Abstract
Toxic heavy metals pollution posed severe health hazards to the environment and biodiversity. Therefore, the development of rapid and non-invasive bioassays is in the race to monitor toxic chemicals using novel approaches. This study isolated and characterized an intense blue luminescence-producing marine bacteria, Vibrio campbellii STF1, for biosensing applications. Species-level identification of this strain was confirmed based on various phenotypic tests and multilocus sequence approach using 16s rRNA, toxR, and luxA gene sequence analysis. Fatty acid methyl ester analysis revealed the presence of three predominant fatty acids C15:0 anteiso (21.73%), C17:0 anteiso (11.27%), and C19:0 anteiso (9.08%) in STF1. Luciferase enzyme from V. campbellii STF1 was extracted, partially purified, and molecular masses (alpha subunit 40 kDa and beta subunit 37 kDa) were determined by SDS-PAGE gel for in vivo assays. MALDI-TOF-MS analysis of V. campbellii cells’ protein extracts showed distinct mass spectral peaks at m/z of 2615, 3948, and 4232 da. V. campbellii STF1 is resistant to heavy metal lead, while other metals such as cadmium, copper, and mercury inhibited its growth and luminescence. Crude ethyl acetate extraction of V. campbellii demonstrated antibacterial activity against Shigella dysenteriae type 5 with a maximum inhibition zone of 27.0±1.0 mm.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42770-021-00471-w.
Keywords: Vibrio campbellii, Multilocus sequence analysis, Biosensor, Heavy metal toxicity, Antibacterial activity
Introduction
Bioluminescent bacteria are one among the known mesmerizing luminous organisms on this tiny blue planet earth [1]. The distribution and diversity of these bacteria in various marine abiotic and biotic samples are widely studied from different environments [2, 3]. Luminescence of these bacteria is regulated by structural and core lux genes [ 4] and by various physicochemical factors [5]. Quorum sensing is a chemical signaling phenomenon in which luminous bacteria release a high concentration of quorum sensing molecules into the growth media, which triggers core genes luxCDABE to produce blue-green luminescence [6]. The emission spectrum of all known bacterial bioluminescence falls within the range of 450-490 nm. Isolation of intense luminescence-producing bacteria is the current research interest in developing luminescence-based rapid bioassays to monitor various pollutants from various biological and environmental samples. On the other hand, the luciferase enzyme is also used as a natural in vitro biosensor to monitor several toxic effluents. Thus, the whole bacteria and enzyme are being used in a myriad of ways, especially in biomedical and toxicological studies [7, 8].
Several recent studies emphasize the increased pollution levels in the atmosphere, hydrosphere, and lithosphere, indicating an alarm to detect and monitor these toxic pollutant levels. In order to monitor and detect pollutants from different environments, studies have been redirected to explore potential novel resources like luminous bacteria. A luminous Vibrio campbellii strain LZ5 isolated from the Pacific oyster, Crassostrea giga, was found to show the potential application in monitoring different heavy metals [9]. For the first time, recent studies observed the bioactive nature of different species of luminous bacteria in the form of antimicrobial activity against various human pathogens [10]. Identification of vibrios based on various phenotypic tests, fatty acid methyl esters analysis, and phylogenetic analyses using multilocus sequence analyzation (MLSA) approach was found to be reliable in current research [11]. Therefore, considering luminous bacteria's important applications, the phenotypic and genotypic characterizations were carried on the potential biosensing and bioactivity showing luminous marine bacteria Vibrio campbellii STF1.
Materials and methods
Study area and sample collection
During low tide time, a live specimen of unscheduled stonefish Synanceia verrucosa was caught carefully using a small hand net from Kodiyaghat coast (Lat. 11°31′47.16″ N; Long 92°43′25.97″ E). Specimen was immediately transferred to a sterile ziplock sample bag and transported to the laboratory within one hour for further microbiological analysis.
Isolation of luminous bacteria
All microbiological tests were performed according to standard aseptic techniques in microbiology [12]. The collected specimen was washed thoroughly with autoclaved sterile seawater to remove epibionts and debris. Using a sterile cotton swab, the specimen’s surface skin was swabbed and spread evenly onto SWC agar media Petri plate. Plate was incubated at 35°C for 24 hours and examined for luminous colonies under dark conditions by straining the eyes for 10 minutes. Using adjustable red light, intense luminous colonies were picked up with sterile toothpicks [3] and purified by streaking on SWC agar plates until pure isolated colonies were obtained. Luminous strain with high intense luminescence was coded as STF1 and deposited in the host institution for further studies.
Phenotypic characterization of luminous bacterium V. campbellii STF1
Morphological study
Overnight-grown pure culture of V. campbellii in the marine broth was used to observe morphology and motility under ZEISS Axiovert 40 CFL inverted microscope. A loop full of broth culture was placed on a clean glass slide and observed under a microscope at 40× magnification using a blue filter.
