LS. High-throughput RNA sequencing from paired lesional- and non-lesional skin reveals major alterations in the psoriasis circRNAome. BMC Med Genomics 2019;12:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Das Mahapatra K, Pasquali L, Sondergaard JN, Lapins J, Nemeth IB, Baltas E, Kemeny L, Homey B, Moldovan LI, Kjems J, Kutter C, Sonkoly E, Kristensen LS, Pivarcsi A. A comprehensive analysis of coding and non-coding transcriptomic changes in cutaneous squamous cell carcinoma. Sci Rep 2020;10:3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hanniford D, Ulloa-Morales A, Karz A, Berzoti-Coelho MG, Moubarak RS, Sanchez-Sendra B, Kloetgen A, Davalos V, Imig J, Wu P, Vasudevaraja V, Argibay D, Lilja K, Tabaglio T, Monteagudo C, Guccione E, Tsirigos A, Osman I, Aifantis I, Hernando E. Epigenetic Silencing of CDR1as Drives IGF2BP3-Mediated Melanoma Invasion and Metastasis. Cancer Cell 2020;37:55–70 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sand M, Bechara FG, Sand D, Gambichler T, Hahn SA, Bromba M, Stockfleth E, Hessam S. Circular RNA expression in basal cell carcinoma. Epigenomics 2016;8:619–632. [DOI] [PubMed] [Google Scholar]
- 104.Wang A, Toma MA, Ma J, Li D, Vij M, Chu T, Wang J, Li X, Xu Landen N. Circular RNA hsa_circ_0084443 Is Upregulated in Diabetic Foot Ulcer and Modulates Keratinocyte Migration and Proliferation. Adv Wound Care (New Rochelle) 2020;9:145–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li M, Wang J, Liu D, Huang H. Highthroughput sequencing reveals differentially expressed lncRNAs and circRNAs, and their associated functional network, in human hypertrophic scars. Mol Med Rep 2018;18:5669–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang J, Wu H, Xiao Z, Dong X. Expression Profiles of lncRNAs and circRNAs in Keloid. Plast Reconstr Surg Glob Open 2019;7:e2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shi J, Yao S, Chen P, Yang Y, Qian M, Han Y, Wang N, Zhao Y, He Y, Lyu L, Lu D. The integrative regulatory network of circRNA and microRNA in keloid scarring. Mol Biol Rep 2020;47:201–209. [DOI] [PubMed] [Google Scholar]
- 108.Pastar I, Wong LL, Egger AN, Tomic-Canic M. Descriptive vs mechanistic scientific approach to study wound healing and its inhibition: Is there a value of translational research involving human subjects? Exp Dermatol 2018;27:551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol 2012;13:436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications - writers that read. EMBO Rep 2015;16:1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem 2009;78:273–304. [DOI] [PubMed] [Google Scholar]
- 112.Rossetto D, Avvakumov N, Cote J. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics 2012;7:1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou W, Wang X, Rosenfeld MG. Histone H2A ubiquitination in transcriptional regulation and DNA damage repair. Int J Biochem Cell Biol 2009;41:12–15. [DOI] [PubMed] [Google Scholar]
- 114.Wong SL, Demers M, Martinod K, Gallant M, Wang Y, Goldfine AB, Kahn CR, Wagner DD. Diabetes primes neutrophils to undergo NETosis, which impairs wound healing. Nat Med 2015;21:815–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fadini GP, Menegazzo L, Rigato M, Scattolini V, Poncina N, Bruttocao A, Ciciliot S, Mammano F, Ciubotaru CD, Brocco E, Marescotti MC, Cappellari R, Arrigoni G, Millioni R, Vigili de Kreutzenberg S, Albiero M, Avogaro A. NETosis Delays Diabetic Wound Healing in Mice and Humans. Diabetes 2016;65:1061–1071. [DOI] [PubMed] [Google Scholar]
- 116.Jorch SK, Kubes P. An emerging role for neutrophil extracellular traps in noninfectious disease. Nat Med 2017;23:279–287. [DOI] [PubMed] [Google Scholar]
- 117.Liu D, Yang P, Gao M, Yu T, Shi Y, Zhang M, Yao M, Liu Y, Zhang X. NLRP3 activation induced by neutrophil extracellular traps sustains inflammatory response in the diabetic wound. Clin Sci (Lond) 2019;133:565–582. [DOI] [PubMed] [Google Scholar]
