Abstract
We studied the expression of Bacillus amyloliquefaciens transglutaminase cloned in Escherichia coli BL21(DE3)pLysS harboring the plasmid pBAD/3C/bTGase, a bicistronic expression system, in bioreactor cultivation. Batch and fed-batch controlled as DO-stat strategies were employed for the production of the recombinant enzyme. In 30 h-batch cultivations using Terrific broth (TB), 6 g/L of biomass and 3.12 U/mgprotein of transglutaminase activity were obtained. DO-stat fed-batch cultivations under the control of oxygen concentration (DO-stat) using TB as medium but fed with glucose allowed the increment in biomass formation (17.5 g/L) and enzyme activity (6.43 U/mgprotein). DO-stat fed-batch using mineral medium (M9) and fed with glucose under the same conditions produced even higher enzymatic activity (9.14 U/mgprotein). The pH effect was investigated, and the best enzymatic activity could be observed at pH 8. In all cultivations, the bicistronic system remained stable, with 100% of plasmid-bearing cells. These results show that E. coli bearing bicistronic plasmid constructs to express recombinant TGase could be cultivated in bioreactors under DO-stat fed-batch using mineral medium and it is a promising strategy in future optimizations to produce this important enzyme.
Keywords: Microbial transglutaminase, Bicistronic plasmid system, Food enzymes, Fed-batch bioreactor, DO-stat, Bacillus amyloliquefaciens
Introduction
Transglutaminase (TGase, protein-glutamine gamma-glutamyltransferase, EC 2.3.2.13) is a multifunctional enzyme that catalyzes the acyl transfer reaction between a γ-carboxamide group of glutamine residue and the ε-amino group of a lysine residue from a variety of primary amines, resulting in the formation of inter- or intramolecular bonds, highly resistant to proteolysis [1–3]. TGases can be found in animal tissues and body fluids and are involved in several biological processes such as blood coagulation, epidermal keratinization, and stiffening of the erythrocyte membrane [4, 5]. It has been suggested that TGases may also be present in eggs and skin of amphibians, in turtle shell, in vegetable tissues of soy, broad beans, and orchard apples. They are also present in fungi and yeasts such as Phytophthora sp., Candida albicans, and Saccharomyces cerevisiae. However, the largest body of studies refers to microbial transglutaminases (mTGase) of bacteria such as Streptomyces and Bacillus [6].
The crosslinking properties of transglutaminases are widely used in industrial processes, especially in the food and pharmaceutical industries. The mTGase is used in several types of food to further improve the quality of final products such as viscosity, firmness, water holding capacity, and elasticity. Important applications in the textile and leather industries are also observed, as well as in regenerative medicine, PEGylation reaction and in the production of antibody conjugates [7].
In general, the most common techniques applied for the production of TGases for industrial uses are extracting and purifying the enzyme from the tissues or body fluids of animals (pigs, fish, cattle); screening for TGase-producing microorganisms (enzyme production by traditional fermentation technologies); and genetic manipulation using host microorganisms (Escherichia coli, Aspergillus, Bacillus) [8–11]. Because of the generally low titer of enzyme concentration in fermentation broths, many researches have been made in the areas of genetic engineering and bioprocess engineering aiming at improving the production of this enzyme on a large scale [11, 12].
E. coli has proved to be the preferred platform for the production of various enzymes and biological products of commercial interest because of its rapid cell growth, reaching high cell densities through simple cultivation procedures and low production costs in addition to the ability to express high concentrations of recombinant proteins [13, 14]. However, this laboratory workhorse bacterium presents several limitations for heterologous protein expression, such as codon bias, formation of inclusion bodies, lack of post-translation modification, and the efficient growth of E. coli has been a challenge for the industry since the early 1970s [15]. Therefore, the optimization of the production bioprocess, such as the modification of media composition, and genetic techniques, such as expression at lower temperatures, co-expression of molecular chaperones, development of new strains, vectors, and markers, have been reported as ways to overcome these limitations [15–18].
High cell density cultivation strategies are necessary to increase microbial biomass and the productivity in bioprocess, and many techniques have been developed for this purpose [19], including optimization of glucose feed rate profile, control of low acetate excretion, and high dry cell weight (DCW), techniques to improve plasmid stability [20–22].
Fed-batch cultures of E. coli strains that host gene-products are often reported in literature, including simple methods of indirect feedback such as DO-stat, pH–stat as well substrate feeding based on glucose uptake rate or demand, predetermined feeding strategies (exponential feeding), among others [23–25]. The DO-stat is a simple scheme that involves the application of glucose feeding pulses and the control of the dissolved oxygen concentration (DO) response to those pulses, a technique that requires only an DO sensor and the control of oxygen concentration and the glucose feeding pump [26].
However, E. coli cultures produce acetate that can inhibit cell growth and product formation, specially under anaerobic or oxygen-limiting conditions, or when carbon flux exceeds biosynthetic demands and the power generation capacity within the cell [27]. As the metabolic flow through glycolysis is closely related to respiratory activity by reducing NAD+ to NADH + H+, the glucose feeding rate is directly linked to oxygen consumption [28]. An increase in acetate (above 5 g/L) leads to a reduction in the growth rate with a reduction in biomass. There are reports in the literature, in which the accumulation of acetate in cultures of recombinant cells may be even greater than in cultures of the wild strain, under the same conditions and therefore must be controlled by special strategies during the fed-batch phase [27].
In a previous work, we constructed a bicistronic plasmid containing the TGase gene fused to the inhibitory Streptomyces caniferus prodomain. We also cloned the 3C protease gene in the same plasmid, in order to make the enzyme active and avoid the need for removal of the prodomain in vitro [8]. The activity of recombinant bTGase was investigated by cross-linking assays of bovine serum albumin (BSA) and by fluorescence, showing a specific activity of 37 mU/mg in shaker cultivations [8].
Based on these considerations, in the present work, we describe the expression of the mTGase from Bacillus amyloliquefaciens cloned in E. coli bearing the bicistronic plasmid pBAD/3C/bTGase, comparing the enzyme expression in batch bioreactors and cultures operated using a DO-stat fed-batch controlled as strategy controlled by oxygen concentration (DO-stat). The objective was to investigate whether we could express this recombinant construct in bioreactor cultures, allowing for future works aiming at optimizing the enzyme production.