Scanning electron microscopic (SEM) analysis
V. campbellii was fixed overnight at 4°C with 2.5% glutaraldehyde in 0.1 M phosphate buffer–0.1 M sucrose (pH 7.4), rinsed thrice with 0.1 M phosphate buffer, and used for the fixative for 5 minutes after each rinse. Sample was postfixed for 1 hour in 1% osmium tetroxide in the same buffer, rinsed thrice with the same buffer for 5 minutes. The sample was dehydrated by adding ethanol solutions in a graded series of 50%, 70%, and 80% for 10 minutes at each concentration, 95% was used twice for 10 minutes, and 100% was used thrice within 15 minutes. The sample was then dried overnight in a fume hood and mounted on aluminum stubs and sputter-coated with gold for observation in a VEGA3 TESCAN SEM.
Biochemical tests
Biochemical tests were performed according to standard techniques [13] and using readymade disks (Himedia labs, Mumbai). All tests were performed according to both the protocols and with an optimum salt level (3% NaCl) used in all these tests. Results were compared with biochemical keys [14].
Fatty acid methyl esters profile analyzation
Extraction of fatty acid methyl esters (FAME) was followed according to the method described by Sasser [15]. Gas chromatography (GC) analysis of extracted fatty acids was carried out in Agilent technologies, model 6890N network with flame ionization detector (FID), and high-resolution gas chromatography column (Agilent Technologies) capillary with sizes 25m × 200μm × 0.33μm was used in this study. The sample was maintained for 21 minutes with injector temperature at 170°C and a detector temperature at 310°C. The carrier gas used was hydrogen with a flow rate of 30 μl/min, and 2μl of sample was used for the GC analysis with a split ratio of about 100:1. Quantification and identification of FAME peaks were determined with a reporting integrator model- Sherlock Microbial Identification System, version 6.0B.
MALDI-TOF mass spectrometry analysis
Protein analyzation of V. campbellii STF1 was performed using matrix-assisted laser desorption-ionization time of flight mass spectrometry (MALDI-TOF MS) following the Tris-EDTA buffer approach method [16]. Fresh culture of V. campbellii STF1 grown on Luria Broth agar (3% NaCl) for 24 hours at 37°C was used. Cells were harvested and transferred into an Eppendorf tube containing 2 ml of deionized water. This was vortexed and centrifuged at 4000 rpm for 5 minutes at room temperature (RT). The supernatant was decanted, and 1.5 ml of deionized water was added to the bacterial pellet. This was vortexed for 5 minutes and centrifuged at 19000 rpm for 30 minutes at RT. The supernatant was decanted, and 1.5 ml of TE buffer was added. Vortex was repeated for 20 minutes at RT and subsequently centrifuged at 19000 rpm for 30 minutes at RT. The pellet obtained was once again suspended with TE buffer and vigorously shaken for 10 minutes. This homogenous bacterial cell suspension of 0.5 μl was loaded in the MALDI target plate.
Growth curve determination
A growth curve study was performed to determine the luminescence activity with varied cell density. A 100 μl of overnight grown culture in seawater broth with an optical density (OD) value of 0.074 was inoculated into 50 ml seawater broth in a 100 ml conical flask and incubated at 35°C on a shaker with 150 rpm. For every one hour, OD values were measured at 600 nm using a spectrophotometer. Same broth without bacterial culture inoculation was used as a standard blank measurement.
Luminescence assay
For luminescence activity, 100 μl of fresh V. campbellii broth culture with an initial OD of 0.018 was inoculated in 50 ml luminescent broth and incubated at 35°C. Broth media without culture inoculation was used as blank. Luminescence readings were recorded every one hour using the luminometer (Hidex).
Effect of pH on growth and luminescence
Tolerance to various pH levels was studied according to Benson [13]. Different pH levels of 2, 5, 7, 9, and 11 were adjusted in 50 ml of seawater broth containing conical flasks using 1M NaOH and 1M HCl. Culture STF1 was inoculated in flasks, incubated in a shaker at 35°C for 24 hours, and arbitrary values were recorded.
Antibiotic sensitivity assay
Luminous bacteria are known to enter easily into aquaculture systems and cause luminous vibriosis disease, which results in dramatic losses in global aquaculture industries. Many studies have proven that these bacteria can resist antibiotics. To understand their resistance profile, different antibiotics were tested (Table S2). Antibiotic susceptibility assay was performed according to Benson [13]. Various antibiotic disks were placed on freshly prepared Muller Hinton agar (3% NaCl) plates swabbed with overnight grown cultures in marine broth. Petri plates were incubated at 35° C for 24 hours, and results observed as inhibition zones were recorded.
Effect of heavy metals on growth and luminescence
In this study, the effect of heavy metals including cadmium chloride, copper sulfate, lead nitrate, and mercuric chloride on growth and luminescence and tolerance to these metals was studied at different concentrations of 0.1 mM, 1 mM, and 10 mM. Heavy metal solutions were prepared according to Gellert [17], and heavy metal sensitivity assay was performed following Well diffusion method [18].