- 118.Kaur T, Dumoga S, Koul V, Singh N. Modulating neutrophil extracellular traps for wound healing. Biomater Sci 2020;8:3212–3223. [DOI] [PubMed] [Google Scholar]
- 119.Hamam HJ, Khan MA, Palaniyar N. Histone Acetylation Promotes Neutrophil Extracellular Trap Formation. Biomolecules 2019;9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hamam HJ, Palaniyar N. Histone Deacetylase Inhibitors Dose-Dependently Switch Neutrophil Death from NETosis to Apoptosis. Biomolecules 2019;9:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kankaanranta H, Janka-Junttila M, Ilmarinen-Salo P, Ito K, Jalonen U, Ito M, Adcock IM, Moilanen E, Zhang X. Histone deacetylase inhibitors induce apoptosis in human eosinophils and neutrophils. J Inflamm (Lond) 2010;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Urdinguio RG, Lopez V, Bayon GF, Diaz de la Guardia R, Sierra MI, Garcia-Torano E, Perez RF, Garcia MG, Carella A, Pruneda PC, Prieto C, Dmitrijeva M, Santamarina P, Belmonte T, Mangas C, Diaconu E, Ferrero C, Tejedor JR, Fernandez-Morera JL, Bravo C, Bueno C, Sanjuan-Pla A, Rodriguez RM, Suarez-Alvarez B, Lopez-Larrea C, Bernal T, Colado E, Balbin M, Garcia-Suarez O, Chiara MD, Saenz-de-Santa-Maria I, Rodriguez F, Pando-Sandoval A, Rodrigo L, Santos L, Salas A, Vallejo-Diaz J, A CC, Rico D, Hernandez-Lopez I, Vaya A, Ricart JM, Seto E, Sima-Teruel N, Vaquero A, Valledor L, Canal MJ, Pisano D, Grana-Castro O, Thomas T, Voss AK, Menendez P, Villar-Garea A, Deutzmann R, Fernandez AF, Fraga MF. Chromatin regulation by Histone H4 acetylation at Lysine 16 during cell death and differentiation in the myeloid compartment. Nucleic Acids Res 2019;47:5016–5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sahakian E, Chen J, Powers JJ, Chen X, Maharaj K, Deng SL, Achille AN, Lienlaf M, Wang HW, Cheng F, Sodre AL, Distler A, Xing L, Perez-Villarroel P, Wei S, Villagra A, Seto E, Sotomayor EM, Horna P, Pinilla-Ibarz J. Essential role for histone deacetylase 11 (HDAC11) in neutrophil biology. J Leukoc Biol 2017;102:475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nassiri S, Zakeri I, Weingarten MS, Spiller KL. Relative Expression of Proinflammatory and Antiinflammatory Genes Reveals Differences between Healing and Nonhealing Human Chronic Diabetic Foot Ulcers. J Invest Dermatol 2015;135:1700–1703. [DOI] [PubMed] [Google Scholar]
- 125.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol 2011;11:750–761. [DOI] [PubMed] [Google Scholar]
- 126.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, Locati M, Mantovani A, Martinez FO, Mege JL, Mosser DM, Natoli G, Saeij JP, Schultze JL, Shirey KA, Sica A, Suttles J, Udalova I, van Ginderachter JA, Vogel SN, Wynn TA. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 2014;41:14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cabanel M, Brand C, Oliveira-Nunes MC, Cabral-Piccin MP, Lopes MF, Brito JM, de Oliveira FL, El-Cheikh MC, Carneiro K. Epigenetic Control of Macrophage Shape Transition towards an Atypical Elongated Phenotype by Histone Deacetylase Activity. PLoS One 2015;10:e0132984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cabanel M, da Costa TP, El-Cheikh MC, Carneiro K. The epigenome as a putative target for skin repair: the HDAC inhibitor Trichostatin A modulates myeloid progenitor plasticity and behavior and improves wound healing. J Transl Med 2019;17:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sathishkumar C, Prabu P, Balakumar M, Lenin R, Prabhu D, Anjana RM, Mohan V, Balasubramanyam M. Augmentation of histone deacetylase 3 (HDAC3) epigenetic signature at the interface of proinflammation and insulin resistance in patients with type 2 diabetes. Clin Epigenetics 2016;8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Teena R, Dhamodharan U, Ali D, Rajesh K, Ramkumar KM. Gene Expression Profiling of Multiple Histone Deacetylases (HDAC) and Its Correlation with NRF2-Mediated Redox Regulation in the Pathogenesis of Diabetic Foot Ulcers. Biomolecules 2020;10:1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zeng Z, Liao R, Yao Z, Zhou W, Ye P, Zheng X, Li X, Huang Y, Chen S, Chen Q. Three single nucleotide variants of the HDAC gene are associated with type 2 diabetes mellitus in a Chinese population: a community-based case-control study. Gene 2014;533:427–433. [DOI] [PubMed] [Google Scholar]