Materials and methods
Bacterial strains, plasmids, cell maintenance, and materials
The strain used in this work was E. coli BL21(DE3)pLysS (Invitrogen) harboring the plasmid pBAD/3C/bTGase, which contains the fragment of bTGase gene from Bacillus amyloliquefaciens DSM7 and the Streptomyces caniferus prodomain beyond 3C protease gene. The construction of this expression vector, cell transformation, and sequencing were described in detail in a previous work [8].
Unless otherwise stated, the chemicals used in this study were of analytical grade or molecular biological grade and purchased from Sigma-Aldrich (Taufkirchen, Germany).
Cultivation media and inoculum preparation
Terrific broth (TB) (tryptone 12 g/L, yeast extract 24 g/L, glycerol 4.5 g/L, KH2PO4 0.17 M, K2HPO4 0.72 M) [29] and modified M9 medium (Na2HPO4 6 g/L, KH2PO4 3 g/L, NH4Cl 1 g/L, NaCl 0.5 g/L, yeast extract 20 g/L, MgSO4.7H2O 0.12 g/L, glucose 5 g/L, trace elements solution 1 mL (FeSO4.7H2O 2.8 g/L, MnCl2.4H2O 2 g/L, CaCl2.6H2O 2 g/L, CuCl2.2H2O 0.26 g/L, ZnSO4.7H2O 0.3 g/L)) [30], both media containing 100 μg/mL carbenicillin and 34 μg/mL chloramphenicol were used for cell growth and recombinant enzyme production. The feed solution used in the fed-batch cultivations were controlled as DO-stat and it was composed of glucose 300 g/L, yeast extract 30 g/L, MgSO4 0.5 g/L, and antibiotics (carbenicillin and chloramphenicol). The inoculum were prepared by transferring a single colony of E. coli BL21(DE3)pLysS transformed with plasmid pBAD/3C/bTGase into 500-mL flasks containing 100 mL of Lysogeny broth (LB) (yeast extract 5 g/L, tryptone 10 g/L, and NaCl 10 g/L) containing 100 μg/mL carbenicillin and 34 μg/mL chloramphenicol and incubated at 37 °C in a rotatory shaker operating at 180 rpm, and allowing cell growth until an OD600 of 1.0 was reached.
Batch bioreactor cultivations
Batch experiments were performed using a bioreactor BIOSTAT® B plus (Sartorius Stedim, Goettingen, Germany), equipped with two 2-L stirred tanks, filled with 1 L of TB medium. The bioreactor was equipped with two Rushton turbines and with aeration, temperature, agitation, and pH controllers. A polarographic electrode (Ingold, Germany) was used to measure the dissolved oxygen concentration in the culture. The pO2, pH, stirrer speed (STIR), base and acid consumption, and aeration rate were measured online and recorded by an external data acquisition and control system (Sartorius Stedim, Germany). The initial pH of the culture was adjusted to 7.2. In order to control the temperature at 20 °C during the expression (induction) of the recombinant proteins, the bioreactor was coupled to a cooling bath (Frigomix® B—B.Braun Biotech International, Germany). The best growth conditions and protein expression were applied to the batch of bioreactors according to conditions previously described in Duarte and collaborators [8]. The culture medium was supplemented with 100 μg/mL carbenicillin and 34 μg/mL of chloramphenicol; a cell suspension of 100 mL with concentration of OD600 of 1.0 was used as inoculum in all cultivations. The cultivation followed with constant air flow rate of 1 vvm (air volume per volume of culture medium per minute), 37 °C, and 300 rpm until reaching an OD600 of 0.4–0.6 (approximately 2 h). At this point in cultivation, the temperature was reduced to 20 °C and continued for another 40 min before induction. Then, the pro-bTGase gene was induced by the addition of 0.4 mM IPTG, followed by 20 h; after this period of time, the 3C protease gene was induced by the addition of L-arabinose 0.2%, and culture followed for another 6 h. Cells were then harvested by centrifugation at 4000 × g for 30 min at 4 °C and stored at − 20 °C. All experiments were performed in duplicate.
DO-stat fed-batch bioreactor cultivations
DO-stat fed-batch bioreactor experiments were performed in the same bioreactor system and its assembly and culture conditions were the same as described for the batch cultivations. All media were supplemented with antibiotics, as described in “Cultivation media and inoculum preparation” section. The pH was controlled at 7.2 using either 5 M NaOH or 5 M H3PO4. Feeding was implemented using an internal peristaltic pump after the first 5 h of operation as batch cultures.
The DO-stat fed-batch process was designed to operate as follows:
-
I
Start as batch cultivation until depletion of the carbon source (glucose), with air supply at a constant rate of 1 vvm and the DO kept at 30% saturation, varying the agitation rate from 300 to 1000 rpm (cascade of agitation);
-
II
After 5 h of feeding, cultivation (DO-stat) was started. Air supply continued at a constant rate of 1 vvm and agitation was fixed to 800 rpm. Using this approach, whenever DO rose above the set value (30%), the pump controlling the feeding supply turned on automatically. As a result, the metabolic activity of E. coli increases again and there is a decline in DO. Likewise, when the DO was lower than the set value, the pump that controlled the feeding supply automatically shut down.
Samples were taken at regular intervals to determine biomass (dry weight), residual sugar, and acetate.
At the end of the exponential growth phase, the temperature was reduced to 20 °C and the cultivation continued for another 40 min before induction. Then, the pro-bTGase gene was induced by the addition of 0.4 mM IPTG for 4 h; after this time, the 3C protease gene was induced by the addition of 0.2% L-arabinose, and the culture continued for another 10 h. At the end of this time, the cells were harvested by centrifugation at 4000 × g for 30 min at 4 °C and stored at − 20 °C. All experiments were performed in duplicate.