Extraction and purification of luciferase and visualization in SDS-PAGE
For more reliability of luciferase elucidation, two different methods have been employed for extractions from luminescent broth cultures [9, 19]. Cell lysates obtained from both methods were centrifuged, 5 ml of the supernatant was run through DEAE-52 column in respective elution buffer described in each method, and fractions were collected at a rate of 0.5 ml/min. The collected fractions from both methods were run in SDS-PAGE to visualize and determined luciferase enzyme molecular weights. The gel was run at 50 V and stained with Coomassie brilliant blue.
Extraction of secondary metabolites
Cultures of V. campbellii STF1 were grown in 30 ml of Marine Broth in 100 mL conical flasks, incubated in a shaker at 220rpm with continuous agitation for a period of 3 days at 28°C [20]. After the incubation period, cultures were centrifuged at 10000 rpm for 15 minutes, and cells were removed. Supernatant of the cultures was extracted with an equal volume of ethyl acetate (EtOAc) and placed on a shaker at 220 rpm with continuous agitation for 12 hours to achieve a homogenous mixture. Subsequently, the organic phases were collected and concentrated with a Rota evaporator under reduced pressure at 37°C to remove solvents. The final crude extract obtained was redissolved in 1 ml EtOAc for antibacterial assay.
Antibacterial activity against human pathogenic bacteria
Antibacterial activity of STF1was screened against 17 human pathogenic bacteria (Table 1). Stringent aseptic conditions were adopted for the preparation of inoculums of these pathogenic bacterial cultures. A loop full of each human pathogenic bacterial culture was inoculated into a 5 ml glass tube containing nutrient broth medium and incubated at 35°C for overnight (12 to 18 hours) and used as inoculums suspension for antibacterial assay.
Table 1.
Antimicrobial activity of V. campbellii STF1 against different human pathogenic bacteria
| S. no | Human pathogenic bacteria | Growth inhibition zones (mean ± standard deviation) |
|---|---|---|
| 1. | Enterotoxigenic Escherichia coli serotype O115 | 7.6±0.5 mm |
| 2. | Enteropathogenic E. coli serotype O114 | 7.6±0.5 mm |
| 3. | Shiga toxin-producing E. coli serotype O157:H7 | 8.6±0.5 mm |
| 4. | Klebsiella pneumoniae | 15.5±0.5 mm |
| 5. | Pseudomonas aeruginosa | - |
| 6. | Salmonella typhi | - |
| 7. | Salmonella typhi C6953 | - |
| 8. | Salmonella typhi B12101 | 8.6±0.5 mm |
| 9. | Staphylococcus aureus | - |
| 10. | Staphylococcus aureus | 11.0±0.5 mm |
| 11. | Shigella flexneri type 2a | 9.0±0.5 mm |
| 12. | Shigella dysenteriae type 5 | 27.0±1.0 mm |
| 13. | Shigella boydii type 1 | 10.5±0.5 mm |
| 14. | Shigella sonnei | 8.6±0.5 mm |
| 15. | Aeromonas hydrophilla | 11.3±0.3 mm |
| 16. | Vibrio cholera O139 | 12.3±0.5 mm |
Antibacterial activity assay was performed according to Kirby-Bauer well diffusion method [ 18]. The inoculum suspensions of pathogenic bacterial cultures were swabbed onto entire surfaces of Mueller-Hinton agar plates. The lids of these swabbed Petri dishes were left ajar for 2 to 5 minutes until excess surface moisture was absorbed. Now, wells were made on these plates with sterile cork borers, and 200 μl of the crude bacterial extract was impregnated into wells. The EtOAc solvent was used as a negative control. After complete diffusion of extract into media, plates were incubated at 35°C for 24 hours, and clear zones of inhibition formed around wells (including well size) were measured in diameters.
GC-MS analysis of crude extractions
Crude extraction was analyzed with GC-MS to determine compounds responsible for antimicrobial activity in this isolate. GC-MS analysis was performed with a Thermo GC – Trace Ultra Ver: 5.0, Thermo MS DSQ II, TR 5 – MS capillary standard non – polar column, injection volume 1 microliter, carrier gas Helium, with a flow rate of 1.0 ml/min.
Statistical analysis
Data results obtained from antibacterial assay and heavy metal sensitivity test were expressed as mean ± standard deviation using Microsoft Excel.
Genotypic characterization of luminous bacterium V. campbellii STF1
Genomic DNA isolation
Genomic DNA was isolated according to Cetyl Trimethyl Ammonium Bromide (CTAB) method [21].
PCR amplification of luxA and toxR genes
For PCR amplification, two sets of primers targeting the luxA 745 bp and toxR 477 bp were employed. PCR was carried out by using corresponding forward primer 5′-CTACTGGATCAAATGTCAAAAGGACG-3′ and reverse primer, 5′-TCAGAACCGTTTGCTTCAAAACC-3′ for luxA gene [22]; and forward primer toxRs 5′-GANCARGGNTTYGARGTNGAYGAYTC-3′ and reverse primer toxRas 5′-TTDKKTTGNCCNCYNGTV GCDATNAC-3′ for toxR gene respectively [23]. PCR of the genomic DNA of V. campbellii STF1 was conducted in a final volume of 25μl. The reaction mixture contained 10× PCR buffer (Himedia, Mumbai), 10 μM DNTP’s, 1 U of Taq DNA polymerase, 10 μM of each forward and reverse oligonucleotide primers, and approximately 20 ng of genomic DNA.