- 132.Yan B, Xie S, Liu Z, Ran J, Li Y, Wang J, Yang Y, Zhou J, Li D, Liu M. HDAC6 deacetylase activity is critical for lipopolysaccharide-induced activation of macrophages. PLoS One 2014;9:e110718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yan B, Xie S, Liu Y, Liu W, Li D, Liu M, Luo HR, Zhou J. Histone deacetylase 6 modulates macrophage infiltration during inflammation. Theranostics 2018;8:2927–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Karnam K, Sedmaki K, Sharma P, Routholla G, Goli S, Ghosh B, Venuganti VVK, Kulkarni OP. HDAC6 inhibitor accelerates wound healing by inhibiting tubulin mediated IL-1beta secretion in diabetic mice. Biochim Biophys Acta Mol Basis Dis 2020;1866:165903. [DOI] [PubMed] [Google Scholar]
- 135.denDekker AD, Davis FM, Joshi AD, Wolf SJ, Allen R, Lipinski J, Nguyen B, Kirma J, Nycz D, Bermick J, Moore BB, Gudjonsson JE, Kunkel SL, Gallagher KA. TNF-alpha regulates diabetic macrophage function through the histone acetyltransferase MOF. JCI Insight 2020;5:e132306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kimball AS, Davis FM, denDekker A, Joshi AD, Schaller MA, Bermick J, Xing X, Burant CF, Obi AT, Nysz D, Robinson S, Allen R, Lukacs NW, Henke PK, Gudjonsson JE, Moore BB, Kunkel SL, Gallagher KA. The Histone Methyltransferase Setdb2 Modulates Macrophage Phenotype and Uric Acid Production in Diabetic Wound Repair. Immunity 2019;51:258–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kittan NA, Allen RM, Dhaliwal A, Cavassani KA, Schaller M, Gallagher KA, Carson WFt, Mukherjee S, Grembecka J, Cierpicki T, Jarai G, Westwick J, Kunkel SL, Hogaboam CM. Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes. PLoS One 2013;8:e78045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kimball AS, Joshi A, Carson WFt, Boniakowski AE, Schaller M, Allen R, Bermick J, Davis FM, Henke PK, Burant CF, Kunkel SL, Gallagher KA. The Histone Methyltransferase MLL1 Directs Macrophage-Mediated Inflammation in Wound Healing and Is Altered in a Murine Model of Obesity and Type 2 Diabetes. Diabetes 2017;66:2459–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Carson WFt, Cavassani KA, Soares EM, Hirai S, Kittan NA, Schaller MA, Scola MM, Joshi A, Matsukawa A, Aronoff DM, Johnson CN, Dou Y, Gallagher KA, Kunkel SL. The STAT4/MLL1 Epigenetic Axis Regulates the Antimicrobial Functions of Murine Macrophages. J Immunol 2017;199:1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xia M, Liu J, Wu X, Liu S, Li G, Han C, Song L, Li Z, Wang Q, Wang J, Xu T, Cao X. Histone methyltransferase Ash1l suppresses interleukin-6 production and inflammatory autoimmune diseases by inducing the ubiquitin-editing enzyme A20. Immunity 2013;39:470–481. [DOI] [PubMed] [Google Scholar]
- 141.Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science 2010;327:1135–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gallagher KA, Joshi A, Carson WF, Schaller M, Allen R, Mukerjee S, Kittan N, Feldman EL, Henke PK, Hogaboam C, Burant CF, Kunkel SL. Epigenetic changes in bone marrow progenitor cells influence the inflammatory phenotype and alter wound healing in type 2 diabetes. Diabetes 2015;64:1420–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Davis FM, denDekker A, Joshi AD, Wolf SJ, Audu C, Melvin WJ, Mangum K, Riordan MO, Kunkel SL, Gallagher KA. Palmitate-TLR4 signaling regulates the histone demethylase, JMJD3, in macrophages and impairs diabetic wound healing. Eur J Immunol 2020;50:1929–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, Allen RM, Carson WFt, Cavassani KA, Li X, Lukacs NW, Hogaboam CM, Dou Y, Kunkel SL. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 2009;114:3244–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, Honma K, Matsuyama T, Yui K, Tsujimura T, Standley DM, Nakanishi K, Nakai K, Akira S. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol 2010;11:936–944. [DOI] [PubMed] [Google Scholar]