Effects of pH on the enzymatic activity of TGase
To evaluate the best buffer to be used in cell disruption, the effects of pH on the enzymatic activity of TGase were studied. Thus, 2 g of frozen cells were resuspended in 20 mL of the following buffers: pH 5 (0.1 M sodium acetate), pH 6 (0.1 M sodium phosphate), pH 7 (0.1 M sodium phosphate), pH 8 (0.1 M Tris–HCl), and pH 9 (0.1 M Tris–HCl). Cell suspensions were completely disrupted by sonication (10 pulses, 10 s each, 60% amplitude) and centrifuged at 13,000 × g for 30 min and the supernatant was collected for analysis of protein and enzyme activity. All experiments were performed in duplicate.
Analytical methods
Determination of protein and enzymatic activity
The determination of protein concentration was carried out following the Bradford method [31] using bovine serum albumin as a standard (Quick Start™ Bradford BIO-RAD).
TGase activity was determined by the colorimetric hydroxamate procedure using N-carbobenzoxy-L-glutaminyl-glycine and hydroxylamine [32]. A calibration curve was prepared using L‐glutamic acid γ‐monohydroxamate. One enzymatic unit of TGase (U) generates 1 µmol hydroxamic acid per minute at 37 °C.
Off-line measurements of cultivation substrates and biomass
The concentrations of glucose and acetate were determined by high-performance liquid chromatography (HPLC) (Shimadzu, Japan) equipped with a refractive index detector (RID-10A, Shimadzu) and Bio‐Rad HPX‐87H column (300 × 7.8 mm) in isocratic mode using as mobile phase a 5 mM H2SO4 solution as eluent with a flow rate of 0.6 mL/min and oven temperature of 45 °C. Biomass was measured as DCW; 10 mL of the culture were collected and centrifuged (3000 × g, 15 min) and oven dried at 80 °C to constant weight using an analytical balance.
Determination of plasmid stability
Because the plasmid pBAD/3C/bTGase was constructed by us in a previous work [8] and had never been tested in bioreactor cultivations, we decided to determine its stability. As the E. coli BL21(DE3)pLysS is resistant to chloramphenicol and, when hosting the plasmid pBAD/3C/bTGase, also resistant to carbenicillin, samples from bioreactor cultivations were appropriately diluted and spread in culturing plates with LB medium containing 100 μg/mL carbenicillin and 34 μg/mL chloramphenicol (selection of cells holding the plasmid) and LB medium containing only 34 μg/mL chloramphenicol (cells that have lost the plasmid), according to methodology described in the work of Thomas and collaborators [33] and compared as colony forming unit (CFU).
Results and discussion
Effects of pH on the enzymatic activity of TGase
Initially, to determine the ideal pH of the expressed enzyme, the cells were disrupted at pH 5, 6, 7, 8, and 9 in the buffers described in “Effects of pH on the enzymatic activity of TGase” section. TGase activity was determined in the supernatant of samples by the colorimetric hydroxamate procedure using N-carbobenzoxy-L-glutaminyl-glycine and hydroxylamine (“Effects of pH on the enzymatic activity of TGase” section) at 37 °C. The graph of the enzymatic activity can be seen in Fig. 1. The best enzymatic activity was observed at pH 8, which is in accordance with the literature. In previous works, it has been reported that the ideal pH for Bacillus transglutaminase is around 8, whereas for Streptomyces mobaraensis, the ideal pH is around 6 to 7 [34–36]. With this result, all further tests were performed at the pH 8.
Fig. 1.
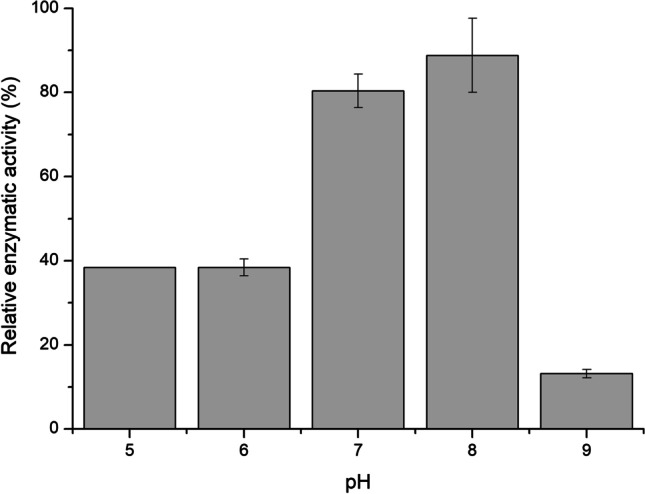
Optimal relative pH activity of the expressed TGase. The cells of E. coli were disrupted using the following buffers: pH 5 (0.1 M sodium acetate), pH 6 (0.1 M sodium phosphate), pH 7 (0.1 M sodium phosphate), pH 8 (0.1 M Tris–HCl), and pH 9 (0.1 M Tris–HCl). The results represent the mean of duplicates, considering the highest activity detected as 100%
Batch bioreactor cultivations
The batch bioreactor cultivations using the TB medium were run under the same induction conditions that were previously used in shaker flask cultivations, which are described in Duarte and collaborators [8]. In that work, cultures in TB medium supplemented with carbenicillin and chloramphenicol were incubated at 37 °C on rotatory shaker at 180 rpm until reaching OD600 of 0.4–0.6. At this point, the temperature was reduced to 20 °C for induction of recombinant pro-bTGase gene, induced with IPTG, and the 3C protease gene was induced by L-arabinose. The results of batch cultures are depicted in Fig. 2A and B and Table 1, comparing with results from the DO-stat fed-batch experiments. Cell concentration reached 5.5 g/L until IPTG was added. After that there was a small increase in cell concentration, up to 6 g/L in 30 h of cultivation, possibly due to the limitation of nutrients and also the necessary energy for the expression of proteins [37]. With 30 h of culture, the specific enzyme activity was 3.12 U/mgprotein.
Fig. 2.