PCR amplification was performed using a GeneAmp PCR system 2720-ThermoCycler, (Applied Biosystem, Foster City, CA, USA). The amplification profile for luxA gene consisted of an initial denaturation at 95°C for 5 min, followed by 35 cycles at 95°C for 1 min, 58°C for 30 sec, 72°C for 30 min, and a final extension step of 72°C for 7 min. For toxR gene, the thermal program consisted of an initial denaturation at 94°C for 3 min, followed by 35 cycles at 94°C for 1 min, 60°C for 1 min, 72°C for 1 min, and a final extension step of 72°C for 7 min. Samples were held at 4°C until further analysis. Further PCR products were resolved in 1.2% agarose gel electrophoresis, gel stained with ethidium bromide, and photographed with gel doc capture system.
Sequencing and phylogenetic analysis of luxA and toxR genes
Sequencing was performed with an automated sequencer (Applied Biosystems, Foster City, USA) determined by the Sanger dideoxynucleotide chain termination method. Prior to sequence, PCR products of toxR and luxA genes were purified with GeNoRime PCR Purification kit (Shrimpex, Chennai) and partially sequenced. The nucleotide sequences obtained were compared with sequences in public gene databases GenBank, EMBL, and DDBJ. Black Box Chimera Check (B2C2) software was used to check chimera formations [24]. Alignment of sequences was carried out using MEGA 6 software [25], and uncertain alignments found at the ends of the sequences were omitted for further analyses. The same software was used to construct a phylogenetic tree. In order to construct and evaluate the reproducibility of the phylogenetic tree, the Neighbor-Joining method, Jukes-Cantor distance matrix, and 1000 bootstrap resamplings were accomplished.
Plasmid DNA isolation
In order to confirm the presence of plasmid in V. campbellii STF1, plasmid DNA extraction was performed with a plasmid DNA extraction kit (Himedia, Mumbai). Plasmid DNAs were visualized in 1.2% agarose gel stained with ethidium bromide and visualized under UV gel doc system.
Results
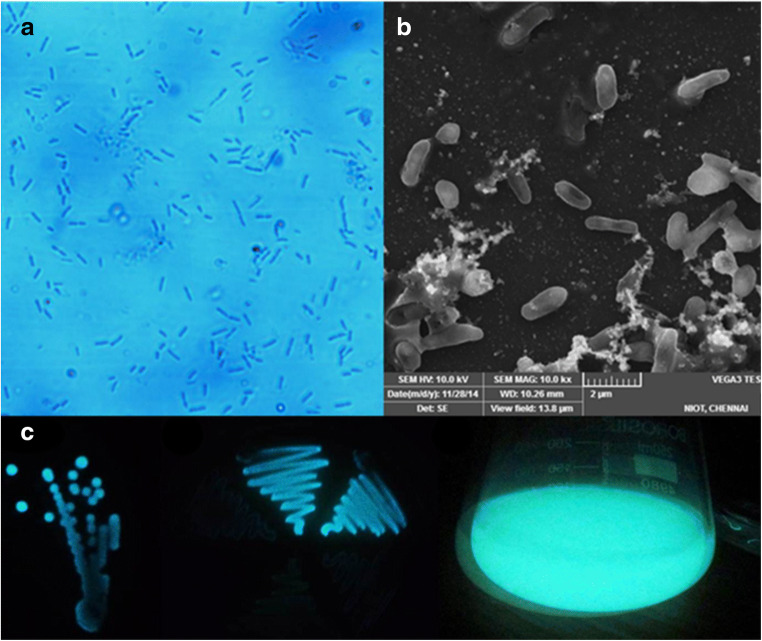
Vibrio campbellii isolated from the surface of stonefish S. verrucosa has shown intense blue luminescence. Luminescence was more intense in broth culture than on the agar plates (Fig. 1).
Fig. 1.
Rod shape morphology of V. campbellii observed under inverted microscope (a) and SEM (b); luminescence on SWC agar plate (left), marine agar (middle), and luminescent broth (right) (c)
Phenotypic characterization of V. campbellii STF1
Morphological observations revealed V. campbellii as short rods and motile (Fig. 1). Colonies are large in size with an entire margin, circular form, and convex elevation. This bacteria is Gram-negative, green in color on TCBS agar, and positive to oxidase, catalase, ONPG, O/129 vibriostatic agent (2,4-diamino-6,7-diisopropyl pteridine sulfate), nitrate reduction, citrate utilization, methyl red test, and positive to various carbohydrates (Table S1). This strain is also positive for dye uptake. The maximum tolerable temperature was 40°C, and it even can tolerate 10% NaCl. Based on the biochemical identification keys, strain STF1 was identified as V. campbellii. This bacterium produces a non-diffusible brownish-orange pigment on marine agar (Fig. S1).