- 146.Alver BH, Kim KH, Lu P, Wang X, Manchester HE, Wang W, Haswell JR, Park PJ, Roberts CW. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun 2017;8:14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pietrzak J, Ploszaj T, Pulaski L, Robaszkiewicz A. EP300-HDAC1-SWI/SNF functional unit defines transcription of some DNA repair enzymes during differentiation of human macrophages. Biochim Biophys Acta Gene Regul Mech 2019;1862:198–208. [DOI] [PubMed] [Google Scholar]
- 148.Lee B, Wu CY, Lin YW, Park SW, Wei LN. Synergistic activation of Arg1 gene by retinoic acid and IL-4 involves chromatin remodeling for transcription initiation and elongation coupling. Nucleic Acids Res 2016;44:7568–7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Campbell L, Saville CR, Murray PJ, Cruickshank SM, Hardman MJ. Local arginase 1 activity is required for cutaneous wound healing. J Invest Dermatol 2013;133:2461–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tracy LE, Minasian RA, Caterson EJ. Extracellular Matrix and Dermal Fibroblast Function in the Healing Wound. Adv Wound Care (New Rochelle) 2016;5:119–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci 2010;123:4195–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hinz B The role of myofibroblasts in wound healing. Curr Res Transl Med 2016;64:171–177. [DOI] [PubMed] [Google Scholar]
- 153.Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol 2007;170:1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pakshir P, Noskovicova N, Lodyga M, Son DO, Schuster R, Goodwin A, Karvonen H, Hinz B. The myofibroblast at a glance. J Cell Sci 2020;133:jcs227900. [DOI] [PubMed] [Google Scholar]
- 155.Stone RC, Chen V, Burgess J, Pannu S, Tomic-Canic M. Genomics of Human Fibrotic Diseases: Disordered Wound Healing Response. Int J Mol Sci 2020;21:8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stone RC, Pastar I, Ojeh N, Chen V, Liu S, Garzon KI, Tomic-Canic M. Epithelial-mesenchymal transition in tissue repair and fibrosis. Cell Tissue Res 2016;365:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Stone RC, Stojadinovic O, Sawaya AP, Glinos GD, Lindley LE, Pastar I, Badiavas E, Tomic-Canic M. A bioengineered living cell construct activates metallothionein/zinc/MMP8 and inhibits TGFbeta to stimulate remodeling of fibrotic venous leg ulcers. Wound Repair Regen 2020;28:164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Glenisson W, Castronovo V, Waltregny D. Histone deacetylase 4 is required for TGFbeta1-induced myofibroblastic differentiation. Biochim Biophys Acta 2007;1773:1572–1582. [DOI] [PubMed] [Google Scholar]
- 159.Jones DL, Haak AJ, Caporarello N, Choi KM, Ye Z, Yan H, Varelas X, Ordog T, Ligresti G, Tschumperlin DJ. TGFbeta-induced fibroblast activation requires persistent and targeted HDAC-mediated gene repression. J Cell Sci 2019;132:jcs233486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Asano Y, Trojanowska M. Fli1 represses transcription of the human alpha2(I) collagen gene by recruitment of the HDAC1/p300 complex. PLoS One 2013;8:e74930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Brinkmann H, Dahler AL, Popa C, Serewko MM, Parsons PG, Gabrielli BG, Burgess AJ, Saunders NA. Histone hyperacetylation induced by histone deacetylase inhibitors is not sufficient to cause growth inhibition in human dermal fibroblasts. J Biol Chem 2001;276:22491–22499. [DOI] [PubMed] [Google Scholar]