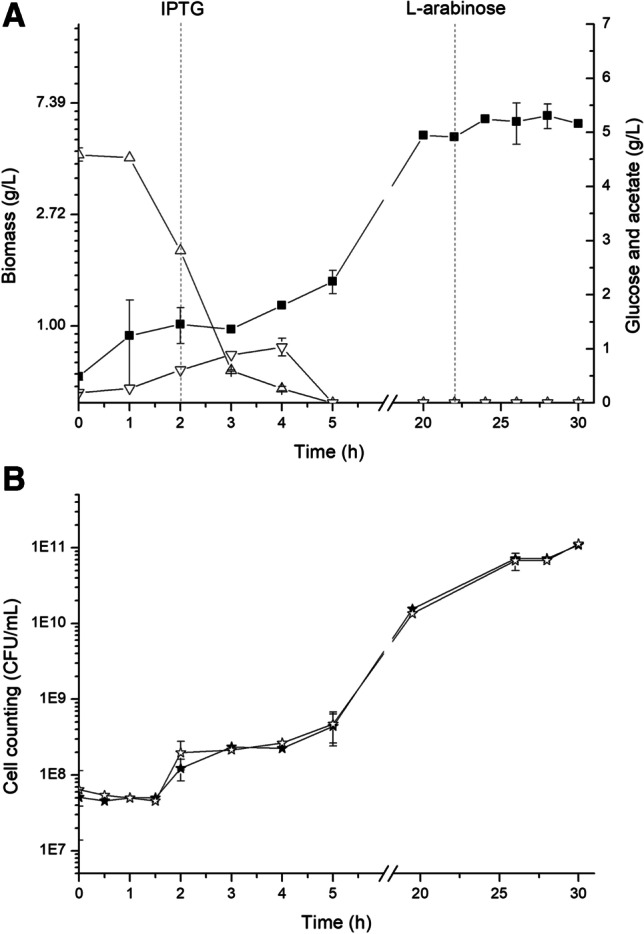
Batch cultures of E. coli BL21(DE3)pLysS (pBAD/3C/bTGase) in TB media. At 2 h, the culture temperature was reduced to 20 °C, the pro-bTGase gene was induced by the addition of 0.4 mM IPTG, and cultivations proceeded for 20 h; gene 3C protease was induced by the addition of 0.2% L-arabinose, followed by 6 h of cultivation. A (■) biomass DCW (g/L); (△) glucose (g/L); (▽) acetate (g/L). The vertical lines mark the time of induction of pro-bTGase and 3C protease. B Cell plasmid stability test: (★) cell count in LB medium containing 34 μg/mL chloramphenicol; (✰) cell count in LB medium containing 100 μg/mL carbenicillin and 34 μg/mL chloramphenicol. All experimental runs were performed in duplicates
Table 1.
Kinetic parameters for mTGase production in different media in 30 h of induction. Results are the mean of duplicates
| Cultivation strategy (culture medium) |
Dry cell weight | Specific activity | Volumetric activity | Enzymatic productivity |
|---|---|---|---|---|
| (g/L) | (U/mgprotein) | (U/L) | Pp (U/L·h) | |
|
Batch (TB) |
6.13 ± 0.10 | 3.12 ± 0.08 | 19.17 ± 0.81 | 0.64 ± 0.03 |
|
Fed-batch (TB then glucose) |
17.56 ± 0.96 | 6.43 ± 0.66 | 19.17 ± 3.21 | 0.64 ± 0.11 |
|
Fed-batch (M9) |
17.07 ± 3.44 | 9.14 ± 0.42 | 29.38 ± 1.54 | 0.98 ± 0.05 |
PP, enzymatic productivity
An important factor in cultures of recombinant microorganisms in industrial processes is plasmid stability, which can be affected by culture conditions such as temperature, aeration, pH, induction factors, among others. A decrease in the concentration of DO in the culture medium as well as elevated temperatures can reduce plasmids stability [38]. In our study, as shown in Fig. 2B, a high stability of plasmid was observed, with practically 100% of cells bearing the plasmid at the end of cultivation. These results demonstrate the viability of using this recombinant strain-plasmid system in order to scale up this process for future industrial applications of TGase cloning and production methodology.
DO-stat fed-batch cultivations
In order to improve the production of the recombinant TGase and achieve high cell densities, it is necessary to adequately control the feeding of glucose and the transfer of oxygen to the culture medium. This control of cellular metabolism can be obtained by adding nutrients in fed-batch mode and appears to be the technology of choice in the bioindustry [27]. Thus, we decided to test a DO-stat fed-batch controlled strategy using an indirect feedback method.
The TB medium was used in the culture medium reservoir and feeding was controlled by a DO-stat feeding strategy. Preliminary bioreactor experiments of cell growth were run to define the best moment to start the feeding, taking into considerations of the conditions used in this research. The DO was kept at 30% saturation by adjusting the agitation rate, starting from 300 rpm. The results showed that in about 5 h of cultivation, carbon source (initial concentration of 4.5 g/L) was depleted, indicating the need to start the feeding. With these data, we set the DO-stat fed-batch cultivations, whose results are depicted in Fig. 3A and B and in Table 1, in comparison with the batch experiments. Previous studies have shown that the induction of plasmids with gene inductions control by IPTG can cause a toxic effect on cells, decreasing their plasmid stability and cell growth [38], leading to reduced production of interest proteins. Transglutaminase, due to its own ability to cross-link proteins, will be toxic to host cells [39], unless its expression is tightly controlled. Therefore, induction of the pro-transglutaminase was done after 16 h of cell growth, at the end of the exponential growth phase and, after 4 h after TGase induction, the 3C protease gene was induced with L-arabinose injection and culture proceeded for another 10 h. In Fig. 3A, it can be seen that the growth of recombinant E. coli was prolonged without accumulation of the carbon source, with control of acetate formation. An increase in the concentration of glucose can also be seen in the same figure in approximately 16 h of culture. This increase may be related to the induction of IPTG, because at that moment there is a change in the metabolism of cell growth by protein induction. At the end of the cultivation, the volume of added medium was 153 mL with 45 g of glucose consumption.
Fig. 3.