Major cellular fatty acids observed were saturated C15:0 anteiso, C17:0 anteiso, C19:0 anteiso, and C13:0 iso 3OH; unsaturated C18:1 ω9c, and summed C18:1 ω6c/C18:1 ω7c. Summed feature 2 comprises C14:0 3OH and/or C16:1 iso I, summed feature 3 comprises C16:1ω6c and/or C16:1 ω7c, summed feature 8 comprises C18:1 ω6c and/or C18:1 ω7c and summed feature 9 comprises C17:1 iso ω9c, and/or C16:0 10-methyl. Various other fatty acids such as C11:0, C12:0, C11:0 3OH, C14:1 iso, C14:0, , C15:0 iso, C14:0 iso 3OH, C16:1ω6c, C16:1 ω7c, C16:1 iso H, C16:0 iso, C16:0, C15:0 iso 3OH, C17:1 iso ω5c, C17:0 iso, C17:1 ω5c, C17:0, C17:0 iso 3OH, C18:0, C18:0 10-methyl, C19:0 iso, and C19:0 cyclo ω8c have also been observed in V. campbellii STF1. MALDI-TOF analysis showed distinct mass spectral peaks at mass-to-charge ratio (m/z) of 2615, 3948, and 4232 Da (Fig. S2). Further analysis is yet to be investigated to characterize the proteins responsible for these distinct peaks.
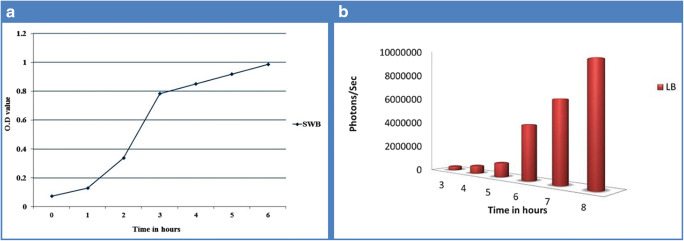
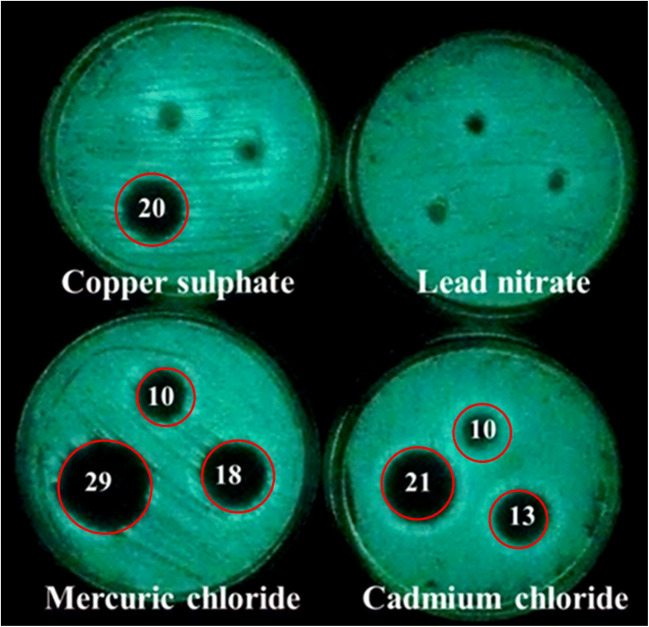
Log phase was observed during 1st hour of incubation and continued till 6th hour. The maximum intensity of luminescence produced by the luciferase enzyme was 8,320,627 photons per second observed at the eighth hour (Figs. 2a, b). Growth and luminescence of V. campbellii were less at pH < 7 and > 11, and optimum was found at pH 9 (data not shown). In vitro, antibiotic sensitivity test showed that this strain is sensitive to most tested antibiotics (Table S2), while resistant to amoxyclave, ampicillin, gentamicin, kanamycin, oxacilin, and penicillin. V. campbellii is sensitive to mercury and cadmium at concentrations of 0.1 mM, 1 mM, and 10 mM with inhibition zones of 10.0±0.0 mm, 18.0±0.0 mm, and 29.0±0.0 mm, and 10.0±0.0 mm, 13.0±0.0 mm, and 21.0±0.0 mm respectively (Fig. 3; Table 2). It is resistant to lead metal. The molecular masses of partially purified luciferase enzyme alpha and beta subunits determined by SDS-PAGE gel were 40 kDa and 37 kDa, respectively (Fig. 4).
Fig. 2.
Growth curve (a) and luminescence count (b) of V. campbellii
Fig. 3.
Sensitivity of STF1 to different heavy metals. Red line rings indicate inhibition zone, and numbers are inhibition zones in diameter
Table 2.