- 162.Guo JR, Yin L, Chen YQ, Jin XJ, Zhou X, Zhu NN, Liu XQ, Wei HW, Duan LS. Autologous blood transfusion augments impaired wound healing in diabetic mice by enhancing lncRNA H19 expression via the HIF-1alpha signaling pathway. Cell Commun Signal 2018;16:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pickard A, Wong PP, McCance DJ. Acetylation of Rb by PCAF is required for nuclear localization and keratinocyte differentiation. J Cell Sci 2010;123:3718–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem 1999;274:1189–1192. [DOI] [PubMed] [Google Scholar]
- 165.Spallotta F, Cencioni C, Straino S, Sbardella G, Castellano S, Capogrossi MC, Martelli F, Gaetano C. Enhancement of lysine acetylation accelerates wound repair. Commun Integr Biol 2013;6:e25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Miao F, Gonzalo IG, Lanting L, Natarajan R. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem 2004;279:18091–18097. [DOI] [PubMed] [Google Scholar]
- 167.de Farias Gabriel A, Wagner VP, Correa C, Webber LP, Pilar EFS, Curra M, Carrard VC, Martins MAT, Martins MD. Photobiomodulation therapy modulates epigenetic events and NF-kappaB expression in oral epithelial wound healing. Lasers Med Sci 2019;34:1465–1472. [DOI] [PubMed] [Google Scholar]
- 168.Spallotta F, Cencioni C, Straino S, Nanni S, Rosati J, Artuso S, Manni I, Colussi C, Piaggio G, Martelli F, Valente S, Mai A, Capogrossi MC, Farsetti A, Gaetano C. A nitric oxide-dependent cross-talk between class I and III histone deacetylases accelerates skin repair. J Biol Chem 2013;288:11004–11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Na J, Lee K, Na W, Shin JY, Lee MJ, Yune TY, Lee HK, Jung HS, Kim WS, Ju BG. Histone H3K27 Demethylase JMJD3 in Cooperation with NF-kappaB Regulates Keratinocyte Wound Healing. J Invest Dermatol 2016;136:847–858. [DOI] [PubMed] [Google Scholar]
- 170.Na J, Shin JY, Jeong H, Lee JY, Kim BJ, Kim WS, Yune TY, Ju BG. JMJD3 and NF-kappaB-dependent activation of Notch1 gene is required for keratinocyte migration during skin wound healing. Sci Rep 2017;7:6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev 2008;22:1865–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li G, Ye Z, Shi C, Sun L, Han M, Zhuang Y, Xu T, Zhao S, Wu X. The Histone Methyltransferase Ash1l is Required for Epidermal Homeostasis in Mice. Sci Rep 2017;7:45401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep 2009;10:881–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shibata S, Kashiwagi M, Morgan BA, Georgopoulos K. Functional interactions between Mi-2beta and AP1 complexes control response and recovery from skin barrier disruption. J Exp Med 2020;217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Indra AK, Dupe V, Bornert JM, Messaddeq N, Yaniv M, Mark M, Chambon P, Metzger D. Temporally controlled targeted somatic mutagenesis in embryonic surface ectoderm and fetal epidermal keratinocytes unveils two distinct developmental functions of BRG1 in limb morphogenesis and skin barrier formation. Development 2005;132:4533–4544. [DOI] [PubMed] [Google Scholar]
- 176.Xiong Y, Li W, Shang C, Chen RM, Han P, Yang J, Stankunas K, Wu B, Pan M, Zhou B, Longaker MT, Chang CP. Brg1 governs a positive feedback circuit in the hair follicle for tissue regeneration and repair. Dev Cell 2013;25:169–181. [DOI] [PubMed] [Google Scholar]
- 177.Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 2011;25:485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kang S, Long K, Wang S, Sada A, Tumbar T. Histone H3 K4/9/27 Trimethylation Levels Affect Wound Healing and Stem Cell Dynamics in Adult Skin. Stem Cell Reports 2020;14:34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Adam RC, Yang H, Ge Y, Infarinato NR, Gur-Cohen S, Miao Y, Wang P, Zhao Y, Lu CP, Kim JE, Ko JY, Paik SS, Gronostajski RM, Kim J, Krueger JG, Zheng D, Fuchs E. NFI transcription factors provide chromatin access to maintain stem cell identity while preventing unintended lineage fate choices. Nat Cell Biol 2020;22:640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Avgustinova A, Benitah SA. Epigenetic control of adult stem cell function. Nat Rev Mol Cell Biol 2016;17:643–658. [DOI] [PubMed] [Google Scholar]