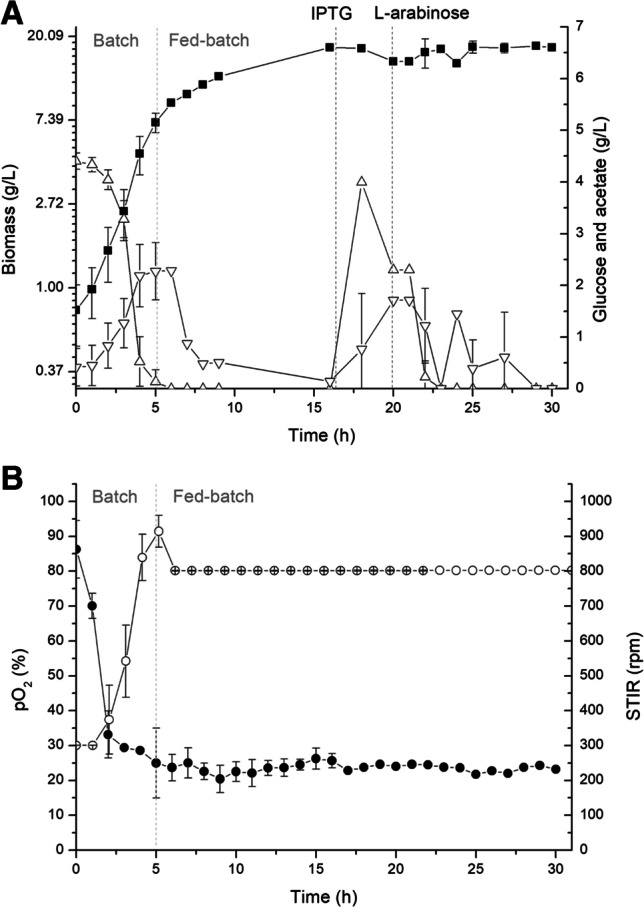
Fed-batch culture by E. coli BL21(DE3)pLysS (pBAD/3C/bTGase) with DO-stat feeding using Terrific broth (TB). In approximately 16 h, the culture temperature was reduced to 20 °C, the pro-bTGase gene was induced by the addition of 0.4 mM IPTG, and cultivations proceeded for 4 h; gene 3C protease was induced by the addition of 0.2% L-arabinose, followed by 10 h of cultivation. A (■) biomass DCW (g/L); (△) glucose (g/L); (▽) acetate (g/L). B (○) STIR (rpm); (●) pO2 (%). All experimental runs were performed in duplicates. The vertical gray lines mark the batch and fed-batch strategy, and the vertical black lines mark the time of induction of pro-bTGase and 3C protease
As expected, the DO-stat fed-batch cultures produced 17.56 g/L of DCW in 30 h, higher than in the batch experiments using the same initial culture medium containing only 4.5 g/L of glycerol as carbon source. The specific enzymatic activity reached 6.43 U/mg protein, approximately 2 times higher than for cells produced in the batch culture.
To further improve the production of enzymes, the modified M9 medium was used, because in addition to be a richer medium in nutrients, a higher level of recombinant proteins was produced in the presence of MgSO4 [40]. Conditions of feeding and controls were the same as for the experiments using TB. At the end of the cultivation, the volume of added medium was 188 mL, with 52 g of glucose consumption. Figure 4A and B and Table 1 show the kinetics of the DO-stat fed-batch using this medium.
Fig. 4.
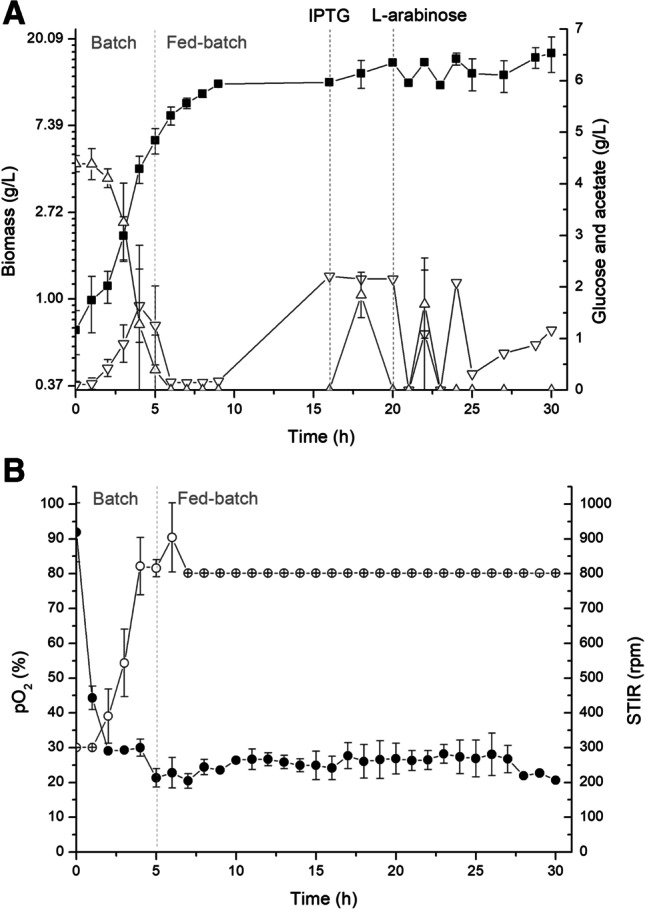
Fed-batch culture by E. coli BL21(DE3)pLysS (pBAD/3C/bTGase) with DO-stat feeding using M9 medium. In approximately 16 h, the culture temperature was reduced to 20 °C, the pro-bTGase gene was induced by the addition of 0.4 mM IPTG, and cultivations proceeded for 4 h; gene 3C protease was induced by the addition of 0.2% L-arabinose, followed by 10 h of cultivation. A (■) biomass DCW (g/L); (△) glucose (g/L); (▽) acetate (g/L). B (○) STIR (rpm); (●) pO2 (%). All experimental runs were performed in duplicates. The vertical gray lines mark the batch and fed-batch strategy, and the vertical black lines mark the time of induction of pro-bTGase and 3C protease
Results for the DO-stat using M9 medium showed a biomass formation of 17.07 g/L in 30 h, similar to that for TB cultivations fed with glucose during the feeding time. However, the specific TGase activity increased to 9.14 U/mg protein, 1.43 times higher than for the medium TB, this being the highest enzyme title achieved in our experiments. The SDS-PAGE showing the expression of recombinant bTGase and 3C protease in E. coli BL21(DE3)pLysS with plasmid pBAD/3C/bTGase with fed-batch culture using feedback-controlled DO-stat in modified M9 medium can be seen in Fig. 5.