Sensitivity of STF1 to different heavy metals (inhibition zones in diameter)
| S. no | Heavy metal | Growth inhibition zones in different concentrations (mean ± standard deviation) | ||
|---|---|---|---|---|
| In 0.1 mM | In 1 mM | In 10 mM | ||
| 1. | Cadmium chloride | 10.0±0.2 mm | 13.0±0.1 mm | 21.0±0.3 mm |
| 2. | Copper sulfate | - | - | 20.0±0.1 mm |
| 3. | Lead nitrate | - | - | - |
| 4. | Mercuric chloride | 10.0±0.1 mm | 18.0±0.2 mm | 29.0±0.4 mm |
Fig. 4.

Visualization of V. campbellii luciferase enzyme alpha (40 kDa) and beta (37 kDa) subunits in SDS-PAGE gel. Lanes: M, marker; 1, method of Hastings et al. (1978); 2, method of Liu et al. (2012)
Crude ethyl acetate extractions stored in -20°C had exhibited good antibacterial activity against various human pathogenic bacteria (Table 1). Maximum zone of inhibition occurred against Shigella dysenteriae type 5 (27.0±1.0 mm), and less activity was observed against E. coli (7.6±0.5 mm). GC-MS chromatogram of crude extraction revealed two important compounds, tetradecanal (RT at 7.55), and 2-tert-Butyl-4-isopropyl-5-methylphenol (RT at 12.74) (Fig. S3).
Genotypic characterization of V. campbellii STF1
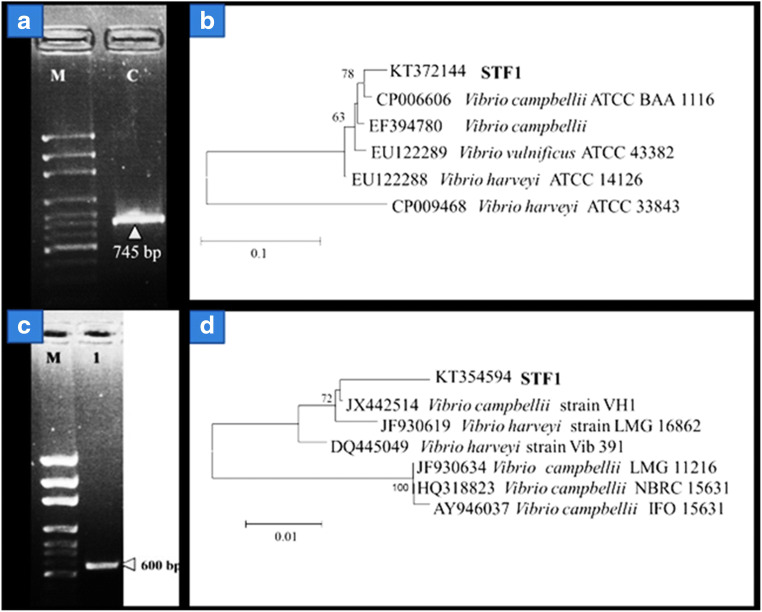
A total length of 745 bp corresponded to luxA gene was successfully amplified by PCR. The amplified toxR gene fragment size was larger (600 bp) than the expected size of 477 bp (Fig. 5). This sequence was aligned with database sequences and trimmed to 477 bp length and analyzed. Nucleotide sequences 16s rRNA, luxA, and toxR genes were deposited in GenBank under the accession numbers of KF961190, KT372144, and KT354594, respectively. While the presence of plasmid indicated that V. campbellii antibiotic resistance is mediated by plasmid DNA.
Fig. 5.
LuxA gene PCR products visualized under gel doc system. Lanes: M, 100 bp marker; C, amplified luxA gene product of 745 bp; (a) NJ tree of luxA gene of STF1, (b) ToxR gene PCR products. (c) Lanes: M, 100 bp marker; 1, amplified toxR gene; (d) NJ tree of toxR gene of STF1
Discussions
Vibrio campbellii strains are known to display morphological and genetic plasticity. Strain STF1 is found to show phenotypic and genotypic plasticity when compared with other strains of V. campbellii isolated previously from the Andaman waters [10]. Brown pigmentations observed in strain STF1 were not produced by other V. campbellii strains isolated in the previous study [10]. Brown pigment production of strain STF1 was similar to V. campbellii ATCC BAA-1116 [26]. A luminous bacterial strain, V. campbellii LZ5 isolated from the meat of Crassostrea gigas was reported to be resistant to two heavy metal ions, cadmium, and mercury [9]. Conversely, in the present study, V. campbellii STF1 was sensitive to both the metals, indicating the intra-species-specific resistance activity as well as the novelty of STF1. An earlier report by Ulitzur and Hastings suggested that tetradecanal is the natural aldehyde substrate involved in luminescence emission [27]. In the present study, also teradecanol was detected from strain STF1; thus, this study is an evidence and supports the previous study observations [27].