- 181.Stojadinovic O, Pastar I, Nusbaum AG, Vukelic S, Krzyzanowska A, Tomic-Canic M. Deregulation of epidermal stem cell niche contributes to pathogenesis of nonhealing venous ulcers. Wound Repair Regen 2014;22:220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Meng H, Cao Y, Qin J, Song X, Zhang Q, Shi Y, Cao L. DNA methylation, its mediators and genome integrity. Int J Biol Sci 2015;11:604–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer 2011;2:607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999;99:247–257. [DOI] [PubMed] [Google Scholar]
- 185.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009;324:930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bian EB, Huang C, Wang H, Wu BM, Zhang L, Lv XW, Li J. DNA methylation: new therapeutic implications for hepatic fibrosis. Cell Signal 2013;25:355–358. [DOI] [PubMed] [Google Scholar]
- 187.Hmadcha A, Bedoya FJ, Sobrino F, Pintado E. Methylation-dependent gene silencing induced by interleukin 1beta via nitric oxide production. J Exp Med 1999;190:1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chen P, Jiang T, Ouyang J, Cui Y, Chen Y. Epigenetic programming of diverse glucocorticoid response and inflammatory/immune-mediated disease. Med Hypotheses 2009;73:657–658. [DOI] [PubMed] [Google Scholar]
- 189.Jozic I, Vukelic S, Stojadinovic O, Liang L, Ramirez HA, Pastar I, Tomic Canic M. Stress Signals, Mediated by Membranous Glucocorticoid Receptor, Activate PLC/PKC/GSK-3beta/beta-catenin Pathway to Inhibit Wound Closure. J Invest Dermatol 2017;137:1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Vukelic S, Stojadinovic O, Pastar I, Rabach M, Krzyzanowska A, Lebrun E, Davis SC, Resnik S, Brem H, Tomic-Canic M. Cortisol synthesis in epidermis is induced by IL-1 and tissue injury. J Biol Chem 2011;286:10265–10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Scotti E, Tontonoz P. Peroxisome proliferator-activated receptor gamma dances with different partners in macrophage and adipocytes. Mol Cell Biol 2010;30:2076–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yan J, Tie G, Wang S, Tutto A, DeMarco N, Khair L, Fazzio TG, Messina LM. Diabetes impairs wound healing by Dnmt1-dependent dysregulation of hematopoietic stem cells differentiation towards macrophages. Nat Commun 2018;9:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.den Dekker A, Davis FM, Kunkel SL, Gallagher KA. Targeting epigenetic mechanisms in diabetic wound healing. Transl Res 2019;204:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Davis FM, Tsoi LC, Wasikowski R, denDekker A, Joshi A, Wilke C, Deng H, Wolf S, Obi A, Huang S, Billi AC, Robinson S, Lipinski J, Melvin WJ, Audu CO, Weidinger S, Kunkel SL, Smith A, Gudjonsson JE, Moore BB, Gallagher KA. Epigenetic regulation of the PGE2 pathway modulates macrophage phenotype in normal and pathologic wound repair. JCI Insight 2020;5:e138443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sawaya AP, Stone RC, Brooks SR, Pastar I, Jozic I, Hasneen K, O'Neill K, Mehdizadeh S, Head CR, Strbo N, Morasso MI, Tomic-Canic M. Deregulated immune cell recruitment orchestrated by FOXM1 impairs human diabetic wound healing. Nat Commun 2020;11:4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yasuda H, Takishita Y, Morita A, Tsutsumi T, Tsuchiya M, Sato EF. DNA demethylation increases NETosis. Arch Biochem Biophys 2020;689:108465. [DOI] [PubMed] [Google Scholar]
- 197.Kohler F, Rodriguez-Paredes M. DNA Methylation in Epidermal Differentiation, Aging, and Cancer. J Invest Dermatol 2020;140:38–47. [DOI] [PubMed] [Google Scholar]