Fig. 5.
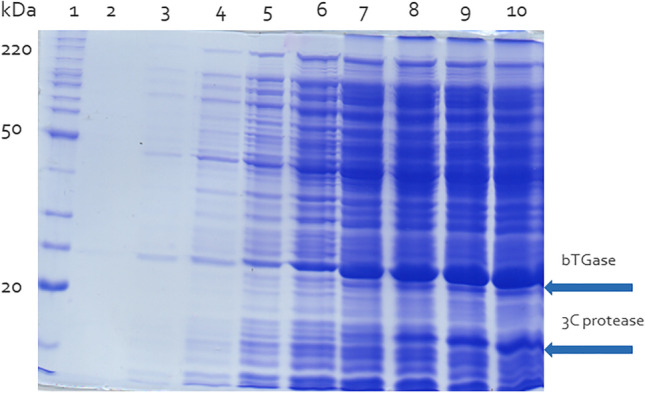
SDS-PAGE analysis of transglutaminase and 3C protease expression in fed-batch controlled as DO-stat cultures. The recombinant E. coli BL21(DE3)pLysS (pBAD/3C/bTGase) cells were cultured in modified M9 medium for 30 h. Feeding started 5 h after the start of batch cultivation and pro-bTGase expression was induced after 16 h of culture by adding 0.4 mM IPTG to the cultures and 3C protease expression was induced after 20 h of culture by adding 0.2% L-arabinose. Lane 1: molecular weight marker, lane 2: pre-inoculum addition, lane 3: 3-h cultivation, lane 4: 5-h cultivation, lane 5: sample collected immediately after induction of IPTG (16 h of culture), lane 6: 18-h cultivation, lane 7: sample collected immediately after induction of L-arabinose (20 h of cultivation), lane 8: 24-h cultivation, lane 9: 27-h cultivation, and line 10: 30-h cultivation
Since 1989, when Ando et al. first reported the production of microbial transglutaminase by Streptoverticillium mobaraense, bioengineering has been striving to improve the production of this enzyme [41]. Efforts to improve the yields of microbial transglutaminase have been made through metabolic optimization, substrate optimization, and environmental control strategies (temperature, pH, agitation, and dissolved oxygen) in cultures of different microorganisms, including S. mobaraense, S. cinnamoneum, S. ladakanum, Streptomyces nigrescens, S. hygroscopicus, S. platensis, Bacillus circulans and B. subtilis [12]. However, after several studies, the activity of transglutaminase in the fermentation broth increased from 2.0 to 6.0 U/mL, the Streptomyces spp. showing higher yields and, therefore, being currently used industrially as a producer of microbial transglutaminase [11].
Presently, the focus for the optimization of transglutaminase production has been the genetic engineering, using the transglutaminase gene from Streptomyces or Bacillus for exogenous expression of the enzyme [12]. In comparison with concentrations of S. mobaraensis native transglutaminase (22.6 U/mgprotein), 23 U/mgprotein was observed for recombinant S. mobaraensis transglutaminase expressed in Corynebacterium ammoniageneses, and 26 U/mgprotein for recombinant S. mobaraensis transglutaminase expressed in Corynebacterium glutamicum [41–44]. Some studies described improved strategies for expressing mTGase, comparing constitutive versus thermo-inducible expression systems, instead of expressing the protein by fusion or wild type protein, for production in E. coli of a recombinant transglutaminase from Streptomyces mobaraensis. The results showed enzymatic activity of the purified mTGase of 24 U/mg in the thermo-inductive expression system, compared to 15 U/mg in the constitutive system [45].
Several authors have reported the production of recombinant enzymes expressed in E. coli with bioreactors using the fed-batch culture approach under DO-stat. Comparison of production of recombinant β-galactosidase in bioreactors by fed-batch culture using feedback-controlled DO-stat and ascendant linear pump feeding showed an increase of approximately 2.5 times in the enzymatic activity using this strategy [46]. The DO-stat strategy proved to be effective in achieving high cellular density and suppressing the accumulation of ethanol in the culture medium, improving the production of interferon-α (pIFN-α) by Pichia pastoris [47]. Higher concentrations and productivities of poly-γ-glutamic acid (γ-PGA) by B. subtilis were achieved after application of the strategy of DO-stat feeding compared with a glucose-feedback feeding strategy [48].
Conclusions
This study presents for the first time the expression of recombinant Bacillus amyloliquefaciens transglutaminase expressed in Escherichia coli under the control of a bicistronic plasmid pBAD/3C/bTGase using batch and fed-batch bioreactor cultivations under the control of DO-stat strategies for the growth of E. coli and the recombinant enzyme. Using a mineral medium (M9) in the DO-stat fed-batch controlled cultivations, it was possible to obtain mTGase with high enzymatic activity, showing to be a promising strategy for the further development of production system of this important enzyme.