Since phenol derivatives are well known as antimicrobial compounds, we speculate that the observed phenol derivative compound 2-tert-Butyl-4-isopropyl-5-methylphenol is likely being responsible for the antibacterial activity of V. campbellii. A recent report revealed the potential antimicrobial activities of luminous bacteria against different human pathogens due to antibacterial compounds such as indole, phenol, 2,4-bis(1,1-dimethylethyl)-,dibutyl phthalate, and 1,2-Benzenedicarboxylic acid, butyl octyl ester [10]. However, such compounds were not found in V. campbellii STF1. Significantly, strain-specific antimicrobial activity was displayed by V. campbellii strains such as BSE1, BSE5, BSECU1, BSECU3, and PEVI1 showed antibacterial activity against various pathogens [10]. Among these strains, BSECU1 displayed maximum inhibition zone of 24 mm against Bacillus subtilis [10]. Whereas the present study strain STF1 showed maximum inhibition zone against Shigella dysenteriae type 5 (27.0±1.0 mm). But strains BSE1, BSE5, BSECU1, BSECU3, and PEVI1 did not show any antibacterial activity against S. dysenteriae [10], indicating the distinct characteristics.
The distinct mass spectral peaks obtained for V. campbellii STF1 were assigned as specific biomarkers, which will serve as species-specific biomarkers for further identification. Multilocus sequence analysis using 16s rRNA [5], toxR, and luxA genes has identified strain STF1 as V. campbellii. This strain BLAST analysis of toxR gene sequence of V. campbellii STF1 revealed 99% sequence identity with V. campbellii VH1 (accession JX442514) isolated from Litopenaeus vannamei shrimp farm [28]. V. campbellii VH1 and V. campbellii STF1 are both haemolytic, and STF1 is different from VH1 as it is resistant to gentamicin and kanamycin. While luxA gene sequence of V. campbellii STF1 showed 99% sequence identity with V. campbellii ATCC BAA-1116 [26]. In NJ tree, also clustering was apparent with these two strains- VH1 and ATCCBAA-116 as similar to BLAST analysis (Fig. 5).
This study shows that luminous V. campbellii STF1 can be readily used as a natural biosensor in vitro to monitor different toxic metals and pollutants from food and agriculture industries. The various concentrations of pollutants can be prepared as similar to our study or earlier study [9] and tested against luminous bacteria to generate luminescence excitation and inhibition profiles associated with each concentration of the interested pollutant. While the extracted luciferase enzyme from the strain STF1 can be employed in luminescence-based assays along with reduced flavin mononucleotide (FMNH2) as luciferin to test pollutants from various environments. Either way, whole bacteria (strain STF1) and its luciferase enzyme would serve as readily available in vitro biosensors to monitor a wide range of pollutants. Natural luminous strain over cloned luminous bacteria is advantageous because natural luminous bacteria produce intense luminescence than cloned bacteria. Developing cloned bacteria with lux genes are cost-effective, time-consuming, and uncertain with the luminescence intensity production. Thus, natural strains with intense luminescence such as strain STF1 are more important in developing rapid toxicological assays. V. campbellii STF1 is also a potential bioactive compound producing marine luminous bacteria for biomedical applications.
Conclusion
The morphological and genetic variations observed within the Vibrio campbellii strains isolated from the Andaman islands suggest that these strains could be novel. Thus, whole-genome analysis is required to understand their genome plasticity in the Andaman waters. The emission patterns of luminescence of V. campbellii STF1 under various toxic heavy metals promises its potential application in developing real-time biosensor in the laboratory. The identified phenol compounds may be extracted for drug application.