- 198.Eaglstein WH. Wound healing and aging. Dermatol Clin 1986;4:481–484. [PubMed] [Google Scholar]
- 199.Gould L, Abadir P, Brem H, Carter M, Conner-Kerr T, Davidson J, DiPietro L, Falanga V, Fife C, Gardner S, Grice E, Harmon J, Hazzard WR, High KP, Houghton P, Jacobson N, Kirsner RS, Kovacs EJ, Margolis D, McFarland Horne F, Reed MJ, Sullivan DH, Thom S, Tomic-Canic M, Walston J, Whitney J, Williams J, Zieman S, Schmader K. Chronic wound repair and healing in older adults: current status and future research. Wound Repair Regen 2015;23:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 2010;463:563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Li J, Jiang TX, Hughes MW, Wu P, Yu J, Widelitz RB, Fan G, Chuong CM. Progressive alopecia reveals decreasing stem cell activation probability during aging of mice with epidermal deletion of DNA methyltransferase 1. J Invest Dermatol 2012;132:2681–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Aguilar C, Gardiner DM. DNA Methylation Dynamics Regulate the Formation of a Regenerative Wound Epithelium during Axolotl Limb Regeneration. PLoS One 2015;10:e0134791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Luo G, Jing X, Yang S, Peng D, Dong J, Li L, Reinach PS, Yan D. DNA Methylation Regulates Corneal Epithelial Wound Healing by Targeting miR-200a and CDKN2B. Invest Ophthalmol Vis Sci 2019;60:650–660. [DOI] [PubMed] [Google Scholar]
- 204.Zhang J, Yang C, Wang C, Liu D, Lao G, Liang Y, Sun K, Luo H, Tan Q, Ren M, Yan L. AGE-induced keratinocyte MMP-9 expression is linked to TET2-mediated CpG demethylation. Wound Repair Regen 2016;24:489–500. [DOI] [PubMed] [Google Scholar]
- 205.Ling L, Ren M, Yang C, Lao G, Chen L, Luo H, Feng Z, Yan L. Role of site-specific DNA demethylation in TNFalpha-induced MMP9 expression in keratinocytes. J Mol Endocrinol 2013;50:279–290. [DOI] [PubMed] [Google Scholar]
- 206.Lam AP, Gottardi CJ. beta-catenin signaling: a novel mediator of fibrosis and potential therapeutic target. Curr Opin Rheumatol 2011;23:562–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Dees C, Schlottmann I, Funke R, Distler A, Palumbo-Zerr K, Zerr P, Lin NY, Beyer C, Distler O, Schett G, Distler JH. The Wnt antagonists DKK1 and SFRP1 are downregulated by promoter hypermethylation in systemic sclerosis. Ann Rheum Dis 2014;73:1232–1239. [DOI] [PubMed] [Google Scholar]
- 208.Altorok N, Tsou PS, Coit P, Khanna D, Sawalha AH. Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann Rheum Dis 2015;74:1612–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wang Y, Fan PS, Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum 2006;54:2271–2279. [DOI] [PubMed] [Google Scholar]
- 210.Dowson C, O’Reilly S. DNA methylation in fibrosis. Eur J Cell Biol 2016;95:323–330. [DOI] [PubMed] [Google Scholar]
- 211.Jones LR, Young W, Divine G, Datta I, Chen KM, Ozog D, Worsham MJ. Genome-Wide Scan for Methylation Profiles in Keloids. Dis Markers 2015;2015:943176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hu B, Gharaee-Kermani M, Wu Z, Phan SH. Epigenetic regulation of myofibroblast differentiation by DNA methylation. Am J Pathol 2010;177:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Mann J, Chu DC, Maxwell A, Oakley F, Zhu NL, Tsukamoto H, Mann DA. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology 2010;138:705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Komatsu Y, Waku T, Iwasaki N, Ono W, Yamaguchi C, Yanagisawa J. Global analysis of DNA methylation in early-stage liver fibrosis. BMC Med Genomics 2012;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zhang W, Qu J, Liu GH, Belmonte JCI. The ageing epigenome and its rejuvenation. Nat Rev Mol Cell Biol 2020;21:137–150. [DOI] [PubMed] [Google Scholar]