Acknowledgements
The authors wish to thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação do Aperfeiçoamento de Pessoal do Ensino Superior (CAPES), Finance Code 001, and FAPERGS for their financial support of this project and scholarships.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Griffin M, Casadio R, Bergamini CM. Transglutaminases: nature’s biological glues. Biochem J. 2002;368:377–396. doi: 10.1042/BJ20021234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang M-T, Chang C-H, Wang JM, Wu TK, Wang Y-K, Chang C-Y, Li TT. Crystal structure and inhibition studies of transglutaminase from Streptomyces mobaraense. J Biol Chem. 2011;286:7301–7307. doi: 10.1074/jbc.M110.203315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yokoyama K, Nio N, Kikuchi Y. Properties and applications of microbial transglutaminase. Appl Microbiol Biotechnol. 2004;64:447–454. doi: 10.1007/s00253-003-1539-5. [DOI] [PubMed] [Google Scholar]
- 4.Aeschlimann D, Paulsson M. Transglutaminases: protein cross-linking enzymes in tissues and body fluids. Thromb Haemost. 1994;71:402–415. doi: 10.1055/s-0038-1642451. [DOI] [PubMed] [Google Scholar]
- 5.Chung SI, Lewis MS, Folk JE. Relationships of the catalytic properties of human plasma and platelet transglutaminases (activated blood coagulation factor XIII) to their subunit structures. J Biol Chem. 1974;249:940–950. doi: 10.1016/S0021-9258(19)43022-6. [DOI] [PubMed] [Google Scholar]
- 6.Mariniello L, Di Pierro P, Giosafatto C, Sorrentino A, Porta R (2008) Transglutaminase in food biotechnology. In: Porta R, Di Pierro P, Mariniello L (eds) Recent research developments in food biotechnology. Enzymes as additives or processing. Research Signpost, Kerala, pp 185–211
- 7.Duarte L, Matte CR, Bizarro CV, Ayub MAZ. Review transglutaminases: part II—industrial applications in food, biotechnology, textiles and leather products. World J Microbiol Biotechnol. 2019;36:11. doi: 10.1007/s11274-019-2792-9. [DOI] [PubMed] [Google Scholar]
- 8.Duarte LS, Barsé LQ, Dalberto PF, da Silva WTS, Rodrigues RC, Machado P, Basso LA, Bizarro CV, Ayub MAZ. Cloning and expression of the Bacillus amyloliquefaciens transglutaminase gene in E. coli using a bicistronic vector construction. Enzyme Microb Technol. 2020;134:109468. doi: 10.1016/j.enzmictec.2019.109468. [DOI] [PubMed] [Google Scholar]
- 9.Sorde KL, Ananthanarayan L. Isolation, screening, and optimization of bacterial strains for novel transglutaminase production. Prep Biochem Biotech. 2019;49:64–73. doi: 10.1080/10826068.2018.1536986. [DOI] [PubMed] [Google Scholar]
- 10.Folk JE, Cole PW. Mechanism of action of guinea pig liver transglutaminase: I. Purification and properties of the enzyme: identification of a functional cysteine essential for activity. J Biol Chem. 1966;241:5518–5525. doi: 10.1016/S0021-9258(18)96373-8. [DOI] [PubMed] [Google Scholar]
- 11.Zhang D, Zhu Y, Chen J. Microbial transglutaminase production: understanding the mechanism. Biotechnol Genet Eng Rev. 2009;26:205–222. doi: 10.5661/bger-26-205. [DOI] [PubMed] [Google Scholar]
- 12.Duarte L, Matte CR, Bizarro CV, Ayub MAZ. Transglutaminases: part I—origins, sources, and biotechnological characteristics. World J Microbiol Biotechnol. 2020;36:15. doi: 10.1007/s11274-019-2791-x. [DOI] [PubMed] [Google Scholar]
- 13.Rosano GL, Morales ES, Ceccarelli EA. New tools for recombinant protein production in Escherichia coli: a 5-year update. Protein Sci. 2019;28:1412–1422. doi: 10.1002/pro.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sørensen HP, Mortensen KK. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J Biotechnol. 2005;115:113–128. doi: 10.1016/j.jbiotec.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:172–172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauer S, Shiloach J. Maximal exponential growth rate and yield of E. coli obtainable in a bench-scale fermentor. Biotechnol Bioeng. 1974;16:933–941. doi: 10.1002/bit.260160707. [DOI] [PubMed] [Google Scholar]
- 17.Khow O, Suntrarachun S. Strategies for production of active eukaryotic proteins in bacterial expression system. Asian Pac J Trop Biomed. 2012;2:159–162. doi: 10.1016/S2221-1691(11)60213-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.VIdalia V-MN, Bernardo F (2017) Escherichia coli como organismo modelo e sua aplicação em biotecnologia. In: Samie A (ed) Escherichia coli - Avanços recentes em fisiologia, patogênese e aplicações biotecnológicas. IntechOpen, London. 10.5772/67306
- 19.Riesenberg D, Guthke R. High-cell-density cultivation of microorganisms. Appl Microbiol Biotechnol. 1999;51:422–430. doi: 10.1007/s002530051412. [DOI] [PubMed] [Google Scholar]
- 20.Shojaosadati SA, Varedi Kolaei SM, Babaeipour V, Farnoud AM. Recent advances in high cell density cultivation for production of recombinant protein. Iran J Biotechnol. 2008;6:63–84. [Google Scholar]
- 21.Dodge TC, Gerstner JM. Optimization of the glucose feed rate profile for the production of tryptophan from recombinant E coli. J Chem Technol Biotechnol. 2002;77:1238–1245. doi: 10.1002/jctb.698. [DOI] [Google Scholar]
- 22.Eiteman MA, Altman E. Overcoming acetate in Escherichia coli recombinant protein fermentations. Trends Biotechnol. 2006;24:530–536. doi: 10.1016/j.tibtech.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Kim BS, Lee SC, Lee SY, Chang YK, Chang HN. High cell density fed-batch cultivation of Escherichia coli using exponential feeding combined with pH-stat. Bioproc Biosystems Eng. 2004;26:147–150. doi: 10.1007/s00449-003-0347-8. [DOI] [PubMed] [Google Scholar]
- 24.Morcelli A, Rech R, Klafke A, Pelegrini R, Ayub MAZ. Exponential fed-batch cultures of Klebsiella pneumoniae under anaerobiosis using raw glycerol as a substrate to obtain value-added bioproducts. J Braz Chem Soc. 2018;29:2278–2286. doi: 10.21577/0103-5053.20180104. [DOI] [Google Scholar]