Supplementary Information
(DOCX 1626 kb)
Acknowledgments
Ramesh thanks the Department of Science and Technology, New Delhi, for providing the INSPIRE fellowship DST/IF120230/2012/280. This is CSIR-NIO’s contribution: 6767.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wilson T, Hastings JW (2013) Bioluminescence: living lights, lights for living. Harvard University Press
- 2.Dunlap PV, Urbanczyk H. Luminous bacteria. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. 4. Berlin, Heidelberg: Springer; 2013. pp. 495–528. [Google Scholar]
- 3.Ramesh CH, Mohanraju R. Genetic diversity of bioluminescent bacteria in diverse marine niches. Indian J Mar Sci. 2017;46(10):2054–2062. [Google Scholar]
- 4.Dunlap PV. Biochemistry and genetics of bacterial bioluminescence. In: Thouand G, Marks R, editors. Bioluminescence: Fundamentals and Applications in Biotechnology. Heidelberg: Berlin; 2014. pp. 37–64. [Google Scholar]
- 5.Ramesh CH, Mohanraju R, Murthy KN, Karthick P, Narayana S. Impact of light, temperature, salinity and glycerol on the intensity of luminescence and growth of marine bioluminescent bacteria Vibrio campbellii (strain STF1) Curr Sci. 2014;106(4):511–513. [Google Scholar]
- 6.Ramesh CH (2016) Studies on marine bioluminescent bacteria from Andaman Islands. PhD Thesis. Pondicherry University, Port Blair Campus
- 7.Shimomura O. Bioluminescence: chemical principles and methods. Singapore: World Scientific; 2006. [Google Scholar]
- 8.Ramesh CH. Terrestrial and marine bioluminescent organisms from the Indian subcontinent: a review. Environ Monit Assess. 2020;192:747. doi: 10.1007/s10661-020-08685-5. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Zhao J, Liang X, Li K, Xiao X, Zhu J, Sun Q, Liang Q. Characterization of luminescent Vibrio campbellii LZ5 and its potential application in the detection of environmental heavy metals. Biotechnol Appl Biochem. 2012;59:405–410. doi: 10.1002/bab.1040. [DOI] [PubMed] [Google Scholar]
- 10.Ramesh CH, Mohanraju R. Antibacterial activity of marine bioluminescent bacteria. Indian J Mar Sci. 2017;46(10):2063–2074. [Google Scholar]
- 11.Gomez-Gil B, Thompson CC, Matsumura Y, et al. Family Vibrionaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thonpson FL, et al., editors. The Prokaryotes. 4. Berlin, Heidelberg: Springer; 2014. pp. 659–747. [Google Scholar]
- 12.WHO . Laboratory biosafety manual. 3. Malta: World Health Organization; 2004. [PubMed] [Google Scholar]
- 13.Benson (2001) Microbiological applications lab manual, 8th edn The McGraw−Hill Companies, New York
- 14.Noguerola I, Blanch AR. Identification of Vibrio spp. with a set of dichotomous keys. J Appl Microbiol. 2008;105:175–185. doi: 10.1111/j.1365-2672.2008.03730.x. [DOI] [PubMed] [Google Scholar]
- 15.Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101, Newark
- 16.Ahmad F, Wu H. High-resolution MALDI-TOF mass spectrometry of bacterial proteins using a Tris-EDTA buffer approach. Microchim Acta. 2012;176:311–316. doi: 10.1007/s00604-011-0714-0. [DOI] [Google Scholar]
- 17.Gellert G. Sensitivity and significance of luminescent bacteria in chronic toxicity testing based on growth and bioluminescence. Ecotoxicol Environ Saf. 2000;45:87–91. doi: 10.1006/eesa.1999.1849. [DOI] [PubMed] [Google Scholar]
- 18.Bauer AW, Kirby WMM, Sherris JS, Turk M. Antibiotic susceptibility by standardized single disc method. Am J Clin Pathol. 1966;45:493–496. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- 19.Hastings JW, Baldwin TO. Nicoli MZ. Bacterial luciferase: assay, purification and properties. Methods Enzymol. 1978;57:135–152. doi: 10.1016/0076-6879(78)57016-X. [DOI] [Google Scholar]
- 20.Wietz M, Mansson M, Gotfredsen CH, Larsen TO. Antibacterial compounds from marine Vibrionaceae isolated on a global expedition. Mar Drugs. 2010;8:2946–2960. doi: 10.3390/md8122946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishiguchi MK, Doukakis P, Egan M, et al. DNA isolation procedure. In: DeSalle R, Giribet G, Wheeler W, et al., editors. Methods and tools in biosciences and medicine techniques in molecular systematics and evolution. Basel: Birkhäuser; 2002. pp. 249–287. [Google Scholar]
- 22.Wimpee C, Nadeau TL, Nealson K. Development of species-specific hybridization probes for marine luminous bacteria by using in vitro DNA amplification. Appl Environ Microbiol. 1991;57:1319–1324. doi: 10.1128/aem.57.5.1319-1324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pascual J, Macian MC, Arahal DR, Garay E, Pujalte MJ. Multilocus sequence analysis of the central clade of the genus Vibrio by using the 16S rRNA, recA, pyrH, rpoD, gyrB, rctB and toxR genes. Int J Syst Evol Microbiol. 2010;60:154–165. doi: 10.1099/ijs.0.010702-0. [DOI] [PubMed] [Google Scholar]
- 24.Gontcharova V, Youn E, Wolcott RD, Hollister EB, Gentry TJ, Dowd SE. Black box chimera check (B2C2): a windows-based software for batch depletion of chimeras from bacterial 16S rRNA gene datasets. Open Microbiol J. 2010;4:47–52. doi: 10.2174/1874285801004010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Lin B, Mostaghim A, et al. Vibrio campbellii hmgA-mediated pyomelanization impairs quorum sensing, virulence, and cellular fitness. Front Microbiol. 2013;4:1–11. doi: 10.3389/fmicb.2013.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ulitzur S, Hastings JW. Evidence for tetradecanal as the natural aldehyde in bacterial bioluminescence. PNAS. 1979;76:265–267. doi: 10.1073/pnas.76.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Chen Y, Huang H, Huang Z, Chen H, Shao Z. Isolation and identification of Vibrio campbellii as a bacterial pathogen for luminous vibriosis of Litopenaeus vannamei. Aquac Res. 2013;46:395–404. doi: 10.1111/are.12191. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 1626 kb)