- 216.Wu D, Hu D, Chen H, Shi G, Fetahu IS, Wu F, Rabidou K, Fang R, Tan L, Xu S, Liu H, Argueta C, Zhang L, Mao F, Yan G, Chen J, Dong Z, Lv R, Xu Y, Wang M, Ye Y, Zhang S, Duquette D, Geng S, Yin C, Lian CG, Murphy GF, Adler GK, Garg R, Lynch L, Yang P, Li Y, Lan F, Fan J, Shi Y, Shi YG. Glucose-regulated phosphorylation of TET2 by AMPK reveals a pathway linking diabetes to cancer. Nature 2018;559:637–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Liu X, Wang Y, Zhang X, Zhang X, Guo J, Zhou J, Chai Y, Ma ZL. MicroRNA-296-5p promotes healing of diabetic wound by targeting sodium-glucose transporter 2 (SGLT2). Diabetes Metab Res Rev 2019;35:e3104. [DOI] [PubMed] [Google Scholar]
- 218.Caskey RC, Zgheib C, Morris M, Allukian M, Dorsett-Martin W, Xu J, Wu W, Liechty KW. Dysregulation of collagen production in diabetes following recurrent skin injury: contribution to the development of a chronic wound. Wound Repair Regen 2014;22:515–520. [DOI] [PubMed] [Google Scholar]
- 219.Icli B, Nabzdyk CS, Lujan-Hernandez J, Cahill M, Auster ME, Wara AK, Sun X, Ozdemir D, Giatsidis G, Orgill DP, Feinberg MW. Regulation of impaired angiogenesis in diabetic dermal wound healing by microRNA-26a. J Mol Cell Cardiol 2016;91:151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Xu J, Wu W, Zhang L, Dorset-Martin W, Morris MW, Mitchell ME, Liechty KW. The role of microRNA-146a in the pathogenesis of the diabetic wound-healing impairment: correction with mesenchymal stem cell treatment. Diabetes 2012;61:2906–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Moura J, Sorensen A, Leal EC, Svendsen R, Carvalho L, Willemoes RJ, Jorgensen PT, Jenssen H, Wengel J, Dalgaard LT, Carvalho E. microRNA-155 inhibition restores Fibroblast Growth Factor 7 expression in diabetic skin and decreases wound inflammation. Sci Rep 2019;9:5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ban E, Jeong S, Park M, Kwon H, Park J, Song EJ, Kim A. Accelerated wound healing in diabetic mice by miRNA-497 and its anti-inflammatory activity. Biomed Pharmacother 2020;121:109613. [DOI] [PubMed] [Google Scholar]
- 223.Zhang J, Sun XJ, Chen J, Hu ZW, Wang L, Gu DM, Wang AP. Increasing the miR-126 expression in the peripheral blood of patients with diabetic foot ulcers treated with maggot debridement therapy. J Diabetes Complications 2017;31:241–244. [DOI] [PubMed] [Google Scholar]
- 224.Tao SC, Guo SC, Li M, Ke QF, Guo YP, Zhang CQ. Chitosan Wound Dressings Incorporating Exosomes Derived from MicroRNA-126-Overexpressing Synovium Mesenchymal Stem Cells Provide Sustained Release of Exosomes and Heal Full-Thickness Skin Defects in a Diabetic Rat Model. Stem Cells Transl Med 2017;6:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lin CJ, Lan YM, Ou MQ, Ji LQ, Lin SD. Expression of miR-217 and HIF-1alpha/VEGF pathway in patients with diabetic foot ulcer and its effect on angiogenesis of diabetic foot ulcer rats. J Endocrinol Invest 2019;42:1307–1317. [DOI] [PubMed] [Google Scholar]
- 226.Ciccacci C, Latini A, Perricone C, Conigliaro P, Colafrancesco S, Ceccarelli F, Priori R, Conti F, Perricone R, Novelli G, Borgiani P. TNFAIP3 Gene Polymorphisms in Three Common Autoimmune Diseases: Systemic Lupus Erythematosus, Rheumatoid Arthritis, and Primary Sjogren Syndrome-Association with Disease Susceptibility and Clinical Phenotypes in Italian Patients. J Immunol Res 2019;2019:6728694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Shimane K, Kochi Y, Horita T, Ikari K, Amano H, Hirakata M, Okamoto A, Yamada R, Myouzen K, Suzuki A, Kubo M, Atsumi T, Koike T, Takasaki Y, Momohara S, Yamanaka H, Nakamura Y, Yamamoto K. The association of a nonsynonymous single-nucleotide polymorphism in TNFAIP3 with systemic lupus erythematosus and rheumatoid arthritis in the Japanese population. Arthritis Rheum 2010;62:574–579. [DOI] [PubMed] [Google Scholar]
- 228.Fitzgerald O’Connor EJ, Badshah II, Addae LY, Kundasamy P, Thanabalasingam S, Abioye D, Soldin M, Shaw TJ. Histone deacetylase 2 is upregulated in normal and keloid scars. J Invest Dermatol 2012;132:1293–1296. [DOI] [PubMed] [Google Scholar]