- 25.Rech R, Ayub MAZ. Fed-batch bioreactor process with recombinant Saccharomyces cerevisiae growing on cheese whey. Braz J Chem Eng. 2006;23:435–442. doi: 10.1590/S0104-66322006000400001. [DOI] [Google Scholar]
- 26.Johnston W, Cord-Ruwisch R, Cooney M. Industrial control of recombinant E. coli fed-batch culture: new perspectives on traditional controlled variables. Bioproc Biosystems Eng. 2002;25:111–120. doi: 10.1007/s00449-002-0287-8. [DOI] [PubMed] [Google Scholar]
- 27.Lee SY. High cell-density culture of Escherichia coli. Trends Biotechnol. 1996;14:98–105. doi: 10.1016/0167-7799(96)80930-9. [DOI] [PubMed] [Google Scholar]
- 28.Krause M, Neubauer A, Neubauer P. The fed-batch principle for the molecular biology lab: controlled nutrient diets in ready-made media improve production of recombinant proteins in Escherichia coli. Microb Cell Fact. 2016;15:110. doi: 10.1186/s12934-016-0513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lessard JC. Growth media for E. coli. Method Enzymol. 2013;533:181–189. doi: 10.1016/B978-0-12-420067-8.00011-8. [DOI] [PubMed] [Google Scholar]
- 30.Harwood CR, Cutting SM. Molecular biological methods for Bacillus. Chichester: Wiley; 1990. [Google Scholar]
- 31.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 32.Grossowicz N, Wainfan E, Borek E, Waelsch H. The enzymatic formation of hydroxamic acids from glutamine and asparagine. J Biol Chem. 1950;187:111–125. doi: 10.1016/S0021-9258(19)50936-X. [DOI] [PubMed] [Google Scholar]
- 33.Thomas P, Sekhar AC, Upreti R, Mujawar MM, Pasha SS. Optimization of single plate-serial dilution spotting (SP-SDS) with sample anchoring as an assured method for bacterial and yeast cfu enumeration and single colony isolation from diverse samples. Biotechnol Rep (Amst) 2015;8:45–55. doi: 10.1016/j.btre.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ragkousi K, Setlow P. Transglutaminase-mediated cross-linking of GerQ in the coats of Bacillus subtilis spores. J Bacteriol. 2004;186:5567–5575. doi: 10.1128/JB.186.17.5567-5575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Huang L, Zheng D, Fu Y, Shan M, Li Y, Xu Z, Jia L, Wang W, Lu F. Characterization of transglutaminase from Bacillus subtilis and its cross-linking function with a bovine serum albumin model. Food Funct. 2018;9:5560–5568. doi: 10.1039/C8FO01503A. [DOI] [PubMed] [Google Scholar]
- 36.Agyare KK, Damodaran S. pH-stability and thermal properties of microbial transglutaminase-treated whey protein isolate. J Agr Food Chem. 2010;58:1946–1953. doi: 10.1021/jf903530d. [DOI] [PubMed] [Google Scholar]
- 37.Harcum SW, Bentley WE. Response dynamics of 26-, 34-, 39-, 54-, and 80-kDa proteases in induced cultures of recombinant Escherichia coli. Biotechnol Bioeng. 1993;42:675–685. doi: 10.1002/bit.260420602. [DOI] [PubMed] [Google Scholar]
- 38.Kumar PKR, Maschke HE, Friehs K, Schügerl K. Strategies for improving plasmid stability in genetically modified bacteria in bioreactors. Trends Biotechnol. 1991;9:279–284. doi: 10.1016/0167-7799(91)90090-5. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi K, Kumazawa Y, Miwa K, Yamanaka S. ϵ-(γ-Glutamyl)lysine cross-links of spore coat proteins and transglutaminase activity in Bacillus subtilis. FEMS Microbiol Lett. 1996;144:157–160. doi: 10.1016/0378-1097(96)00353-9. [DOI] [Google Scholar]
- 40.Mahmoudi S, Abtahi H, Bahador A, Mosayebi G, Salmanian AH, Teymuri M. Optimizing of nutrients for high level expression of recombinant streptokinase using pET32a expression system. Maedica (Buchar) 2012;7:241–246. [PMC free article] [PubMed] [Google Scholar]
- 41.Ando H, Adachi M, Umeda K, Matsuura A, Nonaka M, Uchio R, Tanaka H, Motoki M. Purification and characteristics of a novel transglutaminase derived from microorganisms. Agric Biol Chem. 1989;53:2613–2617. doi: 10.1080/00021369.1989.10869735. [DOI] [Google Scholar]
- 42.Date M, Yokoyama K-i, Umezawa Y, Matsui H, Kikuchi Y. High level expression of Streptomyces mobaraensis transglutaminase in Corynebacterium glutamicum using a chimeric pro-region from Streptomyces cinnamoneus transglutaminase. J Biotechnol. 2004;110:219–226. doi: 10.1016/j.jbiotec.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 43.Itaya H, Kikuchi Y. Secretion of Streptomyces mobaraensis pro-transglutaminase by coryneform bacteria. Appl Microbiol Biotechnol. 2008;78:621–625. doi: 10.1007/s00253-007-1340-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sommer C, Volk N, Pietzsch M. Model based optimization of the fed-batch production of a highly active transglutaminase variant in Escherichia coli. Protein Expr Purif. 2011;77:9–19. doi: 10.1016/j.pep.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 45.Salis B, Spinetti G, Scaramuzza S, Bossi M, Jotti GS, Tonon G, Crobu D, Schrepfer R (2015) High-level expression of a recombinant active microbial transglutaminase in Escherichia coli. BMC Biotechnol:84. 10.1186/s12896-015-0202-4 [DOI] [PMC free article] [PubMed]
- 46.de Andrade BC, Migliavacca VF, Okano FY, Grafulin VY, Lunardi J, Roth G, de Souza CFV, Santos DS, Chies JM, Renard G, Volpato G. Production of recombinant β-galactosidase in bioreactors by fed-batch culture using DO-stat and linear control. Biocatal Biotransfor. 2019;37:3–9. doi: 10.1080/10242422.2018.1493105. [DOI] [Google Scholar]
- 47.Ding J, Gao M, Hou G, Liang K, Yu R, Li Z, Shi Z. Stabilizing porcine interferon-α production by Pichia pastoris with an ethanol on-line measurement based DO-Stat glycerol feeding strategy. J Chem Technol Biotechnol. 2014;89:1948–1953. doi: 10.1002/jctb.4281. [DOI] [Google Scholar]
- 48.Jiang Y, Tang B, Xu Z, Liu K, Xu Z, Feng X, Xu H. Improvement of poly-γ-glutamic acid biosynthesis in a moving bed biofilm reactor by Bacillus subtilis NX-2. Bioresour Technol. 2016;218:360–366. doi: 10.1016/j.biortech.2016.06.103. [DOI] [PubMed] [Google Scholar]


